Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mechanical disassembly of human picobirnavirus like particles indicates that cargo retention is tuned by the RNA–coat protein interaction†
María J.
Rodríguez-Espinosa
 ab,
Javier M.
Rodríguez
b,
José R.
Castón
ab,
Javier M.
Rodríguez
b,
José R.
Castón
 *bd and
Pedro J.
de Pablo
*bd and
Pedro J.
de Pablo
 *ac
*ac
aDepartamento de Física de la Materia Condensada, Universidad Autónoma de Madrid, 28049 Madrid, Spain. E-mail: p.j.depablo@uam.es
bDepartment of Macromolecular Structures, Centro Nacional de Biotecnología (CNB-CSIC), 28049 Madrid, Spain. E-mail: jrcaston@cnb.csic.es
cInstituto de Física de la Materia Condensada (IFIMAC), Universidad Autónoma de Madrid, 28049 Madrid, Spain
dNanobiotechnology Associated Unit CNB-CSIC-IMDEA, Campus Cantoblanco, 28049 Madrid, Spain
First published on 5th October 2023
Abstract
Here we investigate the cargo retention of individual human picobirnavirus (hPBV) virus-like particles (VLPs) which differ in the N-terminal of their capsid protein (CP): (i) hPBV CP contains the full-length CP sequence; (ii) hPBV Δ45-CP lacks the first 45 N-terminal residues; and (iii) hPBV Ht-CP is the full-length CP with a N-terminal 36-residue tag that includes a 6-His segment. Consequently, each VLP variant holds a different interaction with the ssRNA cargo. We used atomic force microscopy (AFM) to induce and monitor the mechanical disassembly of individual hPBV particles. First, while Δ45-CP particles that lack ssRNA allowed a fast tip indentation after breakage, CP and Ht-CP particles that pack heterologous ssRNA showed a slower tip penetration after being fractured. Second, mechanical fatigue experiments revealed that the increased length in 8% of the N-terminal (Ht-CP) makes the virus particles to crumble ∼10 times slower than the wild type N-terminal CP, indicating enhanced RNA cargo retention. Our results show that the three differentiated N-terminal topologies of the capsid result in distinct cargo release dynamics during mechanical disassembly experiments because of the different interaction with RNA.
New conceptsThe idea of using virus-like particles as nanocarriers for heterologous cargo transport and delivery requires controlling the stability of the container–cargo system. In particular, the conditions of the cargo delivery entail tailoring the escape of the molecular payload when the virus particle is disassembled. What happens to the internalized molecules when the nanocage is opened? To this end, it is necessary to control the cargo–container interaction which, in turn, would tune the retention of cargo when the disassembly of the nanocarrier takes place. Thus, it is necessary to develop nanocarrier systems that facilitate the control of the cargo retention conditions as a function of its interaction with the nano-container. We designed three mutants of human picobirnavirus where the RNA–coat protein interaction, observed previously via cryo-electron microscopy, is modified by changing the N-terminal end of the coat protein. Here, we use atomic force microscopy for inducing the mechanical unpacking of the RNA internalized in particles of each mutant. Our experiments crack-opened individual particles in real time to monitor the cargo release. Among other results, we have measured that an increment in the N-terminal length by just 8% increases the cargo retention of partially disrupted particles by a factor of 10 with respect to the wild type. Our study elucidates the interplay between the RNA–coat protein interaction of each mutant and their capacity for cargo retention during disassembly. |
Introduction
Viruses pack, shuttle and deliver nucleic acids to perpetuate themselves.1 These functions require the virus structure to be stable during the extracellular transport to protect the viral genome against physico-chemical assaults,2 but to be labile/flexible enough for uncoating inside the host to release the infectious genome.3–5 In addition, their exquisite programmable architecture makes it possible to develop virus-like particles (VLPs) deprived of infectivity, whose viral origin make them excellent platforms for the development of nanocarriers and functional materials.6 In this context, the application of VLPs and viruses in materials science extends from packing functional enzymes for making nanoreactors7 to transport and delivery of drugs and specific molecules for nanomedicine.8 These viral-based nanocages need to be robust and inert during transport while also releasing their cargo during disassembly, as it happens with many viruses. Genome release is controlled using a variety of strategies. While in the case of bacteriophages, the viral cargo is translocated through the tail by releasing the internal pressure,9 in the case of eukaryotic viruses, the virus disassembles for letting the genome escape from the virus-disrupted cage inside the host. For example, human adenovirus undergoes a programmed disassembly when traveling to the nucleus of the cell,10 where the dsDNA is released through the nuclear pore if the virus particle has undergone sufficient dismantling.11 The dsDNA release depends on the condensation imposed by some histone-like proteins which are present in the adenovirus core.12 The retention of cargo molecules in intact nanocontainers has been investigated for P22 VLPs in large ensembles of particles. In particular, the storage ability of these VLPs depends on ambient conditions and the interaction between the cargo and the shell.13 However, little is known about the retention of heterologous cargo in VLP systems during disassembly and at the nanoscale single particle level. Human picobirnavirus (hPBV) is a dsRNA virus that has been found in healthy respiratory tracts and has infected most of the human population.14 Secondary infections are undetectable due the mildness of hPBV-induced diarrhea.15 Since hPBV VLPs are thus not foreign to our immune system, this virus exhibits an attractive structure for packing heterologous molecules to be used as nanocontainers for cargo delivery and gene therapy.hPBV has a bi-segmented genome, with segment 1 encoding the CP and a protein of unknown function and genome segment 2 encoding the viral polymerase.17 As many dsRNA viruses infecting higher eukaryotes, hPBV probably packages its genome as positive-sense ssRNA segment(s) that nucleate capsid monomers and, once the capsid is assembled, they are replicated into dsRNA in the capsid interior.16
We designed three hPBV VLPs variants morphologically like authentic virions whose CP differs in the N-terminal ends that face the capsid interior.12 These different inner surfaces endorse diverse interactions with heterologous cargo (ssRNA and proteins) packed during assembly.18 The three types of hPBV VLPs offer a model system to directly explore the cargo retention during the capsid disassembly for different RNA–CP interactions controlled by the coat protein topology. In this work, we induce and monitor the mechanical unpacking and disassembly via AFM19,20 of individual virus particles and establish a direct link between cargo retention and the RNA–CP architecture as revealed via cryo-electron microscopy.
Results
Three types of VLPs that differ on the N-terminal pack cargo in different ways
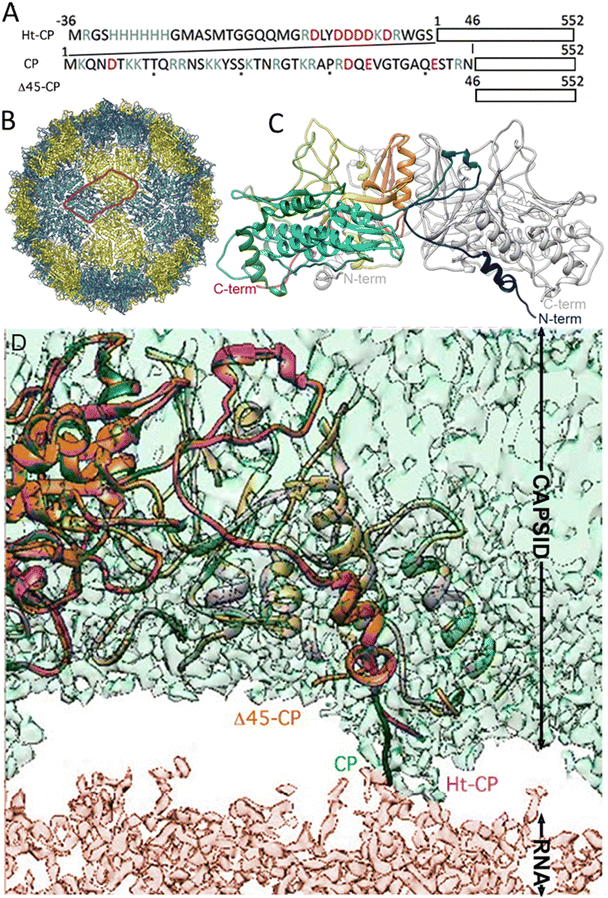
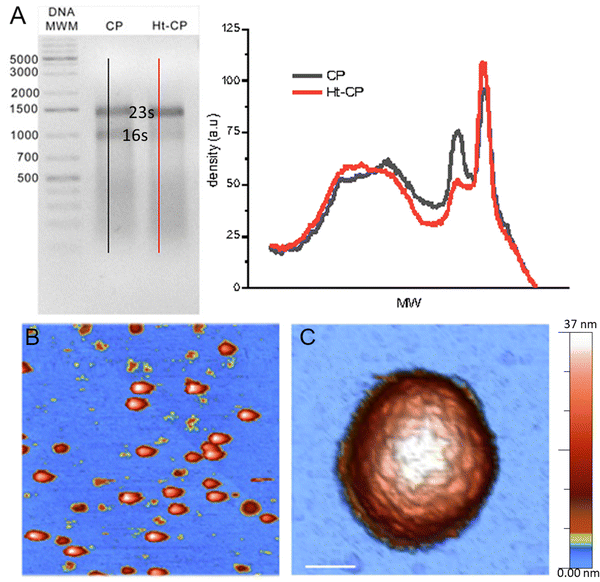
The three hPBV CP variants analyzed in this study are (i) hPBV CP that contains the full-length CP sequence (552 residues, with 13 Arg/Lys at the N-terminal region), (ii) hPBV Δ45-CP that lacks the first 45 N-terminal residues (508 residues), and (iii) hPBV Ht-CP, a full-length CP with an N-terminal 36-residue tag with 10 more basic residues (including the 6-His segment; 588 residues) (Fig. 1(A)). Downstream of the His tag sequence, the pRSETA plasmid (Invitrogen) has several residues bearing targets for different proteases (to remove the His tag). This 36 aa tag occurs when the ORF is cloned in-frame using the plasmid's BamH I restriction site, an approach used to anticipate the possibility that CP would not form capsids in the bacterial system. Proteases were not necessary after CP purification because capsids assembled spontaneously; they were purified by ultracentrifugation, but this 36 aa tag yields a different inner capsid from the wild type (CP) or the Δ45-CP capsids. For the three variants, 3D cryo-EM analysis showed a ∼380 Å diameter T = 1 capsid (Fig. 1(B)) with 60 twisted, elongated protrusions, each bearing two copies of CP monomers (Fig. 1(C)). This quasimetric dimer is the building unit of the capsid. Following established nomenclature for the dimers, the A subunit participates directly in interactions at the five-fold axis (Fig. 1(B), blue) and the B subunit is intercalated between A subunits (Fig. 1(B), yellow). The A subunit N-terminal end extends into the capsid interior, but the B subunit N-terminal end of CP interacts with the inner capsid surface that is highly negative,18 which suggests that protein–RNA interactions are mediated by A subunits (Fig. 1(D)). This scenario is similar for the Ht-CP N-terminal A and B subunits (Fig. 1(D)). While Ht-CP and CP VLP pack bacterial ribosome (rRNA, mostly 23S and 16S) and host proteins (Fig. 2(A)), Δ45-CP VLPs lack nucleic acids and contain only proteins. Mass spectrometry analysis of these hPBV particles showed that they contain diverse host proteins that are probably randomly encapsulated.18 The basic CP N-terminal region is responsible for host rRNA encapsidation, and rRNA binding is probably necessary to neutralize repulsive charge interactions between CP (or Ht-CP), as in hepatitis B virus,21 plant cowpea chlorotic mottle virus (CCMV),22 and brome mosaic virus.23 Thus, in this hPBV coassembly mode, host rRNA emulates genomic nucleic acids (probably as ssRNA segments) acting as a heterologous cargo. Encapsidated bacterial rRNA corresponded to 23S and 16S rRNA of 2905 and 1541 ribonucleotides, respectively. Careful analysis of purified encapsidated rRNA, using ssRNA molecular weight markers, indicated that both segments are not degraded during purification (Fig. S1, ESI†). We have observed that while CP VLPs pack 23S and 16S rRNA, Ht-CP VLPs are enriched in 23S rRNA relative to 16S rRNA (Fig. 2(A)). Nevertheless, except for the differences at the N-terminal region, the three particles are almost identical. A and B subunits of CP lack 42 residues in the N-terminal region (they are disordered and remain invisible in the cryo-EM map).Single indentation assay reveals differences in post breakage spectroscopy and topography of collapsed VLPs
The immobilization of viruses on a solid surface is a sine qua non requisite for manipulating individual VLPs via AFM. We dispersed a droplet of VLPs on highly oriented pyrolytic graphite (HOPG) of a freshly cleaved surface. Virus capsids are adsorbed on the surface through the interaction of the hydrophobic regions of the virus particles and the HOPG.24 The high aspect ratio found by the AFM tip when scanning a virus particle uphill has endorsed jumping mode (JM) as a particularly successful approach for imaging individual icosahedral viruses.25 In the JM, the AFM tip establishes contact with the sample, while the lateral scanning is off. When a predefined cantilever bending (normal force -Fn-) is reached, the topography data of one pixel are recorded and the tip moves back releasing the surface. When the tip and sample are out of contact, separated at a predefined maximum distance, the lateral piezo moves the sample to the next pixel, thus avoiding dragging lateral forces.26 The AFM images reveal some debris and intact virus structures (Fig. 2(B)) whose height indicates an irrelevant deformation (<7%) of the three hPBV structures18,27 (Fig. S2 top left, ESI†). Likewise, high resolution topographies of single VLPs resolve individual capsomers and the adsorption geometry of every capsid18 (Fig. 2(C)). After the localization of an intact and stable VLP, we performed a single indentation assay, which consists of pushing the virus structure using the AFM tip at the very top while recording the cantilever deflection.28 This experiment resulted in a force indentation curve (FIC, Fig. S3, ESI†), which informs about various mechanical parameters of the virus particle, including stiffness, breaking force and brittleness for 25 Δ45-CP, 22 Ht-CP and 32 CP particles18 (Fig. S4, ESI†). The interpretation of FIC data (Fig. S3, ESI†) is as follows. First, the tip approaches the virus showing a flat normal force, until the tip establishes mechanical contact with the protein shell and elastic deformation begins. This almost linear regime indicates a virus deformation of ∼5 nm, whose linear fitting revealed that the spring constant/stiffness of the three hPBV variants almost stays within the standard deviation (Fig. S5, ESI†).18 This mechanical similarity between mutants has been also found previously in Brome Mosaic Virus.29 This linear regime finishes with a jump of the tip downwards, indicating that the tip breaks the protein shell (Fig. S3, ESI†). The coefficient between the indentation at which the particle breaks and the original height (critical strain -εc-) did not present significant differences between the hPBV VLPs variants (Fig. S5, ESI†). Since the basic AFM analysis of viruses30 did not reveal any difference between the three VLPs species, we endeavor new advanced AFM analysis of each hPBV structure that surpasses the classic approaches used before.18 Specifically, the comparison of the AFM topography of the hPBV VLP structure before and after the FIC procedure shows differences in the debris of the collapsed particles (Fig. 3(A)). To deepen in the mechano-structural behavior of VLPs, we have represented the profile data of broken particles for each VLP class (Fig. 3(B)), where FICs have been obtained under the same conditions (50 nm s−1). These data show that Δ45-CP VLPs (that lack packaged cargo) convey collapsed structures with an average maximum height of ∼15 nm (Fig. 3(B), left), while RNA filled VLPs show heights of ∼25 and ∼30 nm for CP and Ht-CP structures, respectively (Fig. 3(B), center and right). However, these height values could be influenced by the irregular topography of the virus particle debris after FIC. Therefore, we analyzed the lost volume by subtracting the resulting virus particle topography after FIC from the value of the intact virus particle before the FIC (Fig. 3(C)). Both profile and volume charts indicate that the presence of ssRNA cargo prevents excessive loss of material during VLP disruption, though Ht-CP VLPs exhibit a lower collapse and volume dopping than CP structures.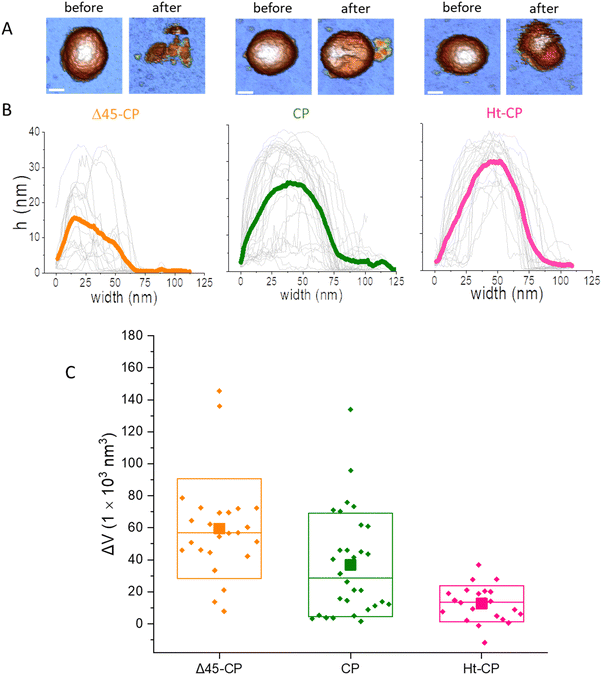 | ||
| Fig. 3 Disruption of VLPs using the single indentation assay. (A) AFM image of an intact VLP before (left) and after (right) being broken with a single indentation for Δ45-CP, CP and Ht-CP variants. Bar, 22 nm. The color of the height scale is the same as shown in Fig. 2. (B) Topographic profiles obtained through each broken particle for the three hPBV variants: 25 Δ45-CP, 32 CP and 22 Ht-CP VLPs (light gray) with their respective averages in thick orange, green and pink. (C) Box plots of the lost material volumes calculated using AFM topographies with the flooding filter of WsXM.39 Average, median and SDs are indicated by solid squares, horizontal lines and boxes, respectively. | ||
Now we focused our study on the breakage spectroscopy of the VLPs beyond the linear deformation of the FIC data, a so far unexplored methodology. The AFM tip reaches a total indentation (δtotal) of ∼25 nm (Fig. S3. ESI†) which accounts for ∼66% of the VLP diameter (Fig. S2 top left, ESI†). The indentation data and topography after indentation (Fig. 3(B)) show that the tip penetrates inside the virus particle, including the capsid and RNA cargo. Therefore, the quantitative analysis of the abrupt decay of the force beyond capsid breakage provides information about the VLP disruption. By grouping all FICs for each hPBV variant (Fig. S4, ESI†), we calculated “thermal” plots where the color indicates the density of FICs points (Fig. 4) that represents the tip position during the indentation experiments for all the VLPs of each variant. These maps are interpreted as the probability of finding the cantilever with a normal force (y axis) at certain indentation (x axis). For instance, it is possible to find the brightest colors around 0 nm until the breaking force because this region corresponds to the elastic deformation of VLPs (Fig. 4(A)–(C)). The dark colors after breakage in RNA-lacking Δ45-CP structures (Fig. 4(A)) illustrate that the AFM tip descends rapidly in free fall from 1 nN at 4 nm of indentation to almost 0 nN at 20 nm of indentation. Afterwards, the tip squeezes the capsid debris without almost bending until the hard surface induces the normal force increment at ∼30 nm of indentation. “Thermal” maps of the RNA-containing VLPs (Fig. 4(B) and (C)) reveal that the indentation of the AFM tip right after breakage happens more gradually than in the Δ45-CP case. From the “thermal” maps, the average force value of the collapsing zone can be defined, i.e. the area that is right after the breaking force up to the indentation at which the FIC slope starts going up (data included between the white dashed lines of Fig. 4(A)–(C)). The charts of these data suggest different behaviors for each VLP variant (Fig. 4(D)): while for Δ45-CP structures, the bending force after breakage peaks at 0.04 nN, this value increases for RNA full particles to 0.25 nN (CP) and 0.32 nN (Ht-CP). We applied the Lilliefors (Kolmogorov–Smirnov) normality test that shows that the data of Fig. 4(D) follow a normal distribution with a 95% significance (P values of 0.064). This indicates the major relevance of highest peaks in comparison with any other lower peak of the plots. Nevertheless, the higher dispersion of the CP chart might indicate a zipper effect during virus indentation originated by the fact that the N-terminal in CP particles is penetrating more perpendicularly into the virus compared to the case of Ht-CP structures (Fig. 1(D)). By using Hook's law with the cantilever spring constant of 0.05 nN nm−1, these peaks of bending values can be converted to cantilever deformations, whose maximum probability after breakage is of 0.5 nm, 4.8 nm and 7 nm for Δ45-CP, CP and Ht-CP VLPs, respectively. These values inform of the amount of material below the tip after the virus particle collapsed, indicating that the Ht-CP variant presents the highest value.
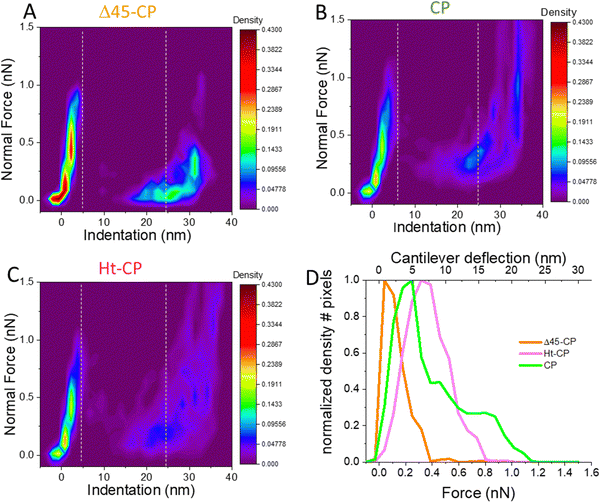 | ||
| Fig. 4 Mechanical unpacking data of the single indentation assay. Density maps of FICs indentation data (Fig. S4, ESI†) obtained for Δ45-CP (A), Ht-CP (B) and CP (C) VLPs. (D) Average density data of the collapsing zone (see text) located between the two dashed vertical lines to obtain the residence time of the tip as a function of the bending force for the three variants. | ||
Mechanical fatigue reveals differentiated cargo retention for each hPBV variant
Cyclic loading of virus particles at low force (∼50 pN) when imaging in JM induces the gradual disruption of capsids31 and reveals additional mechano-structural information related to the proteins of the shell11 and the physical state of the packed nucleic-acids.12,32 Therefore, we studied the response of hPBV VLPs to mechanical fatigue to investigate their disassembly dynamics. Thus, individual VLPs are repeatedly imaged in JM and their topography monitored through the fatigue process. The fatigue-induced disassembly for each VLP variant under ∼50 pN is summarized in three frames of the procedure (Fig. 5(A)). For instance, this process for the CP variant (Movie SM1, ESI†) shows a partial disruption at frame #10, while frame #12 shows the collapsed particle with some debris (capsomers and cargo) spread around (Fig. 5(A), center). Similar experiments performed with Δ45-CP (Movie SM2, ESI†) and Ht-CP (Movie SM3, ESI†) structures show differentiated progression. RNA-empty particles (Δ45-CP) do not present debris on the surface through the disassembly process (Fig. 5(A), left). Thus, we analyzed the area coverage with debris in the vicinity of 10 Δ45-CP, 18 CP and 10 Ht-CP VLPs (Fig. 5(B)) which have been subjected to mechanical fatigue at ∼50 pN with identical JM parameters.31 While Δ45-CP VLPs lacking RNA show a decrease in the coverage (Fig. 5(B), left), CP and Ht-CP VLPs retain a constant area covered with debris (Fig. 5(B); center and right, respectively). These results showed that VLP debris appeared only when RNA was packed inside and suggested that the genome is mediating the binding of these debris to the surface. It is important to remark that the area, height, and volume charts (Fig. 5) indicate a gradual degradation of virus particles, which is not compatible with the sudden detachment of virus structures from the surface. Supplementary Videos (ESI†) clearly indicate the same idea, which demonstrates that the particle does not escape from the surface, but pieces of the particle are released in a progressive fashion instead.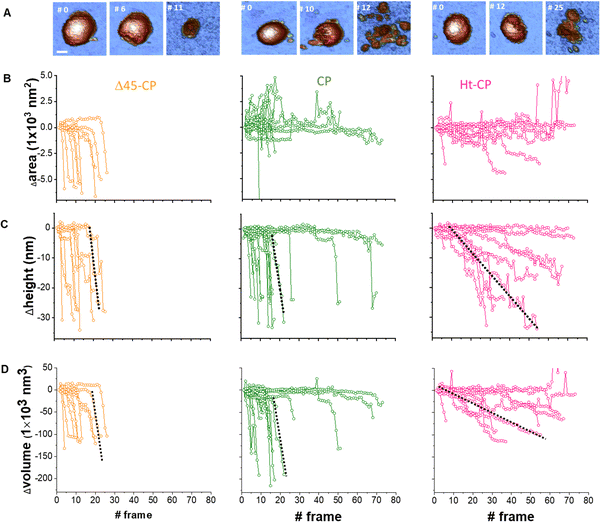 | ||
Fig. 5 Analysis of individual virus particles unpacking under mechanical fatigue. (A) Evolution of individual particles under mechanical fatigue for the three variants: Δ45-CP (left), CP (center) and Ht-CP (right). Scale bar: 25 nm. The initial frame shows the intact particle, the intermediate frame reveals some damage and the last frame is the image of the final collapsed state. The entire fatigue process can be visualized in Movies SM1, SM2 and SM3 (ESI†) for Δ45-CP, CP and Ht-CP, respectively. The color of the height scale is the same as shown in Fig. 2. (B) Evolution of the particle surrounding the covered area with debris for each VLP: 10 Δ45-CP VLPs (left), 18 CP VLPs (center) and 10 Ht-CP VLPs (right). (C) Evolution of the particle height for each virus particle, grouped by the hPBV variant. The dotted dark lines indicate the rate of collapse (RoC) ratio fitting for three cases in Δ45-CP (left, −7 nm per frame), CP (center, −6 nm per frame) and Ht-CP (right, −0.5 nm per frame), see the text. (D) Lost volume for each virus particle, classified by the hPBV variant. The dotted dark lines indicate the RoC ratio fitting for three cases in Δ45-CP (left, −21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3 per frame), CP (center, −21 000 nm3 per frame), CP (center, −21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3 per frame) and Ht-CP (right, −2000 nm3 per frame), see the text. The elapsed time for 1 frame is 120 seconds, resulting in 2 hours 30 minutes for the 80 frames of the x axes. 000 nm3 per frame) and Ht-CP (right, −2000 nm3 per frame), see the text. The elapsed time for 1 frame is 120 seconds, resulting in 2 hours 30 minutes for the 80 frames of the x axes. | ||
A key information to be extracted from fatigue experiments is the size of individual VLPs along the fatigue experiment, whose measurement reports on the amount of the virus structure which remains intact over time.32 It is known that AFM images are affected by tip dilation artifacts that provide wider lateral dimension of the scanned specimens than the real size.33 Since the vertical dimension is not affected by tip dilation, it is possible to use it as an indicator of the virus size through the mechanical fatigue experiments (Fig. 5(C)).31 The analysis of the height indicates that while all cargo lacking Δ45-CP VLPs have lost ∼30 nm before reaching 30 frames (Fig. 5(C) left), 3 out of 10 RNA-loaded CP VLPs surpass 30 frames keeping almost an intact size (Fig. 5(C), center). However, both Δ45-CP and CP VLPs present a drastic loss of height right after disassembly has started (Fig. 5(C), left and center). We can define the rate of collapse (RoC) by fitting linearly the height variation of each particle from the last frame, where Δh ∼ 0 until its minimum value (Fig. 5(C), dotted black). This parameter evaluates how fast the VLPs are unpacked after being crack-opened during the fatigue experiments. RoC median values for Δ45-CP and CP VLPs are found to be −3.7 and −5.7 nm per frame, respectively. These values indicate a high rate of material lost per image. However, Ht-CP data unveil a gradual collapse (Fig. 5(C), right) with a median RoC value of −0.65 nm per frame, indicating a much slower rate of material lost. A complementary volume loss analysis (Fig. 5(D)) indicates medians of −16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 nm3 per frame, −17
900 nm3 per frame, −17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3 per frame and −1800 nm3 per frame for Δ45-CP, CP and Ht-CP, respectively. The comparison of these volume rates, which are equally affected by tip dilation, also indicates a much slower rate of lost material for Ht-CP virus particles.
000 nm3 per frame and −1800 nm3 per frame for Δ45-CP, CP and Ht-CP, respectively. The comparison of these volume rates, which are equally affected by tip dilation, also indicates a much slower rate of lost material for Ht-CP virus particles.
Discussion
Our results demonstrate differences in the retention capacity of cargo for the partially disrupted particles of each hPBV class. This retention can relate to the CP-ssRNA architecture provided by cryo-EM. Specifically, cryo-EM shows that highly disordered RNA is in contact with the capsid at 60 symmetrical positions of N-terminal ends (Fig. 1) in CP and Ht-CP variants.18 These 60 internal locations of the virus shell correspond to the N-terminal of each CP-A subunit (Fig. 1), whose different morphologies determine the cargo of each hPBV class. Biochemistry analyses show that CP and Ht-CP capsids contain bacterial rRNA accounting for 95% (CP) and 99% (Ht-CP) of the encapsidated RNA.18 In contrast, mass spectrometry analysis of Δ45-CP capsids showed that although lacking nucleic acids, they mostly contain diverse host proteins that are probably randomly encapsulated in the crowded cytoplasm.18 Electron microscopy indicates that while purified Ht-CP and CP VLP were homogeneously full, Δ45-CP particles were structurally stable with variable internal content (only 10% are apparently empty).18Previous analysis for virus stiffness and breaking force indicated little differences between the three hPBV variants (Fig. S5, ESI†).18 In this work, we show that data analysis beyond breakage and mechanical fatigue contribute with deeper information that was previously not considered.18 Beyond the VLP stiffness, the single indentation assay provides FIC data whose post-breakage “thermal” maps indicate that RNA-lacking VLPs present an abrupt fall of the AFM tip because of points’ shortage between 10 and 15 nm of indentation (Fig. 4(A)). However, CP and Ht-CP (RNA-loaded) VLPs present more points in the same region (Fig. 4(B) and (C)), indicating a snagged fall of the tip. This result shows that the tip penetrates Δ45-CP VLPs more easily and rapidly compared to CP or HT-CP VLPs that contain heterologous RNA. In addition, the final bending of the cantilever is significantly higher for VLPs containing RNA than for Δ45-CP particles which contain only proteins (Fig. 3(D)). In fact, the FICs of Δ45-CP VLPs show their final deflection accumulated around 0.5 nm, which indicates that very little material remains between the tip apex and the surface (Fig. 4(D)). However, Ht-CP VLPs show a final bending of 7 nm, almost doubling the CP value of 4.5 nm, and about 10 times more than Δ45-CP VLPs. Besides suggesting that RNA-containing VLPs retain more material than RNA empty VLPs after indentation, these data also illustrate that Ht-CP VLPs hold the tip at a higher position than CP particles after particle breakage. This interpretation is supported by the profiles and volumes of the topographies obtained for the disrupted particles after single FICs were carried out under the same conditions. Specifically, virus particles show final sizes of 41%, 70% and 83% of the original height for Δ45-CP, CP and Ht-CP VLPS, respectively (Fig. 3(B)). Although these estimations escape from the tip-sample geometrical dilation artifact,33 the irregular debris appearance of disrupted viruses could not provide a correct value of the height. The 3D topology AFM data allow to estimate the virus volume, although this estimate is prone to the tip-sample geometrical dilation artifact. In fact, the volume of an intact virus, assuming a radius of 19 nm,18 is 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 730 nm3, which is well below the AFM calculated volume of intact virus particles (Fig. S2, right panel, ESI†) of 74
730 nm3, which is well below the AFM calculated volume of intact virus particles (Fig. S2, right panel, ESI†) of 74![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 24
000 ± 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3 (mean ± SD, N = 79). Nevertheless, since the dilation artifact could roughly consider the same for all data, the removed volume (Fig. 3(C)) makes it possible to estimate the remaining material for each virus class. This estimation results in ∼14
000 nm3 (mean ± SD, N = 79). Nevertheless, since the dilation artifact could roughly consider the same for all data, the removed volume (Fig. 3(C)) makes it possible to estimate the remaining material for each virus class. This estimation results in ∼14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3, ∼38
000 nm3, ∼38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3 and ∼61
000 nm3 and ∼61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 nm3 for Δ45-CP, CP and Ht-CP particles, respectively. For the sake of comparison, and removing the dilation artifact, we normalized these values by dividing the above mentioned values by the intact AFM calculated volume (Fig. S2 top right, ESI†) to find the percentage of final size for each virus class as ∼20%, ∼50% and ∼82% for Δ45-CP, CP and Ht-CP particles, respectively. Overall, it can be concluded that both height and volume measurements show the same tendency for each virus type: the capacity of retaining material in crumbled hPBV VLPs is led by Ht-CP, followed by CP and Δ45-CP VLPs. The results of RNA-empty Δ45-CP VLPs in both topography (Fig. 3) and FIC thermal maps (Fig. 4) might indicate in the first glimpse that the ability for retaining material of hPBV collapsed structures relates with the amount of packed RNA.
000 nm3 for Δ45-CP, CP and Ht-CP particles, respectively. For the sake of comparison, and removing the dilation artifact, we normalized these values by dividing the above mentioned values by the intact AFM calculated volume (Fig. S2 top right, ESI†) to find the percentage of final size for each virus class as ∼20%, ∼50% and ∼82% for Δ45-CP, CP and Ht-CP particles, respectively. Overall, it can be concluded that both height and volume measurements show the same tendency for each virus type: the capacity of retaining material in crumbled hPBV VLPs is led by Ht-CP, followed by CP and Δ45-CP VLPs. The results of RNA-empty Δ45-CP VLPs in both topography (Fig. 3) and FIC thermal maps (Fig. 4) might indicate in the first glimpse that the ability for retaining material of hPBV collapsed structures relates with the amount of packed RNA.
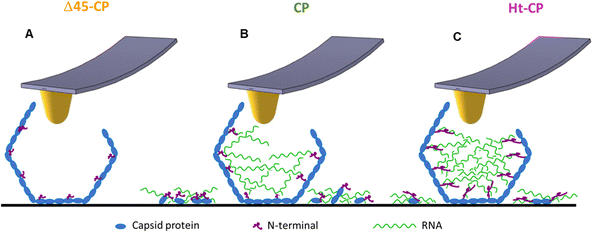
In mechanical fatigue experiments, the AFM tip erodes the VLPs gradually while imaging (Fig. 5(A) and Movies SM1–SM3, ESI†), removing the virus material which can be either adhered by the surface or diffused to the liquid environment. Monitoring the area covered with debris for each hPBV variant reveals that RNA helps to keep the capsomer debris attached to the HOPG surface after being lost from the capsid during the mechanical fatigue procedure (Fig. 5(B)). The hydrophobic nature of HOPG does not capture nucleic acids,34 and charged mica is typically used for this purpose.35 Therefore, we propose that when a capsomer is removed from Δ45-CP VLPs, it escapes from the virus structure diffusing to the liquid environment with low chances of being trapped by the surface (Fig. 6(A)). However, in CP and Ht-CP VLPs, the removed capsomers are held by the RNA around the particle until they are captured by the surface through hydrophobic interactions (Fig. 6(B) and (C)). In addition, the tendency of RNA to acquire a secondary structure might also determine the attachment of this nucleic acid to flat surfaces.36 Thus, the analysis of the covered area around the VLP informs about the presence of RNA but could not report directly about the strength of the CP–RNA interaction. In this vein, the variation of the particle's height and volume over time sheds light on the cargo retention driven by the RNA–capsomer interaction (Fig. 5(C) and (D)). In particular, the collapse undergone by Δ45-CP and CP VLPs of −7.5 and −6 nm per frame, respectively, reveals that even in the case of CP particles containing RNA, virus materials are quickly unpacked once the particles have been crack-opened during mechanical fatigue. Nonetheless, Ht-CP VLPs show a RoC (−0.5 nm per frame) of about one order of magnitude below Δ45-CP and CP VLP values. While this difference could be anticipated with respect to the Δ45-CP RNA empty particles, in the case of the RNA full VLPs, other factors come into play. The genome escapes 10 times slower in Ht-CP during mechanical fatigue. Similar results can be found when analyzing the loss of volume (Fig. 5(D)). Therefore, we pay attention to the differences of the CP–RNA structure in the RNA full VLPs which are determined by the N-terminal structure (Fig. 1(D)). As in many positive ssRNA viruses, the hPBV CP contains an N-terminal region enriched in positively charged residues that face the capsid internal surface (13 basic residues out of 45 residues). The Ht-CP contains 10 additional basic residues in the N-terminal 36-residue tag. Although both basic regions were not resolved in our cryo-EM maps, they determine the differences between mutants in the post-breaking and fatigue AFM analyses. Therefore, the charge distribution in the 36-residue insertion of Ht-CP, with 6 consecutive His in the N-terminal end and 6 Asp at the C-terminal end of the segment, should be optimal for distinguishing cargo retention and unpacking between CP and Ht-CP VLPs. Δ45-CP VLPs do not pack RNA because the N-terminal lacks the first 45 residues and it runs parallel to the capsid, thus precluding RNA trapping during assembly (Fig. 1(D)).18 However, the N-terminal of CP VLPs is complete, and it points to the capsid interior, fomenting the electrostatic interaction of CP with RNA. The N-terminal, which contains numerous basic residues, interacts with the RNA to neutralize itself and allow the assembly of the capsid.21,22 In the Ht-CP case, the N-terminal length has been increased by just ∼8% of the CP value and these residues extend again towards the internal capsid cavity (Fig. 1(D)). This geometry raises their contact area with the RNA and, therefore, makes the RNA–CP interaction stronger with respect to the CP case and increases the cargo retention (Fig. 6(C)). What is the state and nature of the packed genome inside the CP and Ht-CP VLPs? We can estimate the volume occupied by the 36-residue tag of Ht-CP. Assuming a value of 1.33 g cm−3 as the density of protein and considering the molecular weight of the 36-residue peptide of 4147.44 Da (https://web.expasy.org/protparam/), a volume of 5.2 × 103 Å3 (∼5 nm3) is obtained. The internal volume of the capsid is 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 770 nm3 (based on an average internal capsid radius of 140 Å), and the 120 copies of the 36-residue tag would decrease the total available volume in less than 5%. We expect that this small change in volume should not vary the condensation state of packed ssRNA inside the viral cavities between CP and Ht-CP. The analysis of the encapsidated cargo (Fig. 2(A) and Fig. S1, ESI†) indicates that the 16S rRNA secondary and tertiary structures should be less prone than those of 23S rRNA to interact electrostatically with the N-terminal region of Ht-CP. Despite Ht-CP is packing much less 16S rRNA than CP, the cargo retention capacity in Ht-CP is about 10 times higher than in CP (Fig. 4(C)). Hence, the higher specificity of 23S rRNA with the N-terminal Ht-CP increases the RNA–CP interaction. It is worth remarking here that this increased RNA–CP interaction is not apparently helping to pack more RNA in Ht-CP VLPs. Therefore, we conclude that in Ht-CP, the N-terminal VLPs form a stronger interaction with RNA compared to CP VLPs, multiplying their RNA retention capacity by a factor of 10 with an increase of the N-terminal length just by 8% (Fig. 5 and 6). Cargo retention is an important parameter to consider when using virus capsids as molecular containers13 and the findings presented here provide key facts in modifying the N-terminal of protein cages for tuning cargo retention in technological applications. Overall, this work describes for the first time the cargo retention ability of a disrupted nano-container as a function of the capsid–cargo interaction. This is important for the application of protein nanocontainers in materials science and medicine, where it is necessary to control the cargo release.
770 nm3 (based on an average internal capsid radius of 140 Å), and the 120 copies of the 36-residue tag would decrease the total available volume in less than 5%. We expect that this small change in volume should not vary the condensation state of packed ssRNA inside the viral cavities between CP and Ht-CP. The analysis of the encapsidated cargo (Fig. 2(A) and Fig. S1, ESI†) indicates that the 16S rRNA secondary and tertiary structures should be less prone than those of 23S rRNA to interact electrostatically with the N-terminal region of Ht-CP. Despite Ht-CP is packing much less 16S rRNA than CP, the cargo retention capacity in Ht-CP is about 10 times higher than in CP (Fig. 4(C)). Hence, the higher specificity of 23S rRNA with the N-terminal Ht-CP increases the RNA–CP interaction. It is worth remarking here that this increased RNA–CP interaction is not apparently helping to pack more RNA in Ht-CP VLPs. Therefore, we conclude that in Ht-CP, the N-terminal VLPs form a stronger interaction with RNA compared to CP VLPs, multiplying their RNA retention capacity by a factor of 10 with an increase of the N-terminal length just by 8% (Fig. 5 and 6). Cargo retention is an important parameter to consider when using virus capsids as molecular containers13 and the findings presented here provide key facts in modifying the N-terminal of protein cages for tuning cargo retention in technological applications. Overall, this work describes for the first time the cargo retention ability of a disrupted nano-container as a function of the capsid–cargo interaction. This is important for the application of protein nanocontainers in materials science and medicine, where it is necessary to control the cargo release.
Materials and methods
Expression and purification of hPBV capsids
The three CP variants of hPBV were expressed and purified as described.18 Briefly, the sequence of hPBV segment 1 strain Hy005102 (GenBank accession number NC_007026) was ordered from GenScript, produced synthetically, and cloned into plasmid pUC57. HT-CP, CP, and Δ45-CP coding sequences were amplified and the corresponding plasmids were used to transform competent E. coli DH5 cells. Positive recombinant plasmids were transformed into the E. coli BL21(DE3)pLysS expression strain (Invitrogen), and the best clone for each plasmid was selected for preparative scale expression. Bacteria from isolated colonies of the previously selected clones were grown in LB medium with ampicillin and chloramphenicol. This pre inoculum was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in prewarmed LB containing the same antibiotics and then cultured (37 °C) to an A600 of 0.2. The culture was equilibrated to 16 °C, and recombinant protein expression was induced with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside). Cells were then chilled on ice, harvested by centrifugation, and stored at −20 °C until purification. hPBV VLPs were purified from thawed cells by ultracentrifugation on a 20% sucrose cushion and a linear 20–50% sucrose gradient, as described elsewhere.18 Fractions containing purified hPBV Δ45-CP, CP or Ht-CP VLPs were kept in TN buffer (25 mM tris–HCl [pH 7.5], 50 mM NaCl), and analyzed in 12% SDS-PAGE. Purified RNA from CP and Ht-CP VLP was checked on a 1% agarose gel and detected using MidoriGreen.
10 in prewarmed LB containing the same antibiotics and then cultured (37 °C) to an A600 of 0.2. The culture was equilibrated to 16 °C, and recombinant protein expression was induced with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside). Cells were then chilled on ice, harvested by centrifugation, and stored at −20 °C until purification. hPBV VLPs were purified from thawed cells by ultracentrifugation on a 20% sucrose cushion and a linear 20–50% sucrose gradient, as described elsewhere.18 Fractions containing purified hPBV Δ45-CP, CP or Ht-CP VLPs were kept in TN buffer (25 mM tris–HCl [pH 7.5], 50 mM NaCl), and analyzed in 12% SDS-PAGE. Purified RNA from CP and Ht-CP VLP was checked on a 1% agarose gel and detected using MidoriGreen.
Atomic force microscopy
For AFM measurements, a drop of fresh Δ45-CP, CP, or Ht-CP VLP samples (30 μl at 0.1 mg ml−1 in TN buffer) was incubated on a freshly cleaved HOPG piece (ZYA quality) during 15 min. Measurements were performed with an atomic force microscope (Nanotec Electrónica S.L., Madrid, Spain) in Jumping Plus Mode.37For nanoindentation experiments, we used rectangular cantilevers (RC800PSA; Olympus) with nominal spring constants of 0.05 N m−1. The cantilevers were calibrated using Sader's method.38 Critical indentation experiments were carried out on single particles at a constant speed of 50 nm s−1 through a force versus z-piezo displacement (FZ) experiment that is turned into a force-indentation curve (FIC).30 The maximum force applied during each indentation is high enough to induce virus disruption. The indentations were implemented in 25 Δ45-CP, 32 CP VLPs and 22 Ht-CP VLPs and in all cases, a single indentation per particle. Images taken before and after indentation reveal the structural changes induced by the virus deformation (Fig. 3(A)). Images were processed and analyzed using WSxM software,39 where the topographic profile of the viruses was obtained (Fig. 3(B)). Density maps of FICs indentation data (Fig. 4(A)–(C)) show the density of the data points from the indentation curves. In these indentation curves, we focus our analysis on the post-breakage region. The average value of this region between dashed lines (Fig. 4(A)–(C)) was calculated to observe the different post-breakage contours for each VLP species (Fig. 4(D)). These new experiments produce stark differences with the previous AFM classical analysis of spring constant and breaking force.
For mechanical fatigue experiments, rectangular cantilevers (NANOSENSORS™, qp-BioAC AFM probes) with nominal spring constants of 0.05 and 0.1 N m−1 were used. The cantilevers were calibrated using Sader's method. Mechanical fatigue experiments were carried out on single particles, taking consecutive images at a low and constant force of ∼50 pN, below its breaking force. Images were processed and analyzed frame-by-frame using WSxM software39 to obtain additional information about the topography transformation of virus particles during the experiments. The mechanical fatigue experiments were performed in 10 Δ45-CP, 18 CP VLPs and 10 Ht-CP VLPs. Enough frames were obtained until the disruption of the particles took place. The variations over time of the VLPs area and height increments were monitored from the initial value 0 for each frame (Fig. 5).
Author contributions
MJR performed AFM experiments; MJR and JR purified virus samples; MJR, JRC and PJdP analyzed data; PJdP and JRC coordinated the study and wrote the manuscript, with input from all other authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
P. J. d. P. acknowledges the support by grants received from the Spanish Ministry of Science and Innovation projects (FIS2017-89549-R, FIS2017-90701-REDT and PID2021-126608OB-I00) and the Human Frontiers Science Program (HFSPO RGP0012/2018). IFIMAC is the Center of Excellence “María de Maeztu”. J. R. C. acknowledges the Spanish Ministry of Science and Innovation (PID2020-113287RB-I00) and the Comunidad Autónoma de Madrid (P2018/NMT-4389). We thank the Genomics Unit of the Central Scientific-Technical Units of the “Instituto de Salud Carlos III” for its help in characterizing the rRNAs.References
- S. J. Flint, L. W. Enquist, V. R. Racaniello and A. M. Skalka, Principles of virology, ASM Press, Washington D.C., 2004 Search PubMed.
- A. Cordova, M. Deserno, W. M. Gelbart and A. Ben-Shaul, Osmotic Shock and the Strength of Viral Capsids, Biophys. J., 2003, 85, 70–74 CrossRef CAS PubMed.
- M. W. Wien, M. Chow and J. M. Hogle, Poliovirus: new insights from an old paradigm, Structure, 1996, 4, 763–767 CrossRef CAS PubMed.
- J. Reguera, A. Carreira, L. Riolobos, J. M. Almendral and M. G. Mateu, Role of interfacial amino acid residues in assembly, stability, and conformation of a spherical virus capsid, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 2724–2729 CrossRef CAS PubMed.
- M. G. Mateu, Assembly, stability and dynamics of virus capsids, Arch. Biochem. Biophys., 2013, 531, 65–79 CrossRef CAS PubMed.
- S. Nooraei, H. Bahrulolum, Z. S. Hoseini, C. Katalani, A. Hajizade, A. J. Easton and G. Ahmadian, Virus-like particles: preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers, J. Nanobiotechnol., 2021, 19, 59 CrossRef CAS PubMed.
- J. Sharma and T. Douglas, Tuning the catalytic properties of P22 nanoreactors through compositional control, Nanoscale, 2019, 12, 336–346 RSC.
- N. F. Steinmetz, Viral nanoparticles as platforms for next-generation therapeutics and imaging devices, Nanomedicine, 2010, 6, 634–641 CrossRef CAS PubMed.
- M. Hernando-Perez, R. Miranda, M. Aznar, J. L. Carrascosa, I. A. T. Schaap, D. Reguera and P. J. de Pablo, Direct Measurement of Phage phi29 Stiffness Provides Evidence of Internal Pressure, Small, 2012, 8, 2366–2370 CrossRef CAS PubMed.
- U. F. Greber, M. Willetts, P. Webster and A. Helenius, Stepwise dismantling of adenovirus 2 during entry into cells, Cell, 1993, 75, 477–486 CrossRef CAS PubMed.
- N. Martín-González, P. Ibáñez-Freire, Á. Ortega-Esteban, M. Laguna-Castro, C. San Martín, A. Valbuena, R. Delgado-Buscalioni and P. J. de Pablo, Long-Range Cooperative Disassembly and Aging During Adenovirus Uncoating, Phys. Rev. X, 2021, 11, 021025 Search PubMed.
- N. Martín-González, M. Hernando-Pérez, G. N. Condezo, M. Pérez-Illana, A. Šiber, D. Reguera, P. Ostapchuk, P. Hearing, C. San Martín and P. J. de Pablo, Adenovirus major core protein condenses DNA in clusters and bundles, modulating genome release and capsid internal pressure, Nucleic Acids Res., 2019, 47, 9231–9242 CrossRef PubMed.
- K. McCoy, E. Selivanovitch, D. Luque, B. Lee, E. Edwards, J. R. Castón and T. Douglas, Cargo Retention inside P22 Virus-Like Particles, Biomacromolecules, 2018, 19, 3738–3746 CrossRef CAS PubMed.
- S. L. Smits, M. Leeuwen, C. M. E. van, Schapendonk, A. C. Schürch, R. Bodewes, B. L. Haagmans and A. D. M. E. Osterhaus, Picobirnaviruses in the Human Respiratory Tract, Emerg Infect Dis., 2012, 18(9), 1539–1540 Search PubMed.
- C. Gallimore, D. Lewis and D. Brown, Detection and characterization of a novel bisegmented double-stranded RNA virus (picobirnavirus) from rabbit faeces, Arch. Virol., 1993, 133, 63–73 CrossRef CAS PubMed.
- A. Borodavka, U. Desselberger and J. T. Patton, Genome packaging in multi-segmented dsRNA viruses: distinct mechanisms with similar outcomes, Curr. Opin. Virol., 2018, 33, 106–112 CrossRef CAS.
- A. M. Collier, O. L. Lyytinen, Y. R. Guo, Y. Toh, M. M. Poranen and Y. J. Tao, Initiation of RNA Polymerization and Polymerase Encapsidation by a Small dsRNA Virus, PLoS Pathog., 2016, 12, e1005523 CrossRef.
- Á. Ortega-Esteban, C. P. Mata, M. J. Rodríguez-Espinosa, D. Luque, N. Irigoyen, J. M. Rodríguez, P. J. de Pablo and J. Rs Castón, Cryo-electron Microscopy Structure, Assembly, and Mechanics Show Morphogenesis and Evolution of Human Picobirnavirus, J. Virol., 2020, 94 DOI:10.1128/JVI.01542-20.
- M. S. Z. Kellermayer, Z. Vörös, G. Csík and L. Herényi, Forced phage uncorking: viral DNA ejection triggered by a mechanically sensitive switch, Nanoscale, 2018, 10, 1898–1904 RSC.
- A. Ortega-Esteban, G. N. Condezo, A. J. Pérez-Berná, M. Chillón, S. J. Flint, D. Reguera, C. San Martín and P. J. De Pablo, Mechanics of viral chromatin reveals the pressurization of human adenovirus, ACS Nano, 2015, 9, 10826–10833 CrossRef CAS PubMed.
- N. Patel, S. J. White, R. F. Thompson, R. Bingham, E. U. Weiß, D. P. Maskell, A. Zlotnick, E. C. Dykeman, R. Tuma and R. Twarock, et al., HBV RNA pre-genome encodes specific motifs that mediate interactions with the viral core protein that promote nucleocapsid assembly, Nat. Microbiol., 2017, 2, 1–10 Search PubMed.
- D. Luque, A. Escosura, J. de la Snijder, M. Brasch, R. J. Burnley, M. S. T. Koay, J. L. Carrascosa, G. J. L. Wuite, W. H. Roos and A. J. R. Heck, et al., Self-assembly and characterization of small and monodisperse dye nanospheres in a protein cage, Chem. Sci., 2013, 5, 575–581 RSC.
- C. Beren, Y. Cui, A. Chakravarty, X. Yang, A. L. N. Rao, C. M. Knobler, Z. H. Zhou and W. M. Gelbart, Genome organization and interaction with capsid protein in a multipartite RNA virus, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10673–10680 CrossRef CAS PubMed.
- P. J. de Pablo, The application of atomic force microscopy for viruses and protein shells: imaging and spectroscopy, Adv. Virus Res., 2019, 105, 161–187 CAS.
- A. Ortega-Esteban, I. Horcas, M. Hernando-Pérez, P. Ares, A. J. Pérez-Berná, C. San Martín, J. L. Carrascosa, P. J. de Pablo and J. Gómez-Herrero, Minimizing tip–sample forces in jumping mode atomic force microscopy in liquid, Ultramicroscopy, 2012, 114, 56–61 CrossRef CAS PubMed.
- P. J. de Pablo, J. Colchero, J. Gómez-Herrero and A. M. Baró, Jumping mode scanning force microscopy, Appl. Phys. Lett., 1998, 73, 3300–3302 CrossRef CAS.
- C. Zeng, M. Hernando-Perez, B. Dragnea, X. Ma, P. van der Schoot and R. Zandi, Contact Mechanics of a Small Icosahedral Virus, Phys. Rev. Lett., 2017, 119, 038102 CrossRef PubMed.
- P. J. de Pablo, The application of atomic force microscopy for viruses and protein shells: imaging and spectroscopy, Adv. Virus Res., 2019, 105, 161–187 CAS.
- P. Ni, Z. Wang, X. Ma, N. C. Das, P. Sokol, W. Chiu, B. Dragnea, M. Hagan and C. C. Kao, An Examination of the Electrostatic Interactions between the N-Terminal Tail of the Brome Mosaic Virus Coat Protein and Encapsidated RNAs, J. Mol. Biol., 2012, 419, 284–300 CrossRef CAS PubMed.
- I. Ivanovska, P. de Pablo, B. Ibarra, G. Sgalari, F. MacKintosh, J. Carrascosa, C. Schmidt and G. Wuite, Bacteriophage capsids: Tough nanoshells with complex elastic properties, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 7600–7605 CrossRef CAS.
- A. Ortega-Esteban, A. J. Perez-Berna, R. Menendez-Conejero, S. J. Flint, C. S. Martin and P. J. de Pablo, Monitoring dynamics of human adenovirus disassembly induced by mechanical fatigue, Sci. Rep., 2013, 3, 1434 CrossRef CAS PubMed.
- J. Mertens, S. Casado, C. P. Mata, M. Hernando-Perez, P. J. de Pablo, J. L. Carrascosa and J. R. Caston, A protein with simultaneous capsid scaffolding and dsRNA-binding activities enhances the birnavirus capsid mechanical stability, Sci. Rep., 2015, 5, 13486 CrossRef PubMed.
- J. S. Villarrubia, Algorithms for Scanned Probe Microscope Image Simulation, Surface Reconstruction, and Tip Estimation, J. Res. Natl. Inst. Stand. Technol., 1997, 102, 425–454 CrossRef CAS PubMed.
- D. J. Müller, H. Janovjak, T. Lehto, L. Kuerschner and K. Anderson, Observing structure, function and assembly of single proteins by AFM, Prog. Biophys. Mol. Biol., 2002, 79, 1–43 CrossRef PubMed.
- F. Moreno-Herrero, J. Colchero and A. M. Baró, DNA height in scanning force microscopy, Ultramicroscopy, 2003, 96, 167–174 CrossRef CAS PubMed.
- S. Poblete, A. Božič, M. Kanduč, R. Podgornik and H. V. Guzman, RNA Secondary Structures Regulate Adsorption of Fragments onto Flat Substrates, ACS Omega, 2021, 6, 32823–32831 CrossRef CAS PubMed.
- A. Ortega-Esteban, I. Horcas, M. Hernando-Perez, P. Ares, A. J. Perez-Berna, C. San Martin, J. L. Carrascosa, P. J. de Pablo and J. Gomez-Herrero, Minimizing tip-sample forces in jumping mode atomic force microscopy in liquid, Ultramicroscopy, 2012, 114, 56–61 CrossRef CAS PubMed.
- J. E. Sader, J. W. M. Chon and P. Mulvaney, Calibration of rectangular atomic force microscope cantilevers, Rev. Sci. Instrum., 1999, 70, 3967–3969 CrossRef CAS.
- I. Horcas, R. Fernández, J. M. Gómez-Rodríguez, J. Colchero, J. Gómez-Herrero and A. M. Baro, WSXM: A software for scanning probe microscopy and a tool for nanotechnology, Rev. Sci. Instrum., 2007, 78(1), 013705 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nh00195d |
| This journal is © The Royal Society of Chemistry 2023 |