OWL2: a molecular beacon-based nanostructure for highly selective detection of single-nucleotide variations in folded nucleic acids†
Abstract
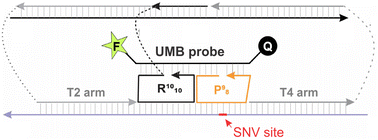
Hybridization probes have been used in the detection of specific nucleic acids for the last 50 years. Despite the extensive efforts and the great significance, the challenges of the commonly used probes include (1) low selectivity in detecting single nucleotide variations (SNV) at low (e.g. room or 37 °C) temperatures; (2) low affinity in binding folded nucleic acids, and (3) the cost of fluorescent probes. Here we introduce a multicomponent hybridization probe, called OWL2 sensor, which addresses all three issues. The OWL2 sensor uses two analyte binding arms to tightly bind and unwind folded analytes, and two sequence-specific strands that bind both the analyte and a universal molecular beacon (UMB) probe to form fluorescent ‘OWL’ structure. The OWL2 sensor was able to differentiate single base mismatches in folded analytes in the temperature range of 5–38 °C. The design is cost-efficient since the same UMB probe can be used for detecting any analyte sequence.
- This article is part of the themed collection: Emerging concepts in nucleic acids: structures, functions and applications


 Please wait while we load your content...
Please wait while we load your content...