Defining the substrate scope of DNAzyme catalysis for reductive amination with aliphatic amines†
Shukun
Yang
 and
Scott K.
Silverman
and
Scott K.
Silverman
 *
*
Department of Chemistry, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, IL 61801, USA. E-mail: sks@illinois.edu; Web: https://scs.illinois.edu/silverman Web: https://twitter.com/sksilverman
First published on 8th February 2023
Abstract
Amines can be alkylated using various reactions, such as reductive amination of aldehydes. In this study, we sought DNAzymes as catalytic DNA sequences that promote reductive amination with aliphatic amines, including DNA-anchored peptide substrates with lysine residues. By in vitro selection starting with either N40 or N20 random DNA pools, we identified many DNAzymes that catalyze reductive amination between the DNA oligonucleotide-anchored aliphatic amino group of DNA-C3-NH2 (C3 = short three-carbon tether) and a DNA-anchored benzaldehyde group in the presence of NaCNBH3 as reducing agent. At pH 5.2, 6.0, 7.5, or 9.0 in the presence of various divalent metal ion cofactors including Mg2+, Mn2+, Zn2+ and Ni2+, the DNAzymes have kobs up to 0.12 h−1 and up to 130-fold rate enhancement relative to the DNA-splinted but uncatalyzed background reaction. However, analogous selection experiments did not lead to any DNAzymes that function with DNA-HEG-NH2 [HEG = long hexa(ethylene glycol) tether], or with short- and long-tethered DNA-AAAKAA and DNA-HEG-AAAKAA lysine-containing hexapeptide substrates (A = alanine, K = lysine). Including a variety of other amino acids in place of the neighboring alanines also did not lead to DNAzymes. These findings establish a practical limit on the substrate scope of DNAzyme catalysis for N-alkylation of aliphatic amines by reductive amination. The lack of DNAzymes for reductive amination with any substrate more structurally complex than DNA-C3-NH2 is likely related to the challenge in binding and spatially organizing those other substrates. Because other reactions such as aliphatic amine N-acylation are feasible for DNAzymes with DNA-anchored peptides, our findings show that the ability to identify DNAzymes depends strongly on both the investigated reaction and the composition of the substrate.
Introduction
DNAzymes (deoxyribozymes, DNA enzymes) are catalytic sequences of DNA, analogous to protein enzymes as catalytic amino acid sequences and ribozymes as catalytic RNA.1–8 Although both protein enzymes and ribozymes evolved naturally, DNAzymes are identified by in vitro selection in the laboratory starting from random sequence populations.9–13 We have sought to expand DNAzyme catalysis to many reactions, including those relevant to peptide modification14–16 such as amine acylation.17 Another such reaction is amine alkylation, where the lysine (Lys) side chain is naturally methylated in many biologically relevant contexts,18,19 and site-selective Lys alkylation is useful for introducing biochemical and biophysical probes.20–23 Because Lys has an aliphatic amine side chain, here we assessed the ability to identify DNAzymes that engage various kinds of aliphatic amines in alkylation reactions.Reductive amination is a broadly useful N-alkylation reaction in which an amine and an aldehyde or ketone condense, and the resulting imine (or iminium ion) is reduced by a hydride donor, often but not always a boron-based compound such as NaBH4 or NaCNBH3.24–26 Biocatalytic reductive amination is increasingly being explored.27 In a previous report, we serendipitously found DNAzymes that catalyze Ni2+-dependent reductive amination between the aromatic N2-amino group of a guanosine nucleobase at the 5′-terminus of an RNA strand and the 3′-dialdehyde at the NaIO4-oxidized ribonucleotide terminus of a DNA strand, with NaCNBH3 as reducing agent.28 Here we reasoned that the greater nucleophilicity of an aliphatic amine should also allow DNAzyme-catalyzed reductive amination, but only if DNAzymes are able to interact with and spatially organize the amine substrate. In a previous study, we showed that substrate reactivity can be more important than structural organization for a DNAzyme-catalyzed aliphatic amine reaction other than reductive amination (N–P bond formation).29
In the present work, it was not possible to predict in advance of experiment whether the substrate reactivity or structural organization consideration would dominate the outcome. Therefore, we used in vitro selection to seek DNAzymes that catalyze reductive amination between DNA-anchored aliphatic amines and a DNA-anchored benzaldehyde reaction partner (Fig. 1), where the latter was chosen as the aldehyde primarily because of its synthetic accessibility. The physical basis of in vitro selection was PAGE shift upon DNAzyme-catalyzed reaction between the amine and benzaldehyde. We identified DNAzymes for our initially evaluated and most structurally organized substrate, DNA-C3-NH2, that presents the aliphatic amino group on a short three-carbon tether at the 3′-terminus of a DNA oligonucleotide anchor strand. However, in subsequent selection experiments, we did not find any DNAzymes when the aliphatic amino group of the substrate DNA-HEG-AAAKAA was part of a Lys-containing hexapeptide connected to a DNA anchor strand by a long hexa(ethylene glycol) tether. We also did not find DNAzymes with the substrates DNA-AAAKAA and DNA-ASXKXS (short non-HEG tether), or DNA-HEG-NH2 and DNA-HEG-ASXKXS (long HEG tether), where S = Ser, and X = Lys (K), Glu (E), or Phe (F) to provide a variety of electrostatic and hydrophobic contexts near the Lys residue. Collectively, these observations reveal a practical limit on the substrate scope of DNAzyme-catalyzed reductive amination of aliphatic amines. The main challenge for this reaction appears to be DNAzyme binding of less structurally organized amine substrates.
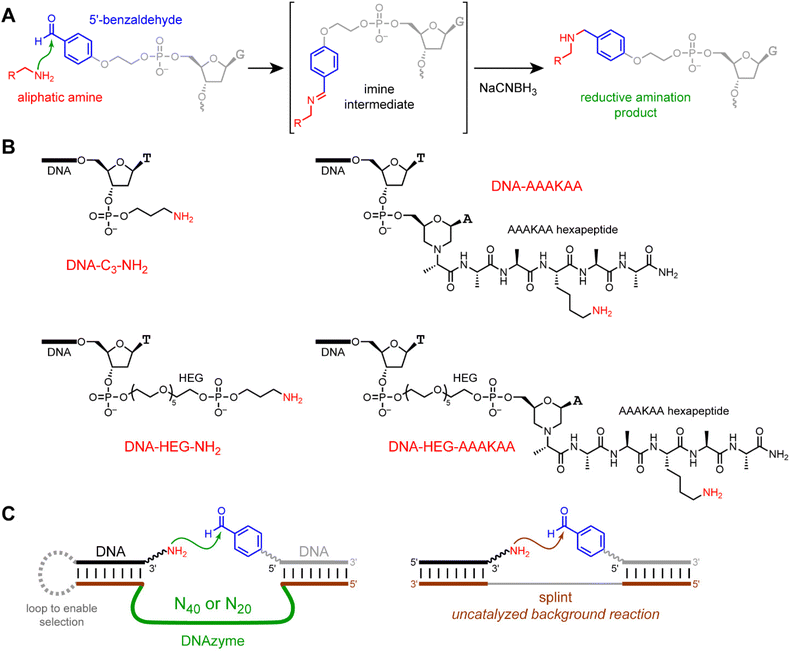 | ||
| Fig. 1 Reductive amination reaction, aliphatic amine substrates, and in vitro selection for DNAzyme-catalyzed reductive amination. (A) N-Alkylation by reductive amination of an aliphatic amine with a 5′-benzaldehyde oligonucleotide. (B) The four aliphatic amine substrates used in this study. HEG = hexa(ethylene glycol). For the two substrates with the hexapeptide, several sequence variants of the AAAKAA peptide were examined. Counting inclusively from the thymidine 3′-O atom to the NH2 group, the tether lengths in atoms are as follows: DNA-C3-NH2 7, DNA-AAAKAA 22, DNA-HEG-NH2 27, and DNA-HEG-AAAKAA 42. (C) The key selection step, in which DNAzyme-catalyzed imine (or iminium ion) formation between the aliphatic amine and the 5′-benzaldehyde reaction partner is followed by NaCNBH3 reduction, enabling PAGE-shift enrichment of the DNA population in catalytically active sequences. At right is shown is the uncatalyzed splinted background reaction. See Fig. 6 in the Experimental section for full nucleotide and primer sequences. | ||
Results
Selection experiments with DNA-C3-NH2 substrate
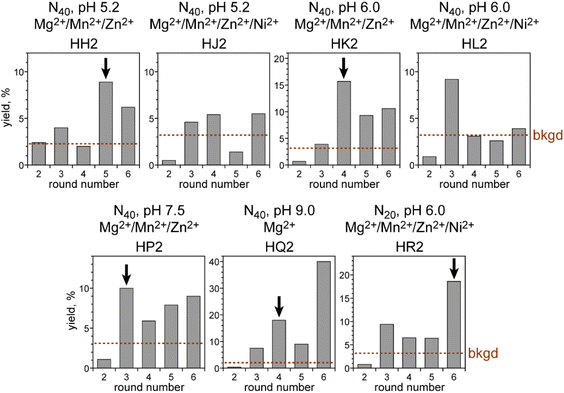
We began by performing in vitro selection using the simplest and shortest DNA-C3-NH2 substrate (Fig. 1B), the 5′-benzaldehyde electrophilic reaction partner that was used throughout the remainder of this study, and one of several incubation conditions chosen on the basis of our experience with other selection experiments.14–16 The three key incubation variables that we explored were random region length, pH, and metal ion cofactors. For the initially random region length, we focused most of our effort on the commonly used N40, i.e., 40 DNA nucleotides in the initially random sequence pool. With N40, sequence space is sampled on the order of 10−10, because we start with 200 pmol ≈ 1014 sequences out of 440 ≈ 1024 possible sequences. The incubation pH was one of 5.2, 6.0, 7.5, or 9.0 (70 mM NaOAc, 70 mM MES, 70 mM HEPES, or 50 mM CHES, respectively). At each of pH 5.2, 6.0, and 7.5 we included all three of 40 mM MgCl2, 20 mM MnCl2, and 1 mM ZnCl2 as common DNAzyme cofactors; an additional selection experiment was performed at each of pH 5.2 and 6.0, providing 20 mM NiCl2 on the basis of our previous report with nucleobase reductive amination28 as well as the other three divalent ions. At pH 9.0, we included only Mg2+, whereas Mn2+ and Zn2+ were excluded because they oxidize and precipitate, respectively, at this higher pH value, and Ni2+ was also omitted. Finally, we performed one N20 selection experiment at pH 6.0 with all four of Mg2+, Mn2+, Zn2+, and Ni2+. With N20, sequence space is ∼102 oversampled, again starting with 200 pmol ≈ 1014 sequences but now with only 420 ≈ 1012 possible sequences. All selection incubations were performed with 30 mM NaCNBH3 at 37 °C for 16 h. The uncatalyzed splinted background yield was 2–3% in all incubation conditions, which allows for a substantial enrichment factor of at least 30-fold in each selection round.Five of the seven selection experiments led to a substantial increase in catalytic activity, i.e., reductive amination yield, as the selection rounds were iterated (Fig. 2). Only the two N40 selections at pH 5.2 and 6.0 that additionally included Ni2+ did not lead to clear emergence of reductive amination activity by round 6 and were set aside. At least one individual DNAzyme was identified from each of the five successful selection experiments, with all sequences in Fig. 3. A wider variety of sequences was found from the two selections at pH 6.0 than from the selections at the other three pH values.
Characterization of individual DNAzymes
DNAzymes from each of the five selections were characterized with regard to their rates and yields, with the highest-yielding DNAzymes from each selection shown in Fig. 4. The N40 and N20 DNAzymes that were identified at pH 6.0 had relatively strong catalytic activity, whereas the N40 DNAzymes found at pH 5.2, 7.5, and 9.0 were less active. All DNAzymes had kobs in the range of 0.07 to 0.12 h−1. The pH 6.0 DNAzymes had yields at 48 h of 50–80%; the DNAzymes at the other pH values had 48 h yields below 30%. The highest rate enhancement value was found for 4HK206 and 6HR205, each with kobs/kbkgd of 130.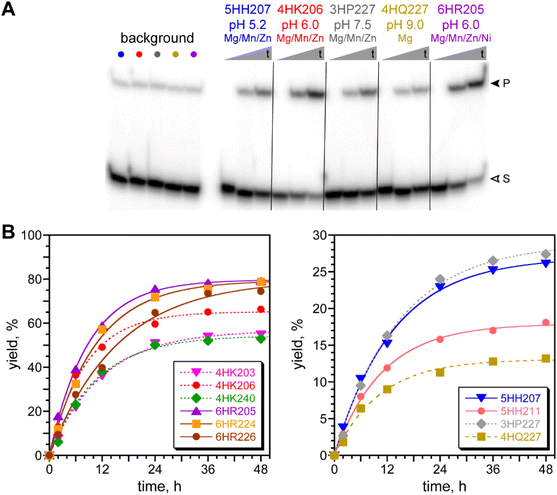 | ||
| Fig. 4 Assays of DNAzymes identified by in vitro selection for reductive amination with the DNA-C3-NH2 substrate. (A) Representative PAGE image for five DNAzymes. All DNAzymes were assayed in the incubation conditions used for their identification, with 100 mM NaCNBH3 to decrease the likelihood of this reagent being rate-limiting. Background = complementary DNA splint in place of DNAzyme, to assess the uncatalyzed background reaction (five background assays, in the same order of incubation conditions as for the five DNAzymes). Shown are representative timepoints (t = 0.5 min, 6 h, 48 h for DNAzymes; 48 h for background; S = substrate, P = product). (B) Kinetic plots for the five representative DNAzymes. kobs values (h−1): 4HK203 0.084; 4HK206 0.122; 4HK240 0.094; 6HR205 0.115, 6HR224 0.096, 6HR226 0.065, 5HH207 0.075, 5HH211 0.092, 3HP227 0.069, 4HQ227 0.098. kbkgd values (left to right in PAGE image, 10−4 h−1): 7.9, 9.2, 8.8, 9.0, 7.3. Additional data for individual DNAzymes not shown in Fig. 4 (five 4HK2, eleven 4HR2) are found in Table S1.† | ||
Mfold predictions30 of the secondary structures of each of the five representative DNAzymes of Fig. 4 are shown in the ESI (Table S2 and Fig. S1†). We have not sought to test experimentally any of these predicted secondary structures. Determining three-dimensional DNAzyme structures31–33 is a separate challenge, also not addressed as part of the present study.
Product validation for individual DNAzymes
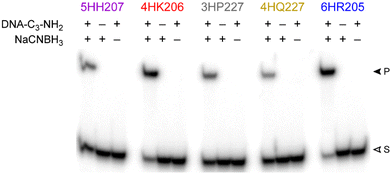
MALDI-TOF mass spectrometry was used to validate product identities for the five representative DNAzymes (Table 1). Negative control experiments confirmed that both the amino group of the DNA-C3-NH2 substrate and the NaCNBH3 were strictly required to observe each DNAzyme's reaction product (Fig. 5), as expected for the reductive amination reaction.| DNAzyme | m/z calcd | m/z found | Δ |
|---|---|---|---|
| 5HH207 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.4 734.4 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735.3 735.3 |
+0.008% |
| 4HK206 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.4 734.4 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.2 734.2 |
−0.002% |
| 3HP227 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.4 734.4 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 735.3 735.3 |
+0.008% |
| 4HQ227 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.4 734.4 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.2 734.2 |
−0.002% |
| 6HR205 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.4 734.4 |
11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 734.7 734.7 |
+0.003% |
Selection experiments with DNA-HEG-AAAKAA substrate
We turned our attention to the DNA-HEG-AAAKAA substrate, seeking to identify DNAzymes for reductive amination with the Lys aliphatic amino group, where the longer HEG tether challenges DNAzymes to function with little structural preorganization of the peptide. For both N40 and N20 initially random pools, we used all four of pH 5.2, 6.0, 7.5, and 9.0, with Mg2+/Mn2+/Zn2+ at pH 5.2, 6.0, and 7.5 and Mg2+ at pH 9.0. We also evaluated N20 at pH 6.0 with Mg2+/Mn2+/Zn2+/Ni2+, which were the same N20 conditions that we used successfully in the first set of selections with DNA-C3-NH2. Unfortunately, in all nine of these selection experiments with DNA-HEG-AAAKAA (30 mM NaCNBH3, 37 °C, 16 h; uncatalyzed splinted background yield 3–5%), no DNAzymes were identified. Each selection experiment was iterated for up to 12 rounds, but no reductive amination activity was observed above the uncatalyzed splinted background level.We performed nine additional selection experiments, this time including 1 mM of 5-methoxyanthranilic acid (5-MA) as a nucleophilic aniline organocatalyst that can form an imine with the 5′-benzaldehyde reaction partner.34 In principle, 5-MA enables an alternative reaction pathway in which the imine from the Lys aliphatic amine and the 5′-benzaldehyde can be formed by transimination from the initially formed 5-MA imine with 5′-benzaldehyde, followed by NaCNBH3 reduction. We evaluated 5-MA with three combinations of substrate and DNA pool: DNA-C3-NH2 substrate and N40 pool, or DNA-HEG-AAAKAA substrate and N40 or N20 pool. For all three combinations, each with all three incubation conditions at pH 6.0, 7.5, and 9.0 with Mg2+/Mn2+/Zn2+ or Mg2+ alone as described above (30 mM NaCNBH3, 37 °C, 16 h), again no DNAzymes that catalyze reductive amination were found.
Additional selection experiments to probe scope of aliphatic amine reactivity
Inspection of Fig. 1B shows that there are two structural differences between DNA-C3-NH2 and DNA-HEG-AAAKAA: the absence or presence of the long HEG tether, and separately, whether the aliphatic amine is part of a Lys residue in a hexapeptide. Given the divergent selection outcomes to this point, we pursued two additional selection experiments at pH 7.5 with the remaining two substrates DNA-AAAKAA (no HEG) and DNA-HEG-NH2 (no peptide). This allowed us to determine which structural aspects of DNA-HEG-AAAKAA contribute to the inability to identify reductive amination DNAzymes for this substrate, even though DNA-C3-NH2 does lead to DNAzymes. In parallel, we considered that the ability of a DNAzyme to interact with the Lys portion of the substrate and enable catalysis may depend on the chemical environment contributed by the nearby peptide side chains and thus the peptide sequence. With this in mind, we performed selections at pH 7.5 with DNA-ASXKXS and DNA-HEG-ASXKXS, where for both hexapeptides, X = K, E, or F (thus, six more selections). For each of the amine substrates of these combined eight new selection experiments, the uncatalyzed splinted background reaction was relatively fast, so we decreased the incubation time in all selection rounds from 16 h to one of 2 h, 1 h, or 0.5 h depending on the substrate, keeping the background yield to 3–6% in each case. None of these eight selection experiments (Mg2+/Mn2+/Zn2+, 100 mM NaCNBH3, 37 °C) led to reductive amination activity above background after 14 rounds. Finally, we tested all 15 of the 4HK2 and 6HR2 DNAzymes that have >40% yield (Fig. 4 and Table S1†) with each of DNA-HEG-NH2 and DNA-HEG-AAAKAA (8 h and 24 h, respectively; DNA-AAAKAA was not tested because of its relatively high uncatalyzed background yield), as well as 5HH207 and 3HP227 with DNA-HEG-AAAKAA (24 h), but no yield above background was observed with these less organized substrates.Discussion
Most of our selection experiments for reductive amination with the DNA-C3-NH2 substrate led to DNAzymes that have substantial rate enhancement above the uncatalyzed splinted background reaction, with the highest activity shown by the DNAzymes identified at pH 6.0 (Fig. 4). We speculate that this pH optimum for emergence of DNAzymes by in vitro selection arises from a balance of pH effects on the multiple steps of imine formation and the subsequent imine reduction, but we have not yet experimentally investigated this interesting mechanistic question. In sharp contrast, no DNAzymes were found with DNA-HEG-NH2, DNA-AAAKAA, or DNA-HEG-AAAKAA, or with any of the evaluated DNA-ASXKXS and DNA-HEG-ASXKXS substrates. The most straightforward interpretation of these outcomes in the context of modifying aliphatic amines by reductive amination is that DNAzymes have substantial difficulty in binding to, and spatially organizing, any of the less structurally organized amine substrates—i.e., anything more structurally complex than DNA-C3-NH2.The 5′-benzaldehyde reaction partner has sufficient intrinsic reactivity to engage in the intended reductive amination reaction, as inferred from the successful DNA-C3-NH2 selections. The DNAzyme kobs values on the order of 0.1 h−1 are modest, perhaps as expected because even the uncatalyzed splinted background reaction is rather slow (Fig. 4), and the selection incubation time of 16 h did not challenge the DNAzymes to perform any better. Merely changing the amine substrate to DNA-HEG-NH2 in selection did not lead to DNAzymes, establishing that the longer HEG tether alone is enough to prevent emergence of DNAzyme catalysis. With this finding, it is consistent that DNA-HEG-AAAKAA was also unsuccessful in selections, including with the alternative imine electrophile formed using 5-MA, because DNA-HEG-AAAKAA should be an even more challenging substrate than DNA-HEG-NH2. The difficulty for DNAzymes to bind short peptide substrates in various DNA-catalyzed reactions has been a recurring theme in our studies. Sometimes DNAzymes for a particular reaction can be found with one peptide substrate sequence but not another.35 In the present situation, changing the peptide sequence from AAAKAA to ASXKXS (X = K, E, or F) did not enable DNAzymes to emerge from selection, despite the increased basis for interaction (i.e., charges and hydrophobic surfaces) of functional groups near the intended nucleophilic Lys residue.
Three caveats on our conclusions are appropriate. First, as the electrophilic reaction partner we evaluated only the 5′-benzaldehyde, not others such as an aliphatic aldehyde. Performing additional experiments is required to say with confidence whether the analogous outcomes would be observed if other aldehydes were used for in vitro selection. Second, we evaluated primarily the N40 random region length, and less comprehensively N20. It is possible that other random region lengths could lead to different outcomes.36 Finally, we have not yet explored the possibility of using directed evolution by reselection with any of the DNAzymes that function with the DNA-C3-NH2 substrate, to determine whether sequence variants can have activity with the less organized amine substrates. There is of course no guarantee that such experiments would be successful, although we have occasionally had favorable results with broadly similar reselections (e.g., ref. 37). The observation that none of the tested DNAzymes have any detectable catalytic activity with substrates less organized than DNA-C3-NH2 is not encouraging in this regard.
In summary, DNAzymes that catalyze N-alkylation by reductive amination were found with the simplest DNA-C3-NH2 substrate, but not with any of the more structurally complex amine substrates of Fig. 1B. These findings establish an experimental limit on the substrate scope of DNAzyme catalysis for reductive amination with aliphatic amines, likely originating in the difficulty of binding the less structurally organized amine substrates. For comparison, our recent work with analogous DNAzymes for amine acylation did lead to peptide acylation (e.g., with DNA-HEG-AAAKAA),17 and therefore the feasibility of DNAzyme catalysis depends on the type of chemical reaction and not solely the substrate. Our earlier study of Lys modification in another context showed that substrate reactivity can be more important than structural organization,29 which is the opposite of our findings here. Taken together, our studies suggest that in the context of DNAzyme catalysis, a rather subtle balance of reactivity and structural effects must be assessed individually for each combination of reaction and substrate.
Experimental section
Oligonucleotide and peptide preparative procedures
DNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA) or prepared by solid-phase synthesis on an ABI 394 instrument using reagents from Glen Research (Sterling, VA). Oligonucleotides were purified by 7 M urea denaturing 20% or 8% PAGE with running buffer 1× TBE (89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3), extracted from the polyacrylamide with TEN buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 300 mM NaCl), and precipitated with ethanol.The Glen Research 5′-benzaldehyde modifier (5′-aldehyde-modifier C2, cat. no. 10-1933) was coupled manually, and the resulting oligonucleotide was deprotected from the 2,2-diethyl-1,3-propanediol acetal to the aldehyde with 80% aqueous acetic acid (1 hour, room temperature) according to the manufacturer's instructions. The 18 nt 5′-benzaldehyde oligonucleotide was purified by HPLC, using a Shimadzu Prominence instrument with a Phenomenex Gemini-NX C18 column (5 μm, 10 × 250 mm), solvent A (20 mM triethylammonium acetate in 50% acetonitrile/50% water, pH 7.0), solvent B (20 mM triethylammonium acetate in water, pH 7.0), gradient of 10% A/90% B at 0 min to 40% A/60% B at 30 min, and flow rate of 3.5 mL min−1, with retention time 18.3 min.
The AAAKAA or ASXKXS hexapeptide was prepared by solid-phase synthesis using Fmoc Rink amide MBHA resin as described.38 Without HPLC purification, the crude peptide was coupled to the DNA anchor oligonucleotide by reductive amination with a periodate-oxidized 3′-terminal rA nucleotide as described.38 After the DNA-anchored hexapeptide was precipitated with ethanol, the Lys(Tfa) protecting group was removed by incubation in 30% aqueous NH4OH at room temperature for 1 h. The sample was dried by SpeedVac and purified by 20% PAGE. Mass spectrometry data for the Lys(Tfa)-protected hexapeptides and DNA-anchored hexapeptides are provided in the ESI.†
In vitro selection details
The key enrichment step of each selection round using the 5′-benzaldehyde oligonucleotide is shown schematically in Fig. 1C, with full nucleotide and primer sequences in Fig. 6. In each round, the ligation step to attach the DNAzyme pool at its 3′-end with the 5′-end of the amine-containing substrate was performed using a DNA splint and T4 DNA ligase. The splint sequence was 5′-ATAGTGAGTCGTATTATCCCCATCAGGATCAGCTTAATACGACTCACTAT-3′. The selection procedures are described in the following paragraphs.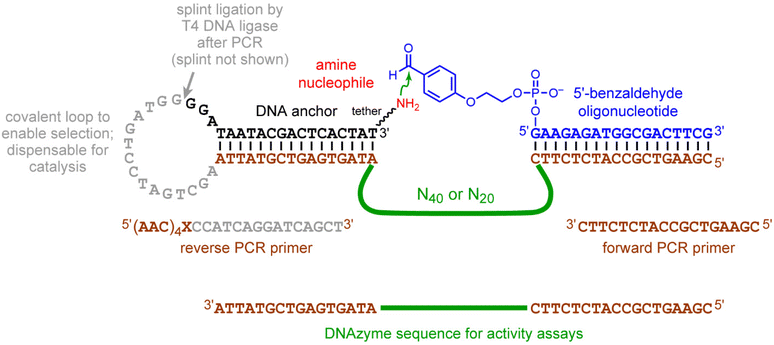 | ||
| Fig. 6 Nucleotide details of the in vitro selection experiments. The complete structures of the DNA-anchored amine substrates are shown in Fig. 1B. | ||
Single-turnover assay procedure for individual DNAzymes
The general single-turnover assay procedure for each DNAzyme (or the background reaction) was as follows. The DNA-anchored amine substrate was 5′-32P-radiolabeled using γ-32P-ATP and T4 polynucleotide kinase. An 8 μL sample containing 0.5 pmol of 5′-32P radiolabeled amine substrate, 10 pmol of DNAzyme (or 10 pmol of DNA splint for the background reaction), and 20 pmol of 5′-benzaldehyde reaction partner was annealed in 5 mM NaOAc, pH 5.2, or 5 mM MES, pH 6.0, or 5 mM HEPES, pH 7.5, or 5 mM CHES, pH 9.0, each with 15 mM NaCl and 0.1 mM EDTA, by heating at 95 °C for 3 min and cooling on ice 5 min. The DNAzyme-catalyzed reaction was initiated by bringing the sample to 20 μL total volume containing 70 mM NaOAc, pH 5.2, 20 mM MnCl2, and 1 mM ZnCl2, or 70 mM MES, pH 6.0, 20 mM MnCl2, 1 mM ZnCl2, and as relevant 20 mM NiCl2, or 70 mM HEPES, pH 7.5, 20 mM MnCl2, and 1 mM ZnCl2, or 50 mM CHES, pH 9.0, each with 40 mM MgCl2, 150 mM NaCl, and 100 mM NaCNBH3. The sample was incubated at 37 °C. At each time point, a 2 μL aliquot was quenched with 7 μL of stop solution (80% formamide, 1× TBE [89 mM each Tris and boric acid and 2 mM EDTA, pH 8.3], 50 mM EDTA, 0.025% bromophenol blue, 0.025% xylene cyanol). Quenched samples were separated by 20% PAGE and quantified using a Phosphorimager. Values of kobs were obtained by fitting the yield versus time data directly to first-order kinetics; i.e., yield = Y(1 − e−kt), where k = kobs and Y is the final yield. For the background reactions, kobs was sufficiently low such that an exponential fit was not meaningful, so the initial points were fit to a straight line, and kobs was taken as the slope of the line.Mass spectrometry assays of DNAzyme products
The products of the five representative individual DNAzymes were analyzed by MALDI-TOF mass spectrometry. For 4HK206 and 6HR205, the product was prepared from a 10 μL sample containing 400 pmol of DNA-C3-NH2 substrate, 480 pmol of DNAzyme, and 600 pmol of 5′-benzaldehyde reaction partner. The sample was annealed in 5 mM MES, pH 6.0, 15 mM NaCl, and 0.1 mM EDTA by heating at 95 °C for 3 min and cooling on ice for 5 min. For 5HH207, 3HP227, and 4HQ227, the product was prepared from a 10 μL sample containing 1.0 nmol of DNA-C3-NH2 substrate, 1.2 nmol of DNAzyme, and 1.5 nmol of 5′-benzaldehyde reaction partner. The sample was annealed in 5 mM NaOAc, pH 5.2 (5HH207), or 5 mM HEPES, pH 7.5 (3HP227), or 5 mM CHES, pH 9.0 (4HQ227), each with 15 mM NaCl and 0.1 mM EDTA, by heating at 95 °C for 3 min and cooling on ice for 5 min. For all five DNAzymes, the DNAzyme-catalyzed reaction was initiated by bringing the sample to 30 μL total volume containing 70 mM NaOAc, pH 5.2, 20 mM MnCl2, and 1 mM ZnCl2 (5HH207), or 70 mM MES, pH 6.0, 20 mM MnCl2, and 1 mM ZnCl2 (4HK206), or 70 mM MES, pH 6.0, 20 mM MnCl2, 1 mM ZnCl2, and 20 mM NiCl2 (6HR205), or 70 mM HEPES, pH 7.5, 20 mM MnCl2, and 1 mM ZnCl2 (3HP227), or 50 mM CHES, pH 9.0 (4HQ227), each with 40 mM MgCl2, 150 mM NaCl, and 100 mM NaCNBH3. The sample was incubated at 37 °C for 48 h. The reductive amination product was purified by 20% PAGE, desalted by Millipore C18 ZipTip, and analyzed by MALDI-TOF mass spectrometry on a Bruker UltrafleXtreme MALDI-TOF mass spectrometer with matrix 3-hydroxypicolinic acid in positive ion mode at the UIUC School of Chemical Sciences Mass Spectrometry Laboratory.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Robert Boyd and Prakriti Das for careful reading of the manuscript.References
- K. Schlosser and Y. Li, Chem. Biol., 2009, 16, 311–322 CrossRef CAS PubMed.
- J. Liu, Z. Cao and Y. Lu, Chem. Rev., 2009, 109, 1948–1998 CrossRef CAS PubMed.
- M. Hollenstein, Molecules, 2015, 20, 20777–20804 CrossRef CAS PubMed.
- M. Liu, D. Chang and Y. Li, Acc. Chem. Res., 2017, 50, 2273–2283 CrossRef CAS PubMed.
- D. Morrison, M. Rothenbroker and Y. Li, Small Methods, 2018, 2, 1700319 CrossRef.
- M. Hollenstein, Curr. Opin. Chem. Biol., 2019, 52, 93–101 CrossRef CAS PubMed.
- L. Ma and J. Liu, iScience, 2020, 23, 100815 CrossRef CAS PubMed.
- M. Cepeda-Plaza and A. Peracchi, Org. Biomol. Chem., 2020, 18, 1697–1709 RSC.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- D. L. Robertson and G. F. Joyce, Nature, 1990, 344, 467–468 CrossRef CAS PubMed.
- G. F. Joyce, Annu. Rev. Biochem., 2004, 73, 791–836 CrossRef CAS PubMed.
- G. F. Joyce, Angew. Chem., Int. Ed., 2007, 46, 6420–6436 CrossRef CAS PubMed.
- S. K. Silverman, Acc. Chem. Res., 2009, 42, 1521–1531 CrossRef CAS PubMed.
- S. K. Silverman, Acc. Chem. Res., 2015, 48, 1369–1379 CrossRef CAS PubMed.
- S. K. Silverman, Trends Biochem. Sci., 2016, 41, 595–609 CrossRef CAS PubMed.
- T. Yao, J. J. Przybyla, P. Yeh, A. M. Woodard, H. J. Nilsson, B. M. Brandsen and S. K. Silverman, Org. Biomol. Chem., 2021, 19, 171–181 RSC.
- D. Husmann and O. Gozani, Nat. Struct. Mol. Biol., 2019, 26, 880–889 CrossRef CAS PubMed.
- K. K. Biggar and S. S.-C. Li, Nat. Rev. Mol. Cell Biol., 2015, 16, 5–17 CrossRef CAS PubMed.
- A. L. Kodal, C. B. Rosen, M. R. Mortensen, T. Tørring and K. V. Gothelf, ChemBioChem, 2016, 17, 1338–1342 CrossRef CAS PubMed.
- M. R. Mortensen, M. B. Skovsgaard, A. H. Okholm, C. Scavenius, D. M. Dupont, C. B. Rosen, J. J. Enghild, J. Kjems and K. V. Gothelf, Bioconjugate Chem., 2018, 29, 3016–3025 CrossRef CAS PubMed.
- M. J. Matos, B. L. Oliveira, N. Martínez-Sáez, A. Guerreiro, P. M. S. D. Cal, J. Bertoldo, M. Maneiro, E. Perkins, J. Howard, M. J. Deery, J. M. Chalker, F. Corzana, G. Jiménez-Osés and G. J. L. Bernardes, J. Am. Chem. Soc., 2018, 140, 4004–4017 CrossRef CAS PubMed.
- M. Chilamari, N. Kalra, S. Shukla and V. Rai, Chem. Commun., 2018, 54, 7302–7305 RSC.
- O. I. Afanasyev, E. Kuchuk, D. L. Usanov and D. Chusov, Chem. Rev., 2019, 119, 11857–11911 CrossRef CAS PubMed.
- E. Podyacheva, O. I. Afanasyev, A. A. Tsygankov, M. Makarova and D. Chusov, Synthesis, 2019, 51, 2667–2677 CrossRef CAS.
- J. Liu, Y. Song and L. Ma, Chem. Asian J., 2021, 16, 2371–2391 CrossRef CAS PubMed.
- F. Hollmann, D. J. Opperman and C. E. Paul, Angew. Chem., Int. Ed., 2021, 60, 5644–5665 CrossRef CAS PubMed.
- O. Wong, A. E. Mulcrone and S. K. Silverman, Angew. Chem., Int. Ed., 2011, 50, 11679–11684 CrossRef CAS PubMed.
- B. M. Brandsen, T. E. Velez, A. Sachdeva, N. A. Ibrahim and S. K. Silverman, Angew. Chem., Int. Ed., 2014, 53, 9045–9050 CrossRef CAS PubMed.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS PubMed.
- A. Ponce-Salvatierra, K. Wawrzyniak-Turek, U. Steuerwald, C. Höbartner and V. Pena, Nature, 2016, 529, 231–234 CrossRef CAS PubMed.
- H. Liu, X. Yu, Y. Chen, J. Zhang, B. Wu, L. Zheng, P. Haruehanroengra, R. Wang, S. Li, J. Lin, J. Li, J. Sheng, Z. Huang, J. Ma and J. Gan, Nat. Commun., 2017, 8, 2006 CrossRef PubMed.
- J. Borggräfe, J. Victor, H. Rosenbach, A. Viegas, C. G. W. Gertzen, C. Wuebben, H. Kovacs, M. Gopalswamy, D. Riesner, G. Steger, O. Schiemann, H. Gohlke, I. Span and M. Etzkorn, Nature, 2022, 601, 144–149 CrossRef PubMed.
- P. Crisalli and E. T. Kool, Org. Lett., 2013, 15, 1646–1649 CrossRef CAS PubMed.
- S. M. Walsh, S. N. Konecki and S. K. Silverman, J. Mol. Evol., 2015, 81, 218–224 CrossRef CAS PubMed.
- T. E. Velez, J. Singh, Y. Xiao, E. C. Allen, O. Wong, M. Chandra, S. C. Kwon and S. K. Silverman, ACS Comb. Sci., 2012, 14, 680–687 CrossRef CAS PubMed.
- O. Wong, P. I. Pradeepkumar and S. K. Silverman, Biochemistry, 2011, 50, 4741–4749 CrossRef CAS PubMed.
- J. Chandrasekar and S. K. Silverman, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5315–5320 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ob00070b |
| This journal is © The Royal Society of Chemistry 2023 |