DOI:
10.1039/D3MA00089C
(Paper)
Mater. Adv., 2023,
4, 3037-3054
Bioinspired green-synthesized silver nanoparticles: in vitro physicochemical, antibacterial, biofilm inhibitory, genotoxicity, antidiabetic, antioxidant, and anticoagulant performance†
Received
26th February 2023
, Accepted 4th June 2023
First published on 6th June 2023
Abstract
Green-synthesized nanobiomaterials, for instance silver nanoparticles (AgNPs), can be engineered as smart nanomedicine platforms for diagnostic and therapeutic purposes of various disorders, including infectious diseases and cancer. In the present study, the Pimpinella anisum aqueous seed extract was applied for the phytofabrication of AgNPs. Several analytical instrumental techniques were applied for the characterization of AgNPs involving UV-visible spectroscopy, field emission scanning electron microscopy (FESEM), dynamic light scattering (DLS), Fourier transmission infrared (FT-IR) spectroscopy, and X-ray diffraction (XRD). The nanoparticles (NPs) were formed with spherical morphology with an average hydrodynamic diameter of 65.40 nm. Furthermore, the biogenic AgNPs exhibited significant antibacterial activity against the reference strain of Escherichia coli (ATCC 25922) and eleven pathogenic E. coli isolates. Interestingly, the minimum inhibitory concentration (MIC) of AgNPs and gentamicin were found to be 4 and 8 μg mL−1 against E. coli (ATCC 25922), respectively. More interestingly, the AgNPs significantly inhibited the biofilm formation against all tested isolates, whereas gentamicin exhibited lower potency for inhibition of bacterial biofilms at the studied concentrations. The interaction of AgNPs with isolated bacterial plasmid and genomic DNA represented the genotoxic effect of these NPs. The AgNPs also exhibited 82.44 ± 1.43% DPPH inhibition, 67.65 ± 4.78% glucose uptake inhibition by Saccharomyces cerevisiae, and 71.43 ± 4.92% alpha-amylase inhibition at a concentration of 1 mg mL−1. Moreover, the AgNPs showed significant anticoagulant activity at 1 mg mL−1 compared to saline (P < 0.05).
Introduction
Nanosized silver particles in the range of 1 to 100 nm have recently attracted significant attention due to their unique physical, chemical, and biological activities.1,2 Interestingly, the AgNPs exhibited significant antibacterial activity against different bacteria including E. coli.3,4 Among different physical, chemical, and biological techniques for the synthesis of AgNPs, the biological approach using algae, yeasts, fungi, bacteria, actinomycetes, and plant extracts has attracted significant attention due to the advantages over traditional physicochemical methods. The biosynthetic approach is a green, eco-friendly, and low-priced process.4,5 Generally, the physical methods require extreme conditions such as high pressure and high temperature, and the chemical methods require hazardous chemicals that may be harmful to the environment.6 Among different natural resources for AgNP fabrication, plant extracts have attracted considerable attention owing to their advantages. The plant-mediated synthesis of AgNPs is an environmentally friendly, economical, simple, and convenient method using the extracts of different plant parts such as the fruit, leaf, root, bark, flower, and rhizome. In this process, no additive chemical reducers and/or stabilizers are involved,7 because the phytoconstituents such as phenols, alkaloids, saponins, flavonoids, terpenoids, proteins, nucleic acids, amino acids, and vitamins that exist in the plant extract play a dual role of reducing and capping agents to form stable colloidal silver nanostructures. Moreover, the phytosynthesis of metal-based NPs is an easy technique and no sophisticated instruments and chemicals are required.8
The World Health Organization (WHO) estimates that multidrug-resistant (MDR) bacterial infections will cause more fatalities than cancers by 2050 worldwide and will cost $100 trillion in healthcare,2 which has a distressingly serious influence on public health and the health economy. The United States Centers for Disease Control has acknowledged that biofilms are responsible for 80% of human bacterial infections.9 The WHO reported a list of antibiotic-resistant biofilm-producing bacteria involving several bacteria such as E. coli that can cause deadly infections.2 Biofilms are around 10 to 1000 fold more resistant to antibiotics than planktonic bacteria.9 Biofilms are a 3-dimensional (3D) complex network composed of proteins, polysaccharides, lipids, and nucleic acids. Pathogenic bacteria tend to aggregate and form biofilms. These biofilms support the bacterial populations from the antibiotics attack and also unfavorable surrounding environments. Biofilms provide an opportunity for bacterial populations to develop resistance to antibiotics through genetic changes.10 An increasing number of MDR biofilm-producing bacteria force the research world to explore alternative antibacterial agents to deal with them.
The antibacterial potential of biologically synthesized AgNPs has been reported. Most of the studies demonstrated the antibacterial activity of the biogenic silver nanostructures against planktonic bacteria. However, few studies focused on the biofilm inhibitory activity of green synthesized nanosized silver particles. Arshad et al. reported the green synthesis of AgNPs using an aqueous root extract of Salvadora persica with spherical and rod shapes and an average particle size of 37.5 nm. The herbal-mediated fabricated AgNPs showed significant antibacterial activity against E. coli (ATCC 11229) and Staphylococcus epidermidis (ATCC 12228). The MIC and MBC of AgNPs were found to be 0.39 and 0.78 μg mL−1 for E. coli, and 0.19 and 0.39 μg mL−1 for S. epidermidis, respectively. The authors stated that the possible antibacterial mechanisms of AgNPs may be attributed to the AgNP cell membrane penetrations, reactive oxygen species (ROS) production as well as ribosome disassembly.11 Besides, Wypij et al. reported the biogenic synthesis of spherical-shaped AgNPs with a size range of 3 to 36 nm with significant antibacterial activity against E. coli (ATCC 8739) and Pseudomonas aeruginosa (ATCC 10145). The MIC and MBC of AgNPs were found to be 8 and 32 μg mL−1 for E. coli, and 8 and 64 μg mL−1 for P. aeruginosa, respectively. The molecular study also revealed the inhibition of ATP synthesis in the AgNP-treated groups which confirmed the inhibition of ATP synthesis as one of the antimicrobial mechanisms of AgNPs.4 In a recent study, the aqueous extract of aerial parts of Zataria multiflora was applied for the green synthesis of AgNPs. The phytosynthesized AgNPs were formed with a spherical morphology and an average hydrodynamic size of 25.5 nm. Then, the antibacterial activity of biogenic AgNPs and commercial AgNPs was compared against Staphylococcus aureus ATCC 25923 and the results showed MICs of 4 and 8 μg mL−1, respectively. Both of the AgNP types exhibited considerable biofilm inhibitory activity at concentrations of 8 μg mL−1 and higher.12
To the best of our knowledge, this is the first study that investigates the biofilm inhibitory activity of P. anisum-derived AgNPs against pathogenic E. coli isolates. Interestingly, we evaluated not only the antibacterial and biofilm inhibitory activity of herbal-mediated synthesized AgNPs against one standard and several pathogenic E. coli, but we also evaluated the antibacterial and biofilm inhibitory activity of standard antibiotic gentamicin for comparison by employing a 96-well microtiter plate assay as a novel biofilm inhibitory activity test. More interestingly, the potential antidiabetic, antioxidant, genotoxicity, and anticoagulant properties of biogenic AgNPs were evaluated.
Materials and methods
Chemical materials, plant specimen, and microbial strain
Salt (AgNO3) was purchased from Merck, Germany. Other chemicals and reagents were purchased from Sigma-Aldrich, USA. The supplied P. anisum seeds from a local market in Tehran (the capital of Iran) were further confirmed by Pharmacognosy Department, School of Pharmacy, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Besides, the microbial strains including E. coli (ATCC 25922), pathogenic E. coli isolates, and Saccharomyces cerevisiae (PTCC 5269) were obtained from the Microbiology Department, Central Research Laboratories, Shahid Beheshti University of Medical Sciences, Tehran, Iran.
Preparation of P. anisum aqueous seed extract
Initially, the P. anisum seeds were ground, and then to obtain the aqueous seed extract, 100 g of the seed powder was soaked in 1000 mL of distilled water for 24 hours at room temperature. Next, a Whatman No-1 filter paper was used to filter the obtained extract. Consequently, the extract was concentrated by using a rotary evaporator (Hei-VAP Value Digital, Heidolph, Germany). Finally, the extract was dried in an oven (PID-C168, Raimand Zist Fanavar, Iran) at 40 °C.
Herbal-mediated synthesis and characterization of AgNPs
Phytosynthesis of AgNPs were conducted according to the method described elsewhere.13 Initially, 100 mL of the aqueous seed extract (0.1%) was added to 100 mL of aqueous silver nitrate solution (1 mM) in an Erlenmeyer flask with a final pH value of 9. Then, the flask was incubated on a rotary shaker (JAL TAJHIZ®, JTSL 40, Iran) at room temperature and 150 rpm for 48 h. Afterward, the color alteration was observed as preliminary evidence for the colloidal synthesis of nanosized silver particles. The plant-mediated synthesized AgNPs were separated from the reaction mixture sample at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min using an ultracentrifuge (Beckman, L90k, US). Deionized double distilled water was used to wash the surface of AgNPs. The AgNPs were washed thrice to improve their purification. The UV-Vis spectroscopy assay was operated by using a double-beam spectrophotometer (Shimadzu, UV-1280, Japan) to confirm the formation of NPs by monitoring a Surface Plasmon Resonance (SPR) peak in the range of 200–800 nm. Besides, the morphology of AgNPs was determined by using the FESEM (Sigma VP, ZEISS, Germany) with an acceleration voltage of 15 kV. In addition, the XRD analysis was carried out for the phytosynthesized AgNPs using PW3050/60 with XPERT-PRO (using CuKα radiation, λ = 1.5406 Å) at 30 kV and 100 mA and recorded from 20 to 80 degrees for 2θ. Moreover, the hydrodynamic diameter and Polydispersity Index (PdI) of AgNPs were recorded using a Zetasizer (Malvern, England) with a scattering angle of 90° at 25 °C. Furthermore, the FT-IR spectroscopy was operated using an FT-IR spectrophotometer (Agilent, Cary 630 model, US) in transmittance mode in the wavelength range of 400 to 4000 cm−1 using the potassium bromide (KBr) pellet method in the ratio of 1
000 rpm for 20 min using an ultracentrifuge (Beckman, L90k, US). Deionized double distilled water was used to wash the surface of AgNPs. The AgNPs were washed thrice to improve their purification. The UV-Vis spectroscopy assay was operated by using a double-beam spectrophotometer (Shimadzu, UV-1280, Japan) to confirm the formation of NPs by monitoring a Surface Plasmon Resonance (SPR) peak in the range of 200–800 nm. Besides, the morphology of AgNPs was determined by using the FESEM (Sigma VP, ZEISS, Germany) with an acceleration voltage of 15 kV. In addition, the XRD analysis was carried out for the phytosynthesized AgNPs using PW3050/60 with XPERT-PRO (using CuKα radiation, λ = 1.5406 Å) at 30 kV and 100 mA and recorded from 20 to 80 degrees for 2θ. Moreover, the hydrodynamic diameter and Polydispersity Index (PdI) of AgNPs were recorded using a Zetasizer (Malvern, England) with a scattering angle of 90° at 25 °C. Furthermore, the FT-IR spectroscopy was operated using an FT-IR spectrophotometer (Agilent, Cary 630 model, US) in transmittance mode in the wavelength range of 400 to 4000 cm−1 using the potassium bromide (KBr) pellet method in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100.
100.
Antibacterial activity of phytosynthesized AgNPs
Well diffusion assay.
To determine the antibacterial potential of the biosynthesized AgNPs, E. coli (ATCC 25922) as the reference strain, and eleven pathogenic strains of E. coli were tested through an agar well diffusion assay for evaluation of the zone of inhibition (ZOI). In this assay, the inoculum of each E. coli isolate was prepared to have a cell density of 108 CFU per mL. A volume of the E. coli inoculum was then spread over the agar for inoculation of the Mueller Hinton Agar (MHA) plate surface. Afterward, different concentrations of AgNPs (128, 256, 512, and 1024 μg mL−1) with a volume of 100 μL were transferred separately through the wells punched aseptically with a diameter of 8 mm. Finally, incubation of the agar plates was performed at 37 °C for 24 h and the ZOI for each concentration of AgNPs was thoroughly estimated.
Broth microdilution assay.
Evaluation of the minimum inhibitory concentration (MIC).
The MIC value of the biogenic AgNPs is considered as the lowest concentration of the content inhibiting the entire growth of the bacteria visibly, for measurement of which twelve wells of a 96-well microtiter plate were poured with 100 μL Mueller Hinton Broth (MHB) medium. Afterward, 100 μL of the biologically fabricated AgNPs was seeded in the first well, 100 μL of the first well composition was seeded in the second well and this technique was similarly performed for the third to the eleventh well. The twelfth well was regarded as the negative control containing no nanomaterials. The next step was the spread of 100 μL E. coli suspension at the concentration of 106 CFU per mL in all of the twelve wells. In a similar procedure, the MIC of antibiotic gentamicin was assessed as the standard and the plates were transferred into the incubator at 37 °C for 24 h once the constituent was mixed.
Evaluation of the minimum bactericidal concentration (MBC).
The determination of the MBC endpoint was through the spread of 50 μL wells contents that lacked any visible bacterial growth over the MHA surface plates. The plates were finally incubated at 37 °C for 48 h and at last, the MBC value, which is defined as the lowest concentration of biofabricated AgNPs in the plates with 99.9% bacteria-free content, was evaluated.
DNA leakage assay.
At first, a reference strain of E. coli (ATCC 25922) with a concentration adjusted to 0.5 McFarland (equivalent to 108 CFU per mL) in the Luria-Bertani (LB) broth was prepared and then treated with AgNPs at the MIC (8 μg mL−1). Afterwards, the sample was incubated at 37 °C and 125 rpm for 24 h. Then, the sample was centrifuged at 8000 rpm for 5 min resulting in the separation of the supernatant. Finally, the optical density (OD) of the supernatant was measured at 260 nm by Thermo Scientific NanoDrop™ 2000 Spectrophotometer.
Protein leakage assay.
After the preparation of a reference strain of E. coli (ATCC 25922) with a concentration of 0.5 McFarland (equivalent to 108 CFU per mL) in the LB broth, the sample was treated with AgNPs at the MIC (8 μg mL−1). Then, the samples were put in the incubator at 37 °C and 125 rpm for 24 h followed by centrifugation at 8000 rpm for 5 min. Finally, the supernatant was separated. For the evaluation of protein leakage, the bicinchoninic acid (BCA) Protein Quantification Kit (Pars Tous Company, Iran; Catalog Number: A101251) was used. According to the method of the manufacturing company, first 25 μL of the supernatant sample was transferred to a 96-well microtiter plate (SPL Life Sciences, Korea). Then, 75 μL of the working reagent solution was added to the samples and was incubated at 60 °C for 1 h. Afterward, the OD of samples at 562 nm was measured using a Hybrid Multimode Reader Cytation 3 (BioTek Company, USA).
Morphological evaluation of the bacteria using electron microscopy
The effect of phytosynthesized AgNPs on the morphological and structural features of E. coli was evaluated using FESEM (Sigma VP, ZEISS, Germany) and transmission electron microscopy (TEM) (EM10C, ZEISS, Germany) to support the findings of cell membrane integrity assays (DNA and protein leakage assays).
Flow cytometry-based quantitative assessment of cell viability
Initially, a reference strain of E. coli (ATCC 25922) with a concentration adjusted to 0.5 McFarland (equivalent to 108 CFU per mL) in the LB broth was prepared and treated with different concentrations of biogenic AgNPs. The standard antibiotic gentamycin also was used as a positive control. Then, the samples were incubated at 37 °C and 125 rpm for 24 h and were centrifuged at 8000 rpm for 1 min. After discarding the supernatant, the plate containing bacteria was washed twice using PBS buffer. Finally, the plate was dissolved in the phosphate-buffered saline (PBS) buffer and after incubation for 20 min with 2 μg mL−1 of propidium iodide (PI) dye, it was assessed using a flow cytometer (BD FACSCalibur™, USA). Data were analyzed using FlowJo software (Tree Star, Inc., Ashland, OR, USA).
Genotoxic activity of phytosynthesized AgNPs
The genotoxic effect of the biogenic AgNPs on DNA (genomic DNA and plasmid) was determined with the agarose gel electrophoresis method described in previous studies with some modifications.14,15
Bacterial genomic DNA damage assay.
The genomic DNA was isolated from a reference strain of E. coli (ATCC 25922) using a DNA Extraction Kit (EX6071, Sinaclon, Iran). 10 μL of genomic DNA (0.5 μg mL−1) was treated with AgNPs at concentrations of 0.125 to 1 mg mL−1 and incubated at room temperature for 24 h. Then, 10 μL of the DNA–AgNPs mixture was mixed with a loading buffer (MM2123, Sinaclon, Iran) and loaded on a 1% agarose gel. A negative control (DNA without treatment by AgNPs) served for comparison. Finally, the electrophoresis was performed at 75 V for 30 min.
Plasmid DNA damage assay.
The plasmid was isolated from a reference strain of E. coli (ATCC 25922) using a Plasmid Extraction Kit (EX6112, Sinaclon, Iran). 10 μL of extracted plasmids (0.5 μg mL−1) were treated with AgNPs at the concentrations of 0.0625 to 1 mg mL−1 and incubated at room temperature for 24 h. Then, 10 μL of the DNA–AgNPs mixture was mixed with a loading buffer (MM2123, Sinaclon, Iran) and loaded on a 1% agarose gel. A negative control (DNA without treatment by AgNPs) served for comparison. Finally, the electrophoresis was performed at 75 V for 30 min.
Biofilm inhibitory potential of phytosynthesized AgNPs
The biofilm inhibitory property of AgNPs was performed according to the method described elsewhere with brief modifications.12,13 A 96-well microplate was used for the measurement of the potential of AgNPs in the inhibition of bacterial biofilm. 200 μL of bacterial isolates were seeded in each well with a concentration of 108 CFU per mL containing Trypticase soy broth (TSB), 2% (w/v) glucose, and samples (Gentamycin as the positive control and biogenic AgNPs) with the concentrations of 0.5, 1, 2, and 4*MIC. Afterward, the plates were put in the incubator at 37 °C for 24 h, and after discarding the content of all the wells, 200 μL of sterile phosphate-buffered saline (PBS, pH 7.4) was seeded into the wells. Once again, the content of the wells was discarded to remove the planktonic. After repeating the removal three times, 200 μL of 95% ethanol was used for 15 min in order to fix the biofilm that existed in the wells. Similarly, the wells' contents were discarded and all the wells were dried. At last, for staining the biofilm, all of the wells containing negative control, positive control, and the sample were seeded with 200 μL of 1% crystal violet dye and set for 5 minutes. Afterward, 200 μL of sterile distilled water was used for the removal of the stain. Afterward, 200 μL of glacial acetic acid (33%) was added to each well in order to solubilize crystal violet dye. At last, evaluation of the optical density (OD570) of the solubilized dye was carried out through analyses by Hybrid Multimode Reader Cytation 3 (BioTek Company, USA) considering that the negative control and positive control (growth control) were wells with sterile TSB alone and bacterial isolates alone, respectively. Knowing that three standard deviations above the mean OD of the negative control were regarded as the cutoff optical density (ODc), the category representing the intensity of the biofilm phenotypes is as follows: OD ≤ ODc (no biofilm), ODc < OD ≤ 2 × ODc (weak biofilm), 2 × ODc < OD ≤ 4 × ODc (moderate biofilm), and 4 × ODc < OD (strong biofilm). Additionally, to further address the data regarding biofilm phenotypes, the percentage of biofilm inhibitory activity of the samples was calculated using the formula [(ODgrowth control − ODsample)/ODgrowth control] × 100.
Antidiabetic activity of phytosynthesized AgNPs
Glucose uptake by S. cerevisiae.
The glucose uptake technique by yeast was performed according to the well-defined method described elsewhere with some modifications.16 After overnight culture of S. cerevisiae (PTCC 5269) in malt extract medium, the medium was centrifuged at 8000 rpm for 10 min and the yeast pellets were collected. The separated pellets with a ratio of 10% w/w were diluted with deionized water containing 20 mM of glucose and different concentrations of AgNPs. The mixture was incubated at 37 °C and 120 rpm for 90 min. The negative control contained no glucose uptake inhibitor. Afterward, the samples were mixed with DNS reagent [3,5 dinitro salicylic acid (96 mM), Na-K tartrate (5.3 M) in 2 M NaOH] in a ratio of 1 to 1 and heated at 80 °C for 15 min. The heat caused the yeast cells to die and stop the absorption of glucose from the solution. Moreover, DNS (yellow) was reduced to 3-amino-5-nitrosalicylic acid (red-brown) in the presence of glucose. Hence, the amount of glucose can be evaluated by spectrophotometry at a wavelength of 540 nm.17 In the next step, the samples were centrifuged at 8000 rpm for 1 min to separate the yeast cells from the solution. Finally, the absorbance of the solution was measured at 540 nm to calculate the amount of used glucose by yeast cells. The glucose uptake percentage was calculated by the following formula:
| Glucose uptake by yeast (%) = (Acontrol − Asample)/Acontrol × 100 |
where Acontrol and Asample correspond to the absorbance of the control and sample, respectively.
In vitro α-amylase inhibition assay.
The α-amylase inhibitory activity of biogenic AgNPs was evaluated according to previous studies with some changes.16,18 Initially, the stocks of acarbose (50 mg mL−1), α-amylase enzyme (2 U mL−1 in 50 mM phosphate buffer with pH 6.8), and potato starch (5 mg mL−1) were prepared. Then, 1 mL of the enzyme was mixed for 30 min with 1 mL of the studied concentrations of AgNPs and standard drug acarbose as well as 1 mL of distilled water as a negative control at ambient temperature. In the next step, 1 mL of potato starch solution (5 mg mL−1) was added to this mixture and shaken for 30 min at 37 °C. After this time, 250 μL of DNS reagent was mixed with 250 μL of each sample and incubated at 80 °C for 15 min before getting cooled to room temperature, and the absorbance value was measured at 540 nm. The positive control was the standard drug acarbose (1 mg mL−1). The inhibition percentage of α-amylase was calculated by the following formula:
α-amylase inhibition (%) = (Acontrol − Asample)/Acontrol × 100where Acontrol and Asample correspond to the absorbance of the control and sample, respectively.
Antioxidant potential of phytosynthesized AgNPs
The antioxidant potential of biofabricated AgNPs was assessed using the 2,2-diphenyl-1-picrylhydrazyl (DPPH) radical scavenging method.13 Firstly, the AgNPs sample (500 μg mL−1) was prepared using ethanol 90%. Afterward, 2.5 mL of the AgNPs sample was mixed with 1 mL of DPPH solution at the concentration of 0.3 mM and incubated at room temperature for 30 min. After evaluation of absorbance value at 518 nm, 5 mg mL−1 of ascorbic acid was assessed for the DPPH radical scavenging activity as a positive control. It is noted that all samples were tested in triplicate. Accordingly, the percentage was obtained using the following formula:
| DPPH scavenging effect (%) = [(Acontrol – (Asample − Ablank)/(Acontrol)] × 100 |
where Acontrol, Asample, and Ablank correspond to the absorbance of the control, sample, and blank sample, respectively.
Anticoagulant activity of phytosynthesized AgNPs
The anticoagulant property of biogenic AgNPs was evaluated according to the previously described method.13 Briefly, a blood sample of a healthy adult male volunteer was prepared and transferred into a tube containing sodium citrate. Afterward, the platelet-poor plasma (PPP) was obtained through centrifugation of the tube at 1500g for 15 min. Then, the activated partial thromboplastin time (aPTT) and prothrombin time (PT) tests were conducted to measure the length of time (in seconds) that it takes for blood coagulation. For the aPTT assay, initially, a mixture of 5 μL of AgNPs, 50 μL of aPTT reagent, and 45 μL of prewarmed PPP was incubated at 37 °C for 180 s. Subsequently, 50 μL of 0.025 M prewarmed calcium chloride was added and the clotting time was measured. In the PT test, after incubation of 5 μL AgNPs and 45 μL prewarmed PPP at 37 °C for 300 s, 100 μL prewarmed thromboplastin-D was added to the mixture. Finally, the clotting time was assessed. aPTT and PT tests were obtained by a coagulation analyzer (Coa DATA 501, LABitec, Germany).
Statistical test
For the assessment of the obtained data, the R statistical software (version 3.4.1) was used applying the ANOVA test. A post hoc test was also performed using Bonferroni correction. All experiments were repeated three times. A p-value < 0.05 was considered statistically significant.
Results and discussion
Herbal-mediated synthesis and characterization of AgNPs
After 48 hours of the addition of silver nitrate solution to the aqueous seed extract of P. anisum, a brown color appeared representing visual evidence for the fabrication of AgNPs. Fig. 1 represents a schematic illustration of the preparation of the aqueous seed extract of P. anisum and phytosynthesis of AgNPs resulting in the stable colloidal nanosized silver particles. It is believed that the bioactive phytochemical constituents in the plant extracts not only have a major role in reducing the silver ions, but also have a significant role in stabilizing the colloidal system by surrounding the AgNPs.12,13 In the next step, the characteristics of AgNPs were evaluated by employing analytical techniques. Particularly, UV-Vis spectroscopy revealed an SPR peak originating from the collective oscillations of the conduction of AgNPs electrons.19 Concretely, Fig. 2A exhibits a maximum absorbance (λmax) at a wavelength of around 420 nm representing the fabrication of nanosized silver particles, whereas the aqueous seed extract of P. anisum exhibited a wavelength of λmax at 207 nm (Fig. 2B). In addition, Fig. 2C represented a Z-average size of 65.40 nm with a PdI of 0.430. According to ISO 22412:2008, the Z-average size can be defined as the “harmonic intensity averaged particle diameter” that is the central and most stable parameter determined by DLS.20 Furthermore, Fig. 2D confirmed the presence of different functional groups on the surface of AgNPs with absorption bands at 3339.69, 2922.23, 1736.94, 1610.21, 1244.93, 1043.65, 879.65, and 656.01 cm−1 in the region of 450 to 4000 cm−1 that could be assigned to the stretching vibrations of N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) H, C–H, C
H, C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–N, C–N, C–H, and C–H, respectively, that are found in the phytochemical constituents in the plant extracts (Fig. S1, ESI†). The FT-IR spectroscopy provided preliminary information and did not characterize the full structure of bioactive phytochemicals surrounding the AgNPs. Hence, HPLC/GCMS analysis can help to characterize the structure of the bioactive molecules that have a role in the bioreduction process. Moreover, the XRD pattern of AgNPs confirmed the fabrication of nanosized silver particles with highly crystalline structures (Ref. code. 01-089-3722 (Ag); 00-006-0480 (AgCl)) (Fig. 3). Besides, Fig. 4 showed the phytofabrication of spherical-shaped AgNPs.
C, C–N, C–N, C–H, and C–H, respectively, that are found in the phytochemical constituents in the plant extracts (Fig. S1, ESI†). The FT-IR spectroscopy provided preliminary information and did not characterize the full structure of bioactive phytochemicals surrounding the AgNPs. Hence, HPLC/GCMS analysis can help to characterize the structure of the bioactive molecules that have a role in the bioreduction process. Moreover, the XRD pattern of AgNPs confirmed the fabrication of nanosized silver particles with highly crystalline structures (Ref. code. 01-089-3722 (Ag); 00-006-0480 (AgCl)) (Fig. 3). Besides, Fig. 4 showed the phytofabrication of spherical-shaped AgNPs.
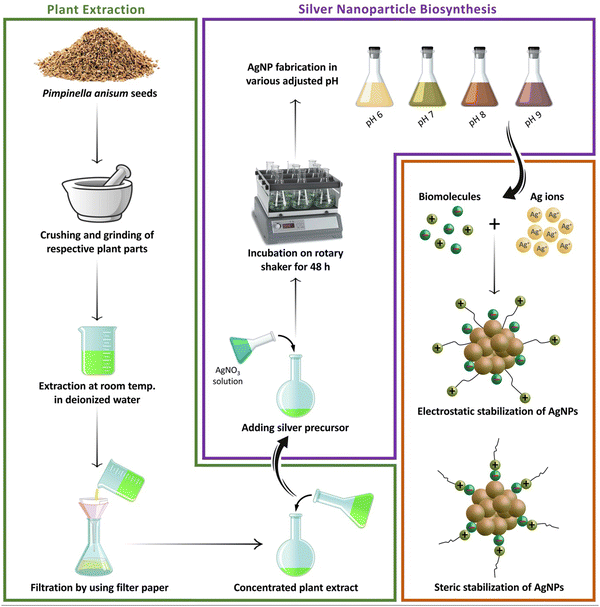 |
| | Fig. 1 A schematic illustration of the process of preparation of the aqueous seed extract of P. anisum and green synthesis of AgNPs resulting in the stable colloidal nanosized silver particles. | |
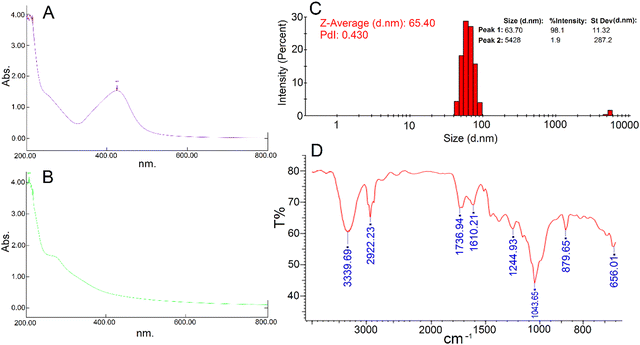 |
| | Fig. 2 (A) Biological synthesized AgNPs UV-vis spectroscopy; (B) P. anisum aqueous extract UV-vis spectroscopy; (C) hydrodynamic particle size of the phytosynthesized AgNPs; (D) FT-IR spectrum of the phytosynthesized AgNPs. | |
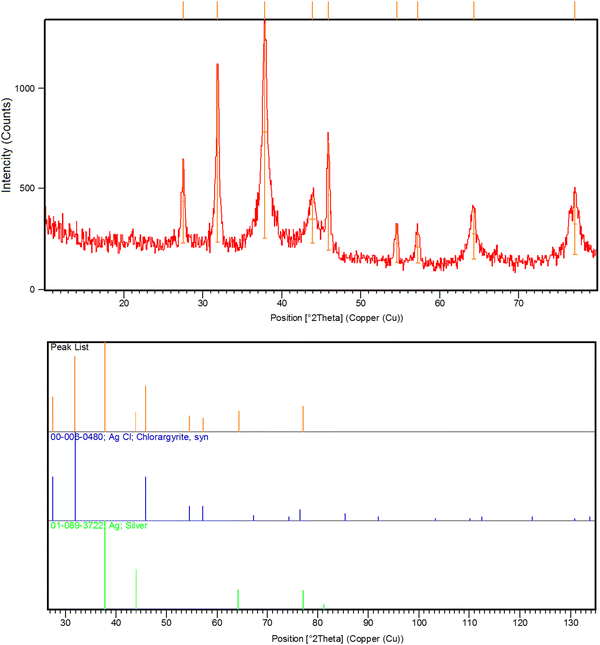 |
| | Fig. 3 XRD spectrum of the phytosynthesized AgNPs. | |
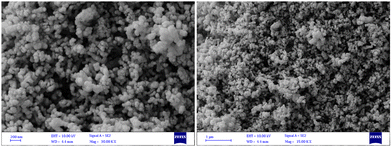 |
| | Fig. 4 FESEM images of the phytosynthesized AgNPs with different magnifications. | |
Antibacterial activity of the phytosynthesized AgNPs
Well diffusion assay.
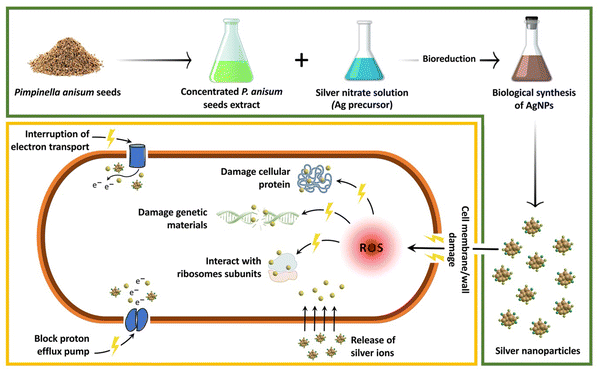
Table 1 shows the results obtained from a well diffusion assay confirming the dose dependent antibacterial activity of biofabricated AgNPs against reference and pathogenic strains of E. coli. Table 1 indicates that 128, 256, 512, and 1024 μg mL−1 of the biogenic AgNPs resulted in dose-dependent antibacterial properties with ZOI of 15, 20, 30, and 40 mm, respectively. In agreement with our findings, Mandal et al. reported the biosynthesis of AgNPs within a spherical shape and size range of 200–300 nm using Cannabis sativa. It was indicated that the biogenic AgNPs could inhibit the growth of E. coli with a ZOI of approximately 16 mm.21 Likewise, in a study, spherical AgNPs were fabricated biologically utilizing seed extracts of jack fruit with the average particle size of 22.12 ± 1.05 nm. The dose-dependent antibacterial activity of the AgNPs against E. coli was determined through the zone inhibition method and the results showed that 4.75, 9.5, and 19 μg of AgNPs led to the ZOI of 14, 14.5, and 15 mm, respectively.22Fig. 5 represents a schematic illustration of the proposed antibacterial mechanisms of AgNPs. The great surface area and opposite charge of AgNPs help them easily attach to the bacterial cell membrane and cause serious damage.23 Moreover, it was suggested that over generation of reactive oxygen species (ROS) and the destructive effect of Ag ions on ATP and DNA were also responsible for the antibacterial effect of AgNPs.24
Table 1 Antibacterial properties of AgNPs against reference and pathogenic strains of E. coli [inhibition zone measured in millimeter (mm).]
|
E. coli code number |
Zone of inhibitions at different concentrations of AgNPs |
| 128 (μg mL−1) |
256 (μg mL−1) |
512 (μg mL−1) |
1024 (μg mL−1) |
| E1 |
22 |
22 |
28 |
30 |
| E2 |
20 |
22 |
25 |
37 |
| E3 |
19 |
26 |
26 |
28 |
| E4 |
18 |
19 |
20 |
23 |
| E5 |
20 |
23 |
25 |
30 |
| E6 |
17 |
19 |
28 |
40 |
| E7 |
14 |
18 |
25 |
27 |
| E8 |
18 |
20 |
21 |
30 |
| E9 |
15 |
15 |
17 |
35 |
| E10 |
15 |
17 |
19 |
32 |
| E11 |
14 |
17 |
18 |
20 |
| ATCC 25922 |
15 |
20 |
30 |
40 |
 |
| | Fig. 5 A schematic illustration of the proposed antibacterial mechanisms of AgNPs. | |
Broth microdilution assay.
As shown in Table 2, the MIC values of standard antibiotic gentamicin and green synthesized AgNPs were found to be 8 and 4 μg mL−1, respectively, against the reference strain of E. coli. Furthermore, MBC endpoints were reported as 8 for the standard drug and 4 μg mL−1 for the AgNPs against the reference strain of E. coli. In agreement with our study, Akter et al. reported the green synthesis of morphologically spherical AgNPs using Sphingobium sp. MAH-11 with a diameter of 7–22 nm. They determined the antibacterial potential of the AgNPs against E. coli successfully with MIC and MBC values of 6.25 and 25 μg mL−1, respectively.25 Similarly, Keshari et al. biofabricated AgNPs with a spherical shape and size of 20 nm on average using leaf extract of Cestrum nocturnum. The MIC of biologically formed AgNPs was reported as 16 μg mL−1 against E. coli.26 Likewise, a study reported the biofabrication of spherical AgNPs using Shewanella sp. ARY1 with an average size of 38 nm. They confirmed the potential of AgNPs for combating E. coli and evaluated the MIC (8–16 μg mL−1) and MBC (32 μg mL−1) accordingly.27 Notably, many factors are effective on the findings of each study. Differences in morphology and size of nanoparticles, surface chemistry, surface charge and bioresources used in the synthesis procedures could lead to a difference in the antibacterial properties of these nanostructures.13
Table 2 MIC and MBC values of phytosynthesized AgNPs and gentamicin against reference and pathogenic strains of E. coli
|
E. coli code number |
Silver nanoparticles |
Gentamicin |
| MIC (μg mL−1) |
MBC (μg mL−1) |
MIC (μg mL−1) |
MBC (μg mL−1) |
| E1 |
4 |
4 |
4 |
4 |
| E2 |
4 |
16 |
4 |
4 |
| E3 |
4 |
4 |
4 |
4 |
| E4 |
4 |
4 |
4 |
4 |
| E5 |
4 |
4 |
4 |
4 |
| E6 |
2 |
2 |
4 |
4 |
| E7 |
2 |
2 |
1024 |
2048 |
| E8 |
2 |
2 |
64 |
64 |
| E9 |
2 |
8 |
32 |
32 |
| E10 |
4 |
16 |
32 |
32 |
| E11 |
4 |
4 |
8 |
8 |
| ATCC 25922 |
4 |
4 |
8 |
8 |
DNA leakage
A DNA leakage assay was performed to evaluate the potency of phytofabricated AgNPs to cause cell membrane damage with further DNA leakage outside the cell. According to the measurement results, a small amount of DNA (OD: 0.283; DNA concentration: 14.2 μg mL−1) was detected in the supernatant of AgNP-treated reference strain E. coli. No DNA was detected in the cultures without any treatment. In a similar study, plant-mediated synthesis of spherical-shaped AgNPs was reported by using the root extract of Momordica dioica with an average size of 13.2 nm. The authors performed a DNA leakage assay and reported the presence of nucleic acids in the supernatant leaked from E. coli after 2 and 8 h of treatment with AgNPs at the MIC with OD values of around 0.09 and 0.22, respectively.28 Alternatively, the green synthesized AgNPs from the mucus of snail Achatina fulica with spherical morphology within the range of 37 to 87 nm caused DNA leakage in S. aureus with a DNA content of 0.136 and 0.159 mg mL−1 after 24 h of treatment with AgNPs at the concentrations of 3 and 6 μg mL−1, respectively.29
Protein leakage
To understand the probable antibacterial mechanisms of phytosynthesized AgNPs, a protein leakage assay was performed for the reference strain of E. coli to evaluate the leakage of proteins through the bacterial membrane into the supernatant by employing a BCA protein assay as a popular technique for colorimetric detection and quantitation of total protein. After overnight incubation of the treated samples with AgNPs at the concentration of MIC (8 μg mL−1), the total protein concentration in the supernatant was found to be 2.423 ± 0.03 mg mL−1, whereas the total protein concentration in the control (cultures without any treatment) was found to be 1.473 ± 0.02 mg mL−1. Hence, the protein leakage via membrane was around 1.64 times higher in the treated sample compared to the control. The results indicated that phytofabricated AgNPs disrupted the bacterial cell membranes. In a study, herbal-mediated synthesized AgNPs showed protein leakage from AgNP treated E. coli. The amount of protein was found to be around 50 and 92 μg mL−1 after treatment for 4 and 8 h.28 Likewise, green synthesized AgNPs from Tectona grandis seed extract exhibited 12.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg mL−1 protein leakage after 6 h of treatment with 1 mg mL−1 of AgNPs.30 Similarly, snail-mucus-mediated fabricated AgNPs in the range of 37 to 87 nm caused protein leakage in S. aureus with a protein content of 3.98 and 4.32 mg mL−1 after 24 h of treatment with AgNPs at the concentrations of 3 and 6 μg mL−1, respectively.29 In a separate study, fungus-mediated synthesized AgNPs with an average size of 16.54 nm increased the protein leakage level from around 75 μg mL−1 (control) to around 140 μg mL−1 after treatment for 8 h with the concentration of 10 mg mL−1 of AgNPs.31
μg mL−1 protein leakage after 6 h of treatment with 1 mg mL−1 of AgNPs.30 Similarly, snail-mucus-mediated fabricated AgNPs in the range of 37 to 87 nm caused protein leakage in S. aureus with a protein content of 3.98 and 4.32 mg mL−1 after 24 h of treatment with AgNPs at the concentrations of 3 and 6 μg mL−1, respectively.29 In a separate study, fungus-mediated synthesized AgNPs with an average size of 16.54 nm increased the protein leakage level from around 75 μg mL−1 (control) to around 140 μg mL−1 after treatment for 8 h with the concentration of 10 mg mL−1 of AgNPs.31
Morphological evaluation of the bacteria using electron microscopy
Fig. 6A–D show the FESEM and TEM images, respectively, representing the morphological changes in the AgNP-treated reference strain of E. coli at the MIC. It was visualized that the phytosynthesized AgNPs led to disturbances in bacterial structure, the formation of rumples, and physiological damage. As shown in Fig. 6, the AgNPs were attached to the surface of bacterial cells and the cell shapes became wrinkled and damaged. Thereby, the morphological studies supported the findings of DNA and protein leakage assays. These results were in agreement with the findings of previous studies.32–34 Moreover, the FESEM represented that the AgNPs were adsorbed on the surface of E. coli cells. In agreement with our results, Tian et al.34 and Ansari et al.35 showed that AgNPs were adsorbed on the surface of bacterial cells and penetrated them leading to cell death.
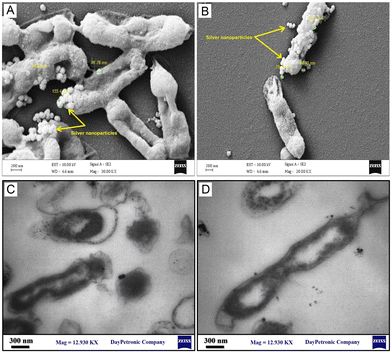 |
| | Fig. 6 Morphological changes of the phytosynthesized AgNP-treated reference strain of E. coli (ATCC 25922) at the MIC determined using electron microscopy: (A and B) FESEM images; (C and D) TEM images. | |
Flow cytometry-based quantitative assessment of cell viability
Bacterial viability of the AgNP-treated reference strain of E. coli was assessed by using flow cytometry employing PI dye to selectively stain dead cells at the MIC (4 μg mL−1) and sub-MIC (2 μg mL−1). Gentamicin also was used as a positive control at the concentration of MIC (8 μg mL−1). PI dye penetrates the bacterial cells with damaged membranes and intercalates to DNA.36 Hence, PI dye was used to identify the percentage of dead cells in a mixed live-dead bacterial population. Fig. 7A exhibited untreated bacterial cells as a control in which 98.23% of cells were live. Fig. 7B and C displayed 33.1% and 24.9% dead cells in the AgNP-treated cells at the concentrations of 4 and 2 μg mL−1, respectively. Besides, representative results of flow cytometric analysis showed 29.0% cell death in the gentamycin-treated cells at the concentration of 8 μg mL−1 (Fig. 7D). In a similar study, the spherical-shaped AgNPs were fabricated by using leaf sap extract of Aloe arborescens in the range of 40 to 50 nm. The phytosynthesized AgNPs exhibited significant antibacterial activity against P. aeruginosa and S. aureus with a MIC of 8 and 12 μg mL−1. The results of the live/dead cell assay by flow cytometry revealed that the percentages of P. aeruginosa and S. aureus dead cells were found to be 66.8% and 25.2%, respectively after 12 h treatment with AgNPs at the concentrations of 8 and 12 μg mL−1, respectively inducing a destructive effect of AgNPs on the bacterial cell membranes.37 Alternatively, flow cytometric analysis was performed for Vatica diospyroides fruit extract-mediated synthesized AgNPs against Bacillus subtilis and S. aureus. The results of flow cytometry-based live/dead cell assay showed that when B. subtilis treated at the concentration of 0.5*MIC (0.37 μg mL−1), MIC (0.74 μg mL−1) and 2*MIC (1.48 μg mL−1) for 12 h, dose-dependent cell death of 17.9, 31.1 and 33.5% was observed, respectively. In addition, when S. aureus was treated at the concentrations of 0.5*MIC (5.915 μg mL−1), MIC (11.83 μg mL−1), and 2*MIC (23.66 μg mL−1) for 12 h, a dose-dependent cell death of 30.37, 54.9 and 59.8% was observed, respectively.38 In a study, flow cytometric analysis showed that plant-mediated fabricated spherical-shaped AgNPs with an average hydrodynamic size of 31 nm killed 89.24% of MDR E. coli cells after 12 h of treatment at the MIC (4 μg mL−1).39 In another study, the authors evaluated the role of different biological sources for the fabrication of AgNPs on the antibacterial activity of AgNPs by using a flow cytometry-based live/dead cell assay. In this study, AgNPs were phytosynthesized by using three plants including Arctium lappa fruit, Solanum melongena leaves, and Taraxacum mongolicum leaves resulting in the synthesis of spherical AgNPs with average diameters of 20.18 nm, 21.00 nm, and 40.08 nm, respectively. Flow cytometric analysis represented 77.95, 72.20 and 62.71% bacterial cell death in Xanthomonas oryzae pv. oryzae, respectively, after 24 h of treatment at the concentration of 20 μg mL−1.34
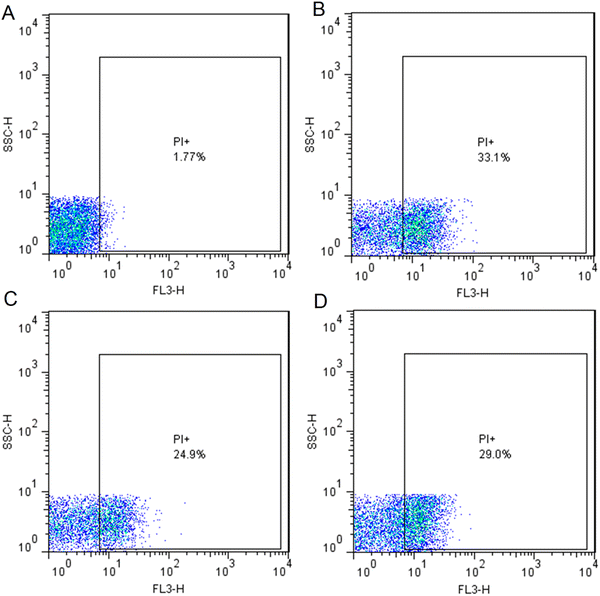 |
| | Fig. 7 Flow cytometry-based quantitative assessment of cell viability. Dot plots representing the cell granularity (side scatter [SSC]) and intensity of fluorescence in the red channel (FL3-H) distributed in logarithm scale (100 to 104). (A) Untreated bacterial cells as the control in which 98.23% cells were live. (B) AgNP-treated bacterial cells at a concentration of 4 μg mL−1 representing 33.1% cell death; (C) AgNP-treated bacterial cells at the concentration of 2 μg mL−1 representing 24.9% cell death; (D) Gentamicin-treated bacterial cells at the concentration of 8 μg mL−1 representing 29.0% cell death. Abbreviation: PI (propidium iodide). | |
Genotoxic activity of phytosynthesized AgNPs
Bacterial genomic DNA damage assay.
Fig. 8A shows the agarose gel image obtained from the incubation of AgNPs in different concentrations (0.125–1 mg mL−1) with isolated genomic DNA of the reference strain of E. coli resulting in dose-dependent DNA damage compared to the untreated DNA. In detail, the brightness of the gel electrophoresis band was weakened with the concentration of AgNPs increasing and in the AgNP-treated sample with the concentration of 1 mg mL−1, a minimal brightness was observed indicating a defamed and damaged DNA compared to the control that exhibited a deep shining band. In agreement with our results, Syed et al. showed the DNA damage activity of plant-mediated synthesized AgNPs against DNA isolated from S. aureus (MTCC 7443) by using an agarose gel-retardation assay. The authors reported bacterial DNA degradation when treated with phytogenic AgNPs at the concentration of 10 mg mL−1 after incubation for 30 min resulting in a weakened band in 1% agarose gel compared to the control.14 Alternatively, in a study, the effect of phytosynthesized AgNPs on E. coli genomic DNA was evaluated by using an agarose gel-retardation assay. After overnight treatment of genomic DNA with AgNPs at concentrations of 2.5 to 10 μg mL−1, the agarose gel represented a smear-like appearance indicating extensive double-strand breaks of DNA with various sizes at all tested concentrations of AgNPs, whereas no breakage was observed in the control.40 Likewise, a study compared the DNA damage activity of chemically and biologically fabricated AgNPs at the concentration of 100 mM against genomic DNA isolated from E. coli (ATCC 25922) by using an agarose gel-retardation assay. The authors used the Datura stramonium leaf extract and sodium citrate for the fabrication of biologically and chemically synthesized AgNPs. After 2 h of treatment, the agarose gel showed higher DNA cleavage of phytosynthesized AgNPs compared to chemically synthesized AgNPs and untreated ones due to the weakened band in agarose gel for the biogenic AgNP-treated sample.41
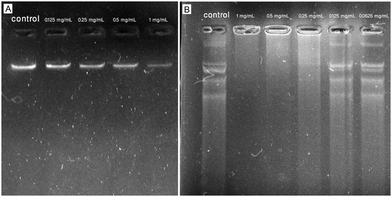 |
| | Fig. 8 Agarose gel images obtained from the incubation of phytosynthesized AgNPs with isolated genomic and plasmid DNA of reference strain of E. coli (ATCC 25922). (A) Interaction of AgNPs with bacterial genomic DNA; (B) interaction of AgNPs with bacterial plasmid DNA. | |
Plasmid DNA damage assay.
Fig. 8B showed the agarose gel image obtained from the incubation of AgNPs in different concentrations (0.0625–1 mg mL−1) with isolated plasmid DNA of the reference strain of E. coli resulting in significant DNA damage at the concentration of 1 mg mL−1 compared to the untreated DNA. The AgNP-treated samples at concentrations of 0.0625 and 0.125 mg mL−1 showed less DNA degradation, but at concentrations of 0.25, 0.5, and 1 mg mL−1 showed a dose-dependent induction of DNA strand break. In agreement with our results, Chowdhury et al. showed the DNA damage activity of fungus-mediated synthesized AgNPs against plasmid pZPY112 (isolated from E. coli DH5α) by using an agarose gel-retardation assay. The authors reported a dose-dependent induction of plasmid degradation due to DNA strand breaks when treated with mycosynthesized AgNPs at the concentrations of 0.51, 1.02, 2.55, 3.57, and 5.1 μg (in a total volume of 100 μL) after 2 h of treatment. The AgNP-treated plasmids showed significant DNA degradation when incubated with 5.1 μg of AgNPs in 1.5% agarose gel compared to the control.42 Alternatively, in a study, the effect of wild edible mushroom-mediated fabricated AgNPs on plasmid DNA (pBR322 isolated from E. coli) was evaluated by using an agarose gel-retardation assay. After 45 min treatment of pBR322 plasmid DNA with AgNPs at concentrations of 50, 100, and 200 μg mL−1, the agarose gel represented extensive plasmid cleavage at all tested concentrations of AgNPs, whereas no breakage was observed in the control.43
Biofilm inhibitory activity of phytosynthesized AgNPs
The ability of bacteria to form biofilms and build resistant structures against conventional drugs has resulted in a major crisis for controlling bacterial infections. Hence, the development of effective antibacterial alternatives for combating such bacteria is of high importance. The present study introduces a novel method for inhibiting biofilm formation by E. coli using plant-mediated fabricated AgNPs. Fig. 9 represents a schematic illustration of different stages of the formation of bacterial biofilm and biofilm inhibitory activity of biosynthesized AgNPs.
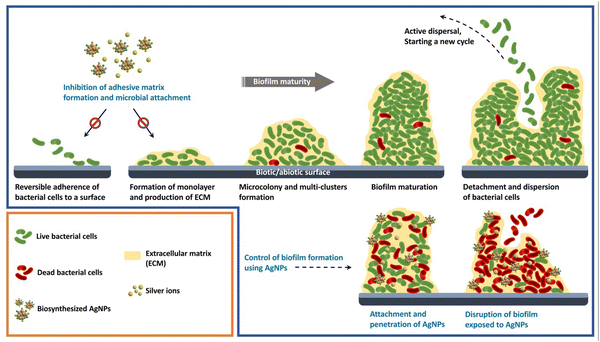 |
| | Fig. 9 A schematic illustration of different stages of the formation of bacterial biofilm and biofilm inhibitory activity of biosynthesized AgNPs. | |
For measurement of the biofilm inhibitory potential of the AgNPs, different concentrations (0.5, 1, 2, and 4*MIC) of the biogenic AgNPs and gentamicin as the standard were prepared. For the case of the AgNPs, the ODc, the mean OD of negative control wells, and the standard deviation were found to be 0.1015, 0.0936, and 0.0026, respectively. The biosynthesized AgNPs successfully hindered the biofilm formation of the reference and pathogenic strains of E. coli at all of the concentrations tested compared to the growth control (positive control) (P < 0.05) as shown in Fig. 10. The data obtained from the phenotypic assay of the biofilm inhibitory effect of biogenic AgNPs at different concentrations against E. coli are exhibited in Table 3. The phenotypic biofilm of weak and negative appeared in the AgNP-treated bacterial strains at the concentration of 4*MIC. Furthermore, the ODc attributed to gentamicin was measured as 0.0914. The mean OD of the negative control and standard deviation of the drug were also found to be 0.0876 and 0.0012, respectively. Fig. 11 shows the inefficiency of gentamicin in inhibiting the biofilm formation in two E. coli strains (strain code: E5 and E6) at all gentamicin-tested concentrations compared to the growth control (P > 0.05). However, some gentamicin-treated strains exhibited biofilm formation inhibition at higher concentrations of AgNPs compared to growth control (P < 0.05). Table 4 shows the data contributing to the phenotypic assay of the biofilm inhibitory effect of gentamicin at different concentrations against E. coli. The percentage of biofilm inhibitory activity of biosynthesized AgNPs and gentamicin at the concentration of 4*MIC against E. coli strains is shown in Table 5 indicating that AgNPs had a higher biofilm inhibitory activity than gentamicin against most of the species of E. coli. In agreement with our findings, in a study, quasi-spherical shaped AgNPs were biosynthesized using Solibacillus isronensis sp. with a diameter of 80–120 nm. A significant biofilm inhibitory effect against E. coli was reported at the concentration of 0.5*MIC (1.56 μg mL−1) of the AgNPs.44 Alternatively, Chandrasekharan et al. biofabricated spherical AgNPs with a size of 36.38 ± 4.75 nm using Gmelina arborea leaf extract. They reported 46.19 ± 1.23% inhibition in the biofilm formation by E. coli when the AgNPs were loaded on hydrogel.45 In a similar study, AgNPs were synthesized biologically with an average size of 72 nm and spherical morphology using Nitzschia palea extract. The biofilm formed by E. coli was significantly inhibited (>80%) by the AgNPs at the concentration of 300 ng mL−1.46 Similarly, Bharathi et al. reported the green fabrication of AgNPs utilizing fruit extract of Cordia dichotoma with a size range of 2–60 nm and spherical structure. The anti-biofilm activity of the biogenic AgNPs at the concentration of 100 μg mL−1 against E. coli was significant (95%) after treatment for 12 h.47 In another study, Singh et al. biosynthesized AgNPs with a crystalline structure and a diameter of 15–30 nm using Rhodiola rosea rhizome extract. They assessed the potential of AgNPs at the concentration range of 1.6–200 μg mL−1 in the inhibition of biofilm formation by E. coli and reported promising results. It was stated that significant anti-biofilm activity was observed at a concentration higher than 6.25 μg mL−1. Furthermore, the authors suggested that the inhibition of expression of genes that are responsible for biofilm formation, could be the mechanism by which AgNPs showed anti-biofilm activities at sub-MIC concentrations.48
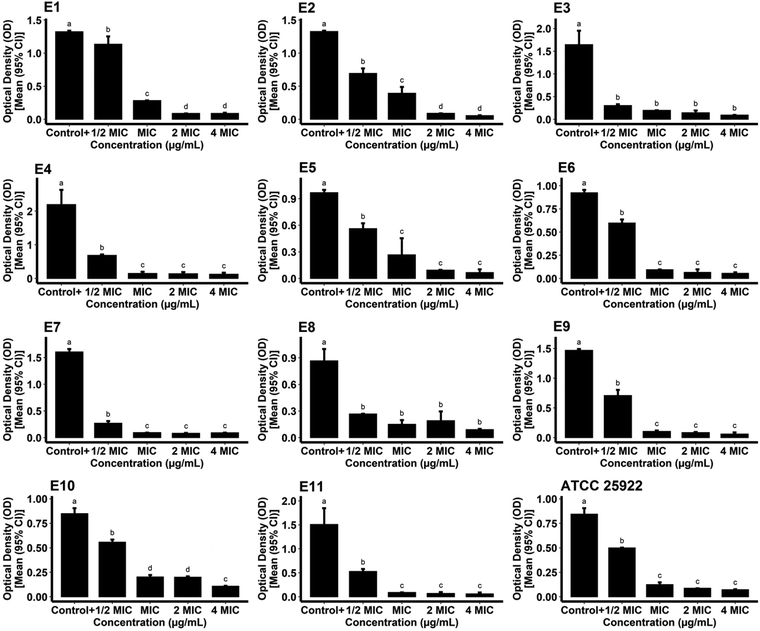 |
| | Fig. 10 Optical densities of phytosynthesized AgNP-treated reference and pathogenic strains of E. coli following the formation of biofilm. | |
Table 3 Biofilm phenotypes and optical densities of phytosynthesized AgNP-treated E. coli species at different concentrations
|
E. coli code number |
Positive control (growth control) |
4*MIC |
2*MIC |
MIC |
0.5*MIC |
| OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
| E1 |
1.3235 |
Strong |
0.0885 |
Negative |
0.0885 |
Negative |
0.2825 |
Moderate |
1.135 |
Strong |
| E2 |
1.327 |
Strong |
0.0545 |
Negative |
0.0895 |
Negative |
0.394 |
Moderate |
0.693 |
Moderate |
| E3 |
1.6445 |
Strong |
0.092 |
Negative |
0.142 |
Weak |
0.1975 |
Weak |
0.302 |
Weak |
| E4 |
2.1965 |
Strong |
0.125 |
Weak |
0.14 |
Weak |
0.1475 |
Weak |
0.6845 |
Moderate |
| E5 |
0.9674 |
Moderate |
0.0655 |
Negative |
0.0955 |
Weak |
0.2655 |
Moderate |
0.5615 |
Moderate |
| E6 |
0.924 |
Moderate |
0.055 |
Negative |
0.065 |
Negative |
0.0945 |
Moderate |
0.597 |
Moderate |
| E7 |
1.604 |
Strong |
0.09 |
Negative |
0.08 |
Negative |
0.094 |
Negative |
0.2685 |
Weak |
| E8 |
0.866 |
Moderate |
0.09 |
Negative |
0.19 |
Weak |
0.15 |
Weak |
0.2655 |
Weak |
| E9 |
1.468 |
Strong |
0.059 |
Negative |
0.084 |
Negative |
0.1045 |
Weak |
0.7085 |
Moderate |
| E10 |
0.848 |
Moderate |
0.1075 |
Weak |
0.1995 |
Weak |
0.2025 |
Weak |
0.5565 |
Moderate |
| E11 |
1.507 |
Strong |
0.061 |
Negative |
0.068 |
Negative |
0.09 |
Negative |
0.5275 |
Moderate |
| ATCC 25922 |
0.843 |
Moderate |
0.072 |
Negative |
0.087 |
Negative |
0.124 |
Negative |
0.4985 |
Moderate |
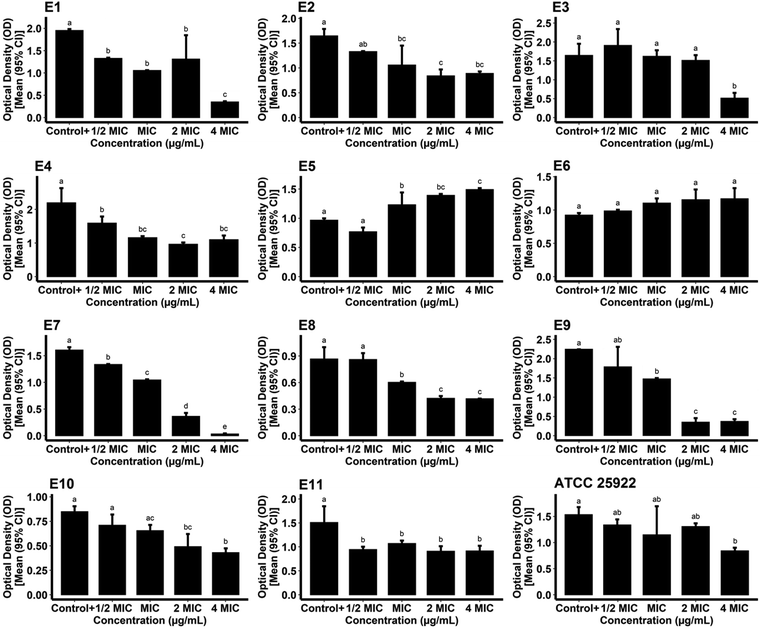 |
| | Fig. 11 Optical densities of gentamicin-treated reference and pathogenic E. coli strains following the formation of biofilm. | |
Table 4 Biofilm phenotypes and optical densities of gentamicin-treated reference and pathogenic strains of E. coli at different concentrations
|
E. coli code number |
Positive control (growth control) |
4*MIC |
2*MIC |
MIC |
0.5*MIC |
| OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
OD |
Biofilm phenotype |
| E1 |
1.951 |
Strong |
0.349 |
Weak |
1.309 |
Moderate |
1.056 |
Moderate |
1.3235 |
Strong |
| E2 |
1.6445 |
Strong |
0.8905 |
Moderate |
0.8405 |
Moderate |
1.0585 |
Moderate |
1.327 |
Strong |
| E3 |
1.6445 |
Strong |
0.5125 |
Weak |
1.5125 |
Strong |
1.619 |
Strong |
1.9065 |
Strong |
| E4 |
2.1965 |
Strong |
1.098 |
Moderate |
0.963 |
Moderate |
1.159 |
Moderate |
1.59 |
Strong |
| E5 |
0.9674 |
Moderate |
1.492 |
Moderate |
1.392 |
Moderate |
1.23 |
Moderate |
0.7685 |
Moderate |
| E6 |
0.924 |
Moderate |
1.169 |
Moderate |
1.154 |
Moderate |
1.104 |
Moderate |
0.986 |
Moderate |
| E7 |
1.604 |
Strong |
0.032 |
Negative |
0.362 |
Weak |
1.045 |
Moderate |
1.335 |
Strong |
| E8 |
0.866 |
Moderate |
0.417 |
Weak |
0.422 |
Weak |
0.602 |
Weak |
0.8595 |
Moderate |
| E9 |
2.2405 |
Strong |
0.37 |
Weak |
0.35 |
Moderate |
1.468 |
Strong |
1.7785 |
Strong |
| E10 |
0.848 |
Moderate |
0.4295 |
Moderate |
0.489 |
Weak |
0.653 |
Weak |
0.709 |
Weak |
| E11 |
1.507 |
Strong |
0.9155 |
Moderate |
0.908 |
Moderate |
1.07 |
Moderate |
0.944 |
Moderate |
| ATCC 25922 |
1.535 |
Strong |
0.843 |
Moderate |
1.3065 |
Moderate |
1.1475 |
Moderate |
1.335 |
Moderate |
Table 5 The percentage of biofilm inhibition of phytosynthesized AgNPs and gentamicin-treated reference and pathogenic strains of E. coli at the maximum tested concentrations
|
E. coli code number |
AgNPs at the concentration of 4*MIC |
Gentamicin at the concentration of 4*MIC |
| Biofilm inhibition percentage (%) |
Biofilm inhibition percentage (%) |
| E1 |
93.31 |
82.11 |
| E2 |
95.89 |
45 |
| E3 |
94.40 |
68.83 |
| E4 |
94.30 |
50.01 |
| E5 |
93.22 |
0 |
| E6 |
94.04 |
0 |
| E7 |
94.38 |
98 |
| E8 |
89.60 |
51.84 |
| E9 |
95.98 |
83.48 |
| E10 |
87.32 |
49.35 |
| E11 |
95.95 |
39.25 |
| ATCC 25922 |
91.45 |
45.08 |
Antidiabetic activity of phytosynthesized AgNPs
Glucose uptake by S. cerevisiae.
The AgNPs exhibited a significant dose-dependent glucose uptake inhibition of 27.31 ± 2.53 and 67.65 ± 4.78% by S. cerevisiae at the concentration of 0.25 and 1 mg mL−1, respectively compared to a negative control (P < 0.05). In agreement with our results, Perumalsamy et al. phytosynthesized the AgNPs by using the hydroethanolic seed extract of Myristica fragrans in the range of 50 to 60 nm with polygonal morphology. The plant-mediated fabricated AgNPs at the concentration of 1 mg mL−1 showed 73.33% glucose uptake inhibition by yeast in 5 mM glucose solution.49 Alternatively, Arsia Tarnam et al. reported the herbal-mediated synthesis of spherical-shaped AgNPs by using ethanolic root extract of Clausena anisate with an average size of 32.75 nm. The phytofabricated AgNPs showed a dose-dependent glucose uptake inhibition by yeast with a maximum glucose uptake of 69.51% (P < 0.001) at the concentration of 2 mg mL−1 of AgNPs in 10 mM glucose solution.50 In another study, the phytosynthesized AgNPs in the range of 30 to 50 nm with spherical morphology showed dose-dependent glucose uptake inhibition by yeast with a maximum glucose uptake of 65.12 ± 0.64 and 63.27 ± 0.34% in 5 and 10 mM glucose solution, respectively at the concentration of 200 μg mL−1 of AgNPs.51
In vitro α-amylase inhibition assay.
Alpha-amylase is a key hydrolyzing enzyme that degrades dietary starch into maltose, a disaccharide cleaved into glucose before absorption. Hence, exploring innovative alpha-amylase inhibitors is a strategy to reduce carbohydrate digestibility to control postprandial hyperglycemia in diabetic patients.52 In the current study, the AgNPs exhibited a significant dose-dependent alpha-amylase inhibition of 44.90 ± 3.87 and 71.43 ± 4.92% at the concentrations of 0.25 and 1 mg mL−1, respectively, compared to a negative control (P < 0.05), whereas the standard drug acarbose showed 57.14 ± 4.32% alpha-amylase inhibition at a concentration of 1 mg mL−1. In agreement with our results, Wilson et al. reported a dose-dependent alpha-amylase inhibition of 31.59 ± 1.02 and 43.96 ± 0.91% at the concentrations of 0.2 and 1 mg mL−1 of phytosynthesized AgNPs, respectively, whereas, at the same concentrations, the standard drug acarbose showed 39.07 ± 0.071 and 55.75 ± 0.71% alpha-amylase inhibition, respectively.51 In a study, the phytofabricated polygonal-shaped AgNPs in the range of 50 to 60 nm showed 52.48% alpha-amylase inhibition at the concentration of 1 mg mL−1.49 Alternatively, the phytofabricated AgNPs from the plant extracts of Zingiber officinale and Mentha longifolia represented 80.52 and 60% alpha-amylase inhibition, respectively, at the concentration of 1 mg mL−1.53 Besides, a study reported the algal synthesis of spherical-shaped AgNPs in the range of 34 to 80 nm by using the seaweed extract of Halymenia poryphyroides. The phycosynthesized AgNPs showed 91.30 ± 0.02% alpha-amylase inhibition at the concentration of 1 mg mL−1, whereas, the standard drug acarbose showed 59.56 ± 0.20% alpha-amylase inhibition at the same concentration.54 Likewise, the mycosynthesized AgNPs in the range of 5 to 20 nm with prominently spherical morphology showed 44.24 ± 0.64% alpha-amylase inhibition at the concentration of 0.5 mg mL−1.55 Alternatively, the biosynthesized spherical-shaped AgNPs from the leaf extract of Solanum khasianum with an average size of 15.96 nm showed 79.56% alpha-amylase inhibition at the concentration of 1 mg mL−1.56 In another study, the spherical-shaped AgNPs were fabricated in the range of 10.34 to 22.12 nm by using the leaf extract of Muntingia calabura. The phytofabricated AgNPs showed 78.16% alpha-amylase inhibition at the concentration of 1 mg mL−1.57
Antioxidant effect of phytosynthesized AgNPs
The antioxidant potential of green synthesized AgNPs and ascorbic acid was evaluated against DPPH free radicals. The results showed that 82.44 ± 1.43% of DPPH free radicals was inhibited by the biofabricated AgNPs at a concentration of 1 mg mL−1. In addition, ascorbic acid (5 mg mL−1) as the positive control hindered 97.89 ± 0.63% of DPPH. Hence, the biosynthesized AgNPs exhibited significant antioxidant potential and were highly capable of inhibiting DPPH free radicals. In agreement with our results, Tyagi et al. utilized Tagetes erecta L. aqueous leaf extract and biosynthesized spherical AgNPs in the size range of 7–35 nm. The best percentage of the inhibition by the biogenic AgNPs at the concentration of 600 μg mL−1 against DPPH radicals was reported to be 62.58 ± 0.55% compared to the percentage of the inhibition of the aqueous leaf extract (38.56 ± 1.12%) at the same concentration. The authors indicated that the antioxidant potential was dose-dependent in both AgNPs and the extract.58 In a study, AgNPs, with spherical morphology and an average size of 22.7 nm, were biosynthesized utilizing Oldenlandia umbellate aqueous extract. They confirmed the antioxidant activity of the biogenic AgNPs by performing three analyses, i.e. DPPH radical scavenging, reducing power, and total antioxidant assays. The percentages of DPPH inhibitory activity of phytofabricated AgNPs were found to be 10.4 ± 0.08 and 54 ± 0.80% at the concentrations of 5 and 100 μg mL−1, respectively. The positive control (ascorbic acid) also showed the DPPH free radical inhibition of 15.1 ± 0.39 and 62.1 ± 0.74% at concentrations of 5 and 100 μg mL−1, respectively. The results of two other analyses were similar to the DPPH radical scavenging method.59 Likewise, Erenler et al. assessed the antioxidant activity of Origanum onites-mediated synthesized AgNPs with an average size of 54.2 nm and spherical structure. The authors reported that the DPPH scavenging effect of the bioengineered AgNPs was significant with the IC50 value of 12.25 μg mL−1 which was higher than the IC50 value of plant extract (IC50: 21.66 μg mL−1).60
Anticoagulant effect of phytosynthesized AgNPs
The interaction of NPs with the proteins in the coagulation pathways could be used to inhibit coagulation. We measured the anticoagulant activity of the biosynthesized AgNPs at the concentration of 1 mg mL−1 in the intrinsic pathway of coagulation through the aPTT test. The mean value was recorded as 89.9 ± 2.83 and 32.2 ± 0.83 s for the biogenic AgNPs and saline as the control, respectively (P < 0.05). The considerable difference between the mean aPTT values probably determines that AgNPs were an effective agent in the intrinsic pathway of blood clotting. Additionally, the anticoagulant activity of the biofabricated AgNPs at the concentration of 1 mg mL−1 in the extrinsic pathway was evaluated by PT analysis. The mean PT values for the phytofabricated AgNPs and saline were found to be 18.2 ± 0.32 and 14.2 ± 0.28 s, respectively (P < 0.05). Hence, it could be suggested that both extrinsic and intrinsic pathways were affected by the biogenic AgNPs in blood clotting. According to the large difference present between the aPTT values, the intrinsic pathway may have a key role in the anticoagulant activity of AgNPs. Hence, our findings revealed that biogenic AgNPs could be an effective agent for the treatment of thrombotic disorders. In a study, Asghar et al. reported the biosynthesis of AgNPs using Sygyzium aromaticum with a spherical shape and size range of 30–40 nm. Then, the authors evaluated the in vivo anticoagulant activity of phytofabricated AgNPs at the concentrations of 0.025 and 0.05 mg kg−1 on healthy white male albino rabbits through oral administration. The results showed that the aPTT (0.025 mg kg−1, 38.3 ± 5.25 s; 0.05 mg kg−1, 41.6 ± 4.77 s) and PT (0.025 mg kg−1, 18.3 ± 3.10 s; 0.05 mg kg−1, 20.5 ± 3.69 s) were increased compared to the control (aPTT: 31.6 ± 4.27 s; PT: 11.2 ± 0.74 s) indicating the effect of the biogenic AgNPs on both intrinsic and extrinsic pathways of coagulation.61 In another study, spherical AgNPs were fabricated biologically with an average diameter of 57.42 nm utilizing Cassia obtusifolia leaf extract as a reducing agent. The significant prolongation of aPTT and PT values exhibited that the biosynthesized AgNPs had anticoagulant activity in both pathways of blood clotting. The authors suggested that the content of the herbal extract was responsible for the anticoagulant potential of the phytosynthesized AgNPs.62
Conclusions
In the current investigation, a sustainable, less expensive, chemical-free, energy-efficient, and scalable approach was used to synthesize AgNPs by employing the aqueous seed extract of P. anisum. This is an appealing technique for the green synthesis of monodispersed AgNPs protecting human health and the environment resulting in safer products as well as lesser waste. The P. anisum-mediated fabricated AgNPs were characterized by using analytical tools, such as UV-visible spectroscopy, DLS, XRD, FT-IR, and FESEM representing the fabrication of nanosized colloidal silver particles with spherical morphology with an average hydrodynamic diameter of 65.40 nm. In the next step, we evaluated the biological properties of phytofabricated AgNPs. The colloidal nanosized silver particles exhibited considerable antibacterial activity against reference and pathogenic strains of E. coli. Impressively, the biogenic AgNPs represented higher biofilm inhibitory properties than standard antibiotic gentamicin. Afterward, the DNA and protein leakage assays were performed to evaluate the cell membrane integrity of the AgNP-treated reference strain of E. coli. The DNA and protein were detected in the supernatant of AgNP-treated bacterial cells. Then, these findings were confirmed by FESEM and TEM representing the disturbances in bacterial structure, formation of rumples, and physiological damage. In addition, the biofabricated AgNPs exhibited bacterial genotoxicity. Furthermore, the herbal-mediated fabricated AgNPs showed antioxidant, antidiabetic, and anticoagulant potential. Notably, despite the significant biological performance of biosynthesized AgNPs, the drawbacks and toxicity of these nanostructures on the human body should be investigated and the safe dosage of the nano-based structures should be addressed in vivo.
Author contributions
HB, HV, FM, and EM supervised the study, provided intellectual input, and contributed to manuscript writing and interpretation of data. HB and FA wrote the first draft of the manuscript. OH, KJ, SSA, FA, AMAA, and SA performed the experiments. NM was involved in the statistical analysis of the study. HB, OH, and EM reviewed and edited the manuscript.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was financially supported by Shahid Beheshti University of Medical Sciences, Tehran, Iran (Grant Number 26134).
References
- J. Osorio-Echavarría, J. Osorio-Echavarría, C. P. Ossa-Orozco and N. A. Gómez-Vanegas, Synthesis of silver nanoparticles using white-rot fungus Anamorphous Bjerkandera sp. R1: influence of silver nitrate concentration and fungus growth time, Sci. Rep., 2021, 11(1), 3842 CrossRef PubMed.
- S. Samuggam, S. V. Chinni, P. Mutusamy, S. C. Gopinath, P. Anbu and V. Venugopal,
et al., Green Synthesis and Characterization of Silver Nanoparticles Using Spondias mombin Extract and Their Antimicrobial Activity against Biofilm-Producing Bacteria, Molecules, 2021, 26(9), 2681 CrossRef CAS PubMed.
- P. C. Mane, S. A. R. Sayyed, D. D. Kadam, D. M. Shinde, A. Fatehmulla and A. M. Aldhafiri,
et al., Terrestrial snail-mucus mediated green synthesis of silver nanoparticles and in vitro investigations on their antimicrobial and anticancer activities, Sci. Rep., 2021, 11(1), 13068 CrossRef CAS PubMed.
- M. Wypij, T. Jędrzejewski, J. Trzcińska-Wencel, M. Ostrowski, M. Rai and P. Golińska, Green Synthesized Silver Nanoparticles: Antibacterial and Anticancer Activities, Biocompatibility, and Analyses of Surface-Attached Proteins, Front. Microbiol., 2021, 12, 888 CrossRef PubMed.
- H. J. C. Barabadi and M. Biology, Nanobiotechnology: A promising scope of gold biotechnology, Cell. Mol. Biol., 2017, 63(12), 3–4 CrossRef.
- D. Wang, B. Xue, L. Wang, Y. Zhang, L. Liu and Y. Zhou, Fungus-mediated green synthesis of nano-silver using Aspergillus sydowii and its antifungal/antiproliferative activities, Sci. Rep., 2021, 11(1), 10356 CrossRef CAS PubMed.
- C. Vanlalveni, S. Lallianrawna, A. Biswas, M. Selvaraj, B. Changmai and S. L. Rokhum, Green synthesis of silver nanoparticles using plant extracts and their antimicrobial activities: a review of recent literature, RSC Adv., 2021, 11(5), 2804–2837 RSC.
- N. Jain, P. Jain, D. Rajput and U. K. Patil, Green synthesized plant-based silver nanoparticles: therapeutic prospective for anticancer and antiviral activity, Micro Nano Syst. Lett., 2021, 9(1), 5 CrossRef.
- B. Skóra, U. Krajewska, A. Nowak, A. Dziedzic, A. Barylyak and M. Kus-Liśkiewicz, Noncytotoxic silver nanoparticles as a new antimicrobial strategy, Sci. Rep., 2021, 11(1), 1–13 CrossRef PubMed.
- P. Singh, S. Pandit, C. Jers, A. S. Joshi, J. Garnæs and I. Mijakovic, Silver nanoparticles produced from Cedecea sp. exhibit antibiofilm activity and remarkable stability, Sci. Rep., 2021, 11(1), 12619 CrossRef CAS PubMed.
- H. Arshad, M. A. Sami, S. Sadaf and U. Hassan, Salvadora persica mediated synthesis of silver nanoparticles and their antimicrobial efficacy, Sci. Rep., 2021, 11(1), 5996 CrossRef CAS PubMed.
- H. Barabadi, F. Mojab, H. Vahidi, B. Marashi, N. Talank and O. Hosseini,
et al., Green synthesis, characterization, antibacterial and biofilm inhibitory activity of silver nanoparticles compared to commercial silver nanoparticles, Inorg. Chem. Commun., 2021, 129, 108647 CrossRef CAS.
- N. Talank, H. Morad, H. Barabadi, F. Mojab, S. Amidi and F. Kobarfard,
et al., Bioengineering of green-synthesized silver nanoparticles: In vitro physicochemical, antibacterial, biofilm inhibitory, anticoagulant, and antioxidant performance, Talanta, 2022, 243, 123374 CrossRef CAS PubMed.
- B. Syed, N. Bisht, P. S. Bhat, R. Nikhil Karthik, A. Prasad and B. L. Dhananjaya,
et al., Phytogenic synthesis of nanoparticles from Rhizophora mangle and their bactericidal potential with DNA damage activity, Nano-Struct. Nano-Objects, 2017, 10, 112–115 CrossRef CAS.
- F. D. Koca and F. Duman, Genotoxic and cytotoxic activity of green synthesized TiO2 nanoparticles, Appl. Nanosci., 2019, 9(5), 815–823 CrossRef CAS.
- F. Saghir, K. Hussain, M. N. Tahir, S. A. Raza, N. Shehzadi and S. Iftikhar,
et al., Antidiabetic Screening, Activity-guided Isolation and Molecular Docking Studies of Flower Extracts of Pongamia pinnata (L.) Pierre, J. Med. Plants By-Prod., 2021, 10(1), 85–92 Search PubMed.
- F. Boshagh, Measurement methods of carbohydrates in dark fermentative hydrogen production- A review, Int. J. Hydrogen Energy, 2021, 46(47), 24028–24050 CrossRef CAS.
- G. Debnath, P. Das and A. K. Saha, Green Synthesis of Silver Nanoparticles Using Mushroom Extract of Pleurotus giganteus: Characterization, Antimicrobial, and α-Amylase Inhibitory Activity, Bionanoscience, 2019, 9(3), 611–619 CrossRef.
- H. Ashraf, T. Anjum, S. Riaz and S. Naseem, Microwave-assisted green synthesis and characterization of silver nanoparticles using melia azedarach for the management of fusarium wilt in tomato, J. Front. Microbiol., 2020, 11, 238 CrossRef PubMed.
- A. Bryła, G. Lewandowicz and W. Juzwa, Encapsulation of elderberry extract into phospholipid nanoparticles, J. Food Eng., 2015, 167, 189–195 CrossRef.
- S. Mandal, S. B. Marpu, R. Hughes, M. A. Omary and S. Q. Shi, Green synthesis of silver nanoparticles using Cannabis sativa extracts and their anti-bacterial activity, Green Sustainable Chem., 2021, 11(1), 38–48 CAS.
- M. Chandhru, R. Logesh, S. K. Rani, N. Ahmed and N. Vasimalai, One-pot green route synthesis of silver nanoparticles from jack fruit seeds and their antibacterial activities with escherichia coli and salmonella bacteria, Biocatal. Agric. Biotechnol., 2019, 20, 101241 CrossRef.
- H. Yousaf, A. Mehmood, K. S. Ahmad and M. Raffi, Green synthesis of silver nanoparticles and their applications as an alternative antibacterial and antioxidant agents, Mater. Sci. Eng., C, 2020, 112, 110901 CrossRef CAS PubMed.
- V. Lakkim, M. C. Reddy, R. R. Pallavali, K. R. Reddy, C. V. Reddy and Inamuddin,
et al., Green Synthesis of Silver Nanoparticles and Evaluation of Their Antibacterial Activity against Multidrug-Resistant Bacteria and Wound Healing Efficacy Using a Murine Model, Antibiotics, 2020, 9(12), 902 CrossRef CAS PubMed.
- S. Akter and M. A. Huq, Biologically rapid synthesis of silver nanoparticles by Sphingobium sp. MAH-11T and their antibacterial activity and mechanisms investigation against drug-resistant pathogenic microbes, Artif. Cells, Nanomed., Biotechnol., 2020, 48(1), 672–682 CrossRef CAS PubMed.
- A. K. Keshari, R. Srivastava, P. Singh, V. B. Yadav and G. Nath, Antioxidant and antibacterial activity of silver nanoparticles synthesized by Cestrum nocturnum, J. Ayurveda Integr. Med., 2020, 11(1), 37–44 CrossRef PubMed.
- A. H. Mondal, D. Yadav, S. Mitra and K. Mukhopadhyay, Biosynthesis of Silver Nanoparticles Using Culture Supernatant of Shewanella sp. ARY1 and Their Antibacterial Activity, Int. J. Nanomed., 2020, 15, 8295–8310 CrossRef CAS PubMed.
- L. S. Naik and C. V. Ramana Devi, Phyto-fabricated silver nanoparticles inducing microbial cell death via reactive oxygen species-mediated membrane damage, IET Nanobiotechnol., 2021, 15(5), 492–504 CrossRef PubMed.
- P. C. Mane, S. A. R. Sayyed, D. D. Kadam, M. D. Shinde, A. Fatehmulla and A. M. Aldhafiri,
et al., Terrestrial snail-mucus mediated green synthesis of silver nanoparticles and in vitro investigations on their antimicrobial and anticancer activities, Sci. Rep., 2021, 11(1), 13068 CrossRef CAS PubMed.
- A. Rautela, J. Rani and M. Debnath, Green synthesis of silver nanoparticles from Tectona grandis seeds extract: characterization and mechanism of antimicrobial action on different microorganisms, J. Anal. Sci. Technol., 2019, 10(1), 5 CrossRef.
- R. Rani, D. Sharma, M. Chaturvedi and J. Yadav, Green synthesis, characterization and antibacterial activity of silver nanoparticles of endophytic fungi Aspergillus terreus, J. Nanomed. Nanotechnol., 2017, 8(4), 1000457 Search PubMed.
- V. Gopinath, S. Priyadarshini, M. F. Loke, J. Arunkumar, E. Marsili and D. MubarakAli,
et al., Biogenic synthesis, characterization of antibacterial silver nanoparticles and its cell cytotoxicity, Arabian J. Chem., 2017, 10(8), 1107–1117 CrossRef CAS.
- S. N. Hazarika, K. Gupta, K. N. A. M. Shamin, P. Bhardwaj, R. Boruah and K. K. Yadav,
et al., One-pot facile green synthesis of biocidal silver nanoparticles, Mater. Res. Express, 2016, 3(7), 075401 CrossRef.
- Y. Tian, J. Luo, H. Wang, H. E. M. Zaki, S. Yu and X. Wang,
et al., Bioinspired Green Synthesis of Silver Nanoparticles Using Three Plant Extracts and Their Antibacterial Activity against Rice Bacterial Leaf Blight Pathogen Xanthomonas oryzae pv. oryzae, Plants, 2022, 11(21), 2892 CrossRef CAS PubMed.
- M. A. Ansari, A. Kalam, A. G. Al-Sehemi, M. N. Alomary, S. AlYahya and M. K. Aziz,
et al., Counteraction of Biofilm Formation and Antimicrobial Potential of Terminalia catappa Functionalized Silver Nanoparticles against Candida albicans and Multidrug-Resistant Gram-Negative and Gram-Positive Bacteria, Antibiotics, 2021, 10(6), 725 CrossRef CAS PubMed.
- P. Stiefel, S. Schmidt-Emrich, K. Maniura-Weber and Q. Ren, Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide, BMC Microbiol., 2015, 15(1), 36 CrossRef PubMed.
- S. S. D. Kumar, N. N. Houreld, E. M. Kroukamp and H. Abrahamse, Cellular imaging and bactericidal mechanism of green-synthesized silver nanoparticles against human pathogenic bacteria, J. Photochem. Photobiol., B, 2018, 178, 259–269 CrossRef CAS PubMed.
- C. Musimun, D. Papiernik, P. Permpoonpattana, P. Chumkaew and T. Srisawat, Synergy of green-synthesized silver nanoparticles and Vatica diospyroides fruit extract in inhibiting Gram-positive bacteria by inducing membrane and intracellular disruption, J. Exp. Nanosci., 2022, 17(1), 420–438 CrossRef CAS.
- B. Das, S. K. Dash, D. Mandal, T. Ghosh, S. Chattopadhyay and S. Tripathy,
et al., Green synthesized silver nanoparticles destroy multidrug resistant bacteria via reactive oxygen species mediated membrane damage, Arabian J. Chem., 2017, 10(6), 862–876 CrossRef CAS.
- M. Singh, Elucidation of biogenic silver nanoparticles susceptibility towards Escherichia coli: an investigation on the antimicrobial mechanism, IET Nanobiotechnol., 2016, 10(5), 276–280 CrossRef PubMed.
- M. Mousavi-Khattat, M. Keyhanfar and A. Razmjou, A comparative study of stability, antioxidant, DNA cleavage and antibacterial activities of green and chemically synthesized silver nanoparticles, Artif. Cells, Nanomed., Biotechnol., 2018, 46(sup3), S1022–S1031 CrossRef CAS PubMed.
- S. Chowdhury, A. Basu and S. Kundu, Green synthesis of protein capped silver nanoparticles from phytopathogenic fungus Macrophomina phaseolina (Tassi) Goid with antimicrobial properties against multidrug-resistant bacteria, Nanoscale Res. Lett., 2014, 9(1), 365 CrossRef PubMed.
- S. Gonca, Investigation of in-vitro biological activities of silver nanoparticles synthesized by green synthesis method using wild edible mushroom Macrolepiota procera, Lokman Hekim J., 2022, 12(1), 198–208 Search PubMed.
- P. Singh, S. Pandit, V. Mokkapati, J. Garnæs and I. Mijakovic, A sustainable approach for the green synthesis of silver nanoparticles from Solibacillus isronensis sp. and their application in biofilm inhibition, Molecules, 2020, 25(12), 2783 CrossRef CAS PubMed.
- S. Chandrasekharan, G. Chinnasamy and S. Bhatnagar, Sustainable phyto-fabrication of silver nanoparticles using Gmelina arborea exhibit antimicrobial and biofilm inhibition activity, Sci. Rep., 2022, 12(1), 156 CrossRef CAS PubMed.
- R. B. Shafreen, S. Seema, A. P. Ahamed, N. Thajuddin and S. Ali Alharbi, Inhibitory Effect of Biosynthesized Silver Nanoparticles from Extract of Nitzschia palea Against Curli-Mediated Biofilm of Escherichia coli, Appl. Biochem. Biotechnol., 2017, 183(4), 1351–1361 CrossRef CAS PubMed.
- D. Bharathi, S. Vasantharaj and V. Bhuvaneshwari, Green synthesis of silver nanoparticles using Cordia dichotoma fruit extract and its enhanced antibacterial, anti-biofilm and photo catalytic activity, Mater. Res. Express, 2018, 5(5), 055404 CrossRef.
- P. Singh, S. Pandit, M. Beshay, V. R. S. S. Mokkapati, J. Garnaes and M. E. Olsson,
et al., Anti-biofilm effects of gold and silver nanoparticles synthesized by the Rhodiola rosea rhizome extracts, Artif. Cells, Nanomed., Biotechnol., 2018, 46(sup3), S886–S899 CrossRef CAS PubMed.
- R. Perumalsamy and L. Krishnadhas, Anti-Diabetic Activity of Silver Nanoparticles Synthesized from the Hydroethanolic Extract of Myristica fragrans Seeds, Appl. Biochem. Biotechnol., 2022, 194(3), 1136–1148 CrossRef CAS PubMed.
- Y. Arsia Tarnam and T. Nargis Begum, Muhammad Ilyas Mohammed H, Shilu M, Archunan G, Ishtiaq Q. Antioxidant and Hypoglycemic Activities of Clausena anisata (Willd.) Hook F. Ex Benth. Root Mediated Synthesized Silver Nanoparticles, Pharmacogn. J., 2016, 8(6), 579–586 CrossRef.
- S. Wilson, S. Cholan, U. Vishnu, M. Sannan, R. Jananiya and S. Vinodhini,
et al., In vitro assessment of the efficacy of free-standing silver nanoparticles isolated from Centella asiatica against oxidative stress and its antidiabetic activity, Der Pharm. Lettre, 2015, 7(12), 194–205 Search PubMed.
- L. Gong, D. Feng, T. Wang, Y. Ren, Y. Liu and J. Wang, Inhibitors of α-amylase and α-glucosidase: Potential linkage for whole cereal foods on prevention of hyperglycemia, Food Sci. Nutr. Diabetes, 2020, 8(12), 6320–6337 CrossRef CAS PubMed.
- S. R. Adil, A. Rizwan, B. Saira and S. Iqra, Bioactivity-guided nanoparticle synthesis from Zingiber officinale and Mentha longifolia, Bioinspired, Biomimetic Nanobiomater., 2021, 10(3), 70–77 CrossRef.
- M. Vishnu Kiran and S. Murugesan, Biogenic silver nanoparticles by Halymenia poryphyroides and its in vitro anti-diabetic efficacy, J. Chem. Pharm. Res., 2013, 5(12), 1001–1008 Search PubMed.
- S. Andleeb, F. Tariq, A. Muneer, T. Nazir, B. Shahid and Z. Latif,
et al., In vitro bactericidal, antidiabetic, cytotoxic, anticoagulant, and hemolytic effect of green-synthesized silver nanoparticles using Allium sativum clove extract incubated at various temperatures, Green Process. Synth., 2020, 9(1), 538–553 Search PubMed.
- P. Chirumamilla, S. B. Dharavath and S. Taduri, Eco-friendly Green Synthesis of Silver Nanoparticles from Leaf Extract of Solanum khasianum: Optical Properties and Biological Applications, Appl. Biochem. Biotechnol., 2023, 195(1), 353–368 CrossRef CAS PubMed.
- S. Vankudoth, S. Dharavath, S. Veera, N. Maduru, R. Chada and P. Chirumamilla,
et al., Green synthesis, characterization, photoluminescence and biological studies of silver nanoparticles from the leaf extract of Muntingia calabura, Biochem. Biophys. Res. Commun., 2022, 630, 143–150 CrossRef CAS PubMed.
- P. K. Tyagi, S. Tyagi, D. Gola, A. Arya, S. A. Ayatollahi and M. M. Alshehri,
et al., Ascorbic acid and polyphenols mediated green synthesis of silver nanoparticles from Tagetes erecta L. aqueous leaf extract and studied their antioxidant properties, J. Nanomater., 2021, 2021, 6515419 Search PubMed.
- P. Subramanian, A. Ravichandran, V. Manoharan, R. Muthukaruppan, S. Somasundaram and B. Pandi,
et al., Synthesis of Oldenlandia umbellata stabilized silver nanoparticles and their antioxidant effect, antibacterial activity, and bio-compatibility using human lung fibroblast cell line WI-38, Process Biochem., 2019, 86, 196–204 CrossRef CAS.
- R. Erenler and B. Dag, Biosynthesis of silver nanoparticles using Origanum majorana L. and evaluation of their antioxidant activity, Inorg. Nano-Met. Chem., 2022, 52(4), 485–492 CAS.
- M. A. Asghar, R. I. Yousuf, M. H. Shoaib and M. A. Asghar, Antibacterial, anticoagulant and cytotoxic evaluation of biocompatible nanocomposite of chitosan loaded green synthesized bioinspired silver nanoparticles, Int. J. Biol. Macromol., 2020, 160, 934–943 CrossRef CAS PubMed.
- L. S. Flaih and N. H. Al-Saadi, Characterization and clinical application of silver nanoparticles synthesized from Cassia Obtusifolia leaves extract, Plant Arch., 2020, 20(2), 1082–1088 Search PubMed.
|
| This journal is © The Royal Society of Chemistry 2023 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Omid
Hosseini
b,
Kamyar
Jounaki
a,
Salar
Sadeghian-Abadi
a,
Fatemeh
Ashouri
a,
Ayat
Mostafa Abdulabbas Alrikabi
a,
Hossein
Vahidi
a,
Salimeh
Amidi
c,
Faraz
Mojab
d,
Neda
Mohammadi
e and
Ebrahim
Mostafavi
*a,
Omid
Hosseini
b,
Kamyar
Jounaki
a,
Salar
Sadeghian-Abadi
a,
Fatemeh
Ashouri
a,
Ayat
Mostafa Abdulabbas Alrikabi
a,
Hossein
Vahidi
a,
Salimeh
Amidi
c,
Faraz
Mojab
d,
Neda
Mohammadi
e and
Ebrahim
Mostafavi
 *fg
*fg
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min using an ultracentrifuge (Beckman, L90k, US). Deionized double distilled water was used to wash the surface of AgNPs. The AgNPs were washed thrice to improve their purification. The UV-Vis spectroscopy assay was operated by using a double-beam spectrophotometer (Shimadzu, UV-1280, Japan) to confirm the formation of NPs by monitoring a Surface Plasmon Resonance (SPR) peak in the range of 200–800 nm. Besides, the morphology of AgNPs was determined by using the FESEM (Sigma VP, ZEISS, Germany) with an acceleration voltage of 15 kV. In addition, the XRD analysis was carried out for the phytosynthesized AgNPs using PW3050/60 with XPERT-PRO (using CuKα radiation, λ = 1.5406 Å) at 30 kV and 100 mA and recorded from 20 to 80 degrees for 2θ. Moreover, the hydrodynamic diameter and Polydispersity Index (PdI) of AgNPs were recorded using a Zetasizer (Malvern, England) with a scattering angle of 90° at 25 °C. Furthermore, the FT-IR spectroscopy was operated using an FT-IR spectrophotometer (Agilent, Cary 630 model, US) in transmittance mode in the wavelength range of 400 to 4000 cm−1 using the potassium bromide (KBr) pellet method in the ratio of 1
000 rpm for 20 min using an ultracentrifuge (Beckman, L90k, US). Deionized double distilled water was used to wash the surface of AgNPs. The AgNPs were washed thrice to improve their purification. The UV-Vis spectroscopy assay was operated by using a double-beam spectrophotometer (Shimadzu, UV-1280, Japan) to confirm the formation of NPs by monitoring a Surface Plasmon Resonance (SPR) peak in the range of 200–800 nm. Besides, the morphology of AgNPs was determined by using the FESEM (Sigma VP, ZEISS, Germany) with an acceleration voltage of 15 kV. In addition, the XRD analysis was carried out for the phytosynthesized AgNPs using PW3050/60 with XPERT-PRO (using CuKα radiation, λ = 1.5406 Å) at 30 kV and 100 mA and recorded from 20 to 80 degrees for 2θ. Moreover, the hydrodynamic diameter and Polydispersity Index (PdI) of AgNPs were recorded using a Zetasizer (Malvern, England) with a scattering angle of 90° at 25 °C. Furthermore, the FT-IR spectroscopy was operated using an FT-IR spectrophotometer (Agilent, Cary 630 model, US) in transmittance mode in the wavelength range of 400 to 4000 cm−1 using the potassium bromide (KBr) pellet method in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100.
100.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) H, C–H, C
H, C–H, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–N, C–N, C–H, and C–H, respectively, that are found in the phytochemical constituents in the plant extracts (Fig. S1, ESI†). The FT-IR spectroscopy provided preliminary information and did not characterize the full structure of bioactive phytochemicals surrounding the AgNPs. Hence, HPLC/GCMS analysis can help to characterize the structure of the bioactive molecules that have a role in the bioreduction process. Moreover, the XRD pattern of AgNPs confirmed the fabrication of nanosized silver particles with highly crystalline structures (Ref. code. 01-089-3722 (Ag); 00-006-0480 (AgCl)) (Fig. 3). Besides, Fig. 4 showed the phytofabrication of spherical-shaped AgNPs.
C, C–N, C–N, C–H, and C–H, respectively, that are found in the phytochemical constituents in the plant extracts (Fig. S1, ESI†). The FT-IR spectroscopy provided preliminary information and did not characterize the full structure of bioactive phytochemicals surrounding the AgNPs. Hence, HPLC/GCMS analysis can help to characterize the structure of the bioactive molecules that have a role in the bioreduction process. Moreover, the XRD pattern of AgNPs confirmed the fabrication of nanosized silver particles with highly crystalline structures (Ref. code. 01-089-3722 (Ag); 00-006-0480 (AgCl)) (Fig. 3). Besides, Fig. 4 showed the phytofabrication of spherical-shaped AgNPs.

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg mL−1 protein leakage after 6 h of treatment with 1 mg mL−1 of AgNPs.30 Similarly, snail-mucus-mediated fabricated AgNPs in the range of 37 to 87 nm caused protein leakage in S. aureus with a protein content of 3.98 and 4.32 mg mL−1 after 24 h of treatment with AgNPs at the concentrations of 3 and 6 μg mL−1, respectively.29 In a separate study, fungus-mediated synthesized AgNPs with an average size of 16.54 nm increased the protein leakage level from around 75 μg mL−1 (control) to around 140 μg mL−1 after treatment for 8 h with the concentration of 10 mg mL−1 of AgNPs.31
μg mL−1 protein leakage after 6 h of treatment with 1 mg mL−1 of AgNPs.30 Similarly, snail-mucus-mediated fabricated AgNPs in the range of 37 to 87 nm caused protein leakage in S. aureus with a protein content of 3.98 and 4.32 mg mL−1 after 24 h of treatment with AgNPs at the concentrations of 3 and 6 μg mL−1, respectively.29 In a separate study, fungus-mediated synthesized AgNPs with an average size of 16.54 nm increased the protein leakage level from around 75 μg mL−1 (control) to around 140 μg mL−1 after treatment for 8 h with the concentration of 10 mg mL−1 of AgNPs.31










