Open Access Article
Open Access ArticleSelf-assembly of glycoprotein nanostructured filaments for modulating extracellular networks at long range†
Roberto
Matassa
 *a,
Marta
Gatti
a,
Martina
Crociati
bc,
Roberto
Brunelli
d,
Ezio
Battaglione
a,
Massimiliano
Papi
*a,
Marta
Gatti
a,
Martina
Crociati
bc,
Roberto
Brunelli
d,
Ezio
Battaglione
a,
Massimiliano
Papi
 ef,
Marco
De Spirito
ef,
Stefania Annarita
Nottola
a and
Giuseppe
Familiari
a
ef,
Marco
De Spirito
ef,
Stefania Annarita
Nottola
a and
Giuseppe
Familiari
a
aDepartment of Anatomical, Histological, Forensic and Orthopaedic Sciences, Section of Human Anatomy, Sapienza University of Rome, Via A. Borelli 50, 00161, Rome, Italy. E-mail: roberto.matassa@uniroma1.it
bDepartment of Veterinary Medicine, University of Perugia, Via San Costanzo, 4, Perugia, 06126, Italy
cCentre for Perinatal and Reproductive Medicine, University of Perugia, 06129 Perugia, Italy
dDepartment of Gynecological-Obstetric and Urologic Sciences, Sapienza University of Rome, Rome, Italy
eDipartimento di Neuroscienze, Università Cattolica del Sacro Cuore, Largo Francesco Vito 1, 00168 Rome, Italy
fFondazione Policlinico Universitario A. Gemelli IRCSS, 00168 Rome, Italy
First published on 11th October 2023
Abstract
The intriguing capability of branched glycoprotein filaments to change their hierarchical organization, mediated by external biophysical stimuli, continues to expand understanding of self-assembling strategies that can dynamically rearrange networks at long range. Previous research has explored the corresponding biological, physiological and genetic mechanisms, focusing on protein assemblies within a limited range of nanometric units. Using direct microscopy bio-imaging, we have determined the morpho-structural changes of self-assembled filament networks of the zona pellucida, revealing controlled levels of structured organizations to join distinct evolved stages of the oocyte (Immature, Mature, and Fertilized). This natural soft network reorganizes its corresponding hierarchical network to generate symmetric, asymmetric, and ultimately a state with the lowest asymmetry of the outer surface roughness, and internal pores reversibly changed from elliptical to circular configurations at the corresponding stages. These elusive morpho-structural changes are regulated by the nanostructured polymorphisms of the branched filaments by self-extension/-contraction/-bending processes, modulated by determinate theoretical angles among repetitive filament units. Controlling the nanoscale self-assembling properties by delivering a minimum number of activation bio-signals may be triggered by these specific nanostructured polymorphic organizations. Finally, this research aims to guide this soft biomaterial into a desired state to protect oocytes, eggs, and embryos during development, to favour/prevent the fertilization/polyspermy processes and eventually to impact interactions with bacteria/virus at multiscale levels.
1. Introduction
The zona pellucida is a dense extracellular network of glycoprotein nanostructured filaments, which supports chemical–mechanical communications between oocyte and granulosa cells through biophysical connections at the nanoscale. This soft extracellular membrane, surrounding all mammalian oocytes, attracts considerable interest due to its ability to control the overall complex system through appropriate self-assembling bio-mechanisms to synchronise the evolved stages of the oocyte.1,2 Distinctive assumptions about the origin, evolution, and coexisting living phases of the zona pellucida (ZP), based on needful experimental/theoretical biological, physiological, and genetic studies, have been correlated with protein assemblies limited to nanometric units. Indeed, they have shown that ZP glycoproteins originate from the late stage of the oogenesis phase through hydrophobic regions rich in proteins anchored to the membrane beginning to grow by continuously secreting glycoproteins.3 This protein accumulation evolved in natural self-assembly nucleation of non-covalently linked domains, favoring only short-range-order structures of oligomeric subdomains of nanometric units (dimers, trimers, etc.).4–6 The presence of the N-terminal in the subdomain increases protein–protein interactions to polymerize the oligomers into high-order filaments. Also, the C-terminal subdomain was associated with protein function, as a unit interaction mediator (i.e., sperm binding). The interactions among protein nanometric units promote the structural transition from a metastable state of the early ZP glycoprotein network to the immature oocyte (I-ZP).7,8 Afterwards, the N-terminal signal peptide (intramolecular disulphide bonds) and the missing C-terminal propeptide should favor the polymerization to reach maturation (M-ZP).9 At the fertilized stage (F-ZP), the oolemma and zona pellucida cooperate by biochemical processes, resulting in cleavage of the N-terminal subdomains, inducing ZP hardening, which is essential to prevent polyspermy.10 Both ZP-N and -C subdomain structures, incorporated into the identified ZP1, ZP2, and ZP3 monomeric units, are regulated by glycoprotein synthesis and secretion processes, modelling the complex cross-linked network (mice case).1,11 Directed electron imaging showed for the first time the existence of individual glycoprotein filaments, not branched, in the ZP, which constituted of globular nanoparticles linked to each other through rods, as observed by Greve and Wassarman.12 Biochemical investigation identified an additional ZP1-like unit, called ZP4, in different mammalian species, including bovine; predicting that it is specialized to create cross-linked networks by assembling long filaments of repetitive –ZP2–ZP3– units clipped by short filaments of –ZP4– units.1,13,14To date, many efforts have been devoted to the emerging and fascinating research of predicting the structure of a protein molecule from its linear amino-acid sequence, right down to the position of each atom at the nanometric scale. Theoretical structural studies coupled with X-ray crystallographic characterization have reported interesting achievements in a complete determination of the glycoprotein nanometric domains, using the crystallization trials method.15,16 Jovine et al. reported relevant prediction information on the multiple functional and folding protein structures on ZP2 and ZP3 filament units and their ZP1/ZP4 inter-unit filament interactions at the atomic scale.17,18 Interesting electron microscopy imaging, confined to the outer/inner surface of the nanometric region, allowed the gathering of information about structural length on polymeric protein filaments without considering the network constraints.19,20 These filaments chemically cross-linked in polymeric protein networks, generating complex bio-structures, have revealed interesting bio-physical surface properties, essential for a variety of cellular processes.21–24 However, these needful outcomes have a crucial lack in matching the persistence length of protein assemblies changed by actual bending/stretching/contraction in the filaments. Thus, understanding how self-assembling processes generate highly organized structures in hierarchical networks for controlling bio-physical properties is essential knowledge.
By increasing the observation scale, morphology studies revealed the smoother outer surface of the I-ZP; while micrometric fenestrations of fibrils of nanostructured filaments with spongy network features in M-ZP were observed.19 Zona pellucida were qualitatively observed in assembled fibrils, forming fibrillar networks at different stages: (i) fibrils organized in a very tight meshed network (I–M–F-ZP); (ii) less condensed mesh networks (I–M–F-ZP); (iii) fibrils assembled in loose or large meshed networks (M–F-ZP); and (iv) fibrils fused together to form a homogeneous structure without a reticular appearance (F-ZP). Nottola et al. observed an outer surface characterized by expanded and flattened fenestration of F-ZP in human cases.25 Instead, bovine I-ZP was found to have a very irregular surface, with an uneven distribution of numerous pores, crevices, and projections.26,27 Interestingly, surface observations of ZP mouse evidenced filaments arranged in aligned beads to form strings connected by globular structures into a long-range network.28 These interesting morphology investigations unfortunately lack of roughness control over a complex surface profile of smaller fibrils affected by a high irregularity of deviations hidden by a dense filamentous network at long range. Thereby, a precise understanding of these morpho-structural changes of filamentous networks at long range that can rearrange dynamically as interactions among the nanostructured filaments changes remains elusive.
To further extend these findings, hierarchical structure analyses of changed glycoprotein networks in the surface/bulk structural properties, governing the oocyte evolution at immature, mature, and fertilized stages, have been reported. To obtain insight into these intriguing and complex soft biomaterial, a combination of microscopy imaging techniques of TEM supported by quantitative imaging analysis (QIA), using statistical quantitative novel approaches, have been exploited to explore the following objectives: (i) morphometric roughness study of both real vertical and spatial features of the outer extracellular surface profile shaped by smaller fibrils of filamentous networks (Fig. 1); (ii) evolution of the internal pores of cross-sectional ZPs (Fig. 2); (iii) polymorphic structural length of self-assembled gly-filaments cross-linked into a network at long range (Fig. 3). Integrating the above scientific objectives in a synergistic manner has been essential for the mechanistic understanding of how hierarchical filaments organizations size/shape the body contours of the zona pellucida, evolving from immature to fertilized stages (Fig. 4).
 | ||
| Fig. 3 TEM images of interconnected glycoprotein filaments from the immature to fertilized stages and analyses of cross-linked network-matrices. (a–c) Portions of cross-sectioned ZP images of Fig. S3,† showing an overlay of simulated colored segments and extracted glycoprotein cross-linked network. (d|) A high magnification TEM image of globular pairing with rod-like structures along glycoprotein filament obtained by a pH-solubilisation procedure and dried referred to Greve et al.12 (d||) Cryo-electron tomography (cryo-ET) image of glycoprotein uromodulin (UMOD) filaments vitrified by a cryogenic method, revealing both a zig-zag backbone and fish-bone shapes refeered to Weiss et al.35 (d|||) A. medium magnification TEM image of interconnected glycoprotein filaments of a ZP matrix fixed and embedded in an epoxydic resin, presented here. (S-abc) Overlapping histograms of I–M–F-ZP showing the length distribution of the branched gly-filaments. (I-a, M-b, F-c) Experimental plots showing the normalized frequency distribution of the gly-filaments lengths extrapolated from panels a–c. Data fitted by Gaussian convolution functions (black line). p < 0.01 statistically significant difference between experimental and theoretical data fitting. (e) A schematic illustration of the estimated glob-rod angles through branched filament segmentation processes. (GA-I-II-III) Theoretical predictions of the glob-rod angles profiles related to the gly-filaments lengths with their corresponding standard deviation (error bars). | ||
2. Results and discussion
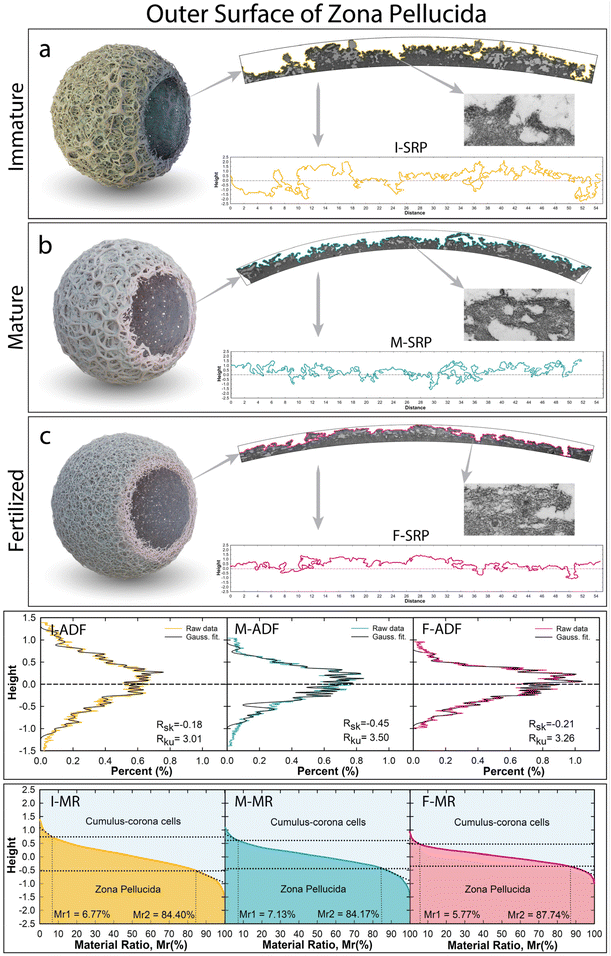
The morpho-structural evolution of the Zona Pellucida has been investigated at immature (I), mature (M), and fertilized (F) stages of bovine oocytes (Fig. S1†). The obtained radial cross-section images represent evidence of the glycoprotein building blocks that guided the hierarchical structures of a specialized networks to assemble into the core–shell shape around the oocyte, which, in turn, opens up the opportunities to perform accurate characterization of the evolving change of the glycoprotein networks at long range. In this regard, the natural dynamic assembly of the complex glycoprotein structures, involved in adapting to changing environmental conditions, has been evaluated quantitatively from the microscopic to nanoscopic scale. The evolution growth of this amazing bio-structured complexity will be clearly visible by observing and quantitatively evaluating the biophysical properties in terms of the outer surface roughness profile and internal porosity biophysical properties, strictly modelled by the self-assembling processes of the glycoprotein filamentous network.2.1. Outer surface proteins driven by a self-assembling filament network
The analyses of the outer surface profiles extracted from cross-sectional specimens are displayed in Fig. 1. Roughness surface properties were then analyzed as topographic contour lines, using similar assessment criteria adopted by surface microscopy techniques (i.e.: SEM, AFM, STM, etc.). Beyond the data resolution enhancement, the TEM cross-sectional profiles allow analysis of the real aspects of the highest complex surface roughness containing forward or backward bending, voids, or other irregular texture profiles, which would not be possible through surface microscopy techniques. Indeed, this new approach used here avoids any shading effect capable of hiding depressions onto the surface generated by irregular bending protrusions shaped by smaller fibrils of filamentous networks along the projection direction (see red-dot-line circles in Fig. S1†). Three different surface roughness profiles (SRP), belonging to each ZP stage, were extrapolated from the cross-sectional TEM image and displayed in Fig. 1-SRP. The high abundance of multidimensional peaks/valleys (protrusions/concavities), shaped by smaller fibrils of filamentous networks, was investigated quantitatively by considering the amplitude distribution function (ADF) extrapolated from the surface profile roughness (SRP). The material ratio (MR) curve, a cumulative density distribution of the ADF, was used to understand the changing surface between the two mediums (Fig. 1-ADF and -MR).29 The I-ZP outer surface profile, extracted from Fig. 1a, clearly shows an irregular micrometric surface with a large maximum vertical distance Rt of 3.17 μm (Fig. 1a-SRP, Table S1†). High fluctuation roughness around the mean line profile in terms of the neighbouring protrusion-concavity pairs exhibited a mean height of Rc = 1.47 μm and a mean width of Rsm = 1.15 μm, having high total peak numbers (Rpc = 8082). The I-SRP has also been evaluated quantitatively in terms of spread and amplitude distributions through shape distribution features of the skewness and kurtosis parameters (Rsk and Rku).30 The estimated Rsk exhibited a value close to zero of −0.18, indicating that the glycoprotein network assembled to shape both protrusions and concavities symmetrical around the mean line (Fig. 1I-SPR). The Rku estimation exhibited a value of 3.01, denoting that the protrusions and concavities have irregular shapesTo gain a deeper understanding of the structural meaning of the glycoprotein surface, the obtained broad I-ADF profile was fitted by convolution of Gaussian functions to assess each height peak position of the single deconvolved Gaussian curve, arising from each protrusion and concavity (Fig. 1I-ADF, Table S2†). The protrusions (positive values) and concavities (negative values) exhibit similar symmetrical positions around the mean line (±0.05 μm, ±0.13 μm, ±0.21 μm, etc.), and the highest amplitudes favoured protrusions rather than concavities. To gain insight into the minimal changing of the network at the surface, the intricate roughness was characterized along the interfacial area ratio between the cumulus-corona cells and the glycoprotein bio-material, the materials ratio curve (Fig. 1I-MR). Among the three different bearing areas, the central portion had a core roughness depth of Rk = 1.24 μm, excluding the asperities contributions of the peaks-valleys. The upper local area exhibited cumulative surface asperities of protruding peaks under the micrometric dimension of Rpk = 0.35 μm with a material portion Mr1 of 6.27% in contact with cumulus-corona cells. Whereas the local minima fluctuation of the jagged deep concavities had a high Rvk = 0.72 μm (Mr2 of 84.40%), evidencing a high friction/adhesion strength between apical extracellular corona cells and the ZP surface.
The mature ZP outer surface is displayed in Fig. 1b and the corresponding extrapolated projected length in Fig. 1M-SPR. The reorganized network shaped the outer surface by decreasing the total height thickness by about 1 μm (Rt = 2.43 μm), reducing the mean height (Rc = 1.17 μm) and increasing the width (Rsm = 1.50 μm) compared to the I-ZP (Table S1†). In this regard, the reorganized cross-linked filaments shaped the protrusions/concavities, involving an asymmetrical distribution over all the surface profile, as established by increasing Rsk (−0.45). A relative increase in Rku (3.50 > 3) concretely supports the enlarging shape for both protrusions/concavities; thereby the bending protrusion humping the surface started to disappear, leading to an increase in the surface porosity. This is evidenced by increasing the bio-active surface area to favour structural interactions with external biological systems. The best fitting of the M-ADF profile, showing a slight shift toward the protrusion formation, exhibited the highest intensity of the Gaussian curve centered at 0.24 and 0.13 μm, losing the external Gaussian curves of the ADF profile (Fig. 1M-ADF). Interestingly, the height distributions of the protrusion and concavities still have similar positions (Table S2†); thereby the reassembled filaments preserve the height positions of the protrusions-concavities formed in the I-ZP, a probably natural sequential order. By evaluating the materials’ ratio curve (Fig. 1M-MR), the roughness core profile slightly decreased (Rk = 0.94 μm), while the local nanometric asperity fluctuations are still higher in the concavities than in the protruding peaks (Rpk = 0.39 μm, Rvk = 0.59 μm). The increasing of the upper material portion (Mr1mature = 7.13% > Mr1immature = 6.27%) indicates that the outer surface has more glycoproteins material to join in the contact or to be bound by sperm through a better load-bearing ability.10
The surface roughness F-ZP profile, displayed in Fig. 1F-SPR, evolved through a contraction in thickness, reducing the mean height (Rc = 0.89 μm). A stretching effect of the surface can be noticed in the increment of the Rsm (1.72 μm) compared to the previous stages. Reducing its previous asymmetry distribution of the M-ZP was achieved by decreasing the concavities width and preserving the flattening shape of the protrusions (Rsk = −0.21, Rku = 3.26). This stretching/flattening effect overall the outer surface is also evident in the height distribution function plotted in Fig. 1F-ADF. Despite the formation of a compact surface, the F-ZP gly-filaments reassembled, preserving their height positions of the surface profile from immature to fertilized stage (Table S2†). Using the shape memory of the glycoprotein networks, the morphological contraction in thickness was also evident in decreasing the lower slope and the less rough surface (Fig. 1F-MR). Further, the core roughness depth still decreased under micrometric dimensions (Rk = 0.62 μm). The bio-action mechanism shaping the protruding peaks/valleys evidenced a decrease in the nanometric asperities, also reaching a roughness balance (Rpk = 220 nm, Rvk = 270 nm). Instead, the peak upper portion showed less glycoproteic material for joining extracellular materials (Mr1fertilized = 5.7% < Mr1mature = 7.13%). Yet from nanometric surface perspectives, the similar estimated Rpk and Rvk, different for the other stage, indicated the presence of a symmetrical nanometric reorganization. This implies that upon contact, the filament units have a tendency to equally share contact, probably to avoid a potential polyspermy, producing a compact and rigid outer surface of a hardened fertilized ZP.23
2.2. Exploring the evolution of the internal pores
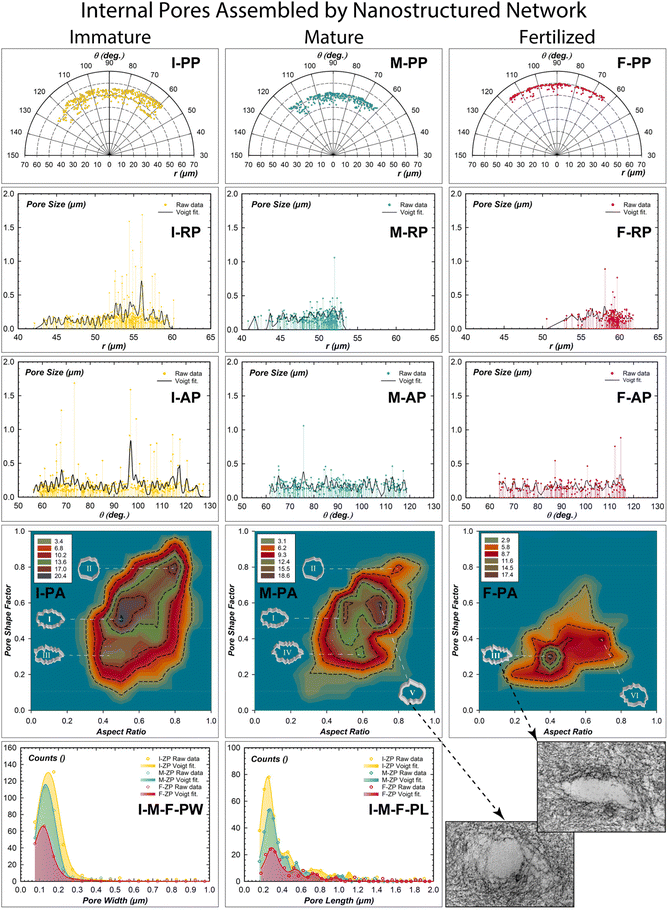
The internal pores of each ZP stage were quantitatively investigated in terms of porosity, pore distribution, pore shape factor, and aspect ratio (Fig. 2). The center of the mass positions of all pores was extrapolated in polar coordinates from the digital images to evaluate their radial and angular distributions (Fig. 2-PP, -RP, -AP, Fig. S2†).31 A total porosity of 5.74% was estimated in I-ZP (Fig. S2-a and Table S3†). An increase in the pore numbers from the inner (40 μm) to the outer (60 μm) surface ranging from 55 to 130 degrees was estimated (Fig. 2I-RP, 2I-AP).Extra-large micrometric pores can be attributed to the strong adhesion of the apical cumulus-corona cells. On the other hand, pores of hundreds of nanometers can be considered to be generated by transzonal or cytoplasmic projections of the cumulus-corona cells; travelling through the thick ZP in search of the oocyte to regulate oocyte activity.32 By fitting the radial profiles, the presence of domains enveloping pore numbers of 20.31 separated by an average spatial frequency of 0.57 μm was revealed (Tables S4 and S5†). The angular pore distribution analysis identified domains of 13.85 pore number separated by an angular frequency of 1.90 degrees (Fig. 2I-AP). Therefore, the cumulus-corona cells communicate with the immature oocyte, generating pore domains distributed every 2 degrees radially separated by half a micron. The frequency distributions of the pore shape factor related to the aspect ratio drawn by 2D contour frequency plots were estimated (Fig. 2I–M–F-PA). The statistical shaping descriptors of multi-shaping pores identified three different shape domains. Synergism between cytoplasmic projections and nanostructured filaments generated a domain pore of high frequency with an elliptical shape and rough walls (0.5, 0.5; labelled I); pores with a quasi-circle shape and low roughness walls (0.8, 0.8; II); and a low-frequency area with a high roughness pore wall and more elongated in shape (0.4, 0.3; III).
Mature ZP exhibited a slight decrease in total porosity of 4.23%, distributed radially from the inner (40 μm) to the outer (54 μm) section and angular from 60° to 120°. The domains of pore numbers are similar radially and decrease angularly (20.43, 10.12), but their repetitive radial center decreases (0.37 μm) and not angularly (1.91°). This changing porosity distribution represents a starting retraction of the cytoplasmic projections. The resulting change of the glycoproteins network by chemical-mechanical action reduces the radial pore separation and the pore number in the angular directions. The low frequency of the three pores shaping the area of the I-ZP (I and II) are still present at the mature ZP, but with low-frequency distribution. The third has a similar roughness pore wall of the area III, but less elongated pores (0.6, 0.3; IV). The main change is revealed by the presence of a high-frequency area, characterizing the pores with circularity and a smooth wall shape (0.6, 0.7; V), similar to the inset of the TEM image (right bottom of Fig. 2). The appearance of a smoother and more circular pore can be considered an action of relaxing/expanding nanostructured filaments to reorganize their long-range network in reaching their maturation purpose. The fertilized ZP exhibited an effective decrease of the total porosity of 2.34% (Table S3†). The radial-angular distribution profiles of the pore size reduced the range from the inner (50 μm, 62°) to the outer (62 μm, 117°). Also, the pore number domains are also reduced in both radial and angular directions (16.64, 6.65); while spatial distribution evidenced a decreasing of pore domains in number without substantially changing in both radial-angular directions (0.36 μm, 1.88°). The resulting pore shaping analysis showed a new formation of an interesting area III (0.4, 0.3), having high-frequency counts, which was already present in the I-ZP with the lowest frequency. There was one more area of low frequency with increasing roughness of the pore wall (0.4, 0.7, VI). Interestingly, the pore analysis shows a preferential pore shape memory established by the notable presence of the area III (0.4, 0.3) detected at the immature stage to finally reappear at the fertilized one as a preferential shape (inset right bottom of Fig. 2). A simple morphometric statistical analysis of the pore dimensions has been reported for pore widths and lengths (Fig. 2I–M–F-PW and -PL). The squeezing effect of the pores shows a moderate change of the width from 0.16 μm (I-ZP) to 0.12 μm (F-ZP); while the estimated pore lengths better evidence an elongation with an increment of 90 nanometers from 0.22 μm (I-ZP) to 0.31 μm (F-ZP).32
2.3. Self-assembling network of plurality glycoprotein filaments
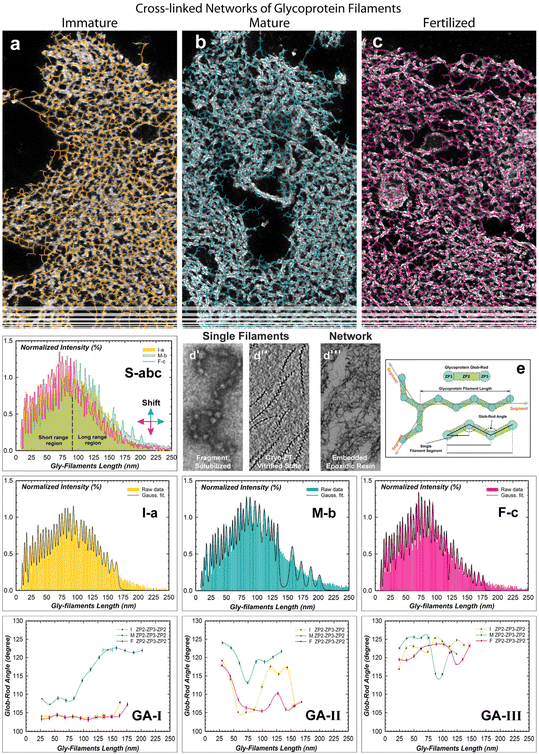
To investigate the evolutionary change of the cross-linked network,15 the super-structural organizations of plurality interconnected gly-filaments have been characterized using high magnification TEM imaging analyses (Fig. 3).33–35 Imaging segmentation process generated total filament segments of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 056 (I-ZP), 11
056 (I-ZP), 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 408 (M-ZP), and 9654 (F-ZP) from the outer to inner surface (as shown in Fig. 3S).36 The estimated branched lengths, plotted in Fig. 3S-abc, exhibited similar lengths and intensities from 9 to 90 nm (the short-range-length region) for the immature and mature ZP networks.37 After that the filament lengths evolved differently (90 to 250 nm, long-range-length regions) with increasing lengths and intensity in the M-ZP network. This could denote a loss of connections among filaments and/or stretching filaments. Instead, the evolved F-ZP networks evidences a contraction effect of lengths, increasing in the short-range region and long-range-region. By quantifying each histogram, the maximum intensities lengths were estimated to be centered around the mean values of 85.67 ± 1.80 nm (I-ZP), 95.37 ± 1.56 nm (M-ZP), and 81.53 ± 2.55 nm (F-ZP).
408 (M-ZP), and 9654 (F-ZP) from the outer to inner surface (as shown in Fig. 3S).36 The estimated branched lengths, plotted in Fig. 3S-abc, exhibited similar lengths and intensities from 9 to 90 nm (the short-range-length region) for the immature and mature ZP networks.37 After that the filament lengths evolved differently (90 to 250 nm, long-range-length regions) with increasing lengths and intensity in the M-ZP network. This could denote a loss of connections among filaments and/or stretching filaments. Instead, the evolved F-ZP networks evidences a contraction effect of lengths, increasing in the short-range region and long-range-region. By quantifying each histogram, the maximum intensities lengths were estimated to be centered around the mean values of 85.67 ± 1.80 nm (I-ZP), 95.37 ± 1.56 nm (M-ZP), and 81.53 ± 2.55 nm (F-ZP).
To elucidate the hidden ordering of the complexity self-assembling mechanism, it is interesting to highlight the morpho-structural meaning of the pronounced discrete bins; thereby, they were fitted by a convolution of Gaussian functions (Fig. 3I-a, 3M-b, and 3F-c).
A first and interesting point is that the estimated centres of each peak exhibited a sequential order in length by pooling information across multiple data sources (e.g. second column of Table S6:† 27.11, 40.12, 53.46, 66.79, etc. have multiple repeat sequences of about 13.33 nm). To evaluate the physical meaning of the repeating unit lengths, past characterizations of morpho-structural analyses of the ZP unbranched filament were considered.11 Greve and Wassarman in 1985 showed glycoprotein units of repeating order given by the globular (ZP3) and rod (ZP2) shape of estimated length (–ZP3–ZP2–ZP3–, Fig. 3d|). Furthermore, cryogenic glycoprotein filaments with zig-zag shaped backbones were also observed (Fig. 3d||).38–41
By considering the results of this experimental imaging, we can assume that the gly-filament length of repeated known units directly depends on the angular variability of the formed glob-rod unit (–ZP3–ZP2–), as shown in the schematic illustration of Fig. 3e. The glob-rod angles were extrapolated by fitting the experimental derived lengths (Fig. 3d|||), using a reiterated estimation control by the RHamilton equation.42,43 The estimated glob-rod unit lengths showed an experimental value of 17.27 and 14.32 nm in accordance with Greve and Wassarman.12 Moreover, the angular prediction among the glob-rod units was constrained by considering X-ray crystallographic protein structure studies (103–126 degree).16,44,45
A new indirect quantitative method of a theoretical prediction, based on experimental data, has been proposed. The resulting matching with the experimental derived lengths generated a polymorphism of distinct oligomeric gly-filaments based on three main glob-rod angular isomorphism (Tables S7 and S8†).39,46 A first angular polymorphism of type-I, with a weight fraction of 49 ± 3%, showed similar structural behaviour for both I- and F-ZP with angles of 104.53 ± 1.43° and 103.99 ± 1.05°, respectively (Fig. 3GA-I). Instead, the mature ZP favours a gly-filament elongation by increasing both the angle and the corresponding standard deviation of 115.10 ± 6.39°; which the latter implies an effect of rising curvature along the filament pathways. This simple parameter may indicate a potential ordering of identical glycoproteic units in a straight conformation by having the lowest standard deviation. Indeed, the F-ZP glob-rod angle showed a low standard deviation, denoting a vanishing curvature effect with a quasi-straight order of the gly-filaments pathway of a contracted conformation. The isomorphism of type-II did not show any relevant change in the angles with a reduced weight of 34 ± 1%. However, the I-ZP glob-rod angle of 111.83 ± 5.64° and the M-ZP of 119.72 ± 4.33° exhibited an increased and slightly reduced curvatures, respectively (Fig. 3GA-II). On the contrary, the fertilized ZP evolved part of the network by decreasing the amount of the filament units still organized in slightly decreasing the curvatures (108.90 ± 3.94°). Further extension lengths were well-estimated in the isomorphism of type-III characterized by the presence of high-angle arrangements for both immature and mature ZP (122.75 ± 2.36°, 121.96 ± 4.05°), detonating a relaxing organization of the gly-filaments with the lowest weight of 29 ± 2%. Similarly, the fertilized ZP also exhibits an increased glob-rod angles distribution (120.59 ± 3.72°) with a slight filament contraction (Fig. 3GA-III). There is one more important implication of the proposed quantitative indirect method, regarding the estimated lengths of the entire distributions (85.67 (I-ZP), 95.37 (M-ZP), and 81.53 (F-ZP) nm). The interconnected filaments favour the structural repeat of six (I- and F-ZP) and seven (M-ZP) filament units; a result close to the previously estimated molar ratio of the ZP1 homodimer to two fibrils of ZP2–ZP3 heterodimers of the mice case at the fertilized stage.12
2.4. Controlling morpho-structural order over the glycoprotein matrix by polymeric self-assembling of interconnected filaments
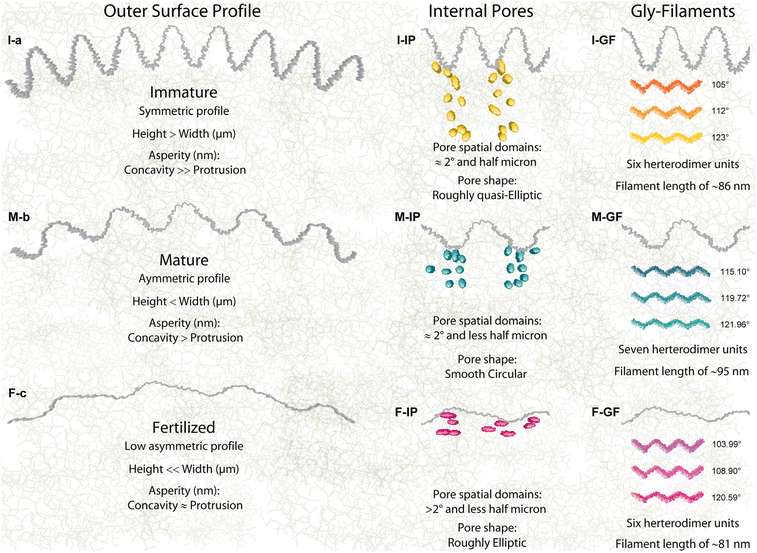
The structural mechanisms have tuned the long-range organizations to generate high and irregular roughness onto the immature ZP outer surface shaped by smaller fibrils of filamentous networks, which are organized in a symmetric distribution of the protrusions-concavities, which has not been shown before (Rsk ≤ 0, Rku ≈ 3, Fig. 4I-a). The unexpected symmetric behavior of an irregular roughness arise from the strong adhesion of the cumulus-corona cells surrounding the outer surface, which are involved in a push–pull action through the penetrating transzonal projections, producing a symmetrical loading over the spherical shell of the zona pellucida. To promote the biological activities of the oocyte development, the transzonal projections penetrated toward the matrix of the 15.6% through concavities with asperities height of 720 nm. The assembled network being not well-packed near the outer surface generated higher heights than widths (Rc = 1.47 μm, Rsm = 1.15 μm). The strong load interactions with the apical cumulus-corona cells generated high nanometric asperities on the concavities, a strain action to favor docking interactions. These findings provide further morpho-structural information not analysed previously by microscopy surface techniques because of the shading effect of the forward or backward bending protrusions. Indeed, compact and smooth external surfaces with few smallest pores of immature ZP mouse oocytes have only been observed at the microscopic scale (SEM).19,28,47 Yet from a roughness surface perspective, the growing protrusions with very irregular and uneven distribution of numerous surface pores were qualitatively observed for ZP bovine, previously.26Glycoprotein frameworks evolve by changing the mature surface in an asymmetrical distribution of the protrusions-concavities both with width and a nearly flattened shape, increasing the surface porosity (Rsk < 0, Rku > 3, Fig. 4M-b). The tight bending protrusion humping the surface started to disappear with decreasing of the jagged concavities and of their local nanometric asperity fluctuation. This is caused by a stretching effect of the border nanostructured filaments with an expansion of the network due to the lower strength of interactions with the cumulus-corona-cells favouring sperm penetration.19,48 After fertilization, the gly-network evolved in shaping the ZP architecture by reducing the protrusion-concavity height through a tangential contraction effect in a flattening shape onto the outer surface. Therefore, the surface profile changed from an asymmetrical distribution (M-ZP) to finally reach an almost symmetric shape (Rsk ≤ 0, Rku > 3, Fig. 4F-c). Interestingly, the gly-network re-shaped the protrusion-concavity by preserving their position heights near to the mean line formed at the immature stages (shaping memory effect, Fig. 1-ADF). A further reduction of the protrusion-concavity nanometric asperities, reaching a balance in the roughness, was cast by densely packed filament contractions. Instead, the absence of symmetric nanometric asperities for I- and M-MR profiles implies unbalanced contact between external contact forces and outer surface compliance. This extraordinary coordination of the glycoproteins network of changing the surface morphology at the micro-nanometric scales represents concrete evidence of their self-assembling ability by following a natural sequential order of the previous stages. Indeed, the reducing nanometric asperities by filament contractions confirms quantitatively the hypothesis of the favored protrusion-concavity to generate disorientation for sperm binding sites after fertilization.49 Moreover, the novel methodology, proposed here, could be applied to other cross-sectional images to understand the surface bio-physical properties of complex multi-layered bio-structures at long range, i.e. cell surface tensions of the actin cortex filaments network.50 The ensemble evolution of the interconnected filaments obviously also involved a changing of internal pores at different ZP stages (Fig. 4I-IP). The internal pores, distributed in pore domains, were spatially assembled with a repeating order every 2 degrees and radially separated by half a micron. This quantitative evidence suggests the independent physiological action of each cumulus-corona cell. Coexisting reorganization of the M-ZP network mainly reduced the spatial separation among pore domains of less than half a micron and the pore number in the angular direction, a resulting action for the starting retraction of the cytoplasmic projections.
The long-range network evolved by relaxing/swelling the branched filaments inducing a spatial expansion able to reduce/close the pores and change their shape from roughly quasi-elliptical to smooth circular (Fig. 4M-IP). This further morphological achievement represents a dual-task of dynamic evolution by near ending of the biological transport mediated by transzonal projections and by promoting the fertilization process, favoring an enlarging framework.50 Effective decrease of total inner porosity is in F-ZP (Fig. 4F-IP). Indeed, it is well known that after maturation, most of these cumulus-corona cells retracted or disintegrated their transzonal projections.33,50 The reduced pore number along both radial-angular directions preserved their center of mass distances detected in M-ZP, prevailing a roughly elliptic shape previously revealed in the I-ZP in minor amounts (region III, Fig. 2I-PA). The applied radial bio-mechanical load for squeezing pores, through a realignment of the contracted filaments,1,51,52 is evident by observing the remaining pores in a roughly elliptical shape quasi-aligned to the outer surface profile (see the outer surface of Fig. S1c†).
The reported findings complements on high variability of both I-ZP sub-micrometric protrusions-concavities and inner pores attributed to the complex network may be attributed to the high hierarchical variability of the estimated lengths and glob-rod angles (Fig. 3 and 4I-GF). These structural differences might be a response to the collective bio-mechanisms of the growing ZP matrix. A structural transition of molecular cooperativity modified the M-ZP architecture through an evident expansion of the network, generating more planar and long filaments by increasing curvature, glob-rod angles, and lengths (Fig. 3 and 4M-GF). This changing structural organization has produced an asymmetrical surface profile of reduced nanometric asperities, and the network expansion reducing/changing the pore. Yet from structural biology perspectives, the expanded network by increasing filament planarity favors the exposition to the finite number of receptor binding sites for its sperm-interacting region.14 The evolved F-ZP hierarchy structure re-assembled the filaments by promoting low glob-rod angles with vanishing curvature to reach a contraction/straight structural conformation, thereby shaping outer surface and closing/squeezing inner pores by decreasing nanometric roughness (104, 109, and 121 degrees; Fig. 3-GA and 4F-GF). Therefore, the mechanistic understanding of the evolved F-ZP morpho-structural architecture confirms quantitatively that the reduced nanometric ensemble is an essential prerequisite for hiding binding sperm interactions to finally reach a hardened cytoskeleton for preventing polyspermy.7 Moreover, the evident filament contraction in the length of the F-ZP network suggests that the bio-mechanical hardening of ZP is not only dependent on modulating the amount of ZP1 or/and ZP4 glycoproteins connecting –ZP3–ZP2–ZP3– oligomers, but also through a structural contractions effect among –ZP3–ZP2^ZP3–ZP2 heterodimers.21
The substantial existence of polymorphic forms capable of organizing bio-filament units (type I, II, and III) confirms the chemically strategic existence of assembling (polymerization) and disassembling (depolymerisation) in response to external stimuli needed for the oocyte bio-evolution, as well as to bio-mechanical force.11,21–24 The lowest weight of the type-III, quite similar for all ZP stages, exhibited a high angle glob-rod, yielding high planarity of the structural conformation. These structural organizations can be related to the steric repulsion among the ZP3 globular species because of the sheer bulk of the proximal peptide groups, generating mutual repulsion and tension in the matrix.27,53 Therefore, this unchanged structural organization could be considered a passive bio-action of the filaments to reach a natural balance adaptation of each evolved stage.
3. Conclusions
Our work shows that hierarchical organizations of interconnected filaments determine the size and shape of the extracellular membrane in response to external biological stimuli driven by basic building blocks in their final self-assembled polymeric network. As shown, we were able to determine for the first time several quantitative morpho-structural features of the ZP evolution from the immature to fertilized stage: (i) the outer surface evidences a symmetrical distribution of the protrusion-concavity with high nanometric asperities (I-ZP) to evolve asymmetrically (M-ZP) and finally reaching a lowest asymmetrical arrangement (F-ZP) with a collective reduction of the symmetrically distributed nano-asperities (Fig. 1); (ii) internal porosity analyses have identified recurring pore domains with preferential radial/angular directions of repeating order distributed in a micrometric region. The pore size-shape evolved from a roughly quasi-elliptic shape (I-ZP) to change into a circular smooth wall (M-ZP) to finally re-organize into a roughly elliptic shape quasi-parallel to the outer surface (F-ZP) (Fig. 2); (iii) structural investigation on the interconnected gly-filaments by using new method for predicting the existence of structural polymorphism (type I, II, and III). This showed push–pull structural actions to generate elongation/bending (M-ZP) and contraction (F-ZP) of the filament lengths by modulating the glob-rod angles among repetitive nano-units (Fig. 3); (iv) the morpho-structural evolution analyses provide the starting point towards network-shape analyses of filaments capable of revealing a hidden organization into a long-range network, which follows a precise morpho-structural memory of the starting stage (Fig. 4).Quantitative understanding of the morpho-structural behaviours is essential to control and exploit the bio-properties of soft biomaterials, wherein the behaviour of the entire network incorporates and responds to stimuli across different multi-scales by following a multiple shaping memory effect. Indeed, the biochemical and mechanical properties of these glycoprotein networks are intimately involved in determining how forces are generated and transmitted in the zona pellucida, aiding human/veterinary reproductive medicine.54,55 The revealed dynamic/reversibility structures and performances of the hierarchical network structures at a long range might also be useful for designing and programming mesoscopic structural networks for new-generation biomaterial devices.56–59 Furthermore, the identification of the major hierarchical structures driven by physiological needs to induce specific complex architectures, not of random organization, guarantees control over the extracellular network of macroscopic structures, especially for the relevant outer profile having the first impact with the external biological identity.60 Furthermore, the novel structural approaches adopted here not only give us the advantage of characterizing self-assembling glycoprotein surface/networks but also the feasibility of applying these morpho-structural studies to any complex system, independently of the dimension scale. Finally, these automated image processing techniques will provide new insights into the extraordinary self-assembling mechanisms of glycoproteins filaments, which may have practical implications for the aggregates’ biochemical and biomechanical properties in controlling human/veterinary reproductive medicine. The revealed capability of assembling glycoprotein with clever dynamic and reversible tuning by changing the external stimuli not only helps us to understand the extraordinary behaviour of life but also aids us in developing new biomaterials with precise functionality in the near future.
4. Experimental section
4.1. Experimental protocol
Bovine immature, in vitro matured and fertilized oocytes (n = 10 each) were collected and prepared by a well-established analytical protocol.22 After that oocytes underwent a chemical extraction, and the extracted samples were prepared for transmission electron microscopy investigation according to an original protocol developed by G. Familiari et al.19 To image a large micrometric area of the ZP network structure, consecutive images were acquired by transmission electron microscopy (ZEISS EM10) operating @ 80 kV.61 TEM images were then merged to obtain a complete view of the outer surface, the internal cross-section, and of the filament network joining the entire path from the outer to the inner ZP surface using Photoshop imaging software (Fig. S1†). Bright Field TEM images were acquired at low magnification (8k×) and were merged to gain a useful area as large as possible to improve the statistical observation/analysis of the morpho-structural changes (Fig. S1†). The different cross-sectional ZP stages, forming shell arcs technically limited by the mash of the TEM holder grid, are shown in Fig. S1.† The reliable and direct measurements of the heights and spatial surface morphology were extrapolated using digital imaging processing of cross-sectional specimens at a high spatial resolution generated by TEM imaging. Moreover, dimensional sampling is another important aspect for quantitatively defining a complex roughness into a usable measure; thereby the roughness surface analysis has been estimated on nine-line profiles each of more than 51 μm in length with incremental steps of 0.01 μm, achieving large datasets for accuracy statistical validation (9215 × 3307 pixels size calibrated for 0.01 μm at 1 pixel).4.2. Surface topographic analysis
Each selected imaging region has been processed to digitalize the outer boundary of the glycoprotein surface in a binary image by extracting and converting pixels in Cartesian coordinates to generate surface profiles (Fiji imaging software).63,64 By considering the curvature surface of the ZPs, being the arc of a sphere, digital processing was used to fit the surface profile with low-order polynomials (3rd order), which is well suited to removing the curvature shape by subtracting the roughness from the average surface (Gwyddion imaging software, Fig. 1-SRP). The natural surface texture clearly exhibits a complex behaviour driven by the finalized self-organization of the glycoprotein filaments (gly-filaments); thereby statistical functions more sensitive to deviations are required for describing and evaluating the irregular geometrical features related to the evolved glycoprotein, protruding from the nanometric to microscopic scale. Several metrology parameters were estimated for characterizing the outer surface profile by using SPIP™ (Version 6.7.8 Premium, Image Metrology, Denmark) software. Each surface profile was processed to determine surface roughness parameters reported in Table S1.† Statistical analysis of the profile heights or height distribution has been evaluated in terms of the fraction of surface heights lying between two assigned heights versus. The roughness parameters were expressed in μm without applying any additional filtering. The entire height profile was plotted to obtain the amplitude distribution function (ADF). The ADF histogram can also be represented as a cumulative distribution function of a large number of random discrete local events derived by plotting the difference in height distribution, following a slope and curvature of a Gaussian distribution. The broadening I-ADF profile was fitted by a convolution of Gaussian functions to assess the single deconvolved Gaussian curves, arising from each protrusion and concavity. The best fitting of the I-ADF profile was deconvolved accurately with 25 Gaussian functions to adequately fit all key features with a standard error of 3.4% (Table S2†). The I-ADF peaks fitting confirmed the behaviour of the estimated Skewness and Kurtosis parameters in terms of symmetric and shape distributions (Fig. 1I-ADF). The best fitting of the M-ADF profile shows a deconvolution constituted by 21 Gaussian functions with a standard error of 5.5% (Fig. 1M-ADF). The best fitting curve with a standard error of 4.4% has been estimated to be deconvolved with 20 fitted Gaussian curves (Fig. 1F-ADF). The glycoprotein material ratio curve describes the ratio between the relative amount of roughness surface for a given height level of the profile divided into three regions: protruding peaks, core roughness, and deep concavities. These curves were useful for investigating surface material with abundancy and dimension of the peaks and valleys able to quantify their amount by giving the peak (A1) and valley area (A2).4.3. Internal pores imaging analysis
By converting the grey image into the binary image, the Cartesian coordinates were generated using Fiji imaging software and displayed in 2D filled pore maps (Fig. S2†).64 This software provided basic image processing tools and the properties of the projected images such as area, center of mass, perimeter, circularity, and Feret's diameter. The resulting imaging analysis provided the pore distribution-size-shaping analyses reported in Table S3.† After that, the centers of mass of each pore were displayed in polar plots to show a perspective view of the local distribution. The best fitting of the cross-sectional I-ZP was generated by 29 Voigt deconvoluted peaks and each fitted Voigt function was convolved to envelop a domain of determinate pore number (Fig. 2I-RP, Tables S4 and S5†). The radial-angular profile fitting was applied to the step-counted intensities by Voigt functions to quantify the main peak positions and full width, showing high probability frequency distributions of pore domains formation. Similarly, the angular distribution analysis of the I-ZP was fitted by 33 Voigt deconvoluted peaks (Fig. 2I-AP). The best fitting of the M-ZP radial-angular profiles was deconvoluted with 22 and 27 Voigt peaks, respectively (Fig. 2M-RP and 2M-AP, Table S4†). The radial pores distribution was estimated with 22 Voigt peaks for F-ZP and the best-fitting curve of the angular pore size distribution was convolved by using 26 Voigt peaks (Fig. 2F-RP and F-AP, Table S4†). Pore shape factor varies with pore wall roughness and the aspect ratio evaluates the pore circularity, both shaping parameters with a value of exactly 1 representing an ideal smooth circle.4.4. Image segmentation analysis of interconnected glycoprotein filaments
Data analysis of individual filament segments was estimated in terms of lengths between two interconnections, and displayed in a histogram of normalized intensity for each ZP stage, exhibiting a broad distribution of branched lengths (Fig. 3S-abc). High magnification TEM imaging analyses merged together to cover a large area of statistical analysis (13.56 × 0.94 μm, Fig. 3, and Fig. S3†).62 Adobe Photoshop CS6-Extended (Adobe, San Jose, CA, USA) was used for the image analysis. The morphometric analyses of the filament meshwork in each ZPs were determined by counting the length from the head to tail of the oligomeric filaments and the amount of knots on the entire image of the merged images. Images at a magnification of 90k× were calibrated according to the original structural finding on the single glycoproteic filaments achieved by J. M. Greve and P. M. Wassarman.12 Segmentation measurements on each TEM image were determined using the public domain Fiji/ImageJ image processing software.64–66 Yet from imaging analysis perspectives, it should be added that the tracing filament segmentation is independent from the edge roughness of the imaged filament (Fig. 3d) since the trace is symmetrically centered and drawn between adjacent edges.32 The best fitting of all the bin sizes provided useful indications on the measures of the main lengths of individual filament segments interconnected to the entire network, as reported in Table S6.†Author contributions
R. M. and G. F. designed the research; R. M. and G. F. performed the research; R. M.: conceptualization, data curation, formal analysis, investigation, methodology, software, validation, and visualization; R. M. wrote the paper and R. M., M. G., M. C., R. B., E. B., M. P., M. D. S., S. N., and G. F. conducted review and editing. All the authors approve of this work.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to the Electron Microscopy Laboratory “Pietro M. Motta” for the provision of their facilities and expertise, Sapienza University of Rome, Italy. This research was supported by grants from Sapienza University of Rome, University of Perugia, and a research project funded by the Italian Ministry for University and Research (MIUR) with the PRIN funding action (N. 2004054995_002, “Dinamica della formazione di strutture sovramolecolari basate su transizioni tiolo-disolfuro catalisi seleno dipendente nella PHGPx”).References
- E. S. Litscher and P. M. Wassarman, Zona Pellucida Proteins, Fibrils, and Matrix, Annu. Rev. Biochem., 2020, 20, 695–715 CrossRef PubMed.
- P. Motta, E. Nesci and Z. Takeva, On the fine structure of the zones of contact between the follicular cells and oocyte during development of the ovarian follicle in mammals, Boll. - Soc. Ital. Biol. Sper., 1970, 46, 257–259 CAS.
- T. Rankin, M. Familari, E. Lee, A. Ginsberg, N. Dwyer, J. Blanchette-Mackie, J. Drago, H. Westphal and J. Dean, Mice homozygous for an insertional mutation in the Zp3 gene lack a zona pellucida and are infertile, Development, 1996, 122, 2903–2910 CrossRef CAS PubMed.
- H. Okumura, T. Sato, R. Sakuma, H. Fukushima, T. Matsuda and M. Ujita, Identification of distinctive interdomain interactions among ZP-N, ZP-C and other domains of zona pellucida glycoproteins underlying association of chicken egg-coat matrix, FEBS Open Bio, 2015, 27, 454–465 CrossRef PubMed.
- H. Serrano and D. Garcia-Suarez, Molecular aspects of mammalian fertilization, Asian J. Androl., 2001, 3, 243–249 CAS.
- N. Egge, A. Muthusubramanian and G. A. Cornwall, Amyloid properties of the mouse egg zona pellucida, PLoS One, 2015, 10, e0129907 CrossRef PubMed.
- P. M. Wassarman, Zona pellucida glycoproteins, J. Biol. Chem., 2008, 283, 24285–24289 CrossRef CAS PubMed.
- L. Jovine, W. G. Janssen, E. S. Litscher and P. M. Wassarman, The PLAC1-homology region of the ZP domain is sufficient for protein polymerisation, BMC Biochem., 2006, 6, 11 CrossRef PubMed.
- L. Jovine, C. C. Darie, E. S. Litscher and P. M. Wassarman, Zona pellucida domain proteins, Annu. Rev. Biochem., 2005, 74, 83–114 CrossRef CAS PubMed.
- P. M. Wassarman, L. Jovine, H. Qi, Z. Williams, C. Darie and E. S. Litscher, Recent aspects of mammalian fertilization research, Mol. Cell. Endocrinol., 2005, 234, 95–103 CrossRef CAS PubMed.
- D. P. Green, Three-dimensional structure of the zona pellucida, Rev. Reprod., 1997, 2, 147–156 CrossRef CAS PubMed.
- J. M. Greve and P. M. Wassarman, Mouse egg extracellular coat is a matrix of interconnected filaments possessing a structural repeat, J. Mol. Biol., 1985, 181, 253–264 CrossRef CAS PubMed.
- S. J. Conner, L. Lefièvre, D. C. Hughes and C. L. Barratt, Cracking the egg: increased complexity in the zona pellucida, Hum. Reprod., 2005, 20, 1148–1152 CrossRef CAS PubMed.
- K. Suzuki, N. Tatebe, S. Kojima, A. Hamano, M. Orita and N. Yonezawa, The Hinge Region of Bovine Zona Pellucida Glycoprotein ZP3 Is Involved in the Formation of the Sperm-Binding Active ZP3/ZP4 Complex, Biomolecules, 2015, 23, 3339–3353 CrossRef PubMed.
- I. Raj, A. L. Sadat, H. Hosseini, E. Dioguardi, K. Nishimura, L. Han, A. Villa, D. de Sanctis and L. Jovine, Structural Basis of Egg Coat-Sperm Recognition at Fertilization, Cell, 2017, 169, 1315–1326 CrossRef CAS PubMed.
- M. Bokhove and L. Jovine, Structure of Zona Pellucida Module Proteins, Curr. Top. Dev. Biol., 2018, 130, 413–442 CAS.
- M. Monné and L. Jovine, A structural view of egg coat architecture and function in fertilization, Biol. Reprod., 2011, 85, 661–669 CrossRef PubMed.
- E. Fahrenkamp, B. Algarra and L. Jovine, Mammalian egg coat modifications and the block to polyspermy, Mol. Reprod. Dev., 2020, 87(3), 326–340 CrossRef CAS PubMed.
- G. Familiari, S. A. Nottola, G. Macchiarelli, G. Micara, C. Aragona and P. M. Motta, Human zona pellucida during in vitro fertilization: an ultrastructural study using saponin, ruthenium red, and osmium-thiocarbohydrazide, Mol. Reprod. Dev., 1992, 32, 51–61 CrossRef CAS PubMed.
- H. Funahashi, H. Ekwall, K. Kikuchi and H. Rodriguez-Martinez, Transmission electron microscopy studies of the zona reaction in pig oocytes fertilized in vivo and in vitro, Reproduction, 2001, 122, 443–452 CAS.
- A. Boccaccio, M. C. Frassanito, L. Lamberti, R. Brunelli, G. Maulucci, M. Monaci, M. Papi, C. Pappalettere, T. Parasassi, L. Sylla, F. Ursini and M. De Spirito, Nanoscale characterization of the biomechanical hardening of bovine zona pellucida, J. R. Soc., Interface, 2012, 9, 2871–2882 CrossRef PubMed.
- M. Papi, R. Brunelli, G. Familiari, M. C. Frassanito, L. Lamberti, G. Maulucci, M. Monaci, C. Pappalettere, T. Parasassi, M. Relucenti, L. Sylla, F. Ursini and M. De Spirito, Whole-depth change in bovine zona pellucida biomechanics after fertilization: how relevant in hindering polyspermy?, PLoS One, 2012, 7, e45696 CrossRef CAS PubMed.
- A. Stracuzzi, J. Dittmann, M. Böl and A. E. Ehret, Visco- and poroelastic contributions of the zona pellucida to the mechanical response of oocytes, Biomech. Model. Mechanobiol., 2021, 20, 751–765 CrossRef PubMed.
- M. Papi, R. Brunelli, L. Sylla, T. Parasassi, M. Monaci, G. Maulucci, M. Missori, G. Arcovito, F. Ursini and M. De Spirito, Mechanical properties of zona pellucida hardening, Eur. Biophys. J., 2010, 39(6), 987–992 CrossRef PubMed.
- S. A. Nottola, S. Makabe, T. Stallone, G. Familiari, S. Correr and G. Macchiarelli, Surface morphology of the zona pellucida surrounding human blastocysts obtained after in vitro fertilization, Arch. Histol. Cytol., 2005, 68, 133–141 CrossRef PubMed.
- K. P. Riddell, D. A. Stringfellow, B. W. Gray, M. G. Riddell, J. C. Wright and P. K. Galik, Structural and viral association comparisons of bovine zonae pellucidae from follicular oocytes, day-7 embryos and day-7 degenerated ova, Theriogenology, 1993, 40, 1281–1291 CrossRef.
- M. Monné, L. Han, T. Schwend, S. Burendahl and L. Jovine, Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats, Nature, 2008, 456, 653–657 CrossRef PubMed.
- G. Macchiarelli, E. Vizza, S. A. Nottola, G. Familiari and P. M. Motta, Cellular and microvascular changes of the ovarian follicle during folliculogenesis: a scanning electron microscopic study, Arch. Histol. Cytol., 1992, 55, 191–204 CrossRef PubMed.
- Q. Zeng, Y. Qin, W. Chang and X. Luo, Correlating and evaluating the functionality-related properties with surface texture parameters and specific characteristics of machined components, Int. J. Mech. Sci., 2018, 149, 62–72 CrossRef.
- E. S. Gadelmawla, M. M. Koura, T. M. A. Maksoud, I. M. Elewa and H. H. Soliman, Roughness Parameters, J. Mater. Process. Technol., 2002, 123, 133–145 CrossRef.
- N. Zoratto, R. Matassa, E. Montanari, G. Familiari, S. Petralito, T. Coviello, C. Di Meo and P. Matricardi, Glycerol as a green solvent for enhancing the formulation of dextran methacrylate and gellan-based semi-interpenetrating polymer networks, J. Mater. Sci., 2020, 55, 9562–9577 CrossRef CAS.
- V. Baena and M. Terasaki, Three-dimensional organization of transzonal projections and other cytoplasmic extensions in the mouse ovarian follicle, Sci. Rep., 2019, 9, 1262 CrossRef PubMed.
- R. Matassa, S. Orlanducci, G. Reina, M. C. Cassani, D. Passeri and M. L. Terranova, Structural and morphological peculiarities of hybrid Au/nanodiamond engineered nanostructures, Sci. Rep., 2016, 31163 CrossRef CAS PubMed.
- R. Li Voti, G. Leahu, C. Sibilia, R. Matassa, G. Familiari, S. Cerra, T. A. Salamone and I. Fratoddi, Photoacoustics for listening to Metal Nanoparticles Super-Aggregates, Nanoscale Adv., 2021, 3, 4692–4701 RSC.
- S. Cerra, T. A. Salamone, F. Sciubba, M. Marsotto, C. Battocchio, S. Nappini, F. A. Scaramuzzo, R. Li Voti, C. Sibilia, R. Matassa, A. M. Beltrán, G. Familiari and I. Fratoddi, Study of the interaction mechanism between hydrophilic thiol capped gold nanoparticles and melamine in aqueous medium, Colloids Surf., B, 2021, 203, 111727 CrossRef CAS PubMed.
- J. C. Russ, The Image Processing Handbook, CRC Press, 5th edn, 2006 Search PubMed.
- R. Matassa, M. Carbone, I. Fratoddi and R. Caminiti, Organometallic Oligomer resolved by Radial Distribution Function of X-ray Diffraction Analysis, J. Phys. Chem. B, 2010, 114, 2359–2364 CrossRef CAS PubMed.
- A. Stsiapanava, C. Xu, M. Brunati, S. Zamora-Caballero, C. Schaeffer, M. Bokhove, L. Han, H. Hebert, M. Carroni, S. Yasumasu, L. Rampoldi, B. Wu and L. Jovine, Cryo-EM structure of native human uromodulin, a zona pellucida module polymer, EMBO J., 2020, 15, e106807 CrossRef PubMed.
- L. Jovine, C. C. Darie, E. S. Litscher and P. M. Wassarman, Zona pellucida domain proteins, Annu. Rev. Biochem., 2005, 74, 83–114 CrossRef CAS PubMed.
- G. L. Weiss, J. J. Stanisich, M. M. Sauer, C. W. Lin, J. Eras, D. S. Zyla, J. Trück, O. Devuyst, M. Aebi, M. Pilhofer and R. Glockshuber, Architecture and function of human uromodulin filaments in urinary tract infections, Science, 2020, 369(6506), 1005–1010 CrossRef CAS PubMed.
- K. R. Porter and I. Tamm, Direct visualization of a mucoprotein component of urine, J. Biol. Chem., 1955, 212(1), 135–140 CrossRef CAS PubMed.
- R. Matassa, M. Carbone, R. Laceri, R. Purrello and R. Caminiti, Supramolecular Structure of Extrinsically Chiral Porphyrin Hetero-Assemblies and Achiral Analogues, Adv. Mater., 2007, 19, 3961–3967 CrossRef CAS.
- R. Matassa, C. Sadun, L. D'Ilario, A. Martinelli and R. Caminiti, Supramolecular Organization of Toluidine Blue Dye in Solid Amorphous Phases, J. Phys. Chem. B, 2007, 111, 1994–1999 CrossRef CAS PubMed.
- R. A. Engh and R. Huber, Accurate Bond and Angle Parameters for X-ray Protein Structure Refinement, Acta Crystallogr., 1991, 47, 392–400 CrossRef.
- P. Conway, M. D. Tyka, F. DiMaio, D. E. Konerding and D. Baker, Relaxation of backbone bond geometry improves protein energy landscape modeling, Protein Sci., 2014, 23, 47–55 CrossRef CAS PubMed.
- R. Kodali and R. Wetzel, Polymorphism in the intermediates and products of amyloid assembly, Curr. Opin. Struct. Biol., 2007, 17, 48–57 CrossRef CAS PubMed.
- F. Báez, A. A. Camargo and G. D. A. Gastal, Ultrastructural Imaging Analysis of the Zona Pellucida Surface in Bovine Oocytes, Microsc. Microanal., 2019, 25, 1032–1036 CrossRef PubMed.
- J. M. Calafell, C. Nogués, M. Ponsà, J. Santaló and J. Egozcue, Zona pellucida surface of immature and in vitro matured mouse oocytes: analysis by scanning electron microscopy, J. Assisted Reprod. Genet., 1992, 9, 365–372 CrossRef CAS PubMed.
- M. Del Collado, J. C. da Silveira, J. R. Sangalli, G. M. Andrade, L. R. da Silva Sousa, L. A. Silva, F. V. Meirelles and F. Perecin, Fatty Acid Binding Protein 3 And Transzonal Projections Are Involved In Lipid Accumulation During In Vitro Maturation Of Bovine Oocytes, Sci. Rep., 2017, 7, 2645 CrossRef PubMed.
- P. Chugh and E. K. Paluch, The actin cortex at a glance, J. Cell Sci., 2018, 19(131), jcs186254 CrossRef PubMed.
- G. Vanroose, H. Nauwynck, A. V. Soom, M. T. Ysebaert, G. Charlier, P. V. Oostveldt and A. de Kruif, Structural aspects of the zona pellucida of in vitro-produced bovine embryos: a scanning electron and confocal laser scanning microscopic study, Biol. Reprod., 2000, 62, 463–469 CrossRef CAS PubMed.
- C. M. Bidan, K. P. Kommareddy, M. Rumpler, P. Kollmannsberger, P. Fratzl and J. W. Dunlop, Geometry as a factor for tissue growth: towards shape optimization of tissue engineering scaffolds, Adv. Healthc. Mater., 2013, 2, 186–194 CrossRef CAS PubMed.
- L. Han, M. Monné, H. Okumura, T. Schwend, A. L. Cherry, D. Flot, T. Matsuda and L. Jovine, Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3, Cell, 2010, 143, 404–415 CrossRef CAS PubMed.
- E. Bianchi, B. Doe, D. Goulding and G. J. Wright, Juno is the egg Izumo receptor and is essential for mammalian fertilization, Nature, 2014, 508, 483–487 CrossRef CAS PubMed.
- L. P. Cheeseman, J. Boulanger, L. M. Bond and M. Schuh, Two pathways regulate cortical granule translocation to prevent polyspermy in mouse oocytes, Nat. Commun., 2016, 19(7), 13726 CrossRef PubMed.
- Z. Gartner and A. Hughes, Bioprinting in space and time, Nature, 2019, 572, 38–39 CrossRef CAS PubMed.
- D. M. Shapiro, G. Mandava, S. E. Yalcin, P. Arranz-Gibert, P. J. Dahl, C. Shipps, Y. Gu, V. Srikanth, A. I. Salazar-Morales, J. P. O'Brien, K. Vanderschuren, D. Vu, V. S. Batista, N. S. Malvankar and F. J. Isaacs, Protein nanowires with tunable functionality and programmable self-assembly using sequence-controlled synthesis, Nat. Commun., 2022, 13(1), 829 CrossRef CAS PubMed.
- Z. Wang, Y. Shang, H. Luo, C. Yang, Z. Yang, C. Ren and J. Liu, Achieving higher hierarchical structures by cooperative assembly of tripeptides with reverse sequences, Nanoscale, 2023, 15(16), 7502–7509 RSC.
- M. Glaser, P. Mollenkopf, D. Prascevic, C. Ferraz, J. A. Käs, J. Schnauß and D. M. Smith, Systematic altering of semiflexible DNA-based polymer networks via tunable crosslinking, Nanoscale, 2023, 15, 7374–7383 RSC.
- A. Kamada, A. Herneke, P. Lopez-Sanchez, C. Harder, E. Ornithopoulou, Q. Wu, X. Wei, M. Schwartzkopf, P. Müller-Buschbaum, S. V. Roth, M. S. Hedenqvist, M. Langton and C. Lendel, Hierarchical propagation of structural features in protein nanomaterials, Nanoscale, 2022, 14, 2502–2510 RSC.
- S. Cerra, V. Dini, T. A. Salamone, F. Hajareh Haghighi, M. Mercurio, A. Cartoni, A. Del Giudice, M. Marsotto, I. Venditti, C. Battocchio, F. A. Scaramuzzo, R. Matassa, S. Nottola, R. Faccini, R. Mirabelli and I. Fratoddi, Acrylates-based hydrophilic co-polymeric nanobeads as nanocarriers for imaging agents, Colloids Surf., A, 2023, 674, 131829 CrossRef CAS.
- G. Reina, E. Tamburri, S. Orlanducci, S. Gay, R. Matassa, V. Guglielmotti, T. Lavecchia and M. L. Terranova, Nanocarbon surfaces for biomedicine, Biomatter, 2014, 4, e28537 CrossRef PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: An open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- R. Matassa, G. Familiari, E. Battaglione, C. Sibilia, G. Leahu, A. Belardini, I. Venditti, L. Fontana and I. Fratoddi, Electron microscopy reveals a soluble hybrid network of individual nanocrystals self-anchored by bifunctional thiol fluorescent bridges, Nanoscale, 2016, 8, 18161–18169 RSC.
- A. Bearzotti, P. Papa, A. Macagnano, E. Zampetti, I. Venditti, R. Fioravanti, L. Fontana, R. Matassa, G. Familiari and I. Fratoddi, Environmental Hg vapours adsorption and detection by using functionalized gold nanoparticles network, J. Environ. Chem. Eng., 2018, 6, 4706–4713 CrossRef CAS.
- Bhushan Modern Tribology Handbook, Two Volume Set Chapter2 Surface Roughness Analysis and Measurement Techniques 2000.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr02644b |
| This journal is © The Royal Society of Chemistry 2023 |