Open Access Article
Open Access ArticleConvergent evolution of animal and microbial rhodopsins
Keiichi Kojima * and
Yuki Sudo
* and
Yuki Sudo *
*
Faculty of Medicine, Dentistry and Pharmaceutical Sciences, Okayama University, Japan. E-mail: keiichikojima@okayama-u.ac.jp; sudo@okayama-u.ac.jp
First published on 13th February 2023
Abstract
Rhodopsins, a family of photoreceptive membrane proteins, contain retinal as a chromophore and were firstly identified as reddish pigments from frog retina in 1876. Since then, rhodopsin-like proteins have been identified mainly from animal eyes. In 1971, a rhodopsin-like pigment was discovered from the archaeon Halobacterium salinarum and named bacteriorhodopsin. While it was believed that rhodopsin- and bacteriorhodopsin-like proteins were expressed only in animal eyes and archaea, respectively, before the 1990s, a variety of rhodopsin-like proteins (called animal rhodopsins or opsins) and bacteriorhodopsin-like proteins (called microbial rhodopsins) have been progressively identified from various tissues of animals and microorganisms, respectively. Here, we comprehensively introduce the research conducted on animal and microbial rhodopsins. Recent analysis has revealed that the two rhodopsin families have common molecular properties, such as the protein structure (i.e., 7-transmembrane structure), retinal structure (i.e., binding ability to cis- and trans-retinal), color sensitivity (i.e., UV- and visible-light sensitivities), and photoreaction (i.e., triggering structural changes by light and heat), more than what was expected at the early stages of rhodopsin research. Contrastingly, their molecular functions are distinctively different (e.g., G protein-coupled receptors and photoisomerases for animal rhodopsins and ion transporters and phototaxis sensors for microbial rhodopsins). Therefore, based on their similarities and dissimilarities, we propose that animal and microbial rhodopsins have convergently evolved from their distinctive origins as multi-colored retinal-binding membrane proteins whose activities are regulated by light and heat but independently evolved for different molecular and physiological functions in the cognate organism.
1. Introduction: rhodopsins as a family of photoreceptive membrane proteins
Rhodopsins are a family of photoreceptive membrane proteins, consisting of 7-transmembrane α-helices and a derivative of vitamin-A, retinal, as a chromophore (Fig. 1).1–3 Retinal is covalently bound to a lysine (Lys) residue located in the seventh helix (called helix VII or helix G) through a Schiff base linkage. Historically, a rhodopsin molecule was firstly identified as a visual reddish pigment isolated from the animal eye (Fig. 1A).4,5 In 1876, Dr Boll recognized that the reddish pigment in frog retina is photosensitive and bleaches to a yellow color when exposed to light.6 Subsequently, Dr Kühne determined that the reddish pigment is the protein expressed in the outer segment of rod photoreceptor cells and named the pigment “visual purple (rhodopsin).” The word rhodopsin originates from the Greek words “rhodo (= rose)” and “opsis (= sight)” based on the bright reddish color of the pigment. Nowadays, a wide variety of rhodopsin-like pigments have been discovered from not only animal's eyes but also from other organs such as the brain, lung and skin.2,7,8 The rhodopsin and rhodopsin-like pigments from metazoans are called animal rhodopsins (or animal opsins).2,7,8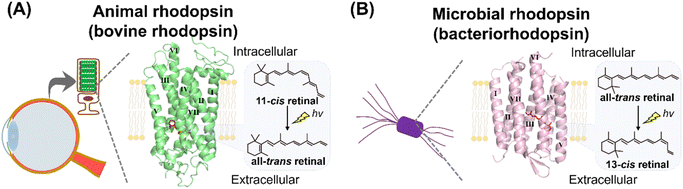 | ||
| Fig. 1 Introduction to animal and microbial rhodopsins. (A) Vertebrate visual rhodopsin, a typical animal rhodopsin (animal opsin), is expressed in the photoreceptor cells of vertebrate retina. The crystal structure of bovine rhodopsin (PDB: 1U19) is shown here. 11-cis retinal is covalently bound to the apoprotein in the dark state as a chromophore and light triggers its isomerization to the all-trans retinal. (B) Bacteriorhodopsin, a typical microbial rhodopsin, is expressed in the cellular membrane of the archaeon H. salinarum. The crystal structure of bacteriorhodopsin (PDB: 1C3W) is shown here. All-trans retinal is covalently bound to the apoprotein in the dark state as a chromophore and light triggers its isomerization to the 13-cis retinal. Transmembrane helix numbers are indicated in the crystal structures. | ||
In 1971, Drs Oesterhelt and Stoeckenius discovered a rhodopsin-like pigment from the purple cellular membrane of the archaeon Halobacterium salinarum (formerly halobium) (Fig. 1B).9 The purple-colored pigment, named bacteriorhodopsin (BR), serves as an outward proton pump. Recently, more than millions of bacteriorhodopsin-like proteins have been identified from not only archaea but also bacteria, eukaryotic microorganisms, and viruses.1,10,11 Bacteriorhodopsin and bacteriorhodopsin-like proteins are called “microbial rhodopsins.” Sequential and phylogenetical analyses revealed that animal and microbial rhodopsins are nonhomologous and taxonomically distinct.12–14 Animal rhodopsins were proposed to evolve from non-retinylidene cAMP receptors in G protein-coupled receptors (GPCRs) having 7-transmembrane structure.15,16 On the other hand, it was proposed that microbial rhodopsins were distantly related to lysosomal cysteine transporters having 7-transmembrane structure and they evolved from a common ancestor.14,17 Thus, it is strongly suggested that animal and microbial rhodopsins emerged from their distinctive origins via convergent evolution despite their structural similarities,12–14 while a few papers proposed that the two rhodopsins emerged from the same origin.18 Before 2000, a few classical rhodopsin molecules (e.g., bovine and squid rhodopsins as animal rhodopsins and bacteriorhodopsin as microbial rhodopsins) have been analyzed in detail and their research indicated that the two rhodopsin families show many distinctive properties. In contrast, recent advances in genomic research revealed a wide variety of animal and microbial rhodopsins in nature and their analysis have shown many similarities in various molecular aspects.1,19 Here, we comprehensively introduce and compare the research of animal and microbial rhodopsins from biological, chemical, and physical perspectives. Furthermore, based on the achievements of the studies and to discuss the nature of rhodopsin, we have summarized the similarities and dissimilarities between animal and microbial rhodopsins.
2. Animal rhodopsins
2.1 Basis of animal rhodopsins
Animal rhodopsins are categorized into a family of G protein-coupled receptors (GPCRs) that act as master regulators of signal transduction cascades in eukaryotes.2,7,8 At the early stages of rhodopsin research, animal rhodopsins (e.g., vertebrate and invertebrate visual rhodopsins and photoisomerase retinochrome) were identified only from eyes; therefore, the rhodopsins were thought to be responsible for mainly visual functions. Subsequently, pinopsin was found in the chicken pineal organ in 1994,20 and a wide variety of animal rhodopsins have been identified from animal photoreceptive (e.g., eye and pineal organ) and non-photoreceptive organs (e.g., lung, liver, skin and skeletal muscle) based on the development of sequencing technology and genomic research.2,7,8 Presently, animal rhodopsins are classified into several subfamilies based on their amino acid sequences (Fig. 2A).2,7 Most of the animal rhodopsins except for two subfamilies, photoisomerases and peropsins, activate cognate heterotrimeric G proteins (e.g., Gs, Gq, Gi, Go, and Gt) to trigger the signal transduction cascades.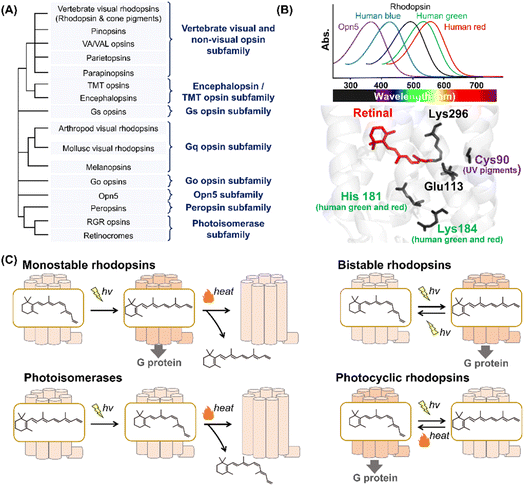 | ||
| Fig. 2 Phylogenetic relationship and the reaction of animal rhodopsins. (A) The phylogenetic tree of animal rhodopsins. Animal rhodopsins are roughly divided into 8 subfamilies.7 (B) Spectral sensitivities of animal rhodopsins, such as vertebrate rhodopsin, human blue-, green-, and red-sensitive cone pigments and Opn5m (upper panel). The crystal structure of bovine rhodopsin around the retinal (lower panel) (PDB: 1U19). The key residues for spectral tuning (His181 and Lys184 in human green- and red-sensitive cone pigments and Cys90 in vertebrate UV-sensitive cone pigments), retinal and Lys296, are highlighted in the structure. (C) The schematics of photoreaction of animal rhodopsins. In monostable rhodopsins, light irradiation induces isomerization from 11-cis to all-trans, thereby forming the active state. The active state bleaches followed by the release of retinal. In the bistable rhodopsins, light irradiation triggers interconversion between the inactive and active state containing 11-cis and all-trans retinal, respectively. Photoisomerases bind to the all-trans retinal exclusively in the dark state. Light irradiation of the dark state induces isomerization from all-trans to 11-cis. The product 11-cis retinal is delivered to regenerate other GPCR-type rhodopsins. The photocyclic rhodopsin Opn5L1 binds to all-trans retinal exclusively in the dark state. Light irradiation induces isomerization from all-trans to 11-cis to form the inactive state that is then thermally reconverted into the active state. | ||
Animal rhodopsins commonly consist of 7-transmembrane helices and an eighth helix (helix VIII) at the intracellular side and parallel to the membrane.2 The chromophore, retinal, is covalently bound to the apoprotein called opsin through a Lys residue (Lys296 in bovine rhodopsin) at the seventh helix (Fig. 1A). While 11-cis retinal is a natural chromophore in the dark inactive state of GPCR-type rhodopsins under physiological conditions, several rhodopsins can bind to 9-cis retinal to form the blue-shifted dark state (called isorhodopsin).21 In addition, mosquito opsin 3 (Opn3) has been reported to bind to 13-cis retinal in the inactive dark state by exogenous addition as a unique feature.22 It is proposed that an ancestral rhodopsin emerged as an all-trans retinal receptor, and then increased the affinity for inverse agonist cis retinals (mainly 11-cis retinal) to act as a photoreceptor during the evolutionary process in GPCRs.2,23 From a chemical point of view, 11-cis retinal is suitable to allow rhodopsins to exhibit various spectral sensitivities from UV- to visible-light region since the rotation around the C6–C7 and C12–C13 single bonds enable the retinal to adopt an infinite number of conformations.24 We speculate that animal rhodopsins mainly selected 11-cis retinal as a natural chromophore to achieve various spectral sensitivities together with the development of the enzymes to supply 11-cis retinal (e.g., RPE65). Many animal rhodopsins absorb visible light in the dark state, while retinal originally absorbs UV light. In visible light-sensitive rhodopsins, the protonation of the Schiff base causes delocalization of π electrons on the polyene chain of retinal, which results in a spectral red-shift in retinal absorbance (called opsin shift).2 The protonated Schiff base is stabilized by the negatively charged counterion (Glu or Asp), such as Glu113, in bovine rhodopsin (Fig. 2B).25–29 In contrast to visible light-sensitive rhodopsins, several types of rhodopsins (e.g., UV-sensitive cone pigments, opsin 5 (Opn5) and parapinopsins) absorb UV light (approx. 350–380 nm) because they have a deprotonated Schiff base in the inactive dark state.30 As humans can see visible light by trichromatic vision (red, green, and blue) using red-, green-, and blue-sensitive cone pigments expressed in the retina, rhodopsins exhibit their characteristic spectral sensitivities (approx. 350–570 nm) (Fig. 2B). Here absorption maximum (λmax) is determined by the energy gap between the ground and excited states of retinal in the protein moiety. The energy gap is attributed to several factors, such as the charge distribution of π electrons and planarity of the polyene chain of the retinal, which is mainly regulated by amino acid residues around the retinal.31,32 For example, human red- and green-sensitive cone pigments have Cl− binding sites (e.g., His181 and Lys184) around the retinal (Fig. 2B).33 The negative charge of Cl− affects the delocalization of π electrons of the retinal, leading to a spectral red-shift. Additionally, Cys90 is responsible for the deprotonation of the Schiff base and ability of UV-absorbance in vertebrate short wavelength-sensitive cone pigments.32 While various types of rhodopsins absorbing UV and visible light have been identified, no report on the existence of infra-red light-sensitive animal rhodopsins exist.
In GPCR-type rhodopsins, light-absorption triggers photoisomerization from 11-cis to all-trans retinal, leading to movements in the helix to form the active state.2,34 In vertebrate visual rhodopsins, the active state containing all-trans retinal is thermally unstable and results in bleaching followed by the release of retinal (Fig. 2C). Illumination does not cause the active state to efficiently convert into the inactive dark state, and thus vertebrate visual rhodopsins are characterized as “monostable rhodopsins.”2,35 Contrastingly, invertebrate visual rhodopsins (e.g., arthropod and mollusc visual rhodopsins) exhibit the thermally stable active state that is convertible into the inactive dark state by illumination (Fig. 2C). This property is observed in other animal rhodopsins (e.g., melanopsins, Go opsins, and parapinopsins) and they are characterized as “bistable rhodopsins.”2,35 Besides, retinal G protein-coupled receptor (RGR) opsins and retinochromes contain all-trans retinal in the dark, and light-absorption triggers photoisomerization from all-trans to 11-cis retinal (Fig. 2C).2,36 Subsequently, the retinal binding proteins shuttle and deliver 11-cis retinal to other GPCR-type rhodopsins.37 Therefore, RGR opsins and retinochromes are characterized as retinal-photoisomerases. Although peropsins show similar photochemical properties to RGR opsins and retinochromes, little experimental evidence exists that explains peropsins to serve as retinal-photoisomerase.38 Thus, the function of peropsins is still controversial. Recently, Opn5L1, a subgroup of opsin 5 (Opn5), was described as a photocyclic rhodopsin, whereas other subgroups of Opn5 (i.e., Opn5m and Opn5L2) were described as having a UV-sensitive bistable property (Fig. 2C).39–41 Opn5L1 binds with all-trans retinal, forming the G protein activating state in the dark. The irradiation induces retinal isomerization from all-trans to 11-cis, forming the G protein inactivation state, and subsequently, a covalent adduct between the retinal and Cys188 residue of Opn5L1 is formed to induce the conversion of the C11![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C12 double bond to a single bond in the retinal. Next, the thermal rotation of the C11–C12 single bond induces the dissociation of the adduct to regenerate the G protein activating dark state. More recently, the bovine rhodopsin, a typical monostable rhodopsin, has been reported to have acquired photocyclic property upon the introduction of Cys at position 188.42 Summarizing the above information, the structural changes between the inactive and active states containing cis- and trans-form of retinal, respectively, is triggered by light and heat in animal rhodopsins.
C12 double bond to a single bond in the retinal. Next, the thermal rotation of the C11–C12 single bond induces the dissociation of the adduct to regenerate the G protein activating dark state. More recently, the bovine rhodopsin, a typical monostable rhodopsin, has been reported to have acquired photocyclic property upon the introduction of Cys at position 188.42 Summarizing the above information, the structural changes between the inactive and active states containing cis- and trans-form of retinal, respectively, is triggered by light and heat in animal rhodopsins.
2.2 Photoactivation of animal rhodopsins
Among various animal rhodopsins, vertebrate rhodopsin (called rhodopsin or rod visual pigment) expressed in rod photoreceptor cells is well studied.2,5 In 2000, the crystal structure of bovine rhodopsin was reported as the first molecule of GPCRs.43 Vertebrate rhodopsin contains 11-cis retinal in the dark, and light absorption results in cis–trans isomerization within 200 fs with a high quantum yield (e.g., 0.65 for bovine rhodopsin).44,45 The efficient and ultrafast conversion of photon energy into chemical energy is attributed to a conical intersection between the potential energy surfaces of the ground and excited electronic states.46–48 After photoisomerization, rhodopsin forms spectroscopically distinctive photointermediates, such as the Photo, Batho, Lumi, MetaI, and MetaII intermediates (Fig. 3A).34,49 MetaII has the deprotonated Schiff base corresponding to the G protein activating state.50,51 In the photoreceptor cells, the photointermediates finally release retinal and bleach into the retinal-unbound opsin state to which 11-cis retinal is supplied to form photoactive pigments. The dark state (i.e., the unphotolyzed state) contains 11-cis retinal that acts as an inverse agonist, and a salt bridge between the protonated Schiff base and a negatively charged counterion (Glu113 in bovine rhodopsin) is present to suppress the constitutive activity before light absorption (Fig. 3B).2,5,34 Additionally, a cytoplasmic hydrogen bonding network exists between the E(D)RY motif (i.e., Glu134, Arg135, and Tyr136 in bovine rhodopsin) on helix III and Glu247 and Glu249 on helix VI in the dark state (Fig. 3B).52 The cytoplasmic salt bridge, which is termed “ionic-lock,” plays important roles in the suppression of constitutive activity of the dark state. The Schiff base is deprotonated by proton transfer reaction to the counterion (Glu113) during the conversion process from MetaI to MetaII, resulting in the disruption of the salt bridge between the protonated Schiff base and the counterion.53 Subsequently, Glu134 in the E(D)RY motif is protonated, resulting in the cytoplasmic ionic-lock disruption.52,54 The accumulated rearrangements of the intrahelical hydrogen bonding network lead to dynamic helix movement, especially the outward movement of the cytoplasmic side of helix VI in MetaII (Fig. 3B).55,56 The open structure of the cytoplasmic side is capable of G protein activation. As described above, many studies on vertebrate rhodopsins (especially bovine rhodopsin) have been conducted to elucidate the structural changes during the photoactivation steps.5 However, the complete imagination of the sequential structural changes remains unsolved, especially the description of how the local structural perturbation around retinal followed by its photoisomerization induces the overall structural changes through intramolecular dynamics. Recently, time-resolved crystallographic techniques using X-ray free-electron lasers (XFEL) have progressively advanced57 and were applied to bovine rhodopsin. Indeed, the techniques enable us to uncover the sequential process of structural changes after photoisomerization.58 Thus, this will provide a three-dimensional overview of the activation process in vertebrate rhodopsins, leading to the implicative information of numerous GPCRs.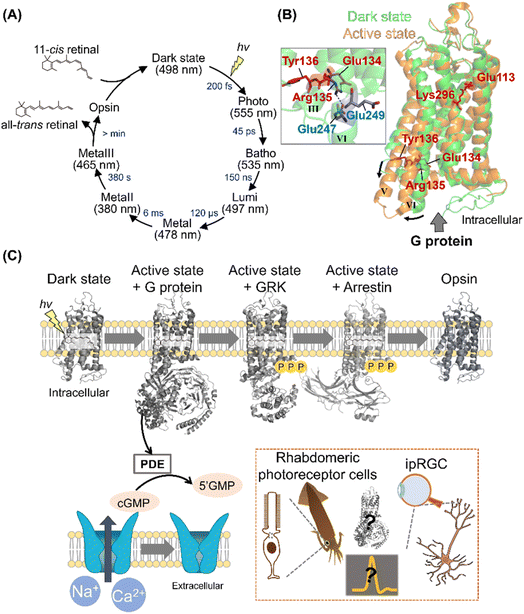 | ||
| Fig. 3 Activation and signal transduction cascades of animal rhodopsins. (A) Schematics of photoreaction and regeneration process of vertebrate visual rhodopsins. The absorption maxima of the intermediates and time constants of the transition are shown here.46 (B) Structural superposition of inactive (PDB: 1U19, denoted in green) and active state of bovine rhodopsin (PDB: 3PXO, denoted in orange). The key residues for the activation process, such as Glu113, Glu134, Arg135, Tyr136, and Lys296 are highlighted in the structure. (Inset) A close-up of E(D)RY motif of inactive state of bovine rhodopsin. The hydrogen-bonds are shown as black dashed lines. (C) The schematics of signal transduction cascades of vertebrate photoreceptor cells. Visual rhodopsins absorb a photon to form the active state, which binds to and activates Gt. The activated Gt binds to and activates PDE, and the PDE then hydrolyses cGMP. The decrease in the concentration of cGMP induces the closure of CNG channels, leading to the hyperpolarization responses of photoreceptor cells. After activating the signaling cascades, vertebrate rhodopsins are rapidly phosphorylated by GRK and then inactivated by arrestin. (Inset) Schematics of the photoresponses in invertebrate rhabdomeric photoreceptor cells and ipRGCs. The structural changes of opsins and overall signal transduction cascades in various visual and non-visual photoreceptive cells will be analysed using the time-resolved techniques. | ||
2.3 Signal transduction by animal rhodopsins
Upon illumination, GPCR-rhodopsins activate G proteins to trigger signal transduction cascades in cells for visual and non-visual physiological responses. Vertebrate retina has two types of photoreceptor cells that are morphologically different, rods and cones for scotopic and photopic vision, respectively.59,60 Rods and cones contain different types of vertebrate visual rhodopsins, rhodopsin (also called rod visual pigment) and cone visual pigments, respectively. In general, vertebrates have several types of cone pigments with different spectral sensitivities (e.g., red-, green-, and blue-sensitive cone pigments for humans) for color discrimination in photopic vision.32 Vertebrate visual rhodopsins activate Gt (transducin), leading to the hydrolysis of cGMP through phosphodiesterase (PDE) activation in the photoreceptor cells. The decrease in cGMP concentration closes cyclic nucleotide-gated (CNG) channels to induce hyperpolarization of the photoreceptor cells as visual signals (Fig. 3C).59–61 After activating the signaling cascades, vertebrate rhodopsins are rapidly inactivated in <100 ms by the cytoplasmic adaptor protein arrestin followed by the phosphorylation of Ser and Thr residues on their C-terminus with G protein receptor kinases (GRKs).60 While the signal transduction cascades in the photoreceptor cells have been analyzed mainly using electrophysiological and biochemical techniques,59–61 the complex structures of vertebrate rhodopsins with G protein, GRK, and arrestin have recently been explored using femtosecond X-ray laser crystallography and cryo-electron microscopy.62–64 These complex structures enable us to visually imagine the snapshots in signal transduction cascades (Fig. 3C). Furthermore, to directly detect the molecular components of the cascade in real-time and within native rod photoreceptor environment, mass spectroscopy has been established.65In contrast to the vertebrate visual signal transduction cascade, arthropod and mollusc visual rhodopsins activate Gq by GDP–GTP exchange reaction in rhabdomeric photoreceptor cells.61 The α subunit of the activated Gq binds to and activates phospholipase C (PLC) that hydrolyzes phosphatidylinositol 4,5-bisphosphatase (PIP2) to 1,4,5-triphosphate (InsP3) and diacylglycerol (DAG). After the activation of PLC-mediated cascades, the transient receptor potential (TRP) channels are finally opened to induce depolarization as visual signals.61 The components and reaction schemes of invertebrate visual signal transduction cascade have been revealed; however, the activation factor of TRP channels remains controversial.61,66
As non-visual phototransduction cascades, melanopsin induces a parallel cascade to the invertebrate visual signal transduction cascade in the intrinsically photosensitive retinal ganglion cells (ipRGCs) of the retina for inducing pupillary light responses and photoentraining circadian rhythm in mammals.61,67 Further analysis using the time-resolved techniques will enable us to capture the sequential overview of the overall signal transduction cascades in various visual and non-visual photoreceptive cells (Fig. 3C, Inset).
2.4 The evolution of animal rhodopsins
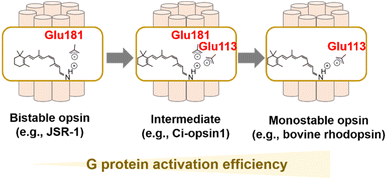
As described, visible light-sensitive rhodopsins have a protonated Schiff base that is stabilized by the negatively charged counterion.25–29 In bovine rhodopsin, the counterion is Glu113 located in helix III.25–27 Glu113 is thought to act as a counterion in vertebrate visual rhodopsins as they are conserved in them. It also accepts proton from the protonated Schiff base during the process of MetaII formation,53 implying the dual important roles of the counterion in visible light-absorption and G protein-activation.28 Most of the other animal rhodopsins except vertebrate visual rhodopsins (e.g., invertebrate visual rhodopsins, Go opsins, photoisomerase opsins, and peropsins) do not have Glu residue at position 113. Drs Terakita and Shichida demonstrated that Glu181 acts as a counterion in Go opsin, photoisomerase opsins, and peropsins by the comprehensive mutational analysis, which suggests that Glu181 has been an ancestral counterion in vertebrate visual rhodopsins.28,29 Furthermore, G protein activation efficiency of Go opsin was approx. 50-fold lower than those of vertebrate visual rhodopsins. The differences in G protein activation efficiencies (i.e., high versus low) and in the photoreaction property (i.e., monostable versus bistable) are consistent with the differences in the counterion position (i.e., Glu113 versus Glu181). Based on the various experimental observation and phylogenetical relationships, vertebrate visual rhodopsins have been proposed to have acquired Glu113 counterion by its replacement from the ancestral position at 181 to increase G protein activation efficiency and to obtain monostable nature during evolutionary process (Fig. 4A).28,68 This counterion displacement is also supported by a previous report that describes an ascidian opsin (Ci-opsin1) showing synergistic counterions of Glu113 and Glu181 and an intermediate property of G protein activation efficiency and photoreaction between monostable and bistable rhodopsins (Fig. 4A).69 Since the efficient G protein activation of vertebrate visual rhodopsins results in the high signal-to-noise ratio in the visual signal transduction cascades, the counterion displacement from Glu181 to Glu113 is thought to be the molecular basis for developing highly functional vertebrate eyes.68 Although the evolutionary scenario is established as described above, understanding the molecular determinant of the differences in G protein activation efficiency and photoreaction in monostable and bistable rhodopsins from a structural aspect remains challenging. The crystal structure of the dark state of the jumping spider rhodopsin-1 (JSR-1) as a bistable rhodopsin containing Glu181 counterion has been resolved, implying a structural difference around the Schiff base and counterion with the bovine rhodopsin (a monostable rhodopsin containing Glu113).70,71 Further structural analysis using time-resolved techniques (e.g., crystallographic techniques, NMR, and spin label electron paramagnetic resonance (EPR)) would be required to investigate the activation process in bistable and monostable rhodopsins.In addition to the counterion displacement process, several evolutionary processes of animal rhodopsins, such as the process of color tuning and adaptation of thermal isomerization rates, have been progressively studied.32,72–75 We expect that further studies on the evolution of animal rhodopsins would reveal their divergence and adaptation and their roles in physiology.
3. Microbial rhodopsins
3.1 Basis of microbial rhodopsins
Historically, research on microbial rhodopsins began with the discovery of BR from H. salinarum in 1971.9 BR works as an outward proton pump that produces a proton concentration gradient between the cellular membranes in the native organism. After the discovery of BR, three microbial rhodopsins were identified from the same archaeon H. salinarum and were named halorhodopsin (HR), sensory rhodopsin-I (SRI), and sensory rhodopsin-II (SRII, also called phoborhodopsin (pR)).76–78 While HR serves as an inward chloride pump, SRI and SRII work as negative/positive and negative phototaxis sensors, respectively. Until 1999, no microbial rhodopsin genes were identified in bacteria and eukaryotic microorganisms, whereas many microbial rhodopsin genes were identified in archaea. Therefore, it was formerly believed that microbial rhodopsins are possessed only by archaea. In 1999, microbial rhodopsin genes were found in the eukaryotic fungus Neurospora crassa.79,80 Since then, more than millions of microbial rhodopsins have been identified from not only archaea but also bacteria, eukaryotic microorganisms, and viruses with a variety of distinct molecular functions,1,10,11 such as outward proton pumps,81–84 inward proton pumps,85–87 cation channels,88–90 anion channels,91,92 inward chloride pumps,93,94 outward sodium pumps,95,96 a sulfate transporter,97 transcriptional regulators,98,99 and enzymes (e.g., histidine kinase and adenylyl/guanylyl cyclases)100,101 (Fig. 5A). The molecular functions of microbial rhodopsins are highly diversified rather than those of animal rhodopsins. In 2018, a rhodopsin family named heliorhodopsin, which shapes a genetically distant group from the classical microbial rhodopsins, was identified by metagenomics.102 The unique feature of heliorhodopsin is that the N-terminus is located on the intracellular side. In other words, the orientation of heliorhodopsin is opposite to that of the animal and classical microbial rhodopsins. The molecular diversity of microbial rhodopsins is still expanding. Other novel rhodopsins beyond expectation are expected to be identified by further analysis.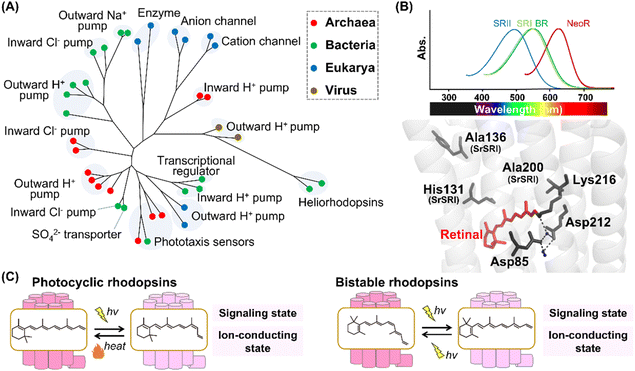 | ||
| Fig. 5 Phylogenetic relationship and reaction of microbial rhodopsins. (A) The phylogenetic tree of microbial rhodopsins. Rhodopsins from archaea, bacteria, eukarya, and viruses are indicated by filled circles colored red, green, blue, and brown, respectively. (B) Spectral sensitivities of microbial rhodopsins, such as BR, SRI, SRII and NeoR (upper panel). Crystal structure of BR around the retinal (lower panel) (PDB: 1C3W). The key residues for spectral tuning (His131, Ala136, and Ala200 in SRI from S. ruber, SrSRI), retinal, Lys216, and the pentagonal cluster (water molecules (blue dots), Asp85 and Asp212), are highlighted in the structure. The hydrogen-bonds of the pentagonal cluster are shown as black dashed lines. (C) The schematics of photoreaction of microbial rhodopsins. In the photocyclic rhodopsins, light irradiation induces isomerization from all-trans to 13-cis to form the signaling and ion-conducting states. The 13-cis retinal-binding form thermally returns to the initial all-trans retinal-binding form. Bistable rhodopsins show the thermally stable 13-cis retinal-binding form that is convertible to the initial all-trans retinal-binding form by illumination. | ||
The microbial and animal rhodopsins are similar such that they share the similar 7-transmembrane α-helical structure and contain a retinal molecule as a chromophore (Fig. 1C).1,10 The enzyme rhodopsins exceptionally consist of 8-transmembrane helices in which the extra helix (named helix 0) and the C-terminal enzyme domain are present at the N-terminus of the transmembrane domain and at the intracellular side, respectively. Furthermore, rhodopsin–bestrophin fusion proteins have been identified in which one rhodopsin domain of 8-transmembrane helices or two rhodopsin domains in tandem are C-terminally fused to a calcium-activated chloride channel (bestrophin).103 Therefore, the molecular structures of apoproteins are highly diversified in microbial rhodopsins compared with animal rhodopsins. In general, microbial rhodopsins contain all-trans retinal through the protonated Schiff base linkage with a Lys residue (Lys216 in BR) located at helix G in the dark state to absorb visible light (approx. 420–600 nm) (Fig. 5B).1,10,11 The binding ability to all-trans retinal is reasonable since the isomer is the most thermally stable.104,105 The protonated Schiff base is generally stabilized by the negatively charged counterion. In BR, two counterion residues (Asp85 and Asp212) and water molecules form the hydrogen-bonding network (called pentagonal cluster) with the protonated Schiff base.1 Similar to animal rhodopsins, the λmax of microbial rhodopsins is determined by the energy gap between the ground and excited states of the retinal in the protein moiety. Like human red- and green-sensitive cone pigments, SRI (λmax = ∼560 nm) has a Cl− binding site composed of a conserved His residue (i.e., His135, His135, and His131 in SRI from H. salinarum, SRI from Haloarcula vallismortis, and SRI from Salinibacter ruber, respectively) and Ala residues (Ala136 and Ala200 in SRI from S. ruber) around the retinal (Fig. 5B).106,107 The negative charge of Cl− induces the delocalization of π electrons of the retinal, leading to the spectral red-shift. In 2020, a microbial rhodopsin from the fungus Rhizoclosmatium globosum (NeoR) was shown to exhibit near-infrared light-sensitivities (λmax = ∼690 nm) (Fig. 5B).108 While there is no report of the existence of UV-sensitive microbial rhodopsins in the dark state containing all-trans retinal, several molecules show a UV-sensitive thermally stable form with 13-cis retinal.101,108
In microbial rhodopsins, light absorption generally triggers the isomerization from all-trans to 13-cis retinal and the photoisomerization induces a sequence of structural changes, resulting in light-dependent molecular functions. Many microbial rhodopsins exhibit the cyclic reaction (called photocycle) in which the 13-cis retinal-binding form thermally returns to the initial all-trans retinal-binding form (Fig. 5C).1 Several microbial rhodopsins (e.g., histidine-kinase rhodopsins and enzyme rhodopsins) exist as bistable rhodopsins, where the thermally stable 13-cis retinal-binding form is convertible into the initial all-trans retinal-binding form by illumination (Fig. 5C).101,108 Thus, similar to animal rhodopsins, the structural changes between the states containing cis- and trans-form of retinal is triggered by light and heat in microbial rhodopsins. From the viewpoint of retinal composition, the conversion between all-trans and 13-cis is observed in most microbial rhodopsins. 13-cis retinal is more thermally stable as compared with other cis isomers (i.e., 7-cis, 9-cis and 11-cis isomers), which implies the low energy barrier of the isomerization from all-trans to 13-cis retinal.104,105 Therefore, we speculate that microbial rhodopsins mainly use C13![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C14 double bond isomerization to absorb light due to its low energy barrier. In 2011, a microbial rhodopsin named middle rhodopsin (MR), which is phylogenetically located at the intermediate between BR and SRII, was found in the archaeon Haloquadratum walsbyi.109 MR uniquely shows the binding ability to 11-cis retinal like animal rhodopsins. More recently, rhodopsin-bestrophin fusion proteins have been reported to show photoisomerization property from all-trans to 11-cis retinal.103 Thus, similar to animal rhodopsins, microbial rhodopsins can bind to various retinal isomers (all-trans, 11-cis, and 13-cis retinal).
C14 double bond isomerization to absorb light due to its low energy barrier. In 2011, a microbial rhodopsin named middle rhodopsin (MR), which is phylogenetically located at the intermediate between BR and SRII, was found in the archaeon Haloquadratum walsbyi.109 MR uniquely shows the binding ability to 11-cis retinal like animal rhodopsins. More recently, rhodopsin-bestrophin fusion proteins have been reported to show photoisomerization property from all-trans to 11-cis retinal.103 Thus, similar to animal rhodopsins, microbial rhodopsins can bind to various retinal isomers (all-trans, 11-cis, and 13-cis retinal).
3.2 Photoactivation of microbial rhodopsins
Among the various microbial rhodopsins, BR has been extensively studied so far. The light absorption of BR results in the isomerization from all-trans to 13-cis retinal within 500 fs and the quantum yield is estimated to be 0.64.110–115 The ultrafast retinal isomerization occurs through the conical intersection in BR as well as bovine rhodopsin.113,114 After photoisomerization, BR forms a variety of photointermediates, such as the J-, K-, L-, M-, N-, and O-intermediates, and subsequently returns to the initial dark state within 10 ms (Fig. 6A).1 During the photocycle, the conformational changes of the protein moiety and continuous pKa changes of charged amino acids, including the protonated Schiff base and its counterion, occur to induce the molecular function (Fig. 6B). The combination of various methods (e.g., UV-visible spectroscopy, vibrational spectroscopy, and X-ray crystallography) elucidates the proton pump mechanism of BR as follows (Fig. 6B).1,116,117 (i) The light absorption triggers the isomerization of the retinal from all-trans to 13-cis form, leading to the K-intermediate formation. (ii) During the transition from the K- to L-intermediate, a proton of the Schiff base is transferred to the primary counterion Asp85, leading to M-intermediate formation. Simultaneously, a proton is released from the extracellular proton releasing group, consisting of Glu194 and Glu204, to the extracellular side. (iii) A proton of Asp96 is transferred to the Schiff base, leading to the Schiff base re-protonation and N-intermediate formation. (iv) A proton is taken up from the intracellular bulk solution to Asp96, leading to Asp96 re-protonation during the transition from N- to O-intermediate. Simultaneously, the retinal is re-isomerized to the all-trans form. (v) Finally, during the transition of the O-intermediate to the initial state, a proton is transferred from the counterion Asp85 to Glu194 of the proton releasing group. Thus, during the single photocycle step, BR vectorially transfers a proton from the intracellular to the extracellular side. As described above, animal rhodopsins show the light-induced movement of helix VI to form the cytoplasmic interaction site with G protein.55,56 A similar helix opening movement (helix F movement) has been observed previously during the N-intermediate formation process in BR.118 The helix movement is speculated to increase the cavity to facilitate the proton transfer reaction during the decay process of M-intermediate. Similar helix F movement is also observed in SRII from Natronomonas pharaonis (NpSRII) that plays important roles in signal transduction to the transducer protein HtrII.119 Thus, animal and microbial rhodopsins commonly show similar helical movements accompanied with the protonation and deprotonation of the charged residues after light absorption.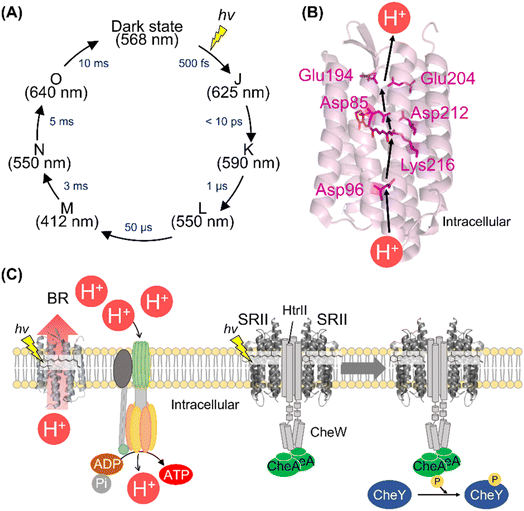 | ||
| Fig. 6 The activation and signal transduction cascades of microbial rhodopsins. (A) Schematics of photoreaction process of bacteriorhodopsin (BR). The absorption maxima of the intermediates and time constants of the transition are shown here.1 (B) Proton transport pathway of BR (PDB: 1C3W). The key residues for proton transport (Asp85, Asp96, Glu194, Glu204, and Asp212), retinal and Lys216, are highlighted in the structure. (C) The schematics of ATP synthesis and signal transduction cascades of H. salinarum. Using light, BR outwardly transports proton to produce the proton gradient across the cellular membrane. ATP synthase uses the proton motive force to produce ATP. SRII activates HtrII and the kinase CheA. Subsequently, CheA activates the response regulator CheY that binds to the flagellar motor, leading to the negative phototaxis of the cell. | ||
In contrast to animal rhodopsins, three-dimensional structures of many kinds of microbial rhodopsin molecules have been solved, which provides valuable insights into the molecular mechanism of the activation of microbial rhodopsins, such as HR and SRII, a cation channel rhodopsin from the algae Chlamydomonas reinhardtii, and an outward sodium pump rhodopsin from Krokinobacter eikastus (KR2).1,120 Recently, time-resolved crystallographic techniques using XFEL was applied to several types of microbial rhodopsins.121–123 This technique allows us to imagine the structural changes of microbial rhodopsins on timescales from fs to ms. In fact, the structures of early photointermediates (e.g., K-, L-, and M-intermediate in BR and K- and L-intermediate in KR2) were trapped.121,122 The accumulation of structural information in various types of microbial rhodopsins reveals essential elements that determine the molecular functions, providing an approach to rational functional conversion and engineering new rhodopsin molecules that are not identified from nature (e.g., Ca2+ and phosphate transporters).
3.3 Signal transduction by microbial rhodopsins
BR and its homologous proteins are abundantly expressed in the halophile archaea such as H. salinarum and outwardly transport a proton to produce the proton concentration gradient between the cellular membrane in a light-dependent manner. The ATP synthase uses the proton motive force to produce adenosine triphosphate (ATP) in the halophile archaea (Fig. 6C). Since the free-energy of hydrolysis of ATP is used as energy source in living organisms, the halophile archaea convert green light into their biological energy source. This similar system composed of outward proton pump rhodopsins such as proteorhodopsins (PRs) is observed in various marine bacteria.81 Genetic and ecological analyses have revealed that the amounts of energy produced by PRs are comparable to those produced by chlorophyll-based photosynthesis, implying the importance of rhodopsin-associated energy production in the survival of marine bacteria.124To efficiently absorb green light for the energy production initiated by BR, the halophilic archaea show attractive response to green light (called positive phototaxis) and avoidance response to harmful blue light (called negative phototaxis) using the green-light sensitive microbial rhodopsin SRI and blue-light sensitive microbial rhodopsin SRII, respectively.125 In the archaeal cells, SRII binds to the cognate transducer protein HtrII with a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry.126 The crystal structure of the SRII and HtrII complex from N. pharaonis (NpSRII and NpHtrII) revealed that NpSRII interact through its helices F and G with NpHtrII.127 After photon absorption, hydrogen bonding between Tyr174 and Thr204 (in NpSRII) located in helices F and G, respectively is altered, resulting in the outward tilting of helix F.128,129 The outward tilting of SRII induces the second helix (TM2) of HtrII to rotate clockwise and the HAMP domain of HtrII from SRII to dissociate, leading to the activation (phosphorylation) of the kinase CheA through the adaptor protein CheW (Fig. 6C).130,131 Finally, CheA activates (phosphorylates) the response regulator CheY, which regulates the rotational direction of the flagellar motor, resulting in the negative phototaxis of the halophilic archaea. The structural changes of the protein and signal transduction cascades after photon absorption of SRII have been analyzed mainly by biochemical and biophysical techniques.120,125 Using the similar strategy with animal rhodopsins,62–64 we expect that the complex structures during signal transduction after light absorption will be visualized by femtosecond X-ray laser crystallography and cryo-electron microscopy. Furthermore, mass spectroscopy technique can be a powerful tool to directly detect the molecular components of the cascade in real-time and within native archaeal and bacterial cells like the previous study using native rod photoreceptor cells.65 In contrast to animal rhodopsins, physiological roles and signal transduction cascades of most microbial rhodopsins (e.g., outward sodium pumps and inward proton pumps) in host organisms are unexplored. The detailed physiological and transcriptome analysis of transgenic animals has revealed the physiological roles and signal transduction cascades of animal rhodopsins.67,132–134 We expect that a similar strategy of microbial rhodopsins will be conducted in the future to reveal their physiological roles.
2 stoichiometry.126 The crystal structure of the SRII and HtrII complex from N. pharaonis (NpSRII and NpHtrII) revealed that NpSRII interact through its helices F and G with NpHtrII.127 After photon absorption, hydrogen bonding between Tyr174 and Thr204 (in NpSRII) located in helices F and G, respectively is altered, resulting in the outward tilting of helix F.128,129 The outward tilting of SRII induces the second helix (TM2) of HtrII to rotate clockwise and the HAMP domain of HtrII from SRII to dissociate, leading to the activation (phosphorylation) of the kinase CheA through the adaptor protein CheW (Fig. 6C).130,131 Finally, CheA activates (phosphorylates) the response regulator CheY, which regulates the rotational direction of the flagellar motor, resulting in the negative phototaxis of the halophilic archaea. The structural changes of the protein and signal transduction cascades after photon absorption of SRII have been analyzed mainly by biochemical and biophysical techniques.120,125 Using the similar strategy with animal rhodopsins,62–64 we expect that the complex structures during signal transduction after light absorption will be visualized by femtosecond X-ray laser crystallography and cryo-electron microscopy. Furthermore, mass spectroscopy technique can be a powerful tool to directly detect the molecular components of the cascade in real-time and within native archaeal and bacterial cells like the previous study using native rod photoreceptor cells.65 In contrast to animal rhodopsins, physiological roles and signal transduction cascades of most microbial rhodopsins (e.g., outward sodium pumps and inward proton pumps) in host organisms are unexplored. The detailed physiological and transcriptome analysis of transgenic animals has revealed the physiological roles and signal transduction cascades of animal rhodopsins.67,132–134 We expect that a similar strategy of microbial rhodopsins will be conducted in the future to reveal their physiological roles.
3.4 The evolution of microbial rhodopsins
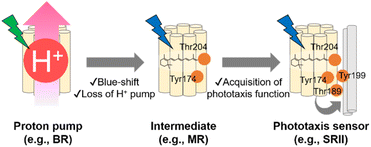
As described in previous sections, the archaeon H. salinarum is attracted to green light for activating BR and avoids shorter wavelength containing harmful blue and near-UV light.125 SRII mediates negative phototaxis and has an approximately 60 nm blue-shifted absorption spectrum (λmax = 498 nm in NpSRII) as compared with BR.135 Phylogenetic analysis suggests that SRII evolved from BR.120,136 This evolutionary scenario is supported by the residual outward proton pump function of SRII in the absence of HtrII.137,138 Moreover, BR can be converted into a SRII-like negative phototaxis sensor by the mutation of only three residues, which strongly suggests the evolutionary relationship between SRII and BR.139 The relationship suggests that SRII acquired the blue-shifted spectral sensitivity and the phototaxis function during the evolutionary process from BR. MR, which is phylogenetically located at the intermediate between BR and SRII, has several key residues responsible for the outward proton pump function (e.g., Asp85, Asp96, Asp212, and Glu204 in BR) and phototaxis function (e.g., Tyr174 and Thr204 in NpSRII), which suggests that MR is like an intermediate state during the evolutionary process from BR to SRII.109 MR shows neither the proton pump function nor phototaxis function, which suggests that MR has lost the proton pump function but has not been fully optimized for the phototaxis function. In fact, a mutant of MR (i.e., A201T/M211Y), having two key residues (Thr189 and Tyr199 in NpSRII) that are responsible for the interaction with HtrII, exhibits phototaxis function similar to that of SRII. In contrast, MR shows blue-light sensitivity (λmax = 485 nm) like SRII, suggesting that MR has blue-shifted spectral sensitivity. Based on the chimeric properties of MR, SRII has been proposed to firstly have blue-shifted its spectral sensitivity and to have lost the outward proton pump function and subsequently acquired the phototaxis function during the evolutionary process from BR (Fig. 7).109,120 While recent advances in genomics have revealed that microbial rhodopsins are highly diversified (Fig. 5A), evolutionary relationships in most microbial rhodopsins are unexplored. Further molecular analysis focusing on the phylogenetic relationship and evolutionary intermediates will be needed to elucidate how molecular properties of microbial rhodopsins have diversified during the evolutionary process.4. Similarities and dissimilarities between animal and microbial rhodopsins
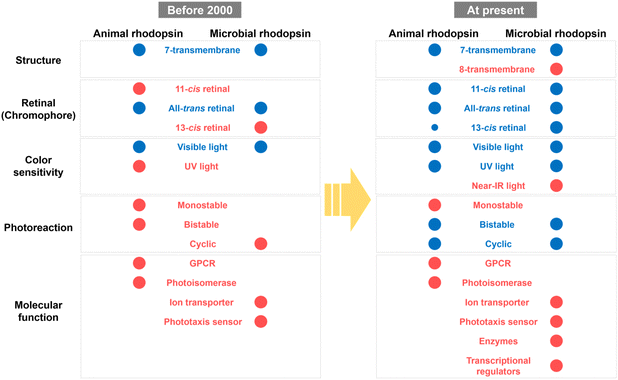
Based on previous studies, the molecular properties of animal and microbial rhodopsins have been summarized as shown in Fig. 8. Before 2000, only a few classical rhodopsin molecules (e.g., vertebrate and invertebrate visual rhodopsins, retinochromes and pinopsin for animal rhodopsins; and BR, HR, SRI, and SRII for microbial rhodopsins) have been identified and analyzed. The common properties between animal and microbial rhodopsins are only a few, such as the 7-transmembrane structure, binding ability to all-trans retinal and visible light sensitivity, whereas any other properties (e.g., the binding ability to 11-cis retinal and bistable photoreaction in animal rhodopsins and the binding ability to 13-cis retinal and cyclic photoreaction nature in microbial rhodopsins) were believed to be their characteristic features. However, a recent comprehensive analysis revealed the high diversity of molecular properties among animal and microbial rhodopsins and that many properties, such as the protein structure, retinal composition, color sensitivity, and photoreaction are common between the two rhodopsins. Firstly, both rhodopsins have 7-transmembrane structures with the eighth helix (helix VIII) at the intracellular side for animal rhodopsins and the extra helix (helix 0) for enzymatic microbial rhodopsins. Secondly, both rhodopsins commonly bind to various isomers of retinal (i.e., 11-cis, 13-cis, and all-trans forms) with various spectral sensitivities from UV- to visible-light region. Thirdly, structural changes between the inactive and active states (i.e., G-protein activating, signaling, and ion conducting states) are triggered by light and heat in both rhodopsins. Based on these similarities, we propose that rhodopsin is defined as the multi-colored retinal-binding membrane proteins whose activities are regulated by light and heat. Since the above three properties (i.e., retinal structure, color sensitivity, and photoreaction) are directly determined by the chemical features of the chromophore, retinal, both rhodopsins are speculated to have diversified their properties through the tuning of the retinal structures by amino-acid substitution mainly around the retinal. Therefore, we expect that the molecular mechanism to regulate the above chemical features in animal rhodopsins can be applied to microbial rhodopsins and vice versa for the production of new rhodopsin molecules with characteristic features.In contrast, molecular functions of animal and microbial rhodopsins are distinctively different (Fig. 8). In general, protein functions are achieved by rationally optimized protein structures and sequences that have been acquired during the long-term evolutionary process for 3.8 billion years on the Earth (Fig. 4 and 7). In fact, animal and microbial rhodopsins have the characteristic important residues that are responsible for individual functions (e.g., cytoplasmic surfaces to interact with G protein in animal rhodopsins and carboxylates to transport the substrate proton in proton pump microbial rhodopsins), and the residues are not common between the two rhodopsins. However, microbial rhodopsins exhibit similar helical opening movement like animal rhodopsins during the photoactivation process, suggesting that both the rhodopsins show a similarity in the global structural changes after photon absorption. Therefore, we speculate that the local structural differences around the functionally important residues are key elements in determining the individual functions that finally leads to the dissimilarity in the functions of the two rhodopsins. Furthermore, protein functions are located at the higher level in the biological hierarchy rather than other molecular properties (e.g., structure, retinal configuration and color sensitivity), and selected for the maintenance of life in host organisms.140 At the moment, the functional diversification imposed by the higher hierarchy disappears in the bottom-up diversification noise. Therefore, it is proposed that the functional diversity of animal and microbial rhodopsins (e.g., GPCRs and photoisomerases for animal rhodopsins versus ion transporters and phototaxis sensors for microbial rhodopsins) have been limited during the evolutionary process, which leads to the dissimilarity in molecular functions (Fig. 8). Based on the similarities and dissimilarities of animal and microbial rhodopsins, we propose that the two rhodopsins have convergently evolved from their distinctive origins as multi-colored retinal-binding membrane proteins whose activities are regulated by light and heat but independently evolved for different molecular and physiological functions in animals and microorganisms. We expect that the understanding of the essential elements related to the individual functions (e.g., GPCRs and photoisomerases for animal rhodopsins versus ion transporters and phototaxis sensors for microbial rhodopsins) using state-of-the-art techniques (e.g., time-resolved mass spectroscopy, cryo-electron microscopy, and XFEL) will enable us to rationally convert the molecular functions across the existing functional dissimilarity among animal and microbial rhodopsins.
Author contributions
K. K. and Y. S. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Our original publications were supported by JSPS KAKENHI with Grant Numbers JP19K16090 and JP21K15054 to KK, JP18H02411, JP19H04727, JP19H05396, JP20K21482, JP21H00404, and JP21H02446 to Y. S., and JST CREST (JPMJCR1656) and AMED (20dm0207060h0004) to Y. S.Notes and references
- O. P. Ernst, D. T. Lodowski, M. Elstner, P. Hegemann, L. S. Brown and H. Kandori, Chem. Rev., 2014, 114, 126–163 CrossRef CAS.
- Y. Shichida and T. Matsuyama, Philos. Trans. R. Soc. London, Ser. B, 2009, 364, 2881–2895 CrossRef CAS PubMed.
- G. Wald and R. Hubbard, Proc. Natl. Acad. Sci. U. S. A., 1950, 36, 92–102 CrossRef CAS PubMed.
- R. Hubbard, Trends Biochem. Sci., 1976, 1, 154–158 CrossRef CAS.
- K. Palczewski, Annu. Rev. Biochem., 2006, 75, 743–767 CrossRef CAS PubMed.
- F. Boll, Arch. Anat. Physiol., 1877, 4–35 Search PubMed.
- A. Terakita, Genome Biol., 2005, 6, 213 CrossRef PubMed.
- S. N. Peirson, S. Halford and R. G. Foster, Philos. Trans. R. Soc. London, Ser. B, 2009, 364, 2849–2865 CrossRef CAS PubMed.
- D. Oesterhelt and W. Stoeckenius, Nat. New Biol., 1971, 233, 149–152 CrossRef CAS.
- E. G. Govorunova, O. A. Sineshchekov, H. Li and J. L. Spudich, Annu. Rev. Biochem., 2017, 86, 845–872 CrossRef CAS.
- K. Kojima, A. Shibukawa and Y. Sudo, Biochemistry, 2020, 59, 218–229 CrossRef CAS.
- E. W. Taylor and A. Agarwal, FEBS Lett., 1993, 325, 161–166 CrossRef CAS.
- J. Soppa, FEBS Lett., 1994, 342, 7–11 CrossRef CAS PubMed.
- N. D. Larusso, B. E. Ruttenberg, A. K. Singh and T. H. Oakley, J. Mol. Evol., 2008, 66, 417–423 CrossRef CAS.
- A. Krishnan, M. S. Almen, R. Fredriksson and H. B. Schioth, PLoS One, 2012, 7, e29817 CrossRef CAS.
- R. Feuda, S. C. Hamilton, J. O. McInerney and D. Pisani, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18868–18872 CrossRef CAS PubMed.
- Y. Zhai, W. H. Heijne, D. W. Smith and M. H. Saier Jr, Biochim. Biophys. Acta, 2001, 1511, 206–223 CrossRef CAS PubMed.
- K. A. Mackin, R. A. Roy and D. L. Theobald, Mol. Biol. Evol., 2014, 31, 85–95 CrossRef CAS.
- T. Nagata and K. Inoue, J. Cell Sci., 2021, 134, jcs258989 CrossRef CAS PubMed.
- T. Okano, T. Yoshizawa and Y. Fukada, Nature, 1994, 372, 94–97 CrossRef CAS.
- R. Hubbard, J. Gen. Physiol., 1958, 42, 259–280 CrossRef CAS.
- M. Koyanagi, E. Takada, T. Nagata, H. Tsukamoto and A. Terakita, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4998–5003 CrossRef CAS.
- H. Tsukamoto, A. Terakita and Y. Shichida, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 6303–6308 CrossRef CAS PubMed.
- K. Nakanishi, Am. Zool., 1991, 31, 479–489 CrossRef CAS.
- T. P. Sakmar, R. R. Franke and H. G. Khorana, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 8309–8313 CrossRef CAS PubMed.
- E. A. Zhukovsky and D. D. Oprian, Science, 1989, 246, 928–930 CrossRef CAS PubMed.
- J. Nathans, Biochemistry, 1990, 29, 9746–9752 CrossRef CAS PubMed.
- A. Terakita, M. Koyanagi, H. Tsukamoto, T. Yamashita, T. Miyata and Y. Shichida, Nat. Struct. Mol. Biol., 2004, 11, 284–289 CrossRef CAS.
- A. Terakita, T. Yamashita and Y. Shichida, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 14263–14267 CrossRef CAS PubMed.
- A. K. Kusnetzow, A. Dukkipati, K. R. Babu, L. Ramos, B. E. Knox and R. R. Birge, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 941–946 CrossRef CAS.
- K. Katayama, S. Sekharan and Y. Sudo, Optogenetics: Light-Sensing Proteins and Their Applications, 2015, ch. 7, pp. 89–107 Search PubMed.
- S. Yokoyama, Annu. Rev. Genomics Hum. Genet., 2008, 9, 259–282 CrossRef CAS.
- Z. Wang, A. B. Asenjo and D. D. Oprian, Biochemistry, 1993, 32, 2125–2130 CrossRef CAS.
- T. Okada, O. P. Ernst, K. Palczewski and K. P. Hofmann, Trends Biochem. Sci., 2001, 26, 318–324 CrossRef CAS PubMed.
- H. Tsukamoto and A. Terakita, Photochem. Photobiol. Sci., 2010, 9, 1435–1443 CrossRef CAS.
- T. Hara and R. Hara, Nature, 1968, 219, 450–454 CrossRef CAS PubMed.
- A. Terakita, R. Hara and T. Hara, Vis. Res., 1989, 29, 639–652 CrossRef CAS PubMed.
- T. Nagata, M. Koyanagi, H. Tsukamoto and A. Terakita, J. Comp. Physiol., A, 2010, 196, 51–59 CrossRef CAS.
- T. Yamashita, H. Ohuchi, S. Tomonari, K. Ikeda, K. Sakai and Y. Shichida, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 22084–22089 CrossRef CAS.
- H. Ohuchi, T. Yamashita, S. Tomonari, S. Fujita-Yanagibayashi, K. Sakai, S. Noji and Y. Shichida, PLoS One, 2012, 7, e31534 CrossRef CAS.
- K. Sato, T. Yamashita, H. Ohuchi, A. Takeuchi, H. Gotoh, K. Ono, M. Mizuno, Y. Mizutani, S. Tomonari, K. Sakai, Y. Imamoto, A. Wada and Y. Shichida, Nat. Commun., 2018, 9, 1255 CrossRef PubMed.
- K. Sakai, Y. Shichida, Y. Imamoto and T. Yamashita, eLife, 2022, 11, e75979 CrossRef CAS PubMed.
- K. Palczewski, T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima, B. A. Fox, I. Le Trong, D. C. Teller, T. Okada, R. E. Stenkamp, M. Yamamoto and M. Miyano, Science, 2000, 289, 739–745 CrossRef CAS.
- J. E. Kim, M. J. Tauber and R. A. Mathies, Biochemistry, 2001, 40, 13774–13778 CrossRef CAS PubMed.
- R. W. Schoenlein, L. A. Peteanu, R. A. Mathies and C. V. Shank, Science, 1991, 254, 412–415 CrossRef CAS PubMed.
- D. Polli, P. Altoe, O. Weingart, K. M. Spillane, C. Manzoni, D. Brida, G. Tomasello, G. Orlandi, P. Kukura, R. A. Mathies, M. Garavelli and G. Cerullo, Nature, 2010, 467, 440–443 CrossRef CAS PubMed.
- P. J. Johnson, A. Halpin, T. Morizumi, V. I. Prokhorenko, O. P. Ernst and R. J. Miller, Nat. Chem., 2015, 7, 980–986 CrossRef CAS PubMed.
- C. Schnedermann, M. Liebel and P. Kukura, J. Am. Chem. Soc., 2015, 137, 2886–2891 CrossRef CAS PubMed.
- Y. Shichida and H. Imai, Cell. Mol. Life Sci., 1998, 54, 1299–1315 CrossRef CAS PubMed.
- D. Emeis, H. Kuhn, J. Reichert and K. P. Hofmann, FEBS Lett., 1982, 143, 29–34 CrossRef CAS PubMed.
- K. P. Hofmann, Biochim. Biophys. Acta, 1985, 810, 278–281 CrossRef CAS PubMed.
- R. Vogel, M. Mahalingam, S. Ludeke, T. Huber, F. Siebert and T. P. Sakmar, J. Mol. Biol., 2008, 380, 648–655 CrossRef CAS PubMed.
- F. Jager, K. Fahmy, T. P. Sakmar and F. Siebert, Biochemistry, 1994, 33, 10878–10882 CrossRef CAS PubMed.
- S. Arnis, K. Fahmy, K. P. Hofmann and T. P. Sakmar, J. Biol. Chem., 1994, 269, 23879–23881 CrossRef CAS.
- H. W. Choe, Y. J. Kim, J. H. Park, T. Morizumi, E. F. Pai, N. Krauss, K. P. Hofmann, P. Scheerer and O. P. Ernst, Nature, 2011, 471, 651–655 CrossRef CAS PubMed.
- J. Standfuss, P. C. Edwards, A. D'Antona, M. Fransen, G. Xie, D. D. Oprian and G. F. Schertler, Nature, 2011, 471, 656–660 CrossRef CAS PubMed.
- R. Neutze and K. Moffat, Curr. Opin. Struct. Biol., 2012, 22, 651–659 CrossRef CAS PubMed.
- T. Gruhl, T. Weinert, M. Rodrigues, C. J. Milne, G. Ortolani, K. Nass, E. Nango, S. Sen, P. J. M. Johnson, C. Cirelli, A. Furrer, S. Mous, P. Skopintsev, D. James, F. Dworkowski, P. Båth, D. Kekilli, D. Ozerov, R. Tanaka, H. Glover, C. Bacellar, S. Brünle, C. M. Casadei, A. D. Diethelm, D. Gashi, G. Gotthard, R. Guixà-González, Y. Joti, V. Kabanova, G. Knopp, E. Lesca, P. Ma, I. Martiel, J. Mühle, S. Owada, F. Pamula, D. Sarabi, O. Tejero, C.-J. Tsai, N. Varma, A. Wach, S. Boutet, K. Tono, P. Nogly, X. Deupi, S. Iwata, R. Neutze, J. Standfuss, G. F. Schertler and V. Panneels, bioRxiv, 2022, preprint, DOI:10.1101/2022.10.14.511948.
- S. Kawamura and S. Tachibanaki, Comp. Biochem. Physiol., Part A: Mol. Integr. Physiol., 2008, 150, 369–377 CrossRef PubMed.
- Y. Fu and K. W. Yau, Pflugers Arch., 2007, 454, 805–819 CrossRef CAS PubMed.
- K. W. Yau and R. C. Hardie, Cell, 2009, 139, 246–264 CrossRef CAS.
- Y. Kang, X. E. Zhou, X. Gao, Y. He, W. Liu, A. Ishchenko, A. Barty, T. A. White, O. Yefanov, G. W. Han, Q. Xu, P. W. de Waal, J. Ke, M. H. Tan, C. Zhang, A. Moeller, G. M. West, B. D. Pascal, N. Van Eps, L. N. Caro, S. A. Vishnivetskiy, R. J. Lee, K. M. Suino-Powell, X. Gu, K. Pal, J. Ma, X. Zhi, S. Boutet, G. J. Williams, M. Messerschmidt, C. Gati, N. A. Zatsepin, D. Wang, D. James, S. Basu, S. Roy-Chowdhury, C. E. Conrad, J. Coe, H. Liu, S. Lisova, C. Kupitz, I. Grotjohann, R. Fromme, Y. Jiang, M. Tan, H. Yang, J. Li, M. Wang, Z. Zheng, D. Li, N. Howe, Y. Zhao, J. Standfuss, K. Diederichs, Y. Dong, C. S. Potter, B. Carragher, M. Caffrey, H. Jiang, H. N. Chapman, J. C. Spence, P. Fromme, U. Weierstall, O. P. Ernst, V. Katritch, V. V. Gurevich, P. R. Griffin, W. L. Hubbell, R. C. Stevens, V. Cherezov, K. Melcher and H. E. Xu, Nature, 2015, 523, 561–567 CrossRef CAS PubMed.
- Y. Kang, O. Kuybeda, P. W. de Waal, S. Mukherjee, N. Van Eps, P. Dutka, X. E. Zhou, A. Bartesaghi, S. Erramilli, T. Morizumi, X. Gu, Y. Yin, P. Liu, Y. Jiang, X. Meng, G. Zhao, K. Melcher, O. P. Ernst, A. A. Kossiakoff, S. Subramaniam and H. E. Xu, Nature, 2018, 558, 553–558 CrossRef CAS PubMed.
- Q. Chen, M. Plasencia, Z. Li, S. Mukherjee, D. Patra, C. L. Chen, T. Klose, X. Q. Yao, A. A. Kossiakoff, L. Chang, P. C. Andrews and J. J. G. Tesmer, Nature, 2021, 595, 600–605 CrossRef CAS PubMed.
- S. Chen, T. Getter, D. Salom, D. Wu, D. Quetschlich, D. S. Chorev, K. Palczewski and C. V. Robinson, Nature, 2022, 604, 384–390 CrossRef CAS PubMed.
- R. C. Hardie and K. Franze, Science, 2012, 338, 260–263 CrossRef CAS PubMed.
- M. W. Hankins, S. N. Peirson and R. G. Foster, Trends Neurosci., 2008, 31, 27–36 CrossRef CAS PubMed.
- T. D. Lamb, Prog. Retinal Eye Res., 2013, 36, 52–119 CrossRef CAS.
- K. Kojima, T. Yamashita, Y. Imamoto, T. G. Kusakabe, M. Tsuda and Y. Shichida, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 6028–6033 CrossRef CAS.
- N. Varma, E. Mutt, J. Muhle, V. Panneels, A. Terakita, X. Deupi, P. Nogly, G. F. X. Schertler and E. Lesca, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 14547–14556 CrossRef CAS.
- T. Nagata, M. Koyanagi, H. Tsukamoto, E. Mutt, G. F. X. Schertler, X. Deupi and A. Terakita, Commun. Biol., 2019, 2, 180 CrossRef PubMed.
- M. Yanagawa, K. Kojima, T. Yamashita, Y. Imamoto, T. Matsuyama, K. Nakanishi, Y. Yamano, A. Wada, Y. Sako and Y. Shichida, Sci. Rep., 2015, 5, 11081 CrossRef CAS PubMed.
- K. Kojima, Y. Matsutani, T. Yamashita, M. Yanagawa, Y. Imamoto, Y. Yamano, A. Wada, O. Hisatomi, K. Nishikawa, K. Sakurai and Y. Shichida, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5437–5442 CrossRef CAS PubMed.
- K. Kojima, Y. Matsutani, M. Yanagawa, Y. Imamoto, Y. Yamano, A. Wada, Y. Shichida and T. Yamashita, Sci. Adv., 2021, 7, eabj1316 CrossRef CAS PubMed.
- M. A. Lienard, G. D. Bernard, A. A. Allen, J. M. Lassance, S. L. Song, R. R. Childers, N. F. Yu, D. J. Ye, A. Stephenson, W. A. Valencia-Montoya, S. Salzman, M. R. L. Whitaker, M. Calonje, F. Zhang and N. E. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2008986118 CrossRef CAS PubMed.
- A. Matsuno-Yagi and Y. Mukohata, Biochem. Biophys. Res. Commun., 1977, 78, 237–243 CrossRef CAS PubMed.
- R. A. Bogomolni and J. L. Spudich, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 6250–6254 CrossRef CAS PubMed.
- T. Takahashi, Y. Mochizuki, N. Kamo and Y. Kobatake, Biochem. Biophys. Res. Commun., 1985, 127, 99–105 CrossRef CAS PubMed.
- J. A. Bieszke, E. L. Braun, L. E. Bean, S. Kang, D. O. Natvig and K. A. Borkovich, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 8034–8039 CrossRef CAS PubMed.
- J. A. Bieszke, E. N. Spudich, K. L. Scott, K. A. Borkovich and J. L. Spudich, Biochemistry, 1999, 38, 14138–14145 CrossRef CAS PubMed.
- O. Beja, E. N. Spudich, J. L. Spudich, M. Leclerc and E. F. DeLong, Nature, 2001, 411, 786–789 CrossRef CAS PubMed.
- D. M. Needham, S. Yoshizawa, T. Hosaka, P. Poirier, C. J. Choi, E. Hehenberger, N. A. Irwin, S. Wilken, C. M. Yung, C. Bachy, R. Kurihara, Y. Nakajima, K. Kojima, T. Kimura-Someya, G. Leonard, R. R. Malmstrom, D. Mende, D. K. Olson, Y. Sudo, S. Sudek, T. A. Richards, E. F. DeLong, P. J. Keeling, A. E. Santoro, M. Shirouzu, W. Iwasaki and A. Z. Worden, Proc. Natl. Acad. Sci. U. S. A., 2019, 16, 20574–20583 CrossRef PubMed.
- S. A. Waschuk, A. G. Bezerra Jr, L. Shi and L. S. Brown, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 6879–6883 CrossRef CAS PubMed.
- T. Tsukamoto, K. Inoue, H. Kandori and Y. Sudo, J. Biol. Chem., 2013, 288, 21581–21592 CrossRef CAS PubMed.
- K. Inoue, S. Ito, Y. Kato, Y. Nomura, M. Shibata, T. Uchihashi, S. P. Tsunoda and H. Kandori, Nat. Commun., 2016, 7, 13415 CrossRef.
- S. Inoue, S. Yoshizawa, Y. Nakajima, K. Kojima, T. Tsukamoto, T. Kikukawa and Y. Sudo, Phys. Chem. Chem. Phys., 2018, 20, 3172–3183 RSC.
- K. Inoue, S. P. Tsunoda, M. Singh, S. Tomida, S. Hososhima, M. Konno, R. Nakamura, H. Watanabe, P. A. Bulzu, H. L. Banciu, A. S. Andrei, T. Uchihashi, R. Ghai, O. Beja and H. Kandori, Sci. Adv., 2020, 6, eaaz2441 CrossRef CAS PubMed.
- G. Nagel, D. Ollig, M. Fuhrmann, S. Kateriya, A. M. Musti, E. Bamberg and P. Hegemann, Science, 2002, 296, 2395–2398 CrossRef CAS PubMed.
- G. Nagel, T. Szellas, W. Huhn, S. Kateriya, N. Adeishvili, P. Berthold, D. Ollig, P. Hegemann and E. Bamberg, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13940–13945 CrossRef CAS.
- N. C. Klapoetke, Y. Murata, S. S. Kim, S. R. Pulver, A. Birdsey-Benson, Y. K. Cho, T. K. Morimoto, A. S. Chuong, E. J. Carpenter, Z. Tian, J. Wang, Y. Xie, Z. Yan, Y. Zhang, B. Y. Chow, B. Surek, M. Melkonian, V. Jayaraman, M. Constantine-Paton, G. K. Wong and E. S. Boyden, Nat. Methods, 2014, 11, 338–346 CrossRef CAS PubMed.
- E. G. Govorunova, O. A. Sineshchekov, R. Janz, X. Liu and J. L. Spudich, Science, 2015, 349, 647–650 CrossRef CAS PubMed.
- J. Oppermann, P. Fischer, A. Silapetere, B. Liepe, S. Rodriguez-Rozada, J. Flores-Uribe, E. Peter, A. Keidel, J. Vierock, J. Kaufmann, M. Broser, M. Luck, F. Bartl, P. Hildebrandt, J. S. Wiegert, O. Beja, P. Hegemann and J. Wietek, Nat. Commun., 2019, 10, 3315 CrossRef PubMed.
- S. Yoshizawa, Y. Kumagai, H. Kim, Y. Ogura, T. Hayashi, W. Iwasaki, E. F. DeLong and K. Kogure, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6732–6737 CrossRef CAS PubMed.
- T. Hasemi, T. Kikukawa, N. Kamo and M. Demura, J. Biol. Chem., 2016, 291, 355–362 CrossRef CAS PubMed.
- K. Inoue, H. Ono, R. Abe-Yoshizumi, S. Yoshizawa, H. Ito, K. Kogure and H. Kandori, Nat. Commun., 2013, 4, 1678 CrossRef PubMed.
- S. P. Balashov, E. S. Imasheva, A. K. Dioumaev, J. M. Wang, K. H. Jung and J. K. Lanyi, Biochemistry, 2014, 53, 7549–7561 CrossRef CAS PubMed.
- A. Niho, S. Yoshizawa, T. Tsukamoto, M. Kurihara, S. Tahara, Y. Nakajima, M. Mizuno, H. Kuramochi, T. Tahara, Y. Mizutani and Y. Sudo, J. Am. Chem. Soc., 2017, 139, 4376–4389 CrossRef CAS PubMed.
- H. Irieda, T. Morita, K. Maki, M. Homma, H. Aiba and Y. Sudo, J. Biol. Chem., 2012, 287, 32485–32493 CrossRef CAS PubMed.
- K. H. Jung, V. D. Trivedi and J. L. Spudich, Mol. Microbiol., 2003, 47, 1513–1522 CrossRef CAS PubMed.
- G. M. Avelar, R. I. Schumacher, P. A. Zaini, G. Leonard, T. A. Richards and S. L. Gomes, Curr. Biol., 2014, 24, 1234–1240 CrossRef CAS PubMed.
- M. Luck, T. Mathes, S. Bruun, R. Fudim, R. Hagedorn, T. M. Tran Nguyen, S. Kateriya, J. T. Kennis, P. Hildebrandt and P. Hegemann, J. Biol. Chem., 2012, 287, 40083–40090 CrossRef CAS PubMed.
- A. Pushkarev, K. Inoue, S. Larom, J. Flores-Uribe, M. Singh, M. Konno, S. Tomida, S. Ito, R. Nakamura, S. P. Tsunoda, A. Philosof, I. Sharon, N. Yutin, E. V. Koonin, H. Kandori and O. Beja, Nature, 2018, 558, 595–599 CrossRef CAS PubMed.
- A. Rozenberg, I. Kaczmarczyk, D. Matzov, J. Vierock, T. Nagata, M. Sugiura, K. Katayama, Y. Kawasaki, M. Konno, Y. Nagasaka, M. Aoyama, I. Das, E. Pahima, J. Church, S. Adam, V. A. Borin, A. Chazan, S. Augustin, J. Wietek, J. Dine, Y. Peleg, A. Kawanabe, Y. Fujiwara, O. Yizhar, M. Sheves, I. Schapiro, Y. Furutani, H. Kandori, K. Inoue, P. Hegemann, O. Beja and M. Shalev-Benami, Nat. Struct. Mol. Biol., 2022, 29, 592–603 CrossRef CAS PubMed.
- R. S. H. Liu and A. E. Asato, Tetrahedron, 1984, 40, 1931–1969 CrossRef CAS.
- K. A. Freedman and R. S. Becker, J. Am. Chem. Soc., 1986, 108, 1245–1251 CrossRef CAS.
- Y. Sudo, Y. Yuasa, J. Shibata, D. Suzuki and M. Homma, J. Biol. Chem., 2011, 286, 11328–11336 CrossRef CAS PubMed.
- J. Yagasaki, D. Suzuki, K. Ihara, K. Inoue, T. Kikukawa, M. Sakai, M. Fujii, M. Homma, H. Kandori and Y. Sudo, Biochemistry, 2010, 49, 1183–1190 CrossRef CAS PubMed.
- M. Broser, A. Spreen, P. E. Konold, E. Peter, S. Adam, V. Borin, I. Schapiro, R. Seifert, J. T. M. Kennis, Y. A. Bernal Sierra and P. Hegemann, Nat. Commun., 2020, 11, 5682 CrossRef CAS PubMed.
- Y. Sudo, K. Ihara, S. Kobayashi, D. Suzuki, H. Irieda, T. Kikukawa, H. Kandori and M. Homma, J. Biol. Chem., 2011, 286, 5967–5976 CrossRef CAS PubMed.
- J. Tittor and D. Oesterhelt, FEBS Lett., 1990, 263, 269–273 CrossRef CAS.
- J. Herbst, K. Heyne and R. Diller, Science, 2002, 297, 822–825 CrossRef CAS PubMed.
- S. Schenkl, F. van Mourik, N. Friedman, M. Sheves, R. Schlesinger, S. Haacke and M. Chergui, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4101–4106 CrossRef CAS PubMed.
- P. Nogly, T. Weinert, D. James, S. Carbajo, D. Ozerov, A. Furrer, D. Gashi, V. Borin, P. Skopintsev, K. Jaeger, K. Nass, P. Bath, R. Bosman, J. Koglin, M. Seaberg, T. Lane, D. Kekilli, S. Brunle, T. Tanaka, W. T. Wu, C. Milne, T. White, A. Barty, U. Weierstall, V. Panneels, E. Nango, S. Iwata, M. Hunter, I. Schapiro, G. Schertler, R. Neutze and J. Standfuss, Science, 2018, 361, eaat0094 CrossRef PubMed.
- G. Nass Kovacs, J. P. Colletier, M. L. Grunbein, Y. Yang, T. Stensitzki, A. Batyuk, S. Carbajo, R. B. Doak, D. Ehrenberg, L. Foucar, R. Gasper, A. Gorel, M. Hilpert, M. Kloos, J. E. Koglin, J. Reinstein, C. M. Roome, R. Schlesinger, M. Seaberg, R. L. Shoeman, M. Stricker, S. Boutet, S. Haacke, J. Heberle, K. Heyne, T. Domratcheva, T. R. M. Barends and I. Schlichting, Nat. Commun., 2019, 10, 3177 CrossRef PubMed.
- S. Tahara, H. Kuramochi, S. Takeuchi and T. Tahara, J. Phys. Chem. Lett., 2019, 10, 5422–5427 CrossRef CAS PubMed.
- H. Kandori, Biochim. Biophys. Acta, 2004, 1658, 72–79 CrossRef CAS PubMed.
- J. K. Lanyi, Annu. Rev. Physiol., 2004, 66, 665–688 CrossRef CAS PubMed.
- T. Oka, N. Yagi, T. Fujisawa, H. Kamikubo, F. Tokunaga and M. Kataoka, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 14278–14282 CrossRef CAS PubMed.
- J. P. Klare, E. Bordignon, M. Engelhard and H. J. Steinhoff, Photochem. Photobiol. Sci., 2004, 3, 543–547 CrossRef CAS PubMed.
- K. Inoue, T. Tsukamoto and Y. Sudo, Biochim. Biophys. Acta, 2014, 1837, 562–577 CrossRef CAS PubMed.
- E. Nango, A. Royant, M. Kubo, T. Nakane, C. Wickstrand, T. Kimura, T. Tanaka, K. Tono, C. Song, R. Tanaka, T. Arima, A. Yamashita, J. Kobayashi, T. Hosaka, E. Mizohata, P. Nogly, M. Sugahara, D. Nam, T. Nomura, T. Shimamura, D. Im, T. Fujiwara, Y. Yamanaka, B. Jeon, T. Nishizawa, K. Oda, M. Fukuda, R. Andersson, P. Bath, R. Dods, J. Davidsson, S. Matsuoka, S. Kawatake, M. Murata, O. Nureki, S. Owada, T. Kameshima, T. Hatsui, Y. Joti, G. Schertler, M. Yabashi, A. N. Bondar, J. Standfuss, R. Neutze and S. Iwata, Science, 2016, 354, 1552–1557 CrossRef CAS PubMed.
- P. Skopintsev, D. Ehrenberg, T. Weinert, D. James, R. K. Kar, P. J. M. Johnson, D. Ozerov, A. Furrer, I. Martiel, F. Dworkowski, K. Nass, G. Knopp, C. Cirelli, C. Arrell, D. Gashi, S. Mous, M. Wranik, T. Gruhl, D. Kekilli, S. Brunle, X. Deupi, G. F. X. Schertler, R. M. Benoit, V. Panneels, P. Nogly, I. Schapiro, C. Milne, J. Heberle and J. Standfuss, Nature, 2020, 583, 314–318 CrossRef CAS PubMed.
- S. Mous, G. Gotthard, D. Ehrenberg, S. Sen, T. Weinert, P. J. M. Johnson, D. James, K. Nass, A. Furrer, D. Kekilli, P. Ma, S. Brunle, C. M. Casadei, I. Martiel, F. Dworkowski, D. Gashi, P. Skopintsev, M. Wranik, G. Knopp, E. Panepucci, V. Panneels, C. Cirelli, D. Ozerov, G. F. X. Schertler, M. Wang, C. Milne, J. Standfuss, I. Schapiro, J. Heberle and P. Nogly, Science, 2022, 375, 845–851 CrossRef CAS PubMed.
- J. Pinhassi, E. F. DeLong, O. Beja, J. M. Gonzalez and C. Pedros-Alio, Microbiol. Mol. Biol. Rev., 2016, 80, 929–954 CrossRef CAS PubMed.
- W. D. Hoff, K. H. Jung and J. L. Spudich, Annu. Rev. Biophys. Biomol. Struct., 1997, 26, 223–258 CrossRef CAS PubMed.
- J. P. Klare, E. Bordignon, M. Doebber, J. Fitter, J. Kriegsmann, I. Chizhov, H. J. Steinhoff and M. Engelhard, J. Mol. Biol., 2006, 356, 1207–1221 CrossRef CAS PubMed.
- H. Luecke, B. Schobert, J. K. Lanyi, E. N. Spudich and J. L. Spudich, Science, 2001, 293, 1499–1503 CrossRef CAS PubMed.
- Y. Sudo, Y. Furutani, H. Kandori and J. L. Spudich, J. Biol. Chem., 2006, 281, 34239–34245 CrossRef CAS PubMed.
- J. P. Klare, V. I. Gordeliy, J. Labahn, G. Buldt, H. J. Steinhoff and M. Engelhard, FEBS Lett., 2004, 564, 219–224 CrossRef CAS PubMed.
- A. A. Wegener, J. P. Klare, M. Engelhard and H. J. Steinhoff, EMBO J., 2001, 20, 5312–5319 CrossRef CAS PubMed.
- A. A. Wegener, I. Chizhov, M. Engelhard and H. J. Steinhoff, J. Mol. Biol., 2000, 301, 881–891 CrossRef CAS PubMed.
- N. Y. Leung and C. Montell, Annu. Rev. Cell Dev. Biol., 2017, 33, 241–264 CrossRef CAS PubMed.
- B. A. Upton, N. M. Diaz, S. A. Gordon, R. N. Van Gelder, E. D. Buhr and R. A. Lang, J. Biol. Rhythms, 2021, 36, 109–126 CrossRef CAS PubMed.
- H. Calligaro, O. Dkhissi-Benyahya and S. Panda, Trends Neurosci., 2022, 45, 200–211 CrossRef CAS PubMed.
- T. Takahashi, B. Yan, P. Mazur, F. Derguini, K. Nakanishi and J. L. Spudich, Biochemistry, 1990, 29, 8467–8474 CrossRef CAS PubMed.
- A. K. Sharma, J. L. Spudich and W. F. Doolittle, Trends Microbiol., 2006, 14, 463–469 CrossRef CAS PubMed.
- Y. Sudo, M. Iwamoto, K. Shimono, M. Sumi and N. Kamo, Biophys. J., 2001, 80, 916–922 CrossRef CAS PubMed.
- G. Schmies, M. Engelhard, P. G. Wood, G. Nagel and E. Bamberg, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 1555–1559 CrossRef CAS PubMed.
- Y. Sudo and J. L. Spudich, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 16129–16134 CrossRef CAS PubMed.
- J. D. Thomas, T. Lee and N. P. Suh, Annu. Rev. Biophys. Biomol. Struct., 2004, 33, 75–93 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |