Open Access Article
Open Access ArticleOptimization and characterization of miRNA-129-5p-encapsulated poly (lactic-co-glycolic acid) nanoparticles to reprogram activated microglia†
Irina
Kalashnikova
 a,
Heather
Cambell
a,
Daniel
Kolpek
a,
Heather
Cambell
a,
Daniel
Kolpek
 a and
Jonghyuck
Park
a and
Jonghyuck
Park
 *ab
*ab
aDepartment of Pharmaceutical Sciences, College of Pharmacy, University of Kentucky, 789 S. Limestone, Lexington, KY 40506, USA. E-mail: jonghyuck.park@uky.edu; Tel: +1-859-257-1850
bSpinal Cord and Brain Injury Research Center, College of Medicine, University of Kentucky, Lexington, KY, USA
First published on 5th May 2023
Abstract
Microglia have become a therapeutic target of many inflammation-mediated diseases in the central nervous system (CNS). Recently, microRNA (miRNA) has been proposed as an important regulator of immune responses. Specifically, miRNA-129-5p has been shown to play critical roles in the regulation of microglia activation. We have demonstrated that biodegradable poly (lactic-co-glycolic acid) (PLGA)-based nanoparticles (NPs) modulated innate immune cells and limited neuroinflammation after injury to the CNS. In this study, we optimized and characterized PLGA-based NPs for miRNA-129-5p delivery to utilize their synergistic immunomodulatory features for activated microglia modulation. A series of nanoformulations employing multiple excipients including epigallocatechin gallate (EGCG), spermidine (Sp), or polyethyleneimine (PEI) for miRNA-129-5p complexation and miRNA-129-5p conjugation to PLGA (PLGA-miR) were utilized. We characterized a total of six nanoformulations through physicochemical, biochemical, and molecular biological methods. In addition, we investigated the immunomodulatory effects of multiple nanoformulations. The data indicated that the immunomodulatory effects of nanoformulation, PLGA-miR with the excipient Sp (PLGA-miR+Sp) and PEI (PLGA-miR+PEI) were significant compared to other nanoformulations including naked PLGA-based NP. These nanoformulations promoted a sustained release of miRNA-129-5p and polarization of activated microglia into a more pro-regenerative phenotype. Moreover, they enhanced the expression of multiple regeneration-associated factors, while alleviating the expression of pro-inflammatory factors. Collectively, the proposed nanoformulations in this study highlight the promising therapeutic tools for synergistic immunomodulatory effects between PLGA-based NPs and miRNA-129-5p to modulate activated microglia which will have numerous applications for inflammation-derived diseases.
1 Introduction
The innate immune cells microglia reside in the central nervous system (CNS) parenchyma and regulate potent inflammatory responses.1,2 Microglia become activated following exposure to pathogen-associated molecular patterns (PAMPs) and/or endogenous damage-associated molecular patterns (DAMPs) after injury to the CNS.2–4 Similar to macrophages/activated monocytes, microglia develop different phenotypes depending on cues in the cells' surrounding environment. While their phenotypes are not binary, microglia can be polarized into distinct functional phenotypes as ranging from classical pro-inflammatory M1 to alternative pro-regenerative M2.5–8 Thus, microglia may exert diverging roles depending on the context of microenvironmental events and signals. Microglia can become activated within minutes after nerve injury, then subsequently recruit neutrophils and force monocyte-derived macrophages (MDMs) to infiltrate into the disease or injury site.9–12 Activated microglia begin to release various factors involved in neuroinflammation, such as cytokines, chemokines, growth factors, reactive oxygen species (ROS), reactive nitrogen species (RNS), and prostaglandins, all of which can magnify inflammatory responses, thereby influencing astrocytes, oligodendrocytes, neurons, and progressive demyelination. Thus, microglia activation is a hallmark of multiple inflammation-mediated diseases on the CNS including multiple sclerosis (MS), Alzheimer's disease, Parkinson's disease, West Nile Virus, and neuropathic pain after spinal cord injury (SCI).7,8,13–15 Therefore, repression of microglial activation may have therapeutic potential for inflammation-derived neuronal deficits with the intent of resolving neuroinflammation. Pharmacologic therapeutic approaches targeting microglia employing small molecules, antioxidants, nanoparticles (NPs), and stem cells may be useful to reprogram activated microglia, yet insufficient data is available to establish guidelines.16–19The microRNA (miRNA) is a type of conserved non-coding RNA which controls the expression of approximately 90% of protein-coded genes.20 Accumulated evidence has demonstrated that dysregulation of miRNA has a significant impact on the regulation of inflammatory responses.21–23 The miRNA-based therapeutic strategies targeting multiple genes result in a broader specific response.21,24 Particularly, mimic miRNA-129-5p (miR-129) has received significant attention as it can suppress cell apoptosis and microglia-induced inflammatory response as well as polarize their phenotype through high-mobility group protein B1 (HMGB1)/toll-like receptor4 (TLR4)/NF-κB and leucine-rich alpha-2-glycoprotein (LRG1)/p38 mitogen-activated protein kinases (MAPK) pathways.25–28 As a well-known maker of inflammatory mediators and cancer therapy, HMGB1 is a direct target of miR-129.29–31 Moreover, HMGB1 is passively released by necrotic and/or stressed cells as well as by immune cells including activated microglia.26–28
Poly (lactic-co-glycolic acid) (PLGA)-based NPs-mediated delivery of miRNA indicated better efficacy on safe miRNA delivery.32–34 Moreover, previous studies demonstrated that the physicochemical properties of PLGA-based NPs influence immune cells reprogramming, specifically targeting pro-inflammatory subsets of immune cells.35–38 These NPs are made of Food and Drug Administration (FDA)-approved material, which is stable at room temperature and can be a widely utilized carrier for drugs, as well as gene transmission. Critically, PLGA-based NPs modulate innate immune cells without the use of active pharmaceutical ingredients that can lead to off-target effects in various inflammation-mediated diseases including West Nile Virus, encephalitis, and SCI.37–40
Although PLGA-based NP for miRNA delivery is promising, they exhibited a burst release with poor encapsulation efficacy.41–43 To overcome these obstacles, different approaches have been applied including utilizing various polymer molecular weights, the ratio of lactic acid to glycolic acid in the copolymers, and PLGA concentration and the volume ratio (water/oil phases), or incorporation of cationic excipients admixing to RNA or cationic excipient conjugation with PLGA.34,44–46 The primary goal of this study was to improve existing approaches of miRNA encapsulation to develop a clinically viable strategy that achieves an effective and sustained release of miRNA with a favorable risk-benefit ratio to limit immune responses for immunomodulation. In particular, aside from miRNA protection by PLGA-based polymer vehicle, we expect to get synergistic anti-inflammatory therapeutic effects using miR-129 and PLGA-based NPs together. In this study, we assessed multiple NP formulations to identify key NP design parameters for mimic miR-129 delivery to provide a clinically feasible immunomodulatory therapy targeting activated microglia. We investigated essential physicochemical features of multiple nanoformulations including cell internalization and cytotoxicity, miR-129 encapsulation efficacy and release kinetics, and immunomodulatory properties as the prioritization criteria for nanoformulation selection.
2 Experimental
2.1 Materials
All reagents were purchased from VWR (Radnor, PA) unless otherwise specified. BV-2 cells were kindly provided by Michael Sanders (Shea lab, University of Michigan). Dulbecco's Modified Eagle's Medium (DMEM/F12) was obtained from Life Technologies (Carlsbad, CA) and was supplemented with 10% heat inactivated fetal bovine serum (Gibco). RNAse free water (non-DEPC treated) was used at all steps of nanoparticles preparation, purification and manipulation. Cytation 7 and Epoch plate reader were purchased from BioTek (Winooski, VT) and used for absorbance and fluorescence intensity measurements, as well as cell imaging. Cell culture 6 and 96 well flat-bottom plates and flasks were purchased from Corning Costar Co. (Corning, NY). Black sided 96 well plates were purchased from Cellvis (Mountain View, CA). Mature miRNA-129-5p (CUUUUUGCGGUCUGGGCUUGC) with modification at 5′ and 3′ ends (FAM dye and amino group, respectively) was purchased from IDT DNA (Coralville, IA) and used in all experiments. Dimethyl sulfoxide (DMSO), sodium hydroxide, lipopolysaccharide (LPS) were purchased from Sigma-Aldrich Chemicals (Atlanta, GA, USA). Trypsin/EDTA, phosphate buffered saline (PBS), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) (MTT), penicillin/streptomycin, BCA assay, Bel-Art™ SP Scienceware™ Flowmi™ cell strainers (400 μm) for 1000 μl pipette tips, and UltraComp compensation beads were purchased from Thermos Fisher (Waltham, MA). 8% paraformaldehyde was from Electron Microscopy Sciences (Hatfield, PA). Mouse TruStain FcX, Zombie Green dye, CD80-PE-Dazzle, CD86-BV786, CD206-BV421, and CD163-PE were purchased from BioLegend (San Diego, CA). IL-4 and IL-13 were purchased from STEM Cells Technologies (Cambridge, MA). All chemicals were of analytical grade and used without further purification.2.2 Nanoparticles fabrication
First, carboxylated 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 PLGA (inherent viscosity = 0.17 dl g−1) purchased from Akina PolySciTech (West Lafayette, IN) was conjugated with Cyanine 5.5 (CY5.5) amine dye (Lumiprobe corporation, Cockeysville, MD) and amine-miRNA-129-5p-FAM using an N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (Sigma-Aldrich, St. Louis, MO)/N-hydroxysuccinimide (NHS) (Thermo Fisher Scientific, Waltham, MA) chemistry. The conjugates were purified by dialysis against DDI and RNAse-free water, respectively and then freeze dried for 72 h. The miRNA PLGA nanoparticles were fabricated using an oil-in-water double emulsion solvent evaporation method. Briefly, 0.01–0.1 mg of miRNA-FAM, its complex with epigallocatechin gallate (EGCG, Sigma-Aldrich, St. Louis, MO), spermidine or polyethyleneimine branched (Sp and PEI, MW = 10
50 PLGA (inherent viscosity = 0.17 dl g−1) purchased from Akina PolySciTech (West Lafayette, IN) was conjugated with Cyanine 5.5 (CY5.5) amine dye (Lumiprobe corporation, Cockeysville, MD) and amine-miRNA-129-5p-FAM using an N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC) (Sigma-Aldrich, St. Louis, MO)/N-hydroxysuccinimide (NHS) (Thermo Fisher Scientific, Waltham, MA) chemistry. The conjugates were purified by dialysis against DDI and RNAse-free water, respectively and then freeze dried for 72 h. The miRNA PLGA nanoparticles were fabricated using an oil-in-water double emulsion solvent evaporation method. Briefly, 0.01–0.1 mg of miRNA-FAM, its complex with epigallocatechin gallate (EGCG, Sigma-Aldrich, St. Louis, MO), spermidine or polyethyleneimine branched (Sp and PEI, MW = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), its conjugate with PLGA (0.01–1 mg) or a complex of its conjugate with spermidine or PEI was mixed with some aliquot of RNAse-free water to get total 100 μl of the aqueous phase. The molar ratio miRNA
000), its conjugate with PLGA (0.01–1 mg) or a complex of its conjugate with spermidine or PEI was mixed with some aliquot of RNAse-free water to get total 100 μl of the aqueous phase. The molar ratio miRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EGCG was 1
EGCG was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, miRNA
2, miRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Sp was 1
Sp was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and miRNA
2 and miRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEI was 1
PEI was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. PLGA polymer and the polymer conjugated with CY5.5 dye (1% w/w) were dissolved in dichloromethane. Next, 1 ml of PLGA mixture was first emulsified with miRNA-contained aqueous phase and secondly, the emulsion obtained was emulsified with 5 ml of 2% poly (ethylene-alt-maleic anhydride) (PEMA, MW = 100–500 kDa, Sigma-Aldrich, St. Louis, MO) solution using a Qsonica Q125 Sonicator Ultrasonic Homogenizer W/Probe 125 W (Cole-Parmer, Vernon Hills, IL). In this study, poly(ethylene-alt-maleic anhydride) was utilized at the last step of NPs fabrication to coat the NPs and provide a strong negative charge to the NPs surface. Control blank NPs with FAM dye loaded (FAM NPs) were prepared in the same way. The emulsion was magnetically stirred at 750 rpm for faster evaporation of the organic solvent. The colloidal suspensions were washed three times with RNAse-free water by centrifugation of 14
1. PLGA polymer and the polymer conjugated with CY5.5 dye (1% w/w) were dissolved in dichloromethane. Next, 1 ml of PLGA mixture was first emulsified with miRNA-contained aqueous phase and secondly, the emulsion obtained was emulsified with 5 ml of 2% poly (ethylene-alt-maleic anhydride) (PEMA, MW = 100–500 kDa, Sigma-Aldrich, St. Louis, MO) solution using a Qsonica Q125 Sonicator Ultrasonic Homogenizer W/Probe 125 W (Cole-Parmer, Vernon Hills, IL). In this study, poly(ethylene-alt-maleic anhydride) was utilized at the last step of NPs fabrication to coat the NPs and provide a strong negative charge to the NPs surface. Control blank NPs with FAM dye loaded (FAM NPs) were prepared in the same way. The emulsion was magnetically stirred at 750 rpm for faster evaporation of the organic solvent. The colloidal suspensions were washed three times with RNAse-free water by centrifugation of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 20 minutes. Obtained NPs samples (Fig. 1) were filtrated using a 0.45 μm syringe filter and stored at 4 °C, which filters the aggregated NPs by multiple centrifugation steps and provides the fine suspension for the study.
000 g for 20 minutes. Obtained NPs samples (Fig. 1) were filtrated using a 0.45 μm syringe filter and stored at 4 °C, which filters the aggregated NPs by multiple centrifugation steps and provides the fine suspension for the study.
2.3 Physicochemical characterization of the NPs
The morphological characterization of NPs was done by scanning electron microscopy (SEM) (Hitachi S-4300) (Fig. 2a), and the size, polydispersity index and zeta potential (Table. S1 in the ESI†) was determined by using a dynamic light scattering (DLS) instrument (Zetasizer, Malvern Nano ZS, Westborough MA). For Fourier-transform infrared spectroscopy (FT-IR) analysis (Fig. 2b and S1a in the ESI†), stock suspension of NPs was freeze-dried for 72 h, and the obtained powder was analyzed by FT-IR spectrometer equipped with an attenuated total reflection (ATR) element (Nicolet iS50 nir, Thermo Scientific, Waltham, MA), operated in the range 4000–400 cm−1 with accumulating 64 scans. Powder X-ray diffraction (pXRD) analysis (Fig. 2c and S1b in the ESI†) was carried out using freeze dried samples as well by placing each one on a low-mass mirrored plate. All samples were shaped into a flat horizontal plane to avoid high intensity spotting. Prepared samples were transferred to a mini flex powder X-ray diffractometer (Rigaku Americas Corporation, TX, USA). Scanning was performed between 2 and 40° 2θ using Cu Kα radiation at 40 kV and 15 mA. The step size was set to 0.020° at a scanning speed of 0.875° min−1. Dynamic vapor sorption analysis (DVS) results are reported in Fig. 3(a) and (b). Isotherms were generated on a DVS Resolution (Surface Measurement Systems, Allentown, PA). Freeze-dried samples were weighed before placed on a sample holder and suspended from a hang-down wire in the sample compartment at 20 °C. Total dry nitrogen gas flow rates of 200 sccm were used for all experiments. Samples were first dried at 20 °C, 0% RH for 170 minutes within the sample chamber. The instrument was programmed to increase the relative humidity (P/P0 vapor pressure) step-wisely, with each step held until equilibrium (Fig. S2 in the ESI†). Equilibrium was said to be achieved when the change in mass with respect to time (dM/dT) was less than 0.002% per min for 10 minutes. The P/P0 stages ranged from 0% up to 90% and back down to 0% in 10% increments. The common sorption and desorption kinetics and isotherms at varying humidity (Fig. S2 in the ESI†) were replotted in water sorption kinetics and isotherm hysteresis (Fig. 3(a) and (b)).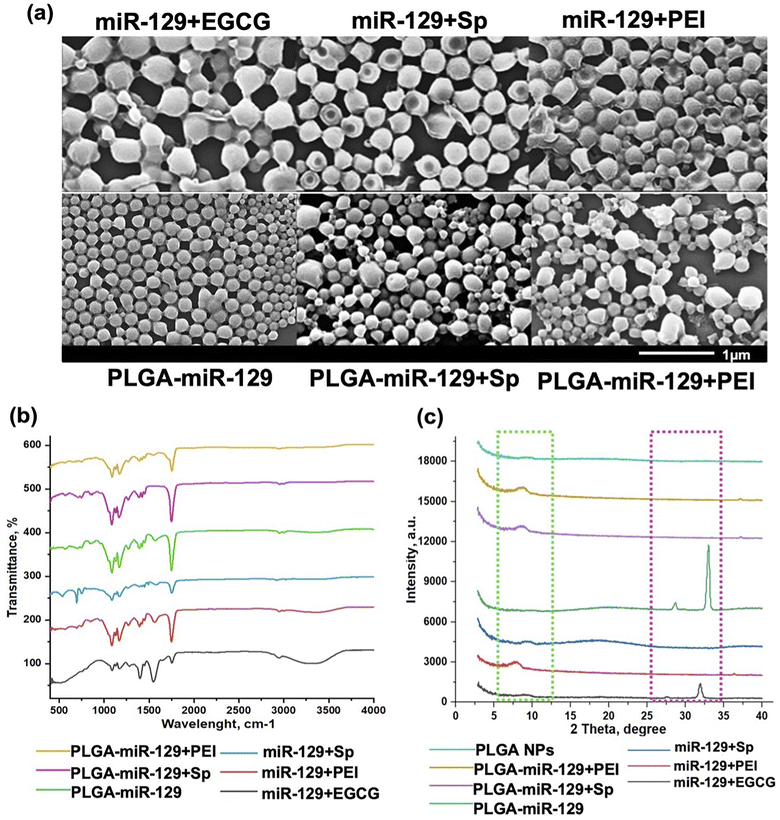 | ||
| Fig. 2 NPs characterization. (a) Scanning electron microscopy (SEM) of NPs (scale bar is 1 μm, ×40k); (b) FTIR; and (c) X-ray diffraction patterns of the nanoformulations (light green dash line: PLGA and magenta dash line: miRNA). The data demonstrated PLGA-incorporated surfactant (PEMA), miR-129, and excipients. The FTIR and pXRD data for controls are presented in Fig. S1.† | ||
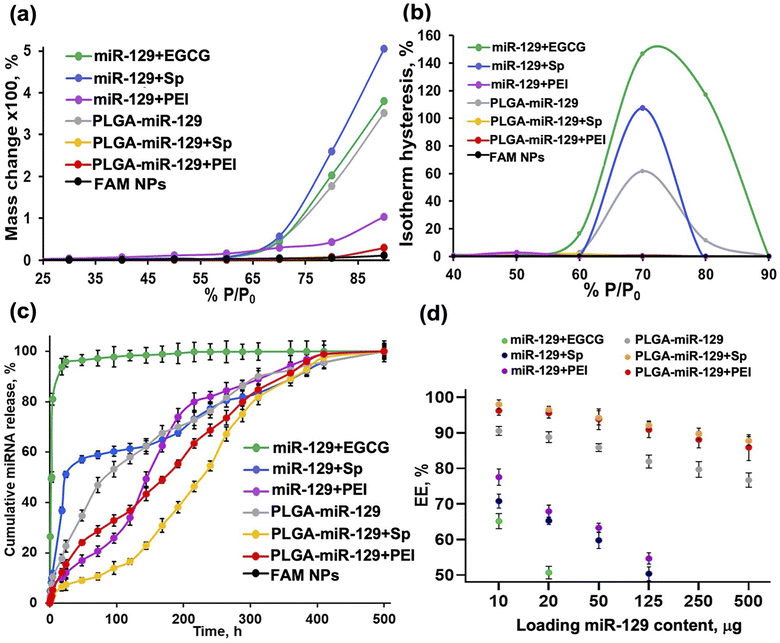 | ||
| Fig. 3 Nanoformulations characterization. Behavior upon exposure to the aqueous phase, miR-129 release kinetics and encapsulation efficacy. DVS results are based on data presented in Fig. S2 in the ESI.† (a) Water sorption kinetics and (b) isotherm hysteresis. (c) miR release kinetics for different nanoformulations at pH 5.5 (conditions: 1 mg freeze-dried NPs (10 μg miR loaded) in 10 ml of MES buffer, at 37 °C using orbital shaker at 80 rpm). (d) Encapsulation efficacy (%) was calculated based on the equation: EE % = (Wf/Wi) × 100%, where Wf is the final total amount of miR-129 in the NPs suspension and Wi is the initial quantity of miR-129 added during preparation. | ||
2.4 miRNA release study
The release profiles of miRNA from all nanoformulations were carried out using Slide-A-Lyzer mini dialysis device (MWCOs = 10 kDa, Thermo Fisher, Waltham, MA). Shortly, 1 mg of freeze dried nanoparticles was distributed in 1 ml of 0.1 M 2-(N-morpholino) ethanesulfonic acid buffer (pH = 5.5) and placed into the mini dialysis device against 5 ml of 0.1 M MES buffer (release medium) to be incubated at 37 °C in an orbital open air shaker (MaxQ, Thermos Fisher, Waltham, MA) at 70 rpm. At designated time points, aliquots of release medium were taken out from the dialysate and replaced by an equal volume of release medium. Content of miRNA released were measured on Cytation 7 plate reader (FAM dye fluorescence intensity) and calculated in cumulative % (Fig. 3c).2.5 Encapsulation efficacy
For loading capacity evaluation, the miRNA amount loaded per unit weight of the nanoparticle was 1 mg. All washes during nanoparticles suspension preparation were collected and analyzed using Cytation 7 plate reader (FAM intensity). Encapsulation efficiency (EE %, Fig. 3d) was calculated using this formula: (EE %) = (Wf/Wi) × 100%, where Wf is the final total amount of miRNA in the NPs suspension and Wi is the initial quantity of miRNA added during preparation. Loading of different quantities of miRNA was tested and EE was examined (Fig. 3d). Size, charge and polydispersity index of the nanoformulations with high content of miRNA loading was verified (Fig. S3 in the ESI†).2.6 Apoptosis assay
BV2 cells were seeded on 6 well plates at a density of 0.5 × 106 cells per well and pre-treated with 10 μg ml−1 LPS for 24 h. Then, the cells were exposed to control NPs and NPs contained miRNA-129 (250 μg ml−1), or PBS for 72 h. After the treatment, the cells were washed twice with cold PBS and resuspended in Annexin V Binding Buffer (ABB) at concentration 1 × 106 cells per ml. Cells were transferred to 5 ml test tubes and stained in 100 μl of ABB with 5 μl of Annexin V-Pacific Blue and 10 μl of propidium iodide (BioLegend, San Diego, CA) for 15 min at RT in the dark. Then 300 μl of ABB was added to each tube and analyzed on a BD FACSymphony A3 (BD Bioscience, San Jose, CA). The cytograms and the summarizing plot are presented in Fig. 4a, b and S4 in the ESI† (controls).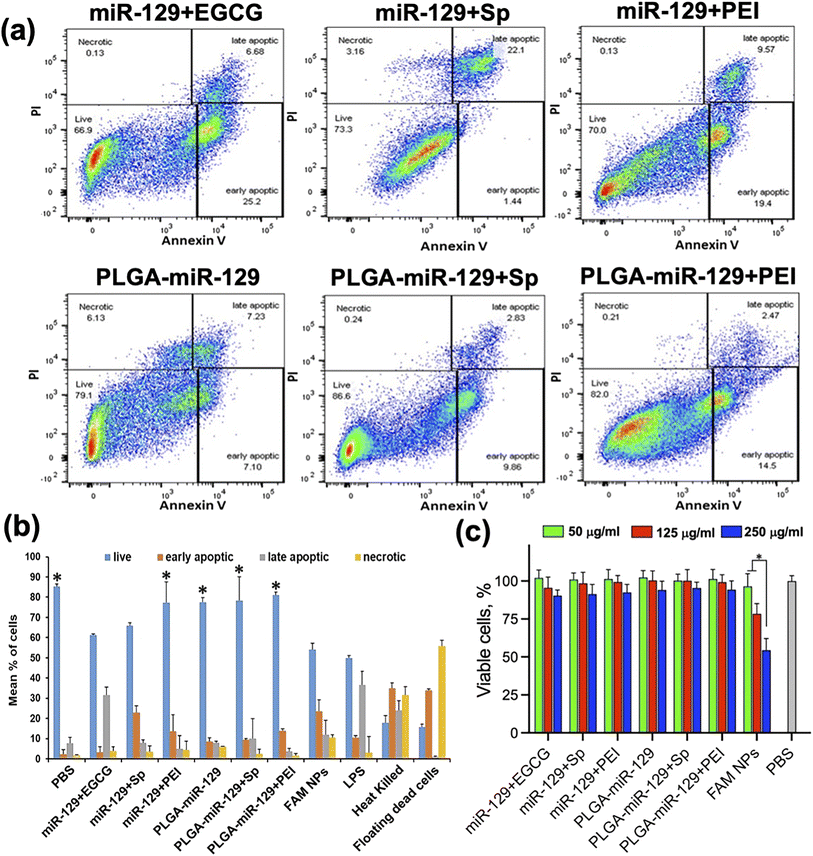 | ||
| Fig. 4 Cytotoxicity analysis. (a and b) The results of Annexin V/PI assay via flow cytometry (a) and summarizing the plot (b). In both cases, BV-2 cells were treated with LPS for 24 h followed by 72 h exposure to all nanoformulations. See flow cytometry controls in Fig. S4 in the ESI.†N = 3/group, error bars = SD, p-value: *p < 0.05 compared to LPS in mean % of live cells. (c) MTT test results; N = 5/group, error bars = SD, p-value: *p < 0.05. | ||
2.7 Cell viability test
BV2 cells were seeded (4000 cells per well, N (wells) = 10) in 96-well plates for 24 h. Then cells were incubated with 100 μl of the NPs samples at different concentrations (5–500 μg ml−1) reconstituted in the cell culture medium for 24–72 h at 37 °C. After, the cells were rinsed twice with PBS and then 100 μl of 5 mg ml−1 MTT reagent in cell culture medium without phenol red was added to each well of the plate and the plate was incubated for 3.5 h at 37 °C. The assay was completed by aspirating the MTT reagent and adding 100 μl of DMSO. A plate was placed on orbital shaker for 15 min at RT to help the formazan crystals dissolve. Optical density (OD) in each well was determined at 550 nm by Epoch plate reader and expressed in % (Fig. 4c).2.8 Cell uptake study
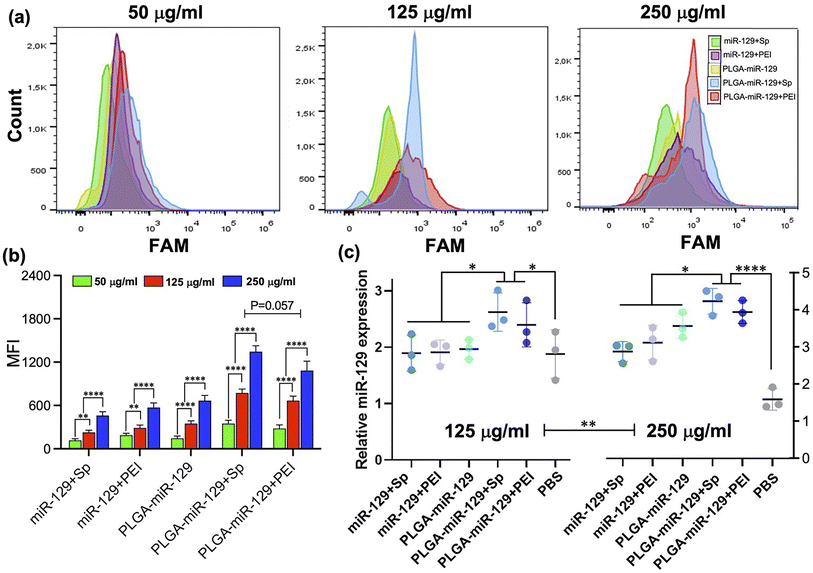
BV2 cells were seeded in 6 well plate at a density of 2 × 105 cells per well and left to adhere overnight. Cell culture medium was replaced on Opti-MEM 1× and cells were then pre-treated with 10 μg ml−1 LPS for 24 h. After that step, the cells were treated with 250 μg NPs per ml of Opti-MEM 1×. At the end of treatment, the cells were washed 3 times with PBS, and 400 μl of 0.5% trypsin/EDTA was added remove cells from the plastic substrate. Cells were collected by adding 500 μl of PBS, centrifuge at 1000 rpm for 5 min at 4 °C. Cells pellets were distributed in 0.3 ml of PBS and analyzed on a BD FACSymphony A3 (BD Bioscience, San Jose, CA). The cytograms obtained and plot in MFI are presented in Fig. 5(a) and (b).2.9 qRT-PCR analysis
RNA was extracted from cells using miRNeasy Mini Kit (Qiagen, Germany) according to the manufacturer's instructions. The RNA quantification was performed using Epoch plate reader (BioTek) and RNA concentration was adjusted to 20 ng μl−1. Reverse transcription for all RNA extracts was performed using the miScript II RT kit (Qiagen, Germany) according to the manufacturer's instructions using the 96-well T100 thermal cycler (Bio-Rad, Hercules, CA). The cDNA was further utilized as a template for real-time PCR with the miScript SYBR Green PCR Kit and miScript Primer Assays (Hs_RNU6-2_11, MMu_miRNA-129-5p; Qiagen Germany). Quantitative RT-PCR (qRT-PCR) was performed using CFX Connect RT system (Bio-Rad, Hercules, CA) with the following parameters:47 95 °C for 15 min, then 40 cycles of 94 °C for 15 s, 55 °C for 30 s and 70 °C for 30 s. The results of RT-PCR are presented in Fig. 5c.2.10 Nitric oxide (Griess) assay
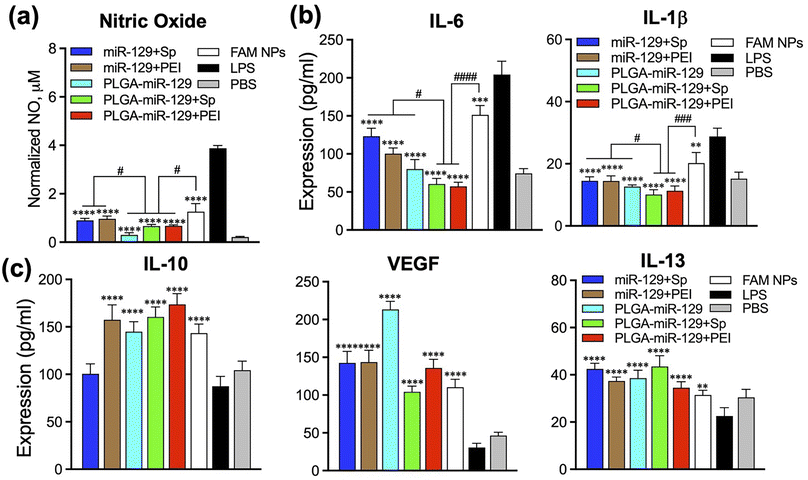
BV2 cells were seeded (500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml) in 96-well plates, incubated overnight, and then pre-treated with 10 μg ml−1 LPS. NPs were loaded to cells after 24 h and kept for 72 h in the incubator at 37 °C. The culture supernatants were collected and processed (N = 5) according to the manufacture protocol. Samples absorbance was measured by Epoch plate reader at 540 nm (±20 nm) and the intensities were normalized to the live cells based on MTT data (Fig. 7a).
000 cells per ml) in 96-well plates, incubated overnight, and then pre-treated with 10 μg ml−1 LPS. NPs were loaded to cells after 24 h and kept for 72 h in the incubator at 37 °C. The culture supernatants were collected and processed (N = 5) according to the manufacture protocol. Samples absorbance was measured by Epoch plate reader at 540 nm (±20 nm) and the intensities were normalized to the live cells based on MTT data (Fig. 7a).
2.11 ELISA assay
Mouse VEGF, IL-1β, IL-10, IL-13, and IL-6 ELISA kits were used (Quantikine ELISA Kit from R&D Systems, Minneapolis, MN) to determine the cytokine and growth factor amounts released by BV2 cells after pre-treatment with 10 μg ml−1 LPS for 24 h and following treatment with control NPs and miRNA-NPs for 72 h. The culture supernatants were collected and processed (N = 5) according to the manufacture's protocol. Samples absorbance was measured by Epoch plate reader at 450 nm (±20 nm) and with the reference wavelength at 540 nm (Fig. 7(b) and (c)).2.12 Flow cytometry
Cells were seeded (300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml) in 6-well plates, incubated overnight, and then treated with NPs at 0.5 μg ml−1 concentration in cell culture medium without phenol red for 24 h. Following, culture medium was aspirated, cells were rinsed twice with PBS, and detached by using 0.25% trypsin/EDTA solution. THP-1 cells were treated similarly, but before exposing them to NPs, they were treated with PMA 20 ng ml−1 for 24 h, then washed and rested for 24 h. The positive controls for M1 and M2 macrophage were prepared by cells treatment either with 40 ng ml−1 of IL-4/IL-13 or combination 10 ng ml−1 LPS with 20 ng ml−1 IFNγ for 24 h. After treatment cell pellets were re-suspended in flow wash buffer (1% BSA + PBS + 0.1% sodium azide) for 10 min at RT, and then cells were stained with anti-mouse or anti-human antibodies (BV421 CD206, AF647 CD209 and APC/Fire750 CD80, Pe-Cy5CD80). After washing with PBS, cells were stained on ice with Zombie NIR at 1
000 cells per ml) in 6-well plates, incubated overnight, and then treated with NPs at 0.5 μg ml−1 concentration in cell culture medium without phenol red for 24 h. Following, culture medium was aspirated, cells were rinsed twice with PBS, and detached by using 0.25% trypsin/EDTA solution. THP-1 cells were treated similarly, but before exposing them to NPs, they were treated with PMA 20 ng ml−1 for 24 h, then washed and rested for 24 h. The positive controls for M1 and M2 macrophage were prepared by cells treatment either with 40 ng ml−1 of IL-4/IL-13 or combination 10 ng ml−1 LPS with 20 ng ml−1 IFNγ for 24 h. After treatment cell pellets were re-suspended in flow wash buffer (1% BSA + PBS + 0.1% sodium azide) for 10 min at RT, and then cells were stained with anti-mouse or anti-human antibodies (BV421 CD206, AF647 CD209 and APC/Fire750 CD80, Pe-Cy5CD80). After washing with PBS, cells were stained on ice with Zombie NIR at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution for 20 min. After staining, cells were washed twice with 1 ml of flow wash buffer (1% BSA + PBS + 0.1% sodium azide) and fixed with 500 μl of 1% paraformaldehyde in PBS for 30 min on ice. After that, 1 ml of flow wash buffer was added to each tube, and cells were centrifuged at 1000 rpm for 5 min at 4 °C. Before analysis, cell pellets were re-suspended in 300 μl of flow wash buffer and filtrated using 70 μm strainer. We performed flow cytometry study using BD FACSymphony A3 (BD Bioscience, San Jose, CA) with FlowJo software (v10.6.2). Three separate flow cytometry studies were done.
500 dilution for 20 min. After staining, cells were washed twice with 1 ml of flow wash buffer (1% BSA + PBS + 0.1% sodium azide) and fixed with 500 μl of 1% paraformaldehyde in PBS for 30 min on ice. After that, 1 ml of flow wash buffer was added to each tube, and cells were centrifuged at 1000 rpm for 5 min at 4 °C. Before analysis, cell pellets were re-suspended in 300 μl of flow wash buffer and filtrated using 70 μm strainer. We performed flow cytometry study using BD FACSymphony A3 (BD Bioscience, San Jose, CA) with FlowJo software (v10.6.2). Three separate flow cytometry studies were done.
2.13 Statistical analysis
We performed a one or two-way ANOVA and Tukey's post hoc test for the multiple comparisons, or Student's t-test were used for the statistical analysis. The Kolmogorov–Smirnov normality test was employed. P-value (alpha, α, 0.05) was used as a cut-off thus P-value larger than 0.05 is considered as passing the normality test. Type II errors (Beta, β) were controlled at 0.2 level for all the statistical tests and α = 0.05 to achieve reasonable statistical power analyses (power = 0.8). Equal variance (ANOVA Model) was validated and assumed for each study. For appropriate sample size, conservative effect size 0.25 was employed for each study. The data were analyzed by G* Power Software,6 Prism 9 (GraphPad Software, La Jolla, CA, USA) and OriginPro (Northampton, MA, USA). Data are showed as mean ± SD and differences of p < 0.05 were considered as statistically significant.3 Results & discussion
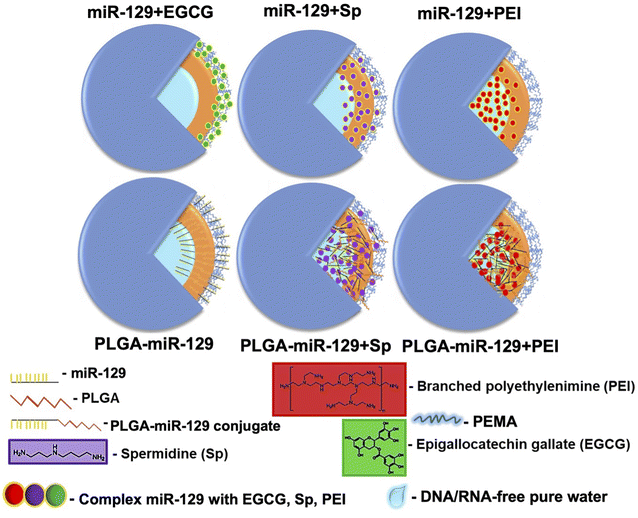
Microglia mediate multiple facets of neuroinflammation, including cytotoxicity, repair, regeneration, and immunosuppression due to their ability to acquire diverse activation states and phenotypes.2–4 We designed six miR-129-PLGA NP formulations (Fig. 1) and tested their immunomodulation properties on LPS-activated immortal microglia cell line (BV-2 cells) modeling neuroinflammation. To improve existing strategies for enhancing miR-129 loading capacity and release profile, we applied three approaches. Initially, the PLGA polymer matrix solution was premixed with PLGA-CY5.5 conjugate and then combined with (1) a preset mixture of miR-129 with one of the following excipients-epigallocatechin gallate (EGCG), spermidine (Sp), or polyethyleneimine (PEI) branched to get miR-129+EGCG, miR-129+Sp, or miR-129+PEI formulations, (2) miR-129 conjugate to PLGA for PLGA-miR-129 formulation, and (3) a preset mixture of PLGA-miR-129 with one of the excipients to get PLGA-miR-129+Sp or PLGA-miR-129+PEI formulations. Our pre-screening investigations and the results in Fig. 3(c) and (d) demonstrate that miR-129+EGCG reveals poor encapsulation efficiency as well as unfavorable release kinetics with about 80% burst release in the first 30 minutes. Therefore, we exclude any alternative variation with EGCG in this study. Previous studies indicate that EGCG can be an effective excipient for complexation with small interference RNA,48,49 yet we were unable to demonstrate such benefits of EGCG for miRNA delivery. All NP formulations were fabricated using the oil-in-water double emulsion solvent evaporation method described previously.33 The excipients were chosen based on the previously reported data of their successful use. It was demonstrated that EGCG facilitated RNA condensation by low-molecular-weight polymers,50,51 whereas Sp conjugated to PLGA46 and PEI anchored into PLGA-based NPs helped with better incorporation of miRNA via electrostatic complexation.33 We characterized all nanoformulations obtained in terms of their morphology, size, zeta potential, physical state and stability, composition, interaction with the aqueous phase, and miR-129 release kinetics. Fig. 1 demonstrates the structure of the NPs based on summarized physicochemical properties. Close to sphere-shaped nanoparticles had a smooth surface with hydrodynamic radius and NPs size determined by Scanning Electron Microscope (SEM) images (Fig. 2a and Table. S1 in the ESI†). All NPs had negative charge due to carboxyl groups of PLGA and PEMA coating at the last step of NPs fabrication since the previous study indicated that negative surface charged NPs enhance cellular binding association through scavenger receptors such as Macrophage Receptor with Collagenous Structure (MARCO) and the class A scavenger receptor SR-AI/II.38,52The results of FTIR and pXRD analysis of the nanoformulations confirmed PEMA, miR-129, and the excipients incorporated into the PLGA matrix (Fig. 2(b) and (c)). The spectral domain (1750–1450 cm−1) shown in Fig. 2b contains absorption bands due to in-plane double bond stretching vibrations of the bases, whereas the spectral domain (800–600 cm−1) is commonly assigned to the out-of-plane vibrations of bases.53 The uracil peak at 1645 cm−1 is observed in the miR-129 spectrum as well as in the miR-129+EGCG (Fig. 2b and S1 in the ESI†). The same band was shifted to 1700 cm−1 in the PLGA-miR-129 conjugate and the conjugate-based nanoformulations. A new band in all nanoformulations and the conjugate (vs. FAM dye loaded blank PLGA based NPs (FAM NPs) on Fig. S1a in the ESI†) can be found at 1550–1564 cm−1 and assigned to the bending vibration of amide II. FAM-NPs were used as a control groups. The sugar band of ν(C–O) ribose (1084 cm−1) in the miR-129 spectrum was shifted to 1087–1090 cm−1 in the PLGA-miR-129 formulation. The weak peak at 1048 cm−1 in miR-129 is assigned to the C–O–C stretching and the combination of the C–O stretching and C–O bending in miR-129. A small shift of this band (1050–1051 cm−1) can be found in PLGA-miR-129 (Fig. 2b). The small bands at 696–704, 806–811, 850–860 cm−1 and 743–749 cm−1 in the nanoformulations correspond to the out-of-plane ring modes of the guanine, ribose-phosphate vibrations, and pyrimidine ring (uracil and/or cytosine) of miR-129, respectively (Fig. 2b).53 The peaks in the NPs spectra at 1397, 1426, 1454, and 1754 cm−1 are ascribed to the bending vibration of carbonyl groups (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of PLGA (Fig. 2b and S1a in the ESI†). The peak at 1474 cm−1 in miR-129+Sp NPs is attributed to bending vibrations of the aminopropyl group of Sp. The peak around 3230 cm−1 indicated the hydroxyl groups of EGCG in miR-129+EGCG (Fig. 2b). As shown in Fig. S1b in the ESI,† the XRD patterns of initial polymers (PLGA, PEMA, and PEI) exhibit no sharp peaks and only wide diffraction ones (5.2 to 7.4°, 12.6 to 19.2°; 16.8 to 20.2°; 10 to 29.4°) that indicates the amorphous state of the polymers was used. As expected, all nanoformulations exist in an amorphous form even in the case of Sp and EGCG, which individually displayed high crystallinity (Fig. 2c and S1b in the ESI†). The latter confirms Sp and EGCG underwent the crystal-to-amorphous transition due to effective entrapment into PLGA mass. However, there are miR-129 peaks (28.7 and 32°) in miR-129+EGCG that imply re-crystallization of miR-129 upon solvent evaporation and its entrapment at the NPs surface. The peak of PEMA (around 18°) was observed in miR-129+Sp and may indicate that a bigger amount of PEMA adsorbed on these NP surfaces compared to other nanoformulations (Fig. 2c). The characteristic crystalline peaks of miR-129 occurred in PLGA-miR-129 at 27.8° and 32.0° as well (Fig. 2c and S1b in the ESI†), however the sharp peaks of miR-129 disappeared in the XRD spectra of the rest of the nanoformulations indicating that miR-129 was successfully encapsulated within them.
O) of PLGA (Fig. 2b and S1a in the ESI†). The peak at 1474 cm−1 in miR-129+Sp NPs is attributed to bending vibrations of the aminopropyl group of Sp. The peak around 3230 cm−1 indicated the hydroxyl groups of EGCG in miR-129+EGCG (Fig. 2b). As shown in Fig. S1b in the ESI,† the XRD patterns of initial polymers (PLGA, PEMA, and PEI) exhibit no sharp peaks and only wide diffraction ones (5.2 to 7.4°, 12.6 to 19.2°; 16.8 to 20.2°; 10 to 29.4°) that indicates the amorphous state of the polymers was used. As expected, all nanoformulations exist in an amorphous form even in the case of Sp and EGCG, which individually displayed high crystallinity (Fig. 2c and S1b in the ESI†). The latter confirms Sp and EGCG underwent the crystal-to-amorphous transition due to effective entrapment into PLGA mass. However, there are miR-129 peaks (28.7 and 32°) in miR-129+EGCG that imply re-crystallization of miR-129 upon solvent evaporation and its entrapment at the NPs surface. The peak of PEMA (around 18°) was observed in miR-129+Sp and may indicate that a bigger amount of PEMA adsorbed on these NP surfaces compared to other nanoformulations (Fig. 2c). The characteristic crystalline peaks of miR-129 occurred in PLGA-miR-129 at 27.8° and 32.0° as well (Fig. 2c and S1b in the ESI†), however the sharp peaks of miR-129 disappeared in the XRD spectra of the rest of the nanoformulations indicating that miR-129 was successfully encapsulated within them.
Characterizing the mechanical stability of nanoformulations at increasing relative humidity is also important in the assessment of storage lifetimes and can predict both polymer degradation and payload release kinetics.54 This analysis was performed by dynamic vapor sorption (DVS). The water adsorption–desorption isotherms by DVS, depicting mass changes as a function of varying relative humidity at constant temperature, are presented in Fig. 3 and S2 in the ESI.† The degree and rate of water sorption can be influenced by encapsulation, controlling particle size, and changing the degree of crystallinity. Higher levels of moisture absorption may cause a plasticizing effect on the NPs, softening and deforming the NPs, filling their porous volume and interfering with the payload interaction with components of the NPs.54,55 Sorption mass change (%) at different % P/P0 displays high water sorption for miR-129+Sp, miR-129+EGCG, and PLGA-miR-129 formulations perhaps due to an allocation of the excipients and miR-129 on or close to the outer surface of NPs increasing its hydrophilicity (Fig. 3a). Other reasons might be a higher PEMA content presenting in the miR-129+Sp and PLGA-miR-129 formulations and higher crystallinity of miR-129+Sp and PLGA-miR-129 formulations (Fig. 2(b) and (c)) that increased water sorption.55 Nanoformulation miR-129+PEI showed a significant decrease in water sorption and nanoformulation, and PLGA-miR-129+Sp and PLGA-miR-129+PEI showed minimal mass change even at the high % P/P0 values (70–90%) (Fig. 3a and in the ESI†). Each excipient being a hydrophilic component may contribute to swelling and mass change as water-moistening enhanced the wettability of the sample. Accumulation of water adsorbed on the NPs surface may lead to the NPs swelling first and then catalyzing PLGA matrix erosion resulting in a generation of some pores into the NPs volume. In general, the adsorption/desorption process is not fully reversible at the same % P/P0, and a larger moisture content is observed during the desorption process; a phenomenon called hysteresis.56 Sorption-induced swelling in various soft structures brings to hysteresis when there is a difference between adsorption and desorption isotherms. The degree of hysteresis in a formulation is a good indicator of the ability to maintain structural integrity. According to DVS results, the least stable samples are PLGA-miR-129, miR-129+Sp, and miR-129+EGCG displaying huge hysteresis (Fig. 3b). The miR-129+PEI has a little hysteresis at a low % P/P0 may be associated with water molecule intercalation in narrow pore spaces leading to irreversible changes in the pore structure.57 Hysteresis isotherms of PLGA-miR-129+PEI and PLGA-miR-129+Sp formulations with only minimal hysteresis indicate that despite being moisture sensitive at high pressure, they are most stable with fewer sorption sites for hydrogen bond formation than other samples. DVS data correlates well with the miR-129 release profile (Fig. 3c). High retention of moisture is considered an unfavorable trait for the maintenance of the structure and therefore a burst release of the entrapped payload and poor sustainability can be expected. It is known that pharmacokinetics (PK) is associated with the degradation rate and the drug release of PLGA-based NPs. Both are influenced by a number of factors such as the initial molecular weight, monomer composition ratio of PLGA matrix (i.e., lactic acid/glycolic acid ratio), payload nature, processing method, polymer crystallinity, polymer end-capping, water absorption, pores, pH, and glass transition temperature.58,59 Different types of payload release patterns can be obtained from PLGA particles, in particular mono-, bi-, or tri-phasic drug release kinetics. Generally, an initial rapid payload release phase (“burst release”) is often met for PLGA-based NPs.58,59 The release curve of the miR-129+EGCG (Fig. 3c) corresponds to the common hydrophilic compound release pattern, the biphasic release that is characterized by the initial burst (80%) and saturation (i.e., zero-order release) which is unfavorable.58 The initial miR-129 burst release phase as in the case of miR-129+EGCG and miR-129+SP might be attributable to the release of oligonucleotide adsorbed onto the surface, to the diffusion of oligonucleotide located close to the surface.58–60 The miR-129+Sp release curve demonstrates combined release kinetics (monophasic/biphasic): small burst release (about 15%), sustained release and short saturation. The 80% miRNA release was observed after 230 h at pH 5.5. Different miRNA patterns are reported in other works with similar approaches using spermidine and PLGA NPs: 80% miRNA released from the NPs after 48 h at pH 5 and 8% of microRNA released in 8 days of incubation at 37 °C under a static condition.61 Apparently, diverse release kinetics in our case may be associated with NPs coating with PEMA and NPs size as well as release study conditions. Like the miR-129+Sp formulation, the PLGA-miR-129 formulation showed combined release kinetics but without any burst release phase (Fig. 3c). Conjugation of miR-129 with PLGA might contribute to solubility/hydrophobicity of miR-129 and distribution within the PLGA matrix that led to a change in diffusion rate, thus improving the release kinetics (Fig. 3c). The PLGA-miR-129+Sp sample displayed fluent sustained release of the miR-129 close to monophasic release from a single homogeneous phase ending by some saturation phase (Fig. 3c). The release curves of miR-129+PEI and PLGA-miR-129+PEI formulations represent the triphasic pattern (i.e., phase I, phase II, and phase III).58 The small initial miR-129 burst release effect is due to the rapid release of surface-bound molecules, the second phase (steady controlled release) is related to diffusion and hydrolysis, and the third phase is a result of bulk erosion.58 Interestingly, Arora et al. showed that PLGA NPs loaded with miR-150 mimics and PEI as a condensing agent could release ∼20% of their cargo within 336 h at pH 7.4.62 At the same release conditions (pH 7.4), it was found double higher % of released miRNA from PLGA-PEI-miR-21i NPs after 60 h.63 These conflicts may be due to different study design parameters including PLGA composition, encapsulation conditions, final NPs size, and release study conditions. Complexation of PLGA-miR-129 conjugate with PEI allowed a heterogeneous distribution of the miR-129 within PLGA nanocarriers, with more concentration at the core area (aqueous phase) and less at peripheries providing slow triphasic release (Fig. 1 and 3c). Thus, three of our six nanoformulations (miR-129+PEI, PLGA-miR-129+Sp, and PLGA-miR-129+PEI) may ensure the prolonged release of miR-129 owing to the optimal distribution of miR-129 within PLGA-based NPs (in high-density areas) (Fig. 3c) and high stability in the aqueous phase (no hysteresis) (Fig. 3b). Encapsulation efficacy is a crucial parameter that permits varying the NPs concentration for in vivo study. The encapsulation efficacy and miR-129 loading content were studied (Fig. 3d and S3a in the ESI†). Encapsulation efficacy was calculated using the equation mentioned above. All nanoformulations displayed miR-129 content-dependent loading capacity (EE, %) with the highest EE found for PLGA-miR-129+Sp and PLGA-miR-129+PEI and the lowest ones for miR-129+EGCG and miR-129+Sp (Fig. 3d). Surprisingly, EE for miR-129+PEI formulation in about 78% matches the value obtained in the study mentioned above using PEI.62 Entrapment of miR-129 as the PLGA-miR-129 conjugate showed a beneficial trend in EE value reaching near 80% at the highest miR-129 content we tested (Fig. 3d). Thereby, the combination of conjugation and complexation approaches allowed reaching 85–98% encapsulation efficacy. The most promising nanoformulations at the highest loading content (125–500 μg of miR-129 per 1 mg of NPs) were characterized using a Zetasizer to measure size, polydispersity index (PdI) and zeta potential (Fig. S3a and b in the ESI†). It can be noted that the nanoformulations' size enlarged slightly (6–8%) as well as PdIs with loading amount growth. In addition, insignificant change was seen for zeta potentials (Fig. S3b in the ESI†).
Cytotoxicity of all nanoformulations was examined with Annexin V/PI assay and MTT test (Fig. 4). Annexin V/PI assay via flow cytometry and summarized numeric data of the Annexin V/PI assay were shown in Fig. 4(a) and (b). All possible controls of Annexin V/PI assay were presented in Fig. S4 in the ESI.† In comparison to LPS only treatment group, all miR-129-contained nanoformulations displayed inhibition of apoptosis of BV-2 cells caused by LPS. It was reported that miR-129 can suppress apoptosis by regulating BCL-2 and caspase-3 expression.26,64 The data indicated that miR-129+EGCG and miR-129+Sp formulations had relatively less anti-apoptotic effects compared to other nanoformulations. MTT test showed the nontoxic behavior of all nanoformulations toward microglia cells indifferently to NPs concentrations tested (Fig. 4c). The exception was observed in the FAM-NPs group which showed a drastic drop in cell viability (up to 58.4%) at 250 μg ml−1.
Subsequently, besides identifying more effective NP designs for mimic miRNA therapy, we investigated the immunomodulatory effects of miR-129 and PLGA NPs for activated microglia. The immunomodulatory effects of each of the nanoformulations were examined on LPS-activated BV-2 cells through in vitro analysis in comparison to control FAM-NPs group. Among the nanoformulations, the miR-129+EGCG formulation demonstrated unfavorable miR-129 release kinetics and the least potency compared to the rest of the nanoformulation based on our NP selection prioritization criteria (Fig. 1–4). Therefore, nanoformulation miR-129+Sp, miR-129+PEI, PLGA-miR-129, PLGA-miR-129+Sp, and PLGA-miR-129+PEI were selected for the following in vitro study. Initially, we investigated the cellular NPs uptake and the expression of miR-129 (Fig. 5). The cellular uptake was measured by FAM signal (miR-129-FAM) using flow cytometry and the most representative cytograms are presented in Fig. 5a. The data indicated that the cellular NP uptake was concentration dependent. In all nanoformulation groups, the cellular NP uptake levels were significantly increased at 250 μg ml−1 and the highest values were collected for PLGA-miR-129+Sp and PLGA-miR-129+PEI formulations (Fig. 5(a) and (b)). Although the cellular NPs uptake levels showed a trend toward greater expression in PLGA-miR-129+Sp formulation compared with PLGA-miR-129+PEI formulation, there were no significant differences between the two nanoformulations (P = 0.057, Fig. 5b). Microimages of the BV-2 cells after NP treatment were taken by Cytation 7 plate reader and presented in Fig. S5 in the ESI.† The data indicated that delivered NPs were taken up by activated BV-2 cells, which were colocalized with miR-129 (yellow). Subsequently, the expression level of miR-129 in BV-2 cells treated with the NPs was analyzed via qRT-PCR (Fig. 5c). All formulations effectively upregulated the expression of miR-129, indicating that our nanoformulations successfully delivered target miRNA-129 to activated cells. We discovered fairly close miR-129 expression patterns for activated BV-2 cells treated with PLGA-miR-129+Sp and PLGA-miR-129+PEI in both concentrations, but the expression levels of miR-129 were concentration-dependent, which was correlated with cellular NP uptake (Fig. 5(b) and (c), P < 0.01 125 μg ml−1vs. 250 μg ml−1 in Fig. 5c).
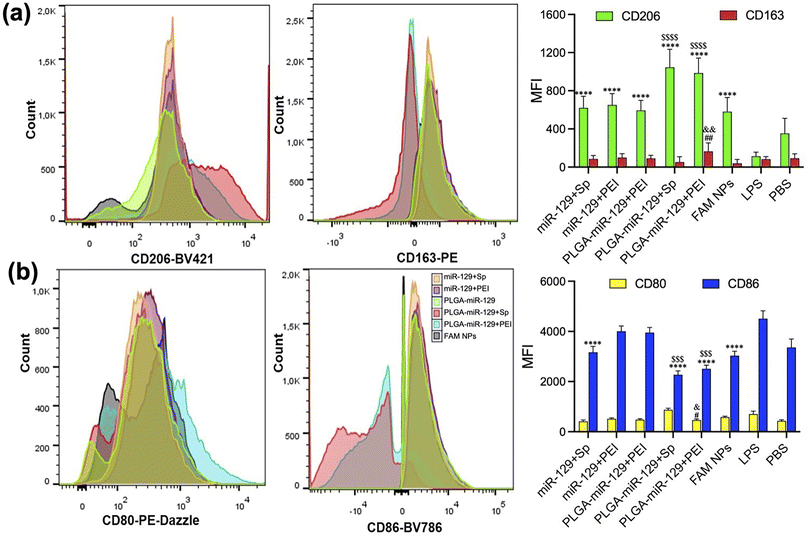
Our last study was devoted to the verification of NPs whether NPs can reprogram the activated microglia by LPS from pro-inflammatory phenotype to anti-inflammatory phenotype. We utilized flow cytometry, Nitric Oxide (NO) assay, and ELISA to examine the NPs-mediated alteration of the main biomarkers from the activated microglia (Fig. 6 and 7). Since the efficacy of NPs was concentration dependent, we employed 250 μg ml−1 of NPs for the following investigations. Initially, LPS-mediated activated microglia indicated pro-inflammatory phenotype (CD80high/CD86high/CD163low/CD206low). However, anti-inflammatory markers, CD206 and CD163, were upregulated in comparison to the LPS group, whereas pro-inflammatory markers, CD80 and CD86, were found to be intensively downregulated in LPS-activated BV2 cells after treatment with PLGA-miR-129+Sp and PLGA-miR-129+PEI formulations (Fig. 6(a) and (b)). Other nanoformulations stimulated the expression of anti-inflammatory factors as well compared to LPS treated group yet were less effective. In addition, NPs significantly attenuated LPS-induced NO production in activated BV-2 cells (Fig. 7a). A decrease in NO production can be observed for all nanoformulations with a concentration-dependent tendency (Fig. 7a and S6 in the ESI†). Almost equal effect on NO production was revealed for three nanoformulations (PLGA-miR-129, PLGA-miR-129+Sp and PLGA-miR-129+PEI) compared to other formulations (P < 0.05). All miR-129 contained nanoformulations were able to regulate the expression of pro-inflammatory and anti-inflammatory factors (Fig. 7b, c and S6 in the ESI†). The expression of pro-inflammatory cytokines including IL-6 and IL-1β was downregulated in LPS-activated microglia after treatment with NPs, while the expression of anti-inflammatory factors vascular endothelial growth factors (VEGF), IL-10 and IL-13 was restored and even exceeded the PBS-treated group. Our previously published study indicated that PLGA-based NPs were internalized by innate immune cells leading to the modulation of immune cells' gene expression and function without active biologics.38 The expression levels of NO and pro-inflammatory factors in PLGA-miR-129+Sp and PLGA-miRNA-129+PEI were significantly reduced compared to the control blank NP group (Fig. 7(a) and (b)), indicating that the synergistic immunomodulatory effects between miR-129 and PLGA-based NPs on activated microglia. Although the expression level of pro-regenerative factors showed a trend toward greater in PLGA-miR-129+Sp and PLGA-miR-129+PEI compared to the control empty NP group, there were no substantial differences between groups. It was confirmed that localized expression of IL-10 can drive microglia and macrophage polarization from pro-inflammatory phenotype to anti-inflammatory phenotype and reduce neutrophil infiltration.65–67 IL-13 has been reported to induce microglia alternative activation and inhibit the expression of pro-inflammatory factors, thereby providing neuroprotection after injury.68 The expression level of VEGF was vastly raised by nanoformulation treatments. VEGF can mediate neuronal restorations through three major mechanisms: stimulating angiogenesis and modulating vascular permeability, direct neuroprotection, and promoting neurogenesis.69,70 In previous work, VEGF down-regulated the expression levels of IL-1β and TNF-α and shifted activated microglia to anti-inflammatory phenotype. Furthermore, VEGF is involved in several signaling pathways, such as PI3K/Akt, Erk/MAPK, VEGF/SphK, JNK, and p38 MAPK pathways in charge of the neuroprotective and nerve regenerative processes.69–71 Therefore, reprogramming of activated microglia post-translationally to attenuate the whole spectrum of anti-regenerative expressions will provide the potential therapeutic opportunity to modulate neuroinflammation after injury and limit its adverse consequences.
Collectively, we selected PLGA-miR-129+Sp and PLGA-miR-129+PEI as the most promising nanoformulations since they demonstrated more effective characterizations for target miRNA delivery and immunomodulation for activated microglia compared to all prepared nanoformulations considering our selection criteria.
4. Conclusion
Microglia play critical role in multiple inflammatory responses-mediated neurological disorders. There is an increasing need for the development of efficient and non-invasive strategies to treat neuroinflammatory aspects by means of microglia activation regulation. The development of clinically viable miRNA-mediated therapy to reprogram the function of activated microglia represents a novel immunotherapeutic strategy for the treatment of multiple inflammation-mediated diseases. We designed effective nanoformulations that allowed for improved target miRNA encapsulation and release kinetics. In this study, we have underscored the synergistic effects of PLGA-based NPs and miR-129 to attenuate inflammatory response in activated microglia. Our selected nanoformulations regained the expression level of miR-129, restored LPS-altered health conditions of microglia, converted the phenotype of microglia, and stimulated the production of multiple pro-regenerative factors. For further study, we will investigate the anti-inflammatory effects of two of the most promising nanoformulations (PLGA-miR-129+Sp and PLGA-miR-129+PEI) through in vivo inflammatory responses-mediated diseases on the CNS models including neuropathic pain after SCI or MS and expect to observe enhanced PK and efficient performance.Author contributions
I. K. and J. P. designed research; I. K., H. C., and D. K. performed research; I. K., D. K., and J P. analyzed data; and I. K., H. C., D. K., and J. P. wrote the paper.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by the Center for Pharmaceutical Research and Innovation (CPRI, NIH P20 GM130456), the National Center for Advancing Translational Sciences (UL1 TR001998), and the University of Kentucky Neuroscience Research Priority Area (NRPA017). The authors thank Larissa Ponomareva for the assistance with the Cytation 7 plate reader and Svitlana Rudik for the graphic abstract.References
- Q. Li and B. A. Barres, Nat. Rev. Immunol., 2018, 18, 225–242 CrossRef CAS PubMed.
- L. Xu, J. Wang, Y. Ding, L. Wang and Y.-J. Zhu, Front. Neurol., 2021, 12, 796704 CrossRef PubMed.
- X. Zhou, X. He and Y. Ren, Neural Regen. Res., 2014, 9, 1787–1795 CrossRef PubMed.
- S. Yoshizaki, T. Tamaru, M. Hara, K. Kijima, M. Tanaka, D. J. Konno, Y. Matsumoto, Y. Nakashima and S. Okada, J. Neuroinflammation, 2021, 18, 12 CrossRef CAS PubMed.
- S. David and A. Kroner, Nat. Rev. Neurosci., 2011, 12, 388–399 CrossRef CAS PubMed.
- J. Park, J. T. Decker, D. R. Smith, B. J. Cummings, A. J. Anderson and L. D. Shea, J. Controlled Release, 2018, 290, 88–101 CrossRef CAS PubMed.
- E. Tatsumi, H. Yamanaka, K. Kobayashi, H. Yagi, M. Sakagami and K. Noguchi, Glia, 2015, 63, 216–228 CrossRef PubMed.
- G. S. Miranpuri, P. Bali, J. Nguyen, J. J. Kim, S. Modgil, P. Mehra, S. Buttar, G. Brown, N. Yutuc, H. Singh, A. Wood, J. Singh and A. Anand, Ann. Neurosci., 2021, 28, 219–228 CrossRef PubMed.
- R. Orihuela, C. A. McPherson and G. J. Harry, Br. J. Pharmacol., 2016, 173, 649–665 CrossRef CAS PubMed.
- Q. M. Pang, S. Y. Chen, Q. J. Xu, M. Zhang, D. F. Liang, S. P. Fu, J. Yu, Z. L. Liu, Q. Zhang and T. Zhang, Int. Immunopharmacol., 2022, 108, 108754 CrossRef CAS PubMed.
- J. Park, B. Muratori and R. Shi, Neural Regen. Res., 2014, 9, 677–683 CrossRef CAS PubMed.
- J. Park, L. Zheng, A. Marquis, M. Walls, B. Duerstock, A. Pond, S. Vega-Alvarez, H. Wang, Z. Ouyang and R. Shi, J. Neurochem., 2014, 129, 339–349 CrossRef CAS PubMed.
- S. J. Rubino, L. Mayo, I. Wimmer, V. Siedler, F. Brunner, S. Hametner, A. Madi, A. Lanser, T. Moreira, D. Donnelly, L. Cox, R. M. Rezende, O. Butovsky, H. Lassmann and H. L. Weiner, Nat. Commun., 2018, 9, 4578 CrossRef PubMed.
- Z. Chen, J. Park, B. Butler, G. Acosta, S. Vega-Alvarez, L. X. Zheng, J. Tang, R. McCain, W. P. Zhang, Z. Ouyang, P. Cao and R. Y. Shi, J. Neurochem., 2016, 138, 328–338 CrossRef CAS PubMed.
- S. Stonedahl, P. Clarke and K. L. Tyler, Vaccines, 2020, 8, 1–14 Search PubMed.
- E. Akhmetzyanova, K. Kletenkov, Y. Mukhamedshina and A. Rizvanov, Front. Syst. Neurosci., 2019, 13, 37 CrossRef CAS PubMed.
- C.-Y. Liu, X. Wang, C. Liu and H.-L. Zhang, Front. Cell. Neurosci., 2019, 13, 514 CrossRef CAS PubMed.
- M. C. Scott, S. S. Bedi, S. D. Olson, C. M. Sears and C. S. Cox, Expert Opin. Ther. Targets, 2021, 25, 365–380 CrossRef CAS PubMed.
- M. Sucksdorff, J. Tuisku, M. Matilainen, A. Vuorimaa, S. Smith, J. Keitila, J. Rokka, R. Parkkola, M. Nylund, J. Rinne, E. Rissanen and L. Airas, Neuroimmunol. Neuroinflammation, 2019, 6, e574 CrossRef PubMed.
- T. Treiber, N. Treiber and G. Meister, Nat. Rev. Mol. Cell Biol., 2019, 20, 5–20 CrossRef CAS PubMed.
- J. A. Slota and S. A. Booth, Noncoding RNAs, 2019, 5(2), 35 CAS.
- A. D. Gaudet, L. K. Fonken, L. R. Watkins, R. J. Nelson and P. G. Popovich, Neuroscientist, 2018, 24, 221–245 CrossRef CAS PubMed.
- I. Khan, K. Preeti, V. Fernandes, D. K. Khatri and S. B. Singh, Cell. Mol. Neurobiol., 2022, 42, 2075–2095 CrossRef CAS PubMed.
- Y. Guo, W. Hong, X. Wang, P. Zhang, H. Körner, J. Tu and W. Wei, Front. Mol. Neurosci., 2019, 12, 125 CrossRef CAS PubMed.
- R. Yang, X. Cai, J. Li, F. Liu and T. Sun, Med. Sci. Monit., 2019, 25, 8281 CrossRef CAS PubMed.
- G. Wan, Y. An, J. Tao, Y. Wang, Q. Zhou, R. Yang and Q. Liang, Biosci. Rep., 2020, 40(3), BSR20193315 CrossRef CAS PubMed.
- X.-Q. Li, F.-S. Chen, W.-F. Tan, B. Fang, Z.-L. Zhang and H. Ma, J. Neuroinflammation, 2017, 14, 1–12 CrossRef PubMed.
- J. Tian, T. Song, W. Wang, H. Wang and Z. Zhang, J. Mol. Neurosci., 2020, 70, 84–93 CrossRef CAS PubMed.
- X. Wu, Y. Tan, J. Zhang, R. Cui, C. Liao and S. Zhang, J. Mater. Chem. B, 2023, 11, 2916–2926 RSC.
- M. Zhang, X. Qin, Z. Zhao, Q. Du, Q. Li, Y. Jiang and Y. Luan, Nanoscale Horiz, 2022, 7, 198–210 RSC.
- J. Tian, T. Song, W. Wang, H. Wang and Z. Zhang, J. Mol. Neurosci., 2020, 70, 84–93 CrossRef CAS PubMed.
- A. Mangraviti, S. Y. Tzeng, K. L. Kozielski, Y. Wang, Y. Jin, D. Gullotti, M. Pedone, N. Buaron, A. Liu, D. R. Wilson, S. K. Hansen, F. J. Rodriguez, G. D. Gao, F. DiMeco, H. Brem, A. Olivi, B. Tyler and J. J. Green, ACS Nano, 2015, 9, 1236–1249 CrossRef CAS PubMed.
- S. W. L. Lee, C. Paoletti, M. Campisi, T. Osaki, G. Adriani, R. D. Kamm, C. Mattu and V. Chiono, J. Controlled Release, 2019, 313, 80–95 CrossRef CAS PubMed.
- C. Cai, Y. Xie, L. Wu, X. Chen, H. Liu, Y. Zhou, H. Zou, D. Liu, Y. Zhao, X. Kong and P. Liu, Sci. Rep., 2017, 7, 46250 CrossRef CAS PubMed.
- D. R. Getts, L. D. Shea, S. D. Miller and N. J. King, Trends Immunol., 2015, 36, 419–427 CrossRef CAS PubMed.
- H. Safari, W. J. Kelley, E. Saito, N. Kaczorowski, L. Carethers, L. D. Shea and O. Eniola-Adefeso, Sci. Adv., 2020, 6, eaba1474 CrossRef CAS PubMed.
- D. R. Getts, R. L. Terry, M. T. Getts, C. Deffrasnes, M. Muller, C. van Vreden, T. M. Ashhurst, B. Chami, D. McCarthy, H. Wu, J. Ma, A. Martin, L. D. Shae, P. Witting, G. S. Kansas, J. Kuhn, W. Hafezi, I. L. Campbell, D. Reilly, J. Say, L. Brown, M. Y. White, S. J. Cordwell, S. J. Chadban, E. B. Thorp, S. Bao, S. D. Miller and N. J. King, Sci. Transl. Med., 2014, 6, 219ra217 Search PubMed.
- J. Park, Y. Zhang, E. Saito, S. J. Gurczynski, B. B. Moore, B. J. Cummings, A. J. Anderson and L. D. Shea, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 14947–14954 CrossRef CAS PubMed.
- E. Saito, R. Kuo, R. M. Pearson, N. Gohel, B. Cheung, N. J. C. King, S. D. Miller and L. D. Shea, J. Controlled Release, 2019, 300, 185–196 CrossRef CAS PubMed.
- E. Saito, R. Kuo, K. R. Kramer, N. Gohel, D. A. Giles, B. B. Moore, S. D. Miller and L. D. Shea, Biomaterials, 2019, 222, 119432 CrossRef CAS PubMed.
- I. Dasgupta and A. Chatterjee, Methods Protoc., 2021, 4, 10 CrossRef CAS PubMed.
- E. Ban, T. H. Kwon and A. Kim, Drug Deliv. Transl. Res., 2019, 9, 1043–1056 CrossRef CAS PubMed.
- M. A. Nguyen, H. Wyatt, L. Susser, M. Geoffrion, A. Rasheed, A. C. Duchez, M. L. Cottee, E. Afolayan, E. Farah, Z. Kahiel, M. Cote, S. Gadde and K. J. Rayner, ACS Nano, 2019, 13, 6491–6505 CrossRef CAS PubMed.
- P. Pantazis, K. Dimas, J. H. Wyche, S. Anant, C. W. Houchen, J. Panyam and R. P. Ramanujam, Methods mol. biol., 2012, 906, 311–319 CrossRef CAS PubMed.
- K. Singha, R. Namgung and W. J. Kim, Nucleic Acid Ther., 2011, 21, 133–147 CrossRef CAS PubMed.
- F. Hu, D. Yang, B. Qian, S. Fan, Q. Zhu, H. Ren, X. Li and B. Zhai, RSC Adv., 2019, 9, 35135–35144 RSC.
- J. Park, M. Farmer, C. Casson, I. Kalashnikova and D. Kolpek, Neurotherapeutics, 2023, 20, 564–577 CrossRef CAS PubMed.
- K. Singha, R. Namgung and W. J. Kim, Nucleic Acid Ther., 2011, 21, 133–147 CrossRef CAS PubMed.
- F. Hu, D. Yang, B. Qian, S. Fan, Q. Zhu, H. Ren, X. Li and B. Zhai, RSC Adv., 2019, 9, 35135–35144 RSC.
- W. Shen, R. Wang, Q. Fan, Y. Li and Y. Cheng, Gene Ther., 2020, 27, 383–391 CrossRef CAS PubMed.
- W. Shen, Q. Wang, Y. Shen, X. Gao, L. Li, Y. Yan, H. Wang and Y. Cheng, ACS Cent. Sci., 2018, 4, 1326–1333 CrossRef CAS PubMed.
- D. R. Getts, L. D. Shea, S. D. Miller and N. J. C. King, Trends Immunol., 2016, 37, 715 CrossRef CAS PubMed.
- F. Geinguenaud, V. Militello and V. Arluison, in RNA Spectroscopy, Springer, 2020, pp. 119–133 Search PubMed.
- G. Degobert and D. Aydin, Pharmaceutics, 2021, 13, 1112 CrossRef CAS PubMed.
- J. Trifol, D. Plackett, P. Szabo, A. E. Daugaard and M. Giacinti Baschetti, ACS Omega, 2020, 5, 15362–15369 CrossRef CAS PubMed.
- X. Ma, W. Shen, X. Li, Y. Hu, X. Liu and X. Lu, Sci. Rep., 2020, 10, 1–12 CrossRef PubMed.
- K. S. Sing and R. T. Williams, Adsorpt. Sci. Technol., 2004, 22, 773–782 CrossRef CAS.
- Y. Xu, C. S. Kim, D. M. Saylor and D. Koo, J. Biomed. Mater. Res. Part B, 2017, 105, 1692–1716 CrossRef CAS PubMed.
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS PubMed.
- Y. Hua, Y. Su, H. Zhang, N. Liu, Z. Wang, X. Gao, J. Gao and A. Zheng, Drug Delivery, 2021, 28, 1342–1355 CrossRef CAS PubMed.
- R. Devulapally, N. M. Sekar, T. V. Sekar, K. Foygel, T. F. Massoud, J. K. Willmann and R. Paulmurugan, ACS Nano, 2015, 9, 2290–2302 CrossRef CAS PubMed.
- S. Arora, S. K. Swaminathan, A. Kirtane, S. K. Srivastava, A. Bhardwaj, S. Singh, J. Panyam and A. P. Singh, Int. J. Nanomedicine, 2014, 9, 2933–2942 CAS.
- Y. Liu, M. Zheng, M. Jiao, C. Yan, S. Xu, Q. Du, M. Morsch, J. Yin and B. Shi, Biomaterials, 2021, 276, 121036 CrossRef CAS PubMed.
- Y. Zhang, N. Yan, X. Wang, Y. Chang and Y. Wang, Biosci. Rep., 2019, 39(12), BSR20192009 CrossRef CAS PubMed.
- C. Porro, A. Cianciulli and M. A. Panaro, Biomolecules, 2020, 10, 1–15 CrossRef PubMed.
- D. J. Margul, J. Park, R. M. Boehler, D. R. Smith, M. A. Johnson, D. A. McCreedy, T. He, A. Ataliwala, T. V. Kukushliev and J. Liang, Bioeng. Transl. Med., 2016, 1, 136–148 CrossRef CAS PubMed.
- J. Park, J. T. Decker, D. J. Margul, D. R. Smith, B. J. Cummings, A. J. Anderson and L. D. Shea, Mol. Ther., 2018, 26, 1756–1770 CrossRef CAS PubMed.
- S. Hamzei Taj, D. Le Blon, C. Hoornaert, J. Daans, A. Quarta, J. Praet, A. Van der Linden, P. Ponsaerts and M. Hoehn, J. Neuroinflammation, 2018, 15, 1–17 CrossRef PubMed.
- K. L. Jin, X. O. Mao and D. A. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 10242–10247 CrossRef CAS PubMed.
- J. Llado, L. Tolosa and G. Olmos, Front. Cell. Neurosci., 2013, 7, 181 CAS.
- J. Li, S. Chen, Z. Zhao, Y. Luo, Y. Hou, H. Li, L. He, L. Zhou and W. Wu, Front. Cell. Neurosci., 2017, 11, 381 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00149k |
| This journal is © The Royal Society of Chemistry 2023 |