Point-of-care (POC) SARS-CoV-2 antigen detection using functionalized aerosol jet-printed organic electrochemical transistors (OECTs)†
Abstract
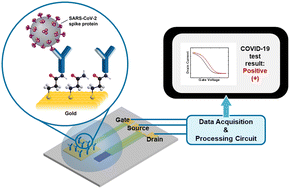
The continuous spread of coronavirus disease 2019 (COVID-19) has highlighted the need for simple and reliable diagnostic technologies for point-of-care (POC) virus detection applications. Here, we report a COVID-19 diagnostic platform based on aerosol jet-printed antibody-functionalized organic electrochemical transistors (OECTs) for rapidly identifying severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) antigens. Selective sensing of SARS-CoV-2 spike S1 protein is achieved in phosphate-buffered saline (PBS) with a detectable range of 1 fg mL−1 to 1 μg mL−1. We used the sensors to detect the antigens in unprocessed patient nasopharyngeal swab samples in universal transport medium (UTM) and achieved an overall accuracy of 70%. In addition, these patient sample tests clearly demonstrate that our OECT threshold voltage shift is correlated with the sample SARS-CoV-2 viral load. Hence, we have demonstrated an accurate POC biosensor for detecting SARS-CoV-2 antigens, which holds great promise towards developing on-site and at-home rapid SARS-CoV-2 infection screening and COVID-19 prognosis.
- This article is part of the themed collections: Nanoscale and Nanoscale Horizons: Nanodevices and Nanomaterials for printed electronics


 Please wait while we load your content...
Please wait while we load your content...