Open Access Article
Open Access ArticleRecent advances in nanoantibiotics against multidrug-resistant bacteria
Mulan
Li†
a,
Ying
Liu†
b,
Youhuan
Gong
a,
Xiaojie
Yan
a,
Le
Wang
 *a,
Wenfu
Zheng
*a,
Wenfu
Zheng
 *cdf,
Hao
Ai
*b and
Yuliang
Zhao
*cde
*cdf,
Hao
Ai
*b and
Yuliang
Zhao
*cde
aCancer Research Center, Jiangxi University of Chinese Medicine, No. 1688 Meiling Avenue, Xinjian District, Nanchang, Jiangxi 330004, P. R. China. E-mail: wangle@jxutcm.edu.cn
bKey Laboratory of Follicular Development and Reproductive Health in Liaoning Province, Third Affiliated Hospital of Jinzhou Medical University, No. 2, Section 5, Heping Road, Jin Zhou, Liaoning 121000, P. R. China. E-mail: fcah@163.com
cCAS Key Lab for Biological Effects of Nanomaterials and Nanosafety, National Center for NanoScience and Technology, No. 11 Zhongguancun Beiyitiao, Haidian District, Beijing, 100190, P. R. China. E-mail: zhengwf@nanoctr.cn; zhaoyl@nanoctr.cn
dThe University of Chinese Academy of Sciences, 19A Yuquan Road, Shijingshan District, Beijing, 100049, P. R. China
eCAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety, Institute of High Energy Physics, Chinese Academy of Sciences, 19B Yuquan Road, Shijingshan District, Beijing, 100049, P. R. China
fCannano Tefei Technology, Co. LTD, Room 1013, Building D, No. 136 Kaiyuan Avenue, Huangpu District, Guangzhou, Guangdong Province 510535, P. R. China
First published on 5th October 2023
Abstract
Multidrug-resistant (MDR) bacteria-caused infections have been a major threat to human health. The abuse of conventional antibiotics accelerates the generation of MDR bacteria and makes the situation worse. The emergence of nanomaterials holds great promise for solving this tricky problem due to their multiple antibacterial mechanisms, tunable antibacterial spectra, and low probabilities of inducing drug resistance. In this review, we summarize the mechanism of the generation of drug resistance, and introduce the recently developed nanomaterials for dealing with MDR bacteria via various antibacterial mechanisms. Considering that biosafety and mass production are the major bottlenecks hurdling the commercialization of nanoantibiotics, we introduce the related development in these two aspects. We discuss urgent challenges in this field and future perspectives to promote the development and translation of nanoantibiotics as alternatives against MDR pathogens to traditional antibiotics-based approaches.
1. Introduction
Bacterial infection has been a major threat to human health leading to high morbidity and mortality since ancient times. Bacterial invasion can cause severe infectious diseases such as sepsis, pneumonia, gastritis and so forth. Since the invention of antibiotics such as penicillin, humans have had powerful weapons to fight against various pathogens. However, the long-term use or abuse of traditional antibiotics has promoted the evolution of bacterial resistance, leading to the emergence of a group of drug-resistant strains, which significantly reduces the therapeutic efficacy of antibiotics. For instance, pathogens such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-intermediate Staphylococcus aureus (VISA) have been spreading worldwide.1,2 Drug-resistant bacteria are classified into three categories based on increasing levels of resistance, multi-drug resistant (MDR, insusceptibility to at least one agent in three or more classes of antibiotics), extensively drug resistant (XDR, insusceptibility to at least one agent in all, except two or fewer classes of antibiotics) and pan-drug resistant (PDR, insusceptibility to all antibiotics from all classes). These drug-resistant pathogens pose a considerable threat to global public health security. Unfortunately, the development speed of new antibiotics is far slower than that of drug resistance (Table 1).3,4 By 2000, only 3 new classes of antibiotics for treatment had been introduced to the market.5,6 Only 1–2 new antibiotics have been approved by the Food and Drug Administration (FDA) for clinical use each year in the 21st century.7 By contrast, after the application of new antibiotics, the corresponding resistant strains appear rapidly, although their working principles are different (Table 1). Thus, it is fundamentally urgent to develop novel antimicrobial strategies for fighting against bacterial infections. In response to the rising demand for new antibiotics, pharmaceutical companies are actively engaged in the research and development of non-traditional drugs. Notably, the nanotechnology industry has emerged as a significant player in this endeavor, directing towards the creation of innovative nanomaterials as promising candidates to replace traditional antibiotics.| Antibiotics | Category | Antibacterial mechanism | Antibiotic development | Antibacterial resistance | Representative cases for resistance generation | Ref. | ||
|---|---|---|---|---|---|---|---|---|
| Dosage | Time | Bacteria | ||||||
| Penicillin | β-Lactam | Inhibit penicillin-binding proteins; inhibit peptidoglycan transpeptidation | 1942 | 1945 | 50–100 mg | 10–14 days | S. pneumoniae | 8 |
| Ceftaroline | 2010 | 2013 | 500 mg | 2 weeks | Neisseria gonorrhoeae | 9–12 | ||
| Streptomycin | Aminoglycosides | Interact with 16S rRNA; positively charged to increase accumulation on bacteria | 1947 | 1947 | 1000 mg | 5 months | Mycobacterium tuberculosis | 13 |
| Gentamicin | 1967 | 1970 | 5 mg | 5 days | S. aureus | 14 and 15 | ||
| Clarithromycin | Tetracycline | Interact with 16S rRNA | 1952 | 1956 | 32 μg | 14 days | Mycobacterium abscessus | 16 |
| Vancomycin | Glycopeptides | Inhibit peptidoglycan synthase | 1958 | 1987 | 40 mg | 30 days | S. aureus | 17 and 18 |
| Linezolid | Oxazolidinone | Interact with 23S rRNA; inhibit 70S subunit | 2000 | 2001 | 600 mg | 5 days | S. aureus | 19 |
| Levofloxacin | Fluoroquinolones | Inhibit DNA synthesis by targeting DNA gyrase | 1994 | 2000 | 1500 mg | 21 days | Mycobacterium tuberculosis | 20 and 21 |
| Azithromycin | Macrolides | Interact with 23S rRNA | 1988 | 1991 | 100 mg | 12 days | S. aureus | 22 and 23 |
| 1 g | 2 weeks | Neisseria gonorrhoeae | 24–26 | |||||
Nanotechnology has emerged as a plausible groundbreaking tool to prioritize the design and develop novel and effective therapeutic options.27 Nanomaterials (NMs) are advantageous as active antibacterial agents due to their exceedingly large surface area relative to their size.28 NMs may provide high bioactivity at an extremely low concentration.29 By adjusting the physicochemical properties of numerous materials, humans can generate various effective antimicrobials that can combat drug-resistant pathogens. Consequently, NMs could serve as an alternative to conventional antibiotics to control bacterial infections.30–32
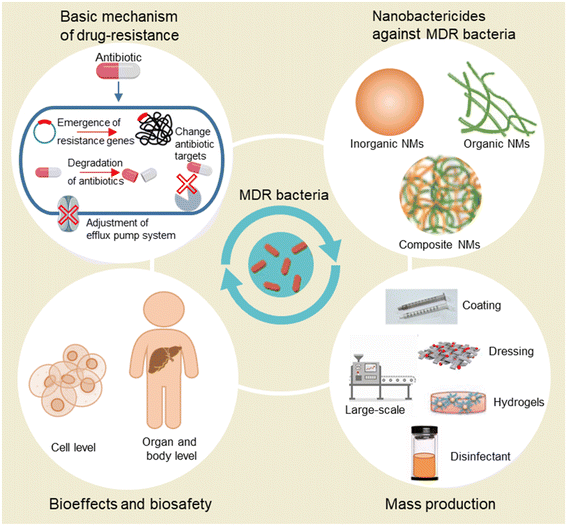
To address the issue of drug resistance, the mechanism of antibiotic resistance should be fully understood. Thus, in this review, we first introduce the basic mechanism of drug resistance including the emergence of resistance genes, change of antibiotic targets, formation of penetration barriers, degradation of antibiotics, and adjustment of the efflux pump system. In the following chapters, we introduce recently developed nanobactericides against MDR bacteria including inorganic and organic NMs. These NMs can not only serve as antibacterial agents but also be used as nanocarriers for loading conventional antibiotics for bypassing the barriers of pathogens and enhancing the antibacterial effects of antibiotics on bacteria. As novel antibacterial agents, NMs may have unexplored toxic effects on the human body. Thus, the evaluation of bioeffects and biosafety of NMs is urgently needed. We also summarize research focusing on the biosafety of antibacterial NMs. Also, the mass production of nanoantibiotics is emphasized for the sustainable development of nanotechnology in the biomedical field. In the last chapter, we discuss the opportunities and challenges for nanoantibiotics and future perspectives in developing novel NMs with better performance in antibacterial effects, biosafety, and mass production (Scheme 1).
2. Antibiotic resistance and the related mechanisms
In the past, antibiotics were considered the primary treatment option for treating bacterial infections and have proven to be effective in many cases. The inappropriate or excessive use of antibiotics has resulted in the emergence of multidrug-resistant (MDR) bacteria, posing a significant global concern that impacts hospitals as well as the natural environment. Thus, it is crucial to comprehend the biochemical and genetic underpinnings of resistance to develop novel antibiotics combating MDR bacteria. This chapter will provide a detailed description of the major mechanisms of antibiotic resistance (Table 2), including tuning resistance genes, changing antibiotic targets, inhibiting penetration barriers, avoiding degradation and adjusting the efflux pump system.33| Mechanism of resistance | Classic example | The affected antibiotic | Examples of bacteria using this mechanism | Ref. |
|---|---|---|---|---|
| Emergence of resistance genes | HGT | β-Lactam | Enterococcus spp | 34 |
| S. aureus | ||||
| K. pneumoniae | ||||
| P. aeruginosa | ||||
| A. baumannii | ||||
| E. coli | ||||
| Aminoglycosides (amikacin) | E. coli | 35 and 36 | ||
| K. pneumoniae | ||||
| Polypeptide (vancomycin) | Enterococcus | 37 | ||
| Fluoroquinolones (ofloxacin, levofloxacin) | Salmonella | 38 | ||
| Biofilm | β-Lactam | S. aureus | 39 | |
| SOS | Quinolones (ciprofloxacin) (fluoroquinolone) | E. coli | 40 | |
| S. pneumoniae | ||||
| Rpos | β-Lactam | E. coli | 41 | |
| Induced drug resistance | β-Lactam | S. pneumonia | 41 | |
| Macrolides | Bacteroideaceae | 41 | ||
| Quinolones (ciprofloxacin) | P. aeruginosa | 42–47 | ||
| Aminoglycosides (tobramycin) | P. aeruginosa | 48 | ||
| Target change or modification | 23S rRNA mutation | Oxazolones (linezolid) | S. aureus | 49–51 |
| Fluoroquinolones (moxifloxacin) | Mycoplasma | 52–55 | ||
| Macrolide (clarithromycin) | Mycoplasma | 53 and 56 | ||
| Helicobacter pylori | ||||
| PBP targeted replacement/targeted bypass replacement | β-Lactam | S. pneumonia | 49 | |
| Penicillin-resistant enzyme penicillin (methicillin) | S. aureus | |||
| Ribosome protective protein | Tetracycline | E. coli | 57 | |
| Formation of penetration barriers | Porin change | β-Lactam (cephalosporins) (carbapenems) | Carbapenem-resistant Enterobacteriins | 58 |
| E. coli | ||||
| K. pneumoniae | ||||
| Wall/membrane change | Polypeptide (vancomycin) | S. aureus | 28 and 37 | |
| Enterococcus | ||||
| Glycopeptides (daptomycin) | Enterococcus | 59 and 60 | ||
| Destroy antibiotics | β-Lactamase | β-Lactam | Enterobacteriaceae | 61 |
| K. pneumoniae | ||||
| A. baumannii | ||||
| Modifying enzyme | Aminoglycosides (amikacin) | Enterococcus | 50 | |
| S. aureus | 62 | |||
| E. coli | 63 | |||
| P. aeruginosa | 64 | |||
| Synercid (quinupristin-dalfopristin) | Enterococcus | 65 | ||
| Enzymatic degradation | Tetracycline | Bacteroides | 66 | |
| Efflux pump system | Efflux pump protein | β-Lactam | K. pneumoniae | 34 |
| A. baumannii | ||||
| P. aeruginosa | ||||
| S. aureus | ||||
| Mycin (chloramphenicol) | E. coli | 67 | ||
| Macrolide (clarithromycin) | Helicobacter pylori | 68–71 | ||
| Nitroimidazoles (metronidazole) | ||||
| Fluoroquinolones | Neisseria gonorrhoeae | 72 | ||
| E. coli | ||||
| Quinolones (ciprofloxacin) | P. aeruginosa | 45 and 73–75 | ||
| Aminoglycosides (tobramycin) | P. aeruginosa | 76 and 77 | ||
| Special efflux pump | Macrolides | S. aureus | 78 | |
| Quinolones (norfloxacin) | E. coli | 79 |
In general, there are two main types of antibiotic resistance. One is intrinsic resistance, where the bacteria are naturally insensitive to some antibiotics. Another is evolutionary resistance which is generated by various mechanisms, including change of antibiotic targets, formation of penetration barriers, degradation of antibiotics, and adjustment of the efflux pump system. The development of drug resistance for most antibiotics belongs to the evolutionary resistance type, which can still be reversed under certain conditions. This provides the opportunity for scientists to take measures to tackle the drug resistance by developing novel drugs or approaches such as nano-drugs or nanotechnology.
2.1 Emergence of resistance genes
Bacteria can develop drug-resistant genes through horizontal gene transfer (HGT), plasmids, transposons, integrons, phages, and others,40,80 which involves the transfer of genetic materials between different bacteria. HGT can occur through various mechanisms such as transformation, transduction, conjugation, or mutation (Fig. 1A).80 Transformation involves the uptake and incorporation of DNA released by dead bacteria into living bacteria, while transduction is the transfer of genetic materials via phages.81 The ssbB gene was induced in response to DNA-damaging agents (streptomycin and norfloxacin), suggesting its involvement in genetic transformation in Streptococcus pneumoniae.82 Conjugation is the direct transfer of DNA from one bacterium to another via a pilus. Mutations can occur during HGT, resulting in the acquisition of antibiotic-resistance genes.80 Specifically, mutations in the topoisomerase genes gyrA and parC have been found to confer fluoroquinolone resistance in Streptococcus pneumoniae.82 Exposure to sublethal doses of antibiotics can also contribute to the development of MDR bacteria. Bacteria activate reactive oxygen species (ROS) defense mechanisms at low antibiotic concentrations, allowing bacteria to survive and proliferate. Over time, these surviving bacteria may accumulate mutations, some of which may confer resistance to multiple antibiotics (Fig. 1B).83 Using low concentrations of ampicillin, Escherichia coli and Staphylococcus aureus can induce resistance.84 In the case of Pseudomonas aeruginosa and Pseudomonas baumannii,34 many strains can acquire MDR genes through HGT, which can reduce the effectiveness of β-lactam81 antibiotics.85 Overuse and misuse of antibiotics have created a strong selective pressure on bacteria, at the same time, HGT provides a rapid pathway for bacteria to develop antibiotic resistance.84 So, proper use of antibiotics and strict control of infectious diseases are essential in preventing the spread of antibiotic resistance in different pathogens.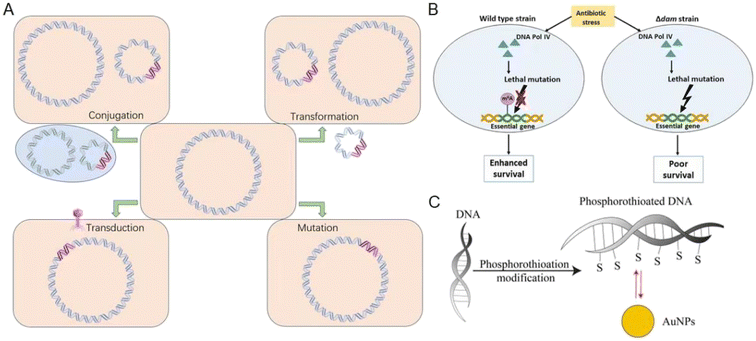 | ||
| Fig. 1 Bacteria develop drug-resistant genes and the related mechanisms. (A) The acquisition of resistance mechanisms involves the transfer of DNA containing antibiotic resistance genes (pink) from the biosphere to a recipient bacterium through conjugation, transformation, transduction, or mutation. (B) Dam-mediated adenine methylation facilitates DNA repair, minimizing deleterious mutations in the bacterial genome and promoting bacterial survival under antibiotic stress. Reproduced from ref. 83 with permission from the American Association for Microbiology, copyright 2020. (C) Bacteria undergo phosphorothioation modification of DNA to withstand oxidative damage. Au NPs show highly specific Au–S bonding interactions with such eDNA. Reproduced from ref. 84 with permission from MDPI, copyright 2020. | ||
Additionally, bacterial biofilms are currently the focus of attention regarding antibiotic resistance.39 Gaining insight into the contribution of biofilms to the emergence of antibiotic resistance is important for developing new strategies to suppress the spread of antibiotic-resistant bacteria.86 Bacteria within biofilms communicate with one another through quorum sensing, allowing them to coordinate their behavior and respond collectively to environmental cues.39 Biofilms are intricate communities of microorganisms that reside within a matrix of extracellular polymeric substances (EPS),39,84 which protect against antibiotics and various other stresses. As a result, persistent infections may be difficult to treat with antibiotics.39 HGT is a key factor in the evolution of antibiotic resistance in biofilms by facilitating the exchange of genetic materials between bacteria. Thus, HGT can help bacteria acquire new resistance genes and adapt to changing environments.85Pseudomonas aeruginosa (Gram-negative bacteria) formed biofilms in the lungs of cystic fibrosis and developed resistance to tobramycin, while Staphylococcus aureus (Gram-positive bacteria) formed biofilms on medical devices and developed resistance to β-lactam antibiotics.87 NMs have been suggested as a potential solution to overcome biofilm resistance.86,88 Metal-based, carbon-based, liposomes, and polymer NMs can increase their interaction with bacterial membranes or biofilm structures. Metal nanoparticles inhibit the development of biofilm resistance by adjusting their surface function and regulating their photo or magnetic properties, so as to increase their effect on destroying the structure of bacterial biofilms.89,90 Phosphorothioation modification of DNA in bacteria has been found to enhance their stability against oxidative damage. Notably, when biofilms containing phosphorothioated DNA were treated with Au NPs, they exhibited significant adsorption due to favorable Au–S chemistry.91 The inhibition of extracellular (eDNA) function is primarily attributed to specific and robust Au–S bonding interactions, supported by electrostatic and hydrophobic interactions (Fig. 1C).84 The interaction between Ag NPs (positive charge) and negatively charged eDNA can potentially disrupt the structural stability of the biofilm matrix, which can make bacteria more vulnerable to environmental antibiotics.86,92
The SOS response and efflux pumps also play a role in antibiotic resistance by promoting asynchronous growth and differential gene expression within the biofilm.39 The SOS response is a regulatory network that assists bacterial cells in managing DNA damage caused by different factors, including drugs, UV radiation, and oxidative stress.34 The regulation of the SOS response involves various genes such as recA, lexA, and umuDC. When the SOS response is triggered, it activates DNA repair mechanisms.93 However, it can also lead to genomic instability or mutations, which may contribute to the emergence of antibiotic resistance in bacterial populations.40 SOS can induce high-pressure resistance by inactivating the mrr gene through spontaneous mutations.88 During the SOS-induced DNA repair process, horizontal transfer of drug-resistant genes can occur, resulting in the development of more resistant strains.41 According to the passage, researchers examined Vibrio anthracis topoisomerase IV that carried the GrlAE85K mutation, and evaluated its activity, susceptibility, and metal ion requirements, suggesting that the Glu85- > Lys mutation in Vibrio anthracis topoisomerase IV can diminish its catalytic activity and contribute to antibiotic resistance.87 Moreover, the mutation of Rpos gene may regulate the antibiotic's effects.86 By comprehending the molecular mechanisms of bacterial stress responses, we can develop new strategies to combat bacterial resistance.94 Mutations induced by β-lactam antibiotics in the Rpos gene can render the drugs ineffective in treating bacterial infections.41 Conversely, Pseudomonas aeruginosa uses the SOS response to stimulate aerobic respiration, which enhances the effectiveness of aminoglycoside antibiotics.61 This mechanism allows the bacteria to survive under low oxygen conditions and resist the effects of the antibiotics.61
2.2 Change of antibiotic targets
Bacteria can reduce their susceptibility to antibiotics by altering target sites via genetic mutations.67,80 The most common way is to change the expression or structure of the target site of antibiotics. For instance, bacteria may exploit point mutations to alter RNA polymerase, making it unresponsive to antibiotics. Similarly, Acinetobacter baumannii developed β-lactam resistance by reducing the binding affinity of penicillin-binding proteins (PBPs). This adaptation enables the bacteria to survive even in the presence of these drugs.95 Although modified PBP retains normal physiological functions, the response to β-lactam antibiotics decreases sharply, resulting in bacterial resistance.96 Bacteria also develop drug resistance through homologous recombination of drug-resistant gene alleles with the target gene. Once antibiotics lose their ability to bind to target sites, the effect of antimicrobial effects is affected.49 Linezolid, used to treat drug-resistant Staphylococcus aureus and coagulase-negative Staphylococcus infections, bonds to 23S ribosomal RNA in the 50S ribosomal subunit (Fig. 2A).97 Resistance to linezolid has been linked to nucleotide mutations in region V of the 23S rRNA gene, as well as mutations in ribosomal L3 and/or L4.98 Additionally, strains carrying the chloramphenicol-fluformone resistance gene have also shown resistance.51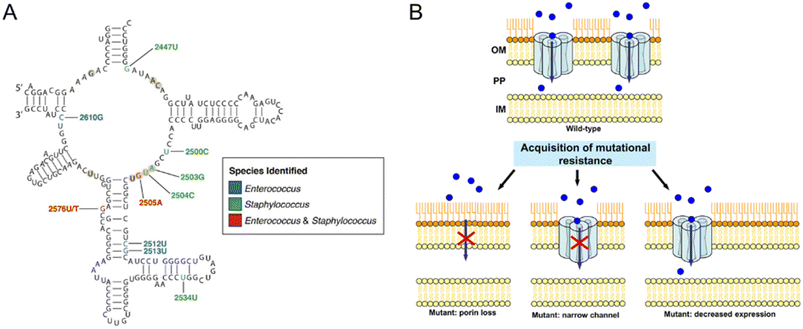 | ||
| Fig. 2 Resistance-associated mutations in bacteria. (A) Secondary structure of the peptidyl transferase loop in domain V of 23S rRNA. Nucleotide positions associated with linezolid resistance are colored according to the species identified, blue for Enterococcus, green for Staphylococcus, and red for both. Reproduced from ref. 97 with the permission from Frontiers Media SA, copyright 2021. (B) Examples of different mechanisms of mutational resistance acquisition associated with porins. The blue circles represent the antibiotic molecules, and the red cross indicates that the antibiotic cannot cross the outer membrane. IM, inner membrane; OM, outer membrane; PP, periplasmic space. Reproduced from ref. 107 with permission from American Association for Microbiology, copyright 2012. | ||
Bacteria also developed antibiotic resistance through the protective mechanism of blocking the targets from antibiotics.49 Qnr protein is a ribosome protection protein that reduces the interaction between bacterial cyclozyme and topological isomerase IV and DNA, thereby reducing the binding sites for quinolones and enhancing quinolone resistance.90,93,99,100 Other ribosome protection proteins such as AAC(6′)-Ib-cr,93 OqxA/OqxB,101 and MfpA94 prevent antibiotics (tetracyclines and macrolide) from reaching their ribosomal target site. To avoid drug resistance, high doses of antibiotics have been used to increase the effectiveness of antibiotics, thereby accelerating the progression of bacterial resistance.66,102
2.3 Formation of penetration barriers
The effectiveness of antibiotics is often reduced by various mechanisms employed by bacteria. Gram-positive bacteria rely on their cell wall as the primary defense barrier against antibiotics, whereas Gram-negative bacteria possess an outer membrane that acts as a protective shield against external antibiotic threats.95 One such mechanism is the alteration of the structure and morphology of the outer membrane, resulting in reduced permeability.103 Protein on the outer membrane acts as a channel to pass small molecules through the membrane, which is directly involved in the permeability regulation. The ring porin structure has high phase velocity and surface charge, which acts as a molecular sieve for small hydrophilic molecules like β-lactam antibiotics. New Delhi metallo-β lactamase enzyme 1 (NDM-1) is predominantly present in Escherichia coli and Klebsiella pneumoniae, which are highly resistant to all antibiotics except tigecycline and myxin.95 However, some bacteria can alter the structure or composition of their outer membrane to reduce the number of porins or change the size of the protein channels.95,104Transport proteins located between the cell wall and the periplasmic intima in Gram-negative bacteria regulate microbial drug resistance by promoting rapid substance transport. β-Lactam enzyme,96,105 aminoglycosidase,98 tetracycline efflux pump,100 oxidative drug efflux pump,100 and other transport proteins are commonly linked to drug resistance in Gram-negative bacteria. The expression level of these transporters in Pseudomonas aeruginosa,98Escherichia coli,102Vibrio cholerae,105 and so forth can affect their sensitivity to various types of antibiotics.99,102Pseudomonas aeruginosa can modify the expression of genes coding for porins or produce efficient variants of these proteins, thereby reducing the transport of antibiotics across the membrane and increasing the contribution to resistance.106 Many carbapenem-resistant bacteria such as Bacillus, Escherichia coli, and Klebsiella pneumoniae also exploit this mechanism to hinder the therapeutic effects of carbapenem antibiotics. When porin is mutated, reduced, or not expressed, these bacteria can evade the effects of antibiotics by diminishing the ability of the drug to penetrate the cell wall or outer membrane (Fig. 2B).107 Substitution or insertion mutation of 1 or 2 amino acids in the L3 ring in the porin OmpC of the Enterobacteriaceae can lead to porin mutants, which change the resulting pore size and charge in the contraction region; this mode greatly reduces the translocation of β-lactams, thereby reducing the sensitivity of bacteria.106,107
2.4 Degradation of antibiotics
Bacteria have evolved various resistance mechanisms against antibiotics, producing enzymes to break down the antibiotic in an efficient way. β-Lactam antibiotics work by inhibiting cell wall synthesis through binding to PBPs, which are special proteins on the bacterial cytosolic membrane.90 The effectiveness of β-lactam antibiotics, including penicillin, cephalosporins, monocytomyces and carbapenems, can be disrupted by antibiotic-modifying enzymes, specifically β-lactamases.108 β-Lactamases break the amide bond in the β-lactam ring, disabling the antibiotics and rendering them ineffective.51 The genes that produce β-lactamase can be controlled by inducible promoters and encoded on chromosomes or plasmids (Fig. 3A).109 These genetic elements may also harbor resistance genes for other antibiotic classes, including aminoglycosides, fluoroquinolones, and tetracycline.66,90,100,102 For example, encoding CTX-M type β-lactam enzyme is usually accompanied by aac(6′)-Ib-cr that codes for aminoglycoside antibiotic resistance, and qnr gene that codes for fluoroquinolone antibiotic resistance.110 This resistance mechanism is a significant problem in clinical settings and highlights the need for careful consideration when prescribing antibiotics to patients. Many bacteria, such as Enterobacteriaceae, Pseudomonas aeruginosa, and Klebsiella pneumonia, produce multiple β-lactamases with varying specificities, resulting in the breakdown of a broad range of β-lactam antibiotics before they can reach their target sites (Fig. 3B).96,105,111 In clinical practice, these bacteria often exhibit multiple drug resistance, posing significant treatment challenges.112 Extended-spectrum beta-lactamases (ESBLs) are a particular type of β-lactamase capable of hydrolyzing a wide range of β-lactam antibiotics, such as penicillin, cephalosporins, and monobactams. Infections caused by ESBL-producing bacteria have emerged as a significant problem in healthcare treatment. Carbapenems, often used as a last resort for MDR bacteria, are susceptible to hydrolysis by carbapenemase enzymes produced by Pseudomonas baumannii, Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumonia, which are associated with high levels of antibiotic resistance.111,113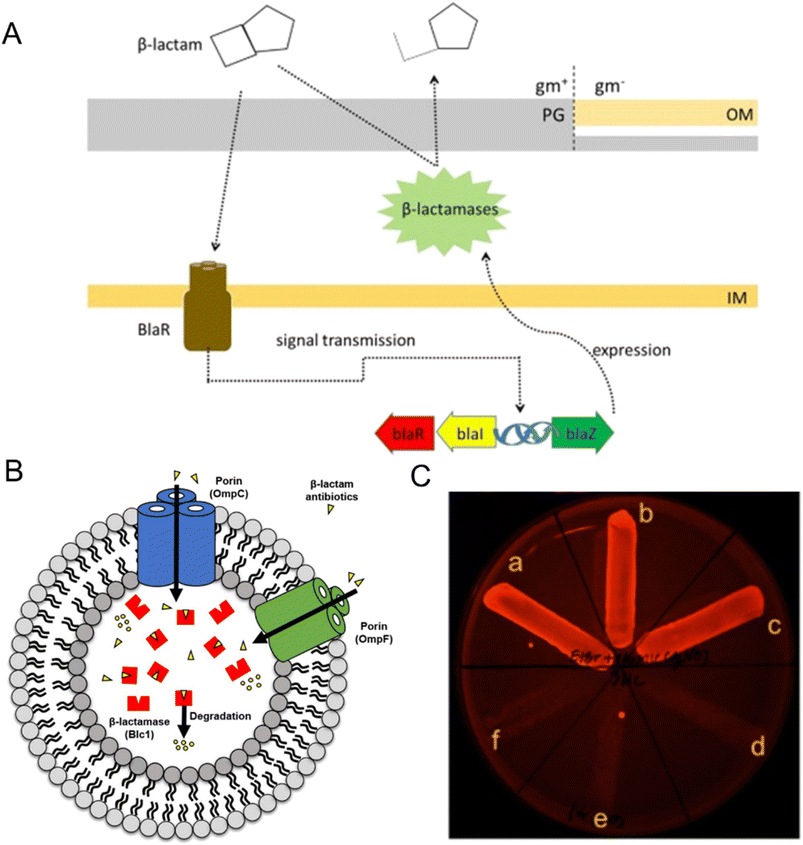 | ||
| Fig. 3 The role in antibiotic resistance of enzyme degradation and efflux pump system. (A) The mechanism of β-lactamase production in bacteria involves related genes (blaR, blaI, and blaZ) that control the expression of β-lactamases. Reproduced from ref. 109 with permission from Elsevier, copyright 2018. (B) Predictive mechanism of β-lactam antibiotics degradation by outer membrane vesicles (OMVs) from β-lactam-resistant Escherichia coli. Porin channels (OmpC and OmpF) in OMVs transport β-lactam antibiotics into their lumen, where the β-lactamase (Blc1) hydrolyzes the confined antibiotics. Reproduced from ref. 105 with permission from MDPI, copyright 2020. (C) Phenotypic efflux pump inhibiting effect of Ag NPs modified with tannic acid. B. pseudomallei treated with 2 μg mL−1 EtBr-agar supplemented with 1/4 MIC of Ag NPs (inset, a–c), compared to the untreated control broth (inset, d–f). Reproduced from ref. 120 with permission from MDPI, copyright 2021. | ||
2.5 Adjustment of the efflux pump system
Efflux pumps are a bacterial defense mechanism, which plays an important role in antibiotic resistance. These pumps actively remove toxic substances, such as antibiotics, from the bacterial cell into the extracellular space.114 In general, Gram-positive bacteria have simpler efflux systems consisting of a single peptide in the plasma membrane, while Gram-negative bacteria typically have more complex efflux pump systems that consist of a three-part combination of an inner membrane component, an outer membrane component, and a periplasmic membrane fusion protein.102,115 Efflux pump genes encode membrane-associated proteins that actively pump drugs out of the cell, leading to lower drug concentrations inside the cell and reducing their effectiveness. The turning on or off of specific genes is dependent on environmental conditions, which make bacteria more or less resistant to certain antibiotics.116Pseudomonas aeruginosa have multiple efflux pump genes, conferring resistance to β-lactam drugs like penicillin and cephalosporins. Similarly, Staphylococcus aureus (including MRSA) can gain resistance through efflux pumps called resistance-nodulation-division (RND) efflux pumps, which also pump out antibiotics from the cell.107 Furthermore, efflux pumps can recognize and remove structurally related compounds, causing cross-resistance to multiple classes of antibiotics.104 The MepA efflux pump of MRSA can recognize and remove not only β-lactam antibiotics but also fluoroquinolones (norfloxacin and ciprofloxacin), making it more difficult to be treated with multiple antibiotics.114,117,118 Overall, the regulation of these genes can lead to increased resistance to antibiotics.Although efflux pump inhibitors (EPIs) have been investigated as a potential strategy for overcoming bacterial resistance, their development is challenging due to specificity issues and potential toxicity. Most antibiotics and their corresponding EPIs have a competitive relationship, and this specificity may lead to a decrease in the clinical efficacy of certain drugs.115 The emergence of antimicrobial NMs is expected to solve this problem.119 Ag NPs modified with tannic acid acted as a long-term EPI. Tannic acid itself does not have antibacterial effects, but it can be used as an EPI in combination with Ag NPs to improve antibacterial activity and decrease the incidence of drug resistance induction (Fig. 3C).120
In summary, bacteria have evolved various resistance mechanisms against antibiotics. How to rebuild the sensitivity of drug-resistant bacteria to the present antibiotics or find alternatives to synthetic antibiotics to combat bacteria is a big challenge to human beings. The rapid development of NMs provides the opportunity to address this tough topic.
3. Nanobactericides against MDR bacteria
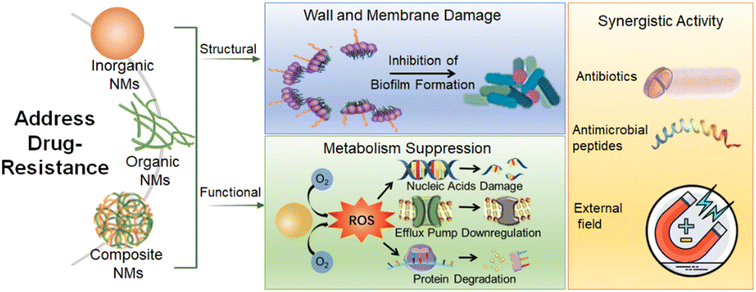
The availability of antibiotics for treating MDR bacteria-caused infections is limited which forces a pressing need to explore new strategies. Among the various ways to solve bacterial resistance, NMs offer a highly promising approach to combat the pathogen due to their high specific surface area, intrinsic physical properties, and simple chemical modification.121,122 The recently developed antibacterial nanobactericides have been summarized in the published review papers.123–125 Thus, in this review, we concisely discuss the related studies focusing on dealing with MDR bacteria and the related infections. In this chapter, we generally divide antibacterial nanobactericides into three categories: inorganic NMs, organic NMs, and composite NMs. The section on inorganic NMs includes metal-based NMs, carbon-based materials, and others. The section on organic NMs includes natural polymers and synthetic polymers. The section on composite NMs includes metal-based NMs, polymer-based materials, and others (Scheme 2).3.1 Inorganic NMs
| Nanomaterials | Strains | Antibacterial effect (MIC) | Mechanisms | Ref. | |
|---|---|---|---|---|---|
| Ag NPs | E. coli | 128 μmol L−1 | Metal ion release | 184 | |
| MDR P. aeruginosa | 1.406–5.625 μg mL−1 | Direct contact, protein degradation | 185 | ||
| B. subtilis | 27 μg mL−1 | Metal ion release; ROS generation | 186 | ||
| E. coli | 31.250 μg mL−1 | ROS generation intracellular substance breakage | 187 | ||
| P. aeruginosa | 15.625 μg mL−1 | ||||
| S. aureus | 31.250 μg mL−1 | ||||
| Au NPs | A-GNCs | MDR E. coli | 8.75 μg mL−1 | Bacterial wall destruction | 188 |
| MDR K. pneumonia | 8.75 μg mL−1 | ||||
| MDR P. aeruginosa | 4.38 μg mL−1 | ||||
| 4AP-Au NPs | E. coli | 7.8 μg mL−1 | Bacterial wall destruction; interaction with 16S rRNA | 189 | |
| A. baumannii | 1.3 μg mL−1 | ||||
| P. aeruginosa | 5.2 μg mL−1 | ||||
| S. aureus | 2.6 μg mL−1 | ||||
| S. enteritidis | 10.5 μg mL−1 | ||||
| E. faecalis | 10.5 μg mL−1 | ||||
| TG-Au NPs | P. aeruginosa | 50 μg mL−1 | Exopolysaccharide secretion inhibition | 190 | |
| Cu NPs | E. coli | 575 μg mL−1 | Metal ion release | 191 | |
| B. subtilis | 40 μg mL−1 | ||||
| ZnO NPs | C. albicans | 128 μg mL−1 | Bacterial membrane disruption | 192 | |
| TiO2 NPs | E. coli | 500 μg mL−1 | ROS generation; bacterial membrane disruption | 193 | |
| B. subtilis | 575 μg mL−1 | ||||
| CuO NPs | C. albicans | 160 μg mL−1 | Bacterial membrane disruption | 194 | |
| Fe2O3 NPs | P. aeruginosa | 17.5 mg mL−1 | ROS generation | 180 | |
| Ag–Cu NPs | S. aureus | 3 μg mL−1 | Metal ion release | 168 | |
| E. coli | 2.5 μg mL−1 | ||||
| GaCur NPs | P. aeruginosa | 82.75 μg mL−1 | Bacterial membrane disruption | 195 | |
Among these metal NMs, silver and gold NMs possess special properties and display great potential in antibacterial applications.
3.1.1.1 Silver nanoparticles (Ag NPs). Silver nanoparticles (Ag NPs) as an effective antibacterial agent have been studied and used for a long period.131–134 Ag NPs could be used as potential alternatives to antibiotics due to their broad-spectrum antimicrobial effects against a variety of bacterial strains, including those that have developed resistance to traditional antibiotics. Ag NPs can interact with the lipid bilayers of bacterial cell walls and affect their stability. Additionally, by capturing amino acids and proteins on the bacterial surface, Ag NPs impede bacterial proliferation (Fig. 4A).135 By inhibiting the expression of cytoskeletal proteins FtsZ and FtsA,89 Ag NPs could impede the growth and biofilm formation of Bacillus subtilis, Salmonella coli, and Salmonella typhimurium.89,90 Releasing silver ions (Ag+) is the major antibacterial mechanism of Ag NPs,136 which changes the membrane structure of bacteria, resulting in increased membrane permeability of bacteria and eventually cell death. Since Ag+ has an affinity for aminos, phosphates, and thiols, they could adhere to the cell surface and enter the bacteria. Ag+ changes mitochondrial function and causes damage to drug-resistant bacteria.137 What is the difference between the antibacterial mechanisms of ionic and colloidal silver at the molecular level? Apart from the effects of Ag+, Ag NPs possess an additional bactericidal property, effectively binding to the bacterial membrane and disrupting its structure, resulting in a notable elevation of intracellular Ag+ concentration. This highlights the advantage of NPs in exhibiting enhanced antibacterial properties.138
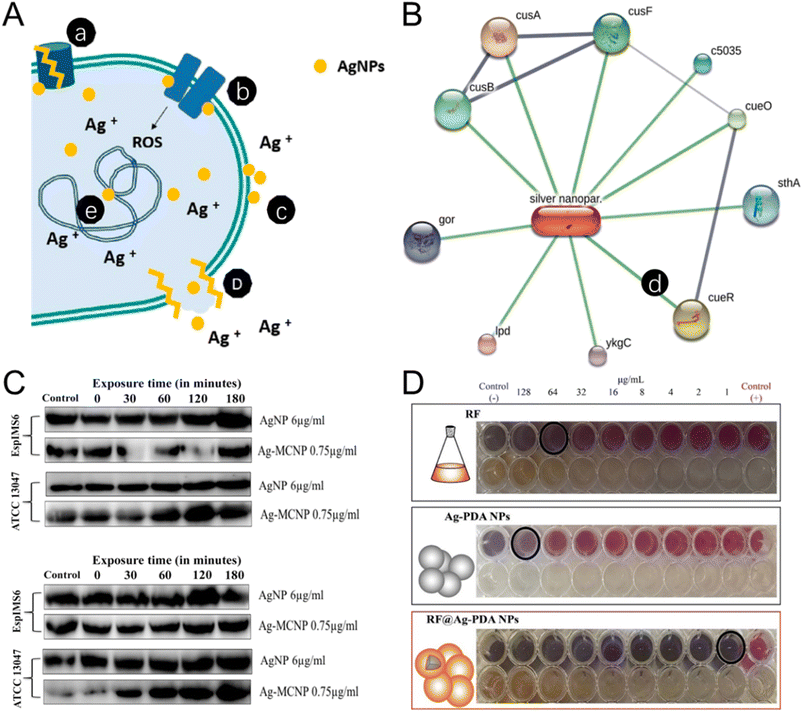 | ||
| Fig. 4 The antibacterial mechanism and antibiotic synergetic effect of the silver NMs. (A) The antibacterial mechanisms of Ag NPs include: (a) efflux pump modification, (b) disruption of the membrane proteins and electron transport chains, (c) accumulation on the membrane to affect membrane permeation, (d) disruption of membrane leading to leakage of intracellular content, (e) DNA damage. Reproduced from ref. 135 with permission from MDPI, copyright 2021. (B) Ag NPs–protein interaction networking of Staphylococcus aureus (MLD4). An individual line between Ag NPs and proteins is illustrated in a networking diagram. Reproduced from ref. 139 with permission from Elsevier, copyright 2022. (C) The expression of AcrA (42 kDa) and AcrB (∼38 kDa) protein in EspIMS6 (top panel) and Enterobacter cloacae ATCC 13047 (bottom panel) is shown to be influenced by Ag and Ag-metal composite nanoparticles. Reproduced from ref. 140 with permission from Frontiers Media SA, copyright 2018. (D) Synergetic effect of rifampin-loaded mussel-inspired Ag NPs with enhanced antibacterial activity against MDR Mycobacterium tuberculosis. Reproduced from ref. 144 with permission from WILEY-VCH, copyright 2021. | ||
Appropriate decoration can stabilize Ag NPs for enhancing their antibacterial activity. Researchers developed glutathione-stabilized silver nanoparticles (GSH-Ag NPs) to treat MDR Campylobacter strains. GSH-Ag NPs exhibited high antibacterial efficacy against all MDR Campylobacter strains, with minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) ranging from 4.92 to 39.4 μg mL−1 and 9.85 to 39.4 μg mL−1 respectively. To decrease the cytotoxicity of Ag, Carica papaya leaf extract was utilized in the biogenic fabrication of chitosan-functionalized silver nanoparticles (Ag-Chito NPs). The MIC of Ag-Chito NPs against Escherichia coli, or Staphylococcus aureus was 12.5 μg mL−1 and 15 μg mL−1 respectively, indicating the highest bacterial sensitivity (Fig. 4B).139 In a recent study, Ag NPs were modified with polysaccharide which showed an inhibitory effect on the expression of the membrane fusion protein AcrA of MDR Enterobacter cloacae isolates, thus, the action of the Ag NPs was not hindered by the efflux protein, a major cause of drug resistance (Fig. 4C).140
By conjugating Ag NPs, conventional antibiotics which are resisted by MDR bacteria could regain their antibacterial activity. A recent work utilized amikacin to functionalize Ag NPs. The MIC of AgNPs@amikacin was no more than 0.5 μg mL−1. Moreover, the rates of Acinetobacter baumannii biofilm metabolic activity were reduced over 50% by AgNPs@amikacin, demonstrating a new strategy to rebuild the antibacterial activity of drugs that have been resisted by MDR bacteria.141 In another study, Ag NPs were functionalized with mercaptopoly(ethylene glycol) carboxylic acid (mPEG-COOH) and amikacin (AK). AgNPs_mPEG_AK was 10-fold more effective than amikacin alone in susceptibility studies, and bactericidal efficacy against 100% of the tested Pseudomonas aeruginosa after 4, 8, 24, or 48 h. AgNPs_mPEG_AK combined with hyperthermia achieved a 75% eradication of planktonic strains and significantly reduced the biofilm formation.142 To tackle the drug-resistance problem of imipenem, imipenem (IMP) was conjugated to silver nanoparticles (IMP-AgNPs) to treat MDR Pseudomonas aeruginosa-infected wounds. A considerable epithelization took place in the IMP-AgNP-treated wounds.143 Tuberculosis (TB) is among the top ten causes of death worldwide. The rise of multidrug-resistant TB poses significant treatment challenges. To ensure the effectiveness of antituberculosis drug rifampin (RF) in the treatment of MDR TB infection, researchers synthesized polydopamine-decorated silver nanoparticles (Ag-PDA NPs) loaded with RF. MIC results showed a synergistic interaction between the Ag-PDA NPs and RF with the optimal antibacterial effect against the MDR M. tuberculosis observed at the mass ratio of 2Ag-PDA NPs:8RF. RF@Ag-PDA NPs showed promise in inhibiting the growth of MDR M. tuberculosis while preserving the potency of RF for clinical application (Fig. 4D).144
In summary, by appropriate modifications, the capability of Ag NPs to inhibit the MDR bacteria could be enhanced. Ag NPs can also conjugate conventional antibiotics to realize synergistic antibacterial effects. As one of the most important kinds of inorganic NMs, the decrease of toxicity and increase of antibacterial activity are the major directions for Ag NPs to be explored. Besides, the stability of Ag NPs is another issue that needs to be resolved.
3.1.1.2 Au NMs. Compared with silver, gold is relatively inert and is safer. Au NMs do not have antibacterial effects themselves, but they have potent antibacterial effects after appropriate surface modifications. Gold is multivalent for binding many types of ligands. Au NMs combat MDR bacteria through several mechanisms including the physical destruction of bacterial structures, disturbing the metabolism of bacteria, and serving as carriers for delivering antibiotics or bioactive molecules. Au NPs can increase the permeability of bacterial cell membranes. For example, 4,6-diamino-2-pyrimidinethiol (DAPT)-capped Au NPs led to enhanced membrane permeability for E. coli (Fig. 5A).145 Au NPs can directly interact with efflux pumps on bacteria. For example, the expression of MexA and MexB efflux pump genes in Pseudomonas aeruginosa was downregulated by Au NPs, leading to a reduction in efflux pump activity.146 Au NPs can also serve as nanocarriers to influence the efflux pump. An example of this is the observed significant synergistic effect of embelin-capped chitosan-Au NPs with ciprofloxacin on the efflux pumps of P. aeruginosa and E. coli.147 To protect antibiotics from enzymatic hydrolysis, researchers functionalized Au NPs with carbapenem through the strong attraction/coordination between their thioether reaction groups. Compared with carbapenem, carbapenem-functionalized Au NPs enhanced the antibacterial effect on Klebsiella pneumoniae and Acinetobacter baumannii, which is a potential way to solve the carbapenem resistance.148 An advantage of Au NPs is their high biosafety. This means that Au NPs themselves are ineffective against any bacteria, however, when Au NPs are modified by some non-antibiotic molecules, the functionalized Au NPs may have potent antibacterial activity. For instance, the growth of Gram-positive bacteria can be effectively and selectively inhibited by multivalent aminosaccharide-capped Au NPs. In particular, aminosaccharide-modified Au NPs showed effectiveness against MRSA.149 Moreover, the antibacterial spectrum of Au NPs can be maximized by controlling their surface ligands. We synthesized Au NPs with tunable antibacterial spectra by small molecule modifications. In the one-step synthesis process, we adjusted the ratio of aminophenylboronic acid (ABA) and thiophenylboronic acid (MBA) to obtain Au NPs (A/M-Au NPs) with different surface ligand densities for different bacterial therapies.150 Remarkably, the A/M-Au NPs demonstrated an exceptionally high median lethal dose (920 mg kg−1), which was approximately 100 times their effective dose (7.2 mg kg−1), indicating their remarkable biosafety.151 Au NPs also can be synthesized in vivo using tetrachloroauric acid and ABA, and the resulting ABA-modified Au NPs exhibited good bacteriostatic effects and high biological safety, indicating their potential to broaden traditional administration (Fig. 5B).152 In addition to functionalization with non-antibiotic molecules, Au NPs can be capped with antibiotics to treat drug-resistant bacteria. Researchers reported kanamycin-capped Au NPs (Kan-Au NPs) which had dose-dependent broad-spectrum activity including kanamycin-resistant bacteria. The underlying mechanism involved the disruption of the bacterial envelope by Kan-Au NPs, leading to the leakage of cytoplasmic contents and subsequent bacterial cell death in a dose-dependent manner.153 Vancomycin-modified Au NPs (Van-Au NPs) can inhibit the proliferation of vancomycin-resistant Enterococci of three different gene types. Compared with sole vancomycin, the efficacy of Van-Au NPs was improved more than 60 times.154 Au NMs could target certain kinds of bacteria by conjugating with phages which can specifically target bacteria and lead to rapid cell lysis. Researchers reported a phage-AuNR bioconjugate carrying Zn2+, which was synthesized by modifying M13-g3p-(Pf1) on Au NRs. The phage nanomaterial exhibited effectiveness against polymyxin-resistant P. aeruginosa, which is typically resistant to last-line antibiotic therapy. In a wound model on mice, under near-infrared light irradiation, the thermal effects resulted in rapid bacterial load reduction and effective Zn2+ release, which facilitated wound healing. The phage-nanomaterial demonstrated no observable toxicity or systemic effects in mice. This finding highlights the potential of phage therapy controlled by NMs as a safe and effective antimicrobial strategy in vivo.155 Reducing the size of Au NPs to a value comparable to the Fermi wavelength of electrons (∼1 nm) can produce the ultrasmall Au nanoclusters (Au NCs) which hold discrete electronic states and characteristic geometric structures and offering them intriguing antibacterial properties. Researchers found that precisely controlling size down to the NC dimension can confer antimicrobial activity to Au NMs. Au NCs could kill both Gram-positive and Gram-negative bacteria. The ultrasmall size of Au NCs allowed them to better interact with bacteria and induce a metabolic imbalance in bacterial cells, leading to an increased production of intracellular ROS which kills bacteria.156 In another work, researchers developed a dual-ligand-functionalized gold nanocluster (Au25(SR1)x(SR2)18−x) and evaluated its bactericidal properties against MDR bacteria. The ligand SR1 (pyridinium ligand) contributed to the bactericidal activity, and the ligand SR2 (zwitterionic ligand) improved its stability and biocompatibility. Through optimization of the ligand ratio, the Au NCs effectively eradicated MDR Gram-positive bacteria by employing multiple antibacterial mechanisms, including aggregation on the bacterial surface, disruption of bacterial membrane integrity, as well as the generation of ROS (Fig. 5C).157 In addition to the direct destruction of bacterial structure, Au NCs can also serve as an adjuvant to improve the antibacterial effect of conventional antibiotics. In a recent study, cell-penetrating peptide (CPP) was modified on Au NCs, the resulting Au NC@CPP did not exhibit antimicrobial activity. But AuNC@CPP was able to eliminate both planktonic persister cells and biofilms when combined with ofloxacin. The mechanism of action of AuNC@CPP involved the disruption of the proton gradient and induction of membrane hyperpolarization. Au NCs can coordinate with organic molecules to form novel structures with antibacterial activity. Researchers developed para-mercaptobenzoic acid (pMBA)-capped Au NCs with adjustable antibacterial activity which is closely related to the protonation level of pMBA ligands in different pH environments. Furthermore, a series of Au NCs-based mixed-metal metal–organic network (MM-MON) films were constructed on titanium disks as antibacterial nanocoatings. By combining robust M4+ (Ti, Zr, Hf)-–bonds and inferior Cu2+–O bonds, the heterobimetallic MM-MON films enabled the controlled dissolution of Cu2+, while the structural integrity is retained. In vitro and in vivo results demonstrated the bacteria-triggered Cu2+ release and contact-killing capability of the MM-MON film nano-coating. This work is insightful for the development of next-generation antibacterial surface modification.158
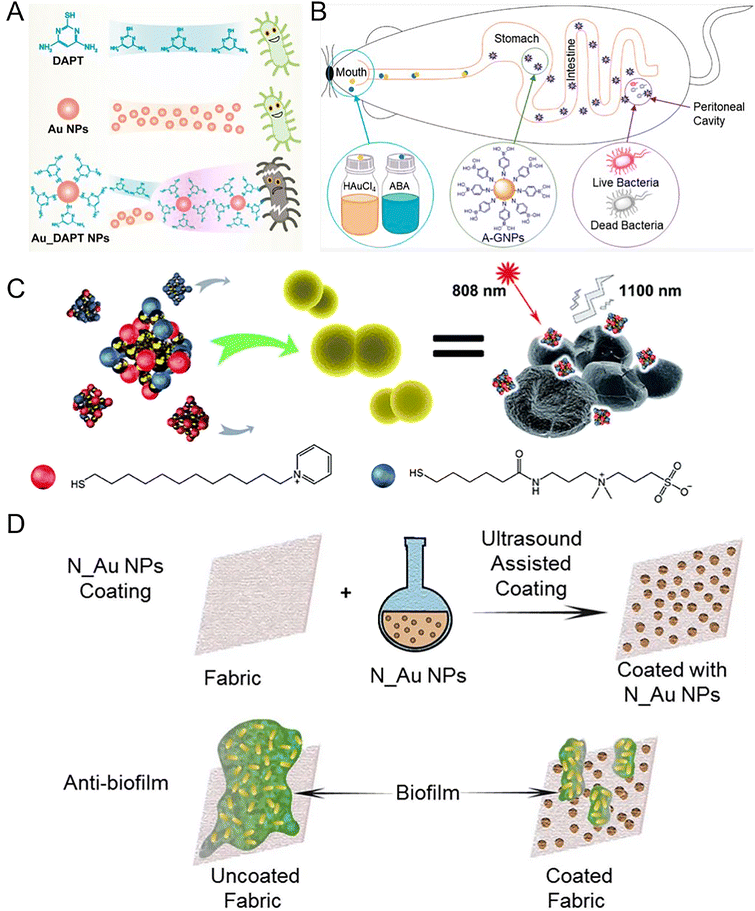 | ||
| Fig. 5 Au nanomaterial-based nano-antibiotics. (A) Schematic diagram of DAPT, Au NPs alone, and Au_DAPT NPs with different antibacterial activities. DAPT or AuNPs individually do not exhibit antibacterial activities, but Au_DAPT NPs have excellent antibacterial activities. Reproduced from ref. 145 with permission from the American Chemical Society, copyright 2022. (B) AuNPs synthesized in vivo. Aminophenyl boronic acid (ABA)-activated AuNPs (A-GNPs) synthesized in vivo using tetrachloroauric acid and ABA, which could be absorbed by the gastrointestinal tract. After oral administration, A-GNPs can reach the peritonitis lesions infected by MDR E. coli in mice. Reproduced from ref. 152 with permission from the American Chemical Society, copyright 2021. (C) P12/C5 modified Au NCs with good biocompatibility interacted with the bacterial cell envelope by aggregating on planktonic bacteria. The special NIR fluorescence of Au NCs facilitates the trace distribution in the body with an 808 nm laser. Golden spheres represent gold atoms, red spheres represent the P12 ligands, and blue spheres represent the C5 ligands. Reproduced from ref. 157 with permission from the Royal Society of Chemistry, copyright 2021. (D) The schematic diagram depicts the process of preparing antibiofilm fabrics coated with various N_Au NPs. Sonochemistry was employed to deposit these Au NPs onto the fabric surface, resulting in outstanding antimicrobial activity against MDR bacteria and remarkable efficacy in inhibiting bacterial biofilms formed by MDR bacteria. Reproduced from ref. 161 with permission from the Royal Society of Chemistry, copyright 2020. | ||
The formation of biofilms, which are communities of bacteria attached to surfaces and enclosed in an extracellular matrix, can significantly enhance bacterial drug resistance and antibiotic tolerance.159 To address this challenge, researchers reported nanocomposites consisting of Au NCs modified by mercaptopropionic acid (denoted as Au18(MPA)14 NCs), and a photosensitizer called protoporphyrin (PpIX), embedded in a chitosan polymer matrix (PpIX-Chito-Au18). The nanocomposite has the ability to generate ROS upon exposure to light. Consequently, it effectively eradicated both Gram-positive and Gram-negative bacteria by damaging their membrane and DNA. Furthermore, the PpIX-Chito-Au18 nanocomposite exhibited the capacity to penetrate and eliminate biofilms formed by S. aureus and P. aeruginosa when activated by light.160 In recent work, we employed ultrasound-assisted coating technology to deposit Au NPs coated with different N-heterocyclic molecules (N_Au NPs) on fabrics, which effectively inhibited the formation of biofilms for addressing the challenge of MDR bacterial infections. Among these Au NPs, mercaptoimidazole (MI)_Au NP-coated fabrics demonstrated significant reductions in the viability of E. coli and S. aureus, with reductions of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) logs and 2
logs and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) logs, respectively (Fig. 5D).161 Although bactericidal Au NMs have been developed, the resistance to these NMs has rarely been reported. A recent study on 4,6-diamino-2-pyrimidine thiol (DAPT)-modified Au NPs (AuDAPTs) showed that a 16-fold increased MIC of E. coli was observed after prolonged exposure (183 days), without developing resistance to conventional antibiotics. Moreover, the resistance was found to be size-specific to Au NPs with the same surface modification. By adjusting the sizes of AuDAPTs without the need for new agents, the antibacterial activities of AuDAPTs against the resistant strain were restored. This unique form of slow and manageable resistance induced by AuDAPTs distinguishes it from traditional antibiotics.162
logs, respectively (Fig. 5D).161 Although bactericidal Au NMs have been developed, the resistance to these NMs has rarely been reported. A recent study on 4,6-diamino-2-pyrimidine thiol (DAPT)-modified Au NPs (AuDAPTs) showed that a 16-fold increased MIC of E. coli was observed after prolonged exposure (183 days), without developing resistance to conventional antibiotics. Moreover, the resistance was found to be size-specific to Au NPs with the same surface modification. By adjusting the sizes of AuDAPTs without the need for new agents, the antibacterial activities of AuDAPTs against the resistant strain were restored. This unique form of slow and manageable resistance induced by AuDAPTs distinguishes it from traditional antibiotics.162
In summary, Au NMs are a group of materials with high safety compared with other metal NMs such as Ag NPs. Moreover, the surface of Au could be functionalized by various molecules that can interact with bacteria through different mechanisms. Moreover, the size, shape, and surface charge of Au NMs could be tuned to maximize the antibacterial capability of the NPs. Thus, Au NMs are promising as next-generation antibacterial agents for dealing with MDR bacteria.
3.1.1.3 Alloy NMs. Alloy NMs usually have stronger antibacterial properties compared with single metal or metal oxide NMs, owing to their special physical and chemical properties, such as surface enlargement effect and grain boundary effect (Fig. 6A).163–166 Copper–silver alloy nanoparticles (Cu–Ag NPs) achieved excellent antibacterial effects against both Gram-negative and Gram-positive bacteria by releasing copper (Cu2+) and silver ions (Ag+),167 which bound to different locations of bacteria, such as sulfides, proteins, and DNA. The MIC against S. aureus and E. coli was 3 and 2.5 mg L−1, respectively.168 Compared with copper and silver, gold and platinum have higher biosafety. The antibacterial effects of AuPt bimetallic NPs were attributed to two mechanisms: (1) the disruption of the inner membrane of bacteria; (2) the increase in ATP levels (Fig. 6B).169 AuPt NPs provide new insights into biological applications and expand the possibilities in medicine. Although alloy nanoparticles have good antibacterial effects, their widespread application is restricted by the high cost and specific pH-dependent dissolution and release.
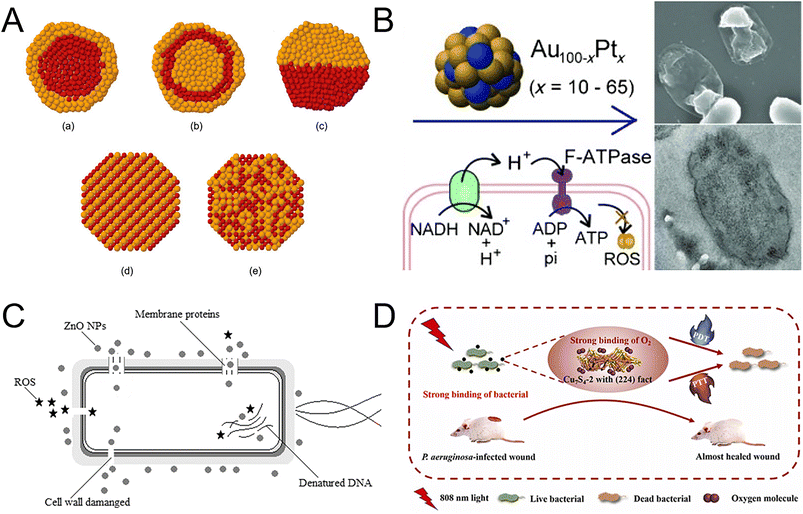 | ||
| Fig. 6 Metal nanocomposites as nanoantibiotics. (A) Structures of bimetallic NPs. (a) Core–shell structure; (b) multi-shell structure; (c) biphase structure; (d) intermetallic structure; (e) nanoalloy. Reproduced from ref. 165 with permission from the American Chemical Society, copyright 2008. (B) The antibacterial properties of AuPt bimetallic NPs vary depending on the metal composition. AuPt bimetallic NPs with 20% Pt (Au80Pt20) exhibit the highest antibacterial activity against Gram-negative bacteria and MDR bacteria. The bactericidal action of AuPt bimetallic NPs involves rupturing the bacterial membrane and enhancing the ATP levels. Reproduced from ref. 169 with permission from WILEY-VCH, copyright 2014. (C) Antibacterial mechanism of ZnO NPs in a cell model. ZnO NPs act on multiple targets to affect bacterial structures. The primary mechanism involves the cytoplasmic membrane, while the effects on other structures are secondary outcomes following membrane rupture. Reproduced from ref. 174 with permission from Springer Nature, copyright 2022. (D) CuS4 with (224) facets exhibited excellent antibacterial efficiency against Gram-positive, Gram-negative and drug-resistant bacteria when exposed to the field of NIR. The antibacterial activity of synergetic photodynamic and photothermal routes provides novel opportunities for designing novel antibacterial NMs. Reproduced from ref. 182 with permission from Elsevier, copyright 2022. | ||
3.1.1.4 Metal oxide NMs and metal sulfide NMs. Metal oxide NMs offer several advantages over metal nanoparticles, such as good biocompatibility, high stability, and strong controllability, making them popular in antibacterial materials in medical treatment,170 food processing,171 and other fields. Among metal oxide NMs, zinc oxide nanoparticles (ZnO NPs), titanium dioxide nanoparticles (TiO2 NPs), iron oxide nanoparticles (Fe2O3 NPs), and copper oxide nanoparticles (CuO NPs) are the most widely studied NMs. Due to their excellent antibacterial activity, low toxicity, and long-term antibacterial effects, ZnO NPs are an ideal agent for medical and biotechnology applications.172,173 ZnO NPs had an excellent antibacterial effect on E. coli and P. aeruginosa, in which the IC100 was 0.6 mM. Furthermore, the efficient antibacterial performance came from the production of Zn2+ and ROS through electrostatic effects (Fig. 6C).174 Different from ZnO NPs, TiO2 NPs had super antimicrobial properties against MDR pathogenic strains based on photocatalytic sterilization.175 When TiO2 NPs absorbed ultraviolet light, they generated strong oxidants, including hydroxyl radicals and superoxide anions, which can destroy the bacterial wall.176 Due to the different cell wall structures and thicknesses of bacteria, TiO2 NPs had greater effectiveness against Gram-positive bacteria than Gram-negative bacteria.177 By combining bismuth, the antimicrobial activity of Bi–TiO2 NPs against Gram-negative bacteria had been significantly enhanced. The hydroxyl group and ROS produced by Bi–TiO2 melted the cell wall, while bacterial phospholipid peroxidation caused bacterial death.178 Compared to ZnO NPs and TiO2 NPs, Fe2O3 NPs have the advantage of low toxicity and high magnetism, which allow for better movement and localization through a magnetic field. This property makes them more effective in killing or inhibiting MDR bacteria.179 Due to the inherent magnetic advantage of Fe2O3 NPs, the diffusion conjugates through the mucin and alginic acid barriers were enhanced. Fe2O3 NPs exhibited inhibitory effects on the P. aeruginosa growth and biofilm formation. The lowest inhibitory concentration against P. aeruginosa was found to be 17.5 mg mL−1.180 Cu-based NMs have been developed as a solution to address antibiotic resistance, due to their superior properties and exceptional biocompatibility. Among these NMs, CuS exhibited peroxidase-like activity, making it a promising candidate for combating bacterial infection by generating hydroxyl radical (OH−) within a specific microenvironment. Researchers created chitosan-oligosaccharide-capped CuS NPs with positive charges (PCuS NPs). Through electrostatic attraction, PCuS NPs effectively bound to bacteria, allowing for direct contact and on-site generation of OH− on the bacterial surface, leading to high antibacterial efficacy in the presence of H2O2.181 By changing surface arrangements of CuS, two Cu7S4 nanosheets with exposed facets, (304) and (224) were obtained. Cu7S4 with (224) exposed facets demonstrated excellent antibacterial activity through synergetic photodynamic and photothermal therapy when exposed to near-infrared light (808 nm). Cu7S4 effectively targeted Gram-positive Bacillus subtilis, Gram-negative Escherichia coli and drug-resistant Pseudomonas aeruginosa and showed a significant therapy effect. Furthermore, Cu7S4 with (224) facets inhibited drug-resistant Pseudomonas aeruginosa in mouse skin. This study demonstrated that properly designed facets can greatly improve the antibacterial efficacy of NMs (Fig. 6D).182 In another study, the potential of ultrasmall copper sulfide nanodots (CuS NDs), known as covellite, for combating drug-resistant pathogens including MRSA and extended-spectrum β-lactamase Escherichia coli was investigated. The CuS NDs exhibited a remarkable photothermal effect, which triggered a potent antibacterial response both in vitro and in vivo. In diabetic mice models infected by MRSA, the application of ultrasmall CuS NDs with photothermal therapy resulted in the eradication of a significant portion of drug-resistant bacteria. Additionally, the released Cu2+ demonstrated the ability to promote fibroblast cell migration and endothelial cell angiogenesis, thus accelerating the wound-healing process. This work offers a helpful antibacterial approach for clinical translation.183
Metal oxide NMs and metal sulfide NMs have unique properties such as photothermal, photodynamic, and magnetic activities, which could be employed to enhance their antibacterial effects. By tuning the size, shape, and surface coating, more metal oxide NMs and metal sulfide NMs could be developed to satisfy the urgent needs of the clinics.
The typical 0 D carbon materials include fullerenes and carbon quantum dots. Fullerenes are carbon-based molecules that have a spherical shape. The smallest fullerene, C20, is made up entirely of pentagons, while the most common fullerene is C60. C60 possesses numerous conjugated double bonds that enable it to absorb light in the ultraviolet and visible regions, generating ROS when exposed to light. The photosensitive properties of fullerene and its derivatives have been applied to antibacterial materials. The bacterial cellulose/C60 composite (BCC60) had antibacterial effects against S. aureus and E. coli under both light and dark conditions. BCC60 had a bacteriostatic rate of only 50% in the dark, while under light, the antibacterial ability of BCC60 improved with an increase in C60 content and the highest bacteriostatic rate reached 95%. In this system, light induced C60 to react with atmospheric oxygen and produce a large amount of ROS, which enhanced the antibacterial effect.197 Carbon quantum dots (CQDs) are semiconductor grains with excellent optical and electrical properties that exhibit promise in the treatment of MDR bacteria through surface modification. Various methods for preparing CQDs from different carbon sources have been outlined to obtain optimal antimicrobial efficacy.198 Researchers modified CQDs with spermine (SPM) and dopamine (DA) mixtures (SPM/DA-CQDs) by a one-step pyrolysis. The MIC of SPM/DA-CQDs was 2–8 μg mL−1, indicating excellent antibacterial activity (Fig. 7A).199 The positively charged surfaces and specific functional groups of CQDs resulted in bacterial agglutination and membrane disruption. The SPM/DA-CQDs exhibited great promise as a coating material to inhibit biofilm formation on contact lenses and safeguard medical devices against contamination. In addition, the one-pot synthesized quaternary ammonium carbon quantum dots (qCQDs) had broad-spectrum antibacterial activity with an MIC range of 12.5 to 50 μg mL−1 for different bacterial strains (S. epidermidis, 12.5 μg mL; S. aureus, 25 μg mL; MRSA, 25 μg mL; E. faecalis, 25 μg mL; E. coli, 50 μg mL−1 and P. aeruginosa, 50 μg mL−1). qCQDs lysed or disintegrated bacterial cells, which destroyed the bacterial integrity, condensed and leaked substances in cells, and led to bacterial death.200
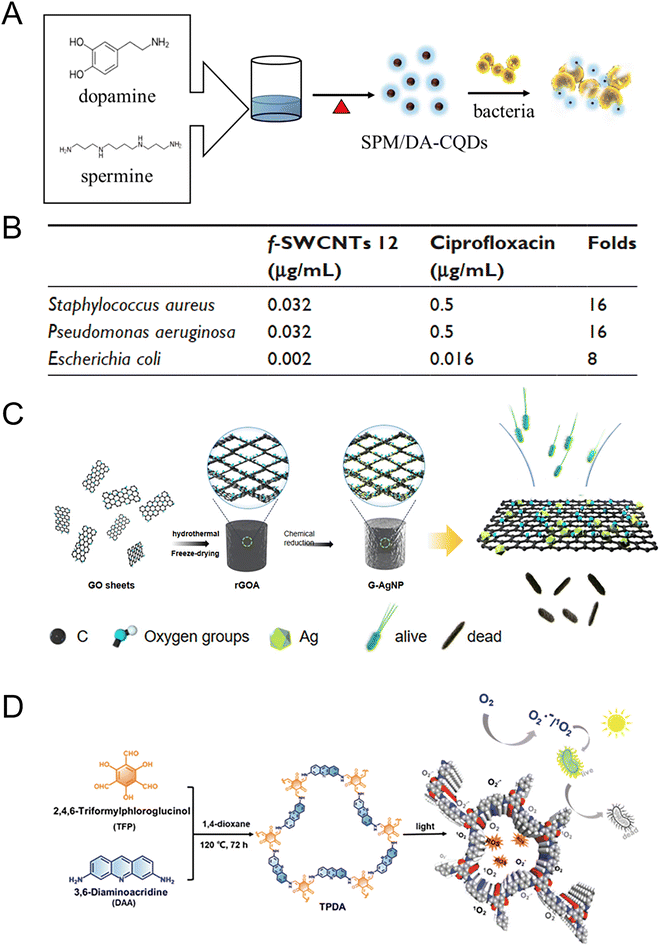 | ||
| Fig. 7 Antibacterial carbon-based NMs. (A) The synthesis of antibacterial carbon quantum dots (CQDs) from spermine and dopamine by a one-step method. Coating contact lenses with CQDs showed the potential to reduce bacterial keratitis development in the injured cornea. Reproduced from ref. 199 with permission from Elsevier, copyright 2020. (B) Compared to ciprofloxacin alone, the functionalized single-walled carbon nanotubes (f-SWCNTs 12) exhibited higher antibacterial activity against S. aureus, P. aeruginosa, and E. coli. The enhanced antibacterial effect is most likely attributed to the aggregation of bacteria with the SWCNTs, leading to increased exposure of bacteria to ciprofloxacin. As a result, the concentration of ciprofloxacin entering into bacteria was raised. Reproduced from ref. 201 with permission from Dove Medical Press Ltd, copyright 2017. (C) The reduced graphene oxide (rGO) with embedded Ag NPs demonstrated a synergistically enhanced antibacterial effect due to the larger specific area and greater number of active sites. This synergistic combination resulted in an antibacterial rate of up to 100% against both E. coli and S. aureus, even at low Ag NP contents. Reproduced from ref. 211 with permission from MDPI, copyright 2022. (D) The obtained COF exhibits high antibacterial activities under light irradiation through ROS generation. Reproduced from ref. 213 with permission from WILEY-VCH, copyright 2021. | ||
The typical 1 D carbon NMs include carbon nanotubes (CNTs), carbon nanofibers, and so forth. Carbon nanotubes exhibit anisotropy, high mechanical strength and elasticity, and excellent electrical and thermal conductivity. The influencing factors of the antibacterial activity of carbon nanotubes include length, diameter, dispersion, and concentration. Researchers modified single-walled carbon nanotubes (SWCNTs) with ciprofloxacin. Compared to ciprofloxacin alone, ciprofloxacin-capped SWCNTs exhibited significantly higher bactericidal activity. The antibacterial effect against Staphylococcus aureus and Pseudomonas aeruginosa was significantly increased by 16-fold, and against E. coli, it was increased by 8-fold (Fig. 7B).201 Carbon nanofibers involve the curling of multilayer graphite sheets to create fibrous materials with high crystalline orientation and impressive electrical and thermal conductivity. Although carbon fibers do not possess inherent antibacterial capabilities, they are infused with antibacterial agents like silver ions or zinc oxide to achieve antibacterial functions. Researchers created Ag@CNT/PA composite nanofibers by combining polyamide (PA), CNTs, and Ag NPs, which exhibited remarkable antibacterial properties and effectively combated drug resistance.202
The typical 2D carbon materials including graphene and MXene have also demonstrated good antibacterial activity. Graphene has garnered extensive attention due to its high antibacterial activity with little resistance and high biocompatibility, which is an ideal carrier for antibacterial substances.203 Graphene-based NMs have antibacterial activities against various microbial species by distinctive mechanisms.204,205 Due to its unique 2D structure, graphene has a high specific surface area and robust physical adsorption capacity, which can adsorb and destroy bacterial cell membranes. Researchers integrated Ag NPs and graphene to prepare a new type of graphene oxide-based nanocomposite and revealed that the antibacterial activity surpassed that of Ag NPs alone.206 At a concentration of 2 mg mL−1, the inhibitory zone for E. coli was 18 mm. In addition, MXene has been identified as a high-potential antibacterial agent with no potential for developing resistance.207 The photothermal ablation of Ti3C2 MXene provided a way to physically eradicate bacteria and biofilms.208 By attracting negatively charged bacteria, it destroyed the structure of the bacterial cell membrane, causing leakage of cytoplasmic contents and eventually killing the bacteria. In terms of its application, 2D Ti3C2Tx MXene-based scaffolds were employed for the treatment of wounds infected by MRSA.209
3D carbon-based NMs have better biocompatibility and higher specific surface area than the aforementioned carbon-based NMs, which show more effective antibacterial effects.210 In addition, they have significant mechanical and chemical stability, which makes them difficult to be destroyed or decomposed and can maintain long-term antibacterial properties (Fig. 7C).211 Thus, 3D carbon-based NMs have great potential in the fields of medicine, environmental sanitation and so forth. Covalent organic frameworks (COFs) and nanodiamonds are two examples of 3D carbon-based NMs with excellent antibacterial properties. COFs are 3D polymers that consist of carbon, hydrogen, and nitrogen. The highly ordered pore structures and tunable surface chemical properties make them effective antibacterial agents in pharmaceuticals and food applications.212 COFs absorb visible light and produce ROS to kill bacteria. Researchers have studied the relationship between the antibacterial rate and the photocatalytic activity of COFs. A synthesized COF-TPDA had a BET specific surface area of 210 m2 g−1, which was favored by oxygen permeation and exposure to catalytically active sites. After 10 minutes of sunlight activation, COF-TPDA had a significant catalytic effect on both E. coli and S. aureus, with a sterilization efficiency of up to 98% (Fig. 7D).213 Similarly, the survival rate of Staphylococcus aureus after the exposure of a porphyrin-COF large-area membrane to visible light for 3 h was nearly 10 times lower than that on exposure for 1.5 h, which verified the effectiveness of photocatalytic sterilization.214 Nanodiamonds possess high mechanical strength and chemical stability, making them capable of crossing cell walls and attacking bacteria.215 Carboxylated nanodiamonds (cNDs) with a size of 5 nm had a significant inhibitory effect on S. mutans by destroying bacterial cell membranes. The MIC was 4 μg mL−1 and the MBC was 16 μg mL−1.216 Nanodiamonds first attacked the outer membrane of bacteria and then gathered around the outer surface and interacted with the cell membrane. The subsequent separation of the outer membrane from the cytoplasmic membrane caused a shift in the outer membrane permeability, allowing nanodiamonds to penetrate deeper into the envelope until the cytoplasmic membrane was destroyed, leading to leakage of bacterial contents and death.217
In summary, the antibacterial effect of carbon-based NMs is a multifaceted process, and the development of biodegradable carbon-based NMs can mitigate their toxicity in antibacterial applications. Further research is needed to improve site specificity for targeted effects. The collaborative interdisciplinary efforts can pave the way for the significant growth of carbon-based NMs in the field of antibacterial applications.
3.2 Organic NMs
Combined with inorganic chemistry, more natural or synthesized organic NMs have broad-spectral antibacterial properties, which could be used in animal husbandry, agriculture, the food packaging industry, preservatives, medical disinfectants, and other fields.218–220 In this chapter, to introduce different organic NMs, we generally divide antibacterial NMs into two categories: natural NMs and synthesized NMs. | ||
| Fig. 8 Preparation of natural polymer- and synthetic polymer-based NMs for antibacterial applications. (A) Acid-functionalized chitosan was chosen as the main structure for the development of antibacterial polypeptide-grafted chitosan-based nanocapsules. These nanocapsules act as “armed” carriers of drugs, exhibiting exceptional antibacterial effectiveness against both Gram-positive and Gram-negative bacteria. Reproduced from ref. 226 with permission from the American Chemical Society, copyright 2013. (B) Schematic of the actuation mechanism of pH-responsive hydrogels. pH-Responsive hydrogels, which belong to the stimulus-responsive hydrogel family, exhibit deformation behaviors such as swelling and shrinking in response to pH changes in the surrounding environment. Reproduced from ref. 229 with permission from Springer Nature, copyright 2019. (C) To treat wounds infected with MDR bacteria, MBA-modified Au NPs modified PCL/gelatin antibacterial wound dressings (PGA) were prepared using a co-electrospinning technique. The PGA electrospun nanofibers facilitate the re-epithelialization process and enhance the rate of healing in the infected wounds. Reproduced from ref. 237 with permission from the American Chemical Society, copyright 2020. (D) The AMP@PLGA-MS@Gln/CS/nHAp composite membrane was fabricated using sequential layer-by-layer electrospinning and electrospraying techniques. This biodegradable membrane was composed of two layers: the barrier layer, consisting of Gln/CS nanofibers, and the osteogenic layer, comprising Gln/CS/nHAp nanofibers. During the electrospinning process, the AMP-loaded PLGA microspheres were electrosprayed alternately during the electrospinning, resulting in their incorporation within the membrane structure. Reproduced from ref. 241 with permission from MDPI, copyright 2018. (E) The Ag NPs/CPH medical hydrogel was synthesized using a specific method and its applications were demonstrated in a new animal model for infected wounds. This innovative hydrogel had excellent conductivity and demonstrated bacteriostatic effects, making it a promising solution for treating severely infected wounds. Reproduced from ref. 243 with permission from Frontiers Media SA, copyright 2021. | ||
In addition, chitosan is another solution for resisting clearance from mucus, which is the key point in the biofilm. The antibiotic resistance of bacteria within biofilms can be up to 1000 times higher compared to bacteria in a free-living state.228 The antimicrobial activity of chitosan inside cells depends on its molecular weight and its ability to penetrate the cell surface. The inhibition of Staphylococci biofilm formation and disruption of biofilm structure have been observed with low molecular weight chitosan. In the concentration range of 400 to 1600 μg mL−1, low molecular weight CS demonstrated significant activity, resulting in 32.9% to 88.7% inhibition for S. aureus V329 biofilm and 95.1% to 98.4% for S. xylosus 1007 biofilm. However, challenges for future applications of CS include its low water solubility and the lack of defined molecular weight.
Due to the insolubility of CS under physiological conditions, several modification strategies of chitosan and its derivatives have been developed to promote their antimicrobial activity. The CS chains stretched out to a greater degree and showed better antibacterial performance under acidic pH.229–231 pH-Sensitive CS hydrogels have emerged as a prominent area of research in recent years (Fig. 8B).229 The pH value of normal skin is approximately 5, while wound tissue typically has a pH of around 7.4. CS hydrogel exhibited pH-responsive behavior, swelling at pH ≤ 5.0 and shrinking at pH ≥ 7.4. The remarkable antimicrobial activity of CS is attributed to the abundance of basic amino groups, resulting in an overall cationic charge at an acidic pH level, which aids in the disruption and lysis of bacterial cells. In comparison to gauze dressing, the application of CS dressing significantly inhibited bacterial growth in the wound during the initial 5 days following surgery.232 Nevertheless, certain challenges remain to be addressed for chitosan-based antibacterial NMs, including issues related to solubility or mechanical properties. It is anticipated that the future development of multifunctional CS NMs will offer promising solutions by combining excellent antibacterial properties with the preservation of their inherent characteristics.
Synthetic polymers are available in the form of nanocapsules and nanospheres and can combine antibacterial agents, growth factors, and nucleic acids to fabricate functional NMs.234,235 Poly(lactic-co-glycolic acid)(PLGA) is a biodegradable polymer extensively employed in the biomedical field. Due to the degradability of PLGA, researchers created a clindamycin delivery system through PLGA-polyethylenimine (PLGA-PEI) nanoparticles (Cly/PPNPs). Cly/PPNPs demonstrated excellent antibacterial effects, resulting in a reduction of more than 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log(99.999% killing) within 24 hours and a speeded wound healing.236 Meanwhile, polymeric film-forming systems (FFSs) are currently generating interest for targeted drug delivery to specific skin sites and improving therapeutic efficacy. Poly(ε-caprolactone) (PCL) is an FDA-approved biomaterial. We incorporated Au_MBA NPs into electrospun PCL nanofibrous membranes (PGA) to fabricate wound dressings. These dressings exhibited remarkable effectiveness in treating MDR S. aureus-infected wounds, offering great potential for widespread clinical use (Fig. 8C).237 Polyethylene glycol (PEG) can resist protein adsorption, which is suitable for changing the chemical properties of the NM surface.238 PEG prevents biofilm formation and cellular adhesion by blocking the adhesion of bacteria on the surface. Researchers combined PEG with chitosan to produce a hydrogel that had a 100% inhibition rate against E. coli and S. aureus.239 PEG can bind to the surface of nylon and prevent the adhesion of S. aureus and P. aeruginosa. Polyethylene oxide (PEO) is a biocompatible and hydrophilic polymer, which is approved by the FDA and soluble in water.240 Researchers produced electrospun and electrospray nanofiber membranes incorporating various concentrations of antimicrobial peptide NP10, chitosan (CS) and PEO.241 CS-PEO nanofibers containing NP10 can accelerate the wound-healing process, displaying excellent wound-healing ability and antibacterial properties (Fig. 8D).241 The conductive polymer with antibacterial monomers makes it possible to develop wound dressings that possess both antibacterial and self-healing capabilities. We utilized an acidic solution of poly(3,4-ethylenedioxythiophene): poly(styrenesulfonate) (PEDOT:PSS), known for its electrical conductivity, to fabricate conductive PEDOT:PSS/guar slime (PPGS) composite, which had a remarkable self-healing capability.242 In the stretchable areas of rats, the GS PPGS group exhibited nearly complete healing, whereas the gauze group only achieved approximately 66% wound closure by day 14 after the operation. The conductive composite materials may enhance the effectiveness of both endogenous and exogenous electrical stimulation, guiding the migration of cells toward the wound site. Researchers created a novel hydrogel with favorable mechanical characteristics and antibacterial effects using PANI as the conductive agent. Combining the Ag NPs in a hydrogel system based on a conductive polymer (CPH), the hydrogel dressing had excellent antibacterial activity against S. aureus-infected wounds for 48 hours, due to the high water-absorption and continuous swelling properties for the antibacterial Ag NP-release (Fig. 8E).243 Additionally, Ag NPs loaded in the conductive hydrogel exchanged with the wound exudate along the concentration gradient, which successfully treated seriously infected wounds, even after a long period of bacterial infection (more than 20 hours).
log(99.999% killing) within 24 hours and a speeded wound healing.236 Meanwhile, polymeric film-forming systems (FFSs) are currently generating interest for targeted drug delivery to specific skin sites and improving therapeutic efficacy. Poly(ε-caprolactone) (PCL) is an FDA-approved biomaterial. We incorporated Au_MBA NPs into electrospun PCL nanofibrous membranes (PGA) to fabricate wound dressings. These dressings exhibited remarkable effectiveness in treating MDR S. aureus-infected wounds, offering great potential for widespread clinical use (Fig. 8C).237 Polyethylene glycol (PEG) can resist protein adsorption, which is suitable for changing the chemical properties of the NM surface.238 PEG prevents biofilm formation and cellular adhesion by blocking the adhesion of bacteria on the surface. Researchers combined PEG with chitosan to produce a hydrogel that had a 100% inhibition rate against E. coli and S. aureus.239 PEG can bind to the surface of nylon and prevent the adhesion of S. aureus and P. aeruginosa. Polyethylene oxide (PEO) is a biocompatible and hydrophilic polymer, which is approved by the FDA and soluble in water.240 Researchers produced electrospun and electrospray nanofiber membranes incorporating various concentrations of antimicrobial peptide NP10, chitosan (CS) and PEO.241 CS-PEO nanofibers containing NP10 can accelerate the wound-healing process, displaying excellent wound-healing ability and antibacterial properties (Fig. 8D).241 The conductive polymer with antibacterial monomers makes it possible to develop wound dressings that possess both antibacterial and self-healing capabilities. We utilized an acidic solution of poly(3,4-ethylenedioxythiophene): poly(styrenesulfonate) (PEDOT:PSS), known for its electrical conductivity, to fabricate conductive PEDOT:PSS/guar slime (PPGS) composite, which had a remarkable self-healing capability.242 In the stretchable areas of rats, the GS PPGS group exhibited nearly complete healing, whereas the gauze group only achieved approximately 66% wound closure by day 14 after the operation. The conductive composite materials may enhance the effectiveness of both endogenous and exogenous electrical stimulation, guiding the migration of cells toward the wound site. Researchers created a novel hydrogel with favorable mechanical characteristics and antibacterial effects using PANI as the conductive agent. Combining the Ag NPs in a hydrogel system based on a conductive polymer (CPH), the hydrogel dressing had excellent antibacterial activity against S. aureus-infected wounds for 48 hours, due to the high water-absorption and continuous swelling properties for the antibacterial Ag NP-release (Fig. 8E).243 Additionally, Ag NPs loaded in the conductive hydrogel exchanged with the wound exudate along the concentration gradient, which successfully treated seriously infected wounds, even after a long period of bacterial infection (more than 20 hours).
The emergence of bacterial infections highlights the need to explore novel polymer-based materials with multifunctional antibacterial properties. Given the intricacy of this problem, future directions may involve a combination of nano-patterned and nano-structured surfaces with adjustable chemical composition to minimize bacterial resistance.
3.3 Composite NMs
Composite NMs represent innovative structures consisting of two or more distinct substances that possess unique physicochemical properties.244 The substantial enhancement and long-lasting antibacterial characteristics of these materials stem from the synergistic effect achieved through combining different types of NMs. Consequently, they have found extensive applications in fields such as healthcare,245,246 food packaging,247 water treatment248,249 and so forth. This chapter aims to provide a comprehensive introduction to various types of composite NMs, which are typically classified into three categories: metal-based nanocomposites, polymer-based nanocomposites, and other nanocomposites.In summary, research on antimicrobial composites is still in its early stages, requiring significant investment in further exploration to effectively enhance the combination of different NMs and optimize their performance.
4. Challenges for nanoantibiotics
NMs have great potential in a wide range of applications due to their excellent ability to deal with drug-resistant pathogens. However, to realize these applications, the development of nanoantibiotics faces major challenges: bioeffects and biosafety as well as large scale manufacturing.Both biosafety and large-scale manufacturing are prerequisites for the wide application of NMs. On one hand, before consideration of large-scale manufacturing, the biosafety of NMs should be evaluated. On the other hand, if large-scale production is not possible, even if the NMs are tested to be safe, they cannot become products. Thus, testing biosafety and mass production in sequence is suggested to be the best strategy for successful commercialization of NMs.
4.1 Bioeffects and biosafety
Nanoantibiotics interact with the human body directly or indirectly. These interactions inevitably pose impacts on human body thus causing beneficial or detrimental bioeffects. Thus, comprehensive evaluation of the bioeffects and biosafety is significant for nanoantibiotics with the potential to be commercialized. Antibacterial NMs interact with the human body at different levels which can reflect the bioeffects and biosafety of NMs from different aspects. So, we discuss this topic by classifying it into three parts: cell level, organ level, and body level.4.1.1.1 Inorganic NMs.
4.1.1.1.1 Ag NMs. Ag NPs are one of the largest groups of manufactured NPs and are widely used in various fields as antimicrobial agents.264 The toxicity of Ag NPs is relevant to their surface charge, size, and surface modification. The number of acidic or basic groups on the surface of nanoparticles can determine the zeta potential of the Ag NPs. The surface of the cell membrane is negatively charged, so the charges on the Ag NPs directly allow the particle to interact with the cell. As a physical parameter, the size affects the performance of Ag NPs. Small-sized Ag NPs are more cytotoxic than large-sized particles.265–267 Ag NPs can penetrate the liver cell membrane and induce the production of ROS, which cause lipid and protein oxidation and DNA damage.268 Ag NPs can ionize in aqueous solution and can be oxidized by O2 and other molecules in the organism, which produces Ag+ and causes potential toxic effects. Ag+ can form stable bonds with S in sulfur-containing proteins and peptides, which can resist damage due to ROS. Moreover, Ag+ ions can interact with NADH dehydrogenase from the respiratory chain and result in the uncoupling of respiration from ATP synthesis.269 Ag NPs can affect the stress-sensing proteins in the endoplasmic reticulum (ER) to restrain unfolded protein from folding, resulting in changes in the homeostasis of the ER and further aggravating ER damage.270 Mitochondria are one of the most sensitive targets for the toxicity of Ag NPs. Ag NPs can penetrate the mitochondrial membrane and damage the ridge structure of mitochondria, and even influence mitochondrial fusion and fission.271,272
4.1.1.1.2 Au NMs. There are three major factors including particle size, surface modification, and particle shape influencing the toxicity of Au NPs. The relationship between toxicity and particle size of Au NPs was studied by different research groups. Among Au NPs ranging from 0.8 nm to 15 nm, 1.4 nm AuNPs were the most cytotoxic, while the 15 nm Au NPs were nontoxic.273 Controversially, another study found that 45 nm Au NPs were more toxic than the smaller ones.274 Thus, the influence of size on the toxicity of Au NPs needs further systematic study. Surface coating on Au NPs directly influences the interaction between the particle and the cells. The modification of molecules with different charges has a great influence on the cellular toxicity of Au NPs. Positively charged NPs were more easily transported into cells due to the electrostatic interaction between negatively charged cell membranes and the particles, which can lead to the breakage of cell membranes. By contrast, the anionic molecules-functionalized Au NPs were relatively less toxic.275 The shape also influences the biocompatibility of Au NPs. Within the 10–100 nm range, Au spheres were taken up more efficiently than Au nanorods.276 The size also influences the biosafety of Au NPs. Au NPs of 5–50 nm size increased the level of ROS, and interfered with mitochondrial membrane potential leading to apoptosis of cells.277 The oxidative stress environment could initiate the autophagic process, which can degrade the Au NPs and thus exert less damage to the cell.278 Au NPs can increase reactive nitrogen species (RNS) levels (NO production) which can eventually result in cell necrosis or apoptosis.279 Au NPs were nontoxic for 24 h in terms of membrane damage, oxidative stress, and cell proliferation inhibition. However, these Au NPs were toxic after 14 days, exhibiting long-term and high-concentration-exposure dependent toxic reactions.280 Researchers demonstrated that Au NPs were toxic to cancer cells and not to normal healthy cells.,281,282 implying that the toxicity of Au NPs is cell-specific.
4.1.1.1.3 CuO NMs. Due to the wide usage of CuO NPs in the antimicrobial field, there is an urgent need to critically assess their toxicity. In vitro, CuO NPs cause cytotoxicity by primarily generating oxidative stress. The size of CuO NPs is associated with their toxicity. The small NPs show more toxicity and are more suspected of cellular internalization than the large ones.283In vitro study demonstrated that CuO NPs induced higher lipid peroxidation and ROS production in A549 cells, and lower antioxidant glutathione (GSH) levels in HepG2 cells, suggesting that oxidative strain might be the key mechanism behind the toxicity of CuO NPs.284 CuO NPs can also cause time- and dose-dependent genotoxicity in A549 cells by inducing lesions and damages that ultimately cause cytotoxicity.285
4.1.1.1.4 ZnO nanoparticles. ZnO is widely applied to the food industry as a Zn supplement and agricultural fertilizer. ZnO is registered as a generally recognized safe (GRAS) material in the United States, suggesting its safety at actual usage levels. The toxicity of ZnO NPs is associated with the released Zn ions and the generation of ROS.286 The cytotoxicity of ZnO concerning particle size has been explored. Size-dependent cytotoxicity in terms of cell viability and ROS generation demonstrated higher toxicity of small-sized ZnO NPs than larger-sized ones.286 Zinc ions disintegrated from ZnO NPs caused dysfunction of mitochondria, activation of caspase and apoptosis of cells.287 ZnO NPs did not enter into human neuronal cells and the nanoparticles in the medium led to cell cycle alterations, apoptosis, micronuclei production, H2AX phosphorylation, and DNA damage-mediated cyto- and genotoxicity.288 ZnO NPs exhibited genotoxic, cytotoxic, clastogenic, and actin depolymerization effects by inducing ROS-mediated oxidative stress responses by macrophages of mice.
4.1.1.1.5 TiO2 nanoparticles. TiO2 NPs have been widely used in many applications including cosmetics industries, textile, plastics, and food packaging. So, the biosafety of TiO2 NPs has been a serious topic. In vitro studies showed that TiO2 NPs promoted the generation of ROS and caused oxidative stress, interfering with cell metabolism and damaging DNA.289 Researchers evaluated the phototoxicity of TiO2 NPs with four different sizes (<25 nm, 31 nm, <100 nm and 325 nm) and two crystal forms (anatase and rutile) toward human skin keratinocytes under ultraviolet radiation. All the TiO2 NPs can generate ROS. By comparison, smaller TiO2 NPs resulted in higher levels of ROS than the big ones.290 Thus, appropriately tuning the size may be a strategy to ameliorate the toxicity of TiO2 NPs.
4.1.1.2 Carbon-based NMs. Carbon-based NMs, such as carbon nanotubes (CNTs), graphene, and fullerenes, show great potential in antibacterial applications. Thus, the evaluation of their toxicity is of significance.
4.1.1.2.1 Carbon nanotubes. Carbon nanotubes (CNTs) including single-walled (SWCNTs) and multi-walled CNTs (MWCNTS) have potent antibacterial activity due to their unique electrical, mechanical, and thermal properties. Due to their lightweight and small size, CNTs can be easily inhaled leading to possible harmful effects on human body. The toxic response of CNTs is dependent on physicochemical characteristics like size, shape, aspect ratio, chemical composition, stability, crystal structure, surface area, surface energy, surface charge, surface roughness, and purity. SWCNTs and MWCNTs can cause DNA damage and inflammatory responses.291 Exposure to SWCNTs and MWCNTs can cause the formation of free radicals, accumulation of peroxidative products and depletion of cell antioxidants in vitro.292,293 CNTs have the potential to induce genotoxicity due to disruption of the chromosomal structure, mutations and double-strand DNA breakage. In vitro exposure of cultured airway epithelial cells (A549) to MWCNTs could increase the DNA repair activity of oxidatively damaged DNA and drive the cells toward replicative senescence, suggesting a low mutagenicity of CNTs in cultured cells.294
4.1.1.2.2 Graphene oxide. The interaction between graphene oxide (GO) and cells is affected by various factors including particle shape, particle size, number of layers, and surface functionalization. Besides, the in vitro cytotoxicity of GO is closely related to incubation conditions such as exposure dose, exposure time, incubation temperature, and cell types.295 Thus, due to the different test conditions, there are contradictory results regarding the biocompatibility of GO. Some research supports the beneficial effects of GO on cells, such as cell adhesion and growth promotion. By contrast, more research suggests the toxic effects of GO on cells, which varies according to the experimental conditions. For example, GO with a smaller size caused more severe oxidative stress and induced more obvious cytotoxicity in A549 cells compared to GO with a larger size.296 Low concentration of GO showed no cytotoxicity on A549 cells,296 but can decrease the metabolic activity of neuronal PC12 cells.297
4.1.1.3 Organic NMs.
4.1.1.3.1 Chitosan-based NPs. Chitosan is a natural linear polysaccharide polymer obtained by the alkaline hydrolysis of chitin which is one of the most abundant natural amino polysaccharides. Chitosan has an abundance of hydroxyl and amine functional groups and is cationic and hydrophilic. Chitosan is biocompatible and biodegradable by certain enzymes into non-toxic oligosaccharides. Chitosan and its derivatives are widely used in constructing nanoparticles for applications in biomedicine, thus, the evaluation of their biosafety is crucial. Researchers developed chitosan-based nanogels as drug-delivery platforms and investigated their potential impacts on human osteocartilaginous cells. Monodisperse nanogels with sizes ranging from 268 to 382 nm neither affected cell proliferation nor induced nitric oxide production in vitro, indicating their good biocompatibility.298 The charge, hydrophobicity and solubility of chitosan are key parameters determining its interaction with cells. Researchers found that positively charged unmodified chitosan did not penetrate cells and was not toxic. Quaternization of chitosan (CHq) led to increased cell penetration, ROS production, cell cycle arrest, and inhibition of cell proliferation. Modification of CHq with hydrophobic residues significantly diminished cytotoxic activity. The negatively charged chitosan derivative succinyl chitosan penetrated the cells efficiently and stimulated cell proliferation.299 Thus, appropriate modification of chitosan is needed to facilitate its safe application.
4.1.1.3.2 Antimicrobial peptides. Antimicrobial peptides (AMPs) have attracted considerable interest in clinical research as potential therapeutic agents against MDR bacteria. AMPs are produced by living organisms to defend against pathogens. AMPs consist of 2–50 amino acids and are generally cationic and rich in hydrophobic amino acid residues.300 To date, more than 3000 AMPs have been officially classified and registered in the AMP database.301 Hydrophobicity, net positive charge and secondary structure are the main physicochemical features that determine the interactions between AMPs and organisms. The electrostatic interaction between the positively charged AMPs and the negatively charged lipid components present in the plasma membrane of microorganisms confers the AMPs with antimicrobial activity. In contrast to microorganisms, the cytoplasmic membrane of mammalian cells consists mainly of zwitterionic phospholipids, which reduce interactions with AMPs, increasing the selectivity of these peptides for microorganisms.302
The unique properties of AMPs, including the broad antibacterial spectrum, low toxicity to mammalian cells, and reduced induction of resistance in target cells make AMPs excellent candidates for the development of a new class of antibacterial agents. Currently, several AMPs such as Nisin and Pediocin PA-1 have been on the market for the food industry.303 In the food industry, many AMPs have been used for the preservation of foods. For instance, nisin has been used in the biopreservation and extending the shelf-life of food items for 40 years in over 50 countries. Natamycin has been approved for use in the food industries in nearly 150 countries.304 ε-Polylysine and pediocins have been added to the GRAS list by the FDA and are allowed to be used as food preservatives. More AMPs including cecropin, defensins, pleurocidin, and enterocin were verified to be non-toxic to mammalian cells and are valuable agents for food preservation.303
4.1.2.1 Inorganic NMs. In this section, we focus on the evaluation of bioeffects and biosafety of metallic nanoparticles, metal oxide nanoparticles, and carbon-based NMs at the organ and body levels.
4.1.2.1.1 Ag NMs. Ag NPs can be absorbed through the respiratory and digestive systems and the skin.305 After inhalation exposure, Ag NPs were detected in the lungs, the blood, and other organs including the liver, kidneys, spleen, heart, and brain,306 and the appearance of multinucleated macrophages was observed in the lungs, indicating inflammation caused by the NPs in rats.307 Orally absorbed Ag NPs can enter into the bloodstream through the intestines and accumulate in other organs such as the duodenum, liver, kidneys, and spleen in mice,308 and evoke an increase in pro-inflammatory cytokines, demonstrating the induction of inflammation.266 Nanocrystal silver dressing-contacting healthy skin showed clusters of silver in the stratum corneum, which confirmed the possibility of Ag NP penetration through intact skin. However, the Ag NPs did not reach systemic circulation and should therefore not have systemic consequences.309 The toxicity of Ag NPs is relevant to their surface charge, size, and surface modification.
4.1.2.1.2 Au NMs. Besides the intravenous administration route, Au NPs can be absorbed through the respiratory system, digestive system, and skin. Inhalation of Au NPs with a size of 6 nm for 90 days caused a decrease in respiratory parameters and infiltration of inflammatory cells in alveoli in rats.310 Inhalation of 20 nm Au NPs led to the retention of the particles in the liver, spleen, kidneys, uterus, and brain. Long-term removal (after 28 days) of Au NPs was dominated by macrophage-mediated transport through interstitial tissue to the larynx and gastrointestinal tract.311 Gold was detected in the urine of volunteers after exposure via inhalation to 4 nm AuNPs, but not in the urine of volunteers exposed to larger particles (34 nm), indicating the size-dependent removal of Au NPs through the kidney.312 Intravenous administration of Au NPs altered in biochemical parameters of glutamic oxaloacetic transaminase (AST), glutamic-pyruvic transaminase (ALT), and blood glucose, implying their impacts on the liver and pancreas. By contrast, the orally administered Au NPs were excreted in feces and entered the blood via the gastrointestinal system.313 Au NPs can penetrate through the skin of the hind paw and the anterior abdominal wall of rats in a size-dependent manner (higher penetration for smaller particles).314 After incubating Au NPs with surgically resected dermal fragments, Au NPs could be detected in the deeper stratum corneum, epidermis, and dermis.315 There are three major factors including particle size, surface modification, and particle shape influencing the toxicity of Au NPs. The relationship between toxicity and particle size of Au NPs was studied by different research groups. Among Au NPs ranging from 0.8 nm to 15 nm, 1.4 nm AuNPs were the most cytotoxic, while the 15 nm Au NPs were nontoxic.273 Controversially, another study found that 45 nm Au NPs were more toxic than the smaller ones.274 Thus, the influence of size on the toxicity of Au NPs needs further systematic study.
Surface coating on Au NPs directly influences the interaction between the particle and the cells. When Au NPs enter the body, proteins in the serum will coat the particle to form a protein corona on the Au NPs. The cellular study demonstrated that the corona played a protective role in the cell membrane.316 Poly(ethyleneglycol) (PEG) is well-known to reduce nonspecific binding of biological molecules to surfaces and avoid macrophage recognition and phagocytosis, thus, PEG coating is a commonly used method to modify Au NPs and has been widely applied to reduce toxicity.317
4.1.2.1.3 CuO NMs. CuO NPs can enter the body through the routes of inhalation, oral administration and skin.318 The size of CuO NPs is associated with their toxicity. In vivo assays indicated a marked increment of the level of Cu in blood and tissues after sub-acute inhalation of CuO NPs in mice, depicting the translocation of Cu into the bloodstream and organs. An elevation in the level of lactate dehydrogenase, total cell counts, macrophages, neutrophils, and inflammatory cytokines was also observed, indicating the inflammatory effects of the NPs on the immune system.319 Oral administration of CuO NPs in mice caused leukocytosis, and increased serum levels of ATL, AST, urea, and creatinine. Increased P53 mRNA and caspase-3 protein expression was also observed in hepatic tissues. Moreover, CuO NPs also caused necrosis in hepatic, renal, and splenic tissues.320 CuO NPs led to a significant decrease in memory and learning in rats321 and showed a reduction in movement distance, velocity and angular velocity of zebrafish larvae.322 Rats exposed to CuO NPs showed a progressive decline of memory and noticeable cognitive and psychiatric disturbances.323
4.1.2.1.4 ZnO NMs. ZnO could be administered via the respiratory and digestive systems, and the skin. Studies demonstrated that ZnO NPs (5 mg mL−1) with constituent particle sizes of 78 nm showed solubility of ∼96% in the simulated gastric fluid (pH ∼1.5),324 implying high absorption through the digestive system. After entering the bloodstream, stable BSA-ZnO NP corona can be formed associated with the conformational change or unfolding of BSA interacting with ZnO NPs, showing the interaction between the body plasma and the NPs.325 Moreover, the ZnO NPs led to severe inflammation and damage to the liver, lungs, and kidneys in mice.326 Totally speaking, the toxicity of ZnO NPs is relatively smaller than those of CuO NPs and Ag NPs. The toxicity of ZnO NPs is dose-dependent and is related to their size, and could be ameliorated by tuning their dose and size.
4.1.2.1.5 TiO2 NMs. Antibacterial TiO2 NPs with an average size of 57.5 nm showed potent activity against human cancer cell lines with IC50 lower than 18.7 μg mL−1. In vivo acute toxicity study in albino male rats showed that the biochemistry and pathology of the liver, kidney, and brain proved the safety of the TiO2 NPs at low doses (10 mg kg−1). However, at a high dose of 1000 mg kg−1, TiO2 NPs were deposited in different organs except the cerebral tissue, suggesting that the dose is a key factor determining the in vivo effects of TiO2. TiO2 nanoparticles can cause pregnancy complications when injected intravenously into pregnant mice.327 TiO2 nanoparticles with diameters of 35 nm were found in the placenta, fetal liver and fetal brain. Mice treated with these nanoparticles had smaller uteri and smaller fetuses than untreated controls.328 In another study, the effect of toxicity of TiO2 nanoparticles was examined on the immunity and antioxidant system of Pacific oysters. The oysters were exposed to different concentrations (0, 5, 10 and 15 μg L−1) of TiO2 for 14 days. TiO2 significantly decreased the total counts of hemocytes and phagocytosis activity, reflecting the adverse effects on immunity. High concentrations of TiO2 (10 and 15 μg L−1) significantly decreased the activity of all antioxidant enzymes including superoxide dismutase, catalase activities, and glutathione peroxidase, implying the toxic effects on antioxidant defense.329 ZnO NPs and TiO2 NPs are often used together in sunscreen products. The joint effects of the two NMs on skin is a critical problem relating to their biosafety. Researchers found that ZnO NPs were toxic to HaCaT cells, which could be rescued with co-exposure to TiO2 NPs. This effect was validated on the epidermal model EpiSkin. The underlying mechanism is that TiO2 NPs restricted the cellular uptake of ZnO NPs and meanwhile decreased the dissociation of Zn2+ from ZnO NPs, performing an antagonistic effect on the cytotoxicity caused by ZnO NPs.330
4.1.2.2 Carbon-based NMs.
4.1.2.2.1 Carbon nanotubes. Due to their lightweight and small size, CNTs can be easily inhaled leading to possible harmful effects in the human body. In vivo experiments on mice indicated accumulation of SWCNTs in the liver, spleen and lung 90 days after a single tail vein injection of SWCNTs (10–30 nm diameter and 2–3 μm length). The decreased GSH levels and increased levels of malondialdehyde in the liver and lung indicated that SWCNTs can induce oxidative damage in vivo.331 Immune-competent nude SKH-1 mice exposed to SWCNTs (5 days, with daily doses of different concentrations of SWCNTs) showed reduction in GSH concentration and oxidation of protein thiols/carbonyls, highlighting the role of SWCNT-mediated oxidative stress in vivo.332 In another study, researchers investigated the acute dermal and acute eye irritation in rabbits by the administration of MWCNTs. A single dose of 5000 mg kg−1 MWCNTs did not cause treatment-related death or toxic signs. Acute oral study revealed that the LD50 of MWCNTs was greater than 5000 mg kg−1. In the 28 day repeated dose study on rats, no significant differences in hematology and clinical biochemistry were detected. However, histopathology results revealed mild periarteriolar lymphoid cell depletion in the spleen and mild tubular cell degeneration on the cortex and medulla of both kidneys of the rats while no remarkable lesions were seen on the other organs.333 The above results indicate that MWCNTs have potential toxicity when the concentration is higher than certain levels.
4.1.2.2.2 Graphene oxide. The natural routes of entry of GO into the body are inhalation, ingestion, and dermal. Besides, GO can be administered by intravenous, intraperitoneal, and subcutaneous injections. Intratracheal administration of GO in mice developed fibrosis in lung tissue,334 whereas intravenous injection of GO could induce the formation of granulomas and pulmonary edema.335 Orally administered GO can be absorbed via the mice's intestine and may pose toxic effects on intestinal villus by reducing their length.336 Also, oral administration of GO in rats showed hepatotoxic effects and induction of oxidative stress.337 After intra-hippocampus injection of GO, patch-clamp recordings of brain slices showed a significant reduction in glutamatergic synaptic activity compared to saline injections, indicating the neurotoxicity of GO.338 However, after intravenous administration of GO in rats for seven days, GO did not affect the locomotor activity and exploratory behavior. The rats treated with GO did not undergo any cerebral cortex changes.339 Thus, the administration routes may affect the toxicity of GO.
4.1.2.3 Organic NMs.
4.1.2.3.1 Chitosan-based NPs. The reported experiments demonstrated controversial results on the toxicity of chitosan-based NPs. Researchers reported that 200 nm and 340 nm chitosan NPs were all toxic to embryos and caused teratogenic phenotypes in zebrafish at very low concentrations (40 mg L−1), although the small NPs were less toxic compared to the large ones.340 Researchers reported that chitosan NPs with sizes of 100–150 nm did not induce teratogenic phenotypes on zebrafish embryos at a concentration of 200 mg L−1. Also, the chitosan NP-treated embryos displayed normal heart physiology at low concentrations and abnormal hyperactivity at high concentrations. Collectively, chitosan NPs are biocompatible biomaterials and are safe under certain concentrations.
4.1.2.3.2 Antimicrobial peptides. Antimicrobial peptides (AMPs) are a promising candidate as potential therapeutic agents against bacterial infections. For clinical application, AMPs are considered to have advantages from the safety perspective compared to small molecule drugs from four aspects:341,342 (1) their degradation products are natural amino acids; (2) the half-life of AMPs is usually short, few AMPs accumulate in tissues; (3) AMPs are generally less immunogenic than recombinant proteins and antibodies; (4) local administration, which is the most common delivery route for AMPs, reduces the risk for any systemic toxicology concerns. Due to the excellent biocompatibility, many AMPs such as plectasin, histatin, ghrelin, murepavadin, and hLF1-11 are in various stages of clinical trials and are promising to be approved as clinical therapeutic agents.343
4.2 Large-scale manufacturing
Although NMs show great potential in fighting against MDR bacteria-causing infections, scaling of successful laboratory-adapted protocols to an industrial level is not easy to achieve. The major reason is that the reaction conditions for the synthesis of monodispersed nanoparticles must be strictly controlled. The window of variability of parameters is usually narrow for the nanoparticle properties to remain the same from laboratory to industry. Thus, how to realize mass production of NMs with high-quality control is a major issue deserving exploration. In this section, we summarize recent developments in methods for large-scale manufacturing of various NMs.4.2.1.1 Ag NPs. Ag NPs have been considered attractive as a new class of antimicrobials offering new solutions for combating a wide range of bacterial pathogens. However, the mass production of Ag NPs faces problems.
One of the factors influencing large-scale production of Ag NPs is the use of toxic reducing agents and organic solvents which suffers from disadvantages such as high energy consumption, low yield and demand for wasteful purifications. Recently developed bioreduction methods based on microorganisms and plant extracts have the potential to resolve this problem. For instance, tangerine extract was used as a solvent, reducing agent and stabilizing agent simultaneously in synthesizing Ag NPs, which were stable over a long period.344 Another study used chitosan as a reducing agent and stabilizer to synthesize Ag NPs based on the one-pot green process in an autoclave. The synthesized Ag NPs had reduced cytotoxicity compared with commercial citrate-stabilized Ag NPs.345
Low productivity per reactor volume is a drawback of conventional methods for synthesizing Ag NPs. For example, the conventional polyol processes require an excess of poly(vinyl pyrrolidone) (PVP) (10–1000 times more than AgNO3). To decrease the reaction volume, researchers used poly(acrylic acid) (PAA) instead of PVP. At a low molar ratio of PAA to AgNO3 (<2), the spherical Ag NPs with the average size of 30 nm were successfully synthesized at 100 gram scale with a high reaction yield of 90%.346
The preparation of NMs with well-controlled size and monodispersed in a small volume is relatively easy to achieve. Based on a small reaction volume, an exponential manufacturing synthesis of Ag NPs was explored. Low concentration of gallic acid was utilized to fabricate small sized (8 ± 4 nm) Ag NPs without any waste byproducts. It is suggested that gallic acid is suitable for scaling up the production of Ag NPs with low volume.347
To achieve large-scale and facile synthesis of Ag NPs, researchers developed a time-saving and efficient method by using microwaves. The obtained Ag NPs homogeneously dispersed in deionized water had a uniform size and average size of 20 nm although the concentration of the silver salt of the precursor solution was as high as 20 wt%. For large-scale industrialization, the synthetic process window was widened to a great extent.348 This study provides a robust way to prepare high-quality Ag NPs with high efficiency.
Compared with the reaction in a liquid state, the solid-state reaction can save solvents and is relatively environmentally friendly. Researchers have presented a one-step strategy for preparing Ag NPs on a large scale based on solid-state reactions at ambient temperature. The synthesis can be achieved by simply grinding AgNO3 and ascorbic acid (AA) for about 30 min without adding any solvent and organic protectors. The size of Ag NPs can be as small as about 10 nm and be readily controlled by adjusting the reaction parameters such as AgNO3/AA molar ratio and reaction time (Fig. 9A).349
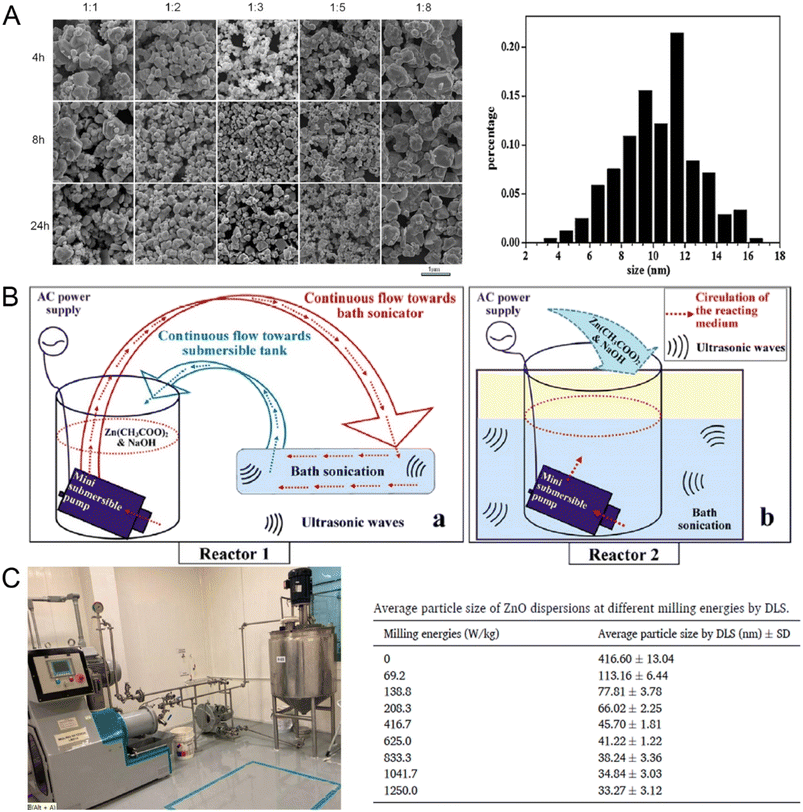 | ||
Fig. 9 Large-scale production of NPs. (A) Left panel, FESEM images of Ag NPs. The columns present samples produced from different molar ratios of AgNO3 to AA, and the rows show materials obtained at different reaction times. Right panel, the size distribution of AgNPs produced by AgNO3 and AA with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 after 4 h. Reproduced from ref. 349 with permission from Elsevier, copyright 2015. (B) Schematic representation of the working of the MSP-assisted sonochemical flow loop reactor (a), and MSP-assisted sonochemical mixing reactor (b). ZnO-NPs and ZnO-NLs are the products from reactors 1 and 2 respectively. Reproduced from ref. 363 with permission from Elsevier, copyright 2019. (C) Left panel, milling mechanism composed of a high-energy ball mill and a dispersion tank for large-scale production of ZnO NPs. Right panel, the average particle size of ZnO dispersions at different milling energies by DLS. Reproduced from ref. 365 with permission from Elsevier, copyright 2023. 3 after 4 h. Reproduced from ref. 349 with permission from Elsevier, copyright 2015. (B) Schematic representation of the working of the MSP-assisted sonochemical flow loop reactor (a), and MSP-assisted sonochemical mixing reactor (b). ZnO-NPs and ZnO-NLs are the products from reactors 1 and 2 respectively. Reproduced from ref. 363 with permission from Elsevier, copyright 2019. (C) Left panel, milling mechanism composed of a high-energy ball mill and a dispersion tank for large-scale production of ZnO NPs. Right panel, the average particle size of ZnO dispersions at different milling energies by DLS. Reproduced from ref. 365 with permission from Elsevier, copyright 2023. | ||
Researchers developed a method to use DNA extracted from salmon milt as a template to mass produce Ag NPs. Spherical Ag NPs with a main diameter of less than 10 nm at a concentration of as high as 5.3 × 10−2 mol L−1 were obtained. This approach offers an alternative route to the mass production of Ag NPs.350 Some wastes could be reused to obtain new materials. Researchers used waste culture after hydrogen production to green synthesize Ag NPs. The waste culture after hydrogen production was autoclaved and centrifuged. The supernatant containing the bacterial cell extracts after autoclaving and the fermentation residual reducing sugars was used for the reduction of silver ions into nanoparticles at 15 psi and 121 °C for only 5 min. Ag NPs with a size range of 5–25 nm and an average nanoparticle size of 15.6 ± 2.46 nm were obtained at an optimum of pH 7 and 10 mM silver nitrate. This one-pot mass-scale green synthesis of Ag NPs provides a good solution for interlinking two technologies and realizing waste recycling and new material production simultaneously.351 Silver is a kind of valuable metal used in various fields such as solar cell wafers. However, the recycling approach of waste solar cell wafers has its disadvantages such as excessive energy consumption and dust emission causing loss of valuable metals. To fulfill the concept of zero waste, researchers presented a sustainable technology for the liberation of silver from waste solar cell wafers and the synthesis of Ag NPs. By using nitric acid and other eco-friendly reagents assisted by ultrasonic treatment, Ag was dissolved and precipitated in the form of NPs with average size of 30 nm and yield of 92%. This is a good example of producing useful materials by recycling of waste.352
Apart from monodisperse nanoparticles, antimicrobial surfaces are also required in many scenarios such as biosensors. Researchers reported a method to deposit Ag NPs on an 8-inch silicon surface by plasma-enhanced atomic layer deposition (PE-ALD). Triethylphosphine-(6,6,7,7,8,8,8-heptafluoro-2,2-dimethyl-3,5-octanedionate)silver(I) [Ag(fod)-(PEt3)C16H25AgF7O2P] was used as the Ag precursor and H2 as the reducing agent. The deposited material was made of polycrystalline pure metallic Ag, indicating the fine control of the Ag NP morphology, which opens the way for interesting applications requiring precise NP dimensions.353
4.2.1.2 Au NPs. Au NPs have great potential as a group of nanoantibiotics for combating drug-resistant bacteria. However, the mass production of Au NPs faces many problems such as strict synthesis conditions, instability for long-term storage, and high cost. To solve these problems, strategies have been proposed. Researchers prepared a nanometer-sized raspberry-like architecture, consisting of an aggregation of repeated sequences of a 3D gold nanoparticle–organic component–Au NP arrangement prepared with a single-step procedure in a biphasic organic/water system. In the biphasic mixture, the reduction of Au3+ by aniline monomers afforded nanoclusters as well as a polyaniline colloid that controls the growth of the clusters into NPs. The reaction parameters (ratio of reactants, time, and temperature) can be varied to control the size of the nanoclusters and nanoparticles from 0.73 nm to 5.5 nm in the polyaniline matrix. Electrochemical degradation of the polyaniline matrix can isolate monodispersed Au nanoclusters and Au NPs.354 This electrochemical system can realize cheap and easy scale-up production of Au NPs by the enlargement of cells and electrodes. To facilitate the mass-production of Au NPs, simplifying the synthetic process is a good solution. In our recent work, we prepared aminophenyl boronic acid (ABA) modified-Au NPs (A-GNPs) by mixing HAuCl4 and ABA solution without any additional reducing agents. The reaction could be performed under ambient conditions or harsh conditions, e.g., acidic conditions. So, we could robustly synthesize Au NPs in vivo by orally administering two starting materials, HAuCl4 and ABA. Compared with other methods for synthesizing Au NPs, our approach has the advantages of simple synthesis conditions, short synthesis time (within 5 min), and comparable size uniformity.152 Our exploration paves the way for mass production of Au NPs by simplifying the reaction conditions.
The development of microfluidics provides an opportunity for the synthesis of NPs continuously. Researchers developed a scalable photochemical flow process for producing Au NPs. In the flowing solution, an acylphosphinate photoinitiator which can be cleaved by exposure to UV irradiation and produce radicals to reduce chloroauric acid into Au(0) was employed to form Au NPs within a short irradiation time (10 s). A productivity of up to 65 g per day of Au NPs was achieved, demonstrating the great potential of this protocol.355
Recently, green synthesis became a promising alternative for the substitution of toxic reducing and capping agents in Au NP synthesis. Plant extracts, microorganisms, and polysaccharides are utilized as reducing agents for eco-friendly synthesis. Also, in situ synthesis of Au NPs by green synthesis protocols is promising for scaling up the fabrication of antibacterial biomaterials. Researchers synthesized AuNPs using a polysaccharide hydrogel as a reducing and stabilizing agent, which enabled the simultaneous synthesis and stabilization of the Au NPs. The reduction of Au from Au3+ to Au0 was mediated by the presence of hydroxyl groups in the polysaccharide chains. The AuNP synthesis took place at the same time that the hydrogel was produced and the prepared AuNP hydrogel could be easily laminated on non-woven fabric to produce antibacterial wound dressing.356
4.2.1.3 Cu and Cu-derivative nanoparticles. Copper and copper derivative materials attract much attention due to their excellent antibacterial performance. Thus, large-scale production of copper and its derivative materials is highly anticipated.
The chemical reduction method is extensively used for synthesizing Cu NPs. However, the current reductants, such as hydrazine hydrate, and borohydride are expensive, and the highest cupric ion concentration is very low, restricting its large-scale production. Researchers presented a method to prepare well-dispersed Cu NPs using diethanolamine (DEA) as a reductant, solvent, as well as a ligand to control the morphology and size of the NPs. This method is succinct and low-cost and can apply to large-scale production of Cu NPs.357 The use of materials like curd, milk, and herbal extracts such as tamarind and lemon juice as agents to prepare Cu NPs is cost-effective, eco-friendly, and easy to scale up. Researchers developed a method to prepare Cu NPs on a large scale from aqueous CuSO4 using capping agents from herbal extracts. The synthesized Cu NPs had sizes ranging from 20 to 50 nm with purity close to 100%.358 To realize mass production, the simplification of the processing procedure is a possible choice. For some inorganic NMs, direct formation of NPs from the raw materials is both economical and time-saving. In a recent study, researchers presented a new approach to producing nanoscale Cu from malachite ore in one step. Malachite ore was first leached with H2SO4 or CH3COOH, partially neutralized using NaOH or NaCO3 respectively and then exposed to nanoscale zerovalent iron (nZVI). The nZVI acted as both a selective and rapid (<240 s) chemical reducing agent but also a magneto-responsive nanoscale Cu recovery vehicle. Up to 31.1 wt% conversion of Cu from ore to discrete Cu/Cu2O NPs was achieved, with nanoparticle purities of up to 81.70 wt% Cu (or 98.59 wt% Cu and O). This work provides a new direct and one-pot in situ nanoparticle mass production route that is conceptually feasible for a wide range of engineered NMs such as Ni, Cr, U, Pb, Ag and Au.359
CuO NPs are known as a potent antibacterial agent with a wide range of applications. To realize large scale production of CuO NPs, a sequential corrosion and detachment strategy was proposed in the growth and dispersion of CuO NPs. The growth of CuO NPs was accompanied by continuous corrosion of Cu powder by ammonia solution and detaching from the Cu powder by sonication. This process was continued up to the complete conversion of Cu powder to CuO NPs. The produced CuO NPs had a size of approximately 6 nm with a high crystallinity. Large-scale production of CuO NPs (120 grams) was achieved which may meet the market criteria for large-scale production of CuO NPs.360 CuS has great potential to be used as an antibacterial agent, so the preparation capability of CuS needs to be increased. Researchers presented a new methodology to synthesize CuS nanoparticles in a large scale at room temperature and pressure using high-concentration Cu complex ion precursors. This methodology was based on the theory that the critical nucleus radius and the critical nucleation free energy decrease as the concentration of the precursor increases to synthesize a large number of nanoparticles by applying low energy. As the precursor concentration increased, the particle size decreased, and the yield improved. The CuS nanoparticles synthesized at the highest concentration had a size of about 17 nm and a yield of about 213.9 g L−1.361
4.2.1.4 ZnO NPs. Zinc oxide is one of the most promising materials for antibacterial applications due to its high biocompatibility and low cost. Various synthesis approaches have been developed in the past few years to fabricate ZnO NPs. For the sol–gel synthesis protocol using zinc acetate dihydrate and potassium hydroxide as precursors, dissolved oxygen plays a key role in the synthesis of NPs. For large-scale synthesis of ZnO NPs, it is favorable to provide sufficient oxygen supply during the reaction. Researchers introduced air bubbles into the reaction system by air flow to promote ZnO NP formation at a large scale. The flow rate of air influences the yield of the NPs. Thus, this method can be easily used for enhancing the production of ZnO NPs.362 In another study, researchers used a mini submersible pump (MSP)-assisted sonochemical flow loop reactor to synthesize ZnO-NPs at gram-scale (11.5 g). The role of the MSP is to provide mechanical mixing in the reactors and to establish a flow loop connection between the submersible tank and bath sonication in the reactor (Fig. 9B). Bare ZnO-NPs and ZnO-NPs-coated cotton fabric showed high antibacterial activity.363 To realize large-scale production of monodispersed spherical ZnO NPs with narrow size distribution, researchers utilized an environmentally friendly wet-chemical route using zinc acetate as a precursor and sodium oleate as a surface capping agent. Zinc acetate was hydrolyzed in the ethanol-2% H2O solution to form the ZnO NPs. The yield of ZnO NPs could be up-scaled to the gram-scale. Moreover, the reaction solution can be recycled which is beneficial for environmental protection.364 Ball milling is one of the most prominent top-down nanotechnology techniques used to prepare a wide spectrum of NMs. In a recent study, the reduction of ZnO particles to NPs was performed by high-energy ball milling. Increasing the milling energy led to a significant reduction of the average particle size of ZnO from 416.60 ± 13.04 nm to 45.70 ± 1.81 nm in the milling energy range of 0–625 W kg−1. ZnO NPs could be produced in large quantities (600 kg) by high energy (Fig. 9C). Thus, the milling energy technique allows for the modification of the properties of ZnO particles in a controlled way for large-scale production.365
4.2.1.5 Carbon NMs. Graphene-based NMs possess potent antibacterial activity and have potential in a wide range of applications. Thus, how to prepare graphene in a large scale is a critical question. Researchers proposed a simple chemical approach for the synthesis of graphene through a mild reduction of graphene oxide (GO) using metal nanoparticles as the catalyst for the hydrolysis reaction of NaBH4 at room temperature. The reaction is conducted under mild conditions and can be easily scaled up and the metal catalyst can be recycled.366 The mass production of high-quality graphene-based NMs in an inexpensive and eco-friendly way is challenging. Researchers reported a facile method for the biosynthesis of graphene by using Allium cepa (onion) extracts as a nontoxic reducing agent in the modified Hummers' method. In this work, graphene oxide was successfully reduced to graphene using onion extract. The resulting graphene showed antibacterial properties against different microorganisms. This work paves the way for a new production method of graphene.367
Ultralong carbon nanotubes (CNTs) are in huge demand in many fields, but their applications are limited by mass production capability. Researchers presented a method to scale up the yield of ultralong CNTs by a substrate interception and direction strategy (SIDS), which couples the advantages of floating-catalyst chemical vapor deposition with the flying-kite-like growth mechanism of ultralong CNTs. The areal density of the ultralong CNT arrays with a length of over 1 cm reached a value of ∼6700 CNTs mm−1, which is 2–3 orders of magnitude higher than the values of traditional methods. This study lays the foundation for the mass production of ultralong CNTs and provides insight for synthesizing other 1D NMs.368
4.2.1.6 Liquid metals. Nanoparticles comprised of liquid metals are useful for biomedical applications. However, the methods for producing liquid metal nanoparticles have disadvantages such as expensive high-power sonication and requirement stabilization of small molecules. In a recent study, researchers presented a microfluidics-enabled platform for mass production of eutectic gallium indium (EGaIn) NPs with tunable size distributions in an aqueous medium. Brushed polyethylene glycol chains with trithiocarbonate end-groups were used as stabilization reagents to negate the requirements for thiol additives while imparting a “stealth” surface layer. Thus, this microfluidic technique is promising as a versatile platform for the rapid production of liquid metal-based nanoparticles.369
4.2.2.1 Polymeric NPs. Polymeric NPs can be prepared through nanoprecipitation by mixing polymers dissolved in organic solvents with anti-solvents. However, due to the inability to precisely control the mixing processes during the synthesis, the mass production of polymeric NPs is limited. Microfluidics enables rapid mixing of reagents to provide homogeneous reaction environments. Fine control of process parameters afforded by microfluidics allows unprecedented optimization of nanoparticle quality. Automation improves the reproducibility and optimization of formulations. Furthermore, the continuous nature of the microfluidic process is inherently scalable, allowing scale-up through process parallelization.370 Moreover, microfluidics can vary the reaction conditions continuously, and allow reagent addition during the progress of a reaction, making it attractive for nanoparticle synthesis. However, the low production rate limits its practical applications. Researchers designed a high-throughput microfluidic device to prepare highly monodisperse methoxyl poly-(ethylene glycol)-poly-(lactic-co-glycolic acid) (MPEG-PLGA) NPs with sizes ranging from 50 to 200 nm regardless of the molecular weight of the polymer and concentration of polymer solution.371 Parallel flow focusing could be realized in the device to yield small nanoparticles with high monodispersity because of predictable control over the mixing process of organic solvents and anti-solvents. Since self-assembly of nanoparticles occurred outside the polydimethylsiloxane (PDMS) microchannels, the precipitating polymer can be isolated from the PDMS wall and thus the possible aggregation on the surface of PDMS wall and clogging of microchannels which was reported previously can be avoided. Researchers designed 100 outlets for the polymer stream in the device, and the production speed of NPs in this device was improved by an order of magnitude as compared to the production efficiency of other studies.372 Moreover, the production speed of NPs could be further improved by two orders of magnitude or more by increasing the number of outlets. This technique can serve as a good base for the large-scale industrial production of polymeric NPs with high monodispersity. To simplify the structure of the microchip, in another study, researchers fabricated hydrodynamic flow-focusing microfluidics for producing poly (lactide-co-glycolide)-b-polyethylene glycol (PLGA-PEG) NPs with uniform sizes ranging from 50 to 150 nm. In contrast to the widely used microfluidic device which has a production rate of 1.8 mg h−1, this simple approach was capable of increasing the production rate of nanoparticles by more than two orders of magnitude up to 288 mg h−1 using a single simple device. This study demonstrated the potential of using simple 2D microfluidic devices for large-scale production of polymeric nanoparticles that could eliminate the need for designing and fabricating complex microfluidic devices.373 To produce a uniform size distribution of polycaprolactone (PCL) NPs with high throughput, five arrangements of microfluidic devices were fabricated. Optimal conditions for rapid production of nanoparticles with a size smaller than 200 nm and polydispersity index (PDI) ≤ 0.31 were obtained at a flow rate ratio (FRR) of 8, and total flow rate (TFR) of 70 mL h−1. Moreover, a higher productivity rate of the PCL NPs with a similar size and lower PDI was achieved on the chip, demonstrating the promise of the device for mass production of PCL NPs.374 The rapid development of microfluidic technologies provides an opportunity to improve the productivity and controllability of polymeric NPs. Researchers presented a swirl mixer on microfluidic to accelerate the translation of NPs from laboratory to clinical application. The swirl mixer provided a high production rate, reproducibility, and precise control of particle size with a low polydispersity index. By changing multiple processing parameters, two different types of nanoformulations: silk nanoparticles (SNPs) and lipid nanoparticles (LNPs) were produced on the microfluidics efficiently with high productivity and allowed for tuning the mean size and size distribution.375
4.2.2.2 Lipid NPs. Liposomes and lipid-based nanoparticles (LNPs) are nanosized vesicles that mainly consist of amphiphilic phospholipids and cholesterol. LNPs have been widely applied in the fields of medicine, cosmetics, and nutrition. In pharmaceutical fields, LNPs are among the most reliable and successful drug-delivery systems for encapsulating various drugs. There are more than 10 approved clinical products, including Doxil376 using LNPs as the carrier. However, the difficulties associated with mass production and the stability of LNPs limit the clinical and commercial applications of LNP technologies. To address this issue, researchers present a simple and industrially accessible method for producing bilayer vesicles. An ethanolic solution dissolving phytantriol, Pluronic F127, and vitamin E acetate was mixed with deionized water. When ethanol was evaporated from the aqueous mixture, vesicles were transformed into liquid NPs with well-defined internal structures such as hexagonal lattices, lined or coiled patterns, and disordered structures, depending on the composition. The production only needs mixing and ethanol evaporation and it is possible to produce about 10 kg per batch of reaction, enabling the large-scale production of lipid NPs for various biomedical applications.377 Another study aimed at developing a large-scale modular production line, which involved a continuous and scalable emulsification and homogenization process to produce nanostructured lipid NPs. The production line exhibited good control over the emulsification and homogenization process and enabled the particle size below 210 nm at a throughput of 25 kg h−1, demonstrating the robustness of this device.378 To realize a continuous and scalable nanoprecipitation synthesis of solid lipid nanoparticles (SLN), researchers developed a platform to mix an acetonic lipid solution with water using static mixers, which exhibited good control over the nanoprecipitation process and enabled the production of SLN below 200 nm at a throughput of 37.5–150 g h−1. This work demonstrates the potential of using the static mixing-nanoprecipitation technique for continuous and large-scale production of SLN.379
Recently, microfluidics has developed as a novel process for simply manufacturing LNPs. In microfluidics, introducing only the aqueous phase and lipids into the organic solvent phase can generate LNPs with the conditioning of flow rate, the ratio between the two phases of liquids, the concentration of lipids and drugs, as well as the mixing modes. In a recent study, a glass-based microfluidic device for mRNA-loaded LNP production with ionizable lipids was used for COVID-19 mRNA vaccines. The chip composed of five-layered microchannels was fabricated by piling up each glass device followed by parallelization (numbering-up) for the mass production of LNPs. This system can produce LNPs with sizes ranging between 20 and 60 nm at a flow rate of 20–50 mL min−1, demonstrating its potential to be commercialized for biomedical applications.380 Microfluidics is a good platform for preparing LNPs. However, the high risk of fouling compromises its reliability for continuous operation. Researchers reported an ultrasound-assisted processing method for the precipitation of LNPs in a microfluidic mixer. Nanoparticles can be produced over several hours without clogging. Thus, this in-process ultrasonication is effective at improving the reliability of microfluidic precipitation and preventing the buildup of precipitates in the channels (Fig. 10A).381 To boost the productivity of LNPs in microfluidics, obtaining a high concentration of LNPs is an alternative strategy. However, high concentration of lipids makes it difficult to control the size and dispersity of LNPs. Researchers developed a microfluidic device named iLiNP in which a simple baffle mixer structure can achieve rapid ethanol dilution. Compared with other microfluidic devices, iLiNP could produce smaller and more concentrated LNPs with good dispersity. This device could be a time- and cost-saving option for the mass production of LNPs for application in nanomedicine and cosmetics (Fig. 10B).382
 | ||
| Fig. 10 Mass-production of lipid nanoparticles on a microfluidic chip. (A) Microchannel structure in the multilamination mixer (a) and picture of the mixing chamber (b). Reproduced from ref. 381 with permission from WILEY-VCH, copyright 2021. (B) Mechanism of LNP formation on a chip. (a) Each initial intermediate forms self-assembled LNPs and results in a large number of LNPs. (b) The initial intermediates fuse with the existing initial intermediates in the vicinity and reduce the number of intermediates and LNPs. Reproduced from ref. 382 with permission from the American Chemical Society, copyright 2022. | ||
4.2.2.3 Lipid–polymer hybrid (LPH) NPs. Lipid–polymer hybrid (LPH) NPs combine the advantages of both liposomes and polymers. Researchers adapted a multi-inlet vortex reactor (MIVR) for the large-scale synthesis of LPH NPs. By tuning parameters such as formulation, polymer concentration, and flow rate, the resulting NPs showed low polydispersity and excellent stability. Using this approach, production rates of greater than 10 g h−1 can readily be achieved, demonstrating that the use of the MIVR is a viable method of producing hybrid nanoparticles in clinically relevant quantities.383 LPH NPs are capable of delivering a wide range of therapeutic compounds in a controlled manner. Microfluidics provides a platform for preparing LPH nanoparticles with high-quality control. However, the low throughput restricts its wide application. Researchers developed a pattern-tunable microvortex platform that allows mass production and size control of LPH NPs with high reproducibility and homogeneity. By varying flow rates, LPH NPs with size in the range of 30–170 nm with high productivity (∼3 g hour−1) and low polydispersity (∼0.1) were obtained. This approach provides a mass production of LPH NPs with a wide range of multicomponents for biomedical applications.384
5. Future research perspectives
This review summarizes the recent developments in the field of antibacterial NMs addressing the issue of drug-resistant pathogens. We reviewed the basic mechanism of the generation and development of drug resistance, which helps guide the exploration of novel antibacterial agents for bypassing or reversing the drug resistance. The recently reported NMs can kill or inhibit bacteria through various mechanisms including destruction of the bacterial structure (membrane, subcellular structures), disturbance of the cellular mechanism, binding to the DNA or protein, and so forth. Moreover, by using external energy sources (laser, acoustics, electricity, and magnetics), NMs can generate heat, ROS, and kinetic energy to enhance their antibacterial effects. Thus, NMs can treat drug-resistant pathogen-induced infections via multiple routes while they can hardly induce drug resistance due to their complex antibacterial mechanism. However, the currently reported NMs need to be improved in several aspects:(1) Specificity. The currently used NMs usually lack specificity to bacteria. Unfortunately, some NMs such as silver nanoparticles are poisonous to the human cells, which limits their wide application. Moreover, NMs cannot discern probiotic bacteria and pathogenic bacteria. Thus, the application of NMs may lead to dysbacteriosis in the human body or ecological environment, which is a more serious situation. Therefore, developing NMs with specific targeting molecules or structures on pathogenic bacteria is meaningful and urgent. To realize this, a deep interdisciplinary collaboration between microbiology and nanomaterial science is needed.
(2) Clear antibacterial mechanism. Although presently reported NMs show potent antibacterial effects, their antibacterial mechanism is still obscure. This situation is not beneficial for further application of nanodrugs due to many uncertainties including the difficulty of profiling their pharmacodynamics and pharmacokinetics, as well as their biosafety. To address this issue, two strategies are suggested: one is to develop antibiotic-loaded NMs in which the nanocarrier is chemically inert and the antibacterial mechanism of the antibiotic is clear. This kind of nanoantibiotics has the advantage of both NMs (nano-effects, do not easily induce drug resistance) and conventional antibiotics (definite antibacterial mechanism) and is promising as the next generation of nanoantibiotics. Another one is to develop core–shell structured NMs on which the core serves as a functional unit that can be actuated by external energy sources such as light, acoustics, and magnetics to elicit bioeffects including thermotherapy or photodynamic therapy and so forth, whereas the shell is composed of chemical groups with certain bioeffects on the bacteria or have definite ligand–receptor interactions with the bacteria. In this kind of nanomaterial, both the nanoeffects and ligand–receptor signaling are employed.
(3) High biosafety. The biggest obstacle restricting the clinical application of nanoantibiotics is their biosafety. So, how to make nanoantibiotics as safe as conventional antibiotics is the point to ponder. To achieve this goal, the composition/structure relationship and the physical and chemical properties of the NMs should be definitely profiled. Moreover, the administration route, biodistribution, biodegradability, and pharmacokinetics/pharmacodynamics of the NMs should be extensively investigated.
In summary, NMs offer a great opportunity to provide a comprehensive solving toolbox to address the issue of antibiotic resistance. To fully explore the potential of this toolbox, interdisciplinary collaborative action is indispensable. We expect that antibacterial NMs will attract attention from scientists in various disciplines to contribute to the development of the next generation of nanoantibiotics fighting against drug-resistant bacteria.
Author contributions
Mulan Li, Ying Liu, Youhuan Gong, Xiaojie Yan: investigation, writing; Le Wang, Wenfu Zheng: investigation, writing and editing; Hao Ai: editing; Yuliang Zhao: editing, supervision, conceptualization.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We thank the Science and Technology Project of Education Department of Jiangxi Province of China (GJJ2200963), Doctor Start-up Fund of Jiangxi University of Chinese Medicine (2022BSZR009), the National Natural Science Foundation of China (22234004, 22304065, and 32071390), and Jie Bang Gua Shuai Project of Department of Science & Technology of Liaoning Province for financial support.References
- J. Carlet, P. Collignon, D. Goldmann, H. Goossens, I. C. Gyssens, S. Harbarth, V. Jarlier, S. B. Levy, B. N'Doye, D. Pittet, R. Richtmann, W. H. Seto, J. W. M. van der Meer and A. Voss, Lancet, 2011, 378, 369–371 CrossRef PubMed.
- P. Nordmann, T. Naas, N. Fortineau and L. Poirel, Curr. Opin. Microbiol., 2007, 10, 436–440 CrossRef CAS PubMed.
- E. Sánchez-López, D. Gomes, G. Esteruelas, L. Bonilla, A. L. Lopez-Machado, R. Galindo, A. Cano, M. Espina, M. Ettcheto, A. Camins, A. M. Silva, A. Durazzo, A. Santini, M. L. Garcia and E. B. Souto, Nanomater., 2020, 10, 292 CrossRef PubMed.
- E. M. Darby, E. Trampari, P. Siasat, M. S. Gaya, I. Alav, M. A. Webber and J. M. A. Blair, Nat. Rev. Microbiol., 2023, 21, 280–295 CrossRef CAS PubMed.
- M. Bassetti, M. Merelli, C. Temperoni and A. Astilean, Ann. Clin. Microbiol. Antimicrob., 2013, 12, 1–5 CrossRef PubMed.
- B. Spellberg, R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett, J. Edwards, Jr. and A. Infectious Diseases Society of, Clin. Infect. Dis., 2008, 46, 155–164 CrossRef PubMed.
- A. J. Huh and Y. J. Kwon, J. Controlled Release, 2011, 156, 128–145 CrossRef CAS PubMed.
- H.- S. Ouangraoua, A. A. Maiga, M. Cairns, I. Zongo, N. Frédéric, R. Serge Yerbanga, B. Tamboura, H. Badji, G. Gore-Langton, I. Kuepfer, H. Tinto, I. Sagara, A. Dicko, S. O. Sow, D. Chandrahoman, B. Greenwood and J. Bosco Ouedraogo, Trop. Med. Int. Health, 2019, 24, 1442–1454 CrossRef PubMed.
- R. Hadad, D. Golparian, I. Velicko, A. K. Ohlsson, Y. Lindroth, E. L. Ericson, H. Fredlund, L. Engstrand and M. Unemo, Front. Microbiol., 2021, 12, 820998 CrossRef PubMed.
- K. Lee, S. I. Nakayama, K. Osawa, H. Yoshida, S. Arakawa, K. I. Furubayashi, H. Kameoka, K. Shimuta, T. Kawahata, M. Unemo and M. Ohnishi, J. Antimicrob. Chemother., 2019, 74, 1812–1819 CrossRef CAS PubMed.
- B. Mlynarczyk-Bonikowska, C. Kowalewski, A. Krolak-Ulinska and W. Marusza, Int. J. Mol. Sci., 2022, 23, 8088 CrossRef CAS PubMed.
- S. López-Argüello, M. Montaner, A. Mármol-Salvador, A. Velázquez-Escudero, F. Docobo-Pérez, A. Oliver and B. Moya, Microbiol. Spectr., 2023, 11(3), e0069223 CrossRef PubMed.
- I. Karaiskos, L. Galani, V. Sakka, A. Gkoufa, O. Sopilidis, D. Chalikopoulos, G. Alivizatos and E. Giamarellou, J. Antimicrob. Chemother., 2019, 74, 1430–1437 CrossRef CAS PubMed.
- T. Ida, R. Okamoto, C. Shimauchi, T. Okubo, A. Kuga and M. Inoue, J. Clin. Microbiol., 2001, 39, 3115–3121 CrossRef CAS PubMed.
- T. Ida, R. Okamoto, M. Nonoyama, K. Irinoda, M. Kurazono and M. Inoue, Antimicrob. Agents Chemother., 2002, 46, 1516–1521 CrossRef CAS PubMed.
- X. H. Li, Y. Y. Huang, L. M. Lu, L. J. Zhao, X. K. Luo, R. J. Li, Y. Y. Dai, C. Qin, Y. Q. Huang and H. Chen, World J. Gastroenterol., 2021, 27, 3595–3608 CrossRef CAS PubMed.
- M. J. Rybak, J. Le, T. P. Lodise, D. P. Levine, J. S. Bradley, C. Liu, B. A. Mueller, M. P. Pai, A. Wong-Beringer, J. C. Rotschafer, K. A. Rodvold, H. D. Maples and B. M. Lomaestro, Pharmacotherapy, 2020, 40, 363–367 CrossRef CAS PubMed.
- J. W. Kim and K. J. Lee, Arch. Microbiol., 2020, 202, 2255–2261 CrossRef CAS PubMed.
- I. Quiles-Melero, R. Gomez-Gil, M. P. Romero-Gomez, A. M. Sanchez-Diaz, M. de Pablos, J. Garcia-Rodriguez, A. Gutierrez and J. Mingorance, J. Clin. Microbiol., 2013, 51, 998–1001 CrossRef CAS PubMed.
- M. Sjolund-Karlsson, R. L. Howie, J. A. Crump and J. M. Whichard, J. Clin. Microbiol., 2014, 52, 877–884 CrossRef CAS PubMed.
- L. Zhao, S. Wang, X. Li, X. He and L. Jian, Antimicrob. Resist. Infect. Control., 2020, 9, 124 CrossRef PubMed.
- Z. H. Gui, H. F. Wang, T. Ding, W. Zhu, X. Y. Zhuang and W. H. Chu, Indian J. Microbiol., 2014, 54, 114–117 CrossRef CAS PubMed.
- A. Bojang, B. Camara, I. J. Cox, C. Oluwalana, K. Lette, E. Usuf, C. Bottomley, B. P. Howden, U. D'Alessandro and A. Roca, Clin. Infect. Dis., 2018, 67, 1191–1197 CrossRef CAS PubMed.
- C. B. Wadsworth, B. J. Arnold, M. R. A. Sater, Y. H. Grad, W. Shafer and M. S. Gilmore, mBio, 2018, 9, e01419-18 CrossRef PubMed.
- M. Olesky, S. Zhao, R. L. Rosenberg and R. A. Nicholas, J. Bacteriol., 2006, 188, 2300–2308 CrossRef CAS PubMed.
- S. A. Chisholm, J. Dave and C. A. Ison, Antimicrob. Agents Chemother., 2010, 54, 3812–3816 CrossRef CAS PubMed.
- C. Blackadar, K.-Y. G. Choi, M. F. Embree, H. M. Hennkens, C. Rodríguez-Rodríguez, R. E. W. Hancock, K. Saatchi and U. O. Häfeli, ACS Appl. Mater. Interfaces, 2022, 14, 26382–26393 CrossRef CAS PubMed.
- N. M. Salah, A. E. Saafan, E. H. Salem, H. A. El Rabey, M. A. Alsieni, F. A. Alatawi, A. I. Alalawy, A. B. Abeer Mohammed and A. Omri, BioMed Res. Int., 2022, 2022, 1–10 CrossRef PubMed.
- L.-J. Luo, T.-Y. Lin, C.-H. Yao, P.-Y. Kuo, M. Matsusaki, S. G. Harroun, C.-C. Huang and J.-Y. Lai, J. Colloid Interface Sci., 2019, 536, 112–126 CrossRef CAS PubMed.
- E. Hodille, L. Delouere, C. Bouveyron, H. Meugnier, M. Bes, A. Tristan, F. Laurent, F. Vandenesch, G. Lina and O. Dumitrescu, Med. Mal. Infect., 2017, 47, 152–157 CrossRef CAS PubMed.
- J. Stojkovska, J. Zvicer and B. Obradovic, Appl. Microbiol. Biotechnol., 2020, 104, 4643–4658 CrossRef CAS PubMed.
- Z. Lin, Y. Zhao, P. K. Chu, L. Wang, H. Pan, Y. Zheng, S. Wu, X. Liu, K. M. C. Cheung, T. Wong and K. W. K. Yeung, Biomaterials, 2019, 219, 119372 CrossRef CAS PubMed.
- J. Davies and D. Davies, Microbiol. Mol. Biol. Rev., 2010, 74, 417–433 CrossRef CAS PubMed.
- L. Narendrakumar, M. Chakraborty, S. Kumari, D. Paul and B. Das, Front. Microbiol., 2022, 13, 1092556 CrossRef PubMed.
- F. Spagnolo, C. Rinaldi, D. R. Sajorda and D. E. Dykhuizen, Antimicrob. Agents Chemother., 2016, 60, 1336–1342 CrossRef CAS PubMed.
- M. E. Tolmasky, R. M. Chamorro, J. H. Crosa and P. M. Marini, Antimicrob. Agents Chemother., 1988, 32, 1416–1420 CrossRef CAS PubMed.
- M.-L. Foucault, F. Depardieu, P. Courvalin and C. Grillot-Courvalin, Proc. Natl. Acad. Sci. U.S.A., 2010, 107, 16964–16969 CrossRef CAS PubMed.
- M. Sjölund-Karlsson, R. L. Howie, J. A. Crump, J. M. Whichard and E. Munson, J. Clin. Microbiol., 2014, 52, 877–884 CrossRef PubMed.
- M. A. Rather, K. Gupta and M. Mandal, Braz. J. Microbiol., 2021, 52, 1701–1718 CrossRef PubMed.
- J. R. Huddleston, Infect. Drug Resist., 2014, 7, 167–176 CrossRef PubMed.
- F. Baquero, J. L. Martinez, V. F. Lanza, J. Rodriguez-Beltran, J. C. Galan, A. San Millan, R. Canton and T. M. Coque, Clin. Microbiol. Rev., 2021, 34, e0005019 CrossRef CAS PubMed.
- S. Hernando-Amado, P. Laborda, J. R. Valverde and J. L. Martinez, Proc. Natl. Acad. Sci. U.S.A., 2022, 119, e2109370119 CrossRef PubMed.
- J. K. Lee, Y. S. Lee, Y. K. Park and B. S. Kim, Int. J. Antimicrob. Agents, 2005, 25, 290–295 CrossRef CAS PubMed.
- X. Feng, Z. Zhang, X. Li, Y. Song, J. Kang, D. Yin, Y. Gao, N. Shi and J. Duan, Infect. Drug Resist., 2019, 12, 261–272 CrossRef CAS PubMed.
- P. Higgins, Infect. Drug Resist., 2003, 21, 409–413 CrossRef CAS PubMed.
- M. R. Pasca, C. Dalla Valle, A. L. De Jesus Lopes Ribeiro, S. Buroni, M. C. Papaleo, S. Bazzini, C. Udine, M. L. Incandela, S. Daffara, R. Fani, G. Riccardi and P. Marone, Microb. Drug Resist., 2012, 18, 23–32 CrossRef CAS PubMed.
- S. Bruchmann, A. Dötsch, B. Nouri, I. F. Chaberny and S. Häussler, Antimicrob. Agents Chemother., 2013, 57, 1361–1368 CrossRef CAS PubMed.
- S. M. Cheer, J. Waugh and S. Noble, Drugs, 2003, 63, 2501–2520 CrossRef CAS PubMed.
- E. Christaki, M. Marcou and A. Tofarides, J. Mol. Evol., 2020, 88, 26–40 CrossRef CAS PubMed.
- W. R. Miller, J. M. Munita and C. A. Arias, Expert Rev. Anti-Infect. Ther., 2014, 12, 1221–1236 CrossRef CAS PubMed.
- I. Quiles-Melero, R. Gomez-Gil, M. P. Romero-Gomez, A. M. Sanchez-Diaz, M. de Pablos, J. Garcia-Rodriguez, A. Gutierrez and J. Mingorance, J. Clin. Microbiol., 2013, 51, 998–1001 CrossRef CAS PubMed.
- K. A. Tagg, N. J. Jeoffreys, D. L. Couldwell, J. A. Donald and G. L. Gilbert, J. Clin. Microbiol., 2013, 51, 2245–2249 CrossRef CAS PubMed.
- P. C. Y. Woo, S. K. P. Lau, C.-m. Chu, K.-h. Chan, H.-w. Tsoi, Y. Huang, B. H. L. Wong, R. W. S. Poon, J. J. Cai, W.-k. Luk, L. L. M. Poon, S. S. Y. Wong, Y. Guan, J. S. M. Peiris and K.-y. Yuen, J. Virol., 2005, 79, 884–895 CrossRef CAS PubMed.
- B. Kaltenboeck, J. Twin, J. S. Jensen, C. S. Bradshaw, S. M. Garland, C. K. Fairley, L. Y. Min and S. N. Tabrizi, PLoS One, 2012, 7, e35593 CrossRef PubMed.
- D. Durukan, T. R. H. Read, G. Murray, M. Doyle, E. P. F. Chow, L. A. Vodstrcil, C. K. Fairley, I. Aguirre, E. Mokany, L. Y. Tan, M. Y. Chen and C. S. Bradshaw, Clin. Infect. Dis., 2020, 71, 1461–1468 CrossRef CAS PubMed.
- X.-H. Li, Y.-Y. Huang, L.-M. Lu, L.-J. Zhao, X.-K. Luo, R.-J. Li, Y.-Y. Dai, C. Qin, Y.-Q. Huang and H. Chen, World J. Gastroenterol., 2021, 27, 3595–3608 CrossRef CAS PubMed.
- K. Yadav, L. Garoff, D. L. Huseby and D. Hughes, J. Antimicrob. Chemother., 2021, 76, 1441–1447 CrossRef CAS PubMed.
- A. Jaffe, Y. A. Chabbert and O. Semonin, Antimicrob. Agents Chemother., 1982, 22, 942–948 CrossRef CAS PubMed.
- C. A. Arias, D. Panesso, D. M. McGrath, X. Qin, M. F. Mojica, C. Miller, L. Diaz, T. T. Tran, S. Rincon, E. M. Barbu, J. Reyes, J. H. Roh, E. Lobos, E. Sodergren, R. Pasqualini, W. Arap, J. P. Quinn, Y. Shamoo, B. E. Murray and G. M. Weinstock, N. Engl. J. Med., 2011, 365, 892–900 CrossRef CAS PubMed.
- K. L. Palmer, A. Daniel, C. Hardy, J. Silverman and M. S. Gilmore, Antimicrob. Agents Chemother., 2011, 55, 3345–3356 CrossRef CAS PubMed.
- K. M. Raible, B. Sen, N. Law, T. E. Bias, C. L. Emery, G. D. Ehrlich and S. G. Joshi, Ann. Clin. Microbiol. Antimicrob., 2017, 16, 75 CrossRef PubMed.
- R. R. Cutler, J. Antimicrob. Chemother., 1983, 11, 263–269 CrossRef CAS PubMed.
- M. H. Perlin and S. A. Lerner, Antimicrob. Agents Chemother., 1979, 16, 598–604 CrossRef CAS PubMed.
- C. Torres, M. H. Perlin, F. Baquero, D. L. Lerner and S. A. Lerner, Int. J. Antimicrob. Agents, 2000, 15, 257–263 CrossRef CAS PubMed.
- L. E. Kehoe, J. Snidwongse, P. Courvalin, J. B. Rafferty and I. A. Murray, J. Biol. Chem., 2003, 278, 29963–29970 CrossRef CAS PubMed.
- S. R. Connell, D. M. Tracz, K. H. Nierhaus and D. E. Taylor, Antimicrob. Agents Chemother., 2003, 47, 3675–3681 CrossRef CAS PubMed.
- J. Kobylka, M. S. Kuth, R. T. Muller, E. R. Geertsma and K. M. Pos, Ann. N. Y. Acad. Sci., 2020, 1459, 38–68 CrossRef CAS PubMed.
- L. B. Kouitcheu Mabeku, B. Eyoum Bille, C. Tepap Zemnou, L. D. Tali Nguefack and H. Leundji, BMC Infect. Dis., 2019, 19, 880 CrossRef PubMed.
- E. I. Kazakos, N. Dorrell, S. A. Polyzos, G. Deretzi and J. Kountouras, World J. Gastroenterol., 2017, 23, 6194–6196 CrossRef CAS PubMed.
- T. Falsafi, A. Ehsani, B. Attaran and V. Niknam, Jundishapur J. Microbiol., 2016, 9, e30726 CrossRef PubMed.
- K. van Amsterdam, A. Bart and A. van der Ende, Antimicrob. Agents Chemother., 2005, 49, 1477–1482 CrossRef CAS PubMed.
- F. Long, C. Rouquette-Loughlin, W. M. Shafer and E. W. Yu, Antimicrob. Agents Chemother., 2008, 52, 3052–3060 CrossRef CAS PubMed.
- T. Köhler, M. Michéa-Hamzehpour, U. Henze, N. Gotoh, L. K. Curty and J. C. Pechère, Mol. Microbiol., 1997, 23, 345–354 CrossRef PubMed.
- N. Masuda, E. Sakagawa, S. Ohya, N. Gotoh, H. Tsujimoto and T. Nishino, Antimicrob. Agents Chemother., 2000, 44, 3322–3327 CrossRef CAS PubMed.
- C. Xu, H. Liu, X. Pan, Z. Ma, D. Wang, X. Zhang, G. Zhu, F. Bai, Z. Cheng, W. Wu and Y. Jin, Front. Microbiol., 2021, 11, 598291 CrossRef PubMed.
- N. Mayer-Hamblett, M. Rosenfeld, R. L. Gibson, B. W. Ramsey, H. D. Kulasekara, G. Z. Retsch-Bogart, W. Morgan, D. J. Wolter, C. E. Pope, L. S. Houston, B. R. Kulasekara, U. Khan, J. L. Burns, S. I. Miller and L. R. Hoffman, Am. J. Respir. Crit. Care Med., 2014, 190, 289–297 CrossRef PubMed.
- S. Hernando-Amado, F. Sanz-García and J. L. Martínez, Sci. Adv., 2020, 6, eaba5493 CrossRef CAS PubMed.
- M. A. Abushaheen, Muzaheed, A. J. Fatani, M. Alosaimi, W. Mansy, M. George, S. Acharya, S. Rathod, D. D. Divakar, C. Jhugroo, S. Vellappally, A. A. Khan, J. Shaik and P. Jhugroo, Disease-a-Month, 2020, 66, 100971 CrossRef PubMed.
- H. H. Lee, M. N. Molla, C. R. Cantor and J. J. Collins, Nature, 2010, 467, 82–85 CrossRef CAS PubMed.
- D. I. Andersson and D. Hughes, Nat. Rev. Microbiol., 2010, 8, 260–271 CrossRef CAS PubMed.
- E. Denamur and I. Matic, Mol. Microbiol., 2006, 60, 820–827 CrossRef CAS PubMed.
- S. K. Henderson-Begg, D. M. Livermore and L. M. Hall, J. Antimicrob. Chemother., 2006, 57, 849–854 CrossRef CAS PubMed.
- D. Ghosh, B. Veeraraghavan, R. Elangovan and P. Vivekanandan, Antimicrob. Agents Chemother., 2020, 64, e02225-19 CrossRef PubMed.
- A. S. Joshi, P. Singh and I. Mijakovic, Int. J. Mol. Sci., 2020, 21, 7658 CrossRef CAS PubMed.
- C. Alves-Barroco, L. Rivas-Garcia, A. R. Fernandes and P. V. Baptista, Front. Microbiol., 2022, 13, 841124 CrossRef PubMed.
- K. J. Aldred, R. J. Kerns and N. Osheroff, Biochemistry, 2014, 53, 1565–1574 CrossRef CAS PubMed.
- K. J. Aldred, S. A. McPherson, C. L. Turnbough, Jr., R. J. Kerns and N. Osheroff, Nucleic Acids Res., 2013, 41, 4628–4639 CrossRef CAS PubMed.
- Z. Baharoglu and D. Mazel, FEMS Microbiol. Rev., 2014, 38, 1126–1145 CrossRef CAS PubMed.
- S. Sanyasi, R. K. Majhi, S. Kumar, M. Mishra, A. Ghosh, M. Suar, P. V. Satyam, H. Mohapatra, C. Goswami and L. Goswami, Sci. Rep., 2016, 6, 24929 CrossRef PubMed.
- X. Li, S. M. Robinson, A. Gupta, K. Saha, Z. Jiang, D. F. Moyano, A. Sahar, M. A. Riley and V. M. Rotello, ACS Nano, 2014, 8, 10682–10686 CrossRef CAS PubMed.
- Y. Cui, Y. Zhao, Y. Tian, W. Zhang, X. Lü and X. Jiang, Biomaterials, 2012, 33, 2327–2333 CrossRef CAS PubMed.
- J. M. V. Makabenta, A. Nabawy, C. H. Li, S. Schmidt-Malan, R. Patel and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed.
- P. Wang, L. Hu and Z. Hao, Infect. Drug Resist., 2020, 13, 749–759 CrossRef CAS PubMed.
- L. Feng, J. E. A. Mundy, C. E. M. Stevenson, L. A. Mitchenall, D. M. Lawson, K. Mi and A. Maxwell, Proc. Natl. Acad. Sci. U.S.A., 2021, 118, e2016705118 CrossRef CAS PubMed.
- K. K. Kumarasamy, M. A. Toleman, T. R. Walsh, J. Bagaria, F. Butt, R. Balakrishnan, U. Chaudhary, M. Doumith, C. G. Giske, S. Irfan, P. Krishnan, A. V. Kumar, S. Maharjan, S. Mushtaq, T. Noorie, D. L. Paterson, A. Pearson, C. Perry, R. Pike, B. Rao, U. Ray, J. B. Sarma, M. Sharma, E. Sheridan, M. A. Thirunarayan, J. Turton, S. Upadhyay, M. Warner, W. Welfare, D. M. Livermore and N. Woodford, Lancet Infect. Dis., 2010, 10, 597–602 CrossRef CAS PubMed.
- N. Padmini, A. A. K. Ajilda, N. Sivakumar and G. Selvakumar, J. Basic Microbiol., 2017, 57, 460–470 CrossRef CAS PubMed.
- A. M. Turner, J. Y. H. Lee, C. L. Gorrie, B. P. Howden and G. P. Carter, Front. Microbiol., 2021, 12, 637656 CrossRef PubMed.
- R. E. Hancock and D. P. Speert, Drug Resistance Updates, 2000, 3, 247–255 CrossRef CAS PubMed.
- X. Z. Li, P. Plesiat and H. Nikaido, Clin. Microbiol. Rev., 2015, 28, 337–418 CrossRef PubMed.
- D. Schnappinger and W. Hillen, Arch. Microbiol., 1996, 165, 359–369 CrossRef CAS PubMed.
- Y. Long, X. Lu, X. Ni, J. Liu, M. Wang, X. Li, Z. Li, H. Zhou, Z. Li, K. Wu, W. Wang, L. Yang, J. Xu, H. Chen and B. Kan, Antibiotics, 2021, 11, 15 CrossRef PubMed.
- J. M. Blair, G. E. Richmond and L. J. Piddock, Future Microbiol., 2014, 9, 1165–1177 CrossRef CAS PubMed.
- M. Huemer, S. Mairpady Shambat, S. D. Brugger and A. S. Zinkernagel, EMBO Rep., 2020, 21, e51034 CrossRef CAS PubMed.
- Q. Zhang, T. Xia and C. Zhang, Environ. Sci. Technol., 2020, 54, 13186–13196 CrossRef CAS PubMed.
- S. W. Kim, J. S. Lee, S. B. Park, A. R. Lee, J. W. Jung, J. H. Chun, J. M. S. Lazarte, J. Kim, J. S. Seo, J. H. Kim, J. W. Song, M. W. Ha, K. D. Thompson, C. R. Lee, M. Jung and T. S. Jung, Int. J. Mol. Sci., 2020, 21, 2822 CrossRef CAS PubMed.
- R. Misra and S. Benson, J. Bacteriol., 1988, 170, 528–533 CrossRef CAS PubMed.
- L. Fernandez and R. E. Hancock, Clin. Microbiol. Rev., 2012, 25, 661–681 CrossRef CAS PubMed.
- C. Rezzoagli, M. Archetti, I. Mignot, M. Baumgartner and R. Kummerli, PLoS Biol., 2020, 18, e3000805 CrossRef CAS PubMed.
- L. Lingzhi, G. Haojie, G. Dan, M. Hongmei, L. Yang, J. Mengdie, Z. Chengkun and Z. Xiaohui, Microbiol. Res., 2018, 215, 126–129 CrossRef CAS PubMed.
- G. Chowdhury, G. P. Pazhani, G. B. Nair, A. Ghosh and T. Ramamurthy, Int. J. Antimicrob. Agents, 2011, 38, 169–173 CAS.
- H. Kotrange, A. Najda, A. Bains, R. Gruszecki, P. Chawla and M. M. Tosif, Int. J. Mol. Sci., 2021, 22, 9596 CrossRef CAS PubMed.
- D. M. Livermore and N. Woodford, Trends Microbiol., 2006, 14, 413–420 CrossRef CAS PubMed.
- B. Khalandi, N. Asadi, M. Milani, S. Davaran, A. J. Abadi, E. Abasi and A. Akbarzadeh, Drug Res., 2017, 67, 70–76 CAS.
- W. Jiang, B. Li, X. Zheng, X. Liu, X. Pan, R. Qing, Y. Cen, J. Zheng and H. Zhou, J. Antibiot., 2013, 66, 339–345 CrossRef CAS PubMed.
- L. J. Piddock, Nat. Rev. Microbiol., 2006, 4, 629–636 CrossRef CAS PubMed.
- D. Pan, Z. Zhan, D. Chen, Z. Wu, A. Pang, Y. Wang and Z. Lin, J. Nanosci. Nanotechnol., 2011, 11, 7621–7626 CrossRef CAS PubMed.
- A. H. I. Faraag, M. W. Shafaa, N. S. Elkholy and L. J. M. Abdel-Hafez, Int. Microbiol, 2022, 25, 427–446 CrossRef CAS PubMed.
- G. W. Kaatz, F. McAleese and S. M. Seo, Antimicrob. Agents Chemother., 2005, 49, 1857–1864 CrossRef CAS PubMed.
- S. Roy, I. Hasan and B. Guo, Coord. Chem. Rev., 2023, 482, 215075 CrossRef CAS.
- O. Srichaiyapol, S. Thammawithan, P. Siritongsuk, S. Nasompag, S. Daduang, S. Klaynongsruang, S. Kulchat and R. Patramanon, Molecules, 2021, 26, 1004 CrossRef CAS PubMed.
- A. M. Diez-Pascual, Int. J. Mol. Sci., 2020, 21, 7533 CrossRef PubMed.
- E. M. Darby, E. Trampari, P. Siasat, M. S. Gaya, I. Alav, M. A. Webber and J. M. A. Blair, Nat. Rev. Microbiol., 2022, 21, 280–295 CrossRef PubMed.
- Y. Liu, L. Q. Shi, L. Z. Su, H. C. van der Mei, P. C. Jutte, Y. J. Ren and H. J. Busscher, Chem. Soc. Rev., 2019, 48, 428–446 RSC.
- A. Gupta, S. Mumtaz, C.-H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC.
- P. V. Baptista, M. P. McCusker, A. Carvalho, D. A. Ferreira, N. M. Mohan, M. Martins and A. R. Fernandes, Front. Microbiol., 2018, 9, 1441 CrossRef PubMed.
- G. V. Vimbela, S. M. Ngo, C. Fraze, L. Yang and D. A. Stout, Int. J. Nanomed., 2017, 12, 3941–3965 CrossRef CAS PubMed.
- N. Goswami, R. Bright, R. M. Visalakshan, B. Biswas, P. Zilm and K. Vasilev, Nanoscale Adv., 2019, 1, 2356–2364 CAS.
- A. Elbourne, V. E. Coyle, V. K. Truong, Y. M. Sabri, A. E. Kandjani, S. K. Bhargava, E. P. Ivanova and R. J. Crawford, Nanoscale Adv., 2019, 1, 203–212 RSC.
- R. Y. Pelgrift and A. J. Friedman, Adv. Drug Delivery Rev., 2013, 65, 1803–1815 CrossRef CAS PubMed.
- D. de Lacerda Coriolano, J. B. de Souza, E. V. Bueno, S. Medeiros, I. D. L. Cavalcanti and I. M. F. Cavalcanti, Braz. J. Microbiol., 2021, 52, 267–278 CrossRef CAS PubMed.
- C. Singh, A. K. Mehata, V. Priya, A. K. Malik, A. Setia, M. N. L. Suseela, Vikas, P. Gokul, Samridhi, S. K. Singh and M. S. Muthu, Molecules, 2022, 27, 7059 CrossRef CAS PubMed.
- P. Slepicka, N. Slepickova Kasalkova, J. Siegel, Z. Kolska and V. Svorcik, Mater., 2019, 13, 1 CrossRef PubMed.
- S. Lee and B.-H. Jun, Int. J. Mol. Sci., 2019, 20, 865 CrossRef CAS PubMed.
- H. Haidari, N. Goswami, R. Bright, Z. Kopecki, A. J. Cowin, S. Garg and K. Vasilev, Nanoscale Adv., 2019, 1, 2365–2371 RSC.
- T. Bruna, F. Maldonado-Bravo, P. Jara and N. Caro, Int. J. Mol. Sci., 2021, 22, 7202 CrossRef CAS PubMed.
- R. K. Rajendran and C. C. Lin, ACS Appl. Mater. Interfaces, 2021, 13, 46233–46246 CrossRef CAS PubMed.
- O. V. Morozova, Int. J. Mol. Sci., 2021, 22, 9928 CrossRef CAS PubMed.
- A. G. Khina and Y. A. Krutyakov, Appl. Biochem. Microbiol., 2021, 57, 683–693 CrossRef CAS.
- S. Samanta, J. Banerjee, B. Das, J. Mandal, S. Chatterjee, K. M. Ali, S. Sinha, B. Giri, T. Ghosh and S. K. Dash, Int. J. Biol. Macromol., 2022, 219, 919–939 CrossRef CAS PubMed.
- M. Mishra, S. Kumar, R. K. Majhi, L. Goswami, C. Goswami and H. Mohapatra, Front. Microbiol., 2018, 9, 823 CrossRef PubMed.
- L. d. O. Camargo, I. Fontoura, T. S. Veriato, L. Raniero and M. L. Castilho, Am. J. Infect. Control, 2023, 51, 871–878 CrossRef CAS PubMed.
- M. Palau, E. Munoz, M. F. Gusta, N. Larrosa, X. Gomis, J. Gilabert, B. Almirante, V. Puntes, R. Texido and J. Gavalda, Microbiol. Spectrum, 2023, 11, e0028023 CrossRef PubMed.
- M. Shahbandeh, A. Eghdami, M. M. Moghaddam, M. J. Nadoushan, A. Salimi, M. Fasihi-Ramandi, S. Mohammadi, M. Mirzaei and R. Mirnejad, J. Biosci., 2021, 46, 1–19 CrossRef.
- D. W. Yu, J. C. Xu, R. N. Li, J. Zhao, F. H. Li, Y. X. Zhai, J. Xue, H. F. Song, F. Yang, P. Xu and Y. Y. Song, Chemistryselect, 2021, 6, 10682–10687 CrossRef CAS.
- H. X. Zhao, H. Tang and X. Y. Jiang, ACS Nano, 2022, 16, 10066–10087 CrossRef PubMed.
- K. Dorri, F. Modaresi, M. R. Shakibaie and E. Moazamian, Pharmacia, 2022, 69, 125–133 CrossRef CAS.
- T. Khare, S. Mahalunkar, V. Shriram, S. Gosavi and V. Kumar, Environ. Res., 2021, 199, 111321 CrossRef CAS PubMed.
- M. A. Shaker and M. I. Shaaban, Int. J. Pharm., 2017, 525, 71–84 CrossRef CAS PubMed.
- X. L. Yang, L. M. Zhang and X. Y. Jiang, Nano Res., 2018, 11, 6237–6243 CrossRef CAS.
- L. Wang, S. Li, J. Yin, J. Yang, Q. Li, W. Zheng, S. Liu and X. Jiang, Nano Lett., 2020, 20, 5036–5042 CrossRef CAS PubMed.
- L. Wang, S. Li, L. Zhong, Q. Li, S. Liu, W. Zheng and X. Jiang, J. Mater. Chem. B, 2021, 9, 3025–3031 RSC.
- L. Wang, J. Yang, S. Li, Q. Li, S. Liu, W. Zheng and X. Jiang, Nano Lett., 2021, 21, 1124–1131 CrossRef CAS PubMed.
- J. N. Payne, H. K. Waghwani, M. G. Connor, W. Hamilton, S. Tockstein, H. Moolani, F. Chavda, V. Badwaik, M. B. Lawrenz and R. Dakshinamurthy, Front. Microbiol., 2016, 7, 9578 Search PubMed.
- H. Gu, P. L. Ho, E. Tong, L. Wang and B. Xu, Nano Lett., 2003, 3, 1261–1263 CrossRef CAS.
- H. Peng, D. Rossetto, S. S. Mansy, M. C. Jordan, K. P. Roos and I. A. Chen, ACS Nano, 2022, 16, 4756–4774 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, D. T. Leong and J. Xie, ACS Nano, 2017, 11, 6904–6910 CrossRef CAS PubMed.
- Z. Pang, Q. Li, Y. Jia, W. Yan, J. Qi, Y. Guo, F. Hu, D. Zhou and X. Jiang, Chem. Sci., 2021, 12, 14871–14882 RSC.
- G. Chu, C. Zhang, Y. Liu, Z. Cao, L. Wang, Y. Chen, W. Zhou, G. Gao, K. Wang and D. Cui, ACS Nano, 2020, 14, 15633–15645 CrossRef CAS PubMed.
- S. Tomane, E. Lopez-Maya, S. Boujday, V. Humblot, J. Marrot, N. Rabasso, J. Castells-Gil, C. Sicard, A. Dolbecq, P. Mialane and A. Vallee, Nanoscale Adv., 2019, 1, 3400–3405 RSC.
- Y. G. Srinivasulu, A. Mozhi, N. Goswami, Q. F. Yao and J. P. Xie, Mater. Chem. Front., 2022, 6, 689–706 RSC.
- L. Wang, M. Natan, W. Zheng, W. Zheng, S. Liu, G. Jacobi, I. Perelshtein, A. Gedanken, E. Banin and X. Jiang, Nanoscale Adv., 2020, 2, 2293–2302 RSC.
- W. Zheng, Y. Jia, Y. Zhao, J. Zhang, Y. Xie, L. Wang, X. Zhao, X. Liu, R. Tang, W. Chen and X. Jiang, Nano Lett., 2021, 21, 1992–2000 CrossRef CAS PubMed.
- H. Akbarzadeh, E. Mehrjouei, M. Abbaspour and A. N. Shamkhali, Top. Curr. Chem., 2021, 379, 22 CrossRef CAS PubMed.
- D. Mitra, E. T. Kang and K. G. Neoh, ACS Appl. Mater. Interfaces, 2020, 12, 21159–21182 CrossRef CAS PubMed.
- R. Ferrando, J. Jellinek and R. L. Johnston, Chem. Rev., 2008, 108, 845–910 CrossRef CAS PubMed.
- J. Hopf, M. Waters, V. Kalwajtys, K. E. Carothers, R. K. Roeder, J. D. Shrout, S. W. Lee and P. D. Nallathamby, Nanoscale Adv., 2019, 1, 4812–4826 RSC.
- X. Fan, L. Yahia and E. Sacher, Biology, 2021, 10, 137 CrossRef CAS PubMed.
- X. Wang, R. Li, Z. Li, R. Xiao, X.-B. Chen and T. Zhang, J. Mater. Chem. B, 2019, 7, 4169–4176 RSC.
- Y. Zhao, C. Ye, W. Liu, R. Chen and X. Jiang, Angew Chem. Int. Ed. Engl., 2014, 53, 8127–8131 CrossRef CAS PubMed.
- A. A. Balhaddad, I. M. Garcia, L. Mokeem, R. Alsahafi, F. M. Collares and M. A. Sampaio de Melo, Bioeng., 2021, 8, 146 CAS.
- S. M. Youn and S. J. Choi, Int. J. Mol. Sci., 2022, 23, 6074 CrossRef CAS PubMed.
- A. Krol, P. Pomastowski, K. Rafinska, V. Railean-Plugaru and B. Buszewski, Adv. Colloid Interface Sci., 2017, 249, 37–52 CrossRef CAS PubMed.
- S. Raha and M. Ahmaruzzaman, Nanoscale Adv., 2022, 4, 1868–1925 RSC.
- C. R. Mendes, G. Dilarri, C. F. Forsan, V. M. R. Sapata, P. R. M. Lopes, P. B. de Moraes, R. N. Montagnolli, H. Ferreira and E. D. Bidoia, Sci. Rep., 2022, 12, 2658 CrossRef CAS PubMed.
- J. Verma, A. S. Khanna, R. Sahney and A. Bhattacharya, Nanoscale Adv., 2020, 2, 4093–4105 RSC.
- K. Chand, D. Cao, D. E. Fouad, A. H. Shah, M. N. Lakhan, A. Q. Dayo, H. J. Sagar, K. Zhu and A. M. A. Mohamed, J. Mol. Liq., 2020, 316, 113821 CrossRef CAS.
- E. Abdolmajid, H. Kharazi, M. Chalaki, M. Khojasteh, S. Haghighat, F. Attar, F. Nemati and M. Falahati, J. Biomol. Struct. Dyn., 2019, 37, 3007–3017 CrossRef CAS PubMed.
- R. H. Al-Serwi, M. El-Sherbiny, T. V. A. Kumar, A. A. Qasim, T. Khattar, Y. Alghazwani, A. Alqahtani, V. Krishnaraju, J. M. Muthu Mohamed and V. P. Sundramurthy, Bioinorg. Chem. Appl., 2022, 2022, 1142727 Search PubMed.
- N. Elahi and M. Rizwan, Artif. Organs, 2021, 45, 1272–1299 CrossRef CAS PubMed.
- L. M. Armijo, S. J. Wawrzyniec, M. Kopciuch, Y. I. Brandt, A. C. Rivera, N. J. Withers, N. C. Cook, D. L. Huber, T. C. Monson, H. D. C. Smyth and M. Osinski, J. Nanobiotechnol., 2020, 18, 35 CrossRef CAS PubMed.
- Z. Li, C. J. Xie, X. W. Ren, Q. Zhang and B. J. Ma, Rare Met., 2023, 42, 1899–1911 CrossRef CAS.
- S. D. Mo, Y. Zhao, J. H. Wen, J. Y. Sun, Z. Zhang, Q. L. Yu, G. C. Wang, X. Chen and M. Y. Liu, J. Hazard. Mater., 2022, 432, 128662 CrossRef CAS PubMed.
- Y. Qiao, Y. Ping, H. B. Zhang, B. Zhou, F. Y. Liu, Y. H. Yu, T. T. Xie, W. L. Li, D. N. Zhong, Y. Z. Zhang, K. Yao, H. A. Santos and M. Zhou, ACS Appl. Mater. Interfaces, 2019, 11, 3809–3822 CrossRef CAS PubMed.
- J. Gouyau, R. E. Duval, A. Boudier and E. Lamouroux, Int. J. Mol. Sci., 2021, 22, 1905 CrossRef CAS PubMed.
- S. Liao, Y. Zhang, X. Pan, F. Zhu, C. Jiang, Q. Liu, Z. Cheng, G. Dai, G. Wu, L. Wang and L. Chen, Int. J. Nanomed., 2019, 14, 1469–1487 CrossRef CAS PubMed.
- S. Gevorgyan, R. Schubert, S. Falke, K. Lorenzen, K. Trchounian and C. Betzel, Sci. Rep., 2022, 12, 14077 CrossRef CAS PubMed.
- Z. Cheng, S. Tang, J. Feng and Y. Wu, Heliyon, 2022, 8, e10010 CrossRef CAS PubMed.
- L. Wang, Q. Hou, W. Zheng and X. Jiang, ACS Nano, 2021, 15, 17885–17894 CrossRef CAS PubMed.
- L. Wang, W. Zheng, S. Li, L. Zhong and X. Jiang, Nano Lett., 2022, 22, 3576–3582 CrossRef CAS PubMed.
- S. G. Ali, M. Jalal, H. Ahmad, K. Umar, A. Ahmad, M. B. Alshammari and H. M. Khan, Molecules, 2022, 27, 8685 CrossRef CAS PubMed.
- A. M. Shehabeldine, B. H. Amin, F. A. Hagras, A. A. Ramadan, M. R. Kamel, M. A. Ahmed, K. H. Atia and S. S. Salem, Appl. Biochem. Biotechnol., 2023, 195, 467–485 CrossRef CAS PubMed.
- A. Miri, N. Mahdinejad, O. Ebrahimy, M. Khatami and M. Sarani, Mater. Sci. Eng., C, 2019, 104, 109981 CrossRef CAS PubMed.
- O. Metryka, D. Wasilkowski, A. Nowak, M. Adamczyk-Habrajska and A. Mrozik, Int. J. Mol. Sci., 2020, 21, 9089 CrossRef CAS PubMed.
- F. Amin, Fozia, B. Khattak, A. Alotaibi, M. Qasim, I. Ahmad, R. Ullah, M. Bourhia, A. Gul, S. Zahoor and R. Ahmad, J. Evidence-Based Complementary Altern. Med., 2021, 2021, 5589703 Search PubMed.
- G. Ramesh, J. E. Kaviyil, W. Paul, R. Sasi and R. Joseph, ACS Omega, 2022, 7, 6795–6809 CrossRef CAS PubMed.
- R. Ren, C. Lim, S. Li, Y. Wang, J. Song, T. W. Lin, B. W. Muir, H. Y. Hsu and H. H. Shen, Nanomater., 2022, 12, 3855 CrossRef CAS PubMed.
- M. Chu, H. Gao, S. Liu, L. Wang, Y. Jia, M. Gao, M. Wan, C. Xu and L. Ren, RSC Adv., 2018, 8, 18197–18203 RSC.
- Y. Wu, C. Li, H. C. van der Mei, H. J. Busscher and Y. Ren, Antibiotics, 2021, 10, 623 CrossRef CAS PubMed.
- H.-J. Jian, J. Yu, Y.-J. Li, B. Unnikrishnan, Y.-F. Huang, L.-J. Luo, D. Hui-Kang Ma, S. G. Harroun, H.-T. Chang, H.-J. Lin, J.-Y. Lai and C.-C. Huang, Chem. Eng. J., 2020, 386, 123913 CrossRef CAS.
- C. Zhao, X. Wang, L. Yu, L. Wu, X. Hao, Q. Liu, L. Lin, Z. Huang, Z. Ruan, S. Weng, A. Liu and X. Lin, Acta Biomater., 2022, 138, 528–544 CrossRef CAS PubMed.
- M. Assali, A. N. Zaid, F. Abdallah, M. Almasri and R. Khayyat, Int. J. Nanomed., 2017, 12, 6647–6659 CrossRef CAS PubMed.
- K. Banihashemi, B. Sobouti, I. Mehregan, R. Bakhtiari and N. Amirmozafari, Rep. Biochem. Mol. Biol., 2020, 9, 89 CrossRef CAS PubMed.
- S. Gurunathan and J. H. Kim, Int. J. Nanomed., 2016, 11, 1927–1945 CrossRef CAS PubMed.
- M. Y. Xia, Y. Xie, C. H. Yu, G. Y. Chen, Y. H. Li, T. Zhang and Q. Peng, J. Controlled Release, 2019, 307, 16–31 CrossRef CAS PubMed.
- K. Yan, C. Mu, L. Meng, Z. Fei and P. J. Dyson, Nanoscale Adv., 2021, 3, 3708–3729 RSC.
- H. Naeem, M. Ajmal, R. B. Qureshi, S. T. Muntha, M. Farooq and M. Siddiq, J. Environ. Manage., 2019, 230, 199–211 CrossRef CAS PubMed.
- A. Z. Warsi, F. Aziz, S. Zulfiqar, S. Haider, I. Shakir and P. O. Agboola, Nanomater., 2022, 12, 713 CrossRef CAS PubMed.
- F. Wu, H. Zheng, W. Wang, Q. Wu, Q. Zhang, J. Guo, B. Pu, X. Shi, J. Li, X. Chen and W. Hong, Sci. China Mater., 2020, 64, 748–758 CrossRef.
- L. Zhou, H. Zheng, Z. Liu, S. Wang, Z. Liu, F. Chen, H. Zhang, J. Kong, F. Zhou and Q. Zhang, ACS Nano, 2021, 15, 2468–2480 CrossRef CAS PubMed.
- V. K. Sharma, T. J. McDonald, H. Kim and V. K. Garg, Adv. Colloid Interface Sci., 2015, 225, 229–240 CrossRef CAS PubMed.
- Y. Lyu, Y. Shi, S. Zhu, Y. Jia, C. Tong, S. Liu, B. Sun and J. Zhang, Sensors, 2022, 22, 7952 CrossRef CAS PubMed.
- F. Mohajer, G. Mohammadi Ziarani, A. Badiei, S. Iravani and R. S. Varma, RSC Adv., 2023, 13, 8136–8152 RSC.
- C. Zhang, J. Guo, X. Zou, S. Guo, Y. Guo, R. Shi and F. Yan, Adv. Healthcare Mater., 2021, 10, e2100775 CrossRef PubMed.
- L. G. Ding, S. Wang, B. J. Yao, F. Li, Y. A. Li, G. Y. Zhao and Y. B. Dong, Adv. Healthcare Mater., 2021, 10, e2001821 CrossRef PubMed.
- S. Szunerits, A. Barras and R. Boukherroub, Int. J. Environ. Res. Public Health, 2016, 13, 413 CrossRef PubMed.
- C. Quan, H. Lin, H. Xiao and J. Zhao, J. Clin. Lab. Anal., 2021, 35, e23872 CrossRef CAS PubMed.
- W. Cao, X. Wang, Q. Li, X. Peng, L. Wang, P. Li, Z. Ye and X. Xing, Mater. Sci. Eng., C, 2018, 92, 307–316 CrossRef CAS PubMed.
- M. Nasrollahzadeh, M. Sajjadi, S. Iravani and R. S. Varma, Carbohydr. Polym., 2021, 251, 116986 CrossRef CAS PubMed.
- N. Morin-Crini, E. Lichtfouse, G. Torri and G. Crini, Environ. Chem. Lett., 2019, 17, 1667–1692 CrossRef CAS.
- L. Liping, L. Kexin, D. Huipu, L. Jia and Z. Jie, Nat. Prod. Commun., 2020, 15, 1934578X20946876 CrossRef.
- U. Garg, S. Chauhan, U. Nagaich and N. Jain, Adv. Pharm. Bull., 2019, 9, 195–204 CrossRef CAS PubMed.
- G. Kravanja, M. Primožič, Ž. Knez and M. Leitgeb, Molecules, 2019, 24, 1960 CrossRef CAS PubMed.
- I. Schlachet, H. Moshe Halamish and A. Sosnik, Molecules, 2020, 25, 4496 CrossRef CAS PubMed.
- M. Wang, Z. Deng, Y. Guo and P. Xu, Nanoscale Adv., 2022, 5, 27–45 RSC.
- R. Zhang, Y. Li, M. Zhou, C. Wang, P. Feng, W. Miao and H. Huang, ACS Appl. Mater. Interfaces, 2019, 11, 26711–26721 CrossRef CAS PubMed.
- C. Zhou, M. Wang, K. Zou, J. Chen, Y. Zhu and J. Du, ACS Macro Lett., 2013, 2, 1021–1025 CrossRef CAS PubMed.
- S. Roy, A. Mondal, V. Yadav, A. Sarkar, R. Banerjee, P. Sanpui and A. Jaiswal, ACS Appl. Bio Mater., 2019, 2, 2738–2755 CrossRef CAS PubMed.
- A. Jolivet-Gougeon and M. Bonnaure-Mallet, Drug Discovery Today: Technol., 2014, 11, 49–56 CrossRef PubMed.
- Q. Shi, H. Liu, D. Tang, Y. Li, X. Li and F. Xu, NPG Asia Mater., 2019, 11, 64 CrossRef CAS.
- D. N. Iqbal, S. Shafiq, S. M. Khan, S. M. Ibrahim, S. A. Abubshait, A. Nazir, M. Abbas and M. Iqbal, Int. J. Biol. Macromol., 2020, 164, 499–509 CrossRef CAS PubMed.
- H. Aksel, F. Mahjour, F. Bosaid, S. Calamak and A. A. Azim, J. Endod., 2020, 46, 1867–1875 CrossRef PubMed.
- C. H. Wang, J. H. Cherng, C. C. Liu, T. J. Fang, Z. J. Hong, S. J. Chang, G. Y. Fan and S. D. Hsu, Int. J. Mol. Sci., 2021, 22, 7067 CrossRef CAS PubMed.
- D. Feldman, J. Macromol. Sci., Part A: Pure Appl.Chem., 2016, 53, 55–62 CrossRef CAS.
- R. Namivandi-Zangeneh, E. H. H. Wong and C. Boyer, ACS Infect. Dis., 2021, 7, 215–253 CrossRef CAS PubMed.
- M. El-Sayed, S. E. Al-Mofty, N. K. Mahdy, W. A. Sarhan and H. M. E. Azzazy, Nanoscale Adv., 2023, 5, 2517–2529 RSC.
- N. Hasan, J. Cao, J. Lee, S. P. Hlaing, M. A. Oshi, M. Naeem, M. H. Ki, B. L. Lee, Y. Jung and J. W. Yoo, Pharmaceutics, 2019, 11, 236 CrossRef CAS PubMed.
- L. Wang, J. Yang, X. Yang, Q. Hou, S. Liu, W. Zheng, Y. Long and X. Jiang, ACS Appl. Mater. Interfaces, 2020, 12, 51148–51159 CrossRef CAS PubMed.
- R. Javed, M. Ahmed, I. u. Haq, S. Nisa and M. Zia, Mater. Sci. Eng., C, 2017, 79, 108–115 CrossRef CAS PubMed.
- P. Sautrot-Ba, N. Razza, L. Breloy, S. A. Andaloussi, A. Chiappone, M. Sangermano, C. Hélary, S. Belbekhouche, T. Coradin and D. L. Versace, J. Mater. Chem. B, 2019, 7, 6526–6538 RSC.
- J. D. Vanza, R. B. Patel, R. R. Dave and M. R. Patel, Pharm. Dev. Technol., 2020, 25, 1169–1187 CrossRef CAS PubMed.
- Y. He, Y. Jin, X. Wang, S. Yao, Y. Li, Q. Wu, G. Ma, F. Cui and H. Liu, Nanomater. , 2018, 8, 327 CrossRef PubMed.
- S. Li, L. Wang, W. Zheng, G. Yang and X. Jiang, Adv. Funct. Mater., 2020, 30, 2002370 CrossRef CAS.
- L. Xiao, F. Hui, T. Tian, R. Yan, J. Xin, X. Zhao, Y. Jiang, Z. Zhang, Y. Kuang, N. Li, Y. Zhao and Q. Lin, Front. Chem., 2021, 9, 787886 CrossRef CAS PubMed.
- I. Armentano, D. Puglia, F. Luzi, C. R. Arciola, F. Morena, S. Martino and L. Torre, Mater., 2018, 11, E795 CrossRef PubMed.
- M. Chao, L. He, M. Gong, N. Li, X. Li, L. Peng, F. Shi, L. Zhang and P. Wan, ACS Nano, 2021, 15, 9746–9758 CrossRef CAS PubMed.
- H. Jin, M. O. G. Nayeem, S. Lee, N. Matsuhisa, D. Inoue, T. Yokota, D. Hashizume and T. Someya, ACS Nano, 2019, 13, 7905–7912 CrossRef CAS PubMed.
- V. K. Pandey, S. N. Upadhyay, K. Niranjan and P. K. Mishra, Int. J. Biol. Macromol., 2020, 157, 212–219 CrossRef CAS PubMed.
- S. Kirti, V. M. Bhandari, J. Jena and A. S. Bhattacharyya, J. Environ. Manage., 2018, 226, 95–105 CrossRef CAS PubMed.
- R. E. Morsi, A. M. Alsabagh, S. A. Nasr and M. M. Zaki, Int. J. Biol. Macromol., 2017, 97, 264–269 CrossRef CAS PubMed.
- J. Troyano, A. Carne-Sanchez, C. Avci, I. Imaz and D. Maspoch, Chem. Soc. Rev., 2019, 48, 5534–5546 RSC.
- P. Walvekar, R. Gannimani, M. Salih, S. Makhathini, C. Mocktar and T. Govender, Colloids Surf., B, 2019, 182, 110388 CrossRef CAS PubMed.
- X. Unamuno, E. Imbuluzqueta, F. Salles, P. Horcajada and M. J. Blanco-Prieto, Eur. J. Pharm. Biopharm., 2018, 132, 11–18 CrossRef CAS PubMed.
- S. Yao, J. J. Chi, Y. T. Wang, Y. J. Zhao, Y. Luo and Y. A. Wang, Adv. Healthcare Mater., 2021, 10, e2100056 CrossRef PubMed.
- M. Chen, Z. Long, R. Dong, L. Wang, J. Zhang, S. Li, X. Zhao, X. Hou, H. Shao and X. Jiang, Small, 2020, 16, 1906240 CrossRef CAS PubMed.
- N. Pal, P. Dubey, P. Gopinath and K. Pal, Int. J. Biol. Macromol., 2017, 95, 94–105 CrossRef CAS PubMed.
- J. A. T. Marin, S. R. Londono, J. Delgado, D. P. N. Porras, M. E. V. Zapata, J. H. M. Hernandez, C. H. Valencia and C. D. G. Tovar, Int. J. Mol. Sci., 2019, 20, 2987 CrossRef CAS PubMed.
- K. S. Venkataprasanna, J. Prakash, S. Vignesh, G. Bharath, M. Venkatesan, F. Banat, S. Sahabudeen, S. Ramachandran and G. D. Venkatasubbu, Int. J. Biol. Macromol., 2020, 143, 744–762 CrossRef CAS PubMed.
- Y. Wang, Y. Lu, J. Zhang, X. Hu, Z. Yang, Y. Guo and Y. Wang, J. Mater. Chem. B, 2019, 7, 538–547 RSC.
- Z. Yang, C. Sun, L. Wang, H. Chen, J. He and Y. Chen, J. Colloid Interface Sci., 2017, 507, 344–352 CrossRef CAS PubMed.
- S. Nadzirah, S. C. B. Gopinath, N. A. Parmin, A. A. Hamzah, M. A. Mohamed, E. Y. Chang and C. F. Dee, Crit. Rev. Anal. Chem., 2022, 52, 637–648 CrossRef CAS PubMed.
- J. Seaberg, H. Montazerian, M. N. Hossen, R. Bhattacharya, A. Khademhosseini and P. Mukherjee, ACS Nano, 2021, 15, 2099–2142 CrossRef CAS PubMed.
- M. Balcioglu, M. Rana and M. V. Yigit, J. Mater. Chem. B, 2013, 1, 6187–6193 RSC.
- M. Mabrouk, R. M. Abd El-Wahab, H. H. Beherei, M. M. Selim and D. B. Das, Int. J. Pharm., 2020, 587, 119658 CrossRef CAS PubMed.
- S. Mishra and H. B. Singh, Appl. Microbiol. Biotechnol., 2015, 99, 1097–1107 CrossRef CAS PubMed.
- M. D. Scherer, J. C. V. Sposito, W. F. Falco, A. B. Grisolia, L. H. C. Andrade, S. M. Lima, G. Machado, V. A. Nascimento, D. A. Goncalves, H. Wender, S. L. Oliveira and A. R. L. Caires, Sci. Total Environ., 2019, 660, 459–467 CrossRef CAS PubMed.
- E. J. Park, E. Bae, J. Yi, Y. Kim, K. Choi, S. H. Lee, J. Yoon, B. C. Lee and K. Park, Environ. Toxicol. Pharmacol., 2010, 30, 162–168 CrossRef CAS PubMed.
- Y. M. Cho, Y. Mizuta, J. I. Akagi, T. Toyoda, M. Sone and K. Ogawa, J. Toxicol. Pathol., 2018, 31, 73–80 CrossRef CAS PubMed.
- Y. Yao, Y. Zang, J. Qu, M. Tang and T. Zhang, Int. J. Nanomed., 2019, 14, 8787–8804 CrossRef CAS PubMed.
- J. Jiravova, K. B. Tomankova, M. Harvanova, L. Malina, J. Malohlava, L. Luhova, A. Panacek, B. Manisova and H. Kolarova, Food Chem. Toxicol., 2016, 96, 50–61 CrossRef CAS PubMed.
- R. Chen, L. Zhao, R. Bai, Y. Liu, L. Han, Z. Xu, F. Chen, H. Autrup, D. Long and C. Chen, Toxicol. Res., 2016, 5, 602–608 CrossRef CAS PubMed.
- J. Li, B. Zhang, X. Chang, J. Gan, W. Li, S. Niu, L. Kong, T. Wu, T. Zhang, M. Tang and Y. Xue, Environ. Pollut., 2020, 256, 113430 CrossRef CAS PubMed.
- M. Akter, M. T. Sikder, M. M. Rahman, A. Ullah, K. F. B. Hossain, S. Banik, T. Hosokawa, T. Saito and M. Kurasaki, J. Adv. Res., 2018, 9, 1–16 CrossRef CAS PubMed.
- Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef CAS PubMed.
- T. Mironava, M. Hadjiargyrou, M. Simon, V. Jurukovski and M. H. Rafailovich, Nanotoxicology, 2010, 4, 120–137 CrossRef CAS PubMed.
- C. M. Goodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Bioconjugate Chem., 2004, 15, 897–900 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- J. G. Nirmala, S. Akila, M. S. A. M. Nadar, R. T. Narendhirakannan and S. Chatterjee, RSC Adv., 2016, 6, 82205–82218 RSC.
- V. Iswarya, J. Manivannan, A. De, S. Paul, R. Roy, J. B. Johnson, R. Kundu, N. Chandrasekaran, A. Mukherjee and A. Mukherjee, Environ. Sci. Pollut. Res. Int., 2016, 23, 4844–4858 CrossRef CAS PubMed.
- H. Y. Jia, Y. Liu, X. J. Zhang, L. Han, L. B. Du, Q. Tian and Y. C. Xu, J. Am. Chem. Soc., 2009, 131, 40–41 CrossRef CAS PubMed.
- M. R. Jo, S. H. Bae, M. R. Go, H. J. Kim, Y. G. Hwang and S. J. Choi, Nanomater., 2015, 5, 835–850 CrossRef CAS PubMed.
- I. Fratoddi, I. Venditti, C. Cametti and M. V. Russo, Toxicol. Res., 2015, 4, 796–800 CrossRef CAS.
- M. V. Brundo, R. Pecoraro, F. Marino, A. Salvaggio, D. Tibullo, S. Saccone, V. Bramanti, M. A. Buccheri, G. Impellizzeri, V. Scuderi, M. Zimbone and V. Privitera, Front. Physiol., 2016, 7, 130 CrossRef PubMed.
- S. Nangia and R. Sureshkumar, Langmuir, 2012, 28, 17666–17671 CrossRef CAS PubMed.
- N. N. Farshori, M. A. Siddiqui, M. M. Al-Oqail, E. S. Al-Sheddi, S. M. Al-Massarani, M. Ahamed, J. Ahmad and A. A. Al-Khedhairy, Biol. Trace Elem. Res., 2022, 200, 5042–5051 CrossRef CAS PubMed.
- P. M. Costa, I. Gosens, A. Williams, L. Farcal, D. Pantano, D. M. Brown, V. Stone, F. R. Cassee, S. Halappanavar and B. Fadeel, J. Appl. Toxicol., 2018, 38, 385–397 CrossRef CAS PubMed.
- T. S. Kang, R. F. Guan, X. Q. Chen, Y. J. Song, H. Jiang and J. Zhao, Nanoscale Res. Lett., 2013, 8, 496 CrossRef PubMed.
- Y. Y. Kao, Y. C. Chen, T. J. Cheng, Y. M. Chiung and P. S. Liu, Toxicol. Sci., 2012, 125, 462–472 CrossRef CAS PubMed.
- V. Valdiglesias, C. Costa, G. Kilic, S. Costa, E. Pasaro, B. Laffon and J. P. Teixeira, Environ. Int., 2013, 55, 92–100 CrossRef CAS PubMed.
- S. Hussain, S. Boland, A. Baeza-Squiban, R. Hamel, L. C. Thomassen, J. A. Martens, M. A. Billon-Galland, J. Fleury-Feith, F. Moisan, J. C. Pairon and F. Marano, Toxicology, 2009, 260, 142–149 CrossRef CAS PubMed.
- J.-J. Yin, J. Liu, M. Ehrenshaft, J. E. Roberts, P. P. Fu, R. P. Mason and B. Zhao, Toxicol. Appl. Pharmacol., 2012, 263, 81–88 CrossRef CAS PubMed.
- K. Yamashita, Y. Yoshioka, K. Higashisaka, Y. Morishita, T. Yoshida, M. Fujimura, H. Kayamuro, H. Nabeshi, T. Yamashita, K. Nagano, Y. Abe, H. Kamada, Y. Kawai, T. Mayumi, T. Yoshikawa, N. Itoh, S.-i. Tsunoda and Y. Tsutsumi, Inflammation, 2010, 33, 276–280 CrossRef CAS PubMed.
- H.-S. Hsieh, R. Wu and C. T. Jafvert, Environ. Sci. Technol., 2014, 48, 11330–11336 CrossRef CAS PubMed.
- C. Ventura, J. F. S. Pereira, P. Matos, B. Marques, P. Jordan, A. Sousa-Uva and M. J. Silva, Nanotoxicology, 2020, 14, 479–503 CrossRef CAS PubMed.
- A. Borghini, M. Roursgaard, M. G. Andreassi, A. Kermanizadeh and P. Moller, Mutagenesis, 2017, 32, 173–180 CrossRef CAS PubMed.
- F. Zhao, H. Meng, L. Yan, B. Wang and Y. Zhao, Sci. Bull., 2015, 60, 3–20 CrossRef CAS.
- Y. L. Chang, S. T. Yang, J. H. Liu, E. Dong, Y. W. Wang, A. N. Cao, Y. F. Liu and H. F. Wang, Toxicol. Lett., 2011, 200, 201–210 CrossRef CAS PubMed.
- Y. Zhang, S. F. Ali, E. Dervishi, Y. Xu, Z. Li, D. Casciano and A. S. Biris, ACS Nano, 2010, 4, 3181–3186 CrossRef CAS PubMed.
- S. Manivong, A. Garcia Ac, S. A. Patten, J. C. Fernandes, M. Benderdour, X. Banquy, F. Moldovan and V. G. Roullin, Nanomaterials, 2022, 12(8), 1337 CrossRef CAS PubMed.
- A. Zubareva, B. Shagdarova, V. Varlamov, E. Kashirina and E. Svirshchevskaya, Eur. Polym. J., 2017, 93, 743–749 CrossRef CAS.
- B. Bechinger and S. U. Gorr, J. Dent. Res., 2017, 96, 254–260 CrossRef CAS PubMed.
- Y. Huan, Q. Kong, H. Mou and H. Yi, Front. Microbiol., 2020, 11, 582779 CrossRef PubMed.
- A. Ebbensgaard, H. Mordhorst, F. M. Aarestrup and E. B. Hansen, Front. Microbiol., 2018, 9, 2153 CrossRef PubMed.
- I. Kamal, U. A. Ashfaq, S. Hayat, B. Aslam, M. H. Sarfraz, H. Yaseen, M. S. R. Rajoka, A. A. Shah and M. Khurshid, Biotechnol. Lett., 2023, 45, 137–162 CrossRef CAS PubMed.
- M. Meena, P. Prajapati, C. Ravichandran and R. Sehrawat, Food Sci. Biotechnol., 2021, 30, 1481–1496 CrossRef CAS PubMed.
- W. Yang, L. Wang, E. M. Mettenbrink, P. L. DeAngelis and S. Wilhelm, Annu. Rev. Pharmacol. Toxicol., 2021, 61, 269–289 CrossRef CAS PubMed.
- E. J. Petersen, M. Mortimer, R. M. Burgess, R. Handy, S. Hanna, K. T. Ho, M. Johnson, S. Loureiro, H. Selck, J. J. Scott-Fordsmand, D. Spurgeon, J. Unrine, N. van den Brink, Y. Wang, J. White and P. Holden, Environ. Sci.: Nano, 2019, 6, 1039 Search PubMed.
- M. S. Jo, J. K. Kim, Y. Kim, H. P. Kim, H. S. Kim, K. Ahn, J. H. Lee, E. M. Faustman, M. Gulumian, B. Kelman and I. J. Yu, Arch. Toxicol., 2020, 94, 773–784 CrossRef CAS PubMed.
- L. Narciso, L. Coppola, G. Lori, C. Andreoli, A. Zjino, B. Bocca, F. Petrucci, A. Di Virgilio, A. Martinelli, A. Tinari, F. Maranghi and R. Tassinari, Nanoimpact, 2020, 18, 100221 CrossRef.
- R. George, S. Merten, T. T. Wang, P. Kennedy and P. Maitz, Australas. J. Dermatol., 2014, 55, 185–190 CrossRef PubMed.
- J. H. Sung, J. H. Ji, J. D. Park, M. Y. Song, K. S. Song, H. R. Ryu, J. U. Yoon, K. S. Jeon, J. Jeong, B. S. Han, Y. H. Chung, H. K. Chang, J. H. Lee, D. W. Kim, B. J. Kelman and I. J. Yu, Part. Fibre Toxicol., 2011, 8, 16 CrossRef CAS PubMed.
- W. G. Kreyling, W. Moeller, U. Holzwarth, S. Hirn, A. Wenk, C. Schleh, M. Schaeffler, N. Haberl, N. Gibson and J. C. Schittny, ACS Nano, 2018, 12, 7771–7790 CrossRef CAS PubMed.
- M. R. Miller, J. B. Raftis, J. P. Langrish, S. G. McLean, P. Samutrtai, S. P. Connell, S. Wilson, A. T. Vesey, P. H. B. Fokkens, A. J. F. Boere, P. Krystek, C. J. Campbell, P. W. F. Hadoke, K. Donaldson, F. R. Cassee, D. E. Newby, R. Duffin and N. L. Mills, ACS Nano, 2017, 11, 4542–4552 CrossRef CAS PubMed.
- M. Bednarski, M. Dudek, J. Knutelska, L. Nowinski, J. Sapa, M. Zygmunt, G. Nowak, M. Luty-Blocho, M. Wojnicki, K. Fitzner and M. Tesiorowski, Pharmacol. Rep., 2015, 67, 405–409 CrossRef CAS PubMed.
- G. Raju, N. Katiyar, S. Vadukumpully and S. A. Shankarappa, J. Dermatol. Sci., 2018, 89, 146–154 CrossRef CAS PubMed.
- R. Fernandes, N. R. Smyth, O. L. Muskens, S. Nitti, A. Heuer-Jungemann, M. R. Ardern-Jones and A. G. Kanaras, Small, 2015, 11, 713–721 CrossRef CAS PubMed.
- L. Wang, J. Li, J. Pan, X. Jiang, Y. Ji, Y. Li, Y. Qu, Y. Zhao, X. Wu and C. Chen, J. Am. Chem. Soc., 2013, 135, 17359–17368 CrossRef CAS PubMed.
- T. Niidome, M. Yamagata, Y. Okamoto, Y. Akiyama, H. Takahashi, T. Kawano, Y. Katayama and Y. Niidome, J. Controlled Release, 2006, 114, 343–347 CrossRef CAS PubMed.
- S. Naz, A. Gul and M. Zia, IET Nanobiotechnol., 2020, 14, 1–13 CrossRef PubMed.
- S. Areecheewakul, A. Adamcakova-Dodd, E. Haque, X. Jing, D. K. Meyerholz, P. T. O'Shaughnessy, P. S. Thorne and A. K. Salem, Part. Fibre Toxicol., 2022, 19, 40 CrossRef CAS PubMed.
- B. E. El Bialy, R. A. Hamouda, M. A. Abd Eldaim, S. S. El Ballal, H. S. Heikal, H. K. Khalifa and W. N. Hozzein, Int. J. Nanomed., 2020, 15, 3827–3842 CrossRef CAS PubMed.
- X. Li, W. Sun and L. An, Toxicol. Ind. Health, 2018, 34, 409–421 CrossRef CAS PubMed.
- Y. Sun, G. Zhang, Z. He, Y. Wang, J. Cui and Y. Li, Int. J. Nanomed., 2016, 11, 905–918 CAS.
- E. I. Hassanen, M. A. Ibrahim, A. M. Hassan, S. Mehanna, S. H. Aljuaydi and M. Y. Issa, Neurochem. Res., 2021, 46, 1264–1279 CrossRef CAS PubMed.
- E.-B. Jung, J. Yu and S.-J. Choi, Nanomaterials, 2021, 11, 2922 CrossRef CAS PubMed.
- A. K. Bhunia, P. K. Samanta, S. Saha and T. Kamilya, Appl. Phys. Lett., 2013, 103, 143701 CrossRef.
- R. Pati, I. Das, R. K. Mehta, R. Sahu and A. Sonawane, Toxicol. Sci., 2016, 150, 454–472 CrossRef CAS PubMed.
- M. T. Hamed, B. A. Bakr, Y. H. Shahin, B. H. Elwakil, M. M. Abu-Serie, F. S. Aljohani and A. A. Bekhit, Bioinorg. Chem. Appl., 2021, 2021, 8171786 Search PubMed.
- K. Yamashita, Y. Yoshioka, K. Higashisaka, K. Mimura, Y. Morishita, M. Nozaki, T. Yoshida, T. Ogura, H. Nabeshi, K. Nagano, Y. Abe, H. Kamada, Y. Monobe, T. Imazawa, H. Aoshima, K. Shishido, Y. Kawai, T. Mayumi, S. Tsunoda, N. Itoh, T. Yoshikawa, I. Yanagihara, S. Saito and Y. Tsutsumi, Nat. Nanotechnol., 2011, 6, 321–328 CrossRef CAS PubMed.
- A. J. Khoei and K. Rezaei, Toxin Rev., 2022, 41, 237–246 CrossRef.
- Y. Liang, A. Simaiti, M. Xu, S. Lv, H. Jiang, X. He, Y. Fan, S. Zhu, B. Du, W. Yang, X. Li and P. Yu, Nanomater., 2022, 12, 2769 CrossRef CAS PubMed.
- S. T. Yang, X. Wang, G. Jia, Y. Gu, T. Wang, H. Nie, C. Ge, H. Wang and Y. Liu, Toxicol. Lett., 2008, 181, 182–189 CrossRef CAS PubMed.
- A. R. Murray, E. Kisin, S. S. Leonard, S. H. Young, C. Kommineni, V. E. Kagan, V. Castranova and A. A. Shvedova, Toxicology, 2009, 257, 161–171 CrossRef CAS PubMed.
- J. P. Reyes, J. R. Celorico, P. A. Deyro, C. N. Ochona, Z. A. Ochona, B. A. Visaya and B. A. Basilia, Mater. Today: Proc., 2022, 66, 3178–3184 CAS.
- M. C. Duch, G. R. Budinger, Y. T. Liang, S. Soberanes, D. Urich, S. E. Chiarella, L. A. Campochiaro, A. Gonzalez, N. S. Chandel, M. C. Hersam and G. M. Mutlu, Nano Lett., 2011, 11, 5201–5207 CrossRef PubMed.
- X. Y. Zhang, J. L. Yin, C. Peng, W. Q. Hu, Z. Y. Zhu, W. X. Li, C. H. Fan and Q. Huang, Carbon, 2011, 49, 986–995 CrossRef CAS.
- C. Fu, T. Liu, L. Li, H. Liu, Q. Liang and X. Meng, Biomaterials, 2015, 40, 23–31 CrossRef CAS PubMed.
- A. K. Patlolla, J. Rondalph and P. B. Tchounwou, Austin J. Environ. Toxicol., 2017, 3, 1021 Search PubMed.
- R. Rauti, M. Medelin, L. Newman, S. Vranic, G. Reina, A. Bianco, M. Prato, K. Kostarelos and L. Ballerini, Nano Lett., 2019, 19, 2858–2870 CrossRef CAS PubMed.
- Y. Li, Y. Wang, L. Tu, D. Chen, Z. Luo, D. Liu, Z. Miao, G. Feng, L. Qing and S. Wang, Int. J. Environ. Res. Public Health, 2016, 13, 1149 CrossRef PubMed.
- Y. L. Hu, W. Qi, F. Han, J. Z. Shao and J. Q. Gao, Int. J. Nanomed., 2011, 6, 3351–3359 CAS.
- M. Mahlapuu, J. Hakansson, L. Ringstad and C. Bjorn, Front. Cell. Infect. Microbiol., 2016, 6, 194 Search PubMed.
- P. Vlieghe, V. Lisowski, J. Martinez and M. Khrestchatisky, Drug Discovery Today, 2010, 15, 40–56 CrossRef CAS PubMed.
- G. Li, Z. Lai and A. Shan, Adv. Sci., 2023, 10, e2206602 CrossRef PubMed.
- A. R. J. Azar and S. Mohebbi, Micro Nano Lett., 2013, 8, 813–815 CrossRef.
- J. Wongpreecha, D. Polpanich, T. Suteewong, C. Kaewsaneha and P. Tangboriboonrat, Carbohydr. Polym., 2018, 199, 641–648 CrossRef CAS PubMed.
- K. J. Lee, Y.-I. Lee, I.-K. Shim, B. H. Jun, H. J. Cho and J. Joung, Solid State Phenom., 2007, 124–126, 1189–1192 CAS.
- D.-Y. Kim, J. S. Sung, M. Kim and G. Ghodake, Mater. Lett., 2015, 155, 62–64 CrossRef CAS.
- Y. G. Cai, X. Q. Piao, W. Gao, Z. J. Zhang, E. Nie and Z. Sun, RSC Adv., 2017, 7, 34041–34048 RSC.
- A. Zhang, Y. Tian, Y. Xiao, Y. Sun and F. Li, Mater. Sci. Eng. B: Solid-State Mater., 2015, 197, 5–9 CrossRef CAS.
- T. Takeshima, L. Sun, Y. Wang, Y. Yamada, N. Nishi, T. Yonezawa and B. Fugetsu, Polym. J., 2014, 46, 36–41 CrossRef CAS.
- F. M. Morsy, Int. J. Hydrogen Energy, 2014, 39, 11902–11912 CrossRef CAS.
- S. Yousef, M. Tatariants, M. Tichonovas and V. Makarevicius, Waste Manage., 2019, 98, 126–134 CrossRef CAS PubMed.
- S. Wack, P. L. Popa, N. Adjeroud, J. Guillot, B. R. Pistillo and R. Leturcq, J. Phys. Chem. C, 2019, 123, 27196–27206 CrossRef CAS.
- H. Shiigi, R. Morita, Y. Muranaka, S. Tokonami, Y. Yamamoto, H. Nakao and T. Nagaoka, J. Electrochem. Soc., 2012, 159, D442–D446 CrossRef CAS.
- P. Bianchi, G. Petit and J.-C. M. Monbaliu, React. Chem. Eng., 2020, 5, 1224–1236 RSC.
- M. S. Marques, K. M. Zepon, J. M. Heckler, F. D. Morisso, M. M. D. Paula and L. A. Kanis, Int. J. Biol. Macromol., 2019, 124, 838–845 CrossRef CAS PubMed.
- Y. Wen, W. Huang, B. Wang, J. Fan, Z. Gao and L. Yin, Mater. Sci. Eng., B, 2012, 177, 619–624 CrossRef CAS.
- A. B. S. Sastry, R. B. K. Aamanchi, C. S. R. L. Prasad and B. S. Murty, Environ. Chem. Lett., 2013, 11, 183–187 CrossRef CAS.
- R. A. Crane and D. J. Sapsford, Miner. Eng., 2023, 196, 108048 CrossRef CAS.
- P. Nazari, B. Abdollahi-Nejand, M. Eskandari and S. Kohnehpoushi, Mater. Res. Express, 2018, 5, 045046 CrossRef.
- H. Y. Jeon, C.-H. Ryu, S. Han, D. H. Lee, J. Byun and Y.-I. Lee, J. Am. Ceram. Soc., 2023, 106, 7278–7287 CrossRef.
- S. Koch, S. Spaeth, A. Shmeliov, V. Nicolosi and K. Mandel, J. Mater. Chem. C, 2015, 3, 12430–12435 RSC.
- A. Gupta and R. Srivastava, Ultrason. Sonochem., 2019, 52, 414–427 CrossRef CAS PubMed.
- Y.-M. Chen and H.-W. Jia, Mater. Lett., 2014, 132, 389–392 CrossRef CAS.
- C. M. M. Palos, A. E. Marino-Gamez, G.-E. Acosta-Gonzalez, M. B. Hernandez, S. Garcia-Villarreal, L. F. Franco, L. Garcia-Ortiz and J. A. Aguilar-Martinez, Phys. B, 2023, 656, 414776 CrossRef.
- Q. Zhuo, J. Gao, M. Peng, L. Bai, J. Deng, Y. Xia, Y. Ma, J. Zhong and X. Sun, Carbon, 2013, 52, 559–564 CrossRef CAS.
- P. N. Khanam and A. Hasan, Int. J. Biol. Macromol., 2019, 126, 151–158 CrossRef PubMed.
- Q. Y. Jiang, F. Wang, R. Li, B. N. Li, N. Wei, N. F. Gao, H. T. Xu, S. M. Zhao, Y. Huang, B. S. Wang, W. S. Zhang, X. K. Wu, S. L. Zhang, Y. L. Zhao, E. Z. Shi and R. F. Zhang, Nano Lett., 2023, 23, 523–532 CrossRef CAS PubMed.
- S. Y. Tang, R. Qiao, S. Yan, D. Yuan, Q. Zhao, G. Yun, T. P. Davis and W. Li, Small, 2018, 14, e1800118 CrossRef PubMed.
- S. Garg, G. Heuck, S. Ip and E. Ramsay, J. Drug Targeting, 2016, 24, 821–835 CrossRef CAS PubMed.
- X. Kang, C. Luo, Q. Wei, C. Xiong, Q. Chen, Y. Chen and Q. Ouyang, Microfluid. Nanofluid., 2013, 15, 337–345 CrossRef CAS.
- M. Rhee, P. M. Valencia, M. I. Rodriguez, R. Langer, O. C. Farokhzad and R. Karnik, Adv. Mater., 2011, 23, H79–H83 CrossRef CAS PubMed.
- T. Baby, Y. Liu, A. P. J. Middelberg and C. X. Zhao, Chem. Eng. Sci., 2017, 169, 128–139 CrossRef CAS.
- F. Heshmatnezhad and A. R. S. Nazar, J. Flow Chem., 2020, 10, 533–543 CrossRef CAS.
- M. A. Tomeh, M. H. Mansor, R. Hadianamrei, W. Sun and X. Zhao, Int. J. Pharm., 2022, 620, 121762 CrossRef CAS PubMed.
- M. E. R. O'Brien, N. Wigler, M. Inbar, R. Rosso, E. Grischke, A. Santoro, R. Catane, D. G. Kieback, P. Tomczak, S. P. Ackland, F. Orlandi, L. Mellars, L. Alland, C. Tendler and C. B. C. S. Grp, Ann. Oncol., 2004, 15, 440–449 CrossRef PubMed.
- D.-H. Kim, S. Lim, J. Shim, J. E. Song, J. S. Chang, K. S. Jin and E. C. Cho, ACS Appl. Mater. Interfaces, 2015, 7, 20438–20446 CrossRef CAS PubMed.
- C. Hu, A. Qian, Q. Wang, F. Xu, Y. He, J. Xu, Y. Xia and Q. Xia, Eur. J. Pharm. Biopharm., 2016, 109, 206–213 CrossRef CAS PubMed.
- Y. Dong, W. K. Ng, S. Shen, S. Kim and R. B. H. Tan, Colloids Surf., B, 2012, 94, 68–72 CrossRef CAS PubMed.
- M. Maeki, Y. Okada, S. Uno, K. Sugiura, Y. Suzuki, K. Okuda, Y. Sato, M. Ando, H. Yamazaki, M. Takeuchi, A. Ishida, H. Tani, H. Harashima and M. Tokeshi, Appl. Mater. Today, 2023, 31, 101754 CrossRef.
- H. Bolze, J. Riewe, H. Bunjes, A. Dietzel and T. P. Burg, Chem. Eng. Technol., 2021, 44, 1641–1650 CrossRef CAS.
- Y. Matsuura-Sawada, M. Maeki, T. Nishioka, A. Niwa, J. Yamauchi, M. Mizoguchi, K. Wada and M. Tokeshi, ACS Appl. Nano Mater., 2022, 5, 7867–7876 CrossRef CAS.
- R. H. Fang, K. N. H. Chen, S. Aryal, C.-M. J. Hu, K. Zhang and L. Zhang, Langmuir, 2012, 28, 13824–13829 CrossRef CAS PubMed.
- Y. T. Kim, B. L. Chung, M. M. Ma, W. J. M. Mulder, Z. A. Fayad, O. C. Farokhzad and R. Langer, Nano Lett., 2012, 12, 3587–3591 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |