Open Access Article
Open Access ArticleSARS-CoV-2 suppression depending on the pH of graphene oxide nanosheets†
Md. Saidul
Islam
 ab,
Masahiro
Fukuda
b,
Md. Jakir
Hossain
cd,
Nurun Nahar
Rabin
ab,
Ryuta
Tagawa
a,
Mami
Nagashima
e,
Kenji
Sadamasu
e,
Kazuhisa
Yoshimura
e,
Yoshihiro
Sekine
ab,
Masahiro
Fukuda
b,
Md. Jakir
Hossain
cd,
Nurun Nahar
Rabin
ab,
Ryuta
Tagawa
a,
Mami
Nagashima
e,
Kenji
Sadamasu
e,
Kazuhisa
Yoshimura
e,
Yoshihiro
Sekine
 f,
Terumasa
Ikeda
*d and
Shinya
Hayami
f,
Terumasa
Ikeda
*d and
Shinya
Hayami
 *abg
*abg
aDepartment of Chemistry, Faculty of Advanced Science and Technology, Kumamoto University, 2-39-1 Kurokami, Kumamoto 860-8555, Japan. E-mail: hayami@kumamoto-u.ac.jp
bInstitute of Industrial Nanomaterials, Kumamoto University, 2-39-1 Kurokami, Chuo-ku, Kumamoto 860-8555, Japan
cDivision of Molecular Virology and Genetics, Joint Research Center for Human Retrovirus Infection, Kumamoto University, 2-2-1 Honjo, Kumamoto 860-0811, Japan
dGraduate School of Medical Sciences, Kumamoto University, Kumamoto 860-0811, Japan
eTokyo Metropolitan Institute of Public Health, Tokyo, Japan
fPriority Organization for Innovation and Excellence, Kumamoto University, 2-39-1 Kurokami, Chuo-ku, Kumamoto 860-8555, Japan
gInternational Research Center for Agricultural and Environmental Biology (IRCAEB), 2-39-1 Kurokami, Chuo-ku, Kumamoto 860-8555, Japan
First published on 10th March 2023
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) inactivation of pH-dependent graphene oxide (GO) nanosheets is presented. The observed virus inactivation using an authentic virus (Delta variant) and different GO dispersions at pH 3, 7, and 11 suggests that the higher pH of the GO dispersion yields a better performance compared to that of GO at neutral or lower pH. The current findings can be ascribed to the pH-driven functional group change and the overall charge of GO, favorable for the attachment between GO nanosheets and virus particles.
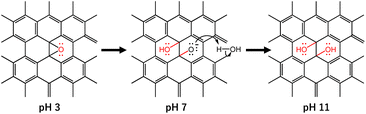
Novel coronavirus disease 2019 (COVID-19) is an infectious disease caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 first appeared at the end of 2019 in Wuhan, China, and still continues to show adverse impacts in terms of quality of life and worldwide economic sustainability.1,2 This virus is believed to spread through small liquid particles ranging from large respiratory droplets to small aerosols coming from the mouth and nose when an infected person coughs, sneezes, speaks, sings, or breathes and its likelihood of spreading increases indoors and in crowded environments. Even though the vaccination process has already been applied to stop the spread of the virus, the virus has continuously evolved with multiple mutations particularly in the spike (S) gene, which contributes to the emergence of new variants and resistance to vaccine-induced immunity. In addition, those who never develop symptoms can also pass the virus on to others, making it more complicated to prevent the virus from spreading. Therefore, the key strategies to prevent or mitigate the spread of the virus include the development of anti-SARS-CoV-2 active materials in the form of face masks and other safety equipment for use in everyday life.3–5 In light of this, recently, our group reported the inactivation of SARS-CoV-2 using graphene oxide (GO) nanosheets along with our understanding of the SARS-CoV-2 inactivation route.6 Particularly, the antiviral mechanism of GO nanosheets is associated with the attachment of virus particles on the GO surface through electrostatic interaction between the negatively charged GO and positively charged virus particles followed by the decomposition of viral proteins. Since the attachment of GO to the virus particles largely depends on the surface functional groups, which can be controlled using different parameters including the pH of the GO dispersion, some fundamental investigations including on the effect of pH on obtaining the optimum efficiency from GO-based anti-SARS-CoV-2 are important.
GO, a carbon-based two-dimensional (2D) material, owing to its exclusive physical and chemical properties, shows impressive potential and possibilities in materials science with a wide area of application including an anti-SARS-CoV-2 response.7–12 The structure of GO possesses a large ratio of oxygen-containing functional groups including epoxy, hydroxyl, and carboxyl groups.13–15 These functional groups of GO are believed to be attached to the positively charged SARS-CoV-2 viral particles and thereafter cause damage to the virus through the deactivation of the S and N (nucleocapsid) protein of the virus.6,12 Interestingly, the functional groups of GO can be altered by changing the pH of the dispersion. In particular, the epoxy groups on the GO surface are quite stable under acidic conditions. However, under alkaline conditions, the ring-opening conversion of epoxy to hydroxyl is caused by a nucleophilic base due to the large strain of the three-membered ring of the epoxy group (Scheme 1).16 In addition, the change in pH of the GO dispersion is associated with the change in the overall negative charge of GO. Therefore, pH-dependent GO is anticipated to show a difference in anti-SARS-CoV-2 activity. In the current work, we demonstrated the effect of pH in the GO dispersion on the inactivation of SARS-CoV-2. We have selected three different GO dispersions at pH 3, pH 7, and pH 11 which represent acidic, neutral, and basic conditions, respectively. We treated the virus stock with the pH dependent GO dispersions while keeping all other conditions unchanged. The virus inactivation was evaluated by plaque assay and enzyme-linked immunosorbent assay (ELISA) against N protein. The current study suggested that GO dispersion at higher pH shows comparatively higher anti-SARS-CoV-2 activity than that at neutral or lower pH.
GO was purchased commercially from Nippon Shokubai Co with an oxidation degree of 33% and used without further purification. The pH of GO was adjusted to the values of 3, 7, and 11 using the appropriate amounts of HCl and ammonia solution. GO at pH 3, 7, and 11 are denoted as GO (3), GO (7) and GO (11), respectively. The prepared GO samples were characterized using FTIR, XPS, and solid-state 13C-NMR analysis. The isolation and titration of the B.1.617.2/Delta variant (strain TKYTK1734; GISAID ID: EPI_ISL_2378732) has been described previously.17 This variant was used to investigate the antiviral activity of each GO sample against SARS-CoV-2. The plaque assay and quantification of N protein by ELISA (Proteintech, cat# KE30007) were conducted in a BSL-3 room as reported previously.6,18,19,23 The details of the experimental procedure are shown in the ESI.†
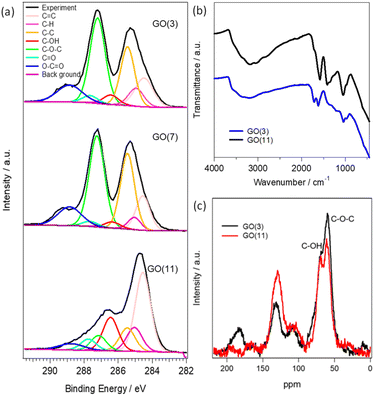
The physical and chemical properties of GO (3), GO (7) and GO (11) are shown in Fig. 1 and S1.† The optical photograph shown in Fig. S1† clearly presents a color change from brown in GO (3) to black in GO (11). The difference in the color of the GO dispersion indicates the alteration of the functional groups in GO. The changes of functional groups in GO (3) and GO (11) are compared using XPS, FTIR, and 13C NMR analysis. Fig. 1a presents the deconvoluted C 1s XPS spectra of GO (3), GO (7), and GO (11). The characteristic peaks for oxygenated functional groups include hydroxyl (C–OH), epoxy (C–O–C), carbonyl (–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and carboxyl (–COOH) groups with peak intensity approximately at 286.4–286.6 eV, 286.8–287.0 eV, 287.8–288.0 eV and 289.0–289.3 eV, respectively.21 Clearly, the peak intensity at 286.8–287.0 eV corresponding to the epoxy group is much higher in GO (3) compared to that of GO (11) indicating the presence of a large number of epoxy (C–O–C) groups in GO (3). On the other hand, the peak intensity in the hydroxyl group at 286.4–286.6 eV is higher in the case of pH (11) compared to that of GO (3). The observation indicates that the conversion of an epoxy to a hydroxyl group occurred during the pH change in GO. The result is further supported by using FTIR in Fig. 1b and 13C NMR analysis in Fig. 1c. The FTIR spectrum of GO samples is difficult to interpret due to the overlapping bands from numerous chemical bonds. However, the characteristic FTIR peaks of GO observed at approximately 1720, 1635, 1290 and 1050 cm−1 correspond to the stretching frequencies of –C
O), and carboxyl (–COOH) groups with peak intensity approximately at 286.4–286.6 eV, 286.8–287.0 eV, 287.8–288.0 eV and 289.0–289.3 eV, respectively.21 Clearly, the peak intensity at 286.8–287.0 eV corresponding to the epoxy group is much higher in GO (3) compared to that of GO (11) indicating the presence of a large number of epoxy (C–O–C) groups in GO (3). On the other hand, the peak intensity in the hydroxyl group at 286.4–286.6 eV is higher in the case of pH (11) compared to that of GO (3). The observation indicates that the conversion of an epoxy to a hydroxyl group occurred during the pH change in GO. The result is further supported by using FTIR in Fig. 1b and 13C NMR analysis in Fig. 1c. The FTIR spectrum of GO samples is difficult to interpret due to the overlapping bands from numerous chemical bonds. However, the characteristic FTIR peaks of GO observed at approximately 1720, 1635, 1290 and 1050 cm−1 correspond to the stretching frequencies of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (carboxyl and carbonyl), –C
O (carboxyl and carbonyl), –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C– (aromatic sp2 carbon), –C–O–C– (epoxy) and –COH (hydroxyl) functional groups, respectively. Moreover, the broad intense band at around 3350 cm−1 indicates the presence of hydroxyl groups or water molecules adsorbed on the hydrophilic functional sites of GO. Clearly, GO (11) shows higher intensities for the peaks at 3350 and 1050 cm−1 corresponding to the –C–OH groups while the peak intensity for the –C
C– (aromatic sp2 carbon), –C–O–C– (epoxy) and –COH (hydroxyl) functional groups, respectively. Moreover, the broad intense band at around 3350 cm−1 indicates the presence of hydroxyl groups or water molecules adsorbed on the hydrophilic functional sites of GO. Clearly, GO (11) shows higher intensities for the peaks at 3350 and 1050 cm−1 corresponding to the –C–OH groups while the peak intensity for the –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and –C–O–C– functional groups at 1720 and 1290 cm−1, respectively, decreases compared to that of GO (3).
O and –C–O–C– functional groups at 1720 and 1290 cm−1, respectively, decreases compared to that of GO (3).
The observed spectra from solid-state 13C NMR of GO (3) and GO (11) are shown in Fig. 1c. The peak intensity at 61 and 70 ppm represents the epoxides and hydroxyl functional groups, respectively.22 The relative peak intensity for the epoxy group is larger in GO (3) compared to that of GO (11). On the other hand, the peak intensity for hydroxyl groups in GO (11) slightly increased compared to that of GO (3). This observation also indicated that the change in pH from 3 to 11 of GO dispersions is associated with the conversion of epoxy to hydroxyl groups. The conversion of epoxy to hydroxyl groups is due to pH change in accordance with some previous reports.16,20 In particular, the three-membered ring of the epoxy groups possesses significant structural strain, and the ring-opening conversion of the epoxy to hydroxyl is caused by a nucleophilic base. The epoxy groups readily undergo nucleophilic attack by OH− thus relieving the strain in the three-membered rings to convert each epoxy group to a trans-vic-diol group via an SN2 reaction.16
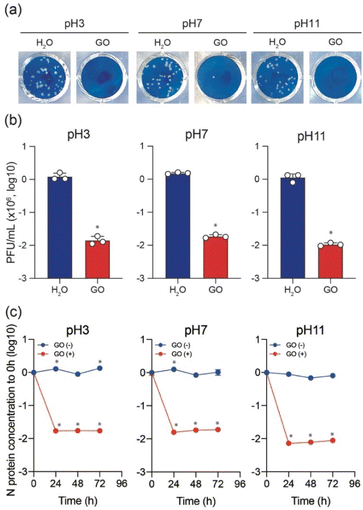
To test the variation of anti-SARS-CoV-2 activity depending on the pH of the GO dispersion, the Delta variant was used to perform plaque assay in the presence and absence of GO while keeping all other conditions unchanged. As shown in Fig. 2a and b, typical cytopathic effects were observed in the absence of GO.6,18,19,23 The treatment of the virus stock with GO (under all pH conditions) significantly decreased the plaque-forming unit (PFU). However, a slight increase in the virus inactivation was observed for both low pH and high pH GO dispersions compared to that at pH 7 (Fig. 2b). The antiviral behaviour shows an increasing order of GO (7) < GO (3) < GO (11). To further ensure the pH effect of the GO dispersion on antiviral activity, ELISA was conducted to cause the destruction of N protein in the presence of GO (3), GO (7), and GO (11) after incubation for 0, 24, 48, and 72 h, respectively. The results are shown on a log scale in Fig. 2c.
Clearly, N protein concentration was stable over time in the absence of GO, and the variations at pH 3, 7, and 11 conditions are insignificant. However, a significant decline in the N protein's concentration is observed in the first 24 h upon treating the virus stock with GO under all pH conditions. As expected, the results also support the increasing order of GO (7) < GO (3) < GO (11) for N protein destruction. It should be noted that in the current work, 1 mg mL−1 GO solutions with different pH (also a solution without GO) were diluted to 0.1 mg mL−1 in a virus dilution buffer [20 mM HEPES, 1% nonessential amino acid, 1% penicillin-streptomycin in DMEM (low glucose)] with the virus stock. Besides, DMEM (low glucose) medium includes phenol red that changes color depending on pH. We always saw the red color of the medium, which indicates no significant change in pH in the virus-GO mixture. After 1 h of incubation, GO was pelleted down in the mixture by centrifugation and the supernatant was used for infection into VeroE6/TMPRSS2 cells to examine the effect of GO on virus inhibition. Without GO (only different pH solutions with the same experimental procedure), SARS-CoV-2 still caused the formation of plaques, showing these experimental conditions to be significant for observing virus infectivity and the effect of GO.
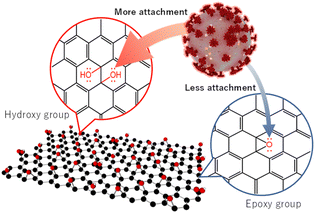
The above observations confirm that GO (11) shows a higher anti-SARS-Cov-2 response than those of GO (3) and GO (7). GO prepared using a modified Hummers oxidation route from a graphite precursor is associated with the appearance of a large number of epoxide functional groups in the basal plane and carboxyl groups at the edge, which is a remarkable feature of GO.24 However, the functional sites and the overall charges of GO can be modified by changing the pH of the GO dispersion. In a lower pH GO dispersion, the epoxy groups are much more stable in the protonated environment. Also, through a molecular dynamics simulation study C.-J. Shih and co-workers suggested that at low pH, the carboxyl groups are protonated such that the GO sheets become less hydrophilic and form a stable GO–water–GO sandwich-like structure.25 On the other hand, at higher pH, the epoxy groups of the GO dispersion are changed to hydroxyl groups. The change of functional groups between epoxide and hydroxyl on the graphene oxide is associated with the bonding nature of linking carbon atoms as well. This results in a significant local structural distortion, including changes in associated bond lengths, altering the electrostatic characteristics of the involved atomic sites and the net surface charge of GO.26 The charges on the O(epoxy) and O(hydroxyl) atoms are −0.36e and −0.70e, respectively.27 Therefore, GO dispersions having a higher percentage of hydroxyls show better elastic properties towards positive particles. In addition, in a low pH environment, the ionized COO− groups are gradually protonated, leading to a decrease in the overall negative charge of the GO. However, at higher pH, carboxyl groups are deprotonated, thus increasing the negative value of zeta potential. The zeta potentials of GO (3) and GO (11) are reported to be −24.90 and −52.20 mV, respectively which represent an overall charge decrease to a negative value, increasing the pH of GO.28 Therefore, better adsorption of positively charged virus particles is expected for GO (11) along with the presence of a large number of surface hydroxyl groups and a much lower net negative charge in the zeta potential, which is reflected in the plaque assay and ELISA of SARS-CoV-2 inactivation.6,12
Furthermore, along with a large number of –OH groups in GO (11), it might have more tendency to show H-bonding which facilitates the attachment of GO (11) and virus particles. Therefore, we propose that the functional sites of GO (11) facilitate the attachment to the virus particles followed by their inactivation compared to those of GO (3)/GO (7), resulting in more virus inactivation (Scheme 2).
In summary, GO nanosheet dispersions at three different conditions including pH 3, 7, and 11 are applied to optimize the suppression of SARS-CoV-2. Both the plaque assay and the quantification of the N protein ELISA suggest that GO (11) has better performance toward SARS-CoV-2 compared to GO (3) and GO (7). The higher SARS-CoV-2 inactivation can be ascribed to the better attachment between the virus particles and GO (11). We believe that the current findings will provide further understanding and important guidelines for the implementation of GO-based infrastructure with/post-COVID-19 society.
Author contributions
S. H., Y. S., and T. I. designed the study and conceived the main conceptual ideas. M. S. I., M. F., and R. T. prepared and characterized the graphene oxide samples. M. N, K. S. and K. Y. provided the resources and collected the data. M. J. H. did the N protein ELISA experiment. M. S. I. and N. N. R. interpreted the results and prepared the original draft manuscript. S. H., Y. S., and T. I. revised the manuscript critically for important intellectual content. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was funded by the Institute of Industrial Nanomaterials (IINa) fusion project fund, Kumamoto University, Japan (to M. S. I), a KAKENHI Grant-in-Aid for Scientific Research (Grant No. JP17H01200) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (to S. H.), JST A-STEP (JPMJTM20SL, to T. I.), a JSPS KAKENHI Grant-in-Aid for Scientific Research C (22K07103, to T. I.), JSPS Leading Initiative for Excellent Young Researchers (LEADER) (to T. I.), Takeda Science Foundation (to T. I.), Shin-Nihon Foundation of Advanced Medical Research (to T. I.), and Waksman Foundation of Japan (to T. I.), an intramural grant from Kumamoto University COVID-19 Research Projects (AMABIE).Notes and references
- COVID-19 Dashboard by the Center for Systems Science and Engineering (CSSE), Johns Hopkins University (JHU), https://coronavirus.jhu.edu/map.html Search PubMed.
- P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li, C.-L. Huang, H.-D. Chen, J. Chen, Y. Luo, H. Guo, R.-D. Jiang, M.-Q. Liu, Y. Chen, X.-R. Shen, X. Wang, X.-S. Zheng, K. Zhao, Q.-J. Chen, F. Deng, L.-L. Liu, B. Yan, F.-X. Zhan, Y.-Y. Wang, G.-F. Xiao and Z.-L. Shi, Nature, 2020, 579, 270–273 CrossRef CAS PubMed
.
- V. Palmieri and M. Papi, Nano Today, 2020, 33, 100883 CrossRef CAS PubMed
.
- A. K. Srivastava, N. Dwivedi, C. Dhand, R. Khan, N. Sathish, M. K. Gupta, R. Kumar and S. Kumar, Mater. Today Chem., 2020, 18, 100385 CrossRef CAS PubMed
.
- G. B. Ramaiah, A. Tegegne and B. Melese, Mater. Today: Proc., 2021, 47, 4357–4363 CAS
.
- M. Fukuda, M. S. Islam, R. Shimizu, H. Nasser, N. N. Rabin, Y. Takahashi, Y. Sekine, L. F. Lindoy, T. Fukuda, T. Ikeda and S. Hayami, ACS Appl. Nano Mater., 2021, 4, 11881–11887 CrossRef CAS
.
- J. Yagyu, M. S. Islam, Y. Shudo, M. Fukuda, H. Ushijima, J. Ohyama, S. Ida, L. F. Lindoy and S. Hayami, ACS Appl. Energy Mater., 2021, 4, 6296–6301 CrossRef CAS
.
- Y. Shudo, M. S. Islam, M. R. Karim, N. N. Rabin, K. Wakata, R. Ohtani, M. Nakamura, L. F. Lindoy and S. Hayami, Global Challenges, 2017, 1, 1700054 CrossRef PubMed
.
- M. S. Islam, J. Yagyu, Y. Sekine, S. Sawa and S. Hayami, Mater. Adv., 2022, 3, 3418–3422 RSC
.
- S. Islam Md, Y. Shudo and S. Hayami, Bull. Chem. Soc. Jpn., 2022, 95, 1–25 CrossRef
.
- J. Yagyu, M. S. Islam, H. Yasutake, H. Hirayama, H. Zenno, A. Sugimoto, S. Takagi, Y. Sekine, S.-I. Ohira and S. Hayami, Bull. Chem. Soc. Jpn., 2022, 95, 862–870 CrossRef CAS
.
- M. A. Unal, F. Bayrakdar, H. Nazir, O. Besbinar, C. Gurcan, N. Lozano, L. M. Arellano, S. Yalcin, O. Panatli, D. Celik, D. Alkaya, A. Agan, L. Fusco, S. Suzuk Yildiz, L. G. Delogu, K. C. Akcali, K. Kostarelos and A. Yilmazer, Small, 2021, 17, 2101483 CrossRef CAS PubMed
.
- H. He, J. Klinowski, M. Forster and A. Lerf, Chem. Phys. Lett., 1998, 287, 53–56 CrossRef CAS
.
- A. Lerf, H. He, M. Forster and J. Klinowski, J. Phys. Chem. B, 1998, 102, 4477–4482 CrossRef CAS
.
- S. Stankovich, R. D. Piner, S. T. Nguyen and R. S. Ruoff, Carbon, 2006, 44, 3342–3347 CrossRef CAS
.
- T. Taniguchi, S. Kurihara, H. Tateishi, K. Hatakeyama, M. Koinuma, H. Yokoi, M. Hara, H. Ishikawa and Y. Matsumoto, Carbon, 2015, 84, 560–566 CrossRef CAS
.
- A. Saito, T. Irie, R. Suzuki, T. Maemura, H. Nasser, K. Uriu and Y. Kosugi,
et al.
, Nature, 2022, 602, 300–306 CrossRef CAS PubMed
.
- R. Suzuki, D. Yamasoba, I. Kimura, L. Wang, M. Kishimoto, J. Ito, Y. Morioka, N. Nao, H. Nasser, K. Uriu, Y. Kosugi, M. Tsuda, Y. Orba, M. Sasaki, R. Shimizu, R. Kawabata, K. Yoshimatsu, H. Asakura, M. Nagashima, K. Sadamasu, K. Yoshimura, H. Sawa, T. Ikeda, T. Irie, K. Matsuno, S. Tanaka, T. Fukuhara and K. Sato, Nature, 2022, 603, 700–705 CrossRef CAS PubMed
.
- D. Yamasoba, I. Kimura, H. Nasser, Y. Morioka, N. Nao, J. Ito, K. Uriu, M. Tsuda, J. Zahradnik and K. Shirakawa,
et al.
, Cell, 2022, 185, 2103–2115 CrossRef CAS PubMed
.
- M. Fukuda, M. S. Islam, Y. Shudo, J. Yagyu, L. F. Lindoy and S. Hayami, Chem. Commun., 2020, 56, 4364–4367 RSC
.
- H. Takehira, M. S. Islam, M. R. Karim, Y. Shudo, R. Ohtani, L. F. Lindoy, T. Taniguchi, M. Osada and S. Hayami, ChemistrySelect, 2017, 2, 6941–6944 CrossRef CAS
.
- A. M. Dimiev, L. B. Alemany and J. M. Tour, ACS Nano, 2013, 7, 576–588 CrossRef CAS PubMed
.
- I. Kimura, D. Yamasoba, T. Tamura, N. Nao, T. Suzuki, Y. Oda, S. Mitoma, J. Ito, H. Nasser, J. Zahradnik, K. Uriu, S. Fujita, Y. Kosugi, L. Wang, M. Tsuda, M. Kishimoto, H. Ito, R. Suzuki, R. Shimizu, M. M. Begum, K. Yoshimatsu, K. T. Kimura, J. Sasaki, K. Sasaki-Tabata, Y. Yamamoto, T. Nagamoto, J. Kanamune, K. Kobiyama, H. Asakura, M. Nagashima, K. Sadamasu, K. Yoshimura, K. Shirakawa, A. Takaori-Kondo, J. Kuramochi, G. Schreiber, K. J. Ishii, T. Hashiguchi, T. Ikeda, A. Saito, T. Fukuhara, S. Tanaka, K. Matsuno and K. Sato, Cell, 2022, 185, 3992–4007 CrossRef CAS PubMed
.
- M. R. Karim, M. S. Islam, K. Hatakeyama, M. Nakamura, R. Ohtani, M. Koinuma and S. Hayami, J. Phys. Chem. C, 2016, 120, 21976–21982 CrossRef CAS
.
- C.-J. Shih, S. Lin, R. Sharma, M. S. Strano and D. Blankschtein, Langmuir, 2012, 28, 235–241 CrossRef CAS PubMed
.
- F. Abbasi, J. Karimi-Sabet, C. Ghotbi, Z. Abbasi, S. A. Mousavi and N. Amini, Sci. Iran., 2017, 24, 3554–3559 Search PubMed
.
- A. J. P. Neto, V. V. Chaban and E. E. Fileti, Phys. Chem. Chem. Phys., 2017, 19, 32333–32340 RSC
.
- B. Konkena and S. Vasudevan, J. Phys. Chem. Lett., 2012, 3, 867–872 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00084b |
| This journal is © The Royal Society of Chemistry 2023 |