Open Access Article
Open Access ArticleSortase A transpeptidation produces seamless, unbranched biotinylated nanobodies for multivalent and multifunctional applications†
Eugene M.
Obeng
 a,
David L.
Steer
b,
Alex J.
Fulcher
c and
Kylie M.
Wagstaff
a,
David L.
Steer
b,
Alex J.
Fulcher
c and
Kylie M.
Wagstaff
 *a
*a
aDepartment of Biochemistry and Molecular Biology, Biomedicine Discovery Institute, Monash University, Clayton, VIC 3800, Australia. E-mail: kylie.wagstaff@monash.edu
bMonash Proteomics and Metabolomics Facility, Monash University, Clayton, VIC 3800, Australia
cMonash Micro Imaging, Monash University, Clayton, VIC 3800, Australia
First published on 15th March 2023
Abstract
Exploitation of the biotin–streptavidin interaction for advanced protein engineering is used in many bio-nanotechnology applications. As such, researchers have used diverse techniques involving chemical and enzyme reactions to conjugate biotin to biomolecules of interest for subsequent docking onto streptavidin-associated molecules. Unfortunately, the biotin–streptavidin interaction is susceptible to steric hindrance and conformational malformation, leading to random orientations that ultimately impair the function of the displayed biomolecule. To minimize steric conflicts, we employ sortase A transpeptidation to produce quantitative, seamless, and unbranched nanobody–biotin conjugates for efficient display on streptavidin-associated nanoparticles. We further characterize the protein–nanoparticle complex and demonstrate its usefulness in optical microscopy and multivalent severe acute respiratory syndrome coronavirus (SARS-CoV-2) antigen interaction. The approach reported here provides a template for making novel multivalent and multifunctional protein complexes for avidity-inspired technologies.
Introduction
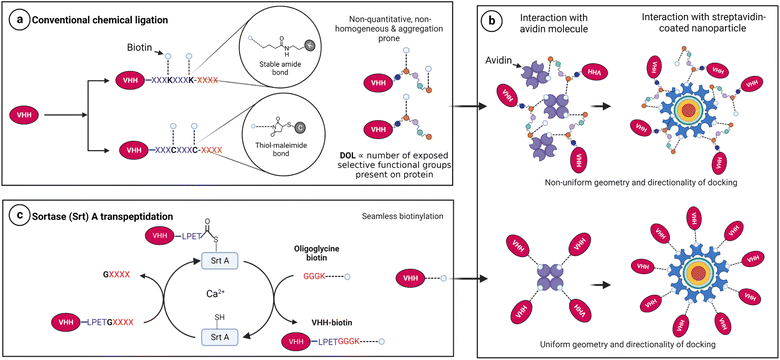
Biotinylation remains an instrumental approach to confer multifunctional properties to proteins via the exploitation of the extremely strong biotin–streptavidin interaction.1 For this purpose, researchers have employed mostly chemical2–4 and enzymatic5–8 approaches to append biotin molecules to proteins of interest for subsequent docking onto streptavidin-associated technologies. Unfortunately, many available chemical conjugation approaches are stochastic and non-orthogonal and often yield heterogeneous products with compromised functionality.9–12 For instance, N-hydroxysuccinimide (NHS)-ester chemistry and thiol–maleimide reactions target surface-exposed, reactive amine groups (e.g., the N-terminal and ε-NH2 side chains of lysine) and the sulfhydryl (–SH) side chain of cysteine residues, respectively.12–14 The caveat here is that proteins with multiple reactive side chains inevitably encounter a non-selective and stochastic degree of labelling (Fig. 1a), leading to products with conformational heterogeneity and malformations.12,15,16Notably, conformational freedom is critical for efficient molecular function, especially for biomolecules with limited flexibility and stringent function-associated orientations.17–20 A typical example is native tetrameric (strept)avidin, which exists as a dimer–dimer, with each subunit contributing to the biotin-binding site of the neighboring monomer.21–23 The biotin-binding site is approachable from one terminal end within the tetrameric architecture.21,22,24 With this complexity, it is known that random biotin orientations significantly affect the docking strength of biotin–streptavidin systems.3,25 Also, single molecule insights suggest that both uniform directionality and center of mass are critical for biotin–streptavidin binding and unbinding.26 Moreover, the forces that hold biotin–streptavidin docking depend on the attachment geometry of biotin.27 These essential requirements are practically challenging to overcome with conventional chemical modification approaches because of their stochastic nature (Fig. 1b).
On the other hand, the use of enzymes such as sortase A (Srt A) guarantees a stoichiometric, seamless, and unbranched biotinylation of proteins of interest near physiological conditions (Fig. 1c). We hereby demonstrate Srt A assisted multivalent protein–nanoparticle complexation via biotin–streptavidin interaction. We perform physical characterization of the chimeric protein–nanoparticle complex and demonstrate the functional usefulness in optical microscopy and multivalent interaction with a severe acute respiratory syndrome coronavirus (SARS-CoV-2) antigen.
Experimental
Bacteria cell culture and protein expression
All nanobody plasmids were synthesized by Genscript. The sortase (Srt) A and mCherry plasmids were kind gifts from David Liu (Addgene plasmid #75144) and Jeanne Stachowiak (Addgene plasmid #113552), respectively. The plasmids were individually transformed into Escherichia coli SHuffle® and grown on terrific broth (TB) agar with kanamycin (50 μg mL−1) and ampicillin (100 μg mL−1) selection for Srt A and mCherry, respectively. A single colony of transformed bacteria was seeded overnight at 30 °C (200 rpm, shaking) and used as an inoculum for protein expression. For protein expression, the inoculum (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) was seeded in TB media with 50 μg mL−1 kanamycin and incubated at 30 °C (200 rpm, shaking) until OD600 of 0.6–0.9 before induction with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Dioxane-Free; Promega). The culture was kept overnight at 16 °C (200 rpm, shaking) post-IPTG induction. The bacteria pellets were harvested via centrifugation (4000×g, 20 min, 10 °C) and stored at −80 °C until ready to use.
30) was seeded in TB media with 50 μg mL−1 kanamycin and incubated at 30 °C (200 rpm, shaking) until OD600 of 0.6–0.9 before induction with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Dioxane-Free; Promega). The culture was kept overnight at 16 °C (200 rpm, shaking) post-IPTG induction. The bacteria pellets were harvested via centrifugation (4000×g, 20 min, 10 °C) and stored at −80 °C until ready to use.
Mammalian cell culture and protein expression
The plasmid containing SARS-CoV-2 spike receptor binding domain (S-RBD; a.a. 333-529) fused to Fc of IgG1 was a gift from Erik Procko (Addgene plasmid #141183). About 50 mg plasmid DNA was transfected into 293F suspension cells (107 total cells in 25 mL media) using polyethyleneimine reagent (PEI MAX; Polysciences). Cells were cultured in Freestyle 293F media at 37 °C (120 rpm, 8% CO2). The total expression volume was 50 mL, which was harvested through centrifugation (4000×g, 20 min, 10 °C) 7 days post transfection. The supernatant was stored at −20 °C until ready to use.Purification of proteins
Nickle affinity chromatography was used for all nanobodies, Srt A and mCherry protein purifications through the ÄKTA Start system (GE Healthcare). The bacteria pellet was resuspended in lysis buffer (40 mM Tris, 0.5 M NaCl, 1 mM EDTA, 0.5% Triton X-100, 20 mM imidazole, pH 8) and sonicated for 15 min (15/45 s ON/OFF pulse, 90% amplitude; QSonica). The lysate was centrifuged at 4000×g (for 30 min, 10 °C), and the supernatant was retained for purification. The supernatant was sterile-filtered prior to loading onto HisTrap FF (1 mL) columns. The purification was performed using binding buffer (40 mM Tris, 0.5 M NaCl, 20 mM imidazole, pH 8) and followed by an optimized step elution process using elution buffer (40 mM Tris, 0.5 M NaCl, 250 mM imidazole, pH 8). The eluted proteins were buffer-exchanged into tris-buffered saline (TBS; 40 mM Tris, 0.15 M NaCl, pH 8) using HiTrap desalting column (5 mL, GE Healthcare) after confirmation using SDS-PAGE. The proteins were concentrated with Amicon® Ultra-4 10 K and stored at −20 °C until ready to use.Protein A affinity chromatography was used for SARS-CoV-2 spike RBD-Fc purification. According to the manufacturer's instruction, the harvested supernatant was conditioned with 3 M NaCl (pH 7.5) and loaded onto HiTrap protein A HP (1 mL, GE Healthcare) columns using binding buffers (20 mM sodium phosphate, 3 M NaCl, pH 7.0) and eluted in elution buffer (100 mM sodium citrate, pH 3.0). The eluate was neutralized with 1 M Tris–HCl (pH 9). The neutralized eluted protein was buffer-exchanged into TBS after confirmation with SDS-PAGE.
Sortase A assisted biotinylation and protein nanoparticle complex synthesis
The Srt A reaction was performed as previously reported.28 In short, 30 μM nanobody, 90 μM Srt A, 2 mM GGGK-biotin peptide and 10 mM CaCl2 were mixed and reacted in TBS. At a total volume of 500 μL, the reaction was incubated for 4 h at 42 °C (800 rpm), followed by separation with magnetic Ni2+–NTA resin. The individual fractions were run on an SDS-PAGE to confirm the completion of the reaction and reasonable product separation. The biotinylated nanobody was buffer-exchanged into TBS and analyzed with electrospray ionization (ESI) mass spectrometry techniques. For anti-mCherry QNC, the biotinylated nanobody was mixed (40 molar excess) with streptavidin-coated Qdot 525 (Invitrogen™). The anti-SARS-CoV-2-S NAC was synthesized by mixing the biotinylated nanobody (5 molar excess) with avidin (Pierce™, Thermo Scientific). The mixtures were incubated at room temperature for 30 min and separated using an Amicon® Ultra-4 100 K.LC-MS and ESI intact protein analysis
Protein samples were analysed by LC-MS using a quadrupole TOF mass spectrometer (MicroTOFq, Bruker Daltonics, Bremen, Germany) coupled online with a 1200 series capillary HPLC (Agilent Technologies, Santa Clara, CA, USA). Samples were injected onto a MabPac SEC-1 5 μm 300 Å 50 × 4 mm (Thermo Scientific) column with 50% acetonitrile 0.05% TFA, 0.05% FA at a flow rate of 50 μL min−1. The protein is eluted over a 20 min run-time monitored by UV detection at 254 nm. The eluant is nebulised and ionised using the Bruker electrospray source with a capillary voltage of 4500 V dry gas at 180 °C, a flow rate of 4 L min−1 and nebuliser gas pressure at 300 mbar. After 20 min, the flow path is switched to infuse low-concentration tune mix (Agilent Technologies, Santa Clara, CA, USA) to calibrate the spectrum post-acquisition. The spectra were extracted and deconvoluted using Data explorer software version 3.4 build 192 (Bruker Daltonics, Bremen, Germany).Transmission electron microscopy (TEM) and dynamic light scattering (DLS) analysis
A negatively stained TEM protein particle analysis was employed to characterize the Qdot samples.29 About 8 μL of sQdot (0.8 μM) and QNC (0.5 μM) were loaded separately onto ultrathin holey carbon film supported by a 300-mesh copper grid (Ted Pella Inc.) and incubated for 5 min. The grids were glow-discharged before the addition of samples. The incubated grids containing the samples were subsequently blot-dried with a clean filter paper, followed by staining with 2% (v/v) uranyl acetate for 1 min to enhance particle contrast, as described previously.29,30 The sQdot and QNC were imaged with FEI Tecnai G2 T20 at 120 kV accelerating voltage using the inbuilt Gatan Orius SC600 (bottom-mounted camera). Further image processing was done using ImageJ software (NIH). Size characterizations were performed on Zetasizer Nano (Malvern Panalytical).Functional assessment of anti-mCherry QNC
The mCherry-coated beads were synthesized by saturating Ni2+–NTA beads with purified His6-mCherry protein, followed by repeated washing with TBS. About 5 μL anti-mCherry QNC (0.5 μM) was added to 50 μL beads in TBS and incubated for 15 min, followed by repeated washing. Glass slides were prepared and imaged using a Zeiss LSM 980 Airyscan 2 microscope (Carl-Zeiss AG, Jena, Germany) with 63× oil C-PlanApo 1.4NA objective. The optimal lasers and filter setup for the QNC and mCherry were 445 and 561 nm, respectively. This choice was made to avoid cross-excitation. Further image processing and analysis were performed using the Zeiss software and ImageJ (NIH).Biolayer interferometry functional assessment of anti-SARS-CoV-2 NAC
The BLItz system (FortéBio) was used for the functional assessment of the anti-SARS-CoV-2-S NAC. First, hydrated anti-human IgG Fc capture (AHC) biosensors were immobilized with purified SARS-CoV-2 spike RBD-Fc followed by equilibration with TBS. Different concentrations of analyte (i.e., anti-SARS-CoV-2-S VHH-biotin or anti-SARS-CoV-2-S NAC) were applied, followed by a dissociation step. The BLItz Pro software was used to fit a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model to determine the binding affinities. The data were replotted with GraphPad Prism Software (v9; Dotmatics).
1 binding model to determine the binding affinities. The data were replotted with GraphPad Prism Software (v9; Dotmatics).
Results and discussion
Sortase A assisted biotinylation of nanobodies
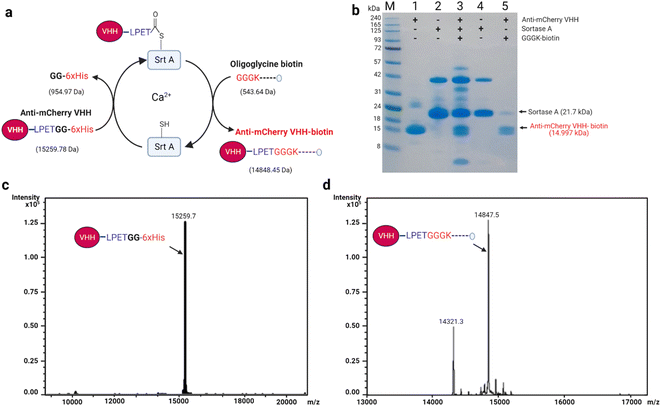
In Srt A transpeptidation (Fig. 2a), the enzyme first recognizes a pentapeptide motif (LPXTG) on any fusion molecule, cleaves between the threonine (T) and glycine (G), and then replaces any sequence C-terminal to the threonine with molecules bearing N-terminal oligoglycine.31,32 The oligoglycine molecule is preferably a tri- or pentaglycine molecule.33 Also, the “X” in the recognition sequence could be any amino acid.34,35 With these in place, and provided that the N-terminus of the oligoglycine is available, many C-terminal decorations such as fluorophores, drugs, lipids, carbohydrates, nucleic acids and biotin are permissible and processible by the enzyme.36–42For demonstration purposes, we chose to utilize nanobodies (VHHs), which are single-domain antigen-specific binders derived from the heavy-chain-only antibodies of camelids.43 VHHs have found enormous and incredible applications in research, diagnostics, and therapeutics over the years.44–48 Specifically, anti-mCherry (a.k.a. LaM-4)49 and anti-SARS-CoV-2-S (a.k.a. Nb15)50 VHHs were subjected in this study to Srt A transpeptidation to yield seamless biotinylated nanobody products.
As depicted in Fig. 2a for anti-mCherry VHH (VHH-LPETG-6xHis; 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259.78 Da, est.), the assistance of Srt A was predicted to capably produce seamless unbranched anti-mCherry VHH-biotin (14
259.78 Da, est.), the assistance of Srt A was predicted to capably produce seamless unbranched anti-mCherry VHH-biotin (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 843.45 Da, est.) via transpeptidation with oligoglycine-biotin molecules (543.64 Da, est.). The composition of each stage of the transpeptidation process is depicted using SDS-PAGE (Fig. 2b), where the last lane shows the purified anti-mCherry VHH-biotin. To confirm the exactness of the estimated (est.) masses, we employed electrospray ionization (ESI) spectrometry to assess the actual (act.) or “intact” masses pre- and post-transpeptidation. As expected, the unmodified anti-mCherry VHH (15
843.45 Da, est.) via transpeptidation with oligoglycine-biotin molecules (543.64 Da, est.). The composition of each stage of the transpeptidation process is depicted using SDS-PAGE (Fig. 2b), where the last lane shows the purified anti-mCherry VHH-biotin. To confirm the exactness of the estimated (est.) masses, we employed electrospray ionization (ESI) spectrometry to assess the actual (act.) or “intact” masses pre- and post-transpeptidation. As expected, the unmodified anti-mCherry VHH (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 259.7 Da, act.) and anti-mCherry VHH-biotin (14
259.7 Da, act.) and anti-mCherry VHH-biotin (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 847.2 Da, act.) were in close agreement with the estimated masses (Fig. 2c and d). Cleaved but unbiotinylated anti-mCherry VHH (14
847.2 Da, act.) were in close agreement with the estimated masses (Fig. 2c and d). Cleaved but unbiotinylated anti-mCherry VHH (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 321.3 Da, act.) was also detected in the modified sample due to incomplete transpeptidation and substrate hydrolysis (Fig. 2d). Also, the transpeptidation yield may likely be influenced by the ability of the enzyme to recognize the conjugated molecule as a new substrate because of the reformation of the LPETG motif.
321.3 Da, act.) was also detected in the modified sample due to incomplete transpeptidation and substrate hydrolysis (Fig. 2d). Also, the transpeptidation yield may likely be influenced by the ability of the enzyme to recognize the conjugated molecule as a new substrate because of the reformation of the LPETG motif.
Similarly, anti-SARS-CoV-2-S VHH (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 121.7 Da, est.; 15
121.7 Da, est.; 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120.30 Da act.) was biotinylated via Srt A transpeptidation into anti-SARS-CoV-2-S VHH-biotin (14
120.30 Da act.) was biotinylated via Srt A transpeptidation into anti-SARS-CoV-2-S VHH-biotin (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 710.37 Da, est.; 14
710.37 Da, est.; 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 707.70 Da act.) (Fig. 3a–d). As was observed for anti-mCherry VHH, cleaved but unbiotinylated anti-SARS-CoV-2-S VHH was detected by ESI (Fig. 3d). The subtle differences between the estimated and actual measurements across these described modifications may potentially be a result of instrument sensitivity. Nevertheless, Srt A-mediated biotinylation was found to be efficient, paving the way for subsequent functional assessments.
707.70 Da act.) (Fig. 3a–d). As was observed for anti-mCherry VHH, cleaved but unbiotinylated anti-SARS-CoV-2-S VHH was detected by ESI (Fig. 3d). The subtle differences between the estimated and actual measurements across these described modifications may potentially be a result of instrument sensitivity. Nevertheless, Srt A-mediated biotinylation was found to be efficient, paving the way for subsequent functional assessments.
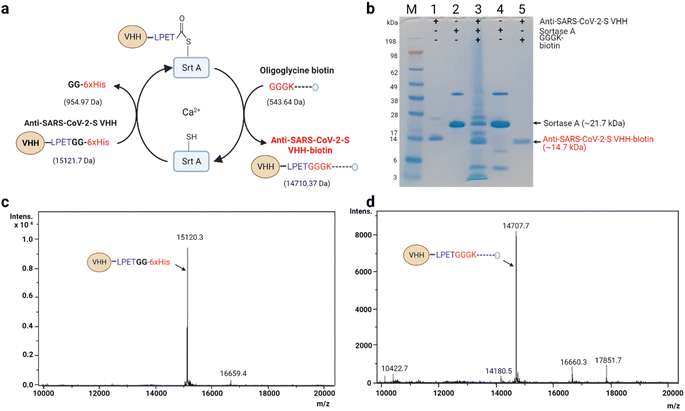 | ||
| Fig. 3 Seamless biotinylation of anti-SARS-CoV-2-S VHH via Srt A transpeptidation. (a) Schematic description of stage-wise molecular weight changes. The details of the reaction are similar to the description in Fig. 2. The estimated molecular weight changes are indicated in brackets. (b) SDS-PAGE analysis of the anti-SARS-CoV-2-S VHH transpeptidation process. The last lane shows the state of the purified anti-SARS-CoV-2-S VHH-biotin. M: molecular weight marker; (1) anti-SARS-CoV-2-S VHH; (2) Srt A protein; (3) reaction mixture containing anti-SARS-CoV-2-S VHH, Srt A and GGGK-biotin; (4) separated Srt A (end of reaction); (5) biotinylated anti-SARS-CoV-2-S VHH. (c) ESI spectra of unmodified anti-SARS-CoV-2-S VHH (note: this sample was not subjected to Srt A transpeptidation). (d) Sample ESI spectra of anti-SARS-CoV-2-S VHH-biotin prepared via Srt A transpeptidation. | ||
Enzymes such as Escherichia coli biotin ligase (BirA) could also be used for protein biotinylation.6,51–55 Like Srt A, BirA recognizes a motif (GLNDIFEAQKIEWHE; Avi-tag) on the protein of interest and tethers a biotin molecule to the lysine (K) within the tag. This reaction can occur irrespective of the tag's position (e.g., N-, C-terminus and internal locations) as long as it remains exposed.5 However, there is no scission of any part of the 15-amino acid recognition motif and, thus, results in a non-seamless connection. Also, the length of the tag may influence the terminal orientation and steric pressure posed in the presence of streptavidin, as discussed earlier. There has also been the practice of fusing streptavidin-binding peptides genetically to proteins for association with streptavidin.56–58 These peptides are long (between 9 and 38 amino acids) and exhibit a moderate (nanomolar) binding behaviour, unlike the femtomolar binding strength of biotin with streptavidin. On the other hand, the Srt A approach reported in this study uses relatively short external fusions to produce seamless unbranched biotinylated products. The seamlessness provided by Srt A ligation has the penchant for preserving the associated binding strength of biotin to streptavidin.
Synthesis and characterization of multivalent protein nanoparticle chimera via biotin–streptavidin interaction
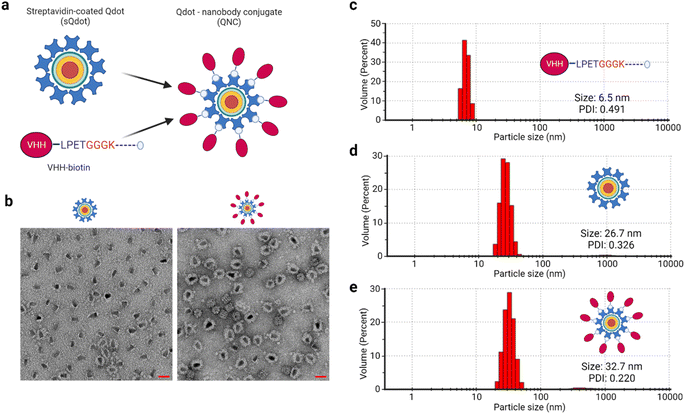
Multivalent nanotechnologies are essential because they minimize off-target consequences during selective targeting applications.59,60 To this end, we synthesized multivalent quantum dot–nanobody chimera (QNC) and nanobody–avidin conjugates (NAC) (Fig. S1†) by exploiting biotin–(strept)avidin non-covalent interaction and tested their application in optical microscopy and SARS-CoV-2-S antigen interaction, respectively. Quantum dots (Qdots) are nanocrystal fluorophores (<10 nm) with exceptional optical, magnetic and electronic capabilities.61,62 They are made with various compounds such as cadmium selenide (CdSe), cadmium sulphite (CdS), cadmium telluride (CdTe), and zinc oxide (ZnO), with core–shell formats available. The smaller size, high quantum efficiency and stability render Qdots suitable candidates for fluorescent probe synthesis. The Qdot used herein is a CdSe/ZnS core–shell type.To synthesize the QNC (Fig. 4a), a 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometric mix of anti-mCherry VHH-biotin and streptavidin-coated quantum dot (sQdot) was made, given that each molecule of quantum dot (Qdot) has more than seven molecules of streptavidin attached. We physically analysed the chimeric protein–nanoparticle complex using negative-stained transmission electron microscopy (TEM), which showed a clear difference in the protein shield between the sQdot and the QNC (Fig. 4b), indicating the attachment of the VHH. To confirm this, a particle size analysis of anti-mCherry VHH-biotin, sQdot and QNC was performed via dynamic light scattering (DLS) measurement, yielding sizes of 6.5, 26.7, and 32.7 nm, respectively (Fig. 4c–e). The incremental size changes from the DLS analysis corroborate the TEM data.
1 stoichiometric mix of anti-mCherry VHH-biotin and streptavidin-coated quantum dot (sQdot) was made, given that each molecule of quantum dot (Qdot) has more than seven molecules of streptavidin attached. We physically analysed the chimeric protein–nanoparticle complex using negative-stained transmission electron microscopy (TEM), which showed a clear difference in the protein shield between the sQdot and the QNC (Fig. 4b), indicating the attachment of the VHH. To confirm this, a particle size analysis of anti-mCherry VHH-biotin, sQdot and QNC was performed via dynamic light scattering (DLS) measurement, yielding sizes of 6.5, 26.7, and 32.7 nm, respectively (Fig. 4c–e). The incremental size changes from the DLS analysis corroborate the TEM data.
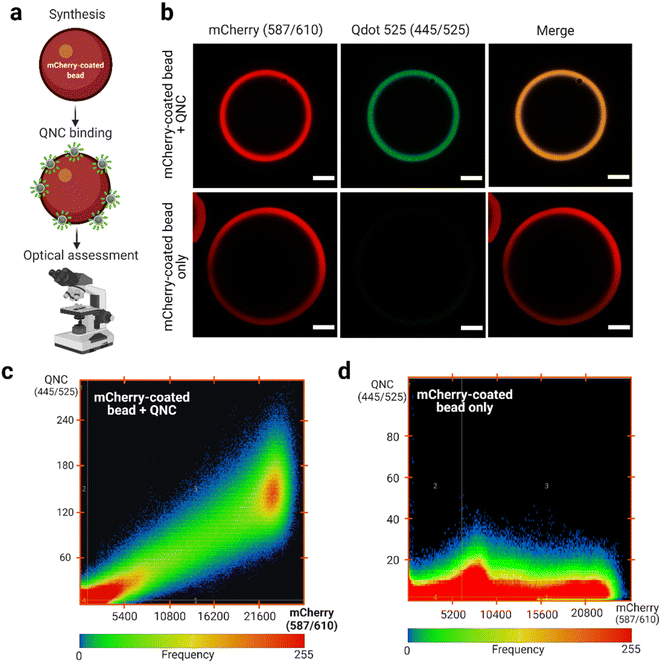
Functional assessment of anti-mCherry QNC
Monomeric anti-mCherry VHH binds to mCherry at high affinity (KD = 0.18 nM).49 According to Shcherbo et al.63 mCherry has excitation and emission wavelengths of 587 and 610 nm, respectively. Therefore, selecting a Qdot with excitation and emission wavelengths of 445 and 525 nm, respectively, provides distinct optical properties for assessing binding (Fig. 5a). It was observed that the anti-mCherry QNC remains functional by interacting with the mCherry-coated bead as determined by confocal laser scanning microscopy (CLSM; Fig. 5b). No signal was observed in the 445/525 channel in the absence of the QNC (Fig. 5b, bottom panels). Analysis of the pixel fluorogram between the test (+QNC) and control (−QNC) samples also revealed a clear molecular colocalization between the anti-mCherry QNC and the mCherry-coated beads.Notably, previous studies have attempted to orient VHHs on Qdots using thiol–maleimide chemistry,64 VHH-rhizavidin fusion65 and genetic code expansion techniques.25,66 However, thiol–maleimide-dependent bioconjugates are sometimes unstable and may implicate variable outcomes.67,68 Contrary, the VHH-rhizavidin fusion system is not well-characterized and, thus, has experienced limited usage. However, the genetic code expansion technique is a developing approach and, unfortunately, experiences critical translational challenges during protein synthesis due to the foreign nature of the amino acids required.69,70 Nonetheless, the biotin–streptavidin approach remains well-established even as the Srt A enzyme complementation addresses the site-specific and orientational requirements. Overall, this QNC synthesis technique reported here may have extended applications in biosensing and single particle tracking of cellular occurrences.
Functional assessment of the anti-SARS-CoV-2-S nanobody–avidin conjugate (NAC)
According to Xu et al.50 monomeric anti-SARS-CoV-2-S VHH (Nb15) is a receptor binding site (RBS) nanobody that interacts with the SARS-CoV-2 spike receptor binding domain (S-RBD) at high affinity (KD = 8.15 nM). A trimeric format derived through genetic linker fusion also showed an improved binding affinity of 1.64 × 10−11 M.50 Considering avidin permits tetravalent protein complex formation,71,72 we anticipated that a tetravalent nanobody–avidin conjugate (NAC) will have enhanced “avidified” S-RBD binding characteristics.73Fig. 6a shows a cartoon structure of a typical RBS nanobody. Using biolayer interferometry analysis (Fig. 6b), we identified that anti-SARS-CoV-2-S VHH-biotin has an interestingly high affinity (KD = 1.017 nM) towards the spike RBD in the same order of magnitude as reported by the source (Fig. 6c).50 The slight binding affinity difference observed between groups may be due to differences in sample preparation and instrumentation. More interestingly, we identified that the NAC binding with S-RBD-Fc was almost irreversible (KD < 1 pM; Fig. 6d) due to its significantly low dissociation rate. The low dissociation rate infers how the complex stays together in actuality, although the association rate is lower than the monomer (Fig. 6c and d). These findings make the NAC worthy of future assessment in terms of actual virus inhibition.
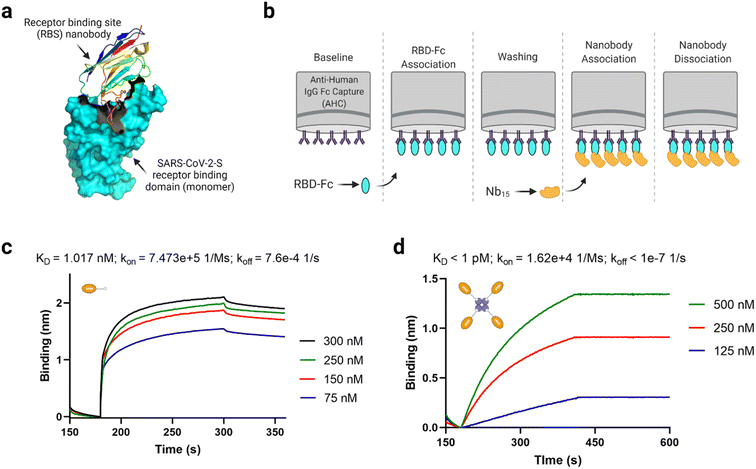 | ||
| Fig. 6 Functional assessment of the anti-SARS-CoV-2-S nanobody–avidin conjugate (NAC). (a) A cartoon structure of a typical receptor binding site (RBS) nanobody (PDB ID: 7B17). (b) Schematic depiction of the biolayer interferometry protocol used to assess binding. (c) Biolayer interferometry analysis of anti-SARS-CoV-2-S VHH-biotin binding with spike RBD-Fc per the description in (b). (d) Biolayer interferometry analysis of NAC binding with spike RBD-Fc per the description in (b). The exact KD value was beyond the detection limit of the BLItz instrument due to the low dissociation rate. It should be noted that one molecule of tetrameric avidin can accommodated up to 4 molecules of biotinylated molecule depending on the availability of exposed binding pockets. | ||
Notably, the irreversible binding characteristic exhibited by the NAC is typical of multivalent and ultrapotent binders, as has been reported for other nanobodies73,74 and angiotensin-converting enzyme 2 (ACE2) decoy receptors.75 There is a closely related example, where an anti-SARS-CoV-2 (Ty1) nanobody was decorated with a click chemistry handle and later fused to a 4-arm polyethylene glycol (PEG) based scaffold via strain-promoted azide–alkyne click chemistry.76 This provided a remarkable enhancement in SARS-CoV-2 pseudovirus neutralization to an IC50 value of about 13 pM. The effectiveness was putatively due to the tetrameric nanobody's ability to cross-link viruses into large immune complexes. The NAC reported in this work may likely achieve a similar result and be useful in future treatment technologies, especially when low or non-immunogenic streptavidin versions are used.77,78 In this way, the NAC may escape adverse immune reactions associated with PEGylated drugs.79
It is worth noting that Srt A-assisted biotinylation has been reported, especially for bio-imaging application.80–84 However, the data in this work further highlights the significance of Srt A-assisted oligomerization centered on biotin–streptavidin interaction. Furthermore, the evidence shows how multivalency substantially improves the binding characteristics of affinity-based molecules such as nanobodies. These finding and considerations may foster future developments in nanomedicine.
Conclusion
In this study, we have demonstrated that Srt A transpeptidation is a versatile approach for generating seamless, unbranched biotinylated biomolecules for multifunctional and multivalent nanotechnologies. The seamless attachment of biotin facilitated a steric-free docking of nanobodies onto (strept)avidin-associated molecules. The QNC synthesized in this study showed incredible labelling of its cognate molecule, making the technique promising for extended applications in biosensing and single-particle tracking of cellular functions. Similarly, the NAC will likely find further applications in avidity-inspired nanomedicine technologies for disease treatment, provided safety requirements are met. Overall, the approach reported here provides a template for making novel multivalent and multifunctional protein complexes.Abbreviations
| AHC | Anti-human IgG Fc capture |
| BLI | Biolayer interferometry |
| DLS | Dynamic light scattering |
| DNA | Deoxyribonucleic acid |
| DOL | Degree of labelling |
| ESI | Electrospray ionization mass spectrometry |
| Fc | Crystallizable fragment of antibody |
| IPTG | Isopropyl β-D-1-thiogalactopyranoside |
| K D | Dissociation constant |
| LB | Luria-Bertani broth |
| LC-MS | Liquid chromatography-mass spectrometry |
| NAC | Nanobody–avidin conjugate |
| NHS | N-Hydroxysuccinimide |
| Ni2+–NTA | Nickel–nitrilotriacetic acid resin |
| OD600 | Optical density at 600 nm |
| PEG | Polyethylene glycol |
| QNC | Quantum dot—nanobody chimera/conjugate |
| SARS-CoV-2 | Severe acute respiratory syndrome coronavirus 2 |
| SARS-CoV-2-S | Severe acute respiratory syndrome coronavirus 2 spike protein |
| SH | Sulfhydryl side chain of cysteine |
| sQdot | Streptavidin-coated quantum dot |
| SRBD | Spike receptor binding domain |
| Srt A | Sortase A |
| TB | Terrific broth |
| TBS | Tris-buffered saline |
| TEM | Transmission electron microscopy |
| VHH | Variable domain of the heavy chain only antibody of camelids |
Data availability
All data are available from the corresponding authors upon reasonable request.Author contributions
E. M. O.: conceptualization, investigation, formal analysis, writing – original draft, writing – review and editing, validation, visualization. D. L. S.: investigation, formal analysis. A. F.: investigation, writing – review and editing, supervision. K. M. W.: writing – review and editing, supervision.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
EMO acknowledges a Monash Graduate Scholarship, a Monash Departmental Scholarship and a Monash Postgraduate Publications Award for his PhD studies. The authors are grateful to Dr Lizhong He for providing the plasmid for anti-mCherry nanobody, Dr Simon R. Corrie for access to the Akta Start purification system, Monash Center for Electron Microscopy, and the Monash Ramaciotti Centre for Cryo-electron Microscopy for access to their TEM facilities. All Confocal Laser Scanning Microscopy was performed at the Monash Micro Imaging Facility. All schematics were drawn and assembled with https://www.biorender.com/.References
- C. M. Dundas, D. Demonte and S. Park, Appl. Microbiol. Biotechnol., 2013, 97, 9343–9353 CrossRef CAS PubMed
.
- A. Muraoka, Y. Matsuura, H. Naitow, M. Ihara and N. Kunishima, Anal. Biochem., 2018, 557, 46–58 CrossRef CAS PubMed
.
- K. Boeneman, J. R. Deschamps, S. Buckhout-White, D. E. Prasuhn, J. B. Blanco-Canosa, P. E. Dawson, M. H. Stewart, K. Susumu, E. R. Goldman, M. Ancona and I. L. Medintz, ACS Nano, 2010, 4, 7253–7266 CrossRef CAS PubMed
.
- Z. Wang, L. Liu, Y. Xu, L. Sun and G. Li, Biosens. Bioelectron., 2011, 26, 4610–4613 CrossRef CAS PubMed
.
-
M. Fairhead and M. Howarth, in Analytical Biochemistry, ed. A. Gautier and M. J. Hinner, Springer New York, New York, NY, 2015, vol. 1266, pp. 171–184 Search PubMed
.
- I. Chen, M. Howarth, W. Lin and A. Y. Ting, Nat. Methods, 2005, 2, 99–104 CrossRef CAS PubMed
.
- D. I. Kim, S. C. Jensen, K. A. Noble, B. Kc, K. H. Roux, K. Motamedchaboki and K. J. Roux, Mol. Biol. Cell, 2016, 27, 1188–1196 CrossRef CAS PubMed
.
- E. de Boer, P. Rodriguez, E. Bonte, J. Krijgsveld, E. Katsantoni, A. Heck, F. Grosveld and J. Strouboulis, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 7480–7485 CrossRef CAS PubMed
.
- N. Jain, S. W. Smith, S. Ghone and B. Tomczuk, Pharm. Res., 2015, 32, 3526–3540 CrossRef CAS PubMed
.
- S. Panowski, S. Bhakta, H. Raab, P. Polakis and J. R. Junutula, MAbs, 2014, 6, 34–45 CrossRef PubMed
.
- K. Tsuchikama and Z. An, Protein Cell, 2018, 9, 33–46 CrossRef CAS PubMed
.
- J. A. Shadish and C. A. DeForest, Matter, 2020, 2, 50–77 CrossRef
.
-
G. T. Hermanson, in Bioconjugate Techniques, Elsevier, 2013, pp. 229–258 Search PubMed
.
-
G. T. Hermanson, in Bioconjugate Techniques, Elsevier, 2013, pp. 127–228 Search PubMed
.
- E. Mahon, A. Salvati, F. Baldelli Bombelli, I. Lynch and K. A. Dawson, J. Control. Release, 2012, 161, 164–174 CrossRef CAS PubMed
.
- L. M. Herda, D. R. Hristov, M. C. Lo Giudice, E. Polo and K. A. Dawson, J. Am. Chem. Soc., 2017, 139, 111–114 CrossRef CAS PubMed
.
- M. C. Childers, C.-L. Towse and V. Daggett, Protein Eng., Des. Sel., 2016, 29, 271–280 CrossRef CAS PubMed
.
- M. Najafi, N. A. Maza and P. D. Calvert, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 203–208 CrossRef CAS PubMed
.
- P. Gattinger, S. Izadi, C. Grünwald-Gruber, S. Kallolimath and A. Castilho, Front. Plant Sci., 2021, 12, 671728 CrossRef PubMed
.
- X. Liu, Y. Zhang, J. Lei, Y. Xue, L. Cheng and H. Ju, Anal. Chem., 2010, 82, 7351–7356 CrossRef CAS PubMed
.
- I. Le Trong, Z. Wang, D. E. Hyre, T. P. Lybrand, P. S. Stayton and R. E. Stenkamp, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2011, 67, 813–821 CrossRef CAS PubMed
.
- B. A. Katz, J. Mol. Biol., 1997, 274, 776–800 CrossRef CAS PubMed
.
- Z. N. Scholl, W. Yang and P. E. Marszalek, ACS Nano, 2015, 9, 1189–1197 CrossRef CAS PubMed
.
- X. Fan, J. Wang, X. Zhang, Z. Yang, J.-C. Zhang, L. Zhao, H.-L. Peng, J. Lei and H.-W. Wang, Nat. Commun., 2019, 10, 2386 CrossRef PubMed
.
- K. W. Yong, D. Yuen, M. Z. Chen, C. J. H. Porter and A. P. R. Johnston, Nano Lett., 2019, 19, 1827–1831 CrossRef CAS PubMed
.
- S. M. Sedlak, L. C. Schendel, M. C. R. Melo, D. A. Pippig, Z. Luthey-Schulten, H. E. Gaub and R. C. Bernardi, Nano Lett., 2019, 19, 3415–3421 CrossRef CAS PubMed
.
- S. M. Sedlak, L. C. Schendel, H. E. Gaub and R. C. Bernardi, Sci. Adv., 2020, 6, eaay5999 CrossRef CAS PubMed
.
- M. D. Witte, C. S. Theile, T. Wu, C. P. Guimaraes, A. E. M. M. Blom and H. L. Ploegh, Nat. Protoc., 2013, 8, 1808–1819 CrossRef PubMed
.
- J. R. Gallagher, A. J. Kim, N. M. Gulati and A. K. Harris, Curr. Protoc. Microbiol., 2019, 54(1), 1–9 Search PubMed
.
- C. K. O. Dzuvor, B. K. Shanbhag, T. Younas, H.-H. Shen, V. S. Haritos and L. He, ACS Appl. Bio Mater., 2022, 5(10), 4993–5003 CAS
.
- S. Tsukiji and T. Nagamune, ChemBioChem, 2009, 10, 787–798 CrossRef CAS PubMed
.
- H. Ton-That, S. K. Mazmanian, K. F. Faull and O. Schneewind, J. Biol. Chem., 2000, 275, 9876–9881 CrossRef CAS PubMed
.
- J. M. Antos, G.-L. L. Chew, C. P. Guimaraes, N. C. Yoder, G. M. Grotenbreg, M. W.-L. L. Popp and H. L. Ploegh, J. Am. Chem. Soc., 2009, 131, 10800–10801 CrossRef CAS PubMed
.
- R. G. Kruger, B. Otvos, B. A. Frankel, M. Bentley, P. Dostal and D. G. McCafferty, Biochemistry, 2004, 43, 1541–1551 CrossRef CAS PubMed
.
- H. Mao, S. A. Hart, A. Schink and B. A. Pollok, J. Am. Chem. Soc., 2004, 126, 2670–2671 CrossRef CAS PubMed
.
- C. P. Guimaraes, J. E. Carette, M. Varadarajan, J. Antos, M. W. Popp, E. Spooner, T. R. Brummelkamp and H. L. Ploegh, J. Cell Biol., 2011, 195, 751–764 CrossRef CAS PubMed
.
- M. W. Popp, J. M. Antos, G. M. Grotenbreg, E. Spooner and H. L. Ploegh, Nat. Chem. Biol., 2007, 3, 707–708 CrossRef CAS PubMed
.
- M. W.-L. Popp and H. L. Ploegh, Angew. Chem., Int. Ed., 2011, 50, 5024–5032 CrossRef CAS PubMed
.
- S. Pritz, O. Kraetke, A. Klose, J. Klose, S. Rothemund, K. Fechner, M. Bienert and M. Beyermann, Angew. Chem., Int. Ed., 2008, 47, 3642–3645 CrossRef CAS PubMed
.
- S. Samantaray, U. Marathe, S. Dasgupta, V. K. Nandicoori and R. P. Roy, J. Am. Chem. Soc., 2008, 130, 2132–2133 CrossRef CAS PubMed
.
- D. J. Williamson, M. A. Fascione, M. E. Webb and W. B. Turnbull, Angew. Chem., Int. Ed., 2012, 51, 9377–9380 CrossRef CAS PubMed
.
- M. Rashidian, L. Wang, J. G. Edens, J. T. Jacobsen, I. Hossain, Q. Wang, G. D. Victora, N. Vasdev, H. Ploegh and S. H. Liang, Angew. Chem., Int. Ed., 2016, 55, 528–533 CrossRef CAS PubMed
.
- C. Hamers-Casterman, T. Atarhouch, S. Muyldermans, G. Robinson, C. Hammers, E. B. Songa, N. Bendahman and R. Hammers, Nature, 1993, 363, 446–448 CrossRef CAS PubMed
.
- T. Che, S. Majumdar, S. A. Zaidi, P. Ondachi, J. D. McCorvy, S. Wang, P. D. Mosier, R. Uprety, E. Vardy, B. E. Krumm, G. W. Han, M.-Y. Lee, E. Pardon, J. Steyaert, X.-P. Huang, R. T. Strachan, A. R. Tribo, G. W. Pasternak, F. I. Carroll, R. C. Stevens, V. Cherezov, V. Katritch, D. Wacker and B. L. Roth, Cell, 2018, 172, 55–67.e15 CrossRef CAS PubMed
.
- H. Kumar, J. S. Finer-Moore, X. Jiang, I. Smirnova, V. Kasho, E. Pardon, J. Steyaert, H. R. Kaback and R. M. Stroud, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 8769–8774 CrossRef CAS PubMed
.
- M. Yamagata and J. R. Sanes, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 2126–2131 CrossRef CAS PubMed
.
- J. C. Y. Tang, T. Szikra, Y. Kozorovitskiy, M. Teixiera, B. L. Sabatini, B. Roska and C. L. Cepko, Cell, 2013, 154, 928–939 CrossRef CAS PubMed
.
- E. Platonova, C. M. Winterflood, A. Junemann, D. Albrecht, J. Faix and H. Ewers, Methods, 2015, 88, 89–97 CrossRef CAS PubMed
.
- P. C. Fridy, Y. Li, S. Keegan, M. K. Thompson, I. Nudelman, J. F. Scheid, M. Oeffinger, M. C. Nussenzweig, D. Fenyö, B. T. Chait and M. P. Rout, Nat. Methods, 2014, 11, 1253–1260 CrossRef CAS PubMed
.
- J. Xu, K. Xu, S. Jung, A. Conte, J. Lieberman, F. Muecksch, J. C. C. Lorenzi, S. Park, F. Schmidt, Z. Wang, Y. Huang, Y. Luo, M. S. Nair, P. Wang, J. E. Schulz, L. Tessarollo, T. Bylund, G.-Y. Chuang, A. S. Olia, T. Stephens, I.-T. Teng, Y. Tsybovsky, T. Zhou, V. Munster, D. D. Ho, T. Hatziioannou, P. D. Bieniasz, M. C. Nussenzweig, P. D. Kwong and R. Casellas, Nature, 2021, 595, 278–282 CrossRef CAS PubMed
.
-
M. Fairhead and M. Howarth, in Methods in Molecular Biology (Clifton, N.J.), 2015, vol. 1266, pp. 171–184 Search PubMed
.
- K. Minamihata, M. Goto and N. Kamiya, Bioconjugate Chem., 2011, 22, 74–81 CrossRef CAS PubMed
.
- M. Li, M. Zhu, C. Zhang, X. Liu and Y. Wan, Toxins, 2014, 6, 3208–3222 CrossRef CAS PubMed
.
- Z. Sun, J. Lv, X. Liu, Z. Tang, X. Wang, Y. Xu and B. D. Hammock, Anal. Chem., 2018, 90, 10628–10634 CrossRef CAS PubMed
.
- A. Noor, G. Walser, M. Wesseling, P. Giron, A.-M. Laffra, F. Haddouchi, J. De Grève and P. Kronenberger, BMC Res. Notes, 2018, 11, 751 CrossRef CAS PubMed
.
- A. D. Keefe, D. S. Wilson, B. Seelig and J. W. Szostak, Protein Expression Purif., 2001, 23, 440–446 CrossRef CAS PubMed
.
- T. G. Schmidt and A. Skerra, Nat. Protoc., 2007, 2, 1528–1535 CrossRef CAS PubMed
.
- T. Lamla and V. A. Erdmann, Protein Expression Purif., 2004, 33, 39–47 CrossRef CAS PubMed
.
- I. R. S. Ribeiro, R. F. da Silva, C. P. Silveira, F. E. Galdino and M. B. Cardoso, Nano Today, 2021, 36, 101012 CrossRef CAS PubMed
.
- L. Woythe, N. B. Tito and L. Albertazzi, Adv. Drug Delivery Rev., 2021, 169, 1–21 CrossRef CAS PubMed
.
- S. Nomura, Y. Segawa, K. Misawa, T. Kobayashi, X. Zhao, Y. Aoyagi and T. Sugano, J. Lumin., 1996, 70, 144–157 CrossRef CAS
.
-
T. Khan, R. S. Vohra and S. Homer-Vanniasinkam, in Biomaterials and Devices for the Circulatory System, Elsevier, 2010, pp. 251–269 Search PubMed
.
- D. Shcherbo, E. M. Merzlyak, T. V. Chepurnykh, A. F. Fradkov, G. V. Ermakova, E. A. Solovieva, K. A. Lukyanov, E. A. Bogdanova, A. G. Zaraisky, S. Lukyanov and D. M. Chudakov, Nat. Methods, 2007, 4, 741–746 CrossRef CAS PubMed
.
- A. Sukhanova, K. Even-Desrumeaux, A. Kisserli, T. Tabary, B. Reveil, J.-M. Millot, P. Chames, D. Baty, M. Artemyev, V. Oleinikov, M. Pluot, J. H. M. Cohen and I. Nabiev, Nanomed. Nanotechnol. Biol. Med., 2012, 8, 516–525 CrossRef CAS PubMed
.
- J. L. Liu, S. A. Walper, K. B. Turner, A. B. Lee, I. L. Medintz, K. Susumu, E. Oh, D. Zabetakis, E. R. Goldman and G. P. Anderson, Biotechnol. Rep., 2016, 10, 56–65 CrossRef PubMed
.
- K. W. Yong, D. Yuen, M. Z. Chen and A. P. R. Johnston, ACS Appl. Mater. Interfaces, 2020, 12, 5593–5600 CrossRef CAS PubMed
.
- S. D. Fontaine, R. Reid, L. Robinson, G. W. Ashley and D. V. Santi, Bioconjugate Chem., 2015, 26, 145–152 CrossRef CAS PubMed
.
- J. F. Ponte, X. Sun, N. C. Yoder, N. Fishkin, R. Laleau, J. Coccia, L. Lanieri, M. Bogalhas, L. Wang, S. Wilhelm, W. Widdison, J. Pinkas, T. A. Keating, R. Chari, H. K. Erickson and J. M. Lambert, Bioconjugate Chem., 2016, 27, 1588–1598 CrossRef CAS PubMed
.
- M. A. Shandell, Z. Tan and V. W. Cornish, Biochemistry, 2021, 60, 3455–3469 CrossRef CAS PubMed
.
- J. J. Porter and R. A. Mehl, Oxid. Med. Cell. Longevity, 2018, 2018, 1–14 CrossRef PubMed
.
-
N. Michael Green, in Methods in Enzymology, ed. M. Wilchek and E. A. Bayer, Elsevier, 1990, vol. 184, pp. 51–67 Search PubMed
.
- H. R. Nordlund, V. P. Hytönen, J. Hörhä, J. A. E. Määttä, D. J. White, K. Halling, E. J. Porkka, J. P. Slotte, O. H. Laitinen and M. S. Kulomaa, Biochem. J., 2005, 392, 485–491 CrossRef CAS PubMed
.
- E. M. Obeng, C. K. O. Dzuvor and M. K. Danquah, Nano Today, 2021, 101350 Search PubMed
.
- M. Schoof, B. Faust, R. A. Saunders, S. Sangwan, V. Rezelj, N. Hoppe, M. Boone, C. B. Billesbølle, C. Puchades, C. M. Azumaya, H. T. Kratochvil, M. Zimanyi, I. Deshpande, J. Liang, S. Dickinson, H. C. Nguyen, C. M. Chio, G. E. Merz, M. C. Thompson, D. Diwanji, K. Schaefer, A. A. Anand, N. Dobzinski, B. S. Zha, C. R. Simoneau, K. Leon, K. M. White, U. S. Chio, M. Gupta, M. Jin, F. Li, Y. Liu, K. Zhang, D. Bulkley, M. Sun, A. M. Smith, A. N. Rizo, F. Moss, A. F. Brilot, S. Pourmal, R. Trenker, T. Pospiech, S. Gupta, B. Barsi-Rhyne, V. Belyy, A. W. Barile-Hill, S. Nock, Y. Liu, N. J. Krogan, C. Y. Ralston, D. L. Swaney, A. García-Sastre, M. Ott, M. Vignuzzi, P. Walter and A. Manglik, Science, 2020, 1479, eabe3255 Search PubMed
.
- K. K. Chan, D. Dorosky, P. Sharma, S. A. Abbasi, J. M. Dye, D. M. Kranz, A. S. Herbert and E. Procko, Science, 2020, 369, 1261–1265 CrossRef CAS PubMed
.
- A. Moliner-morro, D. J. Sheward, V. Karl, L. P. Vidakovics, B. Murrell, G. M. McInerney and L. Hanke, Biomolecules, 2020, 10, 1–11 CrossRef PubMed
.
- K. Yumura, M. Ui, H. Doi, T. Hamakubo, T. Kodama, K. Tsumoto and A. Sugiyama, Protein Sci., 2013, 22, 213–221 CrossRef CAS PubMed
.
- T. Kawato, E. Mizohata, T. Meshizuka, H. Doi, T. Kawamura, H. Matsumura, K. Yumura, K. Tsumoto, T. Kodama, T. Inoue and A. Sugiyama, J. Biosci. Bioeng., 2015, 119, 642–647 CrossRef CAS PubMed
.
- B.-M. Chen, T.-L. Cheng and S. R. Roffler, ACS Nano, 2021, 15, 14022–14048 CrossRef CAS PubMed
.
- M. C. Truttmann, Q. Wu, S. Stiegeler, J. N. Duarte, J. Ingram and H. L. Ploegh, J. Biol. Chem., 2015, 290, 9087–9100 CrossRef CAS PubMed
.
- L. Hanke, D. J. Sheward, A. Pankow, L. P. Vidakovics, V. Karl, C. Kim, E. Urgard, N. L. Smith, J. Astorga-Wells, S. Ekström, J. M. Coquet, G. M. McInerney and B. Murrell, Sci. Adv., 2022, 8, 1–17 Search PubMed
.
- N. Jailkhani, J. R. Ingram, M. Rashidian, S. Rickelt, C. Tian, H. Mak, Z. Jiang, H. L. Ploegh and R. O. Hynes, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 14181–14190 CrossRef CAS PubMed
.
- R. W. Cheloha, F. A. Fischer, A. W. Woodham, E. Daley, N. Suminski, T. J. Gardella and H. L. Ploegh, Nat. Commun., 2020, 11, 2087 CrossRef CAS PubMed
.
- J. Xu, A.-R. Kim, R. W. Cheloha, F. A. Fischer, J. S. S. Li, Y. Feng, E. Stoneburner, R. Binari, S. E. Mohr, J. Zirin, H. L. Ploegh and N. Perrimon, Elife, 2022, 11, 1–28 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na00014a |
| This journal is © The Royal Society of Chemistry 2023 |