Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineering yeast for the production of plant terpenoids using synthetic biology approaches†
Jean-Alexandre
Bureau
 ,
Magdalena Escobar
Oliva
,
Magdalena Escobar
Oliva
 ,
Yueming
Dong
,
Yueming
Dong
 and
Codruta
Ignea
and
Codruta
Ignea
 *
*
Department of Bioengineering, McGill University, Montreal, QC H3A 0C3, Canada. E-mail: codruta.ignea@mcgill.ca
First published on 31st July 2023
Abstract
Covering: 2011–2022
The low amounts of terpenoids produced in plants and the difficulty in synthesizing these complex structures have stimulated the production of terpenoid compounds in microbial hosts by metabolic engineering and synthetic biology approaches. Advances in engineering yeast for terpenoid production will be covered in this review focusing on four directions: (1) manipulation of host metabolism, (2) rewiring and reconstructing metabolic pathways, (3) engineering the catalytic activity, substrate selectivity and product specificity of biosynthetic enzymes, and (4) localizing terpenoid production via enzymatic fusions and scaffolds, or subcellular compartmentalization.
1. Introduction
Plants produce a myriad of small molecules, known as natural products, to respond to biotic and abiotic threats, interact within organismal communities and attract pollinators, adapt to specific habitats, and fight against competitive counterparts.1 These molecules have found applications of utmost importance for human uses2 in the fields of medicine,3 cosmetics,4 food industry,5,6 and agriculture.7,8 Terpenoids (terpenes or isoprenoids) are the largest class of natural products,1,3 with more than 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 known structures. These compounds are synthesized through modular pathways prone to introducing a high degree of diversity.9,10 Terpenoids vary in size, based on a typical 5-carbon unit, from the simplest compound isoprene to large natural polymers (natural rubber), and in structural complexity, from linear (myrcene) to exquisite stereochemistry (taxol).1 Moreover, terpene moieties also occur in hybrid structures, known as meroterpenoids, among which the most notable are monoterpene indole alkaloids (MIAs)11 or cannabinoids.12 This structural diversity bestows a variety of properties to terpenoids. Hence, it is not surprising that their applications are diverse, spanning major economic sectors from industry to agriculture and medicine.
000 known structures. These compounds are synthesized through modular pathways prone to introducing a high degree of diversity.9,10 Terpenoids vary in size, based on a typical 5-carbon unit, from the simplest compound isoprene to large natural polymers (natural rubber), and in structural complexity, from linear (myrcene) to exquisite stereochemistry (taxol).1 Moreover, terpene moieties also occur in hybrid structures, known as meroterpenoids, among which the most notable are monoterpene indole alkaloids (MIAs)11 or cannabinoids.12 This structural diversity bestows a variety of properties to terpenoids. Hence, it is not surprising that their applications are diverse, spanning major economic sectors from industry to agriculture and medicine.
Several valuable terpenoids, specifically those with complex chemical structures required for potent activities, are difficult to obtain in sufficient amounts to meet demand13,14 due to their low abundance in plants. Traditional extraction from native producers or organic synthesis is often inefficient and unfeasible at the industrial level, leading to high costs or shortages of essential products. Thus, the production of these molecules in genetically amenable organisms, such as yeast, is emerging as an attractive alternative.15 Advances in natural products research enabled industrial production of the antimalarial drug artemisinin16,17 or the biofuel farnesene.18 Moreover, multiple approaches (e.g. plant cell culture; summarized in Table S1†) have been developed for the reconstruction of the biosynthetic pathway, successfully achieved for high-value compounds: carnosic acid19,20 forskolin,21 ginsenosides,22 carotenoids,23–27 cannabinoids,28 strictosidine29 or, more recently vinblastine.30
This article will review synthetic biology and metabolic engineering strategies for the production of plant terpenoids using the microbial cell factories yeast Saccharomyces cerevisiae and other non-conventional yeast (Tables S2–S6†). We will discuss rational approaches and key limitations identified through metabolic logic and will illustrate trends to overcome constraints for efficient, scalable, and inexpensive production of the relevant terpenoids. Engineering strategies will be examined at the host, pathway, and enzyme levels. Strategies for intracellular localization of terpenoid biosynthetic pathways in yeast will also be described.
2. Host engineering
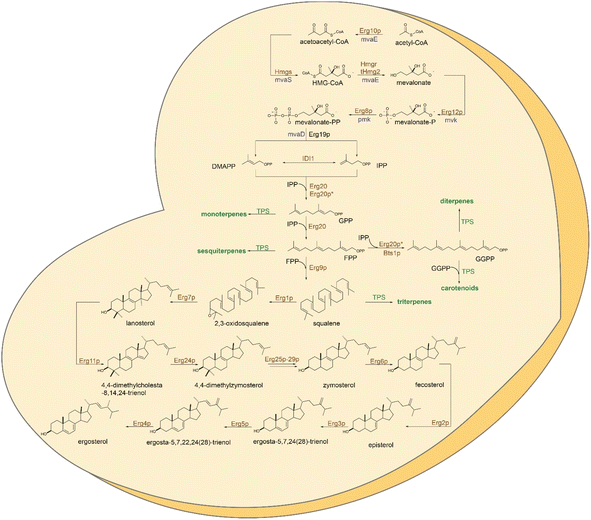
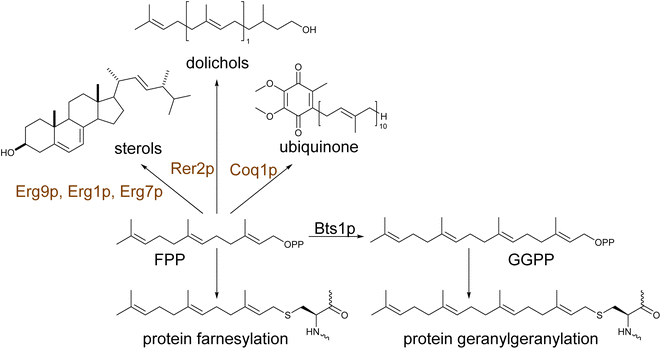
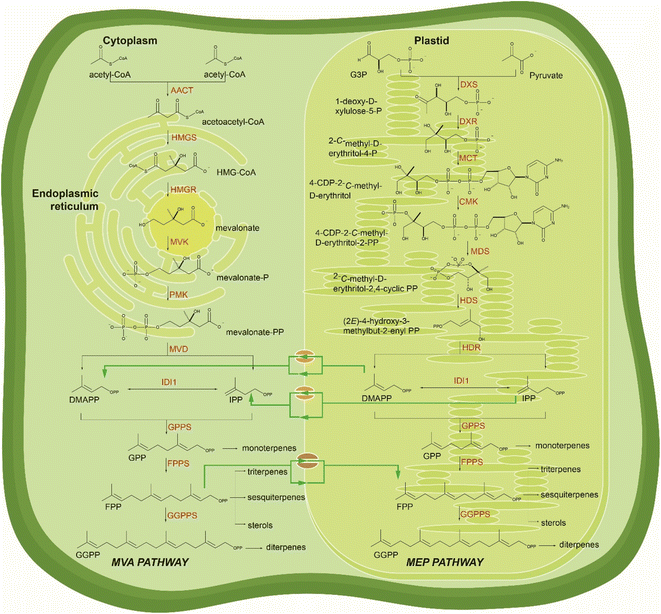
Yeast Saccharomyces cerevisiae has become a workhorse in biotechnology due to its robustness and versatility.31,32 With a well-studied genome and metabolism, yeast combine features of the eukaryotic kingdom that support processes specific to higher organisms, with the simplicity of manipulation conferred by its unicellular makeup.33 Well-established genetic manipulation, fast growth rate, and the ability to be cultivated from simple nutrients combined with high-throughput emerging tools make possible the integration of multiple genome modifications into yeast cells in a short time. Moreover, pathways of central metabolism are conserved and partially shared in yeast and plants enabling the reconstruction of terpenoid pathways into yeast's metabolism.15Yeast naturally synthesizes early precursors of terpenoids, such as isopentenyl diphosphate (IPP), dimethylallyl diphosphate (DMAPP), geranyl diphosphate (GPP), and farnesyl diphosphate (FPP), via the mevalonate pathway (MVA) (Fig. 1). These metabolites are essential components in the primary metabolism of yeast for sterol (Fig. 1), ubiquinone and dolichol biosynthesis.34 Moreover, FPP is used for protein prenylation (Scheme 1). Yeast also produces, albeit in lower amounts, geranylgeranyl diphosphate (GGPP) utilized for post-translational modifications (geranylgeranylation) of proteins.14,35 Compared to other natural products, terpene biosynthesis in higher eukaryotic hosts is significantly active. By contrast, yeast offer a terpeneless background that enables engineering of heterologous pathways with minimal interference from endogenous enzymatic machinery and low production of undesirable by-products. Yeast mainly requires targeted engineering of the central metabolism to minimize ethanol formation and improve glucose conversion to pyruvate and acetyl-CoA, the starting precursor in the MVA pathway.36 Many strategies for engineering the acetyl-CoA metabolism in yeast have been reported and previously summarized in36 and, therefore, not considered here. In this review, we will focus on host engineering strategies (Fig. 2) and achievements to improve the yield of precursors along the MVA pathway and redirect the metabolic flux from sterol to terpene biosynthesis.15 These approaches have targeted the rate limiting steps regulated by product-driven feedback inhibition, favoring conversion rates at divergent metabolic nodes.37–40
2.1 Genome editing
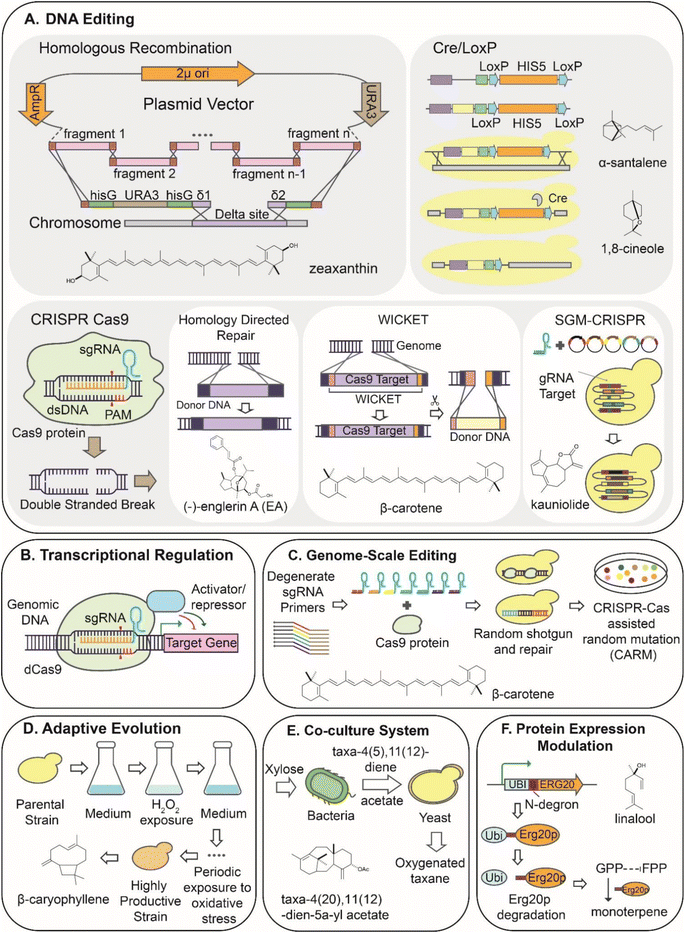
The efficient homologous recombination (HR) system of S. cerevisiae has been instrumental in turning this organism into the workhorse of the biotechnological industry. Early strategies for terpene production employed HR for genomic pathway integration and production of the sesquiterpene amorpha-4,11-diene,41 the carotenoid zeaxanthin42 or products of oxidative cleavage such as β-ionone from β-carotene.43 Coupled with Cre/LoxP system,44 HR allows for selection-free genetic modifications through the use of recyclable cassettes alongside genetic elements of interest (Fig. 2A). This strategy was applied to engineer the yeast genome by promoter exchange, gene integrations, or deletions to produce the monoterpene 1,8-cineole37 and the sesquiterpene α-santalene.45The discovery of the CRISPR/Cas9 system has revolutionized genome editing and has been applied in various organisms.46 Notably, it has been proven an excellent approach for integrating genes of interest in yeast chromosomes due to the highly efficient homology-directed repair (HDR) when double-stranded breaks (DSB) are induced.47,48 CRISPR/Cas9 was used to engineer target genes of MVA pathway to produce the triterpene friedelin49 at 37.07 mg L−1 and sesquiterpene guaia-6,10(14)-diene as the precursor for the semi-synthesis of englerin A.50 Exploiting the high prevalence of the delta sites in the yeast genome, the CRISPR-Cas9 system has also been used to integrate multiple copies of the biosynthetic pathways, achieving a high protein expression in a stable manner.51,52 This strategy was applied to the biosynthesis of santalenes and santalols, reaching production rates of 164.7 and 68.8 mg L−1, respectively.51 Recently, a method (HapAmp), which utilizes haploinsufficiency as an evolutionary-driven force for in vivo gene amplification, was implemented for terpenes production. Using this method, it was possible to integrate up to 47 copies of target genes into the yeast genome in a stable and controlled manner and significantly improved the production of representative terpenes, such as the monoterpene limonene, the sesquiterpene nerolidol, and the tetraterpene lycopene.53
Moreover, CRISPR-based approaches were developed for simultaneous editing of multiple targets, a process known as multiplex gene editing, reviewed by Utomo et al.54 Several of these systems were developed and used for the production of β-carotene, CasEMBLR,55 CrEdit,56 Wicket,57 or astaxanthin, PCR & Go,58 due to ease of visual screening conferred by color (yellow or red, respectively) saturation of productive colonies. Recently, an SGM-CRISPR system employing yeast strains harboring preinstalled synthetic gRNA was used for the production of sesquiterpene lactone, kauniolide.59
CRISPR/Cas9 has also contributed to advances in engineering non-conventional yeast for the production of terpenoids. Yarrowia lipolytica, which naturally produces large amounts of acetyl-CoA, has proven a promising chassis for the production of carotenoids. Accordingly, a standardized CRISPR/Cas9-mediated markerless gene integration strategy was developed for lycopene production,60 and a β-carotene hyper-producing strain was engineered by CRISPR/Cas9-directed overexpression of MVA key genes and multi copies of β-carotene biosynthetic gene.61 Combined with the optimization of bioreactor fermentation, the latest approach enabled 4.5 g L−1 β-carotene production. An alternative method to the labor-intensive HDR-based editing, a CRISPR/Cas9 approach employing the non-homologous end-joining (NHEJ) in a targeted fashion was constructed by optimization of the cleavage efficiency of Cas9 nuclease and through rational regulation of the NHEJ repair pathway and the cell cycle. This attractive, fast engineering was confirmed by the production of 36.1 mg L−1 of canthaxanthin.62
2.2 Transcriptional regulation
Naturally, the MVA pathway is tightly regulated at transcriptional and post-transcriptional levels resulting in a limited supply of terpene precursors.63 Thus, the main direction of metabolic engineering in the production of terpenes is the overexpression of genes in the MVA pathway (described in Section 3.1), leading to the accumulation of target terpenes. However, unbalanced gene expression can cause the accumulation of intermediates that inhibit the activity of enzymes by the feedback mechanism.63 Therefore, combinatorial methods (described in Section 2.3) make it possible to optimize the expression of target genes and, as a result, achieve a balanced metabolic flux, which can improve the production of target terpenoids.76CRISPR-cas9 systems have also been developed for modulation of the transcriptional regulation in different organisms, including yeast.64,65 A deactivated Cas9 (dCas9) fused with an activation or repression domain, CRISPRa or CRISPRi, respectively, can be used to modulate gene expression by targeting the upstream region of the gene of interest.64,65 It has been shown that the use of CRISPRa/i to modulate the expression level of genes involved in the MVA pathway and β-carotene pathway enzymes directly correlates to an increase or decrease in the final yield of the strain65 (Fig. 2B).
2.3 Combinatorial and genome-scale engineering
The modulation of the yeast's genome by the previously described engineering is effective at increasing the yield of terpenoids. The application of a single strategy, however, in most cases, does not unlock the full potential of rewiring metabolic fluxes. Thus, combining multiple approaches has been employed to provide synergistic metabolic adjustment to improve system performance further.66,67 Recently, a CRISPR system (CRISPR-AID) containing three orthogonal CRISPR proteins with different activities was developed for combinatorial metabolic engineering. This system consists of a transcriptional activation domain (CRISPRa), a transcriptional interference domain (CRISPRi), and a catalytically active CRISPR protein for gene deletions (CRISPRd).61 Utilizing β-carotene production as a colorimetric screen, CRISPR-AID was implemented to increase simultaneously the HMG1 expression (CRISPRa), downregulate the ERG9 expression (CRISPRi), and delete ROX1 (CRISPRd). As a result, the application of the CRISPR-AID system improved the final β-carotene yield by 3-fold. CRISPR systems have also been used to develop yeast-based auxin-inducible protein degradation strategies and have also emerged as a strategy to modulate metabolic fluxes for improved terpene production.68 The strategy was developed for mono and sesquiterpenes production chassis, yielding significant improvements in limonene and nerolidol production.68CRISPR-AID strategy is suitable for the metabolic rewiring of biosynthetic pathways where key enzymatic steps or bottlenecks are obvious. However, metabolic pathways are frequently regulated by complex mechanisms that involve hidden interaction.69 Therefore, the modulation of individual genes involved in complex pathways has generally low throughput. To alleviate this issue, a rapid combinatorial method for the assembly of biosynthetic pathways (COMPASS) was developed.67 This approach is a high throughput cloning method of DNA parts of interest (promoters, genes, and synthetic transcription factors) based on the positive selection of correctly assembled pathway fragments.67 The cloning and stable integration of these diversely assembled modules allow high throughput pathway reconstruction while tuning the enzyme expression level to maximize terpenoid production.67 The application of COMPASS to the production pathway of β-ionone led to a 4.2-fold increase over the control strain.67
CRISPR-Cas9 system has been used to make untargeted genome-wide modifications70 to randomly shift the gene expression across the genome to improve the production of a target product. The CRISPR-Cas9 assisted random mutation (CARM) technique was developed to introduce non-oriented genomic modifications into the yeast genome using a random gRNA library70 until acquiring the improvement of a target compound (Fig. 2C). Thus, several rounds of CARM mutagenesis in the BY4741 yeast strain resulted in a 10.5-fold improvement in β-carotene production via altering the expression level of over 2500 distinct genes.70
2.4 Adaptive evolution
A non-directed genomic modification strategy developed for terpene production in yeast is the adaptive laboratory evolution (ALE).71 The ALE strategy is commonly used to adapt industrial strains through selective environmental conditions to produce the specialized metabolites that are often toxic. Using an ALE approach, the natural activity of the terpenoid-type metabolite was exploited to respond to an induced stressor for either increasing the yields of a compound of interest or introducing a desired trait. As a result of an introduced evolutionary pressure, the yeast strain can adapt to stress through acquired mutations and altered gene dosage.72 Exploiting the antioxidant features of carotenoids and intermittent exposure to hydrogen peroxide as an adaptive pressure force, a 3-fold improvement in carotenoid production was estimated in the yeast producer strain.73 Similarly, utilizing oxidative stress as the driving force of ALE, β-caryophyllene production in yeast increased 4-fold in just a few generations74 (Fig. 2D).Using terbinafine as the inhibitor of downstream steps in sterol biosynthesis, a 16.5-fold increase of squalene accumulation over the basic strain was achieved by an ALE approach.75 Moreover, ALE has been employed to alleviate the toxicity of the target product.76 The robustness of a yeast strain producing a blend of highly toxic terpenes (10% cymene, 50% limonene, 40% farnesene) for the production of a jet fuel product was improved by subjecting the strain to repeated rounds of limonene supplementation.76 Following the treatment with the toxic monoterpene, the tolerance towards jet fuel production was improved by 4-fold.76 In addition, the tolerance towards the production of other toxic monoterpenes, β-pinene, and myrcene, was also improved 11- and 8-fold, respectively.76 Similarly, the tolerance of Yarrowia lipolytica to limonene was improved by 8-fold over the base production strain.77 To acquire desirable traits for industrial applications, such as thermotolerance, yeast was ALE-evolved to survive at ≥40 °C by modifying its sterol profile.78
2.5 Co-culture systems
Co-culture systems have also been investigated to improve the production of terpenes in yeast. The co-culture of yeast and bacteria was developed to separate the biosynthetic pathways for terpene production into modules expressed in the two microbes.79 The pathway division alleviated the total metabolic burden placed on either species while engineering a mutually dependent growth to prevent one species from significantly outperforming the other.79 The co-culture system was applied to/for the production of oxygenated taxanes,79 yielding the first microbial-based production of taxa-4(20),11(12)-dien-5α-acetoxy-10β-ol and other terpenes, such as nootkatone and ferruginol79 (Fig. 2E). More recently, a similar system was used for exploring the biosynthesis of strigolactones, plant hormones derived from all-trans-β-carotene.80 Reconstruction of early steps of strigolactone pathways was successfully achieved in E. coli while functionalizing the key intermediate carlactone was only possible in yeast. This bacteria-yeast consortium enabled the functional characterization of CYP722Cs from different plants involved in the oxygenation of carlactone and improved the titer of 5-deoxystrigol by a metabolic engineering approach.803. Reconstruction of biosynthetic pathways
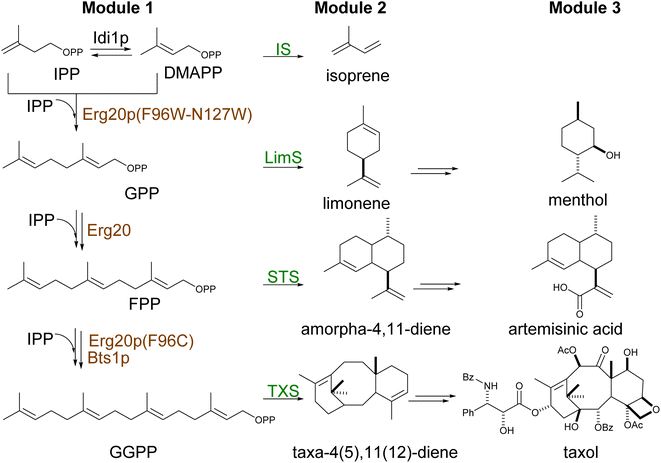
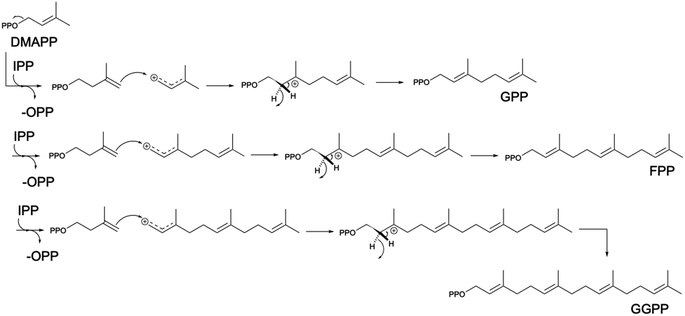
Built from two universal isomer substrates, isoprenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP), terpenoids are synthesized in modular pathways1,81 (Scheme 2). In the first module, prenyltransferases typically catalyze the initial elongation of the diphosphate chain by sequential head-to-tail (1′–4) addition of IPP units,82 producing a handful of building blocks of increasing size by 5 carbon atoms (geranyl diphosphate C10, farnesyl diphosphate C15, geranylgeranyl diphosphate C20) (Scheme 3).In the second module, terpene synthases introduce the first level of diversification through complex reactions of rearrangement and cyclization of the hydrocarbon chain.10 Hence, each building block emerges in distinct classes of thousands of compounds with the same number of carbon atoms, known as monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20), respectively. Triterpene (C30) and carotenoids (C40) are synthesized via a head-to-head addition (Scheme 4) of two molecules of FPP and GGPP, respectively. Subsequent functionalization of terpene scaffold by hydroxylation, reduction, acetylation, methylation, glycosylation, etc., enables, in the third module, synthesis of structurally complex isoprenoids, thus enlarging their chemical variety.
Plants naturally harbor two pathways producing the universal terpene precursors IPP and DMAPP, the MVA in the cytoplasm as the main source of FPP and the specialized methylerythritol phosphate (MEP) pathway83 present in chloroplasts (Fig. 3). MEP coordinates synthesis of GPP and GGPP distinct from the cytosolic formation of FPP, circumventing most of the competition between different metabolic pathways. However, prenyl diphosphates, such as IPP and DMAPP or larger (e.g. FPP), can be shared in between pathways despite their localization in distinct subcellular compartments. Several exceptions of terpenoids produced from building blocks supplied from both plastid and cytosolic pathways have been shown through isotope labeling and feeding studies,84,85 but the crosstalk mechanism and its regulation remains unclear (Fig. 3). Yeast lacking the MEP pathway delivers all terpene building blocks from the cytoplasmic MVA pathway for the reconstruction of downstream biosynthetic pathways of target compounds. While the production of sesquiterpenes derived from the cytoplasmatic FPP is relatively straightforward and industrial yields have been achieved,17,18 the production of GPP- or GGPP-derived terpenes is relatively inefficient and requires extensive pathway engineering and optimization.
3.1 Endogenous pathway modulation
Traditionally, the first steps in the production of terpenes in yeast are aimed at increasing the performance of the MVA pathway (Fig. 1) and downregulating sterol biosynthesis to redirect accumulating prenyl diphosphate precursors to the synthesis of target terpenes.37–40 Engineering hubs can be identified at the HMG-CoA reductase (HMGR), converting HMG-CoA to mevalonic acid, a rate-limiting step of the upstream pathway. Prenyl diphosphate synthesis by the sequential activity of Erg20p and squalene or lanosterol synthesis by Erg9p or Erg7p, respectively, in the downstream pathway, are also key points of intervention according to the target terpene class of interest. The two yeast isozymes, Hmg1p and Hmg2p, are highly regulated by different mechanisms at transcriptional,86 translational,87 and post-translational levels.88 These enzymes are key engineering targets towards lifting the negative feedback inhibition at this step. Overexpression of a truncated version of Hmg1p (tHmg1p) devoid of regulatory sequences after the removal of the transmembrane anchor renders the protein soluble and alleviates an early pathway bottleneck. The truncation of the first 552 amino acids significantly improved the enzyme's catalytic turnover, removing the bottleneck in the pathway.89 This approach has been conventionally used to increase the precursor supply in yeast for the production of various terpenoids,16,90 meroterpenoids,28,30 and even non-natural compounds.91 Targeting HMG2, Ignea and co-workers introduced a stable variant, Hmg2p(K6R), resistant to ubiquitination into the yeast genome to improve monoterpene and sesquiterp ene production.37 They also engineered tandem heterozygous deletions of HMG2 positive genetic interactors that regulate protein turnover and stability.39 It has been demonstrated that the upregulation of other key genes in the pathway was shown to have synergistic effects with HMGR stabilization.92 Specifically, overexpression of ERG10, ERG13, and ERG12, in conjunction with tHmg1, enhanced the metabolic flux through the MVA pathway, increasing the yield of amorpha-4,11-diene 13-fold.92To reroute the carbon flux dedicated to sterol biosynthesis to terpenoid production, diverging points have been rationally engineered according with the first committed substrate for different classes of terpenes.93 FPP, the first dedicated precursor of sterols, is thus the primary endpoint of the pathway.15 Combined with inhibition of HMGR feedback regulation, FPP is efficiently accumulated in yeast and readily converted into sesquiterpenes. This advantageous metabolic flux supported the early breakthroughs for the production of artemisinin precursor16,17 and the highest titers of any terpene reported in yeast to date of 130 g L−1 of farnesene.18 Effective strategies for sesquiterpene production also surround FPP synthesis in yeast and its conversion to squalene by the Erg20 and Erg9 enzymes, respectively.17 Among these, overexpression of the Erg20p and downregulation of ERG9 have become standard strategies that expand beyond the production of sesquiterpenes.38,40 The use of ergosterol-sensitive promoters from ERG11, ERG1, ERG2, and ERG3, was shown to downregulate the expression of Erg9p, increasing the yield of amorpha-4,11-diene between 2-5-fold.94 The best-performing promoter was shown to be ERG1 promoter.94 The methionine-sensitive (MET3) and the copper-regulated CTR3 promoters were also used to downregulate Erg9p.17 The use of both promoters and Erg20p upregulation increased amorpha-4,11-diene production up to 1.4–1.6 g L−1 titers17 (Table S2†).
With the tight regulation of the synthesis of FPP and downstream intermediates in yeast, the availability of other diphosphate precursors, such as GPP and GGPP, for mono- and diterpene or carotenoid biosynthesis, respectively, is limited. The Erg20p yeast prenyl transferase naturally produces GPP. However, it is not released from the enzyme's catalytic site but is immediately used for the subsequent addition of IPP to synthesize FPP.38 This highly efficient substrate channeling of GPP into FPP severely limits the availability of the smaller precursor for efficient synthesis of monoterpenes. Conventional approaches, such as expression of plant GPP synthases95 have only shown little improvements as any excess of GPP accumulated in yeast is efficiently redirected back into FPP synthesis. Overexpression of yeast IPP isomerase Idi1p coupled with Erg20p, aiming to control the IPP/DMAPP ratio, yielded a several-fold increase of 1,8-cineole37 or geraniol.96 More advanced approaches, including engineering a yeast dominant negative GPP synthase,38 N-degron-mediated protein degradation97 (Fig. 2F), de novo synthesis of GPP alternative substrates,98 dynamic control,98 and pathway compartmentalization99 will be discussed in more detail in the next chapters (Table S3†).
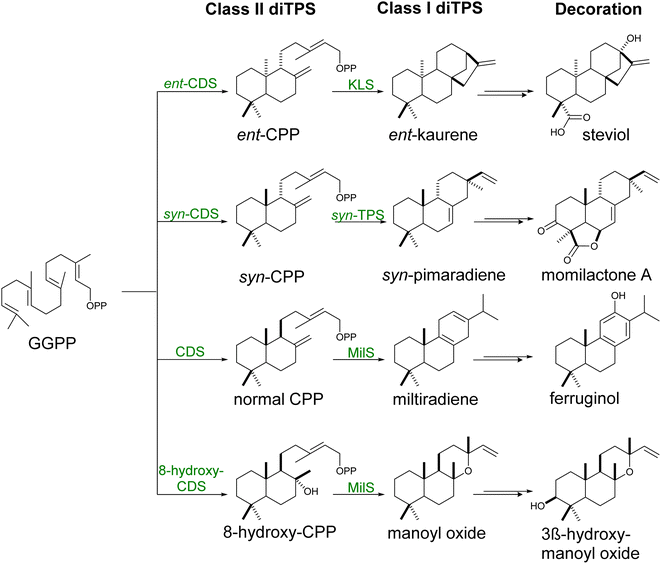
GGPP is naturally produced by yeast by the mitochondrial localized GGPP synthase Bts1p. The metabolite remains, however, a minor by-product of the MVA pathway, being used for the post-translational modification of essential proteins.15,100 Increasing GGPP level via expression of native101 or heterologous GGPP synthases21,90,102 or engineered variants40 and channeling this precursor into the diterpene skeleton formation have been typically used for the efficient production of various diterpenes. Specifically, fusions of biosynthetic enzymes catalyzing the synthesis of cyclic diphosphates (copalyl or 8-hydroxy-copalyl diphosphate; Scheme 5) improved the labdane-type diterpene production including miltiradiene,103 manoyl oxide,102,104 manool,102 sclareol102 among other (Table S4†). The use of glucose-responsive promoter HXT1 and the ERG1 promoter to regulate the expression of Erg20p and Erg9p increased the availability of FPP for its conversion to GGPP,90 leading to an increase in casbene yield over 3-fold.90
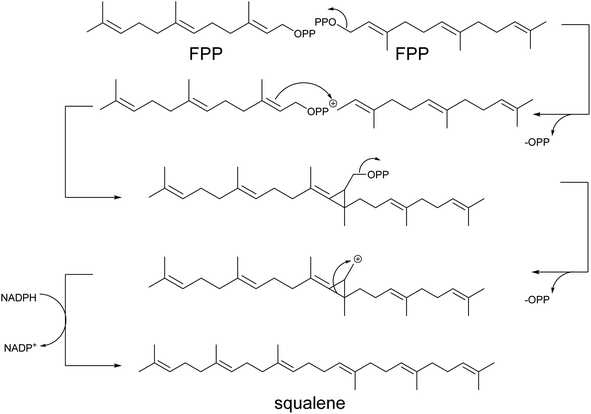
Squalene is the first committed step in triterpene (Scheme 6) and hopanoid synthesis. In yeast, the production of these compounds requires diverging the metabolic flux at the Erg7p step to minimize the conversion of 2,3-oxidosqualene to lanosterol. Consequently, upregulation of the proximal upstream ERG9 and/or ERG1 genes encoding squalene synthase and squalene epoxidase, respectively, has been coupled with repression of ERG7 as standard approaches for the production of triterpene scaffolds. Overexpression of ERG1 resulted in a 10-fold increase in the yield of protopanaxadiol in S. cerevisiae.105 However, joint overexpression of ERG9 and ERG1 in Y. lipolytica only slightly improved betulinic acid production.106 Alternatively, the expression of heterologous squalene epoxidases can be used to boost triterpene production.107 Exchanging the native ERG7 promoter with MET3 (ref. 108) or CTR3 (ref. 109) promoters enabled methionine- or copper-based repression of ERG7, respectively, perhaps due to a deficiency of feedback regulation by ergosterol at this step. As a result, the lupeol content in copper-supplemented engineered cells was increased by 7.6-fold.109 In an attempt to develop a feeding-free procedure for industrial purposes, Zhao et al. engineered the ERG7 promoter using the TetR and TetO-based gene regulation system.110 Nevertheless, no significant increase in protopanaxadiol yield was obtained by this method (Table S5†).
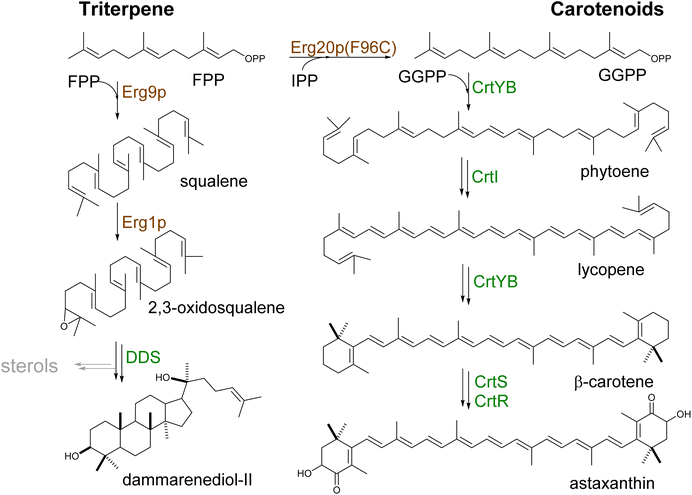 | ||
| Scheme 6 Biosynthesis of triterpenes and carotenoids. Triterpenes and carotenoids (tetraterpenes) are produced by head-to-head addition reactions using two FPP or GGPP molecules, respectively. They are converted to squalene, further oxidized to 2,3-oxidosqualene, or phytoene, which serves as the linear entry scaffolds for the formation of cyclic triterpenes or carotenoids, respectively (Tables S5 and S6†). Yeast enzymes are represented in brown and heterologous enzymes are represented in green. Downregulated steps are indicated in grey. | ||
3.2 Upstream heterologous pathway
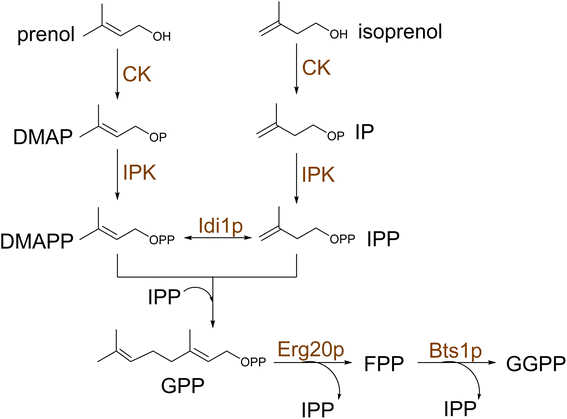
Recent research has shown that the reconstruction of heterologous upstream pathways could achieve efficient synthesis of terpene precursors in yeast. Particularly, the introduction of heterologous enzymes with superior catalytic properties significantly increases metabolic flux through the target pathway. It has been repeatedly demonstrated that the introduction of the mvaE and mvaS genes of Enterococcus faecalis leads to the augmentation of pathway intermediates.92,111,112 The bacteria enzymes have higher catalytic turnover and, in addition, lack the negative feedback regulation of the yeast's MVA pathway,92 resulting in increased metabolic flux and ultimately higher yields of target terpenoid.92,111,112 Furthermore, functional expression of the MEP pathway in yeast cytosol has been successfully achieved113 by assembling an elaborate iron-sulfur cluster (ISC) assembly machinery to functionalize the last two enzymes in the pathway, IspG and IspH. Kirby et al. showed that MEP could maintain cell viability under low aeration conditions in the absence of the MVA pathway, however, at lower growth rates.113 Despite evidence of sustaining isoprenoid synthesis, further incorporation of MEP-derived IPP and DMAPP into other terpene compounds was not reported to date. Recently, a universal terpenoid pathway was engineered by the introduction of a two-step isopentenol utilization (IU) pathway in S. cerevisiae to convert supplemented short alcohols, such as isoprenol and prenol, into terpene precursors IPP and DMAPP114 (Scheme 7). This approach provides a shortcut to accessing prenyl diphosphates in yeast by circumventing the competition with the native sterol pathway that elevated the levels of GPP and GGPP by 147- and 374-fold, providing efficient platforms for mono-and diterpene production.114A highly efficient one-step orthogonal pathway for monoterpene biosynthesis was engineered to synthesize neryl diphosphate (NPP), the cis isomer of GPP.98 Ignea et al. showed that NPP accumulates in yeast as an end-product and can be used to synthesize various cyclic monoterpenes, including limonene, sabinene, 1,8-cineole camphene among others.98 Lacking competition with endogenous metabolism, the approach was combined with protein and metabolic engineering strategies, facilitating a 7-fold increase in monoterpene titers over GPP-based production and reported in vivo monoterpene oxidation for the first time. NPP-derived production of limonene in S. cerevisiae reached titers of 917.7 mg L−1 in fed-batch fermentation,115 and the orthogonal system was also implemented in non-conventional yeasts, such as Rhodosporidium toruloides.116
3.3 Downstream heterologous pathway
The downstream production of terpenes is separated into two distinct biosynthetic modules responsible for terpene scaffold synthesis and decoration. After the removal of the diphosphate group, the prenyl hydrocarbon chain undergoes rearrangements and cyclization by terpene synthases to generate unique terpene scaffolds.10 Functional groups are subsequently attached to the hydrocarbon skeleton by a variety of enzymes, such as dehydrogenases, O-acyl and O-methyltransferases, cytochrome P450s, glycosyltransferases, etc.117 resulting in active terpenoids with high structural complexity. To reconstruct the plant pathways in yeast, the expression of functional biosynthetic enzymes involved in both modules must be achieved. While cyclization of the diphosphate precursors is performed by a single enzyme or, exceptionally, by two (such as class I and class II diterpene synthases in labdane biosynthesis), terpene functionalization typically proceeds through a few to multiple decoration steps.1 Analyzing in detail engineering approaches for overexpression of terpene biosynthetic enzymes in yeast is beyond the scope of this review. Here, we present a short overview and summarize notable examples of TPSs and decorating enzymes (Tables S4–S6†) expressed in yeast for the production of specific compounds.Terpene synthases (TPSs) form a highly intricate group of enzymes in plant-specialized metabolism, catalyzing the most complex reactions found in nature. These enzymes act at the entry points of the terpenoid pathways and are responsible for the first diversification in this large group of natural products. Their activity can dramatically vary from high fidelity to broad promiscuity. Several TPSs precisely synthesize an exclusive product out of the maximum of theoretical possible isomers (limonene synthase from Citrus limon118), but most TPSs are promiscuous enzymes generating multiple products. An extreme example is γ-humulene synthase synthesizing 52 different products.119 Overexpression of TPSs in yeast can be achieved from plasmid-based or chromosomal integration,120–124 and their functionality varies significantly according to the enzyme catalytic activity and the engineering strategy (Tables S2 and S3†). Optimization of TPSs performance is typically accomplished by regulating their expression level, constructing their fusions with proximal biosynthetic enzymes to improve substrate channeling, enzyme stability, and protein engineering, discussed in more detail in Section 4.
The second diversification step in the terpenoid chemical space occurs through the functionalization of terpene skeleton at specific positions, typically in successive reactions catalyzed by different classes of enzymes in stereospecific reactions. Among the most frequent modifications is terpene oxidation, generally catalyzed by cytochrome P450 enzymes and, to a lesser extent, by dioxygenases or dehydrogenases. Terpene-related CYPs belong to CYP71, CYP72, CYP85, and CYP711 clans. These enzymes catalyze the addition of oxygen atoms to terpene structures, which, if sequentially introduced in the same position, can generate terpene alcohols, aldehydes or carboxylic acids (Scheme 8).
Yeast has become an ideal host for the functional expression of CYPs due to their requirement to insert themselves into ER membranes and the low presence of endogenous homologs. These conditions have favored efficient reconstruction of CYP-driven sequential reactions,17,19,20,125,126 CYP library screenings for pathway discovery,19,127 and combinatorial biosynthesis approaches for the production of novel terpenoids.102,128 In fewer instances, CYPs can also orchestrate backbone rearrangements with the formation of new C–C bonds or rings (e.g. lactones).129 Recently, it has been shown that a synergistic combination of Tripterygium wilfordii CYPs from the CYP71BE subfamily catalyzed the formation of the abeo-abietane skeleton (Scheme 9) and subsequent lactone ring present in the structure of triptolide in engineered yeast cells.130 Plant CYPs may frequently catalyze multiple,19,131,132 and/or atypical reactions.133
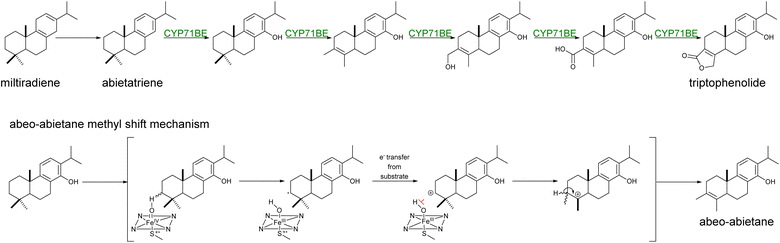 | ||
| Scheme 9 Hypothetical formation of abeo-abietane skeleton via Wagner–Meerwein rearrangement reaction. The combined activity of CYP71BE85 and CYP71BE86 from T. wilfordii was proposed to catalyze the C4 → C3 methyl shift and for the formation of the lactone moiety of triptophenolide.130 The methyl shift of C18 to C3 in the abietane skeleton was proposed to undergo through a Wagner–Meerwein mechanism, where CYP-catalyzed hydrogen abstraction and inhibition of oxygen rebound (shown in red) from the heme bound hydroxyl to the substrate, thus facilitating electron transfer and carbocation formation.130 | ||
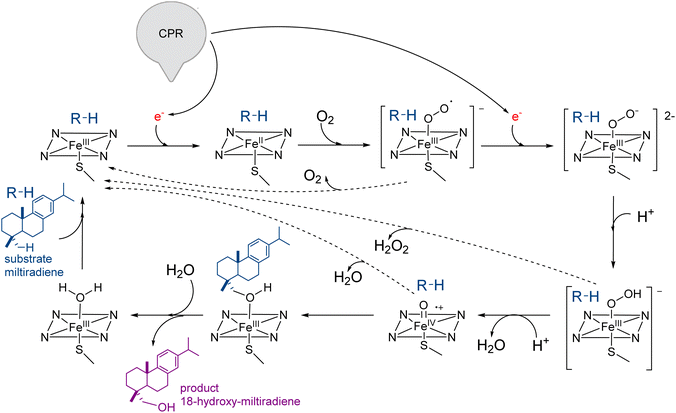
In addition, these catalysts have broad substrate promiscuity that makes this multigene family a major contributor to terpene diversity. A notable example is abietadiene oxidase from Pinus taeda (CYP720B1),134 able to accept as substrate not only different labdane diterpenes102 but also unnatural terpenoids.91 CYPs require the delivery of two electrons from NAD(P)H to the heme moiety by cytochrome P450 reductase (CPR) to catalyze the oxidation of its substrate (Fig. 4). The CYP-CPR pair may also involve an additional component, cytochrome b5 (Cyt b5), which can transfer the second electron and significantly increase the catalytic activity of the paired CYP.135 Due to its low redox potential, cyt b5 can only rarely directly activate the CYPs in the absence of CPR (Scheme 8 and Fig. 4), however it shares the CYP surface binding site with CPR. Thus depending on the expression level and affinity for a specific CYP, cyt b5 can have opposite effects and either negatively impact the reduction of CYP ferric group or enhance the catalytic cycle efficiency in subsequent steps to increase coupling.136 The interaction of CYPs, CPR, and alternative redox partners has been shown critical for modulating titers of oxygenated terpenoids such as ferruginol, 11-hydroxy ferruginol, salviol, pisiferic acid, and carnosic acid.137
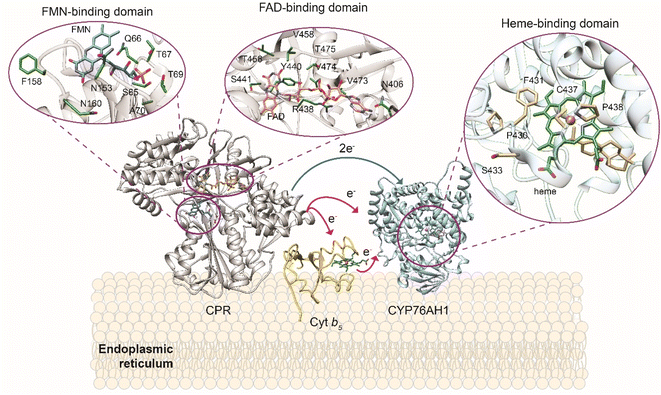 | ||
| Fig. 4 Plant cytochrome P450 electron transfer mechanism and redox system in yeast host. Overview of CYP-CPR interaction and their modulation by cytochrome b5 (Cyt b5) in an ER membrane. Magnifications at the heme ligand in CYP and the FMN and FAD ligands in CPR and residues involved in the substrate binding are shown using X-ray crystal structures of CYP76AH1 from Salvia miltiorrhiza (PDB ID: 7CB9), CPR from Candida tropicalis (PDB ID: 6T1U), and solution NMR structure of Cyt b5 from Oryctolagus cuniculus (PDB ID: 2M33) are used for model generation. CYP76AH1 and CPR crystal structures are truncated in the membrane anchoring domain. Graphics produced by USCF Chimera.138 | ||
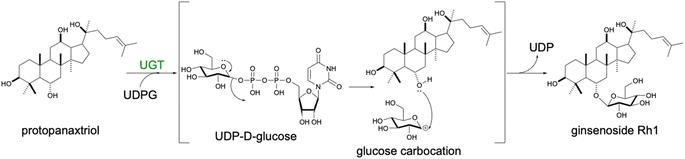
An additional modification directly affecting terpenoid biological activities is glycosylation by transferring uridine diphosphate (UDP) sugars (Scheme 10). Single or multiple sugar attachments are catalyzed by UDP-glycosyltransferases (UGTs), enhancing water solubility and end-product stability. Several UGTs catalyzing the formation of triterpenoid saponins have been expressed in yeast, previously reviewed in.139 Particularly, successful validation of a large number of UGTs has been achieved for the biosynthesis of dammarane-type ginsenosides Rg3 and Rd,140 Rh1 and F1,141 or Rh2 (ref. 142) in yeast. Production of glycosylated terpenoids is generally inefficient. Optimization approaches were aimed at increasing the copy number or the strength of the promoter controlling the level of expression of UGT of interest. By engineering overexpressing URT94 from Panax ginseng capable of using UDP-rhamnose instead of UDP-glucose as a sugar donor in yeast, double and triple glycosylated ginsenoside Rg2 and Re, respectively, were produced.143
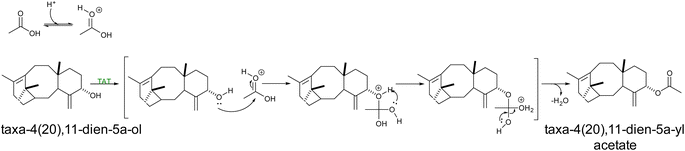
Terpenoid acetylation was also achieved in yeast by overexpression of taxa-4(20),11-dien-5α-ol O-acetyltransferase (TAT) from Taxus sp. producing taxa-4(20),11-dien-5α-yl acetate for the reconstruction of the early steps of taxol pathway79 (Scheme 11). Following two upstream rate-limiting steps in the pathways, the acetylation step remains inefficient, and further optimization is needed. Furthermore, two acetyltransferases from Coleus forskolii were reported to catalyze the conversion of 13R-manoyl oxide to the complex labdane-type diterpenoids. Overexpressing a set of three P450s in combination with a single acetyltransferase resulted in high titers of forskolin in yeast.21 Furthermore, a promiscuous acetyltransferase from Arabidopsis thaliana (THAA2) has been shown to catalyze acetylation of the C-3 hydroxyl group of a wide range of triterpenes in yeast, including thalianol and its derivatives, α-amyrin, β-amyrin, lupeol.144
An important component in the engineering and optimization of terpene functionalization is the availability of donor groups and cofactors, the competition for these metabolites between endogenous and engineered pathways, and the resultant cellular fitness. Heme depletion was identified as a major factor of cellular stress during overexpression of heterologous CYPs.145 Thus, rewiring the heme biosynthetic flux at rate-limiting steps has been employed to alleviate the metabolic burden generated by CYP-expressing yeast platforms (Scheme S1†). Overexpression of HEM3 was reported to favorably support production of various labdane-type diterpenoids.19,104,137 Furthermore, heterologous redox enzymes frequently introduce perturbations in the dynamic utilization and regeneration of NADPH cofactor in yeast and could significantly affect production of reconstructed pathways.146,147 Consequently, a large amount of data, reviewed elsewhere, has been reported for optimization of NADPH levels in yeast for the functionality of CYPs. Rerouting the yeast redox metabolism (Fig. S2†) by overexpression of ZWF1 glucose-6-phosphate dehydrogenase and the mitochondrial NADH kinase POS5, capable of regenerating the NADPH cofactor, enabled increased titers of sesquiterpene lactones parthenolide and costunolide148 and triterpenes ursolic and oleanolic acid.149 Moreover, combining ZWF1 overexpression with replacing ALD2 gene involved in NADH generation with ALD6 responsible for synthesis of NADPH increased production of the aglycone ginsenoside protopanaxadiol by 11-fold.150 Furthermore, to enhance the supply of UDP-glucose in yeast as a sugar donor for UGTs activity, three endogenous genes were targeted and overexpressed. These included HXK1 encoding hexokinase, PGM1 encoding phosphoglucomutase, and UGP1 encoding glucose-1 phosphate uridylyltransferase, the last two acting as rate-limiting steps in the UDP-glucose biosynthetic pathway from glucose.151 Moreover, sucrose synthase (SuSy) was co-expressed with UGT73C5 from A. thaliana to regenerate uridine diphosphate glucose from cheap sucrose resulting in a 10-fold increase in Rh2 production.152 Recently, a UDP-rhamnose donor was made available in yeast by introducing the RHM2 from A. thaliana, which converts UDP-glucose to the desired sugar153 (Scheme S2†).
4. Engineering of terpene biosynthetic enzymes
The biosynthetic enzymes involved in terpenoid biosynthesis are sophisticated biocatalysts responsible for the astounding diversity of chemical structures produced. In native organisms, these enzymes are evolutionary tuned to catalyze specific reactions in biosynthetic pathways governed by a complex regulation. Intricate pathways pose the risks of minimal substrate channeling and product accumulation, functional uncoupling of biosynthetic steps, and synthesis of undesirable by-products. At the enzyme level, we will discuss strategies from rational mutagenesis to directed evolution of biosynthetic enzymes involved in terpene biosynthesis. In plants, the specialized metabolism can dictate the primary end-product of a biosynthetic pathway without inhibiting normal cellular processes. However, the reconstruction of heterologous pathways in yeast stands on the consumption of endogenous metabolites involved in the native metabolism. Specifically, the production of terpenoids is based on isoprene units produced by a central pathway, sterol biosynthesis. As discussed in chapter 2, the availability of the different isoprenoid precursors is based on their respective regulation in essential cellular processes, which in turn influence the varying success of the initial mono-, sesqui, and diterpene production strategies. Protein engineering strategies have been applied to enzymes acting in all three terpenoid biosynthesis modules.4.1 Engineering of prenyl transferase enzymes
To divert the native metabolic flux of the MVA pathway in yeast for the production of carotenoids, protein engineering strategies have been employed on the central enzyme of the pathway ERG20p38 (Fig. 5A). Ignea et al. rationally engineered ERG20p into a dominant negative GPP synthase to increase the downstream production of monoterpenes.38 Several mutations resulted in a decreased affinity for the second reaction substrate, GPP, resulting in its premature release and, subsequently, perturbing synthesis of the final product FPP.38 ERG20(F96W) mutant was identified as the most efficient variant, but the majority of the produced GPP was still converted to FPP38 by the wild-type Erg20p enzyme. The N144 residue of FPS1 from Gallus gallus was previously shown to be an integral part of the active site of the other FPS1 subunit in its homodimer conformation.154 Homology modelling of Erg20p with FPS1 structures (PDB ID: 1UBX, 1UBY), revealed the asparagine in position 127 homologous to N144 in FPS1. Ignea et al. hypothesized that substitution of N127 with a bulkier residue, such as tryptophan, may block the active site of the paired subunit. When expressed in yeast cells, the Erg20p(N127W) mutant abolished the FPP synthesis without affecting GPP production and performed as a dominant negative enzyme that inhibit the FPP synthesis by the wild-type enzyme in the heterodimer conformation with the mutant.38 The double variant ERG20(F96W-N127W) was shown to further increase the synthesis of GPP resulting in an over 343-fold improvement in sabinene production over the base strain.38 Subsequently, this highly efficient biocatalyst has been employed for the production of other monoterpenes155 and meroterpenoids bearing a GPP-derived moiety in their structure, such as cannabinoids28 or vinblastine.30 In a later study, Ignea et al. engineered Erg20p variants to produce geranylgeranyl diphosphate (GGPP), the first dedicated precursor in diterpenoid biosynthesis.40 Bypassing the activity of the native GGPP synthase, BTS1p, enabled the formation of GGPP in successive steps by the same Erg20p variant (Scheme 2). Homology analysis of ERG20p and FPS1 and superposition of E. coli FPP synthase IspA enabled the identification of five residues (Y95, F96, L97, Q168, and Y201) in the ERG20p active site, contributing to the steric hindrance. Rational mutations of these residues enabled the reshaping of the active site cavity and binding of FPP for elongation by the addition of an IPP unit (Fig. 5A). Mutations to the Y95 and F96 residues were associated with chain length termination. Consequently, the ERG20(F96C) mutant achieved a 70-fold improvement in the production of the diterpene sclareol.40 The mutant was also shown not to disrupt ergosterol biosynthesis, maintaining its FPP synthesis capacity to replace the wild-type ERG20p (Table S7†).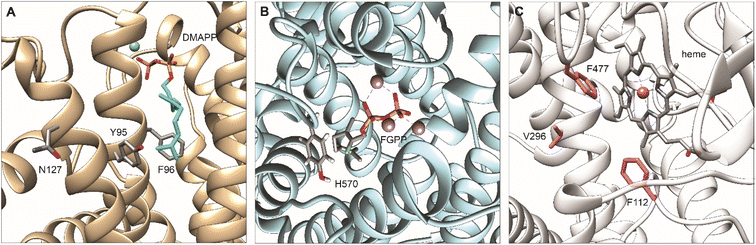 | ||
| Fig. 5 Homology modeling of engineered upstream enzymes for enhanced bioproduction of plant-based terpenoids in yeast with key residues for mutagenesis highlighted. (A) Model of Erg20p complexed with DMAPP showing residues Y95, F96, and N127 that are essential for controlling product length. Homology model produced by PyMod 3 plugin in PyMol160 based on the structure of fanesyl pyrophosphate synthase in Gallus gallus: PDB ID: 1UBY. Adapted with permission from ref. 38 Copyright © 2014, American Chemical Society and ref. 40 Copyright © 2014 International Metabolic Engineering Society, with permision from Elsevier Inc. (B) Model of Citrus limon limonene synthase (ClLimS) complexed with a GPP analogue, 2-fluorogeranyl diphosphate (FGPP); residue H570 influencing substrate selectivity is highlighted. Homology model produced by PyMod 3 plugin in PyMol160 based on the structure of limonene synthase in citrus sinensis: PDB ID: 5UV1. (C) Model of CYP76AH24 from S. pomifera showing heme group and residues F112, V296, F477 that are essential for controlling substrate selectivity and product specificity. Homology model produced by SWISS-MODEL161–163 based on sugiol synthase in S. miltiorrhiza CYP76AH3, PDB ID: 7X2Q. All graphics produced with UCSF Chimera.138 | ||
Another rational protein engineering strategy applied to increase the availability of prenyl diphosphates in yeast consists of the addition of a degradation signal (degron K3K15) at the N-terminus of ERG20p to favor monoterpene production.97 The tagging of degron K3K15 decreased the half-life of GFP to 1 hour. In addition to the use of a sterol-responsive promoter (ERG1 promoter), this signal achieved an improved carbon flux balance between monoterpene synthesis and sterol biosynthesis leading to a 27-fold improvement in monoterpene production.97 However, the reduction in FPP availability increased the doubling time of strain.97 A protein degradation method was applied to the production of sesquiterpenes.156 Although FPP is the main end-product of the MVA pathway, when it is produced in excess, it is converted to squalene by ERG9p.156 The overproduction of squalene is a metabolic burden for cells leading to a reduction in growth rate.156 To avoid squalene accumulation, a protein destabilization technique was developed for ERG9p, localized to the endoplasmic reticulum and ER-derived lipid droplets.156 Thus, a rich in Pro, Glu/Asp, Ser, and Thr (PEST) sequence-dependent endoplasmic reticulum-associated protein degradation (ERAD) system was established for Erg9p.156 The addition of the C-terminal peptide tag to Erg9p enabled recognition of the enzyme as a misfolded ER protein, increasing its degradation rate and reducing its half-life. Following the transcriptional downregulation of Erg9p, the conversion rate of its product (FPP) to the downstream intermediate (squalene) was significantly reduced leading to the availability of FPP in yeast cells as terpene precursor. This mechanism helped divert excess FPP toward nerolidol biosynthesis, resulting in an 86% titer improvement.156
4.2 Terpene synthase engineering
Mechanistically fascinating enzymes, TPSs exhibit great ability to improvise novel or altered functions with a small number of amino acid substitutions, a property referred to as plasticity. Different mutagenesis approaches have been employed to improve the performance of terpene synthases in vitro or in vivo studies. Several studies have demonstrated that a single amino acid residue switch can determine substrate and product specificity in terpene synthases. A conserved residue in plant monoterpene synthases was identified to control isomeric substrate selectivity.98 F571 in 1,8-cineole synthase from Salvia fruticosa (SfCinS) acts as a switch to exert stereochemical control of the enzyme preference in binding the canonical GPP in trans configuration or its cis isomer neryl diphosphate (NPP). Corresponding residues in the other synthases exhibited a similar role in dictating substrate selectivity. Variants of limonene synthase from Citrus lemon (ClLimS) (Fig. 5B), sabinene synthase from Salvia pomifera (SpSabS), and camphene synthase from Solanum elaeangifolium (SeCamS) were able to preferentially convert NPP accumulated in yeast via an orthogonal pathway to produce efficiently a range of monoterpene.98 Sabinene titers by SpSabS(H561F) in yeast reached 113 mg L−1, a 7-fold increase compared with GPP-based synthesis by the wild-type SpSabS. This amount was sufficient to enable the production of trans-sabin-3-ol98 by the sabinene hydroxylase CYP750B1 (ref. 157) from Thuja plicata. Notably, the F571 conserved residue in SfCinS and its homologous counterparts play a role in converting canonical plant monoterpene synthase into unnatural C11 terpene synthases, that specifically catalyze the synthesis of new-to-nature terpenes with 11 carbons in yeast117 (Table S7†). This achievement was demonstrated by engineering variants that preferentially accept the noncanonical substrate 2-methyl-GPP to produce 2-methyllimonene (ClLimS(H570V, L or I)), 2-methyl-α-terpineol (SfCinS1(F571Y)), 2-methyllinalool (ObMyrS(F579V) and PtPinS(F607L)), and 2-methylmyrcene (SfCinS1(N388S-I451A)).117 A conserved amino acid residue was also identified in triterpene synthases from diverse plant species to determine product specificity.158 Mutation of the serine in position 728 in β-amyrin synthase (SAD1) into a phenylalanine (S728F) influenced the cyclization profile of SAD1, enabling a synthesis of tetracyclic (dammarane) instead of pentacyclic triterpene scaffolds. Moreover, when expressed in yeast, SAD1(S728F) mutants showed a preference to cyclize dioxidosqualene rather than SAD1 canonical substrate oxidosqualene. As a result, the production of epoxydammaranes was detected in yeast instead of β-amyrin. Similarly, lupeol synthase from A. thaliana (AtLUP1), mutated at the equivalent amino acid residue (T729F), enabled the synthesis of tetracyclic triterpenes as the major cyclization products and preferentially converted dioxidosqualene to epoxydammaranes in yeast. Remarkably, this residue was demonstrated to be a substrate specificity switch only when expressed in yeast cells, revealing the hidden functional diversity in specialized metabolism.158 More recently, site-directed mutagenesis of santalene synthases revealed a plastic residue F441 that controls the sesquiterpene ratio in sandalwood oil. Introduction of a F441V variant of the santalene synthase for Clausena lansium in engineered yeast resulted in production of a mixture of compounds that match the corresponding ISO standard ratio.159Directed evolution is a powerful protein engineering strategy, utilizing introduced genetic variability in conjunction with a user-defined evolutionary pressure to mutate a target enzyme and select for the desired improvement.164 Directed evolution has been applied to various enzymes, and species in vitro systems and more recently, methods to evolve enzymes in vivo systems, such as yeast, have been developed.165,166 However, in vivo evolution of enzymes related to terpenoids remain in their infancy. Wang et al. applied directed evolution to evolve isoprene synthase (ISPS) and enhance its enzymatic activity for the efficient production of isoprene in yeast. An efficient mutant, ISPSM4(F340L/A570T) with 3-fold activity improvement was identified.167 The team has further improved ISPS activity via saturation mutagenesis considering the two hot spots (F340 and A570) identified by directed evolution and a set of three hydrophobic residues F338–V341–F485 in proximity to the active site. Through combinatorial mutagenesis, an efficient mutant ISPLN was engineered, showing increased isoprene by 4-fold over the strains expressing the wild-type enzyme.168 The expression of the ISPLN enzyme in conjunction with optimization of precursor supply produced from the cytoplasm and a mitochondrial pathway compartmentalization strategy led to the highest isoprene yield (11.9 g L−1) in eukaryotic cells168 (Table S7†).
4.3 Engineering performance of cytochrome P450s
Cytochrome P450s are a large group of plant enzymes involved in specialized metabolism.169 Through evolution, many CYPs acquire a certain degree of promiscuity making these enzymes ideal targets for engineering strategies to improve their catalytic performance or modify their substrate and product specificity. Site-directed mutagenesis was performed on CYP76AH24 from S. pomifera involved in the synthesis of carnosic acid intermediates.19,104,137 The enzyme was shown to accept different substrates and catalyze the synthesis of pathway intermediate ferruginol and 11-hydroxy-ferruginol or by-products, such as 11-keto-miltiradiene. Considering its promiscuity, CYP76AH24 was converted into a surrogate enzyme for the synthesis of forskolin precursor 11β-hydroxy-manoyl oxide.104 Specifically, the reconstruction of the carnosic acid pathway in yeast resulted in the production of minor products, such as pisiferic acid and salviol, proposed to have potent bioactivities yet not well understood. To preferentially produce pisiferic acid and salviol and bypass the natural plasticity of the carnosic acid pathway, site-directed mutagenesis of CYP76AH24 enabled restricting the promiscuity of the enzyme137(Fig. 5C). It has been demonstrated that CYP76AH24 oxidizes miltiradiene, abietatriene, ferruginol, and manoyl oxide. A single amino acid substitution F112L in the enzyme active site rendered the CYP76AH24 into a dedicated ferruginol synthase, increasing the precursor availability for the target minor products that are difficult to isolate from small amounts produced by the wild-type enzyme in plants or microbial hosts. CYP76AH24 (F112L) was shown to increase the production of pisiferic acid and salviol by 24 and 14-fold, respectively, creating a first-of-its-kind dedicated platform for the target products137 (Table S8†). CYP720B1 from P. taeda was also reported as a promiscuous enzyme catalyzing the oxidation of abietadiene as the main precursor, but also of structurally resembling labdane diterpenes, including miltiradiene, manoyl oxide, and manool.102 Recently, CYP720B1 was found to oxidize non-natural terpenoids with 16 carbon and produce several unique molecules not found in nature.91 A rational mutagenesis approach enabled the identification of plastic residues in the proximity of the CYP720B1 active site, which, if mutated, altered product specificity, narrowed substrate selectivity, and improved catalytic efficiency102 of the enzyme. Dedicated biocatalysts were engineered for the production of 18- or 19-hydroxy-miltiradiene. Interestingly, a single residue change (G359A) was sufficient to render the mutant specific to exclusively produce 18-hydroxy-miltiradiene with 2-fold higher efficiency than the originating enzyme. By contrast, mutation of T295 to S had an antagonistic effect resulting in relaxed product specificity. Joint with a second substitution of I223 by G, enabled the conversion of CYP720B1 into a 19-hydroxy-miltiradiene synthase. Screening the activity of a library of single mutants and rationally pairing substitutions at two residues, double mutants were engineered as dedicated 3β-hydroxy-manool synthase (L123V-I223L) and 19-hydroxy-manool synthase (I223G-L466M)102 (Table S8†).5. Targeting localization of terpene biosynthesis in yeast
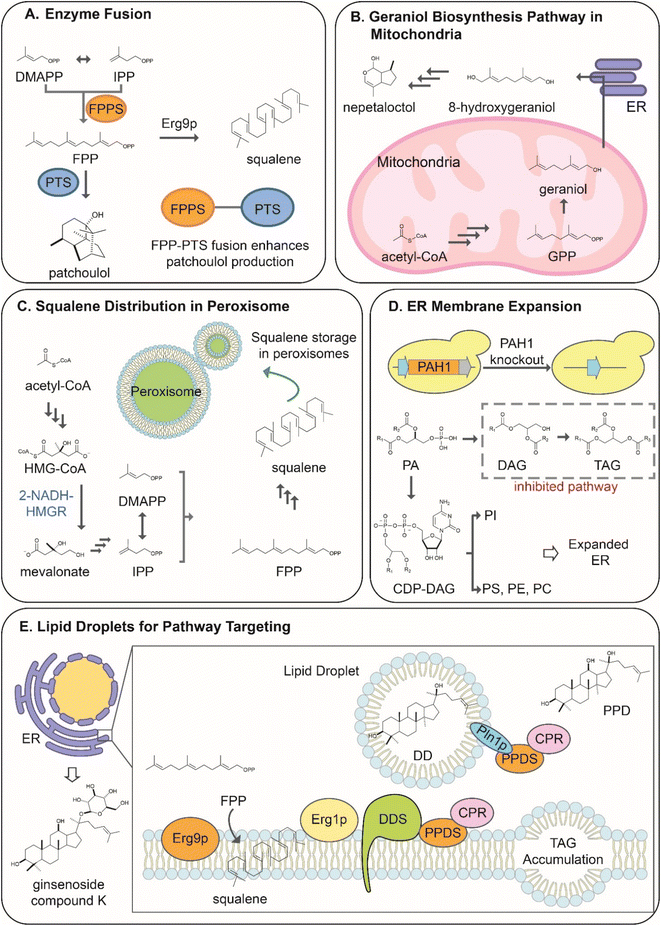
The longer the reconstructed pathway, the greater is the metabolic burden for the cell and the higher possibility of inadvertent crosstalk between the synthetic pathway and native metabolism, pathway deregulation, or even toxicity. To alleviate these limitations in the production of terpenoids in yeast, strategies were developed to increase enzyme proximity, availability of substrates and co-localize biosynthetic enzymes and pathways. Modulating the spatial localization within the cells improves pathway performance by increasing the probability of the desired reactions. This approach deviates from the traditional metabolic flux modulation, enhancing the dedicated carbon flux through the pathway, addressing the metabolic crosstalk between endogenous, and reconstructed pathways and decreasing the competition for substrates with endogenous metabolism. This chapter will discuss strategies for terpene enzyme fusions and scaffolds and compartmentalization of terpene biosynthetic pathways in yeast subcellular organelles.5.1 Enzyme fusions and scaffolds
The simplest strategies investigated to increase the proximity of enzymes and promote substrate channeling are enzyme fusions38,40,102 (Fig. 6A). The linking and subsequent co-localization170 of enzymes allow increased substrate channeling through the pathway. The proximity of the substrate-binding site with the upstream enzyme's catalytic site limits the ability of endogenous enzymes to compete for their common precursors and interfere with the desired reaction.170 Although the rationale behind enzyme fusions is relatively simple, their application is quite complex. The linking of enzymes can result in improper folding and function of the individual linked proteins.1 No standard method has been established to predict the efficacy of the linker strategy, often requiring trial and error to find the optimal length and type of linker used for the fusion strategy.171 Many different linkers are available for fusion strategies, ranging in size and rigidity.171To improve terpenoid production in yeast, strategies have been developed to fuse prenyltransferases acting in successive steps to efficiently synthesize of larger prenyl diphosphates, such as GGPP,40,103 or cyclic labdane-type substrates, such as copalyl diphosphate.40,103 The resulting fusions showed improved synthesis of target terpenes between 2 to 6-fold, according to a specific orientation of enzymes in the fusion. Prenyl building blocks are converted to terpene scaffold at a greater rate by fusing prenyltransferases with the downstream enzyme in the pathway, the terpene synthases. ERG20 fusions were investigated for the production of the sesquiterpene patchoulol.172 The fusion of ERG20 to the patchoulol synthase (PTS) from Pogostemon cablin yielded a two-fold increase of the sesquiterpene compared to the enzymes expressed separately.172 Interestingly, the length and type of linker used were shown to have little effect on the production rate of patchoulol.172 However, the fusion orientation was shown to affect the production. Only the ERG20-PTS fusion was shown to improve production, while the PTS-ERG20 had no advantage over the expression of the separate enzymes, demonstrating the large variability within fusion strategies.172 The ERG20 fusion strategy was also applied to the production of monoterpenes.38 The GPP-producing mutant ERG20(F96W-N127W) was shown to significantly increase the production of monoterpenes when fused to terpene synthases by limiting its conversion to FPP.38 The fusion of the sabinene synthase from S. pomifera (SpSabS1) to ERG20(F96W-N127W) increased the sabinene titer by 3.5-fold.38 Enzyme fusion strategies have also been developed to increase the performance of an integrated pathway. For example, the β-carotene biosynthesis starting from the dedicated precursor GGPP consists of three enzymes: phytoene desaturase (crtI), phytoene synthase (crtB), and lycopene cyclase (crtY)173 (Scheme 6). The expression of enzymes from Xanthophylomyces dendrorhous yielded the best titer of β-carotene in yeast. Although the strategy was the most successful up to that point, there was a buildup of the β-carotene precursor phytoene, indicating that the full potential of the pathway had not yet been unlocked.173 To increase the conversion rate of phytoene to β-carotene, Rabeharindranto et al. investigated tri-domain fusions enzymes with the natural crtYB fusion enzyme. The crtI enzyme fused to the crtYB enzyme (crt(YB)-crtI) with the flexible (EAAK)4 linker was shown to convert phytoene to β-carotene at the highest rate. This resulted in a 2-fold increase in the production over the natural enzyme expression, representing the highest titer of β-carotene up to date.173
In recent work, Kang et al. engineered a multienzyme complex of Idi1p and CrtE by tagging them with RIAD and RIDD short peptides that naturally exhibit an exceptionally strong affinity. The Idi1-CrtE assembly simplified the flux of lycopene biosynthesis in S. cerevisiae, increasing the fermentation titer by 58% over control strains reaching 2.3 g L−1 lycopene.174 Enzyme colocalization through attachment to a synthetic scaffold via noncovalent interactions can also improve production of target compounds in yeast. Using two pairs of anti-idiotypic affibodies, the key enzymes for farnesene formation, FPP synthase and farnesene synthase were tagged to a scaffold,175 which increased farnesene titers up to 120% in fed-batch cultivations. In a different approach, Han et al. spatially organized enzymes involved in the farnesene and farnesol pathway into a multi-enzyme protein body in yeast using the Tya protein176 capable of self-assembly in a shell similarly to virus-like particles (VLPs).177 The authors constructed Tya-fusions at the C- or N-terminus of key enzymes in the sesquiterpene pathway, such as yeast tHMG1, IspA from E. coli, and AFS1 from Malus domestica for farnesene production or yeast DPP1 for farnesol production. The assembly of these enzymes resulted in a synthetic metabolon that channeled the substrates towards the production of target compounds, increasing their titers by 3.1-fold for farnesene and 3.8-fold for farnesol when compared with free enzyme-based production.176
5.2 Converting yeast organelles into terpenoid-producing subcellular compartments
Organelles are defined as subcellular compartments isolated from the cytoplasm by a membrane. The isolation of the intra-organelle environment from the rest of the cytoplasm offers a unique advantage for synthetic biology approaches in yeast.178 The compartmentalization of terpenoid production pathways in organelles provides the ability to sequester a pathway from the endogenous pathways of the cytoplasm178,179 and has been recently shown to facilitate significant yield improvements. In terms of terpenoid synthesis, organelles would most notably limit the overlap of the heterologous pathways with the endogenous MVA pathway and eliminate the competition for isoprenoid precursors8 with immediate benefits for target product yield and host growth, respectively. The compartmentalization strategy also increases substrate concentration to accelerate conversion by heterologous enzymes170 and minimize toxicity effects by hydrophobic intermediates accumulated intracellularly. This leads to a higher probability of substrate to enzyme interaction and reaction, increasing enzyme and pathway performance.170 Differences in organelle structure and metabolic functions confer these compartments a specific cellular environment with unique properties that should be considered for targeting a specific biosynthetic pathway.1,7 Organelles successfully employed for the production of terpenoids are the mitochondria, peroxisome, and endoplasmic reticulum,179 recently reviewed by Yocum et al.170 This review will summarize approaches that apply to the production of plant terpenoids in yeast.The mitochondria, often called the cell's powerhouse, is responsible for the production of the primary energy subunit of the cell, adenosine triphosphate (ATP).180 The organelle is, therefore, rich in acetyl-CoA and ATP.71 In addition, the mitochondria house an abundance of cofactors and a high redox potential beneficial for the activity of specific plant enzymes and pathways.71,180 Moreover, the native GGPP synthase BTS1 is localized in the mitochondria concluding that the mitochondrial membrane is thus permeable to DMAPP and IPP.10 Although the mitochondrial environment is suitable for the activity of plant enzymes and has an abundance of the major substrates for terpenoid production, the importance of the organelle's function to cell viability limits the modification of its native activity.9
The impact of compartmentalizing biosynthetic pathways on the performance of the mitochondria was well demonstrated by the work of Zhu et al., reporting reconstruction of the MVA pathway in the mitochondria for overproduction of squalene.180 MVA enzymes were localized to the mitochondria by adding the mitochondrial localization signal (MLS) of the CoxIV enzyme,9 most commonly used as a targeting signal for this organelle. However, other MLS have also been used, such as Hmil, Coq3, Hsp60, Pda1, Oli1, Cdc9, and Eat1.170 The authors produced IPP and DMAP in the mitochondria by localizing the pathway enzymes Erg10p, Erg13p, tHmg1p, Erg12p, Erg8p, Erg19p and Idi1p in the mitochondria,9 which increased accumulation of squalene in the cytoplasm. However, the strain engineered to compartmentalize MVA in mitochondria exhibited reduced biomass accumulation compared to a wild type-strain.9 The stepwise build-up of the mitochondrial localized MVA pathway allowed for the identification of the toxicity of several pathway intermediates,9 specifically IPP, DMAPP, mevalonate-5-P, and mevalonate-5-PP. Accumulating these phosphorylated intermediates increased the doubling time of the strain, with double phosphorylated compounds being more toxic than the single phosphorylated metabolites.9 The toxicity of the intermediates was attributed to their conversion to ATP analogs inhibiting the normal function of the oxidative phosphorylation pathways.9 The elongation of the MVA pathway in the mitochondria to produce a sequestered GPP pool was achieved by introducing a mutant FPP synthase (mFPS) from Gallus gallus.181 Subsequent localization of the geraniol synthase from Ocimum basilicum was shown to drive geraniol production with a 6-fold increase compared to cytosolic producing strains (Fig. 6B). The high geraniol yield produced in mitochondria supported the cytosolic reconstruction of downstream steps involved in strictosidine biosynthesis catalyzed by geraniol hydroxylase (G8H), geraniol oxidoreductase (GOR), and iridoid synthase (ISY) from Catharanthus roseus. Accumulation of 8-hydroxygeraniol intermediate at a titer of 227 mg L−1 in a fed-batch fermentation enabled further pathway extension, resulting in the first de novo nepetalactol production. In addition, the production of sesquiterpenes in the mitochondria was also investigated by the additional introduction of ERG20p to elongate the mitochondrial MVA pathway to FPP.182 The production of amorpha-4,11-diene and patchoulol was achieved by the localization of the amorpha-4,11-diene synthase (ADS) from Artemisia annua and patchoulol synthase from P. cablin.183 Although the studies successfully produced their target compounds, the production came with a significant reduction in biomass accumulation attributed to the aforementioned toxicity of the pathway intermediates in the mitochondria.181–183
Peroxisomes are small organelles involved in various functions such as energy metabolism.178 In most cases, the organelles are not necessary for the viability of the cell, making it an attractive candidate for the production of terpenoids.178,179 In addition, the beta-oxidation of fatty acids in the peroxisome results in a sizeable acetyl-CoA pool available for the introduced pathway.184,185 The organelle is also permeable to various small molecules due to its simple membrane structure, making it suitable for terpenoid pathway integration.170 Furthermore, the biogenesis of peroxisomes can also be manipulated to increase the total number of organelles present within the cell.178
The localization of pathways to the peroxisome can be achieved by adding peroxisomal localization sequences to the gene sequence. The most commonly used signal is the C-terminus PTS1 localization tag, but N-terminus PTS2 localization signals have also been used.178 Peroxisomal compartmentalization strategy has been investigated, most notably, for the production of monoterpenes, specifically due to tight regulation at the Erg20p steps, as previously mentioned. The production of monoterpenes in the peroxisomes has achieved the highest production titers for geraniol and (R)-(+)-limonene by integrating the MVA pathway up to GPP within the peroxisome with the additional terpene synthases; ObGES and (R)-(+)-limonene synthase from Citrus limon, respectively.99 This localization strategy resulted in a titer of 5.5 g L−1 and 2.6 g L−1 for geraniol and (R)-(+)-limonene, respectively, increasing monoterpene titers up to 125-fold compared to cytosolic production.99 The high monoterpene production in peroxisome is associated with regular biomass accumulation compared with non-compartmentalized strains suggesting that the strategy does not add on to the inherent toxicity of the produced compounds, thus removing the main limitation observed in mitochondrial localization. The significant variation observed between the monoterpenes produced suggests that the variability is due to the GPP conversion rate of the individual terpene synthase. This approach has been, so far, the most efficient strategy to address the current limitations in the availability of GPP for the terpene synthases and cytosolic production of monoterpenes and other GPP-derived compounds.99 Peroxisomal targeting has also been used to produce other classes of terpenoids, such as sesqui- and triterpenes (Fig. 6C). Reconstruction of the MVA pathway up to FPP in Y. lipolytica peroxisomes coupled with localization of the nerolidol synthase from Fragaria ananassa and other metabolic engineering approaches, led to a 11.1 g L−1 yield of the sesquiterpene in a fed-batch fermentation.186 The compartmentalization of the MVA pathway and the squalene producing enzyme (ERG9) led to an increased squalene titer (1.3 g L−1) over the wild-type strain, representing a 138-fold improvement over the native cytosolic pathway.187 When used in conjunction, the peroxisomal and cytosolic pathways achieved a titer of 11 g L−1 squalene in fed-batch culture.187 This approach was further exploited to limit the inhibitory effect of squalene on the β-amyrin synthase from Glycyrrhiza glabra improving β-amyrin production by 2.6 fold.188
5.3 Manipulation of yeast intracellular membrane for colocalization of terpenoid biosynthesis
Synthetic biology approaches related to the engineering of the endoplasmic reticulum mostly aim to manipulate ER-associated proteins and lipid metabolism.8 Particularly, ER membrane is the natural environment for one of the largest classes of enzymes involved in plant-specialized metabolism, the cytochrome P450s. Other biosynthetic enzymes, such as aromatic prenyltransferases or specific cyclases, may also require embedding themselves in the ER membrane for functionalization. Strategies have been developed to increase heterologous protein expression by expanding the endoplasmic reticulum structure.189 This approach led to an increased accumulation of the ER-associated terpene enzymes and consequently improved the production of target compounds.190 The increased size of the endoplasmic reticulum was achieved by overexpressing an ER size regulatory gene, INO2. Consequently, an increased expression level of enzymes involved in squalene and protopanaxadiol synthesis was obtained, leading to a 71-fold and 8-fold titer improvement, respectively.189 The deletion of the phosphatidic acid phosphatase-encoding PAH1 gene also achieved the expansion of the ER190 (Fig. 6D). This resulted in an improved titer of triterpenes such as β-amyrin by 8-fold over the control strain. The PAH1 deletion was also shown to improve the production of other terpenoids, depending on ER association status of the specific enzymes.190 Artemisinic acid production was also increased in strains with enlarged ER membranes, although at a lesser rate than β-amyrin, generating a twofold improvement.190 Furthermore, colocalization of cytosolic enzyme in the ER in conjunction with INO2 overexpression was applied to the biosynthesis of 7-dehydrocholesterol, engineered in yeast through the expression of the Δ24-dehyrocholesterol reductase (DHCR24). Despite the natural compartmentalization of DHCR24 and several other sterol enzymes to the ER, the biosynthesis of 7-dehydrocholesterol was sequestered across cytoplasm, ER and lipid droplets. To further increase the substrate channeling through the pathway, six enzymes (Erg12p, Erg8p, Mvd1p, Idi1p, Erg20p, and Erg7p) were C-terminus fused with the ER-targeting signal peptide CNE1 (ref. 191) and colocalized to ER. This strategy combined with tightly controlled gene expression, conveyed by CRISPR-mediated systems, increased 7-dehydrocholesterol to 2.87 g L−1 in fed-batch cultivation.192Lipid droplets (LD) are small ER-derived organelles involved in lipid metabolism8 that have also been employed to increase the production of terpenoids in yeast. The organelles are attractive for synthetic biology due to their hydrophobic nature, allowing their use as dynamic storage depots and pathway concentrators. Terpenoid pathway enzymes have been targeted to LDs through fusion with the PLN1 protein,193 naturally located in the organelle membrane. This approach allows for the efficient conversion of hydrophobic terpenoid precursors stored in the droplets leading to the prevention of cell saturation and toxicity at high terpenoid concentration and continuous production which overall improves strain performance.193 Targeting protopanaxadiol (PPD) synthase to the lipid droplets resulted in an almost 5-fold increase in its conversion rate of dammarenediol-II substrate193 (Fig. 6E). In conjunction with the further tuning of protein expression, the ginsenoside production reached up to 5 g L−1 in fed-batch fermentations.193 The LD localization of the amyrin synthase (CrAS) and oxidase (CrAO) from C. roseus paired with the CPR from A. thaliana (AtCPR1) was beneficial for the production of ursolic and oleanolic acid triterpenoids. CrAS was shown to naturally localize to LDs. Targeting the fusion CrAO-AtCPR1 to the LD membrane positively impacted ursolic acid and oleanolic acid titers to 1132.9 mg L−1 and 433.9 mg L−1 respectively,149 when combined with MVA pathway engineering and rewiring the fluxes of acetyl-CoA precursor and NADPH cofactor (discussed in Section 3.3). The storage of terpenoids in lipid droplets has also been shown to increase the performance of the strains, as it offers sequestered storage for the lipotoxic compounds.194 In addition, the biogenesis of the lipid droplet can be controlled by tuning the expression of crucial gene regulators.194 For example, the deletion of the FLD1 gene can increase the lipid droplets' size up to 50-fold, increasing the available storage space.194 However, the increase in the size of the lipid droplets should be balanced due to the extensive carbon source usage required for their formation.184 The use of the lipid droplets strategy has been applied to the production of lycopene in S. cerevisiae.194 The overexpression of yeast fatty acid desaturase (OLE1), coupled with the deletion of FLD1, was shown to be beneficial to lycopene accumulation in yeast,194 resulting in the highest titer (2.37 g L−1) achieved in S. cerevisiae. While lycopene production in baker's yeast still remains lower than the yields achieved in Y. lipolytica,194 this could provide a proof-of-concept for increasing production of other lipophilic compounds.
6. Conclusion and future perspectives
Over the last decade, significant advances have been reported for the production of terpenoids in microbial hosts. Due to their eukaryotic yet unicellular organization, comprehensive understanding, and ease of manipulation, yeasts have become prime candidate hosts for terpenoid production. Successful examples are summarized in Tables S2–S6.† Nevertheless, optimizing engineered strains for industrial-scale production is still time-consuming and the acceleration of this process is essential for developing sustainable strategies. The reconstruction of almost the entire pathway of the anticancer drug vinblastine in yeast, comprising 30 biosynthetic steps beyond the yeast metabolism,30 marks a new level in the field. Consequently, fully or partially elucidated biosynthetic pathways of important terpenoids (momilactones,195 triptolide,130 ginkgolides133) are yet to be reconstructed in microbial chassis. Complex and intricate pathways of taxol196 or celastrol,126 are still lacking the knowledge of their full biosynthesis and yeast could provide the appropriate platform to identify the missing steps. Moreover, the discovery of many terpenoid pathways of potent terpenoids is still in its infancy. Such cases include the potent diterpene resiniferatoxin or the antiobesity agent withaferin A. Finally, recent studies have provided proof-of-concept on the enzymatic derivatization of terpenoid compounds to produce non-natural tailored analogs in yeast91,117 that may have improved or broader spectrum of biological activities, increased bioavailability, and reduced side effects when used as therapeutics.To address current limitations, novel synthetic biology approaches must be developed and integrated into the current toolbox for the efficient production of terpenoids in yeast and the intensification of strain optimization. We foresee the development of more advanced high-throughput tools for gene mining, pathway elucidation, and screening of potent variants for specific enzymatic activities or engineering synthetic biology gate systems that read signals resembling the plant's native specialized metabolism. Such strategies will allow further exploration and expansion of the untapped chemical space of terpenoids and add to the natural diversity of an unlimited number of building blocks and scaffolds. Mimicking intracellular plant environments in microbes by manipulating the ER membrane composition197 or engineering plastid-like organelles in yeast will support the optimal performance of plant enzymes and pathways. Rational compartmentalization of pathways will benefit from intraorganellar conditions and address the multilevel toxicity of intermediates to alleviate metabolic burden of the host and bypass enzyme inhibition by substrates or products. It has been shown that specialized metabolism acts at the molecular level in a coordinated fashion through sophisticated metabolic complexes known as metabolons198 to achieve remarkable natural diversity. Reproducing such complexes as customized synthetic metabolons will allow us to control reconstructed pathways with nature's precision and accelerate optimization processes by switching fluxes for the production of target compounds by demand. More recently, membraneless organelles199 have been shown to spontaneously arise from heterogeneous interactions of cellular material and form coacervate drops lacking the typical lipid barrier in a process termed liquid–liquid phase separation (LLPS). Beyond their organizational role, membraneless organelles tune biochemical reactions and improve cellular fitness during stress conditions. Not yet reported in specialized metabolism, coacervate drops may act as intracellular nanobioreactors that, if engineered in a heterologous host, could address our limitations to overcome the intricate natural regulation in terpenoid biosynthesis. Despite becoming an integrated tool manipulating terpene biosynthetic pathways, enzyme engineering remains a very difficult approach. In vivo evolution of biosynthetic enzymes is envisioned to facilitate these efforts assuming an evolutionary pressure is associated. Engineering intracellularly driven selection of the most fitted variants will enable system autotuning and self-optimization. Moreover, the expansion of genetic code offers new horizons to engineer artificial enzymes with tailor activities capable of reshaping protein structures by incorporating isomeric non-natural residues, labeling biosynthetic enzymes with fluorescent signals to monitor enzyme dynamics in vivo systems or introducing intended physicochemical properties in the structures of biosynthetic enzymes to trigger specific interactions. While mastering nature's complexity will remain challenging, novel synthetic biology and metabolic engineering approaches could lead to exciting developments in the field in the coming years.
7. Abbreviations
| IPP | Isopentenyl diphosphate; |
| DMAPP | Dimethylallyl diphosphate |
| GPP | Geranyl diphosphate |
| FPP | Farnesyl diphosphate |
| NPP | Neryl diphosphate |
| MVA | Mevalonate pathway |
| GGPP | Geranylgeranyl diphosphate |
| HR | Homologous recombination |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| HDR | Homology-directly repair |
| DSB | Double-stranded breaks |
| PCR | Polymerase chain reaction |
| NHEJ | Non-homologous end-joining |
| ALE | Adaptive laboratory evolution |
| MEP | Methylerythritol phosphate |
| ISC | Iron sulfur cluster |
| TPSs | Terpene synthases |
| CYPs | Cytochrome P450s |
| UDP | Uridine diphosphate |
| UGTs | UDP-glycosyltransferases |
| LDs | Lipid droplets |
8. Author contributions
Conceptualization: CI; writing original draft: CI, JAB, MEO, YD; manuscript revision and editing: CI, JAB, MEO, YD; figure generation, YD, CI. Funding acquisition: CI. All authors contributed to the article and approved final submission.9. Conflicts of interest
The authors declare no conflict of interest.10. Acknowledgements
We would like to thank to Olga Sofianovich for critical reading of the manuscript and constructive comments and Kristofer Andreas Coolen for discussion on the chemical mechanisms. This work was financially supported by Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grant RGPIN-04360-2022, Canadian Institutes of Health Research (CIHR) Grant 202203PJT-481041-TIR-CFAA-98996, Natural Sciences and Engineering Research Council (NSERC) of Canada Alliance Grant ALLRP 576220-22, Consortium de Recherche et d’Innovation en Bioprocédés Industriels au Québec (CRIBIQ) 2021-013-C77, New Frontiers in Research Fund - Exploration Grant NFRFE-2022-00158, Fonds de recherche du Québec – Nature et technologies (FRQNT) Grant 2024-NCM-326434 and McGill Start Up Fund to Codruta Ignea; Vadasz Scholar McGill Engineering Doctoral Award (MEDA) and Fonds de recherche du Québec – Nature et technologies (FRQNT) Bourses de doctorat en recherche 2023-2024 - B2X - 334275 to Jean-Alexandre Bureau, Consejo Nacional de Ciencia y Tecnología CONACyT Fellowship No. 795399 to Magdalena Escobar Oliva and McGill Engineering Undergraduate Student Masters Award (MEUSMA) to Yueming Dong.11. References
- J. Gershenzon and N. Dudareva, Nat. Chem. Biol., 2007, 3, 408–414 CrossRef CAS PubMed.
- K. C. Nicolaou and T. Montagnon, Molecules that changed the world : a brief history of the art and science of synthesis and its impact on society, Wiley-VCH, Weinheim, 2008 Search PubMed.
- D. Cox-Georgian, N. Ramadoss, C. Dona and C. Basu, Med. Plants, 2019, 333–359 Search PubMed.
- B. Singh and R. A. Sharma, 3 Biotech, 2015, 5, 129–151 CrossRef PubMed.
- L. Caputi and E. Aprea, Recent Pat. Food, Nutr. Agric., 2011, 3, 9–16 CrossRef CAS PubMed.
- X. Lyu, J. Lee and W. N. Chen, J. Agric. Food Chem., 2019, 67, 4397–4417 CrossRef CAS PubMed.
- G. H. Smith, J. M. Roberts and T. W. Pope, Crop Prot., 2018, 110, 125–130 CrossRef CAS.
- V. Ninkuu, L. Zhang, J. Yan, Z. Fu, T. Yang and H. Zeng, Int. J. Mol. Sci., 2021, 22, 5710 CrossRef CAS PubMed.
- A. Vattekkatte, S. Garms, W. Brandt and W. Boland, Org. Biomol. Chem., 2018, 16, 348–362 RSC.
- P. S. Karunanithi and P. Zerbe, Front. Plant Sci., 2019, 10, 1166 CrossRef PubMed.
- S. E. O'Connor and J. J. Maresh, Nat. Prod. Rep., 2006, 23, 532–547 RSC.
- M. Nazir, M. Saleem, M. I. Tousif, M. A. Anwar, F. Surup, I. Ali, D. Wang, N. Z. Mamadalieva, E. Alshammari, M. L. Ashour, A. M. Ashour, I. Ahmed, Elizbit, I. R. Green and H. Hussain, Biomolecules, 2021, 11, 957 CrossRef CAS PubMed.
- L. Chen, Y. Pang, Y. Luo, X. Cheng, B. Lv and C. Li, Eng. Life Sci., 2021, 21, 724–738 CrossRef CAS PubMed.
- S. Moser and H. Pichler, Appl. Microbiol. Biotechnol., 2019, 103, 5501–5516 CrossRef CAS PubMed.
- Y. Zhang, J. Nielsen and Z. Liu, FEMS Yeast Res., 2017, 17(8) CrossRef PubMed.
- D. K. Ro, E. M. Paradise, M. Ouellet, K. J. Fisher, K. L. Newman, J. M. Ndungu, K. A. Ho, R. A. Eachus, T. S. Ham, J. Kirby, M. C. Chang, S. T. Withers, Y. Shiba, R. Sarpong and J. D. Keasling, Nature, 2006, 440, 940–943 CrossRef CAS PubMed.
- C. J. Paddon, P. J. Westfall, D. J. Pitera, K. Benjamin, K. Fisher, D. McPhee, M. D. Leavell, A. Tai, A. Main, D. Eng, D. R. Polichuk, K. H. Teoh, D. W. Reed, T. Treynor, J. Lenihan, M. Fleck, S. Bajad, G. Dang, D. Dengrove, D. Diola, G. Dorin, K. W. Ellens, S. Fickes, J. Galazzo, S. P. Gaucher, T. Geistlinger, R. Henry, M. Hepp, T. Horning, T. Iqbal, H. Jiang, L. Kizer, B. Lieu, D. Melis, N. Moss, R. Regentin, S. Secrest, H. Tsuruta, R. Vazquez, L. F. Westblade, L. Xu, M. Yu, Y. Zhang, L. Zhao, J. Lievense, P. S. Covello, J. D. Keasling, K. K. Reiling, N. S. Renninger and J. D. Newman, Nature, 2013, 496, 528–532 CrossRef CAS PubMed.
- A. L. Meadows, K. M. Hawkins, Y. Tsegaye, E. Antipov, Y. Kim, L. Raetz, R. H. Dahl, A. Tai, T. Mahatdejkul-Meadows, L. Xu, L. Zhao, M. S. Dasika, A. Murarka, J. Lenihan, D. Eng, J. S. Leng, C. L. Liu, J. W. Wenger, H. Jiang, L. Chao, P. Westfall, J. Lai, S. Ganesan, P. Jackson, R. Mans, D. Platt, C. D. Reeves, P. R. Saija, G. Wichmann, V. F. Holmes, K. Benjamin, P. W. Hill, T. S. Gardner and A. E. Tsong, Nature, 2016, 537, 694–697 CrossRef CAS PubMed.
- C. Ignea, A. Athanasakoglou, E. Ioannou, P. Georgantea, F. A. Trikka, S. Loupassaki, V. Roussis, A. M. Makris and S. C. Kampranis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3681–3686 CrossRef CAS PubMed.
- U. Scheler, W. Brandt, A. Porzel, K. Rothe, D. Manzano, D. Bozic, D. Papaefthimiou, G. U. Balcke, A. Henning, S. Lohse, S. Marillonnet, A. K. Kanellis, A. Ferrer and A. Tissier, Nat. Commun., 2016, 7, 12942 CrossRef CAS PubMed.
- I. Pateraki, J. Andersen-Ranberg, N. B. Jensen, S. G. Wubshet, A. M. Heskes, V. Forman, B. Hallstrom, B. Hamberger, M. S. Motawia, C. E. Olsen, D. Staerk, J. Hansen, B. L. Moller and B. Hamberger, eLife, 2017, 6, e23001 CrossRef PubMed.
- L. L. Chu, J. A. V. Montecillo and H. Bae, Front. Bioeng. Biotechnol., 2020, 8, 139 CrossRef PubMed.
- T. Ma, B. Shi, Z. Ye, X. Li, M. Liu, Y. Chen, J. Xia, J. Nielsen, Z. Deng and T. Liu, Metab. Eng., 2019, 52, 134–142 CrossRef CAS PubMed.
- B. Shi, T. Ma, Z. Ye, X. Li, Y. Huang, Z. Zhou, Y. Ding, Z. Deng and T. Liu, J. Agric. Food Chem., 2019, 67, 11148–11157 CrossRef CAS PubMed.
- J. Lopez, V. F. Cataldo, M. Pena, P. A. Saa, F. Saitua, M. Ibaceta and E. Agosin, Front. Bioeng. Biotechnol., 2019, 7, 171 CrossRef PubMed.
- P. Zhou, W. Xie, A. Li, F. Wang, Z. Yao, Q. Bian, Y. Zhu, H. Yu and L. Ye, Enzyme Microb. Technol., 2017, 100, 28–36 CrossRef CAS PubMed.
- V. F. Cataldo, N. Arenas, V. Salgado, C. Camilo, F. Ibanez and E. Agosin, Metab. Eng., 2020, 59, 53–63 CrossRef CAS PubMed.
- X. Luo, M. A. Reiter, L. d'Espaux, J. Wong, C. M. Denby, A. Lechner, Y. Zhang, A. T. Grzybowski, S. Harth, W. Lin, H. Lee, C. Yu, J. Shin, K. Deng, V. T. Benites, G. Wang, E. E. K. Baidoo, Y. Chen, I. Dev, C. J. Petzold and J. D. Keasling, Nature, 2019, 567, 123–126 CrossRef CAS PubMed.
- S. Brown, M. Clastre, V. Courdavault and S. E. O'Connor, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 3205–3210 CrossRef CAS PubMed.
- J. Zhang, L. G. Hansen, O. Gudich, K. Viehrig, L. M. M. Lassen, L. Schrubbers, K. B. Adhikari, P. Rubaszka, E. Carrasquer-Alvarez, L. Chen, V. D’Ambrosio, B. Lehka, A. K. Haidar, S. Nallapareddy, K. Giannakou, M. Laloux, D. Arsovska, M. A. K. Jorgensen, L. J. G. Chan, M. Kristensen, H. B. Christensen, S. Sudarsan, E. A. Stander, E. Baidoo, C. J. Petzold, T. Wulff, S. E. O’Connor, V. Courdavault, M. K. Jensen and J. D. Keasling, Nature, 2022, 609(7926), 341–347 CrossRef CAS PubMed.
- M. Kavšček, M. Stražar, T. Curk, K. Natter and U. Petrovič, Microb. Cell Fact., 2015, 14, 94 CrossRef PubMed.
- M. S. Siddiqui, K. Thodey, I. Trenchard and C. D. Smolke, FEMS Yeast Res., 2012, 12, 144–170 CrossRef CAS PubMed.
- F. David and V. Siewers, FEMS Yeast Res., 2015, 15, 1–14 CrossRef CAS PubMed.
- A. Szkopińska, K. Grabińska, D. Delourme, F. Karst, J. Rytka and G. Palamarczyk, J. Lipid Res., 1997, 38, 962–968 CrossRef.
- B. Dursina, N. H. Thomä, V. Sidorovitch, A. Niculae, A. Iakovenko, A. Rak, S. Albert, A.-C. Ceacareanu, R. Kölling, C. Herrmann, R. S. Goody and K. Alexandrov, Biochemistry, 2002, 41, 6805–6816 CrossRef CAS PubMed.
- J. Nielsen, mBio, 2014, 5, e02153 CrossRef CAS PubMed.
- C. Ignea, I. Cvetkovic, S. Loupassaki, P. Kefalas, C. B. Johnson, S. C. Kampranis and A. M. Makris, Microb. Cell Fact., 2011, 10, 4 CrossRef CAS PubMed.
- C. Ignea, M. Pontini, M. E. Maffei, A. M. Makris and S. C. Kampranis, ACS Synth. Biol., 2014, 3, 298–306 CrossRef CAS PubMed.
- C. Ignea, F. A. Trikka, I. Kourtzelis, A. Argiriou, A. K. Kanellis, S. C. Kampranis and A. M. Makris, Microb. Cell Fact., 2012, 11, 162 CrossRef CAS PubMed.
- C. Ignea, F. A. Trikka, A. K. Nikolaidis, P. Georgantea, E. Ioannou, S. Loupassaki, P. Kefalas, A. K. Kanellis, V. Roussis, A. M. Makris and S. C. Kampranis, Metab. Eng., 2015, 27, 65–75 CrossRef CAS PubMed.
- A. L. Lindahl, M. E. Olsson, P. Mercke, O. Tollbom, J. Schelin, M. Brodelius and P. E. Brodelius, Biotechnol. Lett., 2006, 28, 571–580 CrossRef CAS PubMed.
- Y. Yuan, E. Andersen and H. Zhao, ACS Synth. Biol., 2016, 5, 46–52 CrossRef CAS PubMed.
- J. López, K. Essus, I. K. Kim, R. Pereira, J. Herzog, V. Siewers, J. Nielsen and E. Agosin, Microb. Cell Fact., 2015, 14, 84 CrossRef PubMed.
- U. Güldener, S. Heck, T. Fielder, J. Beinhauer and J. H. Hegemann, Nucleic Acids Res., 1996, 24, 2519–2524 CrossRef PubMed.
- G. Scalcinati, C. Knuf, S. Partow, Y. Chen, J. Maury, M. Schalk, L. Daviet, J. Nielsen and V. Siewers, Metab. Eng., 2012, 14, 91–103 CrossRef CAS PubMed.
- J. A. Doudna and E. Charpentier, Science, 2014, 346, 1258096 CrossRef PubMed.
- F. Storici, C. L. Durham, D. A. Gordenin and M. A. Resnick, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 14994–14999 CrossRef CAS PubMed.
- J. M. Gardner and S. L. Jaspersen, Methods Mol. Biol., 2014, 1205, 45–78 CrossRef CAS PubMed.
- J. Zhou, T. Hu, L. Gao, P. Su, Y. Zhang, Y. Zhao, S. Chen, L. Tu, Y. Song, X. Wang, L. Huang and W. Gao, New Phytol., 2019, 223, 722–735 CrossRef CAS PubMed.
- T. Siemon, Z. Wang, G. Bian, T. Seitz, Z. Ye, Y. Lu, S. Cheng, Y. Ding, Y. Huang, Z. Deng, T. Liu and M. Christmann, J. Am. Chem. Soc., 2020, 142, 2760–2765 CrossRef CAS PubMed.
- Y. Wang, X. Gong, F. Li, S. Zuo, M. Li, J. Zhao, X. Han and M. Wen, Appl. Microbiol. Biotechnol., 2021, 105, 8795–8804 CrossRef CAS PubMed.
- S. Shi, Y. Liang, E. L. Ang and H. Zhao, in Microbial Metabolic Engineering: Methods and Protocols, ed. C. N. S. Santos and P. K. Ajikumar, Springer New York, New York, NY, 2019, pp. 73–91, DOI:10.1007/978-1-4939-9142-6_6.
- B. Peng, L. Esquirol, Z. Lu, Q. Shen, L. C. Cheah, C. B. Howard, C. Scott, M. Trau, G. Dumsday and C. E. Vickers, Nat. Commun., 2022, 13, 2895 CrossRef CAS PubMed.
- J. C. Utomo, C. L. Hodgins and D. K. Ro, Front. Plant Sci., 2021, 12, 719148 CrossRef PubMed.
- T. Jakociunas, A. S. Rajkumar, J. Zhang, D. Arsovska, A. Rodriguez, C. B. Jendresen, M. L. Skjodt, A. T. Nielsen, I. Borodina, M. K. Jensen and J. D. Keasling, ACS Synth. Biol., 2015, 4, 1226–1234 CrossRef CAS PubMed.
- C. Ronda, J. Maury, T. Jakociunas, S. A. Jacobsen, S. M. Germann, S. J. Harrison, I. Borodina, J. D. Keasling, M. K. Jensen and A. T. Nielsen, Microb. Cell Fact., 2015, 14, 97 CrossRef PubMed.
- S. Hou, Q. Qin and J. Dai, ACS Synth. Biol., 2018, 7, 782–788 CrossRef CAS PubMed.
- M. Qi, B. Zhang, L. Jiang, S. Xu, C. Dong, Y. L. Du, Z. Zhou, L. Huang, Z. Xu and J. Lian, Front. Bioeng. Biotechnol., 2020, 8, 613771 CrossRef PubMed.
- S. Baek, J. C. Utomo, J. Y. Lee, K. Dalal, Y. J. Yoon and D. K. Ro, Metab. Eng., 2021, 64, 111–121 CrossRef CAS PubMed.
- C. Schwartz, M. Shabbir-Hussain, K. Frogue, M. Blenner and I. Wheeldon, ACS Synth. Biol., 2017, 6, 402–409 CrossRef CAS PubMed.
- X. K. Zhang, D. N. Wang, J. Chen, Z. J. Liu, L. J. Wei and Q. Hua, Biotechnol. Lett., 2020, 42, 945–956 CrossRef CAS PubMed.
- Z. Cui, H. Zheng, J. Zhang, Z. Jiang, Z. Zhu, X. Liu, Q. Qi and J. Hou, Appl. Environ. Microbiol., 2021, 87(6), e02666 CrossRef CAS PubMed.
- P. Liao, A. Hemmerlin, T. J. Bach and M.-L. Chye, Biotechnol. Adv., 2016, 34, 697–713 CrossRef CAS PubMed.
- E. Cámara, I. Lenitz and Y. Nygård, Sci. Rep., 2020, 10, 14605 CrossRef PubMed.
- E. D. Jensen, R. Ferreira, T. Jakočiūnas, D. Arsovska, J. Zhang, L. Ding, J. D. Smith, F. David, J. Nielsen, M. K. Jensen and J. D. Keasling, Microb. Cell Fact., 2017, 16, 46 CrossRef PubMed.
- J. Lian, M. HamediRad, S. Hu and H. Zhao, Nat. Commun., 2017, 8, 1688 CrossRef PubMed.
- G. Naseri, J. Behrend, L. Rieper and B. Mueller-Roeber, Nat. Commun., 2019, 10, 2615 CrossRef PubMed.
- Z. Lu, B. Peng, B. E. Ebert, G. Dumsday and C. E. Vickers, Nat. Commun., 2021, 12, 1051 CrossRef CAS PubMed.
- J. van Leeuwen, C. Boone and B. J. Andrews, Curr. Opin. Syst. Biol., 2017, 6, 14–21 CrossRef PubMed.
- M. Zhao, M. Gao, L. Xiong, Y. Liu, X. Tao, B. Gao, M. Liu, F. Q. Wang and D. Z. Wei, ACS Synth. Biol., 2022, 11, 1958–1970 CrossRef CAS PubMed.
- L. Duran, J. M. López and J. L. Avalos, FEMS Yeast Res., 2020, 20(6), foaa037 CrossRef CAS PubMed.
- M. Dragosits and D. Mattanovich, Microb. Cell Fact., 2013, 12, 64 CrossRef PubMed.
- L. H. Reyes, J. M. Gomez and K. C. Kao, Metab. Eng., 2014, 21, 26–33 CrossRef CAS PubMed.
- A. Godara and K. C. Kao, Microb. Cell Fact., 2021, 20, 106 CrossRef CAS PubMed.
- K. Paramasivan, A. A, N. Gupta and S. Mutturi, Yeast, 2021, 38, 424–437 CrossRef CAS PubMed.
- T. C. R. Brennan, T. C. Williams, B. L. Schulz, R. W. Palfreyman, J. O. Krömer and L. K. Nielsen, Appl. Environ. Microbiol., 2015, 81, 3316–3325 CrossRef CAS PubMed.
- J. Li, K. Zhu, L. Miao, L. Rong, Y. Zhao, S. Li, L. Ma, J. Li, C. Zhang, D. Xiao, J. L. Foo and A. Yu, ACS Synth. Biol., 2021, 10, 884–896 CrossRef CAS PubMed.
- L. Caspeta, Y. Chen, P. Ghiaci, A. Feizi, S. Buskov, B. M. Hallstrom, D. Petranovic and J. Nielsen, Science, 2014, 346, 75–78 CrossRef CAS PubMed.
- K. Zhou, K. Qiao, S. Edgar and G. Stephanopoulos, Nat. Biotechnol., 2015, 33, 377–383 CrossRef CAS PubMed.
- S. Wu, X. Ma, A. Zhou, A. Valenzuela, K. Zhou and Y. Li, Sci. Adv., 2021, 7, eabh4048 CrossRef CAS PubMed.
- P. M. Dewick, Medicinal Natural Products: A Biosynthetic Approach, Wiley, 3rd edn, 2009 Search PubMed.
- N. V. Bhagavan and C.-E. Ha, in Essentials of Medical Biochemistry, ed. N. V. Bhagavan and C.-E. Ha, Academic Press, San Diego, 2015, 2nd edn, pp. 299–320 Search PubMed.
- M. Rohmer, Nat. Prod. Rep., 1999, 16, 565–574 RSC.
- S. Opitz, W. D. Nes and J. Gershenzon, Phytochemistry, 2014, 98, 110–119 CrossRef CAS PubMed.
- A. Hemmerlin, J. L. Harwood and T. J. Bach, Prog. Lipid Res., 2012, 51, 95–148 CrossRef CAS PubMed.
- M. Thorsness, W. Schafer, L. D'Ari and J. Rine, Mol. Cell. Biol., 1989, 9, 5702–5712 CAS.
- M. Nakanishi, J. L. Goldstein and M. S. Brown, J. Biol. Chem., 1988, 263, 8929–8937 CrossRef CAS PubMed.
- J. L. Goldstein, R. A. DeBose-Boyd and M. S. Brown, Cell, 2006, 124, 35–46 CrossRef CAS PubMed.
- T. Polakowski, U. Stahl and C. Lang, Appl. Microbiol. Biotechnol., 1998, 49, 66–71 CrossRef CAS PubMed.
- R. Callari, Y. Meier, D. Ravasio and H. Heider, Front. Bioeng. Biotechnol., 2018, 6, 160 CrossRef PubMed.
- C. Ignea, M. H. Raadam, A. Koutsaviti, Y. Zhao, Y. T. Duan, M. Harizani, K. Miettinen, P. Georgantea, M. Rosenfeldt, S. E. Viejo-Ledesma, M. A. Petersen, W. L. P. Bredie, D. Staerk, V. Roussis, E. Ioannou and S. C. Kampranis, Nat. Commun., 2022, 13, 5188 CrossRef CAS PubMed.
- J. Yuan and C. B. Ching, Biotechnol. Bioeng., 2014, 111, 608–617 CrossRef CAS PubMed.
- T. Jordá and S. Puig, Genes, 2020, 11(7), 795 CrossRef PubMed.
- J. Yuan and C. B. Ching, Microb. Cell Fact., 2015, 14, 38 CrossRef PubMed.
- J. Zhao, X. Bao, C. Li, Y. Shen and J. Hou, Appl. Microbiol. Biotechnol., 2016, 100, 4561–4571 CrossRef CAS PubMed.
- G. Z. Jiang, M. D. Yao, Y. Wang, L. Zhou, T. Q. Song, H. Liu, W. H. Xiao and Y. J. Yuan, Metab. Eng., 2017, 41, 57–66 CrossRef CAS PubMed.
- B. Peng, L. K. Nielsen, S. C. Kampranis and C. E. Vickers, Metab. Eng., 2018, 47, 83–93 CrossRef CAS PubMed.
- C. Ignea, M. H. Raadam, M. S. Motawia, A. M. Makris, C. E. Vickers and S. C. Kampranis, Nat. Commun., 2019, 10, 3799 CrossRef PubMed.
- S. Dusseaux, W. T. Wajn, Y. Liu, C. Ignea and S. C. Kampranis, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 31789–31799 CrossRef CAS PubMed.
- M. Miaczynska, S. Lorenzetti, U. Bialek, R. M. Benito-Moreno, R. J. Schweyen and A. Ragnini, J. Biol. Chem., 1997, 272, 16972–16977 CrossRef CAS PubMed.
- J. C. Utomo, F. C. Chaves, P. Bauchart, V. J. J. Martin and D.-K. Ro, Metabolites, 2021, 11, 147 CrossRef CAS PubMed.
- C. Ignea, E. Ioannou, P. Georgantea, S. Loupassaki, F. A. Trikka, A. K. Kanellis, A. M. Makris, V. Roussis and S. C. Kampranis, Metab. Eng., 2015, 28, 91–103 CrossRef CAS PubMed.
- Y. J. Zhou, W. Gao, Q. Rong, G. Jin, H. Chu, W. Liu, W. Yang, Z. Zhu, G. Li, G. Zhu, L. Huang and Z. K. Zhao, J. Am. Chem. Soc., 2012, 134, 3234–3241 CrossRef CAS PubMed.
- C. Ignea, E. Ioannou, P. Georgantea, F. A. Trikka, A. Athanasakoglou, S. Loupassaki, V. Roussis, A. M. Makris and S. C. Kampranis, Microb. Cell Fact., 2016, 15, 46 CrossRef PubMed.
- Z. Dai, Y. Liu, X. Zhang, M. Shi, B. Wang, D. Wang, L. Huang and X. Zhang, Metab. Eng., 2013, 20, 146–156 CrossRef CAS PubMed.
- C. C. Jin, J. L. Zhang, H. Song and Y. X. Cao, Microb. Cell Fact., 2019, 18, 77 CrossRef PubMed.
- L. Dong, J. Pollier, J. E. Bassard, G. Ntallas, A. Almeida, E. Lazaridi, B. Khakimov, P. Arendt, L. S. de Oliveira, F. Lota, A. Goossens, F. Michoux and S. Bak, Metab. Eng., 2018, 49, 1–12 CrossRef CAS PubMed.
- T. Moses, J. Pollier, L. Almagro, D. Buyst, M. Van Montagu, M. A. Pedreno, J. C. Martins, J. M. Thevelein and A. Goossens, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1634–1639 CrossRef CAS PubMed.
- J. N. Bröker, B. Müller, N. van Deenen, D. Prüfer and C. Schulze Gronover, Appl. Microbiol. Biotechnol., 2018, 102, 6923–6934 CrossRef PubMed.
- F. Zhao, P. Bai, W. Nan, D. Li, C. Zhang, C. Lu, H. Qi and W. Lu, AIChE J., 2019, 65, 866–874 CrossRef CAS.
- S. Lu, C. Zhou, X. Guo, Z. Du, Y. Cheng, Z. Wang and X. He, Microb. Biotechnol., 2022, 15, 2292–2306 CrossRef CAS PubMed.
- B. Peng, M. R. Plan, A. Carpenter, L. K. Nielsen and C. E. Vickers, Biotechnol. Biofuels, 2017, 10, 43 CrossRef PubMed.
- J. Kirby, K. L. Dietzel, G. Wichmann, R. Chan, E. Antipov, N. Moss, E. E. K. Baidoo, P. Jackson, S. P. Gaucher, S. Gottlieb, J. LaBarge, T. Mahatdejkul, K. M. Hawkins, S. Muley, J. D. Newman, P. Liu, J. D. Keasling and L. Zhao, Metab. Eng., 2016, 38, 494–503 CrossRef CAS PubMed.
- Y. Ma, Y. Zu, S. Huang and G. Stephanopoulos, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2207680120 CrossRef CAS PubMed.
- S. Cheng, X. Liu, G. Jiang, J. Wu, J. L. Zhang, D. Lei, Y. J. Yuan, J. Qiao and G. R. Zhao, ACS Synth. Biol., 2019, 8, 968–975 CrossRef CAS PubMed.
- S. Liu, M. Zhang, Y. Ren, G. Jin, Y. Tao, L. Lyu, Z. K. Zhao and X. Yang, Biotechnol. Biofuels, 2021, 14, 243 CrossRef CAS PubMed.
- C. Ignea, M. Pontini, M. S. Motawia, M. E. Maffei, A. M. Makris and S. C. Kampranis, Nat. Chem. Biol., 2018, 14, 1090–1098 CrossRef CAS PubMed.
- J. Lucker, M. K. El Tamer, W. Schwab, F. W. Verstappen, L. H. van der Plas, H. J. Bouwmeester and H. A. Verhoeven, Eur. J. Biochem., 2002, 269, 3160–3171 CrossRef CAS PubMed.
- C. L. Steele, J. Crock, J. Bohlmann and R. Croteau, J. Biol. Chem., 1998, 273, 2078–2089 CrossRef CAS PubMed.
- S. Mafu, P. S. Karunanithi, T. A. Palazzo, B. L. Harrod, S. M. Rodriguez, I. N. Mollhoff, T. E. O'Brien, S. Tong, O. Fiehn, D. J. Tantillo, J. Bohlmann and P. Zerbe, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 974–979 CrossRef CAS PubMed.
- Y. S. Yeo, S. E. Nybo, A. G. Chittiboyina, A. D. Weerasooriya, Y. H. Wang, E. Góngora-Castillo, B. Vaillancourt, C. R. Buell, D. DellaPenna, M. D. Celiz, A. D. Jones, E. S. Wurtele, N. Ransom, N. Dudareva, K. A. Shaaban, N. Tibrewal, S. Chandra, T. Smillie, I. A. Khan, R. M. Coates, D. S. Watt and J. Chappell, J. Biol. Chem., 2013, 288, 3163–3173 CrossRef CAS PubMed.
- S. Tippmann, G. Scalcinati, V. Siewers and J. Nielsen, Biotechnol. Bioeng., 2016, 113, 72–81 CrossRef CAS PubMed.
- J. Beekwilder, A. van Houwelingen, K. Cankar, A. D. van Dijk, R. M. de Jong, G. Stoopen, H. Bouwmeester, J. Achkar, T. Sonke and D. Bosch, Plant Biotechnol. J., 2014, 12, 174–182 CrossRef CAS PubMed.
- X. Deng, B. Shi, Z. Ye, M. Huang, R. Chen, Y. Cai, Z. Kuang, X. Sun, G. Bian, Z. Deng and T. Liu, Metab. Eng., 2022, 69, 122–133 CrossRef CAS PubMed.
- K. Tamura, H. Seki, H. Suzuki, M. Kojoma, K. Saito and T. Muranaka, Plant Cell Rep., 2017, 36, 437–445 CrossRef CAS PubMed.
- N. L. Hansen, K. Miettinen, Y. Zhao, C. Ignea, A. Andreadelli, M. H. Raadam, A. M. Makris, B. L. Moller, D. Staerk, S. Bak and S. C. Kampranis, Microb. Cell Fact., 2020, 19, 15 CrossRef CAS PubMed.
- W. Yuan, C. Jiang, Q. Wang, Y. Fang, J. Wang, M. Wang and H. Xiao, Nat. Commun., 2022, 13, 7740 CrossRef CAS PubMed.
- N. D. Gold, E. Fossati, C. C. Hansen, M. DiFalco, V. Douchin and V. J. J. Martin, ACS Synth. Biol., 2018, 7, 2918–2929 CrossRef CAS PubMed.
- M. Kwon, J. C. Utomo, K. Park, C. A. Pascoe, S. Chiorean, I. Ngo, K. A. Pelot, C.-H. Pan, S.-W. Kim, P. Zerbe, J. C. Vederas and D.-K. Ro, ACS Catal., 2022, 12, 777–782 CrossRef CAS.
- N. L. Hansen, L. Kjaerulff, Q. K. Heck, V. Forman, D. Staerk, B. L. Møller and J. Andersen-Ranberg, Nat. Commun., 2022, 13, 5011 CrossRef CAS PubMed.
- J. Guo, X. Ma, Y. Cai, Y. Ma, Z. Zhan, Y. J. Zhou, W. Liu, M. Guan, J. Yang, G. Cui, L. Kang, L. Yang, Y. Shen, J. Tang, H. Lin, X. Ma, B. Jin, Z. Liu, R. J. Peters, Z. K. Zhao and L. Huang, New Phytol., 2016, 210, 525–534 CrossRef CAS PubMed.
- J. Guo, Y. J. Zhou, M. L. Hillwig, Y. Shen, L. Yang, Y. Wang, X. Zhang, W. Liu, R. J. Peters, X. Chen, Z. K. Zhao and L. Huang, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12108–12113 CrossRef CAS PubMed.
- V. Forman, D. Luo, F. Geu-Flores, R. Lemcke, D. R. Nelson, S. C. Kampranis, D. Staerk, B. L. Møller and I. Pateraki, Nat. Commun., 2022, 13, 5143 CrossRef CAS PubMed.
- D. K. Ro, G. Arimura, S. Y. Lau, E. Piers and J. Bohlmann, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 8060–8065 CrossRef CAS PubMed.
- S. C. Im and L. Waskell, Arch. Biochem. Biophys., 2011, 507, 144–153 CrossRef CAS PubMed.
- H. Zhang, S. C. Im and L. Waskell, J. Biol. Chem., 2007, 282, 29766–29776 CrossRef CAS PubMed.
- C. Ignea, A. Athanasakoglou, A. Andreadelli, M. Apostolaki, M. Iakovides, E. G. Stephanou, A. M. Makris and S. C. Kampranis, Sci. Rep., 2017, 7, 8855 CrossRef PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- H. Guo, H. Wang and Y.-X. Huo, ACS Synth. Biol., 2020, 9, 2214–2227 CrossRef CAS PubMed.
- S. C. Jung, W. Kim, S. C. Park, J. Jeong, M. K. Park, S. Lim, Y. Lee, W. T. Im, J. H. Lee, G. Choi and S. C. Kim, Plant Cell Physiol., 2014, 55, 2177–2188 CrossRef CAS PubMed.
- W. Wei, P. Wang, Y. Wei, Q. Liu, C. Yang, G. Zhao, J. Yue, X. Yan and Z. Zhou, Mol. Plant, 2015, 8, 1412–1424 CrossRef CAS PubMed.
- P. Wang, W. Wei, W. Ye, X. Li, W. Zhao, C. Yang, C. Li, X. Yan and Z. Zhou, Cell Discovery, 2019, 5, 5 CrossRef PubMed.
- C. Li, X. Yan, Z. Xu, Y. Wang, X. Shen, L. Zhang, Z. Zhou and P. Wang, Commun. Biol., 2022, 5, 775 CrossRef CAS PubMed.
- Y. Bai, P. Fernández-Calvo, A. Ritter, A. C. Huang, S. Morales-Herrera, K. U. Bicalho, M. Karady, L. Pauwels, D. Buyst, M. Njo, K. Ljung, J. C. Martins, S. Vanneste, T. Beeckman, A. Osbourn, A. Goossens and J. Pollier, New Phytol., 2021, 230, 228–243 CrossRef CAS PubMed.
- J. K. Michener, J. Nielsen and C. D. Smolke, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 19504–19509 CrossRef CAS PubMed.
- J. Hou, N. F. Lages, M. Oldiges and G. N. Vemuri, Metab. Eng., 2009, 11, 253–261 CrossRef CAS PubMed.
- G. N. Vemuri, M. A. Eiteman, J. E. McEwen, L. Olsson and J. Nielsen, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2402–2407 CrossRef CAS PubMed.
- Y. Shi, T. Dong, B. Zeng, M. Yao, Y. Wang, Z. Xie, W. Xiao and Y. Yuan, ACS Synth. Biol., 2022, 11, 2473–2483 CrossRef CAS PubMed.
- K. Jin, X. Shi, J. Liu, W. Yu, Y. Liu, J. Li, G. Du, X. Lv and L. Liu, Bioresour. Technol., 2023, 374, 128819 CrossRef CAS PubMed.
- J.-E. Kim, I.-S. Jang, B. H. Sung, S. C. Kim and J. Y. Lee, Sci. Rep., 2018, 8, 15820 CrossRef PubMed.
- Y. Zhuang, G. Y. Yang, X. Chen, Q. Liu, X. Zhang, Z. Deng and Y. Feng, Metab. Eng., 2017, 42, 25–32 CrossRef CAS PubMed.
- Y. Hu, J. Xue, J. Min, L. Qin, J. Zhang and L. Dai, J. Biotechnol., 2020, 309, 107–112 CrossRef CAS PubMed.
- C. Li, X. Yan, Z. Xu, Y. Wang, X. Shen, L. Zhang, Z. Zhou and P. Wang, Commun. Biol., 2022, 5, 775 CrossRef CAS PubMed.
- S. M. Stanley Fernandez, B. A. Kellogg and C. D. Poulter, Biochemistry, 2000, 39, 15316–15321 CrossRef CAS PubMed.
- C. M. Denby, R. A. Li, V. T. Vu, Z. Costello, W. Lin, L. J. G. Chan, J. Williams, B. Donaldson, C. W. Bamforth, C. J. Petzold, H. V. Scheller, H. G. Martin and J. D. Keasling, Nat. Commun., 2018, 9, 965 CrossRef PubMed.
- B. Peng, M. R. Plan, P. Chrysanthopoulos, M. P. Hodson, L. K. Nielsen and C. E. Vickers, Metab. Eng., 2017, 39, 209–219 CrossRef CAS PubMed.
- A. Gesell, M. Blaukopf, L. Madilao, M. M. Yuen, S. G. Withers, J. Mattsson, J. H. Russell and J. Bohlmann, Plant Physiol., 2015, 168, 94–106 CrossRef CAS PubMed.
- M. Salmon, R. B. Thimmappa, R. E. Minto, R. E. Melton, R. K. Hughes, P. E. O'Maille, A. M. Hemmings and A. Osbourn, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E4407–E4414 CrossRef CAS PubMed.
- W. Zha, F. Zhang, J. Shao, X. Ma, J. Zhu, P. Sun, R. Wu and J. Zi, Nat. Commun., 2022, 13, 2508 CrossRef CAS PubMed.
- G. Janson and A. Paiardini, Bioinformatics, 2020, 37, 1471–1472 CrossRef PubMed.
- K. Arnold, L. Bordoli, J. Kopp and T. Schwede, Bioinformatics, 2006, 22, 195–201 CrossRef CAS PubMed.
- M. C. Peitsch, Biochem. Soc. Trans., 1996, 24, 274–279 CrossRef CAS PubMed.
- A. Waterhouse, M. Bertoni, S. Bienert, G. Studer, G. Tauriello, R. Gumienny, F. T. Heer, T. A. P. de Beer, C. Rempfer, L. Bordoli, R. Lepore and T. Schwede, Nucleic Acids Res., 2018, 46, W296–W303 CrossRef CAS PubMed.
- F. H. Arnold, Angew. Chem., Int. Ed. Engl., 2018, 57, 4143–4148 CrossRef CAS PubMed.
- N. Crook, J. Abatemarco, J. Sun, J. M. Wagner, A. Schmitz and H. S. Alper, Nat. Commun., 2016, 7, 13051 CrossRef CAS PubMed.
- A. Ravikumar, G. A. Arzumanyan, M. K. A. Obadi, A. A. Javanpour and C. C. Liu, Cell, 2018, 175, 1946–1957.e1913 CrossRef CAS PubMed.
- F. Wang, X. Lv, W. Xie, P. Zhou, Y. Zhu, Z. Yao, C. Yang, X. Yang, L. Ye and H. Yu, Metab. Eng., 2017, 39, 257–266 CrossRef CAS PubMed.
- Z. Yao, P. Zhou, B. Su, S. Su, L. Ye and H. Yu, ACS Synth. Biol., 2018, 7, 2308–2316 CrossRef CAS PubMed.
- P. R. Ortiz de Montellano, Cytochrome P-450 structure, mechanism, and biochemistry, Plenum Press, New York, 1986 Search PubMed.
- H. C. Yocum, A. Pham and N. A. Da Silva, Front. Bioeng. Biotechnol., 2021, 9 Search PubMed.
- X. Chen, J. L. Zaro and W.-C. Shen, Adv. Drug Delivery Rev., 2013, 65, 1357–1369 CrossRef CAS PubMed.
- L. Albertsen, Y. Chen, L. S. Bach, S. Rattleff, J. Maury, S. Brix, J. Nielsen and U. H. Mortensen, Appl. Environ. Microbiol., 2011, 77, 1033–1040 CrossRef CAS PubMed.
- H. Rabeharindranto, S. Castano-Cerezo, T. Lautier, L. F. Garcia-Alles, C. Treitz, A. Tholey and G. Truan, Metab. Eng. Commun., 2019, 8, e00086 CrossRef PubMed.
- W. Kang, T. Ma, M. Liu, J. Qu, Z. Liu, H. Zhang, B. Shi, S. Fu, J. Ma, L. T. F. Lai, S. He, J. Qu, S. Wing-Ngor Au, B. Ho Kang, W. C. Yu Lau, Z. Deng, J. Xia and T. Liu, Nat. Commun., 2019, 10, 4248 CrossRef PubMed.
- S. Tippmann, J. Anfelt, F. David, J. M. Rand, V. Siewers, M. Uhlén, J. Nielsen and E. P. Hudson, ACS Synth. Biol., 2017, 6, 19–28 CrossRef CAS PubMed.
- J. Y. Han, J. M. Song, S. H. Seo, C. Wang, S.-G. Lee, H. Lee, S.-W. Kim and E.-S. Choi, Biotechnol. Bioeng., 2018, 115, 694–704 CrossRef CAS PubMed.
- N. R. Burns, J. E. Gilmour, S. M. Kingsman, A. J. Kingsman and S. E. Adams, Mol. Biotechnol., 1994, 1, 137–145 CrossRef CAS PubMed.
- S. K. Hammer and J. L. Avalos, Nat. Chem. Biol., 2017, 13, 823–832 CrossRef CAS PubMed.
- A. C. Jaramillo-Madrid, E. Lacchini and A. Goossens, Curr. Opin. Green Sustainable Chem., 2022, 33, 100572 CrossRef CAS.
- Z.-T. Zhu, M.-M. Du, B. Gao, X.-Y. Tao, M. Zhao, Y.-H. Ren, F.-Q. Wang and D.-Z. Wei, Metab. Eng., 2021, 68, 232–245 CrossRef CAS PubMed.
- D. A. Yee, A. B. DeNicola, J. M. Billingsley, J. G. Creso, V. Subrahmanyam and Y. Tang, Metab. Eng., 2019, 55, 76–84 CrossRef CAS PubMed.
- J. Yuan and C. B. Ching, Metab. Eng., 2016, 38, 303–309 CrossRef CAS PubMed.
- X.-Y. Tao, Y.-C. Lin, F.-Q. Wang, Q.-H. Liu, Y.-S. Ma, M. Liu and D.-Z. Wei, Biotechnol. Lett., 2022, 44, 571–580 CrossRef CAS PubMed.
- X. Cao, S. Yang, C. Cao and Y. J. Zhou, Synth. Syst. Biotechnol., 2020, 5, 179–186 CrossRef PubMed.
- N. Kulagina, S. Besseau, N. Papon and V. Courdavault, Front. Bioeng. Biotechnol., 2021, 9 Search PubMed.
- F. Liu, S. C. Liu, Y. K. Qi, Z. Liu, J. Chen, L. J. Wei and Q. Hua, J. Agric. Food Chem., 2022, 70, 15157–15165 CrossRef CAS PubMed.
- G.-S. Liu, T. Li, W. Zhou, M. Jiang, X.-Y. Tao, M. Liu, M. Zhao, Y.-H. Ren, B. Gao, F.-Q. Wang and D.-Z. Wei, Metab. Eng., 2020, 57, 151–161 CrossRef CAS PubMed.
- M. M. Du, Z. T. Zhu, G. G. Zhang, Y. Q. Zhao, B. Gao, X. Y. Tao, M. Liu, Y. H. Ren, F. Q. Wang and D. Z. Wei, J. Agric. Food Chem., 2022, 70, 229–237 CrossRef CAS PubMed.
- H. Davies, G. R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M. J. Garnett, W. Bottomley, N. Davis, E. Dicks, R. Ewing, Y. Floyd, K. Gray, S. Hall, R. Hawes, J. Hughes, V. Kosmidou, A. Menzies, C. Mould, A. Parker, C. Stevens, S. Watt, S. Hooper, R. Wilson, H. Jayatilake, B. A. Gusterson, C. Cooper, J. Shipley, D. Hargrave, K. Pritchard-Jones, N. Maitland, G. Chenevix-Trench, G. J. Riggins, D. D. Bigner, G. Palmieri, A. Cossu, A. Flanagan, A. Nicholson, J. W. C. Ho, S. Y. Leung, S. T. Yuen, B. L. Weber, H. F. Seigler, T. L. Darrow, H. Paterson, R. Marais, C. J. Marshall, R. Wooster, M. R. Stratton and P. A. Futreal, Nature, 2002, 417, 949–954 CrossRef CAS PubMed.
- P. Arendt, K. Miettinen, J. Pollier, R. De Rycke, N. Callewaert and A. Goossens, Metab. Eng., 2017, 40, 165–175 CrossRef CAS PubMed.
- K. Thodey, S. Galanie and C. D. Smolke, Nat. Chem. Biol., 2014, 10, 837–844 CrossRef CAS PubMed.
- X. Xiu, Y. Sun, Y. Wu, K. Jin, L. Qu, Y. Liu, J. Li, G. Du, X. Lv and L. Liu, Bioresour. Technol., 2022, 360, 127572 CrossRef CAS PubMed.
- Y. Shi, D. Wang, R. Li, L. Huang, Z. Dai and X. Zhang, Metab. Eng., 2021, 67, 104–111 CrossRef CAS PubMed.
- T. Ma, B. Shi, Z. Ye, X. Li, M. Liu, Y. Chen, J. Xia, J. Nielsen, Z. Deng and T. Liu, Metab. Eng., 2019, 52, 134–142 CrossRef CAS PubMed.
- R. De La Pena and E. S. Sattely, Nat. Chem. Biol., 2021, 17, 205–212 CrossRef CAS PubMed.
- K. R. Croteau R, R. M. Long, R. Kaspera and M. R. Wildung, Phytochem. Rev., 2006, 5, 75–97 CrossRef PubMed.
- Z. Jin, A. Vighi, Y. Dong, J. A. Bureau and C. Ignea, Biotechnol. Adv., 2023, 64, 108118 CrossRef CAS PubMed.
- B. L. Moller, Science, 2010, 330, 1328–1329 CrossRef PubMed.
- E. Gomes and J. Shorter, J. Biol. Chem., 2019, 294, 7115–7127 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3np00005b |
| This journal is © The Royal Society of Chemistry 2023 |