Open Access Article
Open Access ArticleSwitching between DNA binding modes with a photo- and redox-active DNA-targeting ligand†
Christoph
Dohmen
and
Heiko
Ihmels
 *
*
Department of Chemistry and Biology, University of Siegen, and Center of Micro- and Nanochemistry and (Bio)Technology (Cμ), Adolf-Reichwein-Str. 2, 57068 Siegen, Germany. E-mail: ihmels@chemie.uni-siegen.de
First published on 6th February 2023
Abstract
A disulfide-functionalized bis-benzo[b]quinolizinium is presented that is transformed quantitatively into its cyclomers in a fast intramolecular [4 + 4] photocycloaddition. Both the bis-quinolizinium and the photocyclomers react with glutathione (GSH) or dithiothreitol (DTT) to give 9-(sulfanylmethyl)benzo[b]quinolizinium as the only product. As all components of this reaction sequence have different DNA-binding properties, it enables the external control and switching of DNA association. Hence, the bis-benzo[b]quinolizinium binds strongly to DNA and is deactivated upon photocycloaddition to the non-binding cyclomers. In turn, the subsequent cleavage of the cyclomers with DTT regains a DNA-intercalating benzoquinolizinium ligand. Notably, this sequence of controlled deactivation and recovery of DNA-binding properties can be performed directly in the presence of DNA.
Introduction
Despite considerable progress in tumor treatment, cancer is still a devastating disease and one of the major causes of death.1 In this context, the traditional chemotherapeutic approach, that employs cytostatic, DNA-targeting reagents,2 is among the commonly applied treatments of many types of cancer, albeit this method still has the disadvantage of a low selectivity, which leads to serious side effects on healthy tissue.3 One promising approach to increase the selectivity of chemotherapeutic agents focuses on so-called prodrugs,3a,4 that are intrinsically bioinactive and whose cytotoxicity can be selectively activated by external stimuli, such as light, redox processes, or enzymatic triggers as well as combinations thereof.5 In particular, the reductive cleavage of disulfide bonds under hypoxic conditions by nucleophiles, such as glutathione (GSH),5,6 has been employed in disulfide-containing prodrugs, that change their reactivity towards biological targets once the disulfide is cleaved.5,7 Notably, the disulfide-GSH activation of a substrate has several advantages in drug development8 because GSH is present in the human body in relatively high concentrations and is overproduced in fast-growing, hypoxic cancer cells.5 Furthermore, the disulfide bond increases the propensity of drugs to pass cell membranes.7b,9 Therefore, several drug candidates have been developed that are activated in vivo by the reaction with GSH under hypoxic conditions,5,10 however, to the best of our knowledge, ligands with a disulfide functionality as redox-active unit to control their DNA-binding properties have not been reported, so far. As the controlled modification and/or release of DNA ligands may constitute a useful element in the development of DNA-targeting lead structures11 we investigated whether the functionalization of DNA ligands with a disulfide bond may also be used for such purposes. To this end, we chose the benzo[b]quinolizinium ion as ligand because it has been shown to be a versatile platform for the design of DNA-binding compounds.12 To add to that, this particular DNA ligand is photochromic and can be reversibly transformed in a [4 + 4] photocycloaddition into its dimers.11e,13 The latter do not bind to DNA, so that this system can be used for the controlled release of the DNA-binding monomer upon irradiation of the dimer.11e In order to increase the efficiency of the deactivation of the DNA-binding properties with a photodimerization, it has to be performed in an intramolecular process that does no longer require a diffusion-controlled encounter of the monomers, as realized in the dialkyldisulfide-linked bis-benzo[b]quinolizinium 1 (Fig. 1). Hence, we proposed that the combination of the photochromic properties of benzo[b]quinolizinium derivatives and a redox-active disulfide bond in substrate 1 may enable the modification of its DNA-binding properties by two stimuli, namely by irradiation or reduction, both of which may, in principle, be realized even within a cell. Herein, we show that the disulfide-linked bis-benzo[b]quinolizinium 1 can indeed be switched between different states with distinctly different DNA-binding properties, however, in an unexpected reaction pathway.Results and discussion
Synthesis
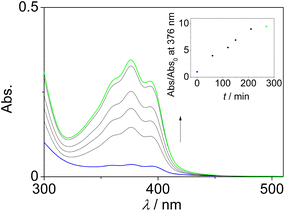
The bis-benzo[b]quinolizinium 1 was synthesized starting from the 9-(bromomethyl)benzo[b]quinolizinium (2), which was obtained by the established cyclodehydration route (cf. ESI, Scheme S1†).14 The reaction of 2 with thioacetic acid resulted in the formation of the corresponding thioacetate 3 in 82% yield (Scheme 1). Subsequent acid-catalyzed ester hydrolysis gave the 9-(sulfanylmethyl)benzo[b]quinolizinium (4), which was immediately oxidized with Br2/HBr to give the product 1 in 91% yield in two steps. All new products were fully characterized and identified by 1D- and 2D-NMR spectroscopy, absorption and emission spectroscopy, and by elemental analysis, except for the thiol 4, which was analyzed from the crude reaction mixture because of its fast oxidation to 1 (cf. ESI, Chapter 3†).It was investigated whether the thiol 4 is also available by the reduction of the disulfide 1. Thus, photometric analysis showed that upon addition of L-glutathione (GSH, pH = 7.8, T = 37 °C) to a solution of 1 (λmax = 380 nm, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 4.17) a new, more intense absorption band formed at 377 nm (log
ε = 4.17) a new, more intense absorption band formed at 377 nm (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε = 4.31) within 3 h, which did not change after additional 16 h (Fig. 2). The absorption was assigned to the thiol 4, which was persistent under these conditions. The formation of this product was also confirmed by the 1H-NMR-spectroscopic investigation of the reaction mixture, which revealed the characteristic signals of the methylene group (4.07 ppm, 2H) and the 6-H proton (10.47 ppm, 1H) of the product 4 (cf. ESI, Fig. S3†). Essentially the same results were obtained with dithiothreitol (DTT) as the reducing agent (cf. ESI, Fig. S2A2, Fig. S3†).
ε = 4.31) within 3 h, which did not change after additional 16 h (Fig. 2). The absorption was assigned to the thiol 4, which was persistent under these conditions. The formation of this product was also confirmed by the 1H-NMR-spectroscopic investigation of the reaction mixture, which revealed the characteristic signals of the methylene group (4.07 ppm, 2H) and the 6-H proton (10.47 ppm, 1H) of the product 4 (cf. ESI, Fig. S3†). Essentially the same results were obtained with dithiothreitol (DTT) as the reducing agent (cf. ESI, Fig. S2A2, Fig. S3†).
 | ||
| Fig. 2 Absorption spectra of 1 (c = 20 μM) before (orange) and after treatment with glutathione (GSH, c = 10 mM) for 3 h (green) and after 16 h (B, black) at 37 °C in phosphate buffer (pH = 7.8). | ||
Photoinduced [4 + 4] cycloaddition of 1
To investigate the photoactivity of bis-benzo[b]quinolizinium 1 it was irradiated with a LED (λex = 365 nm) in aqueous solution which resulted in an almost complete disappearance of the initial absorption bands, along with the development of weaker bands at 340–420 nm (cf. ESI, Fig. S5A†). During the reaction, an isosbestic point at 210 nm was formed, and after 120 s the reaction was completed. These observations indicated the known [4 + 4] photodimerization of the benzoquinolizinium units,13a,b in particular because the absorption spectrum of the product mixture resembled the ones of known photodimers.13a–c,15 Most notably, this photoreaction appeared to be relatively fast as compared with reported examples.13,15a,16 For comparison, under the same conditions the dimerization of the parent benzo[b]quinolizinium15b (cf. ESI, Fig. S4†) took 8 h for completion (cf. ESI, Fig. S5B†), which confirmed the initial proposal that the efficiency of the intramolecular photocycloaddition should significantly increase. To gain further structural information about the photoproducts the reaction was monitored by 1H-NMR spectroscopy in D2O solution (Scheme 2), which indicated full consumption of the starting material and the complete formation of three products, whose structures were assigned by 2D-NMR-spectroscopic analysis, especially based on the characteristic 1H-NMR signals of the bridgehead protons of the photoproducts at 5.7–5.9 ppm (Scheme 2B, cf. ESI, Chapter 3–7†). Thus, the product mixture contained the syn-head-to-head13a (ssh) cycloadduct c1shh (74%), the syn-head-to-tail (sht) product c1sht (18%), and the syn-head-to-head13a photodimer [4]2shh (8%), in which the disulfide bond was cleaved. Additional high-resolution mass spectrometric analysis of the reaction mixture confirmed that all products are dicationic (cf. ESI†).As compared with the intermolecular photodimerization of benzo[b]quinolizinium derivatives,13 the intramolecular photoreaction of the derivative 1 resulted in a different ratio of isomeric cycloadducts, because the linker unit directs the product formation depending on geometric and steric constraints. The isomer [4]2shh is most likely formed as a secondary photoproduct upon cleavage of the disulfide bond of the cycloadduct c1shh because 1H-NMR-spectroscopic monitoring of the reaction showed that the signals of [4]2shh increased with longer irradiation time while the signals of c1shh decreased (cf. ESI, Fig. S7†). However, a photoreaction upon direct absorption of the disulfide and formation of thiyl radicals can be excluded because dialkyldisulfide bonds absorb only below 320 nm.17 Therefore, it is proposed that an intramolecular photoinduced electron transfer (PET) reaction of the photoproduct c1shh, that indirectly or directly involves the disulfide unit, leads to the known photocatalyzed cleavage of the disulfide bond through the formation of intermediate radical ions.18 The observation that only the cycloadduct c1shh is reduced to the dithiol [4]2shh upon further irradiation may indicate that only this substrate absorbs the excitation wavelength or that only in this photoproduct an efficient PET takes place.
The photoproducts turned out to be thermally stable in solution even at slightly elevated temperatures, as indicated by negligible changes of the absorption spectrum of the photoproducts at 55 °C for 1.5 h in aqueous buffer solution (cf. ESI, Fig. S8A†). When the mixture of cycloadducts was irradiated with λex = 270 nm the absorption bands of the cycloadducts disappeared and a broad absorption band with a maximum at 450 nm evolved, which, however, did not fully match the one of compound 1, thus indicating that at least one by-product was formed along with the cycloreversion (cf. ESI, Fig. S8B†). Presumably, a photoinduced cleavage of the disulfide bond was also induced at this excitation wavelength and the resulting thiyl radicals led to alternative reaction pathways.17a
In contrast to the unselective photocleavage of the cycloadducts c1shh, c1sht, and [4]2shh, these substrates were reduced quantitatively to the corresponding thiol 4 upon treatment with DTT (Scheme 2A). Hence, the photometric analysis of this reaction showed a complete disappearance of the cycloadducts and formation of the monomer 4, namely by the increase of the initial absorption over time with formation of the characteristic absorption of 4 at 377 nm (Fig. 3).
In addition, the 1H-NMR-spectroscopic analysis of the reaction mixture confirmed the exclusive formation of the thiol 4 by its characteristic proton spectrum (cf. ESI, Fig. S11†). Although the mechanism of this completely unexpected cycloreversion-reduction reaction is not clear, it may be proposed that it is introduced by the formation of intermediate radical ions, as shown already previously for naphthoquinolizinium dimers.16 Even more important, other than the commonly applied photoinduced release of a benzo[b]quinolizinium unit from its dimer, which requires unfavorable high-energy light,13 this thermally induced dimer cleavage operates under mild, low-temperature conditions and is therefore compatible with the biological conditions in cells.
DNA-binding properties
The DNA-binding properties of the disulfide 1, the corresponding thiol 4, and, for comparison, the thioacetate 3, as well as the ones of the photocycloadducts c1shh, c1sht, and [4]2shh with calf thymus (ct) DNA were investigated with photometric and fluorimetric titrations (Fig. 2, cf. ESI, Chapter 5†). For that purpose, the thiol 4 and the cycloadducts c1shh, c1sht, and [4]2shh were generated from solutions of the disulfide 1 either by reduction with DTT/GSH or by irradiation with λex = 365 nm, as described above, prior to the addition of DNA. The absorption of the thioacetate 3 decreased on addition of DNA while a new red-shifted absorption maximum developed at 415 nm (ΔλAbs = 19 nm, Fig. 4A1). Furthermore, an isosbestic point at 403 nm was formed during the titration. At the same time, the fluorescence of the derivative 3 was quenched with increasing ct DNA concentration (cf. ESI, Fig. S12†). The binding isotherm from the photometric DNA titration of 3 was used to determine the binding constant of Kb = 8.70 × 103 M−1 (cf. ESI, Fig. S17A1†). In the case of the disulfide 1, the absorption firstly decreased upon addition of up to 0.5 molar equiv. of DNA and then increased with a small red shift on further DNA addition (Fig. 4A2). Because of the high photochemical reactivity of disulfide 1 (see above) the association with DNA could not be monitored by fluorescence spectroscopy, as the photodimerization was already induced by the excitation light. Alternatively, the fluorescent indicator displacement (FID) assay was performed as indirect fluorimetric method that allows to monitor the displacement of thiazole orange (TO) from its DNA binding site upon the addition of a competing ligand. The displacement of TO by the analyte is determined by its fluorescence quenching with an excitation at λex = 475 nm, i.e., where 1 does not absorb (cf. ESI, Fig. S13†).19 Thus, upon addition of disulfide 1 to a solution of DNA-bound TO the emission of the latter was significantly quenched with increasing concentration of 1. From this fluorimetric titration, a percentage displacement value at 50% (PD50) was determined at 2.63 μM (cf. ESI, eq. 1†).During the photometric titration of thiol 4 with ct DNA in the presence of DTT the absorption of the ligand decreased and a new weak absorption band at 408 nm (ΔλAbs = 13 nm) and an isosbestic point at 404 nm evolved (Fig. 4A3). The titration in the presence of GSH gave similar results (cf. ESI, Fig. S15A1†). The exact concentration of the ligand within the solution was not known under these conditions. But with the assumption of a quantitative reduction to the thiol 4 (as evidenced by the isosbestic point during subsequent DNA titration) the analysis of the resulting binding isotherms gave binding constants of Kb = 4.0 × 103 M−1 (with DTT) and Kb = 2.5 × 103 M−1 (with GSH), which are essentially in the same range within a supposedly slightly larger error margin under these conditions (cf. ESI, Fig. S17B†).20
The addition of DNA to the mixture of cycloadducts c1shh, c1sht, and [4]2shh did not lead to a significant change of the absorption spectrum (cf. ESI, Fig. S15A2†). Unfortunately, it was not possible to extract binding constants of the cycloadducts and the disulfide 1 from the photometric titration experiments.
To gain further information about the binding modes, circular-dichroism (CD, Fig. 4B, cf. ESI, Chapter 5†) and flow linear-dichroism (LD, Fig. 4C, cf. ESI, Chapter 5†) spectra were recorded at different ligand-DNA ratios (LDR). In the presence of DNA, the CD spectra of compound 3 showed a weak positive and a negative induced CD (ICD) signal in the absorption range of the ligand whose intensity increased with increasing LDR (Fig. 4B1). Conversely, the CD spectra of derivative 1 exhibited exclusively a strong positive ICD band upon association with DNA (Fig. 4B2, cf. ESI, Fig. S16†). Subsequently, the steady formation of this ICD band during titration of DNA to ligand 1 was used to gain a binding isotherm whose fit to the theoretical model gave a binding constant of Kb = 3.2 × 105 M−1 (cf. ESI, Fig. S17A2†). Conversely, the cycloadducts c1shh, c1sht, and [4]2shh only showed a very weak, almost invisible ICD signal (cf. ESI, Fig. S15B2†) in the presence of DNA.
Upon addition of DNA, the thiol 4 and the thioacetate 3 gave a weak negative LD signal in the range of the absorption of the ligand, respectively, which increased with increasing LDR (Fig. 4C1 and C3). Although the disulfide 1 also gave a weak negative LD signal between 340–430 nm, an additional weak positive band was observed between 290–340 nm (Fig. 4C2). Both LD bands increased with increasing LDR. In contrast, only a very weak negative LD signal was observed for the cycloadducts of 1 in the absorption range of the ligand (cf. ESI, Fig. S15C2†).
Overall, both the thioacetate 3 and the thiol 4 showed the characteristic features of a DNA intercalator, namely a positive and a negative ICD signal and a negative LD signal.21 It should be emphasized that the thioacetate 3 was employed as a reference for comparison with the thiol 4 because the latter is a rather sensitive compound. Thus, to exclude that a secondary reaction or decomposition of 4 interfered with the DNA-binding studies the same experiments were performed with the thioacetate 3 as a structurally resembling, but more robust sulfur-containing reference compound, which has the same DNA-binding unit with a resembling sulfur-functionality at the same position. And as both compounds 3 and 4 showed resembling spectroscopic features during the DNA-titrations it can be concluded that the thiol 4 is persistent under these conditions. Notably, however, both binding constants are slightly smaller than the ones of the parent compound and resembling 9-substituted benzo[b]quinolizinium derivatives, which are usually in the range of 104–105 M−1.12b,d This slightly lower binding affinity of 3 and 4 may be explained by the sterically more demanding thiol and thioacetate groups that lead to repulsion within the binding pocket of the DNA.
In contrast, the spectrometric DNA titrations revealed a more complex binding mode for ligand 1 because the CD and LD data indicated both intercalation (negative LD signal) and groove binding (weak positive LD signal and strong positive ICD bands).21 Moreover, these data do not match the characteristic ones obtained from the DNA-intercalating thioacetate derivative 3 that may be used for comparison as a suitable reference compound. And the binding constant of the parent disulfide 1 is also relatively high for a benzo[b]quinolizinium unit.12b,d Hence, considering the particular structure of the tethered bis-benzoquinolizinium derivative 1 these results may indicate a binding mode in which one benzoquinolizinium is intercalated into the DNA, whereas the other one is accommodated in the groove (cf. ESI, Fig. S18†). This binding mode agrees with the changes during photometric titration after addition of 0.5 molar equivalents of DNA, because at the high ligand loading at the beginning of the titration there are not enough binding sites available for this particular binding mode. Furthermore, this binding mode explains why ligand 1 is able to displace TO, that is, a known intercalator (Kb = 3.2 × 105 M−1),22 from its DNA binding site. Notably, the PD50 value at 2.63 μM is relatively low as compared to, e.g., neutral bisquinacridine derivatives with PD50 at 0.4–0.7 μM.19 However, similar values were determined for tetraazoniapentaphenopentaphene with PD50 at >2.5 μM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 and dibenzoazoniachrysenes with PD50 at 1.1–2.5 μM,24 which are annelated derivatives of the parent benzo[b]quinolizinium structure.
23 and dibenzoazoniachrysenes with PD50 at 1.1–2.5 μM,24 which are annelated derivatives of the parent benzo[b]quinolizinium structure.
On the contrary, the cycloadducts of 1 do not show a spectroscopic indication of DNA binding, presumably because the sterically demanding structures can at best bind weakly in a loose backbone association to the DNA.11g
In situ switching of DNA-binding properties
As the disulfide 1 and the cycloadducts c1shh, c1sht, and [4]2shh are responsive to reducing thiols and/or irradiation in a predictable way (Scheme 2A), it was further investigated whether the combination of these properties may be used for the controlled in situ formation of one particular DNA-binding form. Thus, in a control experiment it was first investigated whether the cycloreversion of the cycloadducts c1shh, c1sht, and [4]2shh can be induced thermally in the presence of ct DNA (cf. ESI, Fig. S10†). But like in the absence of DNA (see above), the cycloadducts were persistent also under these conditions. Next, it was examined by absorption and CD spectroscopy whether the controlled locking of the DNA-binding properties of the disulfide 1 by irradiation and subsequent unlocking by reduction of the mixture of c1shh, c1sht, and [4]2shh can be performed in the presence of DNA (Scheme 3). Accordingly, upon irradiation of a mixture of disulfide 1 and ct DNA the strong positive ICD signal of the ligand decreased and eventually vanished, and the absorption also decreased, which indicated the photocycloaddition reaction to the non-binding cycloadducts (Fig. 5). Subsequently, the addition of DTT to this mixture under anaerobic conditions at 37 °C led to the formation of the characteristic absorption and ICD bands of the DNA-bound thiol 4 (Fig. 5), as unambiguously shown by comparison with the spectra of authentic samples (Fig. 4). Hence, these experiments demonstrated that the sequence of deactivation and reactivation of DNA ligands starting with the disulfide 1 is indeed possible directly in the presence of DNA. | ||
| Scheme 3 Photocycloaddition-reduction-sequence of disulfide 1 to thiol 4 in the presence of DNA. The structures of c1sht and [4]shh are omitted for clarity. | ||
Conclusions
In summary, we have established a disulfide-functionalized photochromic system that enables the control of DNA-binding properties in a sequence of initial deactivation of the DNA-binding starting material 1 in a photocycloaddition and subsequent cleavage of the weakly DNA-binding cycloadducts c1shh, c1sht, and [4]2shh by reduction with sulfide to eventually regain the DNA intercalator 4. As the latter conditions resemble the ones in hypoxic cancer cells, this concept of the photo- and redox-triggered activation and deactivation of DNA binders may be a promising starting point for new approaches towards dual-mode switchable prodrugs.Experimental section
Equipment
NMR spectra were recorded on a Jeol JNM-ECZR (1H: 500 MHz, 13C: 125 MHz) or on a Varian VNMR-S 600 (1H: 600 MHz, 13C: 150 MHz). The chemical shifts (δ) are given in ppm and are referenced relative to the sodium salt of trimethylsilylpropionic acid (TMSP-d4, 1H: δ = 0.00 ppm, 13C: δ = 0.00 ppm) or the residual solvent signals (DMSO-d5, 1H: δ = 2.50 ppm, 13C: δ = 39.5 ppm). In general, the HH coupling is specified as 1J, 2J, etc. When the unambiguous assignment was not possible, this information was omitted. Spectra were analysed with the software MestReNova 12.0.0. High-resolution (ESI) mass spectra were acquired with a Thermo Fisher Scientific Exactive MS with Orbitrap mass analyzer (injection flow rate: 10 μL min−1, ESI spray voltage: 4.0 kV). Elemental analyses were measured in-house with a HEKAtech EUROEA combustion analyser (Universität Siegen, Organische Chemie I) or directly at HEKAtech EUROEA. Irradiation experiments were performed with a LED lamp (Roschwege HighPower-LED UV, λex = 365 nm) or a Rayonet (RPR-100) photoreactor equipped with 6 lamps (8 W, λex = 270 nm). Irradiation experiments monitored by NMR spectroscopy were performed in glass tubes (Boro400–5–7).Materials
All commercially available chemicals were used without further purification unless stated otherwise. Chemicals were purchased from the following companies: Acros organics (Geel, BE): Br2, pyridine-2-aldehyd, thiolacetic acid; carbolution (St. Ingberd, DE): 1,4-dithio-DL-threit (DTT); Sigma-Aldrich (Steinheim am Albuch, DE): HPF6 (aq. 60%); fluorochem (Graphite Way, EN): HBr (aq. 48%); Carl Roth GmbH & Co KG (Karlsruhe, DE): L-Glutathione (GSH); Thermo Fisher Scientific Inc (Waltham, MA, USA): BH3-THF. 4-Bromomethylbenzylalcohol was synthesized according to the reported procedures.25Buffer solutions were prepared with biochemistry-grade salts (see below) in deionized water, and the pH values were adjusted with aqueous solutions of NaOH or HCl. Prior to use, the buffer solutions were filtered through a PVDF membrane filter (pore size 0.45 μm). Biphosphate EDTA buffer (BPE buffer): Na2HPO4 (6.00 mM), NaH2PO4 (2.00 mM), = Na2EDTA (1.00 mM). Phosphate-buffered saline (PBS): NaCl (137 mM), KCl (2.68 mM), KH2PO4 (1.98 mM), Na2HPO4 (10.0 mM).
Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) = 395 nm (3.97). – Em. (H2O): λmax [Φfl, relative to anthracene Φfl = 0.28 (EtOH, λex = 340 nm)]26 = 412, (0.30). – 1H-NMR (500 MHz, DMSO-d6): δ = 2.41 (s, 3H, CH3), 4.45 (s, 2H, CH2), 7.89 (dd, 3J = 9 Hz, 4J = 1 Hz, 1H, 8-H), 7.94 (td, 3J = 7 Hz, 4J = 1 Hz, 1H, 3-H), 8.07 (ddd, 3J = 9 Hz, 3J = 7 Hz, 4J = 1 Hz, 1H, 2-H), 8.43 (d, 3J = 9 Hz, 1H, 7-H), 8.28 (s, 1H, 10-H), 8.53 (d, 3J = 9 Hz, 1H, 1-H), 9.17 (s, 1H, 11-H), 9.25 (d, 3J = 7.1 Hz, 1H, 4-H), 10.36 (s, 1H, 6-H). – 13C-NMR (125 MHz, DMSO-d6): δ = 30.3 (CH3), 32.8 (CH2), 122.3 (C3), 124.3 (C11), 124.9 (C6a), 125.6 (C10), 126.8 (C1), 128.6 (C7), 131.2 (2C), 132.3 (C8), 134.3 (C4), 135.3 (C10a), 137.8 (C11a), 139.9 (C6), 145.3 (C8), 194.6 (CO). – MS (ESI+): m/z (%) = 268 (100) [M−Br−]+. – El. Anal. for C16H14BrNOS × 0.5 H2O, calcd (%): C 53.75, H 4.23, N 3.92, S 8.97, found (%): C 54.18, H 3.89, N 3.98, S 9.18.
ε) = 395 nm (3.97). – Em. (H2O): λmax [Φfl, relative to anthracene Φfl = 0.28 (EtOH, λex = 340 nm)]26 = 412, (0.30). – 1H-NMR (500 MHz, DMSO-d6): δ = 2.41 (s, 3H, CH3), 4.45 (s, 2H, CH2), 7.89 (dd, 3J = 9 Hz, 4J = 1 Hz, 1H, 8-H), 7.94 (td, 3J = 7 Hz, 4J = 1 Hz, 1H, 3-H), 8.07 (ddd, 3J = 9 Hz, 3J = 7 Hz, 4J = 1 Hz, 1H, 2-H), 8.43 (d, 3J = 9 Hz, 1H, 7-H), 8.28 (s, 1H, 10-H), 8.53 (d, 3J = 9 Hz, 1H, 1-H), 9.17 (s, 1H, 11-H), 9.25 (d, 3J = 7.1 Hz, 1H, 4-H), 10.36 (s, 1H, 6-H). – 13C-NMR (125 MHz, DMSO-d6): δ = 30.3 (CH3), 32.8 (CH2), 122.3 (C3), 124.3 (C11), 124.9 (C6a), 125.6 (C10), 126.8 (C1), 128.6 (C7), 131.2 (2C), 132.3 (C8), 134.3 (C4), 135.3 (C10a), 137.8 (C11a), 139.9 (C6), 145.3 (C8), 194.6 (CO). – MS (ESI+): m/z (%) = 268 (100) [M−Br−]+. – El. Anal. for C16H14BrNOS × 0.5 H2O, calcd (%): C 53.75, H 4.23, N 3.92, S 8.97, found (%): C 54.18, H 3.89, N 3.98, S 9.18.
Photocycloaddition reactions
A solution of 1 (1.6 mg, 2.6 μmol) in D2O (500 μL) was irradiated in a NMR-tube with a LED (λexc = 365 nm) for 13 min. The sample was placed directly in front of the LED lamp (distance to LED = 3 mm). The reaction mixture was analyzed by NMR spectroscopy and by mass spectrometry and the products were quantified by integration of the CH2-signals c1shh: 74%; c1sht: 18%; [4]2shh: 8%.[4]2shh: 1H-NMR (600 MHz, D2O, 10 °C) δ = 4.10 (s, 4H, 2 × CH2), 5.69 (s, 2H, 11-H), 6.86 (s, 2H, 6-H), 6.98–7.05 (m, 2H, 8-H), 7.13 (s, 2H, 10-H), 7.31 (d, 2H, 3J = 8 Hz, 1H, 7-H), 7.85–7.93 (m, 2H, 3-H), 8.11–8.20 (m, 2H, 1-H), 8.44–8.50 (m, 2H, 2-H), 9.11–9.13 (m, 2H, 4-H). – 13C-NMR (150 MHz, D2O): δ = 40.6 (2 × CH2), 52.4 (2 × C11),75.9 (2 × C6), 130.1 (2 × C3), 132.1 (2 × C1), 132.2 (2 × C8), 133.3 (2 × C7), 134.5 (C6a and C10a), 135.6 (2 × C10), 137.6 (not visible in 13C-NMR spectrum, but in HMBC experiment, C6a and C10a), 144.5 (2 × C9), 149.0 (2 × C4), 151.1 (2 × C2), 157.6 (2 × C11a). – HRMS (ESI+): calcd for: [C28H24N2S2 + 2MeOH + H2O]2+m/z = 267.1000, found: 225.0860 [M + 2MeOH + H2O].
c1shh: 1H-NMR (600 MHz, D2O, 10 °C): δ = 3.57, 3.90 (AX system, 2J = 15 Hz, 2H, 9-CH2), 3.59, 3.83 (AX system, 2J = 14 Hz, 2H, 9-CH2), 5.72 (d, 3J = 11 Hz, 1H, 11-H), 5.82 (d, 3J = 11 Hz, 1H, 11-H), 6.94–6.98 (m, 1H, 7-H or 8-H), 6.98–7.05 (m, 3H, 2 × 6-H and 7-H or 8-H), 7.12 (s, 1H, 10-H), 7.20–7.27 (m, 2H, 7-H and 8-H), 7.55 (s, 1H, 10-H), 7.85–7.93 (m, 2H, 2 × 3-H), 8.16 (t, 3J = 7 Hz, 2H, 2 × 1H), 8.44–8.50 (m, 2H, 2 × 2-H), 9.11 (d, 3J = 6 Hz, 1H, 4-H), 9.13 (d, 3J = 6 Hz, 1H, 4-H). – 13C-NMR (150 MHz, D2O): δ = 43.9 (C9), 45.0 (C9), 53.2 (C11), 53.5 (C11),75.6 (2 × C6),130.0 (C3), 130.2 (C3), 130.4 (C7 or C8), 131.2 (C7 or C8), 131.9 (C1), 132.1 (C1), 132.1 (C7 or C8), 132.7 (C7 or C8), 133.2 (C10), 134.0 (C10), 134.3 (C6a or C10a), 135.0 (C6a), 135.0 (C10a), 137.0 (C6a or C10a), 146.4 (C9), 146.6 (C9), 148.9 (C4), 149.0 (C4), 151.2 (C2), 157.5 (C 11a), 157.7 (C11a). – HRMS (ESI+): calcd for: [C28H22N2S2 ]2+m/z = 225.0607, found: 225.0603 [M].
c1sht: 1H-NMR (600 MHz, D2O, 10 °C) δ = 3.67, 3.72 [AB system, 2J = 13 Hz, 4H, 2 × 9-CH2], 5.85 (d, 3J = 10 Hz, 2H, 2 × 11-H), 6.81 (s, 2H, 2 × 10-H), 6.92 (d, 3J = 10 Hz, 2H, 2 × 6-H), 6.94–6.98 (m, 2H, 2 × 8-H), 7.35 (d, 3J = 8 Hz, 2H, 2 × 7-H), 7.88–7.92 (m, 2H, 2 × 3-H), 8.23 (d, 3J = 8 Hz, 1H, 2 × 1-H), 8.47–8.50 (m, 2H, 2 × 2-H), 9.01 (d, 3J = 6 Hz, 2H, 2 × 4-H). – 13C-NMR (150 MHz, D2O): δ = 41.6 (2 x CH2), 54.1 (2 × C11), 77.5 (2 × C6), 126.8 (2 × C7), 130.3 (2 × C3), 131.5 (2 × C1), 133.6 (2 × C10), 135.5 (2 × C8), 136.3 (2 × C6a, or 2 × C10a), 137.2 (2 × C6a, or 2 × C10a), 137.5 (2 × C9), 149.2 (2 × C4), 151.1 (2 × C2), 154.9 (2 × C11a). – HRMS (ESI+): calcd for: [C28H22N2S2 ]2+m/z = 225.0607, found: 225.0603 [M].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ε) = 396 nm (4.09). – Em. (H2O): λmax = 409 nm. – 1H-NMR (500 MHz, DMSO-d6): δ = 4.22 (s, 2H, 2 × CH2), 7.93–7.96 (m, 2H, 3-H, 3′-H, 8-H, 8′-H), 8.08 (dd, 3J = 9 Hz, 3J = 7 Hz, 1H, 2-H, 2′-H), 8.16 (s, 1H, 10-H, 10′-H), 8.46 (d, 3J = 9 Hz, 1H, 7-H, 7′-H), 8.57 (d, 3J = 9 Hz, 1H, 1-H, 1′-H), 9.20 (s, 1H, 11-H, 11′-H), 9.26 (d, 3J = 7 Hz, 1H, 4-H, 4′-H), 10.41 (s, 1H, 6-H, 6′-H). 1H-NMR (600 MHz, D2O): δ = 4.03 (s, 2H, 2 × CH2), 7.66 (s, 1H, 2H, 10-H, 10′-H), 7.70 (t, 3J = 7 Hz, 1H, 3-H, 3′-H), 7.86 (dd, 3J = 9 Hz, 4J = 2 Hz, 1H, 8-H, 8′-H), 7.90 (t, 3J = 8 Hz, 1H, 2-H, 2′-H), 8.15–8.18 (m, 4H, 1-H, 1′-H, 7-H, 7′-H), 8.45 (s, 1H, 11-H, 11′-H), 8.75 (d, 3J = 7 Hz, 1H, 4-H, 4′-H), 9.63 (s, 1H, 6H, 6′-H). 13C-NMR (125 MHz, DMSO-d6): δ = 40.9 (2 × CH2), 122.3 (C3, C3′), 124.2 (C11, C11′), 125.0 (C6a, C6a’), 126.2 (C10, C10′), 126.7 (C1, C1′), 128.5 (C7, C7′), 131.2 (C2, C2′), 132.6 (C8, C8′), 134.2 (C4, C4′), 135.0 (C10a, C10a’), 137.7 (C11a, C11a’), 139.8 (C6, C-6′), 144.6 (C9, C9′). 13C-NMR (150 MHz, D2O): δ = 47.0 (2 × CH2), 125.7 (C3, C3′), 126.5 (C11, C11′), 127.9 (C6a, C6a’), 128.0 (C10, C10′), 129.3 (C1, C1′), 130.5 (C7, C7′), 134.0 (C2, C2′), 135.7 (C8, C8′), 135.9 (C4, C4′), 137.8 (C10a, C10a’), 139.7 (C11a, C11a’), 140.2 (C6, C6′), 149.6 (C9, C9′). – MS (ESI+): m/z (%) = 225 (100) [M−2Br−]2+. – El. Anal. for C28H22Br2N2S2 × 2 H2O, cald. (%): C 52.02, H 4.05, N 4.33, S 9.92; found (%): C 52.06, H 4.04, N 4.19, S 10.18.
ε) = 396 nm (4.09). – Em. (H2O): λmax = 409 nm. – 1H-NMR (500 MHz, DMSO-d6): δ = 4.22 (s, 2H, 2 × CH2), 7.93–7.96 (m, 2H, 3-H, 3′-H, 8-H, 8′-H), 8.08 (dd, 3J = 9 Hz, 3J = 7 Hz, 1H, 2-H, 2′-H), 8.16 (s, 1H, 10-H, 10′-H), 8.46 (d, 3J = 9 Hz, 1H, 7-H, 7′-H), 8.57 (d, 3J = 9 Hz, 1H, 1-H, 1′-H), 9.20 (s, 1H, 11-H, 11′-H), 9.26 (d, 3J = 7 Hz, 1H, 4-H, 4′-H), 10.41 (s, 1H, 6-H, 6′-H). 1H-NMR (600 MHz, D2O): δ = 4.03 (s, 2H, 2 × CH2), 7.66 (s, 1H, 2H, 10-H, 10′-H), 7.70 (t, 3J = 7 Hz, 1H, 3-H, 3′-H), 7.86 (dd, 3J = 9 Hz, 4J = 2 Hz, 1H, 8-H, 8′-H), 7.90 (t, 3J = 8 Hz, 1H, 2-H, 2′-H), 8.15–8.18 (m, 4H, 1-H, 1′-H, 7-H, 7′-H), 8.45 (s, 1H, 11-H, 11′-H), 8.75 (d, 3J = 7 Hz, 1H, 4-H, 4′-H), 9.63 (s, 1H, 6H, 6′-H). 13C-NMR (125 MHz, DMSO-d6): δ = 40.9 (2 × CH2), 122.3 (C3, C3′), 124.2 (C11, C11′), 125.0 (C6a, C6a’), 126.2 (C10, C10′), 126.7 (C1, C1′), 128.5 (C7, C7′), 131.2 (C2, C2′), 132.6 (C8, C8′), 134.2 (C4, C4′), 135.0 (C10a, C10a’), 137.7 (C11a, C11a’), 139.8 (C6, C-6′), 144.6 (C9, C9′). 13C-NMR (150 MHz, D2O): δ = 47.0 (2 × CH2), 125.7 (C3, C3′), 126.5 (C11, C11′), 127.9 (C6a, C6a’), 128.0 (C10, C10′), 129.3 (C1, C1′), 130.5 (C7, C7′), 134.0 (C2, C2′), 135.7 (C8, C8′), 135.9 (C4, C4′), 137.8 (C10a, C10a’), 139.7 (C11a, C11a’), 140.2 (C6, C6′), 149.6 (C9, C9′). – MS (ESI+): m/z (%) = 225 (100) [M−2Br−]2+. – El. Anal. for C28H22Br2N2S2 × 2 H2O, cald. (%): C 52.02, H 4.05, N 4.33, S 9.92; found (%): C 52.06, H 4.04, N 4.19, S 10.18.
Methods
DNA-binding properties were determined according to published protocols27 (for details, cf. ESI, Chapter 5†).Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support was provided by the University of Siegen. C. D. is grateful to the House of Young Talents (University of Siegen) for a PhD fellowship.References
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, Ca-Cancer J. Clin., 2021, 71, 7 CrossRef PubMed.
- (a) C. Chen, X. Li, H. Zhao, M. Liu, J. Du, J. Zhang, X. Yang, X. Hou and H. Fang, J. Med. Chem., 2022, 65, 3667 CrossRef CAS PubMed; (b) G. Padroni, J. M. Withers, A. Taladriz-Sender, L. F. Reichenbach, J. A. Parkinson and G. A. Burley, J. Am. Chem. Soc., 2019, 141, 9555 CrossRef CAS PubMed; (c) S. Bhaduri, N. Ranjan and D. P. Arya, Beilstein J. Org. Chem., 2018, 14, 1051 CrossRef CAS PubMed.
- (a) W. Wu, Y. Pu and J. Shi, J. Nanobiotechnol., 2022, 20, 4 CrossRef CAS PubMed; (b) Q. Gao, J. Feng, W. Liu, C. Wen, Y. Wu, Q. Liao, L. Zou, X. Sui, T. Xie, J. Zhang and Y. Hu, Adv. Drug Delivery Rev., 2022, 188, 114445 CrossRef CAS PubMed; (c) L. Conti, E. Macedi, C. Giorgi, B. Valtancoli and V. Fusi, Coord. Chem. Rev., 2022, 469, 214656 CrossRef CAS.
- J. K. Patra, G. Das, L. F. Fraceto, E. V. R. Campos, M. P. Del Rodriguez-Torres, L. S. Acosta-Torres, L. A. Diaz-Torres, R. Grillo, M. K. Swamy, S. Sharma, S. Habtemariam and H.-S. Shin, J. Nanobiotechnol., 2018, 16, 71 CrossRef PubMed.
- X. Dong, R. K. Brahma, C. Fang and S. Q. Yao, Chem. Sci., 2022, 13, 4239 RSC.
- W. Xiao and J. Loscalzo, Antioxid. Redox Signal., 2020, 32, 1360 CrossRef PubMed.
- (a) B. Sun, C. Luo, H. Yu, X. Zhang, Q. Chen, W. Yang, M. Wang, Q. Kan, H. Zhang, Y. Wang, Z. He and J. Sun, Nano Lett., 2018, 18, 3643 CrossRef CAS PubMed; (b) R. Zhang, T. Nie, Y. Fang, H. Huang and J. Wu, Biomacromolecules, 2022, 23, 1 CrossRef PubMed.
- (a) C. Luo, J. Sun, D. Liu, B. Sun, L. Miao, S. Musetti, J. Li, X. Han, Y. Du, L. Li, L. Huang and Z. He, Nano Lett., 2016, 16, 5401 CrossRef CAS PubMed; (b) J. Wang, X. Sun, W. Mao, W. Sun, J. Tang, M. Sui, Y. Shen and Z. Gu, Adv. Mater., 2013, 25, 3670 CrossRef CAS PubMed.
- Q. Laurent, R. Martinent, B. Lim, A.-T. Pham, T. Kato, J. López-Andarias, N. Sakai and S. Matile, JACS Au, 2021, 1, 710 CrossRef CAS PubMed.
- M. Zhou, Y. Xie, S. Xu, J. Xin, J. Wang, T. Han, R. Ting, J. Zhang and F. An, Eur. J. Med. Chem., 2020, 195, 112274 CrossRef CAS PubMed.
- (a) M. Lin, S. Zou, T. Li, J. Karges, Y. Chen, Y. Zhao, L. Ji and H. Chao, Chem. Commun., 2022, 58, 4324 RSC; (b) N. A. Simeth, S. Kobayashi, P. Kobauri, S. Crespi, W. Szymanski, K. Nakatani, C. Dohno and B. L. Feringa, Chem. Sci., 2021, 12, 9207 RSC; (c) M. Dudek, M. Deiana, K. Szkaradek, M. J. Janicki, Z. Pokładek, R. W. Góra and K. Matczyszyn, J. Phys. Chem. Lett., 2021, 12, 9436 CrossRef CAS PubMed; (d) M. Deiana, M. Mosser, T. Le Bahers, E. Dumont, M. Dudek, S. Denis-Quanquin, N. Sabouri, C. Andraud, K. Matczyszyn, C. Monnereau and L. Guy, Nanoscale, 2021, 13, 13795 RSC; (e) J. Rodriguez, J. Mosquera, S. Learte-Aymamí, M. E. Vázquez and J. L. Mascareñas, Acc. Chem. Res., 2020, 53, 2286 CrossRef CAS PubMed; (f) H. K. Saeed, S. Sreedharan and J. A. Thomas, Chem. Commun., 2020, 56, 1464 RSC; (g) D. V. Berdnikova, J. Heider, H. Ihmels, N. Sewald and P. M. Pithan, ChemPhotoChem, 2020, 4, 520 CrossRef CAS; (h) M. Linares, H. Sun, M. Biler, J. Andréasson and P. Norman, Phys. Chem. Chem. Phys., 2019, 21, 3637 RSC; (i) H. Chen, R. Li, S. Li, J. Andréasson and J. H. Choi, J. Am. Chem. Soc., 2017, 139, 1380 CrossRef CAS PubMed; (j) T. C. S. Pace, V. Müller, S. Li, P. Lincoln and J. Andréasson, Angew. Chem., Int. Ed. Engl., 2013, 52, 4393 CrossRef CAS PubMed.
- (a) A. Granzhan, H. Ihmels and M. Tian, Arkivoc, 2015, 494 Search PubMed; (b) A. Granzhan and H. Ihmels, Synlett, 2016, 27, 1775 CrossRef CAS; (c) R. M. Suárez, P. Bosch, D. Sucunza, A. M. Cuadro, A. Domingo, F. Mendicuti and J. J. Vaquero, Org. Biomol. Chem., 2015, 13, 527 RSC; (d) H. Ihmels, K. Faulhaber, D. Vedaldi, F. Dall'Acqua and G. Viola, Photochem. Photobiol., 2005, 81, 1107 CrossRef CAS PubMed.
- (a) H. Ihmels and J. Luo, J. Photochem. Photobiol., A, 2008, 200, 3 CrossRef CAS; (b) S. A. Stratford, M. Arhangelskis, D.-K. Bučar and W. Jones, CrystEngComm, 2014, 16, 10830 RSC; (c) T. Wolff, C. Lehnberger and D. Scheller, Heterocycles, 1997, 45, 2036 CrossRef; (d) S. Yamada, Coord. Chem. Rev., 2020, 415, 213601 CrossRef.
- (a) C. K. Bradsher and L. E. Beavers, J. Am. Chem. Soc., 1955, 77, 4812 CrossRef CAS; (b) C. K. Bradsher, Chem. Rev., 1946, 46, 447 CrossRef PubMed.
- (a) H. Ihmels, B. Engels, K. Faulhaber and C. Lennartz, Chem. – Eur. J., 2000, 6, 2854 CrossRef CAS PubMed; (b) C. Bradsher, L. Beavers and J. Jones, J. Org. Chem., 1957, 22, 1740 CrossRef CAS.
- H. Ihmels, C. J. Mohrschladt, A. Schmitt, M. Bressanini, D. Leusser and D. Stalke, Eur. J. Org. Chem., 2002, 2624 CrossRef CAS.
- (a) D. Gupta and A. R. Knight, Can. J. Chem., 1980, 58, 1350 CrossRef CAS; (b) E. Robert-Banchereau, S. Lacombe and J. Ollivier, Tetrahedron, 1997, 53, 2087 CrossRef CAS.
- (a) C. Walling and R. Rabinowitz, J. Am. Chem. Soc., 1959, 81, 1243 CrossRef CAS; (b) W. L. Wallace, R. P. van Duyne and F. D. Lewis, J. Am. Chem. Soc., 1976, 98, 5319 CrossRef CAS; (c) M. You, Z. Zhu, H. Liu, B. Gulbakan Da Han, R. Wang, K. R. Williams and W. Tan, ACS Appl. Mater. Interfaces, 2010, 2, 3601 CrossRef CAS PubMed.
- (a) D. Monchaud, C. Allain and M.-P. Teulade-Fichou, Bioorg. Med. Chem. Lett., 2006, 16, 4842 CrossRef CAS PubMed; (b) W. C. Tse and D. L. Boger, Acc. Chem. Res., 2004, 37, 61 CrossRef CAS PubMed; (c) D. L. Boger, B. E. Fink, S. R. Brunette, W. C. Tse and M. P. Hedrick, J. Am. Chem. Soc., 2001, 123, 5878 CrossRef CAS PubMed.
- F. H. Stootman, D. M. Fisher, A. Rodger and J. R. Aldrich- Wright, Analyst, 2006, 131, 1145 RSC.
- (a) N. C. Garbett, P. A. Ragazzon and J. B. Chaires, Nat. Protoc., 2007, 2, 3166 CrossRef CAS PubMed; (b) T. Šmidlehner, I. Piantanida and G. Pescitelli, Beilstein J. Org. Chem., 2018, 14, 84 CrossRef PubMed; (c) B. Nordén, A. Rodger and T. Dafforn, Linear Dichroism and Circular Dichroism. A Textbook on Polarized-Light Spectroscopy, RSC Publishing, Cambridge, 2010 RSC.
- J. Nygren, N. Svanvik and M. Kubista, Biopolymers, 1998, 46, 39 CrossRef CAS PubMed.
- A. Granzhan, H. Ihmels and K. Jäger, Chem. Commun., 2009, 1249 RSC.
- K. Jäger, J. W. Bats, H. Ihmels, A. Granzhan, S. Uebach and B. O. Patrick, Chem. – Eur. J., 2012, 18, 10903 CrossRef PubMed.
- C. Audin, J.-C. Daran, É. Deydier, É. Manoury and R. Poli, C. R. Chim., 2010, 13, 890 CrossRef CAS.
- K. Suzuki, A. Kobayashi, S. Kaneko, K. Takehira, T. Yoshihara, H. Ishida, Y. Shiina, S. Oishi and S. Tobita, Phys. Chem. Chem. Phys., 2009, 11, 9850 RSC.
- R. Bortolozzi, H. Ihmels, L. Thomas, M. Tian and G. Viola, Chem. – Eur. J., 2013, 19, 8736 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials, methods, experimental procedures, analytical data, and spectra. See DOI: https://doi.org/10.1039/d3ob00013c |
| This journal is © The Royal Society of Chemistry 2023 |