Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
XFCT-MRI hybrid multimodal contrast agents for complementary imaging†
Giovanni Marco
Saladino
 *a,
Carmen
Vogt
a,
Bertha
Brodin
a,
Kian
Shaker
a,
Nuzhet Inci
Kilic
*a,
Carmen
Vogt
a,
Bertha
Brodin
a,
Kian
Shaker
a,
Nuzhet Inci
Kilic
 a,
Kenth
Andersson
a,
Marie
Arsenian-Henriksson
b,
Muhammet Sadaka
Toprak
a,
Kenth
Andersson
a,
Marie
Arsenian-Henriksson
b,
Muhammet Sadaka
Toprak
 a and
Hans Martin
Hertz
*a
a and
Hans Martin
Hertz
*a
aDepartment of Applied Physics, Biomedical and X-Ray Physics, KTH Royal Institute of Technology, SE 10691 Stockholm, Sweden. E-mail: saladino@kth.se
bDepartment of Microbiology Tumor and Cell Biology (MTC), Karolinska Institute, SE 17165 Stockholm, Sweden
First published on 28th December 2022
Abstract
Multimodal contrast agents in biomedical imaging enable the collection of more comprehensive diagnostic information. In the present work, we design hybrid ruthenium-decorated superparamagnetic iron oxide nanoparticles (NPs) as the contrast agents for both magnetic resonance imaging (MRI) and X-ray fluorescence computed tomography (XFCT). The NPs are synthesized via a one-pot polyol hot injection route, in diethylene glycol. In vivo preclinical studies demonstrate the possibility of correlative bioimaging with these contrast agents. The complementarity allows accurate localization, provided by the high contrast of the soft tissues in MRI combined with the elemental selectivity of XFCT, leading to NP detection with high specificity and resolution. We envision that this multimodal imaging could find future applications for early tumor diagnosis, improved long-term treatment monitoring, and enhanced radiotherapy planning.
Introduction
Functional and molecular medical imaging with a single modality presents limitations for early-stage diagnosis due to constraints in tissue penetration depth, sensitivity, specificity, and resolution.1–3 The combination of different imaging techniques with complementary characteristics allows the collection of more comprehensive data and the achievement of more accurate detection. To enable multimodality, nanoparticle- (NP-)based contrast agents with different functionalities may be designed and optimized for use with multiple bioimaging techniques.4–6 Typically, these combine one classical backbone technique – e.g., magnetic resonance imaging (MRI) or positron emission tomography (PET) – with additional optical contrast (e.g., Raman, photoacoustic, fluorescence) providing improved molecular contrast.7–13 Despite their promise, the optical methods suffer from low penetration depth and/or low spatial resolution,14 thereby reducing their applicability for in vivo imaging and often restricting their use to ex vivo tissue analysis.Here, we present complementary multimodal imaging of MRI and X-ray fluorescence computed tomography (XFCT) via a NP dual-mode contrast agent. Both the techniques operate with high penetration depth and exhibit a resolution of ∼100 μm in mice.15,16 MRI is the standard tool for primarily structural and functional in vivo imaging of soft tissue. Superparamagnetic iron oxide NPs (SPIONs) have long been used as negative contrast agents for MRI, although the detection specificity is hampered by several confounding factors.2 XFCT is an emerging in vivo imaging technique with high elemental specificity and multiplexing property, where CT whole-body images are overlaid with the detected X-ray fluorescence (XRF) signal from injected NP contrast agents, enabling functional and molecular imaging.15,17 Recent studies have focused on the design of NP contrast agents for multimodal bioimaging, combining XRF and optical fluorescence for microscopic in vitro and whole-body imaging, where polymer-coated (PVP) Ru NPs were used as XFCT contrast agents, among others.17,18 Although there have been several attempts to design CT-MRI contrast agents to improve the image interpretation,19 both the techniques suffer from low elemental specificity.20 In the present work, we demonstrate complementary XFCT-MRI in preclinical in vivo studies using ruthenium-decorated (Ru-) SPIONs. We demonstrate a similar sensitivity for the two techniques, but the spectral properties of the XFCT signal allows for higher specificity and more accurate localization. Thus, the complementary XFCT-MRI imaging holds promise for high-resolution functional and molecular in vivo imaging for improved diagnostics.21
The key enabler for the XFCT-MRI is the use of Ru-SPIONs as the contrast agent. Generally, the design of dual-mode NP contrast agents includes a multi-step synthesis route, including core formation, decoration, and functionalization with a hydrophilic polymer to improve water dispersibility, which might reduce the reproducibility and unavoidably lead to low-yield reactions.22–25 Furthermore, doping mechanisms and passivation coatings have been demonstrated to directly affect the magnetic performance of MRI contrast agents, due to alternations in the crystal lattice26,27 and reduced proton diffusion and proton-magnetic field interaction,28 respectively. In the present paper, Ru-SPIONs were synthesized in a one-pot polyol synthesis process, resulting in biocompatible NPs, possessing hydrophilic character granted by a polymer (PVP) coating for in vivo XFCT-MRI. The magnetization of the SPIONs was demonstrated to be unaffected by the decoration with Ru NPs. The performance of the contrast agents with the two techniques was evaluated with phantom sensitivity tests and in vivo preclinical studies, highlighting the complementarity of the two techniques.
Results and discussion
Design of the contrast agents
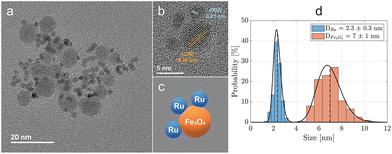
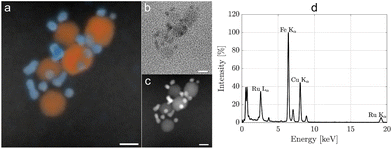
Achieving multimodal XFCT-MRI required a special design of a hybrid nanostructured material presenting an MRI-active magnetic core (SPIONs) and an XRF-active element (Ru) for XFCT, for its absorption edge matches the X-ray source energy. For this scope, Ru-SPIONs were synthesized through a hot-injection polyol route in diethylene glycol (DEG), with PVP as the capping agent (Fig. S1†). Morphological and structural characteristics of the Ru-SPIONs are presented in Fig. 1. Using transmission electron microscopy (TEM), we identified SPIONs and Ru NPs (Fig. 1a), displaying spherical morphology and average diameters of 7 ± 1 nm and 2.3 ± 0.3 nm, respectively (cf. size distributions in Fig. 1d). Ru NPs exhibit a higher contrast in TEM due to their higher atomic number compared to SPIONs (Fe, O). Furthermore, the crystal planes were studied with high resolution- (HR-)TEM in Fig. 1b, where the (002) plane for Ru NPs and (220) plane for SPIONs are identified. In Fig. 2, the energy-dispersive X-ray spectroscopy (EDS) mapping allows the discrimination of Ru NPs and SPIONs. The high-angle annular dark field (HAADF) image underlines the elemental difference between these NPs, due to the Rutherford scattering, yielding brighter intensities for heavier elements (Z-contrast image). To correctly assign the electron diffraction rings, Ru NPs and SPIONs were also synthesized separately using the same procedure (Fig. S2†). SPIONs exhibited similar size distribution, indicating the independence from RuCl3 hot injection. Contrarily, the different size distribution of Ru NPs in the presence or absence of SPIONs confirms a different nucleation mechanism. When Ru3+ was reduced in the presence of SPIONs, it resulted in a faster nucleation kinetics due to the facilitation of heterogeneous nucleation, with reduced surface tension and lower critical radius.29,30 The presence of SPIONs reduced the self-nucleation stage by inducing a faster build-up of supersaturation and, therefore, smaller Ru NPs were obtained. The homogeneous nucleation of Ru NPs in the absence of SPIONs led, instead, to an average diameter of 3.3 nm and a broader size distribution (±0.8 nm). The crystal structures of Ru NPs and SPIONs were analyzed with selected area electron diffraction (SAED), where the main crystal planes are shown (Fig. S3†). Ru NPs displayed hexagonal close-packed (hcp) crystal phase,31,32 while the face-centered cubic (fcc) crystal lattice of SPIONs was confirmed by the observed diffraction rings/peaks of magnetite (or maghemite).33–37 The absence of other diffractions permitted to exclude the existence of other impurity phases (e.g., hematite), which would negatively affect the overall magnetization and relaxivity. Ru-SPIONs exhibited the same crystal phases of the constituents, as the SPION phase and structure are not influenced by the heterogeneous nucleation of Ru NPs.The crystal phase of Ru NPs was mainly dependent on the precursor, rather than the solvent. As previous studies have shown the possibility of obtaining different structures with a polyol synthesis using either RuCl3 in ethylene glycol (hcp) or Ru(acac)3 in triethylene glycol (fcc),31 here we show that RuCl3 in DEG also yields monodispersed Ru NPs with an hcp structure, suggesting that the Ru precursor is the critical factor for the phase selection. The size, morphology, and crystal structure of the SPIONs were not affected by the hot injection process and subsequent decoration with Ru NPs, conserving their magnetite (or maghemite) crystal phase and superparamagnetic performance, thus highlighting some of the advantages of decoration.38 The successful PVP coating was demonstrated with thermogravimetric analysis (TGA, Fig. S4a†), and Fourier transform infrared spectroscopy (FT-IR, Fig. S4b†). FT-IR spectra revealed that the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration band undergoes a shift, from 1673 cm−1 (PVP powder) to 1656 cm−1 in Ru-SPIONs, ascribed to the binding of the carbonyl groups on PVP with the NPs.39 The organic content estimated with TGA is about 33%, in accordance with previous studies.39 Whilst the Ru/Fe ratio is estimated with a combined measurement with XRF and inductively-coupled plasma optical emission spectroscopy (ICP-OES), resulting in a value (∼50% w/w) close to the precursor ratio (∼55% w/w). This difference is attributed to both the unreacted Ru precursor and partial homogeneous nucleation. Magnetic separation was employed in the washing steps, enabling the collection of only the Ru-SPIONs, while excluding unreacted precursors and freestanding Ru NPs in the dispersion (≤10%). The PVP-coated SPIONs formed before the hot injection of RuCl3 allow the heterogeneous nucleation of Ru NPs. Furthermore, the high molecular weight, i.e., longer chain length, of PVP (55 kDa) led to the formation of permanent ensembles of hybrid contrast agents, Ru-SPIONs, as schematically illustrated in Fig. 1c.
O stretching vibration band undergoes a shift, from 1673 cm−1 (PVP powder) to 1656 cm−1 in Ru-SPIONs, ascribed to the binding of the carbonyl groups on PVP with the NPs.39 The organic content estimated with TGA is about 33%, in accordance with previous studies.39 Whilst the Ru/Fe ratio is estimated with a combined measurement with XRF and inductively-coupled plasma optical emission spectroscopy (ICP-OES), resulting in a value (∼50% w/w) close to the precursor ratio (∼55% w/w). This difference is attributed to both the unreacted Ru precursor and partial homogeneous nucleation. Magnetic separation was employed in the washing steps, enabling the collection of only the Ru-SPIONs, while excluding unreacted precursors and freestanding Ru NPs in the dispersion (≤10%). The PVP-coated SPIONs formed before the hot injection of RuCl3 allow the heterogeneous nucleation of Ru NPs. Furthermore, the high molecular weight, i.e., longer chain length, of PVP (55 kDa) led to the formation of permanent ensembles of hybrid contrast agents, Ru-SPIONs, as schematically illustrated in Fig. 1c.
The use of PVP as the capping agent for Ru-SPIONs has outlined its multiple properties in the decoration mechanism, serving as both a passivating biocompatible surface coating, and a scaffold for the formation of covalent ensembles and for the heterogeneous nucleation of Ru NPs on the SPION surface. The magnetic characteristic analysis was performed using a vibrating-sample magnetometer (VSM), to obtain the magnetization curves (Fig. S5a and S5b†), which were then used for the magnetic fitting (Fig. S5c†) to estimate the average size of magnetic cores. The saturation magnetization of the bare SPIONs was estimated as 53.2 emu g−1, in accordance with the literature.35 Ru-SPIONs exhibited a saturation magnetization of 38.4 emu g−1 (normalized to the total inorganic content) and a SPION magnetic diameter of 6.8 nm, with the absence of coercivity (Hc < 1.5 Oe). When normalized to the iron oxide content (Fig. S5b†), the difference in saturation magnetization between SPIONs and Ru-SPIONs is about 2%, which was ascribed to random errors. The magnetic performance of the SPIONs was proven to be unaffected by the Ru decoration, indicating that the hot injection method is preferred over doping co-reduction mechanisms.26 Contrarily to Ru-doped magnetite, the nucleation of Ru NPs on the SPION surface does not induce critical structural modifications which would alter their superparamagnetic characteristic. By comparing the size distribution of SPIONs obtained from the TEM micrographs and magnetic fit, the slightly lower magnetic diameter was ascribed to the magnetic dead layer (MDL) on the NPs.40 Furthermore, the broader magnetic size distribution was in accordance with the previous literature.41 The mathematical derivation of the magnetic diameter can be found in ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42,43
42,43
Imaging performance
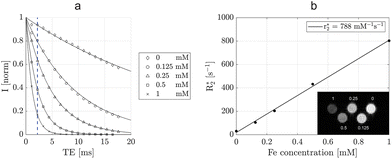
The performance of Ru-SPIONs as MRI contrast agents was evaluated with magnetic relaxivity studies on five samples where Ru-SPIONs or SPIONs were dispersed in agarose with various iron contents (Fig. 3 and S6†).The intensity plots (T2* and T2) as a function of the echo time (TE) at different concentrations of Ru-SPIONs (Fig. 3a and S6a†) exhibited an exponential behavior, with shorter relaxation times at higher concentrations. For example, the concentration of iron –[Fe] is 1 mM, the T2* intensity dropped to zero after 6 ms, due to a strong response to the NPs, compared to agarose ([Fe] = 0 mM).
The five tested concentrations allowed the estimation of the transverse relaxivities (R2* and R2) and demonstrated their linear dependence upon the Fe concentration (Fig. 3b and S6b†).
In particular, the R2 value was estimated as 142 mM−1 s−1, while a value of 788 mM−1 s−1 was found for the transverse relaxivity R2*, more than two folds higher than the commercial contrast agents.44 For this reason, T2*-weighted imaging was pursued for the preclinical studies, to exemplify the use of these contrast agents for complementary imaging. In comparison, the R2* relaxivity of bare SPIONs was estimated as 246 mM−1 s−1, exhibiting a lower value than in Ru-SPIONs (Fig. S7†). This effect was ascribed to the presence of local agglomerations45 caused by the nucleation of Ru NPs on the SPION surface, leading to the increase of local variations in the magnetic field. Contrarily to coating mechanisms (with e.g., silica) on magnetic cores, which then would suffer from diminished MRI performance because of the reduced proton-magnetic field interaction,28,46 the effect of decoration with Ru NPs and subsequent local agglomerations of Ru-SPIONs led to a higher R2* relaxivity.
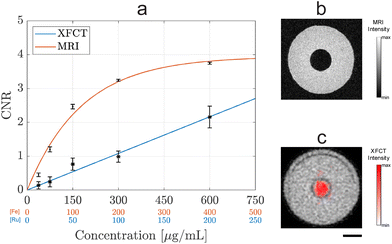
Phantom tests were performed to quantitatively estimate and compare the sensitivity of MRI and XFCT when employing Ru-SPIONs as the contrast agents. To mimic soft tissues, one pipette tip containing Ru-SPIONs in agarose was inserted in a cylindrical holder filled with agarose. In fact, the magnetic relaxation is strongly affected by the molecular environment for MRI imaging; while XFCT is nearly environment-independent, provided with a self-absorption correction algorithm for image processing and reconstruction. Phantoms with different NP concentrations were axially imaged with both MRI and XFCT (Fig. 4 and S8†). The contrast-to-noise ratio (CNR) was estimated within a range of concentrations, for the evaluation of the sensitivity for the two techniques.
Further details on the mathematical derivation of the CNR can be found in ESI,† where a unique definition of CNR was introduced for both MRI and XFCT, permitting to consider the average agarose signal and the Compton scattering, respectively. MRI led to a saturating exponential behavior, with the intensity dropping to zero at sufficiently high contrast agent concentrations.47
On the contrary, XFCT presented a linear response to the NP concentration, within the tested range ([Ru] ≤ 200 μg mL−1). Theoretically, non-linearities might affect higher concentrations – due to XRF signal self-absorption within the NP accumulations – although these were not observed in previous in vivo studies.15 The sensitivity was defined as the minimum contrast agent concentration to yield CNR ≥2, i.e., when the difference between the signal and the background is higher than 95% of the background noise (2σ). The sensitivity in MRI was estimated as [Fe] = 100 μg mL−1, while a concentration of [Ru] = 200 μg mL−1 was the estimation of the XFCT sensitivity for these specific imaging conditions (cf. Experimental section). Nevertheless, the spectral specificity in XFCT isolates the NP signal from false positives (e.g., air pockets in MRI) with structural information provided by the overlaid X-ray absorption data (Fig. 4c). NPs with increased Ru/Fe ratio would lead to a higher XFCT/MRI sensitivity ratio, which can potentially be achieved by increasing the amount of Ru precursor and its injection speed, to tune the Ru NP size and relative number on the SPION surface. Furthermore, the replacement of Ru with, e.g., molybdenum (Mo) as the active XRF element would further increase the XFCT sensitivity due to the lower background from the Compton scattering, as demonstrated in previous studies where sensitivities down to [Mo] = 50 μg mL−1 could be achieved in vivo.15
Biocompatibility
To evaluate the biocompatibility of the synthesized nanostructures, SPIONs, Ru NPs, and Ru-SPIONs were tested in vitro, by exposing RAW 264.7 macrophages, A549 lung carcinoma epithelial cells and primary human mesenchymal stem cells (MSC) to a range of NP concentrations, followed by CellTiter-Glo® (CTG) luminescent cell viability assay, which quantifies cellular ATP levels (Fig. S9†). The tested concentrations were chosen to quantitatively compare the effects of the NPs, in terms of the same metallic elemental composition. In macrophages and mesenchymal stem cells, the bare SPIONs exhibited higher viabilities than the unexposed control cells. The dissolution of iron from SPIONs in endosomal acidic environments after a phagocytic response could bias an over-stimulation by the treatment.48,49 On the contrary, Ru NPs and Ru-SPIONs followed a similar concentration-dependent trend, with above 85% viability at all the tested concentrations. One possible explanation for the absence of overstimulation in Ru-SPIONs is that Ru NPs decorated on the SPION surface would be responsible for a limited and/or slower dissolution rate of the iron oxide. When exposed to A549 cell line, both Ru NPs and SPIONS were slightly cytostatic, with a reduction in cell viability of up to ∼40%. On the contrary, this was not observed in cells exposed to Ru-SPIONs, with viabilities always above 95%. The overall higher nanostructure size yielded a lower cytotoxic effect, as larger NP sizes result in a lower toxicity response, due to reduced surface area (per unit weight) in contact with cells.50Preclinical imaging
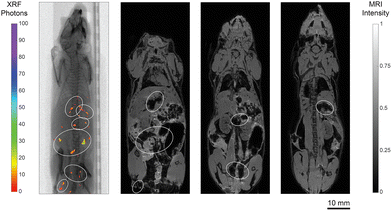
The first part of the animal studies consisted of postmortem full-body imaging of mice to evaluate the performance of Ru-SPIONs as dual-mode contrast agents for correlative imaging. Here, one mouse received Ru-SPIONs through intraperitoneal (IP) injection ([Ru] = 20 mg kg−1, [Fe] = 40 mg kg−1) and was imaged 24 h after the injection. In Fig. 5, the XRF projection image of the mouse highlights local NP accumulations of up to few millimeters in the abdominal area. The XRF projection image is correlated with three selected T2*-weighted multi-gradient-echo 3D (MGE3D) MRI slices, where the Ru-SPIONs were co-localized, and the main agglomerations were highlighted. The spectral specificity of XFCT permits to uniquely localize the XRF active elements, while the effect of SPIONs in lowering the MRI relaxivity is also caused by air-filled bubbles, field inhomogeneity, tissue interfaces, blood clots, endogenous iron accumulation, suppressed fat, among others.2,51 These would unavoidably lead to false positives.Furthermore, the susceptibility artifacts at the air-tissue interface in the lungs and their low proton density limit the use of MRI for related studies.52 Some of these confounding factors could be overcome by using Gd(III)-based positive contrast agents for MRI, although there have been arising concerns over their nephrotoxicity.53,54 Moreover, although a T2 sequence could ameliorate the overall image quality, the T2* sequence did not impede the co-registration of the signal from the contrast agents. On the contrary, XRF signal is not affected by the local chemical environment, as it relies on an inner-shell excitation-relaxation interaction. Thus, it did not lead to limitations in this respect, due to the high spectral specificity of the Ru Kα radiation. In fact, quantitative estimations in organs with XFCT have been shown to accurately match the concentrations obtained via ICP.55
The 3D spatial localization of the features was made possible by the full-body XFCT (Movie S1†), which constituted a complementary analysis to the MRI scan. Finally, XFCT inherently suffers from low soft-tissue contrast, due to the low X-ray absorption, while MRI facilitates the co-registration of NP accumulations and anatomic soft-tissue landmarks. The second animal experiment demonstrated an in vivo complementary XFCT-MRI protocol, using Ru-SPIONs as the contrast agents. Here, four mice were injected with the same NP concentration and imaged sequentially with MRI followed by XRF/XFCT, 24 h (2 mice) or 48 h (2 mice) after injection. Additionally, two other mice were used as untreated controls.
Noticeably, no visible side effects were observed in mice injected with Ru-SPIONs compared to control mice during the experimental period (cf. Experimental section). Furthermore, the presence of 40 cytokines associated with inflammation was investigated with an antibody array, highlighting the absence of cytokine activation in the plasma of injected mice. The corresponding XRF projection images and MRI scans are shown in Fig. S10.† Mice were kept under anesthesia during the scans with the two imaging techniques, and respiratory gating was implemented for MRI to minimize motion artifacts due to the Fourier reconstruction process.56 Images at different times (24 or 48 h) highlighted the absence of macroscopic differences in NP distribution.
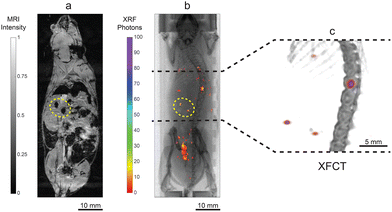
Nevertheless, the lower abdominal region was affected by artifacts due to the presence of gas bubbles in the intestine, decreasing the specificity of MRI in this area. In Fig. 6, we present a combination of in vivo imaging with full-body MRI (Fig. 6a) and XRF projection imaging (Fig. 6b), together with a local XFCT (Fig. 6c and Movie S2†). Here, the correlation is demonstrated focusing on two local accumulations, marked with yellow dashed circles.
The IP injection allowed an optimal comparison between MRI and XFCT, as the NPs randomly distributed in the peritoneal area, where small local accumulations were proven to be ideal for demonstrating the imaging correlation and the complementarity of the techniques. In our study, we did not observe any behavioural or morphological side effects, supported by the high cell viabilities in the in vitro cytotoxicity assay. The transition to intravenous injection is considered a further step, since it represents the most common administration route for drug delivery and diagnosis.57,58
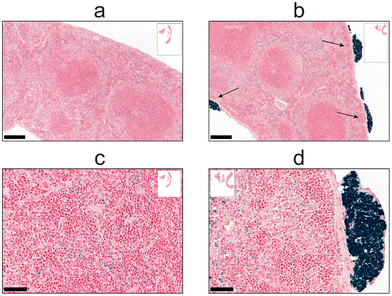
The detected Ru-SPION accumulations could be further confirmed by histochemistry of organ sections from control and injected mice using Prussian blue staining: in the presence of Fe ions, it results in a characteristic blue colour. In mice injected with Ru-SPIONS, we observed particulate and diffused iron in the omentum and diaphragm (Fig. S11†). Major differences between the control and NP-administered mice were also observed in the spleen, where particulate material – accumulations of Ru-SPIONs – surrounded the splenic capsule and diffused into the red pulp as Fe ions, responsible for the scattered blue staining, in proximity of the germinal centres (Fig. 7). The spleen is the major blood filter in mammals with a function in Fe recycling.57 The red pulp is largely composed of chordal macrophages which have the role of regulating the tissue inflammatory response. With their phagocytic function, they were ascribed to be responsible of NP clearance.58,59
While SPIONs are subjected to rapid oxidation and dissolution in lysosomes,60 which might limit their application for long-duration clinical investigations, Ru NPs exhibit a low dissolution rate in acidic environments,61 making them promising for diagnosis and tracking of chronic diseases,62 and long-term monitoring of treatment response.63 Furthermore, the combination with SPIONs might enable a combined Z-enhanced radiotherapy, using Ru NPs as the radiosensitizers and SPIONs for radiation treatment planning.64,65
Conclusions
In this work, we designed multimodal XFCT-MRI contrast agents and demonstrated their potential for complementary imaging. Ru-SPIONs were obtained via a single-pot hot injection polyol synthesis, leading to biocompatible nanostructured contrast agents. The design of multimodal hybrid contrast agents with independent identities through decoration grants minimal interference between the two NP kinds, whereas doping and coating mechanisms unavoidably alter the magnetization properties. Here, the unaffected SPION magnetization granted a high contrast with MRI, while Ru constituted the XRF-active element for XFCT. Phantom tests on the imaging performance, cytotoxicity assay with three different cell lines and in vivo preclinical studies proved the potential of Ru-SPIONs as contrast agents for XFCT-MRI. The detection of MRI contrast agents suffers from several confounding factors, potentially leading to false positives. The combination of a high contrast for soft tissues with MRI with the elemental selectivity and multiplexing property of XFCT could be advantageous for early-tumor diagnosis, where high specificity and sensitivity are a necessity.Experimental
Materials
Ruthenium(III) chloride hydrate (RuCl3·H2O, Ru 40%), iron(III) acetylacetonate (Fe(acac)3, >99.9%), diethylene glycol (DEG, (HOCH2CH2)2O, ≥99%), poly(vinyl pyrrolidone) (PVP, C2H2N(C6H9NO)nC13H10NS2, average MW = 55 kDa), acetone, Agarose with low gelling temperature, Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), lung carcinoma cells (A549, 86012804), and murine macrophages (RAW 264.7, 91062702) were all purchased from Sigma Aldrich (Sweden). Primary human mesenchymal stem cells (MSC) derived from umbilical cord were commercially obtained from PromoCell Mediqip AB (Denmark).Nanoparticle synthesis
Ru-SPIONs were synthesized via a hot injection polyol method. Briefly, Fe(acac)3 (2 mmol) and PVP (5 mmol, repeating units) were dissolved in DEG (20 mL) in a three-neck flask, while magnetically stirring. The solution was heated up to 120 °C and kept for 15 min to evaporate traces of water. Subsequently, the system was brought to the refluxing temperature and let react for 2 h. The dispersion color turned from orange to black. A solution of RuCl3 (0.5 mmol) was prepared in a mix of DEG (4.5 mL) and water (0.5 mL) and introduced to the system via injection (1 mL min−1). The dispersion was kept reacting for 2 h at the refluxing temperature. Subsequently, the system was cooled down to room temperature. The dispersion was then collected and the as-synthesized Ru-SPIONs were washed with acetone and water employing magnetic separation. The prepared stock was stored at 4 °C for further use. Similarly, SPIONs and Ru NPs were independently prepared using the same protocol, but only in the presence of the corresponding precursor and PVP. These were instead washed and collected by repeated centrifugations.Nanoparticle characterization
Ru-SPIONs, SPIONs and Ru NPs were characterized with several techniques to evaluate their morphological and functional characteristics. Size and morphology were estimated with TEM (JEM-2100F, 200 kV, JEOL): 10 μL of diluted samples were drop-casted on copper grids and dried at room temperature. The diameter of at least 200 NPs was measured for evaluation of the size distribution. SAED and HR-TEM were employed for the determination of the crystal structure of the synthesized NPs. STEM-EDS analyses were performed with a FEI Titan Themis 200 with probe corrector and SuperX EDS (Thermo Scientific).FT-IR (ThermoFisher Scientific) and TGA (TGA550, TA Instruments) studies were conducted for the identification and quantification of the organic content on the NPs. Concentration of Fe was determined via ICP-OES (iCAP 6000 series, Thermo Scientific), as 13.9 mg mL−1, after dissolving the SPIONs in HCl. Given the slow dissolution rates of Ru NPs in acids, often requiring microwave digestion processing,61 the Ru concentration, therefore, was estimated with XRF (Ru Kα), by preparing 10× diluted dispersions of Ru NPs, Ru-SPIONs, Ru standard solution (1000 ppm) and water in 2 mL centrifuge tubes. The acquired X-ray spectra were then utilized to estimate the NP concentration in relation to the Ru standard (1000 ppm), leading to a stock concentration of 6.9 mg mL−1.17,18
For the magnetization measurements, a fixed volume (200 μL) with a known concentration ([Fe] + [Ru]) of NPs was dried (35 °C) in 0.2 mL centrifuge tubes. The tubes were directly used in the vibrating-sample magnetometer (VSM, EG&G model 155, Princeton Applied Research), with magnetic fields up to ±8 kOe. The measurement was started and ended at positive saturation, completing a loop. The saturation magnetization was estimated by fitting the experimental results up to infinite field. To obtain the magnetic size distribution, a domain magnetization of 446 emu per cc was used for iron oxide,66 and a lognormal distribution was assumed for the NPs.67 The NP stocks were tested for lipopolysaccharides (LPS) contamination, employing the LAL assay Endosafe-PTS (Charles River) and PTS cartridges (sensitivity of 0.005 EU mL−1). All the stocks used for in vitro and in vivo studies presented LPS values below the maximum admissible limit of 0.1 EU mL−1.
Relaxivity tests
Phantom experiments with syringes filled with agarose containing several concentrations of SPIONs and Ru-SPIONs were prepared. A dispersion with agarose in water (3%) was heated to 50 °C while magnetically stirring. Once the dispersion turned fully transparent, the corresponding amount of Ru-SPIONs was introduced, to obtain a final iron concentration of 0.125, 0.25, 0.5 and 1 mM. One dispersion without NPs was prepared as the control. After 10 min, the warm dispersions were aspirated in 1 mL syringes, which were sealed after cooling down. The MRI relaxivity of the NPs was performed using a preclinical MRI 9.4T system (Bruker). The transverse relaxivities (R2 and R2*) were determined using the spin-echo multi-slice (sems) and gradient-echo multi-slice (gems) sequences, respectively, at different echo times (TE). The relaxivity was plotted as a function of the iron concentration.Sensitivity studies
Cylindrical polylactic acid (PLA) holders were 3D-printed (Ultimaker 2) with a 2 cm diameter (Fig. 6A) to contain a 200 μL pipette tip surrounded by agarose (Fig. 6b). The pipette tip was filled with agarose at several concentrations of Ru-SPIONs. The phantoms were subsequently scanned with MRI and XFCT. T2*-Weighted MRI images of the phantoms were acquired (TE = 1.15 ms) with the multi-gradient echo 3D (mge3d) sequence (∼2 min per slice), with a voxel size of 200 μm. XFCT images with the same voxel size and acquisition times were acquired with our laboratory XFCT arrangement (pencil-beam mode), employing a liquid metal-jet X-ray source coupled to a W–C-coated multilayer Montel mirror. A 3-element photon-counting silicon-drift detector (SDD) was used for XRF signal detection (Ru Kα) and a single SDD for transmitted X-rays.15 For each pixel, a full X-ray spectrum is acquired; photons detected within 19–19.45 keV are associated with Ru Kα radiation. The CNR for both MRI and XFCT was estimated, and an appropriate fit was performed to estimate the sensitivity. A detailed description can be found in the ESI.†Cytotoxicity assays
To assess the in vitro NP biocompatibility, the CellTiter-Glo® luminescent cell viability (CTG) assay was performed on RAW264.7, A549, and MSC cell lines, in quadruplicates (96-well plate). The cells were exposed to three different concentrations of SPIONs, Ru NPs, and Ru-SPIONs for 48 h. Untreated cells were the negative control. The viability estimated was based on the quantification of the ATP, an indicator of metabolically active cells. The luminescence signal was normalized to the control cells.Animal studies
Experiments with mice were approved by the regional animal ethics committee of Northern Stockholm, Sweden (ethical permit numbers 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 579–2020 and 15
579–2020 and 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 099–2021), according to institutional and European guidelines for animal handling and research (EU Directive 2010/63/EU for animal experiments). Six-week-old female albino mice from BALB/c strain were obtained from Taconic Biosciences (Denmark) and housed under controlled temperature (21 ± 1 °C), and humidity (55 ± 5%) conditions, with light–dark cycle, and ad libitum feeding. The general conditions of the mice were assessed prior to and during the study, checking for possible onsets of behavioral and/or morphological changes. Ru-SPIONs were intraperitoneally (IP) injected with 100 μL NP suspension in PBS ([Ru] = 20 mg kg−1, [Fe] = 40 mg kg−1). In the ex vivo study, one mouse was injected and euthanized after 24 h, before imaging, to demonstrate the correlative bioimaging with XFCT and MRI. For the in vivo experiments, four mice were injected and MRI and XFCT scans were performed sequentially under anesthesia with isoflurane (Abbott, Sweden), either 24 or 48 h after the injection. Two mice were used as control for MRI, with no applied treatment. During the imaging sessions, ophthalmic ointment (Oculentum simplex, APL, Sweden) was applied to the eyes for cornea protection; temperature and respiration were also monitored. Extracted plasma from mice exposed to Ru-SPIONs was tested with an inflammation antibody array (ab133999, Abcam).
099–2021), according to institutional and European guidelines for animal handling and research (EU Directive 2010/63/EU for animal experiments). Six-week-old female albino mice from BALB/c strain were obtained from Taconic Biosciences (Denmark) and housed under controlled temperature (21 ± 1 °C), and humidity (55 ± 5%) conditions, with light–dark cycle, and ad libitum feeding. The general conditions of the mice were assessed prior to and during the study, checking for possible onsets of behavioral and/or morphological changes. Ru-SPIONs were intraperitoneally (IP) injected with 100 μL NP suspension in PBS ([Ru] = 20 mg kg−1, [Fe] = 40 mg kg−1). In the ex vivo study, one mouse was injected and euthanized after 24 h, before imaging, to demonstrate the correlative bioimaging with XFCT and MRI. For the in vivo experiments, four mice were injected and MRI and XFCT scans were performed sequentially under anesthesia with isoflurane (Abbott, Sweden), either 24 or 48 h after the injection. Two mice were used as control for MRI, with no applied treatment. During the imaging sessions, ophthalmic ointment (Oculentum simplex, APL, Sweden) was applied to the eyes for cornea protection; temperature and respiration were also monitored. Extracted plasma from mice exposed to Ru-SPIONs was tested with an inflammation antibody array (ab133999, Abcam).
MRI
For the ex vivo study, T2*-weighted MRI full-body images were acquired (TE = 1.4 ms, TR = 50 ms, TI = 100 ms) with the mge3d sequence (200 μm voxel size) for a total scanning time of 4 h, using the preclinical MRI 9.4T system (Bruker). In vivo imaging was performed with the gems sequence (TE = 2.3 ms, TR = 250 ms, TI = 60 ms) with a 16 min scanning time and voxel size of 200 × 200 × 600 μm. Respiratory gating was employed for the imaging.XRF/XFCT imaging
For the ex vivo experiment, a whole-body tomography was obtained in 7.5 h by acquiring XRF projection images with 30 rotation angles (15 min per projection) with pixel size of 200 × 200 μm. For in vivo imaging, the mice previously scanned with MRI were re-anesthetized for a single whole-body XRF projection image and local (1 cm vertical region) XFCT (45 min) with a voxel size of 200 × 200 × 400 μm. Both MRI and XRF/XFCT were performed within 24 h.Histological analysis
At the imaging endpoint, the mice were euthanized. Spleen and diaphragm samples were dissected and fixed in buffered paraformaldehyde (PFA) and embedded in paraffin. Formalin-fixed paraffin embedded (FFPE) sections of ≈ 10 μm were stained with hematoxylin and eosin (H&E) and Prussian Blue.Author contributions
GMS, MST, HMH conceived the ideas. GMS, NIK designed the nanoparticles. GMS synthesized and characterized the nanoparticles. GMS, KS, CV designed the phantom tests. BB, GMS, CV designed, performed, and analyzed the in vitro analyses. GMS, CV, BB, KS, KA performed the in vivo imaging experiments. GMS processed the images. MAH was the holder of the ethical permit and provided expertise in animal experiments. MST, HMH supervised the work. GMS wrote the original draft, assisted by HMH and MST. All authors have reviewed and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project received funding from the Knut and Alice Wallenbergs Foundation (KAW 2016.0057). The MRI scanning was performed at the Department of Comparative Medicine/Karolinska Experimental Research and Imaging Centre at Karolinska University Hospital, Solna, Sweden. We thank Ann-Christin Sandberg Nordqvist, Patrik Jarvoll, and Peter Damberg for the fruitful discussions. The material characterization was made possible by accessing the facilities in Stockholm University (MMK), Uppsala University (Ångström Microstructure Laboratory) and KTH Royal Institute of Technology (Hultgren Laboratory).References
- S. Shaikh, F. U. Rehman, T. Du, H. Jiang, L. Yin, X. Wang and R. Chai, ACS Appl. Mater. Interfaces, 2018, 10, 26056–26063 CrossRef.
- V. I. Mikla, Med. Imaging Technol., 2013, 1–141 Search PubMed.
- L. Fass, Mol. Oncol., 2008, 2, 115 CrossRef PubMed.
- Y.-C. Lin, E. Perevedentseva, Z.-R. Lin, C.-C. Chang, H.-H. Chen, S.-M. Yang, M.-D. Lin, A. Karmenyan, G. Speranza, L. Minati, C. Nebel and C.-L. Cheng, Sci. Rep., 2022, 12, 1–12 CrossRef PubMed.
- S. M. Park, A. Aalipour, O. Vermesh, J. H. Yu and S. S. Gambhir, Nat. Rev. Mater., 2017, 2, 1–20 Search PubMed.
- E. K. H. Chow and D. Ho, Sci. Transl. Med., 2013, 5, 216 Search PubMed.
- X. Li, X. N. Zhang, X. D. Li and J. Chang, Cancer Biol. Med., 2016, 13, 339 CAS.
- M. F. Kircher, A. de La Zerda, J. v. Jokerst, C. L. Zavaleta, P. J. Kempen, E. Mittra, K. Pitter, R. Huang, C. Campos, F. Habte, R. Sinclair, C. W. Brennan, I. K. Mellinghoff, E. C. Holland and S. S. Gambhir, Nat. Med., 2012, 18, 829–834 CrossRef CAS PubMed.
- J. v. Jokerst, C. Khademi and S. S. Gambhir, Sci. Transl. Med., 2013, 5, 177 Search PubMed.
- E. Phillips, O. Penate-Medina, P. B. Zanzonico, R. D. Carvajal, P. Mohan, Y. Ye, J. Humm, M. Gönen, H. Kalaigian, H. Schöder, H. W. Strauss, S. M. Larson, U. Wiesner and M. S. Bradbury, Sci. Transl. Med., 2014, 6, 260 Search PubMed.
- A. Singh and M. M. Amiji, Curr. Opin. Biotechnol, 2022, 74, 241–246 CrossRef CAS PubMed.
- W. Li, X. Sun, Y. Wang, G. Niu, X. Chen, Z. Qian, L. Nie, P. C. Li, C. W. Wei, C. Liao, C. Chen, K. Pao, C. Wang, Y. Wu, D. Shieh, S. K. Balasubramanian, J. Jittiwat, J. Manikandan, C. Ong, L. E. Yu, W. Y. Ong, C. H. Lee, S. H. Cheng, Y. J. Wang, Y. C. Chen, N. T. Chen, J. Souris, C. T. Chen, C. Y. Mou, C. S. Yang, L. W. Lo, S. Ye, R. Yang, J. Xiong, K. K. Shung, Q. Zhou, C. Li and Q. Ren, Biomed. Opt. Express, 2014, 5(8), 2679–2685 CrossRef PubMed.
- Q. Li, K. Chen, W. Huang, H. Ma, X. Zhao, J. Zhang, Y. Zhang, C. Fang and L. Nie, Cancer Lett., 2021, 496, 169–178 CrossRef CAS PubMed.
- P. Singh, S. Pandit, V. R. S. S. Mokkapati, A. Garg, V. Ravikumar and I. Mijakovic, Int. J. Mol. Sci., 2018, 19, 7 Search PubMed.
- K. Shaker, C. Vogt, Y. Katsu-Jimenez, R. v. Kuiper, K. Andersson, Y. Li, J. C. Larsson, A. Rodriguez-Garcia, M. S. Toprak, M. Arsenian-Henriksson and H. M. Hertz, IEEE Trans. Med. Imaging, 2020, 1–1 Search PubMed.
- R. W. Brown, Y.-C. N. Cheng, E. M. Haacke, M. R. Thompson and R. Venkatesan, Magnetic resonance imaging: physical principles and sequence design, John Wiley & Sons, Inc., 2nd edn, 2014 Search PubMed.
- G. M. Saladino, C. Vogt, Y. Li, K. Shaker, B. Brodin, M. Svenda, H. M. Hertz and M. S. Toprak, ACS Nano, 2021, 15, 5085 CrossRef.
- G. M. Saladino, N. I. Kilic, B. Brodin, B. Hamawandi, I. Yazgan, H. M. Hertz and M. S. Toprak, Nanomaterials, 2021, 11, 2165 CrossRef CAS PubMed.
- E. N. M. Cheung, R. D. A. Alvares, W. Oakden, R. Chaudhary, M. L. Hill, J. Pichaandi, G. C. H. Mo, C. Yip, P. M. MacDonald, G. J. Stanisz, F. C. J. M. van Veggel and R. S. Prosser, Chem. Mater., 2010, 22, 4728–4739 CrossRef CAS.
- L. Frullano and T. J. Meade, JBIC, J. Biol. Inorg. Chem., 2007, 12, 939–949 CrossRef CAS.
- J. D. Schiffman, P. G. Fisher and P. Gibbs, Am. Soc. Clin. Oncol. Educ. Book, 2015, 57–65 CrossRef PubMed.
- J. Zhu, J. Wang, X. Wang, J. Zhu, Y. Yang, J. Tian, W. Cui, C. Ge, Y. Li, Y. Pan and H. Gu, J. Mater. Chem. B, 2015, 3, 6905–6910 RSC.
- J. A. Hachtel, S. Yu, A. R. Lupini, S. T. Pantelides, M. Gich, A. Laromaine and A. Roig, Faraday Discuss., 2016, 191, 215–227 RSC.
- J. Zhu, Y. Lu, Y. Li, J. Jiang, L. Cheng, Z. Liu, L. Guo, Y. Pan and H. Gu, Nanoscale, 2013, 6, 199–202 RSC.
- M. Wang, C. Wang, K. L. Young, L. Hao, M. Medved, T. Rajh, H. C. Fry, L. Zhu, G. S. Karczmar, C. Watson, J. S. Jiang, N. M. Markovic and V. R. Stamenkovic, Chem. Mater., 2012, 24, 2423–2425 CrossRef.
- B. T. de Gregorio, R. M. Stroud, L. R. Nittler, H. R. Zebardast, S. Rogak, E. Asselin, M. A. Zayed, S. I. El-dek, M. K. Ahmed and D. H. el Sherbiny, Mater. Res. Express, 2020, 7, 125002 CrossRef.
- C. R. de Silva, S. Smith, I. Shim, J. Pyun, T. Gutu, J. Jiao and Z. Zheng, J. Am. Chem. Soc., 2009, 131, 6336–6337 CrossRef.
- Z. Zhao, M. Li, J. Zeng, L. Huo, K. Liu, R. Wei, K. Ni and J. Gao, Bioact. Mater., 2022, 12, 214–245 CrossRef PubMed.
- N. T. K. Thanh, N. Maclean and S. Mahiddine, Chem. Rev., 2014, 114, 7610–7630 CrossRef PubMed.
- J. Polte, CrystEngComm, 2015, 17, 6809–6830 RSC.
- K. Kusada, H. Kobayashi, T. Yamamoto, S. Matsumura, N. Sumi, K. Sato, K. Nagaoka, Y. Kubota and H. Kitagawa, J. Am. Chem. Soc., 2013, 135, 5493–5496 CrossRef CAS PubMed.
- R. G. Ross and W. Hume-Rothery, J. Less-Common Met., 1963, 5, 258–270 CrossRef CAS.
- G. M. Saladino, B. Hamawandi, C. Vogt, G. K. Rajarao and M. S. Toprak, Appl. Nanosci., 2020, 10, 1861–1869 CrossRef CAS.
- G. M. Saladino, B. Hamawandi, M. A. Demir, I. Yazgan and M. S. Toprak, Colloids Surf., A, 2021, 613, 126086 CrossRef CAS.
- D. Maity, P. Chandrasekharan, F. si-Shen, J. M. Xue and J. Ding, J. Appl. Phys., 2010, 107, 09B310 CrossRef.
- W. H. Bragg, Nature, 1915, 95, 561–561 CrossRef.
- C. Pecharromán, T. González-Carreño and J. E. Iglesias, Phys. Chem. Miner., 1995, 22, 21–29 CrossRef.
- L. León Félix, B. Sanz, V. Sebastián, T. E. Torres, M. H. Sousa, J. A. H. Coaquira, M. R. Ibarra and G. F. Goya, Sci. Rep., 2019, 9, 1–11 CrossRef PubMed.
- Y. Li, G. M. Saladino, K. Shaker, M. Svenda, C. Vogt, B. Brodin, H. M. Hertz and M. S. Toprak, Nanomaterials, 2020, 10, 2129 CrossRef CAS.
- J. Curiale, M. Granada, H. E. Troiani, R. D. Sánchez, A. G. Leyva, P. Levy and K. Samwer, Appl. Phys. Lett., 2009, 95, 043106 CrossRef.
- R. Chantrell, J. Popplewell and S. Charles, IEEE Trans. Magn., 1978, 14, 975–977 CrossRef.
- G. M. Saladino, Superparamagnetic Hybrid Microspheres as a Reliable Platform for Bio-functionalization, M.Sc. Thesis, Dept. Appl. Phys., Roy. Inst. Technol., Stockholm, Sweden, 2019 Search PubMed.
- G. M. Saladino , Zenodo, SPfit-Superparamagnetic Fit.
- D. J. Korchinski, M. Taha, R. Yang, N. Nathoo and J. F. Dunn, Magn. Reson. Insights, 2015, 8, 15 Search PubMed.
- H. T. Ta, Z. Li, Y. Wu, G. Cowin, S. Zhang, A. Yago, A. K. Whittaker and Z. P. Xu, Mater. Res. Express, 2017, 4, 116105 CrossRef.
- H. M. Joshi, M. De, F. Richter, J. He, P. v. Prasad and V. P. Dravid, J. Nanopart. Res., 2013, 15, 1448 CrossRef.
- R. D. A. Alvares, D. A. Szulc and H. L. M. Cheng, Sci. Rep., 2017, 7, 15493 CrossRef.
- I. Iavicoli, V. Leso, L. Fontana and E. J. Calabrese, Int. J. Mol. Sci., 2018, 19, 805 CrossRef PubMed.
- H. van der Woude, G. Alink and I. M. C. M. Rietjens, Crit. Rev. Toxicol., 2008, 35, 603–607 CrossRef PubMed.
- Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef PubMed.
- F. Kiessling, M. E. Mertens, J. Grimm and T. Lammers, Radiology, 2014, 273, 10–28 CrossRef PubMed.
- M. Wielpütz and H. U. Kauczor, Diagn. Interv. Radiol., 2012, 18, 344–353 Search PubMed.
- E. Kanal and M. F. Tweedle, Radiology, 2015, 275, 630–634 CrossRef PubMed.
- J. M. Wild, H. Marshall, M. Bock, L. R. Schad, P. M. Jakob, M. Puderbach, F. Molinari, E. J. R. van Beek and J. Biederer, Insights Imaging, 2012, 3, 345–353 CrossRef CAS PubMed.
- J. C. Larsson, C. Vogt, W. Vågberg, M. S. Toprak, J. Dzieran, M. Arsenian-Henriksson and H. M. Hertz, Phys. Med. Biol., 2018, 63, 164001 CrossRef PubMed.
- M. Zaitsev, J. Maclaren and M. Herbst, J. Magn. Reson. Imaging, 2015, 42, 887 CrossRef.
- B. T. T. Pham, E. K. Colvin, N. T. H. Pham, B. J. Kim, E. S. Fuller, E. A. Moon, R. Barbey, S. Yuen, B. H. Rickman, N. S. Bryce, S. Bickley, M. Tanudji, S. K. Jones, V. M. Howell and B. S. Hawkett, Int. J. Mol. Sci., 2018, 19, 205 CrossRef PubMed.
- M. Cataldi, C. Vigliotti, T. Mosca, M. R. Cammarota and D. Capone, Int. J. Mol. Sci., 2017, 18(6), 1249 CrossRef PubMed.
- K. M. Tsoi, S. A. Macparland, X. Z. Ma, V. N. Spetzler, J. Echeverri, B. Ouyang, S. M. Fadel, E. A. Sykes, N. Goldaracena, J. M. Kaths, J. B. Conneely, B. A. Alman, M. Selzner, M. A. Ostrowski, O. A. Adeyi, A. Zilman, I. D. McGilvray and W. C. W. Chan, Nat. Mater., 2016, 15, 1212 CrossRef CAS PubMed.
- E. J. Guggenheim, J. Z. Rappoport and I. Lynch, Nanotoxicology, 2020, 14, 504–532 CrossRef CAS.
- Y. Yang, C. Sun, Y. Ren, S. Hao and D. Jiang, Sci. Rep., 2014, 4, 1–6 Search PubMed.
- Y. Ma, F. Cai, Y. Li, J. Chen, F. Han and W. Lin, Bioact. Mater., 2020, 5, 732–743 CrossRef.
- M. v. Yezhelyev, X. Gao, Y. Xing, A. Al-Hajj, S. Nie and R. M. O'Regan, Lancet Oncol., 2006, 7, 657–667 CrossRef CAS PubMed.
- P. Retif, S. Pinel, M. Toussaint, C. Frochot, R. Chouikrat, T. Bastogne and M. Barberi-Heyob, Theranostics, 2015, 5, 1030 CrossRef CAS.
- M. A. Schmidt and G. S. Payne, Phys. Med. Biol., 2015, 60, R323 CrossRef CAS.
- L. Maldonado-Camargo, M. Unni and C. Rinaldi, Methods Mol. Biol., 2017, 1570, 47–71 CrossRef CAS.
- C. G. Granqvist and R. A. Buhrman, Solid State Commun., 1976, 18, 123–126 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr05829d |
| This journal is © The Royal Society of Chemistry 2023 |