Open Access Article
Open Access ArticleSynthesis and application of spermine-based amphiphilic poly(β-amino ester)s for siRNA delivery†
Yao
Jin
a,
Friederike
Adams‡
 a,
Anny
Nguyen
a,
Sebastian
Sturm
b,
Simone
Carnerio
a,
Knut
Müller-Caspary
b and
Olivia M.
Merkel
a,
Anny
Nguyen
a,
Sebastian
Sturm
b,
Simone
Carnerio
a,
Knut
Müller-Caspary
b and
Olivia M.
Merkel
 *a
*a
aDepartment of Pharmacy, Ludwig-Maximilians-University Munich, Pharmaceutical Technology and Biopharmaceutics, Butenandtstr. 5-13, 81377 Munich, Germany. E-mail: olivia.merkel@lmu.de
bDepartment of Chemistry and Centre for NanoScience, Ludwig-Maximilians-University Munich, Butenandtstr. 11, 81377 Munich, Germany
First published on 7th September 2023
Abstract
Small interfering RNA (siRNA) can trigger RNA interference (RNAi) to therapeutically silence disease-related genes in human cells. The approval of siRNA therapeutics by the FDA in recent years generated a new hope in novel and efficient siRNA therapeutics. However, their therapeutic application is still limited by the lack of safe and efficient transfection vehicles. In this study, we successfully synthesized a novel amphiphilic poly(β-amino ester) based on the polyamine spermine, hydrophobic decylamine and 1,4-butanediol diacrylate, which was characterized by 1H NMR spectroscopy and size exclusion chromatography (SEC, Mn = 6000 Da). The polymer encapsulated siRNA quantitatively from N/P 5 on as assessed by fluorescence intercalation while maintaining optimal polyplex sizes and zeta potentials. Biocompatibility and cellular delivery efficacy were also higher than those of the commonly used cationic, hyperbranched polymer polyethylenimine (PEI, 25 kDa). Optimized formulations mediated around 90% gene silencing in enhanced green fluorescence protein expressing H1299 cells (H1299-eGFP) as determined by flow cytometry. These results suggest that spermine-based, amphiphilic poly(β-amino ester)s are very promising candidates for efficient siRNA delivery.
Introduction
Small interfering RNA (siRNA) is a double-stranded macromolecule with a length of 19–21 nucleotides per strand used to trigger the innate RNA interference (RNAi) mechanism to degrade complementary mRNA molecules for a subsequent decrease in gene expression.1 RNAi therapy has preclinically been demonstrated promising for the treatment for many diseases, for example, chronic myeloid leukemia,1 non-small cell lung cancer,2 skin melanoma,3 and many others. It also has the potential to treat COVID-19 caused by SARS-CoV-2 virus infections.4,5 Additionally, several siRNA-based products are already approved by the FDA, and many additional siRNA-drugs are in different stages of clinical trials.6 However, the physiochemical properties of siRNAs including their macromolecular characteristics, hydrophilicity, net negative charge, enzymatic degradation by serum endonucleases such as RNases, and rapid renal clearance of siRNA require the combination of bioactive sequences with effective delivery systems.7,8Cationic polymers have shown promise for nucleic acid delivery given their ability to self-assemble with anionic siRNA into condensed nanoparticles (polyplexes) with efficient encapsulation capacity. Various cationic polymers have been developed as nonviral nucleic acid vectors during the past three decades, including polyethylenimine (PEI), chitosan, inulin, poly-L-lysine, and many more.9 Poly(β-amino ester)s (PBAE)s are polymers synthesized from acrylate compounds and amine-containing monomers by Michael addition-based step growth polymerization and have drawn people's attention after their first application for transfection in 2000 due to their inherent biocompatibility, biodegradability, and stimuli-responsiveness.10,11 The first generation of PBAEs contains linear PBAEs (LPAEs), and a variety of LPAEs has been established and proven to be promising for gene delivery. However, the delivery of siRNA bears new challenges in comparison to plasmid DNA because siRNA is linear and short with a rigid structure, which affords limited molecular entanglement points to electrostatically bind to cationic polymers.12 Therefore, high polymer/siRNA weight ratios are reported to be required to fully condense siRNA and enable efficient transfection.13 Hyperbranched and branched PBAEs (HPAEs) have a high density of branching linkers, and the resulting three-dimensional globular shape and multiple chain-end groups have the potential of high siRNA encapsulation efficiency. However, HPAEs with strong nucleic acid binding affinity may restrict the intracellular siRNA release, thus hampering siRNA-mediated gene silencing efficiency.12 Environment-responsive disulfide linkages containing HPAEs were thus designed and provided promising applications for anti-inflammatory siRNA delivery and for efficient cancer gene therapy in approaches of furthering HPAE efficacy.12,14 In this study, brushed PBAEs with spermine side chain were synthesized to overcome some of the limitations reported in the literature.
Spermine is a naturally occurring, biocompatible polycation with primary and secondary amines spaced by methylene groups and has high affinity toward nucleic acids.15,16 The essential role of spermine in eukaryotic cells is to aid in packaging cellular DNA into a compact state.17,18 However, spermine shows a limited siRNA complexation ability and efficiency for transfecting nucleic acids because of its low molecular weight (∼200 Da).18 In addition, spermines yield limited endosomal escape despite of their good proton-buffering capacity.19 The polymerization of spermine to increase its molecular weight can improve the products' buffering capacity as well as enhance the endosomal escape and transfection efficiency of spermine polyplexes.20,21 In this regard, Lote et al. synthesized a variety of oligospermines and found that the molecular weight of the linear oligospermines had a pronounced effect, while the molecular architectures influenced the siRNA encapsulation profiles.15 Duan et al. synthesized a linear polyspermine imidazole-4,5-amide (PSIA), and the resulting PSIA-polyplexes were able to achieve high cellular uptake and efficient luciferase gene silencing.21 Jere et al. synthesized a hydrophilic poly(β-amino ester) containing spermine and PEG for gene delivery and showed higher transfection efficiency than with PEI (25 kDa).16 Liu et al. synthesized a hyperbranched poly(β-amino ester), in which the end-capping with spermine improved protein binding and thus enhanced cytosolic protein delivery.22
In this work, we synthesized a spermine-based poly(β-amino ester) combining the advantages of PBAEs with increased numbers of spermine-units in one polymer. We first protected three amines of spermine to facilitate the reaction with one primary amine to form brushed polymers that still contain primary amines. Inspired by our previous research,23 we found that hydrophobic modification is important for the successful transfection and gene silencing of PBAEs polyplexes. Therefore, decylamine was polymerized together with spermine to give a brush-like poly(β-amino ester) with suitable amphiphilicity.
Results and discussion
In this work, we first converted spermine according to literature procedures to form tri-tert-butyl carbonyl spermine, abbreviated as tri-boc-spermine.24,25 To obtain a pure product, the crude product was purified by column chromatography. The purity of monomers is important for the following steps during polymer synthesis, and the chemical structure of the tri-boc-spermine was therefore characterized by 1H NMR spectroscopy (Fig. S1A†).The tri-boc-spermine and decylamine-based β-amino ester polymer poly(tri-boc-spermine-co-decylamine amino β-amino ester) abbreviated as P(BSpDBAE) was synthesized by the Michael addition-based step-growth polymerization of tri-boc-spermine, decylamine and 1,4-butanediol diacrylate (Scheme 1). As shown in Fig. S1C† the NMR spectrum is in accordance with a polymer consisting of all three monomers, as peaks assigned to all three monomers are visible in the NMR spectrum. The peaks at 1.44 ppm, 1.70 ppm and 3.16 ppm are assigned to tri-boc-spermine; the peaks at 0.88 ppm and 1.26 ppm belong to decylamine; the peaks at 1.70 ppm, 2.46 ppm, 2.82 ppm and 4.11 ppm are assigned to the 1,4-butanediol diacrylate backbone of the polymer. A shift of the peak at 4.18 ppm of 1,4-butanediol diacrylate to 4.11 ppm as well as a decrease in integrals of the acrylate protons indicate the conjugation of the two amine-containing monomers after successful 1,4-Michael-addition. The molar ratio of the two amine monomers was calculated according to the integrations of peaks at 1.70 ppm and 0.88 ppm, and the ratio was approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The reaction was carried out in bulk, as high concentrations of monomers in the absence of solvents can produce PBAEs with higher molecular weights and less intramolecular cyclization while requiring shorter reaction times.26,27
1. The reaction was carried out in bulk, as high concentrations of monomers in the absence of solvents can produce PBAEs with higher molecular weights and less intramolecular cyclization while requiring shorter reaction times.26,27
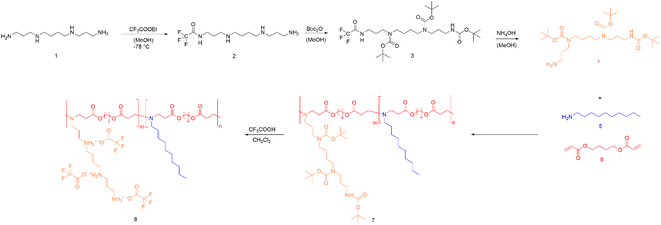 | ||
| Scheme 1 Synthesis route of P(SpDBAE) (8) by reacting tri-boc spermine (4) with decylamine (5) and 1,4-butanediol diacrylate (6) resulting in P(BSpDBAE) (7) followed by deprotection. | ||
The polymer P(BSpDBAE) was then deprotected by treatment with trifluoro acetic acid (TFA) resulting in poly(spermine-co-decylamine amino β-amino ester) P(SpDBAE). As shown in Fig. S1D† the disappearance of the peak at 1.44 ppm indicates that the N-Boc protection groups were removed successfully. The reaction conditions, yield, molecular weight, PDI and protonable unit of the polymers are summarized in Table 1.
| Polymer | Solvent | Time (h) | Yield (%) | M n (Da) | Đ | Protonable unit |
|---|---|---|---|---|---|---|
| a Reaction conditions: no solvent, 120 °C, overnight. b Reaction conditions: 100 mg polymer, 1 mL TFA, 20 mL dichloromethane, room temperature, 2 h. c M n and Đ were tested via SEC (measurement relative to polystyrene and in chloroform at 40 °C). d Molecular weight was calculated on the basis of the Mn of P(BSpDBAE) and the theoretical chemical structure of P(SpDBAE) as TFA salt. | ||||||
| P(BSpDBAE)a | — | 15 | 44 | 6000c | 1.83c | — |
| P(SpDBAE)b | DCM | 2 | 62 | 6200d | — | 220 |
Polyplexes between P(SpDBAE) and siRNA were then prepared as described in the ESI† at different N/P ratios. The hydrodynamic diameter (size) and polydispersity index (PDI) of the polyplexes were measured by dynamic light scattering (DLS) and the zeta potential of the polyplexes was determined by laser doppler anemometry (LDA). As shown in Fig. 1A, the size of the polyplexes depended on the N/P ratios with a minimum size of the polyplexes of 37 nm at N/P 7 with a PDI of 0.18. It was hypothesized that the siRNA was not entirely encapsulated at N/P 1 and 3 as the zeta potential was negative, and the observed sizes were comparably large. For the polyplexes at N/P 10 to 20, an excess of free polymer may cause sedimentation, resulting in a comparably high PDI. Size is an important factor for cellular uptake and transfection. Some reports indicate that nanoparticles with a size below 150 nm are required for the uptake in lung cells by endocytosis,28 while other papers report that spermine-based delivery systems with a larger size have a good transfection efficiency in vivo.29
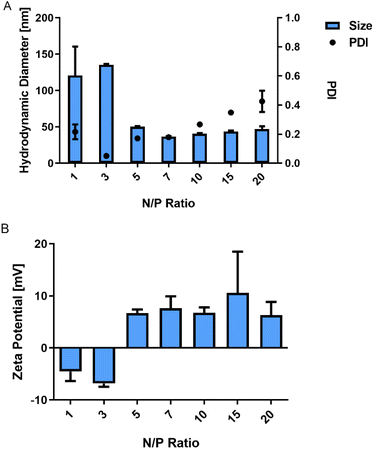 | ||
| Fig. 1 (A) Hydrodynamic diameter (size) and polydispersity index (PDI) of P(SpDBAE) polyplexes and (B) zeta potential of P(SpDBAE) polyplexes at various N/P ratios (mean ± SD, n = 3). | ||
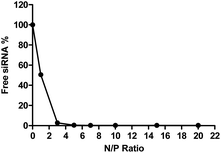
To determine the siRNA encapsulation efficiency of P(SpDBAE), SYBR gold assays were performed. SYBR® gold stain is a high quantum yield proprietary unsymmetrical cyanine dye which can bind to free nucleic acids and exhibits high fluorescence intensity upon binding.30 As shown in Fig. 2, free siRNA was used as a control and its fluorescence intensity was set as 100%. The amount of free siRNA decreased with the increase of N/P ratio, which reflects siRNA encapsulation by the polymer in polyplexes. Almost all the siRNA was encapsulated from N/P 5 on, which indicates that P(SpDBAE) can encapsulate siRNA efficiently. The SYBR gold results also correspond to the size and zeta potential of the polyplexes. As shown in Fig. 2, the siRNA was not fully encapsulated at N/P 1 and 3, and free siRNA attached to the surface of the polyplexes or remained in solution, thus the zeta potentials of the polyplexes were negative (Fig. 1B). The siRNA can be quantitatively encapsulated from N/P 5 on (Fig. 2), which is corresponding to a positive zeta potential of the formed polyplexes (Fig. 1B).
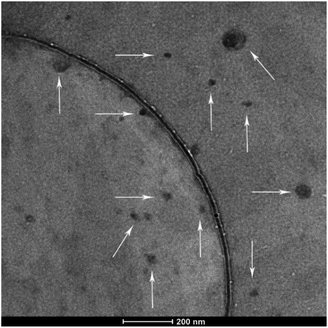
The morphology of the polyplexes was investigated via transmission electron microscopy (TEM) at an FEI TITAN operated at 300 kV. The polyplexes shown in Fig. 3 were mostly spherical. As the white arrows pointed in the figure, particle sizes vary between 20 and 100 nm, which is in accordance with the DLS results (Fig. 1A, N/P 10). Accordingly, the existence of big particles can explain the high PDI of the DLS results.
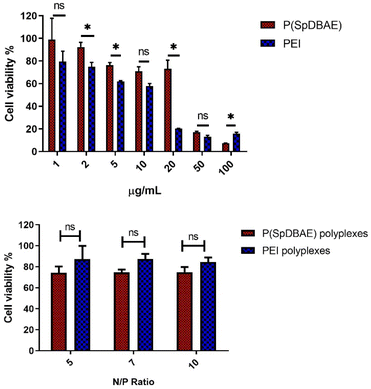
The cell compatibility of P(SpDBAE) free polymer and P(SpDBAE) polyplexes was measured by MTT assay. MTT (dimethylthiazolyl blue diphenyltetrazolium bromide) is a yellow tetrazole, which can be reduced to purple formazan in living cells.31 The method indeed measures the metabolic activity of the cells and reveals the cell viability indirectly. As shown in Fig. 4, the treatment with polymer P(SpDBAE) had a lower effect on the cell viability than hyPEI 25 kDa treatment in general. The IC50 of P(SpDBAE) was 43.5 μg mL−1 and the IC50 of PEI was calculated as 10.5 μg mL−1 (Fig. S2†). The cell viability of P(SpDBAE) polyplexes at N/P 5, 7 and 10 was 79.8%, 75.9% and 64.7% respectively. The cell viability of P(SpDBAE) polyplexes decreased from N/P 5 to 10 because of the increasing amount of excess free polymer. Interestingly, the viability of cells transfected with PEI polyplexes was better than compared with P(SpDBAE) polyplexes. This observation was attributed to the smaller protonable unit of PEI than that of P(SpDBAE), resulting in smaller weight amounts of PEI necessary for preparing the polyplexes than the weight amount of P(SpDBAE) in P(SpDBAE) polyplexes. Besides, the cellular uptake of P(SpDBAE) polyplexes was much higher than that of PEI polyplexes (Fig. 5), which could also lead to stronger cytotoxicity. Although the cell compatibility of P(SpBAE) polyplexes seems not as good as the compatibility of PEI polyplexes, the PBAE is biodegradable. PBAE degradation products are considered to be benign to mammalian cells and were reported to have limited influence on the metabolic activity of healthy cells.32 When applied in vivo, several doses of polyplexes may be required to obtain a therapeutic effect and for improved long-term efficacy.33 Although no animal studies were performed in this initial set of experiments, the characteristics observed here seem favorable for an in vivo follow-up experiment.
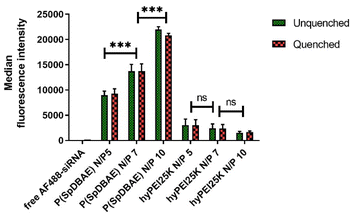
The cellular uptake of P(SpDBAE) polyplexes was quantified by flow cytometry and compared to hyperbranched polyethylenimine (hyPEI, 25 kDa) polyplexes as a positive control and free AF488-siRNA as a negative control. Extracellular fluorescence associated with non-internalized polyplexes on the cell surface was quenched with 0.4% trypan blue.
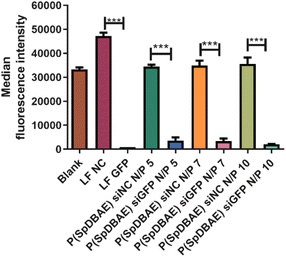
As shown in Fig. 5, the median fluorescence intensities of all the formulations showed no significant difference before and after quenching with trypan blue, which confirms that the determined fluorescence stems from the polyplexes that were internalized into the cells. The uptake of P(SpDBAE) polyplexes at N/P 5 was 59 times higher than that of free AF488-siRNA (negative control) and 3 times higher than that of hyPEI25K polyplexes (positive control); the uptake of P(SpDBAE) polyplexes at N/P 7 was 87 times higher than the negative and 5 times higher than the positive control; the uptake of P(SpDBAE) polyplexes at N/P 10 was 133 times higher than negative and 12 times higher than positive control. The cellular uptake of P(SpDBAE) polyplexes increased from N/P 5 to 10, while the uptake of hyPEI25K polyplexes decreased from N/P 5 to 10.
The cellular uptake of the polyplexes was determined from N/P 5 on, where the siRNA is quantitatively encapsulated according to the SYBR gold assays (Fig. 2). The highest N/P ratio chosen in this experiment was N/P 10 to avoid any possible cytotoxicity at higher concentrations of free polymer. The cellular uptake of P(SpDBAE) polyplexes was in general higher than the PEI polyplexes, which could be favored by the interaction of the hydrophobic C10 chains of decylamine with cell membranes. Hydrophobic modifications have successfully improved the transfection efficiency in the past.34 Interestingly, the cellular uptake of P(SpDBAE) polyplexes increased as the N/P ratio increased from N/P 5 to 10, regardless of the PDI of the polyplexes; while for PEI polyplexes, the cellular uptake decreased from N/P 5 to 10, which may not be attributed to potential cytotoxicity of PEI according to the MTT assay but could indicate that the size of the PEI polyplexes was too disperse at high N/P ratios (Fig. S3†).
To determine the gene silencing efficiency of P(SpDBAE) polyplexes on the protein level, GFP knockdown assays were performed using enhanced green fluorescence protein expressing cells (H1299-eGFP) to quantify the median fluorescence intensity via flow cytometry. Lipofectamine™ 2000 (LF) was used as positive control and LF and P(SpDBAE) were used to encapsulate scrambled siRNA (siNC) as negative control. As shown in Fig. 6, the knockdown efficiency of P(SpDBAE) polyplexes at N/P 5, 7 and 10 was 89.7%, 90.3% and 94.3% respectively. Accordingly, higher cellular uptake of P(SpDBAE) polyplexes led to higher levels of GFP knockdown.
In fact, the cellular uptake and knockdown does not always show a positive correlation. For example, Dosta et al. synthesized a group of PBAEs. Although the cholesterol-modified polymers showed lower siRNA uptake than the unmodified hexylamine and hexadecylamine pendant polymers, the GFP knockdown efficiency of cholesterol-modified polymers was still comparable with the unmodified PBAEs.35
In our case, higher cellular uptake led to more efficient gene silencing efficiency. As far as we understand, the correlation between cellular uptake and gene knockdown mainly depends on the endosomal escape ability of the polyplexes.8 If the polyplexes can escape from the endosomes and lysosomes successfully, high cellular uptake commonly results in efficient gene silencing. However, if the polyplexes have poor ability to escape from endosomes or lysosomes, most of the polyplexes will be entrapped in the latter compartment, and the siRNA will be degraded eventually. In that case, high cellular uptake does not correlate with efficient gene silencing. In addition, the polyplexes should not be too cytotoxic in order to silence gene expression successfully without off-target effects.36 For the same polyplexes with different N/P ratios, the formulations with high cellular uptake usually results in high gene silencing efficiency. However, when comparing polyplexes with and without modification, the modified polyplexes may have low cellular uptake but high gene silencing efficiency.35 Kwon et al. modified PEI with a membrane-lytic peptide called HGP taken from the endodomain of HIV gp41. The PEI-HGP polyplexes showed slightly lower cellular uptake than PEI but the transfection efficiency of PEI-HGP polyplexes was significantly higher than that of unmodified PEI. This effect was explained by the enhanced endosomal escape mediated by the HGP peptide.37
The gene silencing efficacy of polyplexes also strongly depends on the polymers. After the first use of PBAEs for gene delivery by Langer's group,11 a lot of PBAEs were synthesized and researchers tried to understand the relationship between the properties and the chemical structures of PBAEs.38,39 It was found that amine-terminated PBAEs are more suitable for gene delivery, and the transfection potential of PBAEs can also be effected by molecular weight.40 For example, PBAEs with molecular weight less than 11 kDa were unable to form stable polyplexes even at a high 150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer to DNA weight ratio, however, PBAEs at molecular weight more than 13 kDa (Mw = 13
1 polymer to DNA weight ratio, however, PBAEs at molecular weight more than 13 kDa (Mw = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 13
100 and 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) were able to stably complex DNA even at polymer to DNA weight ratio as low as 10
400) were able to stably complex DNA even at polymer to DNA weight ratio as low as 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and showed the highest luciferase expression.40 In addition, low molecular weight PBAEs (Mw < 8 kDa) only poorly transfect DNA compared with higher molecular weight versions, which is the same trend as observed for PEI.39–41 However, we found that for siRNA formulation and delivery investigated here, the molecular weight of PBAEs does not necessarily need to be higher than 8 kDa. Instead, a suitable amphiphilicity may be the determining factor.35
1, and showed the highest luciferase expression.40 In addition, low molecular weight PBAEs (Mw < 8 kDa) only poorly transfect DNA compared with higher molecular weight versions, which is the same trend as observed for PEI.39–41 However, we found that for siRNA formulation and delivery investigated here, the molecular weight of PBAEs does not necessarily need to be higher than 8 kDa. Instead, a suitable amphiphilicity may be the determining factor.35
P(SpDBAE) polyplexes mediated efficient gene silencing at N/P 5 (polymer/siRNA weight ratio 3.5). Compared with polyplexes prepared from cross-linked PEI 800, which achieved gene silencing efficiency of 46% at N/P 115.05,42 and another PBAE polyplexes with 92% GFP knockdown at polymer/siRNA weight ratio 445,43 the N/P ratio of our polyplexes is rather low, and gene silencing is highly efficient. Low N/P ratios are expected to cause less cytotoxicity or side effects when applied in vivo. In the case of intravenous administration, polyplexes encounter a complex environment with subsequent interaction with hundreds of serum proteins. Negatively charged proteins may attach to the polyplexes and form the so-called “protein corona”, which can alter the endocytic pathway of the polyplexes and lower the transfection efficiency due to the reduced endo/lysosomal escape.44,45 P(SpDBAE) polyplexes can be effectively applied in complete cell culture medium regardless of the presence of proteins in serum. Although the serum concentration in cell culture is lower than the serum concentration in full blood, our results still indicate the potential application of the polyplexes in vivo.
Conclusions
Safe and efficient transfection of siRNA has remained a challenge for many years. In this study, we successfully synthesized a novel poly(β-amino ester) composed of decylamine, spermine and 1,4-butanediol diacrylate. The free polymer has higher biocompatibility than the commonly used cationic polymer PEI, and its polyplexes achieved a high cellular uptake and around 90% GFP knockdown at low N/P ratios in complete cell culture medium. These results suggest that this new PBAE P(SpDBAE) is very promising for siRNA delivery and it could be one of the most efficient PBAEs for siRNA delivery reported so far.Author contributions
Conceptualization, Yao Jin, Friederike Adams, and Olivia M. Merkel; methodology and investigation, Yao Jin and Friederike Adams; TEM characterisation, Sebastian Sturm and Müller-Caspary; original manuscript preparation, Yao Jin; review and editing, Friederike Adams and Olivia. M. Merkel; funding acquisition, Olivia M. Merkel.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Yao Jin appreciates the financial support from China Scholarship Council (CSC201906010329). This project was funded by ERC-2014-StG – 637830 to Olivia Merkel.References
- J. Valencia-Serna, C. Kucharski, M. Chen, R. Kc, X. Jiang, J. Brandwein and H. Uludag, J. Controlled Release, 2019, 310, 141–154 CrossRef CAS PubMed.
- A. Mehta, E. Dalle Vedove, L. Isert and O. M. Merkel, Pharm. Res., 2019, 36, 133 CrossRef PubMed.
- M. Z. Wang, J. Niu, H. J. Ma, H. A. Dad, H. T. Shao, T. J. Yuan and L. H. Peng, J. Controlled Release, 2020, 322, 95–107 CrossRef CAS PubMed.
- H. Uludağ, K. Parent, H. M. Aliabadi and A. Haddadi, Front. Bioeng. Biotechnol., 2020, 8, 916 CrossRef PubMed.
- A. Mehta, T. Michler and O. M. Merkel, Adv. Healthcare Mater., 2021, 10, e2001650 CrossRef PubMed.
- C. M. O'Driscoll, A. Bernkop-Schnürch, J. D. Friedl, V. Préat and V. Jannin, Eur. J. Pharm. Sci., 2019, 133, 190–204 CrossRef PubMed.
- M. I. Sajid, M. Moazzam, S. Kato, K. Yeseom Cho and R. K. Tiwari, Pharmaceuticals, 2020, 13, 294 CrossRef CAS PubMed.
- M. Dominska and D. M. Dykxhoorn, J. Cell Sci., 2010, 123, 1183–1189 CrossRef CAS PubMed.
- G. Cavallaro, C. Sardo, E. F. Craparo, B. Porsio and G. Giammona, Int. J. Pharm., 2017, 525, 313–333 CrossRef CAS PubMed.
- J. J. Green, R. Langer and D. G. Anderson, Acc. Chem. Res., 2008, 41, 749–759 CrossRef CAS PubMed.
- D. M. Lynn and R. Langer, J. Am. Chem. Soc., 2000, 122, 10761–10768 CrossRef CAS.
- X. Wang, Q. Liang, Y. Mao, R. Zhang, Q. Deng, Y. Chen, R. Zhu, S. Duan and L. Yin, Biomater. Sci., 2020, 8, 3856–3870 RSC.
- K. L. Kozielski, S. Y. Tzeng, B. A. Hurtado De Mendoza and J. J. Green, ACS Nano, 2014, 8, 3232–3241 CrossRef CAS PubMed.
- S. Liu, Y. Gao, A. Sigen, D. Zhou, U. Greiser, T. Guo, R. Guo and W. Wang, ACS Biomater. Sci. Eng., 2016, 3, 1283–1286 CrossRef PubMed.
- A. R. Lote, V. R. Kolhatkar, T. Insley, P. Král and R. Kolhatkar, ACS Macro Lett., 2014, 3, 829–833 CrossRef CAS PubMed.
- D. Jere, T. H. Kim, R. B. Arote, H. L. Jiang, M. H. Cho, J. W. Nah and C. S. Cho, Pharm. Res., 2007, 25, 875–885 CrossRef PubMed.
- H. Eliyahu, S. Siani, T. Azzam, A. J. Domb and Y. Barenholz, Biomaterials, 2006, 27, 1646–1655 CrossRef CAS PubMed.
- M. Elsayed, V. Corrand, V. Kolhatkar, Y. Xie, N. H. Kim, R. Kolhatkar and O. M. Merkel, Biomacromolecules, 2014, 15, 1299–1310 CrossRef CAS PubMed.
- J. G. Hardy, M. A. Kostiainen, D. K. Smith, N. P. Gabrielson and D. W. Pack, Bioconjugate Chem., 2006, 17, 172–178 CrossRef CAS PubMed.
- M. S. Shim and Y. J. Kwon, Biomaterials, 2011, 32, 4009–4020 CrossRef CAS PubMed.
- S.-Y. Duan, X.-M. Ge, N. Lu, F. Wu, W. Yuan and T. Jin, Int. J. Nanomed., 2012, 7, 3813 CAS.
- X. Liu, Z. Zhao, F. Wu, Y. Chen and L. Yin, Adv. Mater., 2022, 34, 2108116 CrossRef CAS PubMed.
- V. Nadithe, R. Liu, B. A. Killinger, S. Movassaghian, N. H. Kim, A. B. Moszczynska, K. S. Masters, S. H. Gellman and O. M. Merkel, Mol. Pharmaceutics, 2015, 12, 362–374 CrossRef CAS PubMed.
- I. S. Blagbrough and A. J. Geall, Tetrahedron Lett., 1998, 39, 439–442 CrossRef CAS.
- S. K. M. Nalluri, J. Voskuhl, J. B. Bultema, E. J. Boekema and B. J. Ravoo, Angew. Chem., Int. Ed., 2011, 50, 9747–9751 CrossRef CAS PubMed.
- A. Akinc, D. M. Lynn, D. G. Anderson and R. Langer, J. Am. Chem. Soc., 2003, 125, 5316–5323 CrossRef CAS PubMed.
- J. Chen, S.-W. Huang, M. Liu and R.-X. Zhuo, Polymer, 2007, 48, 675–681 CrossRef CAS.
- T. Lebhardt, S. Roesler, M. Beck-Broichsitter and T. Kissel, J. Drug Delivery Sci. Technol., 2010, 20, 171–180 CrossRef CAS.
- H.-L. Jiang, S.-H. Hong, Y.-K. Kim, M. A. Islam, H.-J. Kim, Y.-J. Choi, J.-W. Nah, K.-H. Lee, K.-W. Han and C. Chae, Int. J. Pharm., 2011, 420, 256–265 CrossRef CAS PubMed.
- R. S. Tuma, M. P. Beaudet, X. Jin, L. J. Jones, C.-Y. Cheung, S. Yue and V. L. Singer, Anal. Biochem., 1999, 268, 278–288 CrossRef CAS PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- Y. Liu, Y. Li, D. Keskin and L. Shi, Adv. Healthcare Mater., 2019, 8, e1801359 Search PubMed.
- S. J. Gwak, C. Macks, D. U. Jeong, M. Kindy, M. Lynn, K. Webb and J. S. Lee, Biomaterials, 2017, 121, 155–166 CrossRef CAS PubMed.
- M. Zheng, Y. Zhong, F. Meng, R. Peng and Z. Zhong, Mol. Pharmaceutics, 2011, 8, 2434–2443 CrossRef CAS PubMed.
- P. Dosta, V. Ramos and S. Borrós, Mol. Syst. Des. Eng., 2018, 3, 677–689 RSC.
- L. Parhamifar, A. K. Larsen, A. C. Hunter, T. L. Andresen and S. M. Moghimi, Soft Matter, 2010, 6, 4001–4009 RSC.
- E. J. Kwon, J. M. Bergen and S. H. Pun, Bioconjugate Chem., 2008, 19, 920–927 CrossRef CAS PubMed.
- D. G. Anderson, D. M. Lynn and R. Langer, Angew. Chem., Int. Ed., 2003, 42, 3153–3158 CrossRef CAS PubMed.
- D. G. Anderson, A. Akinc, N. Hossain and R. Langer, Mol. Ther., 2005, 11, 426–434 CrossRef CAS PubMed.
- A. Akinc, D. G. Anderson, D. M. Lynn and R. Langer, Bioconjugate Chem., 2003, 14, 979–988 CrossRef CAS PubMed.
- S. Choosakoonkriang, B. A. Lobo, G. S. Koe, J. G. Koe and C. R. Middaugh, J. Pharmaceut. Sci., 2003, 92, 1710–1722 CrossRef CAS PubMed.
- X. Ge, J. Feng, S. Chen, C. Zhang, Y. Ouyang, Z. Liu and W. Yuan, J. Nanobiotechnol., 2014, 12, 1–8 CrossRef PubMed.
- K. L. Kozielski, S. Y. Tzeng and J. J. Green, Chem. Commun., 2013, 49, 5319–5321 RSC.
- K. E. Wheeler, A. J. Chetwynd, K. M. Fahy, B. S. Hong, J. A. Tochihuitl, L. A. Foster and I. Lynch, Nat. Nanotechnol., 2021, 16, 617–629 CrossRef CAS PubMed.
- D. Zhu, H. Yan, Z. Zhou, J. Tang, X. Liu, R. Hartmann, W. J. Parak, N. Feliu and Y. Shen, Biomater. Sci., 2018, 6, 1800–1817 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00272a |
| ‡ Present address: Chair of Macromolecular Materials and Fiber Chemistry, Institute of Polymer Chemistry, University of Stuttgart, Pfaffenwaldring 55, 70569 Stuttgart, Germany and University Eye Hospital Tübingen, Center for Ophthalmology, Elfriede-Aulhorn-Straße 7, 72076 Tübingen, Germany. |
| This journal is © The Royal Society of Chemistry 2023 |