Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Impact of nucleic acid encapsulated MOF crystal phase on protein corona formation†
Shakil Ahmed
Polash
 ab,
Suneela
Pyreddy
ab,
Amanda N
Abraham
ab,
Suneela
Pyreddy
ab,
Amanda N
Abraham
 ac,
Sanje
Mahasivam
ac,
Sanje
Mahasivam
 ab,
Vipul
Bansal
ab,
Vipul
Bansal
 ab,
Linda
Varadi
ab,
Linda
Varadi
 d,
Gary
Bryant
d,
Gary
Bryant
 a and
Ravi
Shukla
a and
Ravi
Shukla
 *abe
*abe
aSchool of Science, RMIT University, Melbourne, Victoria 3001, Australia. E-mail: ravi.shukla@rmit.edu.au
bSir Ian Potter NanoBiosensing Facility, NanoBiotechnology Research Laboratory (NBRL), RMIT University, Melbourne, Victoria 3001, Australia
cARC Centre of Excellence for Nanoscale BioPhotonics, RMIT University, Melbourne, Victoria 3001, Australia
dSchool of Engineering, RMIT University, Melbourne, Victoria 3001, Australia
eCentre for Advanced Materials and Industrial Chemistry, RMIT University, Melbourne, Victoria 3001, Australia
First published on 21st September 2023
Abstract
Metal–organic frameworks (MOFs) are among the most extensively studied materials for delivering a wide range of therapeutic entities including proteins, carbohydrates, and nucleic acids. Despite significant efforts, the effect of serum proteins on MOFs is still not well understood. The biological identity and fate of therapeutic biocomposites are altered in a biological environment due to the formation of a protein corona around nanomaterials. Although the crystalline property of MOFs controls their biological interactions, the influence of these properties on the MOF-protein interaction is not well understood. Herein, we study the mechanism of interaction of a nucleic acid-encapsulated Zn-based zeolitic imidazolate framework-8 (ZIF-8) and its carbonate-rich variant (ZIF-C) with human serum albumin (HSA). It is found that different crystal topologies of these ZIFs affect their interaction with serum albumin. The key finding is that ZIF-8 has the tendency to interact more strongly with serum proteins compared to ZIF-C. Considering that ZIF-8 tends to spontaneously transform into ZIF-C in an aqueous environment, these findings may have important implications in the rationale design of MOF-based therapeutic agents.
1. Introduction
Metal–organic frameworks (MOFs) are coordination polymers that are made of metal ions coordinated with organic ligands. Their unique physicochemical properties make them attractive candidates for delivering therapeutic biomolecules including proteins, carbohydrates, and nucleic acids.1 It is a distinct class of porous hybrid nanomaterials capable of bio-conjugating, infiltrating, and encapsulating biomolecules.2 Versatility in the choice of metals and linkers define the structure and properties of individual MOFs. Moreover, they are biodegradable and have a high loading and on-demand release of loaded therapeutics, which makes them a promising delivery agent for therapeutic biomolecules.3–5 Zeolitic imidazolate frameworks (ZIFs) are imidazolate-based MOFs where a tetrahedral imidazole (Im) ring is connected with transition metal (M) ions (e.g., Fe, Co, Cu, Zn) via strong coordination bonds (M-Im-M) to form zeolite like isomorphs.6 Among them, ZIF-8 (made of 2-methylimidazole and Zn2+) is the most studied ZIF subclass capable of encapsulating drugs, enzymes, and nucleic acids for biomedical applications.7–11Human blood makes up 45% of the cells and 55% of the plasma. The plasma transports nutrients, hormones, and proteins across the body and removes waste products as well. The majority of plasma is made up of water, the remaining 10% is made up of ions, proteins, dissolved gases, and nutrient molecules.12 Plasma proteins include antibodies, clotting factors, fibrinogen, and albumin. Albumin is the most abundant blood protein. It has a plasma concentration of 35–50 mg mL−1.13 This 66.5 kDa monomeric globular protein is produced in the liver and participates in essential physiological functions such as vitamins, lipids, and hormones transport, pH buffering, and osmotic pressure control.14 Once a therapeutic agent is introduced into the blood, the water-soluble human serum albumin (HSA) tends to non-specifically adsorb onto the surface of drug carriers and biomolecules.15,16 The HSA molecule has metal-dependent (pseudo-)enzymatic properties to play a role for plasma detoxification, pro-drugs activation, nucleic acid recognition and transportation and storage of transition metals.17–19 This can change the molecular identity of the therapeutic agent and so the understanding of the interactions between the HSA and the nanocarrier is a critical factor in designing robust therapeutic and imaging agents. The HSA molecule is made of 585 amino acid residues including all three aromatic amino acids, tyrosine (Try), phenylalanine (Phe), and tryptophan (Trp).20 The aromatic side chains of Trp, Tyr, and Phe have maximum absorption at 280 nm due to the transition of diffused pi electrons from the aromatic ring.21 These aromatic amino acids are intrinsically fluorescent when excited by ultraviolet (UV) light (λ 280 nm) and regulate the optical properties of HSA. However, multiple copies of Try and Phe residues complicate the investigation of biomolecular interactions of the HSA molecule with the nanocarrier. Since HSA has a single copy of the Trp residue, observing the changes in the optical properties of the Trp residue offers a good choice to understand the level of interaction between HSA and a particular nanocarrier. Optical properties such as absorption and fluorescence emission can be used to explore structural changes in HSA and determine complex formations.22
After administration of a nanomaterial, once it is exposed to the biological fluid, the surface of the delivered nanomaterial spontaneously binds to cellular components like proteins, forming a protein corona. This corona formation depends largely on the physicochemical properties of the nanomaterial and the environment.23 The formation of protein corona on inorganic nanoparticles (NPs) has been extensively studied.24–27 For instance, Treuel et al. studied the effect of NP surface chemistry on interactions with serum albumin.28 Walkey et al. studied the effect of particle size, aggregation status and surface chemistry of Au NPs on forming a serum albumin-Au complex.29 The study noted that the adsorption of blood components and subsequent interaction with cells changed the interfacial composition of the nanomaterial by providing it with a distinct biological identity. This study revealed the importance of the rational design of nanomaterial for better cellular uptake and less toxicity. Carnovale et al. further demonstrated the impact of particle morphology on interactions with serum protein.16 Further, an earlier study also supports the idea that cellular uptake is strongly affected by the presence of a protein adsorption layer around therapeutic nanomaterial.30
ZIF-based MOFs have been extensively investigated for delivering therapeutic proteins and nucleic acids.6–8,31 These studies have mainly focused on maximising the loading of biomolecules in MOFs and their release on demand. The formation of protein corona around biomolecule-encapsulated ZIFs has not yet been investigated. Therefore, it is critical to understand the effects of protein corona on MOFs as it can change the inherent properties of MOF particles and influence their biological fate, including the release behaviour of the loaded biomolecules.32 Further, the release rate of biomolecular payloads can be manipulated by varying the corona composition around the carrier molecule.33
To address these fundamental questions on the nature of the protein corona around nucleic acid-encapsulated MOFs, we first utilised a biomimetic mineralisation method to encapsulate a 5.6 kb circular plasmid within the ZIF-8. The resulting biocomposite was washed and purified and then exposed to the HSA. The reciprocal impact of ZIF-8 on the properties of HSA was investigated as a function of incubation time and MOF concentration using a combination of UV-Vis absorption, fluorescence, and circular dichroism (CD) spectroscopies. The ultimate fate of the crystal structure of MOF and its surface properties were analysed via X-ray powder diffraction (XRD) and Fourier-transform infrared spectroscopy (FT-IR). Finally, the protection of loaded nucleic acid after serum treatment was evaluated by agarose gel electrophoresis. The outcomes of this study offer valuable insights for understanding the early molecular interactions and changes that a therapeutic nucleic acid-loaded MOF is expected to face in a biological environment. These outcomes are critical to allow safe translation of therapeutic MOFs for subsequent clinical studies.
2. Experimental section
2.1. Required materials
Zinc acetate dihydrate (Zn(OAc)2·2H2O), 2-methylimidazole (2mIm), and human serum albumin (HSA, ≥97%) were purchased from Sigma-Aldrich (Australia). Propidium iodide and SYBR™ Safe DNA gel stains were purchased from Thermo Fisher Scientific. An engineered plasmid DNA (6.5 kbp) was collected from NanoBiotechnology Research Laboratory (NBRL), RMIT University, Melbourne, Australia. This plasmid carries a gene that codes for a green fluorescent protein (GFP). The plasmid (hereafter referred to as pGFP) itself is non-fluorescent, but when transcribed and translated within cellular machinery, expresses a nontoxic protein, GFP, and emits a strong green fluorescence under blue light excitation.2.2. Nucleic acid encapsulation by ZIF-8
The biomimetic mineralisation method was followed to encapsulate pGFP in ZIF-8.7 In brief, 1000 ng of pGFP was taken in a fresh tube to mix with aqueous 2mIm (160 mM, 20 μl) followed by aqueous ZnOAc (40 mM, 20 μl). The mixture was left undisturbed for 10 minutes at room temperature. The obtained precipitate was recovered by centrifugation at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rpm for 12
rpm for 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) minutes (Eppendorf MiniSpinPlus). The supernatant was separated to remove unreacted precursors and ions. The pellet was washed thrice with ethanol (E) and labelled as pGFP@ZIF-8E. Similarly, a separate batch of the pellet was washed thrice with deionised water (W) and labelled as pGFP@ZIF-8W. Finally, all samples were vacuum dried to remove any residual solvent post-centrifugation.
minutes (Eppendorf MiniSpinPlus). The supernatant was separated to remove unreacted precursors and ions. The pellet was washed thrice with ethanol (E) and labelled as pGFP@ZIF-8E. Similarly, a separate batch of the pellet was washed thrice with deionised water (W) and labelled as pGFP@ZIF-8W. Finally, all samples were vacuum dried to remove any residual solvent post-centrifugation.
2.3. Verification of nucleic acid encapsulation
Encapsulation of pGFP was verified by agarose gel electrophoresis (AGE), UV-Visible (UV-Vis) and fluorescence spectroscopy. Verification of pGFP loading within ZIF-8 was carried out through agarose gel electrophoresis of supernatant and ethylenediaminetetraacetic acid (EDTA)-treated pellet of the pGFP loaded biocomposites. EDTA (20 mM) dissolved the ZIF-8 pellet due to the chelation of the zinc ions.34 Briefly, 1% (w/v) agarose gel was prepared in 100 mL 1× TAE (Tris–acetate–EDTA) buffer. 1× TAE was prepared from a stock of 10× made by dissolving 48.4 g of Tris base, 11.4 mL of glacial acetic acid and 3.7 g of EDTA and bringing the final volume to 1 L. The agarose solution was heated till the solution was completely clear and cooled to roughly 40 °C prior to the addition of SYBR™ Safe DNA stain. It was poured into the gel casting tray with the comb in place. After the gel polymerized, the comb was removed to expose the sample wells and the gel was placed into the tank of the electrophoresis apparatus. Also, 1× TAE was poured to cover the gel and act as a running buffer. The gel was run at 90 V for 60 minutes. Finally, the DNA bands were visualised by placing the gel under a UV light source in a Gel Doc system (BioRad). The pGFP encapsulation was also verified by UV-vis (Lambda365, PerkinElmer) and fluorescence spectroscopy (HORIBA FluoroMax4) by estimating the nucleic acid content in the supernatant and EDTA-digested pellet. Nucleic acids absorb UV light due to the presence of heterocyclic ring structure in nucleotides.35 The fluorescence spectroscopy measurements were performed to reconfirm the successful encapsulation of pGFP in ZIF-8 using propidium iodide (PI), a DNA-binding red fluorescence dye. This dye selectively binds with DNA and emits red fluorescence. In this study, PI (1 μg mL−1) was mixed with an equal volume of samples and incubated in dark for 30 minutes. Finally, the PI mixed solutions were excited at 535 nm and maximum emission at 617 nm was recorded.2.4. Serum protein treatment
HSA solution was prepared in ultrapure water (pH 7, MilliQ). Then, different concentrations of pGFP@ZIF-8E and pGFP@ZIF-8W (5–200 mM) were mixed with HSA (4 μM). The reaction mixtures were incubated for 30 and 60 minutes in ice. The HSA-coated samples were brought to room temperature 10 minutes before the spectroscopic analysis. The HSA-coated pGFP@ZIF-8E and pGFP@ZIF-8W were purified by centrifugation. The HSA-coated pGFP@ZIF-8 pellet was denoted as pGFP@ZIF-8@HSA in subsequent studies.2.5. Characterization of materials
The crystallinity of particles was studied by XRD using Bruker D8 General Area Detector Diffraction System (GADDS). The Cu Kα (λ = 1.54056 Å) radiation at 40 kV and 40 mA was used for the collection of spectra at room temperature. The step size was 0.01° and the collection range (2θ) was set from 5 to 60°. Data was collected in raw file form (.raw) and converted to UXD file format using the File Exchange Program XCH (Ver. 5.0.10, 2004, Bruker AXS, Socabim, Karlsruhe, Germany) before data analysis. As a reference, the calculated ZIF-8 and ZIF-C data were collected from earlier literature.34,36The surface chemical properties of as-prepared pGFP@ZIF-8E and pGFP@ZIF-8W and their HSA-treated derivatives were assessed by FT-IR spectroscopy (Frontier 4000, PerkinElmer, USA). The vacuum-dried pellets were mixed thoroughly with KBr and loaded into the sample holder. The average of 128 scans was recorded within the 4000–400 cm−1 range with a 4 cm−1 resolution. The Spectrum™ software was used to acquire and process the data.
The morphology of particles was visualised by FEI Verios 460L scanning electron microscope (SEM) operated at an accelerating voltage of 3 kV. Before SEM measurements, the samples were diluted with 500 μL deionised water and sonicated for 10 minutes. After that, a 5 μL sample was taken on the clean surface of the silicon wafer substrate, followed by an even 5 nm iridium sputter coating (Leica EM ACE600 Sputter Coater) to enhance the conductivity and vacuum durability in the SEM chamber. The particles were also assessed by transmission electron microscopy (TEM) with the JEOL1010 TEM (Japan) operated at 100 kV. The samples were diluted and sonicated before drop casting on a carbon-coated copper grid. The grid was left to dry overnight under ambient conditions in a dust-free environment.
2.6. Spectroscopic studies of serum protein interaction
Both pGFP@ZIF-8E and pGFP@ZIF-8W were resuspended in water via gentle pipetting and sonication and the emission spectra (from 310 to 450 nm) were recorded at room temperature using a fluorescence spectrophotometer (HORIBA FluoroMax4). Here, water was used as blank and uncoated (HSA-free) pGFP@ZIF-8E and pGFP@ZIF-8W were used for background signal collection. The thermodynamics of interactions between HSA and ZIF particles was also investigated by a temperature-dependent study. The emission spectra of HSA-coated ZIFs at various temperatures (i.e., 20–60 °C) were recorded. From these data, the binding and quenching mechanism was explained using Stern–Volmer plots and van’t Hoff plots.15,16,372.7. CD spectroscopy
Changes in the secondary structure of HSA in the presence of pGFP@ZIF-8E and pGFP@ZIF-8W were investigated using a Jasco J-815 spectropolarimeter (USA). A quartz cell (pathlength 1 mm) was used to collect all spectra at room temperature (25 °C) and ultrapure water was used as a solvent. Baseline correction was performed using ultrapure water. The CD spectra were recorded in the 200 to 260 nm range using a scan speed of 5 nm min−1 under a constant nitrogen flow. For each sample, five scans were acquired and averaged to improve the signal-to-noise ratio. The ellipticity values are expressed in terms of mean residue molar ellipticity (θ) in degree cm2 dmol−1. The baseline-corrected spectra were smoothed with a 9-point Savitzky–Golay smoothing function.2.8. Protection assay
Protection assays, i.e., the ability of ZIFs to protect the pGFP plasmid, were performed using TURBO™ DNase (Invitrogen) following the manufacturer's protocol. HSA-coated pGFP@ZIF-8E and pGFP@ZIF-8W particles were added with 1 μL DNase I (2U μL−1) and incubated at 37 °C. The inactivation reagent was added after 30 minutes to stop DNase I activity, followed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The pellet was re-suspended in 10 μL of aqueous solution and mixed with EDTA (20 mM) to digest the ZIF-8 particles. Agarose gel electrophoresis was then performed in 1× TAE buffer at 90 V for 60 min. Finally, the nucleic acid bands were visualized in GelDoc® (BioRad®, USA).
000 rpm for 10 min. The pellet was re-suspended in 10 μL of aqueous solution and mixed with EDTA (20 mM) to digest the ZIF-8 particles. Agarose gel electrophoresis was then performed in 1× TAE buffer at 90 V for 60 min. Finally, the nucleic acid bands were visualized in GelDoc® (BioRad®, USA).
2.9. Release assay
In vitro release of pGFP from HSA-treated and untreated pGFP@ZIF-8E and pGFP@ZIF-8W was evaluated in a phosphate buffer saline (PBS) solution. Both pGFP@ZIF and pGFP@ZIF@HSA particles were incubated in PBS (pH 5.5 and pH 7.4) at 37 °C and stirred at 100 rpm in a ThermoMixer. The sample was collected at different time points and PI (1 μg mL−1), and a DNA-binding fluorescent dye was added to quantify the released pGFP and the reaction was incubated in the dark. After 10 minutes the fluorescence emission (Ex. λmax = 535 nm and Em. λmax = 617 nm) was recorded to quantify PI labelled pGFP in the solution.3. Results and discussion
3.1. Nucleic acid encapsulation
ZIF MOFs are excellent candidate materials to explore the bio-encapsulation of therapeutically important molecules.34 They enable facile ambient temperature encapsulation of therapeutic biomolecules (e.g., protein, enzymes, carbohydrates, nucleic acids) through their biomimetic mineralisation under aqueous conditions.The process of ZIF-8 growth in the absence of any biomolecules is well studied.38 In aqueous condition, imidazolate ions form bridges with Zn2+ ions via coordination bonds resulting in Zn(2mIm)n2+ units. However, biomimetic mineralization, i.e., the presence of an additional biomolecule in the reaction, triggers the formation of ZIF particles while the biomolecule acts as a nucleating agent, which significantly expedites the reaction. Here, the concentration of biomolecule plays a critical role in nucleation, as without the biomolecule the process is extremely slow.34
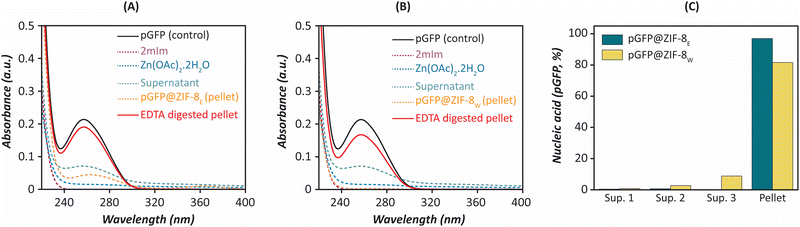
This leads to a several-fold increase in the number of particles formed during biomimetic mineralisation, even when the concentrations of other precursors are relatively lower than under non-biomimetic conditions.7,39 Herein this study, the 2mIm (160 mM) and pGFP (biomolecule) mixture immediately formed a turbid solution soon after the addition of zinc (40 mM). Notably, in the absence of pGFP, the 2mIm and Zn mixture remained transparent while other parameters were consistent. Here, the presence of pGFP induces a synergistic interaction between the MOF precursors and the pGFP. This indicates the pGFP triggered nucleation of ZIF-8 precursors under an aqueous condition followed by rapid particle growth within 10 minutes. UV-Vis and fluorescence spectroscopy were performed to confirm the encapsulation of pGFP in ZIF-8 biocomposites. Nucleic acids have maximum absorption at about 260 nm. In the UV-Vis spectra, no absorption was observed at 260 nm neither in pellet nor supernatant. However, the reappearance of a DNA peak at 260 nm was observed in the EDTA digested pellet similar to the free pGFP solution. In Fig. 1A and B, a small peak at ∼260 nm was observed in the supernatant as not all pGFP was encapsulated inside the ZIF-8E and ZIF-8W, respectively. Similarly, the pGFP@ZIF-8E pellet also showed a tiny peak in that region which might be due to surface deposition on pGFP (Fig. 1A). However, EDTA treatment released the encapsulated pGFP and a sharp peak for nucleic acid appeared similar to a plasmid control (i.e., 1000 ng of pGFP) (Fig. 1A and B). This initial finding confirmed the successful encapsulation of pGFP in both ethanol and water-washed biocomposites.
Agarose gel electrophoresis also confirmed the successful encapsulation of nucleic acid. Here, the MOF pellet was digested with the chelating agent (EDTA, 20 mM) to dissociate the Zn-imidazole bonds in the framework. As EDTA has a high affinity (Kd = 10−16 M) towards zinc, it dissociates the Zn-imidazole bonds of ZIF-8 immediately and releases the plasmid.40 The gel image confirmed the maximum loading of the nucleic acid in the pellet (Fig. S1A, ESI†). The bar graphs represent 96% and 81.5% loading of pGFP in ZIF-8E and ZIF-8W pellets, respectively (Fig. 1C). Furthermore, fluorescence spectroscopy was performed using PI, a common DNA staining dye.41,42 No emission was observed in either supernatant or pellet. However, the reappearance of the PI emission peak in the EDTA digested pellet confirmed the successful encapsulation of pGFP inside the framework (Fig. S1B and C, ESI†). Here, the PI interacted immediately with the pGFP released in the solution after the EDTA treatment, while the relative fluorescent intensities in ZIF-8E and ZIF-8W pellets also corroborate with the absorbance spectroscopy results. This result supports that biomimetic mineralisation allows the encapsulation of nucleic acid inside the framework.7 We studied the encapsulation efficiency of ZIF-C using plasmid and siRNA as a representative of large and small nucleic acid payload.8 Moreover, we and our collaborators studied the encapsulation efficiency of ZIF-based material using different nucleic acid components.43–45 Herein, we also observed that the choice of washing solvents did not significantly compromise the nucleic acid encapsulation efficiency in ZIF biocomposites.
3.2. Characterization
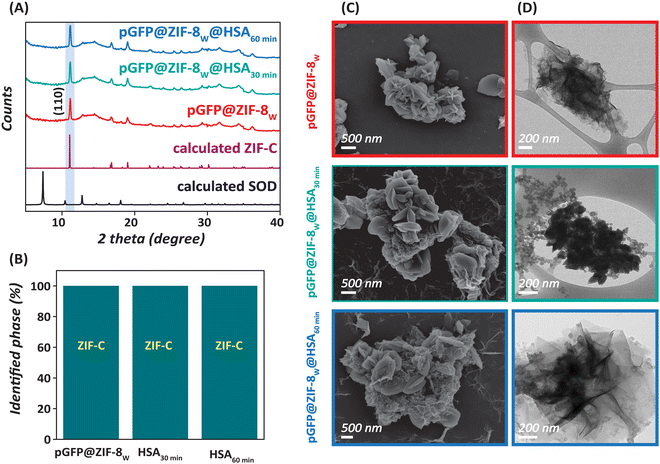
The crystalline properties of as-prepared pGFP@ZIF-8W, pGFP@ZIF-8E and the HSA-treated derivatives were investigated by XRD. The diffraction pattern of water-washed biocomposite (i.e., pGFP@ZIF-8W) matched with carbonate-based ZIF or ZIF-C (Fig. 2A). In particular, the diffracted peaks at 11.03, 14.35, 16.95, 18.31, 22.12, and 23.33° match perfectly with the calculated values.36,46 Moreover, no noticeable change was observed in the diffraction patterns of pGFP@ZIF-8W@HSA (Fig. 2A). This suggests that the crystallinity of pGFP@ZIF-8W was maintained in aqueous HSA for at least up to 60 minutes. A web application, developed at the Graz University of Technology, called “ZIF phase analysis” was used to analyse the crystalline phases in the biocomposites.6 This analysis revealed that the ZIF-C phase was retained completely in the HSA-treated biocomposite (Fig. 2B). Next, the particle morphology was assessed via electron microscopy. The SEM image showed plate-like morphology of pGFP@ZIF-8W particles (Fig. 2C),6,8 which is further evident from a TEM image that shows a thin 2-dimensional (2D) sheet like morphology of these particles (Fig. 2D). Once treated with HSA, a more compact and dense particle morphology was observed in HSA-treated ZIF-8W (Fig. 2C and D). This suggests that while exposure of pGFP@ZIF-8W to HSA does not lead to changes in its crystal structure, it does cause some morphological changes in this material.Conversely, in the case of pGFP@ZIF-8E, the obtained diffraction pattern for pGFP@ZIF-8E showed crystallinity which resembles that of the sodalite (SOD) topology of ZIF-8, typically formed by Zn2+ and 2mIm in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
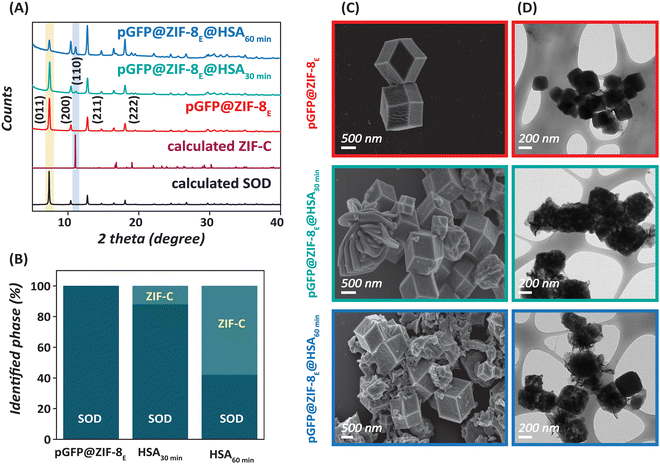
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 stoichiometry (Fig. 3A). The obtained 2θ values of pGFP@ZIF-8E at 7.3, 10.3, 12.7, 14.7, 16.4 and 18° perfectly matched with that of SOD ZIF-8.34,47 However, interestingly, a consistent time-dependent change in crystallinity in pGFP@ZIF-8E was observed after treatment with HSA for 30 and 60 minutes. The signature peak (2θ = 11.07°) for carbonate-based ZIF (ZIF-C) started to appear and increase in intensity with time in pGFP@ZIF-8E@HSA samples. Eventually, the SOD pGFP@ZIF-8E was gradually turned into a mixed-phase biocomposite after HSA treatment (Fig. 3B). It has been noted previously that SOD ZIF-8 is not stable in water under ambient conditions and undergoes a shift to a crystalline, denser substance.48 This was also evident from our data on pGFP@ZIF-8W samples in Fig. 2 that showed a ZIF-C phase instead of a SOD ZIF-8 phase after a water wash step. Moreover, instability of other ZIFs (e.g., ZIF-68 and ZIF-71) has also been observed to result in phase transformation in water.49,50 This indicates a water-mediated phase shift of ZIF structures as most ZIFs are formed on similar coordination chemistry. To understand whether HSA plays any substantial role in the stability of ZIF-8, we performed a control experiment, in which SOD pGFP@ZIF-8E was exposed to water for 1 h in the absence of HSA (Fig. S2, ESI†). The corresponding XRD pattern shows a similar degree of mixed phases having ZIF-8 and ZIF-C that HSA does not significantly influence the stability of ZIF-8 in water. Next, we studied the morphology of the pGFP@ZIF-8E and the HSA-treated derivatives by electron microscopy. The SEM and TEM images of pGFP@ZIF-8E support the synthesis of rhombic dodecahedral morphology of SOD ZIF-8 (Fig. 3C and D).34 However, HSA treatment resulted in a time-dependent change in the morphology, with structural deformations evident at the edges. Consistent with the 2D sheet like morphology of pristine ZIF-C in Fig. 2, pGFP@ZIF-8E@HSA shows hybrid structures containing rhomboids and 2D nanosheets.
2 stoichiometry (Fig. 3A). The obtained 2θ values of pGFP@ZIF-8E at 7.3, 10.3, 12.7, 14.7, 16.4 and 18° perfectly matched with that of SOD ZIF-8.34,47 However, interestingly, a consistent time-dependent change in crystallinity in pGFP@ZIF-8E was observed after treatment with HSA for 30 and 60 minutes. The signature peak (2θ = 11.07°) for carbonate-based ZIF (ZIF-C) started to appear and increase in intensity with time in pGFP@ZIF-8E@HSA samples. Eventually, the SOD pGFP@ZIF-8E was gradually turned into a mixed-phase biocomposite after HSA treatment (Fig. 3B). It has been noted previously that SOD ZIF-8 is not stable in water under ambient conditions and undergoes a shift to a crystalline, denser substance.48 This was also evident from our data on pGFP@ZIF-8W samples in Fig. 2 that showed a ZIF-C phase instead of a SOD ZIF-8 phase after a water wash step. Moreover, instability of other ZIFs (e.g., ZIF-68 and ZIF-71) has also been observed to result in phase transformation in water.49,50 This indicates a water-mediated phase shift of ZIF structures as most ZIFs are formed on similar coordination chemistry. To understand whether HSA plays any substantial role in the stability of ZIF-8, we performed a control experiment, in which SOD pGFP@ZIF-8E was exposed to water for 1 h in the absence of HSA (Fig. S2, ESI†). The corresponding XRD pattern shows a similar degree of mixed phases having ZIF-8 and ZIF-C that HSA does not significantly influence the stability of ZIF-8 in water. Next, we studied the morphology of the pGFP@ZIF-8E and the HSA-treated derivatives by electron microscopy. The SEM and TEM images of pGFP@ZIF-8E support the synthesis of rhombic dodecahedral morphology of SOD ZIF-8 (Fig. 3C and D).34 However, HSA treatment resulted in a time-dependent change in the morphology, with structural deformations evident at the edges. Consistent with the 2D sheet like morphology of pristine ZIF-C in Fig. 2, pGFP@ZIF-8E@HSA shows hybrid structures containing rhomboids and 2D nanosheets.
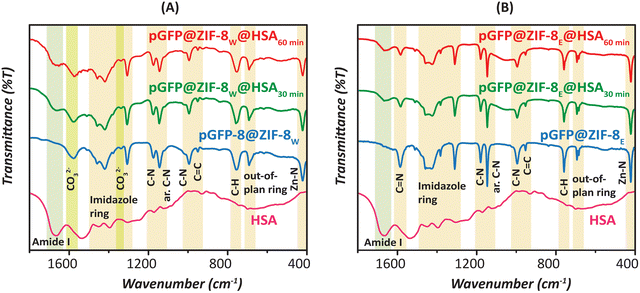
Next, we employed FT-IR spectroscopy to study the chemical composition of ZIFs and confirm the presence of characteristic modes of the peptide backbone of HSA such as the amide I (1600–1750 cm−1) band within the ZIF architecture (Fig. 4).51 A distinct Zn–N band was observed at 423 cm−1 in pGFP@ZIF-8W and the other relevant biocomposites (Fig. 4A).6 The band at 692 cm−1 can be attributed to the out-of-plane bending vibrations of the imidazole ring.52 The peaks at 760 and 996 cm−1 could be assigned to C–H and C–N bending vibrations, respectively.52 The peak at 950 cm−1 refers to in-plane bending.53 The intense peak at 1148 cm−1 is from the aromatic C–N stretching vibrations.53 Moreover, signals between 1300–1450 cm−1 correspond to entire ring stretching.52 The broad peak at 1580 cm−1 and medium intense peak at 1340 cm−1 are attributed to the asymmetric stretching of carbonate.6,46 All these vibrational features, particularly those related to carbonate, support the formation of ZIF-C on exposure to water. The source of carbonate in the water-washed biocomposite could be derived from the CO2 dissolved in the deionized water. Huang et al. studied the material briefly and found no ZIF-C biocomposite formation in an inert atmosphere with degassed water.36 It is not uncommon for imidazole-based ionic liquids to absorb a significant amount of atmospheric CO2, and in the case of ZIF-based MOFs, the imidazole moiety may be presumed to absorb CO2 and convert it to ZIF-C in water.54–56 In addition, after HSA treatment, an additional broad peak around the 1650–1700 cm−1 region is observed, which is due to the amide groups and confirms the presence of HSA on the surface of pGFP@ZIF-8W. Similarly, in pGFP@ZIF-8E, bands corresponding to the ZIF framework are observed at 422 cm−1 (Zn–N stretching), 694 cm−1 (imidazole ring), 753 cm−1 (C–H bending), 952 cm−1 (C–N bending), 1145 cm−1 (aromatic C–N stretching), 1572 cm−1 (C–N stretching), and 1300–1500 cm−1 (imidazole ring stretching) (Fig. 4B).52 However, those corresponding to the carbonate species are missing, supporting the structure of SOD ZIF-8 in ethanol. After treatment with HSA, the carbonate features start to appear in the spectra, and a broad amide peak around the 1650–1750 cm−1 region also emerges signifying the presence of HSA on the biocomposite surface.
3.3. Fluorescence characteristics of HSA-ZIF-8 interactions
Fluorescence quenching is an indication of the variety of interactions such as molecular rearrangement, collisional quenching or ground-state complex formation.57 The Trp-214 residue in HSA is highly sensitive to the changes in its microenvironment, thereby the intrinsic fluorescence emission of Trp is a good indicator of HSA interactions with other species.58 The HSA itself has the maximum emission at 348 nm when excited at 295 nm. By contrast, pGFP@ZIF-8 particles showed no fluorescence at this wavelength. As a result, there were no overlapping fluorescence signals from the ZIF-8 particles (Fig. S3, ESI†). Fig. 5 and 6 show that HSA Trp fluorescence quenching increases with increasing concentrations of both pGFP@ZIF-8E and pGFP@ZIF-8W. A greater extent of quenching was observed at 60 minutes compared to 30 minutes of treatment. Moreover, the pGFP@ZIF-8E showed higher quenching of Trp fluorescence compared to pGFP@ZIF-8W. One of the key factors influencing the efficiency of quenching is the proximity of the quencher to the chromophore.59 Outcomes of the spectrofluorometric study indicate that HSA binds to the ZIF particles at or near its Trp residue, which sits in sub-domain IIA of the HSA molecule. Although both bio-composites have similar surface chemistries, they have different crystal topologies (Fig. 2A and 3A). The pGFP@ZIF-8W particles have plate-like morphology that offers a non-uniform mode of interaction with HSA. As a result, the pGFP@ZIF-8W biocomposites are likely to have fewer interactions, thus exhibiting lower quenching. The water-washed ZIF led to the development of a different crystal phase (ZIF-C) than the typical sodalite phase (Fig. 2 and 3), and this might play a significant role in its interaction with HSA.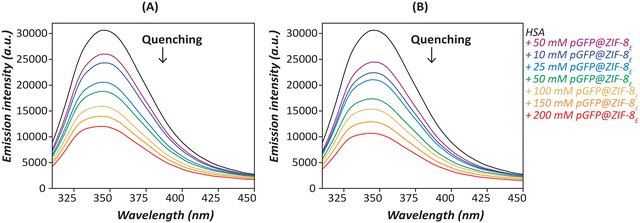 | ||
| Fig. 5 Fluorescence emission spectra depict the interactions of HSA with increasing concentrations of pGFP@ZIF-8E after (A) 30 and (B) 60 min. | ||
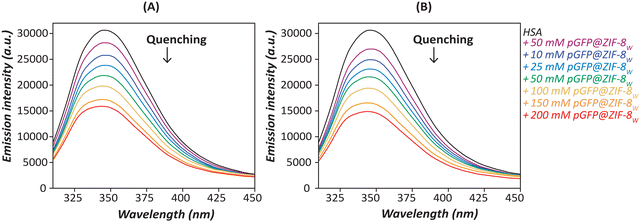 | ||
| Fig. 6 Fluorescence emission spectra depict the interactions of HSA with increasing concentrations of pGFP@ZIF-8W after (A) 30 and (B) 60 min. | ||
3.4. Mechanism of fluorescence quenching
Fluorescence quenching indicates a variety of interactions such as molecular rearrangement, dynamic/collisional quenching or ground-state complex formation.57 Most often, the interaction between a fluorophore and a quencher molecule follows either a dynamic/collisional or static/complex quenching mechanism. Dynamic quenching happens when the excited electron of a fluorophore is deactivated and remains close to the quencher. On the other hand, the formation of a non-fluorescent ground state complex between the quencher and the fluorophore results in static quenching.60,61 These two types of interactions can be differentiated by studying the interactions as a function of temperature. A positive correlation between increasing temperature and molecular diffusion is observed in the dynamic quenching mechanism, whereas a negative correlation is observed for static quenching.62Fluorescence emission spectra of HSA with increasing concentrations of ZIF particles were obtained at temperatures ranging from 20–60 °C. The Stern–Volmer eqn (1) was applied to determine the quenching constant and the quenching mechanism between HSA and ZIF particles.15
| F0/F = 1 + KSV[Q] | (1) |
The Stern–Volmer quenching plots for the pGFP@ZIF-8 are shown in Fig. 7, and the corresponding Stern–Volmer quenching constants are displayed in Tables S1 and S2 (ESI†), respectively for pGFP@ZIF-8E and pGFP@ZIF-8W. A constant decrease in KSV values was observed from 30 to 60 °C for pGFP@ZIF-8E (Table S1, ESI†). This declining trend of KSV values with increasing temperature indicates a reduction in the binding capacity, which suggests predominantly static binding. The number of binding sites (n) and the binding constant (K) between pGFP@ZIF-8 particles and HSA were also calculated to understand the quenching process using eqn (2)
log[(F0 − F)/F] = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K + n K + n![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log[Q] log[Q] | (2) |
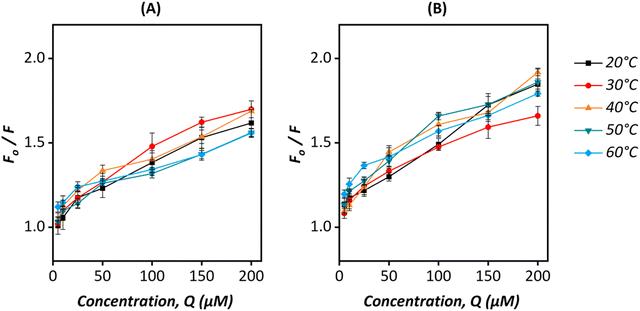 | ||
| Fig. 7 Stern–Volmer plots for (A) pGFP@ZIF-8E@HSA and (B) pGFP@ZIF-8W@HSA at different temperatures (20–60 °C). | ||
A decrease in n with a temperature rise indicates a reduction of binding sites of pGFP@ZIF-8E on HSA. This suggests the instability of the pGFP@ZIF-8E-HSA complex at higher temperatures. The binding sites ‘n’ in the HSA also decreases with temperature (Table S1, ESI†). This feature might suggest that the pGFP@ZIF-8E@HSA complex is unstable.60,63,64 On the other hand, the KSV for pGFP@ZIF-8W showed no consistent trend based on temperature (Table S2, ESI†). The ZIF-C crystal topology of pGFP@ZIF-8W has a relatively higher density than ZIF-8, but a high thermal stability similar to ZIF-8.46 However, the aggregated plate-like form of pGFP@ZIF-8W possibly offers an uneven surface to uniformly bind with protein moieties which results in inconsistency in interacting with HSA at different temperatures. The instability of this interaction is further evident from the number of binding sites (n), which shows less than one binding site is involved in the interaction (Table S2, ESI†). We note that to retain the crystalline and morphological properties of ZIF in a liquid state, water was chosen as a solvent in these studies instead of a buffer. Phosphate ions of the commonly used buffer like PBS tend to degrade ZIF by forming Zn-phosphate precipitate even under physiological conditions.65
3.5. Binding constant and thermodynamic parameters
The double logarithmic regression curves of log[(F0 − F)/F] versus log[Q] were plotted for the pGFP@ZIF-8 (Fig. S4, ESI†). The intercept of this graph gives K, the binding constant. These binding constants are shown in Tables S1 and S2 (ESI†) for pGFP@ZIF-8E and pGFP@ZIF-8W biocomposites, respectively. The K value for pGFP@ZIF-8E showed a gradual decrease with the increase in temperature, indicating that the binding capacity decreases as the temperature rises.The van't Hoff plot (Fig. S5A, ESI†) for pGFP@ZIF-8E showed a linear trend, therefore the thermodynamic parameters were calculated using the following equations.
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = ΔH°/RT + ΔS°/R K = ΔH°/RT + ΔS°/R | (3) |
| ΔG° = ΔH° − TΔS° | (4) |
For pGFP@ZIF-8E (Table S1, ESI†), ΔH° > 0 at lower temperatures indicates an endothermic reaction between pGFP and ZIF-8E, which becomes exothermic at higher temperatures (ΔH° < 0). The ΔS° < 0, suggests that through the interactions between HSA and ZIF-8E, the system becomes more ordered. In this system, ΔH° > 0 and ΔS° < 0 indicating electrostatic forces up to 40 °C, above which both these parameters become <0, indicating that van der Waals forces are responsible for the interaction between HSA and pGFP@ZIF-8E at higher temperatures. Positive ΔG° indicates that these interactions are non-spontaneous across the studied temperature range (Table S1, ESI†). For the pGFP@ZIF-8W biocomposite, the van’t Hoff plot showed polynomial regression at lower temperatures and linear regression at the higher temperature (Fig. S5B, ESI†).15 Since the trend is polynomial from 20–40 °C, polynomial equations were utilized to calculate the thermodynamic parameters.66 The dependence of ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K on 1/T can be reduced to the quadratic form with three parameters (α0, α1, and α2), which is represented in a polynomial equation as follows.
K on 1/T can be reduced to the quadratic form with three parameters (α0, α1, and α2), which is represented in a polynomial equation as follows.
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = α0 + α1/T + (α2/T)2 K = α0 + α1/T + (α2/T)2 | (5) |
| ΔH° = −R(α1 + 2α2/T) | (6) |
| ΔS° = R(α0 – (α2/T)2 | (7) |
3.6. Effect of ZIF-8 on the secondary structure of HSA
CD spectroscopy provides evidence of the conformational change in HSA secondary structure on interaction with ZIF-8. It is a fast and sensitive method to identify the secondary structure, confirmation, and stability of proteins in a solution state.67 Two prominent negative bands of HSA are typically observed at 208 and 222 nm respectively due to the π–π* and n–π* transitions of α-helix peptide bonds.15 The pGFP@ZIF-8 treated HSA retained all the spectral features of HSA, such as the double minima at 222 and 208 nm and the maximum at 190 nm (Fig. 8). However, in the presence of pGFP@ZIF-8, a decrease in the CD signal was observed. This refers to the change in the α-helical content of serum protein. The changes in α-helical content were calculated from mean residue ellipticity (MRE) values at 208 nm position using eqn (8) and (9).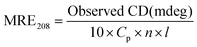 | (8) |
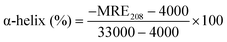 | (9) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 is the MRE value of a pure α-helix.68
000 is the MRE value of a pure α-helix.68
 | ||
| Fig. 8 The CD spectra of (A) pGFP@ZIF-8E and (B) pGFP@ZIF-8W after being incubated with HSA (4 μM) at 30 and 60 min of treatment. | ||
According to this analysis, the pristine α-helix content of HSA (61.8%) is consistent with the reported value.69 The α-helix content in HSA declined to 49% and 43.3%, respectively for 30 min and 60 min treatment with pGFP@ZIF-8E (Fig. 8A). This indicates that the α-helix content of HSA after being incubated with ethanol-washed pGFP@ZIF-8E decreased with the incubation time.70 Additionally, a similar trend was also observed in water-washed biocomposites. However, the pGFP@ZIF-8W particles showed higher α helix content in HSA after being incubated for 30 min (59%) and 60 min (53.3%) (Fig. 8B). This demonstrates higher unfolding of the secondary structure and more conformational changes of the protein in the presence of ethanol-washed pGFP@ZIF-8E compared to water-washed pGFP@ZIF-8W.
3.7. Protection of nucleic acid
The pristine and HSA-treated pGFP@ZIF-8 were treated with DNase I (to cleave pGFP) and then analysed by agarose gel electrophoresis in 1× TAE buffer. Fig. 9 shows the gel image of DNase-treated bio-composites. A clear nucleic acid band appeared in the untreated pGFP (lane 1). However, DNase treatment cleaved the pGFP completely and no band appeared (lane 2). The DNA bands reappeared again in the pGFP@-8 and HSA-treated pGFP@ZIF-8 bio-composites (lanes 3–8). The intensity of nucleic acid bands was quantified by densitometry to determine the content of pGFP after the DNase treatment (Fig. 9). ImageJ analysis showed minor differences in pGFP content in pristine and HSA-treated derivatives (Fig. S6, ESI†) which supports the idea that the protein corona formation around ZIFs does not interfere with their nucleic acid protection ability.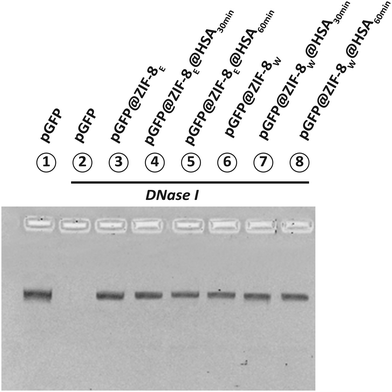 | ||
| Fig. 9 Protection of pGFP by ethanol and water washed ZIF after being treated with HSA. Here, all the particles were treated with DNase I and run in 1% agarose gel. | ||
3.8. The nucleic acid release profile
The % of pGFP released in the solution was quantified by PI-mediated fluorescence assay. Fig. 10 shows the time-dependent release of pGFP from pGFP@ZIF-8E and pGFP@ZIF-8W and their HSA-treated derivatives. In both cases, a steady release of pGFP was found at physiological conditions (pH 7.4). This is because ZIFs are stable at neutral pH.7 However, a fast release of the loaded nucleic acid was observed under acidic conditions (pH 5.5). It is known that the stability of MOFs depends upon the pH, and they can destabilise in highly protonated solutions.71 At acidic pH, the free protons in the solution cleave the Zn–N coordination bonds and break the framework architecture.72,73 About 60% of the pGFP was observed to be released within 30 minutes of exposure to acidic conditions. Though the nucleic acid released trend in HSA treated ethanol washed ZIF-8 was similar, 60 min incubated pGFP@ZIF-8E@HSA showed maximum release (∼87.7%) (Fig. 10A). In pGFP@ZIFW, a higher release was observed in both 30 and 60 minute incubated HSA-treated bio-composites (Fig. 10B). The untreated pGFP@ZIF-8W released 67% pGFP within 30 min and 92% at 5 h. However, both the HSA-treated pGFP@ZIF-8W released ∼97% pGFP within 5 h.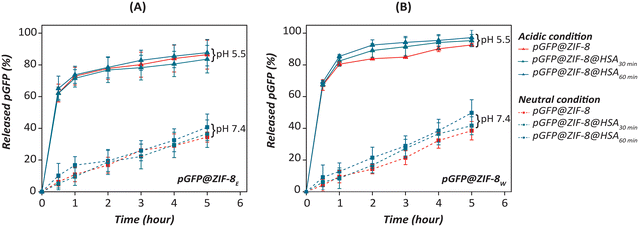 | ||
| Fig. 10 Release of pGFP from (A) pGFP@ZIF-8E@HSA and (B) pGFP@ZIF-8W@HSA at physiological (pH 7.4) and acidic (pH 5.5) at different time points compared to untreated bio-composite (n = 3). | ||
During treatment, the HSA was absorbed on the surface of the ZIFs as confirmed by absorption and fluorescence spectroscopy studied (discussed above). This interaction resulted in structural deformity and more release of loaded nucleic acid after treatment. The crystal phase and morphology of ZIFs played a role in releasing the loaded nucleic acid under acidic conditions. As SOD pGFP@ZIF-8E possesses strong hydrophobicity and is insoluble in water, it takes more time to release the loaded nucleic acid compared to pGFP@ZIF-8W.74 Also phosphate ions in the PBS allow the release of Zn ions from the framework and release the cargo in a time-dependent manner.65
4. Conclusions
The biological fate of a biomolecule carrier can be altered by the formation of a protein corona. From our work, it can be concluded that the crystal phase of the ZIF plays an important role in interacting with HSA which is the most abundant component in human serum. We have demonstrated the impact of different washing solvents on nucleic acid-encapsulated ZIF crystals and their impact on protein corona formation. The results indicate that the porous SOD topology exerts a stronger quenching effect on the inherent fluorescence of HSA compared to the non-porous carbonated ZIF-C phase. A study of the secondary structure of HSA upon its interaction with ZIFs suggests that the ZIF-C phase causes less conformational distortion of serum proteins than the standard sodalite phase. Both these observations from independent experiments are consistent with the observed thermodynamic parameters of interaction, which reveal that the ZIF-C phase interacts less with HSA than the sodalite ZIF-8 phase. It was also confirmed that these interactions do not alter the DNA protection capability of the Zn-based MOFs. These are some very interesting preliminary observations, which we believe, will lay the foundation for extensive studies to thoroughly understand the mechanisms of biomolecular interactions between biomolecule-loaded MOFs of different crystal structures and complex biological fluids.Author contributions
S. A. P.: data curation, formal analysis, investigation, methodology, writing – original draft. S. P.: validation, data curation, writing – review & editing. A. N. A: investigation, validation, data curation, writing – review & editing. S. M.: investigation, validation, writing – review & editing. V. B.: formal analysis, validation, writing – review & editing. L. V.: writing – review & editing. G. B.: supervision, validation, writing – review & editing. R. S.: conceptualization, supervision, validation, project administration, writing – review & editing, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. A. P. and S. M. are thankful to the Australian Government for the RMIT Research Stipend Scholarship (RRSS) to support the PhD program. S. P. is also thankful for RTP Stipend PhD scholarship support. A. A. acknowledges the support of the Australian Research Council (ARC) through its Centre of Excellence for Nanoscale BioPhotonics (CE140100003). R. S. and V. B. acknowledge the Sir Ian Potter Foundation for support in establishing Sir Ian Potter NanoBiosensing Facility. The authors acknowledge the technical support from RMIT Microscopy and Microanalysis Facility (RMMF) towards materials characterisation.References
- M. d J. Velásquez-Hernández, M. Linares-Moreau, E. Astria, F. Carraro, M. Z. Alyami, N. M. Khashab, C. J. Sumby, C. J. Doonan and P. Falcaro, Towards applications of bioentities@MOFs in biomedicine, Coord. Chem. Rev., 2021, 429, 213651 CrossRef.
- R. J. Kuppler, D. J. Timmons, Q. R. Fang, J. R. Li, T. A. Makal, M. D. Young, D. Yuan, D. Zhao, W. Zhuang and H. C. Zhou, Potential applications of metal-organic frameworks, Coord. Chem. Rev., 2009, 253, 3042–3066 CrossRef CAS.
- P. Horcajada, R. Gref, T. Baati, P. K. Allan, G. Maurin, P. Couvreur, G. Ferey, R. E. Morris and C. Serre, Metal–organic frameworks in biomedicine, Chem. Rev., 2012, 112, 1232–1268 CrossRef CAS PubMed.
- P. Horcajada, T. Chalati, C. Serre, B. Gillet, C. Sebrie, T. Baati, J. F. Eubank, D. Heurtaux, P. Clayette, C. Kreuz, J. S. Chang, Y. K. Hwang, V. Marsaud, P. N. Bories, L. Cynober, S. Gil, G. Férey, P. Couvreur and R. Gref, Porous metal-organic-framework nanoscale carriers as a potential platform for drug delivery and imaging, Nat. Mater., 2010, 9, 172–178 CrossRef CAS PubMed.
- S. Li, K. Wang, Y. Shi, Y. Cui, B. Chen, B. He, W. Dai, H. Zhang, X. Wang and C. Zhong, High pulmonary accumulation, favorable biocompatibility, and improved therapeutic outcome, Adv. Funct. Mater., 2016, 26, 2715–2727 CrossRef CAS.
- F. Carraro, M. J. Velásquez-Hernández, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Phase dependent encapsulation and release profile of ZIF-based biocomposites, Chem. Sci., 2020, 11, 3397–3404 RSC.
- A. Poddar, J. J. Conesa, K. Liang, S. Dhakal, P. Reineck, G. Bryant, E. Pereiro, R. Ricco, H. Amenitsch, C. Doonan, X. Mulet, C. M. Doherty, P. Falcaro and R. Shukla, Encapsulation, visualization and expression of genes with biomimetically mineralized zeolitic imidazolate framework-8 (ZIF-8), Small, 2019, 15, e1902268 CrossRef PubMed.
- A. Poddar, S. Pyreddy, F. Carraro, S. Dhakal, A. Rassell, M. R. Field, T. S. Reddy, P. Falcaro, C. M. Doherty and R. Shukla, ZIF-C for targeted RNA interference and CRISPR/Cas9 based gene editing in prostate cancer, Chem. Commun., 2020, 56, 15406–15409 RSC.
- S. K. Alsaiari, S. Patil, M. Alyami, K. O. Alamoudi, F. A. Aleisa, J. S. Merzaban, M. Li and N. M. Khashab, Endosomal escape and delivery of CRISPR/Cas9 genome editing machinery enabled by nanoscale zeolitic imidazolate framework, J. Am. Chem. Soc., 2018, 140, 143–146 CrossRef CAS PubMed.
- M. Z. Alyami, S. K. Alsaiari, Y. Li, S. S. Qutub, F. A. Aleisa, R. Sougrat, J. S. Merzaban and N. M. Khashab, Cell-type-specific CRISPR/Cas9 delivery by biomimetic metal organic frameworks, J. Am. Chem. Soc., 2020, 142, 1715–1720 CrossRef CAS PubMed.
- Y. Li, K. Zhang, P. Liu, M. Chen, Y. Zhong, Q. Ye, M. Q. Wei, H. Zhao and Z. Tang, Encapsulation of plasmid DNA by nanoscale metal-organic frameworks for efficient gene transportation and expression, Adv. Mater., 2019, 31, e1901570 CrossRef PubMed.
- A. Priyam, L. O. Afonso, A. G. Schultz, A. K. Dinda and P. P. Singh, Hemocompatibility of biogenic phosphorus nano-agromaterials at environmentally relevant and supra-environmental concentrations for occupational exposure, Environ. Sci.: Adv., 2023, 2, 313–324 Search PubMed.
- A. M. Merlot, D. S. Kalinowski and D. R. Richardson, Unraveling the mysteries of serum albumin-more than just a serum protein, Front. Physiol., 2014, 5, 299 CrossRef PubMed.
- A. Maji, M. Beg, S. Das, M. N. Aktara, S. Nayim, A. Patra, M. M. Islam and M. Hossain, Study on the antibacterial activity and interaction with human serum albumin of Tagetes erecta inspired biogenic silver nanoparticles, Process Biochem., 2020, 97, 191–200 CrossRef CAS.
- A. N. Abraham, T. K. Sharma, V. Bansal and R. Shukla, Phytochemicals as dynamic surface ligands to control nanoparticle-protein interactions, ACS Omega, 2018, 3, 2220–2229 CrossRef CAS PubMed.
- C. Carnovale, G. Bryant, R. Shukla and V. Bansal, Impact of nanogold morphology on interactions with human serum, Phys. Chem. Chem. Phys., 2018, 20, 29558–29565 RSC.
- G. De Simone, A. di Masi and P. Ascenzi, Serum albumin: A multifaced enzyme, Int. J. Mol. Sci., 2021, 22, 10086 CrossRef CAS PubMed.
- L. I. Alinovskaya, S. E. Sedykh, N. V. Ivanisenko, S. E. Soboleva and G. A. Nevinsky, How human serum albumin recognizes DNA and RNA, Biol. Chem., 2018, 399, 347–360 CAS.
- S. Al-Harthi, J. I. Lachowicz, M. E. Nowakowski, M. Jaremko and Ł. Jaremko, Towards the functional high-resolution coordination chemistry of blood plasma human serum albumin, J. Inorg. Biochem., 2019, 198, 110716 CrossRef CAS PubMed.
- B. Meloun, L. Morávek and V. Kostka, Complete amino acid sequence of human serum albumin, FEBS Lett., 1975, 58, 134–137 CrossRef CAS PubMed.
- N. J. Anthis and G. M. Clore, Sequence-specific determination of protein and peptide concentrations by absorbance at 205 nm, Protein Sci., 2013, 22, 851–858 CrossRef CAS PubMed.
- K. Byadagi, M. Meti, S. Nandibewoor and S. Chimatadar, Investigation of binding behaviour of procainamide hydrochloride with human serum albumin using synchronous, 3D fluorescence and circular dichroism, J. Pharm. Anal., 2017, 7, 103–109 CrossRef PubMed.
- V. H. Nguyen and B. J. Lee, Protein corona: a new approach for nanomedicine design, Int. J. Nanomed., 2017, 12, 3137–3151 CrossRef CAS PubMed.
- J. Bing, X. Xiao, D. J. McClements, Y. Biao and C. Chongjiang, Protein corona formation around inorganic nanoparticles: Food plant proteins-TiO2 nanoparticle interactions, Food Hydrocolloids, 2021, 115, 106594 CrossRef CAS.
- Q. Yu, L. Zhao, C. Guo, B. Yan and G. Su, Regulating protein corona formation and dynamic protein exchange by controlling nanoparticle hydrophobicity, Front. Bioeng. Biotechnol., 2020, 8, 210 CrossRef PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. Toledano Furman, F. Salvatore and E. Tasciotti, The impact of nanoparticle protein corona on cytotoxicity, immunotoxicity and target drug delivery, Nanomedicine, 2016, 11, 81–100 CrossRef CAS PubMed.
- F. S. M. Tekie, M. Hajiramezanali, P. Geramifar, M. Raoufi, R. Dinarvand, M. Soleimani and F. Atyabi, Controlling evolution of protein corona: a prosperous approach to improve chitosan-based nanoparticle biodistribution and half-life, Sci. Rep., 2020, 10, 9664 CrossRef PubMed.
- L. Treuel, M. Malissek, J. S. Gebauer and R. Zellner, The influence of surface composition of nanoparticles on their interactions with serum albumin, ChemPhysChem, 2010, 11, 3093–3099 CrossRef CAS PubMed.
- C. D. Walkey, J. B. Olsen, H. Guo, A. Emili and W. C. Chan, Nanoparticle size and surface chemistry determine serum protein adsorption and macrophage uptake, J. Am. Chem. Soc., 2012, 134, 2139–2147 CrossRef CAS PubMed.
- L. Treuel, S. Brandholt, P. Maffre, S. Wiegele, L. Shang and G. U. Nienhaus, The influence of surface composition of nanoparticles on their interactions with serum albumin, ACS Nano, 2014, 8, 503–513 CrossRef CAS PubMed.
- S. A. Polash, K. Garlick-Trease, S. Pyreddy, S. Periasamy, G. Bryant and R. Shukla, Amino acid-coated zeolitic imidazolate framework for delivery of genetic material in prostate cancer cell, Molecules, 2023, 28, 4875 CrossRef CAS PubMed.
- S. Behzadi, V. Serpooshan, R. Sakhtianchi, B. Müller, K. Landfester, D. Crespy and M. Mahmoudi, Protein corona change the drug release profile of nanocarriers: the “overlooked” factor at the nanobio interface, Colloids Surf., B, 2014, 123, 143–149 CrossRef CAS PubMed.
- A. Cifuentes-Rius, H. de Puig, J. C. Kah, S. Borros and K. Hamad-Schifferli, Optimizing the properties of the protein corona surrounding nanoparticles for tuning payload release, ACS Nano, 2013, 7, 10066–10074 CrossRef CAS PubMed.
- K. Liang, R. Ricco, C. M. Doherty, M. J. Styles, S. Bell, N. Kirby, S. Mudie, D. Haylock, A. J. Hill, C. J. Doonan and P. Falcaro, Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules, Nat. Commun., 2015, 6, 7240 CrossRef CAS PubMed.
- J. Tian, Y. Li and D. Zhao, Basics of Molecular Biology, Molecular Imaging, Springer, 2013, pp. 541–601 Search PubMed.
- Z. Huang, M. Ge, F. Carraro, C. Doonan, P. Falcaro and X. Zou, Can 3D electron diffraction provide accurate atomic structures of metal-organic frameworks?, Faraday Discuss., 2021, 225, 118–132 RSC.
- V. D. Suryawanshi, L. S. Walekar, A. H. Gore, P. V. Anbhule and G. B. Kolekar, Spectroscopic analysis on the binding interaction of biologically active pyrimidine derivative with bovine serum albumin, J. Pharm. Anal., 2016, 6, 56–63 CrossRef PubMed.
- M. Jian, B. Liu, R. Liu, J. Qu, H. Wang and X. Zhang, Water-based synthesis of zeolitic imidazolate framework-8 with high morphology level at room temperature, RSC Adv., 2015, 5, 48433–48441 RSC.
- E. Astria, M. Thonhofer, R. Ricco, W. Liang, A. Chemelli, A. Tarzia, K. Alt, C. E. Hagemeyer, J. Rattenberger, H. Schroettner, T. Wrodnigg, H. Amenitsch, D. M. Huang, C. J. Doonan and P. Falcaro, Carbohydrates@MOFs, Mater. Horiz., 2019, 6, 969–977 RSC.
- J. K. Nyborg and O. B. Peersen, That zincing feeling: the effects of EDTA on the behaviour of zinc-binding transcriptional regulators, Biochem. J., 2004, 381, e3–e4 CrossRef CAS PubMed.
- N. Zhang, Y. Fan, C. Li, Q. Wang, N. Leksawasdi, F. Li and S. Wang, Cell permeability and nuclear DNA staining by propidium iodide in basidiomycetous yeasts, Appl. Microbiol. Biotechnol., 2018, 102, 4183–4191 CrossRef CAS PubMed.
- C. Riccardi and I. Nicoletti, Analysis of apoptosis by propidium iodide staining and flow cytometry, Nat. Protoc., 2006, 1, 1458–1461 CrossRef CAS PubMed.
- S. Pyreddy, A. Poddar, F. Carraro, S. A. Polash, C. Dekiwadia, B. Murdoch, Z. Nasa, T. S. Reddy, P. Falcaro and R. Shukla, Targeting telomerase utilizing zeolitic imidazole frameworks as non-viral gene delivery agents across different cancer cell types, Biomater. Adv., 2023, 149, 213420 CrossRef CAS PubMed.
- R. Tagore, K. Alagarasu, P. Patil, S. Pyreddy, S. A. Polash, M. Kakade, R. Shukla and D. Parashar, Targeted in vitro gene silencing of E2 and nsP1 genes of chikungunya virus by biocompatible zeolitic imidazolate framework, Front. Bioeng. Biotechnol., 2022, 10, 1003448 CrossRef PubMed.
- A. Poddar, S. Pyreddy, S. A. Polash, C. M. Doherty and R. Shukla, A quest for cytocompatible metal organic frameworks in non-viral gene therapy: Relevance of zeolitic imidazolate framework-8, Biomater. Biosyst., 2022, 8, 100065 CrossRef CAS PubMed.
- S. A. Basnayake, J. Su, X. Zou and K. J. Balkus, Jr., Carbonate-based zeolitic imidazolate framework for highly selective CO2 capture, Inorg. Chem., 2015, 54, 1816–1821 CrossRef CAS PubMed.
- A. Schejn, L. Balan, V. Falk, L. Aranda, G. Medjahdi and R. Schneider, Controlling ZIF-8 nano- and microcrystal formation and reactivity through zinc salt variations, CrystEngComm, 2014, 16, 4493–4500 RSC.
- H. Zhang, M. Zhao and Y. Lin, Stability of ZIF-8 in water under ambient conditions, Microporous Mesoporous Mater., 2019, 279, 201–210 CrossRef CAS.
- A. Kasik, X. Dong and Y. Lin, Synthesis and stability of zeolitic imidazolate framework-68 membranes, Microporous Mesoporous Mater., 2015, 204, 99–105 CrossRef CAS.
- M. E. Schweinefuß, S. Springer, I. A. Baburin, T. Hikov, K. Huber, S. Leoni and M. Wiebcke, Zeolitic imidazolate framework-71 nanocrystals and a novel SOD-type polymorph: solution mediated phase transformations, phase selection via coordination modulation and a density functional theory derived energy landscape, Dalton Trans., 2014, 43, 3528–3536 RSC.
- D. Usoltsev, V. Sitnikova, A. Kajava and M. Uspenskaya, Systematic FTIR spectroscopy study of the secondary structure changes in human serum albumin under various denaturation conditions, Biomolecules, 2019, 9, 359 CrossRef CAS PubMed.
- Y. Zhang and Y. Jia, Synthesis of zeolitic imidazolate framework-8 on polyester fiber for PM 2.5 removal, RSC Adv., 2018, 8, 31471–31477 RSC.
- Z. X. Low, J. Yao, Q. Liu, M. He, Z. Wang, A. K. Suresh, J. Bellare and H. Wang, Crystal transformation in zeolitic-imidazolate framework, Cryst. Growth Des., 2014, 14, 6589–6598 CrossRef CAS.
- L. F. Zubeir, D. van Osch, M. A. A. Rocha, F. Banat and M. C. Kroon, Carbon dioxide solubilities in decanoic acid-based hydrophobic deep eutectic solvents, J. Chem. Eng. Data, 2018, 63, 913–919 CrossRef CAS PubMed.
- W. A. N. G. Mei, L. Q. Zhang, L. I. U. Hao, J. Y. Zhang and C. G. Zheng, J. Fuel Chem. Technol., 2012, 40, 1264–1268 CrossRef.
- G. Iijima, T. Kitagawa, A. Katayama, T. Inomata, H. Yamaguchi, K. Suzuki, K. Hirata, Y. Hijikata, M. Ito and H. Masuda, CO2 reduction promoted by imidazole supported on a phosphonium-type ionic-liquid-modified Au electrode at a low overpotential, ACS Catal., 2018, 8, 1990–2000 CrossRef CAS.
- A. Selva Sharma and M. Ilanchelian, Comprehensive multispectroscopic analysis on the interaction and corona formation of human serum albumin with gold/silver alloy nanoparticles, J. Phys. Chem. B, 2015, 119, 9461–9476 CrossRef CAS PubMed.
- T. Sun, L. Liu, Y. Sun, C. Tan, F. Yao, X. Liang, Y. Wang, Y. Yang, X. Hu and J. Fan, Synthesis and characterization of TiO2 nanoparticles: applications in research on the interaction of colloidal TiO2 with human serum albumin by fluorescence spectroscopy, Anal. Sci., 2012, 28, 491–496 CrossRef CAS PubMed.
- S. Mandal, M. Hossain, P. S. Devi, G. S. Kumar and K. Chaudhuri, J. Hazard. Mater., 2013, 248–249, 238–245 CrossRef CAS PubMed.
- J. Mariam, P. M. Dongre and D. C. Kothari, Study of interaction of silver nanoparticles with bovine serum albumin using fluorescence spectroscopy, J. Fluoresc., 2011, 21, 2193–2199 CrossRef CAS PubMed.
- P. K. Sidhu and K. Nehra, Bacteriocin-capped silver nanoparticles for enhanced antimicrobial efficacy against food pathogens, IET Nanobiotechnol., 2020, 14, 245–252 CrossRef PubMed.
- H. C. Ishikawa-Ankerhold, R. Ankerhold and G. P. Drummen, Advanced fluorescence microscopy techniques-Frap, Flip, Flap, Fret and flim, Molecules, 2012, 17, 4047–4132 CrossRef CAS PubMed.
- W. Zhang, Q. Zhang, F. Wang, L. Yuan, Z. Xu, F. Jiang and Y. Liu, Comparison of interactions between human serum albumin and silver nanoparticles of different sizes using spectroscopic methods, Luminescence, 2015, 30, 397–404 CrossRef CAS PubMed.
- S. Ambika and M. Sundrarajan, Green biosynthesis of ZnO nanoparticles using Vitex negundo L. extract: Spectroscopic investigation of interaction between ZnO nanoparticles and human serum albumin, J. Photochem. Photobiol., B, 2015, 149, 143–148 CrossRef CAS PubMed.
- M. d J. Velásquez-Hernández, R. Ricco, F. Carraro, F. T. Limpoco, M. Linares-Moreau, E. Leitner, H. Wiltsche, J. Rattenberger, H. Schröttner, P. Frühwirt, E. M. Stadler, G. Gescheidt, H. Amenitsch, C. J. Doonan and P. Falcaro, CrystEngComm, 2019, 21, 4538–4544 RSC.
- T. Galaon and V. David, Deviation from van't Hoff dependence in RP-LC induced by tautomeric interconversion observed for four compounds, J. Sep. Sci., 2011, 34, 1423–1428 CrossRef CAS PubMed.
- N. J. Greenfield, Using circular dichroism spectra to estimate protein secondary structure, Nat. Protoc., 2006, 1, 2876–2890 CrossRef CAS PubMed.
- Q. Sun, J. He, H. Yang, S. Li, L. Zhao and H. Li, Analysis of binding properties and interaction of thiabendazole and its metabolite with human serum albumin via multiple spectroscopic methods, Food Chem., 2017, 233, 190–196 CrossRef CAS PubMed.
- M. A. Qureshi and S. Javed, Investigating binding dynamics of trans resveratrol to HSA for an efficient displacement of aflatoxin B1 using spectroscopy and molecular simulation, Sci. Rep., 2022, 12, 2400 CrossRef CAS PubMed.
- L. Xu, Y. X. Hu, Y. C. Li, Y. F. Liu, L. Zhang, H. X. Ai and H. S. Liu, Study on the interaction of paeoniflorin with human serum albumin (HSA) by spectroscopic and molecular docking techniques, Chem. Cent. J., 2017, 11, 1–12 CrossRef PubMed.
- K. Leus, T. Bogaerts, J. De Decker, H. Depauw, K. Hendrickx, H. Vrielinck, V. Van Speybroeck and P. Van Der Voort, Systematic study of the chemical and hydrothermal stability of selected “stable” Metal Organic Frameworks, Microporous Mesoporous Mater., 2016, 226, 110–116 CrossRef CAS.
- C. Han, C. Zhang, N. Tyminska, J. R. Schmidt and D. S. Sholl, Insights into the stability of zeolitic imidazolate frameworks in humid acidic environments from first-principles calculations, J. Phys. Chem. C, 2018, 122, 4339–4348 CrossRef CAS.
- H. Nabipour, M. H. Sadr and G. R. Bardajee, Synthesis and characterization of nanoscale zeolitic imidazolate frameworks with ciprofloxacin and their applications as antimicrobial agents, New J. Chem., 2017, 41, 7364–7370 RSC.
- W. Liang, H. Xu, F. Carraro, N. K. Maddigan, Q. Li, S. G. Bell, D. M. Huang, A. Tarzia, M. B. Solomon and H. Amenitsch, Enhanced activity of enzymes encapsulated in hydrophilic metal-organic frameworks, J. Am. Chem. Soc., 2019, 141, 2348–2355 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ma00391d |
| This journal is © The Royal Society of Chemistry 2023 |