Open Access Article
Open Access ArticleDNA nanoparticles targeting FOXO4 selectively eliminate cigarette smoke-induced senescent lung fibroblasts†
Yaopin
Han
a,
Yixing
Wu
a,
Binfeng
He
ab,
Di
Wu
c,
Jianlan
Hua
a,
Hang
Qian
 *c and
Jing
Zhang
*a
*c and
Jing
Zhang
*a
aDepartment of Pulmonary and Critical Care Medicine, Zhongshan Hospital, Fudan University, Shanghai 200032, China. E-mail: zhang.jing@zs-hospital.sh.cn
bDepartment of General Practice, Xinqiao Hospital, Third Military Medical University, Chongqing 400037, China
cInstitute of Respiratory Diseases, Department of Pulmonary and Critical Care Medicine, Xinqiao Hospital, Third Military Medical University, Chongqing 400037, China. E-mail: hqian@tmmu.edu.cn
First published on 5th October 2023
Abstract
The pathogenesis and development of chronic obstructive pulmonary disease (COPD) are significantly related to cellular senescence. Strategies to eliminate senescent cells have been confirmed to benefit several senescence-related diseases. However, there are few reports of senolytic drugs in COPD management. In this study, we demonstrated elevated FOXO4 expression in cigarette smoke-induced senescent lung fibroblasts both in vitro and in vivo. Additionally, self-assembled DNA nanotubes loaded with single-stranded FOXO4 siRNA (siFOXO4-NT) were designed and synthesized to knockdown FOXO4 in senescent fibroblasts. We found that siFOXO4-NT can concentration- and time-dependently enter human lung fibroblasts (HFL-1 cells), thereby reducing FOXO4 levels in vitro. Most importantly, siFOXO4-NT selectively cleared senescent HFL-1 cells by reducing BCLXL expression and the BCL2/BAX ratio, which were increased in CSE-induced senescent HFL-1 cells. The findings from our work present a novel strategy for senolytic drug development for COPD therapy.
Introduction
Chronic obstructive pulmonary disease (COPD) is one of the leading causes of death, disability, and healthcare expenditures worldwide.1 It accounts for more than half of the morbidity associated with chronic respiratory diseases, and its prevalence is projected to rise due to an aging population.2 However, there has been a persistent lack of significant advancements in the types of drugs used to manage COPD. While bronchodilators and mucolytics can alleviate symptoms, they do not effectively halt the progression of the disease.3 Anti-inflammatory drugs, such as inhaled corticosteroids and phosphodiesterase-4 inhibitors, provide benefits only to a small subset of individuals with substantial airway inflammation.4 Therefore, the development of novel therapeutic drugs targeting the underlying pathogenesis of COPD is urgently needed.Repeated cell division or various stressors can lead to an irreversible arrest in the cell cycle, which is known as cellular senescence.5 Since senescent cells often retain metabolic activity, they can impact the local tissue environment by releasing the senescence-associated secretory phenotype (SASP).6 Through autocrine, paracrine, and/or endocrine mechanisms, SASP, in turn, can promote further senescence.7 The development of COPD is now recognized to be closely associated with cellular senescence. Endothelial cells and epithelial cells in the COPD lungs had increased DNA damage foci8 and exhibited various signs of senescence, such as elevated senescence-related-β-galactosidase (SA-β-GAL) activity and increased expression of p21Cip1/Waf1 and p16INK4a.9 Moreover, COPD patients display heightened SASP expression, including IL-1, IL-6, CCL2, and MMP9.10,11
Fibroblasts participate in tissue homeostasis and disease development through the production of extracellular matrices (ECMs).12 Following lung epithelium damage, fibroblasts become activated and locally proliferate. These activated fibroblasts can then migrate to the injury sites and differentiate into myofibroblasts, which possess contractile functions and the ability to produce ECMs, thereby contributing to tissue repair.13 However, studies have revealed that primary fibroblasts derived from the lungs of moderate to severe emphysema patients exhibit significantly higher SA-β-Gal activity, indicating cellular senescence in emphysema patients.14 Furthermore, research has demonstrated that senescent fibroblasts from COPD lungs display impaired migration and repair functions. Additionally, these alterations in fibroblast function were found to be correlated with lung function and emphysema severity.15 Collectively, the senescence of lung fibroblasts may contribute to the pathogenesis and progression of COPD.16,17
Once senescence occurs, cells show resistance to apoptosis. Targeting anti-apoptotic pathways with subsequent clearance by phagocytosis is the major strategy for the elimination of senescent cells, which is also known as senolytic therapy.3 Various drugs, both existing and newly discovered, have been found to reduce the accumulation of senescent cells. They include Dasatinib, Quercetin, BCL2 inhibitors, UBX0101, heat shock protein family inhibitors, and others.18–21 Preclinical studies have demonstrated that these senolytic drugs can effectively treat or improve senescent-associated diseases such as osteoarthritis, Alzheimer's disease, and chronic kidney disease.22–24 Given that the anti-apoptotic pathway patterns of senescent cells vary across different cell types and inducing factors,10 the development of senolytic drugs specifically targeting senescent cells associated with COPD is necessary.
Therapeutic RNA interference (RNAi) molecules possess high selectivity in suppressing pathologically upregulated gene function.25 Compared to traditional small-molecule and monoclonal antibody drugs, RNAi drugs are relatively easy to design and synthesize without the need for cellular expression systems, complex protein purification, or refolding schemes. As a result, their development entails reduced cost and time.26,27 The Watson–Crick base-pairing properties of DNA make it ideal for the design of nanostructures with a predictable size and shape, enabling complete synthesis through self-assembly.28 DNA nanostructures have found widespread application in drug delivery due to their structural stability and biodegradable, nonimmunogenic properties.29 Moreover, the delivery of siRNA using DNA nanostructures has been demonstrated to effectively protect nucleic acids against enzymatic degradation, while offering high biocompatibility and low toxicity.30 Lin et al. reported that tetrahedral DNA nanostructures have a clearing effect on senescent dermal fibroblasts by suppressing the antiapoptotic protein BCL2 and upregulating BAX expression.31 However, it is still unclear how DNA nanostructures interact with human lung fibroblasts and whether DNA nanostructures carrying drugs can effectively clear senescent lung fibroblasts for COPD therapy.
The FOX family is a class of transcription factors, which contains 19 subgroups ranging from FOXA to FOXS.32 As a member of the FOXO subgroup, FOXO4 is of great importance to a number of cellular processes, including oxidative stress, cell cycle, apoptosis, and cell homeostasis.33 FOXO4 is paradoxically involved in apoptosis regulation. It was found that FOXO4 induces apoptosis through up-regulating BIM, BAX and BCL6.33,34 However, the activation of FOXO4 with a low level of oxidative stress could attenuated BCL6 and BIM-mediated cell apoptosis.35 Recently, FOXO4 was proved to interact with p53 in senescent cells and prevent the nuclear exclusion of active p53, thereby inhibiting p53-mediated apoptosis.36
In this study, we conducted a screening of a novel therapeutic target, FOXO4, in senescent fibroblasts induced by cigarette smoke extract (CSE) using RNA-sequencing. We subsequently validated the expression of FOXO4 in both senescent fibroblasts and a mouse model of COPD. Furthermore, a FOXO4 siRNA-loaded self-assembled DNA nanoparticle was designed and synthesized for the selective elimination of senescent lung fibroblasts (Fig. 1). The potential of FOXO4 as a molecular target for senescent fibroblasts in COPD, as well as the efficacy of DNA nanostructures as carriers for siRNA-based gene therapy, was further validated in vitro.
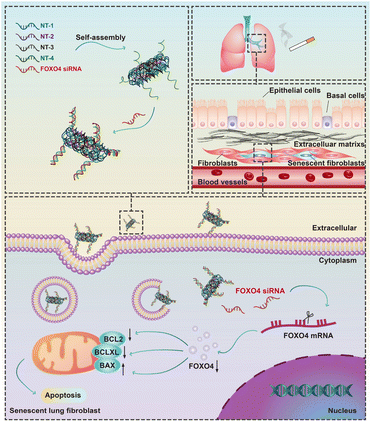 | ||
| Fig. 1 Schematic illustration of the elimination of cigarette smoke induced senescent lung fibroblasts using FOXO4 siRNA-loaded DNA nanoparticles. | ||
Materials and methods
Materials and reagents
DNA oligonucleotides, purified by HPLC, were obtained from Sangon (Shanghai, China). FOXO4 siRNA (5′-UCU UGC UAC AAA GCC UAA GUG-3′) and FOXO4 siRNA with Cy5 modification at the 5′-end were purchased from Tsingke (Beijing, China). Polyacrylamide (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for acrylamide
1 for acrylamide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) bisacrylamide), 10× Tris-Acetic-EDTA (TAE) buffer and Stain-All were purchased from Sangon (Shanghai, China). Lipofectamine™ 3000 were obtained from Thermo Fisher (Springfield Township, NJ, USA). Dulbecco's Modified Eagle Medium (DMEM), Opti-MEM™ Reduced Serum Medium and fetal bovine serum (FBS) were obtained from GIBCO (Shanghai, China). Senescence β-Galactosidase Staining Kit (CST, #9860) was purchased from Cell Signaling Technology (Danvers, MA, USA). Human inflammatory CK CBA kits were obtained from BD bioscience (Shanghai, China). Chlorpromazine, LY294002 and Nystatin were obtained from MedChemExpress (Shanghai, China). The Calcein/PI Assay Kit and Cell counting kit-8 (CCK8) were purchased from Beyotime (Jiangsu, China). RNAiso Plus was obtained from Takara (Beijing, China). The total RNA isolation kit was obtained from Tiangen (Beijing, China). The PrimeScript™ RT reagent Kit was obtained from Takara (Beijing, China). Hieff® qPCR SYBR Green Master Mix was purchased from YEASEN (Shanghai, China).
bisacrylamide), 10× Tris-Acetic-EDTA (TAE) buffer and Stain-All were purchased from Sangon (Shanghai, China). Lipofectamine™ 3000 were obtained from Thermo Fisher (Springfield Township, NJ, USA). Dulbecco's Modified Eagle Medium (DMEM), Opti-MEM™ Reduced Serum Medium and fetal bovine serum (FBS) were obtained from GIBCO (Shanghai, China). Senescence β-Galactosidase Staining Kit (CST, #9860) was purchased from Cell Signaling Technology (Danvers, MA, USA). Human inflammatory CK CBA kits were obtained from BD bioscience (Shanghai, China). Chlorpromazine, LY294002 and Nystatin were obtained from MedChemExpress (Shanghai, China). The Calcein/PI Assay Kit and Cell counting kit-8 (CCK8) were purchased from Beyotime (Jiangsu, China). RNAiso Plus was obtained from Takara (Beijing, China). The total RNA isolation kit was obtained from Tiangen (Beijing, China). The PrimeScript™ RT reagent Kit was obtained from Takara (Beijing, China). Hieff® qPCR SYBR Green Master Mix was purchased from YEASEN (Shanghai, China).
Antibodies are as follows: FOXO4 (Abmart, #T55892), FOXO4 (CST, #9472), p16Ink4a (CST, #18769), p21Cip1/Waf1 (Abmart, #T55088), Vimentin (Abcam, #ab92547) and BAX (Abcam, #ab32503), BCL-XL (Abmart, #T55050), BCL2 (Abmart, #T40056) and β-TUBULIN (Abmart, #M20005). Secondary antibody included goat anti-mouse and rabbit IgG (Abmart, #M21003).
CSE preparation
CSE was prepared by bubbling commercial cigarettes (Shanghai, China) into DMEM medium in accordance with a previous report (draw of 10 mL and 1 cigarette per 3 min), using a vacuum cigarette smoking pump.37 Each of the cigarette contains 12 mg tar and 2.5 mg nicotine. The CSE solution was adjusted to an OD(A320–A540) of 0.9 to 1.2, and a pH of 7.4. Sterilized CSE solution was obtained by filtering with a 0.22 μM filter. One cigarette was first used to prepare a 100% concentration of stock solution, which was then diluted to 2.5% concentration in DMEM medium for 1 h before experiments.Cell culture
Human lung fibroblasts (HFL-1 cells), provided by Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China), were cultured in high glucose DMEM, containing a 1% penicillin and streptomycin mixture and 10% FBS, and specifically used between passages 3 and 5. The medium was changed daily. To establish the CSE-induced senescent cell model, HFL-1 cells were cultured with 2.5% concentration of CSE for 7 consecutive days. The senescent cell model was identified by SA-β-GAL staining, western blot, quantitative real-time PCR (qPCR) and SASP detection.Animal experimental protocol
All the mice (6 week-old, male, C57BL/6) were obtained from the SLAC Laboratory Animals (Shanghai, China), and were bred in the animal facilities at Zhongshan Hospital. In total, 10 mice were randomly selected. The mice model of COPD was established according to the protocol of our previous study.37 Briefly, mice (n = 5) were exposed to 20 commercial cigarettes for 2 h, twice a day, 6 days a week for 6 months. Age-matched mice that were exposed to room air served as non-smoking controls. Lungs of mice were used for western blot, immunohistochemistry (IHC) and immunofluorescence (IF) staining. All animal experiments were conducted in accordance with the “Laboratory animal Guideline for ethical review of animal welfare” (People's Republic of China National Standard GB/T 35892-2018) and the “Regulations on the administration of laboratory animals of Zhongshan Hospital, Fudan University”. All experimental protocols were approved by the Animal Care and Use Committee of Zhongshan Hospital (Approved number: B2021-217A).Preparation of DNA nanoparticles
The computer program SEQUIN was used to design DNA sequences, which are listed in Table S1.† The DNA strand solutions were diluted with nuclease-free water and mixed in 10× TAE/Mg2+ buffer (pH 8.0, 125 mM Mg2+) at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 (NT-1
3 (NT-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NT-2
NT-2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NT-3
NT-3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) NT-4). To form DNA nanotubes, the mixture was cooled as follows: 95 °C/5 min, 65 °C/15 min, 37 °C/15 min, and 22 °C/30 min. We screened out siFOXO4-2, which could most efficiently inhibit FOXO4 expression in HFL-1 cells, from three FOXO4 siRNA with different sequences for subsequent synthesis (Table S2 and Fig. S1†). FOXO4 siRNA (5′-UCU UGC UAC AAA GCC UAA GUG-3′) or Cy5 labeled FOXO4 siRNA was then added to the nanotube solution at a 6
NT-4). To form DNA nanotubes, the mixture was cooled as follows: 95 °C/5 min, 65 °C/15 min, 37 °C/15 min, and 22 °C/30 min. We screened out siFOXO4-2, which could most efficiently inhibit FOXO4 expression in HFL-1 cells, from three FOXO4 siRNA with different sequences for subsequent synthesis (Table S2 and Fig. S1†). FOXO4 siRNA (5′-UCU UGC UAC AAA GCC UAA GUG-3′) or Cy5 labeled FOXO4 siRNA was then added to the nanotube solution at a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio. For the combination of siRNA and DNA nanotubes, the mixture was incubated at 37 °C for more than 10 min.
1 molar ratio. For the combination of siRNA and DNA nanotubes, the mixture was incubated at 37 °C for more than 10 min.
Characterization of DNA nanoparticles
For native polyacrylamide gel electrophoresis (PAGE) characterization, DNA nanoparticles were run on 6% polyacrylamide gels on the electrophoresis unit at 120 V for 60 min in 1× TAE/Mg2+ running buffer. The gels were finally scanned after staining with Stain-All. For serum stability assay, nanoparticles were incubated with 10% FBS for different times, and then detected with 6% native PAGE.Dynamic Light Scattering (Malvern, UK) was used to measure the size distribution and zeta potential right after diluting the DNA nanoparticle solution to a concentration of approximately 100 nM and 1000 μg L−1, respectively.
For Atomic Force Microscope (AFM) imaging, siFOXO4-NT was added dropwise onto a mica surface and was then washed using Mg2+ solution (2 mM) and dried with compressed nitrogen after 2 min of incubation. Subsequently, TAE/Mg2+ buffer was added onto the surface of the mica for AFM imaging in liquid mode. DNA nanoparticles were imaged by using a Multimode 8 AFM system (Bruker, USA). The mode and parameters were set according to our previous research.38
Cell uptake of DNA nanoparticles
Senescent HFL-1 cells were seeded on glass bottom dishes or 6-well dishes overnight. These cells were incubated with different concentrations of siFOXO4-NT-Cy5 in the presence of Lipofectame-3000 for indicated times. For endocytosis pathway detection, senescent HFL-1 cells were pre-exposed with different endocytosis inhibitors for 30 min, including chlorpromazine (10 μg mL−1), LY294002 (20 μg mL−1) and nystatin (10 μg mL−1), and then were treated with or without 100 nM siFOXO4-NT and incubated for 6 h. Cells incubated with siFOXO4-NT in the absence of inhibitors served as the positive control, and cells transfected at 4 °C served as the negative control, while cells incubated without siFOXO4-NT served as the blank control.For Confocal Laser Scanning Microscope (CLSM) analysis, HFL-1 cells were fixed with 4% paraformaldehyde for 15 min. DAPI was used to stain cell nuclei. Images were obtained by CLSM (Olympus, Tokyo, Japan). For the flow cytometry (FACS) analysis, cells were removed by trypsinization and fixed in 200 μL 4% paraformaldehyde. FACS was used to measure the samples. Flow Jo software (BD, Version 10.0) was used to analyse the data.
Live/dead staining assay
Calcein and Propidium Iodide (PI) probes were used to identify live cells (green) and apoptotic cells (red), respectively. Treated senescent cells were stained with calcein/PI for at least 30 min. Images were obtained by using a fluorescence microscope (Olympus, Tokyo, Japan) with 488 and 514 nm excitation channels.Statistical analysis
Data are presented as means ± standard deviation. Statistical analysis was performed with Prism software (Version 8.0.2). Student's t-test was applied for comparisons between two groups. One-way analysis of variance (ANOVA) was used to compare multiple group comparisons. All experiments were performed at least three times. P < 0.05 was statistically significant.Results and discussion
FOXO4 is a potential target for clearing senescent cells
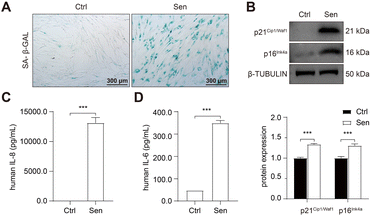
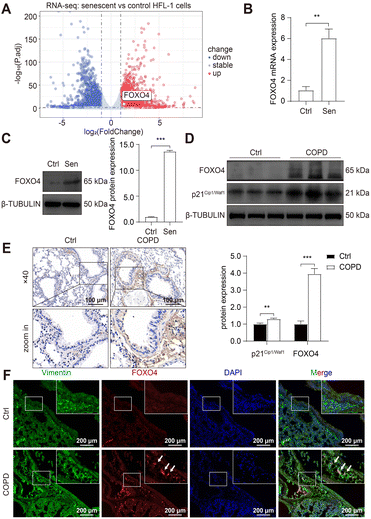
To develop senolytic reagents for COPD therapy, we established CSE-induced senescent fibroblasts by treating human lung fibroblasts 1 (HFL-1 cells) with 2.5% CSE for 7 days.39 The characteristics of lung fibroblast senescence included reduced cell proliferation, flattened and enlarged morphology, and increased p16Ink4a expression and upregulated SA-β-GAL activity.39 In addition, previous studies have confirmed an increase in SASP expression in COPD, including IL-1, IL-6, IL-8, TGF-β, and MMP-9.10,11,16,17 Similarly, we observed increased SA-β-GAL activity (Fig. 2A) and protein expression of p21Cip1/Waf1 and p16Ink4a (Fig. 2B) in senescent HFL-1 cells in our study. Furthermore, IL-6 and IL-8 secreted by CSE-exposed HFL-1 cells were significantly higher compared to the control cells (Fig. 2C and D). These findings provide sustained validation of cellular senescence induced by CSE in HFL-1 cells.Subsequently, RNA sequencing (RNA-seq) was conducted to identify up-regulated anti-apoptotic genes in CSE-induced senescent HFL-1 cells. Through differentially expressed genes (DEGs) analysis, we obtained a total of 2329 upregulated genes and 1963 downregulated genes in the senescent group (Fig. 3A). Interestingly, most of the known senolytic targets, such as BCL2, BCL2L1, BCL2L2, MDM2, USP7, HSP90AA1, BIRC2, and BIRC3, were not significantly upregulated in CSE-induced senescent HFL-1 cells except for FOXO4.40–45 Next, gene ontology analysis was performed on the upregulated DEGs in senescent HFL-1 cells. Unfortunately, none of the enriched GO terms related to “negative regulation of apoptosis” reached the adjusted P-value threshold of <0.05 (Fig. S2†). Interestingly, both the mRNA expression levels of FOXO4 (Fig. 3B) and its protein expression (Fig. 3C) were found to be elevated in CSE-induced senescent HFL-1 cells. DEG analysis of GSE151052 also revealed that FOXO4 expression was upregulated in the lung tissue of COPD patients (Fig. S3†). Consequently, we decided to focus our study on FOXO4.
For further validation, we established a COPD mice model by exposing the mice to cigarette smoke for 6 months, as per our previous study.37 This exposure resulted in disrupted airway epithelial structures and alveolar spaces, confirming COPD development in the mice. We observed elevated p21Cip1/Waf1 expression in smoking-exposed mice lungs, indicating the presence of senescent lung tissues in COPD mice and supporting the potential of senotherapeutic strategies for COPD treatment. Furthermore, compared to the control group, we noticed increased FOXO4 protein expression in the smoking-exposure group (Fig. 3D). Immunohistochemistry (IHC) assay also revealed higher expression of FOXO4 in the lung interstitium of smoking-exposed mice (Fig. 3E). Using dual immunofluorescence staining, we determined that FOXO4 co-localized with vimentin, a cytoplasmic intermediate filament protein expressed in various mesenchymal cells, including fibroblasts.46 This co-localization indicated that FOXO4 was predominantly located in the interstitial area of the smoking-exposed lung (Fig. 3F). These findings strongly support the potential of targeting FOXO4 to eliminate cigarette-induced senescent lung fibroblasts as a treatment approach for COPD.
Design and characterization of FOXO4 siRNA loaded DNA nanotubes
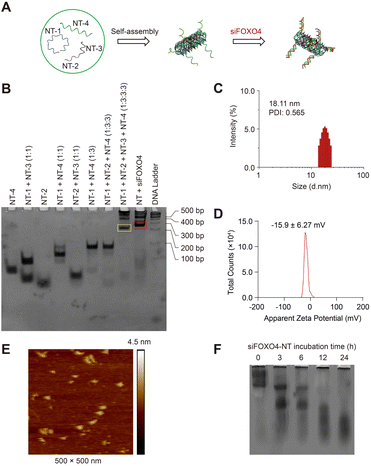
In this study, we utilized single-stranded FOXO4 siRNA (antisense, siFOXO4) loaded onto a DNA self-assembled nanostructure, as previously described.47 Four unique DNA strands, designed using the computer program SEQUIN, were mixed in specific molar ratios to form a six-helix bundle DNA nanotube (NT) through self-assembly. This nanotube was capable of carrying six copies of siFOXO4 through DNA-RNA hybridization (Fig. 4A). The characterization of siFOXO4-NT was performed as follows: the native PAGE gel analysis demonstrated a clear and distinct band at the expected position for both the nanotube and siFOXO4-NT, indicating high assembly yields (Fig. 4B). The siFOXO4-NT nanoparticles exhibited a hydrodynamic diameter of approximately 18.11 nm (Fig. 4C). The surface zeta potential of siFOXO4-NT in TAE/Mg2+ buffer was determined to be −15.6 ± 6.27 mV, which is well consistent with previous research results (Fig. 4D).48 Furthermore, atomic force microscopy (AFM) imaging of the siFOXO4-NT nanoparticles revealed a relatively uniform appearance with good dispersion (Fig. 4E). To evaluate the physiological stability of siFOXO4-NT, the nanoparticles were mixed with 10% FBS and incubated at 37 °C for different time intervals. Analysis using native PAGE showed that siFOXO4-NT remained intact up to 6 h (Fig. 4F). The cytotoxicity of DNA nanotube solution in CSE-induced senescent HFL-1 cells is presented in Fig. S4.† The IC50 of DNA nanotubes was found to be 1457 nM after 62 h incubation, indicating high biosafety.Cellular uptake and effect of siFOXO4-NT on senescent HFL-1 cells
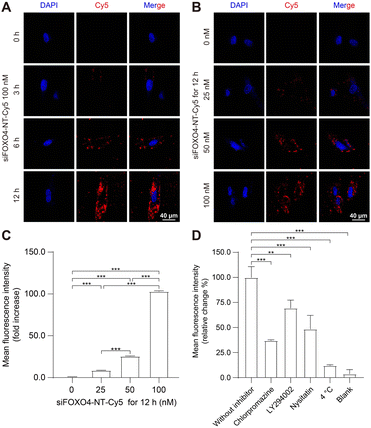
The capacity of nanoparticles to enter cells is one of the main factors influencing the therapeutic effect.49 Nanoparticles can interact with the extracellular matrix or cell membrane, and subsequently enter the cell mainly by endocytosis.50 Previously, a similar six-arm DNA nanotube was utilized to deliver mTOR siRNA. It was shown that this siRNA-loaded nanotube effectively inhibited mTOR expression in pulmonary smooth muscle cells after being internalized through endocytosis.47To assess the cellular uptake of siFOXO4-NT, the FOXO4 siRNA labelled with Cy5 was introduced into the DNA nanotube for drug tracking. The results of cellular uptake observed by CLSM in CSE-induced senescent HFL-1 cells showed a significant enhancement in red fluorescence produced by siFOXO4-NT-Cy5. The fluorescent particles were predominantly located around the cell membrane after 3 hours and 6 hours of incubation. After a 12 hour incubation, a large proportion of red fluorescent particles appeared in the cytoplasmic and perinuclear region (Fig. 5A). Moreover, a significant increase of fluorescence intensity was observed with increasing concentrations of siFOXO4-NT-Cy5 (Fig. 5B). FACS analysis also demonstrated dose-dependent uptake of siFOXO4-NT-Cy5 by senescent HFL-1 cells (Fig. 5C). Endocytosis is an energy-consuming process that can be inhibited at low temperatures. In this study, FACS analysis showed the inhibition of the endocytosis of senescent HFL-1 cells against siFOXO4-NT-Cy5 by 4 °C incubation.30 Additionally, the internalization of siFOXO4-NT was also found to be inhibited by endocytosis inhibitors, including chlorpromazine, LY294002 and nystatin, which inhibit clathrin-mediated endocytosis, macropinocytosis and caveolae-mediated endocytosis, respectively (Fig. 5D).49,51
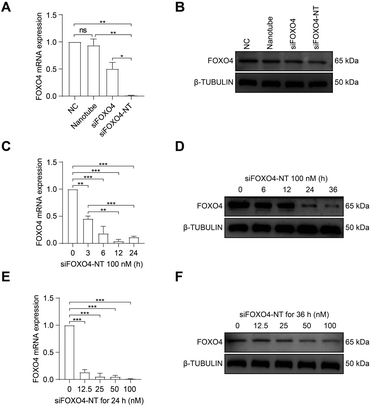
Subsequently, the effect of siFOXO4-NT on FOXO4 expression in CSE-induced senescent HFL-1 cells was evaluated. Both naked FOXO4 siRNA and siFOXO4-NT were able to reduce the expression level of FOXO4, with siFOXO4-NT showing a greater effectiveness than naked FOXO4 siRNA (Fig. 6A and B). As expected, it was also observed that FOXO4 expression decreased with increasing siFOXO4-NT incubation time (Fig. 6C and D) and concentration (Fig. 6E and F).
Taken together, these results confirm that siFOXO4-NT can be efficiently endocytosed by CSE-induced senescent HFL-1 cells mainly through a clathrin-dependent pathway, leading to the intracellular delivery of siRNA and subsequent silencing of FOXO4 expression.
siFOXO4-NT selectively eliminates CSE-induced senescent HFL-1 cells
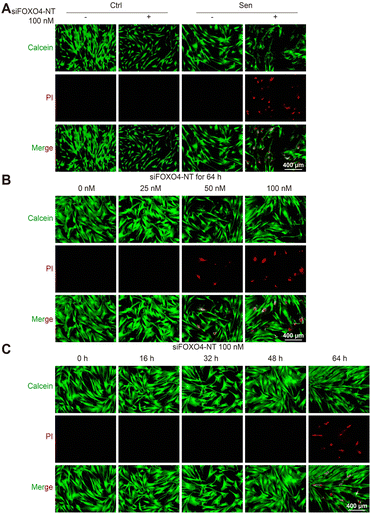
Subsequently, calcein/PI staining was used to detect cell activity. In comparison to the group without siFOXO4-NT treatment, there was an increased number of PI-stained cells observed in CSE-induced senescent HFL-1 cells after 64 hours of siFOXO4-NT incubation. However, siFOXO4-NT had no apparent effect on the cell activity of HFL-1 cells not exposed to CSE (Fig. 7A). Based on the representative images in Fig. 7B and C, it is suggested that siFOXO4-NT can selectively eliminate CSE-induced senescent HFL-1 cells in a dose- and time-dependent manner. CCK8 assay was then used to evaluate the effect of siFOXO4-NT on the growth of CSE-induced senescent HFL-1 cells. The IC50 of siFOXO4-NT was found to be 157.7 nM after 62 h incubation (Fig. S5†).Notably, although the senolytic strategy has been shown to be beneficial in many cases, the elimination of senescent pulmonary endothelial cells through intraperitoneal injection of FOXO4-DRI or oral ABT-263 has been recently confirmed to worsen pulmonary hemodynamics in pulmonary hypertension mouse models.52 These results demonstrate the significant side effects on pulmonary vessels associated with in vivo application of senolytic drugs. For our strategy to control senescent lung fibroblasts in COPD therapy, pulmonary delivery of senolytic drugs may help avoid direct effects of senolytic drugs on pulmonary vessels. Compared to systemic administration, pulmonary administration has been shown to be more acceptable to patients and reduces drug clearance in the liver and kidneys.4,53 Moreover, delivering drugs directly to the lungs can also protect RNAi drugs from degradation by RNA enzymes in the blood.54 Remarkably, senescent cells, which are currently believed to be involved in disease pathogenesis, may have unknown beneficial functions. Thus, the indiscriminate elimination of senescent cells may lead to side effects. Previous studies have revealed the protective influence of senescent endothelial cells against liver and pulmonary vascular remodeling.55,56 Therefore, the roles of senescent cells in COPD pathogenesis, especially the beneficial ones, remain to be elucidated through future studies. Additionally, specific modifications of the drug itself or coated nanocarriers will help identify target cells and enable precise delivery of senolytic drugs, thereby avoiding off-target effects on normal cells or other senescent cells.
siFOXO4-NT selectively eliminates CSE-induced senescent HFL-1 cells through promoting apoptosis
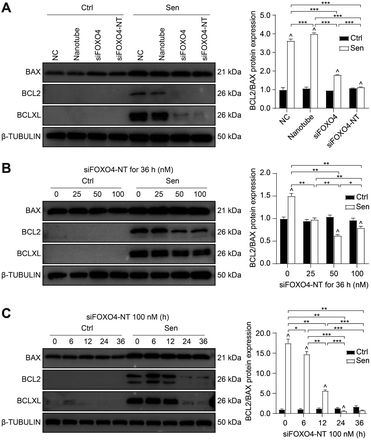
Apoptosis resistance is one of the main characteristics of senescent cells.3 In this study, CSE-induced senescent HFL-1 cells exhibited higher expression of BCLXL and a higher BCL2/BAX ratio compared to HFL-1 cells without CSE treatment. This suggests that senescent HFL-1 cells are resistant to apoptosis (Fig. 8A). Both naked FOXO4 siRNA and siFOXO4-NT significantly decreased BCLXL expression and the BCL2/BAX ratio in senescent HFL-1 cells. However, for HFL-1 cells without CSE treatment, the effect of FOXO4 siRNA and siFOXO4-NT on the BCL2/BAX ratio was negligible. These results indicate that reducing FOXO4 expression through FOXO4 siRNA or siFOXO4-NT can promote apoptosis in CSE-induced senescent HFL-1 cells without affecting normal HFL-1 cells. Furthermore, the ability of siFOXO4-NT to promote apoptosis in senescent HFL-1 cells was dependent on siFOXO4-NT concentration and the duration of incubation (Fig. 8B and C).Conclusions
In summary, our study screened and identified FOXO4 as a therapeutic target for eliminating senescent fibroblasts in COPD therapy. We further demonstrated that silencing FOXO4 promotes apoptosis in HFL-1 cells through the BCLXL-BAX signaling pathway. Additionally, as far as we know, the selective removal of CSE-induced senescent fibroblasts using a self-assembled DNA nanomaterial is reported for the first time. The DNA nanomaterial exhibited excellent properties, including higher cellular uptake efficiencies and specific clearance of senescent cells. This study demonstrates that utilizing DNA nanomaterials for the elimination of senescent lung tissue cells may be a novel strategy for COPD therapy.Author contributions
B. F. He, H. Qian and J. Zhang conceived the idea and designed the experiments. Y. P. Han performed the experiments. D. Wu performed the characterization analysis. Y. X. Wu and J. L. Hua helped in the cell experiments. Y. P. Han analyzed the experimental data and wrote the manuscript. B. F. He, H. Qian and J. Zhang revised the manuscript. J. Zhang provided all experimental resources and supervised the project. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC) (82270039 and 81970035).Notes and references
- C. A. Brandsma, M. de Vries, R. Costa, R. R. Woldhuis, M. Königshoff and W. Timens, Eur. Respir. Rev., 2017, 26, 170073 CrossRef PubMed.
- S. A. Christenson, B. M. Smith, M. Bafadhel and N. Putcha, Lancet, 2022, 399, 2227–2242 CrossRef PubMed.
- J. R. Baker, L. E. Donnelly and P. J. Barnes, Chest, 2020, 158, 562–570 CrossRef CAS PubMed.
- Y. Han, J. Hua, B. He and J. Zhang, J. Transl. Int. Med., 2022, 10, 181–184 CrossRef PubMed.
- J. Birch and J. Gil, Genes Dev., 2020, 34, 1565–1576 CrossRef CAS PubMed.
- P. J. Barnes, J. Baker and L. E. Donnelly, Am. J. Respir. Crit. Care Med., 2019, 200, 556–564 CrossRef CAS PubMed.
- A. Calcinotto, J. Kohli, E. Zagato, L. Pellegrini, M. Demaria and A. Alimonti, Physiol. Rev., 2019, 99, 1047–1078 CrossRef CAS PubMed.
- J. Birch, R. K. Anderson, C. Correia-Melo, D. Jurk, G. Hewitt, F. M. Marques, N. J. Green, E. Moisey, M. A. Birrell, M. G. Belvisi, F. Black, J. J. Taylor, A. J. Fisher, A. De Soyza and J. F. Passos, Am. J. Physiol. Lung Cell Mol. Physiol., 2015, 309, L1124–L1137 CrossRef CAS PubMed.
- J. Birch, P. J. Barnes and J. F. Passos, Pharmacol. Ther., 2018, 183, 34–49 CrossRef CAS PubMed.
- P. J. Barnes, Annu. Rev. Physiol., 2017, 79, 517–539 CrossRef CAS PubMed.
- P. J. Barnes, J. Allergy Clin. Immunol., 2016, 138, 16–27 CrossRef CAS PubMed.
- J. C. Hewlett, J. A. Kropski and T. S. Blackwell, Matrix Biol., 2018, 71–72, 112–127 CrossRef CAS PubMed.
- T. Parimon, M. S. Hohmann and C. Yao, Int. J. Mol. Sci., 2021, 22 Search PubMed.
- K. C. Müller, L. Welker, K. Paasch, B. Feindt, V. J. Erpenbeck, J. M. Hohlfeld, N. Krug, M. Nakashima, D. Branscheid, H. Magnussen, R. A. Jörres and O. Holz, Respir. Res., 2006, 7, 32 CrossRef PubMed.
- S. Togo, O. Holz, X. Liu, H. Sugiura, K. Kamio, X. Wang, S. Kawasaki, Y. Ahn, K. Fredriksson, C. M. Skold, K. C. Mueller, D. Branscheid, L. Welker, H. Watz, H. Magnussen and S. I. Rennard, Am. J. Respir. Crit. Care Med., 2008, 178, 248–260 CrossRef CAS PubMed.
- J. Zhang, L. Wu, J. M. Qu, C. X. Bai, M. J. Merrilees and P. N. Black, J. Cell. Mol. Med., 2012, 16, 1522–1532 CrossRef CAS PubMed.
- J. Zhang, L. Wu, M. X. Feng, P. Sexton, C. X. Bai, J. M. Qu, M. Merrilees and P. N. Black, Respir. Physiol. Neurobiol., 2011, 177, 236–240 CrossRef CAS PubMed.
- L. J. Hickson, L. G. P. Langhi Prata, S. A. Bobart, T. K. Evans, N. Giorgadze, S. K. Hashmi, S. M. Herrmann, M. D. Jensen, Q. Jia, K. L. Jordan, T. A. Kellogg, S. Khosla, D. M. Koerber, A. B. Lagnado, D. K. Lawson, N. K. LeBrasseur, L. O. Lerman, K. M. McDonald, T. J. McKenzie, J. F. Passos, R. J. Pignolo, T. Pirtskhalava, I. M. Saadiq, K. K. Schaefer, S. C. Textor, S. G. Victorelli, T. L. Volkman, A. Xue, M. A. Wentworth, E. O. Wissler Gerdes, Y. Zhu, T. Tchkonia and J. L. Kirkland, EBioMedicine, 2019, 47, 446–456 CrossRef PubMed.
- J. Pan, D. Li, Y. Xu, J. Zhang, Y. Wang, M. Chen, S. Lin, L. Huang, E. J. Chung, D. E. Citrin, Y. Wang, M. Hauer-Jensen, D. Zhou and A. Meng, Int. J. Radiat. Oncol. Biol. Phys., 2017, 99, 353–361 CrossRef CAS PubMed.
- Y. Huang, Y. He, M. J. Makarcyzk and H. Lin, Front. Bioeng. Biotechnol., 2021, 9, 677576 CrossRef PubMed.
- D. S. Hong, U. Banerji, B. Tavana, G. C. George, J. Aaron and R. Kurzrock, Cancer Treat Rev., 2013, 39, 375–387 CrossRef CAS PubMed.
- M. M. Gonzales, S. Krishnamurthy, V. Garbarino, A. S. Daeihagh, G. J. Gillispie, G. Deep, S. Craft and M. E. Orr, Mech. Ageing Dev., 2021, 200, 111589 CrossRef CAS PubMed.
- J. A. Collins, B. O. Diekman and R. F. Loeser, Curr. Opin. Rheumatol., 2018, 30, 101–107 CrossRef CAS PubMed.
- C. Li, Y. Shen, L. Huang, C. Liu and J. Wang, Faseb. J., 2021, 35, e21229 CAS.
- G. M. Traber and A. M. Yu, J. Pharmacol. Exp. Ther., 2023, 384, 133–154 CrossRef CAS PubMed.
- M. Panigaj, M. B. Johnson, W. Ke, J. McMillan, E. A. Goncharova, M. Chandler and K. A. Afonin, ACS Nano, 2019, 13, 12301–12321 CrossRef CAS PubMed.
- Y. Qiu, J. K. Lam, S. W. Leung and W. Liang, Molecules, 2016, 21 Search PubMed.
- A. Rangnekar and T. H. LaBean, Acc. Chem. Res., 2014, 47, 1778–1788 CrossRef CAS PubMed.
- W. Ma, Y. Zhan, Y. Zhang, C. Mao, X. Xie and Y. Lin, Signal Transduct. Target Ther., 2021, 6, 351 CrossRef CAS PubMed.
- Y. Wang, Z. You, J. Du, H. Li, H. Chen, J. Li, W. Dong, B. He, C. Mao and G. Wang, J. Control. Release, 2016, 233, 126–135 CrossRef CAS PubMed.
- C. Mao, W. Pan, X. Shao, W. Ma, Y. Zhang, Y. Zhan, Y. Gao and Y. Lin, ACS Appl. Mater. Interfaces, 2019, 11, 1942–1950 CrossRef CAS PubMed.
- B. Bourgeois, T. Gui, D. Hoogeboom, H. G. Hocking, G. Richter, E. Spreitzer, M. Viertler, K. Richter, T. Madl and B. M. T. Burgering, Cell Rep., 2021, 36, 109446 CrossRef CAS PubMed.
- W. Wang, P. H. Zhou and W. Hu, Mol. Med. Rep., 2016, 13, 2229–2234 CrossRef CAS PubMed.
- T. T. Tang, D. Dowbenko, A. Jackson, L. Toney, D. A. Lewin, A. L. Dent and L. A. Lasky, J. Biol. Chem., 2002, 277, 14255–14265 CrossRef CAS PubMed.
- M. A. Essers, S. Weijzen, A. M. de Vries-Smits, I. Saarloos, N. D. de Ruiter, J. L. Bos and B. M. Burgering, EMBO J., 2004, 23, 4802–4812 CrossRef CAS PubMed.
- H. H. Le, S. S. Cinaroglu, E. C. Manalo, A. Ors, M. M. Gomes, B. Duan Sahbaz, K. Bonic, C. A. Origel Marmolejo, A. Quentel, J. S. Plaut, T. E. Kawashima, E. S. Ozdemir, S. V. Malhotra, Y. Ahiska, U. Sezerman, G. Bayram Akcapinar, J. C. Saldivar, E. Timucin and J. M. Fischer, EBioMedicine, 2021, 73, 103646 CrossRef CAS PubMed.
- B. F. He, Y. X. Wu, W. P. Hu, J. L. Hua, Y. Han and J. Zhang, Free Radic. Biol. Med., 2023, 195, 359–370 CrossRef CAS PubMed.
- Z. You, Q. Huang, L. Xu, X. Liu, J. Fu, B. Li, Y. Yang, S. Li, H. Qian and G. Wang, Chembiochem, 2022, 23, e202200344 CrossRef CAS PubMed.
- T. Nyunoya, M. M. Monick, A. Klingelhutz, T. O. Yarovinsky, J. R. Cagley and G. W. Hunninghake, Am. J. Respir. Cell Mol. Biol., 2006, 35, 681–688 CrossRef CAS PubMed.
- J. Chang, Y. Wang, L. Shao, R. M. Laberge, M. Demaria, J. Campisi, K. Janakiraman, N. E. Sharpless, S. Ding, W. Feng, Y. Luo, X. Wang, N. Aykin-Burns, K. Krager, U. Ponnappan, M. Hauer-Jensen, A. Meng and D. Zhou, Nat. Med., 2016, 22, 78–83 CrossRef CAS PubMed.
- Y. Fang, G. Liao and B. Yu, Acta Pharm. Sin. B, 2020, 10, 1253–1278 CrossRef CAS PubMed.
- O. H. Jeon, C. Kim, R. M. Laberge, M. Demaria, S. Rathod, A. P. Vasserot, J. W. Chung, D. H. Kim, Y. Poon, N. David, D. J. Baker, J. M. van Deursen, J. Campisi and J. H. Elisseeff, Nat. Med., 2017, 23, 775–781 CrossRef CAS PubMed.
- L. Kategaya, P. Di Lello, L. Rougé, R. Pastor, K. R. Clark, J. Drummond, T. Kleinheinz, E. Lin, J. P. Upton, S. Prakash, J. Heideker, M. McCleland, M. S. Ritorto, D. R. Alessi, M. Trost, T. W. Bainbridge, M. C. M. Kwok, T. P. Ma, Z. Stiffler, B. Brasher, Y. Tang, P. Jaishankar, B. R. Hearn, A. R. Renslo, M. R. Arkin, F. Cohen, K. Yu, F. Peale, F. Gnad, M. T. Chang, C. Klijn, E. Blackwood, S. E. Martin, W. F. Forrest, J. A. Ernst, C. Ndubaku, X. Wang, M. H. Beresini, V. Tsui, C. Schwerdtfeger, R. A. Blake, J. Murray, T. Maurer and I. E. Wertz, Nature, 2017, 550, 534–538 CrossRef CAS PubMed.
- H. Fuhrmann-Stroissnigg, Y. Y. Ling, J. Zhao, S. J. McGowan, Y. Zhu, R. W. Brooks, D. Grassi, S. Q. Gregg, J. L. Stripay, A. Dorronsoro, L. Corbo, P. Tang, C. Bukata, N. Ring, M. Giacca, X. Li, T. Tchkonia, J. L. Kirkland, L. J. Niedernhofer and P. D. Robbins, Nat. Commun., 2017, 8, 422 CrossRef PubMed.
- C. Schwarzenbach, L. Tatsch, J. Brandstetter Vilar, B. Rasenberger, L. Beltzig, B. Kaina, M. T. Tomicic and M. Christmann, Cancers, 2021, 13 Search PubMed.
- K. M. Ridge, J. E. Eriksson, M. Pekny and R. D. Goldman, Genes Dev., 2022, 36, 391–407 CrossRef CAS PubMed.
- Z. You, H. Qian, C. Wang, B. He, J. Yan, C. Mao and G. Wang, Biomaterials, 2015, 67, 137–150 CrossRef CAS PubMed.
- S. Zhao, R. Tian, J. Wu, S. Liu, Y. Wang, M. Wen, Y. Shang, Q. Liu, Y. Li, Y. Guo, Z. Wang, T. Wang, Y. Zhao, H. Zhao, H. Cao, Y. Su, J. Sun, Q. Jiang and B. Ding, Nat. Commun., 2021, 12, 358 CrossRef CAS PubMed.
- J. J. Rennick, A. P. R. Johnston and R. G. Parton, Nat. Nanotechnol., 2021, 16, 266–276 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, W. Tao, M. A. Hamaly, M. Y. Alkawareek, E. C. Dreaden, D. Brown, A. M. Alkilany, O. C. Farokhzad and M. Mahmoudi, Chem. Soc. Rev., 2017, 46, 4218–4244 RSC.
- B. Chellan, C. A. Reardon, G. S. Getz and M. A. Hofmann Bowman, Arterioscler. Thromb. Vasc. Biol., 2016, 36, 1101–1113 CrossRef CAS PubMed.
- E. Born, L. Lipskaia, M. Breau, A. Houssaini, D. P. Beaulieu, E. Marcos, R. Pierre, M. Do Cruzeiro, M. Lefevre, G. Derumeaux, D. V. Bulavin, M. Delcroix, R. Quarck, V. Reen, J. Gil, D. Bernard, J. M. Flaman, S. Adnot and S. Abid, Circulation, 2023, 147, 650–666 CrossRef CAS PubMed.
- J. K. Lam, W. Liang and H. K. Chan, Adv. Drug. Deliv. Rev., 2012, 64, 1–15 CrossRef CAS PubMed.
- H. Duan, Y. Liu, Z. Gao and W. Huang, Acta Pharm. Sin. B, 2021, 11, 55–70 CrossRef CAS PubMed.
- N. Mouraret, E. Marcos, S. Abid, G. Gary-Bobo, M. Saker, A. Houssaini, J. L. Dubois-Rande, L. Boyer, J. Boczkowski, G. Derumeaux, V. Amsellem and S. Adnot, Circulation, 2013, 127, 1664–1676 CrossRef CAS PubMed.
- L. Grosse, N. Wagner, A. Emelyanov, C. Molina, S. Lacas-Gervais, K. D. Wagner and D. V. Bulavin, Cell Metab., 2020, 32, 87–99 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00547j |
| This journal is © The Royal Society of Chemistry 2023 |