Open Access Article
Open Access ArticleExploiting the UV excited size-dependent emission of PDMS-coated CdTe quantum dots for in vitro simultaneous multicolor imaging of HepG2 cellular organelles†
Sulaxna
Pandey
ab and
Dhananjay
Bodas
 *ab
*ab
aNanobioscience Group, Agharkar Research Institute, GG Agarkar Road, Pune 411 004, India. E-mail: dsbodas@aripune.org; Tel: +9120 2532 5127
bSavitribai Phule Pune University, Ganeshkhind Road, Pune 411 007, India
First published on 15th March 2023
Abstract
Multicolor bioimaging can be referred to as the imaging method that non-invasively visualizes biological processes using fluorophores. Over the years, this technique has been primarily used in the areas of diagnostic and theranostic applications such as the detection of pathogens, identification of disease-specific markers, and cancer detection. Fluorophores such as organic dyes are widely used in bioimaging studies. However, organic dyes exhibit a major limitation of excitation and emission spectral overlap, especially when multispectral bioimaging is considered. Quantum dots (QDs), in contrast, hold great potential due to their properties such as size-tunable narrow emission, single excitation in UV, photo and chemical stability, high fluorescence lifetime, simple surface modification process, etc. Thus, QDs can be a good alternative to traditional fluorophores for bioimaging applications. Herein, the previously reported poly dimethyl siloxane (PDMS)-coated CdTe QDs (PQDs) are used to simultaneously image cellular organelles such as lysosomes, mitochondria, nucleus, and actin of HepG2. Briefly, blue, green, yellow, and orange PQDs are conjugated to CD68 ab. (Lysosomes), mitochondria ab., nuclear antigen ab. and smooth muscle actin ab. respectively using EDC-NHS chemistry. The intracellular organelle targeting of the conjugated QDs is assessed by colocalization with the commercially available dyes. Finally, PQD-conjugates are used to simultaneously image four cellular organelles of HepG2 at an excitation wavelength of 405 nm. The present study demonstrates the potential of PQDs as a fluorophore in simultaneous multicolor bioimaging.
Introduction
Multicolor bioimaging utilizes several fluorophores with different emission wavelengths to image various moieties of a given sample.1–4 Fluorophores such as organic stains or dyes exhibit an excitation and emission pair for the visualization of biological moieties. The major limitation of organic dyes when used in multicolor imaging is their spectral overlap. Many researchers are working to resolve the spectral overlap of organic dyes for precise multicolor imaging applications.5–8 Bandi et al.7 reported multiplexed two-color and three-color in vivo imaging using per sulfonated indocyanine dyes with absorbance maxima at 872 and 1072![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) nm through catechol-ring and aryl-ring fusion, respectively, onto the nonamthine scaffold. Sharma et al.9 have demonstrated the multicolor imaging of different cancer cell lines using N@FCM and N-P@FCM. Furthermore, Koyama et al.10 have optically diagnosed tumors using three fluorescently labeled antibodies using multiple excitation/emission filters. Wei et al.11 imaged neuronal co-cultures and brain tissues by multiple fluorophores using Raman scattering under the electronic pre-resonance condition. They resolved 24 colors of DNA heterogeneities and protein metabolism.
nm through catechol-ring and aryl-ring fusion, respectively, onto the nonamthine scaffold. Sharma et al.9 have demonstrated the multicolor imaging of different cancer cell lines using N@FCM and N-P@FCM. Furthermore, Koyama et al.10 have optically diagnosed tumors using three fluorescently labeled antibodies using multiple excitation/emission filters. Wei et al.11 imaged neuronal co-cultures and brain tissues by multiple fluorophores using Raman scattering under the electronic pre-resonance condition. They resolved 24 colors of DNA heterogeneities and protein metabolism.
Reports suggest the usage of multiple dyes for bioimaging; however, the techniques are complex and thus deter mass applicability. Moreover, organic dyes exhibit broad excitation and emission spectra, surface non-modifiability, photobleaching, etc. which impedes their pertinence in multicolor bioimaging applications. Owing to the above-mentioned limitations, multicolor imaging using fluorophores is instrument intensive and requires aggressive post-processing. In contrast, quantum dots (QDs) are promising as their size is tunable with a narrow emission spectrum, photo- and chemical-stability, high fluorescence lifetime, and surface modifiability.12–17
Lately, researchers have used QDs for multicolor imaging. Bahari et al.17 demonstrated the multicolor imaging of MDA-MB231 living cancer cells using graphdiyne/graphene quantum dots. Campbell et al.18 reported the multicolor imaging of HeLa, MCF-7 and HEK-293 cells using graphene QDs. The imaging of three different colors of QDs was done at different excitation wavelengths (blue-450 nm), green −535 nm, and NIR −750 nm). Similarly, Chen et al.19 presented the multicolor imaging of HER2 and ER in breast cancer specimens using 605-QD-SA and 545-QD-SA probes. The imaging was done using two excitation wavelengths (blue – 450–480 nm and 545 nm). Furthermore, Panagiotopoulou et al.20 used glucuronic acid (GlcA) and N-acetylneuraminic acid (NANA) conjugated with green and red QDs, respectively, for imaging human keratinocytes. 5 commercially procured dyes were used by Kobayashi et al.21 for lymphatic multicolor imaging. While multicolor imaging of QDs has been widely reported, there is still a need to resolve the multiple emission peaks with a narrow emission wavelength. The simultaneous imaging of multiple targets using QDs at a single excitation wavelength can ease out the complex process of multicolor bioimaging. Multicolor bioimaging application requires high photostability, quantum efficiency, narrow emission spectra, and reduced toxicity of QDs. This would be possible by synthesizing QDs with properties like monodispersity, quantum efficiency, photostability, biocompatibility, and reproducibility.14
Here, the use of size tunable narrow emission wavelength, quantum efficient, photostable, biocompatible, and reproducible PQDs for simultaneous imaging at a single excitation wavelength of four cellular organelles in HepG2 is demonstrated. The synthesis, characterization, optical properties, and bioimaging applications of PQDs used in this work have been reported previously.14 Based on our reported protocol, PQDs of four colors (blue, green, yellow, and orange) are synthesized.14 These PQDs are conjugated to CD68 ab. (lysosomes), mitochondria ab. (mitochondria), nuclear antigen ab. (nucleus) and smooth muscle actin ab. (cytoskeleton) using the reported EDC-NHS chemistry.14,22 Conjugation was confirmed by Fourier Transform Infrared spectroscopy (FTIR). Furthermore, colocalization studies with organelle-specific commercial dyes demonstrate the pertinence of PQD-conjugates. Finally, simultaneous imaging of four cellular organelles of HepG2 with a single excitation (UV-405 nm) using confocal microscopy is demonstrated.
Experimental section
Synthesis and characterization of PQDs
PQDs of 4 colors are synthesized using the method reported in our previous work.14 PQDs were synthesized using the mathematically derived process parameters in conjunction with a microreactor. Briefly, PDMS (polymer) was modified under oxygen plasma to activate hydroxyl groups followed by the addition of APTMS (3-amino propyl trimethoxy silane) to ensure amine functionalization. 100 μL of the amine functionalized solution was diluted with toluene (1 mL). Furthermore, 100 μL of the diluted solution was added into the reaction mixture containing Cd (CdCl2), Te (K2TeO3) as salt precursors, mercaptosuccinic acid (MSA) as a capping agent and sodium borohydride (NaBH4) as a reducing agent. The microreactor was held in between two heaters (Mica) with optimized temperature conditions. The optimized flow rate was maintained using a syringe pump. Finally, synthesized P-QDs of different colors were collected through the outlet of the microreactor and purified using ethanol by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min. The obtained pellet was then suspended in Millipore water and was lyophilized. A stock solution of QDs (1 mg mL−1) was made for further experiments. The synthesized PQDs are characterized for their emission-excitation wavelength by photoluminescence spectroscopy (PL-Hitachi F-2500) and UV-Vis spectrophotometer (Shimadzu 30 UV-Vis spectrophotometer-1801), and size by atomic force microscopy (AFM) and transmission electron microscopy (TEM-Technai-300 KeV).
000 rpm for 20 min. The obtained pellet was then suspended in Millipore water and was lyophilized. A stock solution of QDs (1 mg mL−1) was made for further experiments. The synthesized PQDs are characterized for their emission-excitation wavelength by photoluminescence spectroscopy (PL-Hitachi F-2500) and UV-Vis spectrophotometer (Shimadzu 30 UV-Vis spectrophotometer-1801), and size by atomic force microscopy (AFM) and transmission electron microscopy (TEM-Technai-300 KeV).
Biomarker (cellular organelle-specific antibodies) conjugation of PQDs
For simultaneous multicolor bioimaging using PQDs, cellular organelles of HepG2 such as the nucleus, mitochondria, lysosomes, and cytoskeleton are targeted. CD68 ab. (Lysosomes), Mitochondria ab. (Mitochondria), Nuclear antigen ab. (Nucleus) and Smooth muscle actin ab. (cytoskeleton) are conjugated to blue, green, yellow, and orange PQDs, respectively, using a coupling reagent N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride (EDC) and N-hydroxysuccinimide (NHS) chemistry reported in our previous work.22 For this, 100 μg of PQDs are suspended in 100 μL 1X PBS (10 mM; pH 7.4). PQDs are treated with 0.05 M EDC and 0.01 M NHS and kept for 1 h with gentle rotation to activate the carboxylic groups. Furthermore, the EDC and NHS treated blue, green, yellow, and orange PQDs (100 μg/100 μL) are conjugated with lysosome-specific CD68 ab. (6 μg/100 μL), mitochondria-specific ab. (6 μg/100 μL), nucleus specific nuclear antigen ab. (8 μg/100 μL), and cytoskeleton-specific smooth muscle actin ab. (5 μg/100 μL), respectively. The mixture is incubated overnight at 4 °C with gentle rotation. Furthermore, the conjugated QDs are pelleted at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 45 min. The pellet is washed with 1X PBS (pH 7.4) and re-suspended in 1X PBS for further use. The conjugation is analyzed by Fourier transform Infrared spectroscopy (FTIR-IRAffinity-Shimadzu, USA).
000 rpm for 45 min. The pellet is washed with 1X PBS (pH 7.4) and re-suspended in 1X PBS for further use. The conjugation is analyzed by Fourier transform Infrared spectroscopy (FTIR-IRAffinity-Shimadzu, USA).
Colocalization
HepG2 cells are grown and maintained in Dulbecco's Modified Eagle's Medium (DMEM) for bioimaging. HepG2 cells (104 cells mL−1) are seeded in 6 well plates, and after the cells reach 70% confluency, they are treated with 5% Bovine Serum Albumin (BSA) to avoid non-specific binding. After 1 h incubation and 1X PBS wash, PQD-conjugates are added to a serum-free medium. The conjugates (yellow PQDs-nucleus, green PQDs-mitochondria, blue PQDs-lysosomes, and orange PQDs-cytoskeleton) are added and incubated for 1 h separately in different wells. After the internalization of PQD-conjugates, the medium is removed, and the cells are fixed with 4% paraformaldehyde (PFA). The cells are washed using 1X PBS followed by permeabilization with 0.1% TritonX-100. Furthermore, commercial organelle-specific dyes (Hoechst-Nucleus, Actin Green-Cytoskeleton, Mitotracker Orange-Mitochondria, and Lysotracker Yellow-Lysosomes) are added to the cells and incubated as per the manufacturer's protocol. Finally, the cover glass with PQD and dye treated cells are mounted on a glass slide using 80% glycerol for confocal microscopy (Leica-SP8).Multicolor bioimaging
For multicolor bioimaging, HepG2 cells are treated with PQD-conjugates in a serum-free medium with 5% BSA. The sequence for the treatment of PQD-conjugates is as follows: 1. Green PQDs-Mitochondria ab, 2. Yellow PQDs-Nuclear antigen ab., 3. Orange PQDs-smooth muscle actin ab., and 4. Blue PQDs-CD68 ab., followed by washing with 1X PBS. The incubation time for each treatment is kept at 1 h. Furthermore, the PQD-conjugate treated cells are fixed with 4% PFA followed by washing with 1X PBS. Furthermore, the PQDs samples are mounted on a glass slide using 80% glycerol for confocal microscopy. As a control, HepG2 is treated with organelle-specific commercial dyes (Hoechst-Nucleus, Actin Green-Cytoskeleton, Mitotracker Orange-Mitochondria, and Lysotracker Yellow-Lysosomes). For this, the staining protocol as described by the manufacturer is followed. After treatment, the cells are washed with 1X PBS and mounted on a glass slide using 80% glycerol for confocal microscopy. The excitation and emission wavelengths used in confocal microscopy are given in Table 1.| Organelle | Fluorescent labels | ||||
|---|---|---|---|---|---|
| Dyes | QDs | ||||
| Type | Ex./Em. (nm) | Type | Ex./Em. (nm) | Biomarker | |
| Nucleus | Hoechst | 460/490 | Yellow | 405/585 | Nuclear antigen ab. |
| Cytoskeleton | Actin green | 495/518 | Orange | 405/600 | Smooth muscle actin ab. |
| Mitochondria | Mitotracker Orange | 554/576 | Green | 405/550 | Mitochondria ab. |
| Lysosomes | Lysotracker Yellow | 465/535 | Blue | 405/470 | CD68 ab. |
The bioimaging of cellular organelles of HepG2 using PQDs is done simultaneously at single excitation (UV-405 nm) and multiple emissions are recorded using a spectral density filter (see Table 1). In contrast, the bioimaging of cells using dyes is done sequentially according to the excitation and emission pairs prescribed for each dye (Table 1). The imaging is carried out using a confocal microscope (Leica-SP8), followed by image analysis (LasX software). Moreover, to resolve specific fluorescence signals from PQD treated organelles, high-resolution microscopy (Zeiss LSM 880 with Aery Scan microscope) is performed.
Results
Synthesis and characterization of PQDs
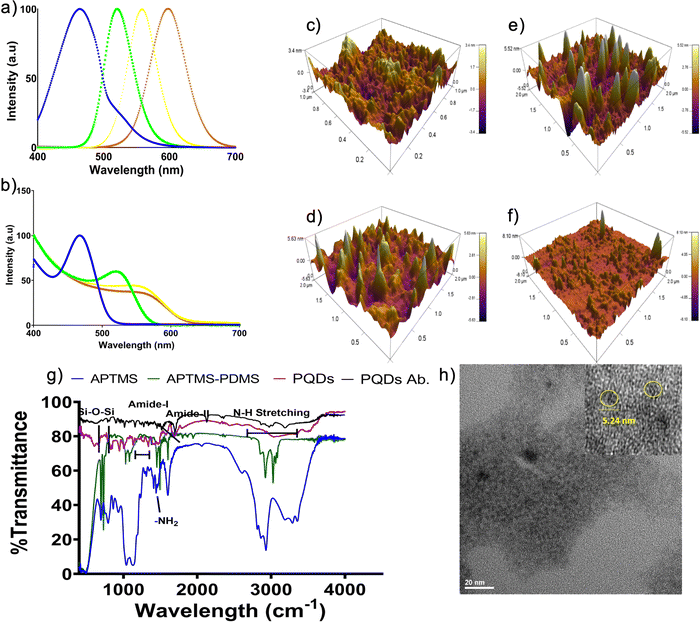
PQDs of 4 different colors are synthesized, and the optical properties are determined for all the PQDs by UV-Vis spectrophotometer and photoluminescence spectrum (PL). The PQDs used in this study are monodisperse (coefficient of variance (CV) ∼2.4%), possess high quantum yield (QY ∼62%), are photostable (44%), and biocompatible.14 These properties of PQDs are characterized in our previous manuscript. Emission maxima of 466 nm, 520 nm, 567 nm, and 598 nm corresponding to blue, green, yellow, and orange PQDs, respectively, are obtained from the PL spectroscopy14 (Fig. 1a). Further, the UV-Vis absorption spectrum (Fig. 1b) reveals the characteristic peak position shift to longer wavelengths (450 nm–580 nm) indicating the synthesis of blue to orange PQDs. An atomic force microscope is used to obtain the size of the PQDs with a cantilever tip size of 1 nm. The size of the PQDs is recorded to be 3.4 nm, 5.52 nm, 5.83 nm, and 8.83 nm for blue, green, yellow, and orange PQDs, respectively (Fig. 1c–f). The transmission electron micrograph (TEM) captured for green PQDs show the size to be ∼5 nm (Fig. 1h) which correlates to that obtained by AFM.Biomarker (cellular organelle-specific antibodies) conjugation of PQDs
The synthesized PQDs are further conjugated with organelle-specific antibodies for their use in bioimaging. The conjugation is done using EDC-NHS chemistry reported in our previous work.14 The peaks obtained from FTIR (Fig. 1g) of ab.-PQDs display all the characteristic peaks of PQDs at 950 cm−1, 1000 cm−1, and a broad peak from 1352–1455 cm−1 corresponding to Si–O–Si, Si–O–CH3, and Si–O–CH2CH3, respectively, indicating the presence of PDMS. A peak at 1598 cm−1, corresponding to NH stretch of primary amines, indicates amine modification of PDMS. Moreover, the characteristic peaks of the amide I band (C–O stretching vibrations between 1700–1650 cm−1) and amide II band (N–H bending vibrations coupled with C–N stretching vibrations between 1500–1400 cm−1) suggest the conjugation of ab. to PQDs via amide bonding.3,23Colocalization
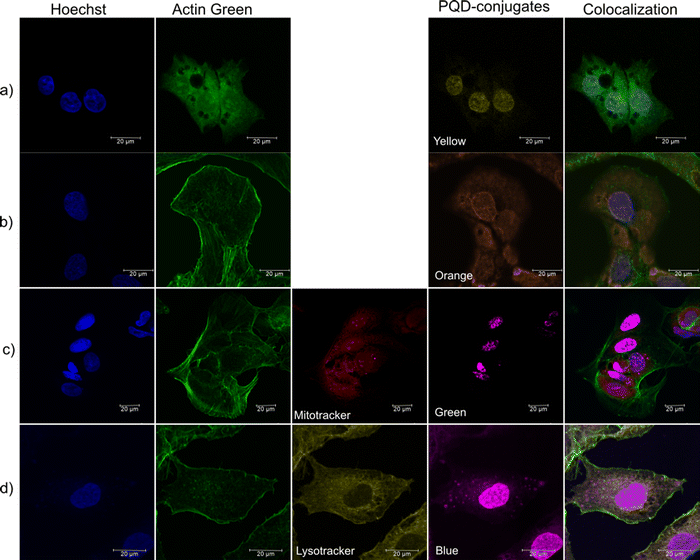
The labeling of organelle-specific antibody-conjugated PQDs is assessed by their colocalization with commercial organelle-specific dyes (Hoechst, Actin Green, Lysotracker Yellow, Mitotracker Orange) in HepG2 cells. These organelle specific commercial dyes are used as a control. As Hoechst and Actin Green dye stain the nucleus and cytoskeleton respectively, they are used in all the samples. Fig. 2a shows the specific uptake of nuclear antigen ab. conjugated yellow PQDs in the nucleus, which can be seen to be colocalized with Hoechst dye (blue). Similarly, in Fig. 2b, the smooth muscle actin ab. conjugated orange PQDs are seen to be explicitly taken up by the cytoskeleton of HepG2, which can be further seen to be colocalized with Actin Green (merge image). Furthermore, in Fig. 2c, the mitochondria ab. conjugated green PQDs (shown in magenta color to avoid overlapping with the green color of Actin Green) are taken up specifically by mitochondria (spherical dots in magenta within cytoskeleton and nucleus) of HepG2, which can be seen colocalized with the commercial dye. Moreover, Fig. 2d shows the specific uptake of CD68 lysosome ab. conjugated blue PQDs (shown in magenta color to avoid the overlapping with the green color of Actin Green dye) by lysosomes, which is further seen to be localized with Lysotracker Yellow. The PQD-conjugates are seen to label their target organelles specifically; however, a few PQDs are internalized non-specifically in the nucleus. This nuclear internalization (passive diffusion) may be attributed to the small size of the PQDs (< 9 nm).14,24,25 Besides, the pathway of cellular colocalization of PQDs may be considered to be a caveolae-dependent pathway based on the previous reports.24,25 Also, the elimination and partial degradation of QDs have been reported to be within 24 h from the cells.26,27 Thus, the PQD-conjugates can stain the cellular organelles specifically and can be used as a bioimaging agent for simultaneous multicolor bioimaging.Simultaneous multicolor bioimaging
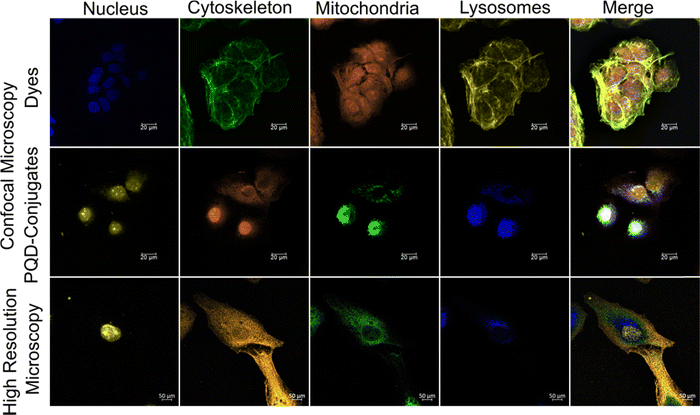
Simultaneous multicolor bioimaging of HepG2 cellular organelles using 4 PQDs at an excitation wavelength of 405 nm is carried out by confocal microscopy (Fig. 3 and Fig. S1 of ESI†). Fig. 3 shows the image of the cellular organelles (nucleus, lysosomes, mitochondria, and actin) of HepG2 treated with commercial dyes and PQD-conjugates. In the case of commercial dyes, the imaging is done sequentially at different excitation and emission wavelengths (see Table 1), whereas the PQD-conjugates are simultaneously imaged at a single excitation wavelength of 405 nm. Fig. 3 (top panel) shows the confocal microscopic images obtained using commercial dyes. An overlap of the signal of green (Actin Green) and yellow (Lysotracker) is evident. However, the middle panel of the PQD-conjugates shows no such overlap with the cellular organelles that are clearly distinguishable. Moreover, the images prove the potential of the organelle-specific PQD-conjugates.To confirm the organelle-specific targeting of the PQD-conjugates, high-resolution microscopic images are obtained from a Zeiss LSM 880 with an Aery Scan microscope (bottom panel). However, some non-specific distribution is seen due to the passive diffusion of small-sized PQDs.14,15,24,28
Discussion
Quantum dots display unique size-tunable emissions when excited by UV. They demonstrate peculiar properties like high quantum efficiency, photostability, monodispersity, biocompatibility, and reproducibility, which could be suitable for bioimaging. They are better, especially in terms of photostability and quantum efficiency, than their commercial counterparts (dyes or stains) and thus, are the ideal choice for bioimaging. Moreover, an exhibition of the visible spectrum at single excitation makes them the first choice for multicolor bioimaging.In the present study, biocompatible polymer-coated QDs are used. These PQDs are quantum efficient, photostable, biocompatible, and reproducible, thus making them fit for simultaneous multicolor imaging. Blue, green, yellow, and orange PQDs were synthesized successfully using the reported protocol.14 Photoluminescence and absorbance spectra demonstrate the narrow emission wavelength of the size-tuned PQDs (Fig. 1a and b). The size of the PQDs is in the range of 3.4 nm to 8.8 nm as revealed from AFM analysis (Fig. 1c–f). The TEM micrograph shows the size of the green color PQDs as 5.24 nm (Fig. 1h). The PQDs are monodisperse with a CV ∼2.4% and reproducible (CV ∼9%).14 Moreover, the PQDs are quantum efficient (∼62%) and photostable (44%). Furthermore, the polymer coating reduces ion leaching and renders them biocompatible.14 Monodispersity ensures a narrow emission spectrum providing a possibility to add more colors by changing the size of the QDs.14,29 Quantum efficiency and photostability will guarantee bright fluorescence with less photobleaching.14,30 Reproducibility in the synthesis process offers commercialization potential to these PQDs.14,31
Thus, blue, green, yellow, and orange PQDs are synthesized and conjugated successfully with organelle-specific biomarkers (see Table 1). The targetability of the PQD-conjugates is assessed by colocalization with the commercially procured organelle-specific dyes. The PQD-conjugates were localized with the commercial dyes to their respective organelles. This result confirms the specificity of organelle-specific targeting of PQD-conjugates. However, a few PQDs are taken up non-specifically owing to passive diffusion because of the small size and efficiency of conjugation.24,28 As PQD-conjugates are seen to colocalize with the commercial dyes, they are deemed suitable for simultaneous multicolor imaging of HepG2 cellular organelles.
Multicolor imaging of cellular organelles treated with dyes is done sequentially at different excitation/emission wavelength pairs (Fig. 3 Dyes panel), whereas the PQD-conjugate treated cellular organelles were imaged simultaneously using a single UV excitation wavelength of 405 nm (Fig. 3 PQD-conjugates panels). The confocal microscopic images (dyes panel of Fig. 3) reveal overlapping spectra for Actin Green (Ex./Em.: 495/518 nm) as well as Lysotracker Yellow (Ex./Em.: 465/535 nm) when imaged sequentially. This is due to the broad excitation and emission spectra associated with the dyes. Moreover, due to the excitation-emission pairs exhibited by the dyes, the selection needs to be highly specific to avoid spectral overlap. The overlapping of the spectra results in a loss of specificity in imaging an organelle of interest. In contrast, PQDs don’t show (PQD-conjugates panel of Fig. 3) any spectral overlap owing to the narrow emission spectra. Moreover, the PQD-conjugate panel shows a bright fluorescence signal. These merits may be attributed to the monodispersity, quantum efficiency, and photostability of the PQDs.14,15,32–34
To further resolve the specific signals from PQD targeted organelles, the samples are seen with the help of a high-resolution Zeiss LSM 880 with an Aery Scan microscope (bottom panel of Fig. 3). Lambda scan is used to observe the HepG2 samples treated with PQD-conjugates. A bright fluorescence due to the lambda scan from the PQDs is visible revealing the targeted organelles. Thus, this result reiterates the superior properties of the QDs.
There are reports available demonstrating the use of QDs for multicolor imaging applications.2,7,16,17,20,35,36 However, very few reports are available for simultaneous imaging of biological moieties at a single excitation wavelength using QDs only. A study demonstrating the simultaneous imaging of tissue targets using streptavidin conjugated commercially available QDs at a single excitation wavelength was reported by Fountain et al.37 Emission fingerprinting was carried out to resolve the emission peaks of the QDs. Out of multiple emission peaks of QDs, few QD emission peaks were resolved, keeping a 20 nm bandgap. This may be credited to the broad emission spectra (polydispersity) of the QDs.38 However, the QDs used in this study were not biocompatible, which is an important property for bioimaging.15,39 In our study, the 4 PQD-conjugates used for simultaneous imaging were resolved clearly without the loss of signal due to their specific labeling. Also, the PQDs synthesized here possess all the desirable properties such as monodispersity, quantum efficiency, photostability, biocompatibility, and reproducibility required for simultaneous multicolor imaging of biological moieties. Thus, the present study establishes the potential of PQDs over commercial dyes for simultaneous multicolor imaging of cellular organelles at a single excitation wavelength. This study opens new possibilities of using QDs for multicolor bioimaging applications to understand several biological processes.
Conclusion
PQDs of 4 colors were synthesized using the reported method and were conjugated successfully to nucleus, cytoskeleton, mitochondria, and lysosome-specific antibodies. Furthermore, a cellular organelle colocalization assay confirmed the specific labeling of the PQD conjugates with organelle-specific commercial dyes in HepG2. Finally, the PQD conjugates were employed successfully for simultaneous multicolor imaging of cellular organelles with a single excitation wavelength using a Leica-SP8 confocal microscope. Moreover, the specific emission signals were successfully resolved by a high-resolution Zeiss LSM 880 with an aery scan microscope. Thus, the developed monodisperse, quantum efficient, photostable, biocompatible, and reproducible PQDs were proved to be an excellent alternative to commercial dyes for in vitro simultaneous multicolor bioimaging.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
SP acknowledges the senior research fellowship from the Indian Council of Medical Research (ISRM/11(73)/2017). The authors highly acknowledge Lt. Dr. Arun Karthik S. for allowing use of the bioimaging facility at National Centre for Cell Science, Pune. The authors thank the help from Dr. Suwarna Datar, Defence Institute of Advanced Technology (DIAT), Pune in AFM analysis.References
- W. S. Kuo, X. C. Shen, C. Y. Chang, H. F. Kao, S. H. Lin, J. Y. Wang and P. C. Wu, ACS Nano, 2020, 14, 11502–11509 CrossRef CAS PubMed
.
- H. Zeng, H. Yang, G. Liu, S. Zhang, X. Zhang and Y. Zhang, J. Microsc., 2020, 277, 32–41 CrossRef CAS PubMed
.
- S. L. Sahoo, C. H. Liu, M. Kumari, W. C. Wu and C. C. Wang, RSC Adv., 2019, 9, 32791–32803 RSC
.
- W. Wang, Z. Liu and X. Lan, Mol. Imaging Biol., 2020, 22, 820–831 CrossRef PubMed
.
- S. Babu, J. H. Cho, J. M. Dowding, E. Heckert, C. Komanski, S. Das, J. Colon, C. H. Baker, M. Bass, W. T. Self and S. Seal, Chem. Commun., 2010, 46, 6915–6917 RSC
.
- Y. Kang, Y.-Z. Wu, X. Hu, X. Xu, J. Sun, R. Geng, T. Huang, X. Liu, Y. Ma, Y. Chen, Q. Wan, X. Qi, G. Zhang, X. Zhao and X. Zeng, Sci. Rep., 2017, 7, 45313 CrossRef CAS PubMed
.
- V. G. Bandi, M. P. Luciano, M. Saccomano, N. L. Patel, T. S. Bischof, J. G. P. Lingg, P. T. Tsrunchev, M. N. Nix, B. Ruehle, C. Sanders, L. Riffle, C. M. Robinson, S. Difilippantonio, J. D. Kalen, U. Resch-Genger, J. Ivanic, O. T. Bruns and M. J. Schnermann, Nat. Methods, 2022, 19, 353–358 CrossRef CAS PubMed
.
- A. N. Butkevich, G. Y. Mitronova, S. C. Sidenstein, J. L. Klocke, D. Kamin, D. N. H. Meineke, E. D’Este, P. Kraemer, J. G. Danzl, V. N. Belov and S. W. Hell, Angew. Chem., Int. Ed., 2016, 55, 3290–3294 CrossRef CAS PubMed
.
- V. Sharma, N. Kaur, P. Tiwari and S. M. Mobin, J. Photochem. Photobiol., B, 2018, 182, 137–145 CrossRef CAS PubMed
.
- Y. Koyama, Y. Hama, T. Barrett, G. Ravizzini, P. L. Choyke and H. Kobayashi, Neoplasia, 2007, 9, 1021–1029 CrossRef CAS PubMed
.
- L. Wei, Z. Chen, L. Shi, R. Long, A. V. Anzalone, L. Zhang, F. Hu, R. Yuste, V. W. Cornish and W. Min, Nature, 2017, 544, 465–470 CrossRef CAS PubMed
.
- A. F. E. Hezinger, J. Teßmar and A. Göpferich, Eur. J. Pharm. Biopharm., 2008, 68, 138–152 CrossRef CAS PubMed
.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Nat. Methods, 2008, 5, 763–775 CrossRef CAS PubMed
.
- S. Pandey, D. Mukherjee, P. Kshirsagar, C. Patra and D. Bodas, Mater. Today Bio, 2021, 11, 100123 CrossRef CAS PubMed
.
- S. Pandey and D. Bodas, Adv. Colloid Interface Sci., 2020, 278, 102137 CrossRef CAS PubMed
.
- Z. G. Wang, S. L. Liu and D. W. Pang, Acc. Chem. Res., 2021, 54, 2991–3002 CrossRef CAS PubMed
.
- D. Bahari, B. Babamiri, A. Salimi and A. Rashidi, J. Lumin., 2021, 239, 118371 CrossRef CAS
.
- E. Campbell, M. T. Hasan, R. Gonzalez Rodriguez, G. R. Akkaraju and A. V. Naumov, ACS Biomater. Sci. Eng., 2019, 5, 4671–4682 CrossRef CAS PubMed
.
- C. Chen, J. Peng, H. Xia, Q. Wu, L. Zeng, H. Xu, H. Tang, Z. Zhang, X. Zhu, D. Pang and Y. Li, Nanotechnology, 2010, 21, 095101 CrossRef PubMed
.
- M. Panagiotopoulou, Y. Salinas, S. Beyazit, S. Kunath, L. Duma, E. Prost, A. G. Mayes, M. Resmini, B. Tse Sum Bui and K. Haupt, Angew. Chem., Int. Ed., 2016, 55, 8244–8248 CrossRef CAS PubMed
.
- H. Kobayashi, M. Ogawa, R. Alford, P. L. Choyke and Y. Urano, Chem. Rev., 2010, 110, 2620–2640 CrossRef CAS PubMed
.
- S. Agrawal, A. Morarka, D. Bodas and K. M. Paknikar, Appl. Biochem. Biotechnol., 2012, 167, 1668–1677 CrossRef CAS PubMed
.
- C. Li, Y. Ji, C. Wang, S. Liang, F. Pan, C. Zhang, F. Chen, H. Fu, K. Wang and D. Cui, Nanoscale Res. Lett., 2014, 9, 1–13 CrossRef PubMed
.
- L. W. Zhang and N. A. Monteiro-Riviere, Toxicol. Sci, 2009, 110, 138–155 CrossRef CAS PubMed
.
- A. K. Narasimhan, S. B. Lakshmi, T. S. Santra, M. S. R. Rao and G. Krishnamurthi, RSC Adv., 2017, 7, 53822–53829 RSC
.
- R. Hardman, Environ. Health Perspect., 2006, 114, 165–172 CrossRef PubMed
.
- E. Yaghini, H. Turner, A. Pilling, I. Naasani and A. J. MacRobert, Nanomedicine, 2018, 14, 2644–2655 CrossRef CAS PubMed
.
- N. S. Awad, M. Haider, V. Paul, N. M. Alsawaftah, J. Jagal, R. Pasricha and G. A. Husseini, Pharmaceutics, 2021, 13, 2073 CrossRef CAS PubMed
.
- S. Chan, M. Liu, K. Latham, M. Haruta, H. Kurata, T. Teranishi and Y. Tachibana, J. Mater. Chem. C, 2017, 5, 2182–2187 RSC
.
- D. A. East, D. P. Mulvihill, M. Todd and I. J. Bruce, Langmuir, 2011, 27, 13888–13896 CrossRef CAS PubMed
.
- U. Resch-Genger, M. Grabolle, S. Cavaliere-Jaricot, R. Nitschke and T. Nann, Nat. Methods, 2008, 5, 763–775 CrossRef CAS PubMed
.
- Y. P. Gu, R. Cui, Z. L. Zhang, Z. X. Xie and D. W. Pang, J. Am. Chem. Soc., 2012, 134, 79–82 CrossRef CAS PubMed
.
- Z. Liang, M. B. Khawar, J. Liang and H. Sun, Front. Oncol., 2021, 11, 1–14 Search PubMed
.
- A. A. H. Abdellatif, H. M. Tawfeek, M. A. Younis, M. Alsharidah and O. Al Rugaie, Int. J. Nanomed., 2022, 17, 1951–1970 CrossRef PubMed
.
- S. Jeong, Y. Jung, S. Bok, Y.-M. Ryu, S. Lee, Y.-E. Kim, J. Song, M. Kim, S.-Y. Kim, G.-O. Ahn and S. Kim, Adv. Healthcare Mater., 2018, 7, 1800695 CrossRef PubMed
.
- S. Hu, S. Zeng, B. Zhang, C. Yang, P. Song, T. J. Hang Danny, G. Lin, Y. Wang, T. Anderson, P. Coquet, L. Liu, X. Zhang and K. T. Yong, Analyst, 2014, 139, 4681–4690 RSC
.
- T. J. Fountaine, S. M. Wincovitch, D. H. Geho, S. H. Garfield and S. Pittaluga, Mod. Pathol., 2006, 19, 1181–1191 CrossRef CAS PubMed
.
- Z. Wan, H. Yang, W. Luan, S. Tu and X. Zhou, Nanoscale Res. Lett., 2009, 5, 130–137 CrossRef PubMed
.
- E. S. Speranskaya, N. V. Beloglazova, P. Lenain, S. De Saeger, Z. Wang, S. Zhang, Z. Hens, D. Knopp, R. Niessner, D. V. Potapkin and I. Y. Goryacheva, Biosens. Bioelectron., 2014, 53, 225–231 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: Supporting data is provided with the manuscript. See DOI: https://doi.org/10.1039/d2ma00964a |
| This journal is © The Royal Society of Chemistry 2023 |