Open Access Article
Open Access ArticleDevelopment of fluorescent carbon nanoparticles from Madhuca longifolia flower for the sensitive and selective detection of Cr6+: a collective experimental–computational approach†
Tuhin
Mandal
 a,
Ashish Kumar
Ghosh
bd,
Shiv Rag
Mishra
ad,
Sarvesh Kumar
Pandey
a,
Ashish Kumar
Ghosh
bd,
Shiv Rag
Mishra
ad,
Sarvesh Kumar
Pandey
 *c and
Vikram
Singh
*c and
Vikram
Singh
 *ad
*ad
aEnvironment Emission and CRM Section, CSIR-Central Institute of Mining and Fuel Research, Dhanbad, Jharkhand 828108, India. E-mail: vikku.010@gmail.com; vikramsingh@cimfr.nic.in
bCoal Quality and Value Addition Division, CSIR-Central Institute of Mining and Fuel Research, Dhanbad, Jharkhand 828108, India
cDepartment of Chemistry, School of Basic Sciences, Manipal University Jaipur, Jaipur – 303007, Rajasthan, India. E-mail: sarvesh.pandey@jaipur.manipal.edu
dAcademy of Scientific and Innovative Research (AcSIR), Ghaziabad, 201002, India
First published on 18th July 2023
Abstract
Herein, blue-emitting carbon nanoparticles (CNPs) were synthesized using the Madhuca longifolia flower for the highly selective and sensitive detection of Cr6+ ions in aqueous media using a simple, green, and cost-effective approach, and computational experiments were also performed. The prepared CNPs were well-dispersed in water with an average diameter of 12 nm and functionalized with carbonyl, hydroxyl and carboxylic acid groups. The decrease in the fluorescence intensity of the CNPs with an increase in the content of Cr6+ provided an important signal for the sensitive and selective detection of Cr6+ in aqueous media. The limit of detection for Cr6+ in an aqueous medium was found to be 103 ppb, which is more sensitive in comparison with the previously reported study. Furthermore, the validation of the proposed higher sensing feature and more selective nature of the CNPs towards Cr6+ was also explained using an in silico approach. The results from the theoretical calculations based on the DFT approach demonstrated that the binding energy (BE) of the CNPs with three transition metal (TM) cations (Cr6+, Fe3+, and Hg2+) follows the order of Cr6+ > Fe3+ > Hg2+, where the Cr6+ TM cation associated with the CNPs possesses the highest valence state, showing the highest sensing feature and highest selectivity among the investigated ions, as expected. The metal ions associated with the CNPs having a higher charge and a smaller radius indicated a higher BE and larger degree of deformation of the CNPs. Moreover, to achieve new insights into the structural, stability/energetics, and electronic features, some useful tools, such as NCI-plot, HOMO–LUMO gap, MESP, and QTAIM analysis were employed, which facilitated noteworthy outcomes.
1. Introduction
Heavy metals are required for normal metabolism at specific concentrations; however, the presence of an excess amount of heavy metal ions causes severe disorders in humans. Chromium is one of the significant heavy metals that is discharged into the environment through industrial and anthropogenic activities.1 Among the prevalent and stable forms of chromium, hexavalent chromium (Cr6+) such as dichromate (Cr2O72−) and chromate (CrO42−) is considered to be the most toxic due to its high mobility and carcinogenic and mutagenic effects in both living organisms and the environment.2,3 The human nervous and cardiovascular systems are susceptible to the accumulation of Cr6+ and it is very difficult to detoxify its toxic effect. In this case, Cr6+-contaminated drinking water and air even at low concentrations cause major health issues, such as lung cancer, edema, pulmonary congestion, gastrointestinal ulceration, dermal irritation and liver damage.4,5 Therefore, the detection of chromium [Cr6+] has received significant attention given that it is one of the most toxic heavy metal ions in recent years, the use of which has been significantly curtailed globally. Its concentration in water used for drinking is rigorously regulated and must not exceed the micromolar range. The US Environmental Protection Agency placed it at a danger threshold of 0.1 mg L−1 (100 ppb) for total Cr and beyond this limit, it causes carcinogenicity and genotoxicity to humans.6–8Therefore, the quantitative and qualitative detection of Cr6+ is essential to protect human health. In this case, various fluorescent sensors have been developed for the convenient and rapid detection of Cr6+ heavy metal in water.9–14 Among the various fluorescent materials, carbon nanoparticles (CNPs) have attracted significant attention owing to their green synthesis, biocompatible nature, water dispersibility, tunable fluorescence, ease of surface functionalization, etc.15–18 Thus, CNPs are considered alternative fluorescent materials to organic dyes and semiconductor quantum dots (QDs). The properties of CNPs have been extensively used in various applications such as biomedicine,19,20 optoelectronics,21,22 fluorescence sensing,23,24 photocatalysts,25,26 water purification,27,28 and visible ink.29,30 Carbon-rich precursors are used for the synthesis of fluorescent CNPs on a feasible scale using the top-down and bottom-up approaches.31
In recent years, vegetables,32 fruits,33 agricultural waste,34 biomass,35 and hydrocarbons36 have been widely used for the development of non-toxic fluorescent CNPs. For example, blue fluorescent, water-soluble, and nitrogen-doped CNPs were prepared via the calcination of diethylenetriaminepentaacetic acid37 and hydrothermal treatment of methyl glycine diacetic acid trisodium salt38 for the determination of Cr6+ ions with a calculated detection limit of 0.15 mM and 2.1 μM, respectively, through turn-off of fluorescence due to the inner filter effect process. X. Guo and co-workers developed a fluorescent aerogel with a 3D net-like structure based on carboxymethylated cellulose nanofibrils and carbon dots as a highly effective adsorbent and sensitive optical sensor of Cr6+ and its limit of detection was found to be 17.6 mg L−1 using the calibration plot. A fluorescent microcline-based nanoprobe was developed by incorporating nitrogenous carbon dots in the natural microcline nanostructure for the sensitive detection of the Cr6+ with a limit of detection of 4.0 μM and it was found that photoinduced electron transfer is responsible for the fluorescence quenching.39 J. Goswami et al. prepared banana stem-derived blue emissive carbon dots using phosphoric acid treatment as a novel sensor for the detection of Cr6+via a photoinduced electron transfer quenching mechanism.40
According to the literature and the above-mentioned examples, it can be observed that green carbon-rich precursors and chemical-free synthetic approaches for the preparation of fluorescent CNPs for the detection of Cr6+ have not been explored to date. Furthermore, the detection limit for Cr6+ using fluorescent CNPs in the above-mentioned reports appears to be slightly high. Therefore, the development of environmentally benign approaches for the sensitive detection of Cr6+ in aqueous medium using natural carbon-rich precursors is necessary. Therefore, in the present study, the Madhuca longifolia flower (a highly rich source of carbohydrates and vitamin C) was chosen,41,42 which mainly contains carbon, hydrogen, and oxygen. The presence of a high content of carbon and oxygen-containing groups in the Madhuca longifolia flower provides high potential for the synthesis of blue fluorescent CNPs in aqueous medium. Herein, Madhuca longifolia flowers were used as a sustainable source of carbon for the preparation of blue-emitting CNPs. Subsequently, the CNPs were used as a nanosensor for the detection of Cr6+ in an aqueous medium via a static quenching mechanism. A detailed experimental investigation was carried out to understand the sensing mechanism for the detection of Cr6+, which was further supported employing an in silico approach.
A few sensing features were established by the non-covalent interactions (NCIs) between the carbon network of the CNPs and external stimuli. Furthermore, useful insights into the sensing features [in terms of the supramolecular approach-based binding energy (BE) or interaction energy (IE)], especially the metal–nonmetal bonding interaction between the metal cation and nonmetals of the associated CNP complex were explored by performing an extensive computational experiment including the involved NCIs and van der Waals (vdW) (extremely weak) interaction(s). To complement the experimental results of the selective sensing of some metal cations (Cr6+/Fe3+/Hg2+), the interaction between the metal ions (Cr6+/Fe3+/Hg2+) and fluorescent CNPs and some other interesting and important features were studied theoretically in the framework of the widely used dense generalized density functional theory (DFT) approach. Quantum theory of atoms in molecules (QTAIM), NCI-plot-based reduced density gradient (RDG), electrostatic surface potential (ESP), and frontier molecular orbital (highest occupied orbital–lowest unoccupied molecular orbital) analyses were also performed to inspect some other useful structural, stability, and electronic features (vide infra). Moreover, in the present work, the quantum mechanical calculations were not only consistent with the experimental findings for the selective sensing of three metal ions (Cr6+, Fe3+, and Hg2+) interacting with a small unit of functionalized coronene as the CNP, but also facilitated a deep understanding and visualization of the CNP complexes associated with the probed metals.
2. Experimental
2.1. Materials
Madhuca longifolia flowers were collected from our institute CSIR-CIMFR Digwadih Campus, Dhanbad. Metal ions including cobaltous chloride, lead acetate trihydrate, manganese chloride, ferric chloride, mercury chloride, chromium oxide, aluminium chloride, zinc acetate, cadmium chloride, nickel chloride, silver nitrate, and magnesium acetate were purchased from Merck India. Whatman 42 filter paper was purchased from Sigma Aldrich.2.2. Instrumentation
The size and morphology of the as-synthesized nanomaterials were determined using a Thermo Fisher Scientific Talos F200X G2 high-resolution transmission electron microscope (HRTEM). The morphology and elemental composition mapping were studied using a Zeiss Merlin VP compact field-emission scanning electron microscope (FESEM) and energy dispersive X-ray spectroscopy (EDS), respectively. The determination of various functional groups was done using a Shimadzu Iraffinity-1s Fourier transform infrared spectrophotometer. X-ray photoelectron spectroscopy for the characterization of surface functionality and elemental composition was carried out using a PHI 5000 versa probe III instrument. The count per second of different functional groups as a function of the BE (eV) was measured using a standard monatomic argon ion gun, which was capable of generating 5 eV to 5 keV Ar ion beams. UV-vis electronic absorption and fluorescence emission spectra of the water-dispersed carbon nanomaterials were analyzed using an Agilent Carry 5000 UV-Vis-NIR and Hitachi F-7000 (150 W xenon lamp) fluorescence spectrophotometer, respectively. The zeta potential measurement was carried out using an Anton Paar Litesizer 500 system.2.3. Preparation of carbon nanoparticles (CNPs) from Madhuca longifolia flower
The collected Madhuca longifolia flowers were washed with water to remove dust particles and allowed to dry for 30 min at 100 °C. Then 5.0 g of Madhuca longifolia flowers was mixed with 30 mL of double distilled water and transferred to a 50 mL Teflon-linked hydrothermal pressure vessel and heated at a temperature of 160 °C for 10 h. After completion of the reaction, the autoclaved was left to cool to room temperature and the reaction mixture was filtered through Whatman 42 filter paper. Further, the filtrate solution was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min. Then, the supernatant solution was purified using a 1 kDa dialysis membrane for 24 h. The purified CNPs [6.0 mg mL−1] were kept at 4 °C for further characterization and application.
000 rpm for 5 min. Then, the supernatant solution was purified using a 1 kDa dialysis membrane for 24 h. The purified CNPs [6.0 mg mL−1] were kept at 4 °C for further characterization and application.
In this green, economical and simple approach, no passivating agent or substrate was used. The CNPs were freely dispersed in an aqueous medium (Scheme 1).
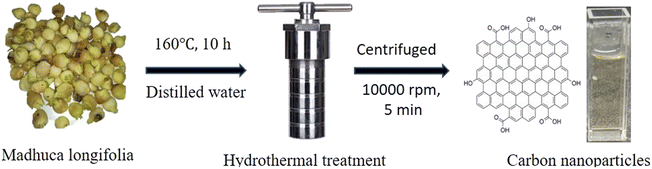 | ||
| Scheme 1 Schematic representation for the one-step synthesis of the fluorescent CNPs via the hydrothermal treatment of Madhuca longifolia. | ||
2.4. Computational methodology
The capability of accurately describing metal–ligand interactions in various fields is demonstrated by the large number of calculations using the generalized DFT (B3LYP) method. The B3LYP functional of the generalized density functional theory was employed to acquire information on the geometrical optimization of the complexes formed by Cr6+, iron (Fe3+) and mercury (Hg2+) ions with the CNPs (i.e., in this work, three types of complexes showing the selective features were examined theoretically), respectively. The initial configuration of all the CNP-based complexes was first passed through the minima (equilibrium) in the potential energy surface (PES), and then their frequencies were analyzed using the same level of theory, which resulted in all positive frequencies and showed the stable configurations of all the optimized complexes. Efforts were made to study the complexes using the dispersion-corrected DFT (DFT-D) approach, but the optimization process did not converge constructively as expected, which can be attributed to the large structure of the complexes constructed from the functionalized coronene-based CNP model associated with the metal cations. Furthermore, to avoid the complexity involved in the DFT-D approach and to make the calculations easier and faster, together with reasonably precise outcomes, the B3LYP/6-31(d) level was employed as an alternative. To obtain the equilibrium structures (minimum energy configurations) of the sensing complexes and their constituting units, the Gaussian 09 package (electronic structure calculations) was used.43 Some computational techniques [for instance, the BE/IE parameter, QTAIM tool, hydrogen bond strength based on the interaction coordinate (HBSBIC) approach, NCI-plot tool, compliance constant (CC), and relaxed force constant (RFC) parameters] to quantify the bond strength (bonding/nonbonding interactions) are frequently used to gain deep insights into the type, nature, and strength of various covalent and weak to moderate to strong NCIs.44–483. Characterization of Madhuca longifolia-derived CNPs
3.1. Morphological characterization
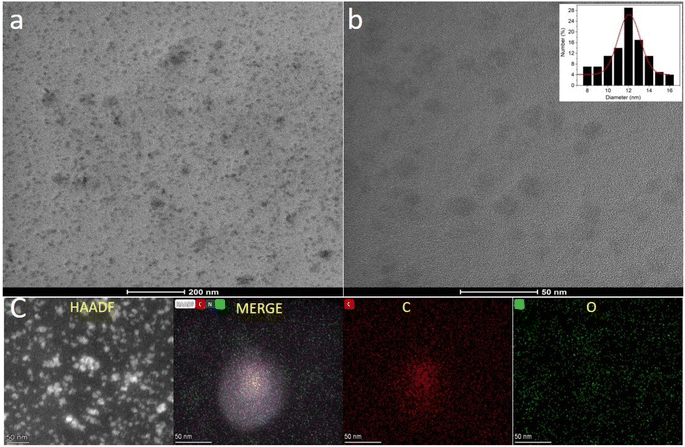
The size and morphology of the CNPs prepared by the hydrothermal treatment of Madhuca longifolia flower were studied via high transmission electron microscopy (TEM). The synthesized CNPs showed a spherical morphology and were uniformly dispersed in the aqueous medium (Fig. 1a and b). The average diameter of the prepared CNPs was determined by the particle size distribution plot (inset, Fig. 1b), which revealed that they have an average core size of 12 nm.Scanning transmission electron microscopy (STEM) and the corresponding energy dispersive X-ray spectroscopy (EDS) images were employed to identify the elemental composition and distribution of the carbon, nitrogen, and oxygen elements in the uniformly dispersed CNPs (Fig. 1c). The obtained result indicates that the CNPs are mainly composed of carbon, nitrogen, and oxygen [Fig. S1a in the ESI†]. The EDS mapping showed that there is a uniform distribution of C, O, and N throughout the CNPs.
X-ray photoelectron spectroscopy (XPS) and FTIR analysis were carried out to understand the nature of the functional groups present on the surface of the as-synthesized CNPs. Three peaks were observed at 284.0 eV, 399.5 eV, and 531.5 eV, which are attributed to C 1s, N 1s and O 1s with the atomic percentages 76.2%, 1.8% and 22.0%, respectively, in the full XPS survey spectrum of CNPs (Fig. 2a).
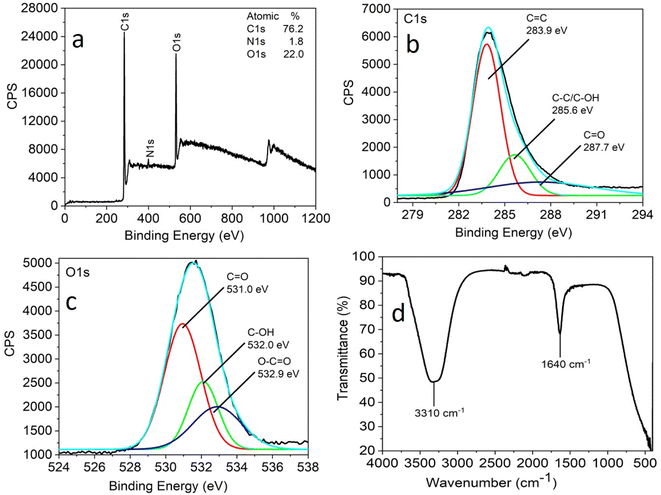 | ||
| Fig. 2 Full scan XPS spectra (a), high-resolution XPS spectra (b) C 1s region and (c) O 1s region and FTIR spectra (d) of CNPs obtained by the hydrothermal treatment of Madhuca longifolia flower. | ||
The full scan (high resolution) spectrum of C 1s and O 1s was obtained to determine the BE values of the various functional groups. The high-resolution scan of the C 1s region shows that carbon is present in three different chemical environments, corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C at 283.9 eV, C–OH/C–C at 285.6 eV, and C
C at 283.9 eV, C–OH/C–C at 285.6 eV, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 287.7 eV (Fig. 2b). The deconvoluted O 1s spectrum shows peaks attributed to C
O at 287.7 eV (Fig. 2b). The deconvoluted O 1s spectrum shows peaks attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 531.0 eV, C–OH at 532 eV and O–C
O at 531.0 eV, C–OH at 532 eV and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O at 532.9 eV (Fig. 2c).
O at 532.9 eV (Fig. 2c).
The FTIR analysis facilitated further insight into the surface functional groups present in the CNPs. This study revealed the presence of functional groups, such as a band at 3310 cm−1 corresponding to the stretching vibrations of C–OH and 1640 cm−1 for the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration (Fig. 2d). The XPS and FTIR analyses suggest that the CNPs were functionalized with carbonyl, hydroxyl, and carboxylic acid groups.
O stretching vibration (Fig. 2d). The XPS and FTIR analyses suggest that the CNPs were functionalized with carbonyl, hydroxyl, and carboxylic acid groups.
The zeta potential (ζ) of CNPs was found to be −17.3 mV at neutral pH, which suggests the presence of acid functionalities on the surface of the synthesized CNPs (Fig. S1b, in the ES I†). The high negative zeta potential value confirms that the dispersion of CNPs in water is very stable. The presence of the aforementioned functional groups on the surface and the zeta potential of the CNPs confirmed their excellent dispersion and stability in water.
3.2. Photophysical study
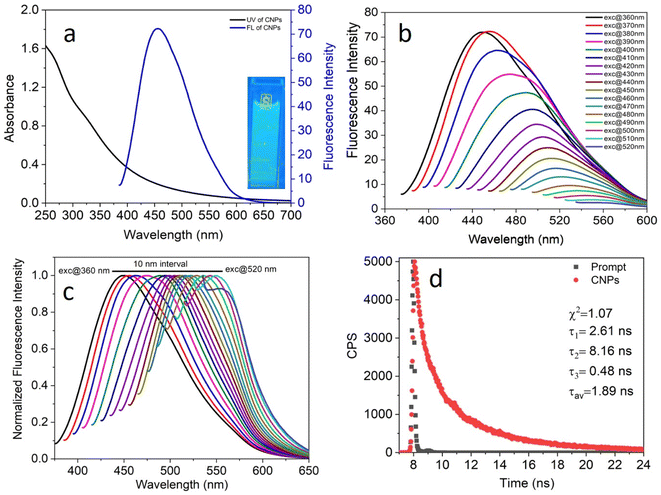
The photophysical properties of the as-synthesized CNPs were studied in an aqueous medium at room temperature. Given that the concentration of the CNPs in terms of molarity could not be calculated accurately, their absorbance was kept in the range of 0.2–0.7 at the excitation wavelength to avoid any inner filter effect. A clear transparent solution of 6.0 mg mL−1 CNPs showed absorption at around 250–550 nm (Fig. 3a, black) with two absorption bands at 271 nm and 330 nm, which can be assigned to the typical absorption of aromatic π systems or the n–π* and π–π* transition of nitrogen or oxygen-based groups and the trapping of excited-state energy by the surface states, respectively. The water-dispersed CNPs exhibited strong blue fluorescence with an emission maximum at 455 nm (Fig. 3a, blue) at 370 nm excitation. Moreover, the CNP solution displayed strong blue light under UV light exposure (Fig. 3a, inset).To explore the excitation-dependent emission property of the water-dispersed CNPs, we scanned the samples at excitation wavelengths in the range of 360 to 520 nm (at 10 nm intervals) and collected their emission response from 350 to 650 nm (Fig. 3b). A prominent red-shift in the emission spectrum with the maximum intensity at 370 nm excitation was observed (Fig. 3b). The observed tunability in the normalized emission spectra (Fig. 3c) may be due to the presence of CNPs with different sizes and carboxylic groups on the surface of the CNPs, and structural inhomogeneity originating from the different surface trap states or surface defects during the hydrothermal synthesis of the CNPs from Madhuca longifolia flower. The presence of different types of aggregates even at a very low concentration may also contribute to the excitation wavelength dependence of the emission spectrum. The tri-exponential function-fitted fluorescence lifetime decay of CNPs is shown in Fig. 3d. The average fluorescence lifetime of CNPs was found to be 1.89 ns at a laser excitation of 405 nm.
4. Experimental results and discussion
4.1. Fluorescence sensing study
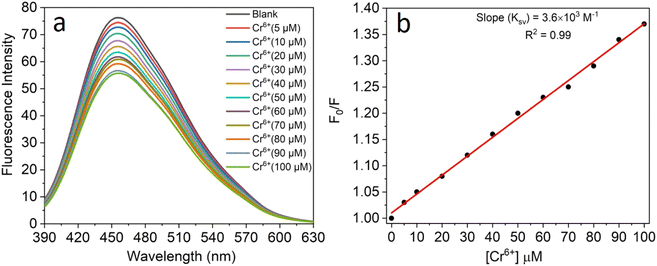
All the experiments were performed at room temperature in double-distilled water for the detection of Cr6+ ions and the concentration of the CNPs was fixed at 2.0 mg mL−1. The fluorescence emission spectra were recorded in the absence and presence of an aqueous solution of Cr6+ (5–100 μM). It was observed that the fluorescence intensity of the CNPs was significantly quenched with an increase in the concentration of Cr6+ ions (Fig. 4a). The formation of a complex between Cr6+ and the CNPs led to quenching of their fluorescence intensity in the aqueous medium. The sensing characteristics or quenching efficiency of the fluorescent CNPs by the Cr6+ ion were best studied using the standard Stern–Volmer equation, as follows:| F0/F = 1 + KSV[Q] | (1) |
To get a complete scenario of the quenching mechanism, the fluorescence lifetime of CNPs with and without Cr6+ was studied because fluorescence lifetime decay is one of the most specific methods to discriminate static and dynamic quenching. In our lifetime experiment, there was almost no change in the average lifetime of the CNPs after the addition of 100 M of Cr6+ (Fig. S2 in the ESI†), which confirms a purely ground-state (static quenching) quenching phenomenon.
4.2 Quenching mechanism
Given that Cr6+ has a high affinity to form a complex with carboxylate groups,49 the formation of a complex between the Cr6+ ion and acidic group of the CNPs may lead to a decrease in the fluorescence intensity of CNPs through charge transfer (CT) quenching. A schematic diagram for the plausible quenching mechanism is depicted in Scheme 2.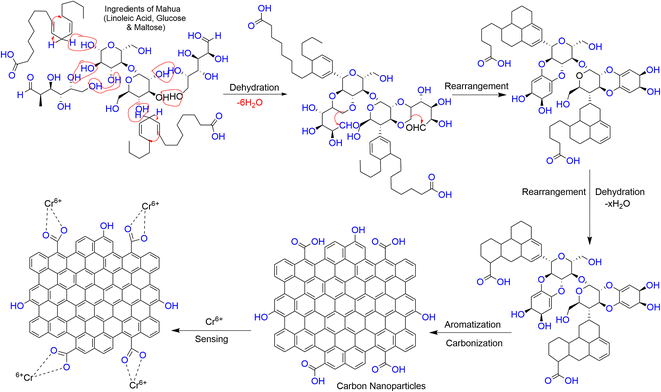 | ||
| Scheme 2 Plausible representation of the formation of CNPs via the hydrothermal treatment of the Madhuca longifolia flower and their formation of a complex with the Cr6+ ion. | ||
4.3 Selective and sensitive detection of the Cr(VI) ion
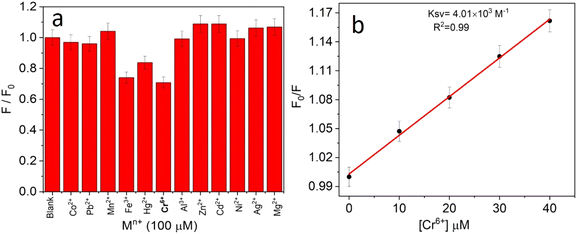
The selectivity towards the detection of the Cr6+ ion was verified by carrying out experiments in the presence of other heavy metal ions (number of samples, n = 5). In this study, a variety of environmentally relevant heavy metal ions was chosen in aqueous medium including Co2+, Pb2+, Mn2+, Fe3+, Hg2+, Cr6+, Al3+, Zn2+, Cd2+, Ni2+, Ag2+, and Mg2+. The fluorescence intensity ratio plot for the CNPs (2.0 mg mL−1) in the presence of 100 μM metal ions in double-distilled water was recorded. Fig. 5a shows the selectivity for Cr6+, where about a 29% decrease in the fluorescence intensity was observed in the presence of Cr6+ (100 μM). However, a slight decrease and increase in the fluorescence intensity ratio were also observed (Fig. 5a) in the presence of other metal ions, mainly Fe3+ (25% reduction), Hg2+, Mn2+, Zn2+, and Co2+, respectively. The p-value is less than 0.05, suggesting that the result of this test is statistically significant. The enhancement in the fluorescence intensity after the addition of Mn2+, Zn2+ and Co2+ may be due to aggregation-induced emission enhancement (AIEE) or coordination-induced aggregation (CIA). When aggregation occurs, the intramolecular motion is restricted, which blocks the nonradiative path and activates radiative decay.There are numerous studies in the literature where a slight variation in the fluorescence intensity was found at the micromolar level, and calculations were made based on the observed variation. Employing the change in the fluorescence intensity in the micromolar range, the limit of detection (LOD) from the standard deviation (S.D.) and the slope (m) of the calibration plot (Fig. 5b) were calculated using eqn (2) (number of samples, n = 5).
The linear plot of the fluorescence intensity ratio versus Cr6+ concentration (Fig. 5b) is useful to calculate the LOD knowing the standard deviation (S.D.) and the slope of the plot.
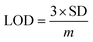 | (2) |
It was found that the Madhuca longifolia-derived fluorescent CNPs have a reasonable LOD (103 ppb), which is almost comparable with the maximum allowed limit of 0.1 mg L−1 (100 ppb) by the U.S. Environmental Protection Agency (EPA). Furthermore, the LOD for Cr6+ sensing in the present study is much better than that in most of the previously reported works in recent years (Table 1).
| S. no. | Nanosensor | LOD (ppb) for Cr6+ | References |
|---|---|---|---|
| 1 | Carbon dots | 109 | Li et al.50 |
| 2 | Carbon dots | 560 | Wang et al.51 |
| 3 | Carbon dots | 120 | Wang et al.52 |
| 4 | N-doped carbon dots | 216.30 | Wang et al.53 |
| 5 | Carbon dots | 520 | Yang et al.54 |
| 6 | N-doped carbon dots | 207 | Bardhan et al.55 |
| 7 | Carbon dots | 181 | Sakaew et al.56 |
| 8 | Carbon dots | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
Yaun et al.57 |
| 9 | Titanate and cellulose nanofibers carbon dots | 8500 | Luo et al.58 |
| 10 | Lignin-based hydrogel with cellulose nanofibers and carbon dots | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
Yaun et al.59 |
| 11 | N-doped carbon dots | 166 | Bogireddy et al.60 |
| 12 | Terylene carbon dots | 150 | Hu et al.61 |
| 13 | Carbon dots | 1700 | Anil et al.62 |
| 14 | N-doped carbon dots | 709.3 | Chen et al.63 |
| 15 | Carbon dot-based fluorescent hydrogel | 147 | Feng et al.64 |
| 16 | Carbon dots | 124 | Goswami et al.40 |
| 17 | N-doped carbon quantum dots | 109 | Wang et al.38 |
| 18 | Carbon nanoparticles (CNPs) | 103 | Our current work |
5. Computational results and discussion
Given that all-inclusive atomistic features (including the atomic/molecular environment) play a dominant role in altering the inter- or intramolecular (metal–nonmetal and nonmetal–nonmetal covalent/ionic bonding, H-bonding, vdW, etc.) interactions, the sensing feature of the probed species was inspected in the framework of the DFT approach. Herein, among the twelve experimentally probed metal cations (Co2+, Pb2+, Mn2+, Fe3+, Hg2+, Cr6+, Al3+, Zn2+, Cd2+, Ni2+, Ag2+, and Mg2+), the three most selective and high sensitivity feature environmentally relevant heavy metal cations (Cr6+, Fe3+, and Hg2+) were chosen for the quantum chemical calculations. A thorough and organized structural, stability/energetic (i.e., diagnostic of the sensitivity feature using the BE parameter), and electronic feature analysis was performed employing the widely used DFT-based quantum mechanical calculation approach. Interestingly, and importantly, a comprehensive theory-based study on the above-mentioned interactions can be viewed in a few previous reports, portraying the structural, energetic/stability, and electronic features of the vastly used polyacrylamide (PAM) and hydroxyapatite (HAP)-based biocomposite models, where the experimental outcomes were supported by the employed in silico approach.65,665.1 Characterization of structural, stability/energetic, and electronic features
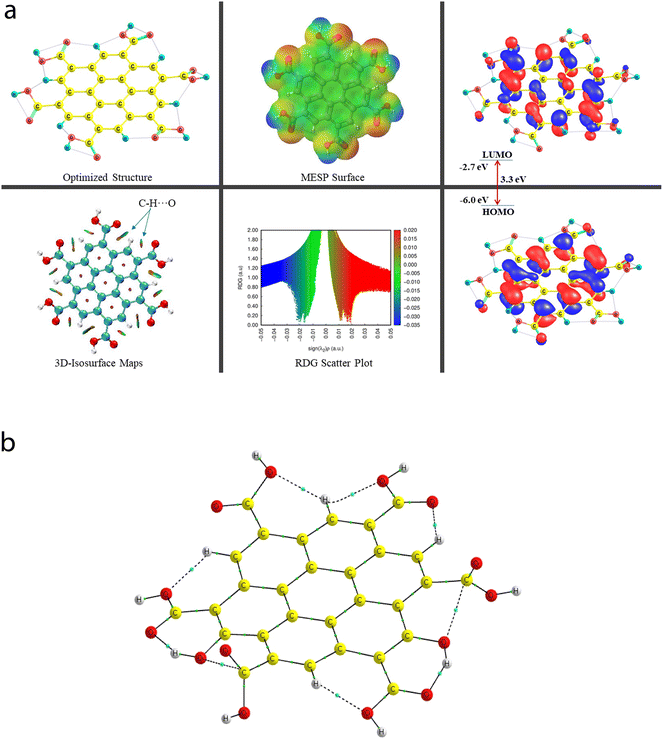
A comprehensive study on the characterization and validation of the structural (optimized parameters such as bond length and bond angle), (HOMO, LUMO, and HOMO–LUMO gap), sensing and selectivity features (in terms of the BEs and visualization of metal–nonmetal and other probable (nonbonding) interactions using the NCI-RDG plot via 3D-isosurface and 2D-scatter plots), and electronic properties (HOMO–LUMO 3D-isosurface maps, MESP, and QTAIM topological picture and parameters) of a free CNP moiety and its associated complexes with the metal ions (Cr, Fe, and Hg cations) is discussed in the following section.5.2 Free functionalized CNP constituent
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond lengths of all six COOH functional groups range from 1.209 Å to 1.239 Å. Importantly, two O–H bonds and four O–H bonds of the COOH group have the bond lengths of 0.976 Å and 0.977 Å, respectively. Because of the presence of multiple COOH groups, many intramolecular H-bonding NCIs were probed using the QTAIM and NCI-plot tools, which show that these (a total of two moderate O–H⋯O and five weak C–H⋯O) interactions take place, and also illustrate that two non-conventional bond paths (NBPs) (C⋯O) stabilize the CNP, as shown in Fig. 6b. The O–H⋯O H-bond distances were calculated to be 1.599 Å and 1.597 Å, respectively. The corresponding bond angles were found to be 147.3° and 147.6°, respectively. The distance between two atoms for one NBP (C29⋯O55) is 2.597 Å, whereas it was computed to be 2.573 Å for the second NBP (C45⋯O53). In seven types of NCIs (excluding both C⋯O NBPs), the O atom acts as a proton acceptor, whereas two O atoms and five C atoms act as proton donors in all the probed NCIs.
O bond lengths of all six COOH functional groups range from 1.209 Å to 1.239 Å. Importantly, two O–H bonds and four O–H bonds of the COOH group have the bond lengths of 0.976 Å and 0.977 Å, respectively. Because of the presence of multiple COOH groups, many intramolecular H-bonding NCIs were probed using the QTAIM and NCI-plot tools, which show that these (a total of two moderate O–H⋯O and five weak C–H⋯O) interactions take place, and also illustrate that two non-conventional bond paths (NBPs) (C⋯O) stabilize the CNP, as shown in Fig. 6b. The O–H⋯O H-bond distances were calculated to be 1.599 Å and 1.597 Å, respectively. The corresponding bond angles were found to be 147.3° and 147.6°, respectively. The distance between two atoms for one NBP (C29⋯O55) is 2.597 Å, whereas it was computed to be 2.573 Å for the second NBP (C45⋯O53). In seven types of NCIs (excluding both C⋯O NBPs), the O atom acts as a proton acceptor, whereas two O atoms and five C atoms act as proton donors in all the probed NCIs.
The energy difference between the two frontier molecular orbitals (FMOs), i.e., highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO), is denoted as the HOMO–LUMO energy gap (Egap). The strength, stability, and chemical behavior of any chemical/molecular species are described by the Egap value. For example, a system having a small Egap value generally shows higher reactivity (i.e., less stability), and thus shows good optical features (i.e., easy photochemical excitation possible) because of the lower energy gap (bandgap) between the valence and conduction bands. Moreover, the electron-accepting and electron-donating capabilities of a particular species are linked to its EHOMO and ELUMO values, respectively. The EHOMO and ELUMO values for the free functionalized CNP species were calculated to be −6.0 eV and −2.7 eV, respectively, and the associated Egap value is 3.3 eV, which are presented in Table 2. Interestingly, Fig. 6a presents the HOMO–LUMO 3D isosurface maps of the free functionalized CNP constituent, where the HOMOs (in the plane) are mainly positioned over almost all the six-membered rings of the free functionalized CNP moiety and the LUMOs follow a similar pattern, but the orbital phases are out-of-plane. This indicates the occurrence of a π to π* transition in the free functionalized CNP framework.
| Species | BE (kcal mol−1) | BSSE complexation energy (kcal mol−1) | HOMO (eV) | LUMO (eV) | E gap (eV) |
|---|---|---|---|---|---|
| Free functionalized CNP | — | — | −6.0 | −2.7 | 3.3 |
| Cr6+@CNPc | −4395.1 | −4759.8 | −23.0 | −21.5 | 1.5 |
| Cr6+@CNPt | −4476.7 | −4688.9 | −23.2 | −22.0 | 1.2 |
| Fe3+@CNPc | −696.5 | −753.1 | −15.1 | −13.8 | 1.3 |
| Fe3+@CNPt | −794.4 | −785.6 | −15.0 | −13.8 | 1.2 |
| Hg2+@CNPt | −183.9 | −226.2 | −11.6 | −11.2 | 0.4 |
The molecular size, shape, and charge distribution are concurrently described by the molecular electrostatic potential surface (MESP) map, which is widely used to understand the NCI(s), ways for the recognition of one molecule by another, relative polarity, and electrophilic and nucleophilic attack, reactive nature, etc. The MESP surface is displayed in Fig. 6a, where the reactive sites such as electron-poor region (high probability of nucleophilic attack) are shown by the absolute positive charge distribution indicated in a blue colour scheme. The neutral regions are exemplified by a green colour scheme, while the strong negative region (shown in the red colour scheme) can be viewed by the absolute negative charge distribution, indicating the electrophilic attack. The positive regions (favorable for nucleophilic attack) are positioned over the H-atoms of the COOH groups, and the negative regions (preferable for electrophilic attack) are located over the O atoms of the COOH groups of the free functionalized CNP constituent.
For instance, the mixed blue-green (light bluish) colour peak (values ranging from −0.02 au to −0.025 au in the 2D scatter plot) shows the H-bonding interactions, which can also be confirmed by the light-bluish (mixed blue-green) disc-shaped 3D-isosurface maps shown in the seven NCIs (two O–H⋯O and five C–H⋯O H-bonding interactions) (see Fig. 6a). These attractive interactions give a vibrant sign of the existence of intramolecular HBs, which indeed play a prime role in stabilizing the functionalized CNP itself. The extremely weak vdW interactions are validated by the 2D scatter plot (green colour peaks lying between −0.015 au and −0.02 au) and the 3D isosurface maps (green colour lentils between some C and O atoms). The low-gradient broad peaks exposed in the red colour scheme and located at the positive side (values ranging from +0.01 au to +0.02 au in the 2D scatter plot) show the existence of a steric effect as an indication of repulsive interaction, which was also confirmed by the 3D isosurface maps. This effect represents the electron density depletion, which is due to the presence of electrostatic repulsion.
To confirm that the NCIs play an important role in the sensing features, the QTAIM tool was employed for all the sensing model complexes together with the free COOH-functionalized CNP constituents. Several parameters such as ρ (electron density), ∇2(ρ) (Laplacian of electron density), V (local potential energy density), and delocalization index (DI) were used to quantify the strength of the NCIs and the NBPs. The bond path lengths (BPLs) acquired from the QTAIM tool appeared to be consistent with the bond length (BL) obtained from the optimized parameters using the Gaussian package, which can be verified in Table 3.
| BP | BL (Å) | BPL (Å) | ρ (au) | ∇2(ρ) (au) | V (au) | DI (A, B) |
|---|---|---|---|---|---|---|
| Noncovalent interaction (NCI) (intramolecular) | ||||||
| O50⋯H54 | 1.597 | 1.622 | 0.0606 | +0.1930 | −0.0565 | 0.131 |
| O34⋯H56 | 1.599 | 1.624 | 0.0604 | +0.1922 | −0.0563 | 0.131 |
| O35⋯H17 | 2.057 | 2.087 | 0.0230 | +0.0878 | −0.0190 | 0.064 |
| O51⋯H18 | 2.064 | 2.088 | 0.0227 | +0.0869 | −0.0188 | 0.066 |
| O42⋯H26 | 2.077 | 2.099 | 0.0233 | +0.0810 | −0.0191 | 0.072 |
| O39⋯H22 | 2.132 | 2.158 | 0.0201 | +0.0743 | −0.0163 | 0.058 |
| O43⋯H22 | 2.234 | 2.411 | 0.0175 | +0.0811 | −0.0140 | 0.04 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Nonconventional bond path (NBP) (intramolecular) | ||||||
| C45⋯O53 | 2.573 | 2.583 | 0.0178 | +0.0687 | −0.0139 | 0.044 |
| C29⋯O55 | 2.597 | 2.609 | 0.0172 | +0.0664 | −0.0133 | 0.041 |
A total of nine [seven NCIs (two O–H⋯O and five C–H⋯O HBs) and two NBPs (C⋯O)] can be visualized in Fig. 6b, which are represented by the bond paths (BPs) shown in dotted/solid lines (black color), where small green color spheres are displayed between two interacting atoms (O and H atoms for the HBs and C and O atoms for the NBPs). A clear QTAIM molecular graph (with proper atomic labeling and numbering) of the free functionalized CNP constituent can be seen in Fig. S3 in the ESI.† As the BLs of the H-bonding interactions increase (ranging from 1.597 Å to 2.234 Å), the corresponding ρ values decrease (range of 0.0175 au to 0.0606 au) as expected. The strongest H-bonding interaction (ρ: 0.0606 au, ∇2(ρ): +0.1930 au, V: −0.0565 au, and DI: 0.131) was detected as the O50⋯H54 bond, whereas the weakest NCI is the O43⋯H22 HB (ρ: 0.175 au, V: −0.0140 au, and DI: 0.04) having the ∇2(ρ) value +0.0811 au, which is greater than that of the second weakest O39–H22 HB. The QTAIM parameters (ρ, ∇2(ρ), V, DI) for two NBPs, C45⋯O53 and C29⋯O55, were computed to be (0.0178, +0.0687, −0.139, 0.044) and (0.172, +0.0664, −0.0133, 0.041) (in au), respectively, which also stabilize the CNP constituent model. These strength estimating parameters correspond to the analyzed BLs and BPLs, as expected.
5.3 Cr6+@CNP, Fe3+@CNP, and Hg2+@CNP complexes
| BP | BL (Å) | BPL (Å) | ρ (au) | ∇2(ρ) (au) | V (au) | DI (A, B) |
|---|---|---|---|---|---|---|
| MNI (intermolecular) | ||||||
| Cr53–C4 | 2.215 | 2.253 | 0.0572 | +0.2194 | −0.0739 | 0.374 |
| Cr53–C9 | 2.298 | 2.337 | 0.0483 | +0.1942 | −0.0570 | 0.311 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| NCI (intramolecular) | ||||||
| O56–H57⋯O31 | 1.467 | 1.490 | 0.0777 | +0.2019 | −0.0766 | 0.159 |
| C23–H26⋯O43 | 2.077 | 2.101 | 0.0213 | +0.0770 | −0.0171 | 0.061 |
| C12–H18⋯O30 | 2.161 | 2.332 | 0.0215 | +0.0966 | −0.0172 | 0.051 |
| C19–H22⋯O42 | 2.195 | 2.402 | 0.0204 | +0.0907 | −0.0162 | 0.049 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| NBP (intramolecular) | ||||||
| C45⋯O54 | 2.689 | 2.704 | 0.0154 | +0.0573 | −0.0109 | 0.042 |
| C45⋯O54 | 2.872 | 2.704 | 0.0154 | +0.0573 | −0.0109 | 0.042 |
| BP | BL (Å) | BPL (Å) | ρ (au) | ∇2(ρ) (au) | V (au) | DI (A, B) |
|---|---|---|---|---|---|---|
| MNI | ||||||
| Cr51–O37 | 1.979 | 2.020 | 0.0768 | +0.4741 | −0.1244 | 0.546 |
| Cr51–O41 | 1.988 | 2.030 | 0.0775 | +0.4408 | −0.1225 | 0.513 |
| Cr51–C18 | 2.268 | 2.400 | 0.0528 | +0.2113 | −0.0643 | 0.318 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| NCI | ||||||
| O48–H57⋯O56 | 1.652 | 1.677 | 0.0515 | +0.1676 | −0.0448 | 0.106 |
| C12–H53⋯O49 | 2.090 | 2.116 | 0.0207 | +0.0789 | −0.0168 | 0.058 |
| C22–H52⋯O40 | 2.221 | 2.259 | 0.0156 | +0.0587 | −0.0116 | 0.045 |
| C6–H17⋯O33 | 2.279 | 2.350 | 0.0149 | +0.0596 | −0.0110 | 0.037 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| NBP | ||||||
| C43⋯O56 | 2.668 | 2.687 | 0.0156 | +0.0587 | −0.0114 | 0.040 |
| O32⋯O54 | 2.743 | 2.853 | 0.0141 | +0.0586 | −0.0114 | 0.055 |
| O28⋯O54 | 1.451 | 2.746454 | 0.275065 | −0.006080 | −0.417653 | 1.135357 |
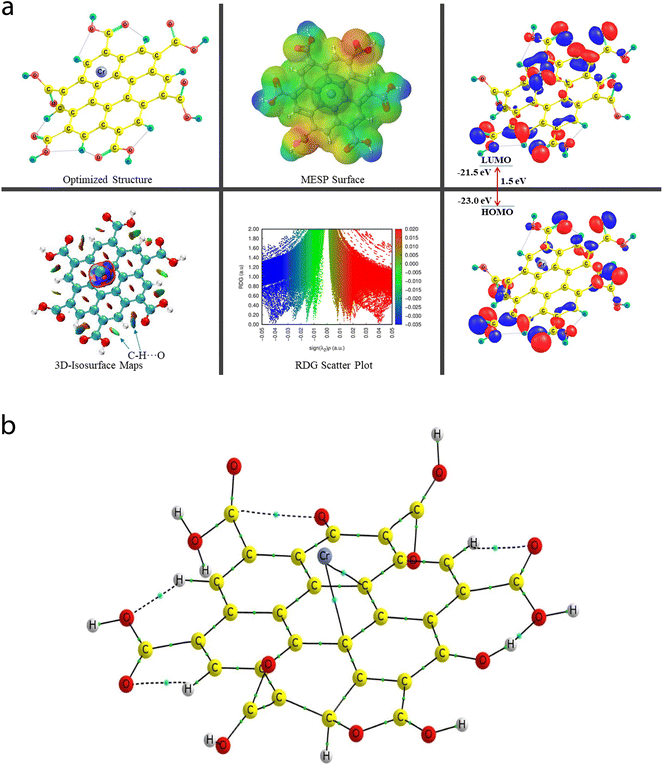
The QTAIM analyses (vide infra) revealed that the Cr metal appears to associate with two C atoms (Cr53–C4 and Cr52–C9) of the central benzene ring of the CNP in the case of the Cr metal ion located at the top of the center of the CNP (Cr6+@CNPc complex) (see Fig. 8b) and the bond distances were detected to be 2.215 Å and 2.298 Å, respectively. Structural changes in the CNP can be seen in the Cr6+@CNPc complex, which is due to incorporation of the Cr6+ ion possessing the highest oxidation state with a smaller size and higher positive charge, thus altering the structural, sensing and selectivity feature (enhanced) and electronic features. A increase in the C–H bond distances in the range of 1.087 Å to 1.09 Å was observed when the Cr metal cation is associated with the CNP (in free functionalized CNP: C–H bond range from 1.077 Å to 1.080 Å). However, interestingly and importantly, another configuration of the Cr metal cation with the CNP (Cr6+@CNPt complex) provides more favorable outcomes because its larger BE than the Cr6+@CNPc complex (vide infra). A total of three MNIs (two Cr–O and one Cr–H) can be discerned in Fig. 7b. The bond distances of the two MNIs, Cr–O were analysed to be 1.979 Å (Cr51–O37) and 1.988 Å (Cr51–O41) and the bond length of the Cr–H bond was computed to be 2.029 Å. It is noteworthy to mention that the higher oxidation state (with highly positive charge) and smaller size of the Cr6+@CNP complex (both cases, Cr6+@CNPc and Cr6+@CNPt) are responsible for the larger degree of deformation of the CNPs.
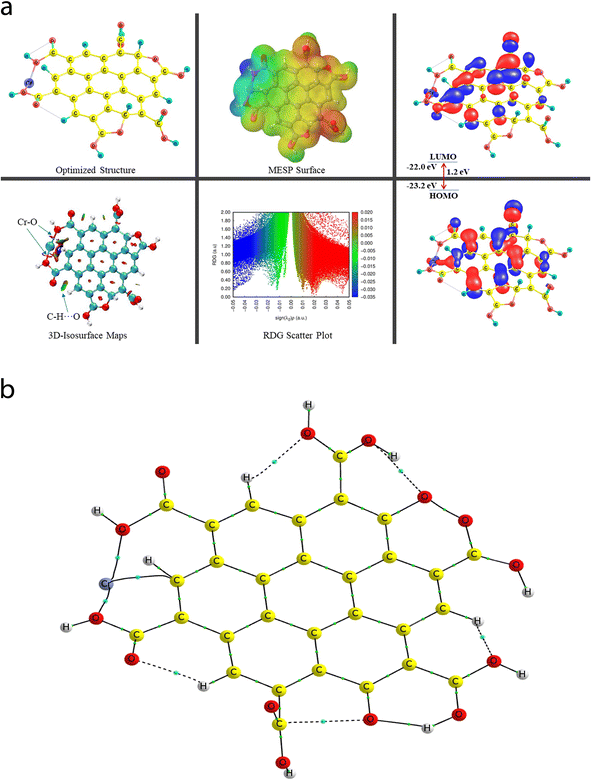
In the case of the Fe3+@CNPc complex, one bond (MNI) between the Fe metal ion and one of the C-atoms of the central benzene ring of the CNP was detected using the QTAIM tool (vide infra), where the bond length of the Fe57–C3 bond was analysed to be 2.336 Å. Moreover, the equilibrium (optimized) structures of the Fe3+@CNPc complex can be seen in Fig. S6† (structural/electronic feature in Fig. S6a† and QTAIM molecular graph in Fig. S6b† for Fe3+@CNPc) and Fig. S7† (geometrical/electronic features in Fig. S7a† and QTAIM molecular graph in Fig. S7b† for Fe3+@CNPt) in the ESI.† Some useful and selected structural [BL and bond path length (BPL)] and QTAIM-based parameters (such as ρ, ∇2(ρ), V, and DI indices) for Fe3+@CNPc and Fe3+@CNPt can be viewed in Tables S1 and S2 of the ESI,† respectively.
However, surprisingly, only one configuration of the Hg2+-related complex, Hg2+@CNPt (Hg2+ at the terminal), can be viewed in the ESI (see Fig. S8a†). Also, the electronic feature-based picture and QTAIM molecular graph of the Hg2+@CNPt complex are displayed in Fig. S8a and b,† respectively. The structural (BL and BPL) and QTAIM-based (ρ, ∇2(ρ), V, and DI) parameters of the Hg2+ ion (probed at the terminal position only) can be seen in Table S3 of the ESI.† The bond distances of two probed MNIs of Hg–O were computed to be 2.943 Å (Hg53–O43) and 3.153 (Hg53–O39), which are much larger than that of the Fe–C bond (2.336 Å) of the Fe3+@CNPc complex.
The calculated BE values for the TM@CNPt complexes associated with the terminal TMs, i.e., Cr6+, Fe3+, and Hg2+ ions, were calculated to be −4476.7 kcal mol−1, −794.4 kcal mol−1, and −183.9 kcal mol−1, whereas the analyzed BEs of the CNP complex with the central Cr6+ and Fe3+ metal ions forming the Cr6+@CNPc and Fe3+@CNPc complexes are −4395.1 kcal mol−1 and −696.5 kcal mol−1, respectively. A high BE difference can be realized among all three cations, which appear to be due to the charge and size of the TM cations. Given that the Cr6+ ion consists of the highest charge and has the lowest size among the three cations, it is expected to show the highest BE with a large difference, revealing that it is a more selective and the most sensitive complex. A comprehensive DFT study on the interaction between different valence metal chromium ions (Cr6+, Cr3+, and Cr2+) and reduced glutathione (GSH) was reported by Jianhua et al. in 2013, where they showed that the BEs for GSH associated with the Cr2+, Cr3+, and Cr6+ metal ions are −834.8 kJ mol−1 (199.5 kcal mol−1), −1162 kJ mol−1 (277.7 kcal mol−1), and −17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 568.6 kJ mol−1 (−4199 kcal mol−1), respectively.67 Here, the emphasis is to compare the selective and sensitivity feature (in terms of the BE value of −4199 kcal mol−1) of the Cr6+ ion interacting with the GSH skeleton (Cr6+@GSH complex), which is 277.7 kcal mol−1 and 196.1 kcal mol−1 smaller than that of the probed Cr6+@CNPt and Cr6+@CNPc complexes, respectively. These findings suggest that the probed Cr6+@CNPt and Cr6+@CNPc complexes reported in this work are much more stable (more selective and higher sensitivity) than the Cr6+@GSH complex.
568.6 kJ mol−1 (−4199 kcal mol−1), respectively.67 Here, the emphasis is to compare the selective and sensitivity feature (in terms of the BE value of −4199 kcal mol−1) of the Cr6+ ion interacting with the GSH skeleton (Cr6+@GSH complex), which is 277.7 kcal mol−1 and 196.1 kcal mol−1 smaller than that of the probed Cr6+@CNPt and Cr6+@CNPc complexes, respectively. These findings suggest that the probed Cr6+@CNPt and Cr6+@CNPc complexes reported in this work are much more stable (more selective and higher sensitivity) than the Cr6+@GSH complex.
Interestingly, the following scenario was seen for the Cr6+- and Fe3+-associated CNP complexes (see Table 2), where it was observed that the calculated BE/IE values suggest that the complexation with both metal ions (Cr6+ and Fe3+) located at the terminal position of the CNP unit is more favorable (i.e., more selective and highly sensitive) (Cr6+@CNPt: −4476.7 kcal mol−1 > Cr6+@CNPc: −4395.1 kcal mol−1) compared to the complexation of the CNP moiety with the TM cations positioned in the center. This can be attributed to the ion–dipole interaction between the metal cation and the COOH-functionalized CNP framework. A similar pattern can be viewed in the case of both Fe3+@CNP complexes (Fe3+@CNPt: −794.4 kcal mol−1 > Fe3+@CNPc: −696.5 kcal mol−1). According to Table 2, the BE results suggest that the TMs located at the terminal of the CNP moiety appear to show much more intense sensing features compared to the TMs located in the center of the CNP framework.
The EHOMO and ELUMO values of the Cr6+@CNPc complex were calculated to be −23 eV and −21.5 eV, respectively, whereas the EHOMO (−23.2 eV) decreased and ELUMO (−22 eV) increased for the Cr6+@CNPt complex. The Egap value of the Cr6+@CNPc complex (1.5 eV) was found to be slightly larger than that of Cr6+@CNPt (1.2 eV), which demonstrates that the latter is more sensitive compared to the former, which is also confirmed by the BE values. The HOMO–LUMO 3D isosurface maps of the Cr6+@CNPc and Cr6+@CNPt complexes can be seen in Fig. 7a and 8a, respectively. In the case of the Cr6+@CNPc complex, its HOMOs appear to be primarily spread over the O atoms of the carbonyl group of the COOH functional groups, whereas the LUMOs seem to be distributed around the O atoms (out-of-plane), which depict an n to π* transition (see the HOMO–LUMO plot in Fig. 7a). In the case of the Cr6+@CNPt complex (see Fig. 8a), the HOMOs are mainly located over the CC bond (in-plane) of the six-membered ring of the CNP framework, while the LUMOs are around the CC bond but in the out-of-plane phase, which describes the π to π* transition. As shown by the 3D MESP surfaces of both chromium-related complexes, it can be seen that the positive regions are positioned over the H atoms of the COOH functional groups, whereas the negative regions are located over the –COO segments of the COOH functional groups in both complexes (Cr6+@CNPc in Fig. 7a and Cr6+@CNPt in Fig. 8a).
The 2D scatter plot and 3D isosurface maps of the Cr6+@CNPt complex are shown in Fig. 8a. It can be seen that three strong MNIs [two Cr–O MNIs: Cr51–O37 and Cr51–O41 and one Cr–C MNI: Cr51–C18] exist in the Cr6+@CNPt complex, which play a major role in stabilizing it. The QTAIM molecular graph of the Cr6+@CNPt complex (see Fig. 8b) and the associated parameters confirmed the presence of these three MNIs. It showed that two Cr–O MNIs (ρ: 0.0768 au and 0.0775; ∇2(ρ): +0.4741 au and +0.4408 au, V: −0.1244 au and −0.1225 au, and DI: 0.546 and 0.513) in the Cr6+@CNPt complex are stronger in comparison to the two Cr–C MNIs present in the Cr6+@CNPc complex, while the Cr–C MNI of the former is weaker than that of the latter and vice versa for the other Cr–C MNI involved in the former. Additionally, the Cr6+@CNPt complex consists of a total of seven intramolecular interactions (one O–H⋯O and three C–H⋯O HBs, as well as one C⋯O and two O⋯O NBPs), which are shown in the 3D isosurface maps and 2D scatter plot. The QTAIM topological parameters for all the NCIs and the NBPs can be seen in Table 5, where the BLs of the NCIs range from 1.652 Å to 2.279 Å. The opposite trend can be observed in the case of the QTAIM-based strength quantifying parameters against their corresponding BLs and BPLs.
The associated QTAIM topological parameters (ρ, ∇2(ρ), and V) were calculated (0.0423 au, +0.196 au, and −0.0454 au) for the Fe57–C3 bond of the Fe3+@CNPc complex, respectively. The QTAIM-based topological features for the various intramolecular NCIs such as six C–H⋯O and two NBPs (C⋯O) involved in the case of the Fe3+@CNPc complex can also be viewed in Table S1 of the ESI.† Alternatively, the Fe3+@CNPt complex demonstrates three MNIs (two Fe–O and one Fe–C), which are slightly stronger than that of the Fe3+@CNPc complex. The (ρ, ∇2(ρ), and V) values for two observed Fe–O intermolecular MNIs, Fe53–O43 and Fe53–O39, of the Fe3+@CNPt complex were determined to be (0.0568 au, +0.3931 au, −0.088 au) and (0.0545 au, +0.399 au, −0.0892 au), while they are (0.0537 au, +0.1918 au, −0.0058 au) for the Fe53–C19 bond, respectively. Alternatively, the five intramolecular NCIs (C–H⋯O) and two NBPs (O⋯O) acquired from the QTAIM technique for the Fe3+@CNPt complex are shown in Table S2 of the ESI.†
The QTAIM topological parameters (ρ, ∇2(ρ), and V) for the Hg53–O43 bond were found to be (0.0163, +0.0491, and −0.0116) (in au), whereas for the Hg53–O39 bond, they were detected to be (0.0108, +0.0305, −0.0067) (in au), respectively, showing that the former is much stronger (approximate one and half times) than the latter. Hence, these MNIs are much weaker than the MNIs (one Fe–C in the Fe3+@CNPc and Fe3+@CNPt complexes, whereas two Fe–O MNIs in only the Fe3+@CNPt complex). The QTAIM parameters for several other intramolecular NCIs (seven C–H⋯O) and NBPs (two O⋯O) taking part in the Hg2+@CNP complex can be seen in Table S3 of the ESI.†
Conclusively considering the sensitivity and selectivity features in all five inspected complexes here in this study, it is noteworthy to mention that the BEs for the COOH-functionalized CNP associated with the TM cations located at the terminal of the framework follow the order of (Cr6+@CNPt: −4476.7 > Fe3+@CNPt: −794.4 > Hg2+@CNPt: −183.9) (kcal mol−1). Importantly and interestingly, this trend is also consistent with the BSSE complexation energy (Cr6+@CNPt: −4688.9 > Fe3+@CNPt: −785.6 > Hg2+@CNPt: −226.2) (kcal mol−1). Table 2 demonstrates that Cr6+@CNPt is the most stable complex (consisting of the largest BE value, i.e., is the most selective and sensitive) among the five computationally examined TM-associated CNP complexes, which assists in portraying the excellent sensitivity and selectivity of the as-prepared CNP for Cr6+. By observing the five analyzed BE findings, the Cr6+@CNPt species is highly preferred in terms of highest selectivity and most sensitive behavior.
6. Conclusions
Herein, a novel, convenient, cost-effective, and green synthetic route for the preparation of CNPs from Madhuca longifolia flower using a hydrothermal approach was proposed in this study. The average diameter of CNPs was determined by TEM and found to be 12.0 nm. The as-synthesized CNPs displayed strong blue fluorescence with an emission maximum of 454 nm at 370 nm excitation, and the prepared CNPs also exhibited excitation-dependent emission properties. The XPS and FTIR studies suggested that the CNPs were functionalized carbonyl, hydroxy and carboxylic acid groups, and the interaction between the carboxylic acid with Cr6+ is responsible for the quenching of fluorescence intensity via the static quenching phenomenon. The fluorescence lifetime decay study also validated the static quenching mechanism given that no change was observed in the fluorescence decay profiles. The LOD was found to be 102 ppb, which is much better in comparison with most of the reported nanosensors. The sensing mechanism for the detection of Cr6+ was well supported by the theoretical study.The experimentally acquired outcomes were strongly supported by the DFT approach. The theoretical findings suggested that the interaction between the functionalized CNP framework and the Cr6+ (located at the terminal) is the strongest among the five studied complexes and demonstrated the highest sensitivity. It should be noted that the bonding features were mainly directed by the MNIs (Cr–C, Fe–C, Fe–O, and Hg–O) in all the investigated complexes, where the Cr6+-associated bonding feature played a dominant role in terms of sensitivity. Moreover, many other NCIs (O–H⋯O and C–H⋯O) and NBPs (C⋯O and O⋯O) taking part in all complexes stabilized the CNP skeleton itself. Given that the quantum chemical calculations provide a good understanding of the structural, stability, and electronic features of the association of three TM cations with the functionalized CNP, these model complexes can facilitate new insights into understanding the sensing features analysed in the field of environmental chemistry.
Author contributions
Methodology, experimental performing, and writing original manuscript (Tuhin Mandal); steady-state fluorescence study (Ashish Kumar Ghosh); reviewing and editing (Shiv Rag Mishra); designing, performing, and analysis of the theoretical study along with the conceptualization and computational methodology (Sarvesh Kumar Pandey); conceptualization, methodology, supervision, analysis of experimental results, writing/editing original manuscript and funding acquisition (Vikram Singh).Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
VS thanks CSIR-Central Institute of Mining and Fuel Research Dhanbad for funding (MLP-155/2021-22) and for providing the facilities. The authors acknowledge the central research facility, Indian Institute of Technology (ISM) Dhanbad for the HRTEM, XPS, and UV-Visible analyses. The authors also acknowledge Prof. Soumit Chatterjee, Chemistry Department and Prof. Ajay Mandal, Petroleum Engineering Department, IIT (ISM) Dhanbad for providing fluorescence lifetime and zeta potential study respectively. SKP acknowledges the Department of Inorganic and Physical Chemistry (IPC), Indian Institute of Science (IISc) Bengaluru for providing the computational facilities.References
- P. Das, M. Maruthapandi, A. Saravanan, M. Natan, G. Jacobi, E. Banin and A. Gedanken, ACS Appl. Nano Mater., 2020, 3, 11777–11790 CrossRef CAS.
- J. Liu, G. Ji, J. Xiao and Z. Liu, Inorg. Chem., 2017, 56, 4197–4205 CrossRef CAS PubMed.
- A. Zhitkovich, Chem. Res. Toxicol., 2011, 24, 1617–1629 Search PubMed.
- M. Jaishankar, T. Tseten, N. Anbalagan, B. B. Mathew and K. N. Beeregowda, Interdiscip. Toxicol., 2014, 7, 60 Search PubMed.
- L. Bu, J. Peng, H. Peng, S. Liu, H. Xiao, D. Liu, Z. Pan, Y. Chen, F. Chen and Y. He, RSC Adv., 2016, 6, 95469–95475 RSC.
- United States Environmental Protection Agency, EPA edition of the drinking water standards and health advisories, 2010 Search PubMed.
- H. A. Maitlo, K.-H. Kim, V. Kumar, S. Kim and J.-W. Park, Environ. Int., 2019, 130, 104748 CrossRef CAS PubMed.
- L. Liu, Y. Leng and H. Lin, Microchim. Acta, 2016, 183, 1367–1373 CrossRef CAS.
- T.-Y. Gu, M. Dai, D. J. Young, Z.-G. Ren and J.-P. Lang, Inorg. Chem., 2017, 56, 4668–4678 CrossRef PubMed.
- X. Wang, Y. Zhang, Z. Shi, T. Lu, Q. Wang and B. Li, ACS Appl. Mater. Interfaces, 2021, 13, 54217–54226 CrossRef CAS PubMed.
- Z. Sun, M. Yang, Y. Ma and L. Li, Cryst. Growth Des., 2017, 17, 4326–4335 CrossRef CAS.
- X. Zhou, Y.-X. Shi, C. Cao, C.-Y. Ni, Z.-G. Ren, D. J. Young and J.-P. Lang, Cryst. Growth Des., 2019, 19, 3518–3528 CrossRef CAS.
- P. Li, X.-M. Yin, L.-L. Gao, S.-L. Yang, Q. Sui, T. Gong and E.-Q. Gao, ACS Appl. Nano Mater., 2019, 2, 4646–4654 CrossRef CAS.
- M. Singh, S. Senthilkumar, S. Rajput and S. Neogi, Inorg. Chem., 2020, 59, 3012–3025 CrossRef CAS PubMed.
- S. Mandal, D. Paul, S. Saha and P. Das, Environ. Sci.: Nano, 2022, 9, 2596–2606 RSC.
- B. Fang, P. Wang, Y. Zhu, C. Wang, G. Zhang, X. Zheng, C. Ding, J. Gu and F. Cao, RSC Adv., 2018, 8, 7377–7382 RSC.
- V. K. Jayswal, A. M. Ritcey and J.-F. Morin, Nanoscale Adv., 2023, 5, 337–343 RSC.
- T. Mandal, S. R. Mishra, K. Singh, H. Agarwalla, R. E. Masto, M. Kumar and V. Singh, J. Nanopart. Res., 2023, 25, 125 CrossRef CAS.
- K. S. Rawat, V. Singh, C. P. Sharma, A. Vyas, P. Pandey, J. Singh, N. M. Gupta, M. Sachdev and A. Goel, J. Imaging, 2023, 9, 19 CrossRef PubMed.
- V. Singh, K. S. Rawat, S. Mishra, T. Baghel, S. Fatima, A. A. John, N. Kalleti, D. Singh, A. Nazir, S. K. Rath and A. Goel, J. Mater. Chem. B, 2018, 6, 3366–3371 RSC.
- G. S. Selopal, M. Mohammadnezhad, L. V. Besteiro, O. Cavuslar, J. Liu, H. Zhang, F. Navarro-Pardo, G. Liu, M. Wang, E. G. Durmusoglu, H. Y. Acar, S. Sun, H. Zhao, Z. M. Wang and F. Rosei, Adv. Sci., 2020, 7, 2001864 CrossRef CAS PubMed.
- V. Singh and A. K. Mishra, J. Mater. Chem. C, 2016, 4, 3131–3137 RSC.
- V. Singh and A. K. Mishra, Sens. Actuators, B, 2016, 227, 467–474 CrossRef CAS.
- D. Bharathi, B. Siddlingeshwar, R. H. Krishna, V. Singh, N. Kottam, D. D. Divakar and A. A. Alkheraif, J. Fluoresc., 2018, 28, 573–579 CrossRef CAS PubMed.
- Z. Khazaee, A. R. Mahjoub and A. H. Cheshme Khavar, Appl. Catal., B, 2021, 297, 120480 CrossRef CAS.
- L. Cao, S. Sahu, P. Anilkumar, C. E. Bunker, J. Xu, K. A. S. Fernando, P. Wang, E. A. Guliants, K. N. Tackett II and Y.-P. Sun, J. Am. Chem. Soc., 2011, 133, 4754–4757 CrossRef CAS PubMed.
- N. Mamidi and R. M. V. Delgadillo, ACS Appl. Mater. Interfaces, 2022, 14, 35789–35801 CrossRef CAS PubMed.
- Y. Zhang, L. Wang, M. Yang, J. Wang and J. Shi, Appl. Surf. Sci., 2019, 466, 515–524 CrossRef CAS.
- V. Singh, B. Gorbel, S. Chatterjee, P. Sen and V. Verma, Mater. Lett., 2022, 309, 131446 CrossRef CAS.
- Y. Wang, Y. Ding, Y. Tan, L. Fu and W. Qing, Inorg. Chem. Commun., 2023, 147, 110220 CrossRef CAS.
- E. Asadian, M. Ghalkhani and S. Shahrokhian, Sens. Actuators, B, 2019, 293, 183–209 CrossRef CAS.
- P. P. Das, S. Pramanik, S. Chatterjee, A. Roy, A. Saha, P. S. Devi and G. Suresh Kumar, ACS Sustainable Chem. Eng., 2018, 6, 10127–10139 CrossRef CAS.
- C. J. Jeong, A. K. Roy, S. H. Kim, J.-E. Lee, J. H. Jeong, I. In and S. Y. Park, Nanoscale, 2014, 6, 15196–15202 RSC.
- V. Singh, S. Chatterjee, M. Palecha, P. Sen, B. Ateeq and V. Verma, Carbon Lett., 2021, 31, 117–123 CrossRef.
- M. Algarra, L. d. Orfãos, C. S. Alves, R. Moreno-Tost, M. S. Pino-González, J. Jiménez-Jiménez, E. Rodríguez-Castellón, D. Eliche-Quesada, E. Castro and R. Luque, ACS Sustainable Chem. Eng., 2019, 7, 10493–10500 CrossRef CAS.
- G. K. Yogesh, E. P. Shuaib, A. Kalai Priya, P. Rohini, S. V. Anandhan, U. M. Krishnan, V. Kalyanavalli, S. Shukla and D. Sastikumar, Opt. Laser Technol., 2021, 135, 106717 CrossRef CAS.
- P. Li, Y. Hong, H. Feng and S. F. Li, J. Mater. Chem. B, 2017, 5, 2979–2988 RSC.
- G. Wang, S. Zhang, J. Cui, W. Gao, X. Rong, Y. Lu and C. Gao, Anal. Chim. Acta, 2022, 1195, 339478 CrossRef CAS PubMed.
- H. Yuan, G. Yang, Q. Luo, T. Xiao, Y. Zuo, X. Guo, D. Xu and Y. Wu, Environ. Sci.: Nano, 2020, 7, 773–781 RSC.
- J. Goswami, S. S. Rohman, A. K. Guha, P. Basyach, K. Sonowal, S. P. Borah, L. Saikia and P. Hazarika, Mater. Chem. Phys., 2022, 286, 126133 CrossRef CAS.
- M. F. Ramadan and E. M. Wagdi Abdel-Hamed, Nuts and Seeds in Health and Disease Prevention, 2nd edn, 2020, pp. 229–237 Search PubMed.
- S. P. Singh, B. Yadav and K. Anupam, in Herbs, Shrubs, and Trees of Potential Medicinal Benefits, CRC Press, 2022, pp. 461–470 Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Revision B.01. Gaussian Inc., Wallingford, CT, 2010 Search PubMed.
- T. A. Keith and T. K. Gristmill, AIMAll (Version 11.05.16), Software, Overland Park KS, 2011, https://aim.tkgristmill.com/ Search PubMed.
- S. K. Pandey, D. Manogaran, S. Manogaran and H. F. Schaefer III, J. Phys. Chem. A, 2017, 121, 6090–6103 CrossRef CAS PubMed.
- J. Contreras-García, E. R. Johnson, S. Keinan, R. Chaudret, J.-P. Piquemal, D. N. Beratan and W. Yang, J. Chem. Theory Comput., 2011, 7, 625–632 CrossRef PubMed.
- J. Baker and P. Pulay, J. Am. Chem. Soc., 2006, 128, 11324–11325 CrossRef CAS PubMed.
- S. K. Pandey, P. Das, P. K. Das, E. Arunan and S. Manogaran, J. Chem. Sci., 2015, 127, 1127–1134 CrossRef CAS.
- M. M. R. Khan, T. Mitra and D. Sahoo, RSC Adv., 2020, 10, 9512–9524 RSC.
- C. Li, W. Liu, X. Sun, W. Pan and J. Wang, Sens. Actuators, B, 2017, 252, 544–553 CrossRef CAS.
- J. Wang, F. Qiu, H. Wu, X. Li, T. Zhang, X. Niu, D. Yang, J. Pan and J. Xu, Spectrochim. Acta, Part A, 2017, 179, 163–170 CrossRef CAS PubMed.
- J. Wang, F. Qiu, X. Li, H. Wu, J. Xu, X. Niu, J. Pan, T. Zhang and D. Yang, J. Lumin., 2017, 188, 230–237 CrossRef CAS.
- H. Wang, S. Liu, Y. Xie, J. Bi, Y. Li, Y. Song, S. Cheng, D. Li and M. Tan, New J. Chem., 2018, 42, 3729–3735 RSC.
- Y. Yang, X. Chen, Y. Wang, M. Wu, Y. Ma and X. Yang, Front. Chem., 2020, 8, 595628 CrossRef CAS PubMed.
- S. Bardhan, S. Roy, D. K. Chanda, S. Ghosh, D. Mondal, S. Das and S. Das, Dalton Trans., 2020, 49, 10554–10566 RSC.
- C. Sakaew, P. Sricharoen, N. Limchoowong, P. Nuengmatcha, C. Kukusamude, S. Kongsri and S. Chanthai, RSC Adv., 2020, 10, 20638–20645 RSC.
- H. Yuan, G. Yang, Q. Luo, T. Xiao, Y. Zuo, X. Guo, D. Xu and Y. Wu, Environ. Sci.: Nano, 2020, 7, 773–781 RSC.
- Q. Luo, X. Huang, Y. Luo, H. Yuan, T. Ren, X. Li, D. Xu, X. Guo and Y. Wu, Chem. Eng. J., 2021, 407, 127050 CrossRef CAS.
- H. Yuan, J. Peng, T. Ren, Q. Luo, Y. Luo, N. Zhang, Y. Huang, X. Guo and Y. Wu, Sci. Total Environ., 2021, 760, 143395 CrossRef CAS PubMed.
- N. K. R. Bogireddy, S. E. Sotelo Rios and V. Agarwal, Chem. Eng. J., 2021, 414, 128830 CrossRef CAS.
- T. Hu, L. Zeng, Y. Li, Y. Wu, Z. Zhu, Y. Zhang, D. Tian, C. Gao and W. Li, Chem. Eng. J., 2022, 432, 134202 CrossRef CAS.
- A. G. Anil, S. Ramachandran, V. Kumar, S. Subramanian and P. C. Ramamurthy, J. Basic Microbiol., 2022, 62, 455–464 CrossRef CAS PubMed.
- X. Chen, Z. Song, B. Yuan, X. Li, S. Li, T. Thang Nguyen, M. Guo and Z. Guo, Chem. Eng. J., 2022, 430, 133154 CrossRef CAS.
- Y. Feng, R. Li, P. Zhou and C. Duan, Microchem. J., 2022, 180, 107627 CrossRef CAS.
- S. Awasthi, S. K. Pandey, J. K. Gaur and C. Srivastava, Mater. Chem. Front., 2022, 6, 3731–3747 RSC.
- S. Awasthi, J. K. Gaur, S. K. Pandey, M. S. Bobji and C. Srivastava, ACS Appl. Mater. Interfaces, 2021, 13, 24505–24523 CrossRef CAS PubMed.
- L. Jianhua, L. Yan and W. Haijun, Chin. J. Appl. Chem., 2013, 30, 963–970 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00289f |
| This journal is © The Royal Society of Chemistry 2023 |