Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Human serum albumin-based supramolecular host–guest boronate probe for enhanced peroxynitrite sensing†
He
Tian Jr.‡
 a,
Chen
Guo‡
a,
Chen
Guo‡
 a,
Xi-Le
Hu
a,
Xi-Le
Hu
 a,
Jing-Bo
Wang
a,
Jing-Bo
Wang
 a,
Yi
Zang
a,
Yi
Zang
 *cf,
Tony D.
James
*cf,
Tony D.
James
 *de,
Jia
Li
*de,
Jia
Li
 *c and
Xiao-Peng
He
*c and
Xiao-Peng
He
 *ab
*ab
aKey Laboratory for Advanced Materials and Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, School of Chemistry and Molecular Engineering, East China University of Science and Technology, 130 Meilong Rd., Shanghai 200237, China. E-mail: xphe@ecust.edu.cn
bNational Center for Liver Cancer, The International Cooperation Laboratory on Signal Transduction, Shanghai 200438, China
cNational Center for Drug Screening, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 189 Guo Shoujing Rd., Shanghai 201203, P. R. China. E-mail: jli@simm.ac.cn
dDepartment of Chemistry, University of Bath, Bath, BA2 7AY, UK. E-mail: t.d.james@bath.ac.uk
eSchool of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang 453007, China
fLingang laboratory, Shanghai 201203, China. E-mail: yzang@simm.ac.cn
First published on 22nd May 2023
Abstract
Peroxynitrite (ONOO−) is an important oxygen/nitrogen reactive species implicated in a number of physiological and pathological processes. However, due to the complexity of the cellular micro-environment, the sensitive and accurate detection of ONOO− remains a challenging task. Here, we developed a long-wavelength fluorescent probe based on the conjugation between a TCF scaffold and phenylboronate; the resulting conjugate is capable of supramolecular host–guest assembly with human serum albumin (HSA) for the fluorogenic sensing of ONOO−. The probe exhibited an enhanced fluorescence over a low concentration range of ONOO− (0–9.6 μM), whist the fluorescence was quenched when the concentration of ONOO− exceeded 9.6 μM. In addition, when human serum albumin (HSA) was added, the initial fluorescence of the probe was significantly enhanced, which enabled the more sensitive detection of low-concentrations of ONOO− in aqueous buffer solution and in cells. The molecular structure of the supramolecular host–guest ensemble was determined using small-angle X-ray scattering.
1. Introduction
Peroxynitrite (ONOO−) is a highly reactive molecule that is formed by the reaction of nitric oxide (NO) and the superoxide anion (O2˙−).1 It is involved in various physiological and pathological processes, including inflammation, oxidative stress, and cell death. ONOO− can react with lipids, proteins, and DNA, leading to cellular damage and dysfunction.2 In disease development, ONOO− has been implicated in a wide range of conditions, including cardiovascular disease, neurodegenerative disorders, and cancer.3 During cardiovascular disease, ONOO− contributes to the development of atherosclerosis by promoting endothelial dysfunction and lipid oxidation,4 and in neurodegenerative disorders, ONOO− is thought to play a role in neuronal damage and death through protein nitration and mitochondrial dysfunction.5 In addition, ONOO− has been shown to promote tumor growth and metastasis by inducing DNA damage and activating signalling pathways that facilitate cell survival and proliferation.Research has indicated that high concentrations of ONOO− are associated with various diseases such as cardiovascular disease, neurodegenerative diseases, inflammation and cancer.6 While low concentrations of ONOO− are required to modulate cellular signalling pathways, a deficit in ONOO− may lead to immune deficiency and increase susceptibility to infection.7 Therefore, the development of analytical tools that are sensitive to the dynamic concentration changes of ONOO− during various biological and pathological processes will facilitate a deeper understanding of ROS/RNS biology and provide tools for disease diagnosis. Conventional techniques for ONOO− detection include microchip electrophoresis,8 chromatographic methods utilizing electron spin resonance spectroscopy and Liquid Chromatography Mass Spectrometry,9 bioluminescent probes,10 photoacoustic probes,11 chemiluminescent probes,12 modified carbon quantum dots,13 and reaction-based fluorescent probes.14,15
The development of fluorescent probes for ONOO− sensing has proven to be effective to help improve our understanding of its intrinsic roles in complicated biological systems.16,17 The probes developed have achieved the visualization of ONOO− in living cells and tissues, allowing researchers to study ONOO− relevant biological events in real-time.18 They can also be used to quantify ONOO− levels in biological samples, thus facilitating the diagnosis of diseases associated with ONOO−. Fluorescent probes targeting ONOO− typically consist of a fluorophore that is coupled to a specific chemical moiety that reacts with ONOO.19,20 Upon reaction with ONOO−, the fluorophore undergoes a change in fluorescence intensity or spectral properties, allowing ONOO− to be detected and quantified. Several types of fluorescent probes have been developed for ONOO− detection, including aryl boronate-based probes, α-ketoamide-based probes and isatin-based probes.21–23 However, because small-molecule probes can be unstable in complicated biological systems, supramolecular strategies to enhance their sensing properties have recently been developed.24
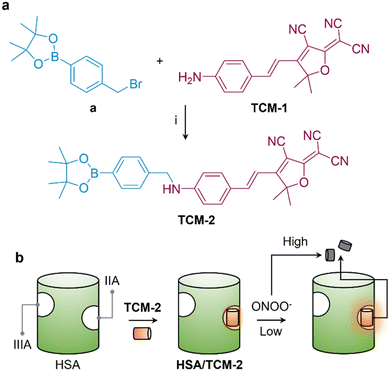
We have demonstrated that human serum albumin (HSA) is an effective, biocompatible host material to enhance the sensitivity of small-molecule fluorescent probes.20,25–27 Here, we extended this strategy to improve the sensing ability of a TCF (2-(3-cyano-4,5,5-trimethylfuran-2(5H)-ylidene) malononitrile)-based long-wavelength ONOO− probe. The probe was synthesized through the conjugation between the TCF scaffold and phenylboronate (Fig. 1a). The resulting conjugate is capable of host–guest assembly with HSA exhibiting an enhanced sensitivity for low-concentrations of ONOO− in aqueous buffer solution and in cells; however, when high concentrations of ONOO− are added, the fluorescence of the probe is quenched due to structural degradation (Fig. 1b). The molecular structure of the supramolecular ensemble between the probe and HSA was determined using small-angle X-ray scattering (SAXS).
2. Results and discussion
The fluorogenic probe was developed based on a known donor–acceptor (D–A) type fluorescent dye, TCM-1 was synthesized by the aldol reaction between 2-(3-cyano-4,5,5-trimethylfuran-2(5H)-ylidene) malononitrile and 4-aminobenzaldehyde (Scheme S1†).28 Then, a phenylboronate, which is a known reactive group for ONOO−,29 was introduced to the amino end of TCM-1 to produce TCM-2 with quenched fluorescence due to inhibited intramolecular charge transfer.30 After reacting with ONOO−, the boronate moiety is removed, resulting in fluorescence recovery of the probe.With the probe in hand, we evaluated the fluorescence changes in the presence of increasing concentrations of ONOO− in an aqueous solution. We found a concentration-dependent fluorescence enhancement of the probe with ONOO− over a concentration range of 0–9.6 μM (Fig. 2a). However, when the concentration continued to increase (9.6–25.6 μM), the fluorescence of the probe gradually decreased (Fig. 2c). In its UV-vis spectrum, we also observed a slightly enhanced absorption peak of TCM-2 at ∼560 nm when the concentration of ONOO− increased from 0–9.6 μM; however, this peak began to decrease gradually when the concentration of ONOO− increased from 9.6–25.6 μM (Fig. S1a†). In contrast, the other absorption peak of the probe at ∼340 nm enhanced gradually over the entire concentration range of ONOO− (0–25.6 μM).
We envisioned that the enhanced fluorescence of TCM-2 with low concentrations of ONOO− was a result of the removal of the phenylboronate group, thus releasing the TCM-1 scaffold. To confirm this hypothesis, mass spectroscopic (MS) analysis was used to determine the mass shift of TCM-2 after treatment with 9.6 μM of ONOO−. As expected, the mass peak assigned to TCM-1 at m/z 302.1169 was detected (Fig. S2†). We further envisioned that when the concentration of ONOO− continues to increase, the structure of TCM-1 degraded. Indeed, using a tandem liquid chromatograph-mass spectrometer, we found the mass peak of a product of TCM-1 with one of the cyano groups having been oxidized (1, Fig. S3†); a further oxidatively cleaved product from TCM-1 was also detected (2, Fig. S3†). In addition, the UV-vis absorption at 512 nm and 350 nm of TCM-1 gradually decreased and increased with increasing concentrations of ONOO− (9.6–25.6 μM), respectively (Fig. S1b†), and the fluorescence of TCM-1 also diminished gradually when 9.6–25.6 μM of ONOO− was added (Fig. S1c†). These findings collectively suggest that the gradually quenched fluorescence of TCM-2 at higher concentrations of ONOO− is the result of structural cleavage. Notably, a previous report also demonstrated the cleavage of the alkenyl bond of TCM-based dyes in the presence of strong oxidants.31
Next, HSA was used for host–guest assembly with TCM-2 under mild sonication (100 W), producing the supramolecular HSA/TCM-2 ensemble. We first determined that increasing the concentration of HSA caused the fluorescence of TCM-2 to enhance gradually (Fig. S4a†), and the fluorescence enhancement is also time-dependent (Fig. S4b†). This observation agrees with previous reports showing that the fluorescence of D–A type fluorogens is enhanced when bound to HSA probably due to a reduction of aggregation-caused quenching and/or an inclusion into the hydrophobic cavity of the protein.20,32–34 The HSA/TCM-2 ensemble exhibited concentration-dependent fluorescence enhancement when low concentrations of ONOO− were added (0–3.2 μM) (Fig. 2b), and the fluorescence began to quench when the concentration exceeded 3.2 μM (Fig. 2c). Interestingly, the fluorescence of HSA/TCM-2 enhanced more dramatically than that of TCM-2 over a smaller concentration range of ONOO− (Fig. 2evs.Fig. 2f). The quenching point of the supramolecular ensemble (3.2 μM) was also 3-fold smaller than that of the probe (9.6 μM). This suggests that the host–guest inclusion of the probe into HSA enhances its reactivity with ONOO− to first remove the phenylboronate group and then induce the structural degradation. Indeed, the limit of detection of HSA/TCM-2 for ONOO− (0.39 nM) was determined to be ∼2-fold smaller than that of TCM-2 (0.84 nM) (Fig. S5†). A thorough comparison to previously developed fluorescent probes for ONOO− detection suggests that our supramolecular system with host and guest assembly is among the most sensitive (Table S1†).
To determine the binding manner of the probe with HSA, SAXS was used. The scattering signals for HSA in the absence and presence of TCM-2 were obtained (Fig. 3a), and then the corresponding interatomic distance distribution functions (P(r)) were determined (Fig. 3b). We found that the maximum dimension of HSA (89.5 Å) increased to 98.4 Å when TCM-2 is included. This suggests a conformational change of the protein after binding with the probe. Subsequently, the 3D structures of HSA and HSA/TCM-2 were simulated and compared to a known crystal structure of HSA (1n5u) extracted from the protein data bank in order to identify the binding site of the probe (Fig. 3c). Shown in Fig. 3d are the superimposed SAXS models of HSA/TCM-2 and HSA (light grey) with the crystal model of HSA (red). We observed a significant conformational change in the IIA domain of the SAXS model of HSA/TCM-2 with respect to the crystalline HSA structure used as control; a 2.067 Å root-mean-square deviation (RMSD) between the two models was determined. In addition, minimal conformational change was seen for the IIIA domain of the SAXS model of HSA, which is also known as a host site for hydrophobic molecules. In contrast, a much greater overlap between the SAXS and crystal model of HSA was seen with a RMSD of 0.743 Å. A subsequent competition assay using known IIIA and IIA binding agents corroborates that the probe is included into the IIA region of HSA (Fig. S6†). The supramolecular HSA/TCM-2 probe also dis-played a wide working pH range of 3–12 (Fig. S7a†), and selectivity over other ROS/RNS tested (Fig. S7b†).
 | ||
| Fig. 3 (a) Small-angle X-ray scattering (SAXS) pattern of HSA and HSA/TCM-2. (b) Stacked interatomic distance distribution function, P(r), of the SAXS patterns of HSA and HSA/TCM-2. (c) Atomic models of HSA/TCM-2 and HSA refined with the scattering data obtained and a known crystal model of HSA (1n5u) extracted from the protein data bank. (d) Superimposed SAXS model of HSA/TCM-2 and HSA (light grey) with the crystal model of HSA (1n5u, red); the IIA and IIIA domains of the superimposed models are enlarged to better illustrate the conformational change of HSA after including TCM-2. | ||
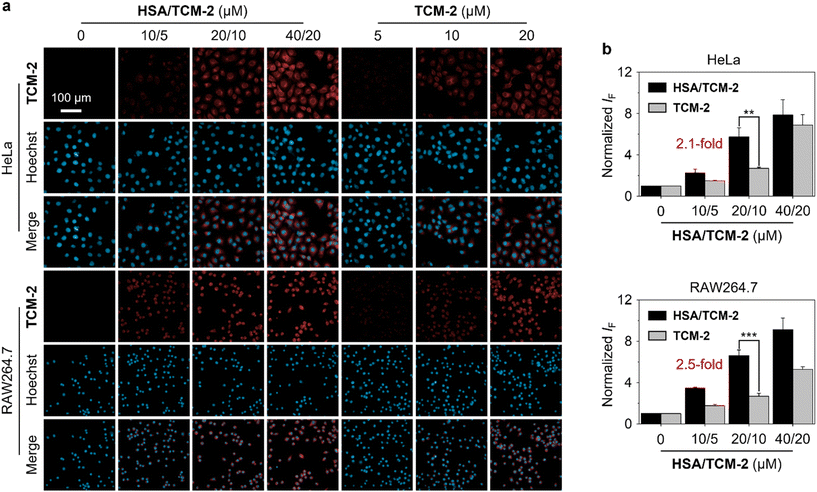
Finally, the supramolecular probe was used for cell imaging. Two cell lines, HeLa (human cervical cancer) and RAW264.7 (mouse macrophage), were used. The cells were pretreated with SIN-1 (3-morpholinosydnonimine, a known ONOO− releasing agent) to upregulate intracellular ONOO− concentration, and were then treated with HSA/TCM-2 and TCM-2 of different concentrations for 30 min. We found that the fluorescence intensities of HSA/TCM-2 were higher than those of TCM-2 over the entire concentration range in both cells imaged (Fig. 4a and b). A remarkable 2.1-fold and 2.5-fold greater fluorescence of the supramolecular ensemble was determined at a TCM-2 concentration of 10 μM in HeLa and RAW264.7 cells, respectively, suggesting the effectiveness of our HSA encapsulation strategy to enhance the sensing properties of fluorescent small-molecule probes in the complicated cellular environment.
The biological applications of small-molecule fluorescent probes are frequently limited by their low fluorescence brightness and insufficient biocompatibility due to molecular aggregation in biological media. Human serum albumin (HSA) is a biocompatible protein carrier suitable as a host for enhancing the imaging capacity of small-molecule probes, and as such a number of fluorescent dyes with HSA binding capacity have been developed.35–38 Besides ONOO− sensing, we have demonstrated the applicability of HSA encapsulation for the enhanced fluorescence imaging of lysosomes based on receptor-directed endocytosis,25 and intracellular glycosidases using super-resolution microscopy.27 In addition, we have shown that this strategy could be extended to targeted fluorescence imaging of triple-negative breast cancer in vivo.26 The use of other biomolecules such as amyloid β peptides to tune the fluorescence properties of small-molecule fluorescent probes has also been recently demonstrated.39
3. Conclusion
We have developed a supramolecular host–guest system formed with HSA, the most abundant protein in the human body, and a long-wavelength fluorogenic probe based on TCF. The probe when bound to a hydrophobic region of HSA exhibited a significantly enhanced sensitivity for ONOO− both in aqueous buffer solution and in cells. SAXS analysis combined with a competition assay corroborates that the probe is included in the IIA region of the HSA. This research offers insight into the simple construction of supramolecular systems with enhanced sensitivity for the detection of biologically relevant reactive species that exist at low concentrations in biological systems.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors thank the National Natural Science Foundation of China (NSFC) (No. 92253306 and 82130099), the Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX03), the Fundamental Research Funds for the Central Universities (222201717003), the Programme of Introducing Talents of Discipline to Universities (B16017) and Open Funding Project of the State Key Laboratory of Bioreactor Engineering of East China University of Science and Technology for financial support. The Research Center of Analysis and Test of East China University of Science and Technology is gratefully acknowledged for assistance in analytical experiments. TDJ wishes to thank the University of Bath and the Open Research Fund of the School of Chemistry and Chemical Engineering, Henan Normal University (2020ZD01) for support.References
- M. Villanueva-Paz, L. Moran, N. Lopez-Alcantara, C. Freixo, R. J. Andrade, M. I. Lucena and F. J. Cubero, Antioxidants, 2021, 10, 390 CrossRef CAS PubMed.
- C. Szabo, H. Ischiropoulos and R. Radi, Nat. Rev. Drug Discovery, 2007, 6, 662–680 CrossRef CAS PubMed.
- R. Radi, J. Biol. Chem., 2013, 288, 26464–26472 CrossRef CAS PubMed.
- F. Wang, Q. Yuan, F. Chen, J. Pang, C. Pan, F. Xu and Y. Chen, Front. Cell Dev. Biol., 2021, 9, 742483 CrossRef PubMed.
- Z. Liu, Z. Ren, J. Zhang, C. C. Chuang, E. Kandaswamy, T. Zhou and L. Zuo, Front. Physiol., 2018, 9, 477 CrossRef.
- J. Li, J. Su, W. Li, W. Liu, B. T. Altura and B. M. Altura, Neurosci. Lett., 2003, 350, 173–177 CrossRef CAS PubMed.
- B. Guo, W. Shu, W. Liu, H. Wang, S. Xing, J. Chen and X. Zhang, Sens. Actuators, B, 2021, 344, 130206 CrossRef CAS.
- M. K. Hulvey, C. N. Frankenfeld and S. M. Lunte, Anal. Chem., 2010, 82, 1608–1611 CrossRef CAS PubMed.
- W. Imaram, C. Gersch, K. M. Kim, R. J. Johnson, G. N. Henderson and A. Angerhofer, Free Radicals Biol. Med., 2010, 49, 275–281 CrossRef CAS PubMed.
- J. B. Li, L. Chen, Q. Wang, H. W. Liu, X. X. Hu, L. Yuan and X. B. Zhang, Anal. Chem., 2018, 90, 4167–4173 CrossRef CAS PubMed.
- X. Qin, F. Li, Y. Zhang, G. Ma, T. Feng, Y. Luo, P. Huang and J. Lin, Anal. Chem., 2018, 90, 9381–9385 CrossRef CAS PubMed.
- B. Wang, Y. Wang, Y. Wang, Y. Zhao, C. Yang, Z. Zeng, S. Huan, G. Song and X. Zhang, Anal. Chem., 2020, 92, 4154–4163 CrossRef CAS.
- Y. Bai, Y. Wang, L. Cao, Y. Jiang, Y. Li, H. Zou, L. Zhan and C. Huang, Anal. Chem., 2021, 93, 16466–16473 CrossRef CAS PubMed.
- S. Wang, L. Chen, P. Jangili, A. Sharma, W. Li, J.-T. Hou, C. Qin, J. Yoon and J. S. Kim, Coord. Chem. Rev., 2018, 374, 36–54 CrossRef CAS.
- W. T. Dou, H. H. Han, A. C. Sedgwick, G. B. Zhu, Y. Zang, X. R. Yang, J. Yoon, T. D. James, J. Li and X. P. He, Sci. Bull., 2022, 67, 853–878 CrossRef CAS PubMed.
- H. H. Han, H. M. Wang, P. Jangili, M. Li, L. Wu, Y. Zang, A. C. Sedgwick, J. Li, X. P. He, T. D. James and J. S. Kim, Chem. Soc. Rev., 2023, 52, 879–920 RSC.
- H. H. Han, H. Tian, Y. Zang, A. C. Sedgwick, J. Li, J. L. Sessler, X. P. He and T. D. James, Chem. Soc. Rev., 2021, 50, 9391–9429 RSC.
- X. Chai, B. Li, C. Chen, W. Zhang, L. Sun, H. H. Han, Y. Zhang, S. Sun, J. Yang, J. Zhang and X. P. He, Anal. Chem., 2023, 95, 5747–5753 CrossRef CAS PubMed.
- M. Weber, H. H. Han, B. H. Li, M. L. Odyniec, C. E. F. Jarman, Y. Zang, S. D. Bull, A. B. Mackenzie, A. C. Sedgwick, J. Li, X. P. He and T. D. James, Chem. Sci., 2020, 11, 8567–8571 RSC.
- H. H. Han, A. C. Sedgwick, Y. Shang, N. Li, T. Liu, B. H. Li, K. Yu, Y. Zang, J. T. Brewster II, M. L. Odyniec, M. Weber, S. D. Bull, J. Li, J. L. Sessler, T. D. James, X. P. He and H. Tian, Chem. Sci., 2019, 11, 1107–1113 RSC.
- Y. Wu, H. H. Han, L. He, L. Li, Y. Zang, J. Li, X. P. He, Y. Ding, W. Cao and T. D. James, Chem. Commun., 2023, 59, 5051–5054 RSC.
- T. Shao, X. Xu, L. Wang, Y. Shen, J. Zhao, H. Li, D. Zhang, W. Du, H. Bai, B. Peng and L. Li, J. Innovative Opt. Health Sci., 2023, 2250039 CrossRef.
- M. Li, H. Han, H. Zhang, S. Song, S. Shuang and C. Dong, Spectrochim. Acta, Part A, 2020, 243, 118683 CrossRef CAS PubMed.
- R. Ramos, J. Bernard, F. Ganachaud and A. Miserez, Small Sci., 2022, 2, 2100095 CrossRef CAS.
- Y. Fu, H. H. Han, J. Zhang, X. P. He, B. L. Feringa and H. Tian, J. Am. Chem. Soc., 2018, 140, 8671–8674 CrossRef CAS PubMed.
- X. L. Hu, Q. Cai, J. Gao, R. A. Field, G. R. Chen, N. Jia, Y. Zang, J. Li and X. P. He, ACS Appl. Mater. Interfaces, 2019, 11, 22181–22187 CrossRef CAS PubMed.
- X. Chai, H. H. Han, A. C. Sedgwick, N. Li, Y. Zang, T. D. James, J. Zhang, X. L. Hu, Y. Yu, Y. Li, Y. Wang, J. Li, X. P. He and H. Tian, J. Am. Chem. Soc., 2020, 142, 18005–18013 CrossRef CAS PubMed.
- A. C. Sedgwick, H. H. Han, J. E. Gardiner, S. D. Bull, X. P. He and T. D. James, Chem. Commun., 2017, 53, 12822–12825 RSC.
- H. Y. Chen, D. Guo, Z. F. Gan, L. Jiang, S. Chang and D. W. Li, Mikrochim. Acta, 2018, 186, 11 CrossRef PubMed.
- X. Wang, L. Wang, T. Jin, K. Sun and J. Yang, Sens. Actuators, B, 2023, 375, 132935 CrossRef CAS.
- D. Li, D. Lei, W. Ren, J. Li, X. Yang, Z. Cai, H. Duan and X. Dou, Org. Biomol. Chem., 2023, 21, 315–322 RSC.
- A. A. Ksenofontov, P. S. Bocharov, K. V. Ksenofontova and E. V. Antina, J. Mol. Liq., 2022, 345, 117031 CrossRef CAS.
- H. Li, Q. Yao, J. Fan, J. Du, J. Wang and X. Peng, Dyes Pigm., 2016, 133, 79–85 CrossRef CAS.
- P. Shen, J. Hua, H. Jin, J. Du, C. Liu, W. Yang, Q. Gao, H. Luo, Y. Liu and C. Yang, Sens. Actuators, B, 2017, 247, 587–594 CrossRef CAS.
- X. Chao, D. Yao, Y. Qi, C. Yuan and D. Huang, Anal. Chim. Acta, 2021, 1188, 339201 CrossRef CAS.
- Y. F. Fan, F. Y. Wang, F. B. Hou, L. Wei, G. H. Zhu, D. F. Zhao, Q. Hu, T. Lei, L. Yang, P. Wang and G. B. Ge, Chin. Chem. Lett., 2023, 34, 1075571–1075574 Search PubMed.
- Y. Ke, J. Cao, J. Gong and N. Fu, Sens. Actuators, B, 2022, 352, 131015 CrossRef CAS.
- D. Yuan, K. Pan, S. Xu and L. Wang, Anal. Chem., 2022, 94, 12391–12397 CrossRef CAS PubMed.
- A. C. Sedgwick, W.-T. Dou, J.-B. Jiao, L. Wu, G. T. Williams, A. T. A. Jenkins, S. D. Bull, J. L. Sessler, X.-P. He and T. D. James, J. Am. Chem. Soc., 2018, 140, 14267–14271 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental section, additional figures and original spectral copies of new compounds. See DOI: https://doi.org/10.1039/d3ob00637a |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2023 |