Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Development of a tissue construct with spatially controllable stiffness via a one-step 3D bioprinting and dual-crosslinking process†
Giorgia
Pagnotta‡
a,
Maila
Becconi‡
 a,
Marco
Malferrari
a,
Marco
Malferrari
 a,
Donatella
Aiello
a,
Donatella
Aiello
 b,
Anna
Napoli
b,
Anna
Napoli
 b,
Luana
Di Lisa
a,
Stefano
Grilli
a,
Stefania
Rapino
b,
Luana
Di Lisa
a,
Stefano
Grilli
a,
Stefania
Rapino
 *ac and
Maria Letizia
Focarete
*ac and
Maria Letizia
Focarete
 *ac
*ac
aDepartment of Chemistry “Giacomo Ciamician” and INSTM UdR of Bologna, University of Bologna, 40126 Bologna, Italy. E-mail: stefania.rapino3@unibo.it; marialetizia.focarete@unibo.it
bDepartment of Chemistry and Chemical Technologies, University of Calabria, Rende 87036, Italy
cHealth Sciences and Technologies - Interdepartmental Center for Industrial Research (HST-ICIR), Alma Mater Studiorum - Università di Bologna, 40064 Ozzano dell’Emilia, Bologna, Italy
First published on 12th July 2023
Abstract
3D bioprinting is a promising technology for manufacturing cellular constructs in which the complex characteristics of real tissues are faithfully represented under in vitro culturing conditions. Many efforts have been made to reproduce the numerous physical, chemical, and mechanical properties that are crucial determinants of cellular biological functions and behavior. Abrupt stiffness changes have been described in tumors, and higher stiffness has been reported for tumor regions within the same tissue. In this study, we developed a green synthetic route for alginate methacrylation with a degree of substitution higher than 60%, which was used for bioink formulations using HeLa cells as a proof of principle cell model. Alginate methacrylate (AlgMa) was characterized by 1H-NMR, FT-IR and MALDI TOF–TOF MS. A dual-step crosslinking procedure was developed to obtain bioprinted constructs with good cellular viability and tunable mechanical properties. A critical and systematic study of the effects of the photoinitiator concentration, photo-crosslinking process, and mechanical properties of the AlgMa-based construct on cell viability was conducted. Extensive rheological characterization of bioink formulations and bioprinted samples that differ in dual-step crosslinking procedures enabled the production of samples characterized by differential and spatially resolved mechanical properties within the same 3D bioprinted construct, approaching the heterogeneous stiffness observed in real tissues.
Introduction
The importance of reproducing and controlling the mechanical properties of hydrogels in 3D cell cultures has been widely reported in the literature.1–3 Cell fate, molecular pathways, proliferation, and apoptosis depend strongly on the constituents of the cell microenvironment and the physico-chemical properties of the Extracellular Matrix (ECM), such as stiffness, cell–matrix interaction, and external directional forces.4–7 ECM stiffness and dysregulation of ECM dynamics have been recognized to play critical roles in cancer progression and malignant cell behavior.8 External mechanical inputs are transduced in cellular systems into molecular outputs (e.g., transcriptional programs and the differential spatial organization of cell molecular determinants) that drive tumor progression,9 epithelial–mesenchymal transition/aggressiveness,10 genomic variation,11 invasion,12 metastatization13 and chemoresistance.14 Tumor and aged tissues are considerably stiffer and usually display a Young's modulus one or two orders of magnitude greater than the corresponding healthy one, depending on the type of tissue and size of the lesion.15,16 Furthermore, pathological tissues, cancer tissues, skin, cartilage, and wounds are characterized by inherent heterogeneity and physical gradients in which stiffness usually increases from the periphery to the core, as in the case of solid tumors.17,18 This enhanced stiffness and heterogeneity make it harder to evaluate all the biological mechanisms related to cell-to-matrix interactions in standard in vitro models that do not recapitulate the reality of human physiological and pathological tissues. Therefore, developing biomaterials that recapitulate the physico-chemical characteristics found in the tissue microenvironment will enable more realistic in vitro studies and better comprehension of cell mechanisms.Hydrogels, highly hydrated macromolecular crosslinked structures, are among the most promising matrices to mimic the ECM of soft tissues. In particular, interest in hydrogels and 3D constructs characterized by controlled stiffness and stiffness gradients has increased significantly over the last few years. As already proven from many scientific works, the structural and mechanical properties of the cell environment in all the three directions influence the diffusion of molecules, generate concentration gradients of soluble factors and mediate cell response to external mechanical inputs.19,20 Many fabrication methods involving the use of thermo-gelling21 and photo-curable polymers have been developed to create mechanical gradients, such as soft-lithography platforms, microgels, flat gradient stiffness surfaces, and microfluidic mixers.21–24 However, these methods present some limitations in terms of (i) cost-effectiveness ratio, (ii) significant technical background or laboratory equipment requirements, and (iii) difficulties in creating desired 3D structures and dimensions. The 3D bioprinting technique has become an attractive tool for the development of 3D in vitro models resembling in vivo conditions. They represent an extremely useful, practical, and easy-to-use tool for the study of cell biology.25–29 In this context, alginate–methacrylate (AlgMa) bioink30,31 is widely recognized as a good candidate for replicating the ECM of 3D constructs because of its high biocompatibility, ability to form hydrogels under mild conditions, and the possibility of modifying and tuning its stiffness.31–34 The mechanical properties of AlgMa are usually modulated by changing the photoinitiator concentration, UV exposure time, and degree of functionalization of the acrylate moieties branched along the polymer chains. These strategies should ensure at the same time the proper cell growth and proliferation and should not alter the printing process along with the replication of the desired mechanical properties reflecting those of the native tissue of interest. Methacrylation is usually reported as a laborious synthesis that involves several pH adjustments, a high molar excess of methacrylic anhydride (MAA), or the use of an organic base. Usually, this leads to a low yield of product and a low degree of substitution (ca. 30%).35,36 In this work, a green synthetic route for the methacrylation of alginate, which leads to a higher degree of substitution compared to those found in the literature, is presented to produce a formulation of printable AlgMa-based bioinks. A dual crosslinking procedure, involving both ionic interactions with divalent cations and the formation of covalent bonds through UV irradiation, has been demonstrated to further stabilize the hydrogel and to improve its mechanical properties.36–38 Usually, this procedure involves the incubation of the AlgMa hydrogel in the crosslinking solution followed by exposure to UV light.36–38 In this work, the AlgMa well-known dual crosslinking procedure was smartly exploited during bioprinting, by starting from a hydrogel solution already containing calcium ions, then controlling both the time and the position of the UV irradiation during the post-printing phase, and, finally, adding a second ionic crosslinking step, placing the printed hydrogel in the calcium containing solution. The designed crosslinking strategy was thoroughly studied using rheology and allowed to obtain a 3D cell culture platform with spatially controllable stiffness, while preserving cell viability. HeLa cells were used for the proof of principle realization of the stiffness heterogeneous tissue-like model.
Materials and methods
Alginate–methacrylate synthesis
Sodium alginate (Alg), derived from brown algae (MW > 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1), was purchased from Carlo Erba (product number 366552). Methacrylic anhydride (MAA) (94%) (product number 102212824), sodium carbonate, and sodium bicarbonate were purchased from Sigma Aldrich. NaOH 5 M was prepared by dissolving pure NaOH pellets in distilled water. A dialysis tube with a cellulose membrane (14 kDa cutoff, avg. flat width of 25 mm) was purchased from Sigma Aldrich. Alg (2% w/v) was dissolved in 0.325 M carbonate (CB) buffer at pH 9. Subsequently, an excess of MAA (1 mL g−1 Alg) was added to the alginate solution under magnetic stirring at 500 rpm and 50 °C. Once the reaction started, a methacrylic acid sub-product was formed, and the pH tended to decrease to below 6. For this reason, NaOH 5 M was added to the reaction until the solution reached pH 9 to promote the methacrylation process. The reaction proceeded for both 24 h and 72 h, at 50 °C, and the pH was stable for the entire process between 7.5 and 8. The solution was then dialyzed for six days at room temperature (RT) using dialysis tubing (cut off 14 kDa) under gentle stirring and protected from light. Then, the dialyzed solutions were filtered through 0.2 μm sterile filters and frozen at −20 °C before lyophilization. Finally, the samples were lyophilized overnight and stored at 4 °C, protected from light, until further use.
000 g mol−1), was purchased from Carlo Erba (product number 366552). Methacrylic anhydride (MAA) (94%) (product number 102212824), sodium carbonate, and sodium bicarbonate were purchased from Sigma Aldrich. NaOH 5 M was prepared by dissolving pure NaOH pellets in distilled water. A dialysis tube with a cellulose membrane (14 kDa cutoff, avg. flat width of 25 mm) was purchased from Sigma Aldrich. Alg (2% w/v) was dissolved in 0.325 M carbonate (CB) buffer at pH 9. Subsequently, an excess of MAA (1 mL g−1 Alg) was added to the alginate solution under magnetic stirring at 500 rpm and 50 °C. Once the reaction started, a methacrylic acid sub-product was formed, and the pH tended to decrease to below 6. For this reason, NaOH 5 M was added to the reaction until the solution reached pH 9 to promote the methacrylation process. The reaction proceeded for both 24 h and 72 h, at 50 °C, and the pH was stable for the entire process between 7.5 and 8. The solution was then dialyzed for six days at room temperature (RT) using dialysis tubing (cut off 14 kDa) under gentle stirring and protected from light. Then, the dialyzed solutions were filtered through 0.2 μm sterile filters and frozen at −20 °C before lyophilization. Finally, the samples were lyophilized overnight and stored at 4 °C, protected from light, until further use.
Alginate–methacrylate chemical characterization
Proton nuclear magnetic resonance (1H-NMR) measurements were carried out on both pristine Alg powder and AlgMa after 24 and 72 h of reaction time using an NMR Varian MR400 coupled with an ATB PFG probe. Both Alg and lyophilized AlgMa were dissolved at 40 mg mL−1 in deuterium oxide, and the chemical shift of each sample was measured at 50 °C to dissolve the materials better and have a higher resolution of the protons peaks associated with the methacrylate groups.Fourier transform infrared (FT-IR) spectroscopy was carried out using a Bruker laser class model ALPHA, equipped with an ATR accessory (PLATINUM-ATR) on both Alg and AlgMa, after 72 h of reaction time, to identify the specific peaks associated with the presence of methacrylate groups. All spectra were recorded between 400 cm−1 and 4000 cm−1 with a resolution of 4 cm−1, accumulation of 16 scans, and step size of 1 cm−1.
Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) was performed on AlgMa after 72 h of reaction time to evaluate the degree of substitution. AlgMa samples were first subjected to partial acidic hydrolysis using a sample of 2% w/v in an acid solution. The partial depolymerization was carried out in trifluoroacetic acid (TFA, 2 M) at a temperature of 90 °C for 5 h to obtain oligosaccharide chains easily detectable by MALDI mass spectrometry.39 5 μL of the hydrolyzed sample was added to 5 μL of NH4OH solution (15%), and then added with 20 μL of MALDI matrix. The best results were obtained using 2,5-dihydroxybenzoic acid (DHB, 10 mg mL−1; H2O/CH3CN, 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60, v-v; 0.1% TFA) as matrix. The dried droplet method was adopted, in combination with a fast-drying protocol.40–43 Then, mass spectrometry analysis was performed on a 5800 MALDI TOF–TOF analyzer (AB SCIEX, Darmstadt, Germany) equipped with an Nd:YLF laser (λ = 345 nm wavelength, <500 ps pulse length up to 1000 Hz repetition rate). The spectra were acquired in reflectron positive mode (HR), with a mass accuracy of 5 ppm and mass range of 300–2500 Da. Each spectrum was recorded, accumulating at least 5000 laser shots with a laser pulse rate of 400 Hz. The sample was spotted three times, and each spot was measured three times to collect nine data points. The spectra were handled using Data Explorer version 4.11 (AB Sciex) to obtain a theoretically calculated distribution and extract the Isotope Cluster Area (ICA) for each ion peak. All signals of interest were detected as singly charged cation adducts or radical cations and used for data analysis (Table S1, ESI†). The Degree of Functionalization (DoF) was calculated as the percentage ratio between the derivatized oligomers vis total oligomers. Therefore, DoF % was determined as the average of three different experiments performed in triplicate (nine data points) and calculated as DoF % = [ΣICAM/ΣICA] × 100, where ICAM are the isotope cluster areas of only ion signals of methacrylate species (Table S2, ESI†). ICA are isotope cluster areas of all the identified oligosaccharides species.
60, v-v; 0.1% TFA) as matrix. The dried droplet method was adopted, in combination with a fast-drying protocol.40–43 Then, mass spectrometry analysis was performed on a 5800 MALDI TOF–TOF analyzer (AB SCIEX, Darmstadt, Germany) equipped with an Nd:YLF laser (λ = 345 nm wavelength, <500 ps pulse length up to 1000 Hz repetition rate). The spectra were acquired in reflectron positive mode (HR), with a mass accuracy of 5 ppm and mass range of 300–2500 Da. Each spectrum was recorded, accumulating at least 5000 laser shots with a laser pulse rate of 400 Hz. The sample was spotted three times, and each spot was measured three times to collect nine data points. The spectra were handled using Data Explorer version 4.11 (AB Sciex) to obtain a theoretically calculated distribution and extract the Isotope Cluster Area (ICA) for each ion peak. All signals of interest were detected as singly charged cation adducts or radical cations and used for data analysis (Table S1, ESI†). The Degree of Functionalization (DoF) was calculated as the percentage ratio between the derivatized oligomers vis total oligomers. Therefore, DoF % was determined as the average of three different experiments performed in triplicate (nine data points) and calculated as DoF % = [ΣICAM/ΣICA] × 100, where ICAM are the isotope cluster areas of only ion signals of methacrylate species (Table S2, ESI†). ICA are isotope cluster areas of all the identified oligosaccharides species.
Cell culture
HeLa cells (ATCC) were used as a cellular model in this study. Both bidimensional and tridimensional cultures were maintained in Dulbecco's Modified Eagle's medium (Microgem, TL1006) supplemented with 10% Fetal Bovine Serum (South America; Microgem, RM10432), 2 mM L-glutamine, 50 U mL−1 penicillin, 50 μg mL−1 streptomycin (Microgem) and cultured at 37 °C, 5% CO2. Bidimensional cultures were passed upon trypsin digestion every two or three days; the growth medium of the 3D constructs was changed every two or three days.AlgMa inks and bioinks preparation
Phosphate buffered saline solution (PBS) was purchased from Microgem (product number TL1006), and Calcium Chloride (product number C8106) and Irgacure 2595 (I2959) (product number 410896) were purchased from Sigma Aldrich. Water-based solutions containing 60 mM CaCl2 were prepared by dissolving the CaCl2 powder in sterile bidistilled water. A mother solution of 10% w/v I2959 was prepared by dissolving Irgacure powder in methanol. Two different AlgMa inks were developed in this study: ink A (IA) and ink B (IB). Both inks were prepared by dissolving 4% w/v of lyophilized AlgMa in PBS 1× at 37 °C. Then, using a mixing system consisting of two syringes linked by a female/female Luer-lock adapter, the AlgMa solution was mixed with a water-based solution of 60 mM CaCl2, at a ratio of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 v/v to obtain the hydrogel. Subsequently, 0.2% w/v and 0.25% w/v of I2959 were added to the pre-crosslinked AlgMa hydrogel formulation to form IA and IB, respectively. Before printing, 100 μL of a suspension of HeLa cells in complete culture medium (1 × 106 cells/100 μL) was gently mixed with 900 μL of each ink formulation to form bioink A (BA) and bioink B (BB), respectively. To make the rheology tests comparable ink A and ink B were adjusted in concentration to compensate for cells dilution.
9 v/v to obtain the hydrogel. Subsequently, 0.2% w/v and 0.25% w/v of I2959 were added to the pre-crosslinked AlgMa hydrogel formulation to form IA and IB, respectively. Before printing, 100 μL of a suspension of HeLa cells in complete culture medium (1 × 106 cells/100 μL) was gently mixed with 900 μL of each ink formulation to form bioink A (BA) and bioink B (BB), respectively. To make the rheology tests comparable ink A and ink B were adjusted in concentration to compensate for cells dilution.
3D bioprinting
Inks and bioinks were printed using an INKREDIBLE+ bioplotter from Cellink (BICO Company). The bioprinter consists of an extrusion-based system controlled by pneumatic pressure. Both bioink formulations (BA and BB) and the corresponding inks without cells (IA and IB) were loaded in UV-shielding cartridges with a 22G conical nozzle (Cellink) and printed at room temperature (RT) by applying a pressure range between 17 kPa and 25 kPa and a printing speed of 10 mm s−1. All the structures were extruded onto separated 35 mm dishes to obtain circular shaped samples with a diameter, ϕ, and height, h, of 10 mm and 1 mm, respectively, which can fit the geometry of the rheometer set up. Furthermore, only bioink A was printed into 3D “fortune cookie” shaped constructs (ϕ = 15 mm × h = 1 mm; printing speed = 10 mm s−1) to obtain the gradient stiffness platform.Dual crosslinking procedure
Different post-printing crosslinking processes were performed on the 3D printed structures to stabilize the hydrogels against water hydrolysis and obtain tunable mechanical properties. The 3D models obtained with bioinks A and B underwent the dual-step crosslinking process shown in Scheme 1 and summarized in Table 1 and described as follows. Step 1: first crosslinking with UV light (intensity: 22–26 mW cm−2, distance from light source: 5 cm, wavelength of UV light: 365 nm) for different irradiation times (60 s, 90 s, 120 s); Step 2: a second crosslinking with CaCl2 solution for 5 min. The acronyms for the 3D models obtained according to the procedure described above are listed in Table 1. In the case of bioink B, three additional 3D models were obtained by crosslinking with only UV light for 60, 90, and 120 s after the printing process. The same protocols were applied to the corresponding models obtained with ink not containing cells (Table 1) to evaluate whether the presence of cells affected the rheological properties of the ink.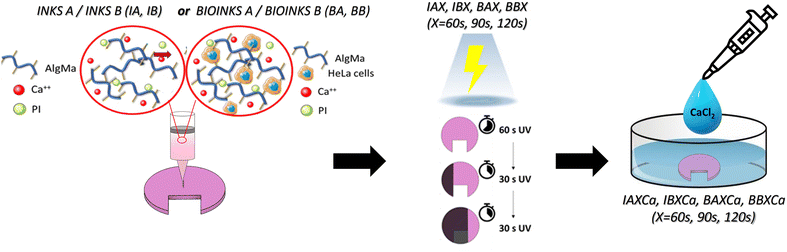 | ||
| Scheme 1 Sketch describing the dual crosslinking process and the preparation of the ink and bioink formulations under investigation. | ||
| Ink | Bioink | Step 1: UV time (s) | Step 2: CaCl2 incubation time (min) | 3D models with cells | 3D models without cell |
|---|---|---|---|---|---|
| IA | BA | 60 | 5 | BA60Ca | IA60Ca |
| 90 | 5 | BA90Ca | IA90Ca | ||
| 120 | 5 | BA120Ca | IA120Ca | ||
| IB | BB | 60 | 5 | BB60Ca | IB60Ca |
| 60 | — | BB60 | IB60 | ||
| 90 | 5 | BB90Ca | IB90Ca | ||
| 90 | — | BB90 | IB90 | ||
| 120 | 5 | BB120Ca | IB120Ca | ||
| 120 | — | BB120 | IB120 |
Rheological measurements
Amplitude sweep, viscosity, and recovery tests were performed using an Anton Paar MCR 102 rheometer in a plate−plate geometry with a diameter of 25 mm and a gap of 0.3 mm. First, amplitude sweep analysis, viscosity, and recovery tests were performed on the Alg and AlgMa solutions, both solubilized in PBS 1×, and on the AlgMa hydrogel not containing I2959, to evaluate how methacrylation and the presence of the CaCl2 pre-crosslinker solution affected rheological parameter and hydrogel printability. All the measurements were carried out at 25 °C to simulate the pre-printing and printing process conditions. Amplitude sweep tests were carried out in a deformation range (γ) from 0.01% to 1000% by using an oscillation mode, keeping constant the frequency at 1 Hz, to characterize G′ and G′′ moduli as a function of the applied strain (%) of the hydrogels and to have a quick evaluation of their printability. Viscosity tests and flow curves were carried out with a shear rate range (![[small gamma, Greek, dot above]](https://www.rsc.org/images/entities/i_char_e0a2.gif) ) from 0.01 s−1 to 1000 s−1 using a rotational mode to assess hydrogel viscosity and shear thinning behavior. The same amplitude sweep analysis performed on Alg and AlgMa solutions and AlgMa hydrogel was carried out also on the 3D bioprinted models (ϕ: 5 mm × h: 1 mm) with and without cells, after the dual-step crosslinking process described above and after 72 h of cell incubation, in a deformation range (γ) from 0.1% to 1000% at 37 °C, to simulate the incubation conditions and to evaluate how the presence of the cells affects the rheological properties of the final 3D printed models.
) from 0.01 s−1 to 1000 s−1 using a rotational mode to assess hydrogel viscosity and shear thinning behavior. The same amplitude sweep analysis performed on Alg and AlgMa solutions and AlgMa hydrogel was carried out also on the 3D bioprinted models (ϕ: 5 mm × h: 1 mm) with and without cells, after the dual-step crosslinking process described above and after 72 h of cell incubation, in a deformation range (γ) from 0.1% to 1000% at 37 °C, to simulate the incubation conditions and to evaluate how the presence of the cells affects the rheological properties of the final 3D printed models.
Biological characterization: biocompatibility and imaging
Live-dead assay with calcein–AM (Thermofisher, c1430) and propidium iodide (Merck, P4864) was performed on the 3D bioprinted constructs to assess the biocompatibility of the formulated hydrogels and the printing process, and how the dual crosslinking procedure affects cell viability in order to choose the best bioink. The assay was performed at 24 h, 48 h and 72 h for all bio ink formulations and at 14 days for the chosen formulation; the assay was adapted for tridimensional constructs from protocols previously described.44 First, the 3D bioprinted samples were washed with PBS supplemented with calcium and magnesium (Gibco 14040) three times for 5 minutes, followed by incubation for 30 minutes at 37 °C with a solution of 3 μM calcein–AM and 4 μM propidium iodide in HBSS (Sigma H8264); before inspection with an optical microscope, the constructs were washed three times for 5 minutes with PBS with calcium and magnesium. The samples were examined with an inverted fluorescence microscope (Nikon Eclipse TiS) using a 10× objective and two fluorescent channels. Propidium iodide signals were measured with a Texas Red HYQ cubic filter (λ excitation = 532–587 nm, λ emission = 608–683 nm), while calcein–AM fluorescence was recorded using a FITC cubic filter (λ excitation = 465–495 nm, λ emission = 512.5–557.5 nm).Confocal laser scanning microscopy
Confocal laser scanning microscopy analyses were performed on the 3D bioprinted samples placed in 35 mm diameter glass bottom microwell dishes (Mattek). Samples stained for the viability assay were observed using a Nikon A1R confocal laser scanning microscope employing two laser lines (489 nm and 561 nm) and two detection GASP channels (525/20 and 595/20 nm) for the green and red false color channels, respectively to measure the fluorescence intensity of calcein–AM and propidium iodide, respectively. Viability percentages were estimated by semi-quantitative analysis using Fiji software:45 live cells (stained green by calcein–AM) and dead cells (stained red by propidium iodide) were manually counted, and the live cell fraction was calculated by dividing the number of live cells by the total number of cells.Statistical analysis
Cell counts are presented as mean ± standard deviation. Statistical analyses were performed using the OriginPro 2019b software. A two-sample t-test was used to determine statistical significance. Differences were considered statistically significant at p < 0.05. Different levels of statistical significance are indicated as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.Results
Alginate methacrylation and assessment of functionalization
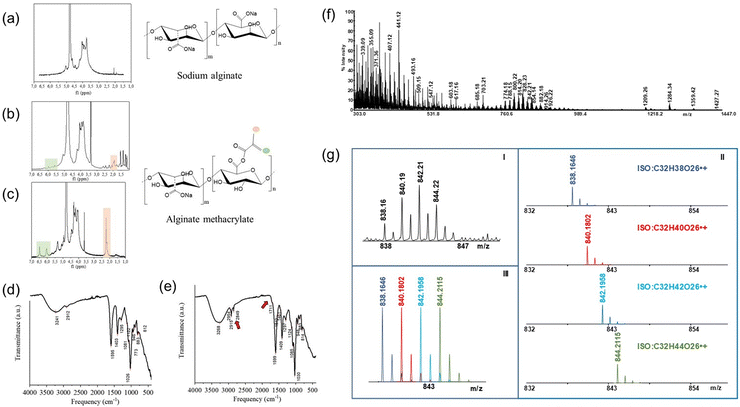
A scheme of the alginate methacrylation reaction is shown in Fig. S1 (ESI†). Pristine alginate was dissolved (2% w/v) in a basic aqueous CB to encourage the deprotonation of the carboxylic groups, making them available for substitution with the methacrylic moieties. 2% w/v was selected as the optimal concentration of alginate to obtain a low viscous solution that can be homogeneously stirred at high rpm (500 rpm) to ensure the collision between the methacrylic anhydride and alginate chains. CB buffer at 0.325 molarity (pH = 9) was selected as an aqueous solvent to carry out the reaction because it could autonomously maintain a pH of approximately eight during the whole reaction time. This choice was made considering its proven better buffering capacity46 with respect to common phosphate-based buffer solutions, which are usually employed for alginate methacrylation and require several pH adjustments with NaOH. A reduced excess of MAA (1 mL g−1 AlgMa) was employed in this synthesis with respect to the molar excess used in traditional AlgMa production, which usually ranges from 15 to 20 mL per gram of AlgMa.47 Under these conditions, only one pH adjustment is required after the solubilization of alginate powder in CB buffer to stabilize the pH between 8 and 9. Finally, two different reaction times were evaluated (24 h and 72 h) to determine the best reaction time duration, considering that in the traditional synthesis of AlgMa, 24 h and 72 h are usually reported as the optimal times to obtain methacrylation.36To assess the presence of methacrylate groups and evaluate the degree of substitution after methacrylation, pristine alginate (Alg) and alginate methacrylate (AlgMa) were characterized by 1H-NMR, FT-IR and MALDI TOF–TOF MS. 1H-NMR spectra reported in Fig. 1(a–c) show the comparison between Alg and AlgMa after 24 h and 72 h of methacrylation. All the spectra display characteristic peaks between 3.50 and 5.20 ppm typical of the saccharide units of the alginate backbone chains.48 In addition, both AlgMa spectra exhibit two distinctive signals of the vinyl in the region ranging from 6.5 ppm to 5.6 ppm and methyl hydrogens peaks of the methacrylate groups at ca. 2.00 ppm, which vary in location depending on the resulting chemical environment. In particular, the AlgMa spectrum obtained after 24 h of reaction time (Fig. 1(b)), shows the vinyl protons signals at 5.6 ppm (singlet) and 6.0 ppm (singlet) and the methyl hydrogens peak at 2.0 ppm (triplet), while the AlgMa obtained after 72 h of reaction time (Fig. 1(c)) shows more intense peaks shifted at 6.0 ppm (doublet) and 6.5 ppm (singlet) for the vinyl hydrogens and 2.23 ppm (triplet) for the methyl ones which integrate for three with respect to the vinyl protons. In summary, the proton signals of AlgMa after 72 h of reaction displayed more intense and defined peaks. Furthermore, Fig. 1(c) shows that the block of peaks associated with the saccharide units of the alginate backbone chain slightly shifted at higher ppm values. This deshielding in the downfield region of the spectrum could be ascribed to the presence of the newly formed ester group, which is directly linked to vinyl functionalization and has a moderate deactivation effect due to its withdrawing nature.
Alg and AlgMa were characterized by FT-IR spectroscopy to further verify the substitution after 72 h of reaction time, which was demonstrated, by 1H-NMR, as the best reaction time to obtain methacrylation. A detailed description of the FT-IR spectra is provided in ESI.† In Fig. 1(e), the differences between the Alg and AlgMa spectra are highlighted by red arrows. The spectrum related to the AlgMa product exhibits more intense peaks at 2980–2850 cm−1 and the presence of a “shoulder” at 1711 cm−1 (Fig. 1(e)) which was not observed in the pristine Alg spectrum (Fig. 1(d)), respectively assigned to the stretching of the –CH of the alkane and alkene bonds contained in the methacrylate functionalization and to the stretching of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group of the esters, both resulting from the grafting of the methacrylate units. Furthermore, a more intense broad band at ca. 3268 cm−1 was observed for AlgMa than for Alg, which might be ascribed to the increased number of –CH groups introduced by methacrylic functionalization. Accordingly, these results, taken together, provide evidence of successful alginate methacrylation. This reaction is assumed to occur in the –OH functional groups since the characteristic peaks of the COO– groups are still present after the reaction and the new band at 1711 cm−1 highlights the formation of the ester bond.
O group of the esters, both resulting from the grafting of the methacrylate units. Furthermore, a more intense broad band at ca. 3268 cm−1 was observed for AlgMa than for Alg, which might be ascribed to the increased number of –CH groups introduced by methacrylic functionalization. Accordingly, these results, taken together, provide evidence of successful alginate methacrylation. This reaction is assumed to occur in the –OH functional groups since the characteristic peaks of the COO– groups are still present after the reaction and the new band at 1711 cm−1 highlights the formation of the ester bond.
MALDI-TOF MS analysis was carried out to quantify the degree of methacrylation of AlgMa after 72 h of reaction. Fig. 1(f) shows representative MALDI-TOF-MS spectra of the product solution obtained by hydrolyzing AlgMa under acidic and thermic conditions (90 °C, 5 h, TFA 2 M). High-intensity readings were observed for several peaks in the mass range of 300–1500 Da, leading to the identification of hexuronic acid (glucuronic/mannuronic acid) chains with two to eight methacrylate units and underivatized. These results confirmed that different oligosaccharides were formed during the experimental treatment. The spectrum complexity is due to the formation of several decomposed products under acid/hydrothermal conditions. Moreover, even and odd species can be observed, which indicate the presence of protonated and radical cation species. All identified oligosaccharide chains are reported in Table S1 in the ESI,† while Fig. 1(g) shows the theoretical distribution of the isotope cluster ions of an exemplificative signal. The composite signal (Fig. 1(g)-III) was derived from the overlapping of the isotope cluster ions of four different species at m/z 838, 840, 842, and 844 (Fig. 1(g)-II). A comparison of the measured experimental isotopic distribution (Fig. 1(g)-I) with the theoretically calculated distribution of the expected summary formula suggests that the ion clusters resulted from the combination of four radical cation species generated as degradation products under the adopted experimental conditions. The same analysis was performed for all the signals to confirm the identity of the observed ion species (Table S1, ESI†). MALDI-MS has major advantages, including high sensitivity and accuracy, applicability to a dynamic mass range,49 fast generation of results,50 and little or no fragmentation of analytes. However, literature reports that MALDI analysis of carbohydrates is affected by salts; therefore, oligo- and polysaccharides often give rise to MALDI spectra of poor quality in the presence of contaminating salts.39,51 In the present study, the choice to perform partial hydrolysis of polysaccharides ensured sodium removal, as confirmed by high-resolution mass measurements (Table S1, ESI†). Moreover, the ammonium hydroxide solution favored the formation of carboxylate/ammonium ion pairs, thus promoting the desorption of oligosaccharides.52 MALDI-TOF-MS measurements allowed calculation the degree of AlgMa methacrylation DoF% of 63.4 ± 3.2%.
Optimization of hydrogel formulation through rheological characterization
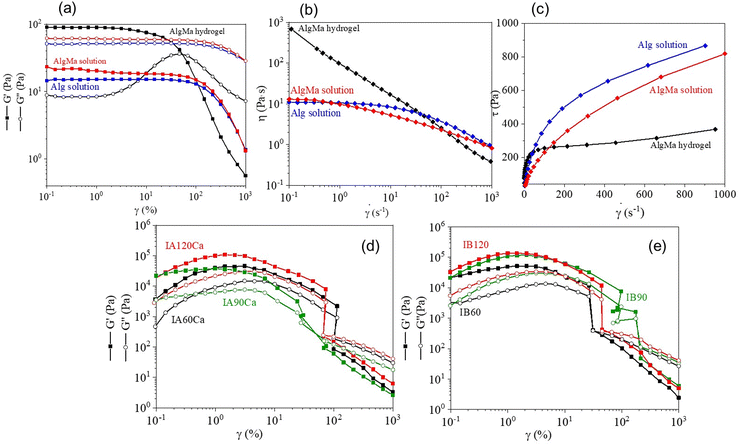
AlgMa obtained after 72 h reaction time was selected to produce hydrogels and bioinks, because of the higher substitution level with respect to the 24 h synthesis, as demonstrated by the previously described chemical characterization. AlgMa solution (4% w/v) was first prepared prior to hydrogel formation by dissolving purified and lyophilized AlgMa in PBS to provide a suitable osmotic concentration for the cells once embedded in the final bioink and to avoid cell dehydration along with cell membrane breakage.The rheological characterization of AlgMa solution was carried out through amplitude sweep and flow curve tests, carried out at 25 °C, and results were compared with those of the pristine Alg solution (4% w/v) to assess the influence of the methacrylation on the rheological properties. Amplitude sweep analysis (Fig. 2(a)) shows the trend of both the storage (G′) and loss (G′′) moduli against the deformation range applied. In particular, pristine Alg and AlgMa solutions display a loss modulus G′′ of 61 Pa and 52 Pa, respectively, higher than the storage modulus G′ of 19 Pa and 15 Pa, respectively, up to 100% of deformation. Above 100% of strain, both moduli drop down without displaying any resistance to the higher deformation. This behavior is typically ascribable to “liquid-like” gels in which the viscous behavior dominates the elastic one. AlgMa and Alg solutions showed a similar trend and values of G′ and G′′ moduli, demonstrating that the methacrylation reaction does not influence the rheological properties of Alg. Flow curves were obtained for both the AlgMa and Alg solutions to verify the shear-thinning behavior of the materials, and the results are shown in Fig. 2(b) and (c). The two solutions display a similar behavior characterized by an initial viscosity plateau (zero-shear viscosity), followed by a linear decrease of the viscosity with an increase in the shear rate; the Alg solution shows a viscosity of 11 Pa s and 0.9 Pa s at a shear rate of at 0.1 s−1 and 1000 s−1 respectively while the AlgMa solution displays a viscosity of 13 Pa s and 0.8 Pa s at a shear rate of at 0.1 s−1 and 1000 s−1 respectively (Fig. 2(b)). Moreover, to better visualize the viscosity behavior of the solutions, the flow curves were also reported by referring to the shear stress (τ) as a function of the shear rate on a linear scale, as shown in Fig. 2(c). The results show the absence of a yield point and confirm the “liquid-like” nature of AlgMa and Alg solutions.
The AlgMa hydrogel was then prepared by mixing AlgMa solution with a 60 mM CaCl2 solution (in a ratio of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 v/v). This specific ratio was used to obtain a printable hydrogel while avoiding instantaneous and inhomogeneous crosslinking between the chains. Rheological measurements carried out on AlgMa hydrogel to evaluate its printability are reported in Fig. 2, together with those of Alg and AlgMa solutions. From the amplitude sweep measurement (Fig. 2(a)), the AlgMa hydrogel shows a clearly different behavior with respect to both Alg and AlgMa solutions by displaying a value of G′ (90 Pa) higher than that of G′′ (8 Pa). Moreover, a linear viscoelastic region (LVE) can be identified from 0.01% of strain up to 3% of strain, which determines the region in which the polymer maintains its elastic behavior and the elastic behavior dominates the viscous one (“gel-like” behavior). By further analyzing the viscosity function of the AlgMa hydrogel formulation through rotational tests (Fig. 2(b) and (c)), it is possible to observe that this material decreases its viscosity with an increase in the applied shear rate (Fig. 2(b)), indicating the ability to flow under an applied shear rate without showing any zero-shear viscosity, typical of the unfilled or uncrosslinked materials, as displayed for Alg and AlgMa solutions. In particular, at a low shear rate, AlgMa hydrogel displays 681 Pa s of viscosity while at a high shear rate, this value decreases to a value of 0.4 Pa s. Furthermore, Fig. 2(c) clearly shows that AlgMa hydrogel displays a well-defined and pronounced yield point around 100 s−1, which defines the point at which a material can flow after the application of external forces that typically characterize hydrogels suitable for 3D bioprinting applications. Rheological results on AlgMa hydrogel, taken all together, show that this material maintains the ability to generate ionic crosslinking between the carboxylate groups and calcium ions even after the methacrylation reaction, and it can be exploited to create the physical entanglements and ionic interactions between polymers chains required for the formulation of the final ink and its printability.
9 v/v). This specific ratio was used to obtain a printable hydrogel while avoiding instantaneous and inhomogeneous crosslinking between the chains. Rheological measurements carried out on AlgMa hydrogel to evaluate its printability are reported in Fig. 2, together with those of Alg and AlgMa solutions. From the amplitude sweep measurement (Fig. 2(a)), the AlgMa hydrogel shows a clearly different behavior with respect to both Alg and AlgMa solutions by displaying a value of G′ (90 Pa) higher than that of G′′ (8 Pa). Moreover, a linear viscoelastic region (LVE) can be identified from 0.01% of strain up to 3% of strain, which determines the region in which the polymer maintains its elastic behavior and the elastic behavior dominates the viscous one (“gel-like” behavior). By further analyzing the viscosity function of the AlgMa hydrogel formulation through rotational tests (Fig. 2(b) and (c)), it is possible to observe that this material decreases its viscosity with an increase in the applied shear rate (Fig. 2(b)), indicating the ability to flow under an applied shear rate without showing any zero-shear viscosity, typical of the unfilled or uncrosslinked materials, as displayed for Alg and AlgMa solutions. In particular, at a low shear rate, AlgMa hydrogel displays 681 Pa s of viscosity while at a high shear rate, this value decreases to a value of 0.4 Pa s. Furthermore, Fig. 2(c) clearly shows that AlgMa hydrogel displays a well-defined and pronounced yield point around 100 s−1, which defines the point at which a material can flow after the application of external forces that typically characterize hydrogels suitable for 3D bioprinting applications. Rheological results on AlgMa hydrogel, taken all together, show that this material maintains the ability to generate ionic crosslinking between the carboxylate groups and calcium ions even after the methacrylation reaction, and it can be exploited to create the physical entanglements and ionic interactions between polymers chains required for the formulation of the final ink and its printability.
Starting from the AlgMa hydrogel, inks and bioink formulations were optimized to find the most suitable photoinitiator concentration and UV time. Mechanical and dimensional stability of the bioprinted constructs under cell culture conditions (i.e., at 37 °C and 5% CO2) was systematically evaluated. In samples showing structural stability over time, HeLa cell viability was evaluated using a live-dead test. Table S3 (ESI†) details all investigated formulations and the obtained viability data. Results showed that when the Irgacure concentration was below 0.2% w/v, the bioprinted sample was not stable and dissolved immediately or after a few hours depending on Irgacure concentration and UV irradiation time. Sample mechanical stability was achieved when Irgacure concentration was equal to or higher than 0.2% w/v. More precisely, with 0.2% w/v Irgacure and UV irradiation time from 1 min to 2 min, the sample was quite stable, while enhancing UV irradiation time up to 5 min, cell mortality reached 100%. Formulations obtained with 0.25% w/v Irgacure showed good mechanical stability and acceptable cell viability (from 58% to 43%), depending on UV irradiation time. Finally, an Irgacure concentration of 0.4% w/v increased cell death, leading to 33% cell viability after 2 min of UV irradiation.
Based on the obtained results, 0.2% w/v and 0.25% w/v were selected as the best Irgacure concentration values to produce constructs from Ink A and Ink B, respectively.
Rheological evaluation of dual crosslinking process
IA and IB were crosslinked using different procedures after the bioprinting process, as detailed in the experimental section (see Table 1). In particular, a double crosslinking process was exploited, involving (i) a photo-crosslinking process through UV irradiation and (ii) an ionic crosslinking by dipping the 3D construct in a calcium chloride solution. This approach has been applied to increase the mechanical resistance and stability of the construct without the need to increase the UV irradiation time, which was found to strongly affect cell viability in relation to the percentage of Irgacure involved in the experiment.Amplitude sweep analyses were performed on printed IA after the dual crosslinking process and printed IB after the single crosslinking process to evaluate how the mechanical properties could be tuned by changing the crosslinking parameters. The results are shown in Fig. 2(d) and (e). All the samples show a ‘gel-like’ behavior, and both moduli increase with the UV exposure time. In fact, IA120Ca showed the highest moduli (21 kPa and 7 kPa for G′ and G′′) while IA90Ca and IA60Ca show 17 kPa and 7 kPa for G′ and G′′ and 5 kPa and 2 kPa for G′ and G′′, respectively (Fig. 2(d)). Similarly, IB120 shows a higher value of G′ (33 kPa) and G′′ (10 kPa) with respect to IB90 (21 kPa for G′ and 6 kPa for G′′) and IB60 (20 kPa for G′ and 4 kPa for G′′) (Fig. 2(e)). The obtained results demonstrate that by introducing the ionic crosslinking process, it is possible to decrease the photoinitiator concentration (from ink B to ink A) and maintain comparable values of G′ and G′′. More interestingly, the results shown in Table 2 indicate that both inks are suitable materials for printing constructs characterized by mechanical properties spanning a range of stiffnesses from 33 to 5 kPa, which can be easily tuned by changing (i) the concentration of the photoinitiator, (ii) the time of UV irradiation, and (iii) the cross-linking method.
| 3D bioprinted construct | UV irradiation time (s) | Time of immersion in CaCl2 (min) | G′ (kPa) | G′′ (kPa) |
|---|---|---|---|---|
| IB120 | 120 | — | 33 | 10 |
| IB90 | 90 | — | 21 | 6 |
| IB60 | 60 | — | 20 | 4 |
| IA120Ca | 120 | 5 min | 21 | 7 |
| IA90Ca | 90 | 5 min | 17 | 7 |
| IA60Ca | 60 | 5 min | 5 | 2 |
Cells influence the stiffness of the 3D models
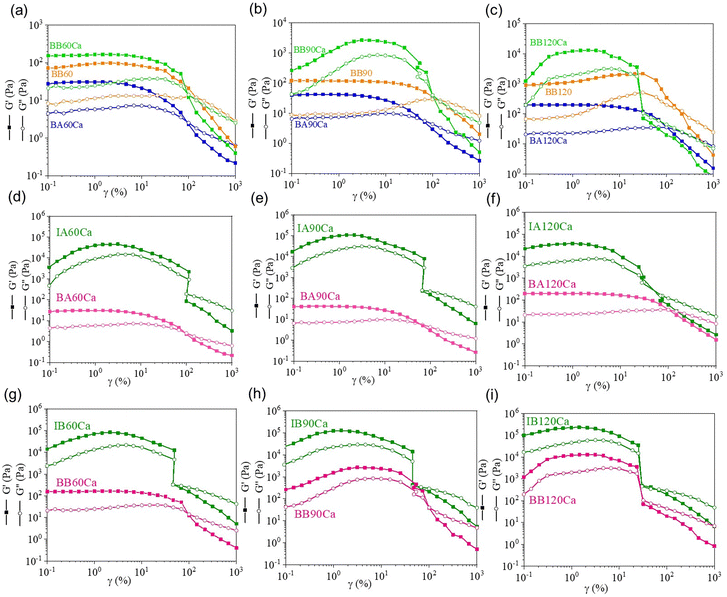
The same post-printing crosslinking processes used for IA and IB were applied to the corresponding bioinks containing HeLa cells (BA, and BB respectively) and amplitude sweep measurements were performed to evaluate whether the presence of the cells affected both G′ and G′′ moduli. The curves obtained are shown in Fig. S2 (ESI†), and the main rheological data are summarized in Table 3. Dual-crosslinked BA scaffolds (Fig. S2(a), ESI†) show a ‘gel-like behavior’ with values of G′ and G′′ moduli consistent with the time of UV irradiation (Table 3). As summarized in Table 3, the LVE range increases at higher strain percentage values with increasing UV irradiation time because of the higher number of crosslinks formed between the polymer chains. A similar rheological behavior was obtained for the constructs printed using bioink BB, which underwent a single photo-crosslinking process (Fig. S2(b), ESI†). Also, in this case, as expected, all the constructs display a ‘gel-like’ behavior with an LVE trend similar to BAXCa constructs (with X indicating, here from, UV time).A dual crosslinking process was also performed on BB 3D printed constructs to (i) compare with the dual crosslinked BA scaffolds, (ii) obtain materials with a wider spectrum of stiffness, and (iii) obtain a stable cell–laden construct while keeping the concentration of the photoinitiator low to reduce possible effects on cell viability. Similar to what was obtained for BAXCa and BBX scaffolds (Fig. S2(a) and (b), ESI†), dual-crosslinked BB scaffolds display a ‘gel-like’ behavior, and BB120Ca shows the highest moduli (Fig. S2(c) and Table 3, ESI†). As the UV exposure time decreased, both G′ and G′′ decreased. However, while BAXCa and BBX constructs showed a clear trend in the LVE range, in the case of the dual crosslinked BBXCa constructs, only BB60Ca highlighted an LVE corresponding to 3% of strain (Table 3). BB90Ca and BB120Ca did not show any G′ plateau in the applied deformation range.
| 3D bioprinted construct | UV irradiation time (s) | Time of immersion in CaCl2 (min) | G′ (kPa) | G′′ (kPa) | LVE (%) |
|---|---|---|---|---|---|
| a n.d.: not detectable. | |||||
| BB120 | 120 | — | 0.900 | 0.390 | 14 |
| BB90 | 90 | — | 0.120 | 0.013 | 10 |
| BB60 | 60 | — | 0.075 | 0.013 | 10 |
| BB120Ca | 120 | 5 | 2 | 0.20 | n.d.a |
| BB90Ca | 90 | 5 | 0.310 | 0.040 | n.d.a |
| BB60Ca | 60 | 5 | 0.120 | 0.020 | 3 |
| BA120Ca | 120 | 5 | 0.200 | 0.020 | 3 |
| BA90Ca | 90 | 5 | 0.041 | 0.007 | 2 |
| BA60Ca | 60 | 5 | 0.030 | 0.002 | 2 |
Interesting evidence was obtained by plotting the overlay of the amplitude sweep curves collected for the BA and BB 3D printed constructs crosslinked with different methods for each UV irradiation time (Fig. 3(a–c)). It was observed that the dual-crosslinked BBXCa constructs always showed higher G′ and G′′ moduli with respect to the dual-crosslinked BAXCa and the single-crosslinked BBX ones, owing to the presence of both ionic crosslinks and the highest number of covalent interchain bonds formed in the presence of the highest concentration of photoinitiator. Moreover, it is possible to identify a correlation between the LVE ranges and the crossover points (G′ = G′′) of BAXCa and single crosslinked BBX constructs because the latter displays a shift of both parameters at higher values of strain with respect to the BAXCa scaffolds. These results might be explained by the fact that in BB, the higher concentration of the Irgacure 2959 photoinitiator leads to a higher number of covalent crosslinks between acrylate moieties compared to those formed in BA. In addition, despite BA constructs underwent a second crosslinking process, they always display lower mechanical properties than BBX constructs because of the weaker ionic crosslinks formed between AlgMa carboxylate groups and Ca2+ ions than the covalent crosslinks formed by photo-crosslinking. Therefore, a higher number of covalent crosslinks in the BBX scaffolds might dominate the sum of both ionic and covalent crosslinks in the BAXCa samples. This ensures higher LVE ranges and higher resistance to deformation before flowing. This correlation was not found for the dual-crosslinked BBXCa constructs because of the strong mechanical properties that lead to a fragile material at high deformations.
Finally, from the comparison between cell-laden and cell-free constructs after the dual crosslinking process, it is possible to observe that the presence of cells strongly affects the mechanical properties of both IA and IB (Fig. 3(d–i)). In particular, the G′ moduli decreased significantly in both the BA and BB samples (see Table S4 in the ESI†). As a general consideration, all moduli after cell loading decreased by approximately two orders of magnitude. This decrease can be explained by considering the steric hindrance of the cells themselves, which usually have dimensions of approximately hundreds of microns; thus, several orders of magnitude higher than the ionic or covalent crosslinks established between polymer chains. In this way, cells might distance AlgMa chains by reducing the statistical probability of interacting with each other to form crosslinks in the presence of calcium ions or when triggered by UV light and a photoinitiator, leading to a decrease in mechanical properties.
Biocompatibility of AlgMa bioinks
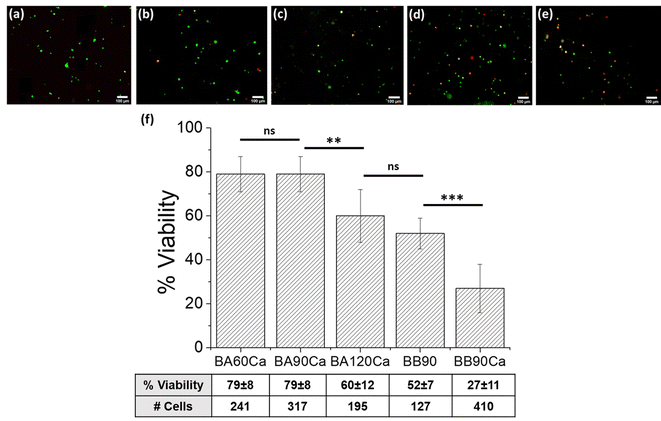
HeLa cell survival in AlgMa hydrogels following 3D bioprinting was evaluated after 3 days of culture using a live/dead assay with calcein–AM and propidium iodide and correlated to the photoinitiator concentration, UV irradiation time, and matrix stiffness. Regarding the duration of UV irradiation, higher mortality is usually observed with increasing UV exposure time. This effect was studied for the BA60Ca, BA90Ca, and BA120Ca samples, and the results are shown in Fig. 4(a–c). Quite high values of cell viability percentages (from 79% for BA60Ca and BA90Ca to 60% for BA120Ca) were obtained (Fig. 4(f)). The 90 seconds irradiation time was selected to compare bioinks A and B and evaluate how the stiffness of the matrix can affect cell behavior by keeping the UV irradiation time fixed. The results showed different viability ranges as a function of bioink formulation and construct characteristics (compare Fig. 4(b), (d) and (e)): starting from BA90Ca, which showed the highest viability (79 ± 8%), cell viability decreased by 52 ± 7% in BB90, while in the more extensively crosslinked sample, BB90Ca, viability percentage slowed down to 27 ± 11%. The high cell mortality rate observed in the latter bioink might be correlated with the higher stiffness of the matrix compared to the other two bioinks, as reported above.Table 4 summarizes the cell viability results for the different materials studied. The data demonstrated that bioprinted samples obtained using bioink A, which contained a lower percentage of Irgacure I2959 (i.e., 0.2%), maintained higher cell viability than all the other samples obtained from bioink B, which was formulated with a higher concentration of Irgacure I2959. The increase in I2959 concentration from 0.2% to 0.25% affected cell viability to an extent of 19% after 60 s of irradiation (viability decreased from 79% to 60%), of 52% after 90 s of irradiation (viability decreased from 79% to 27%), and of 32% after 120 s of irradiation (viability decreased from 60% to 28%). The cell viability decreased when the stiffness and UV irradiation time increased. For example, a change in G′ from 0.120 kPa (BB60Ca) to 2 kPa (BB120Ca) obtained with an increase in UV irradiation time from 60 s to 120 s is related to a decrease in cell viability from 60% to 28%. BB60Ca, BB90Ca, and BB120Ca samples, which have all experienced a dual crosslinking strategy, had higher mortality compared with samples made with the same bioink but that had undergone only photo-crosslinking curing (BB60, BB90, and BB120 samples). In this case, higher mortality is correlated with higher stiffness caused by the calcium chloride crosslinking process. Based on the considerations stated above, it is possible to hypothesize that stiffness plays a role in decreasing the cell viability.
| 3D bioprinted scaffold | UV irradiation time (s) | CaCl2 treatment (min) | G′ (kPa) | Cell viability (%) |
|---|---|---|---|---|
| BB120 | 120 | — | 0.900 | 43 ± 7 |
| BB90 | 90 | — | 0.120 | 52 ± 7 |
| BB60 | 60 | — | 0.075 | 58 ± 7 |
| BB120Ca | 120 | 5 | 2 | 28 ± 17 |
| BB90Ca | 90 | 5 | 0.310 | 27 ± 11 |
| BB60Ca | 60 | 5 | 0.120 | 60 ± 5 |
| BA120Ca | 120 | 5 | 0.200 | 60 ± 7 |
| BA90Ca | 90 | 5 | 0.041 | 79 ± 8 |
| BA60Ca | 60 | 5 | 0.030 | 79 ± 8 |
3D Fabrication of the gradient stiffness platform
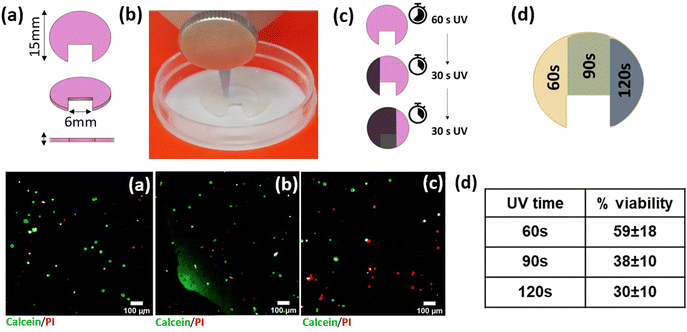
Based on the cell viabilities obtained using the above-described formulations, bioink A was chosen to print the tuneable stiffness platform. A HeLa cell-laden bioprinted platform with a ‘fortune-cookie’-like shape was successfully fabricated, as shown in Fig. 5(a) and (b). The “fortune cookie” shape was chosen for the fabrication of the 3D bioprinted platform with differential stiffness regions (Fig. 5(d), upper panel): the asymmetric shape enables (i) to easily distinguish the different regions during all the experimental phases, (ii) to determine the boundaries of each region, and (iii) to study the cells allocated in the different areas. The constructs were fabricated by printing the same bioink, i.e. BA, for the entire sample. The use of a UV-shielding mask allowed UV rays to pass through only a selected portion of the fortune cookie (Fig. 5(c), upper panel), enabling special control of the UV crosslinking times for the different regions. This strategy allows spatial tuning of the stiffness in the same construct, which is realized using a unique printhead. Live–dead assays directly performed on the ‘fortune cookie’ platform after 14 days of incubation showed a decrease in cell viability compared to that obtained after 3 days of incubation (Fig. 5(a–d), lower panel). Values of 59 ± 18%, 38 ± 10%, and 30 ± 10% in the regions treated with 60 s, 90, and 120 s of UV, respectively, were obtained (Fig. 5(d), lower panel), indicating that cell viability decreased when cell culture was prolonged for two weeks. This result confirmed that we were able to obtain a 3D cell culture with areas characterized by different mechanical properties.Discussion
In this work, a sustainable alginate methacrylation process was accomplished by adapting the gelatin methacryloyl synthesis proposed by Shirahama et al.46 to the alginate polymer chains, which resulted in easier production of alginate methacrylate, both in terms of less complicated procedure and greener synthesis, by reducing the excess MAA used during the whole reaction. This last point is critical in the overall synthesis, as a large excess of MMA induces a decrease in the pH of the reaction bath, requiring frequent additions of NaOH to maintain the pH above the Alg isoelectric point. In the proposed approach, changing the buffering solvent from PB to CB at a specific and well-optimized concentration (0.325 M) produced great self-sustainment of the pH around 8. Moreover, because of the higher buffering ability of the solvent, a lower excess of MAA was needed (1 mL of MAA/g Alg) with respect to previous works in which significant volumes of MAA were involved (from 10 to 20 mL per gram of Alg).471H-NMR spectra confirmed the success of the methacrylation reaction performed for 72 h. It is worth pointing out that, even if 1H-NMR could be useful, in principle, to evaluate the DoF of AlgMa, by integrating the peaks of the protons of the mannuronic and guluronic units with respect to the proton peaks of the vinyl and methyl group of the methacrylate moieties, an accurate quantification of the DoF using 1H-NMR is a challenging task. In particular, the assignment of the peaks and their consequent integration remain highly ambiguous owing to the possible peak shifting caused by the presence of the newly introduced functionalities and the high number of protons contained in the chains. The protons signal of the mannuronic and guluronic units are not fully separated, resulting in an overestimation or underestimation of the DoF if the integration is not considered properly. For this reason, MALDI-MS was considered the best molecular characterization technique for evaluating the DoF of AlgMa. This technique, which is usually applied in the analysis of carbohydrates, can be successfully used to perform an advanced structural investigation of complex polysaccharides and natural complex molecules, exploiting adequate experimental conditions.49–54 The results obtained in this study on AlgMa demonstrate that our synthetic approach can produce AlgMa with 63% of DoF, which is higher with respect to the usual DoF obtained in the literature (around a maximum of 50%).55 Accurate estimation of the DoF is an important parameter that influences the physico-chemical properties of AlgMa-based hydrogels, such as the gelling and rheological behavior of the material.A systematic rheological characterization was carried out in this work as a powerful tool to predict 3D bioprintability and evaluate the stiffness of the obtained constructs. The assessment of printability using rheological tests was first established through oscillation tests (amplitude sweep) that were performed to evaluate the rheological properties of both bioinks A and B, and to assess whether the hydrogels exhibit a ‘solid-like’ (G′ > G′′) or ‘liquid-like’ (G′′> G′) behavior. In fact, ‘solid-like’ features, provided by physical entanglements, crosslinks, and chain interactions, are necessary for a hydrogel to form a continuous and stable filament with good shape fidelity. In our work, this characteristic was achieved only for the AlgMa hydrogel through a pre-crosslinking step with calcium chloride solution to create the proper ionic interactions and ensure suitable rheological properties. The evaluation of the shear-thinning behavior of both AlgMa solution and AlgMa hydrogel, by measuring their viscosity as a function of the shear rate, was a second rheological analysis useful for predicting the printability of the investigated materials. Indeed, to be printable, a material must flow under shear forces by reducing its viscosity with an increase in the shear rate. Even in this case, only the AlgMa hydrogel displayed the right shear-thinning behavior by following the Tscheuschner regression with an exponent p of 0.01 (p < 1), which further quantifies and proves its shear-thinning characteristic.56
Rheology is a useful tool for investigating the stiffness of the obtained 3D models after single and dual crosslinking processes. A comparison of the rheological properties of 3D constructs with and without embedded HeLa cells showed that the presence of cells reduced by two orders of magnitude both G′ and G′′ moduli. The results of this study demonstrate that for both bioinks A and B, it is possible to obtain constructs displaying a wide range of stiffness depending on the post-printing treatment. The mechanical properties obtained herein are summarized in Tables 3 and 4, along with cell viability at 3 days of culture, and can be exploited to reproduce tissues with different stiffness, such as pathological, inflammatory, or tumor tissues.
It is difficult to define a univocal value of rigidity of a tissue, whether physiological or pathological, owing to the intra-heterogeneity of the real tissues. However, AFM measurements performed on cancer biopsies demonstrated that the stiffness of such tissues is described by a bimodal distribution.57 Breast cancer tissue stiffness, for example, shows two prominent peaks at 0.57 ± 0.16 kPa and 1.99 ± 0.73 kPa while healthy specimen reveals a unimodal stiffness distribution of 1.13 ± 0.78 kPa. In the same study, Plodinec et al.57 showed that the core part of breast cancer is mostly a soft area with a specific stiffness peak, whereas the periphery shows a non-homogeneous area with stiff features.
Both physiological and pathological situations can be mimicked with the bioink formulations developed in this study because we were able to reproduce stiffness that ranged from 0.030 kPa to 2 kPa (Table 3). The dual crosslinked samples BB60Ca, BB90Ca, and BB120Ca showed stiffness values ranging from 0.120 kPa to 2 kPa, depending on the UV curing time. These values are very close to those found in tissues characterized by breast cancer (as illustrated above). On the other hand, if we use bioink B and a one-step crosslinking procedure, we obtain tissue models with a stiffness ranging from 0.075 to 0.9 kPa, near to healthy breast tissues. Dual crosslinked bioink A shows stiffness that can mimic softer tissue areas (stiffness range: 0.030–0.2 kPa). However, it should be noted that increased stiffness correlated with decreased HeLa cell viability. Many studies have demonstrated the effects of matrix stiffness on the cellular pathway,15,58,59 which are consistent with the results obtained in this work, but only a few works have deeply analyzed the effects of the photoinitiator concentration and the crosslinking parameters on cell behavior.60,61 It is well recognized that the photoinitiator concentration and UV irradiation time are key parameters that affect cell viability. In this study, all factors related to photo-crosslinking and final construct stiffness were systematically studied and considered in defining the hydrogel formulation. Our results demonstrated that cell viability decreased (i) with increasing UV irradiation time at a fixed photoinitiator concentration, (ii) with an increase in Irgacure I2959 concentration at a fixed UV time, and (iii) with an increase in stiffness at fixed concentration of I2959 and UV time.
The approach proposed in this work is essential to obtain stable cell-laden constructs for two main reasons: (i) because of the strong impact of the presence of cells on the crosslinking efficiency and, at the same time, (ii) to keep the concentration of the photoinitiator and UV irradiation time as low as possible, to avoid adverse effects on cell viability. This method was therefore useful for maintaining good mechanical properties inherent to construct fabrication, limiting cell damage, and, more interestingly, allowing the successful development of a tissue platform with spatially controllable stiffness via one-step 3D bioprinting. HeLa cells viability is approximately 95% in alginate-based bioink, as reported in literature for gelatin/alginate/fibrinogen bioink.62 We aimed to retain the maximum viability of cells, and for this reason, bioink A formulation was chosen to print a ‘fortune-cookie’ platform with tailored mechanical properties, owing to its relatively high viability in comparison to both literature reports and bioink B formulations. Cell viability studies were carried out for two weeks on the 3D printed platform, exploiting the availability of three regions with different properties on the same construct. The results confirmed those obtained on single constructs regarding the effect of UV irradiation time, even though cell viability decreased significantly at 14 days with respect to the 3 days cultures. In this study we classified cell viability as listed in the following: >90% = optimal viability; between 90% and 80%, high viability; between 80% and 60%, good viability; between 60% and 40%, low viability; below 40%, poor viability. Cells that were bioprinted using the bioink A and that underwent dual crosslinking showed from high (80%) to good (60%) viabilities at 72 h after printing (Fig. 4(a–c)) while the viability decreased from good (60%) to poor (30%) at 14 days after printing (Fig. 5). These results indicate that, to a certain extent, tuning matrix mechanical properties affects cell viability; however, the obtained percentages of viability are sufficiently high to enable the study of cellular behavior in an extended range of matrix stiffness values, a feature that is highly relevant in cancer research.
Conclusion
In this study, a 3D bioprinted platform with spatially controllable stiffness was successfully fabricated via a one-step bioprinting and dual-crosslinking process. HeLa cells were used as a proof of principle cell model for platform development because they represent the ‘gold-standard’ model for cancer cell lines. However, the concept of the gradient stiffness platform presented in this study can be used for other cellular systems. A critical and systematic study of the effects of the photoinitiator concentration, photo-crosslinking process, and mechanical properties of the AlgMa-based construct on cell viability was conducted. This study provides fundamental knowledge on the biofabrication of 3D bioprinted models using photo-crosslinkable polymeric matrices, that has attracted increased attention in recent years. Notably, an affordable “greener” synthetic approach to produce highly substituted AlgMa was proposed and the material was thoroughly characterized using spectroscopic techniques (FT-IT, 1H-NMR) and mass spectrometry measurements (MALDI-TOF-MS). A dual crosslinking process was exploited in this study, involving (i) a photo-crosslinking process through UV irradiation, and (ii) an ionic crosslinking, by treating the 3D construct in a CaCl2 solution. The process was optimized to overcome limitations related to (i) low cell viabilities, induced by high photoinitiator concentration and extended UV irradiation time, and (ii) weak mechanical properties inherent to the fabricated constructs. Rheology has been extensively used as a valid support tool for optimizing printable bioinks. Oscillatory amplitude sweeps and rotational tests were employed to demonstrate the modulation of the mechanical properties using the proposed dual-crosslinking strategy and to evaluate the impact of cell steric hindrance on the mechanical properties of the bioink formulations. We demonstrated that ink rheological properties are affected by the presence of cells by approximately two orders of magnitude and that cell viability is strongly dependent (i) on the duration of UV irradiation, (ii) on Irgacure concentration, and (iii) on hydrogel stiffness by itself. By employing the strategy illustrated in our work, it is possible to replicate the stiffness of specific tissues, their variability, and the characteristics of the boundaries between different regions. Our platform easily mimics physiological and pathological situations; for instance, dual-crosslinked bioink B showed a stiffness that ranged from physiological to pathological tissues (from 0.120 kPa to 2 kPa, depending on the UV curing times), while the stiffness of dual-crosslinked bioink A (stiffness range: 0.030 kPa to 0.2 kPa) mimics softer areas of normal tissues. By tuning the duration of UV irradiation, it is possible to obtain a differential stiffness platform that recapitulates the stiffness heterogeneity of specific tissues, such as that existing from the periphery to the core of tumor tissues. Extracellular matrix stiffness regulates several physiological and pathological processes, for example, it is deeply involved in tumor invasion and metastasis. However, the relationship between ECM/tissue stiffness and cancer progression and aggression remains unclear. A differential stiffness platform is an essential tool for studying the interplay between tumor aggressiveness and matrix stiffness. By using the platform described herein, it would be possible to examine cells that feel different mechanical stimuli and observe their interaction and specific behavior in a controlled fashion, paving the way for an understanding of the transduction of mechanical inputs into transcriptional outputs, which drive tumor progression. Moreover, a model that replicates the key features of cancer tissue, including their stiffness, could be used to elucidate the mechanisms of chemoresistance and metastatization. In general, ECM has been proven to directly modulate cellular energetic metabolism;63 consequently, this platform may also be employed to investigate the role of matrix stiffness in pathologies characterized by mitochondrial impairment or defects.64 The 3D platform developed in this work can be used as a model to study cell behavior as a function of mechanical properties of the extracellular microenvironment.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The Italian Ministry for Universities and Research (MUR) is acknowledged. Authors thank National Recovery and Resilience Plan (NRRP), Mission 04 Component 2 Investment 1.5 – NextGenerationEU, Call for tender n. 3277 dated 30/12/2021, Award Number: 0001052 dated 23/06/2022. MB, MM and SR acknowledge “Fondazione AIRC per la Ricerca sul Cancro”, for the MFAG Id. 19044 granted to SR. The PhD scholarship of LDL was funded by the European Union - NextGenerationEU through the Italian Ministry of University and Research under PNRR – Mission 4 Component 2, Investment 3.3 “Partnerships extended to universities, research centres, companies and funding of basic research projects” D.M. 352/2021 – CUP J33C22001330009.Notes and references
- D. Baruffaldi, G. Palmara, C. Pirri and F. Frascella, ACS Appl. Bio Mater., 2021, 4(3), 2233–2250 CrossRef CAS PubMed.
- O. Habanjar, M. Diab-Assaf, F. Caldefie-Chezet and L. Delort, Int. J. Mol. Sci., 2021, 22(22), 12200 CrossRef CAS PubMed.
- R. S. Stowers, A. Shcherbina, J. Israeli, J. J. Gruber, J. Chang, S. Nam, A. Rabiee, M. N. Teruel, M. P. Snyder, A. Kundaje and O. Chaudhuri, Nat. Biomed. Eng., 2019, 3(12), 1009–1019 CrossRef PubMed.
- F. Guilak, D. M. Cohen, B. T. Estes, J. M. Gimble, W. Liedtke and C. S. Chen, Cell Stem Cell, 2009, 5(1), 17–26 CrossRef CAS PubMed.
- R. M. Delaine-Smith and G. C. Reilly, Muscles Ligaments Tendons J., 2012, 2(3), 169–180 Search PubMed.
- R. A. Marklein and J. A. Burdick, Spatially controlled hydrogel mechanics to modulate stem cell interaction, Soft Matter, 2010, 6, 136–143 RSC.
- L. G. Griffith and M. A. Swartz, Nat. Rev. Mol. Cell Biol., 2006, 7(3), 211–224 CrossRef CAS PubMed.
- C. Walker, E. Mojares and A. Del Río Hernández, Int. J. Mol. Sci., 2018, 19(10), 3028 CrossRef PubMed.
- C. Frantz, K. M. Stewart and V. M. Weaver, J. Cell Sci., 2010, 123(24), 4195–4200 CrossRef CAS PubMed.
- R. S. Stowers, A. Shcherbina, J. Israeli, J. J. Gruber, J. Chang, S. Nam, A. Rabiee, M. N. Teruel, M. P. Snyder, A. Kundaje and O. Chaudhuri, Nat. Biomed. Eng., 2019, 3(12), 1009–1019 CrossRef PubMed.
- S. C. Wei, L. Fattet, J. H. Tsai, Y. Guo, V. H. Pai, H. E. Majeski, A. C. Chen, R. L. Sah, S. S. Taylor, A. J. Engler and J. Yang, Nat. Cell Biol., 2015, 17(5), 678–688 CrossRef CAS PubMed.
- C. R. Pfeifer, C. M. Alvey, J. Irianto and D. E. Discher, Curr. Opin. Syst. Biol., 2017, 2, 103–114 CrossRef PubMed.
- I. Acerbi, L. Cassereau, I. Dean, Q. Shi, A. Au, C. Park, Y. Y. Chen, J. Liphardt, E. S. Hwang and V. M. Weaver, Integr. Biol., 2015, 7(10), 1120–1134 CrossRef CAS PubMed.
- S. E. Reid, E. J. Kay, L. J. Neilson, A. Henze, J. Serneels, E. J. McGhee, S. Dhayade, C. Nixon, J. B. Mackey, A. Santi, K. Swaminathan, D. Athineos, V. Papalazarou, F. Patella, A. Román-Fernández, Y. ElMaghloob, J. R. Hernandez-Fernaud, R. H. Adams, S. Ismail, D. M. Bryant, M. Salmeron-Sanchez, L. M. Machesky, L. M. Carlin, K. Blyth, M. Mazzone and S. Zanivan, EMBO J., 2017, 36(16), 2373–2389 CrossRef CAS PubMed.
- J. Schrader, T. T. Gordon-Walker, R. L. Aucott, M. van Deemter, A. Quaas, S. Walsh, D. Benten, S. J. Forbes, R. G. Wells and J. P. Iredale, Hepatology, 2011, 53(4), 1192–1205 CrossRef CAS PubMed.
- E. Azzalini, N. Abdurakhmanova, P. Parisse, M. Bartoletti, V. Canzonieri, G. Stanta, L. Casalis and S. Bonin, Nanomed. Nanotechnol. Biol. Med., 2021, 37, 102452 CrossRef CAS PubMed.
- J. M. Chang, W. K. Moon, N. Cho, A. Yi, H. R. Koo, W. Han, D. Y. Noh, H. G. Moon and S. J. Kim, Breast Cancer Res. Treat., 2011, 129(1), 89–97 CrossRef PubMed.
- L. J. W. Tang, A. Zaseela, C. C. M. Toh, C. Adine, A. O. Aydar, N. G. Iyer and E. L. S. Fong, Adv. Drug Delivery Rev., 2021, 175, 113817 CrossRef CAS PubMed.
- M. Ravi, V. Paramesh, S. R. Kaviya, E. Anuradha and F. D. Paul Solomon, J. Cell. Physiol., 2014, 230(1), 16–26 CrossRef PubMed.
- J. M. Lee, P. Mhawech-Fauceglia, N. Lee, L. C. Parsanian, Y. G. Lin, S. A. Gayther and K. Lawrenson, Lab Investig., 2013, 93(5), 528–542 CrossRef PubMed.
- M. Kuzucu, G. Vera, M. Beaumont, S. Fischer, P. Wei, P. Shastri and A. Forget, ACS Biomater. Sci. Eng., 2021, 7(6), 2192–2197 CrossRef CAS PubMed.
- A. Lavrentieva, T. Fleischhammer, A. Enders, H. Pirmahboub, J. Bahnemann and I. Pepelanova, Macromol. Biosci., 2020, 20(7), 2000107 CrossRef CAS PubMed.
- J. Dou, S. Mao, H. Li and L. Jin-Ming, Anal. Chem., 2019, 92(1), 892–898 CrossRef PubMed.
- C. Liu, M. Li, Z. X. Dong, D. Jiang, X. Li, D. Chen, X. Zou, X. Zhang and G. D. Luker, Acta Biomater., 2021, 131, 326–340 CrossRef CAS PubMed.
- A. M. Bejoy, K. N. Makkithaya, B. B. Hunakunti, A. Hegde, K. Krishnamurthy, A. Sarkar, C. F. Lobo, D. V. S. Keshav, S. Mascarenhas, S. Chakrabarti, S. R. R. D. Kalepu, B. Paul and N. Mazumder, Bioprinting, 2021, 24, e00176 CrossRef.
- I. T. Ozbolat and M. Hospodiuk, Biomaterials, 2016, 76, 321–343 CrossRef CAS PubMed.
- N. Charbe, P. A. McCarron and M. M. Tambuwala, World J. Clin. Oncol., 2017, 8(1), 21–36 CrossRef PubMed.
- P. Datta, M. Dey, Z. Ataie, D. Unutmaz and I. T. Ozbolat, npj Precis Oncol., 2020, 4(1), 18 CrossRef PubMed.
- C. Wang, Z. Tang, Y. Zhao, R. Yao, L. Li and W. Sun, Biofabrication, 2014, 6(2), 022001 CrossRef PubMed.
- E. Axpe and M. L. Oyen, Int. J. Mol. Sci., 2016, 17(12), 1976 CrossRef PubMed.
- M. I. Neves, L. Moroni and C. C. Barrias, Front. Bioeng. Biotechnol., 2020, 8, 665 CrossRef PubMed.
- J. A. Rowley, G. Madlambayan and D. J. Mooney, Biomaterials, 1999, 20(1), 45–53 CrossRef CAS PubMed.
- F. E. Freeman and D. J. Kelly, Sci. Rep., 2017, 7(1), 17042 CrossRef PubMed.
- T. Xia, W. Liu and L. Yang, J. Biomed. Mater. Res., Part A, 2017, 105(6), 1799–1812 CrossRef CAS PubMed.
- F. Araiza-Verduzco, E. Rodríguez-Velázquez, H. Cruz, I. A. Rivero, D. R. Acosta-Martínez, G. Pina-Luis and M. Alatorre-Meda, Materials, 2020, 13(3), 534 CrossRef CAS PubMed.
- M. Hasany, S. Talebian, S. Sadat, N. Ranjbar, M. Mehrali, G. G. Wallace and M. Mehrali, Appl. Mater. Today, 2021, 24, 101150 CrossRef.
- S. I. Somo, K. Langert, C.-Y. Yang, M. K. Vaicik, V. Ibarra, A. A. Appel, B. Akar, M. H. Cheng and E. M. Brey, Acta Biomater., 2018, 65, 53–65 CrossRef CAS PubMed.
- Y. Gao and X. Jin, Mar. Drugs, 2019, 17, 557 CrossRef CAS PubMed.
- A. Jacobs and O. Dahlman, Anal. Chem., 2021, 73(3), 405–410 CrossRef PubMed.
- M. Persike and M. Karas, Rapid Commun. Mass Spectrom., 2009, 23(22), 3555–3562 CrossRef CAS PubMed.
- M. Persike, M. Zimmermann, J. Klein and M. Karas, Anal. Chem., 2010, 82(3), 922–929 CrossRef CAS PubMed.
- N. Bergman, D. Shevchenko and J. Bergquist, Anal. Bioanal. Chem., 2014, 406, 49–61 CrossRef CAS PubMed.
- D. Aiello, F. Lucà, C. Siciliano, P. Frati, V. Fineschi, R. Rongo and A. Napoli, J. Proteome Res., 2021, 20(5), 2607–2617 CrossRef CAS PubMed.
- A. Cantelli, M. Malferrari, A. Solda, G. Simonetti, S. Forni, E. Toscanella, E. J. Mattioli, F. Zerbetto, A. Zanelli, M. Di Giosia, M. Zangoli, G. Barbarella, S. Rapino, F. Di Maria and M. Calvaresi, J. Am. Chem. Soc., 2021, 1(7), 925–935 CAS.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9(7), 676–682 CrossRef CAS PubMed.
- H. Shirahama, B. H. Lee, L. P. Tan and N. J. Cho, Sci. Rep., 2016, 6, 31036 CrossRef CAS PubMed.
- A. D. Rouillard, C. Berglund, J. Y. Lee, W. J. Polacheck, Y. Tsui, L. J. Bonassar and B. J. Kirby, Tissue Eng., Part C, 2011, 17(2), 173–179 CrossRef CAS PubMed.
- B. Stubbe, G. J. Graulus, G. Reekmans, T. Courtin, J. C. Martins, S. Van Vlierberghe, P. Dubruel and P. Adriaensens, A straightforward method for quantification of vinyl functionalized water soluble alginates via 13C-NMR spectroscopy, Int. J. Biol. Macromol., 2019, 134, 722–729 CrossRef CAS PubMed.
- D. Aiello, C. Siciliano, F. Mazzotti, L. Di Donna, C. M. Athanassopoulos and A. Napoli, RSC Adv., 2018, 8(63), 36104–36113 RSC.
- D. Aiello, C. Siciliano, F. Mazzotti, L. Di Donna, C. M. Athanassopoulos and A. Napoli, Food Chem., 2020, 307, 125527 CrossRef CAS PubMed.
- D. J. Harvey, Mass Spectrom. Rev., 2006, 25(4), 595–662 CrossRef CAS PubMed.
- J. Zaia, Mass spectrometry of oligosaccharides, Mass Spectrom. Rev., 2004, 23(3), 161–227 CrossRef CAS PubMed.
- Y. Lin, M. de Kreuk, M. C. M. van Loosdrecht and A. Adin, Water Res., 2010, 44(11), 3355–3364 CrossRef CAS PubMed.
- T. M. Aida, T. Yamagata, M. Watanabe, Jr. and R. L. Smith, Carbohydr. Polym., 2010, 80(1), 296–302 CrossRef CAS.
- H. H. Mishbak, G. Cooper and P. J. Bartolo, Int. J. Bioprint, 2019, 5(2), 189 CAS.
- A. Diab, J. Elastomers Plastics, 2018, 50(4), 354–371 CrossRef CAS.
- M. Plodinec, M. Loparic, C. A. Monnier, E. C. Obermann, R. Zanetti-Dallenbach, P. Oertle, J. T. Hyotyla, U. Aebi, M. Bentires-Alj, R. Y. H. Lim and C. A. Schoenenberger, Nat. Nanotechnol., 2012, 7(11), 757–765 CrossRef CAS PubMed.
- K. H. Vining and D. J. Mooney, Nat. Rev. Mol. Cell Biol., 2017, 18(12), 728–742 CrossRef CAS PubMed.
- J. Xu, S. N. Fu and F. Hug, BMC Musculoskelet Disord, 2021, 22, 829 CrossRef PubMed.
- T. Billiet, E. Gevaert, T. De Schryver, M. Cornelissen and P. Dubruel, Biomaterials, 2014, 35(1), 49–62 CrossRef CAS PubMed.
- S. Piluso, D. Flores Gomez, I. Dokter, L. Moreira Texeira, Y. Li and J. Leijten, J. Mater. Chem. B, 2020, 8(41), 9566–9575 RSC.
- Y. Zhao, R. Yao, L. Ouyang, H. Ding, T. Zhang, K. Zhang, S. Cheng and W. Sun, Biofabrication, 2014, 6(3), 035001 CrossRef CAS PubMed.
- T. Panciera, L. Azzolin, M. Cordenonsi and S. Piccolo, Nat. Rev. Mol. Cell Bio, 2017, 18, 758–770 CrossRef CAS PubMed.
- M. Malferrari, A. Ghelli, F. Roggiani, G. Valenti, F. Paolucci, M. Rugolo and S. Rapino, ChemElectroChem, 2019, 6(3), 627–633 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Scheme of the alginate methacrylation reaction; detailed interpretation of FTIR spectra; mass spectrometry data of AlgMa products after hydrolysis; stability tests and cell viability results; rheological results. See DOI: https://doi.org/10.1039/d3ma00319a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |