On the origins of SARS-CoV-2 main protease inhibitors
Yves L.
Janin

Structure et Instabilité des Génomes (StrInG), Muséum National d'Histoire Naturelle, INSERM, CNRS, Alliance Sorbonne Université, 75005 Paris, France. E-mail: yves.janin@cnrs.fr
First published on 13th October 2023
Abstract
In order to address the world-wide health challenge caused by the COVID-19 pandemic, the 3CL protease/SARS-CoV-2 main protease (SARS-CoV-2-Mpro) coded by its nsp5 gene became one of the biochemical targets for the design of antiviral drugs. In less than 3 years of research, 4 inhibitors of SARS-CoV-2-Mpro have actually been authorized for COVID-19 treatment (nirmatrelvir, ensitrelvir, leritrelvir and simnotrelvir) and more such as EDP-235, FB-2001 and STI-1558/Olgotrelvir or five undisclosed compounds (CDI-988, ASC11, ALG-097558, QLS1128 and H-10517) are undergoing clinical trials. This review is an attempt to picture this quite unprecedented medicinal chemistry feat and provide insights on how these cysteine protease inhibitors were discovered. Since many series of covalent SARS-CoV-2-Mpro inhibitors owe some of their origins to previous work on other proteases, we first provided a description of various inhibitors of cysteine-bearing human caspase-1 or cathepsin K, as well as inhibitors of serine proteases such as human dipeptidyl peptidase-4 or the hepatitis C protein complex NS3/4A. This is then followed by a description of the results of the approaches adopted (repurposing, structure-based and high throughput screening) to discover coronavirus main protease inhibitors.
Introduction
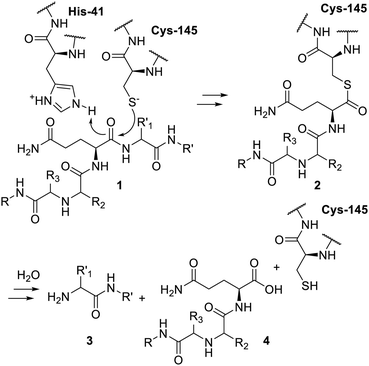
The protein encoded by the nsp5 gene of SARS-CoV-2 virus corresponds to the main protease of this coronavirus. This protein has also been named SARS-CoV-2 Mpro, 3C-like protease/3CLpro (CL for chymotrypsin-like) and C30 endopeptidase. Following N and O-terminal autocleavages, the mature enzyme is, at least,1 able to hydrolyze 9 other conserved sites of the viral polyprotein produced by the host ribosome to generate the corresponding viral proteins. As well reviewed,2–4 the protease features a characteristic cysteine/histidine catalytic dyad. It is the cysteine-145 thiol anion, depicted in Scheme 1, which acts as a nucleophile and the histidine-41 imidazole as a general base in the course of the proteolysis. The peptide 1 cleavage to release 3 and 4 takes place via the occurrence of transient S-acylcysteine (2) which is then hydrolyzed to regenerate the thiol function and release the protein 4. Peptide cleavage-wise, SARS-CoV-2 Mpro has the same hydrolysis selectivity as SARS-CoV-1 Mpro or other proteases from coronaviruses.5,6 A recent report has actually described remarkable structural insights in this sequence recognition process.7 As depicted, the residue R1 of any substrate has to be a glutamine, whereas, upward of the cleavage site, R2 is usually a leucine or another hydrophobic residue, and if R3 can vary, R4 (which is not depicted) remains small and usually aliphatic. Downward, only the residue appears to be governing the cleavage selectivity as it can only be a serine, an alanine or an asparagine.7 Concerning host cell proteins, quite a few8–11 have been reported as substrates of SARS-CoV-2 Mpro, thus providing further insights into the many ways12,13 viruses play havoc in cellular biochemistry and innate immunity. A recent review on the proteins reported as substrates of this protease is also available.14
residue appears to be governing the cleavage selectivity as it can only be a serine, an alanine or an asparagine.7 Concerning host cell proteins, quite a few8–11 have been reported as substrates of SARS-CoV-2 Mpro, thus providing further insights into the many ways12,13 viruses play havoc in cellular biochemistry and innate immunity. A recent review on the proteins reported as substrates of this protease is also available.14
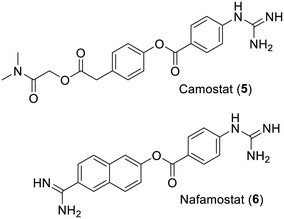
Of note is that comparatively less inhibitors have been reported for the other SARS-CoV-2 protease which is a papain-like cysteine-bearing enzyme. Two papers15,16 and a few reviews17–20 would probably be good starting points for further information on this different viral protease. Moreover, and as far better reviewed recently,21,22 the host cell lysosomal cysteine protease cathepsins B and L, the transmembrane protease serine 2 (TMPRSS2) and the subtilisin-like proprotein convertase furin, a calcium-dependent serine protease, are all capable of cleaving the viral spike protein coating the virus surface and this is an essential step for its cellular entry. Accordingly, in the absence of in vitro selectivity control, concerns have been raised on the true mechanism of observed antiviral effects for some series of unselective protease inhibitors reported,23–25 not to mention the recurrent frequent hitters.26–28 In fact, cathepsins B and L are actually the targets of a selective inhibitor which also displayed an antiviral effect in cellulo.29 Moreover, as for past research against the MERS coronavirus (MERS-CoV),30,31 the importance of these cellular proteases did suggest the use of camostat (5), an approved TMPRSS2 inhibitor, for treatment against COVID-19.32 However, even at high dose, the clinical trials with this anticancer agent pointed out a lack of any benefit.33 Similar reasoning, originally based on MERS-CoV research,34 has also led to trials (NCT04352400) with the nonspecific serine protease inhibitor nafamostat (6). The conclusions of these 2021 human trials against COVID-19 have yet to be published although the short in vivo half-life of this iminoimide will remain of real concern (Fig. 1).35
Two general approaches to discover inhibitors of serine or cysteine-bearing proteases have been used in the past. The main one is to design compounds which can arguably be described as covalent inhibitors. Such compounds will block the enzyme via the occurrence of a covalent bond with their catalytically essential OH or SH residues. Of note is that depending on the chemistry involved, the formation of a covalent bond can sometimes be reversed. The probably more difficult alternative is to discover inhibitors affecting the protease function because of a high and non-covalent affinity for either its catalytic site or another essential component of the enzyme. The latter approach can of course be useful for the former one since a well-placed incorporation of a reactive moiety into a high affinity compound will lead to a possibly more selective covalent inhibitor. Many reports have already listed all the, sometimes related,36 inhibitors reported for their effect on chymotrypsin-like proteases of human rhinovirus,37–40 enterovirus 71,41 SARS and MERS coronaviruses42–46 and then SARS-CoV-2.2–4,6,17,18,47–70 In the present text, the many publications26 solely based on in silico docking approaches71,72 and/or on traditional/ancestral medicine beliefs which only described frequent hitters/pan-assay interference compounds (PAINS)73–77 were ignored. This choice is a bid to discourage such all too obvious pollution of the scientific literature,78,79 not to mention the issue of lack of reproducibility of some data from the academia.80 Aside from these, quite a few virtual-based reports describe modest inhibition of SARS-CoV-2 Mpro by not too obvious frequent hitters which could lead to original series of inhibitors.81–92 However, as for the published results of high throughput or X-ray based screenings,84,93 we chose to wait for some reports focusing on the actual hit to lead progression before including them in this text.
Designing successful covalent inhibitors, a few examples
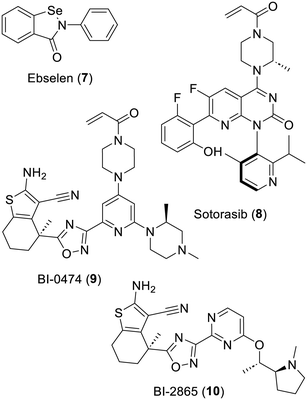
The principle for designing a successful/selective covalent inhibitor in general is to first start with a substance with a degree of specific affinity for the active site of the targeted enzyme. Then, the inclusion of an electrophilic and thus reactive component to the structure of such compounds can lead to a far stronger and possibly more efficient in vivo inhibition effect. A counter example, which unfortunately keeps on attracting undue attention and funding,94 would be the frequent hitter74 ebselen (7). This compound does feature a rather reactive nitrogen–selenium bond but very little else in its structure provides for any target selectivity. Indeed, a selective covalent inhibitor will rely on the principle that, when bound to its target, the reactive component of such a compound is oriented toward a nucleophilic and essential part of the enzyme in order to favor a reaction selectivity. This requires a fine-tuning process not only to improve the affinity of the non-reactive part of the inhibitor for its target but also to secure the best orientation of its reactive component. In other words, ebselen (7) does not comply with such criteria in contrast with, for instance, the recently authorized anticancer drug sotorasib/AMG 510 (8). Indeed, the latter features a Michael accepting acrylamide moiety along with other structural components providing an affinity for its biochemical target. Accordingly, this anticancer substance does preferentially lead to the occurrence of a covalent bond with the oncogenic KRAS (glycine 12 cysteine) mutant.95,96 Of note is that more recent research, starting from BI-0474 (9), another covalent inhibitor of this KRAS mutant, actually led to the non-covalent pan-KRAS inhibitor BI-2865 (10).97 Aside from providing a demonstration of the benefit of long-term research in medicinal chemistry, this illustrates the following facts: (i) in the course of designing inhibitors, it is indeed possible to add a reactive moiety and thus improve an inhibition effect and (ii) it is also sometimes possible to remove such a reactive moiety and, following some more structure-guided design of analogues, reach some very efficient non-covalent inhibitors (Fig. 2).Concerning irreversible inhibitors of serine, cysteine or threonine proteases in general, a few reviews98–101 provide an extensive description of the compounds reported. The following are only illustrations of some of the successes (and failures) of these classes of inhibitors.
Inhibitors of caspase-1/interleukin-1 converting enzyme
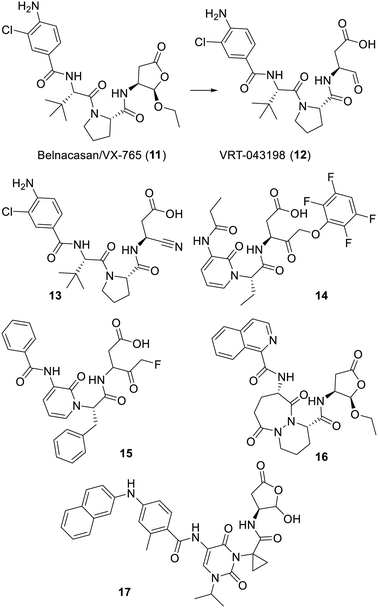
The cysteine protease caspase-1/interleukin-1 converting enzyme (ICE)102 cleaves peptides after an aspartic acid residue, preferably endowed with the sequence Tyr-Val-Ala-Asp. As reviewed,103 research for specific inhibitors started with the aldehyde-bearing peptide Ac-Tyr-Val-Ala-Asp-H and using extensive structure-based insights, proceeded to replace/“deconstruct” most of these residues to reach more metabolically stable inhibitors. Belnacasan/VX-765 (11) is a hemiacetal prodrug, whose first patent application was filed by Vertex in 2000.104 This compound will, upon a hydrolysis, leads to the aldehyde-bearing VRT-043198 (12), a selective covalent inhibitor of ICE. Of note is the nitrile group of analogue 13 which was introduced in 2010 as an alternative to the reactive aldehyde function of compound 11.105 Aside from these, a few other series were also patented by Vertex. As depicted with the structures of the randomly selected N-substituted pyridinones 14 or 15, both “drifted” from the ICE-favored peptidic sequence mentioned above and their central heterocycle provided another type of structural lock to favor a suitable orientation of their reactive ketone.106–108 The possible extent of such a peptide deconstruction approach is also well illustrated with the hemiacetal ICE inhibitor pralnacasan (16), which features a notable bicyclic piperazic component.109 More recent work on the design of ICE inhibitors led another research group to uracil-containing derivatives such as the most advanced compound 17.110,111 Clinic-wise, quite a few trials were conducted with belnacasan (11) for treatments of conditions involving a possible disfunction of ICE, but this class of inhibitors has yet to reach an approved use in human health. Of note is the current trend in addressing the dreadful cytokine storm seen in some SARS-CoV-2 infected patients by inhibiting the many biochemical pathways involved in inflammation. In this regard, a few 2022 patents claimed ICE inhibitors such as belnacasan (11) or the nitrile-bearing analogue 13 for their potential benefit against coronavirus infection.112–114 Similarly, a recent patent115 claims the use of inhibitors of the beta secretase 1 to suppress this storm; however, both cases are quite outside the scope of this review (Fig. 3).Inhibitors of cathepsin K
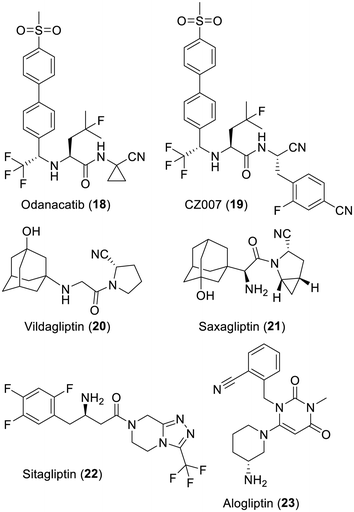
Cathepsin K, which hydrolyses a rather wide range of substrates,116 was selected as a target to treat bone resorption. Extensive research at Merck led to the nitrile-containing inhibitor odanacatib (18)117 and an X-ray based structure further proved its mechanism of inhibition via covalent bonding with the catalytic cysteine residue.118 However, although this inhibitor reached a phase III clinical trial stage, its development was stopped in 2019 because of a stroke risk increase.119 Of note is the closely related analog CZ007 (19) which has been under consideration as a drug against the human parasite Trypanosoma cruzi since it also strongly inhibits cruzipain, a key cysteine protease of this protozoan.120,121Inhibitors of human dipeptidyl peptidase-4/CD26
As well reviewed,122–127 the serine-containing dipeptidyl peptidase-4/CD26, which is a prolyl oligopeptidase, has many physiological roles. These include the proteolysis of glucagon-like peptide 1 or the glucose-dependent insulinotropic polypeptide, which are both key factors in glucose homeostasis. Since 1994, medicinal chemistry efforts have focused on cyanopyrrolidine-containing inhibitors and culminated in the discoveries of vildagliptin (20)128 and saxagliptin (21).129 With their nitrile function, both compounds are covalent inhibitors of this serine protease, although the serine adducts formed130 are slowly reversible.131,132 This class of covalent drugs have been prescribed for years to reduce hyperglycemia in patients with type 2 diabetes mellitus. Interestingly, many non-covalent inhibitors of this protease such as sitagliptin (22)133 or alogliptin (23)134 were also discovered. As demonstrated by X-ray based structures,130,135 these non-covalent inhibitors also target the catalytic site of the protease and their wide structural diversity is a tribute to what can medicinal chemistry do (Fig. 4).Inhibitors of hepatitis C serine protease NS3/4A
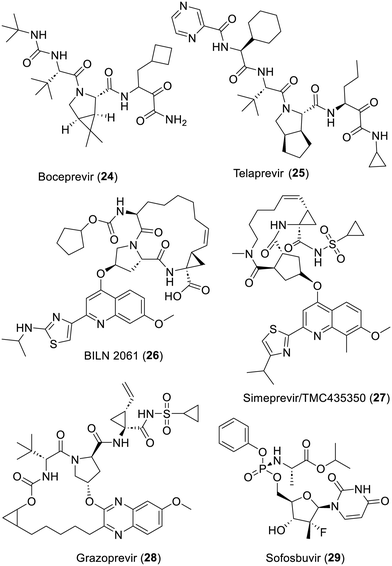
As described in an editorial on the 2020 Nobel Prize award to Harvey J. Alter, Michael Houghton and Charles M. Rice, the quest to first characterize and then discover treatments against hepatitis C infection has been a 30 year-long story.136 Indeed, medicinal chemistry research on the inhibition of a number of viral proteins of hepatitis C virus has today delivered treatment efficacies nearing 95% cure!137 Since the late 90s, many laboratories have focused on the design of covalent or non-covalent inhibitors of the hepatitis C serine protease NS3/4A.138,139 The α-ketoamide-bearing covalent inhibitors boceprevir (24)140–142 and telaprevir (25)143 turned out to be the first clinically approved drugs. Interestingly, a proof of concept was also achieved in patients with the remarkable macrocycle ciluprevir/BILN 2061 (26) which is a non-covalent inhibitor of NS3/4A.144,145 This discovery led to many series of macrocycle-bearing analogues which reached clinical approvals such as simeprevir (27)146 or grazoprevir (28).147 Finally, and as well accounted,148 research on treatment of hepatitis C also focused on discovering inhibitors of its RNA-dependent RNA polymerase NS5B. This was concluded with the prodrug sofosbuvir (29) which is instrumental for reaching the 95% clinical efficacy mentioned above. However, this last achievement also triggered the voluntary withdrawal, or project termination, of quite a few hepatitis C serine protease NS3/4A inhibitors (Fig. 5).Inhibitors of the SARS-CoV-2 main protease
Since the main proteases of SARS-CoV-1 and SARS-CoV-2 share a 96% amino acid sequence identity (but only 50% with the MERS-CoV main protease),2 most if not all research on their inhibition turned out to be useful for the renewed projects focusing on improving such inhibitors. This was also the case for drugs which have been “repurposed” as inhibitors of MERS or SARS-CoV-1, and these include some potential inhibitors of their main protease.Concerning drug repurposing
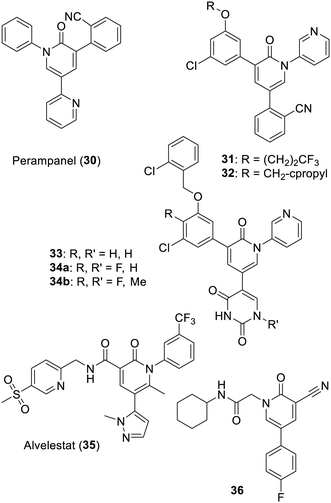
Aside from the excessive number of reports describing in vitro inhibition of SARS-CoV-1(or 2)-Mpro by well-known frequent hitters/PAINS, another approach focused on assessing libraries of drugs already or previously used or tried in humans. The SARS-CoV-2 pandemic thus saw the publication of many papers reporting in vitro inhibition of SARS-CoV-2-Mpro by such compounds. The main result of this approach is an unfortunate illustration of a lack of medicinal chemistry culture in general. Indeed, most often only micromolar level effects were observed in vitro for such drugs. Accordingly, it was more than unlikely that a patient would benefit from a treatment based on them, not to mention the issues of necessary dose increases which would be bound to lead to some side effects including some due to their main biological actions. In medicinal chemistry, compounds effective at the micromolar level against a given target can only be considered as hits (or early leads). Such compounds must undertake rounds of structure–activity relationship and selectivity studies to be further improved before preclinical and clinical trials can be envisaged. Attempts to cut this process short are oblivious to decades of experience in the domain and only slightly more rational than hoping for a miracle. Moreover, even in the rare event of finding a strong level of in vitro SARS-CoV-2-Mpro inhibition for a prescribed drug, it is very likely that it will lack any in vitro or in vivo selectivity. Indeed, quite a few highly reactive compounds are found in the present or past human pharmacopeia. In the following, we describe a few drugs which were reported for their effect on SARS-CoV-2-Mpro. Unfortunately, not all were the focus of some MedChem iterations to improve them before initiating wishful clinical trials which, predictably, led to disappointing results.27,149–152From a screening which identified 14 known drugs, the antiepileptic drug perampanel (30) was found to be an inhibitor of the main protease of SARS-CoV-2.153 Some MedChem helped by X-ray based structures (PDB 7L10 to 7L14) from the same research group provided 3-pyridyl-bearing analogues, such as compounds 31 and 32 (with 32 being much less cytotoxic than 31).154 And further work, helped by nine X-ray based structures in this case, gave improved inhibitors such as pyrimidinediones 33 and 34a,155 as well as the even less cytotoxic N-methylated derivative 34b.156 A more recent report has described analogues featuring a pyridone instead of the uracil moiety of compounds 33 and 34a–b.157 As described in more detail below, the 3-pyridyl component of these improved analogues has actually been known since 2013 for its capability to interact with the histidine-163 of the SARS-CoV-1-Mpro catalytic site.158 Also of note is that this class of inhibitors do have some structural similarities with alvelestat (35), a serine protease (elastase) inhibitor.159 Independent from this work and as stated by its authors, a remarkably lucky de novo-initiated search for inhibitors delivered the somehow related (but not computer-guided) N-substituted pyridinone derivative 36. Further X-ray based crystallography studies are planned in order to improve its relatively modest inhibition level (Fig. 6).160
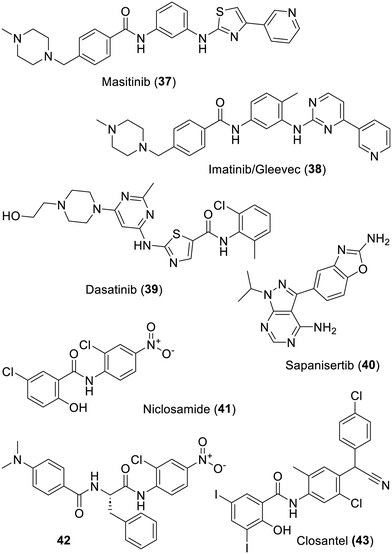
A repurposed curiosity would be the veterinary anticancer drug masitinib (37), it is a tyrosine kinase inhibitor which has also been patented for its modest effect on the replication of SARS-CoV-2.161 Further research actually led to its co-crystallization with SARS-CoV-2-Mpro (PDB 7JU7).162 A note of caution would be that this rather sticky compound, as well as imatinib (38), has also managed to co-crystallize with human deoxycytidine kinase (PDB 5MQL).163 In any case, undertaking clinical trials against COVID-19 (NCT05047783) with compound 37, even in association with the polyphenolic frequent hitter isoquercetin (NCT04622865), is very likely to be disappointing. Indeed, as mentioned for the repurposing of imatinib (38) against COVID-19, which has also been found active in vitro on MERS-CoV,164 clinically achievable doses in humans will not be high enough to be effective against the virus replication.165 One more example of such “sticky” compounds would be dasatinib (39), another tyrosine kinase inhibitor, which was reported to have some effect on SARS-CoV-2-Mpro.166 However, the in cellulo antiviral effects observed for all these amine-bearing compounds are very likely due to drug-induced phospholipidosis.167,168 This cellular-level effect (which impacts to some degree viral replications in cellulo) was unfortunately the cause of a considerable waste of money (as much as 6 billion dollars) when considering all the clinical trials against COVID-19 made with many amine-bearing compounds, especially chloroquine or hydroxychloroquine.150 Concerning other cellular kinases, a recent report has described far stronger in cellulo inhibition of SARS-CoV-2 replication by PI3K/mTOR inhibitors such as sapanisertib (40).169 Future will tell if this translates into an in vivo effect although a precedent would be the inhibitors of cellular dihydroorotate dehydrogenase170 which have yet to translate into effective RNA-based antivirals (including corona) in patients.171–173 In 2004, niclosamide (41) was found to be endowed with a degree of antiviral effect on SARS-CoV-1 although it was not found to inhibit its main protease174 and later on, this anthelminthic drug was also reported for its effect on many viruses, which has been recently reviewed.175 Finally, inhibition of SARS CoV-2 replication was also reported176 and, despite some ongoing structure–activity relationship studies,177 clinical trials were undertaken (NCT04399356 and 04603924)178 with compound 41 and have so far failed.179 Even if mechanism of action-wise, niclosamide (41) should not be mentioned here,180 and some protease inhibitors such as 42 have a puzzling degree of structural similitude to this drug. Compound 42 actually resulted from attempts to prepare protease substrates which would release a fluorescent product. However, it turned out that that these were inhibiting SARS-CoV-2-Mpro and the ensuing structure–activity relationship studies led to this compound.181 More recent work on closely related analogues of compound 42 has reported very modest inhibition of SARS-CoV-2-Mpro.182 Another puzzling similarity is also seen with a patent183 claiming the effect on SARS-CoV-2-Mpro of closantel (43), a rather toxic anthelmintic drug, which shares some degree of similitude to these two amides (Fig. 7).
Covalent or non-covalent inhibitors of SARS-CoV-2-Mpro from previous series
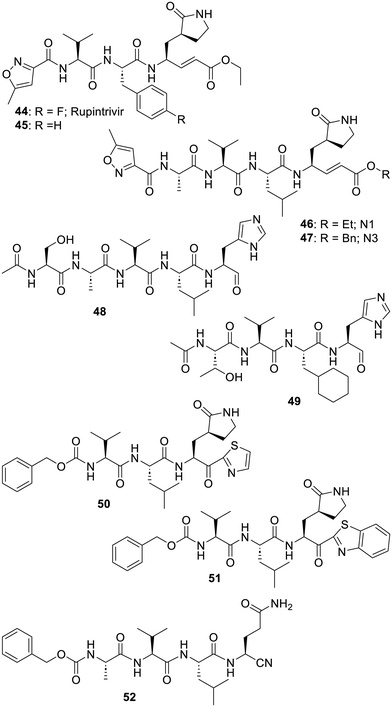
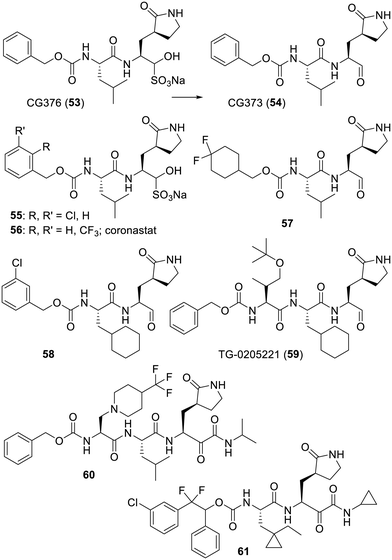
As for the series inhibiting other proteases, one of the approaches to design inhibitors started from the structure of small peptides featuring sequences recognized by the protease (i.e.: Ser-Ala-Val-Leu-Gln) and the addition of various types of warheads next to the cleavage site: downward the glutamine (Gln) residue in the case of SARS-CoV-2-Mpro. It would be beyond the scope of this review to depict all the series of peptide-derived inhibitors made, especially since 2019 an even larger array of warheads has been incorporated in such compounds.184 Indeed, a non-exhaustive list used in the last few decades includes: aldehydes (or the corresponding bisulfite adduct),41,185–202 ketones,203–211 α-ketoamides,153,185,212–214 Michael acceptors,194,215–218 4-iminooxazolidin-2-one,219 reactive halogens,220–224 β-lactam of some penicillins,225,226 phenylsulfide,227 thiocyanate,228 epoxide,24 nitriles229–231 and, last but not least, alkynes.232,233 Interestingly, aside from a 2005 patent234 targeting SARS-CoV-1-Mpro, the design of boron-containing inhibitors, which is a fairly classic approach for protease inhibition,235,236 has not been reported in more recent time. Along with these warheads, intensive efforts were made to modify these peptides and/or replace them with non peptidic spacers. Moreover, compounds designed to block other viral cysteine proteases were of course assayed on the coronaviruses. As an illustration of the multitude of approaches, the Michael-acceptor bearing rupintrivir/AG7088 (44), initially designed in 1999 as a covalent inhibitor of human rhinovirus 3C cysteine protease,237,238 was suggested239 in 2003 to be a starting point to target SARS-CoV-1-Mpro. A strong intensive to evaluate rupintrivir (44) was also the fact that it had previously been the subject of phase 1 and 2 clinical trials against rhinovirus.240,241 However, rupintrivir (44) was “not able to significantly affect virus reduction or moderate disease severity and thus was terminated for clinical development”.242 In any case, if compound 44 was reported inactive in 2005,243 the closely related analogue 45 turned out to modestly inhibit243 SARS-CoV-1-Mpro (or MERS-Mpro)244 and the longer peptides N1 (46) and N3 (47) featuring the more adapted Ala-Val-Leu sequence were even better.245 Although rupintrivir (44) was later found to only be a very modest inhibitor of SARS-CoV-2-Mpro, thanks to its reactive component, it still managed to co-crystallize with this protease (PDB 7L8I).218 Also starting from the Ser-Ala-Val-Leu-Gln sequence, a research group reported in 2011 that the imidazole bearing aldehyde 48 was a modest inhibitor of SARS-CoV-1-Mpro. The ensuing structure-guided iterations of synthesis and evaluation led to the much stronger inhibitor 49 which was also co-crystallized with SARS-CoV-1-Mpro (PDB 3ATW).246 More recent work actually reported related imidazole-bearing peptides.247 Of note is that a glutamine residue is prone to cyclize at the least with ketone or aldehyde warheads.248–250 Accordingly, effort to replace it also stemmed from the search for human rhinovirus 3C cysteine protease inhibitors and provided the bioisosteric lactams depicted in the structures of compounds 44 and 45. As well illustrated below, this bioisosteric replacement was repeatedly used in the structures of other virus protease inhibitors.238,248,251 A recent review has actually described in much more detail this issue of glutamine replacement.252 As for the results reported in 2000 focusing on the inhibition of the rhinovirus 3C protease,203 research on SARS-CoV-1-Mpro, dating from 2009, also explored the incorporation of large components downstream of the warhead. Similar to peptide inhibitors of the rhinovirus 3C protease,203 if the thiazole-2-ketone-bearing peptide 50 was only a modest inhibitor of SARS-CoV-1-Mpro,253 the benzothiazole analogue 51 in which benzene filled a pocket of this protease provided a thousand-fold improvement.254 Also of interest is the nitrile-containing peptide 52 which was reported in 2013 to be a weak inhibitor of SARS-CoV-1 Mpro, but it was established that this function could be used to covalently inhibit this protease (Fig. 8).255The shorter bisulfite prodrug GC376 (53) was reported in 2012 as a norovirus 3CL protease inhibitor.256 As depicted, the bisulfite adduct 53 will release in situ the aldehyde function of the antiviral GC373 (54).251 Interestingly, this compound has demonstrated a degree of in vivo effect against feline enteric coronavirus infection.257,258 Even if it is only modestly effective in vitro217 or in vivo against a mouse model of SARS-CoV-2 infection,259 this pan 3CL virus protease inhibitor has often been used as a positive control in SARS-CoV-2-Mpro assays. Many analogues of GC376 (53) were reported,185,260,261 such as compounds resulting from a “fluorine walk” on the benzyl moiety and/or its replacement by a substituted cyclohexyl, a bulkier adamentyl and even more elaborated substituents.195,199,201,262,263 As an illustration of the “leeway” on this position, the difluorocyclohexyl-bearing analogue 57 was found to be effective against a mouse model of MERS coronavirus infection.188 Moreover, a deuterated derivative of 50 was evaluated on a mouse model of SARS-CoV-2 infection but this analogue showed no real advantage.198 This result may not be too surprising, since a deuteration strategy usually addresses fast metabolic issues but GC376 (53) was reported to be reasonably stable in human plasma (t1/2 > 240 min) or in the presence of human liver microsomes (t1/2 > 80 min) as well as in mice (t1/2, plasma > 240 min, t1/2, microsome > 80).211 Further studies on the protease inhibition selectivity of GC376 (53) as well as analogues EB54 (55) and NK01-63/coronastat (56) pointed out the fact that these compounds are also very strong inhibitors of the host cell cathepsin L.211 Another investigation reported even more analogues of GC376 (53) but also described solubility issues, along with suggestions to administer such bisulfite adducts at higher concentration.264 The cyclohexyl group of compound 49 was also adopted in the design of the related compound 58 targeting the norovirus 3CL protease.265 However, even combined with other types of warheads, this cyclohexyl feature turned out to be associated with cellular toxicity.209,266 Compound TG-0205221 (59) illustrates the structure-based improvements made in 2006 when focusing on the inhibition of SARS-CoV-1-Mpro by this “Boc-derived” series.267 Much later, the O-tert-butyl-threonine component of the tripeptide TG-0205221 (59) was also found to be key to improving the interactions of related analogues with the P3 site of the SARS-CoV-2-Mpro binding pocket and it also provided some cellular potency.268 Interestingly, research at Glaxo, part of it dating from 2018 and focusing on the rhinovirus main protease,269 led to tripeptides such as 60 in which an α-ketoamide warhead was used instead of an aldehyde. However, a modest antiviral activity along with a degree of cellular cytotoxicity probably prevented further development of this series.270 The somehow larger “Boc” derivative 61 is amongst the compounds claimed in four patents by Cocrystal Pharma which has initiated a phase 1 study of the undisclosed coronavirus–norovirus protease inhibitor CDI-988 (NCT05977140) (Fig. 9).271–274
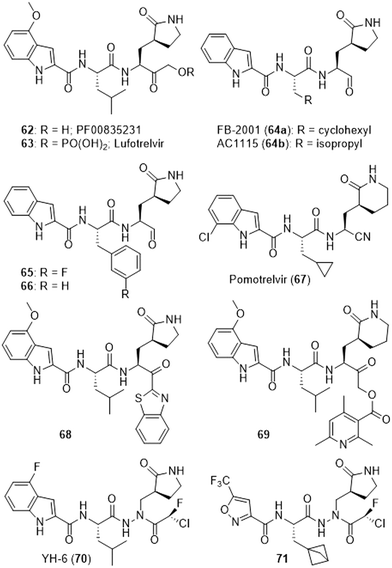
Other series of short peptides were also designed when the world faced the first SARS epidemic. A 2005 patent claimed many isoleucine and lactam-containing inhibitors of SARS-CoV-1-Mpro, including the noteworthy indole-bearing derivative PF00835231 (62).275 In 2020, the corresponding phosphate prodrug PF-07304814/lufotrelvir (63) was then developed by Pfizer to fight COVID-19 infection,205,276 and this intravenous prodrug207 underwent successful phase 1 clinical trials.277,278 However, an additional trial (NCT05780541) was suspended by the FDA and lufotrelvir (63) was then withdrawn. More recent work borrowed the cyclohexane and the indole elements of 62 and led to aldehyde 64a or to the benzyl-bearing analogues 65 and 66.189 Interestingly, FB-2001 (64a) was further evaluated279 and has undergone some clinical trials (NCT04766931). The same research group reported two years later that the non-fluorinated analogue 66 was better suited as a broad-spectrum antiviral since, aside from being active on SARS-CoV-2, it also displayed an effect on entero and rhinoviruses.41 Following extensive structure–activity relationship studies,280 Pardes Biosciences undertook clinical trials (NCT05011812 and NCT05543707) with the nitrile-bearing PBI-0451/pomotrelvir (67) which has also been co-crystallized with SARS-CoV-2-Mpro (8TBE). However, a company statement reported that this compound “did not meet the primary endpoint measured by proportion of participants below the limit of detection for infectious SARS-CoV-2 on day 3 of treatment” and further development was thus suspended.281 The 3-methoxyindole-bearing analogue 68, also featuring the benzothiazole seen in compound 51, was initially designed against SARS-CoV-1-Mpro.282 It was then reported to be efficient against SARS-CoV-2-Mpro and the virus replication.206,210 An extensive study of various ester prodrugs such as compound 69 was reported and also demonstrated that no real difference in antiviral properties was observed between compounds featuring a 5 or a 6-membered lactam.209 In an approach which has been used in the past against rhinovirus 3C proteases283 and which incidentally addressed a racemization risk, the strong aza-bearing SARS-CoV-2-Mpro inhibitor YH-6 (70) featuring a chiral α,α-chlorofluoracetamide warhead was reported.223 Two patents224,284 also claim related α,α-chlorofluoracetamide-containing aza-peptides including the fairly elaborated compound 71 which is effective against a whole panel of coronaviruses (Fig. 10).224
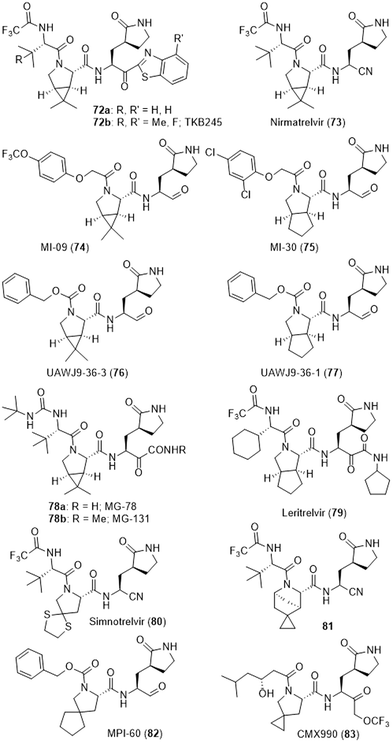
From the end of 2019, the hepatitis C serine protease NS3/4A inhibitors boceprevir (24) and telaprevir (25) depicted above, or the related narlaprevir, were repeatedly reported to be active in SARS-CoV-2-Mpro screening campaigns.153,186,187,285–287 This triggered X-ray based structural studies which pointed out that these reactive compounds did indeed bind to the catalytic site of SARS-CoV-2-Mpro.212,287,288 Of course the bicyclic 3-azabicyclo[3.1.0]hexane component of boceprevir (24), which not only enables the removal of an NH moiety but also probably acts as a conformational lock, provided a lot inspiration for the design of many series of SARS-CoV-2-Mpro inhibitors. In fact, research conducted at Pfizer led, for instance, to the ketone-bearing inhibitor 72a or the nitrile-bearing PF-07321332/nirmatrelvir (73).63,231 A crucial point explaining that nirmatrelvir (73) turned out to be orally active on a mouse model of SARS-CoV-2 infection is the removal of this extra NH moiety with this bicycle which greatly improved its diffusion.289 The ensuing successful clinical trials of nirmatrelvir (73) then provided the first approved SARS-CoV-2-Mpro inhibitor which must be prescribed in association with ritonavir to alter P4503A4-based metabolization and thus improve its pharmacology.290,291 Interestingly, compound 72a was the focus of further optimization which led to the fluorinated derivative TKB245 (72b).292,293 Moreover, an extensive search for an alternative to the trifluoroacetamide moiety of nirmatrelvir (73) was conducted at Ascletis Bioscience.294 The undisclosed protease inhibitor ASC11 probably resulting from this approach is currently undergoing Ascletis-sponsored phase 1 clinical trials, with the co-administration of ritonavir (NCT05718518). The 3-azabicyclo[3.1.0]hexane-bearing compounds MI-09 (74)197 and UAWJ9-36-3 (76)200 or the cyclopenta[c]pyrrole-bearing inhibitors MI-30 (75)197 and UAWJ-9-36-1 (77)200 also stemmed from this “structural lock” idea. Of note is that both 74 and 75 displayed in vivo antiviral effects as well.197 The α-amido ketones MG-78 (78a) and MG-131 (78b) were designed according to similar lines and are very good inhibitors of SARS-CoV-2-Mpro as well.295 Additional α-ketoamide-bearing derivatives featuring such component also had an improved cell permeability.296,297 Moreover, SARS-CoV-2-Mpro structures bound to such compounds have been released by two distinct research groups (PDB 7U92 and 7WQK). The notable cyclopenta[c]pyrrole-bearing SARS-CoV-2 Mpro inhibitor RAY1216/leritrelvir (79) was developed by Raynovent298,299 and granted conditional market approval in China. Interestingly, this strong inhibitor also displayed an improved in vivo half-life in comparison with nirmatrelvir (73) and appeared to have been evaluated on COVID-19 patients with and without ritonavir or other P4503A4 inhibitors (NCT05620160).300,301 A structure of SARS-CoV-2 Mpro bound to this compound has also been released (PDB 8IGN). The design and study of a related series of aldehyde-bearing compounds featuring various conformational locks actually reported that cyclopenta[c]pyrrole was amongst the most efficient pan-corona protease inhibitor in cellulo.302 The dithia-7-azaspiro[4.4]nonane derivative 80 is a strong SARS-CoV-2 Mpro inhibitor which was developed by Simcere Pharmaceutical. In association with ritonavir,303 it has also been granted conditional market approval in China under the name Xiannuoxin/simnotrelvir304–306 and the results of phase 1 clinical trials (NCT05339646) were reported recently.307 The undisclosed SARS-CoV-2 Mpro inhibitor ALG-097558 is developed by Aligos therapeutics, possibly without a co-administration of ritonavir and phase 1 clinical trials have been initiated (NCT05840952).25 Three distinct patents,229,308,309 from this company and the Catholic University of Leuven, describe a large number of compounds featuring a variety of conformational locks. We (randomly) choose to depict the 5-azaspiro[bicyclo[2.2.1]heptane-2,1′-cyclopropane] derivative 81, since this strong SARS-CoV-2 Mpro inhibitor also illustrates what MedChem is about: innovation in chemistry leading to original and thus patentable compounds. The simpler azabicyclo[2.2.1]heptane ring system has also been introduced in the structure of such inhibitors.310 Further insights were provided in the course of an extensive study of boceprevir analogues which reported some key elements to achieve an antiviral effect in cellulo.311 Moreover, the same research group recently reported the azaspiro[4.4]nonane derivative MPI-60 (82), which displays promising antiviral properties312 and a structure of SARS-CoV-2 Mpro bound to this compound was obtained (8STZ). Finally, CMX990 (83), an azaspiro[2,4]heptane-bearing derivative, is also a tribute to the creativity required in MedChem and this compound has reached the stage of phase 1 clinical trials.313,314 This ring system and quite a few others were actually also described in a paper from another research group.315 Moreover, many strongly effective azapeptides, including some α,α-chlorofluoracetamide-containing derivatives284 related to 70 and 71 and featuring such conformational locks, have been claimed.316 Finally, the bisulfite prodrug of the aldehyde derivative AC1115 (64b), a covalent inhibitor of SARS-CoV-2-Mpro and cathepsin L317 has undergone successful clinical trials under the names STI-1558/OVYDSO/Olgotrelvir without the recourse to ritonavir. These trials were sponsored by Sorrento therapeutics/Zhejiang ACEA Pharmaceutical (NCT05716425 and NCT06044233). Interestingly, the corresponding patent is claiming a wide range of SARS-CoV-2-Mpro inhibitors featuring structural elements seen in compounds 62–83 (Fig. 11).318
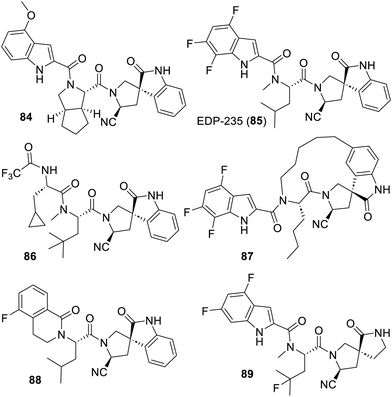
Also of interest, the SARS-CoV-2 Mpro inhibitor EDP-235 (85) from Enanta Pharmaceuticals319 recently met the primary end point of a phase 2 clinical trial without the coadministration of ritonavir (NCT05616728). In this case, 13 patents from this company claim a very large variety of peptides sometimes featuring conformational locks related to the one depicted above. Amongst them, compound 84 (ref. 320) not only features a cyclopenta[c]pyrrole in its structure but its warhead is a cyanopyrrolidine, reminiscent of vildagliptin (20) or saxagliptin (21). This cyanopyrrolidine is also rigidly connected to an indolinone which acts as the bioisoster replacement of the glutamine. Many more compounds featuring such unprecedented spiropyrrolidine along with a variety of warheads were claimed. EDP-235 (85),321 which is also the subject of two process patents for its large-scale production,322,323 or compound 86 (ref. 324) are noteworthy for their N-methyls. The NH homologs of these compounds were also made but their antiviral effects were apparently not deemed of sufficient interest to be mentioned in the corresponding patents.321,324 Interestingly, the structurally very constrained macrocycle 87 (ref. 325) and the simplified compounds 88 (ref. 326) and 89 (ref. 327) were also patented as effective inhibitors of SARS-CoV-2 Mpro (Fig. 12).
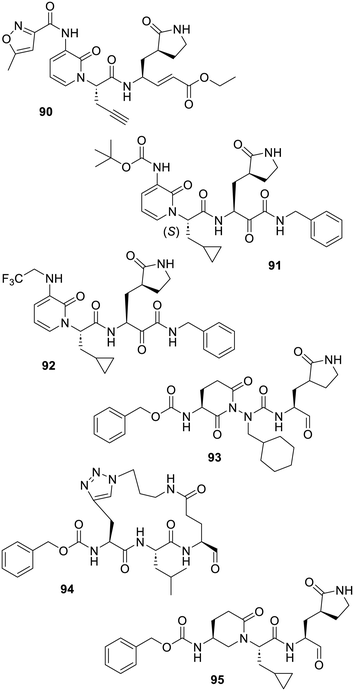
The research on inhibitors of the rhinovirus 3C protease conducted at Agouron/Pfizer at the turn of the century, which provided rupintrivir (44), also led to 2-pyridone-bearing analogues of this peptide.328–330 Amongst them, the orally available inhibitor AG7404 (90) underwent two phase 1 clinical trials,331 the latter, under the drug name V7404, was sponsored by ViroDefense and focused on enterovirus infection.332 As for rupintrivir (44), this alkyne-bearing compound was reported in 2022 to modestly inhibit SARS-CoV-1 and SARS-CoV-2 main proteases and it was co-crystallized with both enzymes (PDB 7ZQW and 7ZQV).333 By following this approach, which consists of rigidifying the P2–P3 amide bond of the protease substrate, the pyridinone derivative (91) was designed and reported to inhibit SARS-CoV-2 Mpro. Moreover, this compound demonstrated an effect on cell-based virus replication assays.212 Interestingly, it was shown that the (S) enantiomer depicted here was the most active although the corresponding (R) enantiomer also had a degree of effect. In fact, even if the (R) isomer was 50 times less efficient as a protease inhibitor, both isomers co-crystallized within the SARS-CoV-2 Mpro catalytic site (PDB 8A4T and 8A4Q) with rather drastically different binding modes.214 This probably illustrates the non-selectivity bias induced by a reactive component in a given molecule. Indeed, if such a compound can reach the nucleophilic part of the biochemical target, it is reasonable to assume that it will react with it, in spite of having a low overall affinity for the binding site. This known bias actually led in 2011 to the following comment: “the stringent substrate specificity of the SARS-CoV Mpro with respect to the P1 and P2 positions can be overruled by the highly electrophilic character of the aldehyde warhead. This constitutes a deviation from the dogma that peptidic protease inhibitors should comprise an amino-acid sequence corresponding to the cleavage specificity of the target enzyme”.334 This comment is also likely valid for a number of aldehyde-bearing compounds which have been reported to be inhibitors of SARS-CoV-2 Mpro.91,196,335 In any case, a patent describes further structure–activity relationship studies around such pyridone derivatives which led to the modest SARS-CoV-2 Mpro inhibitor 92.336 In a 2013 patent, an array of inhibitors, such as the constrained azapeptide 93 featuring a rigidifying 2,6-dioxopiperidine component,337 or macrocyclic-bearing peptides such as 94,338 were claimed for their effects on 3CL proteases of picornaviruses, caliciviruses and coronaviruses. The related 3-amino-6-oxopiperidine spacer was also employed more recently and for instance, compound 95 turned out to be an effective SARS-CoV-2 Mpro inhibitor (Fig. 13).339
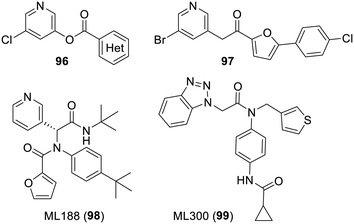
Amongst SARS-CoV-1 Mpro covalent inhibitors of less interest, many 3-pyridyl esters with the general formula 96 were reported between 2004 and 2008.340–343 It was then demonstrated341 by mass spectroscopy that these aryl esters (unsurprisingly) react with the cysteine of the protease. Later on, some compounds were shown to also strongly inhibit SARS-CoV-2 Mpro but again these had a very modest antiviral effect.194,206,344 A more recent report provided an extensive demonstration of their chemical reactivity and further established that these compounds were probably the focus of too much attention in view of their lack of potential in medicinal chemistry.345 Another recent report has actually described covalent inhibitors in which the activated carboxyl was replaced by a sulfone conjugated to a reactive oxadiazole ring system,346 and carmofur analogues also featuring an activated carboxamide side chain were reported.347 Moreover, somehow related thioesters have been reported to similarly react with the catalytic cysteine and displayed rather strong in cellulo antiviral effects.348 It remains to be seen if such compounds will retain some stability in vivo. To address this issue, an attempt was made in 2008 to replace the ester function of compounds 96 by a ketone, as seen in the structure of analogue 97, but the results were disappointing.349 More interestingly, in 2013, a high throughput screening for inhibitors of SARS-CoV-1 Mpro, using the 293![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds of the NIH molecular library, discovered more elaborated 3-pyridyl-bearing compounds such as ML188 (98) or benzotriazole derivatives such as ML300 (99).158,350 One of the reports also included an X-ray based structure of ML188 (98) bound to SARS-CoV-1 Mpro (PDB 3V3M),158 and much later on, both inhibitors were co-crystallized with SARS-CoV-2 Mpro (PDB 7L0D351 PDB 7LME352). Of note is that these inhibitors bind to the catalytic site via key interactions between the imidazole ring of SARS-CoV-2 Mpro histidine-163 and their pyridine or benzotriazole nitrogen. Moreover, for these inhibitors the protease glutamine-166 NH usually interacts with their recurrent carboxamide functions situated four “bonds away” from these nitrogens. As illustrated below, this pattern is recurrent in most of the 3-pyridyl-bearing inhibitors of SARS-CoV main proteases (Fig. 14).
000 compounds of the NIH molecular library, discovered more elaborated 3-pyridyl-bearing compounds such as ML188 (98) or benzotriazole derivatives such as ML300 (99).158,350 One of the reports also included an X-ray based structure of ML188 (98) bound to SARS-CoV-1 Mpro (PDB 3V3M),158 and much later on, both inhibitors were co-crystallized with SARS-CoV-2 Mpro (PDB 7L0D351 PDB 7LME352). Of note is that these inhibitors bind to the catalytic site via key interactions between the imidazole ring of SARS-CoV-2 Mpro histidine-163 and their pyridine or benzotriazole nitrogen. Moreover, for these inhibitors the protease glutamine-166 NH usually interacts with their recurrent carboxamide functions situated four “bonds away” from these nitrogens. As illustrated below, this pattern is recurrent in most of the 3-pyridyl-bearing inhibitors of SARS-CoV main proteases (Fig. 14).
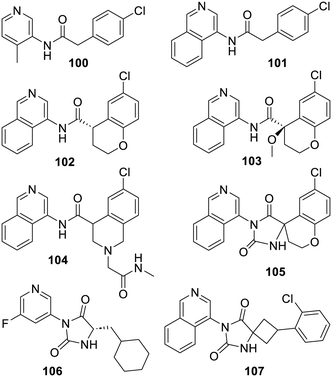
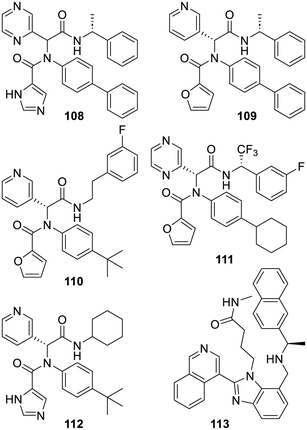
By the end of 2019, these 2013 results became the starting point of research projects in many laboratories. In full collaboration with a large X-ray based facility, many structures of 3-pyridyl-bearing compounds bound to SARS-CoV-2 Mpro were thus made available.93 Such structures were instrumental for the open science COVID Moonshot initiative which undertook a publicly available drug discovery project based on the iterative process required to improve 3-pyridyl-bearing compounds such as ML188 (98).353–357 As described,358,359 in one attempt, the COVID Moonshot initiative went from 3-pyridyl derivative TRY-UNI-714a760b-6 (100) to isoquinoline derivatives such as ADA-UCB-6c2cb422-1 (101), MAT-POS-b3e365b9-1 (102) or PET-UNK-29afea89-2 (103). In fact, these compounds became more and more elaborated not only to strengthen their affinity for the SARS-CoV-2 Mpro catalytic site but also to start addressing the myriad of problems associated with an eventual in vivo use. The next two inhibitors MAT-POS-4223bc15-23 (104) and VLA-UCB-29506327-1 (105) illustrate some more possibilities for further iterations which required extensive profiling before choices were made. Meanwhile, another group also reported some related inhibitors using a docking-based virtual screening along with an X-ray based structure of the derivative X77 (112) bound to SARS-CoV-2 Mpro (PDB 6W63). Interestingly, from the 3-pyridyl derivative 106 discovered with this strategy, the ensuing optimization led to the 300-fold more active inhibitor 107.360 Moreover, out of a virtual screening using an X-ray based structure (PDB 6Y2G) of SARS-CoV-2 Mpro covalently bound to the α-ketoamide inhibitor 91 depicted above, another research group also reported many (modest) hydantoin-bearing inhibitors related to compounds 105–107 (Fig. 15).91
Starting from the less deconstructed ML188 (98), the analogues 108 and 109,361 or the related MAT-POS-f2460aef-1 (110),356 explored some other possibilities and the more elaborated pyrazine-bearing analogue 111 was also reported.362 The design and synthesis of the analogue X77 (112) was never actually reported but this compound was co-crystallized with SARS-CoV-2 Mpro in 2020 and the corresponding X-ray based structure (PDB 6W63) has been repeatedly used for virtual-based approaches. Finally, a search starting with DNA-encoded chemical libraries led to the very strong SARS-CoV-2 Mpro inhibitor 113. As demonstrated by X-ray studies (PDB 7UR9), the isoquinoline moiety of this compound does bind to the protease catalytic site in the same way the series depicted above do (via a hydrogen bond with His-163). However, in one instance in these series, replacing this isoquinoline by a pyridine led to a rather unexpected complete loss of activity (Fig. 16).363
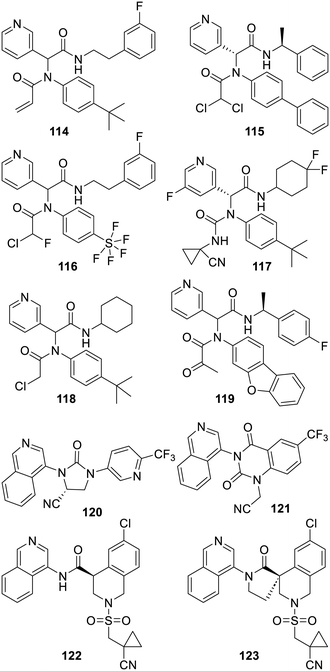
Finally, many research groups resorted to the incorporation of a reactive component in these series.184 As depicted, an acrylamide was used for compound 114,364 and an α,α-dichloroketone for compound 115.221 Of note, the recourse the α,α-chlorofluoracetamide moiety seen in the structures of compounds 70 and 71 was claimed for these series as well,365 and an X-ray based structure of SARS-CoV-2 Mpro covalently bound to the notable Jun10-90-3-C1 (116) has been released by another research group (PDB 8D4P). Out of two large patents from Pardes Biosciences, the randomly chosen α-aminonitrile 117 or even more reactive 1-aminonitriles are additional illustration of this approach.366,367 A report describes a quite systematic introduction of an array of electrophilic moieties leading for instance to the chloroamide 118.368 Even more reactive analogues are described in another publication184 and in a patent.365 Also of interest is the rather large α-ketoamide derivative Y180 (119) which turned out to be an orally available inhibitor of SARS-CoV-2 Mpro endowed with a degree of antiviral effect in vivo.369 In another original approach, two patents from Novartis claim compounds such as the very strong inhibitors 120 (ref. 370) or 121 both combining quite elegantly a reactive nitrile and structural features seen in this class. Concerning the series encompassing compound 121, many analogues claimed in the patent371 do not feature a reactive moiety. Finally, the incorporation of a probably less reactive and hindered nitrile as seen in the structure of MAT-POS-e194df51 (122),356,358 or the more constrained MIK-ENA-5d9157e9 (123), provided some of the most advanced inhibitors of the remarkable COVID Moonshot initiative.359 In fact, compound 122 does not bind covalently to SARS-CoV-2 Mpro as demonstrated by the X-ray based structure code P1788 accessible on the Fragalysis website (Fig. 17).372
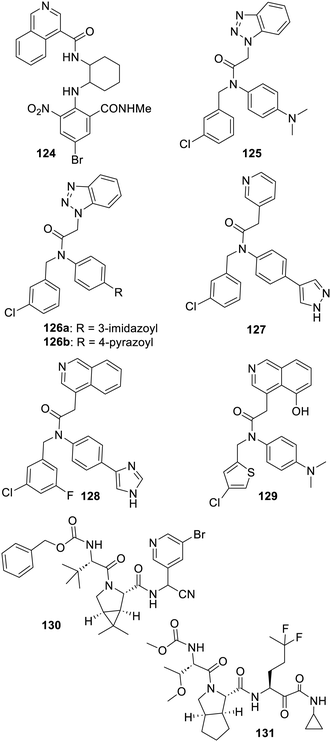
Interestingly, another class of isoquinoline-bearing inhibitors of SARS-CoV-2 Mpro (in which this heterocycle also interacts with His-163, see PDB 7EN8) was found with the combined use of a DNA encoded chemical library and docking-based ranking. This resulted in WU-04 (124) which features a so far required nitro group (for its notable interaction with the backbone CO of the protease Arg188) and is effective in vivo on a mouse model of infection.373 Similar series of nitro-bearing compounds were the focus of extensive structure–activity relationship studies by another research group and were, so far, only illustrated by a recent patent from Qilu Pharmaceuticals.374 Moreover, clinical trials of the undisclosed QLS1128 have been initiated by this company (NCT05458076). The benzotriazole-bearing inhibitor 99 reported350 in 2013 was also the focus of some more research. The MedChem efforts of the COVID Moonshot initiative led to improved analogues such as ALP-POS-6d04362c-2 (125).356 The imidazole-bearing analogue CCF981 (126a) was the fruit of extended SAR studies, although this work also pointed out that strong cytochrome P450 inhibition appeared to be a recurrent issue for these benzotriazole derivatives.352 Another report describes the triazole replacement by a 3-pyridyl moiety, as seen in the structure of compound 127, which led to a less efficient inhibitor in comparison with 126b. But then, the same research group used an isoquinoline and this provided the powerful SARS-CoV-2 Mpro inhibitor 128 (ref. 375), and many more examples, patented by the Cleveland Clinic Foundation, claim a range of inhibitors featuring alternatives to this benzotriazole.376,377 A recent report has also described additional analogues such as compound 129 which combines structural elements of compound 125 as well as 5-hydroxyisoquinoline. The X-ray structure of this inhibitor bound to SARS-CoV-2 Mpro was obtained (PDB 8SXR) and showed that the isoquinoline nitrogen also interacts with the histidine residue 163 of the protease.378 Many strong inhibitors, in which the usual lactams were replaced by a whole array of other bioisosters, were patented by the Global Health Drug Discovery Institute.379 This patent is illustrated by compound 130, which combines peptidic elements and a 3-pyridyl moiety. Moreover, patents from Exscienta or Pardes claim related compounds featuring even more varied heterocycles acting as lactam/3-pyridyl bioisosters.380,381 It is also very likely that an undisclosed derivative of 130 is undergoing clinical trials, sponsored by Jiangsu Hansoh Pharmaceutical, under the name H-10517 (NCT05779579). Finally, a recent patent from Merck382 claims a whole array of inhibitors such as compound 131 which feature a remarkable difluorinated side chain instead of the recurrent lactam. A 2007 paper is actually describing analogues of Boceprivir inhibiting the HCV NS3/4A protease and featuring related difluoromethylene components on the same position.141 In a way, these last two compounds feature most of the hard-won lessons of many structure–activity relationship studies and la boucle est bouclée (the circle is complete). From them, it is actually tempting to suggest to alter the non-covalent 3-pyridyl bearing series of inhibitors by replacing their 3-pyridyl with all the other groups described although it may be a real chemical challenge in some instances (Fig. 18).
Original SARS-CoV-2-Mpro inhibitors
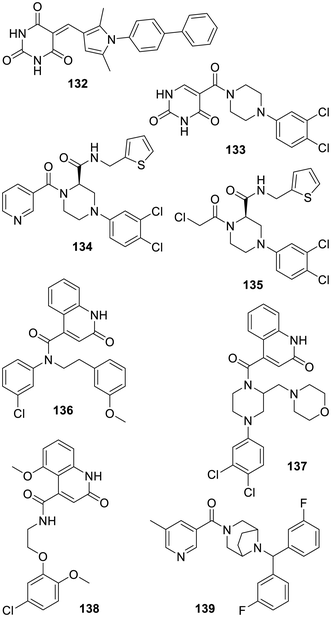
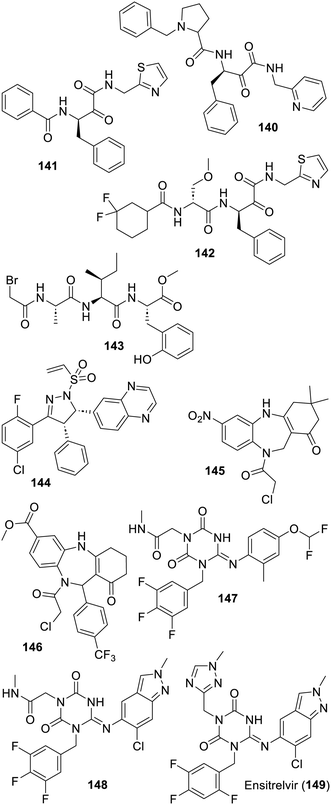
Aside from all the inhibitors described above, which are arguably continuations of the research made on the inhibition of various viral proteases, including SARS-CoV-1 Mpro, one original approach involved the generation of a large number of random peptidic sequences which were then assayed. If a similar but more virtual-based approach failed in an early instance,383 this also led to all D-peptides384,385 or to macrocyclic 11–14-mer peptides featuring modified amino acids386 endowed with affinities for the SARS-CoV-2 Mpro catalytic site. In the latter case, the corresponding X-ray based structure (PDB 7Z4S) may be of future use to deconstruct such rather large peptides into more drug-like compounds. Concerning smaller compounds, a docking-based approach, using half a million bioactive compounds and the structure of SARS-CoV-2 Mpro bound to the peptide derivative 47 (PDB 6LU7), has to be followed by more virtual-based selections, taking into account known inhibitors. This led to the very modest inhibitor 132 featuring a pyrimidinetrione component. A sulfamide derivative was also discovered by the same group but has yet to be developed.387 The related pyrimidinedione 133 was the result of another virtual screening of 6.5 million compounds commercially available using an array of X-ray based structures of SARS-CoV-2 Mpro bound to various inhibitors (compounds 47, X77 (112), telaprevir (25) and masitinib (37); see PDB 7BQY, 6LU7, 6W63, 7C7P, and 7JU7).388 Following this achievement, an X-ray based structure of pyrimidinedione 133 bound to SARS-CoV-2 Mpro was obtained and led to a medicinal chemistry program which has so far provided only modest results.389 Interestingly, another hit to lead project also started with compound 133 and led to the improved GC-14 (134) in which the pyrimidinedione was replaced by the recurrent 3-pyridyl component which interacts with the SARS-CoV-2 Mpro His-163 residue (PDB 8ACL). A replacement with an isoquinoline instead led to the same level of inhibition.390 Unexpectedly, adding instead an α-chloroketone warhead, as depicted for compound 135, had a deleterious impact on the antiviral effect as compared with GC-14 (134).391 However, only a modest antiviral effect was observed in cellulo for compound 134, possibly because of a low cell membrane permeability. From the non-covalent pyridyl-bearing inhibitors listed above, a computer-based “synthesis-directed de novo design model” provided the non-obvious quinolone 136 which has a degree of effect on SARS-CoV-2 Mpro as well as on the seasonal OC43 coronavirus.392 Interestingly, the related inhibitor JZD-07 (137) which gathers features found in 133 and 136 was reported later. In this case, it is the quinolone oxygen which is the bioisosteric replacement for the 3-pyridyl nitrogen (PDB 8GTV), and this inhibitor is endowed with an in vivo efficacy against a mouse model of SARS-CoV-2 infection.393 A related analogue (JZD-26) has been co-crystallized with SARS-CoV-2 Mpro (PDB 8GTW) although a corresponding publication is still expected. The COVID Moonshot initiative also explored related quinolones such as the less rigid but rather modest inhibitor MAT-POS-3b536971-1 (138).356 Another series of 3-pyridyl-bearing inhibitors of SARS-CoV-2 Mpro such as the rather rigid compound 139 were also patented (Fig. 19).394Experimental screenings along with medicinal chemistry approaches were also used to search for SARS-CoV-2 Mpro binding inhibitors. The α-ketoamide 140, with its 2-pyridyl actually interacting with His-163 (PDB 7AEH),395 and compound 141 (ref. 396) were found in the course of a FRET-based screening of a chemical library of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds. The ensuing structure-based (PDB 8HHT) hit to lead progression led to a hundred-fold more active SARS-CoV-2 Mpro inhibitors such as compound SY110 (142).396 A remarkable feature of this original class of peptidic and covalent inhibitors is the absence of the lactam bioisoster of the glutamine residue seen in the many series depicted above. Earlier examples have actually been reported for a modest effect on SARS-CoV-1 (ref. 397) or SARS-CoV-2 multiplication, although in the latter case, these compounds were inhibiting other proteases and had anticancer properties as well.398 Since SY110 (142) displays a robust pan-coronavirus antiviral effect, an original binding mode to the catalytic site (PDB 8HHU), and good preclinical characteristics and is effective in a model animal of COVID-19 infection, further work on this class of inhibitors appears to be warranted. Another recent attempt focused on the variety of sequences found in the substrates of SARS-CoV-2 Mpro including downstream of the cleavage site and the determination of X-ray based structures. This led to the recognition that the peptide Ala-Ile-PheOMe, derived from VKLQAIFR, a larger peptide which has been co-crystallized with SARS-CoV-2 Mpro (PDB 8GWS), retained a degree of affinity for the enzyme. Then, the addition of a warhead onto this peptide and some structure-based improvements led to compounds such as the α-bromoacetamide 143 which is a modest inhibitor of SARS-CoV-2 Mpro and viral replication.399 It is possible that this original work along with the structure of 143 bound to the protease catalytic site reported (PDB 8JPQ) will pave the way for many research projects focusing on the discovery of non peptidic inhibitors targeting this previously unexplored pocket. The strong SARS-CoV-2 Mpro binding inhibitor and vinylsulfoxide-bearing PM-2-071 (144) was reported following a screen of 582 acrylamide or chloroacetamide-bearing compounds and some hit optimization.400 However, the toxicity issue of vinylsulfoxide-bearing compounds should be kept in mind for further development of this type of Michael acceptor.184 The α-chloroamide-bearing benzodiazepine 145 was also found to be a modest inhibitor of SARS-CoV-2 Mpro following the screening of 5000 compounds. The ensuing hit progression has so far provided compound 146 with only a modest 10-fold improvement of activity, despite the obtention of an X-ray based structure (PDB 8JOP).401 Finally and to conclude this part, the truly original and authorized antiviral drug ensitrelvir/S-217622 (149) was discovered in the course of a virtual-based selection using, again, the structure of SARS-CoV-2 Mpro binding X77 (112) (PDB 6W63).402 This selection from hundreds of thousand compounds of an in-house chemical library led to a smaller screening and the identification of compound 147 which has been made years before in the course of a research program focusing on the design of analgesics.403 An X-ray structure (PDB 7VTH) then revealed its binding mode and the ensuing hit to lead program proceeded, via compound 148, to reach ensitrelvir (149). Of note is that as seen in the X-ray based structure, it is the triazole ring which interacts with the His-163 residue for this class of inhibitors (PDB 8DZ0).404 In fact, closely related analogues featuring, amongst other heterocycles, the recurrent 3-pyridyl group instead were claimed later.405 The clinical trials406–408 of ensitrelvir (149), sponsored by Shionogi, turned out to be successful and this is so far the sole non-covalent SARS-CoV-2 Mpro inhibitor approved as an emergency treatment of COVID-19 in Japan. It is currently undergoing phase 3 clinical trials across the world (NCT05605093 and NCT05305547). Two more recent patents have focused on analogues of ensitrelvir (149), the first claims various deuterated analogs which may have improved pharmacological properties,409 and the second has undertaken a rescaffolding of the core 1,3,5-triazine ring and claims analogs featuring a 1,2,4-triazine ring system instead (Fig. 20).410
000 compounds. The ensuing structure-based (PDB 8HHT) hit to lead progression led to a hundred-fold more active SARS-CoV-2 Mpro inhibitors such as compound SY110 (142).396 A remarkable feature of this original class of peptidic and covalent inhibitors is the absence of the lactam bioisoster of the glutamine residue seen in the many series depicted above. Earlier examples have actually been reported for a modest effect on SARS-CoV-1 (ref. 397) or SARS-CoV-2 multiplication, although in the latter case, these compounds were inhibiting other proteases and had anticancer properties as well.398 Since SY110 (142) displays a robust pan-coronavirus antiviral effect, an original binding mode to the catalytic site (PDB 8HHU), and good preclinical characteristics and is effective in a model animal of COVID-19 infection, further work on this class of inhibitors appears to be warranted. Another recent attempt focused on the variety of sequences found in the substrates of SARS-CoV-2 Mpro including downstream of the cleavage site and the determination of X-ray based structures. This led to the recognition that the peptide Ala-Ile-PheOMe, derived from VKLQAIFR, a larger peptide which has been co-crystallized with SARS-CoV-2 Mpro (PDB 8GWS), retained a degree of affinity for the enzyme. Then, the addition of a warhead onto this peptide and some structure-based improvements led to compounds such as the α-bromoacetamide 143 which is a modest inhibitor of SARS-CoV-2 Mpro and viral replication.399 It is possible that this original work along with the structure of 143 bound to the protease catalytic site reported (PDB 8JPQ) will pave the way for many research projects focusing on the discovery of non peptidic inhibitors targeting this previously unexplored pocket. The strong SARS-CoV-2 Mpro binding inhibitor and vinylsulfoxide-bearing PM-2-071 (144) was reported following a screen of 582 acrylamide or chloroacetamide-bearing compounds and some hit optimization.400 However, the toxicity issue of vinylsulfoxide-bearing compounds should be kept in mind for further development of this type of Michael acceptor.184 The α-chloroamide-bearing benzodiazepine 145 was also found to be a modest inhibitor of SARS-CoV-2 Mpro following the screening of 5000 compounds. The ensuing hit progression has so far provided compound 146 with only a modest 10-fold improvement of activity, despite the obtention of an X-ray based structure (PDB 8JOP).401 Finally and to conclude this part, the truly original and authorized antiviral drug ensitrelvir/S-217622 (149) was discovered in the course of a virtual-based selection using, again, the structure of SARS-CoV-2 Mpro binding X77 (112) (PDB 6W63).402 This selection from hundreds of thousand compounds of an in-house chemical library led to a smaller screening and the identification of compound 147 which has been made years before in the course of a research program focusing on the design of analgesics.403 An X-ray structure (PDB 7VTH) then revealed its binding mode and the ensuing hit to lead program proceeded, via compound 148, to reach ensitrelvir (149). Of note is that as seen in the X-ray based structure, it is the triazole ring which interacts with the His-163 residue for this class of inhibitors (PDB 8DZ0).404 In fact, closely related analogues featuring, amongst other heterocycles, the recurrent 3-pyridyl group instead were claimed later.405 The clinical trials406–408 of ensitrelvir (149), sponsored by Shionogi, turned out to be successful and this is so far the sole non-covalent SARS-CoV-2 Mpro inhibitor approved as an emergency treatment of COVID-19 in Japan. It is currently undergoing phase 3 clinical trials across the world (NCT05605093 and NCT05305547). Two more recent patents have focused on analogues of ensitrelvir (149), the first claims various deuterated analogs which may have improved pharmacological properties,409 and the second has undertaken a rescaffolding of the core 1,3,5-triazine ring and claims analogs featuring a 1,2,4-triazine ring system instead (Fig. 20).410
Conclusions
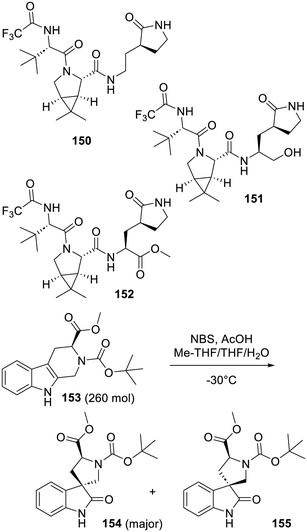
The list of chemicals described above is only representing a portion of all the compounds which have been evaluated around the world on many types of assays targeting SARS-CoV-2 Mpro. In spite of belief-based treatments (using natural or human-made compounds) which unfortunately mobilized a lot of money and claimed an unacceptable number of lives150 and in spite of far too many unexploitable71 computer-involving reports (and patents) which also attracted much funding and raises acute questions on some authors, reviewers (if any), editors and publishers' integrity,79 some approaches did deliver useful drugs. If drug repurposing was, as for the previous coronavirus epidemics, an unmitigated and unfortunately predictable failure,151,152,411,412 past research results on various proteases as well as renewed screenings turned out to be successful starting points. Moreover, as probably not enough emphasized here, the staggering number of X-ray based structures solved greatly helped the resulting hit to lead progressions. A 2014 review on this subject does remain quite relevant.42 As described above, except for ensitrelvir (149), all the most advanced or authorized inhibitors of SARS-CoV-2 Mpro owe their effect to the occurrence of a covalent bond with its catalytic cysteine. Interestingly, in a description of the invention of nirmatrelvir (73),289,413 the choice of the chemically reactive component to be used (nitrile or the ketone function of analogue 72 in this case) remained a difficult one. This was the subject of some investigations,193,414 especially in light of the metabolic and cytotoxicity concerns with aldehyde-bearing covalent inhibitors.268 In one instance,414 a systematic survey was made using the nirmatrelvir (73) structure as a template. From this work which provides precise biochemical and cell-based insights into warhead selection, another quite puzzling result emerged. In fact, if nirmatrelvir (73) or the corresponding ketone-bearing analogue 72b is a really effective inhibitor of SARS-CoV-2 Mpro, the peptides lacking these reactive moieties, such as the decarboxylated derivative 150 or the alcohol 151, are devoid of inhibition effect. This experimental fact is in contrast with what is observed for the KRAS inhibitors 8 and 9 depicted in Fig. 2, since the analogues lacking their acrylamide moiety are still capable of modest inhibition of this kinase. This suggests that, upon the chemical reaction between the warhead of covalent inhibitors and the cysteine of the SARS-CoV-2 Mpro catalytic site, a conformational shift occurs and the newly formed pocket can then bind to some more elements of these inhibitors. Even the ester function of compound 152 which, once this compound settles in the catalytic site, could react with the thiol function, does not. This suggests that 152 never binds to SARS-CoV-2 Mpro and this implies that before a reaction takes place with the cysteine thiol of SARS-CoV-2 Mpro, the catalytic site has a conformation devoid of affinity for the other components of these inhibitors. This could mean that ligands specific to this unknown site conformation have yet to be identified as such. At least one early report on the clustering of various inhibitors of SARS-CoV-2 Mpro according to the shape of the pocket binding to them does point out a degree of flexibility.415 But to account for the complete lack of affinity of compounds 150–152 for the SARS-CoV-2 Mpro catalytic site, one could suspect far larger conformational changes. In fact, one way to detect and characterize such non-obvious conformations remains extensive high throughput screenings for fully original and non-covalent inhibitors, followed by X-ray based structural studies of their interactions with the protease. From the chemistry point of view, many compounds described here are tributes to the creativity of organic chemists. Of note would be the oxidative rearrangement of the tetrahydrocarboline 153 into the spiropyrrolidines 154 and 155.323 As depicted in Scheme 2, it is this synthetic step which paved the way to the design and synthesis of EDP-235 (85) and led to 14 distinct patents claiming many spiropyrrolidine series of SARS-CoV-2 Mpro inhibitors. Interestingly, this rearrangement416 was previously used417 in 1996 to prepare esters and nitrile derivatives somehow related to 154 and 155. History will tell if it was their evaluation on SARS-CoV-2 Mpro which was at the source of these strong inhibitors.Finally, in the following years some viral strains resistant to the currently available SARS-CoV-2 Mpro inhibitors will unfortunately emerge in the population418–423 and these strains will likely compromise the corresponding drug efficacy. For that reason, as well as a price lowering effect, it is very important to have the widest possible range of efficient SARS-CoV-2 multiplication inhibitors so that a lack of cross-resistances between these drugs can be expected. In this regard, the possibility of a general/partial cross-resistance between all the covalent SARS-CoV-2 Mpro inhibitors currently used or developed today is a very relevant issue. In any case, any array of coronavirus-adapted antiviral drugs will be handy to address the next corona epidemic before specific vaccines are designed and mass-produced. Indeed, the recent major progress made with RNA-based vaccines is challenging MedChem and the much longer time usually required to provide a drug-based treatment. Past the next coronavirus pandemics, anticipating such challenges for other zoonotic diseases as well could be a good idea.
Conflicts of interest
There are no conflicts to declare.Notes and references
- S. Yan and G. Wu, Potential 3-chymotrypsin-like cysteine protease cleavage sites in the coronavirus polyproteins pp1a and pp1ab and their possible relevance to COVID-19 vaccine and drug development, FASEB J., 2021, 35, e21573 CAS.
- S. Ullrich and C. Nitsche, The SARS-CoV-2 main protease as drug target, Bioorg. Med. Chem. Lett., 2020, 30, 127377 CrossRef CAS PubMed.
- M. K. Roe, N. A. Junod, A. R. Young, D. C. Beachboard and C. C. Stobart, Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19, J. Gen. Virol., 2021, 102 DOI:10.1099/jgv.0.001558.
- C. C. Chen, X. Yu, C. J. Kuo, J. Min, S. Chen, L. Ma, K. Liu and R. T. Guo, Overview of antiviral drug candidates targeting coronaviral 3C-like main proteases, FEBS J., 2021, 288, 5089 CrossRef CAS.
- A. Hegyi and J. Ziebuhr, Conservation of substrate specificities among coronavirus main proteases, J. Gen. Virol., 2002, 83, 595 CrossRef PubMed.
- M. Xiong, H. Su, W. Zhao, H. Xie, Q. Shao and Y. Xu, What coronavirus 3C-like protease tells us: From structure, substrate selectivity, to inhibitor design, Med. Res. Rev., 2021, 41, 1965 CrossRef CAS PubMed.
- J. Lee, C. Kenward, L. J. Worrall, M. Vuckovic, F. Gentile, A. T. Ton, M. Ng, A. Cherkasov, N. C. J. Strynadka and M. Paetzel, X-ray crystallographic characterization of the SARS-CoV-2 main protease polyprotein cleavage sites essential for viral processing and maturation, Nat. Commun., 2022, 13, 5196 CrossRef CAS.
- J. Wenzel, J. Lampe, H. Müller-Fielitz, R. Schuster, M. Zille, K. Müller, M. Krohn, J. Körbelin, L. Zhang, Ü. Özorhan, V. Neve, J. U. G. Wagner, D. Bojkova, M. Shumliakivska, Y. Jiang, A. Fähnrich, F. Ott, V. Sencio, C. Robil, S. Pfefferle, F. Sauve, C. F. F. Coêlho, J. Franz, F. Spiecker, B. Lembrich, S. Binder, N. Feller, P. König, H. Busch, L. Collin, R. Villaseñor, O. Jöhren, H. C. Altmeppen, M. Pasparakis, S. Dimmeler, J. Cinatl, K. Püschel, M. Zelic, D. Ofengeim, C. Stadelmann, F. Trottein, R. Nogueiras, R. Hilgenfeld, M. Glatzel, V. Prevot and M. Schwaninger, The SARS-CoV-2 main protease Mpro causes microvascular brain pathology by cleaving NEMO in brain endothelial cells, Nat. Neurosci., 2021, 24, 1522 CrossRef CAS PubMed.
- L. Song, D. Wang, G. Abbas, M. Li, M. Cui, J. Wang, Z. Lin and X. E. Zhang, The main protease of SARS-CoV-2 cleaves histone deacetylases and DCP1A, attenuating the immune defense of the interferon-stimulated genes, J. Biol. Chem., 2023, 7(7), 102990 CrossRef.
- J. L. Lu and X. L. Zhou, SARS-CoV-2 main protease Nsp5 cleaves and inactivates human tRNA methyltransferase TRMT1, J. Mol. Cell Biol., 2023, 15, mjad024 CrossRef PubMed.
- K. Zhao, Y. Li, M. Guo, L. Ma and B. Dang, Identification of SARS-CoV-2 PLpro and 3CLpro human proteome substrates using substrate phage display coupled with protein network analysis, J. Biol. Chem., 2023, 299, 104831 CrossRef CAS PubMed.
- A. S. Devasthanam, Mechanisms underlying the inhibition of interferon signaling by viruses, Virulence, 2014, 5, 270 CrossRef.
- H. Li, X. Wang, Y. Wang, Y. Li, Y. Chen, Y. T. Wong, J. He and M. L. He, Secreted LRPAP1 binds and triggers IFNAR1 degradation to facilitate virus evasion from cellular innate immunity, Signal Transduction Targeted Ther., 2023, 8, 374 CrossRef CAS PubMed.
- I. Melano, Y. C. Lo and W. C. Su, Characterization of host substrates of SARS-CoV-2 main protease, Front. Microbiol., 2023, 14, 1251705 CrossRef PubMed.
- Z. Fu, B. Huang, J. Tang, S. Liu, M. Liu, Y. Ye, Z. Liu, Y. Xiong, W. Zhu, D. Cao, J. Li, X. Niu, H. Zhou, Y. J. Zhao, G. Zhang and H. Huang, The complex structure of GRL0617 and SARS-CoV-2 PLpro reveals a hot spot for antiviral drug discovery, Nat. Commun., 2021, 12, 488 CrossRef CAS.
- C. Ma, M. D. Sacco, Z. Xia, G. Lambrinidis, J. A. Townsend, Y. Hu, X. Meng, T. Szeto, M. Ba, X. Zhang, M. Gongora, F. Zhang, M. T. Marty, Y. Xiang, A. Kolocouris, Y. Chen and J. Wang, Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay, ACS Cent. Sci., 2021, 7, 1245 CrossRef CAS.
- A. K. Ghosh, M. Brindisi, D. Shahabi, M. E. Chapman and A. D. Mesecar, Drug Development and Medicinal Chemistry Efforts toward SARS-Coronavirus and Covid-19 Therapeutics, ChemMedChem, 2020, 15, 907 CrossRef CAS.
- V. Anirudhan, H. Lee, H. Cheng, L. Cooper and L. Rong, Targeting SARS-CoV-2 viral proteases as a therapeutic strategy to treat COVID-19, J. Med. Virol., 2021, 93, 2722 CrossRef CAS.
- H. Tan, Y. Hu, P. Jadhav, B. Tan and J. Wang, Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease, J. Med. Chem., 2022, 65, 7561 CrossRef CAS.
- B. C. S. Chia and P. S. Lim, A Patent Review on SARS Coronavirus Papain-Like Protease (PLpro ) Inhibitors, ChemMedChem, 2023, e202300216 CrossRef.
- M. Takeda, Proteolytic activation of SARS-CoV-2 spike protein, Microbiol. Immunol., 2022, 66, 15 CrossRef CAS.
- C. B. Jackson, M. Farzan, B. Chen and H. Choe, Mechanisms of SARS-CoV-2 entry into cells, Nat. Rev. Mol. Cell Biol., 2022, 1, 3 CrossRef.
- C. Ma and J. Wang, Dipyridamole, chloroquine, montelukast sodium, candesartan, oxytetracycline, and atazanavir are not SARS-CoV-2 main protease inhibitors, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2024420118 CrossRef CAS PubMed.
- K. Steuten, H. Kim, J. Widen, B. M. Babin, O. Onguka, S. Lovell, O. Bolgi, B. Cerikan, C. J. Neufeldt, M. Cortese, R. K. Muir, J. M. Bennett, R. Geiss-Friedlander, C. Peters, R. Bartenschlager and M. Bogyo, Challenges for Targeting SARS-CoV2 Proteases as a Therapeutic Strategy for COVID-19, ACS Infect. Dis., 2021, 7, 1457 CrossRef CAS PubMed.
- K. Vandyck, R. Abdelnabi, K. Gupta, D. Jochmans, A. Jekle, J. Deval, D. Misner, D. Bardiot, C. S. Foo, C. Liu, S. Ren, L. Beigelman, L. M. Blatt, S. Boland, L. Vangeel, S. Dejonghe, P. Chaltin, A. Marchand, V. Serebryany, A. Stoycheva, S. Chanda, J. A. Symons, P. Raboisson and J. Neyts, ALG-097111, a potent and selective SARS-CoV-2 3-chymotrypsin-like cysteine protease inhibitor exhibits in vivo efficacy in a Syrian Hamster model, Biochem. Biophys. Res. Commun., 2021, 555, 134 CrossRef CAS.
- C. Ma, Y. Hu, J. A. Townsend, P. I. Lagarias, M. T. Marty, A. Kolocouris and J. Wang, Ebselen, Disulfiram, Carmofur, PX-12, Tideglusib, and Shikonin Are Nonspecific Promiscuous SARS-CoV-2 Main Protease Inhibitors, ACS Pharmacol. Transl. Sci., 2020, 3, 1265 CrossRef CAS.
- C. Ma, H. Tan, J. Choza, Y. Wang and J. Wang, Validation and invalidation of SARS-CoV-2 main protease inhibitors using the Flip-GFP and Protease-Glo luciferase assays, Acta Pharm. Sin. B, 2021, 12, 1636 CrossRef.
- H. Tan, C. Ma and J. Wang, Invalidation of dieckol and 1,2,3,4,6-pentagalloylglucose (PGG) as SARS-CoV-2 main protease inhibitors and the discovery of PGG as a papain-like protease inhibitor, Med. Chem. Res., 2022, 31, 1147 CrossRef CAS PubMed.
- D. M. Mellott, C. T. Tseng, A. Drelich, P. Fajtová, B. C. Chenna, D. H. Kostomiris, J. Hsu, J. Zhu, Z. W. Taylor, K. I. Kocurek, V. Tat, A. Katzfuss, L. Li, M. A. Giardini, D. Skinner, K. Hirata, M. C. Yoon, S. Beck, A. F. Carlin, A. E. Clark, L. Beretta, D. Maneval, V. Hook, F. Frueh, B. L. Hurst, H. Wang, F. M. Raushel, A. J. O'Donoghue, J. L. de Siqueira-Neto, T. D. Meek and J. H. McKerrow, A Clinical-Stage Cysteine Protease Inhibitor blocks SARS-CoV-2 Infection of Human and Monkey Cells, ACS Chem. Biol., 2021, 16, 642 CrossRef PubMed.
- S. Gierer, S. Bertram, F. Kaup, F. Wrensch, A. Heurich, A. Krämer-Kühl, K. Welsch, M. Winkler, B. Meyer, C. Drosten, U. Dittmer, H. Von, T. G. Simmons, H. Hofmann and S. Pöhlmann, The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2 and is targeted by neutralizing anti- bodies, J. Virol., 2013, 87, 5502 CrossRef CAS.
- K. Shirato, M. Kawase and S. Matsuyama, Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2, J. Virol., 2013, 87, 12552 CrossRef CAS PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N. H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, 2020, 181, 271 CrossRef CAS PubMed.
- T. Kinoshita, M. Shinoda, Y. Nishizaki, K. Shiraki, Y. Hirai, Y. Kichikawa, K. Tsushima, M. Sinkai, N. Komura, K. Yoshida, Y. Kido, H. Kakeya, N. Uemura and J. Kadota, A multicenter, double-blind, randomized, parallel-group, placebo-controlled study to evaluate the efficacy and safety of camostat mesilate in patients with COVID-19 (CANDLE study), BMC Med., 2022, 20, 342 CrossRef CAS PubMed.
- M. Yamamoto, S. Matsuyama, X. Li, M. Takeda, Y. Kawaguchi, J. I. Inoue and Z. Matsuda, Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay, Antimicrob. Agents Chemother., 2016, 60, 6532 CrossRef CAS PubMed.
- T. M. Quinn, E. E. Gaughan, A. Bruce, J. Antonelli, R. O'Connor, F. Li, S. McNamara, O. Koch, C. MacKintosh, D. Dockrell, T. Walsh, K. G. Blyth, C. Church, J. Schwarze, C. Boz, A. Valanciute, M. Burgess, P. Emanuel, B. Mills, G. Rinaldi, G. Hardisty, R. Mills, E. G. Findlay, S. Jabbal, A. Duncan, S. Plant, A. D. L. Marshall, I. Young, K. Russell, E. Scholefield, A. F. Nimmo, I. B. Nazarov, G. C. Churchill, J. S. O. McCullagh, K. H. Ebrahimi, C. Ferrett, K. Templeton, S. Rannard, A. Owen, A. Moore, K. Finlayson, M. Shankar-Hari, J. Norrie, R. A. Parker, A. R. Akram, D. C. Anthony, J. W. Dear, N. Hirani and K. Dhaliwal, Randomised controlled trial of intravenous nafamostat mesylate in COVID pneumonitis: Phase 1b/2a experimental study to investigate safety, Pharmacokinetics and Pharmacodynamics, EBioMedicine, 2022, 76, 103856 CrossRef CAS PubMed.
- C. J. Kuo, H. G. Liu, Y. K. Lo, C. M. Seong, K. I. Lee, Y. S. Jung and P. H. Liang, Individual and common inhibitors of coronavirus and picornavirus main proteases, FEBS Lett., 2009, 583, 549 CrossRef CAS.
- A. A. Hernandez and W. R. Roush, Recent advances in the synthesis, design and selection of cysteine protease inhibitors, Curr. Opin. Chem. Biol., 2002, 6, 459 CrossRef CAS PubMed.
- S. R. Shih, S. J. Chen, G. H. Hakimelahi, H. J. Liu, C. T. Tseng and K. S. Shia, Selective human enterovirus and rhinovirus inhibitors: An overview of capsid-binding and protease-inhibiting molecules, Med. Res. Rev., 2004, 24, 449 CrossRef CAS PubMed.
- Q. M. Wanga and S. H. Chen, Human rhinovirus 3C protease as a potential target for the development of antiviral agents, Curr. Protein Pept. Sci., 2007, 8, 19 CrossRef PubMed.
- A. M. De Palma, I. Vliegen, E. De Clercq and J. Neyts, Selective inhibitors of picornavirus replication, Med. Res. Rev., 2008, 28, 823 CrossRef CAS PubMed.
- W. Dai, D. Jochmans, H. Xie, H. Yang, J. Li, H. Su, D. Chang, J. Wang, J. Peng, L. Zhu, Y. Nian, R. Hilgenfeld, H. Jiang, K. Chen, L. Zhang, Y. Xu, J. Neyts and H. Liu, Design, Synthesis, and Biological Evaluation of Peptidomimetic Aldehydes as Broad-Spectrum Inhibitors against Enterovirus and SARS-CoV-2, J. Med. Chem., 2022, 65, 2794 CrossRef CAS PubMed.
- R. Hilgenfeld, From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design, FEBS J., 2014, 4085 CrossRef CAS.
- T. Pillaiyar, M. Manickam, V. Namasivayam, Y. Hayashi and S. H. Jung, An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy, J. Med. Chem., 2016, 59, 6595 CrossRef CAS PubMed.
- P. Thanigaimalai, M. Manickam, V. Namasivayam, Y. Hayashi and S. H. Jung, An Overview of Severe Acute Respiratory Syndrome-Coronavirus (SARS-CoV) 3CL Protease Inhibitors: Peptidomimetics and Small Molecule Chemotherapy, J. Med. Chem., 2016, 59, 6595 CrossRef PubMed.
- R. Liang, L. Wang, N. Zhang, X. Deng, M. Su, Y. Su, L. Hu, C. He, T. Ying, S. Jiang and F. Yu, Development of small-molecule MERS-CoV inhibitors, Viruses, 2018, 10, 721 CrossRef CAS PubMed.
- Y. Liu, C. Liang, L. Xin, X. Ren, L. Tian, X. Ju, H. Li, Y. Wang, Q. Zhao, H. Liu, W. Cao, X. Xie, D. Zhang, Y. Wang and Y. Jian, The development of Coronavirus 3C-Like protease (3CLpro) inhibitors from 2010 to 2020, Eur. J. Med. Chem., 2020, 206, 112711 CrossRef CAS PubMed.
- K. Akaji and H. Konno, Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease, Molecules, 2020, 25, 3920 CrossRef CAS PubMed.
- C. S. Ng, C. C. Stobart and H. Luo, Innate immune evasion mediated by picornaviral 3C protease: Possible lessons for coronaviral 3C-like protease?, Rev. Med. Virol., 2021, 31, 1 CrossRef CAS PubMed.
- R. Banerjee, L. Perera and L. M. V. Tillekeratne, Potential SARS-CoV-2 main protease inhibitors, Drug Discovery Today, 2021, 26, 804 CrossRef CAS PubMed.
- K. Gao, R. Wang, J. Chen, J. J. Tepe, F. Huang and G. W. Wei, Perspectives on SARS-CoV-2 Main Protease Inhibitors, J. Med. Chem., 2021, 64, 16922 CrossRef CAS PubMed.
- A. Citarella, A. Scala, A. Piperno and N. Micale, SARS-CoV-2 M(pro): A Potential Target for Peptidomimetics and Small-Molecule Inhibitors, Biomolecules, 2021, 11, 607 CrossRef CAS PubMed.
- S. A. Amin, S. Banerjee, S. Gayen and T. Jha, Protease targeted COVID-19 drug discovery: What we have learned from the past SARS-CoV inhibitors?, Eur. J. Med. Chem., 2021, 215, 113294 CrossRef CAS PubMed.
- M. Konwar and D. Sarma, Advances in developing small molecule SARS 3CLpro inhibitors as potential remedy for corona virus infection, Tetrahedron, 2021, 77, 131761 CrossRef CAS.
- H. M. Mengist, T. Dilnessa and T. Jin, Structural Basis of Potential Inhibitors Targeting SARS-CoV-2 Main Protease, Front. Chem., 2021, 9, 622898 CrossRef CAS PubMed.
- H. Yang and J. Yang, A review of the latest research on Mpro targeting SARS-COV inhibitors, RSC Med. Chem., 2021, 12, 1026 RSC.
- G. Macip, P. Garcia-Segura, J. Mestres-Truyol, B. Saldivar-Espinoza, G. Pujadas and S. Garcia-Vallvé, A Review of the Current Landscape of SARS-CoV-2 Main Protease Inhibitors: Have We Hit the Bullseye Yet?, Int. J. Mol. Sci., 2022, 23, 259 CrossRef CAS PubMed.
- T. I. Ng, I. Correia, J. Seagal, D. A. DeGoey, M. R. Schrimpf, D. J. Hardee, E. L. Noey and W. M. Kati, Antiviral Drug Discovery for the Treatment of COVID-19 Infections, Viruses, 2022, 14, 961 CrossRef CAS.
- R. Cannalire, C. Cerchia, A. R. Beccari, F. S. Di Leva and V. Summa, Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities, J. Med. Chem., 2022, 65, 2716 CrossRef CAS.
- Q. Hu, Y. Xiong, G. H. Zhu, Y. N. Zhang, Y. W. Zhang, P. Huang and G. B. Ge, The SARS-CoV-2 main protease (Mpro): Structure, function, and emerging therapies for COVID-19, MedComm, 2022, 3, e151 CrossRef CAS PubMed.
- C. S. B. Chia, W. Xu and P. Shuyi Ng, A Patent Review on SARS Coronavirus Main Protease (3CLpro ) Inhibitors, ChemMedChem, 2022, 17, e202100576 CrossRef CAS PubMed.
- L. Agost-Beltrán, S. de la Hoz-Rodríguez, L. Bou-Iserte, S. Rodríguez, A. Fernández-de-la-Pradilla and F. V. González, Advances in the Development of SARS-CoV-2 Mpro Inhibitors, Molecules, 2022, 27, 2523 CrossRef.
- G. La Monica, A. Bono, A. Lauria and A. Martorana, Targeting SARS-CoV-2 Main Protease for Treatment of COVID-19: Covalent Inhibitors Structure-Activity Relationship Insights and Evolution Perspectives, J. Med. Chem., 2022, 65, 12500 CrossRef CAS.
- R. P. Joyce, V. W. Hu and J. Wang, The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations, Med. Chem. Res., 2022, 31, 1637 CrossRef CAS PubMed.
- J. Zhu, H. Zhang, Q. Lin, J. Lyu, L. Lu, H. Chen, X. Zhang, Y. Zhang and K. Chen, Progress on SARS-CoV-2 3CLpro Inhibitors: Inspiration from SARS-CoV 3CLpro Peptidomimetics and Small-Molecule Anti-Inflammatory Compounds, Drug Des., Dev. Ther., 2022, 16, 1067 CrossRef CAS PubMed.
- T. Majerová and J. Konvalinka, Viral proteases as therapeutic targets, Mol. Aspects Med., 2022, 88, 101159 CrossRef PubMed.
- X. Pang, W. Xu, Y. Liu, H. Li and L. Chen, The research progress of SARS-CoV-2 main protease inhibitors from 2020 to 2022, Eur. J. Med. Chem., 2023, 257, 115491 CrossRef CAS PubMed.
- T. Kronenberger, S. A. Laufer and T. Pillaiyar, COVID-19 therapeutics: Small-molecule drug development targeting SARS-CoV-2 main protease, Drug Discovery Today, 2023, 28, 103579 CrossRef CAS PubMed.
- L. Yang and Z. Wang, Bench-to-bedside: Innovation of small molecule anti-SARS-CoV-2 drugs in China, Eur. J. Med. Chem., 2023, 257, 115503 CrossRef CAS PubMed.
- Y. Duan, H. Wang, Z. Yuan and H. Yang, Structural biology of SARS-CoV-2 Mpro and drug discovery, Curr. Opin. Struct. Biol., 2023, 82, 102667 CrossRef CAS PubMed.
- X. Li and Y. Song, Structure and function of SARS-CoV and SARS-CoV-2 main proteases and their inhibition: A comprehensive review, Eur. J. Med. Chem., 2023, 260, 115772 CrossRef CAS PubMed.
- E. N. Muratov, R. Amaro, C. H. Andrade, N. Brown, S. Ekins, D. Fourches, O. Isayev, D. Kozakov, J. L. Medina-Franco, K. M. Merz, T. Oprea, V. Poroikov, G. Schneider, M. Todd, A. Varnek, D. A. Winkler, A. V. Zakharov, A. Cherkasov and A. Tropsha, A critical overview of computational approaches employed for COVID-19 drug discovery, Chem. Soc. Rev., 2021, 50, 9121 RSC.
- G. Macip, P. Garcia-Segura, J. Mestres-Truyol, B. Saldivar-Espinoza, M. J. Ojeda-Montes, A. Gimeno, A. Cereto-Massagué, S. Garcia-Vallvé and G. Pujadas, Haste makes waste: A critical review of docking-based virtual screening in drug repurposing for SARS-CoV-2 main protease (M-pro) inhibition, Med. Res. Rev., 2021, 42, 744 CrossRef PubMed.
- S. L. McGovern, E. Caselli, N. Grigorieff and B. K. Shoichet, A common mechanism underlying promiscuous inhibitors from virtual and high-throughput screening, J. Med. Chem., 2002, 45, 1712 CrossRef CAS PubMed.
- J. Baell and M. A. Walters, Chemical con artists foil drug discovery, Nature, 2014, 513, 481 CrossRef CAS PubMed.
- H. I. Ingolfsson, P. Thakur, K. F. Herold, E. A. Hobart, N. B. Ramsey, X. Periole, D. H. deJong, M. Zwama, D. Yilmaz, K. Hall, T. Maretzky, H. C. Hemmings, Jr., C. Blobel, S. J. Marrink, A. Kocer, J. T. Sack and O. S. Andersen, Phytochemicals perturb membranes and promiscuously alter protein function, ACS Chem. Biol., 2014, 9, 1788 CrossRef CAS PubMed.
- J. Bisson, J. B. McAlpine, J. B. Friesen, S. N. Chen, J. Graham and G. F. Pauli, Can Invalid Bioactives Undermine Natural Product-Based Drug Discovery?, J. Med. Chem., 2016, 59, 1671 CrossRef CAS PubMed.
- J. B. Baell and J. W. M. Nissink, Seven Year Itch: Pan-Assay Interference Compounds (PAINS) in 2017 - Utility and Limitations, ACS Chem. Biol., 2018, 13, 36 CrossRef CAS PubMed.
- J. B. Baell, Screening-Based Translation of Public Research Encounters Painful Problems, ACS Med. Chem. Lett., 2015, 6, 229 CrossRef CAS PubMed.
- J. B. McAlpine, D. Ferreira, N. E. Pauli, S. Gafner and G. F. Pauli, The Ethics of Publishing Biomedical and Natural Products Research, J. Nat. Prod., 2023, 86, 2228–2237 CrossRef CAS PubMed.
- M. C. Parrish, Y. J. Tan, K. V. Grimes and D. Moschly-Rosen, Surviving in the Valley of Death: Opportunities and Challenges in Translating Academic Drug Discoveries, Annu. Rev. Pharmacol. Toxicol., 2019, 59, 405 CrossRef CAS PubMed.
- L. S. Franco, R. C. Maia and E. J. Barreiro, Identification of LASSBio-1945 as an inhibitor of SARS-CoV-2 main protease (MPRO) through in silico screening supported by molecular docking and a fragment-based pharmacophore model, RSC Med. Chem., 2020, 12, 110 RSC.
- L. Zhang, M. Howland, R. Hilgenfeld, M. O. Anderson and S. Eagon, Identification of non-covalent SARS-CoV-2 main protease inhibitors by a virtual screen of commercially available drug-like compounds, Bioorg. Med. Chem. Lett., 2021, 41, 127990 CrossRef CAS PubMed.
- F. Gentile, M. Fernandez, F. Ban, A. Ton, H. Mslati, C. Perez, E. Leblanc, J. Yaacoub, J. Gleave, A. Stern, B. Wong, F. Jean, N. Strynadka and A. Cherkasov, Automated discovery of noncovalent inhibitors of SARS-CoV-2 main protease by consensus Deep Docking of 40 billion small molecules, Chem. Sci., 2021, 12, 15960 RSC.
- S. Günther, P. Y. A. Reinke, Y. Fernández-García, J. Lieske, T. J. Lane, H. M. Ginn, F. H. M. Koua, C. Ehrt, W. Ewert, D. Oberthuer, O. Yefanov, S. Meier, K. Lorenzen, B. Krichel, J. D. Kopicki, L. Gelisio, W. Brehm, I. Dunkel, B. Seychell, H. Gieseler, B. Norton-Baker, B. Escudero-Pérez, M. Domaracky, S. Saouane, A. Tolstikova, T. A. White, A. Hänle, M. Groessler, H. Fleckenstein, F. Trost, M. Galchenkova, Y. Gevorkov, C. Li, S. Awel, A. Peck, M. Barthelmess, F. Schluenzen, X. Lourdu, P. N. Werner, H. Andaleeb, N. Ullah, S. Falke, V. Srinivasan, B. França, M. Schwinzer, H. Brognaro, C. Rogers, D. Melo, J. J. Zaitseva-Doyle, J. Knoska, G. E. Peña-Murillo, A. R. Mashhour, V. Hennicke, P. Fischer, J. Hakanpää, J. Meyer, P. Gribbon, B. Ellinger, M. Kuzikov, M. Wolf, A. R. Beccari, G. Bourenkov, D. von Stetten, G. Pompidor, I. Bento, S. Panneerselvam, I. Karpics, T. R. Schneider, M. M. Garcia-Alai, S. Niebling, C. Günther, C. Schmidt, R. Schubert, H. Han, J. Boger, D. C. F. Monteiro, L. Zhang, X. Sun, J. Pletzer-Zelgert, J. Wollenhaupt, C. G. Feiler, M. S. Weiss, E. C. Schulz, P. Mehrabi, K. Karničar, A. Usenik, J. Loboda, H. Tidow, A. Chari, R. Hilgenfeld, C. Uetrecht, R. Cox, A. Zaliani, T. Beck, M. Rarey, S. Günther, D. Turk, W. Hinrichs, H. N. Chapman, A. R. Pearson, C. Betzel and A. Meents, X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease, Science, 2021, 372, 642 CrossRef PubMed.
- L. Zheng, Y. Chen, J. Bao, L. He, S. Dong, Y. Qi and J. Z. H. Zhang, Discovery of novel inhibitors of SARS-CoV-2 main protease, J. Biomol. Struct. Dyn., 2022, 40, 12526 CrossRef CAS PubMed.
- L. Wang, Z. Yu, S. Wang, Z. Guo, Q. Sun and L. Lai, Discovery of novel SARS-CoV-2 3CL protease covalent inhibitors using deep learning-based screen, Eur. J. Med. Chem., 2022, 244, 114803 CrossRef CAS PubMed.
- D. Deodato, N. Asad and T. M. Dore, Discovery of 2-thiobenzimidazoles as noncovalent inhibitors of SARS-CoV-2 main protease, Bioorg. Med. Chem. Lett., 2022, 72, 128867 CrossRef CAS PubMed.
- J. Glaser, A. Sedova, S. Galanie, D. W. Kneller, R. B. Davidson, E. Maradzike, S. Del Galdo, A. Labbé, D. J. Hsu, R. Agarwal, D. Bykov, A. Tharrington, J. M. Parks, D. M. A. Smith, I. Daidone, L. Coates, A. Kovalevsky and J. C. Smith, Hit Expansion of a Noncovalent SARS-CoV-2 Main Protease Inhibitor, ACS Pharmacol. Transl. Sci., 2022, 5, 255 CrossRef CAS PubMed.
- T. Xu, M. Xu, W. Zhu, C. Z. Chen, Q. Zhang, W. Zheng and R. Huang, Efficient Identification of Anti-SARS-CoV-2 Compounds Using Chemical Structure- and Biological Activity-Based Modeling, J. Med. Chem., 2022, 65, 4590 CrossRef CAS PubMed.
- L. El Khoury, Z. Jing, A. Cuzzolin, A. Deplano, D. Loco, B. Sattarov, F. Hédin, S. Wendeborn, C. Ho, D. El Ahbad, D. T. Jaffrelot Inizan, M. Sturlese, A. Sosic, M. Volpiana, A. Lugato, M. Barone, B. Gatto, M. L. Macchia, M. Bellanda, R. Battistutta, C. Salata, I. Kondratov, R. Iminov, A. Khairulin, Y. Mykhalonok, A. Pochepko, V. Chashka-Ratushnyi, I. Kos, S. Moro, M. Montes, P. Ren, J. W. Ponder, L. Lagardère, J. P. Piquemal and D. Sabbadin, Computationally driven discovery of SARS-CoV-2 Mpro inhibitors: from design to experimental validation, Chem. Sci., 2022, 13, 3674 RSC.
- E. A. Fink, C. Bardine, S. Gahbauer, I. Singh, T. Detomasi, K. White, S. Gu, X. Wan, J. Chen, B. Ary, I. Glenn, J. O'Connell, H. O'Donnell, P. Fajtová, J. Lyu, S. Vigneron, N. J. Young, I. S. Kondratov, A. Alisoltani, L. M. Simons, R. Lorenzo-Redondo, E. A. Ozer, J. F. Hultquist, A. J. O'Donoghue, Y. Moroz, J. Taunton, A. R. Renslo, J. J. Irwin, A. García-Sastre, B. K. Shoichet and C. S. Craik, Large library docking for novel SARS-CoV-2 main protease non-covalent inhibitors, Protein Sci., 2023, 32, e4712 CrossRef CAS PubMed.
- N. Liu, Z. Yang, Y. Liu, X. Dang, Q. Zhang, J. Wang, X. Liu, J. Zhang and X. Pan, Identification of a Putative SARS-CoV-2 Main Protease Inhibitor through In Silico Screening of Self-Designed Molecular Library, Int. J. Mol. Sci., 2023, 24, 11390 CrossRef CAS PubMed.
- A. Douangamath, D. Fearon, P. Gehrtz, T. Krojer, P. Lukacik, C. Owen, E. Resnick, C. Strain-Damerell, A. Aimon, P. Ábrányi-Balogh, J. Brandão-Neto, A. Carbery, G. Davison, A. Dias, T. D. Downes, L. Dunnett, M. Fairhead, J. D. Firth, S. P. Jones, A. Keeley, G. M. Keserü, H. F. Klein, M. P. Martin, M. E. M. Noble, P. O'Brien, A. Powell, R. N. Reddi, R. Skyner, M. Snee, M. J. Waring, C. Wild, N. London, F. von Delft and M. A. Walsh, Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease, Nat. Commun., 2020, 11, 5047 CrossRef CAS PubMed.
- T. M. Rana, Methods and compounds to treat SARS infections, WO2022081984, 2022.
- B. A. Lanman, J. R. Allen, J. G. Allen, A. K. Amegadzie, K. S. Ashton, S. K. Booker, J. J. Chen, N. Chen, M. J. Frohn, G. Goodman, D. J. Kopecky, L. Liu, P. Lopez, J. D. Low, V. Ma, A. E. Minatti, T. T. Nguyen, N. Nishimura, A. Pickrell, A. B. Reed, Y. Shin, A. C. Siegmund, N. A. Tamayo, C. M. Tegley, M. C. Walton, H. L. Wang, R. P. Wurz, M. Xue, K. C. Yang, P. Achanta, M. D. Bartberger, J. Canon, L. S. Hollis, J. D. McCarter, C. Mohr, K. Rex, A. Y. Saiki, T. San Miguel, L. P. Volak, K. H. Wang, D. A. Whittington, S. G. Zech, J. R. Lipford and V. J. Cee, Discovery of a Covalent Inhibitor of KRASG12C (AMG 510) for the Treatment of Solid Tumors, J. Med. Chem., 2020, 63, 52 CrossRef CAS PubMed.
- J. Canon, K. Rex, A. Y. Saiki, C. Mohr, K. Cooke, D. Bagal, K. Gaida, T. Holt, C. G. Knutson, N. Koppada, B. A. Lanman, J. Werner, A. S. Rapaport, T. San Miguel, R. Ortiz, T. Osgood, J. R. Sun, X. Zhu, J. D. McCarter, L. P. Volak, B. E. Houk, M. G. Fakih, B. H. O'Neil, T. J. Price, G. S. Falchook, J. Desai, J. Kuo, R. Govindan, D. S. Hong, W. Ouyang, H. Henary, T. Arvedson, V. Cee and J. R. Lipford, The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity, Nature, 2019, 575, 217 CrossRef CAS PubMed.
- D. Kim, L. Herdeis, D. Rudolph, Y. Zhao, J. Böttcher, A. Vides, C. Ayala-Santos, Y. Pourfarjam, A. Cuevas-Navarro, J. Y. Xue, A. Mantoulidis, J. Bröker, T. Wunberg, O. Schaaf, J. Popow, B. Wolkerstorfer, K. G. Kropatsch, R. Qu, E. de Stanchina, B. Sang, C. Li, D. B. McConnell, N. Kraut and P. Lito, Pan-KRAS inhibitor disables oncogenic signalling and tumour growth, Nature, 2023, 619, 160 CrossRef CAS PubMed.
- J. C. Powers, J. L. Asgian, O. D. Ekici and K. E. James, Irreversible inhibitors of serine, cysteine, and threonine proteases, Chem. Rev., 2002, 102, 4639 CrossRef CAS PubMed.
- B. Turk, Targeting proteases: successes, failures and future prospects, Nat. Rev. Drug Discovery, 2006, 5, 785 CrossRef CAS PubMed.
- Y. Hamada and Y. Kiso, New directions for protease inhibitors directed drug discovery, Biopolymers, 2016, 106, 563 CrossRef CAS PubMed.
- L. Cianni, C. W. Feldmann, E. Gilberg, M. Gütschow, L. Juliano, A. Leitão, J. Bajorath and C. A. Montanari, Can Cysteine Protease Cross-Class Inhibitors Achieve Selectivity?, J. Med. Chem., 2019, 62, 10497 CrossRef CAS PubMed.
- W. Wannamaker, R. Davies, M. Namchuk, J. Pollard, P. Ford, G. Ku, C. Decker, P. Charifson, P. Weber, U. A. Germann, K. Kuida and J. C. Randle, (S)-1-((S)-2-{[1-(4-amino-3-chloro-phenyl)-methanoyl]-amino}-3,3-dimethyl-butanoyl)-pyrrolidine-2-carboxylic acid ((2R,3S)-2-ethoxy-5-oxo-tetrahydro-furan-3-yl)-amide (VX-765), an orally available selective interleukin (IL)-converting enzyme/caspase-1 inhibitor, exhibits potent anti-inflammatory activities by inhibiting the release of IL-1beta and IL-18, J. Pharmacol. Exp. Ther., 2007, 321, 509 CrossRef CAS PubMed.
- M. Poreba, A. Szalek, P. Kasperkiewicz, W. Rut, G. S. Salvesen and M. Drag, Small Molecule Active Site Directed Tools for Studying Human Caspases, Chem. Rev., 2015, 115, 12546 CrossRef CAS PubMed.
- M. W. Wannamaker and R. Davies, Prodrug of an ICE inhibitor, US20020013278, 2002.
- M. B. Boxer, A. M. Quinn, M. Shen, A. Jadhav, W. Leister, A. Simeonov, D. A. Auld and C. J. Thomas, A highly potent and selective caspase 1 inhibitor that utilizes a key 3-cyanopropanoic acid moiety, ChemMedChem, 2010, 5, 730 CrossRef CAS PubMed.
- M. Mortimore, A. Miller, J. Studley and J. D. Charrier, Caspase inhibitors and uses thereof, WO2002094263, 2002.
- G. Brenchley, J. D. Charrier, S. Durrant, R. Knegtel, M. Mortimore and J. R. Studley, Caspase inhibitors and uses therof, WO2004106304, 2004.
- J. D. Charrier, R. Knegtel, M. Mortimore and J. R. Studley, 3[2-(3-acylamino-2-oxo-2H-pyridin-1-yl)-acetylamino]-4-oxo-pentanoic acod derivatives and their use as caspase inhibitors, WO2006057961, 2006.
- G. W. Bemis, M. C. Julian, D. J. Lauffer, M. D. Mullican, M. A. Murcko and D. J. Livingston, Inhibitors of interleukin-1-beta converting enzyme, US5716929, 1998.
- S. Chambon, C. Millois, L. Dumais, R. Pierre, L. Tomas, C. Mathieu, A. L. Ghilini, N. Vanthuyne, K. Reverse, A. Brethon, V. Rodeschini, C. Comino, G. Mouis, G. El-Bazbouz, L. Clary, J. F. Fournier, C. Bouix-Peter, C. S. Harris and L. F. Hennequin, Synthesis and stability evaluation of novel peptidomimetic Caspase-1 inhibitors for topical application, Tetrahedron, 2018, 74, 4805 CrossRef CAS.
- J. F. Fournier, L. Clary, S. Chambon, L. Dumais, C. S. Harris, C. Millois, R. Pierre, S. Talano, E. Thoreau, J. Aubert, M. Aurelly, C. Bouix-Peter, A. Brethon, L. Chantalat, O. Christin, C. Comino, G. El-Bazbouz, A. L. Ghilini, T. Isabet, C. Lardy, A. P. Luzy, C. Mathieu, K. Mebrouk, D. Orfila, J. Pascau, K. Reverse, D. Roche, V. Rodeschini and L. F. Hennequin, Rational Drug Design of Topically Administered Caspase 1 Inhibitors for the Treatment of Inflammatory Acne, J. Med. Chem., 2018, 10, 4030 CrossRef PubMed.
- P. Auberger, P. Chaintreuil, O. Dufies, J. Courjon, L. Boyer and A. Jacquel, Methods and pharmaceutical composition for the treatment of infectious diseases, WO2022008597, 2022.
- E. Jacotot, B. Bung and N. Howard, Treatments of coronavirus infections, cytokine release syndrome, cytokine storm syndrome, or diseases associated with excessive activation of inflammasomes by the use of inhibitors of inflammatory caspases, WO2022074134, 2022.
- O. Alpan, Treatment for diseases caused by RNA viruses, WO2022271877, 2022.
- S. Bao, K. Zhai and Z. Huang, BACE1 inhibitor treatment for suppressing cytokine storm, US20230302009, 2023.
- F. Lecaille, D. Brömme and G. Lalamanach, Biochemical properties and regulation of cathepsin K activity, Biochimie, 2008, 90, 208 CrossRef CAS PubMed.
- J. Gauthier, N. Chauret, W. Cromlish, S. Desmarais, L. T. Duong, J. P. Falgueyret, D. B. Kimmel, S. Lamontagne, S. Léger, T. LeRiche, C. S. Li, F. Massé, D. J. McKay, D. A. Nicoll-Griffith, R. M. Oballa, J. T. Palmer, M. D. Percival, D. Riendeau, J. Robichaud, G. A. Rodan, S. B. Rodan, C. Seto, M. Thérien, V. Truong, M. C. Venuti, G. Wesolowski, R. N. Young, R. Zamboni and W. C. Black, The discovery of Odanacatib (MK-0822), a selective inhibitor of cathepsin K, Bioorg. Med. Chem. Lett., 2008, 18, 923 CrossRef CAS PubMed.
- S. Law, P. M. Andrault, A. H. Aguda, N. T. Nguyen, N. Kruglyak, G. D. Brayer and D. Brömme, Identification of mouse cathepsin K structural elements that regulate the potency of odanacatib, Biochem. J., 2017, 474, 851 CrossRef CAS PubMed.
- M. R. McClung, M. L. O'Donoghue, S. E. Papapoulos, H. Bone, B. Langdahl, K. G. Saag, I. R. Reid, D. P. Kiel, I. Cavallari, M. P. Bonaca, S. D. Wiviott, T. de Villiers, X. Ling, K. Lippuner, T. Nakamura, J. Y. Reginster, J. A. Rodriguez-Portales, C. Roux, J. Zanchetta, C. A. F. Zerbini, J. G. Park, K. Im, A. Cange, L. T. Grip, N. Heyden, C. DaSilva, D. Cohn, R. Massaad, B. B. Scott, N. Verbruggen, D. Gurner, D. L. Miller, M. L. Blair, A. B. Polis, S. A. Stoch, A. Santora, A. Lombardi, A. T. Leung, K. D. Kaufman and M. S. Sabatine, Odanacatib for the treatment of postmenopausal osteoporosis: results of the LOFT multicentre, randomised, double-blind, placebo-controlled trial and LOFT Extension study, Lancet Diabetes Endocrinol., 2019, 7, 899 CrossRef CAS PubMed.
- C. Beaulieu, E. Isabel, A. Fortier, F. Massé, C. Mellon, N. Méthot, M. Ndao, D. Nicoll-Griffith, D. Lee, H. Park and W. C. Black, Identification of potent and reversible cruzipain inhibitors for the treatment of Chagas disease, Bioorg. Med. Chem. Lett., 2010, 20, 7444 CrossRef CAS PubMed.
- M. Ndao, C. Beaulieu, W. C. Black, E. Isabel, F. Vasquez-Camargo, M. Nath-Chowdhury, F. Massé, C. Mellon, N. Methot and D. A. Nicoll-Griffith, Reversible cysteine protease inhibitors show promise for a Chagas disease cure, Antimicrob. Agents Chemother., 2014, 58, 1167 CrossRef.
- A. E. Weber, Dipeptidyl peptidase IV inhibitors for the treatment of diabetes, J. Med. Chem., 2004, 47, 4135 CrossRef CAS PubMed.
- S. L. Gwaltney and J. A. Stafford, Inhibitors of dipeptidyl peptidase, Annu. Rep. Med. Chem., 2005, 40, 149 CAS.
- J. U. Peters, 11 Years of cyanopyrrolidines as DPP-IV inhibitors, Curr. Top. Med. Chem., 2007, 7, 579 CrossRef CAS PubMed.
- S. H. Havale and M. Pal, Medicinal chemistry approaches to the inhibition of dipeptidyl peptidase-4 for the treatment of type 2 diabetes, Bioorg. Med. Chem., 2009, 17, 1783 CrossRef CAS PubMed.
- Y. Liu, Y. Hu and T. Liu, Recent Advances in Non-Peptidomimetic Dipeptidyl Peptidase 4 Inhibitors: Medicinal Chemistry and Preclinical Aspects, Curr. Med. Chem., 2012, 19, 3982 CrossRef CAS PubMed.
- S. Kumar, A. Mittal and A. Mittal, A review upon medicinal perspective and designing rationale of DPP-4 inhibitors, Bioorg. Med. Chem., 2021, 46, 116354 CrossRef CAS PubMed.
- E. B. Villhauer, J. A. Brinkman, G. B. Naderi, B. F. Burkey, B. E. Dunning, K. Prasad, B. L. Mangold, M. E. Russell and T. E. Hughes, 1-[[(3-hydroxy-1-adamantyl)amino]acetyl]-2-cyano-(S)-pyrrolidine: a potent, selective, and orally bioavailable dipeptidyl peptidase IV inhibitor with antihyperglycemic properties, J. Med. Chem., 2003, 46, 2774 CrossRef CAS PubMed.
- D. J. Augeri, J. A. Robl, D. A. Betebenner, D. R. Magnin, A. Khanna, J. G. Robertson, A. Wang, L. M. Simpkins, P. Taunk, Q. Huang, S.-P. Han, B. Abboa-Offei, M. Cap, L. Xin, L. Tao, E. Tozzo, G. E. Welzel, D. M. Egan, J. Marcinkeviciene, S. Y. Chang, S. A. Biller, M. S. Kirby, R. A. Parker and L. G. Hamann, Discovery and Preclinical Profile of Saxagliptin (BMS-477118): A Highly Potent, Long-Acting, Orally Active Dipeptidyl Peptidase IV Inhibitor for the Treatment of Type 2 Diabetes, J. Med. Chem., 2005, 48, 5025 CrossRef CAS PubMed.
- M. Nabeno, F. Akahoshi, H. Kishida, I. Miyaguchi, Y. Tanaka, S. Ishii and T. Kadowaki, A comparative study of the binding modes of recently launched dipeptidyl peptidase IV inhibitors in the active site, Biochem. Biophys. Res. Commun., 2013, 434, 191 CrossRef CAS PubMed.
- T. E. Hughes, M. D. Mone, M. E. Russell, S. C. Weldon and E. B. Villhauer, NVP-DPP728 (1-[[[2-[(5-Cyanopyridin-2-yl)amino]ethyl]amino]acetyl]-2-cyano-(S)-pyrrolidine), a Slow-Binding Inhibitor of Dipeptidyl Peptidase IV, Biochemistry, 1999, 38, 11597 CrossRef CAS PubMed.
- M. Engel, T. Hoffmann, L. Wagner, M. Wermann, U. Heiser, R. Kiefersauer, R. Huber, W. Bode, H.-U. Demuth and H. Brandstetter, The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5063 CrossRef CAS.
- D. Kim, L. Wang, M. Beconi, G. J. Eiermann, M. H. Fisher, H. He, G. J. Hickey, J. E. Kowalchick, B. Leiting, K. Lyons, F. Marsilio, M. E. McCann, R. A. Patel, A. Petrov, G. Scapin, S. B. Patel, R. S. Roy, J. K. Wu, M. J. Wyvratt, B. B. Zhang, L. Zhu, N. A. Thornberry and A. E. Weber, (2R)-4-oxo-4-[3-(trifluoromethyl)-5,6-dihydro[1,2,4]triazolo[4,3-a]pyrazin-7(8H)-yl]-1-(2,4,5-trifluorophenyl)butan-2-amine: a potent, orally active dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes, J. Med. Chem., 2005, 48, 141 CrossRef CAS PubMed.
- J. Feng, Z. Zhang, M. B. Wallace, J. A. Stafford, S. W. Kaldor, D. B. Kassel, M. Navre, L. Shi, R. J. Skene, T. Asakawa, K. Takeuchi, R. Xu, D. R. Webb and S. L. N. Gwaltney, Discovery of alogliptin: a potent, selective, bioavailable, and efficacious inhibitor of dipeptidyl peptidase IV, J. Med. Chem., 2007, 50, 2297 CrossRef CAS PubMed.
- Z. Zhang, M. B. Wallace, J. Feng, J. A. Stafford, R. J. Skene, L. Shi, B. Lee, K. Aertgeerts, A. Jennings, R. Xu, D. B. Kassel, S. W. Kaldor, M. Navre, D. R. Webb and S. L. Gwaltney, Design and synthesis of pyrimidinone and pyrimidinedione inhibitors of dipeptidyl peptidase IV, J. Med. Chem., 2011, 54, 510 CrossRef CAS PubMed.
- N. A. Meanwell, G. I. Georg and S. Wang, The 2020 Nobel Prize in Physiology or Medicine, J. Med. Chem., 2020, 63, 13197 CrossRef CAS PubMed.
- R. D'Ambrosio, E. Degasperi, M. Colombo and A. Aghemo, Direct-acting antivirals: the endgame for hepatitis C?, Curr. Opin. Virol., 2017, 24, 31 CrossRef.
- J. A. McCauley and M. T. Rudd, Hepatitis C virus NS3/4a protease inhibitors, Curr. Opin. Pharmacol., 2016, 30, 84 CrossRef CAS PubMed.
- T. Pillaiyar, V. Namasivayam and M. Manickam, Macrocyclic Hepatitis C Virus NS3/4A Protease Inhibitors: An Overview of Medicinal Chemistry, Curr. Med. Chem., 2016, 29, 3404 CrossRef PubMed.
- S. Venkatraman, S. L. Bogen, A. Arasappan, F. Bennett, K. Chen, E. Jao, Y. T. Liu, R. Lovey, S. Hendrata, Y. Huang, W. Pan, T. Parekh, P. Pinto, V. Popov, R. Pike, S. Ruan, B. Santhanam, B. Vibulbhan, W. Wu, W. Yang, J. Kong, X. Liang, J. Wong, R. Liu, N. Butkiewicz, R. Chase, A. Hart, S. Agrawal, P. Ingravallo, J. Pichardo, R. Kong, B. Baroudy, B. Malcolm, Z. Guo, A. Prongay, V. Madison, L. Broske, X. Cui, K. C. Cheng, Y. Hsieh, J. M. Brisson, D. Prelusky, W. Korfmacher, R. White, S. Bogdanowich-Knipp, A. Pavlovsky, P. Bradley, A. K. Saksena, A. Ganguly, J. Piwinski, V. Girijavallabhan and F. G. Njoroge, Discovery of (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]- 3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (SCH 503034), a selective, potent, orally bioavailable hepatitis C virus NS3 protease inhibitor: a potential therapeutic agent for the treatment of hepatitis C infection, J. Med. Chem., 2006, 49, 6074 CrossRef CAS PubMed.
- A. J. Prongay, Z. Guo, N. Yao, J. Pichardo, T. Fischmann, C. Strickland, J. J. Myers, P. C. Weber, B. M. Beyer, R. Ingram, Z. Hong, W. W. Prosise, L. Ramanathan, S. S. Taremi, T. Yarosh-Tomaine, R. Zhang, M. Senior, R. S. Yang, B. Malcolm, A. Arasappan, F. Bennett, S. L. Bogen, K. Chen, E. Jao, Y. T. Liu, R. G. Lovey, A. K. Saksena, S. Venkatraman, V. Girijavallabhan, F. G. Njoroge and V. Madison, Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization, J. Med. Chem., 2007, 50, 2310 CrossRef CAS PubMed.
- A. Y. Howe and S. Venkatraman, The Discovery and Development of Boceprevir: A Novel, First-generation Inhibitor of the Hepatitis C Virus NS3/4A Serine Protease, J. Clin. Transl. Hepatol., 2013, 1, 22 Search PubMed.
- A. D. Kwong, R. S. Kauffman, P. Hurter and P. Mueller, Discovery and development of telaprevir: an NS3-4A protease inhibitor for treating genotype 1 chronic hepatitis C virus, Nat. Biotechnol., 2011, 29, 993 CrossRef CAS PubMed.
- M. Llinàs-Brunet, M. D. Bailey, G. Bolger, C. Brochu, A. M. Faucher, J. M. Ferland, M. Garneau, E. Ghiro, V. Gorys, C. Grand-Maître, T. Halmos, N. Lapeyre-Paquette, F. Liard, M. Poirier, M. Rhéaume, Y. S. Tsantrizos and D. Lamarre, Structure-activity study on a novel series of macrocyclic inhibitors of the hepatitis C virus NS3 protease leading to the discovery of BILN 2061, J. Med. Chem., 2004, 47, 1605 CrossRef PubMed.
- D. Lamarre, P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bös, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagacé, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R St George, R. B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong and M. Llinàs-Brunet, An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus, Nature, 2003, 426, 186 CrossRef CAS PubMed.
- Å. Rosenquist, B. Samuelsson, P. O. Johansson, M. D. Cummings, O. Lenz, P. Raboisson, K. Simmen, S. Vendeville, H. de Kock, M. Nilsson, A. Horvath, R. Kalmeijer, G. de la Rosa and M. Beumont-Mauviel, Discovery and development of simeprevir (TMC435), a HCV NS3/4A protease inhibitor, J. Med. Chem., 2014, 57, 1673 CrossRef PubMed.
- S. Harper, J. A. McCauley, M. T. Rudd, M. Ferrara, M. DiFilippo, B. Crescenzi, U. Koch, A. Petrocchi, M. K. Holloway, J. W. Butcher, J. J. Romano, K. J. Bush, K. F. Gilbert, C. J. McIntyre, K. T. Nguyen, E. Nizi, S. S. Carroll, S. W. Ludmerer, C. Burlein, J. M. DiMuzio, D. J. Graham, C. M. McHale, M. W. Stahlhut, D. B. Olsen, E. Monteagudo, S. Cianetti, C. Giuliano, V. Pucci, N. Trainor, C. M. Fandozzi, M. Rowley, P. J. Coleman, J. P. Vacca, V. Summa and N. J. Liverton, Discovery of MK-5172, a Macrocyclic Hepatitis C Virus NS3/4a Protease Inhibitor, ACS Med. Chem. Lett., 2012, 3, 332 CrossRef CAS PubMed.
- S. Coats, E. C. Garnier-Amblard, F. Amblard, M. Ehteshami, S. Amiralaei, H. Zhang, L. Zhou, S. R. Boucle, X. Lu, L. Bondada, J. R. Shelton, H. Li, P. Liu, C. Li, J. H. Cho, S. N. Chavre, S. Zhou, J. Mathew and R. F. Schinazi, Chutes and ladders in hepatitis C nucleoside drug development, Antiviral Res., 2014, 102, 119 CrossRef CAS PubMed.
- P. P. Glasziou, S. Sanders and T. Hoffmann, Waste in covid-19 research, BMJ, 2020,(369), m1847 CrossRef PubMed.
- L. K. Boerner, The antivirals that weren't: drug repurposing for COVID-19 produced misleading results, C&En, 2021, vol. 99, (25), p. 7 Search PubMed.
- C. L. Bellera, M. Llanos, M. E. Gantner, S. Rodriguez, L. Gavernet, M. Comini and A. Talevi, Can drug repurposing strategies be the solution to the COVID-19 crisis?, Expert Opin. Drug Discovery, 2021, 16, 605 CrossRef CAS PubMed.
- C. G. Begley, M. Ashton, J. Baell, M. Bettess, M. P. Brown, B. Carter, W. N. Charman, C. Davis, S. Fisher, I. Frazer, A. Gautam, M. P. Jennings, P. Kearney, E. Keeffe, D. Kelly, A. F. Lopez, M. McGuckin, M. W. Parker, C. Rayner, B. Roberts, J. S. Rush and M. Sullivan, Drug repurposing: Misconceptions, challenges, and opportunities for academic researchers, Sci. Transl. Med., 2021, 61, eabd5524 CrossRef PubMed.
- M. M. Ghahremanpour, J. Tirado-Rives, M. Deshmukh, J. A. Ippolito, C. H. Zhang, I. Cabeza de Vaca, M. E. Liosi, K. S. Anderson and W. L. Jorgensen, Identification of 14 Known Drugs as Inhibitors of the Main Protease of SARS-CoV-2, ACS Med. Chem. Lett., 2020, 11, 2526 CrossRef CAS PubMed.
- C. H. Zhang, E. A. Stone, M. Deshmukh, J. A. Ippolito, M. M. Ghahremanpour, J. Tirado-Rives, K. A. Spasov, S. Zhang, Y. Takeo, S. N. Kudalkar, Z. Liang, F. Isaacs, B. Lindenbach, S. Miller, K. S. Anderson and W. L. Jorgensen, Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations, ACS Cent. Sci., 2021, 7, 467 CrossRef CAS PubMed.
- M. G. Deshmukh, J. A. Ippolito, C. H. Zhang, E. A. Stone, R. A. Reilly, S. J. Miller, W. L. Jorgensen and K. S. Anderson, Structure-guided design of a perampanel-derived pharmacophore targeting the SARS-CoV-2 main protease, Structure, 2021, 29, 823 CrossRef CAS PubMed.
- C. H. Zhang, K. A. Spasov, R. A. Reilly, K. Hollander, E. A. Stone, J. A. Ippolito, M. E. Liosi, M. G. Deshmukh, J. Tirado-Rives, S. Zhang, Z. Liang, S. Miller, F. Isaacs, B. D. Lindenbach, K. S. Anderson and W. L. Jorgensen, Optimization of Triarylpyridinone Inhibitors of the Main Protease of SARS-CoV-2 to Low-Nanomolar Antiviral Potency, ACS Med. Chem. Lett., 2021, 12, 1325 CrossRef CAS PubMed.
- L. Jacobs, A. van der Westhuyzen, N. Pribut, D. Cui, M. P. D'Erasmo, P. W. Bartsch, K. Liu, M. C. Cox, S. J. Greenlund, R. K. Plemper, D. Mitchell, J. Marlow, M. K. Andrews, R. E. Krueger, Z. M. Sticher, A. A. Kolykhalov, M. G. Natchus, B. Zhou, S. C. Pelly and D. C. Liotta, Design and Optimization of Novel Competitive, Non-peptidic, SARS-CoV-2 Mpro Inhibitors, ACS Med. Chem. Lett., 2023, 14, 1434 CrossRef CAS PubMed.
- J. Jacobs, V. Grum-Tokars, Y. Zhou, M. Turlington, S. A. Saldanha, P. Chase, A. Eggler, E. S. Dawson, Y. M. Baez-Santos, S. Tomar, A. M. Mielech, S. C. Baker, C. W. Lindsley, P. Hodder, A. Mesecar and S. R. Stauffer, Discovery, synthesis, and structure-based optimization of a series of N-(tert-butyl)-2-(N-arylamido)-2-(pyridin-3-yl) acetamides (ML188) as potent noncovalent small molecule inhibitors of the severe acute respiratory syndrome coronavirus (SARS-CoV) 3CL protease, J. Med. Chem., 2013, 56, 534 CrossRef CAS PubMed.
- F. von Nussbaum and V. M. Li, Neutrophil elastase inhibitors for the treatment of (cardio)pulmonary diseases: Into clinical testing with pre-adaptive pharmacophores, Bioorg. Med. Chem. Lett., 2015, 25, 4370 CrossRef CAS.
- C. Fischer, N. A. Vepřek, Z. Peitsinis, K. P. Rühmann, C. Yang, J. N. Spradlin, D. Dovala, D. K. Nomura, Y. Zhang and D. Trauner, De novo Design of SARS-CoV-2 Main Protease Inhibitors, Synlett, 2021, 33, 458 Search PubMed.
- A. Moussy, S. Tay, N. Drayman, G. Randal and S. Chen, The use of masitinib for the treatment of coronavirus disease 2019 (COVID-19), WO2021205029, 2021.
- N. Drayman, J. K. DeMarco, K. A. Jones, S. A. Azizi, H. M. Froggatt, K. Tan, N. I. Maltseva, S. Chen, V. Nicolaescu, S. Dvorkin, K. Furlong, R. S. Kathayat, M. R. Firpo, V. Mastrodomenico, E. Bruce, M. M. Schmidt, R. Jedrzejczak, M. Á. Muñoz-Alía, B. Schuster, V. Nair, K. Y. Han, A. O'Brien, A. Tomatsidou, B. Meyer, M. Vignuzzi, D. Missiakas, J. W. Botten, C. B. Brooke, H. Lee, S. C. Baker, B. C. Mounce, N. S. Heaton, W. E. Severson, K. E. Palmer, B. C. Dickinson, A. Joachimiak, G. Randall and S. Tay, Masitinib is a broad coronavirus 3CL inhibitor that blocks replication of SARS-CoV-2, Science, 2021, 373, 931 CrossRef CAS PubMed.
- K. Hammam, M. Saez-Ayala, E. Rebuffet, L. Gros, S. Lopez, B. Hajem, M. Humbert, E. Baudelet, S. Audebert, S. Betzi, A. Lugari, S. Combes, S. Letard, N. Casteran, C. Mansfield, A. Moussy, S. De Sepulveda, P. X. Morelli and P. Dubreuil, Dual protein kinase and nucleoside kinase modulators for rationally designed polypharmacology, Nat. Commun., 2017, 8, 1420 CrossRef PubMed.
- J. Dyall, C. M. Coleman, B. J. Hart, T. Venkataraman, M. R. Holbrook, J. Kindrachuk, R. F. Johnson, G. G. J. Olinger, P. B. Jahrling, M. Laidlaw, L. M. Johansen, C. M. Lear-Rooney, P. J. Glass, L. E. Hensley and M. B. Frieman, Repurposing of clinically developed drugs for treatment of Middle East respiratory syndrome coronavirus infection, Antimicrob. Agents Chemother., 2014, 58, 4885 CrossRef PubMed.
- H. Zhao, M. Mendenhall and M. W. Deininger, Imatinib is not a potent anti-SARS-CoV-2 drug, Leukemia, 2020, 34, 3085 CrossRef CAS PubMed.
- J. Loschwitz, A. Jäckering, M. Keutmann, M. Olagunju, R. J. Eberle, M. A. Coronado, O. O. Olubiyi and B. Strodel, Novel inhibitors of the main protease enzyme of SARS-CoV-2 identified via molecular dynamics simulation-guided in vitro assay, Bioorg. Chem., 2021, 111, 104862 CrossRef CAS PubMed.
- A. Sauvat, F. Ciccosanti, F. Colavita, R. Di Rienzo, M. C. Castilletti, M. R. Capobianchi, O. Kepp, L. Zitvogel, G. M. Fimia, M. Piacentini and G. Kroemer, On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2, Cell Death Dis., 2020, 11, 656 CrossRef CAS PubMed.
- T. A. Tummino, V. V. Rezelj, B. Fischer, A. Fischer, M. J. O'Meara, B. Monel, T. Vallet, K. M. White, Z. Zhang, A. Alon, H. Schadt, H. R. O'Donnell, J. Lyu, R. Rosales, B. L. McGovern, R. Rathnasinghe, S. Jangra, M. Schotsaert, J. R. Galarneau, N. J. Krogan, L. Urban, K. M. Shokat, A. Kruse, A. García-Sastre, O. Schwartz, F. Moretti, M. Vignuzzi, F. Pognan and B. K. Shoichet, Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2, Science, 2021, 373, 541 CrossRef CAS PubMed.
- E. J. Fritch, A. L. Mordant, T. S. K. Gilbert, C. I. Wells, X. Yang, N. K. Barker, E. A. Madden, K. H. R. Dinnon, Y. J. Hou, L. V. Tse, I. N. Castillo, A. C. Sims, N. J. Moorman, P. Lakshmanane, T. M. Willson, L. E. Herring, L. M. Graves and R. S. Baric, Investigation of the Host Kinome Response to Coronavirus Infection Reveals PI3K/mTOR Inhibitors as Betacoronavirus Antivirals, J. Proteome Res., 2023, 22, 3159 CrossRef CAS PubMed.
- H. Munier-Lehmann, P.-O. Vidalain, F. Tangy and Y. L. Janin, On dihydroorotate dehydrogenases, their inhibitors and uses, J. Med. Chem., 2013, 56, 3148 CrossRef CAS PubMed.
- M. Wang, Y. Zhao, W. Hu, D. Zhao, Y. Zhang, T. Wang, Z. Zheng, X. Li, S. Zeng, Z. Liu, L. Lu, Z. Wan and K. Hu, Treatment of Coronavirus Disease 2019 Patients With Prolonged Postsymptomatic Viral Shedding With Leflunomide: A Single-center Randomized Controlled Clinical Trial, Clin. Infect. Dis., 2021, 73, e4012 CrossRef CAS PubMed.
- M. J. G. T. Vehreschild, P. Atanasov, K. Yurko, C. Oancea, G. Popov, V. Smesnoi, G. Placinta, H. Kohlhof, D. Vitt, E. Peelen, J. Mihajlović and A. R. Muehler, Safety and Efficacy of Vidofludimus Calcium in Patients Hospitalized with COVID-19: A Double-Blind, Randomized, Placebo-Controlled, Phase 2 Trial, Infect. Dis. Ther., 2022, 6, 2159 CrossRef PubMed.
- I. Kralj-Hans, K. Li, A. Wesek, A. Lamorgese, F. Omar, K. Ranasinghe, M. McGee, K. Brack, S. Li, R. Aggarwal, A. Bulle, A. Kodre, S. Sharma, D. Fluck, I. John, P. Sharma, J. D. Belsey, L. Li, S. R. K. Seshasai, H. L. Li, N. Marczin, Z. Chen and DEFEAT-COVID Investigators, Leflunomide treatment for patients hospitalised with COVID-19: DEFEAT-COVID randomised controlled trial, BMJ Open, 2023, 13, e068179 CrossRef PubMed.
- C. J. Wu, J. T. Jan, C. M. Chen, H. P. Hsieh, D. R. Hwang, H. W. Liu, C. Y. Liu, H. W. Huang, S. C. Chen, C. F. Hong, R. K. Lin, Y. S. Chao and J. T. A. Hsu, Inhibition of Severe Acute Respiratory Syndrome Coronavirus Replication by Niclosamide, Antimicrob. Agents Chemother., 2004, 48, 2693 CrossRef CAS PubMed.
- J. Xu, P. Y. Shi, H. Li and J. Zhou, Broad Spectrum Antiviral Agent Niclosamide and Its Therapeutic Potential, ACS Infect. Dis., 2020, 8, 909 CrossRef PubMed.
- S. Jeon, M. Ko, J. Lee, I. Choi, S. Y. Byun, S. Park, D. Shum and S. Kim, Identification of Antiviral Drug Candidates against SARS-CoV-2 from FDA-Approved Drugs, Antimicrob. Agents Chemother., 2020, 64, e00819 CrossRef CAS PubMed.
- K. Shamim, M. Xu, X. Hu, E. M. Lee, X. Lu, R. Huang, P. Shah, X. Xu, C. Z. Chen, M. Shen, H. Guo, L. Chen, Z. Itkin, R. T. Eastman, P. Shinn, C. Klumpp-Thomas, S. Michael, A. Simeonov, D. C. Lo, G. L. Ming, H. Song, H. Tang, W. Zheng and W. Huang, Application of niclosamide and analogs as small molecule inhibitors of Zika virus and SARS-CoV-2 infection, Bioorg. Med. Chem. Lett., 2021, 40, 127906 CrossRef CAS.
- V. Backer, U. Sjöbring, J. Sonne, A. Weiss, M. Hostrup, H. K. Johansen, V. Becker, D. P. Sonne, T. Balchen, M. Jellingsø and M. O. A. Sommer, A randomized, double-blind, placebo-controlled phase 1 trial of inhaled and intranasal niclosamide: A broad spectrum antiviral candidate for treatment of COVID-19, Lancet Reg. Health Eur., 2021, 100084 CrossRef PubMed.
- D. M. Cairns, D. Dulko, J. K. Griffiths, Y. Golan, T. Cohen, L. Trinquart, L. L. Price, K. R. Beaulac and H. P. Selker, Efficacy of Niclosamide vs Placebo in SARS-CoV-2 Respiratory Viral Clearance, Viral Shedding, and Duration of Symptoms Among Patients With Mild to Moderate COVID-19: A Phase 2 Randomized Clinical Trial, JAMA Netw. Open, 2022, 5(2), e2144942 CrossRef PubMed.
- L. Braga, H. Ali, I. Secco, E. Chiavacci, G. Neves, D. Goldhill, R. Penn, J. M. Jimenez-Guardeño, A. M. Ortega-Prieto, R. Bussani, A. Cannatà, G. Rizzari, C. Collesi, E. Schneider, D. Arosio, A. M. Shah, W. S. Barclay, M. H. Malim, J. Burrone and M. Giacca, Drugs that inhibit TMEM16 proteins block SARS-CoV-2 Spike-induced syncytia, Nature, 2021, 594, 88 CrossRef CAS PubMed.
- J. J. Shie, J. M. Fang, C. J. Kuo, T. H. Kuo, P. H. Liang, H. J. Huang, W. B. Yang, C. H. Lin, J. L. Chen, Y. T. Wu and C. H. Wong, Discovery of potent anilide inhibitors against the severe acute respiratory syndrome 3CL protease, J. Med. Chem., 2005, 48, 4469 CrossRef CAS PubMed.
- S. K. Samrat, J. Xu, X. Xie, E. Gianti, H. Chen, J. Zou, J. G. Pattis, K. Elokely, H. Lee, Z. Li, M. L. Klein, P. Y. Shi, J. Zhou and H. Li, Allosteric inhibitors of the main protease of SARS-CoV-2, Antiviral Res., 2022, 105381 CrossRef CAS PubMed.
- C. Yanyan, Y. Jimbo, W. Zhuoya, Z. Chenyang, Y. Rilei, L. Li and Y. Menglin, Application of closantel or its pharmaceutically acceptable salt in preparation of medicine for preventing and/or treating coronavirus infection, CN112294793, 2020.
- B. Tan, M. Sacco, H. Tan, K. Li, R. Joyce, X. Zhang, Y. Chen and J. Wang, Exploring diverse reactive warheads for the design of SARS-CoV-2 main protease inhibitors, Eur. J. Med. Chem., 2023, 259, 115667 CrossRef CAS PubMed.
- M. D. Sacco, C. Ma, P. Lagarias, A. Gao, J. A. Townsend, X. Meng, P. Dube, X. Zhang, Y. Hu, N. Kitamura, B. Hurst, B. Tarbet, M. T. Marty, A. Kolocouris, Y. Xiang, Y. Chen and J. Wang, Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L, Sci. Adv., 2020, 6, eabe0751 CrossRef CAS PubMed.
- L. Fu, F. Ye, Y. Feng, F. Yu, Q. Wang, Y. Wu, C. Zhao, H. Sun, B. Huang, P. Niu, H. Song, Y. Shi, X. Li, W. Tan, J. Qi and G. F. Gao, Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease, Nat. Commun., 2020, 11, 4417 CrossRef CAS PubMed.
- C. Ma, M. D. Sacco, B. Hurst, J. A. Townsend, Y. Hu, T. Szeto, X. Zhang, B. Tarbet, M. T. Marty, Y. Chen and J. Wang, Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease, Cell Res., 2020, 30, 678 CrossRef CAS PubMed.
- A. D. Rathnayake, J. Zheng, Y. Kim, K. D. Perera, S. Mackin, D. K. Meyerholz, M. M. Kashipathy, K. P. Battaile, S. Lovell, S. Perlman, W. C. Groutas and K. O. Chang, 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice, Sci. Transl. Med., 2020, 12, eabc5332 CrossRef CAS PubMed.
- W. Dai, B. Zhang, X. M. Jiang, H. Su, J. Li, Y. Zhao, X. Xie, Z. Jin, J. Peng, F. Liu, C. Li, Y. Li, F. Bai, H. Wang, X. Cheng, X. Cen, S. Hu, X. Yang, J. Wang, X. Liu, G. Xiao, H. Jiang, Z. Rao, L. K. Zhang, Y. Xu, H. Yang and H. Liu, Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease, Science, 2020, 368, 1331 CrossRef CAS PubMed.
- H. C. Hung, Y. Y. Ke, S. Y. Huang, P. N. Huang, Y. A. Kung, T. Y. Chang, K. J. Yen, T. T. Peng, S. E. Chang, C. T. Huang, Y. R. Tsai, S. H. Wu, S. J. Lee, J. H. Lin, B. S. Liu, W. C. Sung, S. R. Shih, C. T. Chen and J. T. Hsu, Discovery of M Protease Inhibitors Encoded by SARS-CoV-2, Antimicrob. Agents Chemother., 2020, 64, e00872 CrossRef CAS PubMed.
- Y. C. Wang, W. H. Yang, C. S. Yang, M. H. Hou, C. L. Tsai, Y. Z. Chou, M. C. Hung and Y. Chen, Structural basis of SARS-CoV-2 main protease inhibition by a broad-spectrum anti-coronaviral drug, Am. J. Cancer Res., 2020, 10, 2535 CAS.
- W. Vuong, M. B. Khan, C. Fischer, E. Arutyunova, T. Lamer, J. Shields, H. A. Saffran, R. T. McKay, M. J. van Belkum, M. A. Joyce, H. S. Young, D. L. Tyrrell, J. C. Vederas and M. J. Lemieux, Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication, Nat. Commun., 2020, 27, 4282 CrossRef PubMed.
- S. Vankadara, Y. X. Wong, B. Liu, Y. Y. See, L. H. Tan, Q. W. Tan, G. Wang, R. Karuna, X. Guo, S. T. Tan, J. Y. Fong, J. Joy and C. S. B. Chia, A head-to-head comparison of the inhibitory activities of 15 peptidomimetic SARS-CoV-2 3CLpro inhibitors, Bioorg. Med. Chem. Lett., 2021, 48, 128263 CrossRef CAS PubMed.
- S. Iketani, F. Forouhar, H. Liu, S. J. Hong, F. Y. Lin, M. S. Nair, A. Zask, Y. Huang, L. Xing, B. R. Stockwell, A. Chavez and D. D. Ho, Lead compounds for the development of SARS-CoV-2 3CL protease inhibitors, Nat. Commun., 2021, 12, 2016 CrossRef CAS PubMed.
- C. S. Dampalla, A. D. Rathnayake, K. D. Perera, A. M. Jesri, H. N. Nguyen, M. J. Miller, H. A. Thurman, J. Zheng, M. M. Kashipathy, K. P. Battaile, S. Lovell, S. Perlman, Y. Kim, W. C. Groutas and K. O. Chang, Structure-Guided Design of Potent Inhibitors of SARS-CoV-2 3CL Protease: Structural, Biochemical, and Cell-Based Studies, J. Med. Chem., 2021, 64, 17846 CrossRef CAS PubMed.
- S. Chamakuri, S. Lu, M. N. Ucisik, K. M. Bohren, Y. C. Chen, H. C. Du, J. C. Faver, R. Jimmidi, F. Li, J. Y. Li, P. Nyshadham, S. S. Palmer, J. Pollet, X. Qin, S. E. Ronca, B. Sankaran, K. L. Sharma, Z. Tan, L. Versteeg, Z. Yu, M. M. Matzuk, T. Palzkill and D. W. Young, DNA-encoded chemistry technology yields expedient access to SARS-CoV-2 Mpro inhibitors, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2111172118 CrossRef CAS PubMed.
- J. Qiao, Y. S. Li, R. Zeng, F. L. Liu, R. H. Luo, C. Huang, Y. F. Wang, J. Zhang, B. Quan, C. Shen, X. Mao, X. Liu, W. Sun, W. Yang, X. Ni, K. Wang, L. Xu, Z. L. Duan, Q. C. Zou, H. L. Zhang, W. Qu, Y. H. Long, M. H. Li, R. C. Yang, X. Liu, J. You, Y. Zhou, R. Yao, W. P. Li, J. M. Liu, P. Chen, Y. Liu, G. F. Lin, X. Yang, J. Zou, L. Li, Y. Hu, G. W. Lu, W. M. Li, Y. Q. Wei, Y. T. Zheng, J. Lei and S. Yang, SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model, Science, 2021, 371, 1374 CrossRef CAS PubMed.
- C. S. Dampalla, J. Zheng, K. D. Perera, L. R. Wong, D. K. Meyerholz, H. N. Nguyen, M. M. Kashipathy, K. P. Battaile, S. Lovell, Y. Kim, S. Perlman, W. C. Groutas and K. O. Chang, Postinfection treatment with a protease inhibitor increases survival of mice with a fatal SARS-CoV-2 infection, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2101555118 CrossRef CAS PubMed.
- C. S. Dampalla, Y. Kim, N. Bickmeier, A. D. Rathnayake, H. N. Nguyen, J. Zheng, M. M. Kashipathy, M. A. Baird, K. P. Battaile, S. Lovell, S. Perlman, K. O. Chang and W. C. Groutas, Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease, J. Med. Chem., 2021, 64, 10047 CrossRef CAS PubMed.
- Z. Xia, M. Sacco, Y. Hu, C. Ma, X. Meng, F. Zhang, T. Szeto, Y. Xiang, Y. Chen and J. Wang, Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir, ACS Pharmacol. Transl. Sci., 2021, 4, 1048 Search PubMed.
- C. S. Dampalla, A. D. Rathnayake, A. C. Galasiti Kankanamalage, Y. Kim, K. D. Perera, H. N. Nguyen, M. J. Miller, T. K. Madden, H. R. Picard, H. A. Thurman, M. M. Kashipathy, L. Liu, K. P. Battaile, S. Lovell, K. O. Chang and W. C. Groutas, Structure-Guided Design of Potent Spirocyclic Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3C-like Protease, J. Med. Chem., 2022, 65, 7818 CrossRef CAS PubMed.
- H. Wang, R. Pei, X. Li, W. Deng, S. Xing, Y. Zhang, C. Zhang, S. He, H. Sun, S. Xiao, J. Xiong, Y. Zhang, X. Chen, Y. Wang, Y. Guo, B. Zhang and L. Shang, The structure-based design of peptidomimetic inhibitors against SARS-CoV-2 3C like protease as Potent anti-viral drug candidate, Eur. J. Med. Chem., 2022, 238, 114458 CrossRef CAS PubMed.
- P. S. Dragovich, R. Zhou, S. E. Webber, T. J. Prins, A. K. Kwok, K. Okano, S. A. Fuhrman, L. S. Zalman, F. C. Maldonado, E. L. Brown, J. W. R. Meador, A. K. Patick, C. E. Ford, M. A. Brothers, S. L. Binford, D. A. Matthews, R. A. Ferre and S. T. Worland, Structure-based design of ketone-containing, tripeptidyl human rhinovirus 3C protease inhibitors, Bioorg. Med. Chem. Lett., 2000, 10, 45 CrossRef CAS PubMed.
- M. A. T. van de Plassche, M. Barniol-Xicota and S. H. L. Verhelst, Peptidyl Acyloxymethyl Ketones as Activity-Based Probes for the Main Protease of SARS-CoV-2, ChemBioChem, 2020, 21, 3383 CrossRef CAS PubMed.
- R. L. Hoffman, R. S. Kania, M. A. Brothers, J. F. Davies, R. A. Ferre, K. S. Gajiwala, M. He, R. J. Hogan, K. Kozminski, L. Y. Li, J. W. Lockner, J. Lou, M. T. Marra, L. J. J. Mitchell, B. W. Murray, J. A. Nieman, S. Noell, S. P. Planken, T. Rowe, K. Ryan, G. J. R. Smith, J. E. Solowiej, C. M. Steppan and B. Taggart, Discovery of Ketone-Based Covalent Inhibitors of Coronavirus 3CL Proteases for the Potential Therapeutic Treatment of COVID-19, J. Med. Chem., 2020, 63, 12725 CrossRef CAS PubMed.
- S. I. Hattori, N. Higashi-Kuwata, H. Hayashi, S. R. Allu, J. Raghavaiah, H. Bulut, D. Das, B. J. Anson, E. K. Lendy, Y. Takamatsu, N. Takamune, N. Kishimoto, K. Murayama, K. Hasegawa, M. Li, D. A. Davis, E. N. Kodama, R. Yarchoan, A. Wlodawer, S. Misumi, A. D. Mesecar, A. K. Ghosh and H. Mitsuya, A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication, Nat. Commun., 2021, 12, 668 CrossRef CAS PubMed.
- B. Boras, R. M. Jones, B. J. Anson, D. Arenson, L. Aschenbrenner, M. A. Bakowski, N. Beutler, J. Binder, E. Chen, H. Eng, H. Hammond, J. Hammond, R. E. Haupt, R. Hoffman, E. P. Kadar, R. Kania, E. Kimoto, M. G. Kirkpatrick, L. Lanyon, E. K. Lendy, J. R. Lillis, J. Logue, S. A. Luthra, C. Ma, S. W. Mason, M. E. McGrath, S. Noell, R. S. Obach, M. N. O'Brien, R. O'Connor, K. Ogilvie, D. Owen, M. Pettersson, M. R. Reese, T. F. Rogers, R. Rosales, M. I. Rossulek, J. G. Sathish, N. Shirai, C. Steppan, M. Ticehurst, L. W. Updyke, S. Weston, Y. Zhu, K. M. White, A. García-Sastre, J. Wang, A. K. Chatterjee, A. D. Mesecar, M. B. Frieman, A. S. Anderson, C. Allerton and C. Allerton, Preclinical characterization of an intravenous coronavirus 3CL protease inhibitor for the potential treatment of COVID19, Nat. Commun., 2021, 12, 6055 CrossRef CAS PubMed.
- M. A. Redhead, C. D. Owen, L. Brewitz, A. H. Collette, P. Lukacik, C. Strain-Damerell, S. W. Robinson, P. M. Collins, P. Schäfer, M. Swindells, C. J. Radoux, I. N. Hopkins, D. Fearon, A. Douangamath, F. von Delft, T. R. Malla, L. Vangeel, T. Vercruysse, J. Thibaut, P. Leyssen, T. T. Nguyen, M. Hull, A. Tumber, D. J. Hallett, C. J. Schofield, D. I. Stuart, A. L. Hopkins and M. A. Walsh, Bispecific repurposed medicines targeting the viral and immunological arms of COVID-19, Sci. Rep., 2021, 11, 13208 CrossRef CAS PubMed.
- B. Bai, A. Belovodskiy, M. Hena, A. S. Kandadai, M. A. Joyce, H. A. Saffran, J. Shields, M. Khan, E. Arutyunova, J. Lu, S. K. Bajwa, D. Hockman, C. Fischer, T. Lamer, W. Vuong, M. J. van Belkum, Z. Gu, F. Lin, Y. Du, J. Xu, M. Rahim, H. S. Young, J. Vederas, D. L. Tyrrell, M. J. Lemieux and J. A. Nieman, Peptidomimetic α-Acyloxymethylketone Warheads with Six-Membered Lactam P1 Glutamine Mimic: SARS-CoV-2 3CL Protease Inhibition, Coronavirus Antiviral Activity, and in Vitro Biological Stability, J. Med. Chem., 2022, 65, 2905 CrossRef CAS PubMed.
- S. Konno, K. Kobayashi, M. Senda, Y. Funai, Y. Seki, I. Tamai, L. Schäkel, K. Sakata, T. Pillaiyar, A. Taguchi, A. Taniguchi, M. Gütschow, C. E. Müller, K. Takeuchi, M. Hirohama, A. Kawaguchi, M. Kojima, T. Senda, Y. Shirasaka, W. Kamitani and Y. Hayashi, 3CL Protease Inhibitors with an Electrophilic Arylketone Moiety as Anti-SARS-CoV-2 Agents, J. Med. Chem., 2022, 65, 2926 CrossRef CAS PubMed.
- H. Liu, S. Iketani, A. Zask, N. Khanizeman, E. Bednarova, F. Forouhar, B. Fowler, S. J. Hong, H. Mohri, M. S. Nair, Y. Huang, N. E. S. Tay, S. Lee, C. Karan, S. J. Resnick, C. Quinn, W. Li, H. Shion, X. Xia, J. D. Daniels, M. Bartolo-Cruz, M. Farina, P. Rajbhandari, C. Jurtschenko, M. A. Lauber, T. McDonald, M. E. Stokes, B. L. Hurst, T. Rovis, A. Chavez, D. D. Ho and B. R. Stockwell, Development of optimized drug-like small molecule inhibitors of the SARS-CoV-2 3CL protease for treatment of COVID-19, Nat. Commun., 2022, 13, 1891 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors, Science, 2020, 368, 409 CrossRef CAS PubMed.
- L. Zhang, D. Lin, Y. Kusov, Y. Nian, Q. Ma, J. Wang, A. von Brunn, P. Leyssen, K. Lanko, J. Neyts, A. de Wilde, E. J. Snijder, H. Liu and R. Hilgenfeld, α-Ketoamides as Broad-Spectrum Inhibitors of Coronavirus and Enterovirus Replication: Structure-Based Design, Synthesis, and Activity Assessment, J. Med. Chem., 2020, 63, 4562 CrossRef CAS PubMed.
- M. S. Cooper, L. Zhang, M. Ibrahim, K. Zhang, X. Sun, J. Röske, M. Göhl, M. Brönstrup, J. K. Cowell, L. Sauerhering, S. Becker, L. Vangeel, D. Jochmans, J. Neyts, K. Rox, G. P. Marsh, H. J. Maple and R. Hilgenfeld, Diastereomeric Resolution Yields Highly Potent Inhibitor of SARS-CoV-2 Main Protease, J. Med. Chem., 2022, 65, 13328 CrossRef CAS PubMed.
- P. S. Dragovich, S. E. Webber, R. E. Babine, S. A. Fuhrman, A. K. Patick, D. A. Matthews, C. A. Lee, S. H. Reich, T. J. Prins, J. T. Marakovits, E. S. Littlefield, R. Zhou, J. Tikhe, C. E. Ford, M. B. Wallace, J. W. R. Meador, R. A. Ferre, E. L. Brown, S. L. Binford, J. E. Harr, D. M. DeLisle and S. T. Worland, Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure-activity studies, J. Med. Chem., 1998, 41, 2806 CrossRef CAS PubMed.
- P. Li, B. Yang, F. Hao, P. Wang, H. He, L. Huang, X. Zhang, S. Zhang, X. Peng, K. Yin, J. Hu, X. Chen, Z. Gu, L. Wang, L. Shen, G. Hu, N. Li, J. Li, S. Chen, W. Xiao, Z. Wang, Q. Guo, X. Chang, L. Zhang, Q. Cai and T. Lin, Design, synthesis, and biological evaluation of anti-EV71 agents, Bioorg. Med. Chem. Lett., 2016, 26, 3346 CrossRef CAS PubMed.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, J. Yu, L. Wang, K. Yang, F. Liu, R. Jiang, X. Yang, T. You, X. Liu, X. Yang, F. Bai, H. Liu, X. Liu, L. W. Guddat, W. Xu, G. Xiao, C. Qin, Z. Shi, H. Jiang, Z. Rao and H. Yang, Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, 2020, 582, 289 CrossRef CAS PubMed.
- G. J. Lockbaum, M. Henes, J. M. Lee, J. Timm, E. A. Nalivaika, P. R. Thompson, N. Kurt Yilmaz and C. A. Schiffer, Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode, Biochemistry, 2021, 60, 2925 CrossRef CAS PubMed.
- Y. Ma, C. Shang, P. Yang, L. Li, Y. Zhai, Z. Yin, B. Wang and L. Shang, 4-Iminooxazolidin-2-one as a Bioisostere of the Cyanohydrin Moiety: Inhibitors of Enterovirus 71 3C Protease, J. Med. Chem., 2018, 61, 10333 CrossRef CAS PubMed.
- W. Zhu, M. Xu, C. Z. Chen, H. Guo, M. Shen, X. Hu, P. Shinn, C. Klumpp-Thomas, S. G. Michael and W. Zheng, Identification of SARS-CoV-2 3CL Protease Inhibitors by a Quantitative High-Throughput Screening, ACS Pharmacol. Transl. Sci., 2020, 3, 1008 CrossRef CAS PubMed.
- C. Ma, Z. Xia, M. D. Sacco, Y. Hu, J. A. Townsend, X. Meng, J. Choza, H. Tan, J. Jang, M. V. Gongora, X. Zhang, F. Zhang, Y. Xiang, M. T. Marty, Y. Chen and J. Wang, Discovery of Di- and Trihaloacetamides as Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity, J. Am. Chem. Soc., 2021, 143, 20697 CrossRef CAS PubMed.
- J. C. Milligan, T. U. Zeisner, G. Papageorgiou, D. Joshi, C. Soudy, R. Ulferts, M. Wu, C. T. Lim, K. W. Tan, F. Weissmann, B. Canal, R. Fulisawa, T. Deegan, H. Nagara, G. Bineva-Todd, C. Basier, J. F. Curran, M. H. Howell, R. Beale, K. Labib, N. O'Reilly and J. F. X. Diffley, Identifying SARS-CoV-2 Antiviral Compounds by Screening for Small Molecule Inhibitors of nsp5 Main Protease, Biochem. J., 2021, 478, 2499 CrossRef CAS PubMed.
- Y. Hirose, N. Shindo, M. Mori, S. Onitsuka, H. Isogai, R. Hamada, T. Hiramoto, J. Ochi, D. Takahashi, T. Ueda, J. M. M. Caaveiro, Y. Yoshida, S. Ohdo, N. Matsunaga, S. Toba, M. Sasaki, Y. Orba, H. Sawa, A. Sato, E. Kawanishi and A. Ojida, Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease, J. Med. Chem., 2022, 65, 13852 CrossRef CAS PubMed.
- J. Chen, C. Liang, K. Miao, Y. Wu, H. Yun and W. Zhang, Aminocarbamoyl compounds for the treatment of viral infections, WO2022043374, 2022.
- T. R. Malla, A. Tumber, T. John, L. Brewitz, C. Strain-Damerell, C. D. Owen, P. Lukacik, H. T. H. Chan, P. Maheswaran, E. Salah, F. Duarte, H. Yang, Z. Rao, M. A. Walsh and C. J. Schofield, Mass spectrometry reveals potential of β-lactams as SARS-CoV-2 Mpro inhibitors, Chem. Commun., 2021, 57, 1430 RSC.
- T. R. Malla, L. Brewitz, D. G. Muntean, H. Aslam, C. D. Owen, E. Salah, A. Tumber, P. Lukacik, C. Strain-Damerell, H. Mikolajek, M. A. Walsh and C. J. Schofield, Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine, J. Med. Chem., 2022, 65, 7682 CrossRef CAS PubMed.
- E. J. Niesor, G. Boivin, E. Rhéaume, R. Shi, V. Lavoie, N. Goyette, M. E. Picard, A. Perez, F. Laghrissi-Thode and J. C. Tardif, Inhibition of the 3CL Protease and SARS-CoV-2 Replication by Dalcetrapib, ACS Omega, 2021, 6, 16584 CrossRef CAS PubMed.
- P. Ren, H. Li, T. Nie, X. Jian, C. Yu, J. Li, H. Su, X. Zhang, S. Li, X. Yang, C. Peng, Y. Yin, L. Zhang, Y. Xu, H. Liu and F. Bai, Discovery and Mechanism Study of SARS-CoV-2 3C-like Protease Inhibitors with a New Reactive Group, J. Med. Chem., 2023, 66, 12266 CrossRef CAS PubMed.
- K. Vandyck, P. J. M. B. Raboisson, L. Beigelman, V. Serebryany, A. D. Stoycheva, D. A. M.-E. Bardiot, S. Boland and A. D. M. Marchand, Anti-viral compounds for treating coronavirus, picornavirus, and norovirus infections, WO2021252491, 2021.
- B. Bai, E. Arutyunova, M. Khan, J. Lu, M. A. Joyce, H. A. Saffran, J. A. Shields, A. S. Kandadai, A. Belovodskiy, M. Hena, W. Vuong, T. Lamer, H. S. Young, J. C. Vederas, D. L. Tyrrell, M. J. Lemieux and J. A. Nieman, Peptidomimetic nitrile warheads as SARS-CoV-2 3CL protease inhibitors, RSC Med. Chem., 2021, 12, 1722 RSC.
- D. R. Owen, C. M. N. Allerton, A. S. Anderson, L. Aschenbrenner, M. Avery, S. Berritt, B. Boras, R. D. Cardin, A. Carlo, K. J. Coffman, A. Dantonio, L. Di, H. Eng, R. Ferre, K. S. Gajiwala, S. A. Gibson, S. E. Greasley, B. L. Hurst, E. P. Kadar, A. S. Kalgutkar, J. C. Lee, J. Lee, W. Liu, S. W. Mason, S. Noell, J. J. Novak, R. S. Obach, K. Ogilvie, N. C. Patel, M. Pettersson, D. K. Rai, M. R. Reese, M. F. Sammons, J. G. Sathish, R. S. P. Singh, C. M. Steppan, A. E. Stewart, J. B. Tuttle, L. Updyke, P. R. Verhoest, L. Wei, Q. Yang and Y. Zhu, An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19, Science, 2021, 374, 1586 CrossRef CAS PubMed.
- L. Brewitz, L. Dumjahn, Y. Zhao, C. D. Owen, S. M. Laidlaw, T. R. Malla, D. Nguyen, P. Lukacik, E. Salah, A. D. Crawshaw, A. J. Warren, J. Trincao, C. Strain-Damerell, M. W. Carroll, M. A. Walsh and C. J. Schofield, Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine, J. Med. Chem., 2023, 66, 2663 CrossRef CAS PubMed.
- C. Ngo, W. Fried, S. Aliyari, J. Feng, C. Qin, S. Zhang, H. Yang, J. Shanaa, P. Feng, G. Cheng, X. S. Chen and C. Zhang, Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease, J. Med. Chem., 2023, 66, 12237 CrossRef CAS PubMed.
- E. Freire, R. Ottenbrite, Y. Xiao, A. Velazquez-Campoy, S. Leavitt, U. Bacha and J. Barrila, Inhibitors of coronavirus protease and methods of use thereof, WO2005041904, 2005.
- R. Smoum, A. Rubinstein, V. M. Dembitsky and M. Srebnik, Boron containing compounds as protease inhibitors, Chem. Rev., 2012, 112, 4156 CrossRef CAS PubMed.
- S. Song, P. Gao, L. Sun, D. Kang, J. Kongsted, V. Poongavanam, P. Zhan and X. Liu, Recent developments in the medicinal chemistry of single boron atom-containing compounds, Acta Pharm. Sin. B, 2021, 11, 3035 CrossRef CAS PubMed.
- A. K. Patick, S. L. Binford, M. A. Brothers, R. L. Jackson, C. E. Ford, M. D. Diem, F. Maldonado, P. S. Dragovich, R. Zhou, T. J. Prins, S. A. Fuhrman, J. W. Meador, L. Zalman, D. A. Matthews and S. T. Worland, In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease, Antimicrob. Agents Chemother., 1999, 43, 2444 CrossRef CAS PubMed.
- P. S. Dragovich, T. J. Prins, R. Zhou, S. E. Webber, J. T. Marakovits, S. A. Fuhrman, A. K. Patick, D. A. Matthews, C. A. Lee, C. E. Ford, B. J. Burke, P. A. Rejto, T. F. Hendrickson, T. Tuntland, E. L. Brown, J. W. R. Meador, R. A. Ferre, J. E. Harr, M. B. Kosa and S. T. Worland, Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 4. Incorporation of P1 lactam moieties as L-glutamine replacements, J. Med. Chem., 1999, 42, 1213 CrossRef CAS PubMed.
- K. Anand, J. Ziebuhr, P. Wadhwani, J. R. Mesters and R. Hilgenfeld, Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs, Science, 2003, 300, 1763 CrossRef CAS PubMed.
- P. H. Hsyu, Y. K. Pithavala, M. Gersten, C. A. Penning and B. M. Kerr, Pharmacokinetics and safety of an antirhinoviral agent, ruprintrivir, in healthy volunteers, Antimicrob. Agents Chemother., 2002, 46, 392 CrossRef CAS PubMed.
- F. G. Hayden, R. B. Turner, J. M. Gwaltney, K. Chi-Burris, M. Gersten, P. Hsyu, A. K. Patick, G. J. R. Smith and L. S. Zalman, Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers, Antimicrob. Agents Chemother., 2003, 47, 3907 CrossRef CAS PubMed.
- A. K. Patick, Rhinovirus chemotherapy, Antiviral Res., 2006, 71, 391 CrossRef CAS PubMed.
- J. J. Shie, J. M. Fang, T. H. Kuo, C. J. Kuo, P. H. Liang, H. J. Huang, Y. T. Wu, J. T. Jan, Y. S. Cheng and C. H. Wong, Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic alpha,beta-unsaturated esters, Bioorg. Med. Chem., 2005, 13, 5240 CrossRef CAS PubMed.
- S. Tomar, M. L. Johnston, S. E. St John, H. L. Osswald, P. R. Nyalapatla, L. N. Paul, A. K. Ghosh, M. R. Denison and A. D. Mesecar, Ligand-induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus nsp5 Protease (3CLpro), J. Biol. Chem., 2015, 290, 19403 CrossRef CAS PubMed.
- H. Yang, W. Xie, X. Xue, K. Yang, J. Ma, W. Liang, Q. Zhao, Z. Zhou, D. Pei, J. Ziebuhr, R. Hilgenfeld, K. Y. Yuen, L. Wong, G. Gao, S. Chen, Z. Chen, D. Ma, M. Bartlam and Z. Rao, Design of wide-spectrum inhibitors targeting coronavirus main proteases, PLoS Biol., 2005, 3, e324 CrossRef PubMed.
- K. Akaji, H. Konno, H. Mitsui, K. Teruya, Y. Shimamoto, Y. Hattori, T. Ozaki, M. Kusunoki and A. Sanjoh, Structure-Based Design, Synthesis, and Evaluation of Peptide-Mimetic SARS 3CL Protease Inhibitors, J. Med. Chem., 2011, 54, 7962 CrossRef CAS PubMed.
- S. Di Micco, R. Rahimova, M. Sala, M. C. Scala, G. Vivenzio, S. Musella, G. Andrei, K. Remans, L. Mammri, R. Snoeck, G. Bifulco, F. Di Matteo, V. Vestuto, P. Campiglia, J. A. Márquez and A. Fasano, Rational design of the zonulin inhibitor AT1001 derivatives as potential anti SARS-CoV-2, Eur. J. Med. Chem., 2022, 244, 114857 CrossRef CAS PubMed.
- S. E. Webber, K. Okano, T. L. Little, S. H. Reich, Y. Xin, S. A. Fuhrman, D. A. Matthews, R. A. Love, T. F. Hendrickson, A. K. Patick, J. W. Meador, R. A. Ferre, E. L. Brown, C. E. Ford, S. L. Binford and S. T. Worland, Tripeptide aldehyde inhibitors of human rhinovirus 3C protease: design, synthesis, biological evaluation, and cocrystal structure solution of P1 glutamine isosteric replacements, J. Med. Chem., 1998, 41, 2786 CrossRef CAS PubMed.
- H.-Z. Zhang, H. Zhang, W. Kemnitzer, B. Tseng, J. Cinatl, M. Michaelis, H. W. Doerr and S. X. Cai, Design and Synthesis of Dipeptidyl Glutaminyl Fluoromethyl Ketones as Potent Severe Acute Respiratory Syndrome Coronovirus (SARS-CoV) Inhibitors, J. Med. Chem., 2006, 49, 1198 CrossRef CAS PubMed.
- S. I. Al-Gharabli, S. T. Shah, S. Weik, M. F. Schmidt, J. R. Mesters, D. Kuhn, G. Klebe, R. Hilgenfeld and J. Rademann, An efficient method for the synthesis of peptide aldehyde libraries employed in the discovery of reversible SARS coronavirus main protease (SARS-CoV Mpro) inhibitors, ChemBioChem, 2006, 7, 1048 CrossRef CAS PubMed.
- K. C. Tiew, G. He, S. Aravapalli, S. R. Mandadapu, M. R. Gunnam, K. R. Alliston, G. H. Lushington, Y. Kim, K. O. Chang and W. C. Groutas, Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease, Bioorg. Med. Chem. Lett., 2011, 21, 5315 CrossRef CAS PubMed.
- L. A. Stubbing, J. G. Hubert, J. Bell-Tyrer, Y. O. Hermant, S. H. Yang, A. M. McSweeney, G. M. McKenzie-Goldsmith, V. K. Ward, D. P. Furkert and M. A. Brimble, P1 Glutamine isosteres in the design of inhibitors of 3C/3CL protease of human viruses of the Pisoniviricetes class, RSC Chem. Biol., 2023, 4, 533 RSC.
- T. Regnier, D. Sarma, K. Hidaka, U. Bacha, E. Freire, Y. Hayashi and Y. Kiso, New developments for the design, synthesis and biological evaluation of potent SARS-CoV 3CLpro inhibitors, Bioorg. Med. Chem. Lett., 2009, 19, 2722 CrossRef CAS PubMed.
- S. Konno, P. Thanigaimalai, T. Yamamoto, K. Nakada, R. Kakiuchi, K. Takayama, Y. Yamazaki, F. Yakushiji, K. Akaji, Y. Kiso, Y. Kawasaki, S. Chen, E. Freire and Y. Hayashi, Design and synthesis of new tripeptide-type SARS-CoV 3CL protease inhibitors containing an electrophilic arylketone moiety, Bioorg. Med. Chem., 2013, 21, 412 CrossRef CAS PubMed.
- C. P. Chuck, C. Chen, Z. Ke, D. C. Wan, H. F. Chow and K. B. Wong, Design, synthesis and crystallographic analysis of nitrile-based broad-spectrum peptidomimetic inhibitors for coronavirus 3C-like proteases, Eur. J. Med. Chem., 2013, 59, 1 CrossRef CAS PubMed.
- Y. Kim, S. Lovell, K. C. Tiew, S. R. Mandadapu, K. R. Alliston, K. P. Battaile, W. C. Groutas and K. O. Chang, Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses, J. Virol., 2012, 86, 11754 CrossRef CAS PubMed.
- Y. Kim, H. Liu, A. C. Galasiti Kankanamalage, S. Weerasekara, D. Hua, W. C. Groutas, K. O. Chang and N. C. Pedersen, Reversal of the Progression of Fatal Coronavirus Infection in Cats by a Broad-Spectrum Coronavirus Protease Inhibitor, PLoS Pathog., 2016, 12, e1005531 CrossRef PubMed.
- N. C. Pedersen, Y. Kim, H. Liu, A. C. Galasiti Kankanamalage, C. Eckstrand, W. C. Groutas, M. Bannasch, J. M. Meadows and K. O. Chang, Efficacy of a 3C-like protease inhibitor in treating various forms of acquired feline infectious peritonitis, J. Feline Med. Surg., 2018, 20, 378 CrossRef PubMed.
- C. J. Cáceres, S. Cardenas-Garcia, S. Carnaccini, B. Seibert, D. S. Rajao, J. Wang and D. R. Perez, Efficacy of GC-376 against SARS-CoV-2 virus infection in the K18 hACE2 transgenic mouse model, Sci. Rep., 2021, 11, 9609 CrossRef PubMed.
- S. R. Mandadapu, P. M. Weerawarna, M. R. Gunnam, K. R. Alliston, G. H. Lushington, Y. Kim, K. O. Chang and W. C. Groutas, Potent inhibition of norovirus 3CL protease by peptidyl α-ketoamides and α-ketoheterocycles, Bioorg. Med. Chem. Lett., 2012, 22, 4820 CrossRef CAS PubMed.
- K. S. Yang, X. R. Ma, Y. Ma, Y. R. Alugubelli, D. A. Scott, E. C. Vatansever, A. K. Drelich, B. Sankaran, Z. Z. Geng, L. R. Blankenship, H. E. Ward, Y. J. Sheng, J. C. Hsu, K. C. Kratch, B. Zhao, H. S. Hayatshahi, J. Liu, P. Li, C. A. Fierke, C. K. Tseng, S. Xu and W. R. Liu, A Quick Route to Multiple Highly Potent SARS-CoV-2 Main Protease Inhibitors, ChemMedChem, 2021, 16, 942 CrossRef CAS PubMed.
- C. S. Dampalla, H. N. Nguyen, A. D. Rathnayake, Y. Kim, K. D. Perera, T. K. Madden, H. A. Thurman, A. J. Machen, M. M. Kashipathy, L. Liu, K. P. Battaile, S. Lovell, K. O. Chang and W. C. Groutas, Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies, ACS Pharmacol. Transl. Sci., 2023, 6, 181 CrossRef CAS PubMed.
- C. S. Dampalla, M. J. Miller, Y. Kim, A. Zabiegala, H. N. Nguyen, T. K. Madden, H. A. Thurman, A. J. Machen, A. Cooper, L. Liu, K. P. Battaile, S. Lovell, K. O. Chang and W. C. Groutas, Structure-guided design of direct-acting antivirals that exploit the gem-dimethyl effect and potently inhibit 3CL proteases of severe acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) and middle east respiratory syndrome coronavirus (MERS-CoV), Eur. J. Med. Chem., 2023, 254, 115376 CrossRef CAS PubMed.
- W. Vuong, C. Fischer, M. B. Khan, M. J. van Belkum, T. Lamer, K. D. Willoughby, J. Lu, E. Arutyunova, M. A. Joyce, H. A. Saffran, J. A. Shields, H. S. Young, J. A. Nieman, D. L. Tyrrell, M. J. Lemieux and J. C. Vederas, Improved SARS-CoV-2 Mpro inhibitors based on feline antiviral drug GC376: Structural enhancements, increased solubility, and micellar studies, Eur. J. Med. Chem., 2021, 222, 113584 CrossRef CAS PubMed.
- A. C. Galasiti Kankanamalage, Y. Kim, P. M. Weerawarna, R. A. Z. Uy, V. C. Damalanka, S. R. Mandadapu, K. R. Alliston, N. Mehzabeen, K. P. Battaile, S. Lovell, K.-O. Chang and W. C. Groutas, Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure−Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies, J. Med. Chem., 2015, 58, 3144 CrossRef CAS PubMed.
- A. D. Rathnayake, Y. Kim, C. S. Dampalla, H. N. Nguyen, A.-R. M. Jesri, M. M. Kashipathy, G. H. Lushington, K. P. Battaile, S. Lovell, K.-O. Chang and W. C. Groutas, Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease, J. Med. Chem., 2020, 63, 11945 CrossRef CAS PubMed.
- S. Yang, S. J. Chen, M. F. Hsu, J. D. Wu, C. T. Tseng, Y. F. Liu, H. C. Chen, C. W. Kuo, C. S. Wu, L. W. Chang, W. C. Chen, S. Y. Liao, T. Y. Chang, H. H. Hung, H. L. Shr, C. Y. Liu, Y. A. Huang, L. Y. Chang, J. C. Hsu, C. J. Peters, A. H. Wang and M. C. Hsu, Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor, J. Med. Chem., 2006, 49, 4971 CrossRef CAS PubMed.
- Y. Ma, K. S. Yang, Z. Z. Geng, Y. R. Alugubelli, N. Shaabani, E. C. Vatansever, X. R. Ma, C. C. Cho, K. Khatua, J. Xiao, L. R. Blankenship, G. Yu, B. Sankaran, P. Li, R. Allen, H. Ji, S. Xu and W. R. Liu, A multi-pronged evaluation of aldehyde-based tripeptidyl main protease inhibitors as SARS-CoV-2 antivirals, Eur. J. Med. Chem., 2022, 240, 114570 CrossRef CAS PubMed.
- J. Botyanszki, J. G. Catalano, P. Y. Chong, H. Dickson, Q. Jin, A. Leivers, A. Maynard, X. Liao, J. Miller, J. B. Shotwell, V. W. F. Tai and R. K. Thalji, Compounds that inhibit 3C and 3CL proteases and methods of use therof, WO2018042343, 2018.
- J. Botyanszki, Q. Jin, N. D. Pearson and R. K. Thalji,Tripeptides derivatives for treating sars-cov-2 infections, WO2021191827, 2021.
- I. C. Jacobson, S. S. K. Lee and J. C. Pizarro Novoa, Peptidomimetic N5-methyl-N2-(nonanoyl-L-leucyl)-L-glutaminate derivatives, triazaspiro[4.14]nonadecane derivatives and similar compounds as inhibitors of norovirus and coronavirus replication, WO2021188620, 2021.
- I. C. Jacobson, Inhibitors of norovirus and coronavirus replication, WO2021206877, 2021.
- I. C. Jacobson ,Inhibitors of norovirus and coronavirus replication, WO2021206876, 2021.
- I. C. Jacobson, Inhibitors for coronavirus, WO2023014758, 2023.
- R. L. Hoffman, R. S. Kania, J. A. Nieman, S. P. Planken and G. J. Smith, Anticoronaviral Compounds and Compositions, Their Pharmaceutical Uses And Materials For Their Synthesis, WO2005113580, 2005.
- C. Allais, D. Bernhardson, A. R. Brown, G. M. Chinigo, J.-N. Desrosiers, K. J. DiRico, I. Hotham, B. P. Jones, S. A. Kulkarni, C. A. Lewis, R. Lira, R. P. Loach, P. D. Morse, J. J. Mousseau, M. A. Perry, Z. Peng, D. W. Place, A. M. Rane, L. Samp, R. A. Singer, Z. Wang, G. A. Weisenburger, H. G. Yayla and J. M. Zanghi, Early Clinical Development of Lufotrelvir as a Potential Therapy for COVID-19, Org. Process Res. Dev., 2023 DOI:10.1021/acs.oprd.2c00375.
- N. Cheruvu, E. van Duijn, P. A. Spigt, I. M. Barbu, S. S. Toussi, K. Schildknegt, R. M. Jones and R. S. Obach, The metabolism of lufotrelvir, a prodrug investigated for the treatment of sars-cov-2, in humans following intravenous administration, Drug Metab. Dispos., 2023, 51, 1419 CrossRef CAS PubMed.
- P. Robinson, S. S. Toussi, S. Aggarwal, A. Bergman, T. Zhu, F. Hackman, J. G. Sathish, L. Updyke, P. Loudon, G. Krishna, P. Clevenbergh, M. G. Hernandez-Mora, J. M. Cisneros Herreros, T. E. Albertson, M. Dougan, A. Thacker, M. L. Baniecki, H. Soares, M. Whitlock, G. Nucci, S. Menon, A. S. Anderson and M. Binks, Safety, Tolerability, and Pharmacokinetics of Single and Multiple Ascending Intravenous Infusions of PF-07304814 (Lufotrelvir) in Hospitalized Participants With COVID-19, Open Forum Infect. Dis., 2023, 10, ofad355 CrossRef PubMed.
- W. Shang, W. Dai, C. Yao, L. Xu, X. Tao, H. Su, J. Li, X. Xie, Y. Xu, M. Hu, D. Xie, H. Jiang, L. Zhang and H. Liu, In vitro and in vivo evaluation of the main protease inhibitor FB2001 against SARS-CoV-2, Antiviral Res., 2022, 208, 105450 CrossRef CAS PubMed.
- L. D. Arnold, A. Jennings and W. Keung, Inhibitors of cysteine proteases and methods of use thereof, WO2021252644, 2021.
- Pardes Biosciences, https://ir.pardesbio.com/news, (accessed July, 26, 2023).
- P. Thanigaimalai, S. Konno, T. Yamamoto, Y. Koiwai, A. Taguchi, K. Takayama, F. Yakushiji, K. Akaji, S. E. Chen, A. Naser-Tavakolian, A. Schön, E. Freire and Y. Hayashi, Development of potent dipeptide-type SARS-CoV 3CL protease inhibitors with novel P3 scaffolds: design, synthesis, biological evaluation, and docking studies, Eur. J. Med. Chem., 2013, 68, 372 CrossRef CAS PubMed.
- W. M. Kati, H. L. Sham, J. O. McCall, D. A. Montgomery, G. T. Wang, W. Rosenbrook, L. Miesbauer, A. Buko and D. W. Norbeck, Inhibition of 3C protease from human rhinovirus strain 1B by peptidyl bromomethylketonehydrazides, Arch. Biochem. Biophys., 1999, 362, 363 CrossRef CAS PubMed.
- D. Edmonds, C. Liang, H. Yun, B. Zhang and X. Zheng, Antiviral compounds, WO2023104882, 2023.
- J. D. Baker, R. L. Uhrich, G. C. Kraemer, J. E. Love and B. C. Kraemer, A drug repurposing screen identifies hepatitis C antivirals as inhibitors of the SARS-CoV2 main protease, PLoS One, 2021, 16, e0245962 CrossRef CAS PubMed.
- C. Liu, S. Boland, M. D. Scholle, D. Bardiot, A. Marchand, P. Chaltin, L. M. Blatt, L. Beigelman, J. A. Symons, P. Raboisson, Z. A. Gurard-Levin, K. Vandyck and J. Deval, Dual inhibition of SARS-CoV-2 and human rhinovirus with protease inhibitors in clinical development, Antiviral Res., 2021, 187, 105020 CrossRef CAS PubMed.
- Y. Bai, F. Ye, Y. Feng, H. Liao, H. Song, J. Qi, G. F. Gao, W. Tan, L. Fu and Y. Shi, Structural basis for the inhibition of the SARS-CoV-2 main protease by the anti-HCV drug narlaprevir, Signal Transduction Targeted Ther., 2021, 6, 51 CrossRef CAS PubMed.
- D. W. Kneller, S. Galanie, G. Phillips, H. M. O'Neill, L. Coates and A. Kovalevsky, Malleability of the SARS-CoV-2 3CL Mpro Active-Site Cavity Facilitates Binding of Clinical Antivirals, Structure, 2020, 28, 1313 CrossRef CAS PubMed.
- B. Halford, The path to Paxlovid, C&En, 2022, vol. 100, p. 16 Search PubMed.
- B. Halford, Pfizer unveils its oral SARS-CoV-2 inhibitor, C&En, 2021, vol. 99, p. 7 Search PubMed.
- J. Hammond, H. Leister-Tebbe, A. Gardner, P. Abreu, W. Bao, W. Wisemandle, M. Baniecki, V. M. Hendrick, B. Damle, A. Simón-Campos, R. Pypstra and J. M. Rusnak, Oral Nirmatrelvir for High-Risk, Nonhospitalized Adults with Covid-19, N. Engl. J. Med., 2022, 386, 1397 CrossRef CAS PubMed.
- N. Higashi-Kuwata, K. Tsuji, H. Hayashi, H. Bulut, M. Kiso, M. Imai, H. Ogata-Aoki, T. Ishii, T. Kobayakawa, K. Nakano, N. Takamune, N. Kishimoto, S. Hattori, D. Das, Y. Uemura, Y. Shimizu, M. Aoki, K. Hasegawa, S. Suzuki, A. Nishiyama, J. Saruwatari, Y. Shimizu, Y. Sukenaga, Y. Takamatsu, K. Tsuchiya, K. Maeda, K. Yoshimura, S. Iida, S. Ozono, T. Suzuki, T. Okamura, S. Misumi, Y. Kawaoka, H. Tamamura and H. Mitsuya, Identification of SARS-CoV-2 Mpro inhibitors containing P1' 4-fluorobenzothiazole moiety highly active against SARS-CoV-2, Nat. Commun., 2023, 14, 1076 CrossRef CAS PubMed.
- K. Tsuji, T. Ishii, T. Kobayakawa, N. Higashi-Kuwata, K. Shinohara, C. Azuma, Y. Miura, H. Nakano, N. Wada, S. Hattori, H. Bulut, H. Mitsuya and H. Tamamura, Structure-Activity Relationship Studies of SARS-CoV-2 Main Protease Inhibitors Containing 4-Fluorobenzothiazole-2-carbonyl Moieties, J. Med. Chem., 2023, 66, 13516 CrossRef CAS PubMed.
- B. Yang, B. Liang, Y. Lai and J. J. Wu, Inhibitors of cysteine proteases and method of use thereof, WO2023139402, 2023.
- M. Göhl, L. Zhang, H. El Kilani, X. Sun, K. Zhang, M. Brönstrup and R. Hilgenfeld, From Repurposing to Redesign: Optimization of Boceprevir to Highly Potent Inhibitors of the SARS-CoV-2 Main Protease, Molecules, 2022, 27, 4292 CrossRef PubMed.
- M. Z. Lin and M. W. Soerensen, Cell-permeant inhibitors of viral cysteine protease, WO2023114516, 2023.
- M. Westberg, Y. Su, X. Zou, P. Huang, A. Rustagi, J. Garhyan, P. B. Patel, D. Fernandez, Y. Wu, L. Ning, A. Beck, M. Karim, C. Hao, P. Saenkham-Huntsinger, V. Tat, A. Drelich, B. H. Peng, S. Einav, C. T. K. Tseng, C. Blish and M. Z. Lin, Design of SARS-CoV-2 protease inhibitors with improved affinity and reduced sensitivity to mutations, BioRxiv, 2023, preprint, DOI:10.1101/2023.07.19.549739.
- X. Chen, P. Li, J. Huang, Y. Yang, H. Zhang, Z. Wang, Z. Zhu, J. Wang, J. Zhang, K. Chen, H. He, C. Long and S. Chen, Discovery of novel bicyclic[3.3.0]proline peptidyl α-ketoamides as potent 3CL-protease inhibitors for SARS-CoV-2, Bioorg. Med. Chem. Lett., 2023, 90, 129324 CrossRef CAS PubMed.
- X. Chen, X. Huang, Q. Ma, P. Kuzmic, B. Zhou, J. Xu, B. Liu, H. Jiang, W. Zhang, C. Yang, S. Wu, J. Huang, H. Li, C. Long, X. Zhao, H. Xu, Y. J. Sheng, Y. Guo, C. Niu, L. Xue, Y. Xu, J. Liu, T. Zhang, J. Spencer, W. Deng, S.-H. Chen, X. Xiong, Z. Yang and N. Zhong, Inhibition mechanism and antiviral activity of an α-ketoamide based SARS-CoV-2 main protease inhibitor, BioRxiv, 2023, preprint, DOI:10.1101/2023.03.09.53186.
- X. Chen, J. Wang, J. Huang, Z. Liu, C. Long, S. Chen and K. X. Chen, Ketoamide derivatives and application thereof, WO2023036093, 2023.
- B. Wang, H. J. Li, M. M. Cai, Z. X. Lin, X. F. Ou, S. H. Wu, R. H. Cai, Y. N. Wei, F. Yang, Y. M. Zhu, Z. F. Yang, N. S. Zhong and L. Lin, Antiviral efficacy of RAY1216 monotherapy and combination therapy with ritonavir in patients with COVID-19: a phase 2, single centre, randomised, double-blind, placebo-controlled trial, EClinicalMedicine, 2023, 63, 102189 CrossRef PubMed.
- I. Stefanelli, A. Corona, C. Cerchia, E. Cassese, S. Improta, E. Costanzi, S. Pelliccia, S. Morasso, F. Esposito, A. Paulis, S. Scognamiglio, F. Di Leva, P. Storici, M. Brindisi, E. Tramontano, R. Cannalire and V. Summa, Broad-spectrum coronavirus 3C-like protease peptidomimetic inhibitors effectively block SARS-CoV-2 replication in cells: Design, synthesis, biological evaluation, and X-ray structure determination, Eur. J. Med. Chem., 2023, 253, 115311 CrossRef CAS PubMed.
- F. Wang, W. Xiao, Y. Tang, M. Cao, D. Shu, T. Asakawa, Y. Xu, X. Jiang, L. Zhang, W. Wang, J. Tang, Y. Huang, Y. Yang, Y. Yang, R. Tang, J. Shen and H. Lu, Efficacy and safety of SIM0417 (SSD8432) plus ritonavir for COVID-19 treatment: a randomised, double-blind, placebo-controlled, phase 1b trial, Lancet Reg. Health West. Pac., 2023, 38, 100835 CrossRef PubMed.
- X. Jiang, Y. Xu, L. Zhang, H. Su, Q. Zhang, W. Zhao, W. Shang, J. Shen, G. Xiao and H. Jiang, Cyano compound, and preparation method therefor and use thereof, WO2023051657, 2023.
- K. W. Zhu, Deuremidevir and Simnotrelvir−Ritonavir for the Treatment of COVID-19, ACS Pharmacol. Transl. Sci., 2023, 6, 1306 CrossRef CAS PubMed.
- X. Jiang, H. Su, W. Shang, F. Zhou, W. Zhang, W. Zhao, Q. Zhang, H. Xie, L. Jiang, T. Nie, F. Yang, M. Xiong, X. Huang, M. Li, P. Chen, S. Peng, G. Xiao, H. Jiang, R. Tang, L. Zhang, J. Shen and Y. Xu, Structure-based development and preclinical evaluation of the SARS-CoV-2 3C-like protease inhibitor simnotrelvir, Nat. Commun., 2023, 14, 6463 CrossRef CAS PubMed.
- X. M. Yang, Y. Yang, B. F. Yao, P. P. Ye, Y. Xu, S. P. Peng, Y. M. Yang, P. Shu, P. J. Li, S. Li, H. L. Hu, Q. Li, L. L. Song, K. G. Chen, H. Y. Zhou, Y. H. Zhang, F. R. Zhao, B. H. Tang, W. Zhang, X. F. Zhang, S. M. Fu, G. X. Hao, Y. Zheng, J. S. Shen, Y. C. Xu, X. R. Jiang, L. K. Zhang, R. H. Tang and W. Zhao, A First-In-Human Phase 1 Study of Simnotrelvir, a 3CL-like Protease Inhibitor for Treatment of COVID-19, in Healthy Adult Subjects, Eur. J. Pharm. Sci., 2023, 30, 106598 CrossRef PubMed.
- D. A. M.-E. Bardiot, K. Vandyck, S. Boland, A. D. Stoycheva, A. D. M. Marchand, P. J. M. B. Raboisson and L. Beigelman, Anti-viral compounds for treating coronavirus, picornavirus, and norovirus infections, WO2023043816, 2023.
- K. Vandyck, D. A. M.-E. Bardiot, P. J. M. B. Raboisson, L. Beigelman, A. D. Stoycheva, S. Boland and A. D. M. Marchand, Anti-viral compounds, WO2023283256, 2023.
- H. Yang, M. You, X. Shu, J. Zhen, M. Zhu, T. Fu, Y. Zhang, X. Jiang, L. Zhang, Y. Xu, Y. Zhang, H. Su, Q. Zhang and J. Shen, Design, synthesis and biological evaluation of peptidomimetic benzothiazolyl ketones as 3CLpro inhibitors against SARS-CoV-2, Eur. J. Med. Chem., 2023, 257, 115512 CrossRef CAS PubMed.
- Y. R. Alugubelli, Z. Z. Geng, K. Yang, N. Shaabani, K. Khatua, X. R. Ma, E. C. Vatansever, C. C. Cho, Y. Ma, J. Xiao, L. R. Blankenship, G. Yu, B. Sankaran, P. Li, R. Allen, H. Ji, S. Xu and W. R. Liu, A systematic exploration of boceprevir-based main protease inhibitors as SARS-CoV-2 antivirals, Eur. J. Med. Chem., 2022, 240, 114596 CrossRef CAS PubMed.
- Z. Z. Geng, S. Atla, N. Shaabani, V. R. Vulupala, K. S. Yang, Y. R. Alugubelli, K. Khatua, P. C. Chen, J. Xiao, L. R. Blankenship, X. R. Ma, E. C. Vatansever, C. C. Cho, Y. Ma, R. Allen, H. Ji, S. Xu and W. R. Liu, A Systematic Survey of Reversibly Covalent Dipeptidyl Inhibitors of the SARS-CoV-2 Main Protease, J. Med. Chem., 2023, 66, 11040 CrossRef CAS PubMed.
- A. K. Chatterjee, J. J. Chen, E. Nakath, A. Rahimi, A. K. Gupta, G. Grabovyi, K. Wilson, S. Ghorai, A. Nazarian, J. Pedroarena, W. Mazumdar, F. Weiss, L. Song, M. A. Bakowski, L. Riva, K. Wolff, C. W. McNamara, T. F. Rogers, J. Malvin, S. Li, S. Joseph, A. Woods, Y. L. Liu and N. Okwor, Protease inhibitors for the treatment of coronavirus infections, WO2022261473, 2022.
- N. G. R. D. Elshan, K. C. Wolff, L. Riva, A. K. Woods, G. Grabovyi, K. Wilson, A. Rahimi, J. Pedroarena, S. Ghorai, A. K. Gupta, A. Nazarian, F. Weiss, Y. Liu, W. Mazumdar, L. Song, N. Okwor, J. Malvin, M. A. Bakowski, N. Beutler, M. G. Kirkpatrick, A. Gebara-Lamb, E. Huang, V. Nguyen-Tran, V. Chi, S. Li, T. F. Rogers, C. W. McNamara, J. J. Chen, S. B. Joseph, P. G. Schultz and A. K. Chatterjee, Discovery of CMX990: A Potent SARS-CoV-2 3CL Protease Inhibitor Bearing a Novel Covalent Warhead, BioRxiv, 2023, preprint, DOI:10.1101/2023.10.24.563688.
- M. Zhu, T. Fu, M. You, J. Cao, H. Yang, X. Chen, Q. Zhang, Y. Xu, X. Jiang, L. Zhang, H. Su, Y. Zhang and J. Shen, Design, synthesis and biological evaluation of covalent peptidomimetic 3CL protease inhibitors containing nitrile moiety, Bioorg. Med. Chem., 2023, 87, 117316 CrossRef CAS PubMed.
- W. Liu, C. A. Fierke, S. Xu, K. Yang, X. Ma, Y. Ma, Y. R. Alugubelli, E. C. Vatansever, C. C. Cho, Z. Geng and K. Khatua, SARS-COV-2 main protease inhibitors, WO2022020711, 2022.
- Sorrento Therapeutics, https://sorrentotherapeutics.com/research/covid, (accessed July, 26, 2023).
- L. Mao, X. Xu, N. Shaabani and C. Jin, Protease inhibitors as antiviral, WO2022256434, 2022.
- A. Balakrishnan, A. Reyes, R. Shen, N. Bisht, J. Sweeney, R. Levene, N. McAllister, T. Cressey, N. Manalo, M. H. Rhodin, M. Vaine, G. Wang, Y. S. Or and B. Goodwin, Molecular Basis for Antiviral Action of EDP-235: A Potent and Selective SARS-CoV-2 3CLpro Inhibitor for the Treatment of Covid 19, FASEB J., 2022, 36 DOI:10.1096/fasebj.2022.36.S1.0R514.
- G. Wang, R. Shen, J. Ma, Y. He, X. Xing, H. Cao, X. Gao, X. Peng, J. D. Panarese and Y. S. Or, Novel spiropyrolidine derived antiviral agents, WO2022109360, 2022.
- G. Wang, R. Shen, J. Ma, X. Xing, H. Cao, X. Gao, X. Peng, J. Long, W. Li, J. Zhang, J. D. Panarese, N. T. Kenton, S. Bartlett and Y. S. Or, Novel spiropyrolidine derived antiviral agents, WO2022109363, 2022.
- K. Zhu, G. Wang, J. Zhang, H. Cao, R. Shen, Q. Wang, G. Wu and Y. S. Or, Process for the preparation of 4,6,7-trifluoro-1H-indole-2-carboxylic acid, WO2023177854, 2023.
- K. Zhu, G. Wang, X. Zhang, X. Peng, R. Shen, J. Zhang, W. Li, H. Cao, X. Gao, Q. Wang, G. Wu and Y. S. Or, Process for the preparation of substituted spirooxindole derivatives, WO2023177854, 2023.
- R. Shen, Y. He, X. Xing, M. Rhodes, J. D. Panarese, S. Bartlett, W. Li, H. Cao, J. Zhang, X. Peng, G. Wang and Y. S. Or, Novel macrocyclic spiropyrolidine derived antiviral agents, WO2023009187, 2023.
- J. Zhang, X. Peng, B.-C. Suh, J. Kass, X. Gao, H. Cao, W. Li, J. D. Panarese, G. Wang and Y. S. Or, Novel macrocyclic spiropyrolidine derived antiviral agents, WO2022240541, 2022.
- J. D. Panarese, S. Rafferty, J. Thielman, N. T. Kenton, S. Bartlett and Y. S. Or, Heterocyclic antiviral agents, WO2023107417, 2023.
- J. D. Panarese, S. Bartlett and Y. S. Or, Saturated spirocyclics as antiviral agents, WO2023107419, 2023.
- P. S. Dragovich, T. J. Prins, R. Zhou, E. L. Brown, F. C. Maldonado, S. A. Fuhrman, L. S. Zalman, T. Tuntland, C. Lee, A. K. Patick, D. A. Matthews, T. F. Hendrickson, M. B. Kosa, B. Liu, M. R. Batugo, J. P. Gleeson, S. K. Sakata, L. Chen, M. C. Guzman, J. W. R. Meador, R. A. Ferre and S. T. Worland, Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 6. Structure-activity studies of orally bioavailable, 2-pyridone-containing peptidomimetics, J. Med. Chem., 2002, 45, 1607 CrossRef CAS PubMed.
- P. S. Dragovich, T. J. Prins, R. Zhou, T. O. Johnson, E. L. Brown, F. C. Maldonado, S. A. Fuhrman, L. S. Zalman, A. K. Patick, D. A. Matthews, X. Hou, J. W. Meador, R. A. Ferre and S. T. Worland, Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. Part 7: structure-activity studies of bicyclic 2-pyridone-containing peptidomimetics, Bioorg. Med. Chem. Lett., 2002, 12, 733 CrossRef CAS PubMed.
- P. S. Dragovich, T. J. Prins, R. Zhou, T. O. Johnson, Y. Hua, H. T. Luu, S. K. Sakata, E. L. Brown, F. C. Maldonado, T. Tuntland, C. A. Lee, S. A. Fuhrman, L. S. Zalman, A. K. Patick, D. A. Matthews, E. Y. Wu, M. Guo, B. C. Borer, N. K. Nayyar, T. Moran, L. Chen, P. A. Rejto, P. W. Rose, M. C. Guzman, E. Z. Dovalsantos, S. Lee, K. McGee, M. Mohajeri, A. Liese, J. Tao, M. B. Kosa, B. Liu, M. R. Batugo, J. P. Gleeson, Z. P. Wu, J. Liu, J. W. R. Meador and R. A. Ferre, Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 8. Pharmacological optimization of orally bioavailable 2-pyridone-containing peptidomimetics, J. Med. Chem., 2003, 46, 4572 CrossRef PubMed.
- A. K. Patick, M. A. Brothers, F. Maldonado, S. Binford, O. Maldonado, S. Fuhrman, A. Petersen, G. J. R. Smith, L. S. Zalman, L. A. Burns-Naas and J. Q. Tran, In vitro antiviral activity and single-dose pharmacokinetics in humans of a novel, orally bioavailable inhibitor of human rhinovirus 3C protease, Antimicrob. Agents Chemother., 2005, 49, 2267 CrossRef CAS PubMed.
- M. K. Kankam, J. M. Burns, M. S. Collett, M. L. Corrado and J. R. Hincks, A Phase 1 Study of the Safety, Tolerability, and Pharmacokinetics of Single and Multiple Oral Doses of V-7404 in Healthy Adult Volunteers, Antimicrob. Agents Chemother., 2021, 65, e0102921 CrossRef PubMed.
- M. Fàbrega-Ferrer, A. Herrera-Morandé, S. Muriel-Goñi, J. Pérez-Saavedra, P. Bueno, V. Castro, U. Garaigorta, P. Gastaminza and M. Coll, Structure and inhibition of SARS-CoV-1 and SARS-CoV-2 main proteases by oral antiviral compound AG7404, Antiviral Res., 2022, 208, 105458 CrossRef PubMed.
- L. Zhu, S. George, M. F. Schmidt, S. Al-Gharabli, J. Rademann and R. Hilgenfeld, Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease, Antiviral Res., 2011, 92, 204 CrossRef CAS PubMed.
- Z. Shen, Y. Li, J. Zhu, Q. Huang, J. Yin, Y. Xu, A. M. Wu, W. Su and L. Kuai, Virus main protease inhibitor, preparation method therefor, and use, WO2023283831, 2023.
- L. D. Arnold, A. Jennings and W. Keung, Inhibitors of cysteine proteases and methods of use thereof, US11174231, 2021.
- K. O. Chang, Y. Kim and W. C. Groutas, Macrocyclic and peptidomimetic compounds as broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, caliciviruses and coronaviruses, WO2013166319, 2013.
- S. R. Mandadapu, P. M. Weerawarna, A. M. Prior, R. A. Uy, S. Aravapalli, K. R. Alliston, G. H. Lushington, Y. Kim, D. H. Hua, K. O. Chang and W. C. Groutas, Macrocyclic inhibitors of 3C and 3C-like proteases of picornavirus, norovirus, and coronavirus, Bioorg. Med. Chem. Lett., 2013, 23, 3709 CrossRef CAS PubMed.
- L. Wang, C. Ma, M. D. Sacco, S. Xue, M. Mahmoud, L. Calcul, Y. Chen, J. Wang and J. Cai, Development of the Safe and Broad-Spectrum Aldehyde and Ketoamide Mpro inhibitors Derived from the Constrained α, γ-AA Peptide Scaffold, Chemistry, 2023, 29, e202300476 CrossRef CAS PubMed.
- J. E. Blanchard, N. H. Elowe, C. Huitema, P. D. Fortin, J. D. Cechetto, L. D. Eltis and E. D. Brown, High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase, Chem. Biol., 2004, 11, 1445 CrossRef CAS PubMed.
- J. Zhang, H. I. Pettersson, C. Huitema, C. Niu, J. Yin, M. N. James, L. D. Eltis and J. C. Vederas, Design, synthesis, and evaluation of inhibitors for severe acute respiratory syndrome 3C-like protease based on phthalhydrazide ketones or heteroaromatic esters, J. Med. Chem., 2007, 50, 1850 CrossRef CAS PubMed.
- C. Niu, J. Yin, J. Zhang, J. C. Vederas and M. N. James, Molecular docking identifies the binding of 3-chloropyridine moieties specifically to the S1 pocket of SARS-CoV Mpro, Bioorg. Med. Chem., 2008, 16, 293 CrossRef CAS PubMed.
- A. K. Ghosh, G. Gong, V. Grum-Tokars, D. C. Mulhearn, S. C. Baker, M. Coughlin, B. S. Prabhakar, K. Sleeman, M. E. Johnson and A. D. Mesecar, Design, synthesis and antiviral efficacy of a series of potent chloropyridyl ester-derived SARS-CoV 3CLpro inhibitors, Bioorg. Med. Chem. Lett., 2008, 18, 5684 CrossRef CAS PubMed.
- J. Breidenbach, C. Lemke, T. Pillaiyar, L. Schäkel, G. Al Hamwi, M. Diett, R. Gedschold, N. Geiger, V. Lopez, S. Mirza, V. Namasivayam, A. Schiedel, K. Sylvester, D. Thimm, C. Vielmuth, L. Phuong Vu, M. Zyulina, J. Bodem, M. Gütschow and C. E. Müller, Targeting the Main Protease of SARS-CoV-2: From the Establishment of High Throughput Screening to the Design of Tailored Inhibitors, Angew. Chem., Int. Ed., 2021, 60, 10423 CrossRef CAS PubMed.
- R. Wamser, S. Pach, C. Arkona, M. Baumgardt, U. B. A. Aziz, A. C. Hocke, G. Wolber and J. Rademann, A Critical Study on Acylating and Covalent Reversible Fragment Inhibitors of SARS-CoV-2 Main Protease Targeting the S1-Site with Pyridine, ChemMedChem, 2023, 18, e202200635 CrossRef CAS PubMed.
- F. M. Zhang, T. Huang, F. Wang, G. S. Zhang, D. Liu, J. Dai, J. W. Zhang, Q. H. Li, G. Q. Lin, D. Gao, J. Zhao and P. Tian, Discovery of highly potent covalent SARS-CoV-2 3CLpro inhibitors bearing 2-sulfoxyl-1,3,4-oxadiazole scaffold for combating COVID-19, Eur. J. Med. Chem., 2023, 260, 115721 CrossRef CAS PubMed.
- K. M. Kang, Y. Jang, S. S. Lee, M. S. Jin, C. D. Jun, M. Kim and Y. C. Kim, Discovery of antiviral SARS-CoV-2 main protease inhibitors by structure-guided hit-to-lead optimization of carmofur, Eur. J. Med. Chem., 2023, 260, 115720 CrossRef CAS PubMed.
- T. Pillaiyar, P. Flury, N. Krüger, H. Su, L. Schäkel, E. Barbosa Da Silva, O. Eppler, T. Kronenberger, T. Nie, S. Luedtke, C. Rocha, K. Sylvester, M. R. I. Petry, J. H. McKerrow, A. Poso, S. Pöhlmann, M. Gütschow, A. J. O'Donoghue, Y. Xu, C. E. Müller and S. A. Laufer, Small-Molecule Thioesters as SARS-CoV-2 Main Protease Inhibitors: Enzyme Inhibition, Structure-Activity Relationships, Antiviral Activity, and X-ray Structure Determination, J. Med. Chem., 2022, 65, 9376 CrossRef CAS PubMed.
- J. Zhang, C. Huitema, C. Niu, J. Yin, M. N. G. James, L. D. Eltis and J. C. Vederas, Aryl methylene ketones and fluorinated methylene ketones as reversible inhibitors for severe acute respiratory syndrome (SARS) 3C-like proteinase, Bioorg. Chem., 2008, 26, 229 CrossRef PubMed.
- M. Turlington, A. Chun, S. Tomar, A. Eggler, V. Grum-Tokars, J. Jacobs, J. S. Daniels, E. Dawson, A. Saldanha, P. Chase, Y. M. Baez-Santos, C. W. Lindsley, P. Hodder, A. D. Mesecar and S. R. Stauffer, Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamidophenyl carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding, Bioorg. Med. Chem. Lett., 2013, 23, 6172 CrossRef CAS PubMed.
- G. J. Lockbaum, A. C. Reyes, J. M. Lee, R. Tilvawala, E. A. Nalivaika, A. Ali, N. Kurt Yilmaz, P. R. Thompson and C. A. Schiffer, Crystal Structure of SARS-CoV-2 Main Protease in Complex with the Non-Covalent Inhibitor ML188, Viruses, 2021, 13, 174 CrossRef CAS PubMed.
- S. H. Han, C. M. Goins, T. Arya, W. J. Shin, J. Maw, A. Hooper, D. P. Sonawane, M. R. Porter, B. E. Bannister, R. D. Crouch, A. A. Lindsey, G. Lakatos, S. R. Martinez, J. Alvarado, W. S. Akers, N. S. Wang, J. U. Jung, J. D. Macdonald and S. R. Stauffer, Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CL(pro)), J. Med. Chem., 2022, 65, 2880 CrossRef CAS PubMed.
- J. Chodera, A. A. Lee, N. London and F. von Delft, Crowdsourcing drug discovery for pandemics, Nat. Chem., 2020, 12, 581 CrossRef CAS PubMed.
- F. von Delft, M. Calmiano, J. Chodera, E. Griffen, A. Lee, N. London, T. Matviuk, B. Perry, M. Robinson and A. von Delft, A white-knuckle ride of open COVID drug discovery, Nature, 2021, 594, 330 CrossRef CAS PubMed.
- The COVID Moonshot Consortium, J. Chodera, A. Lee, N. London and F. von Delft, Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitors, ChemRxiv, 2021, preprint, DOI:10.33774/chemrxiv.
- H. Achdout, A. Aimon, E. Bar-David, H. Barr, A. Ben-Shmuel, J. Bennett, V. A. Bilenko, M. L. Boby, B. Borden, G. R. Bowman, J. Brun, B. V. N. B. S. Sarma, M. Calmiano, A. Carbery, D. Carney, E. Cattermole, E. Chang, E. Chernyshenko, J. D. Chodera, A. Clyde, J. E. Coffland, G. Cohen, J. Cole, A. Contini, L. Cox, M. Cvitkovic, A. Dias, K. Donckers, D. L. Dotson, A. Douangamath, S. Duberstein, T. Dudgeon, L. Dunnett, P. K. Eastman, N. Erez, C. J. Eyermann, M. Fairhead, G. Fate, D. Fearon, O. Fedorov, M. Ferla, R. S. Fernandes, L. Ferrins, R. Foster, H. Foster, R. Gabizon, A. Garcia-Sastre, V. O. Gawriljuk, P. Gehrtz, C. Gileadi, C. Giroud, W. G. Glass, R. Glen, I. Glinert, A. S. Godoy, M. Gorichko, T. Gorrie-Stone, E. J. Griffen, S. Hassell Hart, J. Heer, M. Henry, M. Hill, S. Horrell, V. D. Huliak, M. F. D. Hurley, T. Israely, A. Jajack, J. Jansen, E. Jnoff, D. Jochmans, T. John, S. De Jonghe, A. L. Kantsadi, P. W. Kenny, J. L. Kiappes, S. O. Kinakh, L. Koekemoer, B. Kovar, T. Krojer, A. Lee, B. A. Lefker, H. Levy, I. G. Logvinenko, N. London, P. Lukacik, H. B. Macdonald, B. MacLean, T. R. Malla, T. Matviiuk, W. McCorkindale, B. L. McGovern, S. Melamed, K. P. Melnykov, O. Michurin, H. Mikolajek, B. F. Milne, A. Morris, G. M. Morris, M. J. Morwitzer, D. Moustakas, A. M. Nakamura, J. Brandao Neto, J. Neyts, L. Nguyen, G. D. Noske, V. Oleinikovas, G. Oliva, G. J. Overheul, D. Owen, R. Pai, J. Pan, N. Paran, B. Perry, M. Pingle, J. Pinjari, B. Politi, A. Powell, V. Psenak, R. Puni, V. L. Rangel, R. N. Reddi, S. P. Reid, E. Resnick, E. G. Ripka, M. C. Robinson, R. P. Robinson, J. Rodriguez-Guerra, R. Rosales, D. Rufa, K. Saar, K. Singh Saikatendu, C. Schofield, M. Shafeev, A. Shaikh, J. Shi, K. Shurrush, S. Singh, A. Sittner, R. Skyner, A. Smalley, B. Smeets, M. D. Smilova, L. J. Solmesky, J. Spencer, C. Strain-Damerell, V. Swamy, H. Tamir, R. Tennant, W. Thompson, A. Thompson, S. Tomasio, I. S. Tsurupa, A. Tumber, I. Vakonakis, R. P. van Rij, L. Vangeel, F. S. Varghese, M. Vaschetto, E. B. Vitner, V. Voelz, A. Volkamer, F. von Delft, A. von Delft, M. Walsh, W. Ward, C. Weatherall, S. Weiss, K. M. White, C. F. Wild, M. Wittmann, N. Wright, Y. Yahalom-Ronen, D. Zaidmann, H. Zidane and N. Zitzmann, Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitor Therapeutics, BioRxiv, 2022, preprint, DOI:10.1101/2020.10.29.339317.
- K. L. Saar, W. McCorkindale, D. Fearon, M. Boby, H. Barr, A. Ben-Shmuel, COVID Moonshot Consortium, N. London, F. Von Delft, J. D. Chodera and A. A. Lee, Turning high-throughput structural biology into predictive inhibitor design, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2214168120 CrossRef CAS PubMed.
- E. Griffen, https://dndi.org/wp-content/uploads/2021/09/COVID_Moonshot_RSC_Cambridge_MedChem_Sept_2021.pdf, (accessed 6th of January 2022).
- M. L. Boby, D. Fearon, M. Ferla, M. Filep, L. Koekemoer, M. C. Robinson, The COVID Moonshot Consortium, J. D. Chodera, A. A. Lee, N. London, A. von Delft and F. von Delft, Open Science Discovery of Potent Non-Covalent SARS-CoV-2 Main Protease Inhibitors, BioRxiv, 2023, preprint, DOI:10.1101/2020.10.29.339317.
- A. Luttens, H. Gullberg, E. Abdurakhmanov, D. D. Vo, D. Akaberi, V. O. Talibov, N. Nekhotiaeva, L. Vangeel, S. De Jonghe, D. Jochmans, J. Krambrich, A. Tas, B. Lundgren, Y. Gravenfors, A. J. Craig, Y. Atilaw, A. Sandström, L. W. K. Moodie, Å. Lundkvist, M. J. van Hemert, J. Neyts, J. Lennerstrand, J. Kihlberg, K. Sandberg, U. H. Danielson and J. Carlsson, Ultralarge Virtual Screening Identifies SARS-CoV-2 Main Protease Inhibitors with Broad-Spectrum Activity against Coronaviruses, J. Am. Chem. Soc., 2022, 144, 2905 CrossRef CAS PubMed.
- N. Kitamura, M. D. Sacco, C. Ma, Y. Hu, J. A. Townsend, X. Meng, F. Zhang, X. Zhang, M. Ba, T. Szeto, A. Kukuljac, M. T. Marty, D. Schultz, S. Cherry, Y. Xiang, Y. Chen and J. Wang, Expedited Approach toward the Rational Design of Noncovalent SARS-CoV-2 Main Protease Inhibitors, J. Med. Chem., 2022, 65, 2848 CrossRef CAS PubMed.
- H. Wang, J. Wen, Y. Yang, H. Liu, S. Wang, X. Ding, C. Zhou and X. Zhang, Identification of highly effective inhibitors against SARS-CoV-2 main protease: From virtual screening to in vitro study, Front. Pharmacol., 2022, 13, 1036208 CrossRef CAS PubMed.
- R. Jimmidi, S. Chamakuri, S. Lu, M. N. Ucisik, P. J. Chen, K. M. Bohren, S. A. Moghadasi, L. Versteeg, C. Nnabuife, J. Y. Li, X. Qin, Y. C. Chen, J. C. Faver, P. Nyshadham, K. L. Sharma, B. Sankaran, A. Judge, Z. Yu, F. Li, J. Pollet, R. S. Harris, M. M. Matzuk, T. Palzkill and D. W. Young, DNA-encoded chemical libraries yield non-covalent and non-peptidic SARS-CoV-2 main protease inhibitors, Commun. Chem., 2023, 6, 164 CrossRef CAS PubMed.
- D. Zaidman, P. Gehrtz, M. Filep, D. Fearon, R. Gabizon, A. Douangamath, J. Prilusky, S. Duberstein, G. Cohen, C. D. Owen, E. Resnick, C. Strain-Damerell, P. Lukacik, C.-M. Consortium, H. Barr, M. A. Walsh, F. von Delft and N. London, An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 Mpro inhibitor, Cell Chem. Biol., 2021, 28, 1 CrossRef PubMed.
- A. Zavoronkovs, Y. A. Ivanenkov and B. Zagribelnyy, SARS-CoV-2 inhibitors having covalent modifications for treating coronavirus infections, WO2021219089, 2021.
- L. D. Arnold, U. Lopatin and W. Keung, Inhibitors of cysteine proteases and methods of use thereof, WO2021212039, 2021.
- L. D. Arnold and W. Keung, Inhibitors of cysteine proteases and methods of use thereof, WO2022221686, 2022.
- J. K. Stille, J. Tjutrins, G. Wang, F. A. Venegas, C. Hennecker, A. M. Rueda, I. Sharon, N. Blaine, C. E. Miron, S. Pinus, A. Labarre, J. Plescia, M. Burai Patrascu, X. Zhang, A. S. Wahba, D. Vlaho, M. J. Huot, T. M. Schmeing, A. K. Mittermaier and N. Moitessier, Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CLpro covalent inhibitors, Eur. J. Med. Chem., 2022, 229, 114046 CrossRef CAS PubMed.
- B. X. Quan, H. Shuai, A. J. Xia, Y. Hou, R. Zeng, X. L. Liu, G. F. Lin, J. X. Qiao, W. P. Li, F. L. Wang, K. Wang, R. J. Zhou, T. T. Yuen, M. X. Chen, C. Yoon, M. Wu, S. Y. Zhang, C. Huang, Y. F. Wang, W. Yang, C. Tian, W. M. Li, Y. Q. Wei, K. Y. Yuen, J. F. Chan, J. Lei, H. Chu and S. Yang, An orally available Mpro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron, Nat. Microbiol., 2022, 7, 716 CrossRef CAS PubMed.
- M. J. Hesse, V. Hornak, S. Joseph, H. E. Moser, J. Papillon, T. Patel, G. Robinson, D. C. Tully and J. Yuan, Compounds and compositions for the treatment of coronaviral related diseases, WO2022224223, 2022.
- D. Bao, F. Guo, M. J. Hesse, V. Hornak, S. Joseph, T. M. Kirrane, B. Liu, H. Lin, Y. Miao, H. E. Moser, J. Papillon, Y. Qu, L. Shi, J. Yuan and T. Zhang, Compounds and compositions for the treatment of coronaviral related diseases, WO2023002409, 2023.
- https://fragalysis.diamond.ac.uk/viewer/react/preview/target/Mpro, (accessed 18th of August 2023).
- N. Hou, L. Shuai, L. Zhang, X. Xie, K. Tang, Y. Zhu, Y. Yu, W. Zhang, Q. Tan, G. Zhong, Z. Wen, C. Wang, X. He, H. Huo, H. Gao, Y. Xu, J. Xue, C. Peng, J. Zou, C. Schindewolf, V. Menachery, W. Su, Y. Yuan, Z. Shen, R. Zhang, S. Yuan, H. Yu, P. Shi, Z. Bu, J. Huang and Q. Hu, Development of Highly Potent Noncovalent Inhibitors of SARS-CoV-2 3CLpro, ACS Cent. Sci., 2023, 9, 217 CrossRef CAS PubMed.
- J. Wang, C. Cheng, B. Chi and D. Sun, 3CL PRO protease inhibitors, WO2023036140, 2023.
- A. Hooper, J. D. Macdonald, B. Reilly, J. Maw, A. P. Wirrick, S. H. Han, A. A. Lindsey, E. G. Rico, T. Romigh, C. M. Goins, N. S. Wang and S. Stauffer, SARS-CoV-2 3CL-protease inhibitors derived from ML300: investigation of P1 and replacements of the 1,2,3-benzotriazole, Res. Sq., 2023 DOI:10.21203/rs.3.rs.
- S. R. Stauffer, S. H. Han, A. Hooper, J. Maw, D. P. Sonawane, M. R. Porter, S. Martinez, J. Alvarado and J. D. Macdonald, Protease inhibitor and method of use, WO2022266203, 2022.
- S. R. Stauffer, J. D. Macdonald, A. Hooper, S. H. Han, D. P. Sonawane, M. R. Porter, J. Maw, S. Martinez and J. Alvarado, Protease inhibitors and method of use, WO2022266368, 2022.
- J. Pérez-Vargas, L. J. Worrall, A. D. Olmstead, A. T. Ton, J. Lee, I. Villanueva, C. A. H. Thompson, S. Dudek, S. Ennis, J. R. Smith, T. Shapira, J. Guzman, S. Gang, F. Ban, M. Vuckovic, M. Bielecki, S. Kovacic, C. Kenward, C. Y. Hong, D. G. Gordon, P. N. Levett, M. Krajden, R. Leduc, P. L. Boudreault, M. Niikura, M. Paetzel, R. N. Young, A. Cherkasov, N. C. J. Strynadka and F. Jean, A novel class of broad-spectrum active-site-directed 3C-like protease inhibitors with nanomolar antiviral activity against highly immune-evasive SARS-CoV-2 Omicron subvariants, Emerging Microbes Infect., 2023, 2246594 CrossRef PubMed.
- R. Liu, Y. Xu, L. Hua, J. Zhou, H. Deng, X. Chu and S. Ding, 3CL protease small-molecule inhibitor for treating or preventing coronavirus infection, and use thereof, WO2023011443, 2023.
- S. Richards, M. Derudas, N. Ahlsten and K. Papachristos, MPro targeting antiviral compounds, WO2023180189, 2023.
- L. D. Arnold, W. Keung and D. V. Kumar, Inhibitors of cysteine proteases and methods of use thereof, WO2023044171, 2023.
- B. T. Campbell, W. Chang, T. J. Hartingh, D. M. Hurzy, M. J. Kelly III, F. M. Klinger, M. E. Layton, J. A. Mccauley, C. C. Nawrat, C. A. Parish, J. J. Perkins, A. J. Roecker, M. De Lera Ruiz, J. D. Schreier, V. W. Shurtleff, J. Su and Q. T. Truong, Protease Inhibitors for Treating or Preventing Coronavirus Infection, WO2023133174, 2023.
- S. Ullrich, V. M. Sasi, M. C. Mahawaththa, K. B. Ekanayake, R. Morewood, J. George, L. Shuttleworth, X. Zhang, C. Whitefield, G. Otting, C. Jackson and C. Nitsche, Challenges of short substrate analogues as SARS-CoV-2 main protease inhibitors, Bioorg. Med. Chem. Lett., 2021, 50, 128333 CrossRef CAS PubMed.
- J. E. Hernández González, R. J. Eberle, D. Willbold and M. A. Coronado, A Computer-Aided Approach for the Discovery of D-Peptides as Inhibitors of SARS-CoV-2 Main Protease, Front. Mol. Biosci., 2022, 8, 816166 CrossRef PubMed.
- R. Eberle, M. Sevenich, I. Gering, L. Scharbert, B. Strodel, N. A. Lakomek, K. Santur, J. Mohrlüder, M. A. Coronado and D. Willbold, Discovery of All-d-Peptide Inhibitors of SARS-CoV-2 3C-like Protease, ACS Chem. Biol., 2023, 18, 315 CrossRef CAS PubMed.
- T. Miura, T. R. Malla, C. D. Owen, A. Tumber, L. Brewitz, M. McDonough, E. Salah, N. Terasaka, T. Katoh, P. Lukacik, C. Strain-Damerell, H. Mikolajek, M. A. Walsh, A. Kawamura, C. J. Schofield and H. Suga, In vitro selection of macrocyclic peptide inhibitors containing cyclic γ2,4-amino acids targeting the SARS-CoV-2 main protease, Nat. Chem., 2023, 15, 998 CrossRef CAS PubMed.
- T. Zhai, F. Zhang, S. Haider, D. Kraut and Z. Huang, An Integrated Computational and Experimental Approach to Identifying Inhibitors for SARS-CoV-2 3CL Protease, Front. Mol. Biosci., 2021, 8, 661424 CrossRef CAS PubMed.
- A. Clyde, S. Galanie, D. W. Kneller, H. Ma, Y. Babuji, B. Blaiszik, A. Brace, T. Brettin, K. Chard, R. Chard, L. Coates, I. Foster, D. Hauner, V. Kertesz, N. Kumar, H. Lee, Z. Li, A. Merzky, J. Schmidt, L. Tan, M. Titov, A. Trifan, M. Turilli, H. Van Dam, S. C. Chennubhotla, S. Jha, A. Kovalevsky, A. Ramanathan, M. S. Head and R. Stevens, High-Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Noncovalent Inhibitor, J. Chem. Inf. Model., 2022, 62, 116 CrossRef CAS PubMed.
- D. W. Kneller, H. Li, S. Galanie, G. Phillips, A. Labbé, K. L. Weiss, Q. Zhang, M. A. Arnould, A. Clyde, H. Ma, A. Ramanathan, C. B. Jonsson, M. S. Head, L. Coates, J. M. Louis, P. V. Bonnesen and A. Kovalevsky, Structural, Electronic, and Electrostatic Determinants for Inhibitor Binding to Subsites S1 and S2 in SARS-CoV-2 Main Protease, J. Med. Chem., 2021, 64, 17366 CrossRef CAS PubMed.
- S. Gao, K. Sylvester, L. Song, T. Claff, L. Jing, M. Woodson, R. H. Weiße, Y. Cheng, L. Schäkel, M. Petry, M. Gütschow, A. C. Schiedel, N. Sträter, D. Kang, S. Xu, K. Toth, J. Tavis, A. E. Tollefson, C. E. Müller, X. Liu and P. Zhan, Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity, J. Med. Chem., 2022, 65, 13343 CrossRef CAS PubMed.
- S. Gao, L. Song, T. Claff, M. Woodson, K. Sylvester, L. Jing, R. H. Weiße, Y. Cheng, N. Sträter, L. Schäkel, M. Gütschow, B. Ye, M. Yang, T. Zhang, D. Kang, K. Toth, J. Tavis, A. E. Tollefson, C. E. Müller, P. Zhan and X. Liu, Discovery and Crystallographic Studies of Nonpeptidic Piperazine Derivatives as Covalent SARS-CoV-2 Main Protease Inhibitors, J. Med. Chem., 2022, 65, 16902 CrossRef CAS PubMed.
- A. Morris, W. McCorkindale, T. C. M. Consortium, N. Drayman, J. D. Chodera, S. Tay, N. London and A. A. Lee, Discovery of SARS-CoV-2 main protease inhibitors using a synthesis-directed de novo design model, Chem. Commun., 2021, 57, 5909 RSC.
- Z. Jiang, B. Feng, Y. Zhang, T. Nie, H. Liu, J. Li, H. Su, L. Zhang, Y. Zang and Y. Zhou, Discovery of novel non-peptidic and non-covalent small-molecule 3CLpro inhibitors as potential candidate for COVID-19 treatment, Signal Transduction Targeted Ther., 2023, 8, 209 CrossRef CAS PubMed.
- J. Sacchettini, N. E. Zhou, M. K. Parai, J. C. Shin, J. L. Wood, I. Krieger, A. C. F. Flores, A. Archarya, Z. Shi, X. Bian, S. Tang, R. Bam and P. K. Jaiswal, Inhibitors of SARS-COV-2, WO2022235813, 2022.
- C. Huang, R. Zeng, J. Qiao, B. Quan, R. Luo, Q. Huang, N. Guo, Y. Li, X. Long, R. Ma, A. Xia, Z. Fang, Y. Wang, Y. Li, Y. Zheng, L. Li, J. Lei and S. Yang, Discovery and structure-activity relationship studies of novel α-ketoamide derivatives targeting the SARS-CoV-2 main protease, Eur. J. Med. Chem., 2023, 259, 115657 CrossRef CAS PubMed.
- C. Huang, H. Shuai, J. Qiao, Y. Hou, R. Zeng, A. Xia, L. Xie, Z. Fang, Y. Li, C. Yoon, Q. Huang, B. Hu, J. You, B. Quan, X. Zhao, N. Guo, S. Zhang, R. Ma, J. Zhang, Y. Wang, R. Yang, S. Zhang, J. Nan, H. Xu, F. Wang, J. Lei, H. Chu and S. Yang, A new generation Mpro inhibitor with potent activity against SARS-CoV-2 Omicron variants, Signal Transduction Targeted Ther., 2023, 8, 128 CrossRef CAS PubMed.
- Y. M. Shao, W. B. Yang, T. H. Kuo, K. C. Tsai, C. H. Lin, A. S. Yang, P. H. Liang and C. H. Wong, Design, synthesis, and evaluation of trifluoromethyl ketones as inhibitors of SARS-CoV 3CL protease, Bioorg. Med. Chem., 2008, 16, 4652 CrossRef CAS PubMed.
- J. Wang, B. Liang, Y. Chen, J. Fuk-Woo Chan, S. Yuan, H. Ye, L. Nie, J. Zhou, Y. Wu, M. Wu, L. S. Huang, J. An, A. Warshel, K. Y. Yuen, A. Ciechanover, Z. Huang and Y. Xu, A new class of α-ketoamide derivatives with potent anticancer and anti-SARS-CoV-2 activities, Eur. J. Med. Chem., 2021, 215, 113267 CrossRef CAS PubMed.
- M. Liu, J. Li, W. Liu, Y. Yang, M. Zhang, Y. Ye, W. Zhu, C. Zhou, H. Zhai, Z. Xu, G. Zhang and H. Huang, The S1'-S3' Pocket of the SARS-CoV-2 Main Protease Is Critical for Substrate Selectivity and Can Be Targeted with Covalent Inhibitors, Angew. Chem., Int. Ed., 2023, e202309657 CAS.
- P. Moon, C. M. Zammit, Q. Shao, D. Dovala, L. Boike, N. J. Henning, M. Knapp, J. N. Spradlin, C. C. Ward, H. Wolleb, D. Fuller, G. Blake, J. P. Murphy, F. Wang, Y. Lu, S. A. Moquin, L. Tandeske, M. J. Hesse, J. M. McKenna, J. A. Tallarico, M. Schirle, F. D. Toste and D. K. Nomura, Discovery of Potent Pyrazoline-Based Covalent SARS-CoV-2 Main Protease Inhibitors, ChemBioChem, 2023, 24, e202300116 CrossRef CAS PubMed.
- F. Wang, R. Zeng, J. Qiao, A. Xia, Y. Li, F. Li, Y. Wu, Y. Liu, X. Zhao, J. Lei and S. Yang, Discovery of benzodiazepine derivatives as a new class of covalent inhibitors of SARS-CoV-2 main protease, Bioorg. Med. Chem. Lett., 2023, 92, 129407 CrossRef CAS PubMed.
- Y. Unoh, S. Uehara, K. Nakahara, H. Nobori, Y. Yamatsu, S. Yamamoto, Y. Maruyama, Y. Taoda, K. Kasamatsu, T. Suto, K. Kouki, A. Nakahashi, S. Kawashima, T. Sanaki, S. Toba, K. Uemura, T. Mizutare, S. Ando, M. Sasaki, Y. Orba, H. Sawa, A. Sato, T. Sato, T. Kato and Y. Tachibana, Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19, J. Med. Chem., 2022, 65, 6499 CrossRef CAS PubMed.
- H. Kai, T. Kameyama, T. Horiguchi, K. Asahi, T. Endoh, Y. Fujii, T. Shintani, K. Nakamura, S. Matsumoto, T. Hasegawa, M. Oohara, Y. Tada, T. Maki and A. Iida, Preparation of triazine derivatives and pharmaceutical compound that contains same and exhibits analgesic activity, WO2012020749, 2012.
- G. D. Noske, E. de Souza Silva, M. O. de Godoy, I. Dolci, R. S. Fernandes, R. V. C. Guido, P. Sjö, G. Oliva and A. S. Godoy, Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease, J. Biol. Chem., 2023, 299, 103004 CrossRef CAS PubMed.
- J. J. Wu, Triazine derivatives and methods of use thereof, US11702406, 2023.
- H. Mukae, H. Yotsuyanagi, N. Ohmagari, Y. Doi, T. Imamura, T. Sonoyama, T. Fukuhara, G. Ichihashi, T. Sanaki, K. Baba, Y. Takeda, Y. Tsuge and T. Uehara, A Randomized Phase 2/3 Study of Ensitrelvir, a Novel Oral SARS-CoV-2 3C-Like Protease Inhibitor, in Japanese Patients with Mild-to-Moderate COVID-19 or Asymptomatic SARS-CoV-2 Infection: Results of the Phase 2a Part, Antimicrob. Agents Chemother., 2022, 66, e0069722 CrossRef PubMed.
- R. Shimizu, T. Sonoyama, T. Fukuhara, A. Kuwata, Y. Matsuo and R. Kubota, Safety, Tolerability, and Pharmacokinetics of the Novel Antiviral Agent Ensitrelvir Fumaric Acid, a SARS-CoV-2 3CL Protease Inhibitor, in Healthy Adults, Antimicrob. Agents Chemother., 2022, 66, e0063222 CrossRef PubMed.
- H. Yotsuyanagi, N. Ohmagari, Y. Doi, T. Imamura, T. Sonoyama, G. Ichihashi, T. Sanaki, Y. Tsuge, T. Uehara and H. Mukae, A phase 2/3 study of S-217622 in participants with SARS-CoV-2 infection (Phase 3 part), Medicine, 2023, 102, e33024 CrossRef CAS PubMed.
- S. Jiang, Y. Xiao, B. Guo, K. Zhang and C. Liu, Triazine compounds or pharamceutically acceptable salt or isomer therof, pharmaceutical composition, and use of thereof, WO2023173708, 2023.
- C. He, H. Xu, Q. Liu, B. Liu, Y. Qi, Z. Zhou, R. Tan, Z. Huang, H. Tan, Z. Chen, Z. Li, Y. Wang, L. Chen, X. Wang, L. Yang, W. Dong, S. J. Lin, X. Zhao and W. Wang, Compounds as SARS-CoV-2 inhibitors, WO2023165459, 2023.
- G. Li, R. Hilgenfeld, R. Whitley and E. De Clercq, Therapeutic strategies for COVID-19: progress and lessons learned, Nat. Rev. Drug Discovery, 2023, 22, 449 CrossRef CAS PubMed.
- U.S. Department of Health & Human Services, https://www.nih.gov/research, (accessed 22/08/ 2023).
- B. Halford, The Path to Paxlovid, ACS Cent. Sci., 2022, 8, 405 CrossRef CAS PubMed.
- S. Vankadara, M. D. Dawson, J. Y. Fong, Q. Y. Oh, Q. A. Ang, B. Liu, H. Y. Chang, J. Koh, X. Koh, Q. W. Tan, J. Joy and C. S. B. Chia, A Warhead Substitution Study on the Coronavirus Main Protease Inhibitor Nirmatrelvir, ACS Med. Chem. Lett., 2022, 13, 1345 CrossRef CAS PubMed.
- N. Pathak, Y. T. Chen, Y. C. Hsu, N. Y. Hsu, C. J. Kuo, H. P. Tsai, J. J. Kang, C. H. Huang, S. Y. Chang, Y. H. Chang, P. H. Liang and J. M. Yang, Uncovering Flexible Active Site Conformations of SARS-CoV-2 3CL Proteases through Protease Pharmacophore Clusters and COVID-19 Drug Repurposing, ACS Nano, 2021, 15, 857 CrossRef CAS PubMed.
- N. Finch, C. W. Gemenden, I. H. S. Hsu, A. Kerr, G. A. Sim and W. I. Taylor, Oxidative Transformations of Indole Alkaloids. III. Pseudoindoxyls from Yohimbinoid Alkaloids and Their Conversion to “Invert” Alkaloids, J. Am. Chem. Soc., 1965, 87, 2229 CrossRef CAS PubMed.
- C. Pelligrini, M. Weber and H. J. Borschberg, Total synthesis of (+)-Elacomine and (−)-Isoelacomine, the two hiterto unamed oxindole alkaloids fro Elaeagnus commutata, Helv. Chim. Acta, 1996, 79, 151 CrossRef.
- S. Iketani, H. Mohri, B. Culbertson, S. Hong, Y. Duan, M. I. Luck, M. K. Annavajhala, Y. Guo, Z. Sheng, A. C. Uhlemann, S. P. Goff, Y. Sabo, H. Yang, A. Chavez and D. D. Ho, Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir, Nature, 2023, 613, 558 CrossRef CAS PubMed.
- M. Kiso, Y. Furusawa, R. Uraki, M. Imai, S. Yamayoshi and Y. Kawaoka, In vitro and in vivo characterization of SARS-CoV-2 strains resistant to nirmatrelvir, Nat. Commun., 2023, 14, 3952 CrossRef CAS PubMed.
- E. Takashita, S. Fujisaki, H. Morita, S. Nagata, H. Miura, M. Nagashima, S. Watanabe, M. Takeda, Y. Kawaoka and H. Hasegawa, Assessment of the frequency of SARS-CoV-2 Omicron variant escape from RNA-dependent RNA polymerase inhibitors and 3C-like protease inhibitors, Antiviral Res., 2023, 216, 105671 CrossRef CAS PubMed.
- M. Kiso, S. Yamayoshi, S. Iida, Y. Furusawa, Y. Hirata, R. Uraki, M. Imai, T. Suzuki and Y. Kawaoka, In vitro and in vivo characterization of SARS-CoV-2 resistance to ensitrelvir, Nat. Commun., 2023, 14, 4231 CrossRef CAS PubMed.
- Y. Hu, E. M. Lewandowski, H. Tan, X. Zhang, R. T. Morgan, X. Zhang, L. M. C. Jacobs, S. G. Butler, M. V. Gongora, J. Choy, X. Deng, Y. Chen and J. Wang, Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir, ACS Cent. Sci., 2023, 9, 1658 CrossRef CAS PubMed.
- B. Tan, R. Joyce, H. Tan, Y. Hu and J. Wang, SARS-CoV-2 Main Protease Drug Design, Assay Development, and Drug Resistance Studies, Acc. Chem. Res., 2023, 56, 157 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |