Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Activity-enhanced DNAzyme for design of label-free copper(II) biosensor†
William
Etheridge
ab,
Frederic
Brossard
b,
Sitan
Zheng
a,
Svenja
Moench
c,
Suraj
Pavagada
 a,
Róisín M.
Owens
a,
Róisín M.
Owens
 a and
Ljiljana
Fruk
*a
a and
Ljiljana
Fruk
*a
aBioNano Engineering Group, Department of Chemical Engineering and Biotechnology, University of Cambridge, West Cambridge Site, Philippa Fawcett Drive CB3 0AS, Cambridge, UK. E-mail: lf389@cam.ac.uk
bHitachi Cambridge Laboratory, Hitachi Europe Ltd, J. J. Thomson Avenue, Cambridge CB3 0HE, UK
cKarlsruhe Institute of Technology (KIT), Institute for Biological Interfaces (IBG 1), Hermann-von-Helmholtz-Platz, D-76344 Eggenstein-Leopoldshafen, Germany
First published on 14th June 2023
Abstract
Metal ion-driven, DNA-cleaving DNAzymes are characterised by high selectivity and specificity. However, their use for metal ion sensing remains largely unexplored due to long reaction times and poor reaction yields relative to RNA-cleaving DNAzymes and other sensing strategies. Herein we present a study demonstrating a significant rate enhancement of a copper-selective DNA cleaving DNAzyme by both polydopamine (PDA) and gold (Au) nanoparticles (NPs). PDA NPs enhance the reaction through the production of hydrogen peroxide, while for AuNPs the enhancement is aided by the presence of citrate surface moeities, both of which drive the oxidative cleavage of the substrate. A 50-fold enhancement for PDA NPs makes the combination of PDA and DNAzyme suitable for a practical application as a sensitive biosensor for Cu(II) ions. Using DNAzyme deposition onto a gold electrode followed by Polydopamine Assisted DNA Immobilisation (PADI), we achieve a cost-effective, label-free and fast (within 15 min) electrochemical biosensor with a limit of detection of 180 nmol (11 ppm), thus opening a route for the rational design of a new generation of hybrid DNAzyme-based biosensors.
1. Introduction
DNAzymes are catalytically active sequences of single stranded DNA (ssDNA) exhibiting a range of functions such as DNA/RNA cleavage, ribonucleotide labelling and catalysis of click chemistry reactions.1,2 Most DNAzyme mediated reactions require metal ion co-factors, and DNAzymes requiring Pb2+, Cu2+, Ag+, Hg2+, Mn2+ and Zn2+ ions have been identified through in vitro selection.3–8 This dependency on metal ions has been exploited in the design of sensitive metal biosensors, mainly utilising RNA-cleaving DNAzymes, which generally exhibit high reaction rates due to the presence of a hydroxyl group within the RNA substrate. However, RNA degrades easily, limiting the application of such sensors in real-life settings such as environmental monitoring.In contrast, despite their inherently higher stability, the use of DNA-cleaving DNAzymes remains largely underexplored due to their long reaction times and poor yields compared to RNA-cleaving counterparts. Thus, to make DNA-cleaving DNAzymes more useful for metal ion sensing, DNAzymes with an improved kinetic profile need to be developed or new strategies enhancing their activity introduced. Herewith, we present a strategy to significantly enhance the activity of a Cu(II)-specific DNA-cleaving DNAzyme to develop a specific and more sensitive electrochemical platform for the detection of Cu(II) ions.
Although in low amounts Cu(II) is beneficial and acts as a metal cofactor of several enzyme classes,9 accumulation of Cu(II) can have numerous detrimental effects. For example, increased amount of Cu(II) in plants due to soil contamination from mining, pesticide production and fossil fuel burning has been shown to have negative effects on germination, growth, and plant development, as well as the photosynthetic cycle.10–13 High levels of Cu(II) in oceans stemming from pollutants such as marine paints and wastewater can lead to metabolic changes and developmental issues in plankton and algae, which are crucial for the health of the oceanic ecosystem.14,15 In medicine, abnormalities in Cu(II) homeostasis have been implicated in an increased risk of developing neurological disorders such as Alzheimer's disease.16 In addition, a genetic disorder known as Wilson's disease is directly related to the inability to process Cu(II) resulting in its accumulation in blood and tissue that can lead to severe tissue damage and death.17 Finally, Cu(II) ingestion through tap water contamination has been linked to increased cases of liver cirrhosis and acute poisoning.18
Clearly, detection and quantification of Cu(II) in water and biological samples is important not only for environmental remediation but also in diagnostics and industrial process monitoring. Commonly, Cu(II) sensor design relies on ion interaction with organic dyes, which is measured by monitoring the resulting change in dye absorbance or emission.19 However, such strategies often require extensive molecular design and the use of components that are often sensitive to environmental conditions.20,21
DNAzymes recently emerged as a suitable alternative to organic dyes, with two Cu-selective RNA-cleaving DNAzymes reported that exhibit a reaction kinetics and sensitivity suitable for point of care sensing.22 These systems utilise fluorescence quenching phenomena and although a limit of detection for Cu(II) of 1.6 nM was achieved, reliance on the use of fluorescent dyes, quenchers, and unstable RNA substrates limits the long-term stability and storage of this sensor.
In addition to RNA-cleaving DNAzymes, six Cu(II) selective DNA-cleaving DNAzymes have been reported, with five of them requiring ascorbate ions alongside Cu(II) for substrate cleavage through the oxidative damage mechanism.23,24 The remaining sixth DNAzyme, named F8, required the addition of Mn(II) ions and metal salts (Na+ and K+), all of which resulted in a low reaction rate of DNA substrate cleavage with Kobs = 0.137 h−1.25
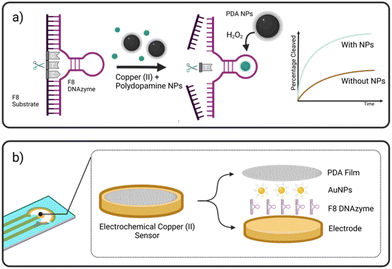
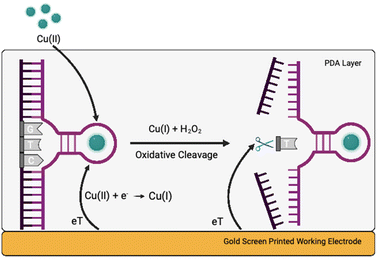
Herewith, we present a simple route to significant improvement of the catalytical activity of Cu(II)-specific F8 DNAzyme by employing nanoparticles (NPs) as additives (Fig. 1a). We further exploit this enhancement to design a new class of sensitive electrochemical sensors for the rapid detection of Cu(II) ions in solution. This was achieved through the deposition of F8 and AuNPs followed by the production of a protective and activity-enhancing polydopamine (PDA) coating to improve the sensitivity, stability, and storage of the sensor (Fig. 1b). Not only does our biosensor results in fast readout and sensitivity, but the use of a label-free electrochemical readout instead of fluorescent labels improves the cost-effectiveness and simplicity of the design. In addition, unlike other DNAzyme sensors, our design does not require complex electrode modification, covalent immobilisation,26 or the use of mediators27 to improve the electrochemical readout and sensitivity.
2. Materials and methods
2.1 DNA sequences
| F8 DNAzyme | 5′ AG TGT TCC GTG GAT GGA GCA ATA GTC TCC CGG GTC CGT ATG GAT CGG CA 3′ |
| F8 substrate | 5′ TGC CGA TCC ATA CTG CGG AAC ACT 3′ |
| W8 non-enzyme DNA | 5′ AG TGT TCC GCA GTA TGG ATC GGC A 3′ |
2.2 Synthesis of polydopamine nanoparticle (PDA NPs)
92 mg of dopamine hydrochloride (Sigma Aldrich) was dissolved in 1 mL ultra-pure water and added to 135 g of Tris Base (Sigma Aldrich) in 200 ml of ultra-pure water. The solution was then stirred at 300 rpm for 30 min and left overnight at room temperature. The solution was centrifuged at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, removing the supernatant and resuspending the pellet in water 3 times yielding the final polydopamine nanoparticles. The nanoparticles were characterised by zeta-potential and DLS measurements (both Malvern Zetasizer Nano ZS) giving a zeta potential of −43.8 mV and a polydispersity index of 0.185. SEM images (ESI, Fig. S1†) were taken and demonstrated a uniform and spherical morphology with an average diameter of 142.4 nm.
000g, removing the supernatant and resuspending the pellet in water 3 times yielding the final polydopamine nanoparticles. The nanoparticles were characterised by zeta-potential and DLS measurements (both Malvern Zetasizer Nano ZS) giving a zeta potential of −43.8 mV and a polydispersity index of 0.185. SEM images (ESI, Fig. S1†) were taken and demonstrated a uniform and spherical morphology with an average diameter of 142.4 nm.
2.3 Screening of F8 DNAzyme activity in presence of nanoparticles
For all assays, 1 μM of both F8 and DNA substrate were annealed in 15 mM NaCl and 50 mM HEPES buffer (pH 7.0–7.6) by heating to 95 °C for 5 min and allowing to cool at room temperature for 30 min. The resulting F8-substrate complex was then exposed to 12.5 μM copper(II) chloride in the absence and presence of NPs (0.01 mg mL−1 of 20 nm citrate capped Au and AgNPs, 25 nm CuNP, 140 nm PDA NPs and 30 nm Fe2O3/Fe3O4). All cleavage reactions were performed at 37 °C for 10 min and 1 hour. After incubation, the cleavage reaction was quenched using 8 M urea and results were analysed using 12% Urea PAGE Gel Electrophoresis at 45 V for 120 min. The resulting gels were imaged using a Syngene G-Box camera and quantified using ImageJ gel analysis software.2.4 Electrochemical sensor design
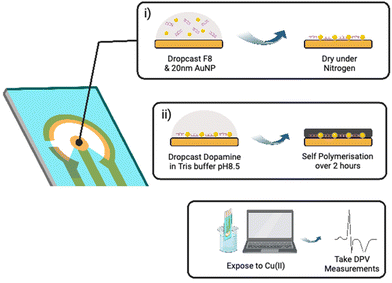
Disposable screen-printed electrodes were purchased from Metrohm UK (DRP C220AT) consisting of a gold working electrode, a gold auxiliary electrode, and a silver reference electrode. They were first electrochemically cleaned by cyclic voltammetry from 0 to 1.5 V in 1 M sulphuric acid for 10 cycles and dried under nitrogen gas. Pre-annealed F8-substrate complex was added to 0.02 mg mL−1 20 nm citrate capped Au NPs and 10 μL was drop cast onto the working electrode. The droplet was dried over 10 min under a light stream of nitrogen gas. 10 μL of 5 mg mL−1 dopamine hydrochloride in 10 mM Tris-buffer pH 8.5 was then drop casted onto the working electrode containing the dried Au NP-doped F8 complex. Dopamine was allowed to polymerise over 2 hours in 100% humidity to prevent drying, and then washed in PBS and water 3 times.2.5 Electrochemical measurements
All electrochemical measurements were taken using a Palmsens 4 potentiostat in PBS. The sensor was clamped in position fully submerged in PBS and allowed to sit for 5 min in stirred solution (100 rpm). Three blank measurements were then taken to generate a stable baseline measurement. The solution was then adjusted to a desired Cu(II) concentration using 2 mM stock solution of CuCl2 solution in ultra-pure water. Differential pulse voltammetry (DPV) measurements were then taken after 15 min. −0.5 V to +0.2 V was used for the voltage scan range with an increment of 0.01 V. The pulse width was 0.02 s with an amplitude of 0.2 V. Peak currents were calculated by subtracting baseline measurement at peak voltage.3 Results and discussion
3.1 NP-mediated rate enhancement of F8 DNAzyme
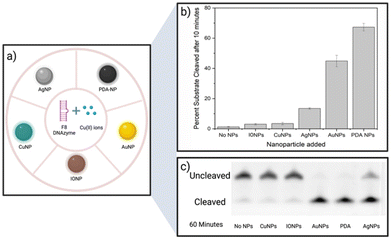
A screening assay was performed to investigate the effect of various classes of commercially available nanoparticles on the cleavage rate of F8 DNAzyme. Initial motivation was to design solid platform for DNAzyme immobilisation to ease the enzyme recovery from the solution. In short, 0.01 mg mL−1 of copper (CuNP), iron oxide (Fe2O3/Fe3O4 NP), polydopamine (PDA NPs), gold (AuNP), or silver (AgNP) nanoparticles were added to the solution of F8 DNAzyme hybridised with F8 DNA sequence spiked with 12.5 μM CuCl2. The reaction was stopped after 10 min and 60 min to assess the percentage of cleaved substrates using gel electrophoresis. Surprisingly, a significant amount of the cleaved substrate was observed already after 10 min in presence of PDA (67.3%), Au (44.9%), and Ag (13.5%) NPs when compared to the control that contain CuCl2 only (1.3%) (Fig. 2). After 60 min, 100% substrate cleavage was obtained in the presence of PDA/AuNP, while control sample showed only 11% (Fig. 2 and complete PAGE gel image in Fig. S2†).The rate enhancement of Au and Ag NPs is attributed to the citrate-coating, with citrate ions directly contributing to the improved cleavage rate as demonstrated by the effect of the citrate ions alone (ESI, Fig. S3 and S4†). On the other hand, we hypothesised that PDA NP-mediated enhancement stems from the direct generation of hydrogen peroxide (H2O2) by PDA NPs in solution. Previously, it was shown that the activity of F8 DNAzyme can be increased by addition of H2O2 to Cu(II) ions.25 In addition, we knew from previous studies that PDA structures can generate H2O2 from molecular oxygen present in water.28 To test our hypothesis that indeed PDA-mediated production of H2O2 impacts the oxidative cleavage of DNA substrate, we measured the concentration of H2O2 generated by PDA NPs. Employing H2O2 screening assay we found that 0.01 mg mL−1 of PDA NPs, the amount used in DNAzyme studies, generates 1.7 μM H2O2 (ESI, Fig. S5†). Due to the smaller enhancement rate, we did not look deeper into the mechanism of IONPs and CuNPs. However, as IONPs can generate H2O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 29 this is a likely reason for observed cleavage enhancement, while in case of CuNPs, the enhancement effect could stem from increased concentration of Cu(II) available to the enzyme through a leaching effect.30
29 this is a likely reason for observed cleavage enhancement, while in case of CuNPs, the enhancement effect could stem from increased concentration of Cu(II) available to the enzyme through a leaching effect.30
3.2 Design of the DNAzyme-based Cu(II) sensor
Considering the remarkable enhancement of the Cu(II)-mediated DNA cleavage in the presence of NPs, we considered potential ways to exploit this effect in the design of an affordable and simple-to-fabricate biosensor for Cu(II) detection. In terms of sensing, electrochemical sensors have numerous advantages such as cost-effectiveness and ease of detection, and we aimed to focus on design of DNAzyme based electrochemical sensing. One of the major challenges in design of electrochemical biosensors is ensuring the deposition and immobilisation of active elements onto electrode surface. To avoid complex immobilisation strategies that would involve additional DNAzyme modifications, we decided to exploit PDA and use it as a protective and activity-enhancing coating. Similar strategy was recently demonstrated in design of electrochemical H2O2 biosensor using heme G-quadruplex DNAzyme, which was entrapped within PDA layer.31Polydopamine assisted DNA immobilisation (PADI) is a simple and effective strategy that does not require any covalent attachment of DNAzymes onto the electrode. However, as PDA is an electrical insulator, it is crucial that the coating step is performed after drop casting of DNAzyme to ensure the current flow to the electrode.
As shown in Fig. 3, the fabrication of our biosensor consists of two steps: (i) drop-casting of DNAzyme/substrate complex onto a gold electrode followed by formation of PDA layer (PADI, ii). PADI proceeds through self-polymerisation of dopamine precursor on top of the deposited biomolecules and any rate-enhancing additives such as NPs.
To ensure that drop casting results in a homogenous and reproducible electrode coating, we investigated drop casting and drying using Cy3-labelled F8 DNAzyme. The coffee ring effect32 was observed when the droplet of DNAzyme/substrate solution was allowed to dry on the electrode under atmospheric conditions causing most of the F8 DNAzyme complex to concentrate in a ring around the edge of the sensing electrode (ESI, Fig. S6a†). Such an effect impairs the reproducibility of the sensor production, and we explored different strategies to avoid it. Finally, homogenous deposition of DNAzyme was achieved by drying deposited solution under a steady stream of nitrogen for 60 seconds (ESI, Fig. S6b†). Such uniform layer of DNAzyme was coated with PDA by exposing the surface to a basic solution of dopamine (5 mg mL−1 Dopamine HCl, 10 mM Tris, pH 8.5).33,34 Significant darkening of the surface was observed after 120 min, which is attributed to the successful formation of a polydopamine film (ESI, Fig. S7†).
To confirm the formation of the coating, impedance spectroscopy measurements of the electrode were taken. Namely, PDA is known to be an electrical insulator and can cause the fouling of electrodes,35 which results in increased impedance as observed with PDA-coated electrodes (ESI, Fig. S8†).
Once the deposition and coating were optimised, the electrochemical performance of the sensor was investigated using differential pulse voltammetry (DPV) before and 15 min after the exposure to Cu(II). DPV was chosen as a read-out method due to its ability to perform fast measurements with high sensitivity.36
3.3 DNAzyme-based electrochemical detection of Cu(II)
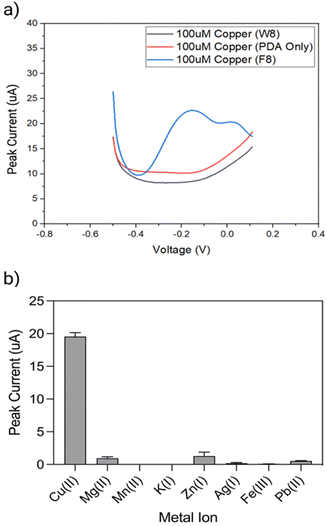
The performance of the biosensor was first explored with F8 DNAzyme and two controls, using DPV measurements before and 15 min after exposure to 10 μM Cu(II) solution (Fig. 4a). The first control comprised of a non-enzymatic, linear strand W8, which was complementary to F8 substrate but did not contain DNAzyme features. The second control was simply an electrode coated with a PDA layer in the absence of F8 DNAzyme. As it can be seen in Fig. 4a, two new cathodic current peaks are observed between −0.2 to 0 V with DNAzyme coated electrode, which are not observed with control samples.To explore the selectivity of F8-DNAzyme electrode towards Cu(II), the electrode was exposed to solutions of other common ions such as Mg(II), Mn(II), Zn(II), and K(I), as well as the pollutants commonly found in soil and water such as Fe(III), Pb(II) and Ag(I) (Fig. 4b). No significant response was observed with those ions indicating high selectivity of our sensor towards Cu(II).
Concerning the mechanism, we hypothesise that Cu(II) ions diffuse through the PDA layer into the active site of the DNAzyme (Fig. 5). The first cathodic peak can be attributed to the reduction of Cu(II) ions into Cu(I), and the second one to H2O2 produced by the PDA layer reacting with Cu(I) causing the cleavage of the substrate via an oxidative radical pathway. Such a mechanism is in line with the results presented here, although there is a possibility that other pathways might be involved.
In addition, the effect of Au NPs on the cleavage rate was evaluated first in the presence of small amount (0.01 mg mL−1) of 20 nm Au NPs. DPV measurements employing PDA entrapped DNAzyme complex with and without citrate capped 20 nm Au NPs in Cu(II) solution are shown in ESI, Fig. S9.† It can be seen that inclusion of the Au NPs resulted in an increase of the peak current. In addition, there is no effect of Au NPs in absence of copper ions. This initial study clearly indicates that the addition of Au NPs to the DNAzyme solution further improves the performance of the sensor without impairing the Cu(II) selectivity. Current work is focused on exploiting this phenomenon further for the design of a sensitive Cu(II) sensor for blood screening.
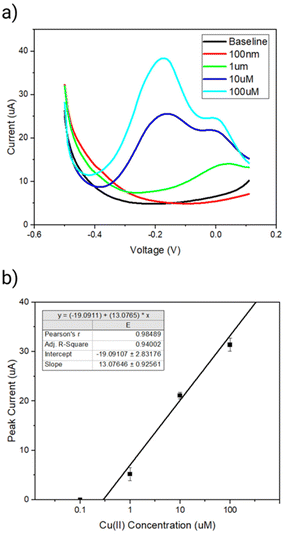
Next, we investigated dependence of the peak current signal between the sensing range of −0.2 to 0.1 V on the Cu(II) concentration. A clear increase in the peak current between a region of interest of −0.2 to 0.1 V in presence of increasing concentration of Cu(II) was observed (Fig. 6a). The linear range of detection was found to be 1 μM to 100 μM copper over a sensing time of 15 min (Fig. 6b). The limit of detection was determined to be 180 nM (11 ppm) Cu(II) using the slope of the concentration curve and a protocol reported by Armbruster et al.37
Finally, an often neglected but important factor when proposing a new biosensor devise is its long-term stability. The stability of our PDA entrapped DNAzyme-based sensor was investigated over two months with the sensor stored under atmospheric conditions (ESI, Fig. S10†). A decay in the peak current was observed after two months, however, this could be improved by storage under dry and/or cold conditions and in the absence of light to minimise PDA and/or electrode degradation.
4. Conclusions
We have shown that the cleavage rate of a Cu(II)-specific DNA-cleaving DNAzyme can be significantly increased in the presence of citrate coated Au (5-fold enhancement in cleavage over 10 min) and polydopamine (PDA) nanoparticles (50-fold enhancement in cleavage over 10 min). Such an increase is directly related to the reducing potential of citrate ions in case the of Au NPs, and intrinsic H2O2 production by PDA. Although their potential was acknowledged, DNAzymes were generally considered too slow to have a huge practical value for metal sensor design. We have shown that nanoparticle additives have significant effect on the rate enhancement making DNAzymes suitable for Cu(II) sensor design. Our electrochemical sensor is cost-effective and simple-to-make with Cu(II) limit of detection of 180 nM (11 ppm). The WHO guideline on the limit of Cu(II) in drinking water is given as 20 μM (1.3 mg ml−1) and the safe limit of copper in serum is regarded to be 2.4 μM.38 As such, our sensor provides a cost effective and efficient alternative to other strategies for monitoring Cu(II) levels in drinking water and agricultural soil, as well as detection of Cu(II) in serum, and continuous monitoring of Cu(II) levels in large bioreactors.Author contributions
W. E. and L. F. contributed equally. They both conceptualised the project with the help of S. P. S. P. and W. E. performed research experiments. S. M., S. P. and S. Z. helped with experimental studies. F. B. and R. O. helped with critical comments, drafting, and revising the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by EPSRC-Hitachi ICase PhD studentship Nr G105536 awarded to W. E. S. P. would like to thank EPSRC Sensor CDT and Cambridge International Trust and TRinity College-Henry Barlow Scholarship.References
- N. Gan, et al., DNAzyme-Catalyzed Click Chemistry for Facilitated Immobilization of Redox Functionalities on Self-Assembled Monolayers, J. Phys. Chem. C, 2020, 124, 19083–19090 CrossRef CAS.
- R. R. Breaker and G. F. Joyce, A DNA enzyme that cleaves RNA, Chem. Biol., 1994, 1, 223–229 CrossRef CAS PubMed.
- P.-J. J. Huang and J. Liu, An Ultrasensitive Light-up Cu 2+ Biosensor Using a New DNAzyme Cleaving a Phosphorothioate-Modified Substrate, Anal. Chem., 2016, 88, 3341–3347 CrossRef CAS PubMed.
- K. Schlosser and Y. Li, Tracing Sequence Diversity Change of RNA-Cleaving Deoxyribozymes under Increasing Selection Pressure during in Vitro Selection, Biochemistry, 2004, 43, 9695–9707 CrossRef CAS PubMed.
- R. Saran and J. Liu, A Silver DNAzyme, Anal. Chem., 2016, 88, 4014–4020 CrossRef CAS PubMed.
- J. Li, In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme, Nucleic Acids Res., 2000, 28, 481–488 CrossRef CAS PubMed.
- R. R. Breaker and G. F. Joyce, A DNA enzyme with Mg2+ -dependent RNA phosphoesterase activity, Chem. Biol., 1995, 2, 655–660 CrossRef CAS PubMed.
- A. K. Brown, J. Li, C. M.-B. Pavot and Y. Lu, A Lead-Dependent DNAzyme with a Two-Step Mechanism, Biochemistry, 2003, 42, 7152–7161 CrossRef CAS PubMed.
- R. Lontie, Copper Proteins and Copper Enzymes, CRC Press, 2018. DOI:10.1201/9781351070911.
- M. Jaishankar, T. Tseten, N. Anbalagan, B. B. Mathew and K. N. Beeregowda, Toxicity, mechanism and health effects of some heavy metals, Interdiscip. Toxicol., 2014, 7, 60–72 CrossRef PubMed.
- H. Ali, E. Khan and M. A. Sajad, Phytoremediation of heavy metals—Concepts and applications, Chemosphere, 2013, 91, 869–881 CrossRef CAS PubMed.
- Z. Shabbir, et al., Copper uptake, essentiality, toxicity, detoxification and risk assessment in soil-plant environment, Chemosphere, 2020, 259, 127436 CrossRef CAS PubMed.
- A. R. Mir, J. Pichtel and S. Hayat, Copper: uptake, toxicity and tolerance in plants and management of Cu-contaminated soil, BioMetals, 2021, 34, 737–759 CrossRef CAS PubMed.
- L. Bat, S. Bilgin and A. Öztekin, Toxicity of copper on marine organisms from the Black Sea, J. Coastal Life Med., 2017, 5, 422–426 CrossRef CAS.
- G. W. Brian, The effects of heavy metals (other than mercury) on marine and estuarine organisms, Proc. R. Soc. London, Ser. B, 1971, 177, 389–410 Search PubMed.
- Z. K. Mathys and A. R. White, Copper and Alzheimer’s Disease, Adv. Neurobiol., 2017, 199–216, DOI:10.1007/978-3-319-60189-2_10.
- P. Ferenci, et al., Diagnostic Value of Quantitative Hepatic Copper Determination in Patients With Wilson's Disease, Clin. Gastroenterol. Hepatol., 2005, 3, 811–818 CrossRef CAS PubMed.
- R. Eife, et al., Chronic poisoning by copper in tap water: I. Copper intoxications with predominantly gastointestinal symptoms, Eur. J. Med. Res., 1999, 4, 219–223 CAS.
- M. Saleem, M. Rafiq, M. Hanif, M. A. Shaheen and S.-Y. Seo, A Brief Review on Fluorescent Copper Sensor Based on Conjugated Organic Dyes, J. Fluoresc., 2018, 28, 97–165 CrossRef PubMed.
- L. Tang, X. Dai, X. Wen, D. Wu and Q. Zhang, A rhodamine–benzothiazole conjugated sensor for colorimetric, ratiometric and sequential recognition of copper(II) and sulfide in aqueous media, Spectrochim. Acta, Part A, 2015, 139, 329–334 CrossRef CAS PubMed.
- X. Chen, et al., New fluorescent and colorimetric chemosensors bearing rhodamine and binaphthyl groups for the detection of Cu2+, Sens. Actuators, B, 2009, 137, 597–602 CrossRef CAS.
- P.-J. J. Huang and J. Liu, An Ultrasensitive Light-up Cu 2+ Biosensor Using a New DNAzyme Cleaving a Phosphorothioate-Modified Substrate, Anal. Chem., 2016, 88, 3341–3347 CrossRef CAS PubMed.
- N. Carmi, L. A. Shultz and R. R. Breaker, In vitro selection of self-cleaving DNAs, Chem. Biol., 1996, 3, 1039–1046 CrossRef CAS PubMed.
- R. Drouin, et al., Cupric ion/ascorbate/hydrogen peroxide-induced DNA damage: DNA-bound copper ion primarily induces base modifications, Free Radicals Biol. Med., 1996, 21, 261–273 CrossRef CAS PubMed.
- M. Wang, et al., In vitro selection of DNA-cleaving deoxyribozyme with site-specific thymidine excision activity, Nucleic Acids Res., 2014, 42, 9262–9269 CrossRef CAS PubMed.
- Y. Tan, et al., An immobilization free DNAzyme based electrochemical biosensor for lead determination, Analyst, 2016, 141, 1121–1126 RSC.
- K. Liu, K. S. Chen, D. Sen and H.-Z. Yu, Ultrasensitive detection of total copper with an electrochemical biosensor built on the in cis coupling of hexynyl CLICK-17 DNAzyme with azido self-assembled monolayers, Electrochim. Acta, 2021, 379, 138125 CrossRef CAS.
- A. Tyo, et al., Development and Characterization of an Antimicrobial Polydopamine Coating for Conservation of Humpback Whales, Front. Chem., 2019, 7, 618 CrossRef CAS PubMed.
- S. I. Nehme, L. Crocker and L. Fruk, Flavin-Conjugated Iron Oxide Nanoparticles as Enzyme-Inspired Photocatalysts for Azo Dye Degradation, Catalysts, 2020, 10, 324 CrossRef CAS.
- M. Raffi, et al., Investigations into the antibacterial behavior of copper nanoparticles against Escherichia coli, Ann. Microbiol., 2010, 60, 75–80 CrossRef CAS.
- A. Gao, Y.-R. Wang, X.-W. He and X.-B. Yin, An Electrochemical Hydrogen Peroxide Biosensor Based on Polydopamine-Entrapped G-Quadruplex-Hemin DNAzyme, Chin. J. Anal. Chem., 2012, 40, 1471–1476 CrossRef CAS.
- R. D. Deegan, et al., Capillary flow as the cause of ring stains from dried liquid drops, Nature, 1997, 389, 827–829 CrossRef CAS.
- A. B. Popov, et al., Size-tuneable and immunocompatible polymer nanocarriers for drug delivery in pancreatic cancer, Nanoscale, 2022, 14, 6656–6669 RSC.
- L. Crocker, et al., Enzyme-inspired flavin–polydopamine as a biocompatible nanoparticle photocatalyst, Nanoscale Horiz., 2019, 4, 1318–1325 RSC.
- J. Szewczyk, D. Aguilar-Ferrer and E. Coy, Polydopamine films: Electrochemical growth and sensing applications, Eur. Polym. J., 2022, 174, 111346 CrossRef CAS.
- B. J. Venton and D. J. DiScenza, Voltammetry, in Electrochemistry for Bioanalysis, Elsevier, 2020, pp. 27–50. DOI:10.1016/B978-0-12-821203-5.00004-X.
- D. A. Armbruster and T. Pry, Limit of blank, limit of detection and limit of quantitation, Clin. Biochem. Rev., 2008, 29(Suppl 1), S49–S52 Search PubMed.
- G. J. Brewer, Wilson's Disease, Springer US, 2001. DOI:10.1007/978-1-4615-1645-3.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr02169f |
| This journal is © The Royal Society of Chemistry 2023 |