Open Access Article
Open Access ArticleA facile one-pot synthesis of water-soluble CQDs for the evaluation of their anti-amyloidogenic propensity
Aniket
Mukherjee
and
Nandini
Sarkar
 *
*
Department of Biotechnology and Medical Engineering, National Institute of Technology Rourkela, Rourkela-769008, Odisha, India. E-mail: sarkarn@nitrkl.ac.in; Fax: 91-661-462281; Tel: 91-661-2462293
First published on 28th March 2023
Abstract
Herein, we present a one-pot microwave-assisted facile synthesis of highly water-soluble CQDs and explore their inhibitory effect towards amyloid fibrillation or their anti-amyloidogenic properties using hen egg-white lysozyme for fibrillation in acidic and neutral pH and explore their potential application as a drug against protein linked neurodegenerative diseases. The average 7 nm sized CQDs prepared using direct carbonization of folic acid and bis-acryl amide under microwave showed long shelf life due to high negative surface charge, which also makes them highly soluble in water, resulting in high photo-stability (pH), high quantum yield (QY), without the need for a surface passivation step after synthesis, thus making the process less cumbersome than the contemporary ones. Upon exploration, the prepared CQDs demonstrated a significant inhibitory effect towards the HEWL amyloid aggregation process in both acidic and neutral pH, as it inhibits up to 60% in acidic pH and up to 50% in neutral pH conditions of the process, which were confirmed using various assays, such as ThT and ANS. Proteins, under certain destabilising conditions, have been seen to unfold and aggregate to form ordered β-sheeted fibrillar aggregates called “amyloids”. These fibrils are the main precursors for protein-linked degenerative diseases, such as Alzheimer's disease, Parkinson's disease, Huntington's disease, and Type II diabetes mellitus. Thus, with the anti-amyloidogenic properties of the CQD as well as their excellent intrinsic characteristics, these CQD have appeared to be good potential candidates for the treatment of protein-linked nuero degenerative diseases.
1. Introduction
Over the past few years, nanomaterials have gained quite a lot of popularity owing to their excellent chemical, physical, and electronic properties that can be harnessed for various chemical, mechanical, biological, and medical applications. Among these nanomaterials, quantum dots are being researched extensively for the last two decades. Quantum dots are particles trapped in an energy state, which have a size of around 2–10 nm and are known to be highly fluorescent and resistant to photobleaching, thus giving them a longer lifetime than regular dyes available.1 There are organic and inorganic quantum dots based on their composition. Inorganic quantum dots are generally semiconductor nanocrystals whose electronic conductivity lies between the bulk and amorphous materials consisting of elements from groups II to VI or III to V in the periodic table and are usually harnessed for their excellent semiconducting properties, optical luminescence, etc. However, they are found to be quite cytotoxic to human cells, thus can not generally be used in any biomedical applications, which is one of the leading fields of research in today's world.Organic quantum dots, which are also known as CQDs, are essentially composed of carbon and most often are non-cytotoxic to human cells and are highly fluorescent in nature. Since their accidental discovery in 2004, CQDs have made a huge impact in the field of chemical, biological, and medical sciences due to their high optical fluorescence, chemical stability, and very low cytotoxicity,2 and have found applications in the field of biosensor development,3 biomedical imaging,4 as well as in electronic devices.5 Carbon quantum dots are quasi-spherical nanoparticles consisting of amorphous as well as nanocrystalline cores with graphitic or turbostratic carbon (sp2 hybridized carbon) that is the carbon crystal whose basal plane has slipped out of alignment or graphene and graphene oxide sheets fused by diamond-like sp3 hybridized carbon insertions.6–8 CQDs are known to provide a greater receptor binding affinity as well as a better cell penetration capability in lieu of their small size.9 Broadly, there are two ways to synthesize CQDs: the top-down approach and the bottom-up approach; CQDs could easily be surface passivated to emit greater fluorescence or enhance its properties of targeted delivery, higher shelf life, etc. The properties of the CQDs such as size quantum yield often essentially depend on the precursor material. The fluorescence quantum yield of the CQDs from cheap sources such as candle soot was found to be around 15%10 while that from citric acid and ethylenediamine was found to be 58%11 at room temperature, which is the highest yet to be recorded. Often to reduce the production cost, chemical processes such as oxidation of candle soot, or nascent carbon by conc. nitric acid or other strong acid10,12 are applied. CQDs have been prepared from various materials having carbon in their molecular backbones such as glucosamine, ascorbic acid, ethanol, citric acid, and saccharides. Recently, self-passivating CQDs are being synthesized by microwave-assisted or hydrothermal methods and high temperature synthesis using carbon precursors.13–16 However, most of these synthesis techniques are multi-stepped and require high post-synthesis purification to obtain the desired product,17 resulting in low quantum yield. Thus, the synthesis of CQDs with high QY and good biocompatibility still remains a challenge.18–20
Protein aggregation and misfolding, which leads to its deposition along human organs and tissue lines, resulting in serious degenerative diseases, is known as amyloidosis. Amyloidosis leads to diseases such as Alzheimer's disease, Spongiform Encephalopathy, and type 2 diabetes and is characterized by long unbranched fibrils.21 They are generally progressive and are associated with a high mortality rate. The destabilization of the native conformations of the proteins leads to the formation of partially unfolded intermediates leading to the assembly and unfolding of proteins forming amyloid fibrils.22,23 Around 20–25 human proteins have been identified over the past few decades, which are known to form amyloid fibril and these proteins are essentially dissimilar having few common traits, which are used to identify and distinguish them.24–26 Hen Egg White lysozyme is among the commonly used model for the fibrillation process and inhibitory effects of small molecules.27–29 Studies have shown that inhibiting the amyloid formation as well as the disintegration of the fibrillar structure can provide an essential therapeutic approach towards the treatment of diseases mentioned in the earlier part.21,30–32 The use of small molecules for suppressing the aggregation process is the most effective way of inhibiting amyloidosis.33 Some naturally found polyphenols have been successful in suppressing the HEWL formation.34,35 MP Taraka Prabhu et al. have shown the inhibitory properties of the CQDs, which were synthesized from naturally occurring spice mix.36 In another study, E. D. Guerrero et al. have shown the inhibitory effect of Na-Citrate CDs on HEWL formation.37 However, despite these reports, the development of effective CQDs-based therapeutics against amyloidosis remains elusive and demands further studies. In this study, we have used folic acid and bis acryl amide (BAA) for the preparation of the carbon dots, folic acid was used in order to have folic acid residues on the surface which can be used for the targeted delivery in the future, and BAA was used as the nitrogen source due to the fact that it has an urea-like structure, also both these sources are available at a cheap rate, which hugely decreases the cost of synthesis, and thus we report a novel, facile, quick one-step microwave-assisted synthesis of water-soluble surface functionalized CQDs with high quantum yield and high fluorescent intensity and average size 7 nm, which show high efficiency in inhibiting the aggregation process of HEWL.
2. Methods and materials
2.1. Materials required
Folic acid (FA) and bis-acrylamide (BAA) were purchased from Himedia, Hen egg white lysozyme (HEWL) (purity ≥90%), guanidine hydrochloride (purity ≥99%), and thioflavin T were purchased from Sigma Aldrich. Glycine (purity ≥99%), hydrogen chloride purity (≥35%) were obtained from Merck. Di-sodium hydrogen phosphate (purity ≥98.5%), sodium di-hydrogenphosphate (purity ≥98%), sodium chloride (purity ≥99.5%) were purchased from HiMedia Laboratory Chemicals (India).2.2. Preparation of CQDs
The CQDs were synthesized using a microwave-assisted quick method. Firstly, 0.2 g FA and 0.2 g BAA were added to 5 mL ultra-pure Miliq water in a glass beaker. The mixture was then stirred without heating at 300 rpm for 15 min. The resulting yellow solution was collected in a conical flask and kept in the microwave at 480 W for 10 min. During this period the solution turned brownish from the yellow colour, which indicated the formation of CQDs. After this process, the resulting solution was taken out and cooled to room temperature. 15 mL of Miliq water was added to the solution. The solution was then transferred to a 50 mL centrifuge tube. After centrifugation at 5000 rpm for 15 min, the supernatant solution was collected and passed through a 0.2 μm filter. The final solution was collected in 15 mL tubes for use in further studies.2.3. CQD characterisation
The mean size of the CQDs was characterized by dynamic light scattering using the nanoparticle size and zeta analyser (Malvern, NanoZS90) and transmission electron microscopy with the FEI, Tecnai G2 TF30-ST model transmission electron microscope. The morphology and the size profile of the CQDs were determined using atomic force microscopy on a Bioscope catalyst scan assist atomic force microscope; 50 μl droplets of CQDs dispersion were drop-cast on a freshly prepared mica surface, and then dried in argon before the examination. The UV-Vis spectroscopy was performed on a UV-Vis NIR spectrophotometer (Horiba Duetta) and the fluorescence behaviour profile of the emission of CQDs at 448 nm was recorded using the same instrument when the CQDs were excited from 270 to 380 nm. The Fourier transform infrared (FTIR) spectroscopy was performed on an FTIR spectrometer (Bruker, Alpha E), which particularly provided information about chemical bonds in the CQDs by providing a transmittance spectrum. XRD pattern was collected on a multipurpose X-Ray diffractometer (Rigaku, Japan & Ultima-IV). Raman spectroscopy was performed using the Witec Alpha 300 Confocal Raman Microscope, with a solid-state LASER of 532 nm with the power of 35 mW, a grating of 600 grooves per mm, magnification was kept at 50×, the scan range was 50–3000 cm−1, and The integration time was kept at 40 s. NMR spectra of CQDs were recorded using a 400 MHz Brucker spectrometer in d6 DMSO. The cytotoxicity assay of the prepared CQDs was performed on the mouse fibroblast cells (L929) for 24 h at a concentration of 400 μg mL−1 in 96 clusters well culture plates at 5% CO2 and 37 °C. Cell viability was measured by the MTT assay.2.4. Quantum yield measurement
The quantum yield measurements were performed by comparing the fluorescence intensities (excitation at 350 nm) and absorbance values of the prepared CQDs and quinine sulphate. Quinine sulphate was dissolved in 0.1 M H2SO4 having a refractive index of 1.33, which is the same for water, which is the solvent for CQDs. The slit widths for both excitation and emission were 5 nm. The quantum yield was calculated using the following equation:38| QY = QYr × (I/Ir) × (Ar/A) × (ηr/η) | (1) |
2.5. Preparation of HEWL amyloid fibril
The HEWL formation was carried out at different pH, i.e. pH 2.7 and pH 7.0, in order to determine the suitable pH for CQDs to work. Amyloid preparation at pH 2.7 was carried out using a modification to the method used by Gazova et al.30 Briefly, 35 μM HEWL was dissolved in a freshly prepared 70 mM Glycine-HCl buffer with pH 2.7. To study the inhibitory activity of CQDs, they were added along with the solution at various concentrations. The solution was then subjected to 65 °C heat and 1200 rpm stirring for 2 h on a Tarsons hot plate magnetic stirrer. Samples were collected in 1.5 mL Eppendorf tubes at 20 min intervals. The final protein concentration was kept 5 μM. For amyloid preparation at neutral pH, the method used by E. Kiran Kumar et al. was followed with some modifications. Briefly, 35 μM HEWL was dissolved in freshly prepared 4 M urea buffer. Similarly, to study the inhibitory effect of CQDs, they were added to the solution at different concentrations. The solution was then placed in a water bath for 12 h. The aliquots were collected at equal intervals of times (2 h) in 1.5 mL Eppendorf tubes. The final protein concentration was maintained at 5 μM.2.6. Thioflavin T(ThT) fluorescence assay
Thioflavin T is a widely used dye to sense amyloid formation or the lack of one by binding with the amyloid structure. Due to its binding, ThT shows an increase in its fluorescence intensity at around 485 nm when excited at 440 nm. As discussed above, the aliquots collected at regular intervals comprised the formed amyloid fibrils both in the presence and absence of CQDs. These aliquots were incubated in the dark for 45 min after the addition of the ThT dye at 20 μM concentration and the protein concentration was maintained at 5 μM. Both the amyloid inhibition assay as well as amyloid disintegration assays were performed. For the amyloid inhibition, CQDs were added at the beginning of the experiment, while for the amyloid disintegration study, the CQDs were added after the HEWL formation. While scanning for a possible change in the fluorescence intensity, the excitation was set a 440 nm, while the emission range was set at 450–700 nm with a slit width of 5 nm and integration time of 0.5 s where the peak value was observed at approximately 485 nm. These readings were recorded in the Horiba Duetta Absorbance and fluorescence spectrophotometer.2.7. ANS binding assay
8-Anilino-1-naphthalenesulfonic acid (ANS) is widely used as a dye to characterize protein conformational states having exposed hydrophobic clusters. A stock solution of 2.5 mM of 8-anilinonaphthalene-1-sulfonic acid (ANS) was prepared by dissolving ANS in double-distilled water. ANS was used at a 100-fold molar excess of HEWL. Samples were excited at 380 nm, and emission spectra were recorded from 390 nm to 600 nm using the Horiba Duetta absorbance and fluorescence spectrophotometer. The emission and excitation slit widths were fixed at 5 nm, and the integration time was set to 1 s.2.8. HEWL fibril characterization
2.9. Equilibrium unfolding study
HEWL was incubated with guanidine hydrochloride at a desired concentration by dissolving them in 50 mM glycine-HCl buffer of pH 2.7 and 4 M urea buffer for approximately 24 h at 25 °C in the presence and absence of CQDs (400 μg mL−1). CQDs were added in order to determine their role in the unfolding of the protein. The final concentration of the protein was maintained at 5 μM and the concentration was varied at 0–6 M. The incubated samples were then subjected to fluorescence spectroscopy for evaluation of the intrinsic fluorescence of the protein. The excitation wavelength was fixed at 290 nm and the emission range was fixed between 300–400 with a slit width of 5 nm. The resulting data were plotted as the protein fraction unfolded against the concentration of Gu-HCl in the presence and absence of CQDs for both the pH.393. Results and discussion
3.1. Synthesis and characterization of CQDs
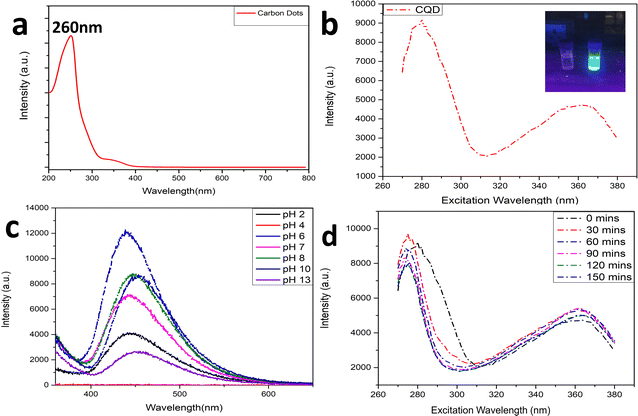
The carbonization process can be visualized from the colour of the solution changing from yellow to brownish solid residues. These residues were then mixed well in 15 mL ultrapure water and the suspension was centrifuged at 5000 rpm for 15 min. The reaction was carried out by firstly condensing the BAA and FA, after which they were carbonized to form CQDs. Fig. 1a shows the UV-Vis absorption spectra of the prepared CQDs. The as-prepared CQD solution showed an absorption peak at 260 nm. The peak at 260 nm might be attributed to the π–π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds, and the n–π* transition of C
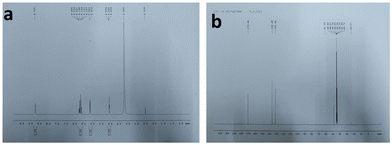
C bonds, and the n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds.40Fig. 1b shows the PL spectra of the as-prepared CQDs. We have provided a profile showing the change in the PL intensities at 448 nm emission wavelength when the aqueous CQDs were excited at different wavelengths of light ranging from 270 nm to 380 nm. It is very clear from the figure that the aqueous solution of the as-prepared CQDs shows excitation dependant emission properties. The CQDs were excited from 270 nm to 380 nm and the strongest emission band was observed at 280 nm excitation, which gradually decreased as the excitation was increased up to 310 nm, after which, the emission intensity gradually increased to 360 nm and again decreased. The exact reason for this behaviour is still unknown. The quantum yield of the sample was found to be very high with a value of 42.8% when referenced with quinine sulphate. The emission was observed at 450 nm, indicating blue light emission. Thus, the CQDs are having excitation and emission values of 280 nm and 450 nm, respectively. This data can be visualized by observing the CQDs and water under a UV illuminator as the CQDs display a strong blue light while the ultra-pure water taken as control did not emit any light. We can also observe the peak shift towards the UV region as we increased the excitation wavelength, which might be due to the surface state/size distribution of the prepared CQDs. The fluorescence stabilities of the CQDs under the effects of different pH were evaluated using the Horiba Duetta spectrophotometer, which is shown in Fig. 1c. Due to the lack of a proper lattice structure, the emission of the CQDs is affected by extreme conditions of pH. As we can infer from Fig. 1c, the best emission was observed when the CQDs were dissolved at pH 6, but the emission hugely decreased at pH 4, and the reason for the same is still unknown. We also performed a photobleaching study. The as-prepared aqueous CQDs were subjected to UV radiation of 302 nm wavelength for 2.5 h and the samples were collected at an interval of 30 min to evaluate the decrease in PL intensity over the range of excitation wavelength. Fig. 1d shows that there is very less photobleaching for almost 2.5 h when excited over a range of 270–380 nm and the emission was measured at 448 nm under environmental room temperature (45° during the day of the experiment), which sheds light on the photostability and the shelf life of the as-prepared CQDs. Fig. 2a shows the typical XRD pattern of the CQD structure with a broad peak located at 24.5°, which suggests the amorphous nature of the CQDs, or a highly disordered structure, which may be due to closely packed carbon atoms around the alkyl chain. Further, Fourier transform infrared spectroscopy was performed to gather information on their surface state. CQDs show many common markers in the transmittance spectra which were taken from 500–4000 cm−1. In Fig. 2b, the broad peaks at 3200–3500 cm−1 are ascribed to the vibrations of O–H/N–H. The carboxyl groups tend to appear at a higher frequency at 1500–1600 cm−1, these carboxylic groups are evidently from the precursors used. The C–H stretching peaks were observed near 2900–3000 cm−1. The dynamic light scattering studies were performed to quantize the size of the CQDs. From Fig. 2c, it can be confirmed that the size of the particle is 7 nm, while some of the particles are coagulated together. The zeta potential of the CQDs was obtained using the nanoparticle size and zeta analyser (Malvern, NanoZS90). Fig. 2d shows a single negative peak at 19.7 mV with a peak width of 5.3 mV. As the zeta potential provides the surface charge of the particle, the presence of a negatively charged moiety on the surface is evident from the figure. The presence of a negatively charged moiety is the reason for the high solubility of CQDs in water and higher shelf life.41,42 A higher negative charge is indicative of higher negative charged moieties on the surface of the CQD, which enables them to easily disperse in the liquid solution for example water, thus preventing aggregation and enabling the retention of the properties for a long term, which in turn increases the shelf life. This in turn is probably the reason for being able to store and use the CQDs at room temperature for around 10 months without observing any signs of turbidity. Raman Spectroscopy is often used to determine the morphological features of carbon-based nanomaterials such as graphene quantum dots, and carbon nanotubes.43 The Raman spectrum of the lyophilized CQDs is shown in Fig. 2e. The characteristic Raman scattering peaks of amide III, alkyl/vinyl/aromatic, and amide I were clearly observed at 1594 and 1446 cm−1, respectively, which corresponds to the reported values from literature, D and G bands were observed at 1249 and 1634 cm−1, the ratio of IG/ID was found to be 1.27.44,45 The IG/ID ratio indicated the amorphous nature of the carbon dots and the ratio of the sp3/sp2 carbon atoms implying the defects in the prepared CQDs. The G band corresponds to the E2g mode of the vibration of the sp2 bonded carbon atom while the smaller D band corresponds to the presence of the sp3 hybridized carbon and some percentage of oxygen in the prepared sample.46 The shift in the peak is observed due to the change in the bond lengths of the carbon atoms. The shortening of the bond lengths causes the Raman peaks to shift towards higher frequency or wavenumber and vice-versa. Another factor that can cause the shifts is the application of stress to the material.47 The morphology of the CQDs was characterized using transmission electron microscopy (TEM) and atomic force microscopy (AFM). The AFM image (Fig. 3a) illustrates that the CQDs formed are of very small size and are well dispersed. Fig. 3b depicts a 3D sectional image of the CQDs obtained from AFM, whose average height was found to be 3–4 nm. Statistically, the average size of the CQD particles was found to be within the range of 1–20 nm. The spherical morphology of the CQDs was observed in the AFM images and was confirmed from TEM images (Fig. 3c) TEM was performed to observe the CQDs, which supports the well-dispersed and the spherical nature of the CQDs, while the high-resolution TEM (HRTEM) image (Fig. 3d) of the CQDs at 10 nm magnification shows the formation of amorphous CQDs with spherical geometry, without any distinctive lattice structure. NMR is an effective tool for distinguishing sp3 and sp2 hybridized carbon atoms. Fig. 4a and b represent the 1H NMR spectra of the CQDs and 13C NMR spectra of the CQDs, respectively. These figures confirm the existence of the sp2 and sp3 hybridized carbon atoms as well as the presence of a carboxylic group and an amide group was also confirmed. In the 13C NMR spectrum, we observed the signals in the range of 23–60 ppm corresponding to the aliphatic carbon (sp3 hybridized) while the signals from 130–170 ppm correspond to the sp2 hybridized carbon atom. The signal at 170 ppm confirms the presence of the carboxyl/amide group in the prepared CQDs.11,481H NMR is used to determine the number of hydrogen bonds present in the compound. 1H NMR spectrum of the CQDs revealed the shift in the proton peaks at signal ranging from 4.55 to 2.5 suggesting the presence of aliphatic protons. Certain low integral peaks were observed between 7 and 5 ppm corresponding to a vinyl proton, and at 8.73 suggesting the presence of an amide group in the CQDs. Thus, it can be estimated that more aliphatic carbons (sp2 hybridized) are present than sp3 hybridized.45 The cytotoxic effect of the CQDs on the mouse fibroblast is shown in Fig. 2f, where we can see that 88% of cells are viable. The cell viability was obtained by the MTT assay.
O bonds.40Fig. 1b shows the PL spectra of the as-prepared CQDs. We have provided a profile showing the change in the PL intensities at 448 nm emission wavelength when the aqueous CQDs were excited at different wavelengths of light ranging from 270 nm to 380 nm. It is very clear from the figure that the aqueous solution of the as-prepared CQDs shows excitation dependant emission properties. The CQDs were excited from 270 nm to 380 nm and the strongest emission band was observed at 280 nm excitation, which gradually decreased as the excitation was increased up to 310 nm, after which, the emission intensity gradually increased to 360 nm and again decreased. The exact reason for this behaviour is still unknown. The quantum yield of the sample was found to be very high with a value of 42.8% when referenced with quinine sulphate. The emission was observed at 450 nm, indicating blue light emission. Thus, the CQDs are having excitation and emission values of 280 nm and 450 nm, respectively. This data can be visualized by observing the CQDs and water under a UV illuminator as the CQDs display a strong blue light while the ultra-pure water taken as control did not emit any light. We can also observe the peak shift towards the UV region as we increased the excitation wavelength, which might be due to the surface state/size distribution of the prepared CQDs. The fluorescence stabilities of the CQDs under the effects of different pH were evaluated using the Horiba Duetta spectrophotometer, which is shown in Fig. 1c. Due to the lack of a proper lattice structure, the emission of the CQDs is affected by extreme conditions of pH. As we can infer from Fig. 1c, the best emission was observed when the CQDs were dissolved at pH 6, but the emission hugely decreased at pH 4, and the reason for the same is still unknown. We also performed a photobleaching study. The as-prepared aqueous CQDs were subjected to UV radiation of 302 nm wavelength for 2.5 h and the samples were collected at an interval of 30 min to evaluate the decrease in PL intensity over the range of excitation wavelength. Fig. 1d shows that there is very less photobleaching for almost 2.5 h when excited over a range of 270–380 nm and the emission was measured at 448 nm under environmental room temperature (45° during the day of the experiment), which sheds light on the photostability and the shelf life of the as-prepared CQDs. Fig. 2a shows the typical XRD pattern of the CQD structure with a broad peak located at 24.5°, which suggests the amorphous nature of the CQDs, or a highly disordered structure, which may be due to closely packed carbon atoms around the alkyl chain. Further, Fourier transform infrared spectroscopy was performed to gather information on their surface state. CQDs show many common markers in the transmittance spectra which were taken from 500–4000 cm−1. In Fig. 2b, the broad peaks at 3200–3500 cm−1 are ascribed to the vibrations of O–H/N–H. The carboxyl groups tend to appear at a higher frequency at 1500–1600 cm−1, these carboxylic groups are evidently from the precursors used. The C–H stretching peaks were observed near 2900–3000 cm−1. The dynamic light scattering studies were performed to quantize the size of the CQDs. From Fig. 2c, it can be confirmed that the size of the particle is 7 nm, while some of the particles are coagulated together. The zeta potential of the CQDs was obtained using the nanoparticle size and zeta analyser (Malvern, NanoZS90). Fig. 2d shows a single negative peak at 19.7 mV with a peak width of 5.3 mV. As the zeta potential provides the surface charge of the particle, the presence of a negatively charged moiety on the surface is evident from the figure. The presence of a negatively charged moiety is the reason for the high solubility of CQDs in water and higher shelf life.41,42 A higher negative charge is indicative of higher negative charged moieties on the surface of the CQD, which enables them to easily disperse in the liquid solution for example water, thus preventing aggregation and enabling the retention of the properties for a long term, which in turn increases the shelf life. This in turn is probably the reason for being able to store and use the CQDs at room temperature for around 10 months without observing any signs of turbidity. Raman Spectroscopy is often used to determine the morphological features of carbon-based nanomaterials such as graphene quantum dots, and carbon nanotubes.43 The Raman spectrum of the lyophilized CQDs is shown in Fig. 2e. The characteristic Raman scattering peaks of amide III, alkyl/vinyl/aromatic, and amide I were clearly observed at 1594 and 1446 cm−1, respectively, which corresponds to the reported values from literature, D and G bands were observed at 1249 and 1634 cm−1, the ratio of IG/ID was found to be 1.27.44,45 The IG/ID ratio indicated the amorphous nature of the carbon dots and the ratio of the sp3/sp2 carbon atoms implying the defects in the prepared CQDs. The G band corresponds to the E2g mode of the vibration of the sp2 bonded carbon atom while the smaller D band corresponds to the presence of the sp3 hybridized carbon and some percentage of oxygen in the prepared sample.46 The shift in the peak is observed due to the change in the bond lengths of the carbon atoms. The shortening of the bond lengths causes the Raman peaks to shift towards higher frequency or wavenumber and vice-versa. Another factor that can cause the shifts is the application of stress to the material.47 The morphology of the CQDs was characterized using transmission electron microscopy (TEM) and atomic force microscopy (AFM). The AFM image (Fig. 3a) illustrates that the CQDs formed are of very small size and are well dispersed. Fig. 3b depicts a 3D sectional image of the CQDs obtained from AFM, whose average height was found to be 3–4 nm. Statistically, the average size of the CQD particles was found to be within the range of 1–20 nm. The spherical morphology of the CQDs was observed in the AFM images and was confirmed from TEM images (Fig. 3c) TEM was performed to observe the CQDs, which supports the well-dispersed and the spherical nature of the CQDs, while the high-resolution TEM (HRTEM) image (Fig. 3d) of the CQDs at 10 nm magnification shows the formation of amorphous CQDs with spherical geometry, without any distinctive lattice structure. NMR is an effective tool for distinguishing sp3 and sp2 hybridized carbon atoms. Fig. 4a and b represent the 1H NMR spectra of the CQDs and 13C NMR spectra of the CQDs, respectively. These figures confirm the existence of the sp2 and sp3 hybridized carbon atoms as well as the presence of a carboxylic group and an amide group was also confirmed. In the 13C NMR spectrum, we observed the signals in the range of 23–60 ppm corresponding to the aliphatic carbon (sp3 hybridized) while the signals from 130–170 ppm correspond to the sp2 hybridized carbon atom. The signal at 170 ppm confirms the presence of the carboxyl/amide group in the prepared CQDs.11,481H NMR is used to determine the number of hydrogen bonds present in the compound. 1H NMR spectrum of the CQDs revealed the shift in the proton peaks at signal ranging from 4.55 to 2.5 suggesting the presence of aliphatic protons. Certain low integral peaks were observed between 7 and 5 ppm corresponding to a vinyl proton, and at 8.73 suggesting the presence of an amide group in the CQDs. Thus, it can be estimated that more aliphatic carbons (sp2 hybridized) are present than sp3 hybridized.45 The cytotoxic effect of the CQDs on the mouse fibroblast is shown in Fig. 2f, where we can see that 88% of cells are viable. The cell viability was obtained by the MTT assay.
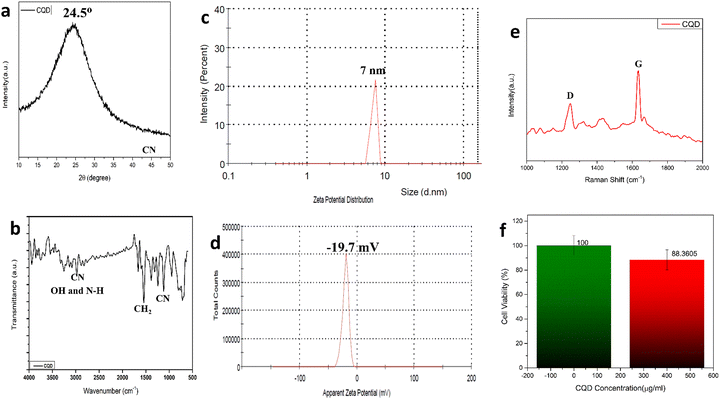 | ||
| Fig. 2 (a)XRD pattern for CQDs; (b) FTIR spectra; (c) dynamic light scattering spectra; (d) zeta potential plot; (e) Raman spectra. | ||
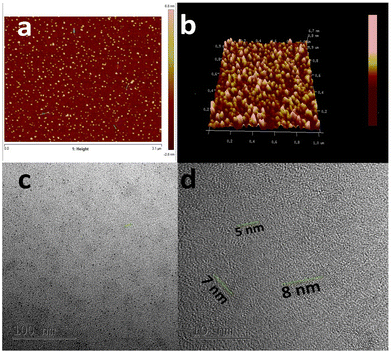 | ||
| Fig. 3 (a) 2D sectional AFM image of CQDs; (b) 3D sectional image of CQDs; (c) TEM image of CQDs at 100 nm; (d) TEM image of CQDs at 10 nm. | ||
3.2. HEWL amyloid fibril size, morphology, and conformation
The HEWL fibrillation process was carried out at different pH to study the inhibitory effect of CQDs in different pHs. The HEWL formation was carried out at an acidic pH (2.7) using Gazova et al.'s method and at a neutral pH (7); it was carried out following the method used by E. Kiran Kumar et al. Dynamic light scattering study of HEWL amyloid formation at pH 7 provided information about the size of the HEWL fibrils in the presence and absence of the CQDs. Fig. 5a–c provide information on the size of the fibrils. Fig. 5a shows the amyloid monomer, whereas a single full-intensity peak at 650 nm was observed, indicating the presence of an amyloid monomer. While, Fig. 5b and c indicate the HEWL amyloid formation in the presence and absence of CQDs, respectively. The process of inhibition can be confirmed by multiple small-sized peaks in Fig. 5c, whereas a large single-intensity peak was observed in the case of HEWL formed without CQDs (Fig. 5b). Thus, we can infer from the figure, that the formed HEWL amyloid fibrils in the absence of CQDs have a larger size indicating the formation of the fibrillar structure while in the presence of CQDs, the fibrils are broken in smaller sizes indicating the inhibitory effect of the CQDs, which is further confirmed in later sections. Far-UV CD spectra and FTIR data were collected to gather an idea about structural changes in protein for the HEWL fibrillation process in the presence and absence of CQDs.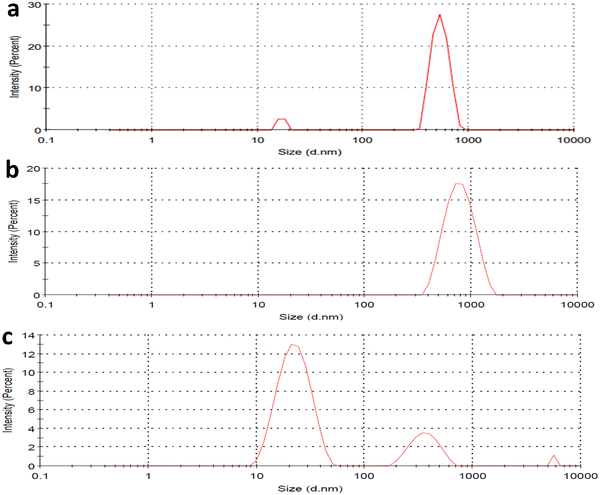 | ||
| Fig. 5 (a) DLS of HEWL monomer; (b) DLS of HEWL amyloid fibril in absence of CQDs; (c) DLS of HEWL amyloid fibril in presence of CQDs; (d) FTIR of amyloid inhibition process. | ||
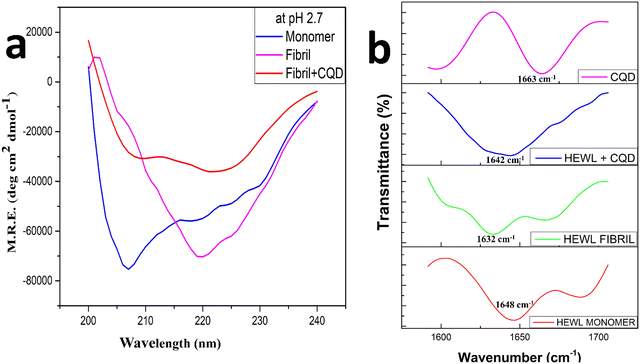
Fig. 6a shows the far UV circular dichroism spectra of the HEWL fibrillation process at pH 2.7, where the analysis of the data shown in Table 1 was performed using the Dichroweb online server.49 There is a peak shift from 209 nm to 219 nm that can be observeed in Fig. 6a as the HEWL amyloid fibrillation proceeds, this can be attributed to the formation of the β-sheets structures in the mature fibril and reduction in the percentage of the α-helix structure, which were more prominent in the monomer stage.50 While in the presence of CQDs, low-intensity peaks can be observed at 210 nm and 223 nm, which are attributed to unrecorded structures or random coiled structures. This observation might be indicative of the theory that the α-helix conformations get reduced in the final solution as well as the final solution does not significantly contain β-sheets while it contains more percentage of random coils, which in turn is indicative of inhibition. These observations correspond to the predicted secondary structural percentage of HEWL in the presence and absence of CQDs.
| α helix (%) | B-sheets (%) | Random coils (%) | |
|---|---|---|---|
| Monomer | 31 ± 1.24 | 12 ± 0.36 | 57 ± 1.9 |
| Fibril | 4 ± 0.18 | 48 ± 1.7 | 48 ± 1.5 |
| Fibril + CQD | 19 ± 0.6 | 25 ± 0.6 | 56 ± 1.5 |
Among other techniques, FTIR (Fourier transform infrared spectra) has been used to characterize amyloid fibrils. Earlier results have shown that FTIR shows an anti-parallel β-sheet structure characterized by two bands at the amide 1 frequency (1600–1700 cm−1), a low-frequency high-intensity band at 1620–1630 cm−1 and a high-frequency low-intensity band at 1685–1695 cm−1.51,52 Later, comparing globular proteins to amyloid fibrils, it was concluded that the amyloid fibrils show their spectral signature clustering around 1630 cm−1 while for the native structure the spectral signature clusters around 1643 cm−1.53 These changes are attributed to the inherent β-sheet properties such as assembling in longer β-sheet conformation or a more planar one or longer sheets.54–57 Thus, in Fig. 6b, we have studied the amide 1 band from 1600–1700 cm−1 for the amyloid fibrillation process both in the presence and the absence of the CQDs. We observed that the peak for the HEWL monomer was at 1648 cm−1, while the peak for the HEWL fibrils in the absence of CQDs was observed at 1632 cm−1, and some random coils were found at 1680 cm−1. In the presence of CQDs, the peaks observed were at 1644 cm−1, which indicates the retainment of the native structure, thus showing an inhibitory effect. The peak for CQDs alone showed a high-intensity band at 1663 cm−1, which is due to the presence of an amide group on the surface of the CQDs.
3.3. HEWL amyloid inhibition and disintegration study
The comparative inhibitory effect of the CQDs on the formation of HEWL amyloid fibrils at different pH was observed using the ThT fluorescence assay and ANS binding assay. To innumerate the amyloid inhibitory effect of CQDs at different pH, two different protocols were followed for the two pH values. The assays were performed both at an acidic (pH 2.7) and a neutral (pH 7) pH. Firstly, for the acidic medium, the glycine-HCl buffer was freshly prepared and pH was adjusted to 2.7 as it is known to promote protein unfolding.58 As mentioned earlier, the method used by Gazova et al. was employed for the HEWL aggregation process. Different concentrations of CQDs (50, 300, 400, and 500 μg mL−1) were added along with the HEWL protein to study amyloid inhibition. The samples were collected at 20 min interval time. The final concentration of the protein was kept at 5 μM and 20 μM and ThT dye was added before incubating for 45 min. Fig. 7a shows the ThT fluorescence assay for the inhibitory effect of CQDs in an acidic medium. As we can infer from the figure, CQDs have shown a concentration-dependent inhibitory effect. A 400 μg mL−1 of CQD concentration showed almost 60% inhibition and so this concentration of CQDs was considered for further studies.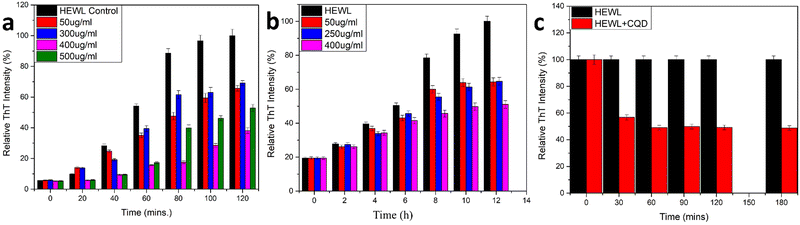 | ||
| Fig. 7 (a) Amyloid inhibition assay using CQDs as inhibitor (pH 2.7); (b) amyloid inhibition assay using CQDs as inhibitor(pH 7); (c) amyloid disintegration study. | ||
Secondly, for the neutral pH, a combination of urea buffer and the temperature was used for the fibril formation process.59 As mentioned earlier, we followed the method used by E. Kiran Kumar et al. with a little modification. Different concentrations of CQDs (50, 250, and 400 μg mL−1) were added to the HEWL solution. Aliquots were collected at regular 2 h intervals. The final concentration of the protein was kept at 5 μM and 20 μM, and ThT dye was added before incubating for 45 min. Fig. 7b shows the ThT fluorescence assay for the inhibitory effect of CQDs for amyloid fibrillation in neutral pH. It is evident from the figure that irrespective of the pH, the inhibitory effect of the CQDs is concentration dependant. Also, we can observe that in neutral pH, 400 μg mL−1 of CQDs has almost 50% of inhibition. So, we can infer that CQDs show a pH and concentration dependant inhibitory effect toward amyloid formation.
In addition to the above studies, we also performed the disintegration of HEWL fibril. In this study, we added 400 μg mL−1 of CQDs to the formed fibrils and incubated them for 180 min initially and periodically (30 min duration for the first 2 h and then 60 min duration for the last hour) and monitored the disintegration using the ThT fluorescence assay. The samples were incubated for 45 min each before taking the fluorescence spectra. Fig. 7c shows the disintegration study for the formed amyloid fibril after the addition of 400 μg mL−1 CQDs to the solution. A sharp decrease in the intensity after the first 30 min of incubation was observed, indicating 40% disintegration of HEWL. As we further incubated for another 30 min, we could see that the HEWL fibrils were already disintegrated more than 50%, which remained almost constant for the rest of the study until 180 min. The same study was performed after 24 h of incubation, which showed 60% disintegration of the HEWL fibrils. Thus, we can say that apart from the inhibitory effect at different pH, the fluorescent CQDs have also shown impressive disintegrative effects towards HEWL. To further study the effects of the addition of CQDs to the amyloid formation process, we added CQDs at different stages of fibrillation. Including, prefibrillar or early stage of aggregation (3 h) and matured fibrillar stage or a later stage of aggregation (7 h) and monitored the HEWL formation by ThT fluorescence assay. Fig. 8a, b, and c show the HEWL formation process as CQDs were added at 0 h, 3 h, and 7 h respectively. It is evident from these figures that as the fibrillation proceeds, the addition is less and less effective with an increase in time. Almost 80% of the fibril was formed when CQDs were added at 7 h while it was only 60% for 3 h addition, and 50% for 0 h addition. This indicates that during the later stages of fibrillation, the process is independent of the surrounding state. We can also infer that in the pre-fibrillar state, the process is quite susceptible to the presence or absence of an inhibitor.
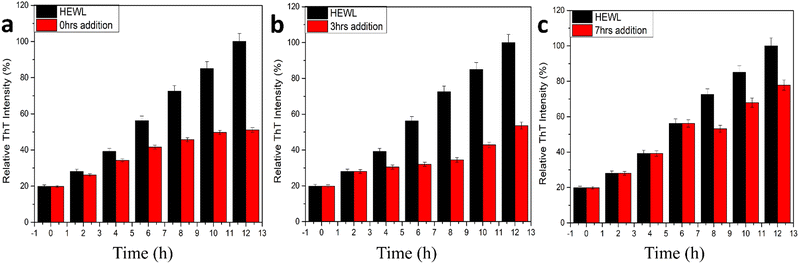 | ||
| Fig. 8 HEWL amyloid inhibition study using ThT fluorescence with CQDs added at (a) 0 h, (b) 3 h, (c) 7 h. | ||
As shown in Fig. 9, an ANS assay was performed to monitor the conformational changes in HEWL in terms of surface hydrophobicity. In the absence of CQDs, an increase in ANS assay was observed followed by a slight decrease. This is in agreement with previous reports, which state that surface hydrophobicity is highest in prefibrillar stages. However, in the presence of the CQDs, a considerable reduction in ANS intensity was observed. This states that the CQDs are interacting with HEWL and altering their conformation, thereby reducing the surface hydrophobicity. Further, we also performed an intrinsic tryptophan fluorescence study for the amyloid formation process in the presence and absence of CQDs at pH 2.7. Fig. 10a–c show the changes in the intrinsic fluorescence over the period of HEWL amyloid formation. Fig. 10a shows the intrinsic fluorescence when the HEWL monomer solution was excited at 280 nm in the presence and absence of CQDs and the emission was recorded at 336 nm, which corresponds to the tryptophan fluorescence. As the fibrillation proceeds similar intrinsic fluorescence was obtained, in which Fig. 10b and c show a gradual increase in the tryptophan fluorescence intensity of the HEWL in the absence of CQDs. Fig. 10b can be identified as the fibrillation phase, while the matured fibril formation can be observed in Fig. 10c in the absence of CQDs. The increase in the tryptophan fluorescence is indicative of the unfolding of the native protein forming matured fibrils within 2 h in the absence of CQDs as the tryptophan residues are on the outside of the fibril structure, thus affecting the fluorescence. However, in the presence of the CQDs, completely different tryptophan emission kinetics were observed, wherein a decrease in emission intensity was observed, followed by a slight increase in λmax value. This suggests that the CQDs on interacting with HEWL, are altering their conformation in such as way that the trp residues are exposed to a more polar environment.
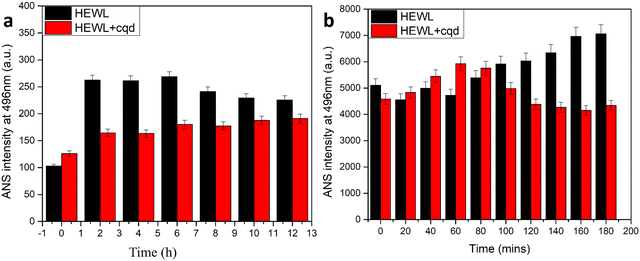 | ||
| Fig. 9 (a) ANS binding assay of amyloid fibrillation at 7 pH; (b) ANS binding of amyloid fibrillation at 2.7. | ||
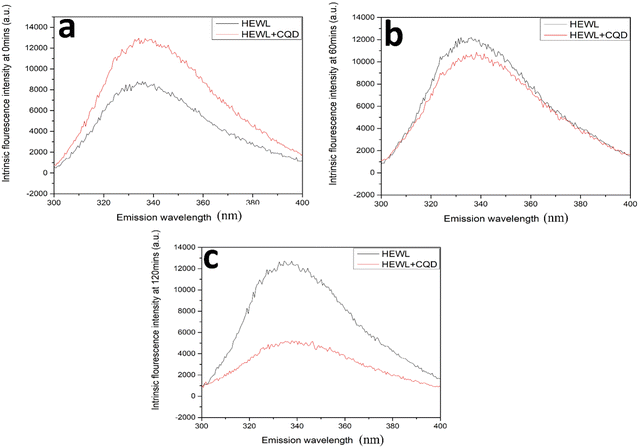 | ||
| Fig. 10 HEWL and CQD interaction study on the basis of intrinsic tryptophan fluorescence measurement in the presence and absence of CQDs at (a) 0 min; (b) 60 min; (c) 120 min. | ||
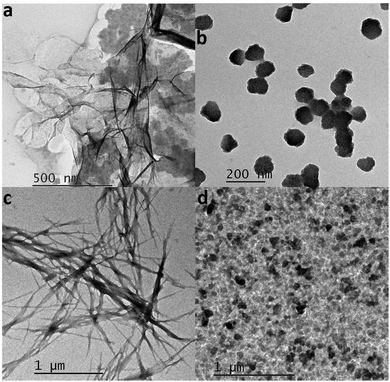
To better visualize the products of the fibrillation process TEM images of the formed fibrils in the presence and absence of CQDs were procured. Fig. 11a and b show the formed fibrils in the absence and presence of CQDs at pH 7, respectively. As we can see in Fig. 11a that the HEWL fibrils have formed as they are characterized by a long-branched structure, while in Fig. 11b, HEWL fibrils are broken into smaller pieces in the presence of CQDs, which confirmed the results of the DLS studies. Fig. 11c and d show the HEWL fibrillation process in the absence and presence of CQDs, respectively, at pH 2.7. Fig. 11c shows the HEWL amyloid in the absence of CQDs while Fig. 10d shows the fibrils in the presence of CQDs, characterized by broken fibrils. The broken fibrils that we can see when the amyloid fibrillation process is carried out in the presence of CQDs, indicate the inhibitory effect of the CQDs toward HEWL formation.
3.4. Equilibrium unfolding study
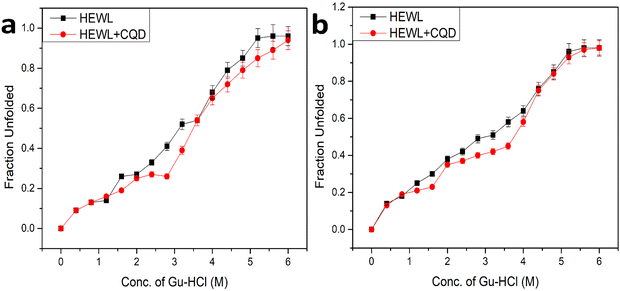
To understand the role of CQDs in amyloid inhibition, we performed an equilibrium unfolding study using guanidine hydrochloride (Gu-HCl) based amyloid (HEWL) inhibition in the presence and absence of CQDs at both the aforementioned pH (pH 2.7 and pH 7). The resulting data were plotted as the fraction unfolded vs. conc. Of Gu-HCl, which is shown in Fig. 12. Fig. 12(a) shows the plot of pH 2.7 where we can observe that in the presence of CQDs, the transition midpoint of unfolding has shown a red shift, which indicates stabilizing the HEWL.36 Similarly, in Fig. 12(b), the data for amyloid inhibition at pH 7 are shown where we can make the same observation that the transition midpoint of the unfolding has a red shift, which essentially indicates the stabilization of the protein and prevents it from aggregation. Further, from the nature of the plot of Fig. 12, we may also infer that the CQDs might be forming some complex with the intermediate state during the unfolding process thus ultimately arresting the entire aggregation process and successfully inhibiting the amyloid formation.4. Conclusion
Summarizing this work, we devised a one-pot facile method for the synthesis of highly fluorescent CQDs using bis acryl amide and folic acid as precursors and a microwave-assisted synthesis for preparing CQDs of an average size of 7 nm. The method is very quick and economical compared to the present methods in use. The prepared CQDs displayed high quantum yield and high fluorescence intensity along with a high inhibitory effect toward HEWL amyloid aggregation. We have also learned that the emission of the prepared CQDs depends on the excitation wavelength and pH, highest at 280 nm and pH 6, respectively. Further, the CQDs show pH as well as concentration dependant inhibitory effects. The emission profile at 448 nm as a function of the excitation wavelength was displayed, which showed the max emission at 280 nm. The photobleaching study has shown that the prepared CQDs can withstand prolonged UV exposure. These CQDs have a high negative surface charge (−19.7 mV), indicating the presence of negatively-charged moieties on the surface. These moieties on the surface make the CQDs highly water soluble and can be stored at room temperature for the long term and used without any turbidity and loss of property (appx. 10 months). Among others, 400 μg mL−1 of concentration of CQDs shows the highest (60%) inhibitory effect. It has also been shown that the CQDs in the above concentration can disintegrate the formed HEWL fibril by 50% within an hour of addition and it can go up to 60% upon incubating the same for 24 h. Also, the role of CQDs in fibril formation or the lack of it has been shown, which indicates that the fibrillation process is dependent on the surroundings (presence of inhibitor/promoter) while they are in the pre-fibrillar state and as the fibrillation progresses the dependence on the surrounding decreases, which in turn sheds light on the high stability of the HEWL amyloid fibrils. From the plots of the equilibrium unfolding studies using Gu-HCl-based amyloid inhibition in the presence and absence of CQDs, we can also conclude that the CQDs seem to form a complex with HEWL during the transition phase of the unfolding process, which is evident from the red shift observed in the transition midpoints at both pHs, thus, ultimately arresting the aggregation process and inhibiting the amyloid formation. Therefore, these CQDs can be used as a promising diagnostic and therapeutic tool for the early detection and cure of protein-linked degenerative diseases in the future.Conflicts of interest
The authors declare the following financial interests/personal relationships, which may be considered as potential competing interests: Dr Nandini Sarkar reports the financial support was provided by the National Institute of Technology Rourkela. Dr Nandini Sarkar reports a relationship with the National Institute of Technology Rourkela that includes employment.Acknowledgements
The authors acknowledge the funding provided to NS by the Science and Engineering Research Board (SERB), Govt. of India (GrantNumber ECR/2017/002081). The infrastructural facilities provided by NIT Rourkela are also acknowledged.References
- M. Fang, C. W. Peng, D. W. Pang and Y. Li, Cancer Biol. Med., 2012, 9, 151–163 CAS.
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- X. Gao, C. Du, Z. Zhuang and W. Chen, J. Mater. Chem. C, 2016, 4, 6927–6945 RSC.
- Z. Peng, E. H. Miyanji, Y. Zhou, J. Pardo, S. D. Hettiarachchi, S. Li, P. L. Blackwelder, I. Skromne and R. M. Leblanc, Nanoscale, 2017, 9, 17533–17543 RSC.
- J. T. Margraf, V. Strauss, D. M. Guldi and T. Clark, J. Phys. Chem. B, 2015, 119, 7258–7265 CrossRef CAS PubMed.
- A. P. Demchenko and M. O. Dekaliuk, Methods Appl. Fluoresc., 2013, 1, 042001 CrossRef CAS PubMed.
- S. N. Baker and G. A. Baker, Angew. Chem., Int. Ed., 2010, 49, 6726–6744 CrossRef CAS PubMed.
- S.-T. Yang, L. Cao, P. G. Luo, F. Lu, X. Wang, H. Wang, M. J. Meziani, Y. Liu, G. Qi and Y.-P. Sun, J. Am. Chem. Soc., 2009, 131, 11308–11309 CrossRef CAS PubMed.
- B. Yu, H. C. Tai, W. Xue, L. J. Lee and R. J. Lee, Mol. Membr. Biol., 2010, 27, 286–298 CrossRef CAS PubMed.
- H. Liu, T. Ye and C. Mao, Angew. Chem., 2007, 119, 6593–6595 CrossRef.
- S. Zhu, Q. Meng, L. Wang, J. Zhang, Y. Song, H. Jin, K. Zhang, H. Sun, H. Wang and B. Yang, Angew. Chem., Int. Ed., 2013, 52, 3953–3957 CrossRef CAS PubMed.
- Z.-A. Qiao, Y. Wang, Y. Gao, H. Li, T. Dai, Y. Liu and Q. Huo, Chem. Commun., 2010, 46, 8812–8814 RSC.
- Z.-C. Yang, M. Wang, A. M. Yong, S. Y. Wong, X.-H. Zhang, H. Tan, A. Y. Chang, X. Li and J. Wang, Chem. Commun., 2011, 47, 11615–11617 RSC.
- P.-C. Hsu and H.-T. Chang, Chem. Commun., 2012, 48, 3984–3986 RSC.
- Y. Yang, J. Cui, M. Zheng, C. Hu, S. Tan, Y. Xiao, Q. Yang and Y. Liu, Chem. Commun., 2012, 48, 380–382 RSC.
- Z. Zhang, J. Hao, J. Zhang, B. Zhang and J. Tang, RSC Adv., 2012, 2, 8599–8601 RSC.
- J. Zhang, W. Shen, D. Pan, Z. Zhang, Y. Fang and M. Wu, New J. Chem., 2010, 34, 591–593 RSC.
- H. Li, F.-Q. Shao, S.-Y. Zou, Q.-J. Yang, H. Huang, J.-J. Feng and A.-J. Wang, Microchim. Acta, 2015, 183, 821–826 CrossRef.
- Q. Y. Cai, J. Li, J. Ge, L. Zhang, Y. L. Hu, Z. H. Li and L. B. Qu, Biosens. Bioelectron., 2015, 72, 31–36 CrossRef CAS PubMed.
- Y. Dong, H. Pang, H. B. Yang, C. Guo, J. Shao, Y. Chi, C. M. Li and T. Yu, Angew. Chem., Int. Ed., 2013, 52, 7800–7804 CrossRef CAS PubMed.
- F. Chiti and C. M. Dobson, Annu. Rev. Biochem., 2006, 75, 333–366 CrossRef CAS PubMed.
- M. Bartolini and V. Andrisano, ChemBioChem, 2010, 11, 1018–1035 CrossRef CAS PubMed.
- M. Bucciantini, E. Giannoni, F. Chiti, F. Baroni, L. Formigli, J. Zurdo, N. Taddei, G. Ramponi, C. M. Dobson and M. Stefani, Nature, 2002, 416, 507–511 CrossRef CAS PubMed.
- S. S. Wang, Y. T. Hung, W. S. Wen, K. C. Lin and G. Y. Chen, Biochem. Biophys. Acta, 1811, 2011, 301–313 Search PubMed.
- K. F. DuBay, A. P. Pawar, F. Chiti, J. Zurdo, C. M. Dobson and M. Vendruscolo, J. Mol. Biol., 2004, 341, 1317–1326 CrossRef CAS PubMed.
- J. Goers, S. E. Permyakov, E. A. Permyakov, V. N. Uversky and A. L. Fink, Biochemistry, 2002, 41, 12546–12551 CrossRef CAS.
- M. R. Krebs, D. K. Wilkins, E. W. Chung, M. C. Pitkeathly, A. K. Chamberlain, J. Zurdo, C. V. Robinson and C. M. Dobson, J. Mol. Biol., 2000, 300, 541–549 CrossRef CAS PubMed.
- R. Swaminathan, V. K. Ravi, S. Kumar, M. V. Kumar and N. Chandra, Adv. Protein Chem. Struct. Biol., 2011, 84, 63–111 CAS.
- Y. Yonezawa, S. Tanaka, T. Kubota, K. Wakabayashi, K. Yutani and S. Fujiwara, J. Mol. Biol., 2002, 323, 237–251 CrossRef CAS PubMed.
- Z. Gazova, K. Siposova, E. Kurin, P. Mučaji and M. Nagy, Proteins, 2013, 81, 994–1004 CrossRef CAS PubMed.
- J. B. Wang, Y. M. Wang and C. M. Zeng, Biochem. Biophys. Res. Commun., 2011, 415, 675–679 CrossRef CAS PubMed.
- M. Masuda, N. Suzuki, S. Taniguchi, T. Oikawa, T. Nonaka, T. Iwatsubo, S. Hisanaga, M. Goedert and M. Hasegawa, Biochemistry, 2006, 45, 6085–6094 CrossRef CAS PubMed.
- M. Mahdavimehr, A. A. Meratan, M. Ghobeh, A. Ghasemi, A. A. Saboury and M. Nemat-Gorgani, PLoS One, 2017, 12, e0187841 CrossRef PubMed.
- M. Stefani and S. Rigacci, Int. J. Mol. Sci., 2013, 14, 12411–12457 CrossRef PubMed.
- Y. Porat, A. Abramowitz and E. Gazit, Chem. Biol. Drug Des., 2006, 67, 27–37 CrossRef CAS PubMed.
- M. P. T. Prabhu and N. Sarkar, Biophys. Chem., 2022, 280, 106714 CrossRef CAS PubMed.
- E. Damian Guerrero, A. M. Lopez-Velazquez, J. Ahlawat and M. Narayan, ACS Appl. Nano Mater., 2021, 4, 2423–2433 CrossRef PubMed.
- X. Wang, C. Xu, H. Song, X. Liu, X. Xie, X. Pang and Y. Zhou, J. Lumin., 2019, 210, 472–478 CrossRef CAS.
- C. Zhang, C. Gao, J. Mu, Z. Qiu and L. Li, BioMed Res. Int., 2013, 2013, 349542 Search PubMed.
- Y. Liu, Y. Zhao and Y. Zhang, Sens. Actuators, B, 2014, 196, 647–652 CrossRef CAS.
- G. Chen, S. Wu, L. Hui, Y. Zhao, J. Ye, Z. Tan, W. Zeng, Z. Tao, L. Yang and Y. Zhu, Sci. Rep., 2016, 6, 19028 CrossRef CAS PubMed.
- A. Dager, T. Uchida, T. Maekawa and M. Tachibana, Sci. Rep., 2019, 9, 14004 CrossRef PubMed.
- M. K. Barman, B. Jana, S. Bhattacharyya and A. Patra, J. Phys. Chem. C, 2014, 118, 20034–20041 CrossRef CAS.
- Z. Ni, Y. Wang, T. Yu and Z. Shen, Nano Res., 2008, 1, 273–291 CrossRef CAS.
- Y. Choi, N. Thongsai, A. Chae, S. Jo, E. B. Kang, P. Paoprasert, S. Y. Park and I. In, J. Ind. Eng. Chem., 2017, 47, 329–335 CrossRef CAS.
- I.-A. Baragau, N. P. Power, D. J. Morgan, T. Heil, R. A. Lobo, C. S. Roberts, M.-M. Titirici, S. Dunn and S. Kellici, J. Mater. Chem. A, 2020, 8, 3270–3279 RSC.
- M. Park, J. S. Choi, L. Yang and H. Lee, Sci. Rep., 2019, 9, 19826 CrossRef CAS PubMed.
- L. Tian, D. Ghosh, W. Chen, S. Pradhan, X. Chang and S. Chen, Chem. Mater., 2009, 21, 2803–2809 CrossRef CAS.
- L. Whitmore and B. A. Wallace, Biopolymers, 2008, 89(5), 392–400 CrossRef CAS PubMed.
- F. Mohammadi, A. Mahmudian, M. Moeeni and L. Hassani, RSC Adv., 2016, 6, 23148–23160 RSC.
- J. Termine, E. Eanes, D. Ein and G. Glenner, Biopolymers, 1972, 11, 1103–1113 CrossRef CAS PubMed.
- G. Glenner, E. Eanes, H. Bladen, R. Linke and J. Termine, J. Histochem. Cytochem., 1974, 22, 1141–1158 CrossRef CAS PubMed.
- G. Zandomeneghi, M. R. H. Krebs, M. G. McCammon and M. Fändrich, Protein Sci., 2004, 13, 3314–3321 CrossRef CAS PubMed.
- J. Kubelka and T. A. Keiderling, J. Am. Chem. Soc., 2001, 123, 12048–12058 CrossRef CAS PubMed.
- J. S. Richardson, Adv. Protein Chem., 1981, 34, 167–339 CrossRef CAS PubMed.
- R. Sarroukh, E. Goormaghtigh, J.-M. Ruysschaert and V. Raussens, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 2328–2338 CrossRef CAS PubMed.
- S. Maity, R. Kumar, S. K. Maity, P. Jana, S. Bera and D. Haldar, MedChemComm, 2013, 4, 530–536 RSC.
- S. Brudar and B. Hribar-Lee, Biomolecules, 2019, 9(2), 65 CrossRef PubMed.
- E. Kiran Kumar and N. P. Prabhu, Phys. Chem. Chem. Phys., 2014, 16(43), 24076–24088 RSC.
| This journal is © The Royal Society of Chemistry 2023 |