Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Profiling rhythmicity of bile salt hydrolase activity in the gut lumen with a rapid fluorescence assay†
Chathuri J.
Kombala
ab,
Neha
Agrawal
a,
Agne
Sveistyte
a,
Ilia N.
Karatsoreos
 c,
Hans P. A.
Van Dongen
c,
Hans P. A.
Van Dongen
 bd and
Kristoffer R.
Brandvold
bd and
Kristoffer R.
Brandvold
 *ae
*ae
aBiological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, USA. E-mail: krbrandvold@gmail.com
bDepartment of Translational Medicine and Physiology, Elson S. Floyd College of Medicine, Washington State University, Spokane, WA 99202, USA
cDepartment of Psychological and Brain Sciences, University of Massachusetts, Amherst, MA 01003, USA
dSleep and Performance Research Center, Washington State University, Spokane, WA 99202, USA
eDepartment of Nutrition and Exercise Physiology, Elson S. Floyd College of Medicine, Washington State University, Spokane, WA 99202, USA
First published on 18th February 2023
Abstract
Diurnal rhythmicity of cellular function is key to survival for most organisms on Earth. Many circadian functions are driven by the brain, but regulation of a separate set of peripheral rhythms remains poorly understood. The gut microbiome is a potential candidate for regulation of host peripheral rhythms, and this study sought to specifically examine the process of microbial bile salt biotransformation. To enable this work, an assay for bile salt hydrolase (BSH) that could work with small quantities of stool samples was necessary. Using a turn-on fluorescence probe, we developed a rapid and inexpensive assay to detect BSH enzyme activity with concentrations as low as 6–25 μM, which is considerably more robust than prior approaches. We successfully applied this rhodamine-based assay to detect BSH activity in a wide range of biological samples such as recombinant protein, whole cells, fecal samples, and gut lumen content from mice. We were able to detect significant BSH activity in small amounts of mouse fecal/gut content (20–50 mg) within 2 h, which illustrates its potential for use in various biological/clinical applications. Using this assay, we investigated the diurnal fluctuations of BSH activity in the large intestine of mice. By using time restricted feeding conditions, we provided direct evidence of 24 h rhythmicity in microbiome BSH activity levels and showed that this rhythmicity is influenced by feeding patterns. Our novel function-centric approach has potential to aid in the discovery of therapeutic, diet, or lifestyle interventions for correction of circadian perturbations linked to bile metabolism.
Introduction
In industrialized nations, many professions (first responders, health care providers, military, etc.) require irregular sleep schedules with extensive nighttime activity, or frequent travel across time zones, both of which lead to misalignment between behavioral cycles and the biological clock.1–5Circadian misalignment between the master biological clock and peripheral organs, such as the gut,6–8 liver,9–11 and pancreas,10–12 leads to a desynchrony that disrupts metabolic processes, which can increase susceptibility to the eventual development of metabolic diseases including obesity,12,13 type 2 diabetes,12,14,15 and cardiovascular disease.15,16 Amelioration of the effects of circadian misalignment would improve personal well-being and decrease cost to society, but the mechanisms through which externally imposed behavioral schedules can dysregulate rhythms in metabolic organs are not currently known. The two main external factors that could produce desynchrony are sleep/wake cycle1,17 and the feeding/fasting schedule.18,19 Sleep deprivation alone does not invoke desynchronization,18,20 so investigating the impact of altered feeding/fasting schedules is particularly appealing. One possibility is that microbes in the gut create signals based on the feeding schedule20–24 which, in turn, regulate peripheral circadian rhythms in the host that are autonomous from the master clock in the brain.25–28 Nutrient availability directly affects microbiome composition28–30 which may act as a timing signal for peripheral circadian rhythms, which is consistent with the observation that certain taxa bloom directly following food ingestion, while others become dominant between meals.31,32 If eating schedules are erratic or not in-sync with the central clock, misalignment could occur. In order to understand why individuals with irregular sleep patterns are predisposed to metabolic disorders33,34 therefore, the microbiome's role in the circadian desynchronization must be appreciated. Promisingly, the gut microbiome is amenable to reshaping, and some characteristics can be rapidly impacted, which makes the gut microbiome an appealing target for therapeutic correction of circadian misalignment.35–38
While sequence-based analyses infer functional rhythmicity based on taxa abundances, direct measurement of microbiome function is under-reported. The extent of functional rhythmicity of the microbiome must be established to understand interactions with host rhythms. We became interested in the possibility of rhythmicity of microbial bile metabolism after we developed a novel assay to quantify the activity of a key metabolic enzyme. Bile is a host-produced fluid that enables the digestion of dietary fats and some lipophilic vitamins, due to the presence of amphipathic bile acid (BA) molecules.39 Microbial BA metabolism directly influence host physiology through changing the capacity to bind host receptors as signaling molecules,40 which impacts the regulation of host dietary lipid absorption,41–43 energy homeostasis,44 cholesterol metabolism,41 and controlling secondary BA level.45,46 Since bile is released from the gall bladder at feeding times, this could act as a “zeitgeber” for peripheral rhythms associated with bile metabolism.
Metabolomics data have already suggested that the bile salt pool experiences daily fluctuations,47 but confounding factors such as changes in bile salt levels due to variations in liver production or excretion in feces could influence the final measurements of metabolomics-based assays. A different approach is needed to investigate rhythmicity in bile metabolism. Multiple structural modifications may occur on a single BA molecule from microbial metabolism, but the first step is always bile salt hydrolysis, which results in free amino acid and BA products (Fig. S1A†). The enzyme responsible for deconjugation, BSH, is therefore thought of as a gatekeeper of microbial BA biotransformation.
Characterization of BSH rhythmicity, and determination of regulatory factors, has been previously untenable due to a lack of tools to specifically report on BSH activity in laboratory-relevant quantities, and issues that plague the study of microbiome functions in general, including inadequate genome annotation. For example, the Unified Human Gastrointestinal Protein catalog reported that there are 204![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 938 nonredundant genomes recorded from human gut microbiota, with 71% of total species having no experimental characterization and 40% no functional annotation whatsoever.48 Therefore, the likelihood of overlooked or improperly annotated BSHs in conventional omics measurements is high. Even in an ideal scenario, with a hypothetically fully annotated microbiome metagenome, transcript measurements do not confirm protein presence, and methods that monitor protein abundance cannot confirm that the enzymes are active, which may be influenced by secondary factors such as co-factor and substrate concentrations, cellular localization, protein–protein interactions, and post-translation modifications. As such, methods for functional measurement must be prioritized.
938 nonredundant genomes recorded from human gut microbiota, with 71% of total species having no experimental characterization and 40% no functional annotation whatsoever.48 Therefore, the likelihood of overlooked or improperly annotated BSHs in conventional omics measurements is high. Even in an ideal scenario, with a hypothetically fully annotated microbiome metagenome, transcript measurements do not confirm protein presence, and methods that monitor protein abundance cannot confirm that the enzymes are active, which may be influenced by secondary factors such as co-factor and substrate concentrations, cellular localization, protein–protein interactions, and post-translation modifications. As such, methods for functional measurement must be prioritized.
We sought to determine unambiguously if BSH activity is rhythmic and provide information on potential regulatory factors through imposition of a feeding schedule that emulates shift work. Direct evaluation of BSH activity in intestinal samples is very challenging due to the complex biological environment. Current methods of assessing BSH activity involve chromatographic techniques such as HPLC which require expensive instrumentation and long assay times.49 A spectrophotometric assay has been developed recently using bioluminescence probes50 which requires complex experimental setup as additional luciferase enzyme or luciferase transfected cells are needed for the assay. We also realized that our previously reported fluorogenic BSH assay,51 which required long incubation periods and large amount of sample, would be impractical in circadian studies due to inherently larger numbers of samples from multiple data collection time points. Therefore, we first sought to develop a novel fluorogenic substrate, which we refer to as ChoRhoS (CholoylRhodamine Substrate), which is a substantial improvement over our previous approach,51,52 because the red-shifted rhodamine fluorophore has lower background noise. In addition, with new assay conditions we reduced the assay time (30× faster) and increased our signal (20-fold reduction in sample size) and only of fraction of a mouse fecal pellet would be needed for analysis. Collectively, these improvements allowed us to definitively characterize the rhythmicity of BSH activity.
Chemical synthesis and spectroscopic characterization of ChoRhoS and related probes
For our BSH assay, we employed an approach commonly used for amide hydrolases that is based on evolution of rhodamine fluorescence. Acylation of rhodamine nitrogen atoms locks the molecule into a closed lactone form that is less fluorescent or nonfluorescent,53 and this enables a variety of fluorogenic enzyme substrates to be built from N,N′-mono or diacylrhodamines, which produce a highly fluorescent free dye upon exposure to an amide cleaving enzyme. Thus, we designed synthetic substrates (Fig. S1B†), which are conjugation products of cholic acid and rhodamine. The designed substrate contains an amide bond that is specifically cleaved by the BSH enzyme (Fig. S1C†), which results in evolution of fluorescence that is proportional to the level of BSH activity (Fig. 1A). We conjugated cholic acid with rhodamine using standard EDC coupling reagents with dimethylformamide (DMF) as a solvent and DIEA as a base (Scheme S1†). The conjugated product was isolated via flash chromatography. Spectroscopic characterization of the conjugated product and free rhodamine showed at least 130-fold less fluorescence for the substrate relative to free rhodamine (Fig. 1B and S1D, Table S1†). This is substantially higher than the 10-fold increase observed with our earlier coumarin-based substrates. Therefore, our probe showed potential for use as a fluorogenic reporter in BSH activity assays.Results
Biochemical characterization of ChoRhoS with purified recombinant BSH enzyme
We first tested our probe with purified, recombinant Lactobacillus plantarum (L. plantarum) BSH. When the mono-cholic conjugate probe (ChoRhoS) was exposed to purified recombinant BSH, rhodamine fluorescence increased significantly (Fig. 1C-green). To confirm the necessity of the cholic acid moiety, we synthesized N-acetylrhodamine as a control substrate, and no fluorescence increase was observed upon incubation with recombinant BSH enzyme (Fig. S1E†). Therefore, the evolution of fluorescence due to co-incubation of BSH with ChoRhoS is not due to non-specific amide cleavage. We also confirmed that the probe was not simply degrading via hydrolysis. No increase in fluorescence was observed when the probe was added just to buffer, indicating the stability of the probe in buffered solutions (Fig. 1C-grey). We were able to detect fluorescence signals from BSH-promoted substrate turnover in the low micromolar concentration range for the substrate, which is a significant improvement over our previous assay with coumarin-derived fluorogenic substrates, which we typically used at a final concentration of 150 μM. We also synthesized di-cholic acid conjugated rhodamine (Di-ChoRhoS; Fig. S1B†) and tested with recombinant protein. Surprisingly, no cleavage was observed for Di-ChoRhoS (Fig. S1E†-gray), which could be due to the steric hindrance from two cholic acid moieties. Due to the negligible activity, Di-ChoRhoS was dropped from consideration, and further probe characterization focused on ChoRhoS.We then quantitatively examined the kinetics of ChoRhoS hydrolysis by recombinant BSH via a standard Michaelis–Menten analysis. Our biochemical characterization of ChoRhoS revealed a low micromolar Michaelis–Menten constant (KM = 7.57 μM; Fig. 1D and S2†), which is substantially lower than reported values54 for native BSH substrates and 8-fold lower than our previously reported synthetic BSH fluorescence probes. The Vmax was observed as 1.07 × 10−4 s−1, and the catalytic efficiency (kcat/Km) for the enzyme was 1.32 M−1 s−1.
Whole-cell characterization of ChoRhoS
After confirming that the ChoRhoS probe is an effective fluorogenic reporter for recombinant BSH, the possibility of analyzing complex biological mixtures was first evaluated with whole-cell suspensions. We tested BSH positive organisms (L. plantarum and L. gasseri) and BSH negative organisms (E. coli Nissle and B. cereus) and observed a correlation between fluorescence evolution and the BSH activity of whole cell monocultures (Fig. 1E and S3, S4†), which suggests that our probe selectively detects activity in BSH expressing organisms (Fig. S3–S5†). Additionally, we observed that fluorescence was proportional to cell density in a dilution series of L. plantarum cells (Fig. S3A†). Finally, we established that ChoRhoS could accurately measure BSH activity in monoculture mixtures (Fig. S3B†), suggesting that the assay could work with more complex samples.Mouse fecal sample analysis using ChoRhoS
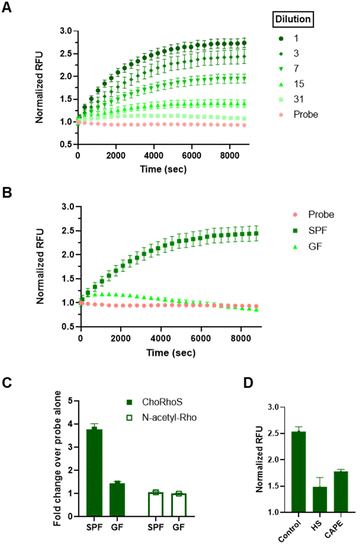
Detecting BSH activity in fecal samples is challenging due to complex and highly heterogeneous sample composition, which is compounded by the limitations of current methods that require extensive sample processing, long assay time, and complex experimental setup.50 Our previous approach, required overnight incubation for any observable signal of native BSH in fecal samples relative to controls. For our new, more rapid approach, we processed fecal samples from conventional (specific pathogen-free; SPF) mice as described previously,50 with minor modifications. After addition of the probe to fecal suspensions, we monitored the kinetics of rhodamine liberation for 3 h. Substantial signal evolution was observed within 1 h, and a substrate turnover plateau was observed within 2 h (Fig. 2A and S6†). In comparison with our previously reported fluorescence assay (cholic acid-conjugated amino coumarin probe; CA-AMCA), which requires 1000 mg of mouse fecal sample for analysis, this assay can be effectively applied to detect BSH activity with 20–50 mg of sample. In addition to the improved characteristics of our probe, we also found that the addition of β-mercaptoethanol, which is commonly added as a reductant to maximize the activity of cysteine hydrolases, including BSH,50 increased assay signal. These conditions improved the activity of our previously published coumarin-based substrates as well (Fig. S7†), but the ChoRhoS assay still performed substantially better under the same conditions. The S/N ratio for ChoRhoS substrate turnover was around 3-fold higher than the cholic acid-conjugated aminocoumarin probe.To demonstrate that the observed signal was the result of microbial activity, we incubated our probe with fecal suspensions of feces from germ-free (GF) mice, with the expectation that we should not see appreciable signal in these samples. As expected, a negligible fluorescence increase was observed in fecal suspensions from GF mice relative to the background fluorescence from the probe alone, which contrasts with the significant increase in fluorescence that was observed within 2 h for fecal suspensions from SPF mice (Fig. 2B and C).
To confirm that the observed activity in SPF samples was from properly folded and active enzymes, we conducted additional experiments in which a significant reduction in evolution of fluorescence was detected when fecal slurries were heat-shocked or incubated with a BSH inhibitor, caffeic acid phenethyl ester (CAPE), prior to addition of the probe (Fig. 2D and S8, S9†). We also confirmed that the observed activity in fecal samples was not due to nonspecific degradation of amides through co-incubation of fecal suspensions with a control probe, N-acetylrhodamine, which, similar to our bile salt probes, has the amine of rhodamine in an amide bond, but does not have the BA element. In contrast to the ChoRhoS probe, no signal increase upon addition to a fecal sample was observed with N-acetylrhodamine (Fig. 2C and S10†); in fact, a slight decrease in signal relative to the buffer was observed. Collectively, our results strongly suggest that the observed signal using ChoRhoS is specifically from active BSH.
The encouraging observations from our mouse fecal sample analysis prompted us to determine if this would additionally translate to the analysis of human fecal samples. In similar experiments, we observed probe fluorescence upon incubation of different dilutions of human fecal suspensions (Fig. S11†). However, although there was a statistically significant difference relative to controls (Fig. S12†), we only observed a 1.04-fold increase in fluorescence for human samples, which is substantially less than the 2.25-fold increase in mouse samples (Fig. S9B†). The lower intensity in human samples could be due to human samples being more heterogeneous as a result of a more complex diet. Thus, further optimization of assay conditions is needed for robust analysis of human samples.
Evaluation of BSH activity in gut luminal content using ChoRhoS
While fecal samples offer a convenient, noninvasive means of characterizing the gut microbiome, microbial activity in the actual gut lumen of a living animal cannot be fully captured by this approach, which is important in the context of BSH activity because this is where free BA products bind host receptors. Microbial activity can vary even within the respective compartments of the gut, as these are substantially different environments and therefore total abundance, and compositional nature can differ. We sought to verify that our assay is compatible with different types of gut lumen content, and intestinal material was collected from the small intestine, cecum, and large intestine tissues isolated from mice.Our assay provided robust kinetic curves for material isolated from the cecum (p = 0.0012) and large intestine (p = 0.0008), but we observed no substantial activity in the small intestine within the timeframe of our assay (Fig. 3A). These data are in good agreement with what is known about the distribution of bacteria in general, and the specific distribution of BSH-expressing organism in the gut.55 We found that these trends were consistent across an examination of four biological replicates (Fig. 3B and S13, S14†). To demonstrate that the evolution of fluorescence was the result of microbial activity, we tested the luminal content of the large intestine in mice treated with an antibiotic cocktail or vehicle control (Fig. 3C) and observed a substantial drop in activity in samples from the antibiotic-treated mice. Additionally, as with fecal samples, we did not observe substantial activity in luminal content from GF mice, but when GF mice received a fecal microbiota transplantation (FMT) from SPF mice, we observed activity levels that were comparable to the control SPF mice (Fig. 3C).
Characterizing fluctuations in BSH activity throughout the day
Finally, after verifying that our assay is compatible with gut lumen content, we sought to determine if BSH activity is rhythmic in this compartment by analyzing samples acquired from mice on a fixed light/dark schedule. We hypothesized that BSH expressing organisms would exist in the highest abundance during the active phase when feeding occurs and bile would be at its highest concentration in the gut. An experiment was designed in which wild-type SPF mice were maintained on a standard 12 h/12 h light/dark cycle. In one condition of the experiment, the mice had access to food ad libitum. Mice are nocturnal animals and are therefore more active and consume more food during the dark phase. We thus expected that we would see the greatest activity with our assay during the dark phase. Mice were sacrificed, in groups of four, at five different time points at 6 h intervals over the course of 24 h, starting at the beginning of the dark phase. Gut lumen content was isolated, and BSH activity was assessed using ChoRhoS. Although there was high variability between biological replicates, the collective data demonstrated rhythmicity in BSH function based on cosinor analysis56 (i.e., nonlinear regression analysis fitting a cosine function to activity values as a function of time) (Fig. 4-green and S15–S19†). Our analysis revealed that BSH activity was highest in the middle of the dark phase when the mice are most active and also eat the most. We further hypothesized that, if this rhythmic pattern was regulated by feeding time, a shift in BSH activity would be observed if the feeding time was restricted to a time when the mice are quiescent and more frequently asleep (light phase), and that we should consequently observe the highest BSH activity being shifted to the phase when food was made available. The experiment, therefore, also included a condition in which the 12 h/12 h light/dark cycle was identical to the first experiment, but mice were only provided chow during the light phase when their activity is typically the lowest, and they sleep the most. To account for any effects due to caloric restriction relative to ad libitum-fed mice, we also had a control condition in which animals were only offered chow during the dark phase. Similar to the ad libitum-fed condition, there was considerable variability between biological replicates for each timepoint, but cosinor analysis demonstrated significantly different rhythms between conditions. In particular, the timing of food availability significantly impacted the timing of the peak in BSH activity (F2,51 = 5.10, p = 0.010). The peak in BSH activity for dark phase-fed mice was about 1.5 h later during the dark phase than that for ad libitum-fed mice, a non-significant difference (p = 0.236). However, the peak in BSH activity for light phase-fed mice was found during the light phase and more than 6 h earlier than that for ad libitum-fed mice, which constituted a significant difference (p = 0.003). Thus, there was a difference of almost 9 h in the peak time of BSH activity between the light phase- and dark phase-fed mice.The peak in BSH activity observed in ad libitum and dark phase-fed groups reflects the normal nocturnal behavior of mice. We speculate that the modestly delayed peak for the dark phase-fed mice relative to the ad libitum-fed mice may be due to the polyphasic nature of rodent sleep, and that ad libitum-fed mice have the opportunity to begin feeding prior to initiation of the dark phase. Conversely, the light phase-fed mice had a BSH activity peak that was much advanced compared to both the dark phase-fed and ad libitum-fed mice, which is consistent with our hypothesis regarding the role of feeding time in the regulation of the functional rhythmicity of BSH. Although other methods have established differences in bile salt concentrations throughout the day, these data constitute the first direct demonstration of diurnal rhythmicity in BSH activity, and this is the first study to directly show that BSH activity levels in the large intestine are strongly influenced by feeding patterns.
Discussion
Bile acid metabolism by the gut microbiome has a substantial influence on human health. Collective BA metabolism in the gut is initiated by BSH, which acts as a crucial player in the coordination of host–microbiome interactions. Development of simple and reliable strategies to detect BSH activity is therefore highly desirable. To specifically characterize BSH as a potential functional oscillator, we developed a simple fluorescence-based tool, ChoRhoS, to directly measure BSH activity rapidly and with high sensitivity. We were able to successfully apply ChoRhoS to the quantification of BSH activity in samples of pure enzyme, microbial monocultures, mixtures of monocultures, and murine feces.We also applied our ChoRhoS assay to directly monitor BSH activity in samples from the mouse gut lumen. Consistent with what is known about the distribution of BSH-expressing organism in the gut, we found the highest BSH activity in the large intestine and cecum, and negligible activity in the small intestine. We also investigated the impact of the daily light/dark cycle and found that ad libitum-fed mice have the highest BSH activity in the large intestine in the dark phase, when the animals are the most active and consume the greatest amounts of food. In addition, we found that we were able to manipulate this trend by restricting the times when food is made available. That is, for mice that received food only during the light period, when they would normally be the least active, peak BSH activity shifted to the light phase. This finding is consistent with a separate study that revealed feeding pattern-based changes in microbiome composition,32 and add valuable information regarding functional significance of such changes. Our data indicate that feeding time is a key regulator of BSH activity, and our ChoRhoS assay allowed for the first functional, direct demonstration of this.
The observation of diurnal rhythmicity of BSH activity has important implications because it directly impacts host signaling processes through modification of the occupancy of host BA receptors by free BAs. The possibility that the observed rhythmicity in BSH function may be modified by other factors as well merits further investigation. For example, significant differences in BSH activity between healthy individuals and patients with atherosclerosis and diabetes have been observed.57 Moreover, an increase in BSH activity has been reported in a mouse model of colitis compared with healthy mice.58 It would be worthwhile to document whether and how these diseases impact the diurnal rhythmicity of BSH activity, and whether alterations in the timing of food intake may have therapeutic potential in this context.
Conclusions
In conclusion, we developed a simple and efficient tool to detect and quantify BSH enzyme activity in complex biological samples. As microbial BSH enzyme is involved in various host functions such as metabolism and immune function, our strategy can be used to assess host physiology and, eventually, to identify and treat BSH-based functional changes and associated diseases. By using our ChoRhoS tool, accurate estimation of BSH activity can be accomplished, with potential for high throughput applications, which will be particularly beneficial for screening of pharmacological modulators or chemical genetic tools for regulating BSH.From a broader perspective, based on our observations with BSH, we believe that a function-focused analysis of diurnal rhythmicity of the gut microbiome in general is needed. As the microbiome displays functional rhythmicity this likely impacts rhythmic processes in the host. Previously observed rapid reversal of metabolite rhythmicity,58,59 exosome-based metabolic signaling,60 and immune parameters60 in humans exposed to simulated night shift work may be mediated by functional microbiome rhythm changes. Desynchronization of host-specific circadian processes between tissues leads to development of a variety of diseases,4 and it is possible that gut microbiome functions play a major role in circadian desynchronization.61 With approximately 20% of the US workforce being exposed to shift work at least some of the time,3,62 mitigation of metabolic disturbances caused by circadian desynchronization associated with such work schedules could significantly reduce the societal health burden. As such, it is crucial that we understand which functions of the microbiome are rhythmic, how these rhythms interact with the host, and which are subject to manipulation, so that we can develop robust intervention strategies. We foresee that function-focused assays, such as our ChoRhoS approach, will be key to driving future discoveries.
Materials and methods
See the ESI† for all the experiment details. All experiments were performed in compliance with the relevant laws and institutional guidelines. All experiments with conventional mice, except for the antibiotic treatment experiment, were conducted at WSU and approved by the WSU IACUC. The experiment conducted with conventional mice involving antibiotic treatment and all experiments conducted with germ-free mice were conducted at PNNL and approved by the PNNL IACUC. Informed consent was obtained for experiments that analyzed human samples.Abbreviations
| BSH | Bile salt hydrolase |
| BA | Bile acid |
| ChoRhoS | CholoylRhodamine substrate |
| BME | Beta-mercaptoethanol |
| CA-AMCA | Cholic acid-conjugated amino coumarin probe |
| CAPE | Caffeic acid phenethyl ester |
| DMF | Dimethylformamide |
| FMT | Fecal microbiota transplantation |
| FS | Fecal suspension |
| GF | Germ free |
| HS | Heat-shocked |
| IACUC | Institutional Animal Care and Use Committee |
| RFU | Relative fluorescence units |
| SPF | Specific pathogen-free |
| ZT | Zeitgeber time |
Author contributions
CJK: conceptualization, methodology, formal analysis, investigation, writing – original draft, writing review & editing, visualization; NA: methodology, investigation; AS: investigation; INK: conceptualization, writing review & editing, HPAVD: conceptualization, formal analysis, writing review & editing, visualization; KRB: conceptualization, methodology, formal analysis, investigation, writing – original draft, writing review & editing, visualization, supervision, project administration, funding acquisition; CJK, NA, IK, HVD, and KRB designed the experiments. CJK, NA, and AS conducted the experiments. CJK, NA, IK, HVD, and KRB analyzed the data. CJK and KRB wrote the manuscript. All authors contributed to revision of the manuscript. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank our following funding sources. Time-restricted feeding experiments were supported by funds provided by the Human Sleep and Cognition Laboratory (HSCL) at Washington State University. CJK was supported by funds provided by the Elson S. Floyd College of Medicine at Washington State University. HVD was supported by USAMRDC grant W81XWH-18-1-0100. KRB was supported in part by funds provided by the HSCL. INK was supported by NIH grant R01 DK119811.Research was also supported in part by the BRAVE and the EBSD SEED Lab Directed Research and Development Programs at Pacific Northwest National Laboratory (PNNL). PNNL is operated by Battelle for the US Department of Energy under contract DE-AC06-76RL01830.
We would also like to thank Aaron T. Wright (PNNL) for his careful reading of the manuscript and helpful editing suggestions. Also, we would like to thank the Integrative Physiology and Neuroscience Behavioral Core at Washington State University and the PNNL Animal Resource Center for technical expertise and execution of mouse experiments. The table of contents graphic was created using Biorender.com.
References
- K. G. Baron and K. J. Reid, Circadian misalignment and health, Int. Rev. Psychiatry, 2014, 26(2), 139–154, DOI:10.3109/09540261.2014.911149
.
- S. L. Chellappa, C. J. Morris and F. A. J. L. Scheer, Circadian misalignment increases mood vulnerability in simulated shift work, Sci. Rep., 2020, 10(1), 18614, DOI:10.1038/s41598-020-75245-9
.
- E. E. Flynn-Evans, L. K. Barger, A. A. Kubey, J. P. Sullivan and C. A. Czeisler, Circadian misalignment affects sleep and medication use before and during spaceflight, npj Microgravity, 2016, 2(1), 15019, DOI:10.1038/npjmgrav.2015.19
.
- A. B. Fishbein, K. L. Knutson and P. C. Zee, Circadian disruption and human health, J. Clin. Invest., 2021, 131(19), e148286, DOI:10.1172/JCI148286
.
- S. M. James, K. A. Honn, S. Gaddameedhi and H. P. A. Van Dongen, Shift, Work: Disrupted Circadian Rhythms and Sleep—Implications for Health and Well-being, Curr. Sleep Med. Rep., 2017, 3(2), 104–112, DOI:10.1007/s40675-017-0071-6
.
- T. D. Butler and J. E. Gibbs, Circadian Host-Microbiome Interactions in Immunity, Front. Immunol., 2020, 11, 1783, DOI:10.3389/fimmu.2020.01783
, review.
- B. A. Matenchuk, P. J. Mandhane and A. L. Kozyrskyj, Sleep, circadian rhythm, and gut microbiota, Sleep Med. Rev., 2020, 53, 101340, DOI:10.1016/j.smrv.2020.101340
.
- S. Mashaqi and D. Gozal, “Circadian misalignment and the gut microbiome. A bidirectional relationship triggering inflammation and metabolic disorders”- a literature review, Sleep Med., 2020, 72, 93–108, DOI:10.1016/j.sleep.2020.03.020
.
- A. J. Davidson, O. Castañón-Cervantes and F. K. Stephan, Daily oscillations in liver function: diurnal vs circadian rhythmicity, Liver Int., 2004, 24(3), 179–186, DOI:10.1111/j.1478-3231.2004.00917.x
.
- J. M. Ferrell and J. Y. L. Chiang, Circadian rhythms in liver metabolism and disease, Acta Pharm. Sin. B, 2015, 5(2), 113–122, DOI:10.1016/j.apsb.2015.01.003
.
- A. Mukherji, S. M. Bailey, B. Staels and T. F. Baumert, The circadian clock and liver function in health and disease, J. Hepatol., 2019, 71(1), 200–211, DOI:10.1016/j.jhep.2019.03.020
.
- M. J. Christopher, J. N. Yang, J. I. Garcia, S. Myers, I. Bozzi, W. Wang, O. M. Buxton, S. A. Shea and F. A. J. L. Scheer, Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(17), E2225–E2234, DOI:10.1073/pnas.1418955112
.
- J. Keller and P. Layer, Circadian pancreatic enzyme pattern and relationship between secretory and motor activity in fasting humans, J. Appl. Physiol., 2002, 93(2), 592–600, DOI:10.1152/japplphysiol.00807.2001
.
- I. C. Mason, J. Qian, G. K. Adler and F. A. J. L. Scheer, Impact of circadian disruption on glucose metabolism: implications for type 2 diabetes, Diabetologia, 2020, 63(3), 462–472, DOI:10.1007/s00125-019-05059-6
.
- J. Qian, C. Dalla Man, C. J. Morris, C. Cobelli and F. A. J. L. Scheer, Differential effects of the circadian system and circadian misalignment on insulin sensitivity and insulin secretion in humans, Diabetes, Obes. Metab., 2018, 20(10), 2481–2485, DOI:10.1111/dom.13391
.
- S. Khan, B. H. Malik, D. Gupta and I. Rutkofsky, The Role of Circadian Misalignment due to Insomnia, Lack of Sleep, and Shift Work in Increasing the Risk of Cardiac Diseases: A Systematic Review, Cureus, 2020, 12(1), e6616–e6616, DOI:10.7759/cureus.6616
.
- F. A. J. L. Scheer, M. F. Hilton, C. S. Mantzoros and S. A. Shea, Adverse metabolic and cardiovascular consequences of circadian misalignment, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(11), 4453–4458, DOI:10.1073/pnas.0808180106
, acccessed 2022/05/22.
- L. Pickel and H.-K. Sung, Feeding Rhythms and the Circadian Regulation of Metabolism, Front. Nutr., 2020, 7, 39, DOI:10.3389/fnut.2020.00039
, review.
- V. D. Longo and S. Panda, Fasting, Circadian Rhythms, and Time-Restricted Feeding in Healthy Lifespan, Cell Metab., 2016, 23(6), 1048–1059, DOI:10.1016/j.cmet.2016.06.001
.
- E. Challet, The circadian regulation of food intake, Nat. Rev. Endocrinol., 2019, 15(7), 393–405, DOI:10.1038/s41574-019-0210-x
.
- E. R. Leeming, A. J. Johnson, T. D. Spector and C. I. Le Roy, Effect, of Diet on the Gut Microbiota: Rethinking Intervention Duration, Nutrients, 2019, 11(12) DOI:10.3390/nu11122862
.
- A. Leshem, E. Segal and E. Elinav, The Gut Microbiome and Individual-Specific Responses to Diet, mSystems, 2020, 5(5), e00665–e00620, DOI:10.1128/mSystems.00665-20
.
- R. K. Singh, H.-W. Chang, D. Yan, K. M. Lee, D. Ucmak, K. Wong, M. Abrouk, B. Farahnik, M. Nakamura and T. H. Zhu,
et al., Influence of diet on the gut microbiome and implications for human health, J. Transl. Med., 2017, 15(1), 73–73, DOI:10.1186/s12967-017-1175-y
.
- R. K. Fritts, A. L. McCully and J. B. McKinlay, Extracellular Metabolism Sets the Table for Microbial Cross-Feeding, Microbiol. Mol. Biol. Rev., 2021, 85(1), e00135–e00120, DOI:10.1128/MMBR.00135-20
.
- M. Murakami and P. Tognini, The Circadian Clock as an Essential Molecular Link Between Host Physiology and Microorganisms, Front. Cell. Infect. Microbiol., 2020, 9, 469, DOI:10.3389/fcimb.2019.00469
, mini review.
- K. Frazier and E. B. Chang, Intersection of the Gut Microbiome and Circadian Rhythms in Metabolism, Trends Endocrinol. Metab., 2020, 31(1), 25–36, DOI:10.1016/j.tem.2019.08.013
, acccessed 2022/05/22.
- E. Zhao, C. Tait, C. D. Minacapelli, C. Catalano and V. K. Rustgi, Circadian Rhythms, the Gut Microbiome, and Metabolic Disorders, Gastro Hep Adv., 2022, 1(1), 93–105, DOI:10.1016/j.gastha.2021.10.008
.
- X. Liang and G. A. FitzGerald, Timing the Microbes: The Circadian Rhythm of the Gut Microbiome, J. Biol. Rhythms, 2017, 32(6), 505–515, DOI:10.1177/0748730417729066
.
- G. Clarke, K. V. Sandhu, B. T. Griffin, T. G. Dinan, J. F. Cryan and N. P. Hyland, Gut Reactions: Breaking Down Xenobiotic–Microbiome Interactions, Pharmacol. Rev., 2019, 71(2), 198, DOI:10.1124/pr.118.015768
.
- A. L. Kau, P. P. Ahern, N. W. Griffin, A. L. Goodman and J. I. Gordon, Human nutrition, the gut microbiome and the immune system, Nature, 2011, 474(7351), 327–336, DOI:10.1038/nature10213
.
- C. A. Thaiss, M. Levy, T. Korem, L. Dohnalová, H. Shapiro, D. A. Jaitin, E. David, D. R. Winter, M. Gury-BenAri and E. Tatirovsky,
et al., Microbiota Diurnal Rhythmicity Programs Host Transcriptome Oscillations, Cell, 2016, 167(6), 1495–1510, DOI:10.1016/j.cell.2016.11.003
.
- C. A. Thaiss, D. Zeevi, M. Levy, G. Zilberman-Schapira, J. Suez, A. C. Tengeler, L. Abramson, M. N. Katz, T. Korem and N. Zmora,
et al., Transkingdom Control of Microbiota Diurnal Oscillations Promotes Metabolic Homeostasis, Cell, 2014, 159(3), 514–529, DOI:10.1016/j.cell.2014.09.048
.
- K. Spruyt, D. L. Molfese and D. Gozal, Sleep Duration, Sleep Regularity, Body Weight, and Metabolic Homeostasis in School-aged Children, Pediatrics, 2011, 127(2), e345–e352, DOI:10.1542/peds.2010-0497
.
- T. Huang and S. Redline, Cross-sectional and Prospective Associations of Actigraphy-Assessed Sleep Regularity With Metabolic Abnormalities: The Multi-Ethnic Study of Atherosclerosis, Diabetes Care, 2019, 42(8), 1422–1429, DOI:10.2337/dc19-0596
.
- E. Milner, B. Stevens, M. An, V. Lam, M. Ainsworth, P. Dihle, J. Stearns, A. Dombrowski, D. Rego and K. Segars, Utilizing Probiotics for the Prevention and Treatment of Gastrointestinal Diseases, Front. Microbiol., 2021, 12, 2121, DOI:10.3389/fmicb.2021.689958
.
- T. Liwinski and E. Elinav, Harnessing the microbiota for therapeutic purposes, Am. J. Transplant., 2020, 20(6), 1482–1488, DOI:10.1111/ajt.15753
.
- D. I. A. Pereira and G. R. Gibson, Effects of Consumption of Probiotics and Prebiotics on Serum Lipid Levels in Humans, Crit. Rev. Biochem. Mol. Biol., 2002, 37(4), 259–281, DOI:10.1080/10409230290771519
.
- S. G. Parkar, A. Kalsbeek and J. F. Cheeseman, Potential Role for the Gut Microbiota in Modulating Host Circadian Rhythms and Metabolic Health, Microorganisms, 2019, 7(2), 41 CrossRef
.
- B. Staels and V. A. Fonseca, Bile Acids and Metabolic Regulation, Diabetes Care, 2009, 32(suppl 2), S237, DOI:10.2337/dc09-S355
.
- N. Molinero, L. Ruiz, B. Sánchez, A. Margolles and S. Delgado, Intestinal Bacteria Interplay With Bile and Cholesterol Metabolism: Implications on Host Physiology, Front. Physiol., 2019, 10, 185, DOI:10.3389/fphys.2019.00185
, mini review.
- C.-C. Tsai, P.-P. Lin, Y.-M. Hsieh, Z.-y. Zhang, H.-C. Wu and C.-C. Huang, Cholesterol-Lowering Potentials of Lactic Acid Bacteria Based on Bile-Salt Hydrolase Activity and Effect of Potent Strains on Cholesterol Metabolism In Vitro and In Vivo, Sci. World J., 2014, 2014, 690752, DOI:10.1155/2014/690752
.
- S. A. Joyce, J. MacSharry, P. G. Casey, M. Kinsella, E. F. Murphy, F. Shanahan, C. Hill and C. G. M. Gahan, Regulation of host weight gain and lipid metabolism by bacterial bile acid modification in the gut, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(20), 7421–7426, DOI:10.1073/pnas.1323599111
.
- B. V. Jones, M. Begley, C. Hill, C. G. M. Gahan and J. R. Marchesi, Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(36), 13580–13585, DOI:10.1073/pnas.0804437105
.
- M. Begley, C. Hill and C. G. M. Gahan, Bile Salt Hydrolase Activity in Probiotics, Appl. Environ. Microbiol., 2006, 72(3), 1729–1738, DOI:10.1128/aem.72.3.1729-1738.2006
.
- M. G. Rooks, P. Veiga, L. H. Wardwell-Scott, T. Tickle, N. Segata, M. Michaud, C. A. Gallini, C. Beal, J. E. T. van Hylckama-Vlieg and S. A. Ballal,
et al., Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission, ISME J., 2014, 8(7), 1403–1417, DOI:10.1038/ismej.2014.3
.
- R. D. Hills Jr., B. A. Pontefract, H. R. Mishcon, C. A. Black, S. C. Sutton and C. R. Theberge, Gut Microbiome: Profound Implications for Diet and Disease, Nutrients, 2019, 11(7), 1613, DOI:10.3390/nu11071613
, review.
- A. A. Adhikari, D. Ramachandran, S. N. Chaudhari, C. E. Powell, W. Li, M. D. McCurry, A. S. Banks and A. S. Devlin, A Gut-Restricted Lithocholic Acid Analog as an Inhibitor of Gut Bacterial Bile Salt Hydrolases, ACS Chem. Biol., 2021, 16(8), 1401–1412, DOI:10.1021/acschembio.1c00192
.
- A. Almeida, S. Nayfach, M. Boland, F. Strozzi, M. Beracochea, Z. J. Shi, K. S. Pollard, E. Sakharova, D. H. Parks and P. Hugenholtz,
et al., A unified catalog of 204,938 reference genomes from the human gut microbiome, Nat. Biotechnol., 2021, 39(1), 105–114, DOI:10.1038/s41587-020-0603-3
.
- A. Knarreborg, R. M. Engberg, S. K. Jensen and B. B. Jensen, Quantitative Determination of Bile Salt Hydrolase Activity in Bacteria Isolated from the Small Intestine of Chickens, Appl. Environ. Microbiol., 2002, 68(12), 6425–6428, DOI:10.1128/aem.68.12.6425-6428.2002
.
- P. V. Khodakivskyi, C. L. Lauber, A. Yevtodiyenko, A. A. Bazhin, S. Bruce, T. Ringel-Kulka, Y. Ringel, B. Bétrisey, J. Torres and J. Hu,
et al., Noninvasive imaging and quantification of bile salt hydrolase activity: From bacteria to humans, Sci. Adv., 2021, 7(6), eaaz9857, DOI:10.1126/sciadv.aaz9857
.
- K. R. Brandvold, J. M. Weaver, C. Whidbey and A. T. Wright, A continuous fluorescence assay for simple quantification of bile salt hydrolase activity in the gut microbiome, Sci. Rep., 2019, 9(1), 1359, DOI:10.1038/s41598-018-37656-7
.
- A. Sveistyte, T. Gibbins, K. J. Tyrrell, C. J. Miller, M. H. Foley, A. E. Plymale, A. T. Wright and K. R. Brandvold, Simple Analysis of Primary and Secondary Bile Salt Hydrolysis in Mouse and Human Gut Microbiome Samples by Using Fluorogenic Substrates, ChemBioChem, 2020, 21(24), 3539–3543, DOI:10.1002/cbic.202000370
.
-
J. B. Grimm, L. M. Heckman and L. D. Lavis, Chapter One - The Chemistry of Small-Molecule Fluorogenic Probes, in Progress in Molecular Biology and Translational Science, ed. M. C. Morris, Academic Press, 2013, vol. 113, pp. 1–34 Search PubMed
.
- C. G. Ha, J. K. Cho, Y. G. Chai, Y. A. Ha and S. H. Shin, Purification and characterization of bile salt hydrolase from Lactobacillus plantarum CK 102, J. Microbiol. Biotechnol., 2006, 16, 1047–1052 CAS
.
- I. Sekirov, S. L. Russell, L. C. M. Antunes and B. B. Finlay, Gut Microbiota in Health and Disease, Physiol. Rev., 2010, 90(3), 859–904, DOI:10.1152/physrev.00045.2009
.
- W. Nelson, Y. L. Tong, J. K. Lee and F. Halberg, Methods for cosinor-rhythmometry, Chronobiologia, 1979, 6(4), 305–323 CAS
. PubMed.
- Z. Song, Y. Cai, X. Lao, X. Wang, X. Lin, Y. Cui, P. K. Kalavagunta, J. Liao, L. Jin and J. Shang,
et al., Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome, Microbiome, 2019, 7(1), 9, DOI:10.1186/s40168-019-0628-3
.
- B. Parasar, H. Zhou, X. Xiao, Q. Shi, I. L. Brito and P. V. Chang, Chemoproteomic Profiling of Gut Microbiota-Associated Bile Salt Hydrolase Activity, ACS Cent. Sci., 2019, 5(5), 867–873, DOI:10.1021/acscentsci.9b00147
.
- J. E. Kyle, L. M. Bramer, D. Claborne, K. G. Stratton, K. J. Bloodsworth, J. G. Teeguarden, S. Gaddameedhi, T. O. Metz and H. P. A. Van Dongen, Simulated, Night-Shift Schedule Disrupts the Plasma Lipidome and Reveals Early Markers of Cardiovascular Disease Risk, Nat. Sci. Sleep, 2022, 14, 981–994, DOI:10.2147/nss.S363437
.
- A. Khalyfa, S. Gaddameedhi, E. Crooks, C. Zhang, Y. Li, Z. Qiao, W. Trzepizur, S. A. Kay, J. Andrade, B. C. Satterfield, D. A Hansen, L. Kheirandish-Gozal, H. P. A Van Dongen and D. Gozal, Circulating Exosomal miRNAs Signal Circadian Misalignment to Peripheral Metabolic Tissues, Int. J. Mol. Sci., 2020, 21(17), 6396, DOI:10.3390/ijms21176396
.
- D. Withrow, S. J. Bowers, C. M. Depner, A. González, A. C. Reynolds and K. P. Wright, Sleep and circadian disruption and the gut microbiome-possible links to dysregulated metabolism, Curr. Opin.
Endocr. Metab. Res., 2021, 17, 26–37, DOI:10.1016/j.coemr.2020.11.009
.
- I. S. Wong, D. Dawson and H. P. A. Van Dongen, International, consensus statements on non-standard working time arrangements and occupational health and safety, Ind. Health, 2019, 57(2), 135–138, DOI:10.2486/indhealth.57_202
.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, biochemical characterization, animal experiment details and spectral data. See DOI: https://doi.org/10.1039/d2ob02257e |
| This journal is © The Royal Society of Chemistry 2023 |