Open Access Article
Open Access ArticleAntibacterial activity of copper pyrazolate coordination polymers†
Corrado
Di Nicola
 *a,
Fabio
Marchetti
*a,
Fabio
Marchetti
 a,
Alessia
Tombesi
a,
Alessia
Tombesi
 b,
Sonila
Xhafa
a,
Patrizio
Campitelli
a,
Marco
Moroni
cd,
Simona
Galli
b,
Sonila
Xhafa
a,
Patrizio
Campitelli
a,
Marco
Moroni
cd,
Simona
Galli
 ce,
Riccardo
Pettinari
ce,
Riccardo
Pettinari
 b and
Claudio
Pettinari
b and
Claudio
Pettinari
 b
b
aChIP Research Center, School of Science and Technology, University of Camerino, via Madonna delle Carceri, 62032 Camerino MC, Italy
bChIP Research Center, School of Pharmacy, University of Camerino, via Madonna delle Carceri, 62032 Camerino MC, Italy
cDipartimento di Scienza e Alta Tecnologia, Università degli Studi dell’Insubria, via, Valleggio 9, 22100 Como, Italy
dDipartimento di Chimica, Università degli Studi di Pavia, via, Taramelli 16, 27100 Pavia, Italy
eConsorzio Interuniversitario Nazionale per la Scienza e Tecnologia dei Materiali, via, Giusti 9, 50121 Firenze, Italy
First published on 21st September 2023
Abstract
Novel copper(II) coordination polymers (CPs) [Cu(μ-4-NO2pz)2]n and [Cu(μ-4-NO2pz)2·DMF H2O]n (4-NO2pz− = 4-nitropyrazolate; DMF = N,N′-dimethylformamide) were synthesized and characterized in the solid state and their thermal behaviour and structural features were investigated. Together with the known CPs [Cu(μ-pz)(μ-OH)]n, [Cu(μ-pz)2(H2O)]n and [Cu(μ-4-Xpz)2(DMF)]n [pz− = pyrazolate; 4-Xpz− = 4-X-pyrazolate, with X = H, Cl, Br, I], they were isolated in the form of very small particles whose dimensions range within 20–300 nm and, as such, they were investigated as potential antibacterial agents. Their antibacterial activity was assessed against two Gram-negative bacteria, namely Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa), and the Gram-positive bacterium Staphylococcus aureus (S. aureus), by monitoring the bacterial growth curve and the antibacterial rate as a function of time. The bactericidal mechanism seems to be related to the ROS production, directly activated by the contact of the CP tiny particles with the bacterial membrane, without a significant release of Cu(II) ions.
Introduction
The need to develop an effective system to fight bacterial infections caused by microbial strains resistant to antibiotics1 has become more and more important in recent years. With this purpose, a huge number of scientific studies have been conducted highlighting the great potential, as antibacterial agents, shown by metals in various forms, such as metal surfaces, metal nanoparticles2,3 and metal-containing compounds.4–6 In this instance, the bactericidal effect is mainly based on the electrostatic interactions arising between the metal ions released by surfaces, nanoparticles or bulk compounds and the negatively charged cell membranes of bacteria. These interactions are able to damage the cell wall membrane, eventually causing the death of the microorganism.7–9 Despite this beneficial effect, numerous metal nanoparticles and metal-based compounds possess the drawback of excessive metal ion release,10,11 that limits their use in food packaging12 and medical devices.13 This critical issue can be solved by using compounds capable of releasing metal ions very slowly and at a low concentration. In this regard, thanks to their stability and low solubility in most of the commonest solvents and, particularly, in water, coordination polymers (CPs)14 and metal–organic frameworks (MOFs)15 can act as slow-releasing metal ion-reservoirs. Moreover, tuneable crystal structures, and with an appropriate choice of linkers and metals, low toxicity, make CPs16,17 and MOFs18 excellent candidates for fighting bacterial antibiotic resistance.In recent years, we have focused our attention on composite materials with antimicrobial properties, obtained by anchoring or incorporating molecular complexes, CPs or MOFs in different matrices, such as cellulose,19 carbon,20 or plastics.21–23 Many other matrices intended for different applications are reported in the literature.24 Among transition metals, bare copper25,26 and Cu(II) compounds27–29 have shown potent bactericidal activity against a wide range of microorganisms.30,31 It is noteworthy that some hydrophobic Cu-MOFs are capable of inducing bacterial death through the electrostatic interactions occurring between the positively charged metal sites on their surface and the negative cell membrane, which provokes a change in its permeability, without any detectable release of copper(II) ions.32,33 In this context, we previously reported several Cu(II) CPs based on the triangular cluster [Cu3(μ3-OH)(μ-pz)3]2+ (pz− = pyrazolate)34–36 or on pyrazolate-bridged 1D chains.37,38 Here, we report the synthesis and solid-state characterization of the novel CPs [Cu(μ-4-NO2pz)2]n and [Cu(μ-4-NO2pz)2·DMF H2O]n (4-NO2pz− = 4-nitropyrazolate; DMF = N,N′-dimethylformamide) (Chart 1) and their antibacterial activity against Gram-positive (S. aureus) and Gram-negative (E. coli and P. aeruginosa) bacterial strains. For comparison purposes, we have extended the biological investigation to six previously described39 Cu(II) pyrazolate CPs, namely [Cu(μ-pz)(μ-OH)]n and [Cu (μ-4-Xpz)2(Solv)]n (4-Xpz− = 4-Xpyrazolate, where X = H, Cl, Br, I and Solv = H2O or DMF) (Chart 1). These materials may successfully combine the antibacterial activity of copper with that of pyrazolate linkers, as many azoles40 and pyrazoles41,42 have shown potential as antibiotics. Moreover, nitro-containing aromatic rings and heterocycles are known for their antibacterial efficiency.43 For example, the 5-nitroimidazolyl ring is a significant moiety in medicinal chemistry, being present in a number of commercial antibiotics including miconazole and metronidazole, whose mechanism of action is likely related to the formation of nitrous and superoxide intermediates, generated by the reduction of nitro group.44 The biological activity of the selected CPs is presented here in terms of the bacteria growth inhibitory effect and antibacterial rate as a function of time. In addition, the role of the bare linkers in the antibacterial activity of the corresponding CPs is investigated. Finally, following the detection of reactive oxygen species and the investigation of the bacterial viability, a mechanism of action involving an electrostatic contact among the CP particles and the bacterial membrane, without leakage of Cu(II) ions, is proposed.
 | ||
| Chart 1 Schematical representation of the 1D chains characterizing the crystal structures of compounds 1, [Cu(μ-pz)(μ-OH)]n, 3, 4, and 6–8, [Cu(μ-4-Xpz)2(Solv)]n (X = H, Cl, Br, and I, respectively; Solv = H2O or DMF),39 as well as the 2D layer characterizing the crystal structure of compound 2, [Cu(μ-4-NO2pz)2]n. | ||
Results and discussion
Synthesis and analytical characterization
Compounds 1, 3, 4 and 6–8 were prepared and characterized according to previous synthetic procedures.39 At variance, the novel compounds 2 and 5 were synthesized at room temperature by combining copper acetate hydrate and 4-nitro-1-H-pyrazole in acetonitrile or DMF, respectively. All the CPs were obtained in the form of very small particles, their dimensions ranging within 20–300 nm. The CPs were separated from the mother liquor by filtration or centrifugation. 1–8 are stable in air and insoluble in all common solvents, water included. The FT-IR spectra of 2 and 5 (Fig. S1, ESI†) display weak bands at 3141 cm−1 and 3136 cm−1, respectively, which can be assigned to the C–H stretching of pyrazolate rings. The –NO2 asymmetric and symmetric stretching vibrations of 2 fall at 1504 cm−1 and 1286 cm−1, respectively, while those of 5 are observed at 1496 cm−1 and 1278 cm−1. The sharp band at 818 cm−1, present in spectra of both 2 and 5, can be ascribed to the C–NO2 stretching vibration.45 In the case of 5, the weak-broad band in the range of 3550–3250 cm−1 can be attributed to the O–H stretching of water molecules. The presence of a H2O molecule is also confirmed by the thermogravimetric analysis (see below). Finally, the presence of DMF molecules in 5 is confirmed by the intense band at 1652 cm−1 due to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, downfield shifted with respect to free DMF (1661 cm−1).
O stretching, downfield shifted with respect to free DMF (1661 cm−1).
Thermal behaviour
As assessed by coupling thermogravimetric analysis and differential thermal analysis, compound 2 is stable up to 563 K (Fig. S2, ESI†). A rapid exothermic decomposition associated with 76% loss of weight occurs in the temperature range of 563–683 K. At the end of the experiment, a black residue is recovered in the crucible, mainly consisting of spongy copper (22.4% residual weight found vs. a theoretical value of 22.1%). Compound 5 exhibits two weight losses before decomposition (Fig. S2, ESI†), highlighting the presence of solvent molecules: the first loss of 4.3% starts at 343 K and ends at 390 K, and can be attributed to the release of one water molecule per formula unit (calculated weight loss of 4.7%, see also the description of the IR spectrum above), whereas the second loss of 19.2% in the temperature range 390 to 485 K is due to the release of one molecule of DMF per formula unit (calculated weight loss 19.2%). The unsolvated compound is stable up to 563 K. Then, a rapid decomposition occurs in the temperature range 563–634 K associated with 46.5% weight loss. In the end, a black residue (23% weight), consisting mainly of spongy copper (calculated weight of 16.7%), is recovered in the crucible. As assessed by IR, TGA, and PXRD (Fig. S1, S2 and S4, ESI†), after heating 5 in an oven at 433 K for one hour, water and DMF loss leads to the formation of 2. The thermal behaviour of 1, 3, 4 and 6–8 was extensively explored previously.39![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , a = 10.1379(7) Å, b = 7.3247(9) Å, c = 10.6400(7) Å, a = 87.78(1)°, b = 112.461(4)°, g = 87.904(9)°, V = 728.5(1) Å3]; see Fig. S5 (ESI†) for the graphical result of the pertinent whole powder pattern refinement). According to the Kempster–Lipson rule,46 the unit cell volume is such that the unit cell can host two Cu(4-NO2pz)(DMF)(H2O) units. On the other hand, the structure determination was successful in the case of 2. The latter crystallizes in the monoclinic space group C2/m. The asymmetric unit features half of a copper(II) ion on an inversion centre (Whyckoff position e) and two halves of 4-NO2pz− ligands on mirror planes (Whyckoff positions i).
, a = 10.1379(7) Å, b = 7.3247(9) Å, c = 10.6400(7) Å, a = 87.78(1)°, b = 112.461(4)°, g = 87.904(9)°, V = 728.5(1) Å3]; see Fig. S5 (ESI†) for the graphical result of the pertinent whole powder pattern refinement). According to the Kempster–Lipson rule,46 the unit cell volume is such that the unit cell can host two Cu(4-NO2pz)(DMF)(H2O) units. On the other hand, the structure determination was successful in the case of 2. The latter crystallizes in the monoclinic space group C2/m. The asymmetric unit features half of a copper(II) ion on an inversion centre (Whyckoff position e) and two halves of 4-NO2pz− ligands on mirror planes (Whyckoff positions i).
The metal ions are hexa-coordinated and show a trans-CuN4O2 octahedral stereochemistry (Fig. 1a; the reader is directed to the figure caption for the main bond distances and angles at the metal ions). μ-Bridging by the nitrogen atoms of both independent pyrazolate rings brings about the formation of 1D chains of collinear metal ions running along the crystallographic b-axis with a pace of 3.80 Å (half of the b-axis) (Fig. 1b).‡‡ The oxygen atoms of the μ-NO2 group of the independent ligand lying approximately on the (100) plane bridge the metal ions of a nearby chain, thus forming 2D slabs extending parallel to this plane and piling, staggered, along the crystallographic a-axis (Fig. 1c). The other independent ligand lies in the (10−1) plane and occupies the empty volume among consecutive slabs (Fig. 1c). Its oxygen atoms are involved in rather weak C–H⋯O non-bonding interactions (C⋯O = 3.2 Å). The crystal structure features a residual empty volume of ca. 12%.
 | ||
| Fig. 2 Growth inhibition curves of E. coli (a) and (b), S. aureus (c) and (d), and P. aeruginosa (e) and (f) in the presence and in the absence (blank) of 1–8. | ||
The number of viable cells was then used to calculate the killing percentage, which is a useful parameter to define if a substance displays bactericidal or bacteriostatic action. The bactericidal activity is defined as the reduction of the number of viable bacterial cells by at least 99.9% within 18–24 h from the inoculum. At variance, the bacteriostatic activity is associated with a reduction in the bacterial growth maintained stable between 90 and 99%, within 18 to 24 h from the inoculum. In general, by analysing the antibacterial activity of the title compounds against each bacterial strain tested (Fig. 3), a homogeneous performance was exhibited by all of them against P. aeruginosa and S. aureus. In contrast, CPs 5 and 8 were less efficient against E. coli in terms of time and rate, as described below. A 95% growth inhibition of P. aeruginosa and S. aureus was reached after 12 h of treatment by all CPs (Fig. 3c–f), thus displaying a clear bacteriostatic activity. Additionally, 1–8 display bactericidal activity against S. aureus and P. aeruginosa within 16 h of treatment, with a 99.99% growth inhibition, that remains stable also after 24 h. The most important differences in terms of performance were observed against E. coli (Fig. 3a and b). In detail, 1–4 and 6 reached a bacteriostatic activity higher than 95% within 4 h, while 5 and 7 reached the same performance after 8 and 12 h, respectively. In terms of bactericidal activity, 99.9% of inhibition was reached very fast by 1–4 and 6, i.e. within 4 h, while 5 and 7 reached it within 12 h. Finally, the worst performance against E. coli, in terms of bactericidal activity, was recorded with 8, which failed to reach the total reduction of viable cells even after 24 h. To evaluate the putative antibacterial activity of the pyrazole ligands used to prepare 1–8, strains of E. coli and S. aureus were chosen, respectively, as models of Gram-negative and Gram-positive bacteria.
 | ||
| Fig. 3 Antibacterial rate of 1–8 as a function of time against E. coli (a) and (b), S. aureus (c) and (d), and P. aeruginosa (e) and (f). | ||
The results showed that none of the ligands, tested under the same conditions applied for 1–8, reached a stable antibacterial activity as a function of time (Fig. S6, ESI†). However, 4-nitropyrazole showed the best performance, probably due to the presence of the nitro moiety, in line with the influence of such a functionality on the biocidal activity of several NO2-containing drugs.43
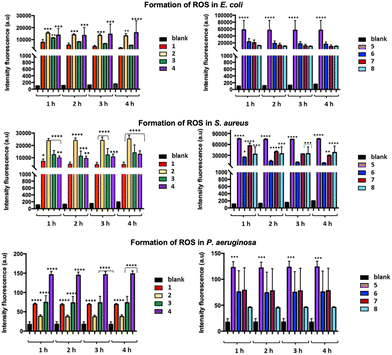
A very fast antibacterial activity of 5 was confirmed for all the tested bacteria by the fluorescence intensity measured after only one hour of treatment. Finally, while 4 showed the highest performance against P. aeruginosa, 1 displayed the lowest fluorescence intensity against all the tested bacteria. The membrane damage caused by 1–8 was investigated using the propidium iodide (PI) assay as an appropriate probe to assess the membrane integrity, as it penetrates only in those bacterial cells with a permeabilized membrane. The red fluorescence emission of bacterial cells exposed to 1–8 and stained with PI was measured at 528 nm vs. bacterial suspensions untreated with 1–8 and contacted with PI. A similar trend was found for the cultured bacterial strains treated with 1–8, with a moderate increase in the fluorescence emission as a function of time (Fig. 5). In summary, 1–8 seem capable of inducing ROS production and formation of highly reactive radicals associated with cellular destruction, with consequent breakdown of the cellular microenvironment. To visually evaluate the overall viability of the bacterial cells, confocal laser scanning microscopy was adopted. According to the LIVE/DEAD® BacLight™ Bacterial Viability Kits,47 the SYTO 9 dye labels all the bacteria in a population, staining them with green color, while PI penetrates into bacteria with disrupted cell membranes, staining them with red color. With an appropriate mixture of both dyes, live bacteria stain fluorescent green, while dead bacteria stain fluorescent red (Fig. 6). This test was carried out contacting 2, arbitrarily chosen, with E. coli and S. aureus for a time of 4 h, under the same conditions as those adopted for the growth inhibition assay. By comparing the image of untreated E. coli (Fig. 6a) with that of the treated one (Fig. 6c), a reduction of live cells (in green) and the appearance of dead cells (in red) is observed. In the case of S. aureus, the treatment effect is even more marked, as only dead cells (in red) are observed in Fig. 6d (vs.Fig. 6b).
These results revealed that after treatment with the title CPs, the cell membrane of both bacterial strains is severely damaged, thus confirming the results shown by the above reported assays. In order to evaluate if the cell membrane damage is related to Cu(II) ion release from the investigated CPs, we verified the occurrence of leakage from 1, 2, 5 and 6, suspending 4 mg of these CPs in 10 mL of physiological solution at 335 K for 3 days: upon analysing the mother liquors by ICP-OES, we detected very low values of 0.010, 0.005, 0.008 and 0.004 ppm of Cu(II), respectively.
In addition, we performed a Cu(II) ion minimum inhibition concentration (MIC) assay using copper nitrate as a source of Cu(II) ions. The data from this assay show bacteriostatic action vs. both E. coli and S. aureus with a concentration of Cu(II) ions above 25 ppm, a higher concentration than that detected by ICP-OES, and a bactericidal effect above 100 ppm (Fig. 7). These results, in accordance with those from the literature,48 lead us to exclude an antibacterial mechanism based on Cu(II) ion release from 1–8. We can therefore hypothesize a mechanism of membrane damage due to electrostatic interactions with the positively charged copper(II) sites on the surface of the CP small particles (size range 20–300 nm) also known as “chelation theory”, as recently found with Cu(II) MOFs with glutarates and bipyridyl linkers,32 and Cu(II) CPs with 4,4′-dicarboxy-2,2′-bipyridine ligands.49
 | ||
| Fig. 7 Copper(II) ions MIC ranging from 0.006 ppm to 100 ppm for E. coli (left) and S. aureus (right). | ||
The cell membrane damage caused by the copper(II) pyrazolates particles was also confirmed by SEM images, which show the morphological alterations of the bacterial cells following treatment with 2. The particle nanometric dimensions of this CP, ranging from 20 to 40 nm (Fig. 8), favor their diffusion and the contact between the bacterial membranes and the coordinatively unsaturated copper(II) centres on the nanoparticle surface, promoting an electrostatic attraction of the nanoparticles to the negatively charged bacterial membrane. In Fig. 8a, a control sample of E. coli shows classical morphology with a rod shape and a smooth surface, while after treatment with 2, Fig. 8b, the contact with the CP nanoparticles changes the regular layer of the bacterial surface. The shrinkage of the bacterial cell and its disruption is magnified in Fig. 8c. In addition, Fig. 8d shows the control sample of S. aureus, having a round appearance with a rigid cell wall. After treatment with 2 (Fig. 8e and magnification in 8f), the cells are surrounded and covered by nanoparticles of 2 and display a vesicular morphology, resulting in the loss of the regular shape and dimensions.
The most interesting aspect of this work lies in the presence of copper(II) pyrazolates which do not release Cu(II) ion (see above), thus excluding a mechanism of action involving a slow and progressive release of copper(II) ions at bactericidal concentrations (above 50/100 ppm). However, it cannot be excluded that local conditions are created in the closeness of the contact area between the bacterial membrane and the CPs which could favour a more significant and targeted release of copper(II) ions. Even so, a “contact kill” mechanism is plausible in the case of 2, given the nanoscale size of its particles, which can approach and intimately join the bacterial membranes through electrostatic interactions between surface unsaturated copper(II) sites and the phospholipidic or peptidoglycan negatively charged cell membranes. Such interactions can deactivate vital membrane enzymes and damage the cell membrane integrity and permeability, as confirmed by the PI tests and SEM images.
Experimental section
Materials and methods
All the chemicals were purchased from commercial suppliers and used without further purification. All the reactions and manipulations were carried out in air, under either ambient or solvothermal conditions. Elemental analyses (% m/m C, H, N) were performed with a Fisons Instruments 1108 CHNS-O elemental analyser. Infrared spectra were recorded from 4000 to 650 cm−1 with a PerkinElmer Spectrum One model FTIR spectrometer in attenuated total reflectance (ATR) mode. In the following, the IR bands are defined as follows: w = weak, m = medium; s = strong; vs = very strong; br = broad. Simultaneous thermal analyses (STAs, simultaneous thermogravimetric analysis and differential thermal analysis) were carried out using a PerkinElmer STA6000 instrument. The temperature program ranged from 303 to 873 K with a heating rate of 10 K min−1 under a flow of N2 (30 mL min−1). The accuracy of the sample temperature measurement was assessed upon measuring the onset fusion temperature of indium (156.6 ± 0.2 K) and tin (232.0 ± 0.2 K) with heating/cooling dynamic segments. Magnetic susceptibilities were measured at 293 K by the Gouy method with a Sherwood Scientific magnetic balance MSB-Auto, using HgCo(NCS)4 as a calibrant, and they were corrected for diamagnetism with the appropriate Pascal constants. The magnetic moments (in Bohr Magnetons, B.M.) were calculated by eqn (1):| μeff = 2.84(χcorrmT)1/2 | (1) |
Synthesis of 1, 3, 4, 6–8
Compounds 1, 3, 4, 6–8 were prepared according to published procedures.39 Their nature and purity were assessed by means of elemental analysis, IR spectroscopy and PXRD.Synthesis of 2, [Cu(μ-4-NO2pz)2]n
4-Nitro-1H-pyrazole (2 mmol, 0.226 g) was dissolved in acetonitrile (CH3CN, 30 mL). Then, copper acetate hydrate, Cu(CH3COO)2·H2O (1.0 mmol, 0.199 g), was added. The resulting reaction mixture was left under magnetic stirring at room temperature for 2 h. A bright purple solid was recovered after centrifugation (three 30 minute-cycles at 6000 rpm), washed twice with acetonitrile, and finally dried in air. Yield: 0.240 g, 83%. Elemental analysis (%) for C6H4CuN6O4 (FW = 287.7 a.m.u.): calculated: C, 25.05; H, 1.40; N, 29.21. Found: C, 25.33; H, 1.47; N, 28.88. IR (ATR, cm−1, Fig. S1, ESI†): 3138w ν(C–Harom), 1504vs νasym(N–O2), 1411s, ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C + C
C + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1286vs νsym(N–O2), 1185m, 1038m, 1008m, 868m, 818m, 754m. STA (Fig. S2, ESI†): 2 starts to decompose at 563 K, leaving a black residue corresponding to 22.4% of the original weight. Room temperature magnetic susceptibility: μeff (293 K) = 1.34 B.M.
N), 1286vs νsym(N–O2), 1185m, 1038m, 1008m, 868m, 818m, 754m. STA (Fig. S2, ESI†): 2 starts to decompose at 563 K, leaving a black residue corresponding to 22.4% of the original weight. Room temperature magnetic susceptibility: μeff (293 K) = 1.34 B.M.
Synthesis of 5, [Cu(μ-4-NO2pz)2·DMF·H2O]n
Copper acetate hydrate, Cu(CH3COO)2·H2O (1.0 mmol, 0.199 g), was added to a 4-nitro-1H-pyrazole (2 mmol, 0.226 g) solution in DMF (25 mL). The obtained mixture was left under magnetic stirring at room temperature for 2 hours. A pale lilac precipitate was obtained. The precipitate was filtered off, washed with fresh DMF and dried in air. Yield: 0.298 g, 79%. Elemental analysis (%) for C9H13CuN7O6 (FW = 378.8 a.m.u.): calculated: C, 28.54; H, 3.46; N, 25.88. Found: C, 28.31; H, 3.39; N, 26.01. IR (ATR, cm−1, Fig. S1, ESI†): 3528br ν(O–H), 3136w ν(C–Harom), 2932wbr ν(C–Halif), 1652vs ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1496s νasym(N–O2), 1408s ν(C
O), 1496s νasym(N–O2), 1408s ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C + C
C + C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1382m, 1278vs νsym(N–O2), 1181m, 1099s, 1033m, 1009m, 867m, 817s, 755s. STA (Fig. S2, ESI†): in the range 343 to 390 K, 5 loses one H2O molecule per formula unit (weight loss found 4.3%, calculated 4.7%), while in the range from 390 to 485 K it loses one DMF molecule per formula unit (weight loss found 19.2%, calculated 19.2%); then it decomposes, leaving a black residue corresponding to 15.3% of the original weight. Room temperature magnetic susceptibility: μeff (293 K) = 1.56 B.M.
N), 1382m, 1278vs νsym(N–O2), 1181m, 1099s, 1033m, 1009m, 867m, 817s, 755s. STA (Fig. S2, ESI†): in the range 343 to 390 K, 5 loses one H2O molecule per formula unit (weight loss found 4.3%, calculated 4.7%), while in the range from 390 to 485 K it loses one DMF molecule per formula unit (weight loss found 19.2%, calculated 19.2%); then it decomposes, leaving a black residue corresponding to 15.3% of the original weight. Room temperature magnetic susceptibility: μeff (293 K) = 1.56 B.M.
Powder X-ray diffraction structure characterization of 2 [Cu(μ-4-NO2pz)2]n and 5 [Cu(μ-4-NO2pz)2·DMF H2O]n
Powdered samples of 2 and 5 (ca. 50 mg) were deposited in the hollow of a 0.2 mm deep silicon zero-background plate. Data acquisitions were performed on a Bruker AXS D8 Advance vertical-scan θ:θ diffractometer equipped with a sealed X-ray tube (Cu Kα, λ = 1.5418 Å), a Bruker Lynxeye linear position-sensitive detector, a filter of nickel located in the diffracted beam and the following optical components: primary- and secondary-beam Soller slits (2.5°), fixed divergence slit (0.5°), and antiscatter slit (8 mm). The generator was operated at 40 kV and 40 mA. After preliminary acquisition to assess the feasibility of the crystal structure determination, performed in a 2θ range of 3–35° with steps of 0.02° and a time/step of 1 s, diffraction data for the structure determination were collected at 2θ values from 5° to 105°, with steps of 0.02°, and a time/step of 10 s. A manual peak search enabled us to estimate the maximum positions of twenty low-to-medium-angle peaks. Working on them, the singular value decomposition algorithm50 implemented in TOPAS-R v. 3.051 provided approximate unit cell parameters for both compounds. The space groups were assigned based on the observed systematic absences. The unit cell parameters and space groups were preliminarily confirmed by whole powder pattern refinements carried out with the Le Bail method,52 as implemented in TOPAS-R v. 3.0. The structure determination was carried out with TOPAS-R v. 3 adopting the Simulated Annealing approach.53 Rigid bodies were used to describe the crystallographically independent portion of the 4-NO2pz− ligand and the DMF molecule, assigning idealized values to bond distances and angles.§ The position of their centre of mass and their orientation (when allowed by symmetry) were varied. The structure determination was successful in the case of 2, while no attempt to disclose the crystal structure of 5 was effective. Structure refinement for 2 was carried out with the so-called Rietveld method54 as implemented in TOPAS-R v. 3.0. The background was modelled through a polynomial function of the Chebyshev type. An isotropic thermal factor (Biso) was attributed to the metal centre, while the other atoms were attributed an isotropic thermal factor amounting to Biso′ = Biso + 2.0 (Å2). The instrumental contribution to the peak profile was modelled by the fundamental parameters approach.55 The anisotropic peak broadening was successfully described using second-order spherical harmonics convoluted to Lorentzian and Gaussian contributions. The final Rietveld refinement plot is shown in Fig. S3 of the ESI.†Crystal data for [Cu(μ-4NO2pz)2]n, 2: C6H4CuN6O4, FW = 287.7 a.m.u., monoclinic, C2/m, a = 20.058(3) Å, b = 7.603(1) Å, c = 7.3757(9) Å, b = 115.018(7)°, V = 1019.2(2) Å3, Z = 8, Z′ = 4, ρ = 1.87 g cm−3, F(000) = 572, RBragg = 0.68, Rp = 1.5% and Rwp = 2.0%, for 4851 data and 41 parameters in the 8.0–105.0° (2θ) range. CCDC no. 2261945.†
Bacterial culture preparation
The antibacterial activity of 1–8 was tested against the two Gram-negative bacteria Escherichia coli (E. coli) ATCC 25922 and Pseudomonas aeruginosa (P. aeruginosa) ATCC 27853, and the Gram-positive bacterium Staphylococcus aureus (S. aureus) ATCC 25923. Bacterial inocula were prepared by adding monocolonies of E. coli, P. aeruginosa or S. aureus to the Tryptone Soy Broth (TSB) medium at 335 K overnight. The enriched cultures (log phase) thus obtained were diluted to a 106 CFU mL−1 concentration.Growth curve observation
To evaluate the trend of bacterial growth as a function of time in untreated bacterial cultures (blanks) and those treated with 1–8, their optical density (OD600) was determined by using a BioTek mQuant MQX200 Microplate spectrophotometer at 600 nm. The bacterial cultures, prepared as detailed above, were treated with 0.4 mg mL−1 powders of 1–8. Then, they were kept on an IKA KS 130 BASIC platform shaker for 24 h at a low speed. The growth inhibitory effect of 1–8 was recorded at 0, 2, 4, 8, 12, 16, 20, and 24 hours.Antibacterial rate (%) evaluation
To evaluate the antibacterial rate (%), for each bacterium 1 μL of inoculum was added to a sterile Eppendorf tube containing 1 mL of autoclaved physiological solution, reaching a concentration of 106 CFU mL−1. 0.4 mg of powders of 1–8 were then added. The Eppendorf tubes were subsequently shaken for 24 h at a speed of 160 rpm. Periodic withdrawals of 10 μL were made at 0, 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, and 24 h. To obtain the bacterial colony count, the supernatant fraction was diluted and included uniformly into Petri dishes containing the Plate Count Agar medium (OXOID PCA). Adopting the same procedure, bacterial strains untreated with 1–8 were used as a control (blank). Growth inhibition was estimated as the antibacterial rate (%) using eqn (2): | (2) |
Detection of reactive oxygen species (ROS)
The production of reactive oxygen species (ROS) was evaluated using an oxidation sensitive probe 2′,7′-dichloro-dihydro-fluorescein diacetate (DCFH-DA). For each bacterium, cells grown aerobically in TSB overnight in phosphonate-buffered saline (PSB) medium to a concentration of 106 CFU mL−1 were incubated for 30 minutes with DCFH-DA at 335 K. 10 μL of this bacterial culture were then added to Eppendorf tubes containing 0.4 mg of 1–8 suspended in 1 mL of physiological solution. For each bacterium, separate Eppendorf tubes were prepared to test ROS production after 1, 2, 3, and 4 h of treatment. Adopting the same procedure, bacterial strains untreated with 1–8 were used as a control (blank). At the end of the treatment time, 100 μL of the sample from each Eppendorf tube were transferred in triplicate into 96 well plates, in the dark. The fluorescence intensity from each sample well was measured using a BMG LABTECH FLUOstar Omega fluorescence cytometer working at 485/20 nm and 528/20 nm for excitation and emission, respectively. The average of the triplicate was calculated and reported as the intensity of fluorescence in arbitrary units (a.u).Propidium iodide uptake
The viability of bacteria treated with compounds 1–8 was assessed using the fluorescence propidium iodide (PI) staining assay. A suspension (106 CFU mL−1) of E. coli, P. aeruginosa or S. aureus in physiological solution treated with 0.4 mg of 1–8 was incubated at 335 K for a period of 48 h. After 4, 8, 24, and 48 h, 100 mL of the content were transferred in triplicate into 96 well plates. 1.5 mL of PI (0.02 mg/1 mL DMSO) were added to each well in the dark. Adopting the same procedure, untreated bacterial strains were used as a control (blank). The fluorescence emitted by the bacterial cells was measured at 528/20 nm upon exciting at 485/20 nm, using a BMG LABTECH FLUOstar Omega fluorescence cytometer.Confocal laser scanning microscopy (CLSM) study
To verify the damaging effect of 1–8 on the cell membranes of the bacterial strains, CLSM analyses were performed with LIVE/DEAD® BacLight™ Bacterial Viability Kits (Invitrogen). Bacterial suspensions with a concentration of 1 × 108 CFU mL−1 (0.3 OD670) for E. coli and 1 × 107 CFU mL−1 (0.15 OD670) for S. aureus were treated with 0.4 mg of 2, arbitrary selected, during the logarithmic growth phase for 4 h. After this incubation period, the suspensions were centrifugated at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10–15 min. Then, after supernatant removal, the resulting pellets were suspended in 0.85% m/m NaCl solution. A mixture of SYTO 9 green-fluorescent stain and PI red-fluorescent stain was prepared and added to all the samples in the dark. After 15 min of incubation, the samples were fixed between a slide and a 18 mm coverslip to observe the fluorescence under a Nikon ECLIPSE Ti confocal microscope. The control assay was conducted without treatment with 2. The adopted excitation/emission wavelengths were 480/500 nm for SYTO 9 and 490/635 nm for PI.
000 rpm for 10–15 min. Then, after supernatant removal, the resulting pellets were suspended in 0.85% m/m NaCl solution. A mixture of SYTO 9 green-fluorescent stain and PI red-fluorescent stain was prepared and added to all the samples in the dark. After 15 min of incubation, the samples were fixed between a slide and a 18 mm coverslip to observe the fluorescence under a Nikon ECLIPSE Ti confocal microscope. The control assay was conducted without treatment with 2. The adopted excitation/emission wavelengths were 480/500 nm for SYTO 9 and 490/635 nm for PI.
Cu(II) ion minimum inhibitory concentration (MIC) test
Cu(II) ions were tested against the Gram-positive bacterium S. aureus and Gram-negative bacterium E. coli. MIC determination was done by the microtitre broth dilution method,56 using a 1000 ppm standard solution of copper nitrate as a source of Cu(II) ion. A loop full of the given bacterial strain was inoculated in 10 mL of N-broth (nutrient broth) and incubated for 24 h at 335 K in order to be activated. The enriched culture was then diluted to reach a concentration of 106 CFU mL−1. A 96 well plate with 100 μL of physiological solution was prepared in triplicate. 100 μL of a solution containing 200 ppm of Cu(II) ions were placed in the first well. From the first well, 100 μL of solution were transferred to the second well. The operation was repeated for 15 dilutions, obtaining a Cu(II) ion final concentration of 0.006 ppm. 20 μL of the bacterial culture were added to each well and incubated at 335 K for 24 hours. To evaluate the trend of growing inhibition, the optical density (OD600) was determined as described above at different time lapses, in order to monitor the bacterial inhibitory activity over time.Copper ion release test
To evaluate the maximum quantity of released copper(II) ions under conditions similar to those of the antibacterial tests, we selected compounds 1, 2, 5, and 6 as representative of analogues 3, 4, 7 and 8 which have very similar structures and properties, and suspended 4 mg of powdered samples in 10 mL of physiological solution. The suspensions were kept under gentle magnetic stirring for 3 days at 335 K. Due to the very small dimensions of the solute grains, filtering by celite pad was necessary. The mother liquors were then analyzed by inductively coupled plasma-optical emission spectrometry (ICP-OES).Bacterial morphological study
The morphology of the bacterial cells after treatment with 2 was determined by SEM, operating at 1–5 kV. For sample preparation, log phase cells of E. coli and S. aureus (106 CFUs) were incubated with 4 mg of compound 2, arbitrarily chosen, for 4 hours at 335 K. After the incubation period, the bacterial strains were centrifugated (2000 rpm, 5 min), washed with physiological solution and fixed on silicon plates with 20 μL of 2.5% (v/v) glutaraldehyde solution for 2 hours. The fixed pellets were immersed for 10 min in physiological solution/ethanol mixtures with increasing concentrations of ethanol [10, 30, 50, 70, 90 and 100% (v/v)]. After drying, the pellets were placed on aluminium stubs using self-adhesive carbon conductive tab and they were chrome coated. Untreated bacterial cells were used as the control.Conclusions
The copper(II) pyrazolate CPs 1–8 showed high antibacterial activity against model Gram-negative and Gram-positive bacteria. The antibacterial mechanism proceeds with ROS production, reasonably activated by the electrostatic interactions between the CP particles and the bacterial membrane, provoking cell membrane disruption. Such an antibacterial mechanism, responsible for the alteration of the bacterial membrane without any relevant Cu(II) ion release, can deactivate vital enzymes and damage the cell membrane integrity and permeability.Author contributions
C. Di Nicola – conceptualization and writing – review & editing; F. Marchetti – resources and writing; A. Tombesi – synthesis and characterization of copper complexes; S. Xhafa – antimicrobial study validation and data curation; P. Campitelli – synthesis and characterization of copper complexes; M. Moroni X-ray crystallographic investigation; S. Galli – X-ray crystallographic study supervision, validation and writing; R. Pettinari – writing – review & editing; C. Pettinari – project administration and ligands and zinc complexes synthesis supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was carried out under the frame of the project Nano4-Fresh – Nanomaterials for environmentally friendly and sustainable handling of perishable products (PRIMA19_00246), which is part of the Partnership on Research and Innovation in the Mediterranean Area (PRIMA) Programme supported by the European Union and funded by the national funding bodies of Participating States (MUR in Italy). Corrado Di Nicola and Fabio Marchetti acknowledge the University of Camerino and the Research Project FAR 2022 PNR “Development of Innovative Antimicrobial Biodegradable Food Packaging” (ActiveFoodPack). Simona Galli and Marco Moroni acknowledge Università degli Studi dell’Insubria for partial funding.Notes and references
- A. Talebi Bezmin Abadi, A. A. Rizvanov, T. Haertlé and N. L. Blatt, Bionanoscience, 2019, 9, 778–788 CrossRef.
- A. I. Ribeiro, A. M. Dias and A. Zille, ACS Appl. Nano Mater., 2022, 5, 3030–3064 CrossRef CAS PubMed.
- S. Gharpure, A. Akash and B. Ankamwar, J. Nanosci. Nanotechnol., 2020, 20, 3303–3339 CrossRef PubMed.
- F. Marchetti, J. Palmucci, C. Pettinari, R. Pettinari, S. Scuri, I. Grappasonni, M. Cocchioni, M. Amati, F. Lelj and A. Crispini, Inorg. Chem., 2016, 55, 5453–5466 CrossRef CAS PubMed.
- Effendy, C. Di Nicola, C. Pettinari, A. Pizzabiocca, B. W. Skelton, N. Somers and A. H. White, Inorganica Chim Acta, 2006, 359(1), 64–80 CrossRef CAS.
- S. Mittapally, R. Taranum and S. Parveen, J. Drug Delivery Ther., 2018, 8, 411–419 CrossRef CAS.
- G. Wyszogrodzka, B. Marszałek, B. Gil and P. Dorożyński, Drug Discov. Today, 2016, 21, 1009–1018 CrossRef CAS PubMed.
- D. Mitra, M. Li, E.-T. Kang and K. G. Neoh, ACS Appl. Mater. Interfaces, 2018, 11, 73–83 CrossRef PubMed.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227 CrossRef CAS PubMed.
- R. Kumar and H. Münstedt, Biomaterials, 2005, 26, 2081–2088 CrossRef CAS PubMed.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4(7), 4317–4323 CrossRef CAS PubMed.
- P.-L. Wang, L.-H. Xie, E. A. Joseph, J.-R. Li, X.-O. Su and H.-C. Zhou, Chem. Rev., 2019, 119, 10638–10690 CrossRef CAS PubMed.
- G. Chedid and A. Yassin, Nanomaterials, 2018, 8, 916 CrossRef PubMed.
- S. R. Batten, S. M. Neville and D. R. Turner, Coordination polymers: design, analysis and application, Royal Society of Chemistry, 2008 Search PubMed.
- S. Kaskel, The Chemistry of metal-organic frameworks, 2 volume set: synthesis, characterization, and applications, John Wiley & Sons, 2016, vol. 1 Search PubMed.
- S. M. Soliman, A. El-Faham and S. E. El Silk, Appl. Organomet. Chem., 2020, 34, e5941 CrossRef CAS.
- F. Marchetti, J. Palmucci, C. Pettinari, R. Pettinari, F. Condello, S. Ferraro, M. Marangoni, A. Crispini, S. Scuri, I. Grappasonni, M. Cocchioni, M. Nabissi, M. R. Chierotti and R. Gobetto, Chem. – Eur. J., 2015, 21, 836–850 CrossRef CAS PubMed.
- C. Pettinari, R. Pettinari, C. Di Nicola, A. Tombesi, S. Scuri and F. Marchetti, Coord. Chem. Rev., 2021, 446, 214121 CrossRef CAS.
- F. Scarpelli, A. Crispini, E. Giorno, F. Marchetti, R. Pettinari, C. Di Nicola, M. P. De Santo, E. Fuoco, R. Berardi and P. Alfano, ChemPlusChem, 2020, 85, 426–440 CrossRef CAS PubMed.
- R. Pettinari, F. Condello, F. Marchetti, C. Pettinari, M. I. Bautista-Toledo, S. Morales-Torres, P. J. Dyson and F. J. Maldonado-Hódar, ACS Appl. Bio Mater., 2018, 1, 153–159 CrossRef CAS.
- C. Di Nicola, F. Marchetti, R. Pettinari, A. Tombesi, C. Pettinari, I. Grappasonni, P. J. Dyson and S. Scuri, Materials, 2020, 13(526), 1–16 Search PubMed.
- S. Scuri, F. Petrelli, I. Grappasonni, L. Idemudia, F. Marchetti and C. Di Nicola, Acta Biomedica., 2019, 90, 370–377 CAS.
- A. Tabacaru, C. Pettinari, F. Marchetti, C. Di Nicola, K. V. Domasevitch, S. Galli, N. Masciocchi, S. Scuri, I. Grappasonni and M. Cocchioni, Inorg. Chem., 2012, 51, 9775–9788 CrossRef CAS PubMed.
- R. Li, T. Chen and X. Pan, ACS Nano, 2021, 15, 3808–3848 CrossRef CAS PubMed.
- M. Vincent, R. E. Duval, P. Hartemann and M. Engels-Deutsch, J. Appl. Microbiol., 2018, 124, 1032–1046 CrossRef CAS PubMed.
- C. E. Santo, D. Quaranta and G. Grass, MicrobiologyOpen, 2012, 1, 46–52 CrossRef CAS PubMed.
- O. Krasnovskaya, A. Naumov, D. Guk, P. Gorelkin, A. Erofeev, E. Beloglazkina and A. Majouga, Int. J. Mol. Sci., 2020, 21, 3965 CrossRef CAS PubMed.
- A. Rauf, J. Ye, S. Zhang, Y. Qi, G. Wang, Y. Che and G. Ning, Dalton Trans., 2019, 48, 17810–17817 RSC.
- G. A. Bowmaker, C. Di Nicola, C. Pettinari, B. W. Skelton, N. Somers and A. H. White, Dalton Trans., 2011, 40, 5102–5115 RSC.
- J. Inkinen, R. Mäkinen, M. M. Keinänen-Toivola, K. Nordström and M. Ahonen, Lett. Appl. Microbiol., 2017, 64, 19–26 CrossRef CAS PubMed.
- X. Sui, R. Xu, J. Liu, S. Zhang, Y. Wu, J. Yang and J. Hao, ACS Appl. Mater. Interfaces, 2018, 10, 36531–36539 CrossRef CAS PubMed.
- J. H. Jo, H.-C. Kim, S. Huh and Y. Kim, Dalton Trans., 2019, 48, 8084–8093 RSC.
- Z. Wang, W. Guo, K. Zhang, Y. Ye, Y. Wang, D. Sui, N. Zhao and F.-J. Xu, Sci. China: Technol. Sci., 2022, 65, 1052–1058 CrossRef CAS.
- C. Di Nicola, E. Forlin, F. Garau, M. Gazzano, A. Lanza, M. Monari, F. Nestola, L. Pandolfo, C. Pettinari, A. Zorzi and F. Zorzi, Cryst. Growth Des., 2013, 13, 126–135 CrossRef CAS.
- C. Di Nicola, F. Garau, M. Gazzano, A. Lanza, M. Monari, F. Nestola, L. Pandolfo and C. Pettinari, Cryst. Growth Des., 2015, 15, 1259–1272 CrossRef CAS.
- E. Forlin, A. Lanza, C. Di Nicola, M. Monari, M. Gazzano, F. Nestola, C. Pettinari and L. Pandolfo, Inorganica. Chim. Acta., 2018, 470, 385–392 CrossRef CAS.
- A. Cingolani, S. Galli, N. Masciocchi, L. Pandolfo, C. Pettinari and A. Sironi, J. Am. Chem. Soc., 2005, 127, 6144–6145 CrossRef CAS PubMed.
- A. Bencini, M. Casarin, D. Forrer, L. Franco, F. Garau, N. Masciocchi, L. Pandolfo, C. Pettinari, M. Ruzzi and A. Vittadini, Inorg. Chem., 2009, 48, 4044–4051 CrossRef CAS PubMed.
- C. Di Nicola, A. Tombesi, M. Moroni, R. Vismara, F. Marchetti, R. Pettinari, L. Nardo, G. Vesco, S. Galli, S. Casassa, L. Pandolfo and C. Pettinari, CrystEngComm, 2020, 22, 3294–3308 RSC.
- H.-Z. Zhang, L.-L. Gan, H. Wang and C.-H. Zhou, Mini Rev. Med. Chem., 2017, 17, 122–166 CrossRef CAS PubMed.
- M. A. Alam, Future Med. Chem., 2022, 14, 343–362 CrossRef PubMed.
- F. K. Keter and J. Darkwa, Biometals, 2012, 25, 9–21 CrossRef CAS PubMed.
- S. Noriega, J. Cardoso-Ortiz, A. López-Luna, M. D. R. Cuevas-Flores and J. A. Flores De La Torre, Pharmaceuticals, 2022, 15, 717 CrossRef CAS PubMed.
- E. Ghobadi, Z. Ghanbarimasir and S. Emami, Eur. J. Med. Chem., 2021, 223, 113669 CrossRef CAS PubMed.
- A. Regiec, H. Mastalarz and P. Wojciechowski, J. Mol. Struct., 2014, 1061, 166–174 CrossRef CAS.
- C. J. E. Kempster and H. Lipson, Acta Crystallogr. B, 1972, 28, 3674 CrossRef CAS.
- L. Boulos, M. Prevost, B. Barbeau, J. Coallier and R. Desjardins, J. Microbiol. Methods, 1999, 37, 77–86 CrossRef CAS PubMed.
- L. Fowler, H. Engqvist and C. Öhman-Mägi, Materials, 2019, 12, 3798 CrossRef CAS PubMed.
- K. Wang, Z. Geng, Y. Yin, X. Ma and Z. Wang, CrystEngComm, 2011, 13, 5100–5104 RSC.
- A. A. Coelho, J. Appl. Crystallogr., 2003, 36, 86–95 CrossRef CAS.
- TOPAS v. 3.0; Bruker AXS, Karlsruhe, Germany, 2005 Search PubMed.
- A. Le Bail, H. Duroy and J. L. Fourquet, Mater. Res. Bull., 1988, 23, 447–452 CrossRef CAS.
- A. A. Coelho, J. Appl. Crystallogr., 2000, 33, 899–908 CrossRef CAS.
- R. A. Young, IUCr Monograph N. 5, Oxford University Press, New York, 1981 Search PubMed.
- R. W. Cheary and A. Coelho, J. Appl. Crystallogr., 1992, 25, 109 CrossRef CAS.
- I. Wiegand, K. Hilpert and R. E. W. Hancock, Nat. Protoc., 2008, 3, 163–175 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: IR spectra of 2 and 5, TGA curves of 2 and 5, PXRD pattern of 2 and antibacterial activity of the pyrazole ligands against E. coli and S. aureus. CCDC 2261945. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d3nj02378h |
| ‡ The existence of a crystallographic axis of ca. 7.32 Å suggests that a 1D motif featuring pyrazolate bridged Cu(II) ions with a pace of ca. 3.65 Å is present also in 5. The existence of a polymeric motif in 5 is supported also by its marked insolubility. |
§ Bond distances and angles for the rigid body describing the ligand: endocyclic C–C and C–N bonds = 1.36 Å; exocyclic C–N bond = 1.40 Å; N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds = 1.25 Å; C–H bonds = 0.95 Å; pyrazolate ring internal and external bond angles = 108° and 126°, respectively; angles at the nitrogen atom of the NO2 group = 120°. Bond distances and angles for the rigid body describing the DMF molecule: C–N bonds = 1.40 Å; C O bonds = 1.25 Å; C–H bonds = 0.95 Å; pyrazolate ring internal and external bond angles = 108° and 126°, respectively; angles at the nitrogen atom of the NO2 group = 120°. Bond distances and angles for the rigid body describing the DMF molecule: C–N bonds = 1.40 Å; C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond = 1.25 Å; C–H bonds = 0.95 Å; angles at the nitrogen and carbonyl carbon atoms = 120°; and angles at the methyl carbon atoms = 109.5°. O bond = 1.25 Å; C–H bonds = 0.95 Å; angles at the nitrogen and carbonyl carbon atoms = 120°; and angles at the methyl carbon atoms = 109.5°. |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2023 |