Open Access Article
Open Access ArticleCharacterizing polyproline II conformational change of collagen superhelix unit on adsorption on gold surface†
Yuntao
Li
a,
Jinrong
Yang
 *a and
Xiao
He
*a and
Xiao
He
 *ab
*ab
aShanghai Engineering Research Center of Molecular Therapeutics and New Drug Development, Shanghai Frontiers Science Center of Molecule Intelligent Syntheses, School of Chemistry and Molecular Engineering, East China Normal University, Shanghai, 200062, China. E-mail: jryang@chem.ecnu.edu.cn
bNew York University–East China Normal University Center for Computational Chemistry, New York University Shanghai, Shanghai, 200062, China. E-mail: xiaohe@phy.ecnu.edu.cn
First published on 22nd August 2023
Abstract
The dynamic process of protein binding onto a metal surface is a frequent occurrence as gold nanoparticles are increasingly being used in biomedical applications, including wound treatment and drug transport. Collagen, as a major component of the extracellular matrix, has potentially advantageous biomedical applications, due to its excellent biocompatibility and elasticity properties. Therefore, a mechanistic comprehension of how and which species in collagen interact with gold nanoparticles is a prerequisite for collagen–gold complexes in clinical application. However, the dynamic behavior of collagen with the polyproline II (PPII) conformation on gold sheets at the molecular level is too complex to capture under current experimental conditions. Here, using molecular dynamics simulations, we investigate the adsorption process and conformational behavior of the tripeptide Gly–Pro–Hyp with the repetitive unit of the collagen superhelix on the gold surface as a function of number of repeating units from 1 to 10. The different numbers of repeating units all prefer to approach the gold surface and adsorb via charged residues at the C-terminal or N-terminal ends, tending to form arch structures on the gold surface. Compared with the various tripeptide units in solution still retaining the native PPII conformation, the presence of the gold surface affects the formation of hydrogen bonds between the protein and water molecules, thus disrupting the PPII conformation of collagen. Specifically, the interaction between the gold surface and HYP limits the rotation of the dihedral angle of collagen, resulting in a tendency for the PPII conformation of the gold surface to transform to the β-sheet conformation. The results provide an indication of how to improve the interaction between the terminal groups and the gold surface for the design of a bioavailable protein–gold material for medicinal purposes.
Introduction
Adsorption processes between proteins and inorganic solid surfaces are a common phenomenon when nanoparticles are applied for biology, materials science, and industrially. Gold nanoparticles, due to their good biocompatibility, ease of synthesis, and shape control,1,2 have been extensively investigated for various biomedical applications, such as antibiotics,3 drug delivery4,5 wound treatment,1 and clinical diagnostics.6 In particular, gold nanoparticles can be used as biosensors to detect the presence and concentration of biomolecules by interacting with specific proteins. In addition, gold nanoparticles can be used to enhance the sensitivity and specificity of certain clinical diagnostics. The interaction of gold nanoparticles with proteins can also help to develop more precise drug delivery systems, as well as for the treatment of specific types of cancer and other diseases. Therefore, it is essential to investigate thoroughly how gold nanoparticles interact with components of the biological environment, but mainly with proteins.The interaction of proteins with gold surfaces is complex, involving different molecular forces, such as the solid–water interface and the protein–solid surface, leading to the investigation of the molecular mechanisms still being at a preliminary stage. Since Vroman's7 prominent research on the adsorption of proteins and peptides on liquid or solid surfaces in the 1960s, numerous experimental studies have been carried out.8–10 Protein adsorption at such interfaces is inevitable and largely irreversible, except in limited cases. Thereafter, various protein adsorption models were proposed to elucidate the molecular forces that determine the interaction, including steric repulsion,11 hydration theory12,13 and others to explain common protein adsorption phenomena such as structural rearrangement and protein aggregation, but debates surrounding these models still continue.14 However, owing to limitations caused by the scale of a protein, experimental studies cannot provide further information at the molecular level and the explanation of the protein adsorption mechanism remains unclear.15–18
Due to the limitations of experimental techniques, computational methods have become widely used to investigate the adsorption of proteins and peptides on solid surfaces.19 All-atom molecular dynamics simulations have become the primary tool to describe the interaction of solid surfaces with proteins,20–22 peptides8,9,14,23,24 and amino acids7,10,25,26 due to their balance between accuracy and computational feasibility. Earlier studies focused on the study of peptide adsorption states on solid surfaces. Penna et al.27 started to focus on the process of proteins or peptides as they move away from the interface to the adsorbed state, proposing a three-phase adsorption mechanism and, by simulating peptides of over 240 length, proposed a generic peptide adsorption mechanism at the molecular level. This has provided theoretical support for subsequent studies on the adsorption of proteins onto solid surfaces at the molecular level.
Collagen is the most common protein in the extracellular matrix and is widely used in cancer treatment28 and tissue engineering;29–31 therefore, investigation of the adsorption mechanism and the conformational changes of collagen on a gold surface is particularly important. Tang et al.32,33 investigated the adsorption mechanism of the human type I collagen triple helix (CMS) on a gold surface by molecular dynamics simulation and explored the adsorption conformation of collagen on surfaces and nanoparticles, which revealed the unfolding mechanism of the collagen triple helix on the gold surface at the atomic level. However, there is a question of whether this adsorption process is also applicable to single-chain collagen. Furthermore, the adsorption conformation of different chain lengths of collagen on gold surfaces needs to be further investigated.
In this research, we aim to investigate the interaction between collagen and gold surfaces at a smaller scale by constructing peptide chains containing 1–10 PRO–HYP–PRO (PPG) units and investigating the adsorption process and conformational changes of collagen adsorption on gold surfaces and in water solutions, respectively. The results show that the different chain lengths of the PPG peptide chains follow a three-step adsorption process with biased adsorption, anchoring, and lockdown; furthermore, the structural restriction of the gold surface to the collagen becomes stronger as the chain length increases, which provides relevant data to support the application of collagen in biomedical applications.
Computational models and methodology
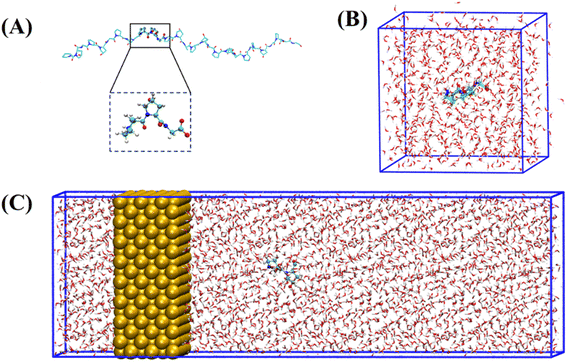
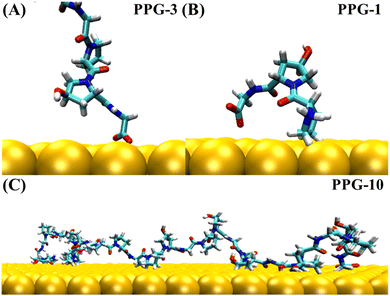
The collagen model was downloaded from the RCSB Protein Data Bank (PDB ID: 1CAG).34 This peptide has a triple helix structure composed of three peptide chains, where each peptide chain consists of ten Pro–Hyp–Gly (PPG) structures (Fig. 1(A)). On this basis, we constructed structures containing one to ten PPG repeat units to investigate the effect of chain length on the adsorption of PPG on the gold surface. In previous studies, the adsorption of collagen-like proteins on titanium-based material surfaces35 and single-walled carbon nanotubes36 has been investigated. The gold surfaces of the 6-atom-thick face-centered cubic (1,0,0) surfaces were constructed. The size of the gold surfaces depends on the length of the PPG cells, ensuring that they interact adequately (Table 1).| Model information | Gold surface system | Water system |
|---|---|---|
| Water box dimension (Å × Å × Å) | 150 × 65 × 65 | 48 × 48 × 48 |
| Number of gold atoms | 3072 | Null |
| Number of water molecules | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 533 533 |
3587 |
| Water box type | Triclinic | Triclinic |
To investigate the molecular behavior of PPG structures of different lengths on gold surfaces, molecular dynamics simulations of PPG structures on gold surfaces were performed, as shown in Fig. 1(C). For comparison, molecular dynamics simulations were also carried out in explicit solvents for different chain lengths in Fig. 1(B). The details of the gold atoms and water molecules for these two species systems are shown in Table S1.† In each system, we added a sufficient amount of TIP3P water molecules to ensure that the system has a distance of 10 Å from the boundary. During the simulation, the density of water in each system remained around 1.0 g cm−3. To investigate the adsorption behavior of different lengths of PPG on the gold surface, three molecular dynamics simulations were carried out for the presence and absence of gold surfaces, and each started with the same structure without initial atomic velocity. All molecular dynamics simulations were performed using GROMACS version 2021.3 (ref. 37) and the AMBER FF99SB38 force field parameter sets. We used the interfacial force fields developed by Heinz et al.39 as the parameters for the gold–solution interactions. In this force field the equilibrium nonbonding distance and the equilibrium nonbonding energy are r0 = 2.951 Å and ε0 = 5.29 kcal mol−1,39 respectively. In order to eliminate too close atomic contacts, all systems were simulated separately with 5000 steps of energy minimization using the steepest descent method. This was followed by 100 picoseconds of equilibrium simulations under NVT and NPT ensembles, in order to equilibrate the solvent and the whole system. Finally, molecular dynamics simulations were performed for 200 ns under the NPT ensemble using 2 femtosecond time steps for integration calculations at a temperature of 300 K and a pressure of one atmosphere. The temperature and pressure controllers used were a velocity rescaling (V-rescale) thermostat and Parrinello–Rahman, respectively. The LINCS algorithm was used to constrain the bonds including hydrogen atoms. A standard cutoff point of 1.0 nm was chosen for neighborhood generation as well as for Coulomb and Leonard-Jones interactions. The long-range electrostatic interactions were calculated using the particle mesh Ewald (PME) method. The time step was 2 femtoseconds and the trajectory coordinates were saved every 10 picoseconds for subsequent analysis. Trajectories were processed and further analyzed using GROMACS37 software. VMD40 software was used for trajectory visualization.
Results and discussion
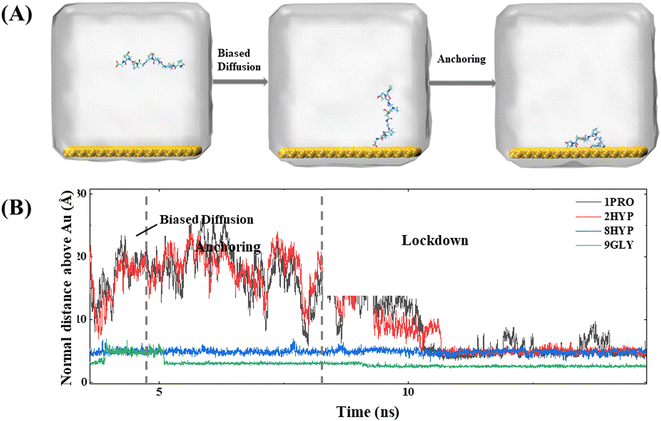
In simulations of collagen with different chain lengths, proteins containing different numbers of PPG units all follow the three steps proposed by Penna et al. for the adsorption of short peptides on hydrophilic surfaces:27 (1) PPG diffuses in a biased way from the bulk phase towards the water–gold surface; (2) PPG is anchored at the interface by forming hydrogen bonds with the water layer on the metal surface or by direct hydrophobic interactions with the metal surface; (3) PPG residues progressively displace water molecules from the gold surface and are adsorbed on the gold surface. For further explanation, we take the dynamic trajectory containing three PPG unit as an example to be illustrated in Fig. 2.The initial distance on the polypeptide closest to the gold surface is set as approximately 30 Å larger than the interaction cutoff range between the peptide and the solid surface. At the beginning of the simulation, the polypeptide adapts its conformation and fluctuates up and down over the gold surface until the C-terminal residue approaches the gold surface, which allows the distance between the polypeptide and the gold surface to be smaller than the cutoff range for the interaction between the peptide and the solid surface for dispersion. At approximately 2 ns from the beginning of the simulation, the polypeptide starts to approach the metal surface rapidly, which is observed in all simulations. The average force potential (PMF) of PPG is shown in Fig. S1,† indicating that the system is in a high energy state close to the gold surface. In contrast, the polypeptide in the water solution is consistently fluctuating, which indicates that biased diffusion occurs only in the presence of gold surfaces. This suggests that there appears to be a longer range of forces at play between the surface and the peptide. Due to the directional ordering of water near the solid surface, the solid surface is covered with a charged water layer, which allows long-range electrostatic interactions between the peptide and the solid surface to operate.
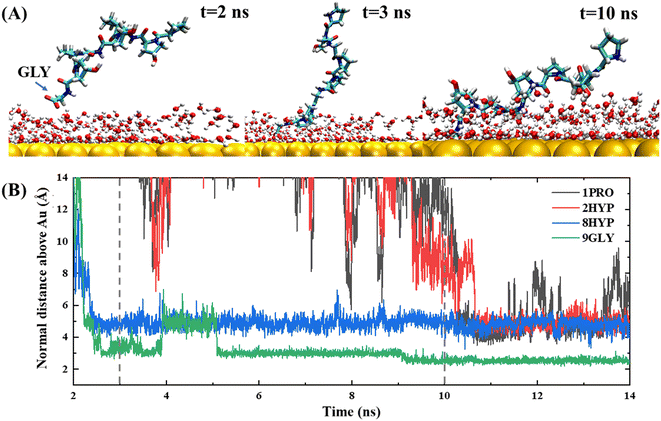
The anchoring phase is the process by which the polypeptide interacts with the water on the gold surface and progressively enters the second water layer at the interface. Although collagen experiences fluctuations in water at distances far away from the gold surface during the initial diffusion phase, the subsequent simulation demonstrates that the C-terminal carboxyl group of the polypeptide is able to anchor to the gold surface, thereby forming a stable anchoring pattern. This suggests that the traction provided by the carboxyl group is a critical factor in the polypeptide's approach to the gold surface, consistent with the work of Hoefling and co-workers.41,42 The anchoring process of PPG on the gold surface is reversible, as illustrated by the exemplary trajectory in Fig. 2(B), where residue 3GLY entered the second water layer at about 2.5 ns, followed by drifting and re-anchoring 3 times before entering the second water layer.
The hydrophilic groups and their interactions with interfacial water play a key role in the anchoring phase of the peptide.27 In simulations of PPG for different chain lengths, all show that the C-terminal region initiated the first anchoring event of the polypeptide, which demonstrates that the charged terminal groups play an important role in anchoring. As the collagen molecule peptide is slightly bent in water, some of the polar groups also undergo anchoring events during subsequent simulations. The carbon terminal can quickly pass through the second water layer into the first water layer as a consequence of the directional ordering of adjacent water on the solid surface, which results in a charged layer on the solid surface, thereby allowing long-range electrostatic interactions between the peptide and the gold surface to function. Residues 8HYP and 7PRO near the C-terminus approach the second water layer under the pull of GLY by the C-terminus. Interestingly, HYP is anchored steadily at the interface after approaching the second water layer and remains without leaving, which is related to the formation of hydrogen bonds between HYP and the surface-bound water layer, as illustrated in Fig. 3. Interaction of the hydrophilic group with the intermediate water indicates that the hydroxyl group also serves as an anchor and can be anchored to the gold surface. Afterwards, PRO enters the second water layer with the pulling action of two other amino acids. In comparison, PRO drifts at about 1 ns near the interface and finally enters the second water layer after three anchoring events.
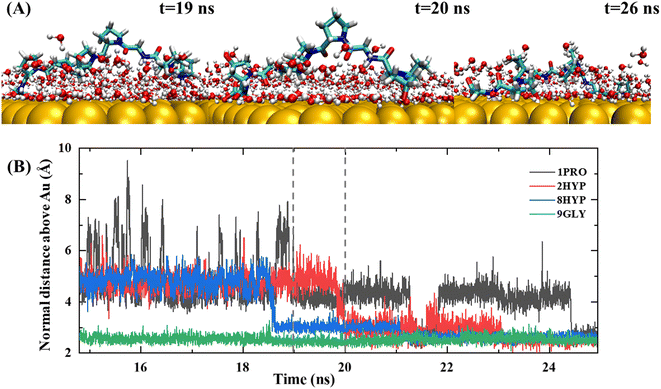
The lockdown phase forms when the anchored peptides are gradually rearranged into the second water layer on the gold surface and then eventually into the water layer immediately adjacent to the solid surface, whereby the peptide is adsorbed onto the gold surface. Insertion of the peptide group into the second water layer of the gold surface is rapid; however, the insertion of the peptide group into the first water layer close to the gold surface is far slower. The anchored collagen has to restructure itself and then wait until it is able to insert groups into the first intimately bound water layer of the gold surface. The anchoring process is generally gradual and protracted, typically lasting more than 10 ns, as shown by the trajectory in Fig. 4. The carbon terminal of the PPG first approaches the gold surface and is anchored to the interface at approximately 5 ns. After a further 5 ns of simulation, the center of mass of the PPG approaches the gold surface, showing that the entire PPG has been adsorbed onto the gold surface.
Penna and co-workers27 showed that after a peptide group has been adsorbed on a gold surface, the probability of adsorption of subsequent groups is related exponentially to their distance from the initially adsorbed group. Nevertheless, our trajectory demonstrated a different result. Fig. 4 shows an exemplary trajectory of a peptide containing three PPG units on a gold surface, where the C-terminal carboxyl group initiates the adsorption onto the gold surface first, followed by the adsorption of the entire C-terminal GLY onto the gold surface and then after 15 ns of simulation residue 8HYP is adsorbed onto the gold surface. Next, residue 2HYP at the N-terminus is adsorbed at approximately 20 ns, and residue 1PRO is finally adsorbed on the gold surface after another 10 ns of simulation. This is consistent with the simulation results of Tang et al.32 for a three-chain collagen-like peptide, determined by the slightly bent conformation of the collagen peptide in water. Consequently, the order of adsorption of polypeptide groups on the gold surface is related not only to the distance of the initially adsorbed groups, but also to the structural properties of the polypeptide and the protein–surface contact pattern.
The trajectories of collagen containing between 1 and 10 PPG units on the gold surface show that PPG polypeptides of different chain lengths all follow a three-step adsorption process; representative snapshots of each dynamic trajectory are shown in Fig. S2–S11.† Both the C-terminal and N-terminal ends are likely to act first as anchors into the second and first water layers of the gold surface and lock onto the gold surface, eventually forming an arch structure on the gold surface. The statistics for the number of anchoring occurrences at the C-terminal and the N-terminal of the 30 trajectories simulated on the gold surface are shown in Table S2.† As the chain length grows, residues in intermediate positions also interact with the gold surface to form multiple arch structures, as illustrated in Fig. 5(C).
After conducting molecular dynamics simulations of various chain lengths for 200 nanoseconds, we observe a clear interaction between the gold surface and all PPG molecules. To gain further insight into the impact of the gold surface on the structure of PPG, we examine the conformational changes of PPG units of different chain lengths during simulations both in the presence and the absence of the gold surface. We analyzed the geometrical parameters of the protein, including Ramachandran angle and mean radius, to identify conformational changes. Our analysis reveals that the Au(1,0,0) surface interacts with PPG, thereby inhibiting the formation of the PPII structure. This phenomenon raises concerns about the potential risks of using bare gold in therapeutic and biological applications.
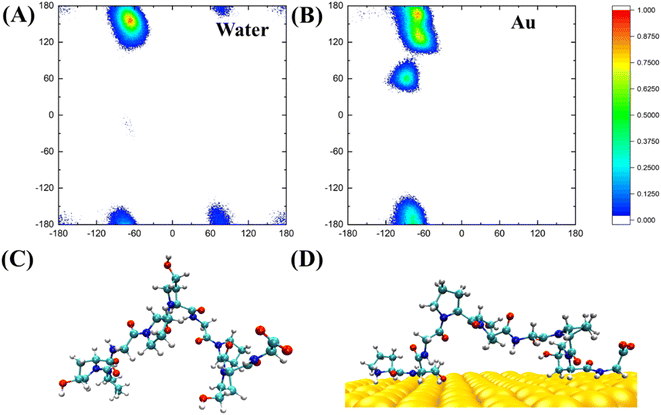
PPG-type collagen peptides have a typical polyproline II secondary structure in which the dihedral angle of the backbone of sequential residues is roughly (−75, 150°). The polyproline II secondary structure of PPG is essential for collagen to form a triple helix structure. Consequently, investigating the Ramachandran plot of PPG units of different lengths contributes to a better understanding of the effect of the gold surface on the PPG structure. Fig. 6 shows the Ramachandran plot (φ, ψ) and representative structures for PPG-3 in two systems. The trajectories used are averaged from the last 50 ns of simulated trajectories for the two systems of gold presence and gold absence. As can be seen from the plot, the Ramachandran plot for the PPG residues in the water solution is concentrated mainly in the polyproline II region, except for the terminal residues; however, the PPG residues on the gold surface have mostly deviated from the position expected for the formation of polyproline II. This suggests that the presence of the gold surface disrupts the important PPII structure required to form the triple helix structure of collagen, which in turn allows the polypeptide to unpeel from the gold surface.
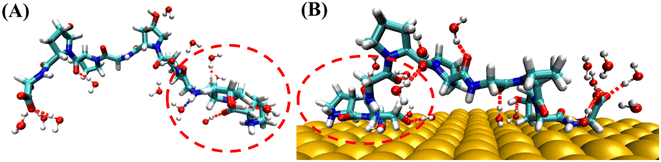
Hydrogen bonding analysis between PPG and water molecules on the gold surface and in water solution was conducted, which showed that the number of hydrogen bonds between PPG and water molecules on the gold surface is significantly less than that between PPG and water molecules in the water solution. As shown in Fig. 7, the number of hydrogen bonds formed between the N-terminal amino acids of PPG-3 and water molecules in water solution is about 4; however, it is reduced to 2 on the gold surface due to the interaction of PPG with the gold surface, which disrupts the hydrogen bonding sites between PPG and water molecules, consequently changing the structure of the polypeptide. This is also demonstrated by the RDF of water molecules with the gold surface, as shown in Fig. S12.†
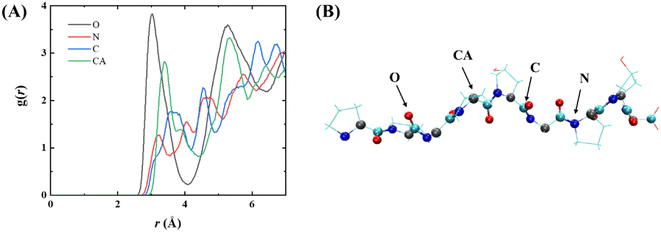
To explore the interaction of PPG with the gold surface at the atomic level, a radial distribution function (RDF) of the gold surface with the main chain atoms of PPG was investigated, as illustrated in Fig. 8, showing the RDF for PPG-3; RDF plots for the other chains are shown in Fig. S13.† Each of the main-chain atoms has a distinct peak in the RDF. The first peak for all main-chain atoms is less than 3.5 Å, indicating that the polypeptide interacts directly with the gold surface after 200 ns of dynamic simulation. Furthermore, the peak of the RDFs for the carbonyl oxygen in Fig. 8 is sharper and higher than the peaks for the other main-chain atoms. This suggests that the gold surface forms a stronger interaction with the carbonyl oxygen of the adsorbed polypeptide than with the other main-chain atoms. This also demonstrates the important role of the carbonyl oxygen in the adsorption process. In conclusion, the gold surface interacts directly with the main-chain atoms of the PPG, which limits the formation of hydrogen bonds between them and water molecules, leading to an apparent unfolding of the triple helix structure of the PPG.
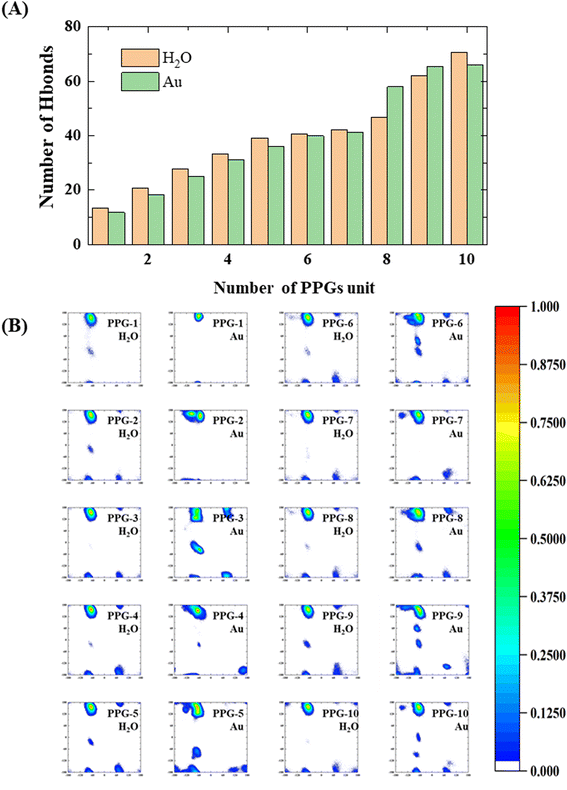
During the analysis performed on the structures of collagen peptides with different chain lengths on the gold surface and in water solution, we find that the number of hydrogen bonds between the collagen and the water molecules on the gold surface is significantly less than in water solution, with significant implications for the stability and conformation of the collagen in this environment, as shown in Fig. 9(A). Interestingly, the results also show that for peptides containing 8 or 9 PPG units, the folding of collagen in water solution results in the opposite situation where the number of hydrogen bonding sites to water molecules decreases as the chain length grows. This observation may be related to the conformational flexibility of PPG, which allows the peptide chain to adopt a range of conformations depending on the local environment. The findings suggest that the conformational preferences of collagen on gold surfaces may differ from those in water solution. Specifically, shorter collagen chains may be more likely to adopt an extension conformation on gold surfaces, which may expose more hydrophobic residues and reduce the number of hydrogen bonding sites with water molecules. Conversely, longer collagen chains may be more likely to fold and adopt a compact conformation, which may enhance their interactions with water molecules and increase the number of hydrogen bonds. These findings suggest that the water environment plays a key role in determining the conformational behavior of collagen on gold surfaces, while a balance between hydrophobic and hydrophilic interactions is a key factor in driving collagen–gold interactions.
To further investigate the conformational behavior of collagen in water and on the gold surface, a Ramachandran plot analysis was carried out, as shown in Fig. 9(B). From the Ramachandran plots, the collagen on the gold surface is more concentrated in the PPII region at shorter chain lengths, while as the chain length increases, the probability of ψ appearing in the (−60°, 60°) range becomes greater at a φ of approximately −60°, which indicates that the interaction between the gold surface and the collagen leads to a restricted dihedral angle rotation of the PPG as the chain length increases. As a result, the interaction between collagen and the gold surface is highly dependent on chain length and dihedral angle. Shorter chain lengths exhibit greater collagen concentrations in the PPII region, demonstrating unrestricted rotation of the PPG. As chain length increases, the ability of collagen molecules to rotate becomes more restricted, with a higher probability of dihedral angles falling in regions other than the PPII. Furthermore, the trends observed in the Ramachandran plots suggest that the conformation of collagen on the gold surface is influenced not only by chain length, but also by the properties of the interactions between the surface and the polypeptide. Previous research has shown that polypeptide adsorption on surfaces is dictated by a complex interaction of various forces, including van der Waals, electrostatic and hydrophobic interactions. These investigations have implications for the design of collagen-based materials for biomedical applications. The ability to control the conformation and orientation of collagen at the surface may affect the mechanical properties of the material, as well as how it interacts with cells and other biomolecules. Further investigation of the mechanisms underlying this interaction is necessary to fully understand and exploit the potential of collagen as a biomaterial.
Conclusion
By simulating PPG with different chain lengths on the gold surface and in water, we find that for different lengths of collagen polypeptide chains on the gold surface, the adsorption process is consistent with the previously mentioned adsorption mechanism, which is based on three stages: biased adsorption, anchoring and lockdown. The C-terminal and N-terminal ends of the polypeptide are more likely to act as anchors to adsorb on the gold surface at first. This is caused by the charged layer formed by the orientated arrangement of water molecules on the gold surface. Furthermore, we discussed the changes in the adsorption conformation of PPG polypeptides of different chain lengths on the gold surface, demonstrating that the gold surface breaks down the PPII structure of collagen to various degrees. The results obtained may provide theoretical support for the development of protein and inorganic surface interactions for biomedical applications and the development of composites designed for collagen and gold.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was supported by the National Key R&D Program of China (Grant No. 2019YFA0905200), the National Natural Science Foundation of China (Grant No. 22273023, 22203032, and 21922301), Shanghai Municipal Natural Science Foundation (Grant No. 23ZR1418200), Shanghai Frontiers Science Center of Molecule Intelligent Syntheses, and the Fundamental Research Funds for the Central Universities. We thank the Supercomputer Center of East China Normal University (ECNU Multifunctional Platform for Innovation 001) for providing computer resources.References
- Y. Li, Y. Tian, W. Zheng, Y. Feng, R. Huang, J. Shao, R. Tang, P. Wang, Y. Jia, J. Zhang, W. Zheng, G. Yang and X. Jiang, Small, 2017, 13, 1700130 CrossRef PubMed.
- R. Shukla, V. Bansal, M. Chaudhary, A. Basu, R. R. Bhonde and M. Sastry, Langmuir, 2005, 21, 10644–10654 CrossRef CAS PubMed.
- Y. Feng, W. Chen, Y. Jia, Y. Tian, Y. Zhao, F. Long, Y. Rui and X. Jiang, Nanoscale, 2016, 8, 13223–13227 RSC.
- Y. Lei, L. Tang, Y. Xie, Y. Xianyu, L. Zhang, P. Wang, Y. Hamada, K. Jiang, W. Zheng and X. Jiang, Nat. Commun., 2017, 8, 15130 CrossRef PubMed.
- S. Deyev, G. Proshkina, A. Ryabova, F. Tavanti, M. C. Menziani, G. Eidelshtein, G. Avishai and A. Kotlyar, Bioconjugate Chem., 2017, 28, 2569–2574 CrossRef CAS PubMed.
- E. K. Lim, T. Kim, S. Paik, S. Haam, Y. M. Huh and K. Lee, Chem. Rev., 2015, 115, 327–394 CrossRef CAS PubMed.
- L. Vroman, Nature, 1962, 196, 476–477 CrossRef CAS PubMed.
- Q. Li, K. H. A. Lau, E. K. Sinner, D. H. Kim and W. Knoll, Langmuir, 2009, 25, 12144–12150 CrossRef CAS PubMed.
- O. Mermut, D. C. Phillips, R. L. York, K. R. McCrea, R. S. Ward and G. A. Somorjai, J. Am. Chem. Soc., 2006, 128, 3598–3607 CrossRef CAS PubMed.
- W. Norde, Adv. Colloid Interface Sci., 1986, 25, 267–340 CrossRef CAS PubMed.
- I. Szleifer, Biophys. J., 1997, 72, 595–612 CrossRef CAS PubMed.
- P. Harder, M. Grunze, R. Dahint, G. M. Whitesides and P. E. Laibinis, J. Phys. Chem. B, 1998, 102, 426–436 CrossRef CAS.
- R. L. C. Wang, H. J. Kreuzer and M. Grunze, J. Phys. Chem. B, 1997, 101, 9767–9773 CrossRef CAS.
- M. Rabe, D. Verdes and S. Seeger, Adv. Colloid Interface Sci., 2011, 162, 87–106 CrossRef CAS PubMed.
- C. Pinholt, R. A. Hartvig, N. J. Medlicott and L. Jorgensen, Expert Opin. Drug Delivery, 2011, 8, 949–964 CrossRef CAS PubMed.
- I. Fenoglio, B. Fubini, E. M. Ghibaudi and F. Turci, Adv. Drug Delivery Rev., 2011, 63, 1186–1209 CrossRef CAS PubMed.
- M. R. Duff and C. V. Kumar, J. Phys. Chem. B, 2009, 113, 15083–15089 CrossRef CAS PubMed.
- J. J. Gray, Curr. Opin. Struct. Biol., 2004, 14, 110–115 CrossRef CAS PubMed.
- M. M. Orosco, C. Pacholski and M. J. Sailor, Nat. Nanotechnol., 2009, 4, 255–258 CrossRef CAS PubMed.
- M. Ozboyaci, D. B. Kokh and R. C. Wade, Phys. Chem. Chem. Phys., 2016, 18, 10191–10200 RSC.
- R. Gopalakrishnan, E. R. Azhagiya Singam, J. Vijaya Sundar and V. Subramanian, Phys. Chem. Chem. Phys., 2015, 17, 5172–5186 RSC.
- M. Hoefling, S. Monti, S. Corni and K. E. Gottschalk, PLoS One, 2011, 6, e20925 CrossRef CAS PubMed.
- H. Chen, L. Yuan, W. Song, Z. K. Wu and D. Li, Prog. Polym. Sci., 2008, 33, 1059–1087 CrossRef CAS.
- M. Morra, J. Biomater. Sci., Polym. Ed., 2000, 11, 547–569 CrossRef CAS PubMed.
- W. Norde, Colloids Surf., B, 2008, 61, 1–9 CrossRef CAS PubMed.
- S. Graslund, P. Nordlund, J. Weigelt, J. Bray, B. M. Hallberg, O. Gileadi, S. Knapp, U. Oppermann, C. Arrowsmith, R. Hui, J. Ming, S. Dhe-Paganon, H. W. Park, A. Savchenko, A. Yee, A. Edwards, R. Vincentelli, C. Cambillau, R. Kim, S. H. Kim, Z. Rao, Y. Shi, T. C. Terwilliger, C. Y. Kim, L. W. Hung, G. S. Waldo, Y. Peleg, S. Albeck, T. Unger, O. Dym, J. Prilusky, J. L. Sussman, R. C. Stevens, S. A. Lesley, I. A. Wilson, A. Joachimiak, F. Collart, I. Dementieva, M. I. Donnelly, W. H. Eschenfeldt, Y. Kim, L. Stols, R. Wu, M. Zhou, S. K. Burley, J. S. Emtage, J. M. Sauder, D. Thompson, K. Bain, J. Luz, T. Gheyi, F. Zhang, S. Atwell, S. C. Almo, J. B. Bonanno, A. Fiser, S. Swaminathan, F. W. Studier, M. R. Chance, A. Sali, T. B. Acton, R. Xiao, L. Zhao, L. C. Ma, J. F. Hunt, L. Tong, K. Cunningham, M. Inouye, S. Anderson, H. Janjua, R. Shastry, C. K. Ho, D. Y. Wang, H. Wang, M. Jiang, G. T. Montelione, D. I. Stuart, R. J. Owens, S. Daenke, A. Schutz, U. Heinemann, S. Yokoyama, K. Bussow, K. C. Gunsalus, C. Struct Genomics, M. Architecture Fonction, C. Berkeley Struct Genomics, C. China Struct Genomics, F. Integrated Ctr Struct, C. Israel Struct Proteomics, G. Joint Ctr Struct, G. Midwest Ctr Struct, X. R. C. New York Struct Genomi, N. E. S. G. Consortium, F. Oxford Prot Prod, F. Prot Sample Prod, M. Max Delbruck Ctr Mol, R. S. G. Proteomics and S. Complexes, Nat. Methods, 2008, 5, 135–146 CrossRef PubMed.
- M. J. Penna, M. Mijajlovic and M. J. Biggs, J. Am. Chem. Soc., 2014, 136, 5323–5331 CrossRef CAS PubMed.
- V. Raeesi and W. C. Chan, Nanoscale, 2016, 8, 12524–12530 RSC.
- X. Wang, T. Coradin and C. Helary, Biomater. Sci., 2018, 6, 398–406 RSC.
- J. Yang, X. Yang, L. Wang, W. Zhang, W. Yu, N. Wang, B. Peng, W. Zheng, G. Yang and X. Jiang, Nanoscale, 2017, 9, 13095–13103 RSC.
- S. Udhayakumar, K. G. Shankar, S. Sowndarya and C. Rose, Biomater. Sci., 2017, 5, 1868–1883 RSC.
- M. Tang, N. S. Gandhi, K. Burrage and Y. Gu, Langmuir, 2019, 35, 4435–4444 CrossRef CAS PubMed.
- M. Tang, N. S. Gandhi, K. Burrage and Y. Gu, Phys. Chem. Chem. Phys., 2019, 21, 3701–3711 RSC.
- J. Bella, M. Eaton, B. Brodsky and H. M. Berman, Science, 1994, 266, 75–81 CrossRef CAS PubMed.
- H. Matsuno, A. Yokoyama, F. Watari, M. Uo and T. Kawasaki, Biomaterials, 2001, 22, 1253–1262 CrossRef CAS PubMed.
- R. Gopalakrishnan, K. Balamurugan, E. R. Singam, S. Sundaraman and V. Subramanian, Phys. Chem. Chem. Phys., 2011, 13, 13046–13057 RSC.
- H. J. C. Berendsen, D. van der Spoel and R. van Drunen, Comput. Phys. Commun., 1995, 91, 43–56 CrossRef CAS.
- J. Wang, P. Cieplak and P. A. Kollman, J. Comput. Chem., 2000, 21, 1049–1074 CrossRef CAS.
- H. Heinz, T. J. Lin, R. K. Mishra and F. S. Emami, Langmuir, 2013, 29, 1754–1765 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14(33–38), 27–38 Search PubMed.
- M. Hoefling, F. Iori, S. Corni and K. E. Gottschalk, Langmuir, 2010, 26, 8347–8351 CrossRef CAS PubMed.
- M. Hoefling, F. Iori, S. Corni and K. E. Gottschalk, ChemPhysChem, 2010, 11, 1763–1767 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed information and snapshots of representative trajectories of polypeptide simulation systems containing different PPG units. See DOI: https://doi.org/10.1039/d3na00185g |
| This journal is © The Royal Society of Chemistry 2023 |