Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
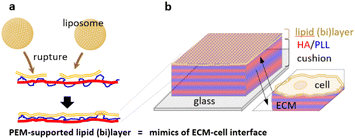
A lipid membrane supported on an artificial extracellular matrix made of polyelectrolyte multilayers: towards nanoarchitectonics at the cellular interface†
Anna
Vikulina
ab,
Alena
Wulf
a,
Guy
Guday
a,
Rawil
Fakhrullin
 *c and
Dmitry
Volodkin
*c and
Dmitry
Volodkin
 *d
*d
aFraunhofer Institute for Cell Therapy and Immunology, Branch Bioanalytics and Bioprocesses (Fraunhofer IZI-BB), Am Mühlenberg 13, 14476 Potsdam, Germany
bFriedrich-Alexander-Universität Erlangen-Nürnberg (FAU), Bavarian Polymer Institute, Dr.-Mack-Straße 77, 90762 Fürth, Germany
cKazan Federal University, Institute of Fundamental Medicine and Biology, Kreml uramı 18, Kazan, Republic of Tatarstan 420008, Russian Federation. E-mail: kazanbio@gmail.com
dNottingham Trent University, School of Science and Technology, Clifton Lane, Nottingham NG11 8NS, UK. E-mail: dmitry.volodkin@ntu.ac.uk
First published on 19th December 2022
Abstract
To implement a specific function, cells recognize multiple physical and chemical cues and exhibit molecular responses at their interfaces – the boundary regions between the cell lipid-based membrane and the surrounding extracellular matrix (ECM). Mimicking the cellular external microenvironment presents a big challenge in nanoarchitectonics due to the complexity of the ECM and lipid membrane fragility. This study reports an approach for the assembly of a lipid bilayer, mimicking the cellular membrane, placed on top of a polyelectrolyte multilayer cushion made of hyaluronic acid and poly-L-lysine – a nanostructured biomaterial, which represents a 3D artificial ECM. Model proteins, lysozyme and α-lactalbumin, (which have similar molecular masses but carry opposite net charges) have been employed as soluble signalling molecules to probe their interaction with these hybrids. The formation of a lipid bilayer and the intermolecular interactions in the hybrid structure are monitored using a quartz crystal microbalance and confocal fluorescence microscopy. Electrostatic interactions between poly-L-lysine and the externally added proteins govern the transport of proteins into the hybrid. Designed ECM–cell mimicking hybrids open up new avenues for modelling a broad range of cell membranes and ECM and their associated phenomena, which can be used as a tool for synthetic biology and drug screening.
1. Introduction
Fabrication of mammalian cell-like functional materials from the nano- to macroscopic scale is regarded as the primary and most ambitious goal of nanoarchitectonics.1 The construction of artificial cells opens up avenues for many potential applications, such as nano- and micromotors and chemical synthesis, e.g., the polymerase chain reaction, RNA polymerization, and protein synthesis.1–3 Simplified mimics of biological cells provide another application, which is widely employed for addressing fundamental questions related to cell and molecular biology.1,4 A few remarkable examples, which represent recent progress in the fabrication of artificial cells, are the use of giant liposomes,5 self-assembly of cells,6 hydrogel microspheres encapsulating exosomes,7 porous polymer capsules,8 artificial reaction promoters,9etc. Proposed approaches enable the reconstitution of many cellular features and functions, e.g. release of specific exosomes7 or acetyl-transfer reactions in mitochondria.9 In terms of dimensionality, one can distinguish between 3D vesicle-based approaches for the fabrication of artificial cells and nanoarchitectonics with 2D materials proposed as a tool to build liquid–liquid interfaces.4It is noted that in the mimicking systems mentioned above and many others, cell–cell interactions are disregarded as they are not required in order for the functions to be reproduced. However, the cells do not exist in isolation. The communication between cells remains a key feature of life, and advanced mimics of artificial cells should take into consideration how they conduct dialog and behave at a population level.10 For instance, cell–cell junctions govern the final tissue pattern11 and self-healing12 of epithelium. Mimicking the cellular interface has been addressed in several studies.12–15 Although native cell–cell junctions appear in a 3D context and are often spatially orthogonal to the matrix plane, traditionally, artificial cell–cell junctions are produced in a 2D configuration (e.g. patterned proteins on a glass slide as reviwed in ref![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). Possibly, the first approaches proposed towards a more realistic model system to mimic cellular interfaces have been focused on the recapitulation of the asymmetry of the biological membrane, which has a different environment from those of the inner and outer sides of the cell. These approaches are reviewed in ref. 16. Recently, the assembly of a spatial network of multiple adjacent vesicles has been proposed to mimic cell–cell communication.17–19 For instance, a double membrane was formed at the interface between two adjacent giant vesicles using optical tweezers.17 However, to the best of our knowledge, none of these studies takes into account the role of the ECM in the intercellular communication.
10). Possibly, the first approaches proposed towards a more realistic model system to mimic cellular interfaces have been focused on the recapitulation of the asymmetry of the biological membrane, which has a different environment from those of the inner and outer sides of the cell. These approaches are reviewed in ref. 16. Recently, the assembly of a spatial network of multiple adjacent vesicles has been proposed to mimic cell–cell communication.17–19 For instance, a double membrane was formed at the interface between two adjacent giant vesicles using optical tweezers.17 However, to the best of our knowledge, none of these studies takes into account the role of the ECM in the intercellular communication.
A polyelectrolyte layer-by-layer (LbL) assembly allows the formulation of polymer-based films or so-called polyelectrolyte multilayers (PEMs) with a high level of control over film properties such as thickness, mechanical properties and composition, which includes controlled loading of bioactive functional molecules.20 PEMs provide unique control over the nanoarchitecture of surface polymer-based structures and therefore are considered as one of the most promising structures for the reconstitution of an artificial ECM. Special attention has been paid to a biopolymer pair of poly-L-lysine (PLL) and hyaluronic acid (HA). The interest in these biopolymers has arisen because HA/PLL films are typically biocompatible, biodegradable, and can be grown in a so-called exponential regime, which results in films reaching micrometre dimensions in thick PEM stacks to provide an exclusively high loading capacity for biomolecules.21,22 A few previous studies investigated the integration of liposomes into polyelectrolyte multilayers (PEMs) made of PLL and HA.23–25 Therein, large unilamellar vesicles remained intact and served as carriers for drug encapsulation, protection and localization.
Herein, we proposed an approach for the formation of a lipid bilayer on the surface of HA/PLL multilayers as the mimic of a cell–ECM interface (Fig. 1). The lipid coating was deposited on an HA/PLL multilayer cushion from the stock of a liposome suspension. In this hybrid layered structure, the layer of lipid mimics the membrane of the cell while the PEM mimics the ECM. This study specifically focuses on control over liposome rupture when being supported on the PEM cushion, the architecture of the lipid layer, and the inward transport of two model proteins, lysozyme and α-lactalbumin, through the lipid (bi)layer into the PEMs.
2. Experimental
2.1. Materials
Sodium hyaluronate (HA, Lifecore Biomedical, 360 kDa), poly-L-lysine HBr (PLL, Sigma), PLL of 15–30 kDa (PLL28, where the average Mw is 28 kDa) and PLL of 150–300 kDa (PLL280) were used. Poly(ethylenimine) (PEI, Sigma), 2-oleoyl-1-palmitoyl-sn-glycero-3-phosphocholine (POPC, Avanti Polar Lipids, Mw: 785.6), 2-oleoyl-1-palmitoyl-sn-glycero-3-phospho-rac-(1-glycerol) sodium salt (POPG, Avanti Polar Lipids, Mw: 771.0), N-(NBD-aminododecanoyl)-1,2-ditetradecanoyl-sn-glycero-3-phosphoethanolamine sodium salt (NBD-DMPE, Avanti Polar Lipids, Mw: 816.0), cholesterol (Chol, Sigma, Mw: 386.7), lysozyme (chicken egg white, CalBioChem, Mw: 14.3 kDa), and α-lactalbumin (bovine milk, type II, calcium depleted, Sigma, Mw: 14.2 kDa) were also used. 10 mM Tris buffer solution containing 15 mM NaCl at pH 7.4 and 0.1 M carbonate buffer solution at pH 9.0 were prepared using distilled water and chemicals purchased from Sigma. Other chemicals and reagents were of analytical grade.Chemical structures of the lipids and polymers used in this study are provided in Fig. S1.†
2.2. Deposition of polyelectrolyte multilayers
PEMs were prepared via the LbL technique using a dipping robot as described elsewhere.26 Deposition was performed on 12 mm round microscopy coverslips (Menzel, Germany) cleaned with a 2% Hellmanex solution followed by 2× rinsing with 1 M HCl and 2× washing with distilled water at 60 °C for 15 min in each step. Subsequently, coverslips were immersed in 1 mg ml−1 PEI in Tris buffer solution for 10 min followed by 24 polyanion and polycation cycles over 10 minutes to deposit layers from 0.5 mg ml−1 polyelectrolyte solution at room temperature. After each step of polymer deposition, the coverslips were 3× washed with Tris buffer solution (3 min each step). The prepared PEMs were stored in Tris buffer solution at 4 °C and used within one week. PEMs used in this study were composed of 24 bilayers of PEI–(HA/PLL15–30)24 and PEI–(HA/PLL150–300)24 (further denoted as HA/PLL28 and HA/PLL280 corresponding to 15–30 kDa and 150–300 kDa PLL median molecular weights of 28 and 280 kDa, respectively, as determined in a previous study).27 HA/PLL280 films were deposited on 100 μm diameter fiber glass following the same procedure as described above but at a temperature of 37 °C as described in ref. 26.2.3. Liposome fabrication
Unilamellar liposomes were prepared as described previously28 with slight modifications. Briefly, 10.5 mg of lipids were dissolved in 1 ml of a chloroform/ethanol mixture 1/1v in a 50 ml round bottom flask. The solvent was evaporated at 40 °C with intensive rotation under 75 mbar for 2 hours. The obtained thin lipid film was maintained under vacuum overnight at room temperature. Then, the lipid film was hydrated at 52–57 °C by the addition of 2 ml of Tris buffer solution. The obtained suspension was subjected to 11 freeze–thaw cycles – quenching in liquid nitrogen for 8 min followed by heating in water at 52–57 °C for 15 min. Finally, liposomes were extruded 10 times through 400 nm polycarbonate membranes and afterwards 10 times through 100 nm polycarbonate membranes using an Avanti mini-extruder at 52–57 °C. The liposome suspension was stored at 4 °C. The two types of prepared liposomes were denoted POPC/POPG/Chol/NBD-DMPE 7/2/1/0.5w and POPC/Chol/NBD-DMPE 9/1/0.5w and further referred to as PC–PGNBD and PCNBD liposomes, respectively.2.4. Lipid deposition on top of PEMs
PEMs supported on glass (plane and fiber) were exposed to 300 μl of liposome suspension in Tris buffer solution. Total lipid concentrations of 20, 50, 100 and 200 μM in PC–PG liposome suspensions were used for the preparation of HA/PLL28 films. The 50 μM concentration was chosen for further deposition of PC–PG and PC liposomes on HA/PLL280 films. Quantitative assessments of adsorbed lipids was performed by measurement of a decrease in the fluorescence of the supernatant by means of fluorescent spectroscopy using excitation and emission wavelengths at 474 and 532 nm, respectively.2.5. Protein labelling
Lysozyme and α-lactalbumin were conjugated with fluorescein isothiocyanate (FITC) in a molar dye![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein ratio of 1
protein ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. The labelling reaction was carried out for 4 hours using 2 mg ml−1 of protein dissolved in carbonate buffer solution. Dialysis against water (2×) and Tris buffer solution (1×) was carried out to purify the proteins. The labelling ratio was calculated from the absorbance of the FITC–protein conjugates at 280 and 495 nm.
5. The labelling reaction was carried out for 4 hours using 2 mg ml−1 of protein dissolved in carbonate buffer solution. Dialysis against water (2×) and Tris buffer solution (1×) was carried out to purify the proteins. The labelling ratio was calculated from the absorbance of the FITC–protein conjugates at 280 and 495 nm.
2.6. Protein interaction with the lipid membrane supported on PEMs
HA/PLL280 films were coated with lipids by incubating them in 50 μM PC–PGNBD liposome suspension for 1 minute and subsequently washing with Tris buffer solution. Uncoated and lipid-coated PEMs were incubated with 0.5 g l−1 FITC-lysozyme or α-lactalbumin solution in Tris buffer solution for 30 min.2.7. Dynamic light scattering (DLS)
The hydrodynamic radii of the liposomes were determined by DLS using a Zetasizer Nano ZS (Malvern Instruments Limited, Worcestershire, UK) from 10 parallel measurements using intensity weighted profiles.2.8. Confocal laser scanning microscopy (CLSM)
CLSM experiments were performed using a Zeiss LSM 510 Meta installation (Zeiss, Germany) equipped with an oil-immersion 63×/NA1.4 objective to record optical transmittance and fluorescence images. Standard filter settings for the excitation and emission of FITC were applied for laser sources with a wavelength of 488 nm. All CLSM experiments were performed at room temperature. Analysis of the CLSM images has been completed by means of ImageJ and an LSM Image Browser.To monitor lipid deposition, fluorescence has been monitored by means of z-stack CLSM. The diffusion coefficients of fluorescently labelled lipids were estimated from fluorescence recovery after photobleaching.
Both lipids and proteins were conjugated with fluorophores having green fluorescence. Therefore, in order to monitor protein interactions, first the fluorescence originating from lipids supported on PEMs was bleached over a large region of interest (ROI), 150 × 150 μm2, by short-time exposure to a 488 nm laser at 100% intensity. Uncoated PEMs were subjected to the same procedure. Then, the protein solution was added to uncoated/coated PEMs and the fluorescence originating from FITC–proteins was monitored by z-stack CLSM. The signal was calculated at a distance of at least 100 μm from the unbleached region of the PEM, which allowed the diffusion of fluorescently labelled lipids to be neglected.
2.9. Quartz crystal microbalance (QCM) measurements
For QCM measurements, PEMs were assembled in a temperature-controlled eCell T microchamber (Hettich, Germany) connected to an eQCM 10M (Gamry, USA). All solutions were introduced into the microchamber with an nEMESYS 290N syringe pump (Cetoni GmbH, Germany). Depositions were started with a pre-treatment step using PEI for 12 minutes, followed by a repetitive cycle of rinsing with Tris buffer (8 min), polyanion adsorption (12 min), rinsing with Tris buffer (8 min), and polycation adsorption (12 min). All depositions were performed at 25 °C, with a net flowrate of 50 μl min−1. QCM results were collected using Gamry Resonator software. For the treatment of data, the Sauerbrey equation was used to determine the deposited mass based on the parallel frequency (fp).3. Results and discussion
3.1. Factors controlling PEM–liposome interactions
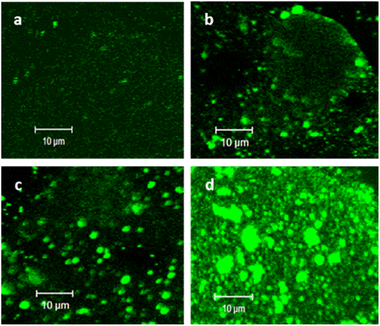
PC–PG liposomes labelled with cap-conjugated NBD-phosphoethanolamine were prepared using an extrusion method. The hydrodynamic radii of the obtained liposomes were measured by DLS (56 ± 17 nm). (HA/PLL)24 was assembled using the LbL technique on round glass slides and the accessible surface of the PEMs was ca. 63.6 mm2. The total amount of liposomes for the formation of an HA/PLL-supported lipid bilayer can be roughly estimated at ca. 0.6 nmol (total lipids), which corresponds to the concentration of liposomes of 2 μM under the settings of experiments in this study. Herein, the surface area of the POPC head group in the bilayer was taken to be ∼0.7 nm2.29 The range of liposome concentrations has been chosen based on this estimation. PC–PGNBD liposomes were adsorbed on (HA/PLL)24 PEMs assembled from high-molecular-weight HA and low-molecular-weight PLL (PLL28) from 20, 50, 100 and 200 μM PC–PGNBD suspensions, corresponding to a total amount of lipids of 6, 15, 30 and 60 nmol, respectively (Fig. 2). As controls, PEM films were scratched with a sharp needle to show an area with no PEMs, which is clearly visible under transmitted light (Fig. S2†).These results suggest that liposomes aggregate by their interaction with PLL on the top of the PEM cushion, and the degree of aggregation is enhanced with an increase of liposome concentration. Apparently, the aggregation originates from the electrostatic attraction of PC–PGNBD to PLL. A similar aggregation occurred upon the addition of PLL to the suspension of PC–PGNBD, while no aggregation in the presence of HA was observed (data not shown). Liposome–PLL aggregates are predominantly formed on the surface of the PEMs.
However, one can observe the formation of liposome–PLL aggregates far from the surface of the PEMs (Fig. S3†), which points to the diffusion of PLL from the PEM to the surrounding medium. PLL28 is highly diffusive (mobile) in the HA/PLL multilayers and HA (360 kDa) is immobile.30
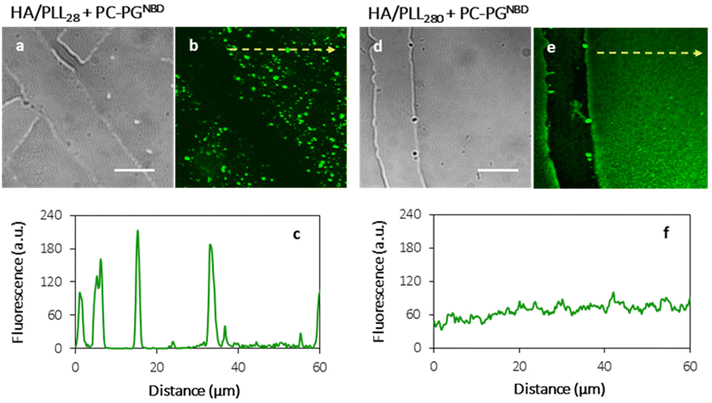
In order to prevent the aggregation of liposomes with PLL and the consequent depletion of (HA/PLL)24 films caused by the diffusion of PLL out of the PEM, two approaches have been proposed and verified (Fig. 3). Apparently, the interplay between the affinity of PLL to HA and PLL to the liposomes could be altered either by the PEM or by the liposome composition. In the first scenario, mobile PLL28 has been replaced with the longer polymer, i.e., PLL280 (Fig. 3). This allows reduction of the polymer molecular mobility, preserving the chemical nature of interactions in the system. A clear suppression of (PC–PGNBD)/PLL aggregation results in the formation of a homogeneous lipid layer on top of the HA/PLL280 cushion. These results suggest that polymer molecular mobility is a key factor influencing liposome adsorption. In its turn, replacing anionic PC–PGNBD liposomes with PCNBD liposomes, which have the same radii (64 ± 19 nm, measured by DLS) but contain no anionic phospholipids, did not provide additional improvement of the lipid layer morphology (Fig. S4†). Taking into account the negative surface charge of natural cell membranes, modulation of liposome–PEM interactions by means of polymer mobility seems to be more favourable than by means of liposome composition.
3.2. Spatial distribution of the (HA/PLL)24-supported lipid layer
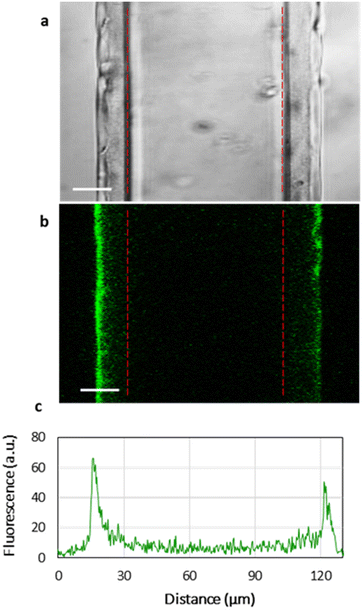
Fig. 3 shows the homogeneous distribution of the lipid layer supported on PEMs in the xy-plane (a top view of the lipid-coated PEM). However, even CLSM resolution in the z-plane is insufficient to visualize the localization of lipids in 3D, since the thickness of the HA/PLL280 film is ca. 1 μm, and it remains unclear whether the lipids penetrate into the entire depth of the PEMs or are retained on top of the multilayers. In order to investigate the spatial distribution of lipids, HA/PLL280 films were deposited on glass fiber instead of flat glass slides (Fig. 4). In this case, HA and PLL form thicker films due to both the cylindrical geometry of the glass support and the elevated temperature upon deposition.26 Orthogonal distribution of lipids is then well resolved in the xy-plane. It is clearly seen that lipids are solely localized on the top of PEMs, and do not penetrate inside the multilayers.In order to confirm the interconnectivity of the lipid coating supported on top of the PEM, the apparent lateral diffusion coefficient (D) of fluorescently labelled lipids in the PC–PGNBD coating was measured based on the profiles of fluorescence recovery after photobleaching. The D value of NBD-PE was found to be (1.3 ± 0.4) × 10−2 μm2 s−1. Fig. S5† shows normalized fluorescence profiles just after the bleaching and after 7 minutes of recovery. This lateral diffusion coefficient is significantly lower than the expected value for the fluid lipid bilayer (10 μm2 s−1). However, it is within the range of 10−2–10−1 μm2 s−1 reported for a lipid bilayer deposited over poly(styrenesulfonate)/poly(allylamine) hydrochloride (PSS/PAH) multilayers.31 This comparison supports the integrity of the lipid coating.
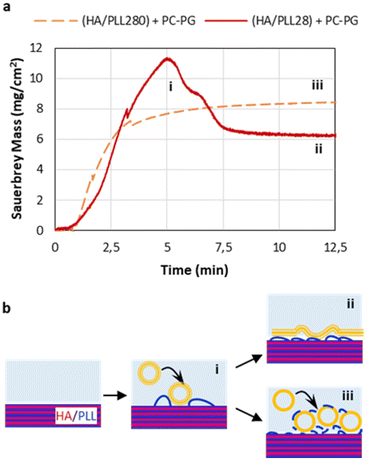
3.3. Liposome rupture
The rupture of liposomes and the formation of a liposome layer on top of the PEMs were further investigated by means of a QCM. For this, short-chain HA/PLL28 and long-chain HA/PLL280 were deposited on a gold-coated quartz crystal and exposed to a 50 μM (total lipids) PC–PGNBD liposome suspension. The adsorbed mass was calculated according to the Sauerbrey equation (Fig. 5a).It was found that the kinetics of liposome adsorption on short-chain HA/PLL28 multilayers obeys the monotonic function. No loss of the adsorbed mass indicates that the liposomes remain intact and do not rupture. Most likely, this is due to the stabilisation of liposomes by means of their aggregation with PLL28, which is known in the literature.28 In contrast, the peak clearly observed for liposome adsorption on HA/PLL280 points to liposome rupture, which is accompanied by a loss of water enclosed in the liposome cavities. Plausibly, the mobility of the PLL molecules plays a role in liposome rupture. Indeed, the diffusion coefficient of the mobile fraction of PLL28 is ca. 0.2 μm2 s−1, and the immobile fraction (D below 10−3 μm2 s−1) is below 30% as determined by fluorescence recovery after photobleaching of HA/PLL28 multilayers.32 Meanwhile, PLL280 is almost completely immobile (>90%).32 The lower diffusivity of PLL280 molecules compared with PLL28 favours PLL retention inside HA/PLL multilayers and hinders the liberation of PLL from the multilayer stack. In this case, the stabilisation of liposomes by PLL does not occur and they rupture on the surface of the PEM cushion. This also supports the results presented in Fig. 3.
It might be useful to compare molecular mobility of PLL in the PEMs with that in the ECM. The majority of work in this field is devoted to the diffusion of guest molecules (mainly proteins). For instance, ref. 33 reports the values of ca. 101–102 μm2 s−1 for the interstitial mobility of albumin and immunoglobulin in different tumour tissues. Diffusion coefficients in the range of 1–10 μm2 s−1 of various macromolecules in the brain extracellular space were also reported.34 To the best of our knowledge, no data on the mobility of ECM components themselves have been reported. It is highly likely that they can be considered as immobile in comparison with the guest molecules, as in the case of an HA/PLL280 system.
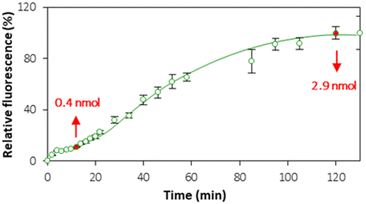
3.4. Lipid bilayer or lipid multilayers?
Although the QCM provided useful information about liposome rupture, the deposition of lipid layers from ruptured liposomes can lead to the formation of either a lipid bilayer or lipid multilayers. In order to establish the settings of bilayer formation that are favorable for cell interface mimics, the kinetics of lipid layer formation was monitored in real time by means of CLSM (Fig. S6†). Fig. 6 shows the kinetics of accumulation of the fluorescence signal originating from PC–PGNBD liposomes at the PEM interface. This kinetic curve includes two plateaus observed at approximately 15–20 and 100–120 minutes.Quantitative analysis of the fluorescence in the PC–PGNBD liposome suspension allowed us to calculate the depletion of the suspensions of 0.4 ± 0.1 and 2.9 ± 0.4 nmol (total lipids) at 15 and 120 minutes of incubation, respectively. This depletion is caused by the adsorption of the lipid layer on top of HA/PLL280 multilayers. It is worth noting the good correlation between the fluorimetry data and the fluorescence signal measured using confocal microscopy (Fig. 6) where the ratio between confocal fluorescence signals at the points of 15 and 120 minutes is ca. 6.2 ± 0.7, while the ratio between PC–PGNBD liposome amounts calculated from the bulk fluorescence values is 7.2 ± 2.7.
Bearing in mind that the amount of lipids required for the formation of a lipid bilayer under the settings of this experiment was estimated as 0.6 nmol, one can anticipate the formation of a lipid bilayer after 15–20 minutes, followed by further adsorption of lipids and the formation of lipid multilayers. After 100–120 minutes this process stops, likely due to a full compensation of the surface charge.
A further remark is that a blue-shift towards lower wavelengths in emission spectra of PC–PGNBD liposomes is observed for both 15 and 120 minutes of incubation with PEMs when compared with control PC–PGNBD liposomes not incubated with the multilayers (Fig. S7†). This blue-shift most likely results from an interaction of PC–PGNBD liposomes with the PEMs.
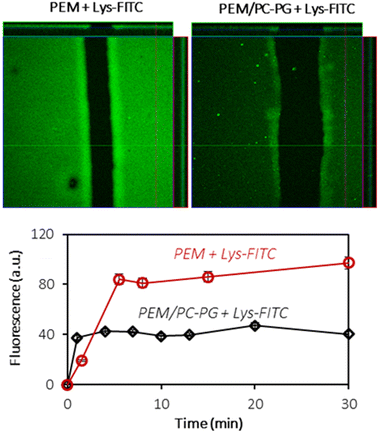
3.5. Proteins interacting with the (HA/PLL)24-supported lipid bilayer
In order to investigate the interactions of the (HA/PLL280)24-supported lipid bilayer with macromolecules, two proteins have been chosen: lysozyme (Lys) and α-lactalbumin (αLA). Both proteins carry the opposite charge at neutral pH: positive and negative net charges for Lys (pI 11) and αLA (pI 4.2), respectively. At the same time both proteins have similar globular structures and sizes (hydrodynamic radius ∼ 2 nm). Protein solutions have been added to PEM–lipid bilayer hybrids and protein inward transport was monitored in real time (Fig. 7 and Fig. S8†).Lys-FITC is able to penetrate into both uncoated and lipid bilayer-coated PEMs. One can reasonably assume that the transport of the protein occurs via the defects in the lipid coating structure.
The equilibrium is reached within less than 10 minutes (Fig. 7). The lipid coating does not prevent the transport of Lys-FITC into the multilayers and the protein is homogeneously distributed within the entire volume of PEMs. However, the supporting lipid bilayer reduces the capacity of PEMs for Lys by ca. 2 times without significant influence on Lys-FITC adsorption kinetics (Fig. 7). The mechanism of Lys transport into the PEM should further be investigated in more detail. It can be supposed that the presence of a lipid coating increases the adsorption energy required for Lys to anchor the binding sites in the PEM due to the partial occupation of these binding sites with lipid molecules.
Additional experimental data are required to support this hypothesis. This will be considered in our further studies. Such changes in the macromolecule hosting properties of PEMs in direct contact with the lipid membrane are important in view of their potential use as artificial ECMs.
In contrast, the lipid coating possesses no barrier properties against the transport of αLA (Fig. S8†). αLA forms μ-sized aggregates on top of HA/PLL280 multilayers regardless of the presence of a lipid bilayer. It can be assumed that negatively charged αLA has a strong affinity to positively charged PLL. Such patterns of αLA interaction with a PEM-supported lipid bilayer points to potential limitations of the proposed PEM–lipid hybrids in both mimics of the ECM and the cellular interface.
The above observations prove again that electrostatically attractive interactions between the highly mobile polycation PLL as presented in multilayers and externally added molecules, such as proteins or lipids, govern the transport of molecules into multilayers. An enhanced interaction of externally added molecules with a mobile PLL results in a strong interaction and the formation of aggregates (either on the multilayer surface or even in solution), which therefore hinders diffusion into multilayers. Positively charged external molecules are freely able to diffuse inside multilayers that may be driven by the compensation of oppositely charged groups of HA. However, the mechanism of diffusion and main driving forces should be further assessed and will be under the focus of an ongoing study.
4. Conclusions
This paper investigates the possibility of supporting a lipid bilayer on a soft biopolyelectrolyte-made multilayer cushion to mimic the cellular interface. It has been demonstrated that the adsorption of small liposomes and their rupture and fusion to form a lipid bilayer can be controlled by the diffusivity of polyelectrolyte constituting the PEMs. Specifically, the higher mobility of PLL in HA/PLL multilayers facilitates its escape from the multilayers in favor of interacting with anionic liposomes. Suppressing PLL mobility without changing its chemical structure (i.e. using higher molecular weight PLL) allows liposome rupture and the formation of bi- or multiple layers of lipids on top of PEMs. The prepared assemblies possess some barrier properties in the case of Lys-FITC transport from the surrounding media into PEMs. The capacity of mobile and positively charged PLL to electrostatically bind the molecules added to multilayers also governs the diffusion of externally added macromolecules – positively charged protein Lys can diffuse into multilayers and fill them homogeneously despite the negatively charged protein αLA forming aggregates with PLL on top of the multilayers.Protein transport analysis revealed the main drawback of this study. Although the structure of the PEM-supported lipid layer can be controlled via polymer mobility, the fabrication of an “ideal” lipid bilayer remains a challenge. Perhaps this can be principally overcome by an opposite design of experiments, i.e., by coating free-standing lipid bilayers (e.g. giant unilamellar vesicles) with PEM assemblies. Apparently, the role of polymer mobility revealed in this study should play a crucial role in such a system too. This will be investigated in our further studies.
This study demonstrates how polymer diffusion within PEMs governs the assembly and architecture of sandwich-like PEMs/lipid bilayer stacks. This technically simple and elegant tool can be used to control the formation of the lipid membrane on top of the PEM as advanced ECM mimics, which opens up new avenues for nanoarchitectonics at the cellular interface.
Author contributions
A. V. and D. V.: conceptualisation and data curation; A. V., A. W., and G. G.: formal analysis and investigation; A. V., A. W., and D. V.: methodology; R. F., A. V., and D. V.: project administration, supervision, and validation; A. V.: writing – original draft; and all authors: visualization and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. V. acknowledges financial support from the Staedtler Foundation in the frames of the project “Function by Design: Cellular Hybrids”. R. F. acknowledges the support of the Kazan Federal University Strategic Academic Leadership Program (PRIORITY-2030).References
- M. Aono and K. Ariga, The Way to Nanoarchitectonics and the Way of Nanoarchitectonics, Adv. Mater., 2016, 28(6), 989–992 CrossRef CAS PubMed.
- K. Ariga, M. Nishikawa, T. Mori, J. Takeya, L. K. Shrestha and J. P. Hill, Self-assembly as a key player for materials nanoarchitectonics, Sci. Technol. Adv. Mater., 2019, 20(1), 51–95 CrossRef CAS PubMed.
- K. Ariga and R. Fakhrullin, Materials Nanoarchitectonics from Atom to Living Cell: A Method for Everything, Bull. Chem. Soc. Jpn., 2022, 95(5), 774–795 CrossRef CAS.
- X. C. Shen, J. W. Song, C. Sevencan, D. T. Leong and K. Ariga, Bio-interactive nanoarchitectonics with two-dimensional materials and environments, Sci. Technol. Adv. Mater., 2022, 23(1), 199–224 CrossRef PubMed.
- Y. Shimane and Y. Kuruma, Rapid and Facile Preparation of Giant Vesicles by the Droplet Transfer Method for Artificial Cell Construction, Front. Bioeng. Biotechnol., 2022, 10, 873854 CrossRef PubMed.
- R. F. Fakhrullin and V. N. Paunov, Fabrication of living cellosomes of rod-like and rhombohedral morphologies based on magnetically responsive templates, Chem. Commun., 2009, 2511–2513 RSC.
- P. Chen, X. D. Ning, W. R. Li, Y. X. Pan, L. Wang, H. K. Li, X. L. Fan, J. X. Zhang, T. T. Luo, Y. B. Wu, C. W. Ou and M. S. Chen, Fabrication of T beta 4-Exosome-releasing artificial stem cells for myocardial infarction therapy by improving coronary collateralization, Bioact. Mater., 2022, 14, 416–429 CrossRef CAS PubMed.
- C. Contini, W. Y. Hu and Y. Elani, Manufacturing polymeric porous capsules, Chem. Commun., 2022, 58(28), 4409–4419 RSC.
- Y. Shindo, H. Komatsu, K. Hotta, K. Ariga and K. Oka, An Artificial Reaction Promoter Modulates Mitochondrial Functions via Chemically Promoting Protein Acetylation, Sci. Rep., 2016, 6, 29224 CrossRef CAS PubMed.
- D. J. Cohen and W. J. Nelson, Secret handshakes: cell-cell interactions and cellular mimics, Curr. Opin. Cell Biol., 2018, 50, 14–19 CrossRef CAS PubMed.
- H. Togashi, K. Kominami, M. Waseda, H. Komura, J. Miyoshi, M. Takeichi and Y. Takai, Nectins establish a checkerboard-like cellular pattern in the auditory epithelium, Science, 2011, 333(6046), 1144–1147 CrossRef CAS PubMed.
- D. J. Cohen, M. Gloerich and W. J. Nelson, Epithelial self-healing is recapitulated by a 3D biomimetic E-cadherin junction, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(51), 14698–14703 CrossRef CAS PubMed.
- B. Ladoux, E. Anon, M. Lambert, A. Rabodzey, P. Hersen, A. Buguin, P. Silberzan and R. M. Mège, Strength dependence of cadherin-mediated adhesions, Biophys. J., 2010, 98(4), 534–542 CrossRef CAS PubMed.
- M. Lambert, F. Padilla and R. M. Mège, Immobilized dimers of N-cadherin-Fc chimera mimic cadherin-mediated cell contact formation: contribution of both outside-in and inside-out signals, J. Cell Sci., 2000, 113(12), 2207–2219 CrossRef CAS PubMed.
- Q. Li, Y. Zhang, P. Pluchon, J. Robens, K. Herr, M. Mercade, J. P. Thiery, H. Yu and V. Viasnoff, Extracellular matrix scaffolding guides lumen elongation by inducing anisotropic intercellular mechanical tension, Nat. Cell Biol., 2016, 18(3), 311–318 CrossRef CAS PubMed.
- D. Marquardt, B. Geier and G. Pabst, Asymmetric lipid membranes: towards more realistic model systems, Membranes, 2015, 5(2), 180–196 CrossRef CAS PubMed.
- G. Bolognesi, M. S. Friddin, A. Salehi-Reyhani, N. E. Barlow, N. J. Brooks, O. Ces and Y. Elani, Sculpting and fusing biomimetic vesicle networks using optical tweezers, Nat. Commun., 2018, 9(1), 1882 CrossRef PubMed.
- T. Y. D. Tang, D. Cecchi, G. Fracasso, D. Accardi, A. Coutable-Pennarun, S. S. Mansy, A. W. Perriman, J. L. R. Anderson and S. Mann, Gene-Mediated Chemical Communication in Synthetic Protocell Communities, ACS Synth. Biol., 2018, 7(2), 339–346 CrossRef CAS PubMed.
- Q. Yang, Z. Guo, H. Liu, R. Peng, L. Xu, C. Bi, Y. He, Q. Liu and W. Tan, A Cascade Signaling Network between Artificial Cells Switching Activity of Synthetic Transmembrane Channels, J. Am. Chem. Soc., 2021, 143(1), 232–240 CrossRef CAS PubMed.
- A. S. Vikulina, A. G. Skirtach and D. Volodkin, Hybrids of Polymer Multilayers, Lipids, and Nanoparticles: Mimicking the Cellular Microenvironment, Langmuir, 2019, 35(26), 8565–8573 CrossRef CAS PubMed.
- C. Uzum, J. Hellwig, N. Madaboosi, D. Volodkin and R. von Klitzing, Growth behaviour and mechanical properties of PLL/HA multilayer films studied by AFM, Beilstein J. Nanotechnol., 2012, 3, 778–788 CrossRef PubMed.
- D. Volodkin, R. von Klitzing and H. Moehwald, Polyelectrolyte Multilayers: Towards Single Cell Studies, Polymers, 2014, 6(5), 1502–1527 CrossRef.
- D. Volodkin, Y. Arntz, P. Schaaf, H. Moehwald, J. C. Voegel and V. Ball, Composite multilayered biocompatible polyelectrolyte films with intact liposomes: stability and temperature triggered dye release, Soft Matter, 2008, 4(1), 122–130 RSC.
- M. Malcher, D. Volodkin, B. Heurtault, P. Andre, P. Schaaf, H. Mohwald, J. C. Voegel, A. Sokolowski, V. Ball, F. Boulmedais and B. Frisch, Embedded silver ions-containing liposomes in polyelectrolyte multilayers: Cargos films for antibacterial agents, Langmuir, 2008, 24(18), 10209–10215 CrossRef CAS PubMed.
- D. V. Volodkin, P. Schaaf, H. Mohwald, J. C. Voegel and V. Ball, Effective embedding of liposomes into polyelectrolyte multilayered films: the relative importance of lipid-polyelectrolyte and interpolyelectrolyte interactions, Soft Matter, 2009, 5(7), 1394–1405 RSC.
- N. Velk, K. Uhlig, A. Vikulina, C. Duschl and D. Volodkin, Mobility of lysozyme in poly(L-lysine)/hyaluronic acid multilayer films, Colloids Surf., B, 2016, 147, 343–350 CrossRef CAS PubMed.
- D. Volodkin, H. Mohwald, J.-C. Voegel and V. Ball, Coating of negatively charged liposomes by polylysine: Drug release study, J. Controlled Release, 2007, 117(1), 111–120 CrossRef CAS PubMed.
- D. Volodkin, V. Ball, P. Schaaf, J.-C. Voegel and H. Mohwald, Complexation of phosphocholine liposomes with polylysine. Stabilization by surface coverage versus aggregation, Biochim. Biophys. Acta, 2007, 1768(2), 280–290 CrossRef CAS PubMed.
- J. F. Nagle and S. Tristram-Nagle, Structure of lipid bilayers, Biochim. Biophys. Acta, 2000, 1469(3), 159–195 CrossRef CAS PubMed.
- L. Jourdainne, S. Lecuyer, Y. Arntz, C. Picart, P. Schaaf, B. Senger, J.-C. Voegel, P. Lavalle and T. Charitat, Dynamics of Poly(L-lysine) in Hyaluronic Acid/Poly(L-lysine) Multilayer Films Studied by Fluorescence Recovery after Pattern Photobleaching, Langmuir, 2008, 24(15), 7842–7847 CrossRef CAS PubMed.
- T. Cassier, A. Sinner, A. Offenhäuser and H. Möhwald, Homogeneity, electrical resistivity and lateral diffusion of lipid bilayers coupled to polyelectrolyte multilayers, Colloids Surf., B, 1999, 15(3), 215–225 CrossRef CAS.
- V. Z. Prokopović, A. S. Vikulina, D. Sustr, C. Duschl and D. Volodkin, Biodegradation-Resistant Multilayers Coated with Gold Nanoparticles. Toward a Tailor-made Artificial Extracellular Matrix, ACS Appl. Mater. Interfaces, 2016, 8(37), 24345–24349 CrossRef PubMed.
- P. A. Netti, D. A. Berk, M. A. Swartz, A. J. Grodzinsky and R. K. Jain, Role of Extracellular Matrix Assembly in Interstitial Transport in Solid Tumors, Cancer Res., 2000, 60(9), 2497–2503 CAS.
- E. Syková and C. Nicholson, Diffusion in Brain Extracellular Space, Physiol. Rev., 2008, 88(4), 1277–1340 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr05186a |
| This journal is © The Royal Society of Chemistry 2023 |