Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evidence of Cytolysin A nanopore incorporation in mammalian cells assessed by a graphical user interface†
Florian Leonardus Rudolfus
Lucas‡
 *a,
Rocio K.
Finol-Urdaneta‡
*a,
Rocio K.
Finol-Urdaneta‡
 *bcd,
Toon
Van Thillo
*bcd,
Toon
Van Thillo
 a,
Jeffrey R.
McArthur
bd,
Nieck Jordy
van der Heide
a,
Jeffrey R.
McArthur
bd,
Nieck Jordy
van der Heide
 e,
Giovanni
Maglia
e,
Giovanni
Maglia
 e,
Peter
Dedecker
e,
Peter
Dedecker
 a,
Olaf
Strauss
f and
Carsten
Wloka‡
a,
Olaf
Strauss
f and
Carsten
Wloka‡
 *f
*f
aLab for Nanobiology, Department of Chemistry, KU Leuven, Belgium. E-mail: florian.lucas@kuleuven.be
bIllawarra Health and Medical Research Institute, Wollongong, NSW 2522, Australia. E-mail: rfinolu@uow.edu.au
cElectrophysiology Facility for Cell Phenotyping and Drug Discovery, Wollongong, NSW 2522, Australia
dFaculty of Science, Medicine and Health, University of Wollongong, Wollongong, NSW 2522, Australia
eGroningen Biomolecular Sciences and Biotechnology Institute, University of Groningen, Groningen, 9747 AG, Groningen, The Netherlands
fExperimental Ophthalmology, Department of Ophthalmology, Charité - Universitätsmedizin Berlin, A Corporate Member of Freie Universität, Humboldt-University, The Berlin Institute of Health, Berlin, Germany. E-mail: carsten.wloka@charite.de
First published on 10th October 2023
Abstract
Technologies capable of assessing cellular metabolites with high precision and temporal resolution are currently limited. Recent developments in the field of nanopore sensors allow the non-stochastic quantification of metabolites, where a nanopore is acting as an electrical transducer for selective substrate binding proteins (SBPs). Here we show that incorporation of the pore-forming toxin Cytolysin A (ClyA) into the plasma membrane of Chinese hamster ovary cells (CHO-K1) results in the appearance of single-channel conductance amenable to multiplexed automated patch-clamp (APC) electrophysiology. In CHO-K1 cells, SBPs modify the ionic current flowing though ClyA nanopores, thus demonstrating its potential for metabolite sensing of living cells. Moreover, we developed a graphical user interface for the analysis of the complex signals resulting from multiplexed APC recordings. This system lays the foundation to bridge the gap between recent advances in the nanopore field (e.g., proteomic and transcriptomic) and potential cellular applications.
Introduction
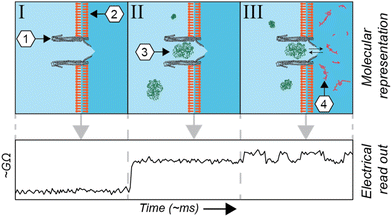
The metabolome of a single cell is one of the major driving forces of its phenotype, representing its physiological, and potential malignant, status. Mass spectrometry1,2 and NMR-based3,4 technologies are extraordinarily sensitive and can quantify and qualify hundreds of metabolites from single cells. The measurement of metabolites in living cells, however, is a challenging endeavour for those technologies. Among other reasons, they lack the temporal resolution required to quantify the dynamic change of metabolite concentrations, which can fluctuate on the timescale of seconds. A potential solution is fluorescent-based probes,5 which are often used to assess subcellular metabolite concentrations in living cells. There are, however, relatively few fluorescent probes available and alternatives have proven labour-intensive to engineer.6 Equally, Raman spectroscopy is a non-invasive proxy for fluorescence, but it is currently limited by low signal intensities. Alternatively, it is possible to perform single-cell magnetic resonance imaging (MRI); however, molecules can only be detected based on their spin polarization, limiting the range of compounds that can be imaged.A complementary technique, derived from voltage-clamp recordings of single channels, utilizes pore-forming toxins to create nanometre-sized pores in membranes (nanopores). Commercially used for DNA sequencing, this technique can be applied to the quantification of metabolites7–12 and detection and study of proteins and peptides13–26 at the single-molecule level. In brief, in this technique an electrical current is applied across a non-conductive membrane and the conductance across the nanopore is measured. The conductance is altered whenever an analyte modifies the ionic current flowing through a nanopore (Fig. 1). Advantageously, voltage-clamp recordings have been used for several decades to investigate membrane-associated channels. The combination of nanopore spectrometry with traditional electrophysiology therefore represents the next logical step.
Recently, it has been shown that an engineered nanopore derived from Mycobacterium smegmatis porin A (MspA) can be inserted into the membrane of HEK293 cells, allowing the stochastic detection of externally applied glutamine.27 Due to the stochastic nature, this approach lacks the ability to measure analytes selectively. Functional synthetic ion channels have been investigated using the patch-clamp technique,28,29 and we have recently shown that an engineered nanopore derived from S. typhi, Cytolysin A (ClyA), is particularly well-suited as an electrical transducer for non-stochastic measurements. ClyA is a comparably large nanopore, able to trap proteins in the range of 20 kDa to 50 kDa in size. The sometimes long-lasting (order of minutes) capture of a single protein is achieved by balancing electroosmotic and electrophoretic forces, which can be tuned by the applied voltage. In this system, a substrate-specific binding protein, or domain, (SBP/SBD) is captured inside the vestibule of the ClyA nanopore,12 allowing non-stochastic quantification of metabolites. Whenever the SBP/SBD binds to its cognate analyte, it undergoes a conformational change that induces a change in conductance across the nanopore. The environmental tolerance of the ClyA nanopore and SBPs/SBDs provide robust readouts in complex matrices such as urine,7 blood,11 and sweat,11 making it thus potentially suitable to measure intracellular metabolites.
Here we establish suitable conditions for ClyA nanopore incorporations into mammalian (CHO-K1) cells that allow the confinement of SBPs specific for glucose and thiamine. The ion channel activity resulting from nanopores inserted in mammalian cells is recorded by octoplexed automated patch-clamp (APC) electrophysiology. Using fluorescence microscopy for a calcium influx assay, we confirm incorporation into the same cell line. Finally, we develop “HoleyPatch” as a customizable, open-source, graphical user interface (GUI) that enables the visualization and analysis of complex data derived from nanopore-coupled APC systems with scalable throughput.
Results and discussion
Experimental setup for the recording of nanopore incorporation in living mammalian cells
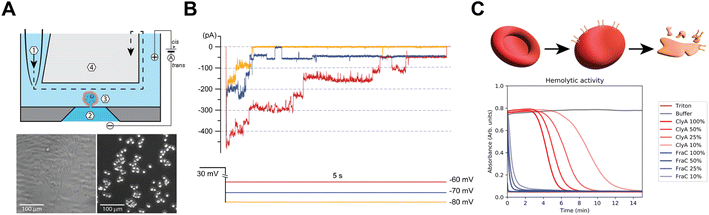
To study the functional properties of Cytolysin A-AS nanopores inserted in mammalian cells, we used an octoplexed automated patch-clamp (APC) electrophysiology system (Patchliner Octo, Nanion) as indicated in Fig. S1A.† We chose Chinese hamster ovary K1 cells (CHO-K1, Fig. 2A) for our study, because they have low endogenous ion channel activity and are commonly used in electrophysiological studies of ion channels. We recorded CHO-K1 cells in suspension at 298 K using standard APC solutions, which included the following buffers: intracellular/trans 60 mM KF, 60 mM KCl, 7 mM NaCl, 10 mM HEPES, 5 mM EGTA, and extracellular/cis 140 mM NaCl, 4 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 10 mM HEPES. Fig. S1B† shows a typical whole-cell voltage-clamp recording of eight different naïve CHO-K1 cells subjected to a standard current–voltage (IV) stimulation protocol. The protocol consisted of a 5-second pulse to −80 mV followed by step depolarizations from −80 mV to 50 mV (5 s, Δ10 mV) delivered at 0.1 Hz and recorded from a holding potential (Vh) of −80 mV. In the absence of exogenously incorporated pore-forming proteins, CHO-K1 cells displayed negligible endogenous ionic currents at applied transmembrane potentials between 0 mV and −80 mV (Fig. S1B†). Our results indicate that CHO-K1 cells and the established APC recording conditions are well-suited for the proof-of-principle detection of ClyA activity in mammalian cell lines. We refer to this system as APC-CHO.The monomeric form of ClyA can spontaneously assemble into artificial lipid bilayers (or bilayer lipid membranes, BLMs), creating membrane-spanning conducting channels with different stoichiometries. First, whole-cell patch-clamped CHO-K1 cells were exposed to monomeric ClyA (mClyA). To achieve this, mClyA was diluted in extracellular recording solution and full bath exchanges were performed on the cells. The cells were then stimulated constantly for 10 minutes at 0.1 Hz, with 5-second square pulses to −80 mV from Vh of −40 mV. During these recordings, conducting events were observed that were consistent with both single and multiple channel events. This attests to the insertion and assembly of functional ClyA nanopores in CHO-K1 cells (Fig. 2B). We found that membrane depolarization enhanced ClyA nanopore activity, as evidenced by the appearance of longer lasting channel events as well as more channel incorporations at −70 mV and −60 mV (Fig. 2B).
The amount of mClyA suspension applied to the extracellular compartment correlated with the incorporation of ion conducting channels, with higher doses of mClyA resulting in a faster and more abundant appearance of ClyA nanopores in patch-clamped CHO-K1 cells. However, at high doses, the on-cell oligomerization of mClyA occurred rapidly, resulting in the overload of nanopores on the recorded cell and subsequent loss of seal and/or cell death. By titrating mClyA, recordings up to approximately 10 minutes after the first appearance of nanopore conducting events were possible. Nevertheless, all experiments were invariably terminated by the cumulative and catastrophic incorporation of too many nanopores into the plasma membrane of CHO-K1 cells.
During these experiments, mClyA oligomerization on-cell was observed, which is consistent with hemolytic activity assays that were performed by exposing ovine (Ovis aries) red blood cells to purified mClyA (Fig. 2C). Therefore, the avid incorporation of mClyA into CHO-K1 cells required modifications to the pulsing regime in order to control the number of channels present during recordings. Specifically, under standard asymmetrical (Ki/Nao) “quasi-physiological” ionic conditions, episodic stimulation consisting of pulses 5–10 seconds long to −80 mV (Vh −40 mV) was implemented going forward in order to capture the nanopores at different applied potentials. In summary, mClyA forms functional nanopores in CHO-K1 cells that can be adequately controlled to enable the characterization of ClyA in mammalian cells.
ClyA nanopores, consisting of 12–14 monomers, are functional in BLMs made of diphytanoyl-sn-glycero-3-phosphocholine (DPhPC). However, only the dodecameric (12 mer) complex has been well studied and confirmed to effectively capture target proteins within the range of 20 kDa to 50 kDa, with dwell times ranging from seconds to minutes, making it suitable for metabolite quantification. To trigger the oligomerization process, the monomeric ClyA solution was supplemented with 0.2% n-dodecyl β-D-maltoside (DDM), and the resulting different oligomeric forms were separated using blue native polyacrylamide gel electrophoresis (BN-PAGE). We attempted to incorporate oligomeric ClyA into CHO-K1 cells for on-cell measurements using 12 mers extracted from the corresponding gel band via overnight incubation in 50 μl of extracellular solution supplemented with 0.2% DDM. The resulting extraction was then diluted in extracellular buffer (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), and increasing volumes of this dilution were applied to the extracellular side of the APC chamber of whole-cell patch-clamped CHO-K1 cells. The electrical activity of individual cells was tracked for up to 30 minutes after sequential and cumulative addition of extracted ClyA 12 mers to the cis chamber bathing CHO-K1 cells. However, due to cell lysis caused by the cumulative addition of detergent-laden eluate, the amount of oligomeric ClyA that could be applied to these cells was curtailed. As a result, under the established recording conditions, exposure to oligomeric ClyA failed to reveal functional ClyA nanopores in APC recordings of CHO-K1 cells (not shown). Therefore, all experiments reported here were conducted using monomeric ClyA.
200), and increasing volumes of this dilution were applied to the extracellular side of the APC chamber of whole-cell patch-clamped CHO-K1 cells. The electrical activity of individual cells was tracked for up to 30 minutes after sequential and cumulative addition of extracted ClyA 12 mers to the cis chamber bathing CHO-K1 cells. However, due to cell lysis caused by the cumulative addition of detergent-laden eluate, the amount of oligomeric ClyA that could be applied to these cells was curtailed. As a result, under the established recording conditions, exposure to oligomeric ClyA failed to reveal functional ClyA nanopores in APC recordings of CHO-K1 cells (not shown). Therefore, all experiments reported here were conducted using monomeric ClyA.
To confirm the incorporation of the nanopores into the cellular membranes, we performed calcium influx assays. In brief, we transfected CHO-K1 cells with a plasmid carrying a genetically encoded fluorescent calcium sensor (GCaMP6s)30 and observed the influx of calcium into the cell from the external medium (1.25 mM Ca2+) by an increase in fluorescence. As a control, we added 0.2% DDM, and its effect was observed as a faint haze on the fluorescent images due to GCaMP6s diffusion (Fig. 3A, left column). The secondary control utilizes a mixture of ionomycin and saponin to translocate calcium across the cell membranes. Cells started to fluoresce without changing the shape of the CHO-K1 cells (Fig. 3A, middle column).31 When ClyA-AS monomers were added to the CHO-K1 cells, we observed a steep increase in fluorescence after several minutes of incubation (Fig. 3A, right column).
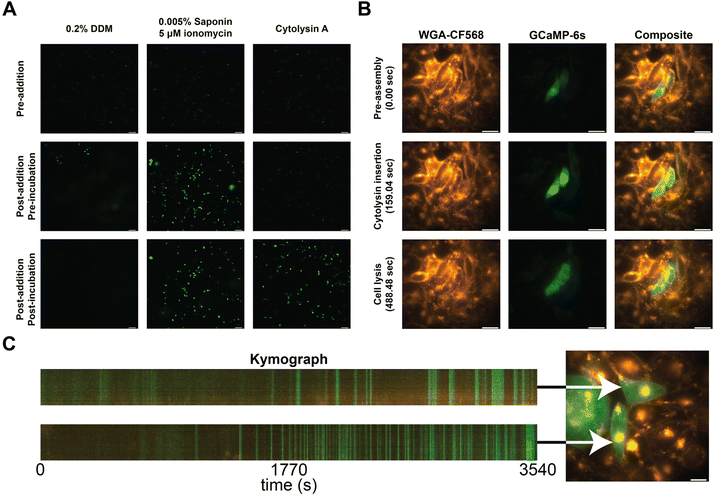 | ||
| Fig. 3 Calcium influx assay in CHO-K1 cells. (A) Calcium influx assay measured using epifluorescence imaging in the presence of 0.2% DDM, 0.005% Saponin and 5 μM ionomycin, and Cytolysin A monomers. Cells prior to addition are shown on the first row, directly after addition in the middle row, and after waiting approximately 60 seconds on the bottom row. (B) Calcium influx of two cells measured using epifluorescence imaging. The first column shows the stained cells using WGA-CF568, the middle column shows GCaMP6s, and the final column shows the composite of both channels. Each row represents an image taken at 0, 159, and 488 seconds after addition of Cytolysin A monomers. (C) Kymographs of two cells (Movie S2†), exposed to 10 times diluted ClyA, showing fluorescence intensity across a virtual line in the cell over time. Selected cells are indicated using an arrow. | ||
To further investigate the insertion of the nanopores, we examined several cells on a high resolution microscope. Herein we stained the cellular membrane with WGA-CF568, allowing us to monitor cell shape (Movie S1†). We observed that there was influx of calcium after several minutes (Fig. 3B). However, shortly after, we observed the outflux of calcium bound dye indicating that the membrane started to disintegrate once a higher concentration ClyA had incorporated. To test this hypothesis, we also monitored the ClyA incorporation at a 10-fold dilution (Fig. 3C and Movie S2†). We observed short bursts of calcium influx, followed by (near) complete recovery to the base fluorescence.
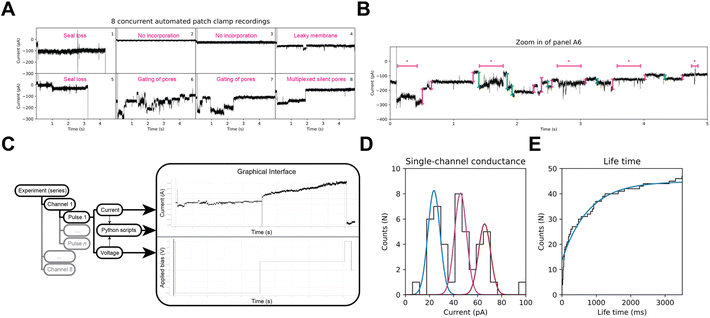
Development of a graphical user interface
In BLM experiments, the composition of the bilayer as well as the cis/trans solutions are tightly controlled, and under these conditions ClyA displays little gating. However, biological membranes have a complex and variable composition, and during APC-CHO whole-cell recordings, different components of the cell cytoplasm may be dialyzed or retained based on their physicochemical properties. As shown in Fig. 4A and B, ClyA nanopores in CHO-K1 cells display complex behaviours that pose substantial challenges for the currently available analysis tools. Therefore, we developed an open-source and flexible alternative to existing public and proprietary software to potentially extract quantitative information from octoplexed APC nanopore signals in mammalian cells. We created an adjustable Python library, named HoleyPatch (Fig. 4C), along with a graphical user interface (GUI) framework that allows rapid display and analysis of electrophysiology traces. HoleyPy,32 our single-channel recordings library, was extended to create HoleyPatch, which we used to assess the nanopore activity of all eight cells, both individually and as an ensemble in Fig. 4.Using HoleyPatch, we investigated the ionic current observed from single-nanopore openings in ClyA during repeated 5–10 s pulses to −80 mV (Vh −40 mV). Our results revealed three distinct distributions of current amplitudes in ClyA openings, measuring 22 ± 3 pA, 45 ± 9 pA, and 64 ± 5 pA, respectively (n = 47, as shown in Fig. 4D). Consistent with these, mClyA incorporation into giant plasma membrane vesicles (GPMVs) recorded by manual patch-clamp displayed similar current amplitudes (22.8 ± 0.5 pA, 45.7 ± 0.3 pA, and 73.3 ± 0.3 pA) as depicted in Fig. S2.† The different current amplitudes or sub-conductance observed in CHO-K1 cells and GPMVs may be attributed to various factors beyond the scope of this study, such as the binding of cytoplasmic components and a highly complex membrane lipid composition and distribution33,34 that are known to affect the function of many ion channels.35–37
ClyA nanopores in CHO-K1 cells displayed an apparent mean lifetime of 759 ± 18 ms (n = 47, Fig. 4E). However, it should be noted that the latter value is skewed by the relatively short stimulation pulses used to preserve cell viability.
ClyA nanopores in APC-CHO cells couple to substrate-binding proteins
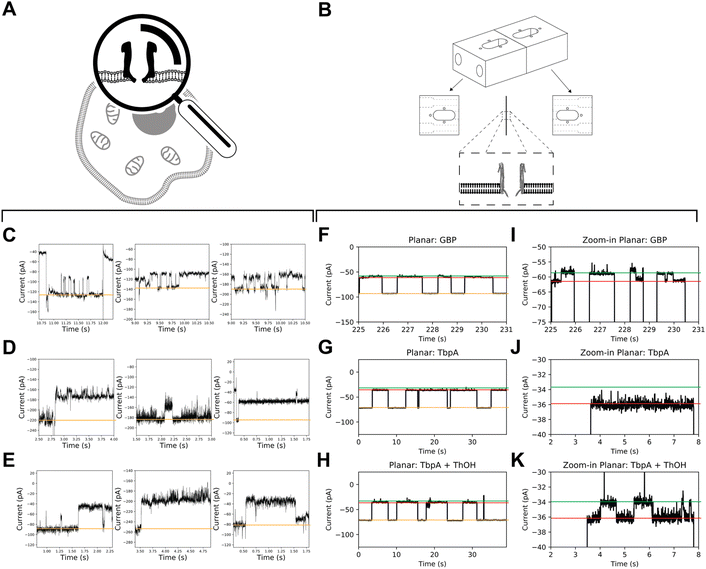
A comparison between recordings obtained from mClyA incorporation into BLMs (Fig. 5) and recordings of nanopore incorporation into mammalian cells reveals that the signals obtained from the latter are more complex. Specifically, the nanopore conductance levels show high frequency, low amplitude fluctuations (Fig. 5C–E). It can be inferred from these findings that the fluctuation in nanopore currents recorded in CHO-K1 cells is, at least in part, caused by brief incursions of endogenous cytoplasmic molecules into the nanopores.Glucose-binding protein (GBP) and the Y27A mutant of thiamine-binding protein (TbpA) are well-suited SBPs due to their well-known characteristics, and fast (un)binding.7,11 To acquire on-cell data, we used mClyA to assemble ClyA nanopores on CHO-K1 cells under nominally glucose/thiamine and GBP/TbpA-free conditions. We then perfused the extracellular buffer with ∼500 nM of either GBP or TbpA and observed nanopore blockades that indicated capture of GBP and TbpA by ClyA (Fig. 5C–E).
To account for potential differences of SBP activity in APC-CHO solutions, we conducted control measurements to observe the incorporation of GBP and TbpA in BLMs under APC conditions (Fig. 5). We found that the ionic asymmetry did not affect the intrinsic closure of GBP (Fig. 5F and I), nor the ability of TbpA to bind to its substrate (Fig. 5G–K), confirming that both GBP and TbpA remain active under APC conditions. In BLMs, proteins are captured by 12 mer ClyA for extensive periods, and unlikely to be captured with similar dwell time in alternate oligomeric forms. Therefore, the long-lasting capture of TbpA and GBP indirectly confirms the insertion of 12 mer oligomeric ClyA. However, it appears that the residence time of the SBPs is shorter than expected from BLMs. This discrepancy may be explained by diverging salt concentrations, as the concentration inside the CHO-K1 cells may differ locally and it is well-known that the localization and residence time of the captured protein are related to the electrostatic conditions inside the nanopore.38,39
In contrast to the stereotypical changes in current levels observed in BLM recordings of GBP coupled to ClyA (Fig. 5F and I), the addition of glucose to the extracellular side of patch-clamped CHO-K1 cells did not result in measurable changes in conductance levels between glucose-free and glucose-supplemented conditions, nor did it cause apparent intrinsic opening and closure of GBP. Similarly, the addition of thiamine did not induce observable changes in the blocked current. Under the conditions used, the baseline variations in APC-CHO traces were in the same order of magnitude as TBP/GBP transitions in planar lipid bilayer experiments.
Conclusion
In this contribution, we verified the feasibility of studying nanopore function in real time in mammalian cells using automated patch-clamp (APC) electrophysiology. Comparison between recordings obtained from mClyA incorporation into CHO-K1 cells and BLMs highlight the higher complexity of the signals derived from the electrical recording of nanopores in mammalian cells. This complexity is evidenced by presence of high frequency, low amplitude fluctuations in the nanopore current levels. We acknowledge that other factors may be of significance, however, we identify the signal-to-noise ratio as the main contributor that disallows the observation of SBP dynamics in CHO-K1 cells. It can be speculated that the complex fluctuation of nanopore currents recorded in CHO-K1 cells may, at least in part, be attributed to short-lived incursions of endogenous cytoplasmic molecules into the nanopores. Additionally, discrepancies in ClyA nanopore behaviours between BLM and APC systems could be attributed to the difference between the membranes: biological membranes are composed of heterogeneous mixtures of lipids and membrane-associated proteins that are often subjected to biochemical modifications. In contrast, the BLMs we employ consist only of DPhPC lipids.While this contribution represents a stepping stone, demonstrating functional insertion of the nanopore sensor, capture of binding proteins, and the software that is required for its analysis, it remains challenging to detect metabolites in live cells. The detection, and by extension quantification, of analytes inside the cell is subject to several challenges. Firstly, the ability of the analyte to diffuse and/or translocate towards the area of the membrane in which the nanopore is incorporated. Secondly, the puncture for patch-clamping combined with the toxicity of the nanopore eventually kills the cells allowing only a brief window of measurement, currently disallowing the continuous monitoring of metabolites. However, the kinetics of assembly of nanopores may be controlled by modification of the nanopores.40 Moreover, other methods, such as optical single channel recording,41 may replace the need for patch-clamping. We envision that the approach presented in this work will eventually enable extended monitoring of metabolites during different physiological scenarios.
Materials and methods
Protein expression, purification, and oligomerization
The ClyA-AS gene with a C-terminal histidine-tag was transformed into the E. cloni ®EXPRESS BL21(DE3) cells via electroporation. The transformed cells were inoculated into 200 mL 2xYT medium supplemented with 100 μg mL−1 ampicillin. The cell culture was grown at 37 °C and 200 rpm until an OD600 of ∼0.8–1.0 was obtained. Protein expression was induced by addition of 0.5 mM Isopropyl β-D-thiogalactopyranoside (IPTG) and subsequent culturing overnight at 25 °C and 200 rpm.The bacterial cells were harvested by centrifugation at 6500 rpm for 20 minutes at 4 °C and subjected to three freeze–thaw cycles at −80 °C to increase susceptibility to cell lysis. The cell pellets were resuspended in 20 mL buffer A (15 mM Tris-HCl pH: 7.5, 150 mM NaCl, 10 mM Imidazole, 1 mM MgCl2, 10 μg mL−1 lysozyme, 0.1 U mL−1 DNase) and incubated for 20 minutes at 4 °C while rotating at 25 rpm. Cell lysis was further improved via probe sonication (Brandson) for 2 minutes at 25% output power. The lysate was centrifuged at 6500 rpm for 30 minutes at 4 °C to remove cellular debris. The supernatant was mixed with 200 μL Ni-NTA resin (Qiagen) in buffer B (15 mM Tris-HCl pH: 7.5, 150 mM NaCl, 10 mM Imidazole) and incubated for 15 minutes at 4 °C while rotating at 10 rpm. The incubated Ni-NTA resin was loaded onto a gravity flow column (Bio-Rad) pre equilibrated with buffer B and washed with 20 mL of buffer B. The monomeric ClyA-AS was eluted from the Ni-NTA resin with three elution steps of 200 μL buffer C (15 mM Tris-HCl pH: 7.5, 150 mM NaCl, 300 mM Imidazole). The protein concentration was measured with the Nanodrop and diluted to 1 mg mL−1 for oligomerization.
The monomeric ClyA-AS was oligomerized by addition of 0.2% n-dodecyl β-D-maltoside (DDM) and incubated for 30 minutes at 37 °C. The different oligomeric forms of ClyA-AS where separated by Blue Native polyacrylamide gel electrophoreses (BN-PAGE) on 4–20% native gels (Bio-Rad). The lowest band on the gel, corresponding to type I ClyA-AS oligomers, was cut out of the gel and resuspended in buffer E (15 mM Tris-HCl pH: 7.5, 150 mM NaCl, 2 mM EDTA, 0.02% DDM).
Haemolytic activity assay
Defibrinated sheep blood (Fisher Scientific) was washed and diluted in buffer D (15 mM Tris-HCl pH: 7.5, 150 mM NaCl) to an OD650 of 0.8–0.9. 100 μL of diluted blood was added to each well of a 96-well plate containing several solutions of 10 μL with different concentrations of pore-forming toxin and the controls 1% TritonX (100% lysis) and buffer D (0% lysis). Immediately after adding the diluted blood the haemolytic activity was measured for 30 minutes by monitoring the decrease in OD650 using the Synergy H1 Hybrid-Multimode reader (BioTek).Planar lipid bilayers recordings
We prepare planar lipid bilayers following the Montal–Mueller method (Fig. 1A) as described recently in extended method.42Thiamine-Y27A and glucose binding protein
We prepared Thiamine-Y27A (TbpA-Y27A) and Glucose binding protein (GBP) as described in recently extended methods.7,11Automated patch-clamp recordings of ClyA nanopores in CHO-K1 cells
CHO-K1 cells were cultured in DMEM-F12 Media (Life Technologies, Australia) supplemented with 10% FBS (Bovigen, Australia), 2 mM GlutaMAX (Life Technologies) and 1% penicillin/streptomycin (Pen/Strep, Life Technologies) in a humidified incubator at 37 °C, 5% CO2. On the day of recording, CHO-K1 cells were detached from T25 flasks using TrypLE (ThermoFisher Scientific), re-suspended in cold external recording solution, and kept in suspension at 4 °C until recording.Automated patch-clamp was performed as previously reported.44,45 Briefly, whole-cell recordings of naïve CHO-K1 were generated by automated patch clamp (APC) electrophysiology using a Patchliner Octo HT (Nanion Technologies GmbH, München, Germany) using single-hole planar NPC-16 chips with resistance of ∼5 MΩ. Chip and whole-cell capacitance were fully compensated with >50% series resistance compensation. The intracellular solution, IS, contained (in mM): 60 KF, 80 KCl, 10 EGTA, 10 glucose, and 10 HEPES (pH 7.2, 285 mOsmol kg−1). The extracellular solution, ES, contained (in mM): 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, 10 glucose, and 10 HEPES (pH 7.4, 298 mOsmol kg−1).
Currents were recorded at room temperature (21–23 °C) using pulse protocol that consisted of 1 s at −80 mV, 1 s at 30 mV and 9 s to −80 mV (or as otherwise stated) from a holding potential (Vh) of −40 mV. Pulses delivered at 0.1 Hz (non-recorded time ∼1 s). The current voltage (IV) stimulation protocol consisted of sequential depolarizations following 5 seconds pre-pulses to −80 mV (10 mV steps, Vh −40 mV).
Calcium influx assay
CHO-K1 cells were cultured in Ham's F12K (Kaighn's) medium (ThermoFisher Scientific) supplemented with 10% FBS (ThermoFisher Scientific) and 10 μg ml−1 gentamicin (ThermoFisher Scientific) in a humidified incubator at 37 °C, 5% CO2. The cells were split twice per week at a confluency of 80 to 100%, and washed twice using Dulbecco's Phosphate-Buffered Saline (DPBS, ThermoFisher Scientific). CHO-K1 cells were detached from T25 flasks using trypsin-EDTA 0.5% (ThermoFisher Scientific) and resuspended in phosphate-buffered saline (PBS). After 5 minutes of incubation (37 °C, 5% CO2), the trypsination was stopped by addition of Ham's F12K (Kaighn's) medium supplemented with 10% FBS and 10 μg ml−1 gentamicin. Cells were counted using a Luna II counter (Logos biosystems) to determine cell viability and concentration (cells per mL). Subsequently, 15 K CHO-K1 cells were put in a 8-well dish (ibidi GmbH) and left to grow for 20 hours at 37 °C and 5% CO2. Each well was transfected with cDNA containing the GCaMP6s insert by addition of a mixture containing FuGENE®6 (Promega) and 250 ng plasmid DNA, prepared according to manufacture recommendation. Transfected cells were left to grow for 20 h at 37 °C and 5% CO2.Immediately before imaging, CHO-K1 cells were washed thrice with Hanks’ Balanced Salt Solution containing calcium, magnesium and glucose (HBSS; ThermoFisher Scientific). After baseline fluorescence was measured, and immediately before imaging, the cells were treated with either 0.2% DDM, 0.005% saponin + 5 μM ionomycin, or a dilution of purified ClyA-AS monomers. The cells were kept at 37 °C throughout the experiment with a stagetop incubator (Okolabs).
Large field-of-view (FOV) images were obtained on an Olympus IX71 microscope (10× UplanSApo) equipped with a Spectra X light engine (Lumencor), a ZT488RDC dichroic mirror (Chroma) and an ET525/30em emission filter (Chroma). Images were acquired with an ORCA-Flash4.0-LT+ sCMOS camera (Hamamatsu) with a 33 ms exposure time under cyan light illumination.
For individual cell imaging, cells were washed thrice with HBSS, similar to earlier described method.43 The membrane of CHO cells were then stained with a 2 μg mL−1 solution of wheat germ agglutinin CF568 conjugate (WGA-CF568; Biotium) in HBSS for 15 minutes, whereafter the triple washing step was repeated.
Individual cell imaging was performed on a Nikon Ti2-e inverted widefield microscope (CFI Apo TIRF 100XC) equipped with an L6Cc laser combiner (Oxxius), a ZET405/488/561/640xv2 excitation filter (Chroma) and a ZT405/488/561/640rpcv2 dichroic mirror (Chroma). GCaMP6s and WGA-CF568 were excited with LBX-488 and LCX-561S laser lines and their fluorescence emission was separated with an ET525/50m (Chroma) and a FF01-593/46 (Semrock) filter, respectively. Images were acquired with a PCO edge 4.2 (PCO GmbH) camera set to a 20 ms exposure time, alternating between the two fluorescence channels.
Data availability
HoleyPatch code description and availability – Python scripts are available from DOI: https://dx.doi.org/10.5281/zenodo.8395352.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Dist. Prof. David J. Adams (IHMRI-UOW) for access to manual patch-clamp facilities, and Iris Govaerts for culturing CHO-K1 cell lines for fluorescent experiments. This research was supported by the Australian Research Council's Linkage Infrastructure, Equipment and Facilities (LE180100066) grant. F. L. R. L. was partially supported by the Research Foundation – Flanders (12B7523N). P. D. was supported by the Research Foundation – Flanders (G086222N). T. V. T. was supported by the Research Foundation – Flanders (1SD1322N). C. W. was supported by a NWO Veni (722.017.010). This work was partly supported by a Rebecca Cooper Foundation for Medical Research Project Grant (PG2019396) to J. R. M., R. K. F.-U. and J. R. M. are grateful to S. Nüsch and E. Nunivut for editing assistance and constant support. N. J. H. and F. L. R. L. prepared the mClyA, oligmeric ClyA, TbpA, and GBP solutions. J. R. M. and R. K. F.-U. performed the APC and GPMV experiments. T. V. T., F. L. R. L., and P. D. performed the analysis and experiments for fluorescent imaging. F. L. R. L. and C. W. developed and performed the data analysis. C. W. and F. L. R. L. performed BLM experiments and analysis. O. S. gave crucial insights to development of a functional system. F. L. R. L., C. W., and R. K. F.-U. wrote the manuscript with contributions from all authors.References
- B. Shrestha, Single-Cell Metabolomics by Mass Spectrometry, Methods Mol. Biol., 2020, 2064, 1–8 CrossRef CAS PubMed.
- R. Zenobi, Single-cell metabolomics: analytical and biological perspectives, Science, 2013, 342(6163), 1243259 CrossRef CAS PubMed.
- R. Kumar, M. Ghosh, S. Kumar and M. Prasad, Single Cell Metabolomics: A Future Tool to Unmask Cellular Heterogeneity and Virus-Host Interaction in Context of Emerging Viral Diseases, Front. Microbiol., 2020, 11, 1152 CrossRef PubMed.
- A.-H. Emwas, R. Roy, R. T. McKay, L. Tenori, E. Saccenti, G. A. N. Gowda, D. Raftery, F. Alahmari, L. Jaremko, M. Jaremko and D. S. Wishart, NMR Spectroscopy for Metabolomics Research, Metabolites, 2019, 9, 123 CrossRef CAS PubMed.
- S. Benson, A. Fernandez, N. D. Barth, F. de Moliner, M. H. Horrocks, C. S. Herrington, J. L. Abad, A. Delgado, L. Kelly, Z. Chang, Y. Feng, M. Nishiura, Y. Hori, K. Kikuchi and M. Vendrell, SCOTfluors: Small, Conjugatable, Orthogonal, and Tunable Fluorophores for In Vivo Imaging of Cell Metabolism, Angew. Chem., Int. Ed., 2019, 58(21), 6911–6915 CrossRef CAS PubMed.
- T. Van Thillo, V. Van Deuren and P. Dedecker, Smart genetically-encoded biosensors for the chemical monitoring of living systems, Chem. Commun., 2023, 59(5), 520–534 RSC.
- F. L. R. Lucas, T. R. C. Piso, N. J. van der Heide, N. S. Galenkamp, J. Hermans, C. Wloka and G. Maglia, Automated Electrical Quantification of Vitamin B1 in a Bodily Fluid using an Engineered Nanopore Sensor, Angew. Chem., Int. Ed., 2021, 60(42), 22849–22855 CrossRef CAS PubMed.
- S. Zernia, N. J. van der Heide, N. S. Galenkamp, G. Gouridis and G. Maglia, Current Blockades of Proteins inside Nanopores for Real-Time Metabolome Analysis, ACS Nano, 2020, 14(2), 2296–2307 CrossRef CAS PubMed.
- N. S. Galenkamp and G. Maglia, Single-Molecule Sampling of Dihydrofolate Reductase Shows Kinetic Pauses and an Endosteric Effect Linked to Catalysis, ACS Catal., 2022, 12(2), 1228–1236 CrossRef CAS PubMed.
- N. S. Galenkamp, A. Biesemans and G. Maglia, Directional conformer exchange in dihydrofolate reductase revealed by single-molecule nanopore recordings, Nat. Chem., 2020, 12(5), 481–488 CrossRef CAS PubMed.
- N. S. Galenkamp, M. Soskine, J. Hermans, C. Wloka and G. Maglia, Direct electrical quantification of glucose and asparagine from bodily fluids using nanopores, Nat. Commun., 2018, 9(1), 4085 CrossRef PubMed.
- M. Soskine, A. Biesemans and G. Maglia, Single-Molecule Analyte Recognition with ClyA Nanopores Equipped with Internal Protein Adaptors, J. Am. Chem. Soc., 2015, 137(17), 5793–5797 CrossRef CAS PubMed.
- F. L. R. Lucas, K. Sarthak, E. M. Lenting, D. Coltan, N. J. van der Heide, R. C. A. Versloot, A. Aksimentiev and G. Maglia, The Manipulation of the Internal Hydrophobicity of FraC Nanopores Augments Peptide Capture and Recognition, ACS Nano, 2021, 15(6), 9600–9613 CrossRef CAS PubMed.
- F. L. R. Lucas, R. C. A. Versloot, L. Yakovlieva, M. T. C. Walvoort and G. Maglia, Protein identification by nanopore peptide profiling, Nat. Commun., 2021, 12(1), 5795 CrossRef CAS PubMed.
- G. Huang, A. Voet and G. Maglia, FraC nanopores with adjustable diameter identify the mass of opposite-charge peptides with 44 dalton resolution, Nat. Commun., 2019, 10(1), 835 CrossRef PubMed.
- C. Wloka, V. Van Meervelt, D. van Gelder, N. Danda, N. Jager, C. P. Williams and G. Maglia, Label-Free and Real-Time Detection of Protein Ubiquitination with a Biological Nanopore, ACS Nano, 2017, 11(5), 4387–4394 CrossRef CAS PubMed.
- H. Brinkerhoff, A. S. W. Kang, J. Liu, A. Aksimentiev and C. Dekker, Multiple rereads of single proteins at single-amino acid resolution using nanopores, Science, 2021, 374(6574), 1509–1513 CrossRef CAS PubMed.
- H. Ouldali, K. Sarthak, T. Ensslen, F. Piguet, P. Manivet, J. Pelta, J. C. Behrends, A. Aksimentiev and A. Oukhaled, Electrical recognition of the twenty proteinogenic amino acids using an aerolysin nanopore, Nat. Biotechnol., 2020, 38(2), 176–181 CrossRef CAS PubMed.
- S. Straathof, G. Di Muccio, M. Yelleswarapu, M. Alzate Banguero, C. Wloka, N. J. van der Heide, M. Chinappi and G. Maglia, Protein Sizing with 15 nm Conical Biological Nanopore YaxAB, ACS Nano, 2023, 17(14), 13685–13699 CrossRef CAS PubMed.
- X. Li, K. H. Lee, S. Shorkey, J. Chen and M. Chen, Different Anomeric Sugar Bound States of Maltose Binding Protein Resolved by a Cytolysin A Nanopore Tweezer, ACS Nano, 2020, 14(2), 1727–1737 CrossRef CAS PubMed.
- J. C. Foster, B. Pham, R. Pham, M. Kim, M. D. Moore and M. Chen, An Engineered OmpG Nanopore with Displayed Peptide Motifs for Single-Molecule Multiplex Protein Detection, Angew. Chem., Int. Ed., 2023, 62(7), e202214566 CrossRef CAS PubMed.
- M. Ahmad, J. H. Ha, L. A. Mayse, M. F. Presti, A. J. Wolfe, K. J. Moody, S. N. Loh and L. Movileanu, A generalizable nanopore sensor for highly specific protein detection at single-molecule precision, Nat. Commun., 2023, 14(1), 1374 CrossRef CAS PubMed.
- N. Cardozo, K. Zhang, K. Doroschak, A. Nguyen, Z. Siddiqui, N. Bogard, K. Strauss, L. Ceze and J. Nivala, Multiplexed direct detection of barcoded protein reporters on a nanopore array, Nat. Biotechnol., 2022, 40(1), 42–46 CrossRef CAS PubMed.
- R. C. A. Versloot, F. L. R. Lucas, L. Yakovlieva, M. J. Tadema, Y. Zhang, T. M. Wood, N. I. Martin, S. J. Marrink, M. T. C. Walvoort and G. Maglia, Quantification of Protein Glycosylation Using Nanopores, Nano Lett., 2022, 22(13), 5357–5364 CrossRef CAS PubMed.
- G. Huang, A. Voorspoels, R. C. A. Versloot, N. J. van der Heide, E. Carlon, K. Willems and G. Maglia, PlyAB Nanopores Detect Single Amino Acid Differences in Folded Haemoglobin from Blood, Angew. Chem., Int. Ed., 2022, 61(34), e202206227 CrossRef CAS PubMed.
- R. C. A. Versloot, S. A. P. Straathof, G. Stouwie, M. J. Tadema and G. Maglia, beta-Barrel Nanopores with an Acidic-Aromatic Sensing Region Identify Proteinogenic Peptides at Low pH, ACS Nano, 2022, 16(5), 7258–7268 CrossRef CAS PubMed.
- X. Zhang, L. Dou, M. Zhang, Y. Wang, X. Jiang, X. Li, L. Wei, Y. Chen, C. Zhou and J. Geng, Real-time sensing of neurotransmitters by functionalized nanopores embedded in a single live cell, Mol. Biomed., 2021, 2(1), 6 CrossRef PubMed.
- R. Wesolowski, A. Sommer, H. D. Arndt, U. Koert, P. Reiss, S. Wimmers and O. Strauss, Functional studies of synthetic gramicidin hybrid ion channels in CHO cells, ChemBioChem, 2007, 8(5), 513–520 CrossRef CAS PubMed.
- P. Fidzinski, A. Knoll, R. Rosenthal, A. Schrey, A. Vescovi, U. Koert, M. Wiederholt and O. Strauss, Electrophysiological response of cultured trabecular meshwork cells to synthetic ion channels, Chem. Biol., 2003, 10(1), 35–43 CrossRef CAS PubMed.
- T. W. Chen, T. J. Wardill, Y. Sun, S. R. Pulver, S. L. Renninger, A. Baohan, E. R. Schreiter, R. A. Kerr, M. B. Orger, V. Jayaraman, L. L. Looger, K. Svoboda and D. S. Kim, Ultrasensitive fluorescent proteins for imaging neuronal activity, Nature, 2013, 499(7458), 295–300 CrossRef CAS PubMed.
- A. J. Morgan and R. Jacob, Ionomycin enhances Ca2+ influx by stimulating store-regulated cation entry and not by a direct action at the plasma membrane, Biochem. J., 1994, 300(Pt 3), 665–672 CrossRef CAS PubMed.
- F. L. R. Lucas, K. Willems, M. J. Tadema, K. M. Tych, G. Maglia and C. Wloka, Unbiased Data Analysis for the Parameterization of Fast Translocation Events through Nanopores, ACS Omega, 2022, 7(30), 26040–26046 CrossRef CAS PubMed.
- T. Kobayashi and A. K. Menon, Transbilayer lipid asymmetry, Curr. Biol., 2018, 28(8), R386–R391 CrossRef CAS PubMed.
- N. Rojko and G. Anderluh, How Lipid Membranes Affect Pore Forming Toxin Activity, Acc. Chem. Res., 2015, 48(12), 3073–3079 CrossRef CAS PubMed.
- P. Sathyanarayana, S. Maurya, A. Behera, M. Ravichandran, S. S. Visweswariah, K. G. Ayappa and R. Roy, Cholesterol promotes Cytolysin A activity by stabilizing the intermediates during pore formation, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(31), E7323–E7330 CrossRef CAS PubMed.
- R. K. Finol-Urdaneta, J. R. McArthur, P. F. Juranka, R. J. French and C. E. Morris, Modulation of KvAP unitary conductance and gating by 1-alkanols and other surface active agents, Biophys. J., 2010, 98(5), 762–772 CrossRef CAS PubMed.
- A. G. Lee, Interfacial Binding Sites for Cholesterol on Kir, Kv, K(2P), and Related Potassium Channels, Biophys. J., 2020, 119(1), 35–47 CrossRef CAS PubMed.
- J. Houghtaling, C. Ying, O. M. Eggenberger, A. Fennouri, S. Nandivada, M. Acharjee, J. Li, A. R. Hall and M. Mayer, Estimation of Shape, Volume, and Dipole Moment of Individual Proteins Freely Transiting a Synthetic Nanopore, ACS Nano, 2019, 13(5), 5231–5242 CrossRef CAS PubMed.
- E. C. Yusko, B. R. Bruhn, O. M. Eggenberger, J. Houghtaling, R. C. Rollings, N. C. Walsh, S. Nandivada, M. Pindrus, A. R. Hall, D. Sept, J. Li, D. S. Kalonia and M. Mayer, Real-time shape approximation and fingerprinting of single proteins using a nanopore, Nat. Nanotechnol., 2017, 12(4), 360–367 CrossRef CAS PubMed.
- N. L. Mutter, J. Volaric, W. Szymanski, B. L. Feringa and G. Maglia, Reversible Photocontrolled Nanopore Assembly, J. Am. Chem. Soc., 2019, 141(36), 14356–14363 CrossRef CAS PubMed.
- S. Huang, M. Romero-Ruiz, O. K. Castell, H. Bayley and M. I. Wallace, High-throughput optical sensing of nucleic acids in a nanopore array, Nat. Nanotechnol., 2015, 10(11), 986–991 CrossRef CAS PubMed.
- C. Wloka, N. S. Galenkamp, N. J. van der Heide, F. L. R. Lucas and G. Maglia, Strategies for enzymological studies and measurements of biological molecules with the cytolysin A nanopore, Methods Enzymol., 2021, 649, 567–585 CAS.
- F. Bierbuesse, A. C. Bourges, V. Gielen, V. Monkemoller, W. Vandenberg, Y. Shen, J. Hofkens, P. Vanden Berghe, R. E. Campbell, B. Moeyaert and P. Dedecker, Absolute measurement of cellular activities using photochromic single-fluorophore biosensors and intermittent quantification, Nat. Commun., 2022, 13(1), 1850 CrossRef CAS PubMed.
- R. K. Finol-Urdaneta, Multitarget nociceptor sensitization by a promiscuous peptide from the venom of the King Baboon spider, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(5) DOI:10.1073/pnas.211093211.
- R. K. Finol-Urdaneta, Brevetoxin versus Brevenal Modulation of Human Nav1 Channels, Mar. Drugs, 2023, 21(7) DOI:10.3390/md21070396.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr01977b |
| ‡ Contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |