Open Access Article
Open Access ArticleInterfacial stabilization of aqueous two-phase systems: a review
Caitlyn
Fick
a,
Zara
Khan
b and
Samanvaya
Srivastava
 *bcd
*bcd
aDepartment of Chemistry and Biochemistry, University of California, Los Angeles, Los Angeles, CA 90095, USA
bDepartment of Chemical and Biomolecular Engineering, University of California, Los Angeles, Los Angeles, CA 90095, USA
cCalifornia NanoSystems Institute, University of California, Los Angeles, Los Angeles, CA 90095, USA
dInstitute for Carbon Management, University of California, Los Angeles, Los Angeles, CA 90095, USA. E-mail: samsri@ucla.edu
First published on 11th September 2023
Abstract
Aqueous two-phase systems (ATPS) are useful in various applications, from purification and separation of biomolecules to wastewater treatment. While they have great utility on their own, there is great interest in discovering how their emulsions, comprising droplets of one aqueous phase dispersed in the other aqueous phase, might be stabilized to enhance their functionality and applications. There are several examples of these systems, but the two most common systems found in the literature are PEG–dextran and complex coacervate ATPS. In this Review, we discuss these systems, their utility, and many different approaches for stabilizing their water/water (w/w) emulsions. We highlight examples wherein interfacial stabilizers such as liposomes, polymers of diverse architectures, colloids of varied shapes and morphologies, and even whole cells have been employed. These stabilization approaches for both PEG–dextran and complex coacervate ATPS are discussed. We conclude with a discussion of the applications of these ATPS and how they can benefit from the creation of corresponding w/w emulsions with stabilized droplets.
Introduction
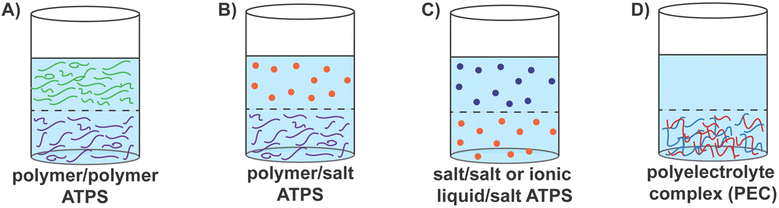
Aqueous two-phase systems (ATPS), consisting of two immiscible aqueous solutions, are routinely encountered in biology, chemistry, chemical engineering, and food science.1–5 One of the first demonstrations of the utility of these systems was using dextran and methylcellulose for the separation of proteins by Albertsson in 1958.6 Since then, ATPS have been utilized for the separation and purification of biomolecules,1,5,6 in drug delivery,7–9 as protocells and bioreactors,10,11 detection of drug residues in food, drink, and water samples,12,13 sequestration of precious metals,14–16 and even for wastewater treatment.1,3,17–19 These systems provide benign, biocompatible, and cheaper alternatives to oil–water systems, making them attractive materials systems for environmentally friendly manufacturing and processing techniques.1,20–24The two phases in ATPS can consist of two immiscible aqueous polymer solutions, aqueous solutions of a polymer and a salt, two aqueous salt solutions, or a complex coacervate phase comprising oppositely charged polyelectrolytes in equilibrium with a polyelectrolyte-lean supernatant phase.4,25–27 ATPS can be categorized as segregative or associative systems, depending on the thermodynamic drivers of phase separation.11,25–27 In segregative systems, the incompatibility between the aqueous solutions of the (macro)molecular solutes results in aqueous phases enriched in one solute (macro)molecule or the other.1–3,25 A common, widely employed system is the mixture of polyethylene glycol (PEG) and dextran, which form a segregative ATPS with two phases comprising either PEG or dextran chains (Fig. 1A).5,28 In contrast, associative ATPS form upon attractive interactions-driven complexation of (macro)molecules. Common examples of such systems include polyelectrolyte complex coacervates (Fig. 1D) and membraneless cellular organelles.29–32
Segregative polymer–polymer ATPS have proven effective for separating many biological compounds and thus has become a familiar and frequently-used method in biotechnological settings.6,21,33–37 The utility of ATPS for extracting biomacromolecules has been enhanced further by influencing the biomacromolecule to partition into either phase.38,39 This can be done by affinity partitioning,38–46 where the polymers themselves may be modified with affinity groups that are specific for biomacromolecules of interest39–42,44,45 or by modulating the molecular weight of the polymers.4,6,46–49 The physicochemical properties of the polymers, such as superficial electrochemical charge and hydrophobicity, can also affect the partitioning of biomacromolecules.3,44,47,50 Polymer–polymer ATPS have been utilized not only for the separation and purification but also for the eventual extraction and encapsulation of proteins,51 nucleic acids,52 viruses,53 antibodies,39,54 and even whole cells.43,45,55–60 These examples describe the diverse utility of polymer–polymer aqueous two-phase systems.
Polymer–salt systems are another common way to create segregative ATPS, wherein salts with high ionic strengths, such as phosphates, sulfates, and citrates, are most often used in combination with PEG and result in coexisting salt-rich and polymer-rich phases (Fig. 1B).4,34,61 These systems are often used for cell encapsulation using microfluidic platforms, where droplet sizes and behaviors can be precisely manipulated.57 Polymer–salt systems have also been used to separate proteins,62–64 DNA,65 and virus-like particles66,67 in systems such as PEG–sodium sulfate or phosphate, in addition to the encapsulation of cells57 and purification of antibodies and biopharmaceutical products.23,35,37,68 Salt–salt systems or ionic liquid–salt ATPS have also emerged in recent years as segregative ATPS (Fig. 1C).4,61,69,70 These systems are formed upon the co-dissolution of a salt with strongly charged ions with another salt or ionic liquid comprising low-charge density ions.4,61,70 Greater tunability of polarity enables these systems to overcome some of the issues, such as slow phase separation, found in polymer–polymer ATPS.4,61 Therefore, extraction and purification abilities can be enhanced in salt–salt ATPS, encouraging their use for extraction and purification of biomolecules when in solution with secondary compounds or contaminants.61,71,72
Associative ATPS, in contrast, are driven by the complexation of (macro)molecules via attractive interactions and include systems such as polyelectrolyte complex coacervates and membraneless cellular organelles.29–32,73,74 In the former, oppositely charged macroions (typically polyelectrolytes) undergo complexation, creating a macroion-rich complex coacervate phase separating from a macroion-lean supernatant phase (Fig. 1D).29–32 In the latter, intrinsically disordered proteins (IDPs), or proteins with intrinsically disordered regions (IDRs), undergo phase separation in the cytoplasmic milieu, forming membraneless organelles such as nucleoli, stress granules, and P-bodies.74,75 While attractive electrostatic interactions and the entropy gains from counterion release are the primary drivers for complex coacervation, the formation of membraneless organelles also benefit from hydrophobic collapse, hydrogen-bonding, π–π stacking, van der Waals interactions, and other short-range attractive intermolecular interactions to drive phase separation.73,74,76–78
Complex coacervates comprising oppositely charged polyelectrolytes are fascinating systems with immense potential for many different applications ranging from their use as biomaterials for cartilage mimics, adhesives for wound healing, and drug delivery vehicles to their use as colloidal bioreactors in cell-free biocatalysis,79,80 encapsulants in cosmetics,81,82 and sorbents in wastewater treatment processes.17,83,84 Concomitantly, a significant body of research investigating the thermodynamics of complexation as well as on the influence of diverse intrinsic properties (such as the polyelectrolyte chemistry,85,86 length,31,87 architecture,32,86 charge density,88–90etc.) and extrinsic properties (ionic strength,32,87 pH,32,86,91 solvent quality,85 temperature,92etc.) on the composition and properties of the coacervate and supernatant phases has emerged.93 The crowded interiors of the coacervates and their ability to sequester charged (bio)macromolecules from their surroundings have promoted their utility as encapsulants and stabilizers in biological,79,94 industrial,95–97 environmental,17,19,83 and cosmetic applications.81,82
Stabilization of the aqueous two-phase systems, including those such as polymer/polymer, water/ionic liquid,98,99 and polyelectrolyte complex systems,80 at the micrometer length scale or smaller, can be expected to enhance their utility even further by enabling the implementation of ATPS in a host of novel applications.1,10,25,26,100 The canonical two-phase mixture of oil and water derives its significant utility from the stabilized mixtures in the form of emulsions and micellar solutions. These have inspired efforts for stabilizing water–water (w/w) interfaces, resulting in progressively increasing research attention. While the design of the interfacial stabilizer depends on the nature of the w/w interface and the composition of the two aqueous phases, a few broad themes can still be identified. As such, the energetically favorable coalescence (that reduces the interfacial area) must be superseded to stabilize droplets of one phase in another. This is typically accomplished by adding a third component that accumulates at the interface, reduces the interfacial area, and stabilizes the droplets.101,102 The energy barrier that stabilizes the droplets is estimated as the product of interfacial tension and the area taken up by the stabilizing components and is referred to as the trapping energy. In w/w interfaces, the interfacial tension is low, and correspondingly the trapping energy is significantly lower as compared to oil–water systems. Therefore, unique design approaches that go beyond employing surfactant-like molecules (with one anchoring point at the interface) for stabilizing the w/w interfaces must be developed. At the same time, the design criteria need to encode an affinity for both phases of the specific ATPS in the different parts of the stabilizer to promote its interfacial assembly.
The effectiveness of typical w/w interfacial stabilizers is typically ascertained by microimaging (to monitor the timespans over which droplets resist coalescence) and turbidimetry (to monitor the temporal evolution of the emulsion turbidity as a proxy for emulsion stability). The temporal evolution of the droplet size distribution can also be monitored to ascertain droplet stability. It can also provide insights into how the size distribution may affect the ability of the droplets to be stabilized. However, it is more often that researchers consider and modify the volumes of polymer or salt solutions or the size of stabilizing particles instead, which can ultimately affect the size of the droplets.80,103,104 For example, it has been found that native proteins are generally too small for stabilization purposes (radius of ∼2 nm).101 It has also been reported that droplets stabilized by triblock polymers, discussed in detail in the following section, can experience changes in stability based on droplet size. In this case, smaller droplets with diameters of a few micrometers are more stable than those with sizes of the order of tens of micrometers.105 It may be noted that the coalescence time varies significantly for different systems. The coalescence time can be affected by Brownian diffusion or the height of the energy barrier to form a hole in the stabilizing shell, which is influenced by the extent to which the droplet is covered.106 Additionally, while the interfacial energy is minimized in these systems, it is not zero. Thus, the droplets will eventually coalesce to reach their lowest energy state.
In this Review, we assess recent progress in creating and stabilizing ATPS (micro)emulsions, specifically focusing on PEG–dextran and complex coacervate (micro)emulsions. We highlight different stabilization methods for PEG–dextran and complex coacervate (micro)emulsions and their prospective applications. These systems provide unique avenues for separation and encapsulation, using compounds that are commonly used, are generally cheap, and are environmentally friendly. As a result, they will likely continue to expand and become ubiquitous materials in research and industry.
Stabilization strategies for PEG–dextran emulsions
Aqueous mixtures of PEG and dextran are one of the most common systems that undergo segregative liquid–liquid phase separation to produce aqueous two-phase systems (ATPS) and the corresponding water–water (w/w) interfaces (Fig. 1). Numerous examples of this system being particularly useful in separating and purifying biomolecules have appeared in the literature.35,39,107,108 The incompatible phases both generally contain 70–95% water, and the interfacial tension between them is low (10−4–10−1 mN m−1); significantly lower than oil–water interfaces (∼25 mN m−1) or alkane–water interfaces (∼50 mN m−1).100,109–112 This ultimately leads to high mass transfer and facilitates the partitioning of sensitive biomolecules, often into the dextran phase, in a non-destructive manner.59,100,109,113Proteins, in particular, can be extracted and purified using PEG–dextran systems. A notable benefit of this approach is that protein denaturation can be diminished in these systems, as polymers typically used in these systems (PEG, dextran) can stabilize both the structure and the activity of proteins and enzymes.28,114,115 Additionally, depending on the protein, it is possible to separate a protein into either the PEG or dextran phase, and this partitioning can be modulated based on the molecular weight of and modifications to the polymers and the proteins’ hydrophobicity and surface properties.3,39,46,68,116
These systems are not only gentle enough to maintain protein structure and function, but they can also support enzyme function in bacterial cells. Dextran, in particular, can have stabilizing effects on microbial cells.60,107,117 At the same time, the partitioning of cells can be modulated by altering the molecular weights and concentration of the polymers.118 For example, if the molecular weight of dextran is lowered, more cells partition into the dextran-rich phase.119 As such, PEG–dextran ATPS have been employed to partition complex cell lines, such as hybridoma cells (mouse/mouse hybridoma cell line BIF6A7), as well as other cell types and cell particles.28,58–60,107,120 These hybridoma cells were supported for long-term growth.59 Additionally, different cell types can be separated from each other using ATPS43,45 or sequestered.56 Thus, this system allows for a wide range of biological applications and affords excellent opportunities for optimizing the system for specific proteins and other biomacromolecules of interest.52,118,119
Stabilizing the water–water interface against coalescence and subsequent emulsification and formation of PEG-in-dextran or dextran-in-PEG emulsions can further enhance the utility of PEG–dextran ATPS. These emulsions have both been investigated extensively, but there is no clear consensus on which is more stable.104,121 However, in either form, these emulsions are particularly exciting because they allow for compartmentalization, typically achieved in oil–water emulsions, without using organic solvents and hydrophobic fluids. The emulsion droplets can be stabilized in several ways by employing materials such as block copolymers, colloids, and liposomes.1,25,26,100 These different methods and some specific examples are highlighted below.
The use of block polymers containing a hydrophobic middle block and two hydrophilic ends stands out as one of the most intuitive approaches to successfully stabilize PEG–dextran interfaces. Self-assembly of an ABC triblock copolymer, where A is hydrophilic and has a specific affinity for one polymer phase, B is hydrophobic, and C is hydrophilic and has a specific affinity for the second polymer phase, in water can form a polymersome (Fig. 2A).105 Such polymersomes have been effective in stabilizing PEG/dextran ATPS, in some cases, for over eight months.105 In particular, it has been reported that more stable systems tend to have smaller droplets and vice versa; increasing the hydrophobic block length correlates linearly with droplet stability against coalescence.105 Jin and coworkers have simplified this system by using two block copolymers to form polymersomes that stabilize PEG–dextran ATPS, where two amphiphilic diblock copolymers co-assemble to form polymersomes with an aqueous cavity. In this case, one block copolymer has a hydrophilic block that has an affinity for one polymer phase and a hydrophobic block, and the other block copolymer has a hydrophilic block that has a specific affinity for the second polymer phase and a hydrophobic block (Fig. 2B).122 These polymersomes self-assemble and enable facile modifications and tunability, presenting a versatile method for creating PEG–dextran microemulsions.
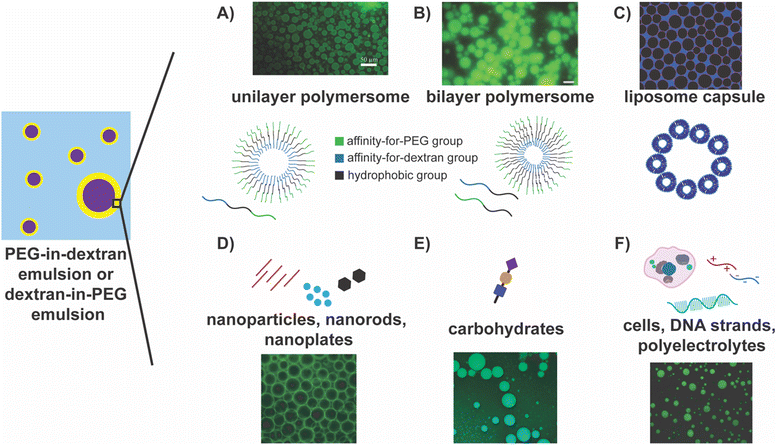 | ||
| Fig. 2 Highlights of approaches for stabilizing PEG–dextran ATPS emulsions. In the schematic on the left, the PEG and the dextran phases are depicted by the blue and the purple regions, respectively, or vice versa, and the yellow region depicts the interface between them. (A) Unilayer105 and (B) bilayer polymersomes122 composed of tri- and diblock copolymers, respectively, stabilizing dextran-in-PEG emulsions. Reprinted (adapted) with permission from D. M. A. Buzza, P. D. I. Fletcher, T. K. Georgiou and N. Ghasdian, Langmuir, 2013, 29, 14804–14814. Copyright 2013 American Chemical Society. Reprinted from Journal of Controlled Release: Official Journal of the Controlled Release Society, 147, Yulong Zhang, Feik Wu, Weien Yuan, and Tuo Jin, Polymersomes of asymmetric bilayer membrane formed by phase-guided assembly, 413–419, Copyright 2010, with permission from Elsevier. (C) Liposome capsules stabilizing dextran droplets in PEG phase.123 Reprinted with permission from Soft Matter, RSC. (D) Nanomaterials, including nanoparticles, nanorods, and nanoplates, stabilize the w/w interface by assembling at the interface. The fluorescent image depicts PEG-in-dextran emulsions stabilized by nanoplatelets, where dextran is labelled with fluorescein.128 (E) Carbohydrates, namely chitosan, assemble at the interface to stabilize the w/w interface. The fluorescent image shows chitosan stabilizing dextran-in-PEG emulsions.103 Reprinted with permission from Carbohydrate Polymers, Elsevier. (F) Cells, DNA strands, and polyelectrolytes as stabilizers of w/w interfaces. The fluorescent image depicts DNA stabilizing the dextran-in-PEG interface.102 Reprinted (adapted) with permission from Y. Wang, J. Yuan, S. Dong and J. Hao, Langmuir, 2022, 38, 4713–4721. Copyright 2022 American Chemical Society. | ||
Liposomes, commonly consisting of phosphatidyl glycerol and phospholipids, form capsules that can accumulate at the PEG–dextran w/w interface, stabilizing the PEG–dextran phases (Fig. 2C).123,124 They have also been employed to stabilize PEG/Ficoll and PEG/sulfate aqueous two-phase systems.123 Moreover, lipids can be modified such that the liposomes achieve selectivity for the transport of different solutes across the liposome membrane.124 These modified lipids are combined with unmodified lipids to form multifunctional liposomes. Additionally, liposome concentration has been shown to influence the size of the droplet, with greater liposome concentrations leading to smaller droplet sizes. Permeability of these liposome capsules has also been shown to be tunable, where only molecules of specific sizes can pass through, and this has been successful using a (sub)monolayer coverage. Here, only part of the droplet interface is stabilized by liposomes, meaning there are gaps in between, and excitingly, both transport and stabilization can occur simultaneously.124 This dual utility expands upon the initial goal of stabilization, and the opportunities for modification provide greater tunability for a specific system or compound of interest. Combined with their biodegradability and biocompatibility, this approach presents an exciting avenue for using ATPS microemulsions in biomedicine and biotechnology.
Similar to polymersomes, colloids can assemble at the PEG–dextran interface to form colloidosomes, which have also been investigated as stabilization methods. Colloidal particles stabilize PEG–dextran emulsions as they are readily trapped at the interface and lower the interfacial energy, forming Pickering emulsions.25,101,103,109,121,125–131 For instance, microgel particles, comprising a crosslinked polymer network, can serve as interfacial stabilizers while imparting stimuli responsiveness to the stabilization.121,126,129 In these systems, droplets have been shown to remain stable for at least one week. At the same time, a dramatic increase in microgel diameter was noted as the pH of the system approached the pKa of the particles.121 Thus, droplets were quickly destabilized upon raising the pH from 7.2 to 8.0, exemplifying the system's sensitivity to pH changes.121 Latex particles have also been employed as they can be trapped at the interface. Alternatively, colloidosomes can be formed from multilayered protein fibrils that form fibrillosomes.127,132,133 Similarly, amphoteric protein particles (created by heat treating and salt exposure), as opposed to native, intact proteins, can form a monolayer that maintains the droplet form while lowering the interfacial tensions significantly.104,125 Nanoparticles, such as nanocrystals, nanorods, nanoplates, and metal (gold and silver) nanospheres and nanowires, some equipped with DNA, have all been shown to stabilize the PEG–dextran w/w interface successfully (Fig. 2D).14,128,134–137 Once self-assembled, they readily form a layer around the droplet. In fact, nanorods and nanoplates can be more effective stabilizers, as they can lie flat along the interface and increase the covered surface area.128 Complexation of oppositely charged polyelectrolytes (PEs) or nanoparticles to form a shell around the droplet that maintains high rigidity and permeability can also serve as a stabilization mechanism.56,130,138–140 This approach accomplishes both goals of droplet stability and permeability while using inorganic materials.
Biomolecules, such as proteins, carbohydrates, DNA, and even whole cells, have also been employed to stabilize PEG–dextran emulsions. Carbohydrates, specifically chitosan, have found great success, and in some cases, the resultant droplets remained stable for at least one week (Fig. 2E).103 In this setting, increasing the PEG and dextran concentration increased the viscosity of the phase-separating polymer solutions, ultimately allowing for tuning of the stability and size of a droplet. Chitosan has also been used in conjunction with cellulose nanocrystals to form a solid, membranous interfacial layer via electrostatic complexation, stabilizing the interface.134 Carbohydrates can also be combined with protein microgels to form complexes that spontaneously form a layer around the droplets.129 DNA (both single and double-stranded) of different lengths have been shown to stabilize droplets and modulate droplet size (Fig. 2F).102 Different cells have also been used as stabilizers, including red blood cells (RBC), NAMRU mouse mammary gland epithelial cells (NMuMG cells), and BIF6A7 hybridoma cells (Fig. 2F).58,60 Under varying proportions of PEG and dextran solutions, different cells could be seen at the interface, and in cases where the interface had a higher tension/surface energy, RBCs, in particular, helped to reduce the surface energy.58 The ability to use live cells as stabilizing agents will be helpful moving forward, opening avenues for novel biological applications.
Overall, while these techniques vary in their approach and chemistries, they all serve and seek to stabilize the w/w interfaces between PEG–dextran solutions, maintain their droplet form, and mitigate droplet coalescence. These systems have immense potential, as they can be used for various applications, imparting little to no detrimental effects on biological compounds and the ability to separate and purify a wide range of compounds from many different (biological, chemical, environmental) settings. It is also important to note the ability of these strategies to simultaneously lower the interfacial tension and stabilize the droplets, often by self-assembly, meaning that little to no input of energy or pre-treatment is required. The opportunities for simultaneous modulation and tuning of the stability of these systems and controlling the transport of molecules across the stabilizing membranes124 make them vital assets that will undoubtedly lead to exciting discoveries in the future.
Stabilization strategies for complex coacervate emulsions
Complex coacervation of oppositely charged polyelectrolytes (polyanions and polycations) is attributed to the increase in entropy of the counterions and water when they are released from the vicinity of their corresponding polyelectrolyte chains upon the latter's complexation as well as the gains in the attractive electrostatic energy of the two polyelectrolytes.32,141,142 The subsequent phase separation results in the formation of a polymer-rich complex coacervate phase which remains in equilibrium with the polymer-lean supernatant phase.29,31,32 These phases exhibit very low interfacial tension (∼100 mN m−1), nearly 1000 times lower than water.27,32,143–146 The lack of any membrane at the interface also allows for rapid transport of small molecules between the two phases. However, this lack of a stabilizing membrane also suggests that the droplets of one phase in another can grow magnanimously via coalescence or Ostwald ripening until they macro-phase-separate.147Due to the low interfacial tension and the ability for rapid transport, these systems have found great utility as cellular mimics,148–152 bioreactors,80,134,150,152–160 and carriers.95,97 Because these systems mimic membraneless organelles, they have been used to study cellular processes outside of a cellular environment, not least of which are enzymatic pathways and cascades, as well as synthetic processes such as nanoparticle catalysis.152,155–159,161 They have also been used to encapsulate scents for perfumes81,82 and hold great promise for other encapsulation strategies and applications.
Stabilization strategies for coacervate droplets have gained increasing research attention as the applications for these systems have become increasingly diverse. While the unstable coacervate systems have broad utility, some of which are listed above, it is in their stabilized droplet forms that their potential uses can be greatly expanded. Recent attempts to stabilize complex coacervates have been inspired by strategies adopted to stabilize hydrophobic phases in water. Conjugation of the polyelectrolytes with a neutral block restricts the coarsening of the coacervates at the nanoscale, resulting in complex coacervate micelles that have been successfully employed as delivery vehicles for charged biomacromolecules, including nucleic acids and proteins.29,162,163 However, their small size and relatively low loading capacity for biomolecules have limited the use of coacervate micelles as viable bioreactors. At the same time, efforts to stabilize micrometer-sized coacervate droplets have relied on the introduction of additional interfaces around the droplets comprising amphiphilic fatty acids,164 amphiphilic terpolymers,94,149,150,155,165,166 and phospholipid vesicles.167,168
ABC triblock copolymers, or terpolymers, have been employed to create unilamellar polymersomes to stabilize coacervate droplets (Fig. 3A).94,149,150,155,165,166 The vast design space wherein each block can be specifically designed for droplet stabilization is a key advantage of this approach.166 The first block of the triblock polymer extends toward the aqueous phase. Its length can be modulated but must avoid internalization by the complex coacervate and ensure successful assembly on the surface.166 This is also conjugated with a hydrophobic middle block, forming the membrane around the droplet.166 The third block of the polymer is often charged at neutral pH and thus provides anchoring to the charged surface of the coacervate microdroplets.166 Following successful demonstrations of the prevention of the coalescence of coacervate droplets, the structure of these polymers has been further explored through various analytical techniques, including MALS, TEM, and confocal microscopy, and refined to enable the transport of selective molecules such as proteins, RNA, and small molecule substrates through the membrane.155
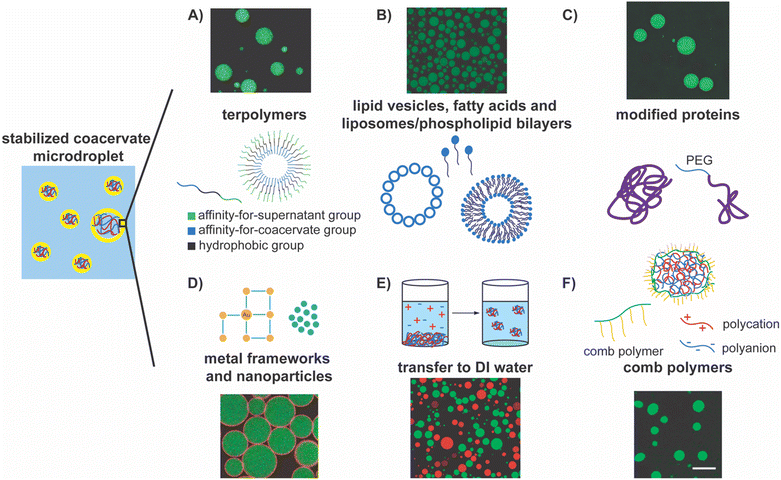 | ||
| Fig. 3 Highlights of the approaches for stabilizing complex coacervate droplets. In the schematic on the left, polyanions are depicted as blue chains, polycations are depicted as red chains, the polymer-lean supernatant phase is depicted by the blue region, and the yellow region depicts the interface between the coacervate and the supernatant phases. (A) Terpolymer, or triblock copolymers, form polymersomes that stabilize the coacervate droplets. In the schematic, the green block has an affinity for the supernatant group, the blue block has an affinity for the coacervate, and black blocks is hydrophobic. The fluorescent image shows stabilization of droplets using this kind of terpolymers.166 Reprinted with permission from Methods in Enzymology, Elsevier. (B) Different self-assemblies of lipids, including lipid vesicles, fatty acids, liposomes, and phospholipid bilayers can serve as stabilizers of coacervate droplets. The fluorescent image shows stabilization of coacervate droplets by phospholipid vesicles.154 Reprinted (adapted) with permission from F. Pir Cakmak, A. T. Grigas and C. D. Keating, Langmuir, 2019, 35, 7830–7840. Copyright 2019 American Chemical Society. (C) Modified proteins can serve as stabilizers for the coacervate/water interface. The fluorescent image shows stabilization of coacervate microdroplets by mPEG-modified BSA accumulated at the interface.173 Reprinted with permission from Small, Wiley. (D) Nanoparticles and metal frameworks can assemble at the coacervate/water interface and stabilize them. The fluorescent image shows gold/PEG nanoparticle stabilization of the coacervate/water interface.177 Copyright 2022 American Chemical Society. Available under the Creative Commons Attribution 4.0 International Public License. No modification to the material was made. (E) Transfer of coacervate droplets from supernatant phase to DI water as a method of stabilizing coacervate droplets. The fluorescent image shows stable coacervate microdroplets in DI water.178 Copyright 2022 A. Agrawal, J. F. Douglas, M. Tirrell, and A. Karim. Available under the Creative Commons Attribution 4.0 International Public License. No modification to the material was made. (F) Comb polymer stabilization of polyelectrolyte complexes via interfacial assembly. The fluorescent image shows stable coacervate droplets.80 Reprinted (adapted) with permission from S. Gao and S. Srivastava, ACS Macro Lett., 2022, 11, 902–909. Copyright 2022 American Chemical Society. | ||
Similarly, unilamellar lipid vesicles have been employed to stabilize coacervate droplets as they readily accumulate at the coacervate–supernatant interfaces, ultimately forming liposomes (Fig. 3B).154,169 Unfortunately, in the earliest studies with coacervates composed of a modified RNA (RNA (polyuridylic acid)) and short polyamines (spermine), such vesicles were not shown to provide sufficient stabilization against coalescence due to the apparent neutralization of adsorbed spermine.169 Subsequently, lipid vesicles were modified with PEG groups and were negatively charged to prevent aggregation. They have even been made pH-responsive such that the coacervate dispersion's stability can be reversibly tuned.170 These modifications have improved droplet stabilization while retaining their biodegradability and biocompatibility, as they can now remain stable for at least one day.170
In a similar vein, fatty acids and phospholipid bilayers have been used as stabilizers, as these molecules can readily self-assemble at the coacervate–water interface (Fig. 3B).167 Drawing inspiration from bilayer cell membranes, lipids that have been extracted from erythrocyte cells via hemolysis have been used to encapsulate coacervate protocells, in some instances providing stabilization for at least 90 minutes.79 Similarly, yeast cell wall fragments have also been shown to assemble at the interface and stabilize the coacervates droplets.171 The biocompatibility of these stabilizers provides exciting avenues to explore for biological and biomedical applications.
Researchers have taken nature-inspired stabilization a step further and modified enzymes such that they decorate the surface of coacervate droplets.172 For example, proteins and enzymes have been modified with PEG and cationized to promote assembly at the interface (Fig. 3C).173 As such, proteins are known to assemble at the interface as self-assembled filaments (actin filaments) or as unfolded globules (e.g., bovine serum albumin), though neither were explicitly shown to stabilize the interface.157,174 Huang and coworkers capitalized on the coacervates’ negatively charged surface to motivate the migration of proteins to the interface and deemed the subsequent shell formation a proteinaceous membrane.173 Other groups have explored this encapsulation method, resulting in proteinosome vesicles.175 This biocompatibility expands our toolbox, offering new stabilization methods and the opportunity to broaden the scope of applications.
Polyoxometalate frameworks have also been used to form and stabilize coacervate droplets (Fig. 3D).176 This method successfully prevented the coalescence of coacervate droplets, and the resulting emulsions could even be lyophilized and rehydrated without losing their overall structure. Similarly, tannic acid–protected gold (Au) nanoparticles have been used together with thioctic acid–modified PEG to slow coalescence.10,177 Conjugation of modified PEG with the tannic acid–Au surface has been accomplished at the droplet interface by harnessing the affinity of the modified PEG and tannic acid–Au particles’ affinity for the continuous and coacervate phases, respectively.177 These conjugate particles pack closely and form a cage around the microdroplets, stabilizing them and creating Pickering emulsions. Using inorganic materials provides a new lens through which the stabilization of coacervate microdroplets can be viewed, and novel ways to synthesize and employ stabilizing materials can be envisioned.
Recently, a unique approach that harnesses the coupling of screening of electrostatic interaction by salt ions with the composition of the coacervate phase has been employed to garner stability to coacervate droplets by creating an elastic coacervate shell around them via interfacial crosslinking. When the coacervate droplets were transferred from the supernatant phase (moderate to high salinity) to DI water (low salinity), an interfacial viscoelastic region formed owing to a reduced screening of the interaction among the oppositely charged polyelectrolytes, rendering stability against coalescence (Fig. 3E).178 Even after vigorous mixing, these droplets with elastic shells remain intact. At the same time, fluorescent proteins (dye-labeled bovine serum albumin) were readily transported into the droplets, confirming the porosity of the elastic shells. Droplets stabilized using this method were shown to be stable for up to a month in standard laboratory conditions.178
Unfortunately, while stabilizing droplets, many of these strategies may result in a loss of functionality by introducing a hydrophobic or elastic gel-like region around the droplets that hinder the transport of small, hydrophilic molecules across them. So far, very few viable strategies for the long-term stabilization of complex coacervate droplets while maintaining their membraneless interface have been demonstrated, slowing the development of complex coacervate-based enzymatic reactors. Recently, we introduced a new approach for stabilizing complex coacervate microdroplets while maintaining the coacervate–water interfaces by using comb polyelectrolytes as interfacial stabilizers (Fig. 3F). The anionic backbones of the comb polyelectrolytes are posited to anchor to the coacervate–water interface via electrostatic interactions while the neutral sidechains extend into the aqueous surroundings and provide steric repulsion between the droplets, minimizing their coalescence and providing long-term stability to these microdroplets. In effect, membraneless complex coacervate emulsions that are stable for at least four months have been created.80 Moreover, this stabilization approach allowed for the successful spontaneous sequestration of proteins and enzymes into the coacervate microdroplets. A combination of macromolecular crowding in the coacervate environment and successful transport of small molecules facilitated by the large area of the stabilized microdroplet interfaces enhanced enzyme-mediated bioreaction rates substantially. Notably, the bioreaction rate enhancements persisted over extended periods.80
Given the increased interest and potential applications for coacervate microdroplets, it is no surprise that a wide range of stabilization techniques and methods have been explored. Although they differ in approach, they follow a common thread in that they all strive to stabilize coacervate microdroplets against coalescence. A future can be envisioned wherein such droplets are utilized as delivery vehicles for hydrophilic cargo, such as pesticides, scents, or drugs, and diverse ways in which their modularity can be harnessed and manipulated to suit the intended applications. In stabilized form, these droplets have the potential to profoundly influence approaches for targeted delivery in medicine, in consumer products, and in the environment.
Applications and utility of stabilized W/W emulsions
Stabilizing water–water interfaces introduces feasible paths to several exciting applications in biology, chemistry, chemical engineering, food science, and various other fields, examples of which are discussed in Fig. 4A–E.37 For instance, extractive bioconversions involving the synthesis and separation of products from their biocatalyst soon after they are synthesized can be facilitated using PEG/dextran ATPS. The bioconversion can be run in one aqueous phase, while the product can be extracted into the other phase.33,107,108,124 ATPS are particularly useful in such a case because they provide a mild environment for the biocatalysts and allow for the extraction of hydrophilic species, such as proteins.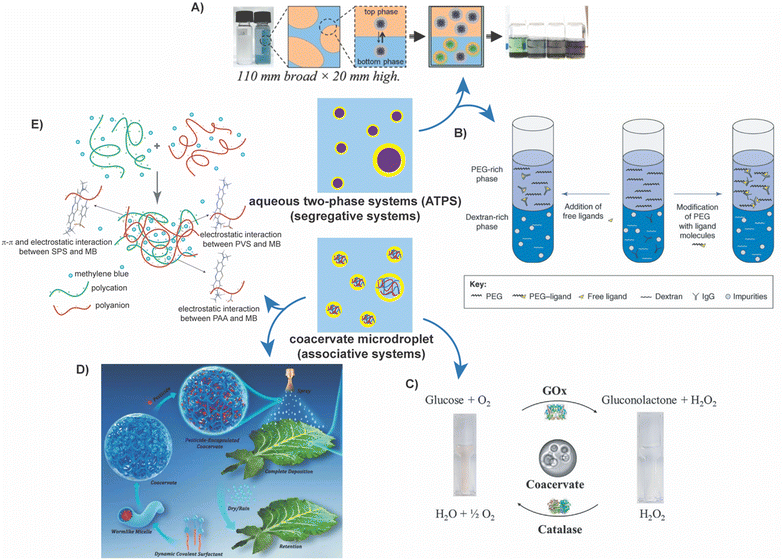 | ||
| Fig. 4 Applications of stabilized ATPS and coacervate microdroplets. (A) ATPS separation of metal particles from solution. The figure shows the general procedure for isolating single semiconducting and metallic single-wall carbon nanotubes from a synthetic solution using a PEG/dextran ATPS.185 Reprinted with permission from Advanced Materials, Wiley. (B) ATPS separation of impurities from antibodies. The figure shows methods by which an antibody can be influenced to partition into the PEG-rich phase, by introduction of free ligands or modification of PEG. In this case, a PEG/dextran ATPS was used.35 Reprinted with permission from Trends in Biotechnology, Cell Press. (C) Enzymatic cascade reactions occurring in coacervate microdroplets. The figure shows the general cascade reaction that was successfully implemented in coacervate droplets.115 Reprinted with permission from Journal of Colloid and Interface Science, Elsevier. (D) Pesticide application and release via sequestration and release from coacervate microdroplets. The figure shows the process of the pesticide, Buprofezin, being encapsulated in coacervates, sprayed on a leaf, and eventual deposition in a controlled fashion.95 Reprinted with permission from Advanced Functional Materials, Wiley. (E) Sequestration of environmental contaminants in coacervate microdroplets. This figure portrays the encapsulation of methylene blue in coacervate microdroplets.19 Reprinted with permission from Macromolecular Rapid Communications, Wiley. | ||
These processes can also be scaled up to industrial levels, which is restrictive with in situ product recovery techniques. ATPS, as a separation technique, has been taken a step further in its use for growing animal cells (mouse/mouse hybridoma cell line BIF6A7), as it similarly provides a mild environment for cell growth.59,60 Additionally, it has been used to partition out hybridoma PFU-83 and BIC-2 CHO cell lines into the same phase and, in other cases, to selectively separate and isolate different cell lines.55,60 It can also be used as a cell encapsulation technique in its stabilized form, where cell viability could be tested.56 ATPS have been used to facilitate the growth of corneal tissue for applications in tissue repair, where stabilization of the interface led to a greater accumulation of cells at the interface and more uniform distribution across the interface.179 This accumulation of cells at the interface can also be used to form tissue constructs that provide convenience in constructing tissue models and may prove advantageous in regenerative medicine.180 ATPS have also proven useful in immunostaining, circumventing common issues such as needing large amounts of expensive antibodies and providing the ability to detect multiple antigens in one sample.181
Enhanced and precise separation of ligands, conjugated with protein, has also been demonstrated by employing ATPS. Many works have cited ATPS to partition and purify proteins. In some cases, large molecular weight proteins have even been employed to selectively alter the partitioning of an affinity ligand, such as horseradish peroxidase (HRP) and human IgG, respectively.38 In this specific example, this affinity ligand conjugated with HRP could then promote the successful recovery of rabbit anti-human IgG.38 Additionally, these ligands may be chemically altered to influence which phase they partition into, ligands can be immobilized, and alterations can be made to the polymers that make up the aqueous solutions to influence partitioning. Stabilization of these systems may lead to better partitioning results.
Similarly, ATPS can isolate specific synthetic products from leftover reactants or side products in chemical syntheses or other reaction mixtures.18,35,39–44,48,49,100,182–184 PEG/Na2CO3 ATPS can extract heavy metals from a sample containing many other compounds.182 A more involved example is the isolation of carbon nanotubes from a complex synthetic mixture.185 This PEG/dextran ATPS technique, demonstrated by Zheng and coworkers, was reported to be rapid and robust, with scope for further tunability. Similarly, PEG/dextran ATPS has been used to select the most promising and ordered DNA-SWCNT (single-wall carbon nanotubes) structures of the many that are available, as the partitioning is strongly dependent on the DNA sequence and SWCNT structure.186 This system can even separate SWCNTs based on chirality and subsequently purified from a synthetic mixture.186,187
Likewise, coacervate droplets provide new ways of pursuing drug and other small molecule delivery.188 They can encapsulate proteins and have been shown to enhance the rates of enzymatic reactions occurring within the coacervate droplets.80 Coacervate droplets can also be used as protocells and, potentially, as bioreactors.79,148,158,168 If stabilized, these reactors and protocells could be used for longer periods and enable the evolution of bioreactions and the accomplishment of enzymatic cascades.
Sequestration and sustained release of cancer drugs and other therapeutics within coacervate systems have been demonstrated.8,9 As such, coacervates have garnered much attention in this realm and continue to be pursued as they also show enhanced drug solubility and protein stability.7 They have been proposed as vehicles to deliver protein-based therapeutics essential for overall function189 through oral delivery routes. Though the cited examples saw little to no need for a stabilizing agent, using a stabilizer could lead to greater selectivity and longer stabilization during delivery.
Proteins have also been shown to complex with polyelectrolytes to form coacervate droplets, especially after supercharging.190,191 These protein–polyelectrolyte complexes are essential for the formation and function of membrane-less organelles.32,163,192 In addition, the encapsulation of proteins often enhances their stability and activity and can help to maintain enzyme activity over time, providing a prime opportunity to use these droplets as protocells or synthetic cells and potentially provide information surrounding the origin of life.80,115,174,193 Researchers have also created coacervates from peptides to understand their compartmentalization properties and allow for greater tunability of phase separation and selectivity for guest molecules. Other biomolecules have also been used, including ribonucleotide monomers and longer nucleic acids such as polyribonucleotides and tRNA, which can remain stable under varying pH and temperature changes.169,194,195 Coacervate droplets were also used to study ribozyme catalysis, and faster ribozyme reactions (up to 12-fold enhancements) when paired with a polyanion were noted.196 Enzymatic cascade reactions have also been performed in coacervate droplets, often deemed enzymatic reactors, where reaction rates have similarly been increased. If the coacervate droplets are stabilized, many more enzymatic cascades and pathways can be studied.
Coacervates have also been used in environmental applications to treat wastewater and extract potentially detrimental compounds from solutions.17,19,84 The corresponding polyelectrolytes can also be modified such that they specifically sequester a dye or contaminant of interest. These contaminants are often organic compounds, such as dyes, that are regularly and readily released and can travel through water sources. Thus, coacervates provide an effective technique to clean these water supplies. They have also been used as soil stabilizers, as they can work to prevent erosion and desertification by strengthening the polymer–soil crust and forming a protective layer. They have even been used as pesticide applicators, where the pesticide particles are successfully encapsulated inside the coacervate droplets. Successful and enhanced deposition of the droplet, excellent erosion prevention by rainwater, and controlled release of the pesticide from the droplet have been reported. Stabilization may include precisely controlled release methods, as the stabilizers could be tuned to respond to only certain external stimuli or target specific plant areas.
They also find uses in cosmetics and can be used specifically in perfumes to help stabilize the scent for extended periods.81,82 Protein/polysaccharide and other coacervate complexes are often used in food systems to enhance gellifying, viscosifying, foaming, and emulsification properties. In practical applications, this can lead to greater perceptions of creaminess in ice cream and sherbet and affect the structure of yogurts. By stabilizing these droplets, perfume scents would have the potential to last longer. Similarly, droplet stabilization may lead to longer-lasting emulsification in foods.
Conclusions
ATPS with water/water interfaces provide a unique materials platform that can be utilized in diverse applications. Thus, considerable research efforts have endeavored to find new routes for maintaining these phases in their droplet form by stabilizing their w/w interfaces against coalescence. In this Review, several of these approaches have been examined for stabilizing the two most common ATPS: aqueous PEG–dextran mixtures and complex coacervates. In both systems, stabilization approaches have witnessed steady development and optimization in the past decade. These approaches can be broadly classified into two categories, comprising either the formation of a hydrophobic membrane, in effect converting the water/water interface into a water/hydrophobe/water interface, accompanied by a crowded hydrophilic shell that prevents coalescence by steric hindrance or by localization of colloids or polymers, synthetic or bioderived, at the water/water interface, thus creating Pickering-like emulsions that minimize interfacial energy and dramatically slow down coalescence. Modular synthesis of terpolymers and modifications of different particles, biological compounds, and inorganic materials have allowed for even greater tunability, with the ability to selectively partition molecules of interest (such as proteins, nanoparticles, small molecules, etc.) or to regulate droplet stabilization.Yet, there are significant gaps in our understanding of the composition and interfacial properties of the water–water interface. Moreover, investigations of how the stabilizers that have already been demonstrated as effective emulsifiers interact with and influence the interface as well as the thermodynamic equilibrium between the two aqueous phases are critical for the development of the next generation of interfacial stabilizers and expanding the utility of w/w ATPS emulsions. We envision that this field of research will continue to evolve as newer strategies for stabilizing ATPS emulsions and their applications in novel areas are investigated.
Author contributions
CF, ZK, and SS conceived the idea and contributed to the literature survey and writing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
CF acknowledges helpful discussion with Holly Senebandith and Divya Iyer. This research was supported by National Science Foundation under Grant No. DMR-2048285.Notes and references
- Y. Chao and H. Cheung Shum, Chem. Soc. Rev., 2020, 49, 114–142 RSC.
- R. Hatti-Kaul, Mol. Biotechnol., 2001, 19, 269–277 CrossRef CAS PubMed.
- M. Iqbal, Y. Tao, S. Xie, Y. Zhu, D. Chen, X. Wang, L. Huang, D. Peng, A. Sattar, M. A. B. Shabbir, H. I. Hussain, S. Ahmed and Z. Yuan, Biol. Proced Online, 2016, 18, 18 CrossRef PubMed.
- J. F. B. Pereira and J. A. P. Coutinho, Liquid-Phase Extraction, Elsevier, 2020, pp. 157–182 Search PubMed.
- A. G. Teixeira, R. Agarwal, K. R. Ko, J. Grant-Burt, B. M. Leung and J. P. Frampton, Adv. Healthcare Mater., 2018, 7, 1701036 CrossRef PubMed.
- P.-Å. Albertsson, Nature, 1958, 182, 709–711 CrossRef CAS PubMed.
- V. Bourganis, T. Karamanidou, O. Kammona and C. Kiparissides, Eur. J. Pharm. Biopharm., 2017, 111, 44–60 CrossRef CAS PubMed.
- M. Kundu, D. L. Morris, M. A. Cruz, T. Miyoshi, T. C. Leeper and A. Joy, ACS Appl. Bio Mater., 2020, 3, 4626–4634 CrossRef CAS PubMed.
- U. Park, M. S. Lee, J. Jeon, S. Lee, M. P. Hwang, Y. Wang, H. S. Yang and K. Kim, Acta Biomater., 2019, 90, 179–191 CrossRef CAS PubMed.
- N. Gao and S. Mann, Acc. Chem. Res., 2023, 56, 297–307 CrossRef CAS PubMed.
- N. Martin, ChemBioChem, 2019, 20, 2553–2568 CrossRef CAS PubMed.
- J. Han, Y. Wang, C. Yu, Y. Yan and X. Xie, Anal. Bioanal. Chem., 2011, 399, 1295–1304 CrossRef CAS PubMed.
- C.-X. Li, J. Han, Y. Wang, Y.-S. Yan, X.-H. Xu and J.-M. Pan, Anal. Chim. Acta, 2009, 653, 178–183 CrossRef CAS PubMed.
- M. R. Helfrich, M. El-Kouedi, M. R. Etherton and C. D. Keating, Langmuir, 2005, 21, 8478–8486 CrossRef CAS PubMed.
- G. D. Rodrigues, M. do, C. H. da Silva, L. H. M. da Silva, F. J. Paggioli, L. A. Minim and J. S. dos Reis Coimbra, Sep. Purif. Technol., 2008, 62, 687–693 CrossRef CAS.
- R. D. Rogers, A. H. Bond, C. B. Bauer, J. Zhang and S. T. Griffin, J. Chromatogr. B: Biomed. Sci. Appl., 1996, 680, 221–229 CrossRef CAS PubMed.
- J. K. Bediako, J.-H. Kang, Y.-S. Yun and S.-H. Choi, ACS Appl. Polym. Mater., 2022, 4, 2346–2354 CrossRef CAS.
- F. Ruiz-Ruiz, J. Benavides, O. Aguilar and M. Rito-Palomares, J. Chromatogr. A, 2012, 1244, 1–13 CrossRef CAS PubMed.
- M. Zhao and N. S. Zacharia, Macromol. Rapid Commun., 2016, 37, 1249–1255 CrossRef CAS PubMed.
- A. Basaiahgari and R. L. Gardas, Curr. Opin. Green Sustainable Chem., 2021, 27, 100423 CrossRef CAS.
- W. N. Phong, P. L. Show, Y. H. Chow and T. C. Ling, J. Biosci. Bioeng., 2018, 126, 273–281 CrossRef CAS PubMed.
- K. S. M. S. Raghavarao, T. V. Ranganathan, N. D. Srinivas and R. S. Barhate, Clean. Technol. Environ. Policy, 2003, 5, 136–141 CrossRef CAS.
- P. A. J. Rosa, I. F. Ferreira, A. M. Azevedo and M. R. Aires-Barros, J. Chromatogr. A, 2010, 1217, 2296–2305 CrossRef CAS PubMed.
- P. A. J. Rosa, A. M. Azevedo, S. Sommerfeld, W. Bäcker and M. R. Aires-Barros, Biotechnol. Adv., 2011, 29, 559–567 CrossRef CAS PubMed.
- E. Dickinson, Trends Food Sci. Technol., 2019, 83, 31–40 CrossRef CAS.
- J. Esquena, Curr. Opin. Colloid Interface Sci., 2016, 25, 109–119 CrossRef CAS.
- V. M. Prabhu, Curr. Opin. Colloid Interface Sci., 2021, 53, 101422 CrossRef CAS.
- P. A. Albertsson, Adv. Protein Chem., 1970, 24, 309–341 CrossRef CAS PubMed.
- J. van der Gucht, E. Spruijt, M. Lemmers and M. A. Cohen, Stuart, J. Colloid Interface Sci., 2011, 361, 407–422 CrossRef CAS PubMed.
- A. M. Rumyantsev, N. E. Jackson and J. J. de Pablo, Annu. Rev. Condens. Matter Phys., 2021, 12, 155–176 CrossRef.
- C. E. Sing and S. L. Perry, Soft Matter, 2020, 16, 2885–2914 RSC.
- S. Srivastava and M. V. Tirrell, Advances in Chemical Physics, John Wiley & Sons, Ltd, 2016, vol. 161, pp. 499–544 Search PubMed.
- E. Andersson and B. Hahn-Hägerdal, Enzyme Microb. Technol., 1990, 12, 242–254 CrossRef CAS PubMed.
- J. A. Asenjo and B. A. Andrews, J. Chromatogr. A, 2011, 1218, 8826–8835 CrossRef CAS PubMed.
- A. M. Azevedo, P. A. J. Rosa, I. F. Ferreira and M. R. Aires-Barros, Trends Biotechnol., 2009, 27, 240–247 CrossRef CAS PubMed.
- M. S. M. Mendes, M. E. Rosa, F. Ramalho, M. G. Freire and F. A. E. Silva, Sep. Purif. Technol., 2023, 317, 123875 CrossRef CAS.
- Y. K. Yau, C. W. Ooi, E.-P. Ng, J. C.-W. Lan, T. C. Ling and P. L. Show, Bioresour. Bioprocess., 2015, 2, 49 CrossRef.
- H.-M. Park, S.-W. Lee, W.-J. Chang and Y.-M. Koo, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 856, 108–112 CrossRef CAS PubMed.
- P. A. J. Rosa, A. M. Azevedo, I. F. Ferreira, J. de Vries, R. Korporaal, H. J. Verhoef, T. J. Visser and M. R. Aires-Barros, J. Chromatogr. A, 2007, 1162, 103–113 CrossRef CAS PubMed.
- B. A. Andrews, D. M. Head, P. Dunthorne and J. A. Asenjo, Biotechnol. Tech., 1990, 4, 49–54 CrossRef CAS.
- L. Ekblad, J. Kernbichler and B. Jergil, J. Chromatogr. A, 1998, 815, 189–195 CrossRef CAS PubMed.
- L. Ekblad, B. Jergil and J. P. Gierow, J. Chromatogr. B: Biomed. Sci. Appl., 2000, 743, 397–401 CrossRef CAS PubMed.
- A. Kumar, M. Kamihira, I. Y. Galaev, B. Mattiasson and S. Iijima, Biotechnol. Bioeng., 2001, 75, 570–580 CrossRef CAS PubMed.
- M. Lu and F. Tjerneld, J. Chromatogr. A, 1997, 766, 99–108 CrossRef CAS PubMed.
- K. A. Sharp, M. Yalpani, S. J. Howard and D. E. Brooks, Anal. Biochem., 1986, 154, 110–117 CrossRef CAS PubMed.
- G. Tubio, B. Nerli and G. Picó, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2004, 799, 293–301 CrossRef CAS PubMed.
- P.-Å. Albertsson, A. Cajarville, D. E. Brooks and F. Tjerneld, Biochim. Biophys. Acta, Gen. Sub., 1987, 926, 87–93 CrossRef CAS PubMed.
- A. D. Diamond and J. T. Hsu, AIChE J., 1990, 36, 1017–1024 CrossRef CAS.
- S. D. Flanagan and S. H. Barondes, J. Biol. Chem., 1975, 250, 1484–1489 CrossRef CAS PubMed.
- A. L. Grilo, M. Raquel Aires-Barros and A. M. Azevedo, Sep. Purif. Rev., 2016, 45, 68–80 CrossRef.
- A. S. Schmidt, A. M. Ventom and J. A. Asenjo, Enzyme Microb. Technol., 1994, 16, 131–142 CrossRef CAS.
- K. Suga, H. Tomita, S. Tanaka and H. Umakoshi, Int. J. Biol. Sci., 2012, 8, 1188–1196 CrossRef CAS PubMed.
- E. C. J. Norrby and P. Å. Albertsson, Nature, 1960, 188, 1047–1048 CrossRef CAS PubMed.
- A. M. Azevedo, A. G. Gomes, P. A. J. Rosa, I. F. Ferreira, A. M. M. O. Pisco and M. R. Aires-Barros, Sep. Purif. Technol., 2009, 65, 14–21 CrossRef CAS.
- J. M. S. Cabral, in Cell Separation: Fundamentals, Analytical and Preparative Methods, ed. A. Kumar, I. Y. Galaev and B. Mattiasson, Springer, Berlin, Heidelberg, 2007, pp. 151–171 Search PubMed.
- S. D. Hann, T. H. R. Niepa, K. J. Stebe and D. Lee, ACS Appl. Mater. Interfaces, 2016, 8, 25603–25611 CrossRef CAS PubMed.
- M. Mastiani, N. Firoozi, N. Petrozzi, S. Seo and M. Kim, Sci. Rep., 2019, 9, 15561 CrossRef PubMed.
- H. Sakuta, T. Fujimoto, Y. Yamana, Y. Hoda, K. Tsumoto and K. Yoshikawa, Front. Chem., 2019, 7, 44 CrossRef CAS PubMed.
- G. M. Zijlstra, C. D. de Gooijer, L. A. van der Pol and J. Tramper, Enzyme Microb. Technol., 1996, 19, 2–8 CrossRef.
- G. M. Zijlstra, M. J. Michielsen, C. D. de Gooijer, L. A. van der Pol and J. Tramper, Biotechnol. Prog., 1996, 12, 363–370 CrossRef CAS PubMed.
- L. McQueen and D. Lai, Front. Chem., 2019, 7, 135 CrossRef CAS PubMed.
- D. Balasubramaniam, C. Wilkinson, K. Van Cott and C. Zhang, J. Chromatogr. A, 2003, 989, 119–129 CrossRef CAS PubMed.
- R. A. Hart, P. M. Lester, D. H. Reifsnyder, J. R. Ogez and S. E. Builder, Nat. Biotechnol., 1994, 12, 1113–1117 CrossRef CAS PubMed.
- A. Veide, A.-L. Smeds and S.-O. Enfors, Biotechnol. Bioeng., 1983, 25, 1789–1800 CrossRef CAS PubMed.
- S. C. Ribeiro, G. A. Monteiro, J. M. S. Cabral and D. M. F. Prazeres, Biotechnol. Bioeng., 2002, 78, 376–384 CrossRef CAS PubMed.
- J. Benavides, J. A. Mena, M. Cisneros-Ruiz, O. T. Ramírez, L. A. Palomares and M. Rito-Palomares, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2006, 842, 48–57 CrossRef CAS PubMed.
- C. Ladd Effio, L. Wenger, O. Ötes, S. A. Oelmeier, R. Kneusel and J. Hubbuch, J. Chromatogr. A, 2015, 1383, 35–46 CrossRef CAS PubMed.
- J. A. Asenjo, A. S. Schmidt, F. Hachem and B. A. Andrews, J. Chromatogr. A, 1994, 668, 47–54 CrossRef CAS.
- D. Degoulange, R. Pandya, M. Deschamps, D. A. Skiba, B. M. Gallant, S. Gigan, H. B. de Aguiar and A. Grimaud, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2220662120 CrossRef CAS PubMed.
- M. G. Freire, A. F. M. Cláudio, J. M. M. Araújo, J. A. P. Coutinho, I. M. Marrucho, J. N. Canongia Lopes and L. P. N. Rebelo, Chem. Soc. Rev., 2012, 41, 4966–4995 RSC.
- C. He, S. Li, H. Liu, K. Li and F. Liu, J. Chromatogr. A, 2005, 1082, 143–149 CrossRef CAS PubMed.
- S. P. M. Ventura, F. A. e Silva, M. V. Quental, D. Mondal, M. G. Freire and J. A. P. Coutinho, Chem. Rev., 2017, 117, 6984–7052 CrossRef CAS PubMed.
- G. L. Dignon, R. B. Best and J. Mittal, Annu. Rev. Phys. Chem., 2020, 71, 53–75 CrossRef CAS PubMed.
- A. A. Hyman, C. A. Weber and F. Jülicher, Annu. Rev. Cell Dev. Biol., 2014, 30, 39–58 CrossRef CAS PubMed.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS PubMed.
- A. V. Fonin, I. A. Antifeeva, I. M. Kuznetsova, K. K. Turoverov, B. Y. Zaslavsky, P. Kulkarni and V. N. Uversky, Essays Biochem., 2022, 66, 831–847 CrossRef CAS PubMed.
- K. K. Turoverov, I. M. Kuznetsova, A. V. Fonin, A. L. Darling, B. Y. Zaslavsky and V. N. Uversky, Trends Biochem. Sci., 2019, 44, 716–728 CrossRef CAS PubMed.
- S. Liu, Y. Zhang, M. Li, L. Xiong, Z. Zhang, X. Yang, X. He, K. Wang, J. Liu and S. Mann, Nat. Chem., 2020, 12, 1165–1173 CrossRef CAS PubMed.
- S. Gao and S. Srivastava, ACS Macro Lett., 2022, 11, 902–909 CrossRef CAS PubMed.
- S. Leclercq, K. R. Harlander and G. A. Reineccius, Flavour Fragrance J., 2009, 24, 17–24 CrossRef CAS.
- D. R. Perinelli, G. F. Palmieri, M. Cespi and G. Bonacucina, Molecules, 2020, 25, 5878 CrossRef CAS PubMed.
- B. Liu, W. Zhao, Y. Shen, Y. Fan and Y. Wang, Langmuir, 2021, 37, 5993–6001 CrossRef CAS PubMed.
- W. Zhao, Y. Fan, H. Wang and Y. Wang, Langmuir, 2017, 33, 6846–6856 CrossRef CAS PubMed.
- L. Li, A. M. Rumyantsev, S. Srivastava, S. Meng, J. J. de Pablo and M. V. Tirrell, Macromolecules, 2021, 54, 105–114 CrossRef CAS.
- D. Priftis, X. Xia, K. O. Margossian, S. L. Perry, L. Leon, J. Qin, J. J. de Pablo and M. Tirrell, Macromolecules, 2014, 47, 3076–3085 CrossRef CAS.
- L. Li, S. Srivastava, M. Andreev, A. B. Marciel, J. J. de Pablo and M. V. Tirrell, Macromolecules, 2018, 51, 2988–2995 CrossRef CAS.
- L.-W. Chang, T. K. Lytle, M. Radhakrishna, J. J. Madinya, J. Vélez, C. E. Sing and S. L. Perry, Nat. Commun., 2017, 8, 1273 CrossRef PubMed.
- J. Huang, F. J. Morin and J. E. Laaser, Macromolecules, 2019, 52, 4957–4967 CrossRef CAS.
- A. E. Neitzel, Y. N. Fang, B. Yu, A. M. Rumyantsev, J. J. de Pablo and M. V. Tirrell, Macromolecules, 2021, 54, 6878–6890 CrossRef CAS PubMed.
- L. Li, S. Srivastava, S. Meng, J. M. Ting and M. V. Tirrell, Macromolecules, 2020, 53, 7835–7844 CrossRef CAS.
- S. Ali, M. Bleuel and V. M. Prabhu, ACS Macro Lett., 2019, 8, 289–293 CrossRef CAS.
- M. Muthukumar, Macromolecules, 2017, 50, 9528–9560 CrossRef CAS PubMed.
- A. F. Mason, N. A. Yewdall, P. L. W. Welzen, J. Shao, M. van Stevendaal, J. C. M. van Hest, D. S. Williams and L. K. E. A. Abdelmohsen, ACS Cent. Sci., 2019, 5, 1360–1365 CrossRef CAS PubMed.
- B. Liu, Y. Fan, H. Li, W. Zhao, S. Luo, H. Wang, B. Guan, Q. Li, J. Yue, Z. Dong, Y. Wang and L. Jiang, Adv. Funct. Mater., 2021, 31, 2006606 CrossRef CAS.
- I. G. Panova, D. D. Khaydapova, L. O. Ilyasov, A. B. Umarova and A. A. Yaroslavov, Colloids Surf., A, 2020, 590, 124504 CrossRef CAS.
- L. Zhang, J. Wang, Y. Fan and Y. Wang, Adv. Sci., 2023, 10, 2300270 CrossRef CAS PubMed.
- S. G. Birrer, P. Quinnan and L. D. Zarzar, Langmuir, 2023, 39, 10795–10805 CrossRef CAS PubMed.
- H. Monteillet, M. Workamp, X. Li, B. Schuur, J. M. Kleijn, F. A. M. Leermakers and J. Sprakel, Chem. Commun., 2014, 50, 12197–12200 RSC.
- C. Wang, Z. Zhang, Q. Wang, J. Wang and L. Shang, Trends Chem., 2023, 5, 61–75 CrossRef CAS.
- T. Nicolai and B. Murray, Food Hydrocolloids, 2017, 68, 157–163 CrossRef CAS.
- Y. Wang, J. Yuan, S. Dong and J. Hao, Langmuir, 2022, 38, 4713–4721 CrossRef CAS PubMed.
- W. Cui, C. Xia, S. Xu, X. Ye, Y. Wu, S. Cheng, R. Zhang, C. Zhang and Z. Miao, Carbohydr. Polym., 2023, 303, 120466 CrossRef CAS PubMed.
- B. T. Nguyen, T. Nicolai and L. Benyahia, Langmuir, 2013, 29, 10658–10664 CrossRef CAS PubMed.
- D. M. A. Buzza, P. D. I. Fletcher, T. K. Georgiou and N. Ghasdian, Langmuir, 2013, 29, 14804–14814 CrossRef CAS PubMed.
- S. Arditty, C. P. Whitby, B. P. Binks, V. Schmitt and F. Leal-Calderon, Eur. Phys. J. E: Soft Matter Biol. Phys., 2003, 11, 273–281 CrossRef CAS PubMed.
- J.-P. Chen and M.-S. Lee, Enzyme Microb. Technol., 1995, 17, 1021–1027 CrossRef CAS.
- H. S. Ng, C. W. Ooi, M. N. Mokhtar, P. L. Show, A. Ariff, J. S. Tan, E.-P. Ng and T. C. Ling, Bioresour. Technol., 2013, 142, 723–726 CrossRef CAS PubMed.
- B. P. Binks, Langmuir, 2017, 33, 6947–6963 CrossRef CAS PubMed.
- L. R. Fisher, E. e Mitchell and N. S. Parker, J. Food Sci., 1985, 50, 1201–1202 CrossRef CAS.
- R. Hans Tromp, in Soft Matter at Aqueous Interfaces, ed. P. Lang and Y. Liu, Springer International Publishing, Cham, 2016, pp. 159–186 Search PubMed.
- S. Zeppieri, J. Rodríguez and A. L. López de Ramos, J. Chem. Eng. Data, 2001, 46, 1086–1088 CrossRef CAS.
- G. M. Zijlstra, C. D. de Gooijer and J. Tramper, Curr. Opin. Biotechnol, 1998, 9, 171–176 CrossRef CAS PubMed.
- J. L. Cleland, C. Hedgepeth and D. I. Wang, J. Biol. Chem., 1992, 267, 13327–13334 CrossRef CAS PubMed.
- R. Toor, L. Hourdin, S. Shanmugathasan, P. Lefrançois, S. Arbault, V. Lapeyre, L. Bouffier, J.-P. Douliez, V. Ravaine and A. Perro, J. Colloid Interface Sci., 2023, 629, 46–54 CrossRef CAS PubMed.
- B. A. Andrews, A. S. Schmidt and J. A. Asenjo, Biotechnol. Bioeng., 2005, 90, 380–390 CrossRef CAS PubMed.
- J. Sinha, P. K. Dey and T. Panda, Appl. Microbiol. Biotechnol., 2000, 54, 476–486 CrossRef CAS PubMed.
- S. Singh and H. Tavana, Front. Chem., 2018, 6, 379 CrossRef PubMed.
- E. Atefi, R. Joshi and H. Tavana, MRS Adv., 2017, 2, 2415–2426 CrossRef CAS.
- H. Walter, E. J. Krob and D. E. Brooks, Biochemistry, 1976, 15, 2959–2964 CrossRef CAS PubMed.
- B. T. Nguyen, W. Wang, B. R. Saunders, L. Benyahia and T. Nicolai, Langmuir, 2015, 31, 3605–3611 CrossRef CAS PubMed.
- Y. Zhang, F. Wu, W. Yuan and T. Jin, J. Controlled Release, 2010, 147, 413–419 CrossRef CAS PubMed.
- A. T. Rowland and C. D. Keating, Soft Matter, 2021, 17, 3688–3699 RSC.
- D. C. Dewey, C. A. Strulson, D. N. Cacace, P. C. Bevilacqua and C. D. Keating, Nat. Commun., 2014, 5, 4670 CrossRef CAS PubMed.
- G. Balakrishnan, T. Nicolai, L. Benyahia and D. Durand, Langmuir, 2012, 28, 5921–5926 CrossRef CAS PubMed.
- F. Dumas, J.-P. Benoit, P. Saulnier and E. Roger, J. Colloid Interface Sci., 2021, 599, 642–649 CrossRef CAS PubMed.
- A. Gonzalez-Jordan, T. Nicolai and L. Benyahia, Langmuir, 2016, 32, 7189–7197 CrossRef CAS PubMed.
- M. Inam, J. R. Jones, M. M. Pérez-Madrigal, M. C. Arno, A. P. Dove and R. K. O’Reilly, ACS Cent. Sci., 2018, 4, 63–70 CrossRef CAS PubMed.
- H. Khemissi, H. Bassani, A. Aschi, I. Capron, L. Benyahia and T. Nicolai, Langmuir, 2018, 34, 11806–11813 CrossRef CAS PubMed.
- Q. Ma, Y. Song, J. W. Kim, H. S. Choi and H. C. Shum, ACS Macro Lett., 2016, 5, 666–670 CrossRef CAS PubMed.
- J. Zhang, PhD thesis, Universität Potsdam, 2020.
- Y. Song, T. C. T. Michaels, Q. Ma, Z. Liu, H. Yuan, S. Takayama, T. P. J. Knowles and H. C. Shum, Nat. Commun., 2018, 9, 2110 CrossRef PubMed.
- Q. Ma, Y. Song, W. Sun, J. Cao, H. Yuan, X. Wang, Y. Sun and H. C. Shum, Adv. Sci., 2020, 7, 1903359 CrossRef CAS PubMed.
- D. Lin, T. Liu, Q. Yuan, H. Yang, H. Ma, S. Shi, D. Wang and T. P. Russell, ACS Appl. Mater. Interfaces, 2020, 12, 55426–55433 CrossRef CAS PubMed.
- L. Bai, S. Huan, B. Zhao, Y. Zhu, J. Esquena, F. Chen, G. Gao, E. Zussman, G. Chu and O. J. Rojas, ACS Nano, 2020, 14, 13380–13390 CrossRef CAS PubMed.
- K. R. Peddireddy, T. Nicolai, L. Benyahia and I. Capron, ACS Macro Lett., 2016, 5, 283–286 CrossRef CAS PubMed.
- E. Ben Ayed, R. Cochereau, C. Dechancé, I. Capron, T. Nicolai and L. Benyahia, Langmuir, 2018, 34, 6887–6893 CrossRef CAS PubMed.
- S. D. Hann, D. Lee and K. J. Stebe, Phys. Chem. Chem. Phys., 2017, 19, 23825–23831 RSC.
- S. D. Hann, K. J. Stebe and D. Lee, ACS Appl. Mater. Interfaces, 2017, 9, 25023–25028 CrossRef CAS PubMed.
- W. Mendez-Ortiz, K. J. Stebe and D. Lee, ACS Nano, 2022, 16, 21087–21097 CrossRef CAS PubMed.
- S. Chen and Z.-G. Wang, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2209975119 CrossRef CAS PubMed.
- Z. Ou and M. Muthukumar, J. Chem. Phys., 2006, 124, 154902 CrossRef PubMed.
- S. Ali and V. M. Prabhu, Macromolecules, 2019, 52, 7495–7502 CrossRef CAS PubMed.
- D. Priftis, R. Farina and M. Tirrell, Langmuir, 2012, 28, 8721–8729 CrossRef CAS PubMed.
- J. Qin, D. Priftis, R. Farina, S. L. Perry, L. Leon, J. Whitmer, K. Hoffmann, M. Tirrell and J. J. de Pablo, ACS Macro Lett., 2014, 3, 565–568 CrossRef CAS PubMed.
- E. Spruijt, J. Sprakel, M. A. C. Stuart and J. van der Gucht, Soft Matter, 2010, 6, 172–178 RSC.
- K. K. Nakashima, M. H. I. van Haren, A. A. M. André, I. Robu and E. Spruijt, Nat. Commun., 2021, 12, 3819 CrossRef CAS PubMed.
- J.-P. Douliez, N. Martin, C. Gaillard, T. Beneyton, J.-C. Baret, S. Mann and L. Beven, Angew. Chem., Int. Ed., 2017, 56, 13689–13693 CrossRef CAS PubMed.
- M. H. M. E. van Stevendaal, L. Vasiukas, N. A. Yewdall, A. F. Mason and J. C. M. van Hest, ACS Appl. Mater. Interfaces, 2021, 13, 7879–7889 CrossRef CAS PubMed.
- W. J. Altenburg, N. A. Yewdall, D. F. M. Vervoort, M. H. M. E. van Stevendaal, A. F. Mason and J. C. M. van Hest, Nat. Commun., 2020, 11, 6282 CrossRef CAS PubMed.
- T.-Y. D. Tang, D. van Swaay, A. deMello, J. L. R. Anderson and S. Mann, Chem. Commun., 2015, 51, 11429–11432 RSC.
- E. Sokolova, E. Spruijt, M. M. K. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. S. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- Y. Zhang, Y. Chen, X. Yang, X. He, M. Li, S. Liu, K. Wang, J. Liu and S. Mann, J. Am. Chem. Soc., 2021, 143, 2866–2874 CrossRef CAS PubMed.
- F. Pir Cakmak, A. T. Grigas and C. D. Keating, Langmuir, 2019, 35, 7830–7840 CrossRef CAS PubMed.
- N. A. Yewdall, B. C. Buddingh, W. J. Altenburg, S. B. P. E. Timmermans, D. F. M. Vervoort, L. K. E. A. Abdelmohsen, A. F. Mason and J. C. M. van Hest, ChemBioChem, 2019, 20, 2643–2652 CrossRef CAS PubMed.
- P. Gobbo, L. Tian, B. V. V. S. Pavan Kumar, S. Turvey, M. Cattelan, A. J. Patil, M. Carraro, M. Bonchio and S. Mann, Nat. Commun., 2020, 11, 41 CrossRef CAS PubMed.
- P. M. McCall, S. Srivastava, S. L. Perry, D. R. Kovar, M. L. Gardel and M. V. Tirrell, Biophys. J., 2018, 114, 1636–1645 CrossRef CAS PubMed.
- B. Drobot, J. M. Iglesias-Artola, K. Le Vay, V. Mayr, M. Kar, M. Kreysing, H. Mutschler and T.-Y. D. Tang, Nat. Commun., 2018, 9, 3643 CrossRef PubMed.
- R. R. Poudyal, R. M. Guth-Metzler, A. J. Veenis, E. A. Frankel, C. D. Keating and P. C. Bevilacqua, Nat. Commun., 2019, 10, 490 CrossRef CAS PubMed.
- N.-N. Deng and W. T. S. Huck, Angew. Chem., Int. Ed., 2017, 56, 9736–9740 CrossRef CAS PubMed.
- S. Koga, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2011, 3, 720–724 CrossRef CAS PubMed.
- H. Cabral, K. Miyata, K. Osada and K. Kataoka, Chem. Rev., 2018, 118, 6844–6892 CrossRef CAS PubMed.
- S. Gao, A. Holkar and S. Srivastava, Polymers, 2019, 11, 1097 CrossRef CAS PubMed.
- T.-Y. Dora Tang, C. Rohaida Che Hak, A. J. Thompson, M. K. Kuimova, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2014, 6, 527–533 CrossRef CAS PubMed.
- A. F. Mason, B. C. Buddingh, D. S. Williams and J. C. M. van Hest, J. Am. Chem. Soc., 2017, 139, 17309–17312 CrossRef CAS PubMed.
- A. F. Mason, W. J. Altenburg, S. Song, M. van Stevendaal and J. C. M. van Hest, in Methods in Enzymology, ed. C. D. Keating, Academic Press, 2021, vol. 646, pp. 51–82 Search PubMed.
- F. Pir Cakmak, A. M. Marianelli and C. D. Keating, Langmuir, 2021, 37, 10366–10375 CrossRef CAS PubMed.
- M. H. van Haren, K. K. Nakashima and E. Spruijt, J. Syst. Chem., 2020, 8, 107–120 Search PubMed.
- W. M. Jr. Aumiller, F. Pir Cakmak, B. W. Davis and C. D. Keating, Langmuir, 2016, 32, 10042–10053 CrossRef PubMed.
- C. Love, J. Steinkühler, D. T. Gonzales, N. Yandrapalli, T. Robinson, R. Dimova and T.-Y. D. Tang, Angew. Chem., Int. Ed., 2020, 59, 5950–5957 CrossRef CAS PubMed.
- C. Zhao, J. Li, S. Wang, Z. Xu, X. Wang, X. Liu, L. Wang and X. Huang, ACS Nano, 2021, 15, 10048–10057 CrossRef CAS PubMed.
- L.-H. Xue, C.-Y. Xie, S.-X. Meng, R.-X. Bai, X. Yang, Y. Wang, S. Wang, B. P. Binks, T. Guo and T. Meng, ACS Macro Lett., 2017, 6, 679–683 CrossRef CAS PubMed.
- J. Li, X. Liu, L. K. E. A. Abdelmohsen, D. S. Williams and X. Huang, Small, 2019, 15, 1902893 CrossRef PubMed.
- N. Martin, M. Li and S. Mann, Langmuir, 2016, 32, 5881–5889 CrossRef CAS PubMed.
- W. Mu, Z. Ji, M. Zhou, J. Wu, Y. Lin and Y. Qiao, Sci. Adv., 2021, 7, eabf9000 CrossRef CAS PubMed.
- D. S. Williams, A. J. Patil and S. Mann, Small, 2014, 10, 1830–1840 CrossRef CAS PubMed.
- N. Gao, C. Xu, Z. Yin, M. Li and S. Mann, J. Am. Chem. Soc., 2022, 144, 3855–3862 CrossRef CAS PubMed.
- A. Agrawal, J. F. Douglas, M. Tirrell and A. Karim, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2203483119 CrossRef CAS PubMed.
- L. T. Hung, S. H. L. Poon, W. H. Yan, R. Lace, L. Zhou, J. K. W. Wong, R. L. Williams, K. C. Shih, H. C. Shum and Y. K. Chan, ACS Biomater. Sci. Eng., 2022, 8, 1987–1999 CrossRef CAS PubMed.
- J. P. Frampton, B. M. Leung, E. L. Bingham, S. C. Lesher-Perez, J. D. Wang, H. T. Sarhan, M. E. H. El-Sayed, S. E. Feinberg and S. Takayama, Adv. Funct. Mater., 2015, 25, 1694–1699 CrossRef CAS.
- J. P. Frampton, M. Tsuei, J. B. White, A. T. Abraham and S. Takayama, Biotechnol. J., 2015, 10, 121–125 CrossRef CAS PubMed.
- A. Hamta and M. R. Dehghani, J. Mol. Liq., 2017, C, 20–24 CrossRef.
- E. Laboureau and M. A. Vijayalakshmi, J. Mol. Recognit., 1997, 10, 262–268 CrossRef CAS PubMed.
- G. A. Gomes, A. M. Azevedo, M. R. Aires-Barros and D. M. F. Prazeres, Sep. Purif. Technol., 2009, 65, 22–30 CrossRef CAS.
- J. A. Fagan, C. Y. Khripin, C. A. Silvera Batista, J. R. Simpson, E. H. Hároz, A. R. Hight Walker and M. Zheng, Adv. Mater., 2014, 26, 2800–2804 CrossRef CAS PubMed.
- G. Ao, J. K. Streit, J. A. Fagan and M. Zheng, J. Am. Chem. Soc., 2016, 138, 16677–16685 CrossRef CAS PubMed.
- G. Ao, C. Y. Khripin and M. Zheng, J. Am. Chem. Soc., 2014, 136, 10383–10392 CrossRef CAS PubMed.
- F. Zhao, G. Shen, C. Chen, R. Xing, Q. Zou, G. Ma and X. Yan, Chem. – Eur. J., 2014, 20, 6880–6887 CrossRef CAS PubMed.
- S. Barthold, S. Kletting, J. Taffner, C. de, S. Carvalho-Wodarz, E. Lepeltier, B. Loretz and C.-M. Lehr, J. Mater. Chem. B, 2016, 4, 2377–2386 RSC.
- A. C. Obermeyer, C. E. Mills, X.-H. Dong, R. J. Flores and B. D. Olsen, Soft Matter, 2016, 12, 3570–3581 RSC.
- C. S. Cummings and A. C. Obermeyer, Biochemistry, 2018, 57, 314–323 CrossRef CAS PubMed.
- J. M. Horn, R. A. Kapelner and A. C. Obermeyer, Polymers, 2019, 11, 578 CrossRef PubMed.
- M. Abbas, W. P. Lipiński, J. Wang and E. Spruijt, Chem. Soc. Rev., 2021, 50, 3690–3705 RSC.
- W. M. Aumiller and C. D. Keating, Nat. Chem., 2016, 8, 129–137 CrossRef CAS PubMed.
- D. Porschke, Biophys. Chem., 1979, 10, 1–16 CrossRef CAS PubMed.
- R. R. Poudyal, C. D. Keating and P. C. Bevilacqua, ACS Chem. Biol., 2019, 14, 1243–1248 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |