Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Beyond iron: metal-binding activity of the Pseudomonas quinolone signal-motif†
Dávid
Szamosvári
a,
Viktoriia
Savchenko
ab,
Natalie
Badouin
c and
Thomas
Böttcher
 *a
*a
aFaculty of Chemistry, Institute for Biological Chemistry & Centre for Microbiology and Environmental Systems Science, Department of Microbiology and Ecosystems Science, University of Vienna, Josef-Holaubek-Platz 2 (UZA II), 1090 Vienna, Austria. E-mail: thomas.boettcher@univie.ac.at
bVienna Doctoral School in Chemistry (DoSChem), University of Vienna, Währinger Str. 42, 1090 Vienna, Austria
cUniversity of Konstanz, 78457 Konstanz, Germany
First published on 30th May 2023
Abstract
The Pseudomonas quinolone signal (PQS) is an important quorum sensing signal controlling virulence of the human pathogen Pseudomonas aeruginosa. PQS also exhibits multiple additional biological functions for P. aeruginosa, including the trapping of ferric iron. The PQS-motif has proven as privileged structure with great potential which is why we here explored the synthesis of two different types of crosslinked dimeric PQS-motifs as potential iron chelators. These compounds indeed chelated ferric iron and produced colorful and fluorescent complexes also with other metal ions. Inspired by these results, we revisited the metal ion binding capabilities of the natural product PQS and were able to detect further metal complexes beyond ferric iron and confirm the complex stoichiometry by mass spectrometry.
Introduction
The opportunistic, nosocomial human pathogen Pseudomonas aeruginosa is known for its biosynthesis of a vast variety of small molecules as secondary metabolites and virulence factors that contribute to its survival and pathogenicity in the human host.1 A prime example of a small molecule with such diverse biological functions is the main quorum sensing molecule of the Pseudomonas quinolone signaling system (2-heptyl-3-hydroxy-4(1H)-quinolone, PQS). While its biosynthetic precursor 2-heptyl-4(1H)-quinolone (HHQ) is primarily acting as a quorum sensing signal, PQS is a multifunctional molecule with many additional functions beyond signaling. For example, PQS is involved in outer membrane vesicle formation and can trap ferric iron as a bidentate ligand in the vicinity of cells of P. aeruginosa. Iron trapping eventually induces the production of siderophores via iron scarcity (Fig. 1).2 Chemical modifications of the PQS scaffold already proved PQS to be a promising structural starting point for the generation of compounds with novel bioactive properties.3 Here we report the synthesis of metal chelating compounds with two covalently linked PQS motifs that led to the identification of metal binding capabilities of PQS beyond ferric iron. In addition to the previously known ferric iron complexes of PQS, we could identify so far undescribed PQS–molybdate and PQS–aluminum complexes by LC-HRMS as well as a suspected vanadate complex. The identification of PQS as a versatile metallophore reveals unprecedented insights into the multifunctional roles of PQS for the survival and metal homeostasis of P. aeruginosa. | ||
| Fig. 1 Structures of the quinolone quorum sensing signals HHQ and PQS of Pseudomonas aeruginosa and congeners with different chain lengths. PQS is a bidentate ligand for ferric iron. | ||
Results and discussion
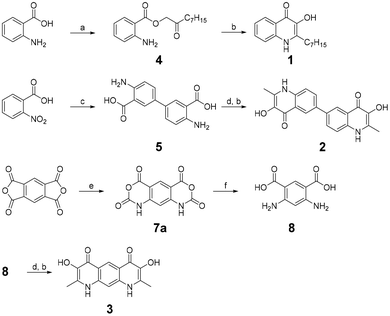
As one of our research interests revolves around the synthesis of PQS derivatives and analogs to study their bioactivity and influence on bacterial population behavior,3,4 we envisioned to synthetically explore structures with a joined dimeric PQS motif. Therefore, we designed scaffolds that represent PQS analogs connected either by a carbon–carbon bond at position 6 (2) or by a shared aromatic system (3). As PQS and derivatives are known to exhibit a generally poor solubility we decided to replace the hydrophobic heptyl side chains of the linked dimeric PQS analogs with methyl groups to improve solubility properties.5 The synthesis of these undescribed compounds was adapted from the synthesis of PQS molecules as reported by Hradil et al.6 where 2-aminobenzoic acid is esterified to 2-oxononyl 2′-aminobenzoate (4) and further cyclized to PQS (1) (Scheme 1). Similarly, we started with the synthesis of compound 2 by the generation of 4,4′-diamino-[1,1′-biphenyl]-3,3′-dicarboxylic acid (5) via reduction of 2-nitrobenzoic acid and subsequent benzidine rearrangement.7 The 2-aminobenzoic acid motifs thus obtained were esterified with chloroacetone and cyclized to the PQS analog 3,3′-dihydroxy-2,2′-dimethyl-[6,6′-biquinoline]-4,4′(1H,1′H)-dione (2). The synthesis of compound 3 started with a double nitrogen insertion in pyromellitic dianhydride by its reaction with trimethylsilyl azide (TMSA) as described for phthalic anhydrides.8 The resulting complex mixture of reaction products required extensive purification efforts and only one regioisomer, 1,9-dihydro-2H,4H-benzo[1,2-d:5,4-d′]bis([1,3]oxazine)-2,4,6,8-tetraone (7a), could be obtained reliably and in practical amounts. Following ring-opening by hydrolysis resulted in 4,6-diaminoisophthalic acid (8) which was esterified and cyclized to 3,7-dihydroxy-2,8-dimethylpyrido[3,2-g]quinoline-4,6(1H,9H)-dione (3) as described for compound 2 (Scheme 1). Since PQS is poorly water soluble and in its native environment secreted in the form of membrane vesicles, we used DMSO as organic solvent system to increase solubility. Qualitative investigations on the metal ion complexing properties of 2 and 3 revealed that both compounds form colorful complexes with Fe3+ ions in DMSO. Similar to PQS, compound 2 formed a blue complex whereas compound 3 formed a greenish-brown color. Upon these initial observations, we investigated the behavior of 2 and 3 with other di- and trivalent metal ions (Al3+, Mn2+, Cu2+, Co2+, Zn2+, Ni2+) and the oxoanions molybdate (MoO42−) and vanadate (VO43−). UV absorption spectroscopy showed that compound 2 changes its UV-absorption spectra when incubated with an excess of Fe3+, Al3+, Cu2+, MoO42− and VO43− although a change in color of the, otherwise colorless, solution of 2 was only observed for Fe3+ and MoO42− (Fig. 2A and C). Complex formation of 2 with Fe3+ and MoO42− was also observed by a change in the fluorescence under UV light of 365 nm (Fig. 2C and S8†). In contrast, the otherwise yellow solution of compound 3, with maximum absorption at 445 nm and bright yellow fluorescence under 365 nm, showed a rapid change in color when incubated with the same ions that form a complex with compound 2 (Fig. 2B and D) and a much more pronounced change in the fluorescence was observed when 3 was incubated with Fe3+, Al3+, MoO42− and VO43− (Fig. 2D). Complexes with other metal ions were not detected by UV absorption spectroscopy. These results inspired us to revisit the metal chelating capabilities of PQS of P. aeruginosa in more detail. Surprisingly no other metal ions or oxoanions of metals, apart from ferric iron, have been described to form complexes with PQS. Therefore, we preselected ions capable to form complexes with 2 and 3 (Fe3+, Al3+, Cu2+, MoO42− and VO43−), and investigated potential complex formation with PQS.Comparable to 2, a change in color of the otherwise colorless PQS solution was only observed for Fe3+ and MoO42− while minor changes in fluorescence were observable for all ions tested (Fig. 3F). To identify and confirm possible PQS-complexes, we subsequently used LC-HRMS analysis of the PQS complex formation experiments (Fig. 3). Analysis of the ferric iron spiked PQS sample revealed the m/z-values of two distinct PQS–Fe3+ complexes (572.2335 Da and 831.3920 Da) that can be assigned to Fe(PQS)2+ and Fe(PQS)3 + H+ (Fig. 3A and B). These complexes have been reported before by LR-MS9 and according ferric iron complexes are known for the methyl derivative of PQS (MPQS).5 Interestingly, the PQS–Al3+ sample showed a similar 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 3
1 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric ratio of PQS and Al3+. Here, we detected the m/z-values of 543.2797 Da and 802.4307 Da corresponding to Al(PQS)2+ and Al(PQS)3 + H+ (Fig. 3C and D). In the case of MoO42−, we could detect a mass of 647.2013 Da that represents the MoO2(PQS)2 + H+ complex (Fig. 3E). Although, the measurement of PQS in a mixture with VO43− ions clearly shows the detection of unique mass signals likely corresponding to PQS–vanadate complexes, an unambiguous assignment of the m/z-values to a complex structure was not possible (Fig. S7†). Surprisingly, even though measurements of the UV-Vis spectra of 2 and 3 with Cu2+ ions indicated complex formation by a significant change in the absorption spectra (Fig. 2), we could not detect the presence of a PQS–Cu2+ complex by LC-HRMS. Our results hence demonstrate that PQS is capable of forming complexes with metals beyond ferric iron, including Al3+, MoO42−, and likely VO43−.
1 stoichiometric ratio of PQS and Al3+. Here, we detected the m/z-values of 543.2797 Da and 802.4307 Da corresponding to Al(PQS)2+ and Al(PQS)3 + H+ (Fig. 3C and D). In the case of MoO42−, we could detect a mass of 647.2013 Da that represents the MoO2(PQS)2 + H+ complex (Fig. 3E). Although, the measurement of PQS in a mixture with VO43− ions clearly shows the detection of unique mass signals likely corresponding to PQS–vanadate complexes, an unambiguous assignment of the m/z-values to a complex structure was not possible (Fig. S7†). Surprisingly, even though measurements of the UV-Vis spectra of 2 and 3 with Cu2+ ions indicated complex formation by a significant change in the absorption spectra (Fig. 2), we could not detect the presence of a PQS–Cu2+ complex by LC-HRMS. Our results hence demonstrate that PQS is capable of forming complexes with metals beyond ferric iron, including Al3+, MoO42−, and likely VO43−.
Transition metal elements, including Co, Cu, Fe, Mg, Mn, Mo, Ni, and Zn ions, are essential micronutrients for all organisms.10 As cofactors in enzymes, they facilitate catalytic reactions, contribute to the structural integrity and stability of proteins, and are crucial for electron transport systems.
The low abundance and bioavailability of these trace elements forced bacteria to develop a multitude of acquisition mechanisms like metallophores, metal-recruiting proteins, and metal-complex transporters, to secure a sufficient supply of these nutrients from the environment.11 Especially during a bacterial infection, the host can sequester essential trace metal ions and thereby actively restrict their availability to the bacteria, which is known as nutritional immunity. Overcoming this limitation is decisive for the survival and virulence of the pathogen and the course of the infection.12 In P. aeruginosa, especially the intake mechanisms of ferric and ferrous iron are well characterized.13 Thereby, PQS plays an important role by acting as a chelating agent for ferric iron in a proposed 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 or 3
1 or 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex which we confirmed by our LC-HRMS analysis. Contrary to siderophores, PQS serves as an iron trap since its ferric iron complexes cannot be taken up into the bacteria directly but instead accumulate in the cell envelope and outer membrane vesicles of the bacteria.2a,5,14 By trapping ferric iron, PQS can activate an iron starvation response in P. aeruginosa and induce the biosynthesis of the siderophores pyochelin and pyoverdine,5 and affect the Rhl quorum-sensing system.9 The acquisition mechanisms and roles of other important transition metals in P. aeruginosa are known to a lesser extent10b and the chelation by PQS was so far only described for ferric iron. Applying our dimeric PQS analogs 2 and 3 as qualitative chromogenic and fluorogenic PQS–metal ion binding probes allowed us to identify the PQS-motif being capable of complexing MoO42−, Al3+, and likely VO43−. The existence of PQS-complexes with these ions could finally be confirmed and their stoichiometry elucidated by LC-HRMS as MoO2(PQS)2 as well as Al(PQS)2+ and Al(PQS)3 complexes. In the environment of the bacteria, the trace element molybdenum is mainly present as the oxyanion molybdate (MoO42−) and essential for the growth of P. aeruginosa under anaerobic and microaerophilic conditions. As complex with its ligand molybdopterin, molybdate plays an important role as molybdenum cofactor in the periplasmic nitrate reductase NarGHI that facilitates the reduction of NO3− to NO2−.15 A lack of MoO42− was shown to significantly reduce the growth of P. aeruginosa under anaerobic conditions.16 To facilitate molybdenum uptake, P. aeruginosa uses a high-affinity ATP-binding cassette permease, ModABC to recruit and import molybdate from the environment.16,17 Importantly, the molybdate transporter ModABC was found to be essential for anaerobic growth in an in vivo animal model of chronic lung infection.18 ModA was shown to be a molybdate-binding protein and important for bacterial competition and virulence of P. aeruginosa in a mouse model.19 However, the exact mode of molybdate sequestration from the environment is still unclear and the PQS–MoO42− complex could represent a missing link in this transport chain.
1 complex which we confirmed by our LC-HRMS analysis. Contrary to siderophores, PQS serves as an iron trap since its ferric iron complexes cannot be taken up into the bacteria directly but instead accumulate in the cell envelope and outer membrane vesicles of the bacteria.2a,5,14 By trapping ferric iron, PQS can activate an iron starvation response in P. aeruginosa and induce the biosynthesis of the siderophores pyochelin and pyoverdine,5 and affect the Rhl quorum-sensing system.9 The acquisition mechanisms and roles of other important transition metals in P. aeruginosa are known to a lesser extent10b and the chelation by PQS was so far only described for ferric iron. Applying our dimeric PQS analogs 2 and 3 as qualitative chromogenic and fluorogenic PQS–metal ion binding probes allowed us to identify the PQS-motif being capable of complexing MoO42−, Al3+, and likely VO43−. The existence of PQS-complexes with these ions could finally be confirmed and their stoichiometry elucidated by LC-HRMS as MoO2(PQS)2 as well as Al(PQS)2+ and Al(PQS)3 complexes. In the environment of the bacteria, the trace element molybdenum is mainly present as the oxyanion molybdate (MoO42−) and essential for the growth of P. aeruginosa under anaerobic and microaerophilic conditions. As complex with its ligand molybdopterin, molybdate plays an important role as molybdenum cofactor in the periplasmic nitrate reductase NarGHI that facilitates the reduction of NO3− to NO2−.15 A lack of MoO42− was shown to significantly reduce the growth of P. aeruginosa under anaerobic conditions.16 To facilitate molybdenum uptake, P. aeruginosa uses a high-affinity ATP-binding cassette permease, ModABC to recruit and import molybdate from the environment.16,17 Importantly, the molybdate transporter ModABC was found to be essential for anaerobic growth in an in vivo animal model of chronic lung infection.18 ModA was shown to be a molybdate-binding protein and important for bacterial competition and virulence of P. aeruginosa in a mouse model.19 However, the exact mode of molybdate sequestration from the environment is still unclear and the PQS–MoO42− complex could represent a missing link in this transport chain.
Aluminum, in its form as Al3+ ion, on the other hand, lacks biological function as a cofactor. Even though the bioavailability of aluminum in an infected host is low, it was shown that aluminum can interfere with the iron acquisition of P. aeruginosa by occupying pyoverdine and accumulation in the periplasm of the bacteria.20 Chelation of Al3+ by PQS could therefore serve as a detoxification strategy by sequestration of the toxic metal ion to prevent interference with metabolic processes. Similar protection mechanisms by P. aeruginosa against antimicrobial and antibiofilm gallium (Ga3+) were already observed for its siderophore pyoverdine.21 Although a small number of bacteria are known to utilize vanadium as a cofactor in their enzymes, such as vanadium nitrogenases or vanadate-dependent haloperoxidases,22 vanadate (VO43−) is generally considered cytotoxic for bacteria due to the structural similarity to the phosphate ion (PO43−).23 Vanadate was shown to substitute for phosphate in enzymes such as phosphatases and kinases. Vanadium, in its vanadyl and vanadate forms, was shown to inhibit the growth of P. aeruginosa, especially in an iron-limited medium and it was suggested that vanadium can affect different functions in the bacteria depending on the iron content of the cell.24 Interestingly, the siderophores pyochelin and pyoverdine of P. aeruginosa have been found to complex vanadium. Even though the underlying mechanisms are still unclear, exposure of P. aeruginosa cells to vanadium was shown to cause an increase in reactive oxygen species and, in response, increase superoxide dismutase (SOD) activity in P. aeruginosa.24 Although we do not at present have a structure of the PQS–VO43− complexes it is conceivable that PQS serves again as a detoxification agent by complexation of vanadate, similar to aluminum.
Experimental Section
General information: Chemicals and solvents for the synthesis were purchased from Sigma-Aldrich, TCI, Carl Roth or VWR Chemicals and were used without further purification. For silica gel chromatography, distilled technical grade solvents and silica gel 60 A (Carl Roth) were used. Thin layer chromatography (TLC) was performed using aluminum sheets “TLC silica gel 60 F254” from Merck Millipore® and analyzed with UV-light or by permanganate staining. NMR spectra were obtained on Bruker Avance-III 400 and Bruker Avance-III 600 NMR spectrometers at ambient temperature. Multiplicities are given as follows: s – singlet, d – doublet, t – triplet, q – quartet, quint. – quintet, m – multiplet. Chemical shifts (δ) are given in parts per million (ppm) relative to the solvent residual signal with CDCl3δH = 7.26 ppm and δC = 77.16 ppm, DMSO-d6δH = 2.50 ppm and δC = 39.52 ppm.25 The data obtained were processed and analyzed with Bruker Topspin 3.5 software. High resolution mass spectrometry data for the synthetic compounds were obtained on an ESI-Orbitrap (Thermo Scientific, LTQ Orbitrap Velos) by direct injection and analyzed with Xcalibur (Thermo Scientific) software. UV-Vis spectroscopy was performed on a TECAN infinite® M200 PRO plate reader using i-control™ software using 96-well plates from Sarstedt (Microtest plates 96- well, flat-bottom, without lid).
Preparation of the metal ion complex solutions: For the qualitative analysis of a complexation event (Fig. 2C, 2D, and 3F), 25 μL of a 2 mM stock solution of PQS (1), compound 2 or compound 3 in DMSO was added to 25 μL of a 4 mM stock solution of a metal salt solution in DMSO (the stock solutions for Na3VO4 and Na2MoO4 were prepared in DMSO and DMSO/H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), respectively). For the measurements of the UV-Vis absorption spectra (Fig. 2A and 2B) 50 μL of a 0.4 mM stock solution of compound 2 or compound 3 in DMSO was added to 50 μL of a 0.8 mM stock solution of a metal salt solution and incubated for 1 h at room temperature before measurement. For the measurements of the HRMS spectra (Fig. 3A–F and Fig. S1–7†), the metal-PQS complexes were obtained by incubating salt solutions (30 μM final) with proportional to molar ratio amounts of PQS solution (60–180 μM final) in 1 mL of Methanol (or Acetone) (Table S1†). Resulting mixtures were kept for at least one hour at +4 °C before subjection to HRMS measurements.
1), respectively). For the measurements of the UV-Vis absorption spectra (Fig. 2A and 2B) 50 μL of a 0.4 mM stock solution of compound 2 or compound 3 in DMSO was added to 50 μL of a 0.8 mM stock solution of a metal salt solution and incubated for 1 h at room temperature before measurement. For the measurements of the HRMS spectra (Fig. 3A–F and Fig. S1–7†), the metal-PQS complexes were obtained by incubating salt solutions (30 μM final) with proportional to molar ratio amounts of PQS solution (60–180 μM final) in 1 mL of Methanol (or Acetone) (Table S1†). Resulting mixtures were kept for at least one hour at +4 °C before subjection to HRMS measurements.
Identification of PQS-metal ion complexes via HRMS: High-resolution mass spectrometry data were obtained on a Hybrid FT Mass Spectrometer LTQ Orbitrap Velos (Thermo Scientific) in positive ESI mode. The structures of PQS-metal complexes were proposed based on their full HRMS spectra and the coordination numbers of metal ions. Detected m/z and their distribution for PQS and complexes were compared to the calculated values (ChemDraw Professional 20.0.0.41).
Conclusion
While PQS is mostly considered a quorum sensing molecule of P. aeruginosa, its properties as iron chelating molecule indicate further relevance for the pathogen beyond quorum sensing. Our identification of PQS as metallophore of Al3+, MoO43−, and VO43− in addition to Fe3+ suggests that PQS serves a crucial and more complex role in the metal ion homeostasis of P. aeruginosa beyond ferric iron.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Malin Bein and Prof. Dr Tanja Gaich for access to mass spectrometry. We gratefully acknowledge funding by the Emmy Noether program of the Deutsche Forschungsgemeinschaft (DFG).References
- (a) I. Jurado-Martín, M. Sainz-Mejías and S. McClean, Int. J. Mol. Sci., 2021, 22, 3128 CrossRef PubMed; (b) M. E. Mattmann and H. E. Blackwell, J. Org. Chem., 2010, 75(20), 6737 CrossRef CAS PubMed.
- (a) J. Lin, J. Cheng, Y. Wang and X. Shen, Front. Cell. Infect. Microbiol., 2018, 8, 230 CrossRef PubMed; (b) G. D. Vrla, M. Esposito, C. Zhang, Y. Kang, M. R. Seyedsayamdost and Z. Gitai, PLoS Pathog., 2020, 16(9), 1 Search PubMed.
- (a) D. Szamosvári, V. F. Reichle, M. Jureschi and T. Böttcher, Chem. Commun., 2016, 52, 13440 RSC; (b) D. Szamosvári, T. Schuhmacher, C. R. Hauck and T. Böttcher, Chem. Sci., 2019, 10, 6624 RSC; (c) J. Hodgkinson, S. D. Bowden, W. R. J. D. Galloway, D. R. Spring and M. Welch, J. Bacteriol., 2010, 192(14), 3833 CrossRef CAS PubMed.
- M. Prothiwa, D. Szamosvári, S. Glasmacher and T. Böttcher, Beilstein J. Org. Chem., 2016, 12, 2784 CrossRef CAS PubMed.
- S. P. Diggle, S. Matthijs, V. J. Wright, M. P. Fletcher, S. R. Chhabra, I. L. Lamont, X. Kong, R. C. Hider, P. Cornelis, M. Cámara and P. Williams, Chem. Biol., 2007, 14(1), 87 CrossRef CAS PubMed.
- P. Hradil, J. Hlaváč and K. Lemr, J. Heterocycl. Chem., 1999, 36, 141 CrossRef CAS.
- M. Kurihara and N. Yoda, Bull. Chem. Soc. Jpn., 1967, 40(10), 2429 CrossRef CAS.
- (a) T. Nagasaka and Y. Koseki, J. Org. Chem., 1998, 63, 6797 CrossRef CAS PubMed; (b) J. I. Sarmiento-Sánchez, J. Montes-Avila, A. Ochoa-Terán, F. Delgado-Vargas, V. Wilson-Corral, S. P. Díaz-Camacho, F. García-Páez and P. Bastidas-Bastidas, Quim. Nova, 2014, 37(8), 1297 Search PubMed; (c) O. Maiatska, J. Omeis and H. Ritter, Macromolecules, 2016, 49, 737 CrossRef CAS.
- F. Bredenbruch, R. Geffers, M. Nimtz, J. Buer and S. Häussler, Environ. Microbiol., 2006, 8(8), 1318 CrossRef CAS PubMed.
- (a) S. S. Merchant and J. D. Helmann, Adv. Microb. Physiol., 2012, 60, 91 CrossRef CAS PubMed; (b) I. J. Schalk and O. Cunrath, Environ. Microbiol., 2016, 18(10), 3227 CrossRef CAS PubMed.
- (a) S. C. Andrews, A. K. Robinson and F. Rodríguez-Quiñones, FEMS Microbiol. Rev., 2003, 27, 215 CrossRef CAS PubMed; (b) P. Chandrangsu, C. Rensing and J. D. Helmann, Nat. Rev. Microbiol., 2017, 15(6), 338 CrossRef CAS PubMed.
- (a) M. I. Hood and E. P. Skaar, Nat. Rev. Microbiol., 2012, 10, 525 CrossRef CAS PubMed; (b) C. C. Murdoch and E. P. Skaar, Nat. Rev. Microbiol., 2012, 10, 657 Search PubMed; (c) G. Núñez, K. Sakamoto and M. P. Soares, J. Immunol., 2018, 201(1), 11 CrossRef PubMed.
- (a) P. Cornelis and J. Dingemans, Front. Cell. Infect. Microbiol., 2013, 14(3), 75 Search PubMed; (b) F. Minandri, F. Imperi, E. Frangipani, C. Bonchi, D. Visaggio, M. Facchini, P. Pasquali, A. Bragonzi and P. Visca, Infect. Immun., 2016, 21(84), 2324 CrossRef PubMed.
- (a) J. Lin, W. Zhang, J. Cheng, X. Yang, K. Zhu, Y. Wang, G. Wei, P. Y. Quin, Z. Q. Luo and X. Shen, Nat. Commun., 2017, 8, 14888 CrossRef CAS PubMed; (b) C. Schwechheimer and M. Kuehn, Nat. Rev. Microbiol., 2015, 13, 605 CrossRef CAS PubMed.
- (a) M. Bertero, R. Rothery, M. Palak, C. Hou, D. Lim, F. Blasco, J. H. Weiner and N. C. J. Strynadka, Nat. Struct. Mol. Biol., 2003, 10, 681 CrossRef CAS PubMed.
- V. G. Pederick, B. A. Eijkelkamp, M. P. Ween, S. L. Begg, J. C. Paton and C. A. McDevitt, Appl. Environ. Microbiol., 2014, 80(21), 6843 CrossRef PubMed.
- E. A. Maunders, D. H. Y. Ngu, K. Ganio, S. I. Hossain, B. Y. J. Lim, M. G. Leeming, Z. Luo, A. Tan, E. Deplazes, B. Kobe and C. A. McDevitt, Front. Microbiol., 2022, 13, 903146 CrossRef PubMed.
- S. Périnet, J. Jeukens, I. Kukavica-Ibrulj, M. M. Ouellet, S. J. Charette and R. C. Levesque, BMC Res. Notes, 2016, 12(9), 23 CrossRef PubMed.
- T. Wang, X. Du, L. Ji, Y. Han, J. Dang, J. Wen, Y. Wang, Q. Pu, M. Wu and H. Liang, Cell Rep., 2021, 35(2), 108957 CrossRef CAS PubMed.
- J. Greenwald, G. Zeder-Lutz, A. Hagege, H. Celia and F. Pattus, J. Bacteriol., 2008, 190(20), 6548 CrossRef CAS PubMed.
- Y. Kaneko, M. Thoendel, O. Olakanmi, B. E. Britigan and P. K. Singh, J. Clin. Invest., 2007, 117, 877 CrossRef CAS PubMed.
- J. C. Pessoa, E. Garribba, M. F. A. Santos and T. Santos-Silva, Coord. Chem. Rev., 2015, 301–302, 49 CrossRef.
- D. Rehder, Metallomics, 2015, 7(5), 730 CrossRef CAS PubMed.
- C. Baysse, D. De Vos, Y. Naudet, A. Vandermonde, U. Ochsner, J. M. Meyer, H. Budzikiewicz, M. Schäfer, R. Fuchs and P. Cornelis, Microbiology, 2000, 146, 2425 CrossRef CAS PubMed.
- G. R. Fulmer, A. J. M. Miller, N. H. Sherden, H. E. Gottlieb, A. Nudelman, B. M. Stoltz, J. E. Bercaw and K. I. Goldberg, Organometallics, 2010, 29, 217 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ob00710c |
| This journal is © The Royal Society of Chemistry 2023 |