Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Kinetic analysis of highly effective triplex formation between a small molecule–peptide nucleic acid conjugate probe and the influenza A virus RNA promoter region at neutral pH†
Chioma Uche
Okeke
 ,
Hiromasa
Miura
,
Yusuke
Sato
,
Hiromasa
Miura
,
Yusuke
Sato
 * and
Seiichi
Nishizawa
* and
Seiichi
Nishizawa
 *
*
Department of Chemistry, Graduate School of Science, Tohoku University, Sendai 980-8578, Japan. E-mail: seiichi.nishizawa.c8@tohoku.ac.jp; yusuke.sato.a7@tohoku.ac.jp
First published on 29th March 2023
Abstract
In order to overcome the pH limitations of triplex-forming peptide nucleic acid (PNA) in binding to double-stranded RNA (dsRNA), we have recently proposed a new design of triplex-forming PNA-based fluorogenic probes that work at neutral pH for sensing the panhandle structure of the influenza A virus (IAV) RNA promoter region. Our strategy is based on the conjugation of a small molecule (DPQ) capable of selectively binding to the internal loop structure with the triplex-forming forced intercalation of thiazole orange (tFIT) probe with natural PNA nucleobases. In this work, the triplex formation of tFIT–DPQ conjugate probes with IAV target RNA at neutral pH was examined by means of a stopped-flow technique UV melting and fluorescence titration experiments. The obtained results revealed that (i) the conjugation strategy is responsible for the observed strong binding affinity due to a very fast association rate constant and a slow dissociation rate constant; (ii) the binding follows a pattern of the DPQ unit binding first to the internal loop region, followed by the tFIT unit binding to the complementary dsRNA region. Our results emphasize the importance of both the tFIT and the DPQ components of the conjugate probe design and revealed an association mechanism for the tFIT–DPQ probe–dsRNA triplex formation towards the IAV RNA at neutral pH.
Introduction
The influenza A virus (IAV) is a major human and animal pathogen responsible for seasonal epidemics and occasional pandemics. It is very infectious and belongs to the Orthomyxoviridae virus family. The genome of IAV consists of eight single-stranded RNA-negative segments. It is highly variable as viral strains accumulate genetic mutations, which results in novel possible pandemic strains.1 A typical example is the 2009 swine flu pandemic.2 Considering the rising resistance of new influenza virus strains to the current anti-influenza medications, finding conserved regions in this virus that are less prone to mutations became critically important.It was found that 13 nucleotides at the 5′ terminus and 12 nucleotides at the 3′ terminus of each RNA segment were conserved among various human IAV strains (cf.Fig. 1A). These termini interact to form double-stranded structures,3 also called panhandle-like structures that act as promoters for viral transcription and replication.4 Mutations in the conserved sequences negatively affect viral replication efficiency. Thus, the resulting structure has emerged as a new attractive target for the development of anti-influenza drugs as the sequences are not involved in the gene variations associated with pathogenesis and antiviral resistance.5,6
NMR spectroscopy results revealed that the RNA promoter of the influenza A virus adopted an A-form helix with an (A·A)-U internal loop structure (cf.Fig. 1A), which is an important requirement for specific interaction with RNA-dependent RNA polymerase (RdRp).3 It is worthy of note that internal loop regions create binding pockets for small molecules.7 Armed with this structural knowledge of the IAV RNA promoter, screening for small molecules that target the (A·A)-U internal loop structure became a possible route for finding hits that possess anti-influenza replication activity.5 These small molecule binders would be prospective anti-influenza drug candidates for various IAV human strains.
In an initial attempt, aminoglycoside antibiotics were shown to work as useful binders, with neomycin showing the highest binding affinity for the (A·A)-U internal loop (dissociation constant Kd = 2.7 μM, pH = 7.5).8 However, given some inherent problems of aminoglycosides such as promiscuous binding to various RNA structures9a and the risk of adverse effects9b such as nephrotoxicity (the rapid deterioration of kidney function) and ototoxicity (affecting the ear), their clinical use would be severely limited. This made it important to screen for new RNA binders with non-aminoglycoside scaffolds.10,11 Actually, through screening a fragment library of 4279 compounds by NMR spectroscopy, DPQ (6,7-dimethoxy-2-(1-piperazinyl)-4-quinazolinamine, cf.Fig. 1C) has been recently identified by Varani and co-workers as a non-aminoglycoside ligand. DPQ has binding selectivity towards this internal loop (Kd = 50.5 μM).11 This study highlighted the great potential of DPQ as a new anti-influenza drug candidate as it could inhibit the replication of the influenza virus in cell-based assays. However, the anti-influenza virus activity is moderate compared to those of approved drugs such as amantadine and ribavirin.11
On the other hand, a triplex-forming peptide nucleic acid (PNA)-based probe, named IR-1, has been recently developed by the Chen group for targeting the double-stranded RNA (dsRNA) region in the panhandle structure of IAV RNA for inhibiting IAV replication.12a PNA is a synthetic DNA analogue with a pseudo-peptide backbone composed of N-(2-aminoethyl)glycine units where nucleobases are connected to the glycine nitrogen via carbonyl methylene linkers (cf.Fig. 1C).13 Significantly, the Rozners group has discovered that homopyrimidine PNAs tightly bind to homopurine tracts within dsRNA by means of sequence-selective Hoogsten base-pairing to form a triplex at acidic pH (Fig. 1B), and that generally, triplex forming PNAs exhibit higher binding affinity for dsRNA than for dsDNA.14a This finding has opened a novel dsRNA-targeting strategy for biochemical and therapeutic applications, facilitating the design of artificial nucleobases to overcome both pH and sequence limitations of triplex-forming PNAs (TFP) in binding to dsRNA.12,15–18 In the case of IR-1 reported by the Chen group,12a they utilized two artificial nucleobases, the cytosine analogue thio-pseudoisocytosine (L)18a for G and guanidine-modified 5-methylcytosine (Q)18b for the C–G pair, in combination with the natural T nucleobase. They demonstrated that IR-1 (NH2-Lys-TLTTTQTLLL-CONH2) exhibited strong and selective binding to the panhandle structure with a Kd of 0.3 μM under physiological conditions (pH 7.5, 200 mM NaCl). They also demonstrated that the conjugation of IR-1 with an amino sugar, neamine (probe named IR-1b), enhanced the cellular uptake, and IR-1b caused a significant reduction of viral replication.12a Furthermore, they designed an IR-1-based fluorescent probe by the incorporation of 5-benzothiophene uracil (btU) as a fluorescence signalling unit into the TFP sequence, and the resulting probe called IR-1X (NH2-Lys-TLTbtUTQTLLL-CONH2) was applicable to the fluorescence detection of IAV RNA (detection sensitivity for total RNA from IAV-infected cells: ∼3600 ng mL−1).12b While further efforts are essential, their study provided the foundation for the development of panhandle RNA structure-targeting antiviral TFPs. In addition, TFP would be a promising candidate for the detection of IAV RNA based on the direct sensing of the IAV RNA promoter region. Considering the importance of detecting virus RNA, a wide variety of methods should be developed as complementary methods to PCR19 or immunochromatography.20
In this context, our group has proposed a fluorogenic TFP probe containing thiazole orange (TO) as a base surrogate, which we call the tFIT (triplex-forming forced intercalation of thiazole orange) probe.21–25 Similar to parent FIT probes for single-stranded RNA (ssRNA),26,27 the TO base surrogate of the tFIT probe is forcibly intercalated into the triplex structure, resulting in a significant light-up response because of the restriction of rotation around the methine bridge between the two heterocycles.28 Importantly, the TO base surrogate also functions as a universal base for any base-pair opposite the TO unit, allowing its binding to pyrimidine bases (C and U) in the target dsRNA. We have also proposed a new design of TFPs with natural nucleobases that do work at neutral pH based on the conjugation of tFIT probes with a small molecule capable of selectively binding to the internal loop structure of the target dsRNA.24,25 Indeed, by using DPQ as the small molecule for the (A-A)-U internal loop binding, we have recently designed a TFP-based fluorogenic probe for the panhandle structure of the IAV RNA promoter.25 The sequence of this 8-mer tFIT with natural nucleobases (C and T) was designed to be complementary to the sequence in the terminal stem region (from A2 to A9, Fig. 1A), in which the TO base surrogate faces towards U4 when the triplex is formed. DPQ was linked to the PNA backbone with Dap (2,3-diaminopropinic acid), and a lysine residue was also involved in the C-terminus of the tFIT structure for increasing the solubility as well as the binding affinity through electrostatic interaction. Significantly, the resulting conjugate named tFIT–DPQ (Dap) (NH2-TC(TO-C1)TCTTT-Lys-Dap(DPQ)-CONH2) was able to bind to the panhandle structure even at neutral pH (pH 7.0, 100 mM NaCl), and the binding affinity (Kd) reached 107 ± 9.4 nM. tFIT–DPQ (Dap) was indeed applicable to the fluorescence detection of IAV RNA with considerable sensitivity (for the total RNA from IAV-infected cells: ∼60 ng mL−1). tFIT–DPQ (Dap) also works as a sensitive indicator for screening test compounds targeting the IAV RNA promoter region in the fluorescence indicator displacement assay.10,29 Together with our previous work on targeting the bacterial rRNA A-site,24 this study reveals that the conjugation of a small molecule does indeed promote an effective triplex formation of a PNA with natural nucleobases. Because the internal loop binding of DPQ does proceed at neutral pH,10a,11 it could serve as an anchor for the conjugate probe to the target RNA, resulting in shifts in the local pKa value of cytosine that facilitate protonation at neutral pH. We believe that the conjugation with an anchoring unit (small molecule) would be a useful approach for overcoming the pH limitation of TFPs, and this can be a complementary approach to the use of artificial cytosine analogues such as thiopseudoisocytosine (L)18a or 2-aminopyridine (M).16d The use of the TO base surrogate is also key to overcoming sequence limitations of TFPs considering the challenge in recognizing the pyrimidine nucleobases (C and U) due to the limited availability of hydrogen bonding donors/acceptors on the Hoogsteen edges compared to the purine nucleobases (A and G). Following this background, we noted that data explaining the binding mechanism of these tFIT–DPQ conjugate probes towards the IAV RNA promoter region are lacking. This is important to fully understand how this conjugation strategy promotes the effective triplex formation of PNA with natural nucleobases even at neutral pH. We expect that a much deeper understanding of the triplex formation of tFIT–DPQ conjugate probes would provide a crucial clue for the advanced design of TFPs targeting the panhandle structure of IAV RNA.
In this work, the focus was on carrying out kinetic analysis of the binding of tFIT–DPQ conjugate probes for the model RNA containing the IAV RNA promoter region. This was done at pH 7.0 (100 mM NaCl, 25 °C) using the stopped-flow technique, together with UV melting experiments and fluorescence titration experiments. In addition to a control probe that lacks the DPQ small molecule unit (NH2-TC(TO-C1)TCTTT-Lys-Lys(Ac)-CONH2, Fig. S1†),25 we examined two types of 8-mer tFIT–DPQ conjugate probes with a lysine residue as the linker between DPQ and the PNA backbone (Fig. 1C). One is the previously reported tFIT–DPQ (Lys) (NH2-TC(TO-C1)TCTTT-Lys-Lys(DPQ)-CONH2) with a Kd of 185 ± 31 nM,25 here we call probe 1. Another is a newly designed one (named probe 2), in which the TO base surrogate was attached to the PNA backbone with a propyl linker (H2N-TC(TO-C3)TCTTT-Lys-Lys(DPQ)-CONH2). This is based on the previous finding by our group,21b which revealed that the TO base surrogate linker significantly affected both the binding affinity and the fluorescence response upon triplex formation with the target dsRNA. It was established that the TO base surrogate connected through the propyl linker in the tFIT probes was the best. Probe 2 was indeed found to work as a stronger binder than any other tFIT–DPQ probes,25 and the binding affinity (Kd) reached 52 ± 16 nM. UV melting experiments confirm that the binding is not based on duplex- or triplex-invasion of tFIT–DPQ probes when binding to the panhandle structure of IAV RNA. Kinetic analysis reveals that the conjugation strategy is responsible for the observed strong binding affinity because of the very fast association rate constant and slow dissociation rate constant. Based on the principle of microscopic reversibility,30 it is highly likely that the rate-limiting step of the association reaction is the DPQ binding to the internal loop region of the target RNA: the binding of the conjugate probe follows a pattern of the DPQ unit binding first to the internal loop region, followed by the tFIT unit binding next to the complementary dsRNA to form the triplex. We discuss these results to better understand the binding event of tFIT–DPQ probes towards the IAV RNA promoter region.
Experimental
All probes were synthesized according to our reports.21b,25 Briefly, the tFIT component of the conjugate probe was synthesized using a Biotage Initiator + microwave peptide synthesizer (Biotage Uppsala, Sweden) based on the standard Fmoc-based solid phase synthesis31 with a Rink-Amide-Chem Matrix resin (Biotage), followed by the attachment of the TO base surrogate and the DPQ small molecule. The probe sequence is complementary to the sequence of the target RNA (5′-A2G3U4A5G6A7A8A9-3′, Fig. 1A), with the G30 * U4 base pair at the opposite position of the TO unit when the triplex is formed. The crude product was purified using a reverse phase HPLC system (Fig. S2†) and characterized by MALDI-TOF-MS (Fig. S3, Table S1†). For experimental details on synthesis and characterization, see the ESI.†Unless otherwise mentioned, all measurements were performed at 25 °C in 10 mM sodium acetate buffer solution (pH 5.5) or sodium phosphate buffer solution (pH 7.0) containing 100 mM NaCl and 1.0 mM EDTA, according to our previous reports.21–25,32 As for the kinetic investigation, we utilized the stopped-flow technique,33 which has been used to quantitatively determine the association rate constant (Kon) of triplex formation by TFPs with dsRNA.21,23,32 The fluorescence quantum yield (Φ) of probes was determined relative to fluorescein in 0.1 M NaOH (Φ = 0.93).34 The details are also given in the ESI.†
Results and discussion
Fluorescence response of the probes and UV melting measurements
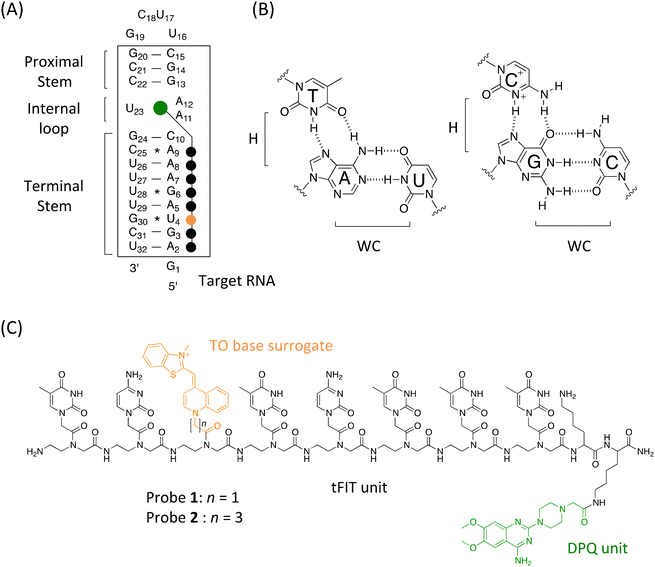
Initially, we examined the fluorescence response of the TO base surrogate in the tFIT–DPQ probes for the target RNA containing the IAV RNA promoter region. As shown in Fig. 2, at acidic pH, the probes showed negligible fluorescence in the absence of RNA because of the free rotation in the TO unit around the monomethine bond causing non-radiative energy decay.21 However, upon the addition of 100 nM target RNA, there was a significant light-up response because of the restricted rotation of TO base surrogates due to the intercalation of the probe into the RNA during triplex formation.21 As observed in our previous studies using simple tFIT probes,21–23 these results indicated that triplex formation indeed induced a light-up response of the probes.It has been recognized that acidic pH is a pre-requisite for the protonation of PNA cytosine (pKa = 4.5) to enhance the G–C+ Hoogsteen base pairs in the target dsRNA during triplex formation.14 Indeed, all the simple tFIT probes with natural nucleobases developed previously by our group worked effectively only at acidic pH.21–23 On the other hand, amazingly, these tFIT–DPQ conjugate probes showed an obvious light-up response even at pH 7.0. In solutions buffered to pH 7.0 at 25 °C, the fluorescence quantum yield Φ of the free probes was determined to be <0.01 and upon binding to target RNA, both probes exhibited an obvious increase (Fig. 2). While the light-up response is dependent on the linker length of the TO base surrogate as previously reported,21b the changes in probe 1 in the absence (free) and presence (bound) of target RNA were found to be Φbound/Φfree = >94 (Φbound = 0.094),25 and the changes in probe 2 were found to be Φbound/Φfree = >5.1 (Φbound = 0.049). Although with lower fluorescence intensity than that at pH 5.5, this result indicates that the binding of the tFIT oligomer to the target RNA occurs even at neutral pH. The possibility that tFIT–DPQ conjugate probes form the duplex-invasion complex with target RNA through Watson–Crick base pairing would be ruled out because probe 2 showed a much weaker response for the complementary single-stranded RNA (5′-r(GAGUAGAAACAAGG)-3′) (Fig. S4†). This is also true for probe 1 as reported previously,25 indicating that the off–on signalling becomes less effective if the probes form the duplex with the target RNA. It is therefore highly likely that triplex formation is indeed taking place at neutral pH between the tFIT–DPQ conjugate probes and the target IAV RNA.
Additional evidence for the absence of duplex or triplex invasion was obtained by UV melting (Tm) experiments to assess the thermal stability of the target IAV RNA in the absence or presence of tFIT–DPQ probes (Fig. 3 and Table 1). For comparison, the effect of a simple tFIT probe without the DPQ moiety (control probe) was also examined (Fig. S5†). Here, the absorbance change at 260 nm was monitored, which is a typical method for thermal melting experiments for duplexes and triplexes.14
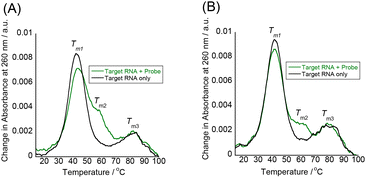 | ||
| Fig. 3 UV melting curves of the target IAV RNA (3.0 μM) recorded at 260 nm in the presence and absence of the probes (3.0 μM) at pH 7.0; (A) probe 1 and (B) probe 2. | ||
| T m1/°C | T m2/°C | T m3/°C | |
|---|---|---|---|
| a Values obtained in solutions buffered to pH 7.0 (10 mM sodium phosphate) containing 100 mM NaCl and 1.0 mM EDTA. [Probe] = 3.0 μM and [RNA] = 3.0 μM. | |||
| None (RNA only) | 42 | ND | 81 |
| Probe 1 | 43 | 58 | 82 |
| Probe 2 | 41 | 61 | 79 |
| Control probe (no DPQ unit) | 44 | ND | 80 |
The model target IAV RNA has two melting temperatures (Tm1 and Tm3) in the absence of tFIT–DPQ probes. Compared to the previous paper that reports the UV melting of a promoter duplex (42.5 °C),35 the lower Tm1 at 42 °C is due to the breaking of the Watson crick base pairs in the terminal stem region while the higher Tm3 at 81 °C is likely to come from the breaking of the duplex in the proximal stem region of the target RNA (cf.Fig. 1A). Significantly, even in the presence of tFIT–DPQ probes, the Tm1 clearly appeared, and almost no changes in melting temperatures were observed (ΔTm1 = ±1 °C). This result reveals that the model target IAV RNA does retain its original duplex structure even after binding with tFIT–DPQ probes. If the binding of tFIT–DPQ probes was based on duplex or triplex invasion, the original Tm1 should have disappeared and/or a significant change in melting behaviour should have been observed as it is known that invasion distorts the original RNA structure.36
Here, it should be noted that a new melting (Tm2) appeared around 60 °C in the presence of tFIT–DPQ probes (Fig. 3), whereas this was absent in the case of the control probe (Fig. S5†). Consistently, a clear melting around 60 °C was also observed when we monitored the change in absorbance at 520 nm due to the TO moiety of tFIT–DPQ probes (Fig. S6†). Although the change at 520 nm is complex and hard to fully interpret, tFIT–DPQ probes seem to finally dissociate from the target RNA around 60 °C. Thus, the Tm2 should be related to probe binding/dissociation. Because the duplex structure in the terminal stem region is already broken above Tm1 (41–43 °C), this should not be due to the triplex formation. Instead, it is reasonable to consider a very simple hybridization between the probe and the complementary single-strand counterpart of the melted target RNA terminal stem in this temperature range. That is, Tm2 corresponds to the melting transition of an RNA–PNA duplex. The DPQ unit may facilitate this hybridization by maintaining the binding to the loop region since the Tm2 was not clearly observed in the case of the control probe (Fig. S5†).
Binding affinity and kinetics
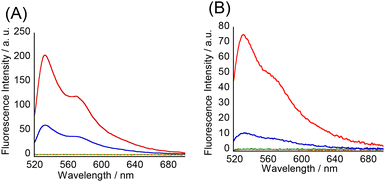
Subsequently, fluorescence titration experiments at pH 7.0 were performed to assess the binding affinity of the tFIT–DPQ conjugate probes towards the target RNA. The light-up response of the TO dye increased as the concentration of the target RNA increased for probe 2 (Fig. 4), in a very similar manner to that of probe 1 previously reported.25 The obtained concentration dependence of the fluorescence response was then used to assess the binding affinity of the probes. The resulting titration curve was fitted using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding isotherm24 (Fig. 4, Inset),‡ which gave a dissociation constant of 52 ± 16 nM for probe 2 (Table 2), which can be observed to be about 3.5 times smaller than that for probe 1 (Kd = 185 ± 31 nM).25 This is as expected and confirms our previous report about the effect of TO linker length on improving binding affinity with such simple modifications.21b Significantly, these tFIT–DPQ conjugate probes showed stronger binding affinity by more than 2 orders of magnitude than DPQ (Kd = 50.5 μM).11,25 Clearly, the binding affinity of tFIT–DPQ conjugate probes drastically improved compared to that of just the DPQ small molecule. This implies that the conjugation of the tFIT oligomer with DPQ leads to an improved binding affinity for the IAV target RNA, for which the triplex formation of the tFIT unit should be responsible. Furthermore, the affinity of tFIT–DPQ conjugate probes is much stronger than that of the control probe without the DPQ unit (Kd = 1.54 ± 0.34 μM, Fig. S7†). This indicates that triplex formation becomes more effective by conjugation with the DPQ unit.
1 binding isotherm24 (Fig. 4, Inset),‡ which gave a dissociation constant of 52 ± 16 nM for probe 2 (Table 2), which can be observed to be about 3.5 times smaller than that for probe 1 (Kd = 185 ± 31 nM).25 This is as expected and confirms our previous report about the effect of TO linker length on improving binding affinity with such simple modifications.21b Significantly, these tFIT–DPQ conjugate probes showed stronger binding affinity by more than 2 orders of magnitude than DPQ (Kd = 50.5 μM).11,25 Clearly, the binding affinity of tFIT–DPQ conjugate probes drastically improved compared to that of just the DPQ small molecule. This implies that the conjugation of the tFIT oligomer with DPQ leads to an improved binding affinity for the IAV target RNA, for which the triplex formation of the tFIT unit should be responsible. Furthermore, the affinity of tFIT–DPQ conjugate probes is much stronger than that of the control probe without the DPQ unit (Kd = 1.54 ± 0.34 μM, Fig. S7†). This indicates that triplex formation becomes more effective by conjugation with the DPQ unit.
| Probe 1 | Probe 2 | Control probe (no DPQ unit) | |
|---|---|---|---|
| a Values obtained in solutions buffered to pH 7.0 (10 mM sodium phosphate) containing 100 mM NaCl and 1.0 mM EDTA at 25 °C. Errors are standard deviations obtained from three independent experiments. b K off values were calculated from Koff = Kd × Kon. c Values from the literature.25 d Re-examined in this study. | |||
| K d [μM] | 0.185 ± 0.031c | 0.052 ± 0.02 | 1.54 ± 0.34d |
| K on [106 M−1 S−1] | 0.65 ± 0.09 | 0.66 ± 0.24 | 0.21 ± 0.15 |
K
off![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b [S−1] b [S−1] |
0.12 | 0.034 | 0.32 |
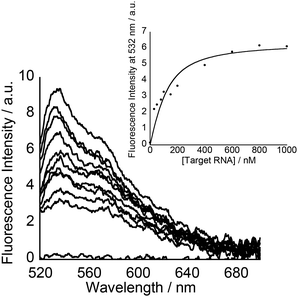
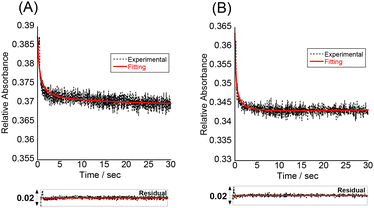
To understand more details about how the tFIT–DPQ conjugate probe binds to the target IAV RNA, we further characterized the binding kinetics by stopped-flow experiments at pH 7.0 (Fig. 5). The absorbance at 260 nm was monitored upon mixing the probes with the target RNA. We observed a decrease in the absorbance at 260 nm upon triplex formation as soon as mixing the PNA and RNA solutions at the same concentrations, which enables the evaluation of the association rate constant (Kon) by a non-linear least squares regression analysis.23,32 We then calculated the dissociation rate constant (Koff) using the equation (Koff = Kd × Kon), where we assumed two-state binding for the bimolecular complexes; referring to the kinetic analysis of triplex formation based on the nucleation-zipping model,32,38 the absorbance changes at 260 nm were analyzed assuming that the only absorbing species are the separated PNA and the target RNA. The obtained kinetic parameters are summarized in Table 2.
The association rate constants of probes 1 and 2 for RNA were determined as 0.65 ± 0.09 × 106 and 0.66 ± 0.24 × 106 M−1 S−1, respectively, which indicates that the conjugate probes bind rapidly to the target RNA as the reaction proceeds. These values are even larger than that of conventional 9-mer PNA-dsRNA triplex formation at pH 5.5 (Kon; 0.28 × 106 M−1 S−1)32a where a total of 6 protonated cytosines are involved in the binding event (H2N-Lys-TCTCCTCCC-CONH2). The conjugation with the DPQ small molecule is therefore highly likely to be important for the observed rapid association for the binding of the conjugate probe to target RNA at neutral pH. This was also confirmed by the comparison of the Kon values to that of the control probe with no DPQ unit (Table 2, Fig. S8†). The control probe showed Kon = 0.21 ± 0.15 × 106 M−1 S−1, and this means probes 1 and 2 are three times faster than the control probe. Furthermore, the Koff value of the control probe (0.32 S−1) is larger than that of the conjugate probes (probe 1, 0.12 S−1; probe 2, 0.033 S−1). These results reveal that the conjugate probes show improved binding affinity compared to the control probe because the association to the target RNA becomes faster while the dissociation becomes slower. The conjugation of the tFIT oligomer with the DPQ unit is thus responsible for the improved binding kinetics. In addition, the stronger binding of probe 2 than that of probe 1 can be explained by the much slower dissociation from the target RNA. The Koff value of probe 2 (0.033 S−1) was indeed one order of magnitude different from that of probe 1 (0.12 S−1), for which the TO-C3 unit should be responsible.
Further analysis of the kinetic data for the tFIT–DPQ probes provided more information about the binding pathway of the probes towards the IAV target RNA. According to the principle of microscopic reversibility,30 the reaction necessarily follows the same pathway in the forward and reverse directions. Since both reactions pass through a common intermediate, the rate-limiting step is the same. Taking into consideration the obtained kinetics, we could propose the association and dissociation mechanism for the probes with target IAV RNA (Fig. 6). From Table 2, it can also be seen that even though probe 2 showed an about 3.5 times increase in binding affinity by exploring TO with the propyl linker, it still has a very similar association rate constant value to probe 1. However, the association rate constant of the control probe is about 3 times less than those of probes 1 and 2. Thus, the rate-limiting step of the association reaction in the formation of the conjugate probe-RNA complex would be the DPQ binding to the internal loop region of the target RNA. Because, if the first associative step involves the rate-limiting triplex formation with the dsRNA region, we should have observed comparable Kon values to that of conventional PNA–dsRNA triplex formation (0.28 × 106 M−1 S−1 at pH 5.5)32a or the control probe without the DPQ unit (0.21 ± 0.15 × 106 M−1 S−1). Also, we should have observed very different Kon values for probe 1 and probe 2. Therefore, we reasoned that the internal loop recognition by the DPQ ligand occurred first and the triplex formation of the tFIT unit with the dsRNA region followed next during the binding to target IAV RNA (Fig. 6).
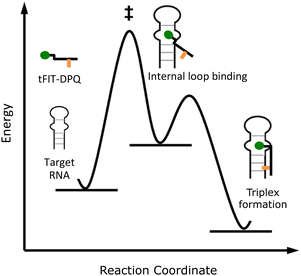 | ||
| Fig. 6 Estimated pathway of the binding between the tFIT–DPQ probes and the target RNA.§ | ||
To further confirm this estimated binding pathway, we again performed a stopped-flow experiment for another set of tFIT–DPQ conjugate probes (Fig. S9, Fig. S10†), in which in place of the lysine used in probes 1 and 2, Dap (2,3-diaminopropionic acid) was utilized as the linker that introduces the DPQ small molecule into the tFIT unit. One of the probes was the previously reported tFIT–DPQ (Dap) (H2N-TC(TO-C1)TCTTT-Lys-Dap(DPQ)-CONH2), here named DPQ-Dap. Compared to probe 1 with the lysine linker, DPQ-Dap with the Dap linker showed an improved binding affinity of 107 nM towards the target IAV RNA.25 We used another probe that is similar to DPQ-Dap but with a mismatch in the tFIT sequence (here all the cytosine residues were replaced with thymine: H2N-TT(TO-C1)TTTTT-Lys-Dap(DPQ)-CONH2). We named this probe DPQ-Dap(pT), and this probe showed a lower binding affinity of 509 nM which is about 5 times weaker than that of the original DPQ-Dap probe (Fig. S11†).
As summarized in Table 3, we observed that although the DPQ-Dap(pT) probe has a weaker binding affinity for the target IAV RNA because of the mismatch, it still has a very similar association rate constant (Kon) value to the DPQ-Dap probe that showed stronger binding affinity. Furthermore, the Kon values of both DPQ-Dap probes (Kon/106 M−1 S−1; DPQ-Dap, 0.89 ± 0.05, DPQ-Dap(pT), 0.81 ± 0.03) are much larger than that of the control probe without the DPQ unit (0.21 ± 0.15 × 106 M−1 S−1). Here, interestingly, the Kon values of both DPQ-Dap probes are a little larger than those of probes 1 and 2 with the lysine linker, and each set of probes has a very similar Kon value depending on the linker (Dap or lysine). Apparently, the association rate constant is governed by the DPQ unit binding to the loop region, not by the tFIT unit for triplex formation with the dsRNA region. Taken together, when tFIT–DPQ conjugate probes bind to the target IAV RNA, it is most likely that the DPQ unit binds first to the internal loop region as proposed in Fig. 6.
| DPQ–Dap probe | DPQ–Dap(pT) probe | |
|---|---|---|
| a Values obtained in solutions buffered to pH 7.0 (10 mM sodium phosphate) containing 100 mM NaCl and 1.0 mM EDTA at 25 °C. Errors are standard deviations obtained from three independent experiments. b K off values were calculated from Koff = Kd × Kon. c Values from the literature.25 | ||
| K d [μM] | 0.107 ± 0.009c | 0.509 ± 0.12 |
| K on [106 M−1 S−1] | 0.89 ± 0.05 | 0.81 ± 0.03 |
K
off![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b [S−1] b [S−1] |
0.095 | 0.41 |
Conclusions
In summary, the triplex formation of tFIT–DPQ probes with the RNA promoter region of the influenza A virus was examined by means of the stopped-flow technique, UV melting and fluorescence titration experiments. We prepared and examined two sets of tFIT–DPQ conjugate probes with different binding affinities. The comparison with the control probe without DPQ clearly revealed that DPQ plays a crucial role in the observed very fast association rate constant in all tFIT–DPQ probes. Importantly, it is highly likely that the binding of the DPQ unit to the internal loop region takes place first, which facilitates the following binding of the tFIT unit to the complementary dsRNA. We believe that the conjugation is indeed the key factor to realize the effective triplex formation of PNA with natural nucleobases at neutral pH. The overall binding affinity of this kind of conjugate probe, on the other hand, was influenced by both DPQ and tFIT units, and was highly sensitive to the slight changes in the probe structure such as the linker between DPQ and tFIT units (lysine or Dap), and the linker length attaching the TO base surrogate to the PNA backbone (C1 or C3). Significantly, probe 2 works as a stronger binder (Kd = 52 nM) than the previously designed DPQ-Dap probe (Kd = 107 nM),25 indicating the further possibility of designing triplex-forming PNA probes with improved binding affinity based on the conjugate probe strategy.We hope that this work inspires the adaptation of the conjugate probe strategy and could provide further insights into the advanced design of fluorogenic probes capable of binding to the IAV RNA promoter with a view towards the sensing of IAV RNA as well as the screening of novel anti-influenza drugs. We also believe that the obtained results would provide valuable insights into molecular interactions for the development of anti-influenza drug candidates targeting the IAV RNA promoter. This concept would be adopted to fit other RNA targets at neutral pH, as has been demonstrated in our previous work that targeted the bacterial rRNA A-site.24 The combined use of excellent artificial PNA nucleobases such as the cytosine analogue 2-aminopyridine (M)16d is expected to further make the probe functions solid. We are now undertaking further studies in these directions.
Author contributions
C. U. O. conceived the study. C. U. O. and H. M. synthesized the probes and characterized their functions. All authors analysed the data. C. U. O., Y. S. and S. N. wrote the paper. Y. S. and S. N. supervised the research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by JSPS KAKENHI Grant Number JP20H02761.References
- N. M. Bouvier and P. Palese, Vaccine, 2008, 26, 49–53 CrossRef PubMed.
- J. S. M. Peiris, L. L. M. Poon and Y. Guan, J. Clin. Virol., 2009, 45, 169–173 CrossRef PubMed.
- S. H. Bae, H. K. Cheong, J. H. Lee, C. Cheong, M. Kainosho and B. S. Choi, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 10602–10607 CrossRef CAS PubMed.
- E. Noble, D. H. Mathews, J. L. Chen, D. H. Turner, T. Takimoto and B. Kim, J. Biol. Chem., 2011, 286, 22965–22970 CrossRef CAS PubMed.
- A. Bottini, S. K. De, B. Wu, C. Tang, G. Varani and M. Pellecchia, Chem. Biol. Drug Des., 2015, 86, 663–673 CrossRef CAS PubMed.
- T. Brown, Harv. Bus. Rev., 2008, 86, 84–92 Search PubMed.
- J. R. Thomas and P. J. Hergenrother, Chem. Rev., 2008, 108, 1171–1224 CrossRef CAS PubMed.
- H. Kim, M. K. Lee, J. Ko, C.-J. Park, M. Kim, Y. Jeong, S. Hong, G. Varani and B.-S. Choi, Mol. Biosyst., 2012, 8, 2857–2859 RSC.
- (a) P. N. Asare-Okai and C. S. Chow, Anal. Biochem., 2011, 408, 269–276 CrossRef CAS PubMed; (b) D. Perez-Fernandez, D. Shcherbakov, T. Matt, N. C. Leong, I. Kudyba, S. Duscha, H. Boukari, R. Patak, S. R. Dubbaka, K. Lang, M. Meyer, R. Akbergenov, P. Freihofer, S. Vaddi, P. Thommes, V. Ramakrishnan, A. Vasella and E. C. Bottger, Nat. Commun., 2014, 5, 3112 CrossRef PubMed.
- (a) Y. Sato, Y. Aiba, S. Yajima, T. Tanabe, K. Higuchi and S. Nishizawa, ChemBioChem, 2019, 20, 2752–2756 CrossRef CAS PubMed; (b) Y. Sato, S. Yajima, A. Taguchi, K. Baba, M. Nakagomi, Y. Aiba and S. Nishizawa, Chem. Commun., 2019, 55, 3183–3186 RSC.
- M.-K. Lee, A. Bottini, M. Kim, M. F. Bardaro, Z. Zhang, M. Pellecchia, B.-S. Choi and G. Varani, Chem. Commun., 2014, 50, 368–370 RSC; A. Bottini, S. K. De, B. Wu, C. Tang, G. Varani and M. Pellecchia, Chem. Biol. Drug Des., 2015, 86, 663–673 CrossRef CAS PubMed.
- (a) J. Kesy, K. M. Patil, S. R. Kumar, Z. Shu, H. Y. Yong, L. Zimmermann, A. A. L. Ong, D.-D. K. Toh, M. S. Krishna, L. Yang, J.-L. Decout, D. Luo, M. Prabakaran, G. Chen and E. Kierzek, Bioconjugate Chem., 2019, 30, 931–943 CrossRef CAS PubMed; (b) M. S. Krishna, D.-F. Toh, Z. Meng, A. A. L. Ong, Z. Wang, Y. Lu, K. Xia, M. Prabakaran and G. Chen, Anal. Chem., 2019, 91, 5331–5338 CrossRef CAS PubMed.
- P. E. Nielsen, M. Egholm, R. H. Berg and O. Buchardt, Science, 1991, 254, 1497–1500 CrossRef CAS PubMed.
- (a) M. Li, T. Zengeya and E. Rozners, J. Am. Chem. Soc., 2010, 132, 8676–8681 CrossRef CAS PubMed; (b) K. Aupeix, R. Le Tinévez and J.-J. Toulmé, FEBS Lett., 1999, 449, 169–174 CrossRef CAS PubMed.
- V. Kotikam, S. D. Kennedy, J. A. Mackay and E. Rozners, Chem. – Eur. J., 2019, 25, 4367–4372 CrossRef CAS PubMed; V. Kumar, N. Brodyagin and E. Rozners, ChemBioChem, 2020, 21, 3410–3416 CrossRef PubMed.
- (a) P. Gupta, T. Zengeya and E. Rozners, Chem. Commun., 2011, 47, 11125–11127 RSC; (b) T. Zengeya, M. Li and E. Rozners, Bioorg. Med. Chem. Lett., 2011, 21, 2121–2124 CrossRef CAS PubMed; (c) P. Gupta, O. Muse and E. Rozners, Biochemistry, 2012, 51, 11125–11127 CrossRef PubMed; (d) T. Zengeya, P. Gupta and E. Rozners, Angew. Chem., Int. Ed., 2012, 51, 12593–12596 CrossRef CAS PubMed; (e) O. Muse, T. Zengeya, J. Mwaura, D. Hnedzko, D. W. McGee, C. T. Grewer and E. Rozners, ACS Chem. Biol., 2013, 8, 1683–1686 CrossRef CAS PubMed; (f) I. Kumpina, N. Brodyagin, J. A. MacKay, S. D. Kennedy, M. Katkevics and E. Rozners, J. Org. Chem., 2019, 84, 13276–13298 CrossRef CAS PubMed.
- T. Endoh, D. Hnedzko, E. Rozners and N. Sugimoto, Angew. Chem., Int. Ed., 2016, 55, 899–903 CrossRef CAS PubMed; T. Endoh, C. Annoni, D. Hnedzko, E. Rozners and N. Sugimoto, Phys. Chem. Chem. Phys., 2016, 18, 32002–32006 RSC.
- (a) G. Devi, Z. Yuan, Y. Lu, Y. Zhao and G. Chen, Nucleic Acids Res., 2014, 42, 4008–4018 CrossRef CAS PubMed; (b) D.-F. K. Toh, G. Devi, K. M. Patil, Q. Qu, M. Maraswami, Y. Xiao, T. P. Loh, Y. Zhao and G. Chen, Nucleic Acids Res., 2016, 44, 9071–9082 CAS; (c) A. A. L. Ong, D.-F. K. Toh, K. M. Patil, Z. Meng, Z. Yuan, M. S. Krishna, G. Devi, P. Haruehanroengra, Y. Lu, K. Xia, K. Okamura, J. Sheng and G. Chen, Biochemistry, 2019, 58, 1319–1331 CrossRef CAS PubMed.
- S. Jeong, D.-M. Kim, S.-Y. An, D.-H. Kim and D.-E. Kim, Anal. Biochem., 2018, 561–562, 66–69 CrossRef CAS PubMed; H. Lee, D.-M. Kim and D.-E. Kim, Analyst, 2020, 145, 8002–8007 RSC.
- S.-T. Yu, C. T. Bui, D. T. H. Kim, A. V. T. Nguyen, T. T. T. Trinh and S.-J. Yeo, Sci. Rep., 2018, 8, 13468 CrossRef PubMed.
- (a) T. Sato, Y. Sato and S. Nishizawa, J. Am. Chem. Soc., 2016, 138, 9397–9400 CrossRef CAS PubMed; (b) T. Sato, Y. Sato and S. Nishizawa, Chem. – Eur. J., 2017, 23, 4079–4088 CrossRef CAS PubMed.
- T. Chiba, T. Sato, Y. Sato and S. Nishizawa, Org. Biomol. Chem., 2017, 15, 7765–7769 RSC; Y. Yoshino, Y. Sato and S. Nishizawa, Anal. Chem., 2019, 91, 14254–14260 CrossRef CAS PubMed.
- (a) T. Tanabe, T. Sato, Y. Sato and S. Nishizawa, RSC Adv., 2018, 8, 42095–420990 RSC; (b) Y. Sato, Y. Takahashi, T. Tanabe and S. Nishizawa, Org. Biomol. Chem., 2020, 18, 4009–4013 RSC.
- E. T. T. Lee, Y. Sato and S. Nishizawa, Chem. Commun., 2020, 56, 14976–14979 RSC; Y. Sato, M. Rokugawa, S. Ito, S. Yajima, H. Sugawara, N. Teramae and S. Nishizawa, Chem. – Eur. J., 2018, 24, 13862–13870 CrossRef CAS PubMed.
- Y. Sato, H. Miura, T. Tanabe, C. U. Okeke, A. Kikuchi and S. Nishizawa, Anal. Chem., 2022, 94, 7814–7822 CrossRef CAS PubMed.
- O. Köhler, D. V. Jarikote and O. Seitz, ChemBioChem, 2005, 6, 69–77 CrossRef PubMed; F. Hövelmann, L. Bethge and O. Seitz, ChemBioChem, 2012, 13, 2072–2081 CrossRef PubMed; F. Hövelmann, I. Gaspar, A. Ephrussi and O. Seitz, J. Am. Chem. Soc., 2013, 135, 19025–19032 CrossRef PubMed; F. Hövelmann, I. Gaspar, S. Loibl, E. A. Ermilov, B. Röder, J. Wengel, A. Ephrussi and O. Seitz, Angew. Chem., Int. Ed., 2014, 53, 11370–11375 CrossRef PubMed; F. Hövelmann, I. Gaspar, J. Chemiolo, M. Kasper, J. Steffen, A. Ephrussi and O. Seitz, Chem. Sci., 2016, 7, 128–135 RSC; F. Hövelmann and O. Seitz, Acc. Chem. Res., 2016, 49, 714–723 CrossRef PubMed.
- N. Kolevzon, D. Hashoul, S. Naik, A. Rubinstein and E. Yavin, Chem. Commun., 2016, 52, 2405–2407 RSC; I. Peled and E. Yavin, ACS Omega, 2018, 3, 3813–3818 CrossRef CAS PubMed; D. Hashoul, R. Shapira, M. Falchenko, O. Tepper, V. Paviov, A. Nissan and E. Yavin, Biosens. Bioelectron., 2019, 137, 271–278 CrossRef PubMed; O. Tepper, H. Zheng, D. H. Appella and E. Yavin, Chem. Commun., 2021, 57, 540–543 RSC; O. Tepper, I. Peled, Y. Fastman, A. Heinberg, V. Mitesser, R. Dzikowski and E. Yavin, ACS Sens., 2022, 7, 50–59 CrossRef PubMed.
- V. Karunakaran, J. L. P. Lustres, L. Zhao, N. P. Ernsting and O. Seitz, J. Am. Chem. Soc., 2006, 128, 2954–2962 CrossRef CAS PubMed.
- S. L. Wicks and A. E. Hargrove, Methods, 2019, 167, 3–14 CrossRef CAS PubMed.
- R. L. Burwell and R. G. Pearson, J. Phys. Chem., 1966, 70, 300–302 CrossRef CAS.
- D. V. Jarikote, O. Köhler, E. Socher and O. Seitz, Eur. J. Org. Chem., 2005, 2005, 3187–3195 CrossRef.
- (a) T. Sato, N. Sakamoto and S. Nishizawa, Org. Biomol. Chem., 2018, 16, 1178–1187 RSC; (b) T. Sato, Y. Sato and S. Nishizawa, Biopolymers, 2021, e23474 Search PubMed.
- S. Wang, A. E. Friedman and E. T. Kool, Biochemistry, 1995, 34, 9774–0784 CrossRef CAS PubMed.
- R. Sjoback, J. Nygren and M. Kubista, Spectrochim. Acta, Part A, 1995, 51, L7–L21 CrossRef.
- E. Noble, D. H. Mathews, J. L. Chen, D. H. Turner, T. Takimoto and B. Kim, J. Biol. Chem., 2011, 286, 22965–22970 CrossRef CAS PubMed.
- P. E. Nielsen, Chem. Biodivers., 2010, 7, 786–804 CrossRef CAS PubMed.
- P. Wittung, P. Nielsen and B. Nordén, Biochemistry, 1997, 36, 7973–7979 CrossRef CAS PubMed.
- (a) M. Rougée, B. Faucon, J. L. Mergny, F. Barcelo, C. Giovannangeli, T. Garestier and C. Hélène, Biochemistry, 1992, 31, 9269–9278 CrossRef PubMed; (b) P. Alberti, P. B. Arimondo, J.-L. Mergny, T. Garestier, C. Hélène and J.-S. Sun, Nucleic Acids Res., 2002, 30, 5407–5415 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, fluorescence response, melting curves, and stopped-flow kinetics trace. See DOI: https://doi.org/10.1039/d3ob00262d |
‡ It is reported that thymine-rich homopyrimidine PNA oligomers prefer binding to double-stranded DNA by triplex invasion (PNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DNA = 2 DNA = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1).37 While the target here is double-stranded RNA, there is a posibility that probe 2 binds to the target RNA by triplex invasion under the condition of [probe 2] > [RNA]. 1).37 While the target here is double-stranded RNA, there is a posibility that probe 2 binds to the target RNA by triplex invasion under the condition of [probe 2] > [RNA]. |
| § Here we just assume the internal loop binding as a reaction intermediate with lower energy, referring to the model of triplex formation, the so-called nucleation-zipping model (the activation energy is negative in this case!).32,38 More in-depth research should be performed to draw the exact energy diagram. |
| This journal is © The Royal Society of Chemistry 2023 |