Open Access Article
Open Access ArticleInconsistent hydrogen bond-mediated vibrational coupling of amide I†
Suranjana Chakrabarty and
Anup Ghosh *
*
a, Department of Condensed Matter of Physics and Materials Sciences, S. N. Bose National Centre for Basic Sciences, JD Block, Sector-III, Salt Lake City, Kolkata – 700 106, India. E-mail: anupg86@gmail.com; anup.ghosh@bose.res.in
First published on 5th January 2023
Abstract
Using infrared spectroscopy and density functional theory (DFT) calculations, we scrutinized an amide (dimethylformamide) as a “model” compound to interpret the interactions of amide 1 with different phenol derivatives (para-chlorophenol (PCP) and para-cresol (CP)) as “model guest molecules”. We established the involvement of amide I in vibrational coupling with symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of different phenolic derivatives and how their coupling was dependent upon different guest aromatic phenolic compounds. Interestingly, substitution of phenol perturbed the pattern of vibrational coupling with amide I. The symmetric and asymmetric C
C modes of different phenolic derivatives and how their coupling was dependent upon different guest aromatic phenolic compounds. Interestingly, substitution of phenol perturbed the pattern of vibrational coupling with amide I. The symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of PC were coupled significantly with amide 1. For PCP, the symmetric C
C modes of PC were coupled significantly with amide 1. For PCP, the symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode coupled significantly, but the asymmetric mode coupled negligibly, with amide I. Here, we reveal the nature of vibrational coupling based on the structure of a guest molecule hydrogen-bonded with amide I. Our conclusions could be valuable for depiction of the unusual dynamics of coupled amide-I modes as well as the dependency of vibrational coupling on altered factors.
C mode coupled significantly, but the asymmetric mode coupled negligibly, with amide I. Here, we reveal the nature of vibrational coupling based on the structure of a guest molecule hydrogen-bonded with amide I. Our conclusions could be valuable for depiction of the unusual dynamics of coupled amide-I modes as well as the dependency of vibrational coupling on altered factors.
Introduction
Hydrogen bonds are pervasive in protein molecules, and are involved in biological processes such as molecular association, catalysis, and signal transmission.1–15 How biologically active, small organic molecules interact with other molecules to elicit different biological effects is an important research area.16–20 However, identifying the vibrational modes of biologically active molecules (e.g., proteins) is difficult. To overcome such difficulties, the structural and environmental properties of biomolecules have been investigated using “vibrational probes”.21–26Many biomolecules contain “amide I”, “amide II”, “amide III”, and “amide A” modes of vibration. However, the amide-I mode is studied widely as a vibrational probe.27,28 The amide bond is present in many organic molecules and biomolecules.29–32 Most importantly, as an infrared (IR) probe, amide I is employed extensively because of its sensitivity to the native electric field, solvation, and large molar extinction coefficient.33–38 In particular, vibrational spectroscopic measurements of the amide-I band are used to monitor shifts in the transition frequency, which is sensitive to the local electric fields as well as interactions with specific “guest molecules”.24–27 Many studies have focused on the relationship between vibrational couplings and conformational dynamics of proteins/peptides.34–41 Various theoretical methodologies and multidimensional IR-spectroscopy methods have been employed to investigate the vibrational coupling and structural details of biological systems.42–49 Vibrational coupling and the interactions between different vibrational modes have been investigated.50 The vibrational coupling between hydrogen bonds associated with amide-A and amide-I/II modes within the same amide component for several dipeptides has been studied using two-dimensional IR spectroscopy.51 The hydrogen bonding between amide I and phenol derivatives, dimethylformamide (DMF), and dimethyl acetamide has been considered.52–54
Investigation of the amide-I vibrational mode is very complex because it is delocalized in biomolecules. However, to study the molecular perceptions and sensitivity of the amide-I mode in the presence of intermolecular hydrogen bonds, we used DMF as a “model” molecule. We measured the IR absorbance of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode involved in vibrational coupling during intermolecular hydrogen bonding with amide I. Correlations between the hydrogen bond-induced vibrational coupling of C
C mode involved in vibrational coupling during intermolecular hydrogen bonding with amide I. Correlations between the hydrogen bond-induced vibrational coupling of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O transitions with different factors were investigated by employing linear IR spectroscopy. We revealed how pervasive formation of hydrogen bonds in the presence of phenolic compounds (hydrogen-bond contributors) could disturb the amide-I transition and symmetric/asymmetric C
O transitions with different factors were investigated by employing linear IR spectroscopy. We revealed how pervasive formation of hydrogen bonds in the presence of phenolic compounds (hydrogen-bond contributors) could disturb the amide-I transition and symmetric/asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C transition of guest molecules. Hydrogen-bond formation as well as the dependency of vibrational coupling upon different orientations between coupled modes were also investigated in our work.
C transition of guest molecules. Hydrogen-bond formation as well as the dependency of vibrational coupling upon different orientations between coupled modes were also investigated in our work.
We employed linear IR spectroscopy and density functional theory (DFT) calculations as theoretical approaches. The frequency gap between symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching of phenol derivatives and the C
C stretching of phenol derivatives and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrational mode of DMF, as well as the enhancement factor in IR absorption during vibrational coupling, were monitored in the presence of different donor molecules. Vibrational coupling in biomolecules is important to understand the many biological interactions and processes at the microscopic level, so the coupling of amide I must be investigated. Overall, this structural evidence of vibrational coupling can be used to elucidate many biological and chemical effects.
O vibrational mode of DMF, as well as the enhancement factor in IR absorption during vibrational coupling, were monitored in the presence of different donor molecules. Vibrational coupling in biomolecules is important to understand the many biological interactions and processes at the microscopic level, so the coupling of amide I must be investigated. Overall, this structural evidence of vibrational coupling can be used to elucidate many biological and chemical effects.
Experimental section
Para-chlorophenol (PCP; 99.9% purity), para-cresol (PC; 99.9% purity), and DMF (99.9% purity) were purchased from MilliporeSigma and used without additional purification. The chemical structure of PCP and PC are drawn in Scheme 1. A solution of DMF (0.1 M) in carbon tetrachloride (CCl4) was used for linear IR spectroscopy. The sample was placed in a homemade Fourier transform infrared (FTIR) sample cell with CaF2 windows and a Teflon™ spacer (60 μm). Linear IR absorption spectroscopy was undertaken using an FTIR spectrometer (JASCO-FTIR-6300). The background of the solvent (CCl4) was measured and subtracted from all spectra of interactions between DMF and phenol derivatives. The Beer–Lambert law was validated by plotting the area of IR absorbance for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode vs. concentration (Fig. S1, ESI†).
C mode vs. concentration (Fig. S1, ESI†).
Theoretical section
We wished to gain detailed knowledge about the IR absorption spectra of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode in DMF and C
O mode in DMF and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of different phenol substitutions, so we undertook DFT calculations employing Gaussian 09. In a preliminary manner, all the initial geometries of DMF and different phenolic complexes were optimized by the B3LYP/6-311G+ (D, P) level of theory. Then, calculations to determine the frequency of IR absorption were done for all DMF–phenol hydrogen-bonded complexes.
C mode of different phenol substitutions, so we undertook DFT calculations employing Gaussian 09. In a preliminary manner, all the initial geometries of DMF and different phenolic complexes were optimized by the B3LYP/6-311G+ (D, P) level of theory. Then, calculations to determine the frequency of IR absorption were done for all DMF–phenol hydrogen-bonded complexes.
Results and discussion
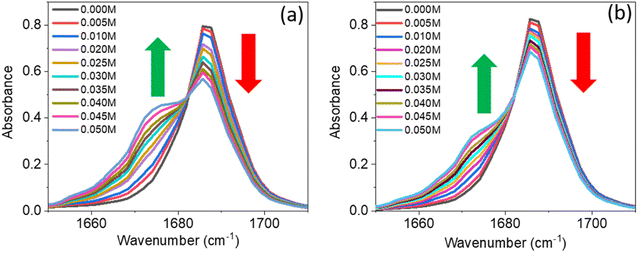
A series of linear IR spectra of DMF solution (0.1 M) in CCl4 were taken with increasing concentrations of PC and PCP from 0 M to 0.05 M (Fig. 1). Preliminarily, the IR absorption frequency of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode was shown to be 1686 cm−1 (black single peak) in the absence of phenolic compounds (0.00 M). With gradual addition of PC or PCP, the IR absorbance of amide 1 decreased progressively and the frequency shifted towards a lower-wavenumber region (Fig. 1a and b, respectively). Fig. 1 reveals that increasing the concentration of PC and PCP led to hydrogen-bond formation of C
O mode was shown to be 1686 cm−1 (black single peak) in the absence of phenolic compounds (0.00 M). With gradual addition of PC or PCP, the IR absorbance of amide 1 decreased progressively and the frequency shifted towards a lower-wavenumber region (Fig. 1a and b, respectively). Fig. 1 reveals that increasing the concentration of PC and PCP led to hydrogen-bond formation of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and gradual shifting of the peak position of IR absorption. However, in PC (0.1 M) and PCP (0.1 M), with a gradual increase in the DMF concentration, the IR absorption spectra for symmetric and asymmetric C
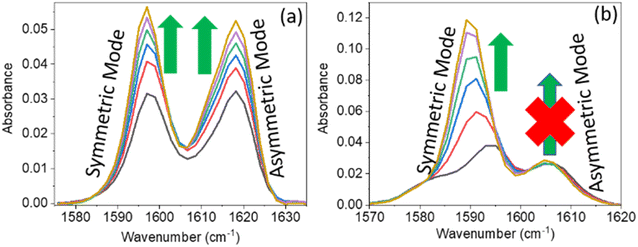
O and gradual shifting of the peak position of IR absorption. However, in PC (0.1 M) and PCP (0.1 M), with a gradual increase in the DMF concentration, the IR absorption spectra for symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes showed anomalous behaviours (Fig. 2). For PC, the IR absorption peaks for symmetric and asymmetric C
C modes showed anomalous behaviours (Fig. 2). For PC, the IR absorption peaks for symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes were at 1597.5 cm−1 and 1618.5 cm−1 (Fig. 2a) whereas, for PCP they were at 1594.0 cm−1 and 1606.5 cm−1, respectively (Fig. 2b). For PC and PCP, with an increasing concentration of DMF (0.000–0.10 M), a significant difference in IR absorbance was observed between symmetric and asymmetric C
C modes were at 1597.5 cm−1 and 1618.5 cm−1 (Fig. 2a) whereas, for PCP they were at 1594.0 cm−1 and 1606.5 cm−1, respectively (Fig. 2b). For PC and PCP, with an increasing concentration of DMF (0.000–0.10 M), a significant difference in IR absorbance was observed between symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes (Fig. 2). For PC, with a gradual increase in the DMF concentration from 0.000 M to 0.10 M, IR absorbance for symmetric and asymmetric C
C modes (Fig. 2). For PC, with a gradual increase in the DMF concentration from 0.000 M to 0.10 M, IR absorbance for symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes was enhanced significantly. However, in contrast with PCP, though IR absorbance for symmetric C
C modes was enhanced significantly. However, in contrast with PCP, though IR absorbance for symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching was enhanced, IR absorbance for asymmetric C
C stretching was enhanced, IR absorbance for asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching was altered negligibly throughout the experiment (Fig. 2). The IR-absorbance enhancement ratio for the symmetric and asymmetric C
C stretching was altered negligibly throughout the experiment (Fig. 2). The IR-absorbance enhancement ratio for the symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of PC was 1.35 and 1.40 whereas, for PCP, it was 5.97 and 1.00, respectively (Table 1), as calculated from Fig. 2. The transition dipole moment of the symmetric and asymmetric modes of PC and PCP changed accordingly (Table 1). These unusual phenomena focused our attention on the intermolecular interactions between amide (C
C modes of PC was 1.35 and 1.40 whereas, for PCP, it was 5.97 and 1.00, respectively (Table 1), as calculated from Fig. 2. The transition dipole moment of the symmetric and asymmetric modes of PC and PCP changed accordingly (Table 1). These unusual phenomena focused our attention on the intermolecular interactions between amide (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) and phenolic compounds (PC and PCP).
O) and phenolic compounds (PC and PCP).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode and the ratio of IR absorption area for the hydrogen-bonded C
C mode and the ratio of IR absorption area for the hydrogen-bonded C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode and free C
C mode and free C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of phenol derivatives. The enhancement factor and transition dipole moment ratio were calculated from the experimental data shown in Fig. 2
C mode of phenol derivatives. The enhancement factor and transition dipole moment ratio were calculated from the experimental data shown in Fig. 2
| Phenol derivatives | Stretching mode | Frequency (cm−1) | IR enhancement ratio (R) | Transition dipole moment ratio |
|---|---|---|---|---|
| PC | Symmetric | 1597.5 | 1.35 | 1.16 |
| Asymmetric | 1618.5 | 1.44 | 1.20 | |
| PCP | Symmetric | 1694.0 | 5.97 | 2.44 |
| Asymmetric | 1606.5 | 1 | 1 |
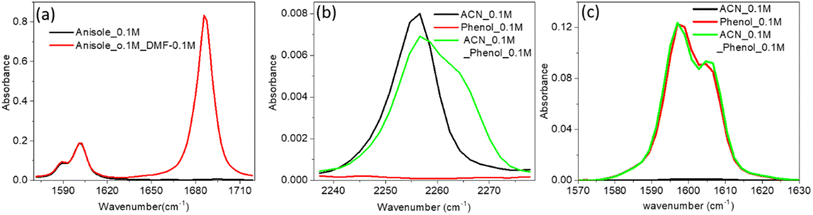
The increase in IR absorbance of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode could have been due to the altered hydrogen bonding with vibrational modes or n–π* bonding of PC and the PCP ring with the C
C mode could have been due to the altered hydrogen bonding with vibrational modes or n–π* bonding of PC and the PCP ring with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode of DMF (Fig. 2a and b). To elucidate the precise reason underpinning the enhancement, we undertook IR spectroscopy of a 1
O mode of DMF (Fig. 2a and b). To elucidate the precise reason underpinning the enhancement, we undertook IR spectroscopy of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMF
1 DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anisole mixture. The unaltered enhancement of IR absorbance for the C
anisole mixture. The unaltered enhancement of IR absorbance for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of the DMF
C mode of the DMF![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) anisole (1
anisole (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) mixture signified no interaction of the non-bonded electron of the oxygen atom of DMF with the π electron cloud of anisole. We did not know whether the enhancement was due to the altered electron density of the phenolic ring (C
1) mixture signified no interaction of the non-bonded electron of the oxygen atom of DMF with the π electron cloud of anisole. We did not know whether the enhancement was due to the altered electron density of the phenolic ring (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode) after hydrogen bonding with DMF, so we undertook IR spectroscopy of a mixture of acetonitrile (ACN) and phenol at a ratio of 1
C mode) after hydrogen bonding with DMF, so we undertook IR spectroscopy of a mixture of acetonitrile (ACN) and phenol at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 3b). The shifting to a higher IR absorbance frequency of CN signified formation of a hydrogen bond between ACN with phenolic OH (Fig. 3b). An absence of enrichment of IR absorbance of C
1 (Fig. 3b). The shifting to a higher IR absorbance frequency of CN signified formation of a hydrogen bond between ACN with phenolic OH (Fig. 3b). An absence of enrichment of IR absorbance of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C explained the non-involvement of hydrogen bonds. Hence, these results suggested that the enhancement in IR absorbance of the C
C explained the non-involvement of hydrogen bonds. Hence, these results suggested that the enhancement in IR absorbance of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode was not due to n–π* bonding or hydrogen bonding. Therefore, the enhancement was probably due to vibrational coupling between the C
C mode was not due to n–π* bonding or hydrogen bonding. Therefore, the enhancement was probably due to vibrational coupling between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of amide (DMF) and phenol derivatives (PC and PCP).
O modes of amide (DMF) and phenol derivatives (PC and PCP).
To check that our hypotheses on vibrational coupling were robust, we carried out DFT calculations (b3lyp, 6311 G (d, p)) for PC and PCP hydrogen-bonded with DMF, and the videos are provided in ESI† (AV1, AV2, AV3, AV4). Vibrational couplings were visualized between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C of phenol derivatives and C
C of phenol derivatives and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of DMF. Interestingly, our experimental observations aligned with the videos for DFT calculations. In AV1 and AV2, the symmetric and asymmetric C
O of DMF. Interestingly, our experimental observations aligned with the videos for DFT calculations. In AV1 and AV2, the symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of PC were coupled significantly with amide 1 of DMF; for PCP, the symmetric C
C modes of PC were coupled significantly with amide 1 of DMF; for PCP, the symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode was coupled significantly (AV3) but the asymmetric mode was not (AV4). PC and PCP are phenol derivatives and form hydrogen bonds with amide 1 of DMF, but they showed different vibrational coupling. Hence, phenolic substitution altered the pattern of vibrational coupling.
C mode was coupled significantly (AV3) but the asymmetric mode was not (AV4). PC and PCP are phenol derivatives and form hydrogen bonds with amide 1 of DMF, but they showed different vibrational coupling. Hence, phenolic substitution altered the pattern of vibrational coupling.
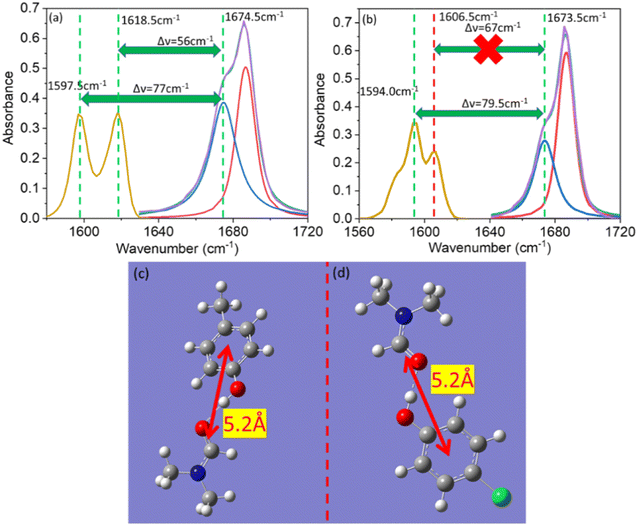
Vibrational coupling is dependent upon the frequency gap of IR absorption, the distance between two vibration modes, and orientation. The frequency gap between the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of PC and C
C mode of PC and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching mode of DMF was smaller (56 cm−1) than that of the symmetric C
O stretching mode of DMF was smaller (56 cm−1) than that of the symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (76 cm−1) modes of PC (Fig. 4a). However, the intensity of asymmetric and symmetric C
O (76 cm−1) modes of PC (Fig. 4a). However, the intensity of asymmetric and symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes was enhanced simultaneously. The frequency gap was smaller for the asymmetric C
C modes was enhanced simultaneously. The frequency gap was smaller for the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode (67 cm−1) than symmetric C
C mode (67 cm−1) than symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode (79.5 cm−1) of PCP with amide 1 of DMF. Significant enhancement in IR absorbance was observed for the symmetric C
C mode (79.5 cm−1) of PCP with amide 1 of DMF. Significant enhancement in IR absorbance was observed for the symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode, but negligible enhancement was observed for the asymmetric C
C mode, but negligible enhancement was observed for the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode (Fig. 4b). The distance between the C
C mode (Fig. 4b). The distance between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of phenol derivatives and DMF according to DFT calculations are shown in Fig. 4c and d. An identical distance (5.2 Å) between C
O modes of phenol derivatives and DMF according to DFT calculations are shown in Fig. 4c and d. An identical distance (5.2 Å) between C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O could not explain the disparity in vibrational coupling. Hence, we assumed that a different transition dipole angle between the C
O could not explain the disparity in vibrational coupling. Hence, we assumed that a different transition dipole angle between the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode and amide-I mode was the cause of this difference in vibrational coupling. To ascertain the reason for this anomalous coupling behaviour, we exposed different orientations between vibration modes (C
C mode and amide-I mode was the cause of this difference in vibrational coupling. To ascertain the reason for this anomalous coupling behaviour, we exposed different orientations between vibration modes (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
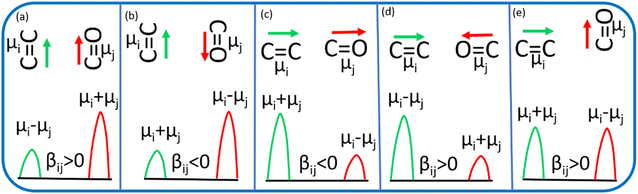
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (Fig. 5). The sign of the coupling constant (βij) was dependent upon the geometry of the hydrogen-bonded DMF and phenol derivatives (eqn (1)).55 If C
O) (Fig. 5). The sign of the coupling constant (βij) was dependent upon the geometry of the hydrogen-bonded DMF and phenol derivatives (eqn (1)).55 If C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes are parallel (Fig. 5a) and βij is positive, the intensity of the C
O modes are parallel (Fig. 5a) and βij is positive, the intensity of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrational mode will be enhanced, with a transition dipole moment μi + μj. In contrast, the intensity of the C
O vibrational mode will be enhanced, with a transition dipole moment μi + μj. In contrast, the intensity of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibrational mode will be weaker, with a transition dipole moment μi − μj. In the case of antiparallel C
C vibrational mode will be weaker, with a transition dipole moment μi − μj. In the case of antiparallel C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes (Fig. 5b) and negative coupling constant βij, the C
O modes (Fig. 5b) and negative coupling constant βij, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode will be weaker and correspondingly the C
C mode will be weaker and correspondingly the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode will be stronger, with a transition dipole moment of μi + μj and μi − μj, respectively. In the head-to-tail (Fig. 5c) or head-to-head orientation (Fig. 5d) of C
O mode will be stronger, with a transition dipole moment of μi + μj and μi − μj, respectively. In the head-to-tail (Fig. 5c) or head-to-head orientation (Fig. 5d) of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, the lower frequency mode of C
O, the lower frequency mode of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C will carry more oscillator strength compared with the C
C will carry more oscillator strength compared with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode. An unperturbed intensity will result if the C
O mode. An unperturbed intensity will result if the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode and C
C mode and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode are perpendicular to each other (Fig. 5e). The geometries of the DMF–PC complex and DMF–PCP complex are in-between the limits we modelled in Fig. 5. The symmetric and asymmetric C
O mode are perpendicular to each other (Fig. 5e). The geometries of the DMF–PC complex and DMF–PCP complex are in-between the limits we modelled in Fig. 5. The symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of PC and symmetric C
C modes of PC and symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of PCP are coupled in the manner shown in Fig. 5c and d. Probably, the asymmetric C
C mode of PCP are coupled in the manner shown in Fig. 5c and d. Probably, the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of PCP was coupled with the C
C mode of PCP was coupled with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O mode of DMF as shown in Fig. 5e or they were coupled very weakly according to the model shown in Fig. 5a–d.
O mode of DMF as shown in Fig. 5e or they were coupled very weakly according to the model shown in Fig. 5a–d.
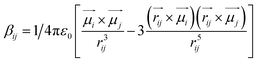 | (1) |
We wished to validate our hypotheses of coupling of the symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of PC and PCP with amide 1. Hence, we carried out DFT calculations of para-ethylphenol and para-nitrophenol. Surprisingly, we observed the same results as PC for para-ethylphenol (AV5, AV6) and as PCP for para-nitrophenol (AV7, AV8). We calculated the vibrational coupling for meta-chlorophenol and ortho-chlorophenol complexes with DMF. The symmetric and asymmetric C
C modes of PC and PCP with amide 1. Hence, we carried out DFT calculations of para-ethylphenol and para-nitrophenol. Surprisingly, we observed the same results as PC for para-ethylphenol (AV5, AV6) and as PCP for para-nitrophenol (AV7, AV8). We calculated the vibrational coupling for meta-chlorophenol and ortho-chlorophenol complexes with DMF. The symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of meta-chlorophenol were coupled with amide I (AV9, AV10). The symmetric C
C modes of meta-chlorophenol were coupled with amide I (AV9, AV10). The symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of ortho-chlorophenol was coupled with amide I, but the asymmetric C
C mode of ortho-chlorophenol was coupled with amide I, but the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode was not (AV11, AV12). Hence, the C
C mode was not (AV11, AV12). Hence, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C symmetric and asymmetric modes coupled with amide 1 for para-electron-promoting phenolic compounds. Only the C
C symmetric and asymmetric modes coupled with amide 1 for para-electron-promoting phenolic compounds. Only the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C symmetric mode coupled with amide 1 for para- and ortho-electron-withdrawing-substituted phenolic compounds. The symmetric and asymmetric C
C symmetric mode coupled with amide 1 for para- and ortho-electron-withdrawing-substituted phenolic compounds. The symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes of electron-withdrawing meta-substituted phenolic compounds were coupled like para-electron-promoting phenolic compounds.
C modes of electron-withdrawing meta-substituted phenolic compounds were coupled like para-electron-promoting phenolic compounds.
Conclusions
Employment of FTIR spectroscopy and DFT calculations revealed the anomalous vibrational coupling between amide 1 and the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode of phenol derivatives. For PC, IR absorbance was enhanced markedly for symmetric and asymmetric C
C mode of phenol derivatives. For PC, IR absorbance was enhanced markedly for symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes upon gradual addition of DMF. For PCP, the symmetric C
C modes upon gradual addition of DMF. For PCP, the symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode was enhanced significantly, whereas the asymmetric C
C mode was enhanced significantly, whereas the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C mode was not. Even though the frequency gap was less for the asymmetric C
C mode was not. Even though the frequency gap was less for the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C transition compared with that for the C
C transition compared with that for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O transition, and the distance between C
O transition, and the distance between C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O was constant for symmetric and asymmetric C
O was constant for symmetric and asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C modes for PC and PCP, distinctive behaviour was observed for different phenol derivatives in the presence of DMF. Theoretical and experimental observations revealed that, with alteration of phenol substituents, the coupling pattern changed. The area of IR absorbance was enhanced ≈5.97 times for the C
C modes for PC and PCP, distinctive behaviour was observed for different phenol derivatives in the presence of DMF. Theoretical and experimental observations revealed that, with alteration of phenol substituents, the coupling pattern changed. The area of IR absorbance was enhanced ≈5.97 times for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C symmetric mode, whereas it was altered negligibly for the asymmetric mode, in the case of PCP. For PC, the area of IR absorbance for symmetric and asymmetric modes was enhanced significantly (≈1.4 times). Thus, we revealed the nature of vibrational coupling based on the structure of a guest molecule hydrogen-bonded with amide I. Our conclusions could be valuable for depiction of the unusual dynamics of coupled amide-I modes as well as the dependency of vibrational coupling on altered factors.
C symmetric mode, whereas it was altered negligibly for the asymmetric mode, in the case of PCP. For PC, the area of IR absorbance for symmetric and asymmetric modes was enhanced significantly (≈1.4 times). Thus, we revealed the nature of vibrational coupling based on the structure of a guest molecule hydrogen-bonded with amide I. Our conclusions could be valuable for depiction of the unusual dynamics of coupled amide-I modes as well as the dependency of vibrational coupling on altered factors.
Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
AG thanks SNBNCBS, Kolkata, India, for instrumental facilities and financial support from the DST-India. SC thanks DST India for a fellowship.References
- R. E. Dickerson and I. Geis, The Structure and Action of Proteins, Harper and Row, New York, Evanston, London, 1969 Search PubMed.
- D. L. Nelson and M. M. Cox, Lehninger Principles of Biochemistry, W. H. Freeman and Company, New York, 2005 Search PubMed.
- J. M. Berg, J. L. Tymoczko and L. Stryer, Biochemistry, W. H. Freeman and Company, New York, 2007 Search PubMed.
- S. Krimm and J. Bandekar, Adv. Protein Chem., 1986, 38, 181–364 CrossRef CAS.
- A. Barth and C. Zscherp, Q. Rev. Biophys., 2002, 35, 369–430 CrossRef CAS.
- S. Woutersen and P. Hamm, J. Phys.: Condens. Matter, 2002, 14, R1035–R1062 CrossRef CAS.
- P. Hamm, M. Lim and R. M. Hochstrasser, J. Phys. Chem. B, 1998, 102, 6123–6138 CrossRef CAS.
- J. Ma, I. M. Pazos, W. Zhang, R. M. Culik and F. Gai, Annu. Rev. Phys. Chem., 2015, 66, 357–377 CrossRef CAS.
- R. Adhikary, J. Zimmermann and F. E. Romesberg, Chem. Rev., 2017, 117, 1927–1969 CrossRef CAS.
- S. D. Fried and S. G. Boxer, Acc. Chem. Res., 2015, 48, 998–1006 CrossRef CAS PubMed.
- H. Kim and M. Cho, Chem. Rev., 2013, 113, 5817–5847 CrossRef CAS PubMed.
- W. Huan, Z. Yanfei, Z. Fengtao, K. Zhengang, H. Buxing, H. Junfeng, W. Zhenpeng and L. Zhimin, Sci. Adv., 2021, 7(22), 1–9 Search PubMed.
- A. Ghosh, B. Cohn, A. K. Prasad and L. Chuntonov, J. Chem. Phys., 2018, 149(18), 184501 CrossRef.
- S. Chakrabarty, A. Barman and A. Ghosh, J. Phys. Chem. B, 2022, 126, 5490–5496 CrossRef CAS.
- S. Chakrabarty, S. H. Deshmukh, A. Barman, S. Bagchi and A. Ghosh, J. Phys. Chem. B, 2022, 126, 4501–4508 CrossRef CAS PubMed.
- M. K. Gilson and H. X. Zhou, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 21–42 CrossRef CAS.
- R. U. Lemieux, Acc. Chem. Res., 1996, 29, 373–380 CrossRef CAS.
- Y. Levy and J. N. Onuchic, Annu. Rev. Biophys. Biomol. Struct., 2006, 35, 389–415 CrossRef CAS.
- D. L. Mobley, E. Dumont, J. D. Chodera and K. A. Dill, J. Phys. Chem. B, 2007, 111, 2242–2254 CrossRef CAS.
- T. H. Plumridge and R. D. Waigh, J. Pharm. Pharmacol., 2002, 54, 1155–1179 CrossRef CAS.
- N. S. Myshakina, Z. Ahmed and S. A. Asher, J. Phys. Chem. B, 2008, 112(38), 11873–11877 CrossRef CAS.
- J. Ma, I. M. Pazos, W. Zhang, R. M. Culik and F. Gai, Annu. Rev. Phys. Chem., 2015, 66, 357–377 CrossRef CAS.
- R. Adhikary, J. Zimmermann and F. E. Romesberg, Chem. Rev., 2017, 117, 1927–1969 CrossRef CAS PubMed.
- S. D. Fried and S. G. Boxer, Acc. Chem. Res., 2015, 48, 998–1006 CrossRef CAS PubMed.
- H. Kim and M. Cho, Chem. Rev., 2013, 113, 5817–5847 CrossRef CAS.
- A. I. Ahmed and F. Gai, Protein Sci., 2017, 26(2), 375–381 CrossRef.
- S. Chakrabarty, S. Maity, D. Yazhini and A. Ghosh, Langmuir, 2020, 36, 11255–11261 CrossRef CAS.
- A. Ghosh, A. K. Prasad and L. Chuntonov, J. Phys. Chem. Lett., 2019, 10, 2481–2486 CrossRef CAS.
- D. G. Brown and J. Bostrom, J. Med. Chem., 2016, 59, 4443–4458 CrossRef CAS.
- V. R. Pattabiraman and J. W. Bode, Nature, 2011, 480, 471–479 CrossRef CAS.
- A. B. Hughes, Origins and Synthesis of Amino Acids, Amino Acids, Peptides and Proteins in Organic Chemistry, Wiley-VCH, Weinheim, Germany, 2009, vol. 1 Search PubMed.
- A. A. Kaspar and J. M. Reichert, Drug Discovery Today, 2013, 18, 807–817 CrossRef CAS PubMed.
- R. B. Dyer, F. Gai, W. H. Woodruff, R. Gilmanshin and R. H. Callender, Acc. Chem. Res., 1998, 31, 709–716 CrossRef CAS.
- S. Woutersen, R. Pfister, P. Hamm, Y. Mu, D. S. Kosov and G. Stock, J. Chem. Phys., 2002, 117, 6833–6840 CrossRef CAS.
- N. Demirdöven, C. M. Cheatum, H. S. Chung, M. Khalil, J. Knoester and A. Tokmakoff, J. Am. Chem. Soc., 2004, 126, 7981–7990 CrossRef.
- M. M. Waegele, R. M. Culik and F. Gai, J. Phys. Chem. Lett., 2011, 2, 2598–2609 CrossRef CAS.
- A. L. Serrano, M. M. Waegele and F. Gai, Protein Sci., 2012, 21, 157–170 CrossRef CAS PubMed.
- H. Kim and M. Cho, Chem. Rev., 2013, 113(8), 5817–5847 CrossRef CAS PubMed.
- P. Hamm, and R. M. Hochstrasser, Ultrafast Infrared, and Raman Spectroscopy, 2001 Search PubMed.
- A. M. Cunha, E. Salamatova, R. Bloem, S. J. Roeters, S. Woutersen, M. S. Pshenichnikov and T. L. C. Jansen, J. Phys. Chem. Lett., 2017, 8, 2438–2444 CrossRef CAS.
- H. Li, R. Lantz and D. Du, Molecules, 2019, 24, 186 CrossRef.
- W. M. Zhang, V. Chernyak and S. Mukamel, J. Chem. Phys., 1999, 110, 5011–5028 CrossRef CAS.
- C. Scheurer, A. Piryatinski and S. Mukamel, J. Am. Chem. Soc., 2001, 123, 3114–3124 CrossRef CAS PubMed.
- A. Moran and S. Mukamel, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 506–510 CrossRef CAS.
- E. G. Buchanan, W. H. James, S. H. Choi, L. Guo, S. H. Gellman, C. W. Müller and T. S. Zwier, J. Chem. Phys., 2012, 137, 094301 CrossRef.
- M. Lima, R. Chelli, V. V. Volkov and R. Righini, J. Chem. Phys., 2009, 130, 204518 CrossRef.
- S. Krimm and Y. Abe, Proc. Natl. Acad. Sci. U. S. A., 1972, 69, 2788–2792 CrossRef CAS PubMed.
- S. Roy, T. L. C. Jansen and J. Knoester, Phys. Chem. Chem. Phys., 2010, 12, 9347–9357 RSC.
- C. Liang, M. Louhivuori, S. J. Marrink, T. L. C. Jansen and J. Knoester, J. Phys. Chem. Lett., 2013, 4, 448–452 CrossRef CAS PubMed.
- J. Schaefer, H. G. Ellen, Y. N. Backus and M. Bonn, J. Phys. Chem. Lett., 2016, 7, 4591–4595 CrossRef CAS PubMed.
- I. V. Rubtsov, J. Wang and R. M. Hochstrasser, J. Phys. Chem. A, 2003, 107, 3384–3396 CrossRef CAS.
- J. V. Hatton and R. E. Richards, Mol. Phys., 1960, 3, 253–263 CrossRef CAS.
- M. Malathi, R. Sabesan and S. Krishnan, Curr. Sci., 2002, 86, 838–842 Search PubMed.
- A. Ghosh, J. Phys. Chem. B, 2019, 123, 7771–7776 CrossRef CAS.
- P. Hamm and M. Zanni, Concepts and Methods of 2D Infrared Spectroscopy, Cambridge University Press, New York, 2011 Search PubMed.
Footnote |
† Electronic supplementary information (ESI) available: We have provided videos showing coupling of the: symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of PC and DMF (AV1); asymmetric C O modes of PC and DMF (AV1); asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of PC and DMF (AV2); symmetric C O modes of PC and DMF (AV2); symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of PCP and DMF (AV3); asymmetric C O modes of PCP and DMF (AV3); asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of PCP and DMF (AV4); symmetric C O modes of PCP and DMF (AV4); symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of para-ethylphenol and DMF (AV5); asymmetric C O modes of para-ethylphenol and DMF (AV5); asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of para-ethylphenol and DMF (AV6); symmetric C O modes of para-ethylphenol and DMF (AV6); symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of para-nitrophenol and DMF (AV7); asymmetric C O modes of para-nitrophenol and DMF (AV7); asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of para-nitrophenol and DMF (AV8); symmetric C O modes of para-nitrophenol and DMF (AV8); symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of meta-chlorophenol and DMF (AV9); asymmetric C O modes of meta-chlorophenol and DMF (AV9); asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of meta-chlorophenol and DMF (AV10); symmetric C O modes of meta-chlorophenol and DMF (AV10); symmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of ortho-chlorophenol and DMF (AV11); asymmetric C O modes of ortho-chlorophenol and DMF (AV11); asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O modes of ortho-chlorophenol and DMF (AV12). See DOI: https://doi.org/10.1039/d2ra07177k O modes of ortho-chlorophenol and DMF (AV12). See DOI: https://doi.org/10.1039/d2ra07177k |
| This journal is © The Royal Society of Chemistry 2023 |