Open Access Article
Open Access ArticleUse of a bioresource nanocomposite as a heterogeneous base catalyst for the green synthesis of novel bioactive pyrazoles: antibacterial evaluation using molecular docking†
Sarah
Aldulaijan
 ab,
Sara
Nabil
ab,
Salha
Alharthi
ab,
Sara
Nabil
ab,
Salha
Alharthi
 ab,
Bushra AL
Abdullatif
ab and
Abir S.
Abdel-Naby
ab,
Bushra AL
Abdullatif
ab and
Abir S.
Abdel-Naby
 *ab
*ab
aChemistry Department, College of Science, Imam Abdulrahman Bin Faisal University, P.O. Box 1982, 31441, Dammam, Saudi Arabia. E-mail: aabdelnaby@iau.edu.sa
bWater Treatment Unit, Basic & Applied Scientific Research Centre (BASRC), Imam Abdulrahman Bin Faisal University, P.O. Box 1982, 31441, Dammam, Saudi Arabia
First published on 28th June 2023
Abstract
Chitosan modified with diallylamine (CS-DAA) was synthesized and mixed with nano magnesium oxide (in various percentages) to prepare CS-DAA–MgO nanocomposites for use as a heterogeneous base catalyst for the synthesis of novel pyrazole derivatives. 13C NMR spectroscopic analysis confirmed the synthesis of the new functionalized chitosan CS-DAA. The nanocomposite containing 5% MgO exhibited the highest thermal stability as shown in the thermogravimetry curves. The X-ray diffraction pattern and scanning electron microscopy confirmed the formation of the nanocomposite CS-DAA–MgO. The ecofriendly, biodegradable nanocomposite was used as a heterogeneous base catalyst for the synthesis of four novel pyrazole derivatives. Pyrazolone was condensed with thiocarbohydrazide in two different ratios to afford the corresponding hydrazones (1 and 2). These hydrazones were treated with α-haloesters or hydrazonoyl halides in the presence of the heterogeneous nanocomposite catalyst, resulting in the formation of thiadiazines (3) and thiazolidin-4-one (4). The catalytic effect of the nanocomposite showed higher catalytic efficiency compared to the traditional homogeneous base catalysts Et3N and NaOH. The four pyrazole structures were confirmed using elemental analysis, 1H NMR and 13C NMR. A molecular docking study was used to predict the inhibition potential for the four pyrazole derivatives as anti-bacterial drugs for Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus and Staphylococcus epidermidis. The results showed that all the compounds exhibit good inhibition potential as anti-bacterial drugs, especially compound (3) and compound (4).
1. Introduction
Recently, as is environmentally necessary, scientific attention has been directed towards greener synthesis methods with the aim of avoiding the use of toxic catalysts and solvents.1–4 Thus, heterogeneous catalysts have been suggested as replacements for homogeneous catalysts. Heterogeneous catalysts have the advantage of being easily isolated from the reaction medium by simple filtration, enabling them to be reused. Polymeric nanocomposites have been used for base-catalyzed reactions.5–7 Chitosan (CS) possesses excellent properties: it is biodegradable, non-toxic and insoluble in most solvents. Despite these advantages, chitosan is unable to form a film or membrane, as it forms a gel when cast.In our previous study, aluminium oxide nanopowder was added to chitosan to increase its basicity and enhance the formation of the composite film.7 Moreover, to increase the base characteristics of chitosan, magnesium oxide and copper oxide nanocomposites have been used as efficient catalysts for the synthesis of triazoles.8,9
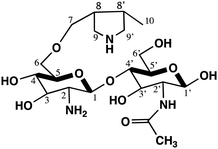
In the present study, CS was allowed to react with diallylamine (DAA) to reduce the hydrophilicity of the polymer, thus enhancing its ability to be cast into a film. This also increases its basicity due to the presence of DAA moieties, as well as pyrrolidine rings as branches, on the CS main chains.
While CS-DAA exhibits some base characteristics, the CS-DAA–MgO nanocomposite enhances the formation of the films due to its nano size, in addition to the basicity of MgO.
Pyrazole is considered to be one of the most important heterocyclic scaffolds, as it exhibits a wide range of physiological and pharmacological properties,10 including antibacterial,11 anti-fungal,12 anti-inflammatory,13 anti-tubercular,14 antiviral,15 anti-HIV,16 anti-tumoral,17 anti-leishmanial,18 anti-cancer19 and anti-depressant20 activities. An increase in the inhibitory characteristics of pyrazole derivatives could be accomplished through the presence of nitrogen and sulfur heteroatoms in their molecular structure, as well as through introducing benzene moieties. Furthermore, few studies have suggested pyrazole carbothioamide derivatives as corrosion inhibitors. When compared with other families of organic chemicals, pyrazoles have low toxicity and exhibit high solubility in acidic conditions, which makes them greener corrosion inhibitors. In recent decades, the synthesis of thiadiazine derivatives, as well as studies of their chemical and biological activity, have become more important, as they have been explored for their antimicrobial,21 anti-diabetic,22 diuretic,23 antioxidant,24 anti-leishmanial,25 and antiviral26 activities.
A synthesis of thidazines has been reported using an ultrasonic technique at 60 °C with a moderate yield.27 Thus, our efforts have been directed towards the synthesis of a new series of thiadiazines using heterogeneous catalysts, aiming to reduce the reaction time and improve the yield percentage. Moreover, the introduction of thiadiazine to the pyrazole moiety could increase its anti-bacterial activity.28
Bacterial diseases can lead to death, and bacterial infections are known for their multidrug resistance properties; thus, new compounds that act as antibacterial drugs are increasingly being sought.29 Pyrazole scaffolds have been explored as bioactive moieties. Therefore, in this study we aimed to develop novel pyrazole derivatives that would be expected to exhibit antibacterial efficiency.
The inhibition mechanisms of antibacterial drugs involve interactions with targets of specific proteins that are responsible for a specific activity such as protein synthesis or cell wall synthesis.30,31 The interactions between the antibacterial drugs and the proteins lead to blocking of the active site of the protein, preventing the synthesis of a new protein and destroying the bacteria.31
Molecular docking is a very popular tool in drug discovery.32,33 Therefore, in this work, it has been used to predict the inhibition potential of novel synthesized compounds as anti-bacteria drugs by evaluating their binding with Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus and Staphylococcus epidermidis. Moreover, the molecular docking results help to understand the types of interactions between these compounds and the active sites of their receptors in these bacteria. The proteins used in this study are the peptide deformylase (PDF)34–37 and transcriptional regulator (TcaR),38 which are commonly used in designing antibacterial drugs.
In this study, a novel synthetized CS-DAA–MgO nanocomposite will be investigated as a heterogenous base catalyst and a green method to synthetize four novel pyrazole derivatives. Thus, we could predict the inhibition potentials of these compounds as antibacterial drugs.
2. Materials and methods
2.1. Materials
3-Methyl-1-phenyl-pyrazol-5-one, acetone, ethanol, ethyl bromoacetate, diallylamine, magnesium oxide (MgO) nanopowder and chitosan of medium molecular weight (85% deacetylation) were used to obtain a high percentage of NH2 groups over the amide groups. All chemicals were purchased from Sigma Aldrich, USA. Triple-distilled water was used in the preparation of all solutions.2.2. Instruments
2.3. Methods
The catalyst was added as a film (by weight) to the reactants in absolute ethanol as a solvent.
The synthesis of each of the following compounds was first attempted using the traditional method. The products took a long time (up to 6 h) to formed and had very poor yield percentages.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and 1260 (C
N) and 1260 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S); 1H NMR: 1.35 (s, 3H, CH3), 2.05 (s, 2H, CH2), 4.05 (s, 1H, NH), 4.24 (s, 2H, NH2), 7.16 (s, 1H, NH), 7.17–8.05 (m, 5H, Ar-H). 13C NMR: 12.96 (CH3), 17.14 (CH2), 124.42–133.19 (C-aromatic), 147.27 (C
S); 1H NMR: 1.35 (s, 3H, CH3), 2.05 (s, 2H, CH2), 4.05 (s, 1H, NH), 4.24 (s, 2H, NH2), 7.16 (s, 1H, NH), 7.17–8.05 (m, 5H, Ar-H). 13C NMR: 12.96 (CH3), 17.14 (CH2), 124.42–133.19 (C-aromatic), 147.27 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 149.85 (CH3–C
N), 149.85 (CH3–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 180.99 (C
N), 180.99 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) (Fig. S2 and S3, ESI†).
S) (Fig. S2 and S3, ESI†).
Anal. calcd for C11H13N6S (262.10): C, 50.36; H, 5.38; N, 32.04; S, 12.22%; found: C, 50.33; H, 5.40; N, 32.06; S, 12.20%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) and 1282 (C
N) and 1282 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S); 1H NMR: 1.60 (s, 6H, 2CH3), 2.45 (s, 4H, 2CH2), 7.32–7.85 (m, 10H, Ar-H), 7.86 (s, 2H, 2NH). 13C NMR: 16.56 (2CH3), 18.12 (CH2), 25.27 (2CH2), 117.86–130.15 (C-aromatic), 159.08 (2 N–C
S); 1H NMR: 1.60 (s, 6H, 2CH3), 2.45 (s, 4H, 2CH2), 7.32–7.85 (m, 10H, Ar-H), 7.86 (s, 2H, 2NH). 13C NMR: 16.56 (2CH3), 18.12 (CH2), 25.27 (2CH2), 117.86–130.15 (C-aromatic), 159.08 (2 N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 161.56 (2C
N), 161.56 (2C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 188.39 (C
N), 188.39 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) (Fig. S4 and S5, ESI†).
S) (Fig. S4 and S5, ESI†).
Anal. calcd for C21H22N8S (418.17): C, 60.27; H, 5.30; N, 26.77; S, 7.66%; found: C, 60.30; H, 5.28; N, 26.78; S, 7.65%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1590 (C
O), 1590 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H NMR: 1.69 (s, 2H, CH2), 2.04 (s, 3H, CH3), 3.79 (s, 3H, CH3), 6.71–8.27 (m, 10H, Ar-H), 11.69 (s, 1H, NH). 13C NMR: 12.97 (CH3), 14.38 (CH3), 22.92 (CH2), 116.28–133.19 (C-aromatic), 142.52 (S–C
N). 1H NMR: 1.69 (s, 2H, CH2), 2.04 (s, 3H, CH3), 3.79 (s, 3H, CH3), 6.71–8.27 (m, 10H, Ar-H), 11.69 (s, 1H, NH). 13C NMR: 12.97 (CH3), 14.38 (CH3), 22.92 (CH2), 116.28–133.19 (C-aromatic), 142.52 (S–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 146.41 (C
N), 146.41 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 153.82 (N–C
N), 153.82 (N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 159.70 (S–C
N), 159.70 (S–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 164.90 (CH2–C
N), 164.90 (CH2–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) (Fig. S6 and S7, ESI†).
N) (Fig. S6 and S7, ESI†).
Anal. calcd for C20H20N8S (404.15): C, 59.39; H, 4.98; N, 27.70; S, 7.93%; found: C, 59.36; H, 4.97; N, 27.73; S, 7.95%.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1590 (C
O), 1590 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N). 1H NMR: 1.28 (s, 6H, 2CH3), 1.60 (s, 4H, 2CH2), 4.17 (s, 2H, CH2 thiazoline), 7.37–7.80 (m, 10H, Ar-H). 13C NMR: 12.28 (2CH3), 22.40 (CH2), 26.52(CH), 28.40 (S–CH2), 116.26–133.19 (C-aromatic), 157.27 (N–C
N). 1H NMR: 1.28 (s, 6H, 2CH3), 1.60 (s, 4H, 2CH2), 4.17 (s, 2H, CH2 thiazoline), 7.37–7.80 (m, 10H, Ar-H). 13C NMR: 12.28 (2CH3), 22.40 (CH2), 26.52(CH), 28.40 (S–CH2), 116.26–133.19 (C-aromatic), 157.27 (N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 159.60 (S–C
N), 159.60 (S–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 168.90 (2 N
N), 168.90 (2 N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–CH2), 180.99 (N–C
C–CH2), 180.99 (N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 181.26 (C
N), 181.26 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (Fig. S8 and S9, ESI†).
O) (Fig. S8 and S9, ESI†).
Anal. calcd for C23H22N8OS (458.16): C, 60.24; H, 4.84; N, 24.44; S, 6.99%; found: C, 60.25; H, 4.85; N, 24.45; S, 6.96%.
3. Results and discussion
3.1. Synthesis of chitosan-diallyl amine (CS-DAA)
The synthetized (CS-DAA) was characterized using 13C NMR spectroscopy, which revealed the following:(1) A peak at 70 ppm indicated the formation of a bond between the hydroxyl group at the C6 position of chitosan and the DAA moiety.
(2) New peaks at 22, 30, and 40 ppm indicated the formation of a pyrrolidine ring through an intramolecular cyclopolymerization process49 (Table 1).
Chitosan is unable to be cast as a film and usually forms a gel, due to its high hydrophilicity. The reaction of chitosan with DAA leads to the formation of some pyrrolidine rings through the chitosan matrix, which decreases its hydrophilicity, thus enhancing its casting into a film. Moreover, pyrrolidine rings exhibit basic characteristics due to the free lone pair of electrons belonging to the nitrogen atom, which increases the basicity of the chitosan polymeric matrix.
3.2. Fabrication of the CS-DAA–MgO nanocomposite
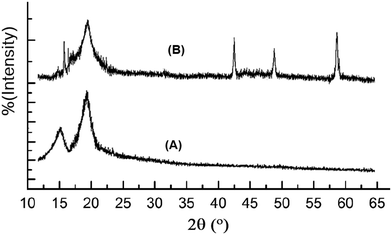
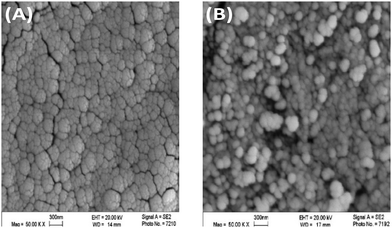
The XRD pattern for the nanocomposite is shown in Fig. 1. The main broad reflection at 2θ = 18–21° is characteristic of the polymer hydrated crystalline structure of modified chitosan. The peaks at 2θ ≤ 15 and the sharp peaks from 2θ = 40 to 60° are associated with MgO coordination bonds with the polymer main chains, as the MgO nanoparticles exhibit two characteristic peaks at 2θ = 43° and 60°.50Fig. 2 shows the difference in the morphology of the nanocomposite (A) compared to that of MgO nanoparticles.
Fig. 3 (A) Shows the EDX image of CS-DAA–MgO, in which all the nanocomposite constituents are present, as compared to that of nano-MgO (B). The yellow color indicates carbon atoms, the pale blue nitrogen atoms, the green color oxygen atoms, and the blue color magnesium atoms. Fig. 3(A) shows the EDX image indicating the uniform distribution of the nano-MgO through the nanocomposite. The EDX analysis showing the atomic percentages in the nanocomposite is presented in Fig. 3(C).
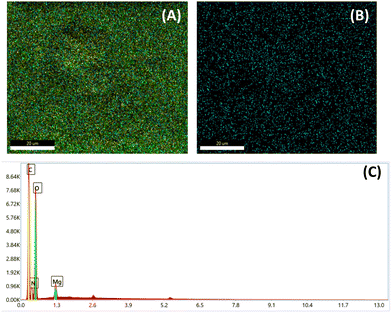 | ||
| Fig. 3 EDX image of the CS-DAA–MgO (A) and nano-MgO (B), and EDX analysis of the CS-DAA–MgO nanocomposite (C). | ||
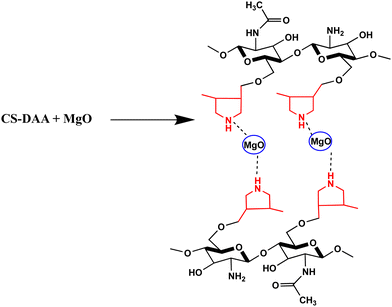
From all the data mentioned above, a schematic representation of the CS-DAA–MgO nanocomposite could be prepared (Fig. 4).
3.3. Thermal studies
The thermal stability of the nanocomposite was investigated prior to suggesting it as a heterogenous catalyst for the synthesis of pyrazole derivatives. The results revealed that the composite exhibited higher thermal stability than the parent chitosan, as shown from the initial decomposition temperature (T), which was the temperature at which the polymer started to lose parts of its matrix, and the weight loss percentage at 500 °C. Moreover, increasing the percentage of nano-MgO powder in the nanocomposite increased its thermal stability. The 5% MgO composite exhibited a higher T value than that of the composite containing 2% (Table 2 and Fig. 5).| Polymeric material | T (°C) | Weight loss % at 500 °C |
|---|---|---|
| Chitosan | 235 | 45 |
| CS-DAA | 220 | 36 |
| CS-DAA–MgO (2%) | 280 | 25 |
| CS-DAA–MgO (5%) | 295 | 15 |
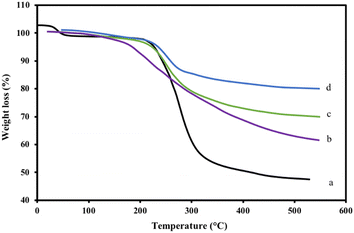 | ||
| Fig. 5 TGA of the CS-DAA–MgO nanocomposites with (c) 2% nano-MgO (d) and 5% nano-MgO, along with those of (b) CS-DAA and (a) CS. | ||
3.4. CS-DAA–MgO nanocomposite as base catalyst for the synthesis of pyrazole derivatives (1), (2), (3), and (4)
Green methods are widely used in heterocyclic chemistry. The base characteristics of the synthetized nanocomposite enable its use as a heterogeneous biodegradable catalyst for the synthesis of four pyrazole derivatives (Scheme 1).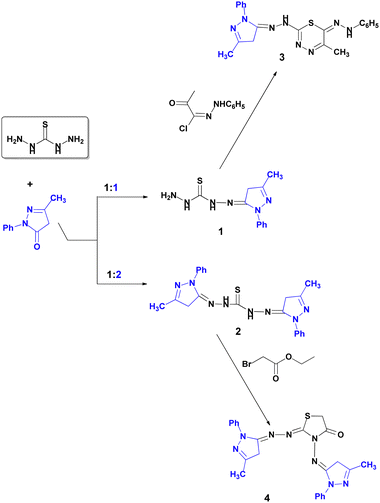 | ||
| Scheme 1 Synthesis of pyrazole derivatives 1–4 using the heterogeneous base nanocomposite CS-DAA–MgO. | ||
The use of this catalyst minimized the reaction time and increased both the percentage yield and the purity of the products as compared with the traditional method. According to the literature, the removal of water is the rate-determining step.51 Therefore, the catalyst was thought to engage in this step, as presented in Scheme 2.
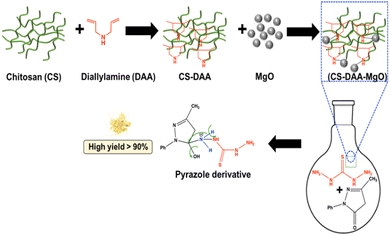 | ||
| Scheme 2 Schematic representation of the role of the CS-DAA–MgO catalyst in the synthesis of compound (1). | ||
The composite exhibited base characteristics because it consisted of pyrrolidine rings, the constructed branches, and the nano-sized base MgO. Thus, the composite could enhance the formation of the products in a short time with a high percentage yield. The branches of the pyrrolidine rings covered the chitosan main chains, surrounding the reactant molecules, thus enhancing their reaction.
Thus, sono-condensation of the pyrazolone derivative with thiocarbohydrazide at 70 °C in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratios afforded the corresponding starting mono- and bis-thiocarbohydrazone mono- and dihydrazones (1, 2) in higher yields than the traditional methods (Scheme 1).
1 ratios afforded the corresponding starting mono- and bis-thiocarbohydrazone mono- and dihydrazones (1, 2) in higher yields than the traditional methods (Scheme 1).
The structures of these hydrazones were elucidated via spectral data. The IR spectrum of (1) showed absorption peaks in the regions 3390, 3360, and 3290 cm−1, which were attributed to NH2 and the N–H functional groups. Additionally, the peak at 1260 cm−1 confirmed the presence of a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S group. Furthermore, 1H NMR (DMSO) of (1) showed signals at δ 4.05 and 7.16 related to the NH2 and 2NH protons. The 13C NMR spectrum (DMSO) revealed signals at 181.93 (C
S group. Furthermore, 1H NMR (DMSO) of (1) showed signals at δ 4.05 and 7.16 related to the NH2 and 2NH protons. The 13C NMR spectrum (DMSO) revealed signals at 181.93 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S). Meanwhile, the 1H NMR spectrum of dihydrazone (2) showed characteristic signals at 7.86 ppm for 2NH. The 13C NMR spectrum (DMSO) exhibited signals at 188.39 (C
S). Meanwhile, the 1H NMR spectrum of dihydrazone (2) showed characteristic signals at 7.86 ppm for 2NH. The 13C NMR spectrum (DMSO) exhibited signals at 188.39 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S).
S).
In the present study, we achieved our targets: yields of 85% to 92% at room temperature. Moreover, the sono-reaction of (1) with hydrazonoyl halides afforded thiadiazine (3), as shown in Scheme 1. The 1H NMR spectrum of (3) exhibited a signal at δ 11.69 ppm, indicating NH groups. The 13C NMR spectrum (DMSO) exhibited a signal at 158.64 ppm, which confirmed the formation of a thiadiazine ring.
Finally, the cyclocondensation of bis-hydrazone (2) with ethyl bromoacetate afforded thiazolidin-4-one. The spectroscopy of compound (4) elucidated its structure. The IR spectrum showed a peak at 1690 cm−1, which corresponds to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O. The 1H NMR spectrum of (4) showed a singlet signal at 4.17 (s, 2H, CH2 thiazoline). Its 13C NMR spectrum exhibited a signal at 181.93 (C
O. The 1H NMR spectrum of (4) showed a singlet signal at 4.17 (s, 2H, CH2 thiazoline). Its 13C NMR spectrum exhibited a signal at 181.93 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).
O).
3.5. Reusability of the heterogeneous of CS-DAA–MgO nanocomposite
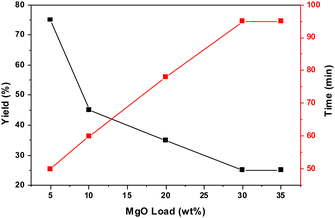
To estimate the appropriate catalyst loading percentage, the synthesis of derivative (2) was tested using various catalyst loading percentages under the same reaction conditions. The results revealed that the optimum loading percentage is 30 wt% (Fig. 6).The reusability of the catalyst was investigated by extensively washing the used catalyst with ethanol and allowing the ethanol traces to evaporate at room temperature. The results revealed that the catalyst could be reused four times without a significant loss of its catalytic activity (Fig. S10, ESI†).
3.6. Antibacterial activity
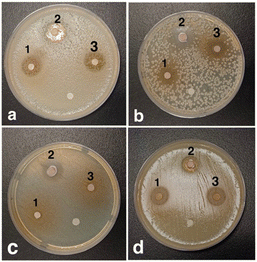 | ||
| Fig. 7 Clearance zones of compounds (1), (2), and (3) for various bacteria: Pseudomonas (a), Staphylococcus epidermidis (b), Escherichia coli (c), and Staphylococcus aureus (d). | ||
From the lab antibacterial test, it was observed that the synthetized pyrazole derivatives exhibited high efficiencies. The following molecular docking calculations will evaluate and confirm these anti-bacterial efficiencies.
3.6.2.1. Escherichia coli. The docking results of the four novel compounds were investigated and compared with the docking results of the original ligand in the active site of the protein PDB file as a reference. All four compounds present better performance than the original ligand.
Table 3 shows that compound (1) and compound (2) have binding energies of −7.2 kcal mol−1 and −7.7 kcal mol−1, respectively.
| Compound | Binding affinity (kcal mol−1) | No. of H-bonds | H-bonding residues |
|---|---|---|---|
| 1 | −7.2 | 2 | GlyA45, GluA133 |
| 2 | −7.7 | 1 | GluA42 |
| 3 | −8.7 | 1 | GluA95 |
| 4 | −9.2 | 1 | ArgA97 |
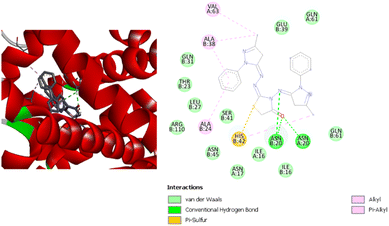
Both compounds have several types of interactions with the residues in the active site of the receptor. Compound 1 has two hydrogen bonds with GlyA45 and GluA133, alkyl and pi–alkyl interactions with the three residues IleA44, CysA129 and IleA128, and a pi–sigma interaction with HisA132. Compound 2 has alkyl and pi–alkyl interactions with the four residues IleA91, CysA90, ArgA97 and CysA129, and one hydrogen bond with GluA42. Moreover, compound (2) has a pi–sigma interaction with IleA44, and a pi–pi interaction with HisA132. Compound (3) has the second-best binding energy for this receptor. Fig. 8 shows the interactions between the compound and the residues in the active site. It has one hydrogen bond with GluA95, and alkyl and pi–alkyl interactions with five residues: LeuA91, IleA86, IleA44, CysA129 and ProA94. In addition, there is a pi–anion interaction with GluA133 and pi–sulfur interaction with CysA90. These interactions exhibit a binding energy equal to −8.7 kcal mol−1.
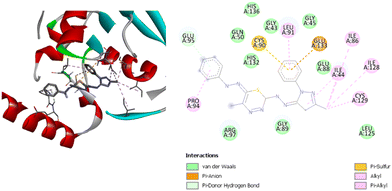 | ||
| Fig. 8 3D (left) and 2D (right) images of the binding interactions of compound (3) with the active site of Escherichia coli. | ||
Fig. 9 shows the interactions between compound (4) and the residues in the active site. This compound has a binding energy of −9.2 kcal mol−1, which is the highest affinity of all the compounds to the Escherichia coli active site based on the docking results.
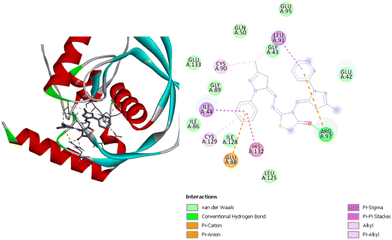 | ||
| Fig. 9 3D (left) and 2D (right) images of the binding interactions of compound (4) with the active site of Escherichia coli. | ||
Compound (4) has one hydrogen bond with ArgA97, as well as several other interactions including alkyl and pi–alkyl interactions with CysA90, GlyA43, and the CysA129; pi–sigma interactions with IleA44 and LeuA91; a pi–pi interaction with HisA132; and pi–cation and pi–anion interactions with GluA88 and ArgA97. All the compounds present several van der Waals interactions with different residues of the active site of the receptor. This is shown in Fig. 8 and 9 for compounds (3) and (4), respectively, and in Fig. S11 and S12 (ESI†) for compounds (1) and (2), respectively. Notably, all four compounds exerted van der Waals interactions with GlnA50 and GlyA89.
3.6.2.2. Pseudomonas aeruginosa. The docking results of the four novel compounds with the active site of the Pseudomonas aeruginosa protein are shown in Table 4. The most promising compounds are compound (3) and compound (4).
| Compound | Binding affinity (kcal mol−1) | No. of H-bonds | H-bonding residues |
|---|---|---|---|
| 1 | −7.0 | 2 | ValA95, CysA92 |
| 2 | −7.7 | — | — |
| 3 | −8.0 | 2 | TyrA99, TyrA88 |
| 4 | −8.7 | 3 | TyrA99, TyrA88, GlyA91 |
Compound (1) has the lowest binding affinity with the Pseudomonas aeruginosa active site, with a binding energy of −7.0 kcal mol−1. This compound has two hydrogen bonds with ValA95 and CysA92. This compound also has two alkyl interactions with LeuA83 and IleA45, and one pi–pi interaction with TyrA99. On the other hand, compound (2) shows a better binding infinity than compound (1) at −7.7 kcal mol−1. This compound did not show as many types of interactions as the other compounds. It has only alkyl and pi–alkyl interactions with the three residues IleA45, CysA131, and LeuA93. Fig. 10 shows the interactions between compound (3) and the residues in the active site. This compound has a very good binding energy. It has a binding energy of −8.0 kcal mol−1. It has one hydrogen bond with TyrA99 and one pi-doner hydrogen bond with TyrA88. In addition, alkyl and pi–alkyl interactions with LeuA127, IleA45, LeuA93, CysA131 and a pi–cation interaction with HisA134. Compound (4) has the highest affinity with the Pseudomonas aeruginosa active site. It has a binding energy of −8.7 kcal mol−1. Fig. 11 shows the interaction between the compound and the residues in the active site. It has three hydrogen bonds with TyrA99, TyrA88, and GlyA91, one pi-donor hydrogen bond with GlyA91, three pi–alkyl interactions with ProA43, TyrA99, and IleA45, and an unfavorable donor–donor interaction with IleA45. Several van der Waals interactions between these compounds have been identified from Fig. 10 and 11 for compounds (3) and (4), and from Fig. S13 and S14 (ESI†) for compounds (1) and (2). It was found that all these compounds exhibit van der Waals interactions with the two residues GluA90 and GlnA89.
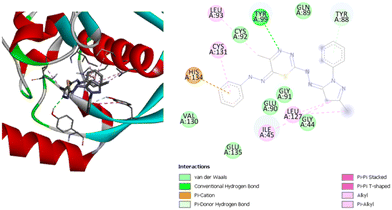 | ||
| Fig. 10 3D (left) and 2D (right) images of the binding interactions of compound (3) with the active site of Pseudomonas aeruginosa. | ||
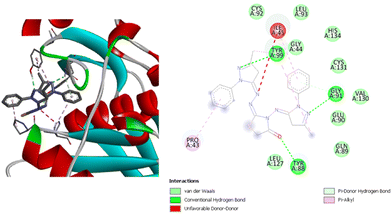 | ||
| Fig. 11 3D (left) and 2D (right) images of the binding interactions of compound (4) with the active site of Pseudomonas aeruginosa. | ||
3.6.2.3. Staphylococcus aureus. Table 5 shows the binding energies of the four novel compounds with the active site of Staphylococcus aureus protein. The most promising compounds are compound (3) and compound (4).
| Compound | Binding affinity (kcal mol−1) | No. of H-bonds | H-bonding residues |
|---|---|---|---|
| 1 | −7.4 | 1 | SerA57 |
| 2 | −7.2 | 2 | TypA147, GlyA110 |
| 3 | −9.4 | 1 | ValA59 |
| 4 | −8.9 | 1 | ArgA56 |
Compound (1) has a binding energy of −7.4 kcal mol−1. It was found that there is one hydrogen bond with SerA57, pi–anion and pi–cation interactions with HisA154 and GluA109, and hydrophobic interactions with ValA151, LeuA112, and CysA111. Compound (2) has a binding energy equal to −7.2 kcal mol−1. It has two hydrogen bonds with TypA147 and GlyA110, and alkyl and pi–alkyl interactions with the four residues LeuA105, ValA183, ValA151 and TypA147. Fig. 12 shows the interactions between compound (3) and the residues in the active site of the receptor. It has a binding energy of −9.4 kcal mol−1, which is the highest binding score for this protein.
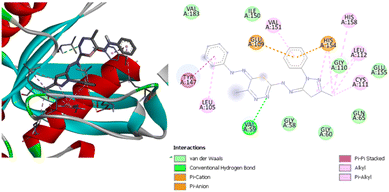 | ||
| Fig. 12 3D (left) and 2D (right) images of the binding interactions of compound (3) with the active site of Staphylococcus aureus. | ||
It was found that there is one hydrogen bond with ValA59, two pi–cation and pi–anion interactions with GluA109 and HisA154, one pi–pi stacked interaction with TyrA147, and alkyl and pi–alkyl interactions with the four residues ValA151, HisA158, LeuA112 and CysA111. Compound (4) has a binding energy of −8.9 kcal mol−1, which is the second highest binding score for this protein. Fig. 13 shows the interactions between this compound and the active site of the receptor. There is one hydrogen bond with ArgA56, one pi–anion interaction with GluA155, and one pi–sulfur interaction with CysA111. In addition, there is also an unfavorable donor–donor interaction with GlyA110, alkyl and pi–alkyl interactions with ValA151, HisA154, IleA150, LeuA112 and LeuA105. Fig. 12 and 13 depict the van der Waals interactions of compounds 3 and 4, while Fig. S15 and S16 (ESI†) show the van der Waals interactions of compounds 1 and 2. It has been found that all these compounds exhibit van der Waals interactions with the residue GlyA58.
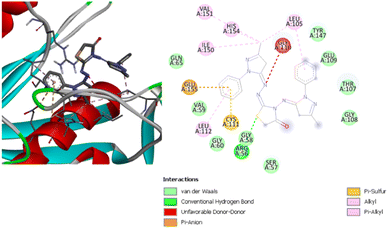 | ||
| Fig. 13 3D (left) and 2D (right) images of the binding interactions of compound (4) with the active site of Staphylococcus aureus. | ||
3.6.2.4. Staphylococcus epidermidis. Table 6 shows the binding energies of the four novel compounds with the active site of Staphylococcus epidermidis protein. The most promising compounds are compound (3) and compound (4). They perform better than the reference ligand.
| Compound | Binding affinity (kcal mol−1) | No. of H-bonds | H-bonding residues |
|---|---|---|---|
| 1 | −7.0 | — | — |
| 2 | −7.9 | 4 | LysB28, GluB39, GlnA61 |
| 3 | −8.9 | 1 | HisB42 |
| 4 | −9.2 | 3 | AsnA20, AsnB20 |
Based on the docking results, compound (1) is shown as the least active compound with Staphylococcus epidermidis active site. It has a binding energy of −7.0 kcal mol−1. It only has one type of interaction with the active site of the receptor. Two pi–alkyl interactions are identified with HisB42 and IleA16. On the other hand, compound (2) has a binding energy of −7.9 kcal mol−1. It has four hydrogen bonds with LysB28, GluB39 and GlnA61, a pi–sigma interaction with AlaB38, and a pi–cation interaction with LysB28, as well as alkyl and pi–alkyl interactions with LysB25, AlaB24, and HisB42. Fig. 14 shows the interaction between compound (3) and the residues in the active site, with a binding energy of −8.9 kcal mol−1. Compound (3) has one hydrogen bond with HisB42, and alkyl and pi–alkyl interactions with AlaB24, IleA16, and LeuB27. Compound (4) has a binding energy equal to −9.2 kcal mol−1, which is the highest binding score for this protein.
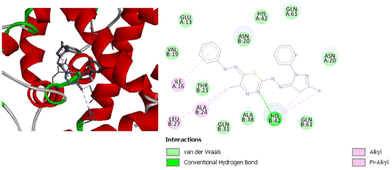 | ||
| Fig. 14 3D (left) and 2D (right) image of the binding interactions of compound (3) with the active site of Staphylococcus epidermidis. | ||
Fig. 15 shows the interactions between this compound and the active site of the receptor. It was found that there are three hydrogen bonds with AsnA20 and AsnB20, one pi–sulfur interaction with HisB42, and alkyl and pi–alkyl interactions with ValA63, AlaB38, AlaB24, and HisB42. For this protein, it has been found from Fig. 14 and 15 for compounds (3) and (4), and from Fig. S17 and S18 (ESI†) for compounds (1) and (2) that all compounds have van der Waals interactions with the ThrB23 and GlnB31 residues.
4. Conclusions
CS-DAA–MgO nanocomposite films were prepared with various percentages of nano-MgO via a simple solution casting method. The structure of CS-DAA was confirmed using 13C NMR. The dispersed nanopowder was characterized using SEM; the whole composite film was characterized using XRD. The nanocomposite containing 5% MgO exhibited the highest thermal stability, as shown from the TGA curves. The nanocomposite (5% MgO) was used as a heterogeneous base catalyst for the green synthesis of four novel pyrazole derivatives. The optimal catalyst loading was 30%, and the catalyst was easily separated from the reaction medium after the formation of the product. The catalyst was effectively reusable up to four times without significant loss of its activity.Under the effect of CS-DAA–MgO, pyrazolone was converted into pyrazolyl derivatives bearing hydrazones, thiadiazines, and thiazolidin-4-one moieties. The four novel pyrazole derivatives were synthetized in the minimum time and in high percentage yield. The structures of the new derivatives were elucidated via spectral data and elemental analysis. Moreover, their bioactivity has been explored, and they are expected to be powerful antibacterial agents, as shown from laboratory tests and confirmed by molecular docking evaluations.
The results of the molecular docking study showed that all ligands had a binding energy inhibition potential between −7.0 and −9.4 kcal mol−1. The most promising compounds were compound (3) and compound (4), where the binding energy inhibition potential values were between −8.0 and −9.4 kcal mol−1, which are better than the binding energy inhibition potentials of the reference ligands for Escherichia coli and Staphylococcus epidermidis. However, biological activity studies and more advanced investigations are necessary before the application of these compounds as antibacterial drugs.
Author contributions
Conceptualization, Abir S. Abdel-Naby; Sara Nabil; Sarah Aldulaijan; Salha Alharthi; data curation, Sara Nabil; Salha Alharthi; methodology, S. Abdel-Naby; Sara Nabil; Sarah Aldulaijan; Lab antibacterial test was conducted by Bushra AL Abdullatif; formal analysis, S. Abdel-Naby; Sara Nabil; Sarah Aldulaijan; software, Sarah Aldulaijan; Salha Alharthi; writing-original draft preparation, Abir S. Abdel-Naby; Sara Nabil; Sarah Aldulaijan; supervision, Abir S. Ab-del-Naby; writing-review and editing, Abir S. Abdel-Naby; Sara Nabil; Sarah Aldulaijan; Salha Alharthi. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work is financially supported by institutional fund by Ministry of Education, Kingdom of Saudi Arabia, project no. IF 20-2020.References
- K. D. Khalil, E. I. Ibrahim and F. A. Al-Sagheer, A Novel, Efficient, and Recyclable Biocatalyst for Michael Addition Reactions and Its Iron (Iii) Complex as Promoter for Alkyl Oxidation Reactions, Catal. Sci. Technol., 2016, 6, 1410–1416 RSC.
- K. Khalil, H. Al-Matar and M. Elnagdi, Chitosan as an Eco-Friendly Heterogeneous Catalyst for Michael Type Addition Reactions. A Simple and Efficient Route to Pyridones and Phthalazines, Eur. J. Chem., 2010, 1, 252–258 CrossRef CAS.
- K. D. Khalil and H. M. Al-Matar, Chitosan Based Heterogeneous Catalyses: Chitosan-Grafted-Poly (4-Vinylpyridne) as an Efficient Catalyst for Michael Additions and Alkylpyridazinyl Carbonitrile Oxidation, Molecules, 2013, 18, 5288–5305 CrossRef CAS.
- B. Mallesham, D. Raikwar and D. Shee, The Role of Catalysis in Green Synthesis of Chemicals for Sustainable Future, Advanced functional solid catalysts for biomass valorization, Elsevier, 2020, pp. 1–37 Search PubMed.
- M. Madkour, K. D. Khalil and F. A. Al-Sagheer, Heterogeneous Hybrid Nanocomposite Based on Chitosan/Magnesia Hybrid Films: Ecofriendly and Recyclable Solid Catalysts for Organic Reactions, Polymers, 2021, 13, 3583 CrossRef CAS PubMed.
- S. M. Riyadh, K. D. Khalil and A. Aljuhani, Chitosan-MgO Nanocomposite: One Pot Preparation and Its Utility as an Ecofriendly Biocatalyst in the Synthesis of Thiazoles and [1, 3, 4] Thiadiazoles, Nanomaterials, 2018, 8, 928 CrossRef PubMed.
- A. S. Abdel-Naby, S. Nabil, S. Aldulaijan, I. M. Ababutain, A. I. Alghamdi, S. Almubayedh and K. D. Khalil, Synthesis, Characterization of Chitosan-Aluminum Oxide Nanocomposite for Green Synthesis of Annulated Imidazopyrazol Thione Derivatives, Polymers, 2021, 13, 1160 CrossRef CAS PubMed.
- P. K. Sahu, P. K. Sahu, S. K. Gupta and D. D. Agarwal, Chitosan: An Efficient, Reusable, and Biodegradable Catalyst for Green Synthesis of Heterocycles, Ind. Eng. Chem. Res., 2014, 53, 2085–2091 CrossRef CAS.
- K. D. Khalil, S. M. Riyadh, S. M. Gomha and I. Ali, Synthesis, Characterization and Application of Copper Oxide Chitosan Nanocomposite for Green Regioselective Synthesis of [1, 2, 3] Triazoles, Int. J. Biol. Macromol., 2019, 130, 928–937 CrossRef CAS PubMed.
- F. E. Bennani, L. Doudach, Y. Cherrah, Y. Ramli, K. Karrouchi and M. E. A. Faouzi, Overview of Recent Developments of Pyrazole Derivatives as an Anticancer Agent in Different Cell Line, Bioorg. Chem., 2020, 97, 103470 CrossRef CAS PubMed.
- T. Omara, B. Musau and S. Kagoya, Frugal Utilization of Flue-Cured Virginia Nicotiana Tabacum Leaf Wastes as a Vicissitudinous Substrate for Optimized Synthesis of Pyridine-3-Carboxylic Acid, Amer. J. Hetero Chem., 2018, 4, 49–54 Search PubMed.
- S. A. Khanum, S. Shashikanth, S. G. Sathyanarayana, S. Lokesh and S. A. Deepak, Synthesis and Antifungal Activity of 2-azetidinonyl-5-(2-benzoylphenoxy) Methyl-1, 3, 4-oxadiazoles against Seed-borne Pathogens of Eleusine Coracana (L.) Gaertn, Pest Manag. Sci. Former. Pestic. Sci., 2009, 65, 776–780 CrossRef CAS PubMed.
- M. J. N. Khadri, A. B. Begum, M. K. Sunil and S. A. Khanum, Synthesis, Docking and Biological Evaluation of Thiadiazole and Oxadiazole Derivatives as Antimicrobial and Antioxidant Agents, Results Chem., 2020, 2, 100045 CrossRef.
- S. G. Kini, A. Bhat, Z. Pan and F. E. Dayan, Synthesis and Antitubercular Activity of Heterocycle Substituted Diphenyl Ether Derivatives, J. Enzyme Inhib. Med. Chem., 2010, 25, 730–736 CrossRef CAS PubMed.
- V. A. S. Pardeshi, N. S. Chundawat, S. I. Pathan, P. Sukhwal, T. P. S. Chundawat and G. P. Singh, A Review on Synthetic Approaches of Benzimidazoles, Synth. Commun., 2021, 51, 485–513 CrossRef CAS.
- M. Zhu, H. Zhou, L. Ma, B. Dong, J. Zhou, G. Zhang, M. Wang, J. Wang, S. Cen and Y. Wang, Design and Evaluation of Novel Piperidine HIV-1 Protease Inhibitors with Potency against DRV-Resistant Variants, Eur. J. Med. Chem., 2021, 220, 113450 CrossRef CAS PubMed.
- T. Legigan, E. Migianu-Griffoni, M. A. Redouane, A. Descamps, J. Deschamp, O. Gager, M. Monteil, F. Barbault and M. Lecouvey, Synthesis and Preliminary Anticancer Evaluation of New Triazole Bisphosphonate-Based Isoprenoid Biosynthesis Inhibitors, Eur. J. Med. Chem., 2021, 214, 113241 CrossRef CAS PubMed.
- M. A. S. Abdelwahid, T. Elsaman, M. S. Mohamed, S. A. Latif, M. M. Mukhtar and M. A. Mohamed, Synthesis, Characterization, and Antileishmanial Activity of Certain Quinoline-4-Carboxylic Acids, J. Chem., 2019, 2019, 1–9 CrossRef.
- F. K. Keter and J. Darkwa, Perspective: The Potential of Pyrazole-Based Compounds in Medicine, Biometals, 2012, 25, 9–21 CrossRef CAS.
- N. A. Abdel-Latif, Synthesis and Antidepressant Activity of Some New Coumarin Derivatives, Sci. Pharm., 2005, 73, 193–216 CrossRef.
- D. S. Arora and J. Kaur, Antimicrobial Activity of Spices, Int. J. Antimicrob. Agents, 1999, 12, 257–262 CrossRef CAS.
- M. C. Sabu and R. Kuttan, Anti-Diabetic Activity of Medicinal Plants and Its Relationship with Their Antioxidant Property, J. Ethnopharmacol., 2002, 81, 155–160 CrossRef CAS PubMed.
- P. Goya, J. A. Paez, I. Alkorta, E. Carrasco, M. Grau, F. Anton, S. Julia and M. Martinez-Ripoll, N-Substituted Pyrazino [2, 3-c][1, 2, 6] Thiadiazine 2, 2-Dioxides. A New Class of Diuretics, J. Med. Chem., 1992, 35, 3977–3983 CrossRef CAS PubMed.
- N. J. Miller, J. Sampson, L. P. Candeias, P. M. Bramley and C. A. Rice-Evans, Antioxidant Activities of Carotenes and Xanthophylls, FEBS Lett., 1996, 384, 240–242 CrossRef CAS PubMed.
- C. A. Moreno-Camacho, J. R. Montoya-Torres, A. Jaegler and N. Gondran, Sustainability Metrics for Real Case Applications of the Supply Chain Network Design Problem: A Systematic Literature Review, J. Clean. Prod., 2019, 231, 600–618 CrossRef.
- A. Chauhan, P. K. Sharma and N. Kaushik, Pyrazole: A Versatile Moiety, Int. J. ChemTech Res., 2011, 3, 11–17 Search PubMed.
- S. N. Shabaan, B. S. Baaiu, A. Abdel-Aziem and M. S. Abdel-Aziz, Ultrasound-Assisted Green Synthesis and Antimicrobial Assessment of 1, 3-Thiazoles and 1, 3, 4-Thiadiazines, Green Chem. Lett. Rev., 2021, 14, 679–688 CrossRef CAS.
- X. Zhong, X. Wang, L. Chen, X. Ruan, Q. Li, J. Zhang, Z. Chen and W. Xue, Synthesis and Biological Activity of Myricetin Derivatives Containing 1, 3, 4-Thiadiazole Scaffold, Chem. Cent. J., 2017, 11, 1–9 CrossRef PubMed.
- G. J. Gabriel, A. Som, A. E. Madkour, T. Eren and G. N. Tew, Infectious Disease: Connecting Innate Immunity to Biocidal Polymers, Mater. Sci. Eng., R, 2007, 57, 28–64 CrossRef PubMed.
- R. B. Ghooi and S. M. Thatte, Inhibition of Cell Wall Synthesis—Is This the Mechanism of Action of Penicillins?, Med. Hypotheses, 1995, 44, 127–131 CrossRef CAS PubMed.
- M. Hussain, T. Qadri, Z. Hussain, A. Saeed, P. A. Channar, S. A. Shehzadi, M. Hassan, F. A. Larik, T. Mahmood and A. Malik, Synthesis, Antibacterial Activity and Molecular Docking Study of Vanillin Derived 1, 4-Disubstituted 1, 2, 3-Triazoles as Inhibitors of Bacterial DNA Synthesis, Heliyon, 2019, 5, e02812 CrossRef PubMed.
- B. K. Shoichet, S. L. McGovern, B. Wei and J. J. Irwin, Lead Discovery Using Molecular Docking, Curr. Opin. Chem. Biol., 2002, 6, 439–446 CrossRef CAS PubMed.
- L. G. Ferreira, R. N. Dos Santos, G. Oliva and A. D. Andricopulo, Molecular Docking and Structure-Based Drug Design Strategies, Molecules, 2015, 20, 13384–13421 CrossRef CAS PubMed.
- C. M. Apfel, H. Locher, S. Evers, B. Takács, C. Hubschwerlen, W. Pirson, M. G. P. Page and W. Keck, Peptide Deformylase as an Antibacterial Drug Target: Target Validation and Resistance Development, Antimicrob. Agents Chemother., 2001, 45, 1058–1064 CrossRef CAS.
- T. Agarwal, P. Gupta, S. Asthana and A. Khursheed, In Silico Analysis to Access the Antibacterial Effect of Thiazides on PDfs: Molecular Docking Approach, Int. J. Pharm. Pharm. Sci., 2014, 6, 387–391 Search PubMed.
- P. Lin, T. Hu, J. Hu, W. Yu, C. Han, J. Zhang, G. Qin, K. Yu, F. Götz and X. Shen, Characterization of Peptide Deformylase Homologues from Staphylococcus Epidermidis, Microbiology, 2010, 156, 3194–3202 CrossRef CAS.
- D. J. Beecher and A. C. Wong, Identification of Hemolysin BL-Producing Bacillus Cereus Isolates by a Discontinuous Hemolytic Pattern in Blood Agar, Appl. Environ. Microbiol., 1994, 60, 1646–1651 CrossRef CAS PubMed.
- J.-C. Palomino, A. Martin, M. Camacho, H. Guerra, J. Swings and F. Portaels, Resazurin Microtiter Assay Plate: Simple and Inexpensive Method for Detection of Drug Resistance in Mycobacterium Tuberculosis, Antimicrob. Agents Chemother., 2002, 46, 2720–2722 CrossRef CAS PubMed.
- D. Mazel, S. Pochet and P. Marliere, Genetic Characterization of Polypeptide Deformylase, a Distinctive Enzyme of Eubacterial Translation, EMBO J., 1994, 13, 914–923 CrossRef CAS PubMed.
- Y.-M. Chang, W.-Y. Jeng, T.-P. Ko, Y.-J. Yeh, C. K.-M. Chen and A. H.-J. Wang, Structural Study of TcaR and Its Complexes with Multiple Antibiotics from Staphylococcus Epidermidis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 8617–8622 CrossRef CAS PubMed.
- J.-P. Guilloteau, M. Mathieu, C. Giglione, V. Blanc, A. Dupuy, M. Chevrier, P. Gil, A. Famechon, T. Meinnel and V. Mikol, The Crystal Structures of Four Peptide Deformylases Bound to the Antibiotic Actinonin Reveal Two Distinct Types: A Platform for the Structure-Based Design of Antibacterial Agents, J. Mol. Biol., 2002, 320, 951–962 CrossRef CAS PubMed.
- A. Kreusch, G. Spraggon, C. C. Lee, H. Klock, D. McMullan, K. Ng, T. Shin, J. Vincent, I. Warner and C. Ericson, Structure Analysis of Peptide Deformylases from Streptococcus Pneumoniae, Staphylococcus Aureus, Thermotoga Maritima and Pseudomonas Aeruginosa: Snapshots of the Oxygen Sensitivity of Peptide Deformylase, J. Mol. Biol., 2003, 330, 309–321 CrossRef CAS.
- H. M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T. N. Bhat, H. Weissig, I. N. Shindyalov and P. E. Bourne, The Protein Data Bank, Nucleic Acids Res., 2000, 28, 235–242 CrossRef CAS PubMed.
- BIOVIA and D. S. BIOVIA, Discovery Studio Visualizer. Available Online: https://Discover.3ds.Com/Discovery-Studio-Visualizer-Download (Accessed on 1 May 2021).
- Acd/Chemsketch, ChemSketch Chemical Structure Drawing Software, Available Online: https://Www.Acdlabs.Com/Products/Chemsketch/(Accessed on 1 Januray 2021). Toronto, ON, Canada 2021.
- PyMOL by Schrödinger, Available online: https://pymol.org/2/ (accessed on 1 Januray 2021).
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading, J. Comput. Chem., 2010, 31, 455–461 CAS.
- A. A. Al-Ghamdi, S. N. Al-Harthi, A. M. EL-Sharif and A. S. Abdel-Naby, Cyclocopolymerization of N, N-Diallylammonium and N, N-Diallylguanidinium Acetate with Acrylonitrile Characterization, Thermal and Morphological Properties, Arab. J. Sci. Eng., 2019, 44, 6303–6311 CrossRef CAS.
- G. J. Safaei, S. Zahedi, M. Javid and M. A. Ghasemzadeh, MgO Nanoparticles: An Efficient, Green and Reusable Catalyst for the Onepot Syntheses of 2, 6-Dicyanoanilines and 1, 3-Diarylpropyl Malononitriles under Different Conditions, J. Nanostruct., 2015, 5, 153–160 CrossRef.
- U. P. Singh, H. R. Bhat, M. K. Kumawat and R. K. Singh, Utilisation of Home Laundry Effluent (HLE) as a Catalyst for Expeditious One-Pot Aqueous Phase Synthesis of Highly Functionalised 4-Thiazolidinones, SpringerPlus, 2013, 2, 1–11 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nj05513a |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2023 |