Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Replication of synthetic recognition-encoded oligomers by ligation of trimer building blocks†
Diego
Núñez-Villanueva‡
 and
Christopher A.
Hunter
and
Christopher A.
Hunter
 *
*
Yusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK. E-mail: herchelsmith.orgchem@ch.cam.ac.uk
First published on 25th October 2023
Abstract
The development of methods for replication of synthetic information oligomers will underpin the use of directed evolution to search new chemical space. Template-directed replication of triazole oligomers has been achieved using a covalent primer in conjunction with non-covalent binding of complementary building blocks. A phenol primer equipped with an alkyne was first attached to a benzoic recognition unit on a mixed sequence template via selective covalent ester base-pair formation. The remaining phenol recognition units on the template were then used for non-covalent binding of phosphine oxide oligomers equipped with an azide. The efficiency of the templated CuAAC reaction between the primer and phosphine oxide building blocks was investigated as a function of the number of H-bonds formed with the template. Increasing the strength of the non-covalent interaction between the template and the azide lead to a significant acceleration of the templated reaction. For shorter phosphine oxide oligomers intermolecular reactions compete with the templated process, but quantitative templated primer elongation was achieved with a phosphine oxide 3-mer building block that was able to form three H-bonds with the template. NMR spectroscopy and molecular models suggest that the template can fold, but addition of the phosphine oxide 3-mer leads to a complex with three H-bonds between phosphine oxide and phenol groups, aligning the azide and alkyne groups in a favourable geometry for the CuAAC reaction. In the product duplex, 1H and 31P NMR data confirm the presence of the three H-bonded base-pairs, demonstrating that the covalent and non-covalent base-pairs are geometrically compatible. A complete replication cycle was carried out starting from the oligotriazole template by covalent attachment of the primer, followed by template-directed elongation, and hydrolysis of the the ester base-pair in the resulting duplex to regenerate the template and liberate the copy strand. We have previously demonstrated sequence-selective oligomer replication using covalent base-pairing, but the trimer building block approach described here is suitable for replication of sequence information using non-covalent binding of the monomer building blocks to a template.
Introduction
Replication of chemical information is the molecular foundation for the evolution of living organisms.1 Sequence information transfer takes place via template-directed synthesis from nucleic acid templates. This process, optimised by millions of years of evolution, has been exploited in the development of breakthrough techniques, namely PCR and SELEX.2–6 Directed evolution is a powerful method for tailoring biomolecules and has led to advances in the search for novel therapeutic agents and more efficient and sustainable manufacturing processes.7–9 To date, the transfer of information from a parent template to a daughter oligomer in such techniques relies exclusively on nucleic acids.10–12 However, there is potential to extend molecular evolution techniques to synthetic systems and access new regions of unexplored chemical space.13,14 The physicochemical properties of such synthetic oligomers will depend on the chemical structure of the backbone and the functional groups that encode information, allowing exploitation in different environments from nucleic acids, i.e. in organic solvents, low temperature, etc.15–17 The functional properties of these synthetic oligomers would be defined by the sequence of the monomer units in the chain, which could be optimised by evolution.18–20 A key requirement for the use of synthetic oligomers in molecular evolution is an efficient method to copy the encoded sequence information into a daughter strand.We recently described the use of covalent templating to achieve efficient sequence information transfer between synthetic triazole oligomers (Fig. 1, top channel).21,22 The base-pairing system involves kinetically inert ester bonds between phenol and benzoic acid side-chains, and copper catalyzed azide alkyne cycloaddition (CuAAC) is used for oligomerization of the backbone of the daughter strand. The resulting covalent duplex can be cleaved by ester hydrolysis to regenerate the initial template along with the complementary copy. The use of covalent base-pairs overcomes the issue of incomplete loading of monomers onto the template when weaker non-covalent interactions are used. It also enables reprogramming of the information transfer process via the use of traceless linkers to attach monomers to the template, so that direct replication, reciprocal replication and mutation are all accessible.23–25
We have extended the use of covalent base-pairs to develop an alternative replication method, inspired by the polymerase chain reaction (PCR) of nucleic acids. A key element in PCR is the use of primers, which define the region of the single-stranded template to be elongated. In non-enzymatic nucleic acid replication, a primer is also essential for template-directed synthesis of the complementary strand. The primer must have a high affinity for the template to avoid non-templated ligation reactions, which requires the design of long primers for effective annealing or the use of excess template to ensure that the primer is fully template bound.26,27
With the aim of developing efficient replication methods for synthetic oligomers, we recently described a hybrid template-directed primer extension method, combining covalent and non-covalent base-pairing (Fig. 1, bottom channel).28 A cleavable covalent ester base-pair was used for efficient primer loading, via a benzoic acid unit at one end of the template. This kinetically stable attachment enables the use of short and simple primers, in contrast to nucleic acid replication. The resulting primed template has phenol H-bond donors that form strong non-covalent interactions with phosphine oxide H-bond acceptor monomers. The primer is equipped with an alkyne for CuAAC elongation with azide monomers. Phenol-phosphine oxide H-bonding interactions lead to the pre-ZIP complex, which templates the CuAAC reaction of the monomer with the primer. Hydrolysis of the template-primer ester bond in the resulting duplex releases the complementary copy from the template.
Fig. 2 shows the key parameters determining the efficiency of the non-covalent template-directed primer elongation when a 3-mer template is used with different types of azide monomer.28 The template has one benzoic acid unit (blue), used for covalent attachment of the primer, and two phenol groups (red) for H-bonding with phosphine oxides (purple). When a phosphine oxide 1-mer is used (Fig. 2, top channel), the first step involves binding to the template (K1 ≈ 200 M−1 in dichloromethane). The rate of the templated CuAAC reaction is given by the product of the rate constant for the untemplated reaction (k) and the kinetic effective molarity for the intramolecular process (EM†). The rate constant for a competing untemplated reaction with a phenol 1-mer is k. The success of the templating process is therefore related to the product of K1 and EM†. The value of EM† depends on the geometric compatibility of the template-monomer complex with the transition state for the CuAAC reaction and on backbone flexibility and is therefore rather difficult to predict or control. In contrast, the binding affinity of the azide monomer (K1) for the template can be manipulated in a straightforward manner.
We have shown previously that the use of a phosphine oxide 2-mer leads to a significant acceleration of the templated reaction (Fig. 2 bottom channel), because K2 is one order of magnitude higher than K1 (K2 ≈ 1500 M−1 in dichloromethane). Similar phenomena have been described in non-enzymatic nucleic acid replication, where the use of activated oligomers instead of monomers significantly increases the efficiency of primer extension.29 Here we show how this approach can be developed further to achieve near quantitative template-directed primer extension of an oligotriazole.
Results and discussion
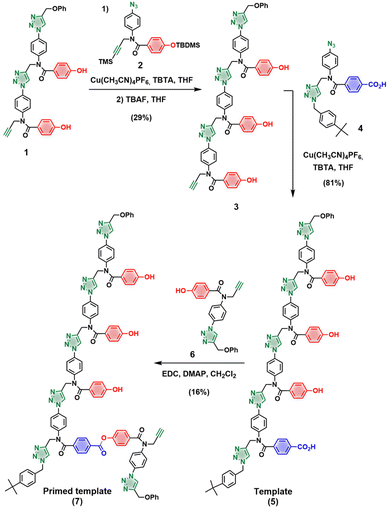
The oligotriazole architecture highlighted in Fig. 1 is ideal for further optimization of the templating process, because the binding affinity between two oligomers increases by one order of magnitude for each H-bonded base-pair added to the chain. Increasing the length of the monomer building blocks therefore provides a convenient tool for improving the efficiency of the templated CuAAC reaction by increasing the stability of the pre-ZIP complex. We report here the output of the templating process for a 4-mer template and an azide-functionalised phosphine oxide 3-mer used to elongate the covalent primer.Synthesis of the 4-mer template containing three H-bonding phenol units and the subsequent incorporation of the covalent primer is shown in Scheme 1. CuAAC coupling between the phenol 2-mer 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 21 and the protected azide 2,21 followed by TBAF-mediated removal of the silyl groups afforded phenol 3-mer 3. The terminal alkyne in 3 was reacted with capped benzoic acid 1-mer 4 to yield template 5 in good yield. The primer was loaded onto the template by ester coupling with an excess of phenol 1-mer 6, affording primed template 7. Scheme 2 shows the 3-mer phosphine oxide azide 8 and the phenol azide 9 used to assess the template effect in competition reactions.28
21 and the protected azide 2,21 followed by TBAF-mediated removal of the silyl groups afforded phenol 3-mer 3. The terminal alkyne in 3 was reacted with capped benzoic acid 1-mer 4 to yield template 5 in good yield. The primer was loaded onto the template by ester coupling with an excess of phenol 1-mer 6, affording primed template 7. Scheme 2 shows the 3-mer phosphine oxide azide 8 and the phenol azide 9 used to assess the template effect in competition reactions.28
 | ||
| Scheme 2 (a) Phosphine oxide 3-mer azide, prepared by oligomerization of a phosphine oxide 1-mer bearing an alkyne and an azide in the presence of a 4-tert-butylbenzyl azide end-capping group. (b) Phenol 1-mer azide.28 | ||
The primed template 7 was used in a competition reaction between phosphine oxide 3-mer 8 and phenol 9 to assess the template effect (Fig. 3). All reactions were performed in dry and degassed dichloromethane using Cu(CH3CN)4PF6, TBTA and an equimolar excess of each azide. Fig. 4c shows the UPLC trace of the crude reaction mixture after 2 days. The templated product (11, green peak) is formed almost exclusively, with only traces of untemplated product (12, red peak). As a control, the same reaction was performed with a simple alkyne instead of the primed template in order to quantify any difference in the intrinsic reactivity of the two azides (see ESI† for details). In this case, 9 was found to be twice as reactive as 8, which means that the efficiency of the templating process is even higher than Fig. 4c suggests.
Fig. 4a and b show the corresponding results for template-directed primer elongation with a phosphine oxide 1-mer and 2-mer respectively. When the azide makes one H-bond with the template, there are no significant differences between the amounts of the templated (green) and untemplated (red) products (Fig. 4a). When the azide makes two H-bonds with the template, there is a significant acceleration in the rate of the templated pathway and an increase in the yield of the templated product (green) relative to the untemplated product (red). However, substantial amounts of the untemplated product are formed in Fig. 4b. It is only with three H-bonds that the off-template pathway is almost completely supressed, and the template-directed primer elongation product is formed efficiently (Fig. 4c).
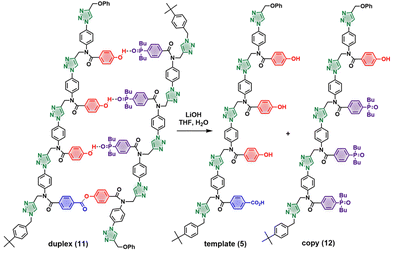
Evidence for the H-bonding interactions involved in templating the CuAAC reaction between the primed template 7 and 3-mer 8 can be obtained from NMR spectra. Fig. 5a shows the 1H NMR spectra of primed template 7 and duplex 11 in CDCl3. The signals corresponding to the OH groups of 7 appear at 8–9 ppm, which suggest that phenols are involved in H-bonds.28 There is no self-association at these concentrations, so there is probably some folding of the primed template due to intramolecular H-bonding between the phenol groups and the carbonyl H-bond acceptors on the backbone. In the duplex 11, the three OH signals are clearly resolved and appear as sharp singlets at 10 ppm, which indicates that they are fully H-bonded to the three phosphine oxide acceptors on the copy strand. Additional evidence for these base-pairing interactions was obtained from 31P NMR spectra (Fig. 5b). In duplex 11, the three signals due to the phosphine oxide groups show downfield shifts of 2–4 ppm compared with the 3-mer phosphine oxide 8 where no H-bonding is possible. Thus, the NMR data suggest that the H-bonding interactions that lead to acceleration of the rate of the templated reaction live on in the product, demonstrating that the covalent and non-covalent base-pairs can be simultaneously accommodated within the duplex structure.
The NMR data suggest that the templated pathway may be affected by competition with folding of the primed template. Molecular mechanics calculations were performed to obtain further insight into the conformational properties of the template and the duplex (Fig. 6).30,31 For the primed template 7, the lowest energy conformation in chloroform indicates that the phenol groups are able to form intramolecular H-bonds with the carbonyl groups of the covalent primer.30,31 This structure is consistent with the 1H NMR spectrum of 7, which indicates partially H-bonded phenol groups.
 | ||
| Fig. 6 Conformational equilibria involved in assembly of the pre-ZIP complex between 7 and 8. The primed template 7 is partially folded. Complexation with the complementary phosphine oxide 3-mer 8 opens up the template to give the pre-ZIP complex. CuAAC reaction leads to irreversible formation of the duplex 11. Lowest energy structures from conformational searches using molecular mechanics are shown for the primed template and the duplex (MMFFs force field with chloroform solvation with H-bonds as green dotted lines).30,31 | ||
The lowest energy structure for the duplex in chloroform is an extended conformation with three phenol-phosphine oxide H-bonds, which is again consistent with the NMR data. The H-bond acceptor parameters for the ester and amide groups (β = 5.3 and 7.8) are much lower than the corresponding parameter for phosphine oxide (β = 10.7), so H-bonding interactions between the phenol groups and the backbone carbonyl groups do not compete with the base-pairing interactions in the duplex.32 These observations suggest that although the primed template is in equilibrium between a folded and extended conformation, when the phosphine oxide 3-mer is added, the pre-ZIP complex is fully assembled with three H-bonds between the phosphine oxide and phenol groups.
These results show that it is possible to tune the efficiency of a templating process using the strength of the interaction between the monomer and the template. The 3-mer phosphine oxide that makes three H-bonds with the primed template is therefore suitable to carry out the full replication cycle illustrated in Fig. 1. Fig. 7 shows the UPLC results for each step of the pathway starting from template 5. The first step involves covalent attachment of the primer 6 by ester coupling to give the primed template 7 (Fig. 7b). The template-directed CuAAC reaction was then carried out using a mixture of the complementary phosphine oxide 3-mer 8 and competing phenol 9 (Fig. 7c shows the UPLC trace of the crude reaction mixture). Column chromatography was used to separate duplex 11 from the excess azides (Fig. 7d). The final step is cleavage of the covalent ester base-pair to release the template and the copy strand (Scheme 3 and Fig. 7e). The two oligomers were separated by column chromatography (Fig. 7f), and the pure copy 12 was fully characterized (see ESI†).
Conclusions
Current directed evolution methodologies rely exclusively on nucleic acids for the transfer of sequence information, limiting the scope to systems that interface with DNA. The use of synthetic polymers in evolutionary searching would give access to a vast and unexplored region of chemical structure space, opening the way to develop synthetic systems that could rival the functional properties of biopolymers. However, methods for the template-directed replication of synthetic oligomers are required before evolutionary optimization of functional polymers becomes possible. Here we describe an efficient process for template-directed replication of triazole oligomers using a combination of covalent and non-covalent base-pairing interactions to transfer information from the template to the daughter strand. A key feature is the use of a covalent primer, which is attached to the template via ester base-pairing, and functionalized with an alkyne group. The template has phenol recognition units able to form non-covalent base-pairs with phosphine oxide monomers via H-bonding. These phosphine oxide monomers are functionalized with an azide group, so the templated elongation of the primer is viable through CuAAC. Competition reactions between azides equipped with phosphine oxide recognition units and azides equipped with phenol recognition units were used to assess the template effect.The efficiency of the templated reaction can be optimized by tuning a single parameter: the binding strength of the azide-functionalized phosphine oxide monomer. When the number of H-bonds between the template phenol units and the phosphine oxide monomer is increased, there is a significant acceleration of the templated reaction. Thus, a phosphine oxide 2-mer leads to a higher rate acceleration than a phosphine oxide 1-mer, because it binds the template with an order of magnitude higher affinity. When a 3-mer phosphine oxide strand was used, the higher affinity for the template leads to quantitative templated primer elongation.
Evidence for the phenol-phosphine oxide H-bonding involved in the templated pathway was obtained from NMR spectroscopy and is supported by molecular models. These data also suggest that the primed template is in equilibrium between a folded and extended conformation. When the phosphine oxide 3-mer is added, the template-substrate complex is fully assembled with three H-bonds between phosphine oxide and phenol groups, enabling the alignment of the azide and alkyne groups in a favourable geometry for the CuAAC reaction. The results show that the covalent and non-covalent base-pairing motifs used here are geometrically compatible and can be successfully deployed in combination.
This system was used to carry out a full cycle of templated-directed primer elongation. Starting from the mixed sequence template, the primer was covalently loaded, then templated oligomer synthesis provided the hybrid covalent/non-covalent duplex. Cleavage of the ester base-pair in the duplex regenerated the template and released the sequence-complementary copy oligomer. The behaviour of this synthetic system is in line with observations on non-enzymatic nucleic acid replication, where the rate and fidelity of primer extension can be improved by using activated oligomers as the building blocks instead of monomers.29
We have previously demonstrated sequence-selective oligomer replication using covalent base-pairing.21 Covalent base-pairs are formed in a quantitative manner, guaranteeing full loading of monomers onto a template, and they also provide a mechanism for irreversible cleavage of the product duplex. However, replication cycles using this approach require six chemical steps. The dynamic nature of non-covalent base-pairs means that base-pair formation and cleavage can be carried out avoiding many of these chemical steps, but incomplete binding to the template lead to low yields of templated product.28 The 3-mer building blocks introduced here form three cooperative non-covalent base-pairs, which leads to full loading of monomers onto the template and high yields of templated product. When used in conjunction with a covalent primer, the product duplex can then be cleaved to irreversibly release the daughter strand from the template. These components therefore set the scene for sequence-selective information transfer between synthetic polymers using non-covalent base-pairing.
Author contributions
The manuscript was written through contributions of all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank the Engineering and Physical Sciences Research Council (EP/P027067/1), the European Research Council (ERC-2020-AdG-101018984-InfoMols) and the Herchel Smith Fund for funding. D. N.-V. acknowledges the funding support provided by the Fundación General CSIC's ComFuturo programme which has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No. 101034263.References
- R. R. Sinden, DNA Structure and Function, Academic Press, 1994 Search PubMed.
- R. K. Saiki, S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich and N. Arnheim, Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia, Science, 1985, 230, 1350 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, In vitro selection of RNA molecules that bind specific ligands, Nature, 1990, 346, 818 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Systematic Evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 1990, 249, 505 CrossRef CAS PubMed.
- K. Chen and F. H. Arnold, Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 5618 CrossRef CAS PubMed.
- G. F. Joyce, Forty years of in vitro evolution, Angew. Chem., Int. Ed., 2007, 46, 6420 CrossRef CAS PubMed.
- M. Famulok, G. Mayer and M. Blind, Nucleic acid aptamers-from selection in vitro to applications in vivo, Acc. Chem. Res., 2000, 33, 591 CrossRef CAS PubMed.
- N. J. Turner, Directed evolution drives the next generation of biocatalysts, Nat. Chem. Biol., 2009, 8, 567 CrossRef PubMed.
- U. T. Bornscheuer, B. Hauer, K. E. Jaeger and U. Schwaneberg, Directed evolution empowered redesign of natural proteins for the sustainable production of chemicals and pharmaceuticals, Angew. Chem., Int. Ed., 2019, 58, 36 CrossRef CAS PubMed.
- J. W. Chin, Expanding and reprogramming the genetic code, Nature, 2017, 550, 53 CrossRef CAS PubMed.
- D. L. Usanov, A. I. Chan, J. P. Maianti and D. R. Liu, Second-generation DNA-templated macrocycle libraries for the discovery of bioactive small molecules, Nat. Chem., 2018, 10, 704 CrossRef CAS PubMed.
- J. C. W. Willis and J. W. Chin, Mutually orthogonal pyrrolysyl-tRNA synthetase/tRNA pairs, Nat. Chem., 2018, 10, 831 CrossRef CAS PubMed.
- L. E. Orgel, Unnatural selection in chemical systems, Acc. Chem. Res., 1995, 28, 109 CrossRef CAS PubMed.
- S. Samokhvalova and J.-F. Lutz, Macromolecular information transfer, Angew. Chem., Int. Ed., 2023, 62, e202300014 CrossRef CAS PubMed.
- E. Yashima, K. Maeda and Y. Furusho, Single- and double-stranded helical polymers: synthesis, structures, and functions, Acc. Chem. Res., 2008, 41, 1166 CrossRef CAS PubMed.
- K. R. Strom and J. W. Szostak, Folding and duplex formation in sequence-defined aniline benzaldehyde oligoarylacetylenes, J. Am. Chem. Soc., 2022, 144, 18350 CrossRef CAS PubMed.
- G. Iadevaia and C. A. Hunter, Recognition-encoded synthetic information molecules, Acc. Chem. Res., 2023, 56, 712 CrossRef CAS PubMed.
- H. Ito, Y. Furusho, T. Hasegawa and E. Yashima, Sequence- and chain-length-specific complementary double-helix formation, J. Am. Chem. Soc., 2008, 130, 14008 CrossRef CAS PubMed.
- B. Gong, Molecular duplexes with encoded sequences and stabilities, Acc. Chem. Res., 2012, 45, 2077 CrossRef CAS PubMed.
- A. E. Stross, G. Iadevaia, D. Núñez-Villanueva and C. A. Hunter, Sequence-selective formation of synthetic H-bonded duplexes, J. Am. Chem. Soc., 2017, 139, 12655 CrossRef CAS PubMed.
- D. Núñez-Villanueva, M. Ciaccia, G. Iadevaia, E. Sanna and C. A. Hunter, Sequence information transfer using covalent template-directed synthesis, Chem. Sci., 2019, 10, 5258 RSC.
- M. Ciaccia, D. Núñez-Villanueva and C. A. Hunter, Capping strategies for covalent template-directed synthesis of linear oligomers using CuAAC, J. Am. Chem. Soc., 2019, 141, 10862 CrossRef CAS PubMed.
- D. Núñez-Villanueva and C. A. Hunter, Molecular replication using covalent base-pairs with traceless linkers, Org. Biomol. Chem., 2019, 17, 9660 RSC.
- D. Núñez-Villanueva and C. A. Hunter, Controlled mutation in the replication of synthetic oligomers, Chem. Sci., 2021, 12, 4063 RSC.
- D. Núñez-Villanueva and C. A. Hunter, Replication of sequence information in synthetic oligomers, Acc. Chem. Res., 2021, 54, 1298 CrossRef PubMed.
- T. Walton, W. Zhang, L. Li, C. P. Tam and J. W. Szostak, The mechanism of nonenzymatic template copying with imidazole-activated nucleotides, Angew. Chem., Int. Ed., 2019, 58, 10812 CrossRef CAS PubMed.
- A. S. Tupper and P. G. Higgs, Rolling-circle and strand-displacement mechanisms for non-enzymatic RNA replication at the time of the origin of life, J. Theor. Biol., 2021, 527, 110822 CrossRef CAS PubMed.
- D. Núñez-Villanueva and C. A. Hunter, H-bond templated oligomer synthesis using a covalent primer, J. Am. Chem. Soc., 2022, 144, 17307 CrossRef PubMed.
- L. Zhou, D. K. O'Flaherty and J. W. Szostak, Template-directed copying of RNA by non-enzymatic ligation, Angew. Chem., Int. Ed., 2020, 59, 15682 CrossRef CAS PubMed.
- Schrödinger Release 2022-1: MacroModel, Schrödinger, LLC, New York, NY, 2022 (version 13.1.141) Search PubMed.
- The PyMOL Molecular Graphics System (open-source PyMOL). Version 2.3.0a0. Schrödinger, LLC Search PubMed.
- C. A. Hunter, Quantifying intermolecular interactions: guidelines for the molecular recognition toolbox, Angew. Chem., Int. Ed., 2004, 43, 5310 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and methods, detailed synthetic procedures, 1H and 13C NMR spectra of all compounds and UPLC traces for the ZIP reactions. See DOI: https://doi.org/10.1039/d3qo01717f |
| ‡ Present address: ComFuturo Fellow, Instituto de Química Médica (IQM-CSIC), Juan de la Cierva 3, 28006 Madrid, Spain. |
| This journal is © the Partner Organisations 2023 |