Point-of-care nucleic acid tests: assays and devices
Abstract
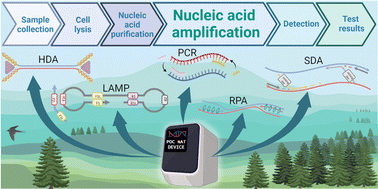
The COVID-19 pandemic (caused by the SARS_CoV_2 virus) has emphasized the need for quick, easy-to-operate, reliable, and affordable diagnostic tests and devices at the Point-of-Care (POC) for homes/fields/clinics. Such tests and devices will contribute significantly to the fight against the COVID-19 pandemic and any future infectious disease epidemic. Often, academic research studies and those from industry lack knowledge of each other's developments. Here, we introduced DNA Polymerase Chain Reaction (PCR) and isothermal amplification reactions and reviewed the current commercially available POC nucleic acid diagnostic devices. In addition, we reviewed the history and the recent advancements in an effort to develop reliable, quick, portable, cost-effective, and automatic point-of-care nucleic acid diagnostic devices, from sample to result. The purpose of this paper is to bridge the gap between academia and industry and to share important knowledge on this subject.
- This article is part of the themed collections: Recent Review Articles and Emerging concepts in nucleic acids: structures, functions and applications


 Please wait while we load your content...
Please wait while we load your content...