Open Access Article
Open Access ArticleStar-shaped poly(L-lysine) with polyester bis-MPA dendritic core as potential degradable nano vectors for gene delivery†
Smiljana
Stefanovic
 a,
Katie
McCormick
a,
Katie
McCormick
 bc,
Sarinj
Fattah
bc,
Sarinj
Fattah
 b,
Ruiari
Brannigan
d,
Sally-Ann
Cryan
b,
Ruiari
Brannigan
d,
Sally-Ann
Cryan
 bce and
Andreas
Heise
bce and
Andreas
Heise
 *ace
*ace
aDepartment of Chemistry, RCSI University of Medicine and Health Sciences, Dublin 2, Ireland. E-mail: andreasheise@rcsi.ie
bSchool of Pharmacy and Biomolecular Sciences, RCSI University of Medicine and Health Sciences, Dublin 2, Ireland
cAMBER, The SFI Advanced Materials and Bioengineering Research Centre, RCSI, Dublin 2, Ireland
dSchool of Chemical Sciences, Dublin City University, Collins Avenue, Whitehall, Dublin 9, Ireland
eScience Foundation Ireland (SFI) Centre for Research in Medical Devices (CURAM), RCSI, Dublin 2, Ireland
First published on 17th May 2023
Abstract
Cationic star-shaped poly(L-lysine) are efficient nanovectors for the delivery of nucleic acids (gene delivery). They are commonly obtained by the ring-opening polymerisation of L-lysine N-carboxyanhydrides (NCA) from multifunctional amine initiators such as dendrimers. To date, commonly reported dendritic precursors are non-degradable which can cause dose dependant cytotoxicity in vivo. Herein, we present a new synthetic route for the preparation of star-shaped poly(L-lysine) from ammonium terminated 2,2-bis-(hydroxymethyl)propionic acid (bis-MPA) dendrimers. A range of well-defined star polypeptides with exceptionally low dispersities (ĐM < 1.05) was obtained by varying the dendrimer generation and adjusting the monomer to initiator feed ratio. The ability of efficient dendrimer core degradation was confirmed under physiological and basic conditions. Moreover, preliminary pDNA complexation studies demonstrate successful polyplex formation between the gene cargo and the star poly(L-lysine). The results suggest that the bis-MPA star poly(L-lysine) platform is a promising fully degradable alternative to leading non-degradable polymeric vectors.
Introduction
Gene therapy has long been seen as a potential solution to previously unmet clinical needs.1 To realise the immense possibilities presented by nucleic acid-based therapeutics, the development of effective and safe delivery vehicles (vectors) is critical.2 Nucleic acids are large, negatively charged molecules and are rapidly cleared by circulating nucleases in the body. Suitable vectors must not only protect the gene cargo against hydrolysis but also efficiently deliver them intracellularly. Positively charged polymers are poised to form stable complexes (polyplexes) with nucleic acids and have been widely investigated as non-viral gene vectors.1 Cationic polymers including polyamidoamine (PAMAM) dendrimers and poly(ethylene imine) (PEI) have seen some success for in vitro gene delivery but underperform in vivo, which is a barrier to clinical translation.3–7 One critical drawback of these synthetic polyamines is their dose dependent cytotoxicity.8–10 While improved stability and easier scale-up is recognised as a benefit of polymeric vectors, it is indeed surprising that their major advantage, i.e. the greater flexibility in structural design and the ability to easily modify them, has not been utilised to any large extend to overcome these drawbacks.We recently began to investigate star-shaped poly(L-lysine) (PLL) as gene delivery vectors.11,12 Well-defined star-shaped PLL was readily prepared via controlled ring opening polymerisation (ROP) of amino acid-derived N-carboxyanhydrides (NCAs) by initiation from the primary amino groups of poly(propylene imine) (PPI) dendrimers.13 Our research has shown that the adaptability of the macromolecular architecture is a critical advantage when optimising delivery performance to best meet the nature of the gene cargo and the clinical target. Star-shaped PLL has since been successfully applied for the delivery of advanced gene therapeutics in pre-clinical in vitro and in vivo models. The versatility in design enabled easy tailoring of the technology for delivery of gene therapeutics with different physicochemical properties. For example, a star PLL system was used to form gene nanomedicines with plasmid DNA (pDNA) to effectively transfect mesenchymal stem cells as well as for siRNA and pDNA delivery to epithelial cells to treat conditions such as cystic fibrosis (CF).11,14 Incorporated into gene activated scaffolds and implanted into a rat calvarian defect model (bone defect), star PLL nanomedicines were found to significantly enhance bone regeneration after just four weeks.15 These materials were found to be more efficient at delivery of gene cargoes to difficult-to-transfect cells, while exhibiting enhanced biocompatibility when compared to leading commercial vectors.16 One concern of the current star PLL delivery vector technology is the incorporation of dendritic PPI. PPI dendrimers show a low degree of hydrolytic degradation which could result in prolonged circulation in the blood stream and potential bioaccumulation in the liver.17 Moreover, PPI dendrimer synthesis is tedious, requiring high temperature and pressure, which limits easy access to the material.18 Other multivalent polymers have been used as a core in the preparation of star-shaped polypeptides, including polyamidoamine (PAMAM) dendrimers but all of them suffer from similar drawbacks as PPI.19–24
Conversely, 2,2-bis-(hydroxymethyl)propionic acid (bis-MPA) based dendrimers are an economical polyester-based alternative to PPI dendrimers, because of their non-demanding and inexpensive preparation. A main advantage is their proven biodegradability, biocompatibility, and low toxicity. They undergo pH triggered hydrolytic degradation resulting in low molecular weight products that can easily be eliminated from the body.25 Recently, Garcia-Gallego et al. proposed a concept of fluoride promoted esterification chemistry to prepare bis-MPA dendrimers.26 That concept is based on a straightforward synthetic method, where 1,1′-carbinyldiimidazole (CDI) is used as a coupling agent for activation of acetonide-protected bis-MPA and caesium-fluoride (CsF) as a catalyst, allowing full conversion of the building blocks. Bis-MPA dendrimers thus combine a number of benefits, which makes them interesting alternatives as core materials for the star PLL platform. However, their feasibility as initiators for NCA ROP has never been investigated. Here we present a systematic study into the design of fully degradable star PLL based on a bis-MPA dendritic core. We demonstrate the flexibility in the design of well-defined star polypeptides with different number of arms and molecular weights. Moreover, preliminary data on degradation and plasmid DNA complexation are presented.
Experimentals
Materials and methods
All chemicals and solvents were purchased from Sigma Aldrich and used as received unless otherwise noted. Syntheses of Lysine NCA and alanine bis-MPA initiators were carried out following literature procedures (details provided in the ESI†).26–28 Amberlyst A21 resin was purified following literature.27 1,1-Carbonyldiimidazole (CDI) was stored in a desiccator due to moisture sensitivity. Attenuated total reflection (ATR) FT-IR spectroscopy was performed using a PerkinElmer Spectrum 100 in the spectral region of 650–4000 cm−1. Proton (1H) and carbon (13C) nuclear magnetic resonance (NMR) spectra as well as diffusion ordered (DOSY) spectra were recorded using a Bruker Avance 400 (400 MHz) spectrometer at room temperature in d-TFA, CDCl3, CD3OD, and D2O as solvents. All chemical shifts of 1H and 13C NMR spectra are reported in parts per million (ppm) while diffusion coefficients are reported in cm2 s−1. The obtained spectra were analysed in MestReNova 6.02 software. DOSY data were processed using the Bayesian transform method. Diffusion coefficients were calculated from intensity of methylene signal of L-lysine side chain at 2.81 ppm using a mono-exponential decay equation, which is simplified version of Stejskal–Tanner function: I = I0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−DZ) and Z = γ2G2δ(Δ − d/3).29I is the intensity of integral (methylene signal at 2.81 ppm); I0 is the maximum intensity, G is the gradient strength in G cm−1, δ is the gradient duration in seconds and Δ is the echo delay in seconds. Z represents the argument of the exponent and it is calculated from the arrayed experimental data. Molecular weight dispersities (Đ) and number average molecular weights (Mn) of the polymers were determined by gel permeation chromatography (GPC). GPC was conducted in 1,1,1,3,3,3-hexafluoro-2-propanol (HFiP) using a PSS SECurity GPC system equipped with a PFG 7 μm 8 × 50 mm pre-column, a PSS 100 Å, 7 μm 8 × 300 mm and a PSS 1000 Å, 7 μm 8 × 300 mm column in series and a differential refractive index (dRI) detector at a flow rate of 1.0 mL min−1. The system was calibrated against Agilent Easi-Vial linear poly(methyl methacrylate) (PMMA) standards and analysed by PSS winGPCUniChrom. All GPC samples were prepared using a concentration of 2 mg mL−1, and were filtered through a 0.2 μm millipore filter prior to injection.
exp(−DZ) and Z = γ2G2δ(Δ − d/3).29I is the intensity of integral (methylene signal at 2.81 ppm); I0 is the maximum intensity, G is the gradient strength in G cm−1, δ is the gradient duration in seconds and Δ is the echo delay in seconds. Z represents the argument of the exponent and it is calculated from the arrayed experimental data. Molecular weight dispersities (Đ) and number average molecular weights (Mn) of the polymers were determined by gel permeation chromatography (GPC). GPC was conducted in 1,1,1,3,3,3-hexafluoro-2-propanol (HFiP) using a PSS SECurity GPC system equipped with a PFG 7 μm 8 × 50 mm pre-column, a PSS 100 Å, 7 μm 8 × 300 mm and a PSS 1000 Å, 7 μm 8 × 300 mm column in series and a differential refractive index (dRI) detector at a flow rate of 1.0 mL min−1. The system was calibrated against Agilent Easi-Vial linear poly(methyl methacrylate) (PMMA) standards and analysed by PSS winGPCUniChrom. All GPC samples were prepared using a concentration of 2 mg mL−1, and were filtered through a 0.2 μm millipore filter prior to injection.
Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600, Mw = 14
600, Mw = 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, Đ = 1.03.
000, Đ = 1.03.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching) at 1793 cm−1 and a peak from L-Lys fingerprint region (C–H wags), at 1255 cm−1 which remain unchanged during the polymerisation (Fig. S21†). Quantification of the NCA conversion was calculated using equations:
O stretching) at 1793 cm−1 and a peak from L-Lys fingerprint region (C–H wags), at 1255 cm−1 which remain unchanged during the polymerisation (Fig. S21†). Quantification of the NCA conversion was calculated using equations:| Conversion, % = 100 − Rt/R0 × 100; |
| Rt = It(1786)/I(1255), R0 = I0(1786)/I(1255) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphate (N/P) ratios including, 0.5, 1, 2, 5, 10 and 20 (N/P represents the molar ratio of positively charged nitrogen (N) in the polyLys arms to negatively phosphate (P) group present in the pGlu molecules). The concentration of pGlu remained constant at 1 μg μl−1. Gel retardation assays were used to assess the encapsulation efficiency across N/P ratios in the range 0.5–20. Briefly, a 1% agarose gel (Fisher Scientific) was prepared in 1× TBE buffer (Invitrogen, USA) containing SYBR® safe nucleic acid stain (Invitrogen, USA). The gel and tray were placed in an electrophoresis tank containing 1× TBE running buffer. 20 μl of each polyplex sample containing 6× gel loading dye was carefully loaded into each well alongside an appropriately sized DNA ladder (1Kb DNA Plus Ladder, Invitrogen, Ireland). The samples were then electrophoresed at 80 V for 40 minutes then visualised using Amersham Imager RGB 600 (GE Healthcare). To explore polyplex size and zeta potential, polyplexes were prepared in 50 μl volumes and diluted to 1 ml with molecular grade water. The polyplexes were assessed for diameter (nm) using nanoparticle tracking analysis (NTA) utilising a Nanosight NS3000 (Malvern, UK), while a zeta potential measurement (mV) was performed using a Malvern Zetasizer ZS 3000 (Malvern, UK). All techniques have been previously described.31
phosphate (N/P) ratios including, 0.5, 1, 2, 5, 10 and 20 (N/P represents the molar ratio of positively charged nitrogen (N) in the polyLys arms to negatively phosphate (P) group present in the pGlu molecules). The concentration of pGlu remained constant at 1 μg μl−1. Gel retardation assays were used to assess the encapsulation efficiency across N/P ratios in the range 0.5–20. Briefly, a 1% agarose gel (Fisher Scientific) was prepared in 1× TBE buffer (Invitrogen, USA) containing SYBR® safe nucleic acid stain (Invitrogen, USA). The gel and tray were placed in an electrophoresis tank containing 1× TBE running buffer. 20 μl of each polyplex sample containing 6× gel loading dye was carefully loaded into each well alongside an appropriately sized DNA ladder (1Kb DNA Plus Ladder, Invitrogen, Ireland). The samples were then electrophoresed at 80 V for 40 minutes then visualised using Amersham Imager RGB 600 (GE Healthcare). To explore polyplex size and zeta potential, polyplexes were prepared in 50 μl volumes and diluted to 1 ml with molecular grade water. The polyplexes were assessed for diameter (nm) using nanoparticle tracking analysis (NTA) utilising a Nanosight NS3000 (Malvern, UK), while a zeta potential measurement (mV) was performed using a Malvern Zetasizer ZS 3000 (Malvern, UK). All techniques have been previously described.31
Results and discussion
Synthesis and characterisation of star PLL from bis-MPA dendrimers
Three generations of bis-MPA hydroxy dendrimers were prepared via esterification of acetonide protected bis-MPA (Scheme S1†) and pentaerythritol (PERT) as a tetrafunctional core following a literature procedure.27 The divergent growth was accomplished by addition of bis-MPA monomer ‘layer-by-layer’. Due to the unique 1H NMR resonances of both bis-MPA and PERT, it was possible to confirm the successful synthesis and purity of three generations of hydroxy terminated dendrimers corresponding to 8, 16 and 32 terminal hydroxy groups (Fig. S5–S10†). Typically, controlled NCA ROP polymerisation is initiated by nucleophiles, mostly primary amines. This offers straightforward control over molecular weight through the initiator (amine) to monomer (NCA) ratio.32 Initiation from weaker nucleophiles such as hydroxy groups is rarely reported due to their low nucleophilicity relative to primary amines. Indeed, when the direct polymerisation of ε-carbobenzyloxy-L-lysine N-carboxyanhydride (ZLys NCA) from the native 8-arm bis-MPA dendrimer was attempted, reactions were slow producing polymers with relatively high dispersity (ĐM = 1.35) implying an uncontrolled polymerisation (Fig. S11†). This result was not unexpected for the initiation from a weak nucleophile, although two recent reports disclosed some success for the initiation from hydroxy groups using 1,1,3,3-tetramethylguanidine (TMG) as a catalyst for N-substituted and unsubstituted NCAs.33,34 In the present study, a different strategy is proposed by converting the bis-MPA dendrimer hydroxy groups into amino groups. Following Stenstorm's protocol, the functionalisation of hydroxyl-terminated bis-MPA dendrimers was performed by addition of imidazole-activated tert-butyloxycarbonyl (Boc) protected β-alanine (Scheme S2†).28 Subsequent deprotection by trifluoroacetic acid (TFA) yielded the three generations of amino terminated dendrimers with 8, 16, and 32 amino end groups as ammonium trifluoroacetate salts (denoted as G1-(8), G2-(16) and G3-(32)). To compare the efficiency of chemically identical, but architecturally different initiators, a commercially available second generation of bis-MPA hyper-branched polyester was purchased and post-functionalised using the same synthetic strategy as for the dendrimers (G2HB-(16)). 1H and 13C NMR spectra of the dendrimers and hyper-branched polymer (Fig. S12–S19†) confirmed the presence of both β-alanine and bis-MPA moieties in all samples verifying quantitative functionalisation with amino acid units. Specifically, for the G1-(8) the β-alanine methylene proton resonance at 2.81 and 3.24 ppm are clearly distinguishable from the methyl signals of bis-MPA segment at 1.32 ppm and the PERT methylene groups at 4.22 ppm. GPC results (Fig. S20†) confirmed monodisperse molecular weight distributions with increasing Mn as a function of dendrimer generation (Table S1†).It was hypothesised that the ROP polymerisation of ZLys NCA can be directly initiated from the ammonium trifluoroacetate terminated dendrimers (Scheme 1). This was based on previous reports on NCA initiation from tertiary amino salts. The concept was first demonstrated by Dimirtov and Schlaad using hydrochloride salt of ω-amino terminated polystyrene.35 The ammonium chloride chain ends were found to be a dormant species, while their dissociation affords the ROP of NCA through the nucleophile attack by the primary amines. The authors later proposed further modification of the procedure by introducing tertiary amines as the catalysts.36,37 Additionally, ammonium hydrochloride salts were used in the preparation of block copolymers.38 Tetrafluoroborate salts were also used successfully for the initiation of γ-benzyl-L-glutamate from different amines.39,40 However, to date, the concept of using ammonium salts for the initiation of the ROP of NCAs has not been applied in the synthesis of complex polypeptides architectures. Moreover, ammonium trifluoroacetate salts have not been investigated for their ability to initiate NCA polymerisation hitherto. Successful initiation would open a straightforward route from readily obtained β-alanine functionalised bis-MPA dendrimers to star polypeptides omitting any neutralisation of the ammonium groups after Boc deprotection.
 | ||
| Scheme 1 Synthesis of PLL star polypeptides by NCA polymerisation using three generations bis-MPA dendrimers as the initiator. | ||
All NCA polymerisations were performed at 40 °C due to the increased dissociation rate of the ammonium salts at elevated temperatures.41 Initially the synthesis of G1-(8)-PZLL5 (8 arms; 5 ZLys units per arm) was carried out (Scheme 1). The monomer conversion was monitored by FT-IR spectroscopy following the NCA signature bands at 1852 and 1786 cm−1 attributed to the anhydride C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group (Fig. S21†). By comparison of the 1786 cm−1 band intensity with that of an unchanging band at 1255 cm−1, the monomer conversion was estimated. As can be seen in Fig. 1 after an initial conversion of about 30% within the first 2 h, the NCA conversion increased linearly to reach quantitative conversion after 72 h for all three samples. GPC traces of samples taken within the 72 h time interval confirmed a steady increase in molecular weight and highly symmetrical monomodal distributions verifying a well-controlled polymerisation process (Fig. 1). Extending the reaction time beyond 72 h did not result in significantly further molecular weight increase corroborating the FT-IR spectroscopic analysis. Similar results were observed for the reactions with second and third generation dendrimers and the second generation of the hyper-branched bis-MPA polymer when targeting 5 (Z)Lys units per arm. However, increasing the target degree of polymerisation (DP) per arm to 10 or more required longer reaction times.
O group (Fig. S21†). By comparison of the 1786 cm−1 band intensity with that of an unchanging band at 1255 cm−1, the monomer conversion was estimated. As can be seen in Fig. 1 after an initial conversion of about 30% within the first 2 h, the NCA conversion increased linearly to reach quantitative conversion after 72 h for all three samples. GPC traces of samples taken within the 72 h time interval confirmed a steady increase in molecular weight and highly symmetrical monomodal distributions verifying a well-controlled polymerisation process (Fig. 1). Extending the reaction time beyond 72 h did not result in significantly further molecular weight increase corroborating the FT-IR spectroscopic analysis. Similar results were observed for the reactions with second and third generation dendrimers and the second generation of the hyper-branched bis-MPA polymer when targeting 5 (Z)Lys units per arm. However, increasing the target degree of polymerisation (DP) per arm to 10 or more required longer reaction times.
 | ||
| Fig. 1 Top: Conversion of ZLys NCA initiated from β-alanine ammonium trifluoroacetate bis-MPA dendrimers. Conversion was calculated from FTIR reduction of the 1786 cm−1 NCA band relative to the unchanged band at 1255 cm−1 (Fig. S21†). Bottom: GPC traces taken at different time points of the ZLys NCA polymerisation from the G1-(8) β-alanine ammonium trifluoroacetate bis-MPA dendrimer. | ||
Next, a set of star-shaped (Z)PLL with different degrees of branching and arm lengths was synthesised, whereby FT-IR spectroscopy was used to determine full monomer conversion. Three generations of (Z)PLL with 8, 16 and 32 arms and 5 Lys units per arm were targeted, denoted as G1-(8)-PLL5, G2-(16)-PLL5 G3-(32)-PLL5 and G2HB-(16)-PLL5, as well as a star-shaped polypeptide with 32 arms and 10 L-lysine units per arm, G3-(32)-PLL10. GPC analysis (Fig. 2) demonstrated that all resulting star-shaped polypeptides possess a monomodal and narrow molecular weight distribution (ĐM = 1.03–1.04). For the samples with constant degree of polymerisation (DP = 5) per arm, their molecular weight (MGPCn) gradually increased with increasing number of arms from 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 to 35
600 to 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 g mol−1 (Table 1). Similarly, for the two samples with constant number of arms (32), a shift to higher molecular weight was seen when the DP per arm was increased from 5 (MGPCn = 35
300 g mol−1 (Table 1). Similarly, for the two samples with constant number of arms (32), a shift to higher molecular weight was seen when the DP per arm was increased from 5 (MGPCn = 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 g mol−1) to 10 (MGPCn = 58
300 g mol−1) to 10 (MGPCn = 58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 g mol−1). Moreover, for all samples a clear shift of the traces from initiator to star polypeptide was observed (Fig. 2). The GPC trace of hyper-branched polypeptide followed the same trend, showing monodisperse molecular weight distribution (Đ = 1.15) even though the initiator was less structurally uniform than the corresponding dendrimers (Fig. S24†). 1H NMR spectra were in agreement with the polymer structures displaying signature peaks of the dendritic initiator and the (Z)PLL (Fig. S22†). Overall, the results emphasize a highly controlled polymerisation from the ammonium trifluoroacetate terminated multifunctional initiators giving rise to well-defined star (Z)PLL in which the molecular weight and number of arms can be readily controlled similar to our previously reported PPI core.12 It is hypothesised that the TFA anions behave as dormant species during the polymerisation of (Z)LL NCA as previously proposed in the literature.35,38,39
400 g mol−1). Moreover, for all samples a clear shift of the traces from initiator to star polypeptide was observed (Fig. 2). The GPC trace of hyper-branched polypeptide followed the same trend, showing monodisperse molecular weight distribution (Đ = 1.15) even though the initiator was less structurally uniform than the corresponding dendrimers (Fig. S24†). 1H NMR spectra were in agreement with the polymer structures displaying signature peaks of the dendritic initiator and the (Z)PLL (Fig. S22†). Overall, the results emphasize a highly controlled polymerisation from the ammonium trifluoroacetate terminated multifunctional initiators giving rise to well-defined star (Z)PLL in which the molecular weight and number of arms can be readily controlled similar to our previously reported PPI core.12 It is hypothesised that the TFA anions behave as dormant species during the polymerisation of (Z)LL NCA as previously proposed in the literature.35,38,39
| Sample | Arms | M theon (g mol−1) |
M
GPCn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (g mol−1) a (g mol−1) |
Đ
M![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b b |
DPtheo | DPNMR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b b |
|---|---|---|---|---|---|---|
| a GPC using HFiP at a flow rate of 1 mL min−1 (PMMA calibration) of the protected (Z)PLL. b Degree of polymerisation per arm obtained by 1H-NMR integration ratio between –CH2 of alanine at 2.70 ppm and PLL at 3.06 ppm after deprotection. | ||||||
| G1-(8)-PLL5 | 8 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 670 670 |
13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.03 | 5 | 5 |
| G2-(16)-PLL5 | 16 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640 640 |
22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
1.03 | 5 | 5 |
| G2HB-(16)-PLL5 | 16 | 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 694 694 |
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
1.15 | 5 | 6 |
| G3-(32)-PLL5 | 32 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 665 665 |
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
1.04 | 5 | 6 |
| G3-(32)-PLL10 | 32 | 89![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 636 636 |
58![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 400 |
1.03 | 10 | 11 |
The final synthetic step was the deprotection of the (Z)PLL arms with TFA and HBr (33 wt% in acetic acid), which was confirmed by the disappearance of the characteristic protecting group proton signals at 7.28 and 5.08 ppm (Fig. S22†). The degree of polymerisation (DP) was obtained from the 1H NMR spectra of the deprotected star PLL by the calculating the integrated peak area of the –CH2 signal from β-alanine at 2.70 ppm and the PLL signal at 3.06 ppm (Fig. 3). It was found that the calculated DPs were in good agreement with the targeted ones (Table 1). Notably, 1H NMR analysis did not suggest any degradation under the applied conditions. The results confirm that the proposed synthetic route gives access to well defined and structurally distinct PLL star polymers.
 | ||
| Fig. 3 1H NMR spectra of deprotected star PLL with arm length 5 and 10 in D2O. Peaks used for the calculation of the degree of polymerisation (DP) are highlighted. | ||
Core degradation
The motivation for replacing the PPI dendrimer with the bis-MPA dendrimer in the star PLL platform was to impart core degradability, while retaining the ability to form polyplexes with a gene cargo. In a recent study we have demonstrated the degradability of the PLL arms on PPI 8-arm star polypeptide by the proteolytic enzyme trypsin.42 In light of that, here we exclusively focused on investigating hydrolytic degradation of the bis-MPA core. Previously reported results of degradation studies on hydroxy terminated bis-MPA dendrimers showed fast degradation at physiological and basic pH with a first degradation fraction at pH 7.5 appearing after only 6 h.25 All degradation studies were standardised and carried out on G3-(32)-PLL10 in aqueous media at a concentration 0.3 mM for 6 days at 37 °C. The star PLL was subjected to hydrolytic degradation under acidic (pH 4.5), basic (pH 9.5) and physiological (pH 7.4) conditions. At the end of the degradation experiment aliquots of the reaction solutions were removed and lyophilised. The obtained solids were subject to diffusion ordered spectroscopy (DOSY) and GPC analysis. DOSY measures diffusion coefficients of individual components in a solution mixture.43,44 Due to diffusion being a function of molecular weight and hydrodynamic volume, we found DOSY measurements a convenient method to track the changes in molecular weight through changes in the diffusion coefficient. The DOSY contour plot of the starting sample and the materials isolated after degradation under the applied conditions is shown in Fig. 4A with diffusion coefficients listed in Table 2. The diffusion coefficient of freshly made G3-(32)-PLL10 was 2.55 × 10−7 cm2 s−1. An increase of diffusion coefficient of the initial star polymer was recorded for the sample subjected to degradation under physiological (9.10 × 10−7 cm2 s−1) and basic conditions (8.70 × 10−7 cm2 s−1) suggesting the formation of lower molecular weight molecules due to hydrolysis. When exposed to acidic hydrolysis a smaller increase in the diffusion coefficient (4.99 × 10−7 cm2 s−1) was seen. It is hypothesised that buffering by the Lys amines protonation causes the higher stability at low pH.| Sample |
D
NMR![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) a (10−7 cm2 s−1) a (10−7 cm2 s−1) |
M
GPCn prim.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b (g mol−1) b (g mol−1) |
M
GPCn secd.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (g mol−1) c (g mol−1) |
|---|---|---|---|
| a Diffusion coefficient in D2O calculated from intensity of methylene signal of L-lysine side chain using a mono-exponential decay equation. b Number-average molecular weight of primary GPC trace. c Number-average molecular weight of secondary GPC trace. d Diffusion coefficient in D2O calculated from intensity of methylene signal of β-alanine moiety of the dendrimer using a mono-exponential decay equation. e Diffusion coefficient in D2O calculated from intensity of methylene signal of bis-MPA moiety of the dendrimer using a mono-exponential decay equation. | |||
| G3-(32)-PLL10 | 2.55 | 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 630 630 |
— |
| pH 4.5 | 4.99 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
5500 |
| pH 7.4 | 9.10 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
5000 |
| pH 9.5 | 8.70 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 510 510 |
5150 |
| G3-(32)-NH3+ | 10.87d | 6530 | — |
| G3-(32)-OH | 16.86e | 5310 | — |
GPC measurements corroborate the DOSY results. Fig. 4B depicts a clear breakdown of the G3-(32)-PLL10 starting sample evident from the second lower molecular weight maximum after degradation under basic and physiological conditions, while absent under acid conditions. Previous studies on β-alanine functional bis-MPA dendrimers showed that the bond between the β-alanine and the dendrimer is preferably hydrolysed as it is the most accessible labile bond.45 It is reasonable that this is also the case for the star polypeptides as the PLL arms are stable under the hydrolysis conditions. While at this point somewhat speculative, this would result in the initial detachment of PLL arms which are then susceptible to further proteolytic degradation and full in vivo clearance.
Pharmaceutical characterisation and pDNA loading assessment
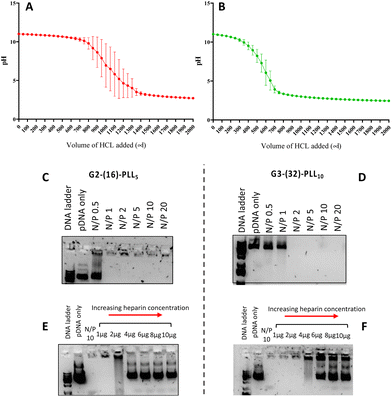
The buffering capacities of selected star PLL were investigated to determine their suitability as gene delivery vectors. The buffering capacity is a good indicator of the material's ability to facilitate a “proton sponge” effect upon cell internalisation by endocytosis.46 From a range of polypeptides synthesised, G2-(16)-PLL5 and G3-(32)-PLL10, were selected as the reference star polypeptide materials in the following studies. As shown in Fig. 5A and B, G2-(16)-PLL5 demonstrated higher buffering capacity than G3-(32)-PLL10. G2-(16)-PLL5 required 1187 ± 171 μl of 0.1 N HCL to reduce the pH to 4 versus G3-(32)-PLL10 which required 681 ± 48 μl of 0.1 N HCl to reduce the pH to 4. The potential capacity of cationic polymers to facilitate gene delivery is primarily attributed to the presence of titratable amine groups within the pH range of 5–7. The presence of these amines in the material structure promotes a “proton sponge effect” in addition to the polymer's ability to volumetrically expand at lower pH 5–6 via the so-called “umbrella hypothesis”. Together, these effects lead to endosome rupture and the prompt release of internalised polyplexes.47 Thus the result obtained herein would suggest that G2-(16)-PLL5 is more likely to function as better proton sponge in the acidic lysosomal environment once taken up by cells compared to G3-(32)-PLL10.To investigate the suitability of the bis-MPA star PLL polypeptides as gene delivery vectors the materials were complexed with plasmid DNA (pGLu) across a range of N/P ratios to form star PLL-pGLu polyplexes and the physicochemical properties of the polyplexes characterised. N/P represents the molar ratio of positively charged nitrogen (N) in the PLL arms to negatively phosphate (P) group present in the pGlu molecules. G2-(16)-PLL5 and G3-(32)-PLL10 polymers formed negatively charged polyplexes with pDNA at N/P 1 but positively charged polyplexes with pDNA when complexed at N/P 10 indicating complete condensation of the negatively charged pGlu at the higher N/P ratio. In addition, all polyplexes had a particle size less than 150 nm in diameter as measured with Nanoparticle Tracking Analysis (NTA) which is widely quoted as being optimal for effective nanoparticle uptake via endocytosis (Table 3).48
| Sample | N/P ratioa | Sizeb (nm) | Zeta potential (mV) |
|---|---|---|---|
| a N/P ratio is the molar ratio of positively charged nitrogen (N) arms present in the bis-MPA-PLL polymer to negatively phosphate (P) group present in the pGlu molecules. b Polyplex diameter (nm) measured using nanoparticle tracking analysis (NTA). | |||
| G2-(16)-PLL5 | 1 | 129 ± 45 | −13 ± 3 |
| G2-(16)-PLL5 | 10 | 120 ± 36 | 31 ± 3 |
| G3-(32)-PLL10 | 1 | 125 ± 54 | −14 ± 3 |
| G3-(32)-PLL10 | 10 | 142 ± 55 | 33 ± 4 |
The encapsulation efficiency of pGLu was further confirmed using gel retardation assays. The G2-(16)-PLL5 polymer achieved complete complexation of the anionic pGluc from N/P 1 (though not at N/P 0.5) as indicated by clear lanes on the agarose gel for all N/P ratios from 1–20 (Fig. 5C). The polyplexes formulated with G3-(32)-PLL10 achieved full complexation of the plasmid above N/P 2 but free (uncomplexed) pDNA was still evident in the gel at N/P 0.5 and N/P 1 (Fig. 5D). These results demonstrate complete complexation of pDNA at relatively low N/P ratios. It can be concluded that star bis-MPA-PLLs exhibit a similar pDNA complexation ability as the star PPI-PLLs with a similar structural composition.11 To date, very few studies have investigated the capacity of bis-MPA dendrimers to complex with genetic materials. One study by Movellan et al. demonstrated for the first time the ability of bis-MPA dendrimers to condense pDNA.49 However, very high N/P ratios were required to achieve full complexation with the dendrimer alone. In the study, a generation 2 bis-MPA with 16 terminal amino groups required a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80
80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 w/w ratio, which is reported as a N/P ratio of 16
000 w/w ratio, which is reported as a N/P ratio of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 161, to fully condense the pDNA. On the other hand, the ionic analogue, ammonium-carboxylate bis-MPA, was capable of complexing pDNA at 1
161, to fully condense the pDNA. On the other hand, the ionic analogue, ammonium-carboxylate bis-MPA, was capable of complexing pDNA at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 w/w ratio that corresponds to N/P 100. In both cases, this is a striking difference in terms of N/P ratio versus our results and substantiates the significant role that the PLL arms likely play when it comes to nucleic acid complexation and condensation. In addition, results by Stenstrom et al. showed that second to fourth generations of ammonium trifluoroacetate bis-MPA dendrimers, with β-alanine moiety, at the periphery of the macromolecule, were able to complex small interfering RNA (siRNA) at N/P 1.5–3, which is similar to our results with pDNA and again, emphasises the importance of post-functionalisation of the bis-MPA structure for gene complexation.50 The polyplex integrity, i.e. stability and ability of pDNA to dissociate from polyplexes, was further examined in the presence of a competing polyanion (heparin). Heparin is a large anionic polysaccharide found in the extracellular matrix that can compete with nucleic acid binding and disrupt polyplex stability.51 As shown in Fig. 5E, pDNA-G2-(16)-PLL5 polyplexes were destabilised in the presence of heparin at all tested concentrations, however, complete dissociation was only observed at heparin dose 2 μg or higher. While pDNA-G3-(32)-PLL10 polyplexes were stable up to 4 μg heparin (Fig. 5F). These findings indicate that pDNA was effectively condensed by bis-MPA star polymers and that the stability of the polyplexes in the presence of a physiologically relevant molecule was dependent on the molecular structure of the bis-MPA star polypeptide. Polyplexes formed with bis-MPA star polypeptides synthesised with a greater number of arms i.e. G3 (32) were more stable as evidenced by the increased dose of heparin required to dissociate pDNA from these polyplexes. Similar studies were performed with PPI-based star polypeptides complexed with pDNA polyplexes and found that dissociation of pDNA was evident around the 4 μg dose of heparin.14
10 w/w ratio that corresponds to N/P 100. In both cases, this is a striking difference in terms of N/P ratio versus our results and substantiates the significant role that the PLL arms likely play when it comes to nucleic acid complexation and condensation. In addition, results by Stenstrom et al. showed that second to fourth generations of ammonium trifluoroacetate bis-MPA dendrimers, with β-alanine moiety, at the periphery of the macromolecule, were able to complex small interfering RNA (siRNA) at N/P 1.5–3, which is similar to our results with pDNA and again, emphasises the importance of post-functionalisation of the bis-MPA structure for gene complexation.50 The polyplex integrity, i.e. stability and ability of pDNA to dissociate from polyplexes, was further examined in the presence of a competing polyanion (heparin). Heparin is a large anionic polysaccharide found in the extracellular matrix that can compete with nucleic acid binding and disrupt polyplex stability.51 As shown in Fig. 5E, pDNA-G2-(16)-PLL5 polyplexes were destabilised in the presence of heparin at all tested concentrations, however, complete dissociation was only observed at heparin dose 2 μg or higher. While pDNA-G3-(32)-PLL10 polyplexes were stable up to 4 μg heparin (Fig. 5F). These findings indicate that pDNA was effectively condensed by bis-MPA star polymers and that the stability of the polyplexes in the presence of a physiologically relevant molecule was dependent on the molecular structure of the bis-MPA star polypeptide. Polyplexes formed with bis-MPA star polypeptides synthesised with a greater number of arms i.e. G3 (32) were more stable as evidenced by the increased dose of heparin required to dissociate pDNA from these polyplexes. Similar studies were performed with PPI-based star polypeptides complexed with pDNA polyplexes and found that dissociation of pDNA was evident around the 4 μg dose of heparin.14
Overall, these results would suggest that different generations of star bis-MPA-PLL hybrids have the ability to effectively complex pDNA into nanoparticles that have properties suitable for cellular transfection.52
Conclusions
In this work, the synthesis of well-defined star-shaped PLL by ring opening polymerisation of (Z)LL NCA from three generations of bis-MPA ammonium terminated dendrimers as well as a second generation of bis-MPA hyper-branched polymer was demonstrated. The ability of efficient core degradation was confirmed under physiological and basic conditions. Moreover, preliminary pDNA complexation studies demonstrate successful polyplex formation between the gene cargo and the star PLL. The results suggest that the bis-MPA star PLL platform is a promising fully degradable alternative to leading non-degradable polymeric vectors. Additional studies in cell transfection are underway and will further inform the potential of this platform as gene vectors.Author contributions
S. S.: investigation, methodology, writing original draft, formal analysis. K. McC.: investigation, formal analysis. S. F.: investigation, formal analysis. R. B.: supervision, writing – review and editing. S.-A. C.: conceptualisation, funding acquisition, supervision, writing – review and editing. A. H.: conceptualisation, funding acquisition, supervision, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work received funding from the Science Foundation Ireland (SFI) Investigators Program under grand code 13/IA/1840 and Science Foundation Ireland (SFI) Centre for Advanced Materials and BioEngineering Research (AMBER) under grant code SFI 12/RC/2278_P2.References
- E. Papanikolaou and A. Bosio, Front. Genome Ed., 2021, 3, 618346 CrossRef PubMed.
- K. Gersbach and C. A. Bulaklak, Nat. Commun., 2020, 11, 5820 CrossRef PubMed.
- M. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259 CrossRef CAS PubMed.
- A. S. Chauhan, Molecules, 2018, 23, 938 CrossRef PubMed.
- D. Luo, K. Haverstick, N. Belcheva, E. Han and W. M. Saltzman, Macromolecules, 2002, 35, 3456 CrossRef CAS.
- S. Taranejoo, J. Liu, P. Verma and K. Hourigan, J. Appl. Polym. Sci., 2015, 132, 42096 CrossRef.
- A. Kargaard, J. P. G. Sluijter and B. Klumperman, J. Controlled Release, 2019, 316, 263 CrossRef CAS PubMed.
- N. Shao, Y. Su, J. Hu, J. Zhang, H. Zhang and Y. Cheng, Int. J. Nanomed., 2011, 6, 3361 CAS.
- A. Janaszewska, J. Lazniewska, P. Trzepiński, M. Marcinkowska and B. Klajnert-Maculewicz, Biomolecules, 2019, 9, 330 CrossRef CAS PubMed.
- E. Haladjova, S. Halacheva, V. Posheva, E. Peycheva, M.-V. Doumanova, T. Topouzova-Hristova, J. Doumanova and S. Rangelov, Langmuir, 2015, 31, 10017 CrossRef CAS PubMed.
- M. Byrne, D. Victory, A. Hibbitts, M. Lanigan, A. Heise and S.-A. Cryan, Biomater. Sci., 2013, 1, 1223 RSC.
- M. Byrne, R. Murphy, A. Kapetanakis, J. Ramsey, S.-A. Cryan and A. Heise, Macromol. Rapid Commun., 2015, 36, 1862 CrossRef CAS PubMed.
- M. Byrne, P. D. Thornton, S.-A. Cryan and A. Heise, Polym. Chem., 2012, 3, 2825 RSC.
- D. P. Walsh, R. D. Murphy, A. Panarella, R. M. Raftery, B. Cavanagh, J. C. Simpson, F. J. O'Brien, A. Heise and S.-A. Cryan, Mol. Pharm., 2018, 15, 1878 CrossRef CAS PubMed.
- D. P. Walsh, A. Heise, R. M. Raftery, R. Murphy, G. Chen, F. J. O'Brien and S.-A. Cryan, Biomater. Sci., 2021, 9, 4984 RSC.
- D. P. Walsh, R. M. Raftery, I. Mencía Castañob, R. Murphy, B. Cavanag, A. Heise, F. J. O'Brien and S.-A. Cryan, J. Controlled Release, 2019, 304, 191 CrossRef CAS PubMed.
- H. Kobayashi, S. Kawamoto, T. Saga, N. Sato, A. Hiraga, T. Ishimori, Y. Akita, M. H. Mamede, J. Konishi, K. Togashi and M. W. Brechbiel, Magn. Reson. Med., 2001, 46, 795 CrossRef CAS PubMed.
- E. M. M. de Brabander-van den Berg and E. W. Meijer, Angew. Chem., Int. Ed. Engl., 1993, 32, 1308 CrossRef.
- J. Li, J. Li, S. Xu, D. Zhang and D. Liu, Colloids Surf., B, 2013, 110, 183 CrossRef CAS PubMed.
- K. Inoue, S. Horibe, M. Fukae, T. Muraki, E. Ihara and H. Kayama, Macromol. Biosci., 2003, 3, 26 CrossRef CAS.
- H. A. Klok, J. R. Hernández, S. Becker and K. Müllen, J. Polym. Sci., Part A: Polym. Chem., 2001, 39, 1572 CrossRef CAS.
- D. Ma, Z. H. Liu, Q. Q. Zheng, X. Y. Zhou, Y. Zhang, Y. F. Shi, J. T. Lin and W. Xue, Macromol. Rapid Commun., 2013, 34, 548 CrossRef CAS PubMed.
- S. J. Lam, A. Sulistio, K. Ladewig, E. H. H. Wong, A. Blencowe and G. G. Qiao, Aust. J. Chem., 2014, 67, 592 CrossRef CAS.
- S. J. Lam, E. H. H. Wong, N. M. O'Brien-Simpson, N. Pantarat, A. Blencowe, E. C. Reynolds and G. G. Qiao, ACS Appl. Mater. Interfaces, 2016, 8, 33446 CrossRef CAS PubMed.
- N. Feliu, M. V. Walter, M. I. Montañez, A. Kunzmann, A. Hult, A. Nyström, M. Malkoch and B. Fadeel, Biomaterials, 2012, 33, 1970 CrossRef CAS PubMed.
- S. García-Gallego, D. Hult, J. V. Olsson and M. Malkoch, Angew. Chem., Int. Ed., 2015, 54, 2416 CrossRef PubMed.
- P. Stenstrom, O. C. Andren and M. Malkoch, Molecules, 2016, 21, 366 CrossRef PubMed.
- P. Stenstrom, E. Hjorth, Y. Zhang, O. C. J. Andren, S. Guette-Marquet, M. Schultzberg and M. Malkoch, Biomacromolecules, 2017, 18, 4323 CrossRef PubMed.
- P. Groves, Polym. Chem., 2017, 8, 6700 RSC.
- J. Shi, J. G. Schellinger, R. N. Johnson, J. L. Choi, B. Chou, E. L. Anghel and S. H. Pun, Biomacromolecules, 2013, 14, 1961 CrossRef CAS PubMed.
- D. P. Walsh, A. Heise, F. J. O'Brien and S.-A. Cryan, Gene Ther., 2017, 24, 681 CrossRef CAS PubMed.
- A. R. Mazo, S. Allison-Logan, F. Karimi, N. Jun-An Chan, W. Qiu, W. Duan, N. M. O'Brien-Simpson and G. G. Qiao, Chem. Soc. Rev., 2020, 49, 4737 RSC.
- B. A. Chan, S. Xuan, M. Horton and D. Zhang, Macromolecules, 2016, 49, 2002 CrossRef CAS.
- K. Li, Z. Li, Y. Shen, X. Fu, C. Chen and Z. Li, Polym. Chem., 2022, 13, 586 RSC.
- I. Dimitrov and H. Schlaad, Chem. Commun., 2003, 2944 RSC.
- C. D. Vacogne and H. Schlaad, Chem. Commun., 2015, 51, 15645 RSC.
- C. D. Vacogne and H. Schlaad, Polymer, 2017, 124, 203 CrossRef CAS.
- J. F. Lutz, D. Schütt and S. Kubowicz, Macromol. Rapid Commun., 2005, 26, 23 CrossRef CAS.
- I. Conejos-Sánchez, A. Duro-Castano, A. Birke, M. Barz and M. J. Vicent, Polym. Chem., 2013, 4, 3182 RSC.
- A. Duro-Castano, R. M. England, D. Razola, E. Romero, M. Oteo-Vives, M. A. Morcillo and M. J. Vicent, Mol. Pharm., 2015, 12, 3639 CrossRef CAS PubMed.
- Y. Knobler, S. Bittner and M. Frankel, J. Chem. Soc., 1964, 3941 RSC.
- D. Skoulas, S. Fattah, D. Wang, S.-A. Cryan and A. Heise, Macromol. Biosci., 2022, 2200175 CrossRef CAS PubMed.
- W. Li, H. Chung, C. Daeffler, J. A. Johnson and R. H. Grubbs, Macromolecules, 2012, 45, 9595 CrossRef CAS PubMed.
- P. Groves, Polym. Chem., 2017, 8, 6700 RSC.
- E. F. Durán-Lara, J. L. Marple, J. A. Giesen, Y. Fang, J. H. Jordan, W. T. Godbey, A. Marican, L. S. Santos and S. M. Grayson, Biomacromolecules, 2015, 16, 3434 CrossRef PubMed.
- J.-P. Behr, CHIMIA Int. J. Chem., 1997, 51, 34 CrossRef CAS.
- J. Nguyen and F. C. Szoka, Acc. Chem. Res., 2012, 45, 1153 CrossRef CAS PubMed.
- D. J. Gary, N. Puri and Y. Y. Won, J. Controlled Release, 2007, 121, 64 CrossRef CAS PubMed.
- J. Movellan, R. Gonzalez-Pastor, P. Martin-Duque, T. Sierra, J. M. de la Fuente and J. L. Serrano, Macromol. Biosci., 2015, 15, 657 CrossRef CAS PubMed.
- P. Stenstrom, D. Manzanares, Y. Zhang, V. Cena and M. Malkoch, Molecules, 2018, 23, 2028 CrossRef PubMed.
- J. McCarthy, M. J. O'Neill, L. Bourre, D. Walsh, A. Quinlan, G. Hurley, J. Ogier, F. Shanahan, S. Melgar, R. Darcy and C. M. O'Driscoll, J. Controlled Release, 2013, 168, 28 CrossRef CAS PubMed.
- J. Rejman, V. Oberle, I. S. Zuhorn and D. Hoekstra, Biochem. J., 2004, 377, 159 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: bis-MPA dendrimer synthesis and characterisation, additional polymer NMR and GPC analysis. See DOI: https://doi.org/10.1039/d3py00346a |
| This journal is © The Royal Society of Chemistry 2023 |