Open Access Article
Open Access ArticlePreferential adhesion of bacterial cells onto top- and bottom-mounted nanostructured surfaces under flow conditions†
S. W. M. A. Ishantha
Senevirathne
 *ab,
Asha
Mathew
ab,
Yi-Chin
Toh
*ab,
Asha
Mathew
ab,
Yi-Chin
Toh
 ab and
Prasad K. D. V.
Yarlagadda
ab and
Prasad K. D. V.
Yarlagadda
 c
c
aQueensland University of Technology, Faculty of Engineering, School of Mechanical, Medical, and Process Engineering, Brisbane, QLD 4000, Australia. E-mail: s2.senevirathne@qut.edu.au
bQueensland University of Technology, Centre for Biomedical Technologies, Brisbane, QLD 4000, Australia
cSchool of Engineering, University of Southern Queensland, Springfield Campus, Springfield Central, QLD 4300, Australia
First published on 12th October 2023
Abstract
The bactericidal effect of biomimetic nanostructured surfaces has been known for a long time, with recent data suggesting an enhanced efficiency of the nanostructured surfaces under fluid shear. While some of the influential factors on the bactericidal effect of nanostructured surfaces under fluid shear are understood, there are numerous important factors yet to be studied, which is essential for the successful implementation of this technology in industrial applications. Among those influential factors, the orientation of the nanostructured surface can play an important role in bacterial cell adhesion onto surfaces. Gravitational effects can become dominant under low flow velocities, making the diffusive transport of bacterial cells more prominent than the advective transport. However, the role of nanostructure orientation in determining its bactericidal efficiency under flow conditions is still not clear. In this study, we analysed the effect of surface orientation of nanostructured surfaces, along with bacterial cell concentration, fluid flow rate, and the duration of time which the surface is exposed to flow, on bacterial adhesion and viability on these surfaces. Two surface orientations, with one on the top and the other on the bottom of a flow channel, were studied. Under flow conditions, the bactericidal efficacy of the nanostructured surface is both orientation and bacterial species dependent. The effects of cell concentration, fluid flow rate, and exposure time on cell adhesion are independent of the nanostructured surface orientation. Fluid shear showed a species-dependent effect on bacterial adhesion, while the effects of concentration and exposure time on bacterial cell adhesion are independent of the bacterial species. Moreover, bacterial cells demonstrate preferential adhesion onto surfaces based on the surface orientation, and these effects are species dependent. These results outline the capabilities and limitations of nanostructures under flow conditions. This provides valuable insights into the applications of nanostructures in medical or industrial sectors, which are associated with overlaying fluid flow.
Introduction
Bacteria are omnipresent and can colonise on surfaces, such as human tissues and metallic, organic, or polymeric surfaces. Adhered bacteria can form biofilms at solid-to-liquid, solid-to-air, and liquid-to-air interfaces.1,2 When biofilms are formed, bacteria become highly resistant to anti-septic agents, antibiotics, and immune killing.1,3 Biofilms affect different fields, including, but not limited to, healthcare and engineering. Infections,4 blocked filtration mechanisms,5 blocked aviation fuel systems,6 reduced heat exchanger efficiency,7 and increased fuel consumption of marine vessels due to higher drag resistance caused by biofouling8 are some of the examples of adverse effects of bacterial colonisation.Nanostructures found on several natural surfaces, such as cicada wings,9–11 dragonfly wings,12 and gecko skin,13,14 were revealed to be bactericidal. Such nanostructures were able to kill bacteria by bursting the cell membrane via a non-chemical-based mechanism.10,15,16 On average, the bactericidal insect wing surfaces have nanofeatures with 200–300 nm height, 40–100 nm diameter at the tip, and 100–200 nm spacing between the pillars.9,12,17 The discovery of these bactericidal nanostructured bio-surfaces lead to the development of alternative methods to alleviate bacterial colonisation–associated issues, such as infections and biofouling.
Interestingly, the bactericidal efficacy (BE) of nanostructured surfaces has been reported to be more pronounced on motile bacteria that are capable of active motion by self-propulsion.17,18 Recent reports on increased bactericidal effects of nanostructured surfaces under flow conditions opened a broad spectrum of potential applications of these surfaces, such as on catheters, ship hulls, and pipelines.19 Moreover, these nanostructures can be fabricated on 3-dimensional surfaces without compromising the bactericidal effect.20 These findings further broaden the scope of applications of antibacterial nanostructured surfaces. However, some aspects have not yet been fully explained. Bacterial deposition on a surface under flow conditions is influenced by gravity,21 as well as flow has caused increase in dead cell count and detachment of cells.19,22 Moreover, there are contradicting reports on the effect of gravity on bacterial adhesion,23–26 and importantly, there were no studies on the effect of surface orientation under flow conditions on the bactericidal properties of nanostructured surfaces. Bacterial motion in a flowing media is dictated by the flow velocity of such media.27,28 As the flow velocity increases, the advective transport of bacterial cells becomes predominant over the diffusive transport, as the effect of gravitational force is reduced by the higher drag forces acting on the cells.29–32 Understanding the impact of surface orientation would assist the translation of antibacterial nanostructured surfaces into industrial applications.
The objective of this study is to determine the effect of nanostructure orientation on bactericidal efficiency. The effects of bacterial cell concentration in the flowing fluid, the fluid flow rate, and the length of time the surface was in contact with the flow on cell adhesion, viability, and bactericidal efficacy on surfaces mounted at the top and bottom of the fluid channel was analysed. In addition, factors triggering differential adhesion and bactericidal efficacy between top and bottom surfaces were also examined.
Methodology
Materials
The nutrient broth for cell culturing was prepared by dissolving 26 g of nutrient broth powder (Oxoid, USA) in 1 l distilled water to make 1 l solution, followed by autoclave sterilisation at 121 °C for 20 minutes. Phosphate buffered saline (PBS) was prepared by dissolving one tablet of 1 X PBS (Oxoid, USA) in 500 ml of distilled water followed by autoclave sterilisation at 121 °C for 20 minutes.The two bacterial species used for the experiments were, Gram-positive S. aureus (ATCC 25923) and Gram-negative P. aeruginosa (ATCC 27853). A bacterial suspension was prepared by inoculating a colony of bacteria in 5 ml of nutrient broth and incubating in a shaking incubator at 37 °C and 200 RPM for 16 hours. Then, the bacterial suspension was centrifuged at 5250 RCF for 5 minutes and the separated pellet was resuspended in PBS. A required volume of suspension was made by adding centrifuged pellets to the measured volume of PBS, until the target turbidity level was achieved. The turbidity of the suspension was measured using a BioPhotometer (Eppendorf, Germany) with OD600 scale.
Nanostructure fabrication
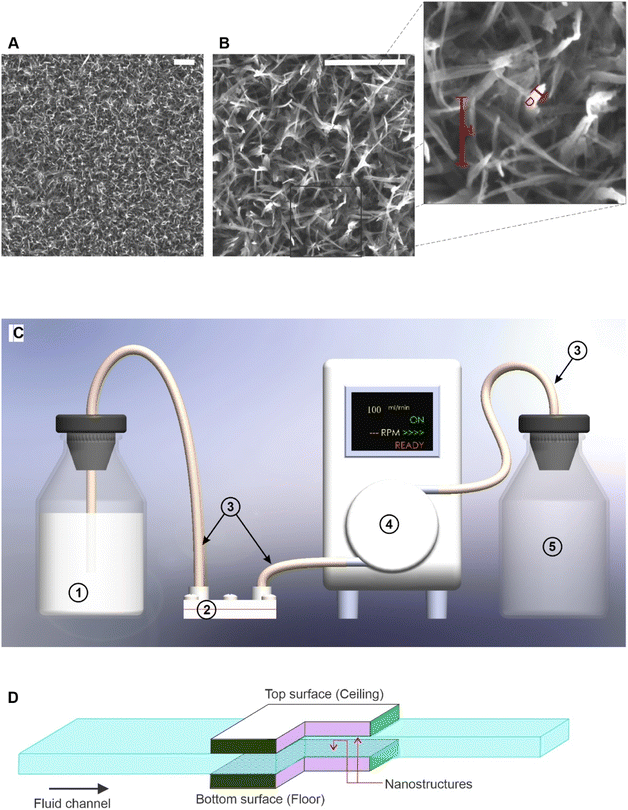
Nanostructures on Titanium (Ti–6Al–4V grade-5) substrates sized 7 × 10 mm were fabricated via a hydrothermal process. Two sets of substrates were used for the experiment. All the substrates were polished into a surface roughness of 0.04 μmRa by electro polishing. One half of the substrates were left untreated and used as the control experiment, while the rest were subjected to a hydrothermal process. These surface-modified substrates were reacted in 1.0 M sodium hydroxide (NaOH) at 180 °C for 2 hours to form nanowire structures33,34. This set of fabrication parameters result in nanowire structures on the titanium substrate with an average wire diameter of 50 nm and a height of 300 nm,34 and the modified surface was hydrophilic.35 The scanning electron microscopic (SEM) images of the fabricated nanowires are shown in Fig. 1A and B. This nanowire structure on a Ti–6Al–4V alloy has been extensively studied by previous researchers for antibacterial properties34,36 and eukaryotic cell proliferation.35 The images acquired using a field emission scanning electron microscope (FEG-SEM) (TESCAN Mira 3) confirmed the nanostructure formation on the substrates. Polished substrates without hydrothermal treatment were the second type of substrates used for the experiment, referred to as untreated substrates. All substrates were soaked in 80% ethanol for 15 minutes, washed with a stream of sterile PBS, and exposed to UV light for 20 minutes, before incubating the bacterial cells on them.Experiment design and procedure
The aim of the experiment was to investigate the effects of bacterial cell concentrations, flow rate, and flow duration on the bactericidal properties of the nanostructured surface under flow conditions, and the effect of surface orientation on bacterial adhesion and viability on nanostructured surfaces under flow conditions. Bacterial cells in a suspension were pumped through a custom designed microfluidic device that hold two nanostructured surfaces at the top and bottom of the flow channel. The cells were quantified using an epifluorescence microscope following staining with a epifluorescence dye kit. First, using factorial analysis, the main effects of cell concentration in the flowing suspension, flow rate of the cell-suspended media, and duration of the flow on cell adhesion and bactericidal effect of the surface were analysed. In the second part of the analysis, using ANOVA, cell counts and bactericidal efficacy were compared between top and bottom locations under each combination of factors. A partial factorial experiment designed using the MiniTab software (version 18), with two levels for each factor was used. The standard order of trials is shown in Table 1, and the low and high levels of each factor are given in Table 2.| Standard order | Concentration | Flowrate | Exposure time |
|---|---|---|---|
| 1 | Low | Low | Low |
| 2 | Low | Low | High |
| 3 | Low | High | Low |
| 4 | High | Low | Low |
| 5 | High | Low | High |
| 6 | High | High | Low |
| 7 | High | High | High |
| Factor | Low level | High level | |
|---|---|---|---|
| Concentration (OD 600) | S. aureus | 0.0660 ± 0.0079 | 0.1942 ± 0.0148 |
| P. aeruginosa | 0.0559 ± 0.0027 | 0.1670 ± 0.0040 | |
| Flowrate (ml min−1) | 0.12 | 12.00 | |
| Flowtime (h) | 1.0 | 3.0 | |
Microfluidic device and flow experiment
The components of the microfluidic system are illustrated in Fig. 1C. Two sterilised substrates were exposed to the flow on ceiling and floor of the channel, as shown in Fig. 1D, to investigate if the adhesion of bacterial cells is preferential. The fluid dynamics of MFD was computed as described in ref. 19. The MFD, its connectors, and tubing were autoclaved before conducting each trial. The number of live cells and the number of dead cells were the two response variables of the experiment. The bacterial cells were suspended in PBS and the cell concentration was adjusted to the targeted turbidity measured in OD600 scale within tolerance of ±0.01. Full details of cell preparation are presented in another section. The prepared bacterial suspension was pumped using a peristaltic pump. In each trial, two nanostructured substrates and two flat (control) substrates were exposed to the bacterial suspension flow with one on the ceiling (top) and the other on the floor (bottom) of the flow channel. Two identical MFDs were used for nanostructured and flat (control) substrates and a bacterial suspension from the same reservoir was pumped through both devices. The two devices were coupled to the same peristaltic pump with two channels of tubing from the same reservoir containing the bacterial suspension. The schematic illustration of this experimental setup with only one microfluidic device is shown in Fig. 1C. The MFD has two metal plates, a 0.2 mm metallic sheet with a flow channel machined on it, two rubber O-rings, and stainless-steel screws to assemble the device. All the metallic parts are made of medical grade AISI315L stainless steel. Before each trial, the MFD was autoclaved at 121 °C for 20 minutes without the substrates and allowed to cool down to room temperature. The MFD was kept in a sealed bag during autoclaving and only opened inside a biosafety cabinet. All substrates were washed with 70% v/v ethanol and exposed to UV light for 20 minutes before use for bacterial incubation. These sterilised substrates were inserted into the MFD inside a running biosafety cabinet. Each of the two substrates is exposed to the fluid flow in an area of 3 × 10 mm on the side facing the flow. The channel is 3 mm wide and 0.2 mm high with a total length of 30 mm. The inlet and outlet channels are 10 mm in length. A peristaltic pump (Ismatec ISM915A with CA-8 cassette, Germany) was used to flow the bacterial suspension at a controlled flow rate through the microfluidic device. The pump was calibrated by manual measurements, by pumping sterilised water using a Ø0.89 isoprene tube (Ismatec, Germany) at flow rates of 1 to 10 ml min−1 for 10 minutes and at a flow rate of 0.1 ml min−1 for 60 minutes, and then the volume of dispensed water was measured using a measuring cylinder.Fluorescence imaging and cell count quantification
After the flow, substrates were immediately retrieved from the MFD and dyed using a Live/Dead BacLight kit (Invitrogen detection technologies, L7012), a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of SYTO9 and propidium iodide (PI) was used to stain bacterial cells in a dark room. Then, 5 μl aliquot of each dye component was mixed in 1 ml of phosphate buffered saline (PBS) to make the dye mixture. The preparation and storage of the dye stock and mixtures was done in the darkness. The dye stock and mixture were stored in a −4 °C freezer. Then, 3 μl of dye mixture was pipetted onto a glass-bottomed microslide (Ibidi, Germany), and the substrate was placed on the top of the dye droplet with the cell-adhered side facing down. After staining, incubation was allowed for 15 minutes before imaging the surfaces. Following the incubation, samples placed face down on the glass-bottomed microslide were immediately imaged using an inverted fluorescence microscope (Nikon Eclipse TiS, Japan) with a FITC filter for live cells and a Cy3 filter for dead cells. SYTO9 got excited at 480 nm wavelength and emitted at 500 nm wavelength, while PI got excited at 490 nm and emitted at 635 nm. Objective lens with 40× magnification and 0.6 numerical aperture was used for imaging surfaces with 100 ms exposure time. An area of 206.40 × 165.12 μm2 in the substrate was captured in each image with a resolution of 1280 × 1024 pixels. Therefore, 1 μm2 is equivalent to 38.5 pixels. Each point was imaged using both filters. The minimum number of measurements was determined for 90% confidence interval with 0.25 margin of error. Two controls with 100% dead cells and 100% live cells were used to confirm the dye functionality. Then, 5 μl of bacterial suspension was pipetted into two microwells, and one of them was added with the same volume of 70% v/v ethanol. This was used as the 100% dead cell control and the other was used as the 100% live cell control. SYTO9 function was confirmed by receiving signals through a FITC filter from 100% live control, and the PI function was confirmed by receiving signals through a CY3 filter with 100% dead control.
1 mixture of SYTO9 and propidium iodide (PI) was used to stain bacterial cells in a dark room. Then, 5 μl aliquot of each dye component was mixed in 1 ml of phosphate buffered saline (PBS) to make the dye mixture. The preparation and storage of the dye stock and mixtures was done in the darkness. The dye stock and mixture were stored in a −4 °C freezer. Then, 3 μl of dye mixture was pipetted onto a glass-bottomed microslide (Ibidi, Germany), and the substrate was placed on the top of the dye droplet with the cell-adhered side facing down. After staining, incubation was allowed for 15 minutes before imaging the surfaces. Following the incubation, samples placed face down on the glass-bottomed microslide were immediately imaged using an inverted fluorescence microscope (Nikon Eclipse TiS, Japan) with a FITC filter for live cells and a Cy3 filter for dead cells. SYTO9 got excited at 480 nm wavelength and emitted at 500 nm wavelength, while PI got excited at 490 nm and emitted at 635 nm. Objective lens with 40× magnification and 0.6 numerical aperture was used for imaging surfaces with 100 ms exposure time. An area of 206.40 × 165.12 μm2 in the substrate was captured in each image with a resolution of 1280 × 1024 pixels. Therefore, 1 μm2 is equivalent to 38.5 pixels. Each point was imaged using both filters. The minimum number of measurements was determined for 90% confidence interval with 0.25 margin of error. Two controls with 100% dead cells and 100% live cells were used to confirm the dye functionality. Then, 5 μl of bacterial suspension was pipetted into two microwells, and one of them was added with the same volume of 70% v/v ethanol. This was used as the 100% dead cell control and the other was used as the 100% live cell control. SYTO9 function was confirmed by receiving signals through a FITC filter from 100% live control, and the PI function was confirmed by receiving signals through a CY3 filter with 100% dead control.
Fluorescence images were enhanced for contrast, and binarized using ImageJ (version 1.53f51). The number of illuminated pixels of the binarized image was counted using a BioFilmAnalyzer,37 and the number of pixels was taken as the unit of cell surface coverage. An area of 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080.1 μm2 on the substrate was captured by each image, and therefore, this was taken as a unit area and is equivalent to 1
080.1 μm2 on the substrate was captured by each image, and therefore, this was taken as a unit area and is equivalent to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310
310![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 pixels in the image. All cell counts were presented as the number of pixels in an area of 1
720 pixels in the image. All cell counts were presented as the number of pixels in an area of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 310
310![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 720 pixels. The threshold level for binarizing images was established by a trial-and-error method, by comparing the final processed image with the original for discrepancies in cell map. This process was done for each batch of images taken together.
720 pixels. The threshold level for binarizing images was established by a trial-and-error method, by comparing the final processed image with the original for discrepancies in cell map. This process was done for each batch of images taken together.
Data analysis
Factorial analysis was performed using the MiniTab software (version 18). A confidence interval of 95% was used for all statistical analyses. ANOVA was used to find the significance of differences between cell counts. The following group comparisons were made with ANOVA using the GraphPad Prism software (version 9):(1) Live versus dead cell count on the same surface.
(2) Live, dead, and total cell count on nanostructured surfaces versus the control surface.
(3) Live, dead, and total cell count on the top surface (ceiling) versus the bottom surface (floor) of the same kind.
Main effects of cell concentration, flow rate, and flow time (exposure time) were examined by the factorial analysis using factorial plots. This analysis shows the effect of each independent variable on dependent variables averaged across all levels of the other two independent variables. The effect on top and bottom surfaces were analysed separately, and the main effect of each factor on live cell count, dead cell count, and total cell adhesion were analysed. One-way and two-way interactions of the factors for each condition were also studied using the factorial analysis. Pareto charts were used to identify the order of magnitude of main effects. The effects of bacterial cell concentration in the suspension, flow rate of the suspension, and duration of the flow (3-independent variables) on total adhesion and bactericidal effect (2-response variables) were examined. Based on this analysis, the three independent variables were ranked considering their overall influence on response variables. ANOVA was used to assess the significance of differences in group mean values between the groups using the GraphPad Prism software. A confidence interval of 95% (p ≤ 0.05) was used to test the statistical significance. Statistical significance with ANOVA is shown by ns: P > 0.05, *: P ≤ 0.05, **: P ≤ 0.01, ***: P ≤ 0.001, and ****: P ≤ 0.0001.
Results
Variations in cell adhesion and viability on nanostructured surfaces when placed at the top and bottom of the flow channel
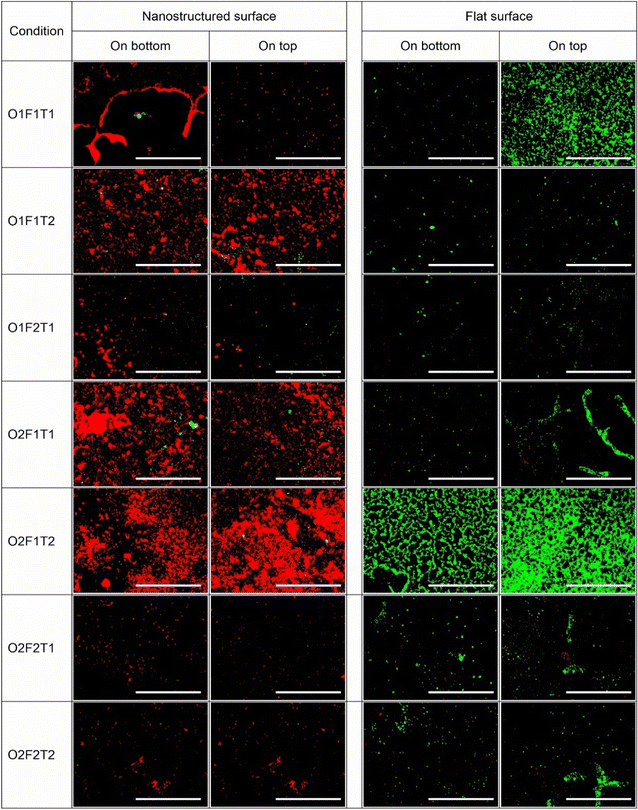
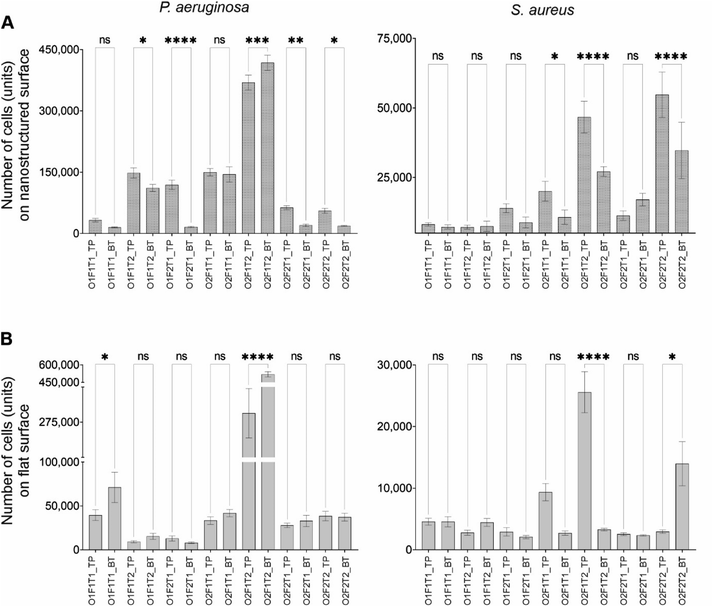
Fluorescence images of substrates stained with SYTO9 and PI were qualitatively analysed for adhesion and viability on surfaces. Fig. 2 and 3 show the fluorescence microscopic images of P. aeruginosa and S. aureus species on nanostructured and flat surfaces located at top and bottom under each trial in the factorial experiment. The most prominent observation was the higher surface coverage of dead cells shown in red colour on the nanostructured surface than that of the flat surface, indicating more cell death on nanostructured surfaces. This observation was consistent throughout the different conditions investigated. Comparatively, P. aeruginosa adhesion on both types of surfaces was noted to be drastically higher than that of S. aureus on the corresponding surface. Under specific conditions, an abnormal increase in cell adhesion was observed. Only exposure time at high levels (O1F1T2), only concentration at high levels (O2F1T1), and both concentration and time at high levels (O2F1T2) resulted in unusual coverage of P. aeruginosa on the nanostructured surface, and only one condition (O2F1T2) gave the same results on the flat surface as well (Fig. 2). When all factors were at low levels (O1F1T1), adhesion on the bottom nanostructured surface was higher than that on one top, but the top flat surface had higher adhesion than the bottom flat surface. Notably, higher adhesion on the nanostructured surface than on the flat surface was observed with either species of bacteria. However, P. aeruginosa showed a higher preference for forming large clusters of cells, whereas S. aureus was forming smaller clusters. Three conditions, namely, (O2F1T1), (O2F1T2), and (O2F2T2) resulted in notably higher adhesion of S. aureus cells on the bottom nanostructured surface.Effects of bacterial cell concentration, flow rate, or exposure time on bacterial adhesion onto nanostructured surfaces are independent of surface orientation
The main effect of each of the three factors (cell concentration, flow rate, and exposure time) on bacterial attachment had no variation with the placement of the nanostructured surface in the fluid channel (data shown in ESI Fig. S1†). The increase in cell concentration in the flowing fluid and exposure time resulted in an increase in cell attachment onto the nanostructured surface without showing any difference in trends with respect to the orientation of the surface. This observation was valid for both species. However, the effect of the flow rate on cell attachment was species dependent. P. aeruginosa attachment on either top-mounted or bottom-mounted surfaces decreased with the increase in flow rate. Nevertheless, the flow rate had no effect on S. aureus cell attachment onto the top-mounted surface, while the attachment onto the bottom-mounted surface was increased with the increase in flow rate.Some of the factors had varying effects on the top- and bottom-mounted flat surface (control) (data shown in Fig. S1†). Notably, those effects were species dependent as well. Primarily, the effects of the three factors on P. aeruginosa adhesion were independent of the surface placement. The increasing cell concentration resulted in an increase in adhesion, while the increase in the flow rate resulted in a decrease in cell adhesion. Nevertheless, this observation was made on both top- and bottom-mounted surfaces. While exposure time increment caused a slight reduction in the adhesion, that on the bottom-mounted surface was not statistically significant. However, S. aureus adhesion on the flat surface saw differential effects by the three factors between top- and bottom-mounted surfaces for all factors.
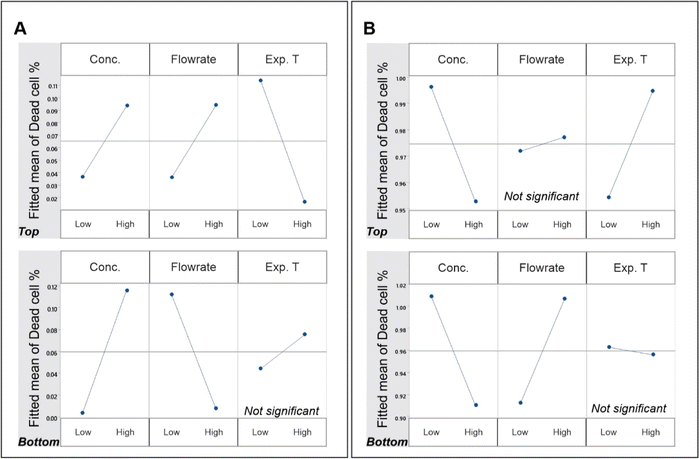
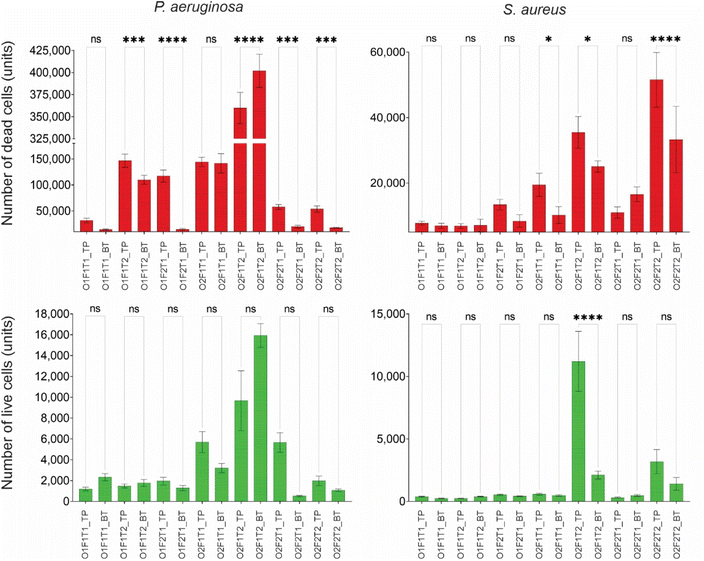
Unlike the total cell count, the dead cell count on the nanostructured surface experienced a complicated pattern of effects from the 3 independent variables, as shown in Fig. 4. Moreover, these effects were species-dependent and orientation-dependent. The effect of cell concentration was positive on P. aeruginosa, while it was negative on S. aureus. However, this was not orientation dependent. Despite the variation with bacterial species, the effect of cell concentration was the same on both top and bottom surfaces for both species. The flow rate and exposure time had species- and orientation-dependent effects on dead cell count. While the flow rate caused a positive effect on dead P. aeruginosa cell count on the top surface, opposing effects were observed on the bottom surface. Meanwhile, the S. aureus dead cell count saw drastically different patterns to this. While the dead cell count on top surface had no effect from flow, there was a positive effect of it on the bottom surface. The flow duration (Exp. T) also showed no specific pattern. While P. aeruginosa dead cell count on the top-mounted nanosurface had a negative effect from flow duration, the same on S. aureus saw a positive effect. Notably, the flow duration had no effect on dead cell count on the bottom-mounted surface for either bacterial species.
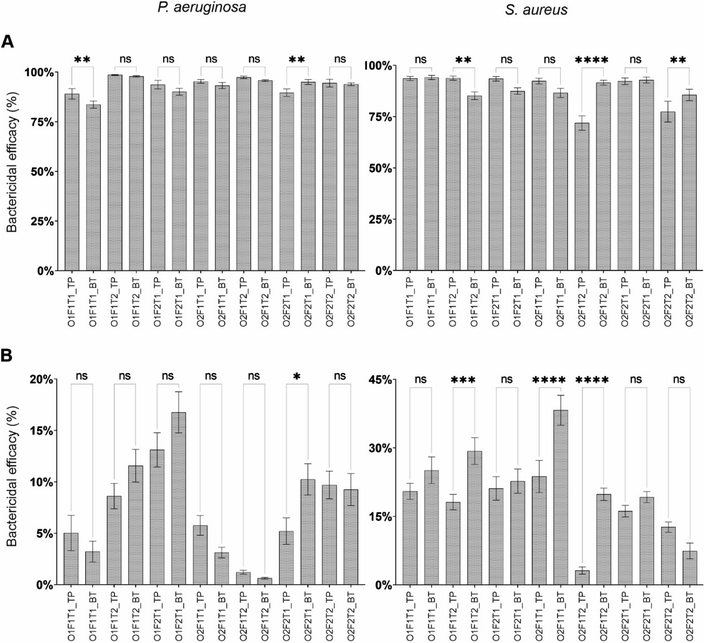
Species- and orientation-dependent effect of fluid flow on bactericidal efficacy
Factorial analysis was carried out to examine the effects of cell concentration, flow rate, and exposure time on the bactericidal efficacy of the nanostructured surfaces placed at the top and bottom, as presented in Fig. 4. The effects of concentration, flow rate and exposure time on bactericidal efficacy was species-dependent in addition to the location dependency. The effect of cell concentration on the bactericidal property was different on the two species, but there was no difference between the top and bottom surfaces. While the efficacy against P. aeruginosa was increased with the increase in cell concentration, that against S. aureus was decreased. However, the observations for each species had no difference between the two locations. Apart from that, there were no other similarity observed between locations or species. The flow rate had opposing effects on bactericidal efficacy against P. aeruginosa on the top and bottom surfaces. The same had drastically different effects on S. aureus. While the bottom surface showed an increase in bactericidal effects with the increase in flow, the top surface had no effect. The exposure time had no effect on the bottom surface bactericidal efficacy with either species of bacteria. Although the top surface had significant effects from exposure time, the effect was species dependent.Species-specific preference in adhesion onto top- and bottom-mounted nanostructured surfaces
Two locations tested in this work were (a) nanostructured surfaces placed on the top surface of the flow channel and (b) nanostructured surfaces placed at the bottom of the flow channel. Cell adhesion and viability of those adherent cells (by percentage of dead cells) were compared using ANOVA to investigate for the differences between the two surface orientations for both nanostructured and flat (control) surfaces. The difference in cell adhesion between top and bottom was observed on the nanostructured surface only under specific combinations of factors, with P. aeruginosa and S. aureus species (Fig. 5), despite no recognisable trend. However, compared to the nanostructured surface, the flat surface saw only few instances of differential adhesion between the top and bottom surfaces. Five of the seven conditions tested showed difference in the adhesion of P. aeruginosa on the top- and bottom-mounted nanostructured surface.Except at high concentrations and exposure time periods (O2F1T2), P. aeruginosa preferred adhering to the bottom-mounted nanostructured surface. Only under two conditions, where all factors were at low level (O1F1T1) and at high concentration and exposure time (O2F1T2), P. aeruginosa showed differential adhesion between top- and bottom-mounted control (flat) surfaces with preferential adhesion onto the top mounted surface. In comparison under five conditions, the adhesion of P. aeruginosa on the bottom nanostructured surface was significantly different from that of the bottom control surface. However, when all three factors at low level (O1F1T1), and when only exposure time at low level (O2F2T1), an insignificant difference (P > 0.05) in adhesion between nanostructured and flat surfaces was observed. Three instances saw an insignificant difference in adhesion between the top-nanostructured and top-flat surface (data not shown). Notably, when all the three factors were at their highest level (O2F2T2), adhesion was similar between the two top surfaces and between the two bottom surfaces. Interestingly, when either, or both, the flow rate or the flow duration is at high levels (F1T2, F2T1, and F2T2), P. aeruginosa cell adhesion on the nanostructured surface showed a significant difference between top and bottom surfaces. The comparison of S. aureus adhesion onto top- and bottom-mounted surfaces is shown in Fig. 5. Only three out of the seven instances showed significant difference in S. aureus adhesion onto top- and bottom-mounted nanostructured surfaces. When only the concentration was at high levels (O2F1T1), when only the flow rate was at low levels (O2F1T2), and when all the three factors were at high levels (O2F2T2), differential adhesion of S. aureus on top and bottom nanostructured surfaces was observed. In comparison, the same on flat surface had two instances of differential adhesion. When only the flow rate was at low levels (O2F1T2) and when all the three factors were at high levels (O2F2T2), S. aureus had differential adhesion between the top and bottom flat surfaces. S. aureus preferred the bottom-mounted nanostructured surface over the top-mounted of the same type for adhesion. The same trend was demonstrated for the flat surface as well. This was in contrast to P. aeruginosa adhesion, which showed preference to the top-mounted nanostructured surface.
These observations suggest that cell adhesion onto nanostructured surfaces depends on cell concentration, flow rate, and flow duration. S. aureus cell adhesion on top and bottom nanostructured surfaces was different only on three of the seven conditions studied. Two of those three conditions had caused significant difference in cell count between top and bottom flat surfaces as well. Compared to this, P. aeruginosa species had five of seven conditions with significantly different cell adhesion on top and bottom nanostructured surfaces. Out of those five conditions, only one had the same observation on the flat (control) surface pair.
Insignificant difference in live cell count between top and bottom nanostructured surfaces
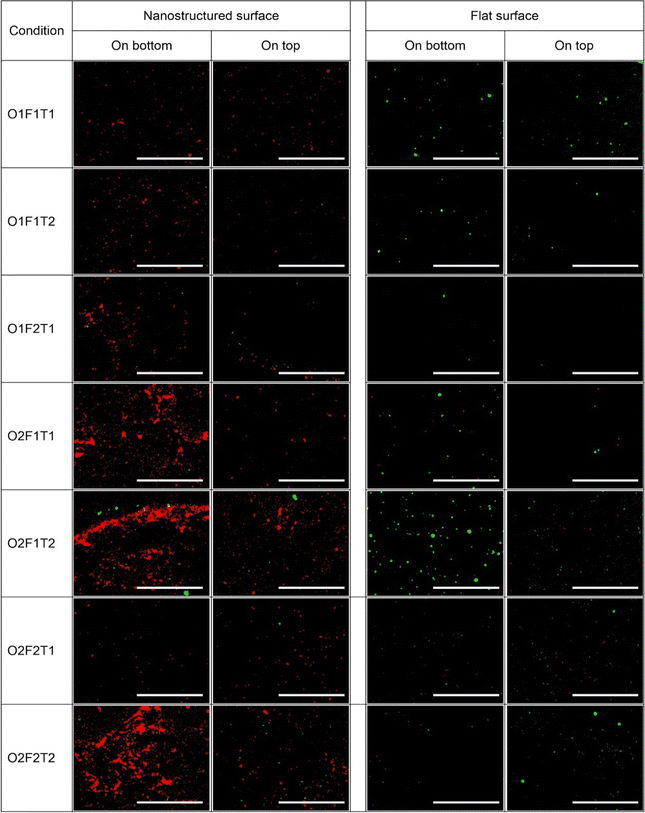
Fig. 6 shows the live and dead cell counts of P. aeruginosa and S. aureus species on top and bottom nanostructured surfaces. While the dead cell count of P. aeruginosa showed differences between the top and bottom nanostructured surfaces, the live cell count did not show any difference between the two locations. Similarly, variation of dead S. aureus cells between the two locations was significant under some of the conditions, while the live cell count was different under only one condition. Under one condition when only the flow rate was at low levels (O2F1T2), there was a significant variation in the live S. aureus cell count between the locations, whereas S. aureus dead cell counts were significantly different under three conditions. However, the P. aeruginosa dead cell count was not different only under two conditions. When all three factors were at low levels (O1F1T1), and only concentration was at high levels (O2F1T1), no difference was observed in dead cell counts between the top and bottom surfaces. Compared to this, the live cell count showed no variations between the two locations under any condition.The dead and live cell counts of P. aeruginosa and S. aureus species on the top- and bottom-mounted flat surfaces are compared as shown in Fig. S3.† The dead cell count had no difference between the top and bottom flat (control) surfaces, but had significant differences between top and bottom nanostructured surfaces under five different conditions for both species. In contrast, the live cell count on the flat surface showed a statistically significant difference between top and bottom locations under some of the tested conditions. P. aeruginosa had only two conditions with differential live cell counts between top and bottom, while S. aureus had three conditions. When only the flow rate was at low levels (O2F1T2), both species showed differential live cell counts between the top and bottom surfaces but not under other conditions. It is evident that primarily only the dead cell count varies with the surface orientation for nanostructured surfaces, and only the live cell count varies with the surface orientation for flat surfaces. Notably, when only the flow rate was at low levels (O2F1T2), the S. aureus live cell count on the nanostructured surface, as well as the flat surface, showed difference between top and bottom. Under such condition, the S. aureus live cell count on the top-mounted surface was significantly higher than that on the bottom-mounted surface.
Bactericidal efficacy varies between the top and bottom surfaces under specific conditions
Fig. 7 shows the bactericidal efficacy of (A) nanostructured and (B) flat surfaces compared between top and bottom locations for P. aeruginosa and S. aureus species. Compared to the bactericidal efficacy demonstrated by the nanostructured surface, the efficacy of the flat surface was drastically low on both bacterial species. Under all the tested conditions, the bactericidal efficacy of the nanostructured surface was significantly higher than that of the flat surface under the same condition (P ≤ 0.0001). The comparison of dead cell percentage of P. aeruginosa and S. aureus between the nanostructured and control surfaces is presented in Fig. S2.†Except under two conditions, the bactericidal efficacy of the nanostructured surface against P. aeruginosa showed no statistically significant difference in between top and bottom orientations. When all factors were at low levels (O1F1T1) and the concentration and flow rate at high levels (O2F2T1), differential bactericidal efficacies between the two locations were observed. Three conditions resulted in dissimilar bactericidal efficacies between top and bottom locations against S. aureus on nanostructured surfaces, and none of the condition was the same as that of P. aeruginosa. When only exposure time was at high levels (O1F1T2), only flow rate at low levels (O2F1T2), and all three factors at high levels (O2F2T2), dissimilar bactericidal efficacies on top and bottom nanostructured surfaces were observed. The bactericidal efficacy of the nanostructured surface was between 83% and 99% on P. aeruginosa, and 72% to 94% on S. aureus. The flat surface demonstrated a relatively low bactericidal efficacy where it was 0.6% to 17% against P. aeruginosa, and 3.1% to 38% against S. aureus. Notably, a dissimilar efficacy between top and bottom flat surfaces was observed under several conditions. Under the condition when only the exposure time was at low levels (O2F2T1), the efficacy against P. aeruginosa was higher on the bottom surface. However, under three conditions, the bottom flat surface demonstrated a higher efficacy than the top surface against S. aureus.
Discussion
The nanowires lyse bacterial cells by piercing the bacterial cell membrane. The attraction forces between the cell and the surface causes the cells to move towards the surface, and thus, the cells get pierced by nanowires upon contact. There are several theories presented by scientists to explain the mechanism of cell membrane failure under mechanical stress caused by sharp nanofeatures. The hydrodynamic forces by the flowing liquid acting on cells cause passive motion of cells. As a consequence, more cells collide with sharp nanowires, resulting in an increased cell lysis. The random orientation of nanowires (Fig. 1A and B) plays a vital role in increasing the bactericidal effect of the surface. Since the nanowires are omni directed, some of the sharp tips are directed against the direction of the fluid flow. This increases the collision of bacterial cells onto sharp tips, leading to increased lysis of the cells. Further, this will cause deeper piercings of cells, which will avoid covering the sharp tip of nanowire that may hinder subsequent cell lysis.Due to the difference in shape, the fluid dynamic forces acting on the cells differ largely. While the spherical shape of S. aureus experiences relatively stable motion under the flow, rod-shaped P. aeruginosa will experience unstable motion. Due to the shape of the cell, the forces acting on it are dependent on the orientation of cell relative to the flow direction. Therefore, P. aeruginosa cells will experience rotations, where the other species may not experience this.
Apart from the differences between bacterial species, the cell concentration in the fluid, flow rate, and flow time will make the adhesion on surfaces differ from one type to another. While lower cell concentrations in the suspension allow cells to move freely, under higher cell concentrations, the cells will experience higher attractive or repulsive forces from neighbouring cells in the suspension. Similarly, the flow rate has an influence on bacterial attachment and death on nanostructured surfaces. The hydrodynamic forces such as drag and lift that act on the cells will cause the cells to gain high inertia. When those cells with high inertia collide with surfaces, they will get pierced by the nanostructures, causing the cells to get lysed and pinned onto the surface. However, this may affect the two bacterial species differently as the cell membrane strength is different between the species. Gram-positive bacteria have a layer of peptidoglycan in their cell wall making it thicker and stronger than that of their Gram-negative counterparts.38,39 Moreover, the higher flow rates can cause deeper piercing of bacterial cells. This will prevent the dead bacterial cell covering the sharp tip of the nanostructure from hindering subsequent lysis of cells. Therefore, a higher flow rate can further increase the cell death on nanostructures. In addition, a higher flow rate will force the cells to have advective transport and will hinder the diffusion of cells in the flow.29 However, lower flow rates will allow the cells to diffuse in lateral directions to the flow, enabling more cells to reach the surface and thereby adhere to the surface. In addition, higher flow rates will cause some of the adhered cells to get detached from the surface, causing variation in the adhered cell count. Moreover, it has been reported that, under flow conditions, it is more likely for the live bacterial cells to get detached from the nanostructured surface than dead cells on the same surface. The flow duration, which is the third factor investigated in this study, will also have an influence on cell attachment. Longer durations will enable cells to get attracted to already adhered cells on the surface, causing an increase in cell adhesion. Moreover, longer duration will allow cells to proliferate, resulting in an increase in the cell count on the surface. However, the same phenomenon can cause higher detachment of cells from the surfaces as well. Species such as S. aureus form 3-dimensional biofilms.40 This increase in size will result in an increase in fluid shear on the biofilm, which can lead to its removal from the surface. However, this removal will depend on the adhesion strength of the two species, which is notably different from each other.
Higher cell concentrations caused an increase in cell adhesion, which is quite intuitive. More cells in the flowing fluid indicate more population of cells near the solid surfaces, which causes more cells to get attracted to the surfaces. This phenomenon was observed across the two types of surfaces and the two bacterial species. However, the effect of flow rate on adhesion onto nanostructured surfaces was species dependent, but it was location independent at the same time. The flow rate increment caused an increase in P. aeruginosa cell adhesion on both locations. However, S. aureus cell adhesion on top mounted surface was not impacted by the flow rate, but increased adhesion on the bottom-mounted surface. Compared to S. aureus cells, P. aeruginosa cells are larger in size. S. aureus cells are spherical in shape and have an average of 1 μm diameter,41 while P. aeruginosa cells are rod shaped with 1 μm diameter and 5 μm length.42 Therefore, S. aureus has an average volume of 0.52 μm2, and that of P. aeruginosa is 3.93 μm2, which is approximately 7.5 times larger than that of the S. aureus cell. Similarly, while S. aureus has a projected area of 0.79 μm2, the maximum projected area of a P. aeruginosa cell is 5 μm2. Therefore, due to the size and shape of the cells, P. aeruginosa cells have higher fluid dynamic drag exerted on them. This explains the decrease in P. aeruginosa attachment with the increase in fluid flow rate. Longer duration of exposure caused higher adhesion of cells onto the nanostructured surface, which was independent of location or species. This is due to more cells passing the nanostructured surface, and hence, the number of cells getting adhered onto nanostructures increased over time. Dead bacterial cells had been demonstrated to get pinned onto nanostructures under flow conditions, while live cells getting detached and taken away by the flow.22 The effects of the three factors were mostly independent of the location. The only exception was with the flow rate on S. aureus species. However, differential impacts on live and dead cells were observed. Particularly, while exposure time increased the number of dead P. aeruginosa cells, it had no impact on the number of live P. aeruginosa cells. Similarly, while the flow rate increased the number of dead S. aureus cells on the bottom surface, it had increased the number of live cells on the same surface. This agrees with the previous reports on the differential effect of fluid flow based on the viability of bacterial cells.19,22
Although variation in the cell count of either dead or live cells had similar effects from the three factors, the ratio between dead and live cells had varying influence by some of the factors based on the location of the surface. The effect of cell concentration was dependent on the bacterial species, but it was consistent between the two locations. Higher cell concentrations caused the nanostructures to show less bactericidal efficacy against S. aureus but opposing effect on P. aeruginosa. Lower bactericidal effects against S. aureus than P. aeruginosa were reported under static conditions as well as fluid flow conditions.19,22,43,44 The effect of flow rate or exposure time had no similarity between the two locations or species. The increase in flow rate resulted in a decrease in both live and dead cells but increased the percentage of dead cells on the top surface. It was shown that live cells get removed from the nanostructured surface more easily than the dead cells under flow conditions.22 Therefore, it can be expected that higher removal of live cells causes increased dead cell percentage. However, the dead cell percentage on the bottom surface was increased by flow rate. A lesser number of dead P. aeruginosa cells were observed on the bottom surface in most of the instances (Fig. 6). The higher removal of live cells from the top surface under flow may result in more cells being settled on the bottom surface. However, the bactericidal efficacy of the top surface against S. aureus had no effect from the flow rate, while that of the bottom surface was increased. Unlike P. aeruginosa, S. aureus is a non-motile species. This prevents them from swimming away from the surface as P. aeruginosa cells do. Therefore, more cells can be expected to accumulate on the bottom surface and get lysed. Higher cell concentrations flowing at slow pace over a long time period may have given motile P. aeruginosa an opportunity to move to the top surface.
The O2F1T2 condition resulted in an increase in P. aeruginosa cell count on the flat surface. The group O2F1T2 has a higher concentration of bacteria sent at a lower flow rate for a longer time, which allowed settling of a higher number of P. aeruginosa cells on the flat surface. Moreover, the lower flow rate results in less fluid drag forces on cells. This will allow the cells to have more diffusive transport under the flow. Due to the lower hydrodynamic forces acting on the cells, the removal of adhered cells from the surface will also be reduced. The increase in cell count on the flat sample can be due to the combined effect of the above.
The bottom-mounted flat surface under the O1F1T1 condition saw an increase in P. aeruginosa live cells on it. This had lower cell concentrations with slower flow for a shorter duration. The slow flow allows the cells to diffuse more in directions lateral to the flow and adhere to the surfaces. Moreover, the lower concentration would allow cells to proliferate more, as ample nutrients are available in the media. Further, the slow flow rate would cause less removal of adhered cells from the surface.
Dissimilar adhesion between the top and bottom locations was observed only under specific conditions. Both species demonstrated preferential adherence to the top-mounted surface over the bottom-mounted surface. P. aeruginosa is a motile species of bacteria that have shown to be responsive to external environmental factors such as flow.45 Several forces are acting on a bacterial cell flowing in a fluid. The weight of the cell, buoyancy force, drag force, and hydrodynamic forces are acting on a cell in passive motion in a fluid. The Peclet number calculated for the two types of bacterial cells shows that advection is prominent for both fluid velocities considered. Higher advection suggested lesser settling of cells on the bottom surface. Only when both the flow rate and flow duration were at low levels (irrespective of the concentration), P. aeruginosa had no difference in adhesion between the top- and bottom-mounted nanostructured surfaces. When either, or both, flow rate or duration was high, it showed significantly different adhesion between the top- and bottom-mounted nanostructured surfaces. When the flow rate is high, P. aeruginosa cells may be experiencing higher lift force acting on the cells due to the high flow velocity causing more cells to reach the top surface. Moreover, it is not uncommon for motile species of bacteria to swim against gravity.46,47 Therefore, when the duration is high, then the cells will have more time to swim against the gravity.
Conclusion
The effects of cell concentration, fluid flow rate, and exposure time on cell adhesion are not dependent on the orientation of the nanostructured surface in the fluid channel. While the concentration and exposure time affected the adhesion independent of the bacterial species, fluid shear has a species-dependent effect on bacterial adhesion. Notably, the effect of fluid flow on the bactericidal efficacy of the nanostructured surface is both orientation and bacterial species dependent. The preference of adhesion onto top- or bottom-mounted nanostructured surfaces is species specific. This choice of adhesion is influenced by the cell concentration, fluid flow rate, and exposure time. Under certain conditions, preferential adhesion on the flat surface also occurs.The difference in cell adhesion is due to the differences in dead cell counts on the nanostructured surface. The nanostructured surface had only subtle difference in bactericidal efficacy against P. aeruginosa on top- and bottom-mounted surfaces. In contrast, the same against S. aureus was pronounced. The bactericidal efficacy of the top-mounted surface was higher when the cell concentration is high. Prolonged exposure resulted in higher bactericidal efficacy against S. aureus on the top-mounted surface.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
Authors wish to acknowledge support from following individuals and institutions. Centre for Biomedical Technologies (CBT), Queensland University of Technology (QUT), Australia. Dr K. A. S. N. Gunarathne for support in designing experiments with microbes. Dr Christina Theodoropoulos for supporting for fluorescence microscopy. Funding was received for this research from AHEAD project (Grant: AHEAD/PhD/R2/ENG/TECH/161), University Grants Commission of Sri Lanka, and Queensland University of Technology. APC was supported by ECR Seed Grant (2023) from School of Mechanical, Medical, and Process Engineering, Queensland University of Technology.References
- H.-C. Flemming, Biofouling in water systems – cases, causes and countermeasures, Appl. Microbiol. Biotechnol., 2002, 59(6), 629–640, DOI:10.1007/s00253-002-1066-9.
- J. W. Costerton, Introduction to biofilm, Int. J. Antimicrob. Agents, 1999, 11(3–4), 217–221, DOI:10.1016/S0924-8579(99)00018-7.
- D. Davies, Understanding biofilm resistance to antibacterial agents, Nat. Rev. Drug Discovery, 2003, 2(2), 114–122, DOI:10.1038/nrd1008.
- M. Ribeiro, F. J. Monteiro and M. P. Ferraz, Infection of orthopedic implants with emphasis on bacterial adhesion process and techniques used in studying bacterial-material interactions, Biomatter, 2012, 2(4), 176–194, DOI:10.4161/biom.22905.
- J. S. Vrouwenvelder, D. A. Graf von der Schulenburg, J. C. Kruithof, M. L. Johns and M. C. M. van Loosdrecht, Biofouling of spiral-wound nanofiltration and reverse osmosis membranes: A feed spacer problem, Water Res., 2009, 43(3), 583–594, DOI:10.1016/j.watres.2008.11.019.
- L. M. Brown, et al., Community dynamics and phylogenetics of bacteria fouling Jet A and JP-8 aviation fuel, Int. Biodeterior. Biodegrad., 2010, 64(3), 253–261, DOI:10.1016/j.ibiod.2010.01.008.
- L. F. Melo and T. R. Bott, Biofouling in water systems, Exp. Therm. Fluid Sci., 1997, 14(4), 375–381, DOI:10.1016/S0894-1777(96)00139-2.
- M. Salta, L. Capretto, D. Carugo, J. A. Wharton and K. R. Stokes, Life under flow: A novel microfluidic device for the assessment of anti-biofilm technologies, Biomicrofluidics, 2013, 7(6), 064118, DOI:10.1063/1.4850796.
- E. P. Ivanova, et al., Natural Bactericidal Surfaces: Mechanical Rupture of Pseudomonas aeruginosa Cells by Cicada Wings, Small, 2012, 8(16), 2489–2494, DOI:10.1002/smll.201200528.
- S. Pogodin, et al., Biophysical model of bacterial cell interactions with nanopatterned cicada wing surfaces, Biophys. J., 2013, 104(4), 835–840, DOI:10.1016/j.bpj.2012.12.046.
- J. Hasan, et al., Selective bactericidal activity of nanopatterned superhydrophobic cicada Psaltoda claripennis wing surfaces, Appl. Microbiol. Biotechnol., 2013, 97(20), 9257–9262, DOI:10.1007/s00253-012-4628-5.
- C. D. Bandara, et al., Bactericidal Effects of Natural Nanotopography of Dragonfly Wing on Escherichia coli, ACS Appl. Mater. Interfaces, 2017, 9(8), 6746–6760, DOI:10.1021/acsami.6b13666.
- H. Lee, B. P. Lee and P. B. Messersmith, A reversible wet/dry adhesive inspired by mussels and geckos, Nature, 2007, 448(7151), 338–341, DOI:10.1038/nature05968.
- G. S. Watson, et al., A gecko skin micro/nano structure - A low adhesion, superhydrophobic, anti-wetting, self-cleaning, biocompatible, antibacterial surface, Acta Biomater., 2015, 21, 109–122, DOI:10.1016/j.actbio.2015.03.007.
- F. Dundar Arisoy, K. W. Kolewe, B. Homyak, I. S. Kurtz, J. D. Schiffman and J. J. Watkins, Bioinspired Photocatalytic Shark-Skin Surfaces with Antibacterial and Antifouling Activity via Nanoimprint Lithography, ACS Appl. Mater. Interfaces, 2018, 10(23), 20055–20063, DOI:10.1021/acsami.8b05066.
- V. K. Truong, M. Al Kobaisi, K. Vasilev, D. Cozzolino and J. Chapman, Current perspectives for engineering antimicrobial nanostructured materials, Curr. Opin. Biomed. Eng., 2022, 23, 100399, DOI:10.1016/j.cobme.2022.100399.
- T. Diu, et al., Cicada-inspired cell-instructive nanopatterned arrays, Sci. Rep., 2014, 4(1), 7122, DOI:10.1038/srep07122.
- K. Jindai, et al., Adhesion and bactericidal properties of nanostructured surfaces dependent on bacterial motility, RSC Adv., 2020, 10(10), 5673–5680, 10.1039/C9RA08282D.
- S. W. M. A. I. Senevirathne, A. Mathew, Y. Toh and P. K. D. V. Yarlagadda, Bactericidal Efficacy of Nanostructured Surfaces Increases under Flow Conditions, ACS Omega, 2022, 7(45), 41711–41722, DOI:10.1021/acsomega.2c05828.
- A. Jaggessar, S. W. M. A. I. Senevirathne, A. Velic and P. K. D. V. Yarlagadda, Antibacterial activity of 3D versus 2D TiO2 nanostructured surfaces to investigate curvature and orientation effects, Curr. Opin. Biomed. Eng., 2022, 100404, DOI:10.1016/j.cobme.2022.100404.
- D. R. Korber, J. R. Lawrence, L. Zhang and D. E. Caldwell, Effect of gravity on bacterial deposition and orientation in laminar flow environments, Biofouling, 1990, 2(4), 335–350, DOI:10.1080/08927019009378155.
- S. W. M. A. I. Senevirathne, Y.-C. Toh and P. K. D. V. Yarlagadda, Fluid Flow Induces Differential Detachment of Live and Dead Bacterial Cells from Nanostructured Surfaces, ACS Omega, 2022, 7(27), 23201–23212, DOI:10.1021/acsomega.2c01208.
- G. Chen, Y. Hong and S. L. Walker, Colloidal and bacterial deposition: Role of gravity, Langmuir, 2010, 26(1), 314–319, DOI:10.1021/la903089x.
- D. R. Walt, J. B. Smulow, S. S. Turesky and R. G. Hiller, The Effect of Gravity on Initial Microbial Adhesion, J. Colloid Interface Sci., 1985, 107(2), 334–336, DOI:10.1016/0021-9797(85)90185-7.
- V. Carniello, B. W. Peterson, H. C. van der Mei and H. J. Busscher, Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth, Adv. Colloid Interface Sci., 2018, 261, 1–14, DOI:10.1016/j.cis.2018.10.005.
- K. Yang, et al., Bacterial anti-adhesion surface design: Surface patterning, roughness and wettability: A review, J. Mater. Sci. Technol., 2022, 99, 82–100, DOI:10.1016/j.jmst.2021.05.028.
- N. Figueroa-Morales, et al., Living on the edge: transfer and traffic of E. coli in a confined flow, Soft Matter, 2015, 11(31), 6284–6293, 10.1039/C5SM00939A.
- J. Cremer, et al., Effect of flow and peristaltic mixing on bacterial growth in a gut-like channel, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(41), 11414–11419, DOI:10.1073/pnas.1601306113.
- S. W. M. A. I. Senevirathne, J. Hasan, A. Mathew, M. Woodruff and P. K. D. V. Yarlagadda, Simulation of Bacterial Motion Under Flow Inside Micro Channel Using CFD and DPM, in Lecture Notes in Networks and Systems, 2022, pp. 95–108 Search PubMed.
- V. Tokárová, et al., Patterns of bacterial motility in microfluidics-confining environments, Proc. Natl. Acad. Sci. U. S. A., 2021, 1108(17), e2013925118, DOI:10.1073/pnas.2013925118/-/DCSupplemental.
- E. Secchi, et al., The effect of flow on swimming bacteria controls the initial colonization of curved surfaces, Nat. Commun., 2020, 11(1), 2851, DOI:10.1038/s41467-020-16620-y.
- I. Tuval, L. Cisneros, C. Dombrowski, C. W. Wolgemuth, J. O. Kessler and R. E. Goldstein, Bacterial Swimming and Oxygen Transport Near Contact Lines, 2005, [Online], Available: http://www.pnas.orgcgidoi10.1073pnas.0406724102 Search PubMed.
- A. Jaggessar and P. K. D. V. Yarlagadda, Modelling the growth of hydrothermally synthesised bactericidal nanostructures, as a function of processing conditions, Mater. Sci. Eng. C, 2020, 108, 110434, DOI:10.1016/j.msec.2019.110434.
- A. Jaggessar, T. Tesfamicheal, H. Wang, C. Yan and P. K. D. V. Yarlagadda, Investigation of mechanical properties and morphology of hydrothermally manufactured titanium dioxide nanostructured surfaces, Procedia Manuf., 2019, 30, 373–379, DOI:10.1016/j.promfg.2019.02.052.
- A. Jaggessar, A. Mathew, T. Tesfamichael, H. Wang, C. Yan and P. K. K. D. V. K. Yarlagadda, Bacteria Death and Osteoblast Metabolic Activity Correlated to Hydrothermally Synthesised TiO2 Surface Properties, Molecules, 2019, 24(7), 1201, DOI:10.3390/molecules24071201.
- R. Bright, et al., Bio-Inspired Nanostructured Ti-6Al-4V Alloy: The Role of Two Alkaline Etchants and the Hydrothermal Processing Duration on Antibacterial Activity, Nanomater., 2022, 12(7), 1140, DOI:10.3390/nano12071140.
- M. I. Bogachev, et al., Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images, PLoS One, 2018, 13(5), 1–24, DOI:10.1371/journal.pone.0193267.
- J. A. F. Sutton, O. T. Carnell, L. Lafage, J. Gray, J. Biboy and J. F. Gibson, et al., Staphylococcus aureus cell wall structure and dynamics during host-pathogen interaction, PLoS Pathog., 2021, 17(3), e1009468, DOI:10.1371/journal.ppat.1009468.
- M. Wang, G. Buist and J. M. van Dijl, Staphylococcus aureus cell wall maintenance – the multifaceted roles of peptidoglycan hydrolases in bacterial growth, fitness, and virulence, FEMS Microbiol. Rev., 2022, 46(5), 1–19, DOI:10.1093/femsre/fuac025.
- M. Idrees, S. Sawant, N. Karodia and A. Rahman, Staphylococcus aureus biofilm: Morphology, genetics, pathogenesis and treatment strategies, Int. J. Environ. Res. Public Health, 2021, 18(14), 7602, DOI:10.3390/ijerph18147602.
- J. M. Monteiro, et al., Cell shape dynamics during the staphylococcal cell cycle, Nat. Commun., 2015, 6, 8055, DOI:10.1038/ncomms9055.
- S. P. Diggle and M. Whiteley, Microbe profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat, Microbiol., 2020, 166(1), 30–33, DOI:10.1099/mic.0.000860.
- D. P. Linklater, S. Juodkazis, R. J. Crawford and E. P. Ivanova, Mechanical inactivation of Staphylococcus aureus and Pseudomonas aeruginosa by titanium substrata with hierarchical surface structures, Mater., 2019, 5, 100197, DOI:10.1016/j.mtla.2018.100197.
- J. Hasan, S. Jain and K. Chatterjee, Nanoscale Topography on Black Titanium Imparts Multi-biofunctional Properties for Orthopedic Applications, Sci. Rep., 2017, 7, 1–13, DOI:10.1038/srep41118.
- R. Rosenzweig, K. Perinbam, V. K. Ly, S. Ahrar, A. Siryaporn and A. F. Yee, Nanopillared Surfaces Disrupt Pseudomonas aeruginosa Mechanoresponsive Upstream Motility, ACS Appl. Mater. Interfaces, 2019, 11(11), 10532–10539, DOI:10.1021/acsami.8b22262.
- S. M. Vater, et al., Swimming behavior of Pseudomonas aeruginosa studied by holographic 3D tracking, PLoS One, 2014, 9(1), e87765, DOI:10.1371/journal.pone.0087765.
- A. J. T. M. Mathijssen, N. Figueroa-Morales, G. Junot, É. Clément, A. Lindner and A. Zöttl, Oscillatory surface rheotaxis of swimming E. coli bacteria, Nat. Commun., 2019, 10(1), 3434, DOI:10.1038/s41467-019-11360-0.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00581j |
| This journal is © The Royal Society of Chemistry 2023 |