Programming rigidity into size-defined wireframe DNA nanotubes†
Abstract
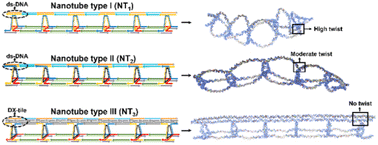
Nanotubes built from DNA hold promise for several biological and materials applications, due to their high aspect ratio and encapsulation potential. A particularly appealing goal is to control the size, shape, and dynamic behaviour of DNA nanotubes with minimal design alteration, as nanostructures of varying morphologies and lengths have been shown to exhibit distinct cellular uptake, encapsulation behaviour, and in vivo biodistribution. Herein, we report a systematic investigation, combining experimental and computational design, to modulate the length, flexibility, and longitudinal patterns of wireframe DNA nanotubes. Subtle design changes govern the structure and properties of our nanotubes, which are built from a custom-made, long, and size-defined template strand to which DNA rungs and linkers are attached. Unlike DNA origami, these custom-made strands possess regions with repeating sequences at strategic locations, thereby reducing the number of strands necessary for assembly. Through strand displacement, the nanotubes can be reversibly altered between extended and collapsed morphologies. These design concepts enable fine-tuning of the nanotube stiffness and may pave the way for the development of designer nanotubes for a variety of applications, including the study of cellular internalization, biodistribution, and uptake mechanisms for structures of varied shapes and sizes.
- This article is part of the themed collection: Emerging concepts in nucleic acids: structures, functions and applications


 Please wait while we load your content...
Please wait while we load your content...