Open Access Article
Open Access ArticleSilver N-heterocyclic carbene complexes are potent uncompetitive inhibitors of the papain-like protease with antiviral activity against SARS-CoV-2†
Maria
Gil-Moles
 ab,
Cillian
O'Beirne
a,
Igor V.
Esarev
a,
Petra
Lippmann
a,
Matthias
Tacke
c,
Jindrich
Cinatl
Jr.
d,
Denisa
Bojkova
d and
Ingo
Ott
ab,
Cillian
O'Beirne
a,
Igor V.
Esarev
a,
Petra
Lippmann
a,
Matthias
Tacke
c,
Jindrich
Cinatl
Jr.
d,
Denisa
Bojkova
d and
Ingo
Ott
 *a
*a
aInstitute of Medicinal and Pharmaceutical Chemistry, Technische Universität Braunschweig, Beethovenstr. 55, 38106 Braunschweig, Germany. E-mail: ingo.ott@tu-bs.de
bDepartamento de Química, Universidad de La Rioja, Centro de Investigación de Síntesis Química (CISQ), Complejo Científico Tecnológico, 26004 Logroño, Spain
cSchool of Chemistry, University College Dublin, Belfield, Dublin 4, Ireland
dInstitute of Medical Virology, Universitätsklinikum Frankfurt, Paul-Ehrlich-Str. 40, 60596 Frankfurt, Germany
First published on 16th May 2023
Abstract
The ongoing SARS-CoV-2 pandemic has caused a high demand for novel innovative antiviral drug candidates. Despite promising results, metal complexes have been relatively unexplored as antiviral agents in general and in particular against SARS-CoV-2. Here we report on silver NHC complexes with chloride or iodide counter ligands that are potent inhibitors of the SARS-CoV-2 papain-like protease (PLpro) but inactive against 3C-like protease (3CLpro) as another SARS-CoV-2 protease. Mechanistic studies on a selected complex confirmed zinc removal from a zinc binding domain of PLpro as relevant factor of their activity. In addition, enzyme kinetic experiments revealed that the complex is an uncompetitive inhibitor and with this rare type of inhibition it offers great pharmacological advantages in terms selectivity. The silver NHC complexes with iodide ligands showed very low or absent host cell toxicity and triggered strong effects on viral replication in cells infected with SARS-CoV-2, making them promising future antiviral drug candidates.
Introduction
Since the discovery of the antitumoral properties of the platinum complex cisplatin, which has become the iconic lead compound for metallodrugs in general, there have been increasing efforts and many success stories regarding the development of metal-based cytostatics.1,2 Among few other therapeutic indications, infectious diseases have emerged as an additional relevant therapeutic field for new metallodrug candidates, with antibacterial agents as the most intensively studied class.2,3Due to the ongoing SARS-CoV-2 pandemic, the spotlight has been shifted towards viral infections, an area that has been relatively underexplored regarding metal-based bioactive substances.4–6 However, a slowly increasing number of reports on metal complexes with activity in SARS-CoV-2 infection models confirms the potential of metal complexes in this area.7–10
We have recently reported on gold and other metal complexes as inhibitors of the SARS-CoV-2 papain-like protease PLpro and the interaction of the viral spike protein with the host cell ACE2 receptor.8,11 In particular for the cysteine protease PLpro, which is essential for viral replication in infected cells, very promising enzyme inhibitory activity has been observed with various metal complexes and the triggering of antiviral effects against SARS-CoV-2 has been confirmed in infected cells. Among the promising hit compounds were some silver complexes, of which an example with a N-heterocyclic carbene (NHC) ligand displayed very promising activity as a PLpro inhibitor and antiviral agent against SARS-CoV-2 (see Fig. 1).8 Inhibitors of PLpro have recently attracted increasing attention in SARS-CoV-2 drug discovery projects.12
 | ||
| Fig. 1 Previously reported antiviral silver complexes.4,8,16 | ||
The anti-infective properties of silver NHC complexes have been well studied going back to a first report by Youngs et al. in 2004 on antibacterial activity.13 To date, many reports provide strong evidence on the antibacterial properties of silver NHC complexes (e.g. complex SBC3) and their efficacy has been proven both in vitro and in vivo.14,15
As for the antiviral activity of silver complexes, the number of reports is comparably limited, despite that silver nitrate and other silver compounds showed efficacy against herpes simplex and other viruses.4 Interestingly, a macrocyclic silver complex with a NHC ligand displayed strong antiviral activity against HIV-1.16
Based on the above screening results, we have extended our studies on silver NHC complexes as possible antiviral agents against SARS-CoV-2. A series of complexes, including the antibacterial SBC3 as reference, was prepared, characterized and evaluated as inhibitors of the target enzymes PLpro and 3CLpro. Non-toxic examples were selected for antiviral studies in SARS-CoV-2 infected cells. The results of this study are presented in this paper.
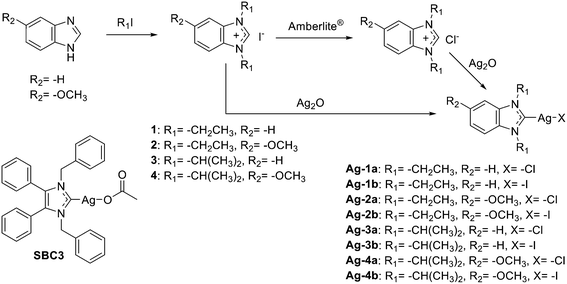
Chemistry
The target silver NHC complexes could be obtained by an established procedure (see Scheme 1) going back to the method reported by Wang and Lin in 1998.17 In the first step, the respective (benz)imidazoles were alkylated leading to the respective (benz)imidazolium iodides 1–4. To obtain the silver NHC chlorido complexes Ag-1a–Ag-4a, the iodide counterion of 1–4 was replaced with chloride followed by treatment with silver oxide. The silver NHC iodido complexes Ag-1b–Ag-4b could be generated directly from 1–4 by reaction with silver oxide. All silver NHC complexes could be prepared in high purity as evidenced by elemental analysis measurements (maximum deviation of the theoretical values: ≤0.5%).The disappearance of the hydrogen signal at position 2 of the (benz)imidazolium cations at 10.72–11.86 ppm in the 1H-NMR spectra of the silver NHC complexes Ag-1a/b–Ag-4a/b is a clear confirmation of formation of the metal carbon bond. Comparing the 13C-NMR spectra of the chlorido complexes Ag-1a to Ag-4a with those of the respective iodido analogues Ag-1b to Ag-4b, shows that the C2 signals of the chlorido complexes are shifted approx. 2–3 ppm towards the upper field compared with the analogous iodido complexes (see ESI† Fig. S1). An increase in the σ-donor ability or, in other words, a lower electronegativity of the halogene ligand should cause a decrease in the Lewis acidity of the metal, which results in the observed increasing 13C chemical shift of C2.18
Mass spectrometry (ESI) measurements showed biscarbene formation with the respective [M–X + NHC] signals in the positive and the AgX2− counterions in the negative mode.
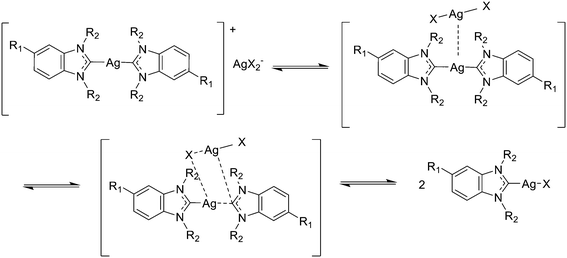
In solution silver NHC complexes exist in a dynamic equilibrium between the neutral monomeric [(NHC)AgX] form and a cationic dimeric [(NHC)2Ag]+ form with a AgX2− counter ion (see Fig. 2).17,19–21 In the solid state various bonding motifs can exist, while in solution also the monomeric form can be dominant. The lack of a 13C–107,109Ag coupling in the 13C-NMR spectra of complexes Ag-1a/b–Ag-4a/b is indicative of such fluxional behaviour with a fast equilibrium in solution indicating dominance of the monomeric form.17,21 In addition, 109Ag-NMR spectra taken exemplarily for complexes Ag-4a/b show a single narrow signal, also indicative of a predominant and likely monocarbenic form. The observed chemical shift for Ag-4a is 633.6 ppm. There is only one example of 109Ag-NMR of a monomeric [(NHC)AgCl] complex in the literature, where the signal was detected at 597 ppm.22 An additional and even more interesting aspect is the difference in the chemical shifts observed for Ag-4a (chloride) and Ag-4b (iodide). Compared with Ag-4a, the signal for compound Ag-4b is strongly shifted towards low field and it appears at 709.7 ppm. To the best of our knowledge, this is the first report of a 109Ag-NMR signal for a [(NHC)AgI] complex. Although, in the literature there are some examples of 109Ag-NMR for silver biscarbenes, [(NHC)2Ag]+, where the signals appear at 642 ppm (ref. 23) and at 727 ppm,24 the latter for a complex with ferrocenyl side chains. The data obtained with both 13C-NMR and 109Ag-NMR overall confirm significant differences in the electronic environments of the silver ions in [(NHC)AgCl] vs. [(NHC)AgI] complexes.
Conductivity measurements can be used to further distinguish between neutral and charged complexes in solution. Here we measured the conductivity of the silver NHC complexes in DMSO in comparison to the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 electrolyte AgNO3 after different periods up to 48 h (see Table 1). All silver NHC complexes generally showed much lower conductivity than AgNO3 (S mol−1 cm2 values of 2.15 to 9.70 for the silver NHC complexes vs. 38 for AgNO3), which again supports that the organometallics exist predominantly in the neutral monocarbene form in DMSO solution. The values remained very stable over 48 h with only marginal increases in some cases, confirming that the equilibrium was formed immediately after dissolution and remained unaltered over the course of the experiment.
1 electrolyte AgNO3 after different periods up to 48 h (see Table 1). All silver NHC complexes generally showed much lower conductivity than AgNO3 (S mol−1 cm2 values of 2.15 to 9.70 for the silver NHC complexes vs. 38 for AgNO3), which again supports that the organometallics exist predominantly in the neutral monocarbene form in DMSO solution. The values remained very stable over 48 h with only marginal increases in some cases, confirming that the equilibrium was formed immediately after dissolution and remained unaltered over the course of the experiment.
| Start | 24 h | 48 h | |
|---|---|---|---|
| AgNO3 | 38 | 38 | 38 |
| SBC3 | 2.15 | 2.25 | 2.31 |
| Ag-1a | 3.00 | 3.09 | 3.31 |
| Ag-1b | 8.44 | 9.31 | 9.70 |
| Ag-2a | 2.94 | 3.13 | 3.37 |
| Ag-2b | 9.19 | 9.25 | 9.82 |
| Ag-3a | 2.34 | 2.70 | 2.67 |
| Ag-3b | 6.67 | 7.00 | 6.69 |
| Ag-4a | 2.15 | 2.25 | 2.40 |
| Ag-4b | 6.72 | 6.92 | 7.32 |
Complexes Ag-3a (chlorido), Ag-3b (iodido) and SBC3 (acetato) were selected as examples with different secondary ligands for conductometry experiments in diluted solutions (see ESI† Fig. S2). In these experiments the molecular conductivity values of Ag-3b only slightly increased upon decreasing the concentrations. In contrast, the values for Ag-3a and SBC3 showed roughly a 2–4 fold increase upon 10-fold dilution, indicating that for the complexes with chloride or acetate ligands the contribution of the charged (biscarbene) form could be increased at lower concentrations.
Enzymatic inhibition studies with SARS-CoV-2 PLpro and 3CLpro
The inhibition of SARS-CoV-2 PLpro and 3CLpro as two relevant SARS-CoV-2 proteases was studied by enzymatic FRET assays and IC50 values were determined for the active inhibitors (see Table 2). Complexes Ag-1a/b–Ag-4a/b, SBC3 and silver nitrate were evaluated in concentrations up to 50 μM. Disulfiram (PLpro) and GC376 (3CLpro) served as positive reference inhibitors of the enzymes.| 3CLpro (SARS-CoV-2) | PLpro (SARS-CoV-2) | PLpro (SARS-CoV) | |
|---|---|---|---|
| Disulfiram | n.d. | 1.05 ± 0.31 | 5.52 ± 0.10 |
| GC376 | 0.90 ± 0.20 | n.d. | n.d. |
| 4 | n.d. | 105 ± 2 | 103 ± 2 |
| AgNO3 | >50 (97 ± 6%) | 0.70 ± 0.27 | 12.05 ± 0.87 |
| SBC3 | >50 (70 ± 4%) | 0.26 ± 0.03 | n.d. |
| Ag-1a | >50 (106 ± 2%) | 0.21 ± 0.03 | n.d. |
| Ag-1b | >50 (110 ± 0%) | 0.24 ± 0.11 | n.d. |
| Ag-2a | >50 (98 ± 15%) | 0.18 ± 0.02 | n.d. |
| Ag-2b | >50 (115 ± 4%) | 0.43 ± 0.06 | n.d. |
| Ag-3a | >50 (116 ± 5) | 0.21 ± 0.02 | n.d. |
| Ag-3b | >50 (111 ± 14%) | 0.19 ± 0.04 | n.d. |
| Ag-4a | >50 (115 ± 4%) | 0.22 ± 0.11 | n.d. |
| Ag-4b | >50 (116 ± 2%) | 0.39 ± 0.07 | 13.71 ± 1.02 |
Against 3CLpro, none of the silver complexes reached more than 50% enzyme inhibition at the highest applied dosage of 50 μM. At this comparably high concentration only SBC3 showed some moderate inhibition of the enzymatic activity, while all other silver NHC complexes were completely inactive.
All silver NHC complexes were potent inhibitors of SARS-CoV-2 PLpro with IC50 values in a rather narrow range of 0.2–0.4 μM, which is in excellent agreement with the results obtained with the silver NHC complex of our previous report.8 Whereas structure–activity-relationships regarding the NHC or the halide ligands could not be identified, it should be noted that all silver NHC complexes exceeded the activity of the reference compounds silver nitrate and disulfiram as SARS-CoV-2 PLpro inhibitors.
Complex Ag-4b was selected for additional mechanistic studies on the nature of the inhibition of PLpro. Initially, the related metal free (benz)imidazolium iodide 4 was exemplarily tested as reference and was inactive (IC50 value >100 μM), which confirmed that the silver ions are essential for enzymatic inhibition. Next, complex Ag-4b was studied as inhibitor of SARS-CoV PLpro and showed an IC50 value of 13.71 μM. Taken together with the inactivity against 3CLpro, it can be concluded that the silver NHC complexes are selective inhibitors of PLpro from SARS-CoV-2.
Time dependent studies on the inhibition of SARS-CoV-2 PLpro by Ag-4b were done at a fixed concentration of 0.75 μM (see Fig. S3†) and showed that the enzymatic turnover was reduced to approx. 60% of the untreated control immediately at the beginning of the experiment up to 30 min of exposure. After 60 min the activity was decreased to approx. 40%. These data clearly indicate that the enzymatic inhibition occurs immediately with some increase over extended exposure.
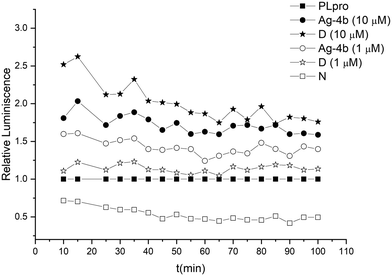
SARS-CoV-2 PLpro harbours a zinc binding domain that is responsible for the structural integrity of the enzyme and where the metal is coordinated by cysteine residues.25 In our previous paper we had reported on the Zn ejection from PLpro by gold complexes using an assay that detects free Zn ions with a fluorescent sensor molecule.11,26 As Zn replacement from cysteines is also feasible for the soft Lewis acidic silver compounds, we investigated complex Ag-4b for Zn ejection properties in the same assay with the Zn-ejector disulfiram as a reference (see Fig. 3). In fact, the complex was effective in removing Zn from SARS-CoV-2 PLpro with a similar efficacy as disulfiram (low efficacy at 1.0 μM and high efficacy at 10 μM).
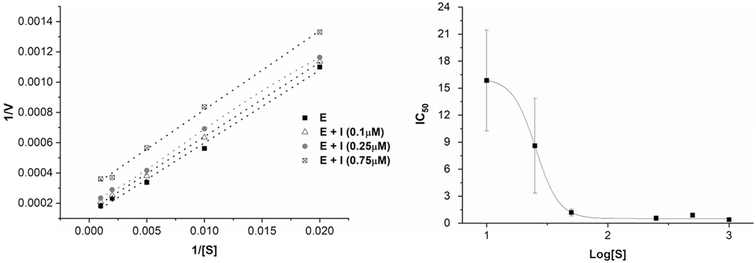
An enzyme kinetic analysis was performed exemplarily for Ag-4b. With Km values between 411 and 176 μM (for Ag-4b = 0, 0.1, 0.25, 0.75 μM, respectively) that are much higher than the Ki value (0.48 ± 0.22 μM), as well as the parallel lines in the Lineweaver–Burk representation (see Fig. 4, left), a clear pattern of an uncompetitive enzyme inhibitor was obtained. As such Ag-4b interacts only with the enzyme-substrate complex, but not with the enzyme alone.27 Accordingly, increasing substrate concentrations will lead to a “paradox” decrease of IC50 values. This was further evaluated for Ag-4b and in fact the IC50 values at low substrate levels were much higher than those at high levels (see Fig. 4, right). A more detailed analysis of the Michaelis–Menten enzyme kinetics of Ag-4b is presented in the ESI.†
Cytotoxicity evaluation against CaCo-2 cell layers
Toxicity of test compounds against host cells can lead to false positive results in cell-based antiviral assays. In order to exclude toxic compounds and to evaluate in which concentration range the complexes could be applied in subsequent antiviral assays, almost confluent cell layers of CaCo-2 cells were grown and challenged for 24 h or 48 h with the complexes in concentrations between 25 μM and 500 μM. Fig. 5 exemplarily shows the effect of 100 μM of the test compounds against Caco-2 cells. At this concentration the iodido silver NHC complexes Ag-1b to Ag-4b are not cytotoxic, whereas all other complexes showed strong effects against the cells. Results with all other applied concentrations are given in the supporting info (see Fig. S6†). Notably, complex Ag-4b remained non-toxic up to 500 μM in these experiments.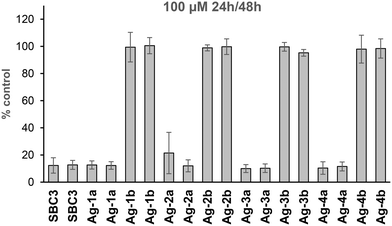 | ||
| Fig. 5 Cytotoxicity of 100 μM of silver NHC complexes against almost confluent cell layers of Caco-2 cells (as % control of untreated cells). Left bars: 24 h, right bars: 48 h. | ||
Antiviral activity against SARS-CoV-2
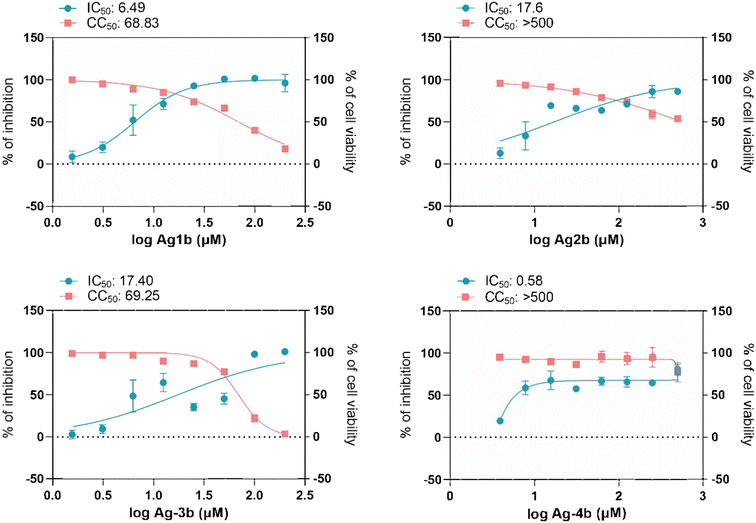
Based on their favourable low or absent host cell cytotoxicity complexes Ag-1b to Ag-4b were selected for further evaluation of antiviral activity against authentic SARS-CoV-2 infection in Caco-2-F03 cells. The complexes were added simultaneously with the virus and the infection rate was determined by staining of spike protein after 24 h (see Fig. 6). All silver NHC complexes were effective antiviral agents that suppressed the infection at least by 50% starting at low micromolar non-toxic concentrations of roughly 0.5–20 μM. Only complexes Ag-1b and Ag-3b completely terminated viral activity at higher dosages, which however overlapped with toxicity in a parallel toxicity counter screen. Missing toxicity was again noted for Ag-4b and Ag-2b was moderately toxic only at very high dosage (CC50 values for both complexes >500 μM). Ag-1b to Ag-3b (IC50 values 6–18 μM) were effective antiviral agents and the by far most active compound was Ag-4b with an IC50 value of 0.58 μM.Discussion and conclusions
Silver NHC complexes were prepared and evaluated as antiviral metallodrug candidates against SARS-CoV-2. Enzymatic inhibition studies confirmed a strong and selective inhibition of SARS-CoV-2 PLpro with IC50 values below 0.5 μM. A rough comparison with IC50 values reported for different types of SARS-CoV-2 PLpro inhibitors shows that the silver NHC complexes of this report are among the most effective inhibitors.28 In contrast, their activity against SARS-CoV PLpro was low and the complexes were inactive in experiments with SARS-CoV-2 3CLpro, confirming that the complexes display a preference/selectivity for PLpro from SARS-CoV-2 over other related proteases.In our previous reports we could confirm activity against SARS-CoV-2 PLpro also for gold and rhodium complexes as well as polyoxometalates.8,11 Strong inhibition of this enzyme has also been reported for bismuth compounds10 or zink pyrithione.29 Inhibition of different SARS-CoV-2 enzymatic targets has been demonstrated for a small number of metal complexes, with helicase for bismuth9,10 compounds, the spike/ACE2 interaction for titanocene dichloride and polyoxometalates,8 3CLpro (Mpro) for bismuth,10 rhenium,30 zinc,31 gold,32 mercury33 and ferrocene34 complexes, and nsp14 N7-methyltansferase for gold and mercury complexes.35
PLpro is a cysteine protease and in addition it hosts a zinc binding domain, in which the zinc is coordinated by four cysteine residues. Therefore, targeting the catalytic cysteine or the cysteines in the zinc binding domain is a promising strategy to inhibit the activity of this enzyme as it has been demonstrated with gold and other thiol-reactive probes, including disulfiram and ebselen.11,25,26 Silver as a soft Lewis acid has a high affinity to soft bases such as thiolates. Zinc ejection experiments performed exemplarily with Ag-4b in fact confirmed efficacy comparable to that of disulfiram. Metal displacement has been suggested as a promising antiviral strategy and has been studied in detail for bismuth compounds with the enzyme helicase.6,9,10 In general, zinc finger domains are considered as therapeutic targets for metal based compounds.36 Also for silver compounds, although little explored in this aspect, zinc replacement from zinc finger binding sites has been reported.37 Interestingly, Sun and colleagues recently confirmed zinc replacement from the colistin-resistance protein mcr-1 by silver ions in connection with the re-sensitisation of resistant bacteria to colistin.38 It can therefore be speculated that zinc replacement could be a general mechanism of high relevance for silver-based anti-infectives.
Kinetic studies proved that Ag-4b exhibits an uncompetitive inhibition mechanism. This comparably rare mechanism of enzymatic inhibition explains that the inhibitor does not coordinate with the active centre of the enzyme, interacts with a different binding site, and binds only when the substrate has bound. Such mechanism is therefore also supportive of Ag-4b and other silver NHC complexes binding to the Zn binding domain of SARS-CoV-2 PLpro. It is important to note that, as part of the uncompetitive binding mechanism, the affinity of Ag-4b for the preformed enzyme–substrate complex (Ki = 0.48 ± 0.22 μM) was approx. 4 orders of magnitude higher than the affinity for the enzyme before formation of the enzyme–substrate complex (Km values: 411–176 μM). The development of inhibitors that show an uncompetitive mechanism brings significant pharmacological advantages. Due to fact that the inhibitor interacts only with the enzyme–substrate complex, it is only active when the enzyme is coordinated to the substrate, which provides high specificity and selectivity. The uncompetitive binding mechanism confirmed for Ag-4b might also explain the selectivity for PLpro from SARS-CoV-2 over that from SARS-CoV (see Table 2). Similar selectivity had also been observed with other silver complexes in our previous report and it could therefore be speculated that uncompetitive PLpro inhibition could be a general feature of silver complexes.8
A preliminary toxicity screening showed very low to absent toxicity for the iodido complexes Ag-1b–Ag-4b, while the chlorido analogues Ag-1a–Ag-4a were quite cytotoxic. This could be attributed to the higher stability (lower reactivity) of the iodido silver NHC complexes compared with the chlorido analogues.19 Based on their favourable toxicity profile, Ag-1b–Ag-4b were selected for antiviral studies in SARS-CoV-2 infected cells. All complexes effectively suppressed SARS-CoV-2 viral replication at non-toxic low micromolar dosages with Ag-4b as the most active and completely non-toxic example.
Taken together, silver NHC complexes represent a promising novel type of SARS-CoV-2 PLpro inhibitors with a rare uncompetitive binding mechanism and efficient antiviral efficacy at non-toxic concentration.
Experimental
General
Unless noted otherwise, all reagents were obtained from Sigma-Aldrich, VWR and Fluorochem and used as supplied. The purity of the synthesized compounds was proven by elemental analysis (Flash EA 1112, Thermo Quest) and differed by less than 0.5% from the predicted values. NMR spectra were recorded on a Bruker DRX-400 AS, AV III HD 500 or a AVII 600 NMR spectrometer; positive-ion ESI (electrospray ionization) mass spectra were recorded a expressionL CMS spectrometer (Advion). Complexes SBC3 (ref. 15) and Ag-1a (ref. 39) have been reported previously. Compounds 1,3-diethylbenzimidazolium iodide (1),40 1,3-di(isopropyl)benzimidazolium iodide (3),41 1,3-diethylbenzimidazolium chloride (1a)42 and 1,3-di(isopropyl)benzimidazolium chloride (3a)43 were reported in the literature already.109Ag-NMR spectroscopy
109Ag-NMR spectra were recorded on a AV 300 MHz spectrometer and chemical shifts were referenced with respect to an external solution of 4.00 M silver nitrate in deuterium oxide (defined as δ 0 ppm). 100 mg (Ag-4a) or 120 mg (Ag-4b) were dissolved in 600 μL of d6-DMSO. The AV 300 spectrometer was equipped with a PABBO BB-1H/DZ-GRD Z828401/0054, pulse sequence zg30, receiver gain 203.0, relaxation delay 20 s.Synthesis
General procedure for synthesis of NHC–Ag–X complexes
A flask was charged with the corresponding (benz)imidazolium precursor and silver oxide (0.6 equivalents) and stirred in dried dichloromethane for 2 h (for Ag-1b, Ag-2a, Ag-2b, Ag-3b) or 18 h (for Ag-1a, Ag-3a, Ag-4a, Ag-4b) at 40 °C in the dark. The reaction mixture was filtered through a bed of celite, the filtrate was reduced to 5 mL and 50 mL of pentane were added to precipitate a solid. The solid was isolated by filtration under reduced pressure and dried.Conductometry
Conductivity measurements were performed using an Apera Instruments EC9500 conductivity meter. Solutions (1.0 mM, or as indicated) of all compounds were prepared using freshly distilled DMSO with a relative electric conductivity of ≤3 × 10−8 Ω cm−1. The conductivities were measured at 25 ± 2 °C immediately after complete dissolution, after 24 h, and after 48 h.SARS-CoV-2 and SARS-CoV PLpro inhibition
The inhibition of PLpro was determined as previously described.11 The inhibitor compounds were prepared as stock solutions in DMSO and diluted hundredfold with HEPES buffer (50 mm HEPES, pH 7.5, 0.1 mg mL−1 bovine serum albumin, 0.1% Triton-X100) to micromolar concentrations. Volumes of 50 μL of 200 nM SARS-CoV-2 PLpro (Elabscience)/His6-SARS-CoV-1 PLpro (South Bay Bio) in HEPES buffer or blank HEPES buffer (negative control) were added to the wells of a black 96-well microtiter plate (Greiner Bio One). Volumes of 50 μL of the inhibitor solutions or 1% DMSO in HEPES buffer (positive control) were added. The resulting solutions (100 nm SARS-CoV-2 PLpro/His6-SARS-CoV-1 PLpro, 0.5% DMSO, 0.1–100 μm test compound or blank HEPES buffer) were mixed and incubated at 37 °C for one hour. A volume of 100 μL of 100 mm Z-Arg-Leu-Arg-Gly-Gly-AMC (Bachem) was added to all wells. The resulting solutions were mixed and the fluorescence emission was measured immediately every 30 s for 10 min (λexc = 355 nm; λem = 460 nm) at 37 μC using a VictorTM X4 Perkin Elmer 2030 multilabel reader. The increase of emission over time followed a linear trend (r2 >0.97) and the enzymatic activities were calculated as the slope thereof. The IC50 values were calculated as the concentration of the inhibitor that was required to decrease the enzymatic activity to 50% of the positive control. The wells containing the negative control were used to confirm the absence of false positive results by reaction of the inhibitor compound with the fluorogenic substrate.Zn-Ejection assay SARS-CoV-2 PLpro inhibition
To determine if the inhibitors are Zn-ejecting agents, the presence of free Zn2+ in solution was measured as previously described.11 The inhibitor compounds were prepared as stock solutions in DMSO and diluted hundredfold with HEPES buffer (50 mM HEPES, pH 7.5) to 2 μM and 20 μM concentrations. Volumes of 50 μL of 1 μM SARS-CoV-2 PLpro (Elabscience) in HEPES buffer or blank HEPES buffer (negative control) were added to the wells of a black 96-well microtiter plate (Greiner Bio One). Volumes of 50 μL of the inhibitor solutions or 1% DMSO in HEPES buffer (positive control) were added. The resulting solutions (500 nM PLpro SARS-CoV-2, 0.5% DMSO, 1 μM or 10 μM test compound or blank HEPES buffer) were mixed. A volume of 100 μL of 2.0 μM of zinc-specific fluorophore FluoZinTM-3 (Invitrogen/Life Technologies) was added to all wells. The resulting solutions were mixed and the fluorescence emission was measured immediately every 5 min for 100 min (λexc = 485 nm; λem = 535 nm) at 37 °C using a Victor™ X4 Pekin Elmer 2030 multilabel reader. The wells containing the negative control were used to confirm the absence of false positive results by reaction of the inhibitor compound with the zinc-specific fluorophore.Michaelis–Menten kinetics with SARS-CoV-2 PLpro
The inhibitor compounds were prepared as stock solutions in DMSO and diluted hundredfold with HEPES buffer (50 mm HEPES, pH 7.5, 0.1 mg mL−1 bovine serum albumin, 0.1% Triton-X100) to micromolar concentrations. Volumes of 50 μL of 200 nM SARS-CoV-2 PLpro (Elabscience) in HEPES buffer were added to the wells of a black 96-well microtiter plate (Greiner Bio One). Volumes of 50 μL of the inhibitor solutions or 1% DMSO in HEPES buffer (positive control) were added. The resulting solutions (100 nm SARS-CoV-2 PLpro 0.5% DMSO, 0.1–0.75 μm test compound or blank HEPES buffer) were mixed and incubated at 37 °C for 10 min. A volume of 100 μL of 20–8000 μm of substrate (Z-Arg-Leu-Arg-Gly-Gly-AMC, Bachem) was added to all wells. The resulting solutions were mixed and the fluorescence emission was measured immediately every minute for 1 h (λexc = 355 nm; λem = 460 nm) at 37 °C using a Victor™ X4 Perkin Elmer 2030 multilabel reader. The enzyme activity of PLpro was represented by the Michaelis Menten equation (eqn (1)) and the Km and Vmax values were calculated with Lineweaver–Burk equation (eqn (2)) where the slope is Km/Vmax, the intersection with the Y-axis corresponds to 1/Vmax, and intersection with the X-axis corresponds to −1/Km.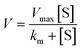 | (1) |
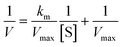 | (2) |
SARS-CoV-2 3CLpro inhibition
The inhibition of 3CLpro was determined according to reported protocols with modifications.44 The inhibitor compounds were prepared as stock solutions in DMSO and diluted hundredfold with HEPES buffer (50 mM HEPES, pH 7.5, 0.1 mg mL−1 bovine serum albumin, 0.1% Triton-X100, 0.1 mM DTT) to micromolar concentrations. Volumes of 50 μL of 200 nM MBP-tag SARS-CoV-2 3CLpro (Bioscience) in HEPES buffer or blank HEPES buffer (negative control) were added to the wells of a black 96-well microtiter plate (Greiner Bio One). Volumes of 50 μL of the inhibitor solutions or 1% DMSO in HEPES buffer (positive control) were added. The resulting solutions (100 nM SARS-CoV-2 3CLpro, 0.5% DMSO, 100 μm test compound or blank HEPES buffer) were mixed and incubated at 37 °C for one hour. A volume of 100 μL of 100 μM DABCYL-KTSAVLQSGFRKME-EDANS (Bachem) was added to all wells. The resulting solutions were mixed and the fluorescence emission was measured immediately every 3 min for 75 min (λexc = 355 nm; λem = 460 nm) at 37 °C using a Victor™ X4 Perkin Elmer 2030 multilabel reader. The increase of emission over time followed a linear trend (r2 >0.97) and the enzymatic activities were calculated as the slope thereof. The wells containing the negative control were used to confirm the absence of false positive results by reaction of the inhibitor compound with the fluorogenic substrate.Toxicity against host cells (CaCo-2)
Caco-2 cells were grown as almost confluent monolayers in 96 well plates. The complexes were dissolved as stock solutions in DMSO (0.2%) diluted with DMEM cell culture medium, which was supplemented with 1% fetal calf serum and 50 mg L−1 gentamicin. The cell layers were incubated with the drug containing media for 24 h and 48 h at 37 °C/5% CO2 in an incubator. The cell viability was determined by crystal violet staining and cell viability was calculated as percentage of an untreated control. Results were obtained in three independent experiments.Antiviral effects against SARS-CoV-2
Virus isolation and preparation
SARS-CoV-2 B.1 (SARS-CoV-2/FFM7, MT358643) was isolated from naso-pharyngeal swabs of COVID-19 patients using subline Caco-2-F03. SARS-CoV-2 stocks used in the experiments had undergone two passages and were stored at −80 °C. Virus titers were determined as TCID50 per mL in confluent cells in 96-well microtiter plates. The isolate was authenticated and the sequence is publicly available at https://GISAID.org.Antiviral activity of silver compounds
Confluent layers of Caco-2-F03 were infected with SARS-CoV-2 B.1 at a MOI of 0.01. The tested compounds were added simultaneously with virus and incubated in MEM supplemented with 1% FBS. Immunohistochemistry of viral spike protein was performed after 24 h post infection to determine antiviral activity against SARS-CoV-2 infection.Immunohistochemistry
Cells were fixed with acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (40
methanol (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) solution and immunostaining was performed using a monoclonal antibody directed against the S protein of SARS-CoV-2 (1
60) solution and immunostaining was performed using a monoclonal antibody directed against the S protein of SARS-CoV-2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1500, Sinobiological), which was detected with a peroxidase-conjugated anti-rabbit secondary antibody (1
1500, Sinobiological), which was detected with a peroxidase-conjugated anti-rabbit secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, Dianova), followed by addition of AEC substrate. The S positive area was scanned and quantified by automated plate reader. The results were expressed as percentage of infection relative to virus control which received no drug.
1000, Dianova), followed by addition of AEC substrate. The S positive area was scanned and quantified by automated plate reader. The results were expressed as percentage of infection relative to virus control which received no drug.
Cell viability assay
Cell viability was measured by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) dye reduction assay. 25 μL of MTT solution (2 mg mL−1 in PBS) were added per well, and the plates were incubated at 37 °C for 4 h. After this, the cells were lysed using 100 μL of a buffer containing 20% sodium dodecylsulfate and 50% N,N-dimethylformamide with the pH adjusted to 4.7 at 37 °C for 4 h. Absorbance was determined at 560 nm (reference wavelength 620 nm) using a Tecan infinite M200 microplate reader (TECAN).Conflicts of interest
There is no conflict of interest to declare.Acknowledgements
M. G.-M. acknowledges support by a Margarita-Salas-Fellowship (European Union, NextGenerationEU), I. V. E. acknowledges support by a Georg-Christoph-Lichtenberg-Fellowship by the Ministry of Science and Culture of the State of Lower-Saxony within the graduate program iCA (“Drug Discovery and Cheminformatics for New Anti-Infectives”). We thank Kerstin Euler, Sebastian Grothe and Lena Stegmann for their technical support.References
- (a) I. Ott and R. Gust, Arch. Pharm., 2007, 340, 117 CrossRef CAS PubMed; (b) G. Gasser, I. Ott and N. Metzler-Nolte, J. Med. Chem., 2011, 54, 3–25 CrossRef CAS.
- K. D. Mjos and C. Orvig, Chem. Rev., 2014, 114, 4540–4563 CrossRef CAS.
- A. Frei, J. Zuegg, A. G. Elliott, M. Baker, S. Braese, C. Brown, F. Chen, C. G. Dowson, G. Dujardin, N. Jung, A. P. King, A. M. Mansour, M. Massi, J. Moat, H. A. Mohamed, A. K. Renfrew, P. J. Rutledge, P. J. Sadler, M. H. Todd, C. E. Willans, J. J. Wilson, M. A. Cooper and M. A. T. Blaskovich, Chem. Sci., 2020, 11, 2627–2639 RSC.
- R. E. F. de Paiva, A. Marçal Neto, I. A. Santos, A. C. G. Jardim, P. P. Corbi and F. R. G. Bergamini, Dalton Trans., 2020, 49, 16004–16033 RSC.
- (a) T. Marzo and L. Messori, ACS Med. Chem. Lett., 2020, 11, 1067–1068 CrossRef CAS PubMed; (b) J. Karges and S. M. Cohen, ChemBioChem, 2021, 2600–2607 CrossRef CAS PubMed; (c) K. Ioannou and M. C. Vlasiou, BioMetals, 2022, 35(4), 639–652 CrossRef CAS PubMed.
- H. Li, S. Yuan, X. Wei and H. Sun, Chem. Commun., 2022, 58, 7466 RSC.
- (a) H. A. Rothan, S. Stone, J. Natekar, P. Kumari, K. Arora and M. Kumar, Virology, 2020, 547, 7 CrossRef CAS PubMed; (b) D. Cirri, T. Marzo, I. Tolbatov, A. Marrone, F. Saladini, I. Vicenti, F. Dragoni, A. Boccuto and L. Messori, Biomolecules, 2021, 11, 1858 CrossRef CAS PubMed; (c) C. Chuong, C. M. DuChane, E. M. Webb, P. Rai, J. M. Marano, C. M. Bernier, J. S. Merola and J. Weger-Lucarelli, Viruses, 2021, 13, 980 CrossRef CAS PubMed.
- M. Gil-Moles, S. Türck, U. Basu, A. Pettenuzzo, S. Bhattacharya, A. Rajan, X. Ma, R. Büssing, J. Wölker, H. Burmeister, H. Hoffmeister, P. Schneeberg, A. Prause, P. Lippmann, J. Kusi-Nimarko, S. Hassell-Hart, A. McGown, D. Guest, Y. Lin, A. Notaro, R. Vinck, J. Karges, K. Cariou, K. Peng, X. Qin, X. Wang, J. Skiba, Ł. Szczupak, K. Kowalski, U. Schatzschneider, C. Hemmert, H. Gornitzka, E. R. Milaeva, A. A. Nazarov, G. Gasser, J. Spencer, L. Ronconi, U. Kortz, J. Cinatl, D. Bojkova and I. Ott, Chem. – Eur. J., 2021, 27, 17928–17940 CrossRef CAS PubMed.
- S. Yuan, R. Wang, J. F.-W. Chan, A. J. Zhang, T. Cheng, K. K.-H. Chik, Z.-W. Ye, S. Wang, A. C.-Y. Lee, L. Jin, H. Li, D.-Y. Jin, K.-Y. Yuen and H. Sun, Nat. Microbiol., 2020, 5, 1439–1448 CrossRef CAS.
- R. Wang, J. F.-W. Chan, S. Wang, H. Li, J. Zhao, T. K.-Y. Ip, Z. Zuo, K.-Y. Yuen, S. Yuan and H. Sun, Chem. Sci., 2022, 13, 2238–2248 RSC.
- M. Gil-Moles, U. Basu, R. Büssing, H. Hoffmeister, S. Türck, A. Varchmin and I. Ott, Chem. – Eur. J., 2020, 26, 15140–15144 CrossRef CAS PubMed.
- (a) C. Ma, M. D. Sacco, Z. Xia, G. Lambrinidis, J. A. Townsend, Y. Hu, X. Meng, T. Szeto, M. Ba, X. Zhang, M. Gongora, F. Zhang, M. T. Marty, Y. Xiang, A. Kolocouris, Y. Chen and J. Wang, ACS Cent. Sci., 2021, 7, 1245–1260 CrossRef CAS PubMed; (b) Z. Shen, K. Ratia, L. Cooper, D. Kong, H. Lee, Y. Kwon, Y. Li, S. Alqarni, F. Huang, O. Dubrovskyi, L. Rong, G. R. J. Thatcher and R. Xiong, J. Med. Chem., 2022, 65, 2940–2955 CrossRef CAS.
- (a) A. Melaiye, R. S. Simons, A. Milsted, F. Pingitore, C. Wesdemiotis, C. A. Tessier and W. J. Youngs, J. Med. Chem., 2004, 47, 973–977 CrossRef CAS PubMed; (b) K. M. Hindi, M. J. Panzner, C. A. Tessier, C. L. Cannon and W. J. Youngs, Chem. Rev., 2009, 109, 3859 CrossRef CAS PubMed.
- (a) C. L. Cannon, L. A. Hogue, R. K. Vajravelu, G. H. Capps, A. Ibricevic, K. M. Hindi, A. Kascatan-Nebioglu, M. J. Walter, S. L. Brody and W. J. Youngs, Antimicrob. Agents Chemother., 2009, 53, 3285–3293 CrossRef CAS PubMed; (b) K. M. Hindi, A. J. Ditto, M. J. Panzner, D. A. Medvetz, D. S. Han, C. E. Hovis, J. K. Hilliard, J. B. Taylor, Y. H. Yun, C. L. Cannon and W. J. Youngs, Biomaterials, 2009, 30, 3771 CrossRef CAS; (c) N. A. Johnson, M. R. Southerland and W. J. Youngs, Molecules, 2017, 22, 1263 CrossRef PubMed; (d) J. O'Loughlin, S. Napolitano, F. Alkhathami, C. O'Beirne, D. Marhöfer, M. O'Shaughnessy, O. Howe, M. Tacke and M. Rubini, ChemBioChem, 2021, 22, 1093–1098 CrossRef; (e) W. Streciwilk, J. Cassidy, F. Hackenberg, H. Müller-Bunz, F. Paradisi and M. Tacke, J. Organomet. Chem., 2014, 749, 88 CrossRef CAS; (f) S. A. Patil, S. A. Patil, R. Patil, R. S. Keri, S. Budagumpi, G. R. Balakrishna and M. Tacke, Future Med. Chem., 2015, 7, 1305–1333 CrossRef CAS PubMed; (g) C. O'Beirne, H. T. Althani, O. Dada, J. Cassidy, K. Kavanagh, H. Müller-Bunz, Y. Ortin, X. Zhu and M. Tacke, Polyhedron, 2018, 149, 95–103 CrossRef.
- S. Patil, A. Deally, B. Gleeson, H. Müller-Bunz, F. Paradisi and M. Tacke, Metallomics, 2011, 3, 74–88 CrossRef CAS PubMed.
- O. Sánchez, S. González, Á. R. Higuera-Padilla, Y. León, D. Coll, M. Fernández, P. Taylor, I. Urdanibia, H. R. Rangel, J. T. Ortega, W. Castro and M. C. Goite, Polyhedron, 2016, 110, 14–23 CrossRef.
- H. M. J. Wang and I. J. B. Lin, Organometallics, 1998, 17, 972–975 CrossRef CAS.
- (a) M. V. Baker, P. J. Barnard, S. K. Brayshaw, J. L. Hickey, B. W. Skelton and A. H. White, Dalton Trans., 2005, 37–43 RSC; (b) H. V. Huynh, Y. Han, R. Jothibasu and J. A. Yang, Organometallics, 2009, 28, 5395–5404 CrossRef CAS.
- J. C. Garrison and W. J. Youngs, Chem. Rev., 2005, 105, 3978–4008 CrossRef CAS.
- (a) L. Busetto, M. C. Cassani, C. Femoni, A. Macchioni, R. Mazzoni and D. Zuccaccia, J. Organomet. Chem., 2008, 693, 2579–2591 CrossRef CAS; (b) P. de Frémont, N. M. Scott, E. D. Stevens, T. Ramnial, O. C. Lightbody, C. L. B. Macdonald, J. A. C. Clyburne, C. D. Abernethy and S. P. Nolan, Organometallics, 2005, 24, 6301–6309 CrossRef.
- U. Hintermair, U. Englert and W. Leitner, Organometallics, 2011, 30, 3726–3731 CrossRef CAS.
- T. Ramnial, C. D. Abernethy, M. D. Spicer, I. D. McKenzie, I. D. Gay and J. A. C. Clyburne, Inorg. Chem., 2003, 42, 1391–1393 CrossRef CAS.
- A. J. Arduengo, H. V. R. Dias, J. C. Calabrese and F. Davidson, Organometallics, 1993, 12, 3405–3409 CrossRef CAS.
- B. Bildstein, M. Malaun, H. Kopacka, K. Wurst, M. Mitterböck, K.-H. Ongania, G. Opromolla and P. Zanello, Organometallics, 1999, 18, 4325–4336 CrossRef CAS.
- B. K. Maiti, ACS Pharmacol. Transl. Sci., 2020, 3, 1017–1019 CrossRef CAS.
- K. Sargsyan, C.-C. Lin, T. Chen, C. Grauffel, Y.-P. Chen, W.-Z. Yang, H. S. Yuan and C. Lim, Chem. Sci., 2020, 11, 9904–9909 RSC.
- T. P. Kenakin, Enzymes as Drug Targets, Pharmacology in Drug Discovery, ed. T. P. Kenakin, Academic Press, 2012, ch. 6, pp. 105–124 Search PubMed.
- H. Tan, Y. Hu, P. Jadhav, B. Tan and J. Wang, J. Med. Chem., 2022, 65, 7561–7580 CrossRef CAS PubMed.
- J. Kladnik, A. Dolinar, J. Kljun, D. Perea, J. Grau-Expósito, M. Genescà, M. Novinec, M. J. Buzon and I. Turel, J. Enzyme Inhib. Med. Chem., 2022, 37, 2158–2168 CrossRef CAS.
- (a) J. Karges, M. Kalaj, M. Bembicky and S. M. Cohen, Angew. Chem., Int. Ed., 2021, 60, 10716–10723 CrossRef CAS; (b) J. Karges and S. M. Cohen, J. Med. Chem., 2023, 66, 3088–3105 CrossRef CAS; (c) J. Karges, M. A. Giardini, O. Blacque, B. Woodworth, J. L. Siqueira-Neto and S. M. Cohen, Chem. Sci., 2023, 14, 711–720 RSC.
- (a) L. Panchariya, W. A. Khan, S. Kuila, K. Sonkar, S. Sahoo, A. Ghoshal, A. Kumar, D. K. Verma, A. Hasan, M. A. Khan, N. Jain, A. K. Mohapatra, S. Das, J. K. Thakur, S. Maiti, R. K. Nanda, R. Halder, S. Sunil and A. Arockiasamy, Chem. Commun., 2021, 57, 10083 RSC; (b) X. Tao, L. Zhang, L. Du, K. Lu, Z. Zhao, Y. Xie, X. Li, S. Huang, P.-H. Wang, J.-A. Pan, W. Xia, J. Dai and Z.-W. Mao, J. Inorg. Biochem., 2022, 231, 111777 CrossRef CAS PubMed.
- J. D. Baker, R. L. Uhrich, G. C. Kraemer, J. E. Love and B. C. Kraemer, PLoS One, 2021, 16, e0245962 CrossRef CAS.
- (a) C. Coelho, G. Gallo, C. B. Campos, L. Hardy and M. Würtele, PLoS One, 2020, 15, e0240079 CrossRef CAS; (b) J. Chen, Y. Zhang, D. Zeng, B. Zhang, X. Ye, Z. Zeng, X.-K. Zhang, Z. Wang and H. Zhou, Biochem. Biophys. Res. Commun., 2022, 591, 118–123 CrossRef CAS.
- D. Graf, N. Farn, J. Klopf, M. Hojjati and U. Schatzschneider, Metallomics, 2023, 15, mfad023 CrossRef PubMed.
- R. Kasprzyk, T. J. Spiewla, M. Smietanski, S. Golojuch, L. Vangeel, S. de Jonghe, D. Jochmans, J. Neyts, J. Kowalska and J. Jemielity, Antiviral Res., 2021, 193, 105142 CrossRef CAS PubMed.
- C. Abbehausen, Metallomics, 2019, 11, 15–28 CrossRef CAS PubMed.
- (a) K. Kluska, M. D. Peris-Díaz, D. Płonka, A. Moysa, M. Dadlez, A. Deniaud, W. Bal and A. Krężel, Chem. Commun., 2020, 56, 1329–1332 RSC; (b) K. Kluska, G. Veronesi, A. Deniaud, B. Hajdu, B. Gyurcsik, W. Bal and A. Krężel, Angew. Chem., Int. Ed., 2022, 61, e202116621 CrossRef CAS PubMed; (c) G. Park, Z. N. Amaris, M. K. Eiken, K. V. Baumgartner, K. A. Johnston, M. A. Williams, J. G. Markwordt, J. E. Millstone, K. E. Splan and K. E. Wheeler, Environ. Sci.: Nano, 2019, 6, 2367–2378 RSC.
- Q. Zhang, R. Wang, M. Wang, C. Liu, M. Koohi-Moghadam, H. Wang, P.-L. Ho, H. Li and H. Sun, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2119417119 CrossRef CAS PubMed.
- T. Rehm, M. Rothemund, A. Bär, T. Dietel, R. Kempe, H. Kostrhunova, V. Brabec, J. Kasparkova and R. Schobert, Dalton Trans., 2018, 47, 17367–17381 RSC.
- H. M. J. Wang, C. Y. L. Chen and I. J. B. Lin, Organometallics, 1999, 18, 1216–1223 CrossRef CAS.
- Y. Han, H. V. Huynh and L. L. Koh, J. Organomet. Chem., 2007, 692, 3606–3613 CrossRef CAS.
- X. Ling, N. Schaeffer, S. Roland and M.-P. Pileni, Langmuir, 2013, 29, 12647 CrossRef CAS PubMed.
- H. V. Huynh, T. T. Lam and H. T. T. Luong, RSC Adv., 2018, 8, 34960–34966 RSC.
- W. Zhu, M. Xu, C. Z. Chen, H. Guo, M. Shen, X. Hu, P. Shinn, C. Klumpp-Thomas, S. G. Michael and W. Zheng, ACS Pharmacol. Transl. Sci., 2020, 3, 1008–1016 CrossRef CAS PubMed.
- D. Bojkova, P. Reus, L. Panosch, M. Bechtel, T. Rothenburger, J. Kandler, A. Pfeiffer, J. U. Wagner, M. Shumliakivska, S. Dimmeler, R. Olmer, U. Martin, F. Vondran, T. Toptan, F. Rothweiler, R. Zehner, H. Rabenau, K. L. Osman, S. T. Pullan, M. Carroll, R. Stack, S. Ciesek, M. N. Wass, M. Michaelis and J. Cinatl, Identification of novel antiviral drug candidates using an optimized SARS-CoV-2 phenotypic screening platform, iScience, 2023, 26, 105944 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3md00067b |
| This journal is © The Royal Society of Chemistry 2023 |