Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efficient filter-in-centrifuge separation of low-concentration bacteria from blood†
Kaiyang
Zeng‡
,
Mohammad
Osaid‡
 and
Wouter
van der Wijngaart
and
Wouter
van der Wijngaart
 *
*
Division of Micro and Nanosystems, KTH Royal Institute of Technology, Stockholm, Sweden. E-mail: wouter@kth.se
First published on 15th September 2023
Abstract
Separating bacteria from infected blood is an important step in preparing samples for downstream bacteria detection and analysis. However, the extremely low bacteria concentration and extremely high blood cell count make efficient separation challenging. In this study, we introduce a method for separating bacteria from blood in a single centrifugation step, which involves sedimentation velocity-based differentiation followed by size-based cross-flow filtration over an inclined filter. Starting from 1 mL spiked whole blood, we recovered 32 ± 4% of the bacteria (Escherichia coli, Klebsiella pneumonia, or Staphylococcus aureus) within one hour while removing 99.4 ± 0.1% of the red blood cells, 98.4 ± 1.4% of the white blood cells, and 90.0 ± 2.6% of the platelets. Changing the device material could further increase bacteria recovery to >50%. We demonstrated bacterial recovery from blood spiked with 10 CFU mL−1. Our simple hands-off efficient separation of low-abundant bacteria approaches clinical expectations, making the new method a promising candidate for future clinical use.
Introduction
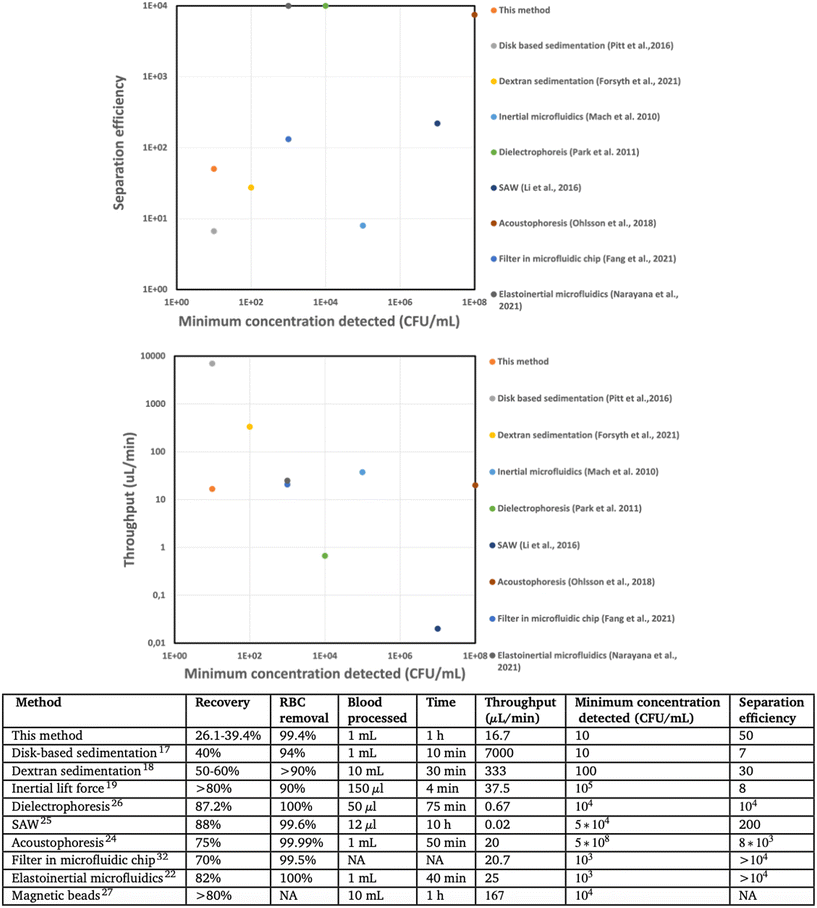
Severe bloodstream infections represent a significant global healthcare challenge, resulting in high mortality rates and medical expenses. Sepsis alone is responsible for over 11 million deaths annually worldwide, accounting for one in every five deaths, with an estimated cost to the US economy of approximately $20 billion.1–5 However, the current clinical diagnosis of BSI relies on a positive blood culture, which is a slow process that can take several days.6 As the mortality rate of untreated patients increases by 8% per hour,7 clinicians often resort to broad-spectrum antibiotics. Unfortunately, in approximately 30% of cases, this approach is incorrect and can lead to more fatalities.8 Furthermore, the use of broad-spectrum antibiotics promotes the growth of antimicrobial resistance.9Rapid pathogen identification and antimicrobial susceptibility testing (AST) are preferred methods for diagnosing sepsis.10,11 Various rapid phenotypic and genotypic methods have been developed for this purpose.12–16 However, the bottleneck in sepsis diagnosis lies in sample preparation, particularly in separating bacteria from whole blood, given that the concentration of bacteria is as low as 1–10 CFU mL−1, whereas blood cells have concentrations in the range of 4–6 billion per mL for red blood cells (RBCs) and 5–10 million per mL for white blood cells (WBCs). Various methods have been investigated for bacterial separation, including sedimentation velocity-based differentiation,17,18 inertial19–21 and elastoinertial22 microfluidics, acoustophoresis,23,24 surface acoustic waves (SAW),25 dielectrophoresis (DEP),26 and magnetic beads based separation.27 However, these methods suffer from low throughput (DEP, SAW), low separation efficiency (density-based separation, inertial microfluidics), or require high bacterial concentration for efficient separation (inertial and elastoinertial microfluidics, DEP, SAW, acoustophoresis) or are selective to specific bacterial strains (magnetic beads) (Fig. 5).
Size-based filtration is a simple, inexpensive, and highly selective method with successful applications in separating WBCs28 or tumor cells29,30 from blood and in blood-serum separation.31 WBCs have typical size 10–20 μm, RBCs 6.2–8.2 μm, platelets 2.0–4.0 μm, and bacteria around 1.5 μm.17 Nonetheless, filter caking can impede filtration, particularly during dead-end filtration, where abundant RBCs accumulate on the filter surface to form a clogging layer that blocks bacteria and limits volumetric throughput.17 A solution to filter caking during bacteria filtration is resuspension of RBCs, allowing near-surface bacteria to pass through the filter, albeit at limited volumetric throughput, impeding detection of low CFU concentrations.32 Cross-flow filtration is another solution to filter caking, although we found no reports of cross-flow filtration of bacteria from blood. Concatenating centrifugation and filtration in two subsequent manual steps has been proposed.33
In this study, we developed a filter-based centrifugal device for efficiently separating low-abundant bacteria from whole blood at relevant throughput in a single step.
Results
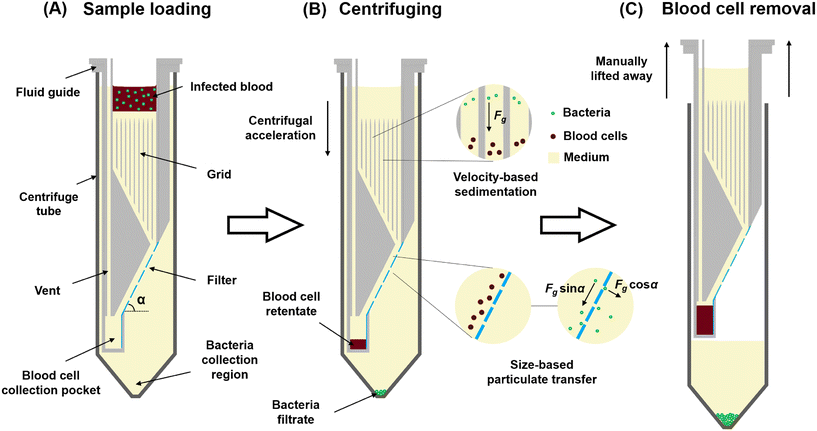
We developed a separation device, based on the difference in cell size and terminal velocity between blood cells and bacteria,17 that utilizes centrifuge-driven cross-filter particle transport (Fig. 1 and S1†). The device comprises a 3D-printed fluid guide inserted into a 50 mL centrifuge tube. The fluid guide incorporates a filter, inclined at angle α with respect to the centrifugal acceleration, and has pores smaller than blood cells but larger than bacteria. The region of the fluid guide above the filter features a grid of vertical walls of thickness 0.4 mm placed in a square mesh with a pitch of 1.6 mm to prevent liquid convection induced by the Coriolis effect during centrifuge acceleration (Fig. S4†).34 At the lower side of the filter, the fluid guide includes a blood cell collection pocket for the filter retentate, while the filtrate is collected in the lower region of the centrifuge tube. An air vent in the fluid guide mitigates air trapping during liquid priming.Operating the device proceeds in four steps.
1. Fill the fluid guide with density medium (a mixture of 75% Lymphoprep and 25% broth) until it reaches the top of the grid. Centrifuge the device for a short duration to remove any air bubbles that may be trapped.
2. Load the blood sample onto the top of the medium.
3. Centrifuge the device, during which the larger blood cells sediment at ∼30× higher velocity than the bacteria (according to Stoke's law17). Blood cells move along the filter surface before the bacteria and sediment in the blood cell collector at the bottom edge of the filter. Bacteria that land on the solid fraction of the filter experience a force of Fg·sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α tangential to the filter surface, where Fg is the artificial gravity acting on the bacteria. Bacteria that reach the filter pores are dragged into the pore and across the filter with force Fg·cos
α tangential to the filter surface, where Fg is the artificial gravity acting on the bacteria. Bacteria that reach the filter pores are dragged into the pore and across the filter with force Fg·cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α, after which they sediment into the bacteria collection region.
α, after which they sediment into the bacteria collection region.
4. Lift the fluid guide containing the blood cells away from the device, leaving the bacteria in 18 mL of Lymphoprep–broth medium in the centrifuge tube. The sample is now ready for downstream processing.
We first investigated the relative recovery (CFU in filtrate/CFU in filtrate and retentate) of bacteria for the following operation parameters: i) Lymphoprep![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) broth mixture ratios 100
broth mixture ratios 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 75
0, 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25, 50
25, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and 0
50 and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
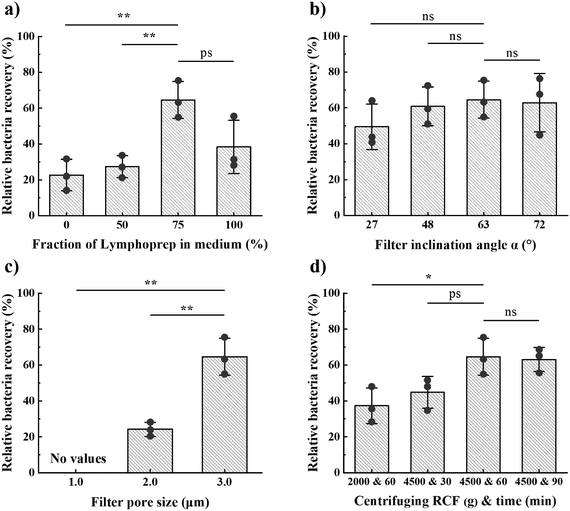
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, ii) centrifuging time between 30 and 90 minutes at an RCF in the range 2000–4500g and with approximately 300 g s−1 acceleration and deceleration, iii) filter pore sizes of 1.0, 2.0 or 3.0 μm, and iv) filter angles of 27°, 48°, 63° or 72°. We found the highest mean relative recovery for 3.0 μm pore size, a medium of 75% Lymphoprep and 25% broth, 63° filter angle, and centrifuging for 60 min at 4500g (from hereon “optimal design”; Fig. 2). During this parameter optimisation study, we used Klebsiella pneumonia (K. pneumonia) spiked in healthy patient blood at 104 CFU mL−1.
100, ii) centrifuging time between 30 and 90 minutes at an RCF in the range 2000–4500g and with approximately 300 g s−1 acceleration and deceleration, iii) filter pore sizes of 1.0, 2.0 or 3.0 μm, and iv) filter angles of 27°, 48°, 63° or 72°. We found the highest mean relative recovery for 3.0 μm pore size, a medium of 75% Lymphoprep and 25% broth, 63° filter angle, and centrifuging for 60 min at 4500g (from hereon “optimal design”; Fig. 2). During this parameter optimisation study, we used Klebsiella pneumonia (K. pneumonia) spiked in healthy patient blood at 104 CFU mL−1.
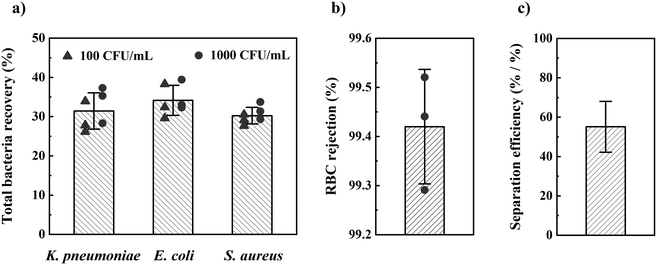
Once the optimal design parameters were established, we tested our method for the most prevalent Gram− and Gram+ sepsis-causing bacteria: Escherichia coli (E. coli), K. pneumonia, and Staphylococcus aureus (S. aureus). Using these optimal design parameters, we separated these bacteria from 1 mL of spiked blood in 1 h with 32 ± 4% (sd, n = 18) total bacteria recovery (CFU in filtrate/CFU in blood) and 99.4 ± 0.1% (sd, n = 3) RBC rejection (Fig. 3). We found no significant difference in bacteria recovery between 100 and 1000 CFU mL−1 concentration of spiked blood, nor between different bacterial species. Blood spiked with 10 CFU mL−1 of the three bacteria species resulted in positive overnight culture of the filtrate for all tests (n = 3 for each bacterial species). In contrast, overnight culture of filtrate from non-spiked blood (negative control) became positive for only two out of nine tests (contamination).
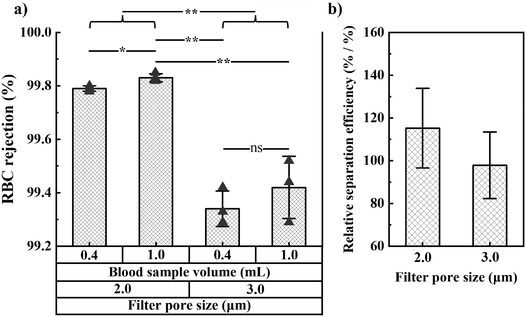
We also studied RBC rejection for filters with 2.0 and 3.0 μm pore size and found 2.0 μm pores to provide the highest RBC rejection and relative separation efficiency (relative bacteria recovery/[1 − RBC rejection]). We investigated WBC and platelet rejection for a filter with 3.0 μm pore size (Fig. S8†).
Further experimental details are provided in the Methods section, and all raw data are in ESI.†
Discussion
Our approach constitutes a simple, hands-off, relevant-throughput, and label-free method to efficiently isolate bacteria from whole blood in clinically relevant concentrations. We successfully tested our method for the most prevalent Gram− and Gram+ sepsis-causing bacteria: E. coli, K. pneumonia, and S. aureus. Despite differences in size and shape (rod-like or round) among these bacteria, we found no significant differences in recovery (Fig. 3). The remainder of this section discusses first the physical phenomena and parameters involved in the operation of the device, thereafter an analysis of the results, next a comparison with state-of-the-art, and finally, future steps required to realize the potential impact of the approach in the clinical setting.Lymphoprep is a sterile and endotoxin-tested density gradient medium used to isolate cells based on their cell density. Mixing Lymphoprep with broth creates different densities of medium. The optimal medium mixture was found to be 75% Lymphoprep in broth, which has a density slightly above that of blood (Table 1), thus promoting both efficient layered loading (Fig. S2c†) and sedimentation of particles through the filter pores (Fig. S2a†). In contrast, using a lower-density mixture (0 or 50% Lymphoprep) prevented layered loading and resulted in the uncontrollable sinking of blood (Fig. S2a and b†), causing cells to be transported downward by convection rather than sedimentation. This led to lower bacteria recovery and compromised the efficiency of the device. On the other hand, using a higher density mixture (100% Lymphoprep) slowed sedimentation and reduced the artificial gravity (Fg) that drives the bacteria through the filter pores, which also resulted in lower bacteria recovery. Overall, our findings highlight the importance of choosing the right density for the medium to optimize performance and improve efficiency.
| Liquid/particles | Average density of mass relative to DIW |
|---|---|
| RBCs | 1.086–1.122 |
| Bacteria | 1.080–1.100 |
| WBCs | 1.057–1.092 |
| 100% Lymphoprep–0% broth | 1.077 |
| Platelets | 1.072–1.077 |
| 75% Lymphoprep–25% broth | 1.058 |
| Whole blood | 1.043 to 1.060 (ref. 35) |
| 50% Lymphoprep–50% broth | 1.039 |
| Blood plasma | 1.035 |
| 0% Lymphoprep–100% broth | 1.000 |
The blood cell collection pocket was designed for up to 0.5 mL of blood cells, limiting the blood volume that could be processed to 1 mL per tube. Designing for larger blood sample volumes would require more space in the fluid guide, which would also require longer centrifuge tubes. Typical centrifuges can hold several centrifuge tubes, allowing for the straightforward handling of larger blood volumes through parallelization.
The upper section of the fluid guide is designed for terminal velocity-based particle separation. However, the Coriolis force causes a flow vortex during centrifuge acceleration, which can cause the mixing of the separating particles and reduce the bacteria recovery (Fig. S4†). The vertical grid walls effectively block horizontal flow components, strongly diminishing the effect of the Coriolis vortex. By blocking the horizontal flow component using the vertical wall grid, we do not impede the vertical sedimentation transport of the bacteria and blood cells. The size of the grid used was sufficient to prevent the mixing.
To ensure effective performance of the device, it is important that the bacteria have enough time to sediment to and through the filter. The sedimentation length is directly proportional to the product of the artificial gravity and centrifugation time. Our observations revealed that most RBCs sediment to the collection pocket within 1–2 minutes at 4500 g (Fig. S3†). Given that RBCs have an estimated sedimentation velocity 30 times larger than that of bacteria, we expect bacteria to typically sediment in 30–60 min. Consistent with this expectation, we found that extending the centrifugation time beyond 60 min did not significantly increase bacteria recovery (Fig. 2d). Our optimal conditions for centrifugation were therefore 60 min at 4500g, which provided a relative bacteria recovery of 65%.
Filter caking was not observed, which we attribute to two design elements. The first design feature is the effect of artificial gravity, which propels blood cells along the inclined filter surface, preventing them from stagnating at pores. The second design feature is velocity-based differentiation, which leads to blood cells passing over the filter before the bacteria arrive, eliminating any obstruction to the bacteria's interaction with the filter.
Our approach to filtration utilizes sedimentation-driven particle movement through the size-based filter, which differs fundamentally from pressure-driven filtration. In pressure-driven filtration, particle movement results from fluid drag. Flow lines bend along the surface and through the pores, actively driving particles towards and through the pores with a force controlled by the liquid velocity. In contrast, in sedimentation-driven filtration, particle movement is caused by the downward artificial gravity on particles in a stationary liquid, in which no force component actively drives the particles toward the pores. Here, the angular velocity and medium density control the force driving particles through the pores. Our method employs a hanging bucket centrifuge, wherefore the method does not depend on the placement of the device in the centrifuge. Previous work with focus on cancer diagnostics has investigated slanted rail filters incorporated in lab-on-a-disk centrifugal devices for cancer cell separation from blood after extensive sample preprocessing.36 In contrast, this work focuses on infectious disease diagnostics, where a slanted filter in a centrifuge tube is concatenated with terminal velocity-based separation, which uniquely allows the separation of bacterial cells (which are more than an order of magnitude smaller in size than cancer cells), processing of liquid volumes that are more than one magnitude order larger, and using whole blood as the input sample without the need for sample preparation.
As expected, the fraction of bacteria and RBCs in the filtrate both increase with the filter pore size (Fig. 2c and 4a). The optimal pore size depends on what parameter one wants to optimize. Small pores are beneficial if maximal blood cell rejection is targeted. When maximal bacteria recovery is targeted, large pores provide a better result. For maximal separation efficiency, we observed that 2 μm pores gave the best results. In order to prioritize bacteria recovery, given the very low bacteria concentration in patient samples, and with the understanding that several downstream processes can tolerate a small fraction of red blood cells, we chose to use 3.0 μm pores as the standard pore size in our experiments.
Filters at 63° angle resulted in the highest average bacterial recovery, wherefore we chose this angle for our optimal design (Fig. 2b). However, the influence of filter inclination angle on bacteria recovery for the range 27° < α < 72° was not significant. The total filter length, along which particles move, scales with 1/cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α, whereas the force driving particles through the pores scales with cos
α, whereas the force driving particles through the pores scales with cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α. We hypothesize that these two effects cancel each other.
α. We hypothesize that these two effects cancel each other.
During design parameter optimization, we optimized the relative bacteria recovery because this measure is less prone to differences in bacterial growth during experimental handling. Of the number of bacteria spiked in the blood, typically only 60% are found in the filtrate and retentate (comparing total bacteria recovery (Fig. 3) and relative bacteria recovery values (Fig. 2)). We believe this discrepancy could be attributed to either the immune response of the healthy donor blood attacking the bacteria or bacteria sticking to the rough 3D-printed surface of the fluid guide. The former is in line with observations that recovery of bacteria spiked in blood diminishes with the duration between spiking and bacteria quantitation. We experimentally tested the effect of the 3D-printed surface and found a bacterial recovery reduction of 40% when centrifuging in 3D-printed tubes compared to commercial centrifuge tubes of polypropylene (Fig. S6†). We therefore speculate that injection molding our fluid guides in polypropylene could increase the total bacterial recovery to 53%.
Our method uniquely provides the combined separation of bacteria at low concentration, high separation efficiency and relevant throughput, which are the three most important performance parameters from a clinical application perspective (Fig. 5). Other sedimentation-based methods have enabled a low bacterial concentration detection but feature limited RBC17 or WBC17,18 removal, affecting potential downstream sample processing. A separation efficiency of 100 or above is typically desirable to avoid interference of blood cells during microfluidic handling (clogging) or optical readout. Several methods report high separation efficiency22,24–26 or high throughput,19,22,24,27 but these methods have been shown only for high bacteria concentration, 1000 CFU mL−1 or above, which would necessitate time-consuming bacterial culture of the clinical sample prior to processing with these methods. To allow rapid sepsis diagnostics necessitates a sample processing throughput of at least 10 mL blood (considering the low bacterial count) in one hour (considering the need for rapid results). Our approach allows reaching this requirement through parallelization, e.g., by dividing a 10 mL blood sample into 10 aliquots in different devices during the same centrifugation. The downstream processing requires blood cell removal to avoid interference with the blood cells in the identification of pathogens and AST. Achieving high RBC, WBC, and platelet removal would be significant for implementing easy downstream processing.37 Our approach achieves 99.4 ± 0.1% RBC removal, 98.4 ± 1.4% WBC removal, and 90.0 ± 2.6% platelet removal, positively addressing the clinical need.
Our method shows promising results that approach clinical expectations, making it a potential candidate for future clinical use. To translate these findings into the clinical setting, several aspects will require further investigation. First, our method was tested only with blood from healthy individuals that was spiked with bacteria. Septic patients could have higher levels of white blood cells, inflammatory proteins, and other substances that could potentially interfere with the separation process. Clinical studies would be necessary to investigate the potential impact of these effects. Second, even though we here evaluate the three most prevalent sepsis-causing bacterial species, the method should be tested with a broader panel of bacteria to evaluate its efficacy in identifying a clinically relevant range of bacterial species. Third, the clinical impact of the method is contingent on its ability to concatenate with rapid downstream identification of bacteria or AST. To determine whether additional sample preparation steps are required prior to downstream analysis, further investigation is necessary.
Methods
Device fabrication
The fluid guide was 3D-printed with a Form 3+ (Formlabs, USA) using clear V4 resin, cleaned in Form Wash (Formlabs, USA) using isopropanol (IPA) in an ultrasonic bath for 30 min, and cured in Form Cure (Formlabs, USA) with ultraviolet light for 40 min at 60 °C. Fluid guide dimensions are provided in ESI.† A Nuclepore track-etched polycarbonate membrane filter (Whatman, Cytiva, UK) with pore sizes 1.0, 2.0, or 3.0 μm was inserted in a guiding structure of the fluid guide and glued using double-sided tape (Scotch, USA). 50 mL centrifuge tubes (Corning, Mexico) were used for the experiments.Medium preparation
Broth was prepared by dissolving Luria low salt powder (L3397, Sigma-Aldrich, USA) in deionized water (DIW) at 25 g L−1 concentration followed by autoclaving. The density of broth was confirmed by weighing 10 mL volume on an electronic lab scale (n = 3). Density medium was prepared by mixing Lymphoprep (STEMCELL Technologies, Canada) and broth.Preparation of spiked blood
Fresh blood in EDTA tubes from healthy anonymous donors was purchased from the blood bank (Blodcentralen, Stockholm, Sweden) and used for experiments no later than one week after purchase. E. coli ATCC 11775 and K. pneumonia ATCC 13883 were obtained from Uppsala University, Uppsala, and S. aureus, is a clinical isolate from Karolinska Hospital, Stockholm. Bacteria were long-term stored at −80 °C in standard glycerol solution. The bacteria were grown overnight in LB broth media at 37 °C and reached the final concentration of around 109 CFU mL−1. The fresh bacteria culture was then diluted to 104, 103, or 102 CFU mL−1. 50–100 μL of the diluted culture (104, 103, or 102 CFU mL−1) was spiked into 3.5 mL of whole blood, acquiring bacteria concentrations of 103, 102, or 10 CFU mL−1 respectively. The concentration of the spiking solution was quantified by plate counting (n = 3).Post-centrifuge analysis
Filter retentate was prepared by placing the fluid guide, after removal from the centrifuge tube, into a clean centrifuge tube and resuspending the cells by pipetting. Filtrate was prepared by resuspending the cells in the liquid remaining in the centrifuge tube. The volume of the filtrate, Vfil, and retentate, Vret, were determined using a 10 mL glass pipette. Bacteria and RBC quantitation were performed on aliquots of the filtrate and retentate (n = 3).Bacteria quantitation
Agar plates were prepared by dissolving LB broth with agar (Miller) (Sigma-Aldrich, USA) in DIW at 40 g L−1 concentration, followed by autoclaving and transferring on clean plates. Bacteria quantitation was performed after plating the bacteria on the agar and overnight incubation at 37 °C.For the 10 CFU mL−1 spiked sample, we resorted to overnight culture in a culture bottle to obtain a yes/no answer on bacteria recovery because quantifying low bacteria concentration in the 18 mL sample of the bacteria collection tube using plate culture is cumbersome. For the experiments of recovering bacteria at 10 CFU mL−1 concentration, the fluid guide was discarded after centrifuging, and the broth was added to the remaining centrifuge tube, followed by overnight culture at 37 °C. The cultured tube would then be inoculated on agar plates to observe bacterial presence. Controls consisted of the overnight culture of the filtrate for non-spiked blood samples, demonstrating the potential for contamination.
RBC quantitation
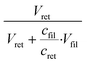
Both filter retentate and filtrate were added to 96-well plates (Avantor, VWR, USA) and diluted by DIW in the wells. The optical absorbance of hemoglobin (Hb) in each well was read by a SpectraMax 340PC384 Microplate Reader (Molecular Devices, USA) by setting the detection endpoint at 410 nm and the background at 500 nm.38 The ratio of RBC concentration in the filtrate over RBC concentration in the retentate, cfil/cret, was obtained from the Hb absorbance values (see Fig. S7† for details). RBC rejection from the bacteria collection tube was determined asWBC and platelet quantification
After centrifuging for 1 h at 4500g, the supernatant of the filtrate liquid in the centrifuge tube was removed by pipetting, and blood cells sedimented to the bottom of the centrifuge tube were concentrated to 1 mL suspension.17 The WBC and platelets counting of the whole blood and the filtrate was then performed using hematology analyzer Swelab Alfa Plus (Boule Diagnostics, Sweden). The rejection of WBCs and platelets by the filter was quantified by comparing the concentrations (WBC or platelet) in the filtrate with the concentrations (WBC or platelet) in whole blood (n = 3).Conclusions
We developed a centrifuge tube containing an inclined filter to separate bacteria from whole blood in a single centrifuging step. Combining centrifugation and size-based filtration addresses the current limitations of both methods, specifically, the limited density differentiation between bacteria and blood cells in end-point centrifugation and caking in blood filtration. As a result, we achieved relevant-throughput bacterial isolation with a unique combination and adaptation of well-known techniques. Within one hour, we recovered 32 ± 4% of E. coli, K. pneumonia, or S. aureus from 1 mL spiked blood, from concentrations as low as 100 CFU mL−1, while rejecting 99.4 ± 0.1% of the red blood cells, 98.4 ± 1.4% of the white blood cells, and 90.0 ± 2.6% of the platelets, and recovering bacteria at 10 CFU mL−1 concentration with positive results. Our technique thus uniquely provides efficient separation of low-concentration bacteria. Findings indicate that a shift in fluid guide material may further improve bacterial isolation by 67%.Our simple hands-off efficient separation of low-abundant bacteria and rejection of blood cells could form a great tool for sample preparation in conjunction with downstream (microfluidic) analysis techniques. The bacterial isolation from blood here demonstrated approaches clinical expectations, making the new method a promising candidate for future clinical use. Future work should investigate the performance of injection molded fluid guides, device performance for a broader panel of pathogens, handling of blood from infected patients, as well as the potential of combining this technique with downstream bacterial identification or AST, to enable staking out a road for translation to the clinical setting. Our results could thus form an important step to the rapid diagnosis of bloodstream infections.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded through the Swedish Foundation for Strategic Research (SSF) via the Agenda 2030 Research Centre “Ultra rapid antibiotic susceptibility determination” ARC19-0016.Notes and references
- K. E. Rudd, S. C. Johnson, K. M. Agesa, K. A. Shackelford, D. Tsoi, D. R. Kievlan, D. V. Colombara, K. S. Ikuta, N. Kissoon and S. Finfer, et al. , Lancet, 2020, 395, 200–211 CrossRef PubMed.
- R. S. Hotchkiss, L. L. Moldawer, S. M. Opal, K. Reinhart, I. R. Turnbull and J.-L. Vincent, Nat. Rev. Dis. Primers, 2016, 2, 1–21 Search PubMed.
- K. Thompson, B. Venkatesh and S. Finfer, Intern. Med. J., 2019, 49, 160–170 CrossRef PubMed.
- H. Arefian, S. Heublein, A. Scherag, F. M. Brunkhorst, M. Z. Younis, O. Moerer, D. Fischer and M. Hartmann, J. Infect., 2017, 74, 107–117 CrossRef PubMed.
- W. H. Organization, et al., Global report on the epidemiology and burden of sepsis: current evidence, identifying gaps and future directions, 2020 Search PubMed.
- P. Barman, S. Chopra and T. Thukral, J. Lab. Physicians, 2018, 10, 260–264 CrossRef PubMed.
- D. J. Castillo, R. F. Rifkin, D. A. Cowan and M. Potgieter, Front. Cell. Infect. Microbiol., 2019, 9, 148 CrossRef CAS PubMed.
- M. W. Pletz, N. Wellinghausen and T. Welte, Intensive Care Med., 2011, 37, 1069–1076 CrossRef CAS PubMed.
- L. S. Roope, R. D. Smith, K. B. Pouwels, J. Buchanan, L. Abel, P. Eibich, C. C. Butler, P. S. Tan, A. S. Walker and J. V. Robotham, et al. , Science, 2019, 364, eaau4679 CrossRef CAS PubMed.
- L. Evans, A. Rhodes, W. Alhazzani, M. Antonelli, C. M. Coopersmith, C. French, F. R. Machado, L. Mcintyre, M. Ostermann and H. C. Prescott, et al. , Intensive Care Med., 2021, 47, 1181–1247 CrossRef PubMed.
- S. A. Asner, F. Desgranges, I. T. Schrijver and T. Calandra, J. Infect., 2021, 82, 125–134 CrossRef CAS PubMed.
- T. Oeschger, D. McCloskey, V. Kopparthy, A. Singh and D. Erickson, Lab Chip, 2019, 19, 728–737 RSC.
- A. Kothari, M. Morgan and D. A. Haake, Clin. Infect. Dis., 2014, 59, 272–278 CrossRef PubMed.
- K. C. Tjandra, N. Ram-Mohan, R. Abe, M. M. Hashemi, J.-H. Lee, S. M. Chin, M. A. Roshardt, J. C. Liao, P. K. Wong and S. Yang, Antibiotics, 2022, 11, 511 CrossRef CAS PubMed.
- Ö. Baltekin, A. Boucharin, E. Tano, D. I. Andersson and J. Elf, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 9170–9175 CrossRef PubMed.
- M. Osaid, Y.-S. Chen, C.-H. Wang, A. Sinha, W.-B. Lee, P. Gopinathan, H.-B. Wu and G.-B. Lee, Lab Chip, 2021, 21, 2223–2231 RSC.
- W. G. Pitt, M. Alizadeh, G. A. Husseini, D. S. McClellan, C. M. Buchanan, C. G. Bledsoe, R. A. Robison, R. Blanco, B. L. Roeder and M. Melville, et al. , Biotechnol. Prog., 2016, 32, 823–839 CrossRef CAS PubMed.
- B. Forsyth, P. Torab, J.-H. Lee, T. Malcom, T.-H. Wang, J. C. Liao, S. Yang, E. Kvam, C. Puleo and P. K. Wong, Biosensors, 2021, 11, 288 CrossRef CAS PubMed.
- A. J. Mach and D. Di Carlo, Biotechnol. Bioeng., 2010, 107, 302–311 CrossRef CAS PubMed.
- H. W. Hou, R. P. Bhattacharyya, D. T. Hung and J. Han, Lab Chip, 2015, 15, 2297–2307 RSC.
- X. Lu, J. J. M. Chow, S. H. Koo, B. Jiang, T. Y. Tan, D. Yang and Y. Ai, Lab Chip, 2021, 21, 2163–2177 RSC.
- S. Narayana Iyengar, T. Kumar, G. Mårtensson and A. Russom, Electrophoresis, 2021, 42, 2538–2551 CrossRef CAS PubMed.
- P. Dow, K. Kotz, S. Gruszka, J. Holder and J. Fiering, Lab Chip, 2018, 18, 923–932 RSC.
- P. Ohlsson, K. Petersson, P. Augustsson and T. Laurell, Sci. Rep., 2018, 8, 9156 CrossRef PubMed.
- S. Li, F. Ma, H. Bachman, C. E. Cameron, X. Zeng and T. J. Huang, J. Micromech. Microeng., 2016, 27, 015031 CrossRef PubMed.
- S. Park, Y. Zhang, T.-H. Wang and S. Yang, Lab Chip, 2011, 11, 2893–2900 RSC.
- J. H. Kang, M. Super, C. W. Yung, R. M. Cooper, K. Domansky, A. R. Graveline, T. Mammoto, J. B. Berthet, H. Tobin and M. J. Cartwright, et al. , Nat. Med., 2014, 20, 1211–1216 CrossRef CAS PubMed.
- V. Liu, M. Patel and A. Lee, 17th International Conference on Miniaturized Systems for Chemistry and Life Sciences, 2013, pp. 27–31 Search PubMed.
- A. Lee, J. Park, M. Lim, V. Sunkara, S. Y. Kim, G. H. Kim, M.-H. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 11349–11356 CrossRef CAS PubMed.
- T.-H. Kim, M. Lim, J. Park, J. M. Oh, H. Kim, H. Jeong, S. J. Lee, H. C. Park, S. Jung and B. C. Kim, et al. , Anal. Chem., 2017, 89, 1155–1162 CrossRef CAS PubMed.
- K. D. Lenz, S. Jakhar, J. W. Chen, A. S. Anderson, D. C. Purcell, M. O. Ishak, J. F. Harris, L. E. Akhadov, J. Z. Kubicek-Sutherland and P. Nath, et al. , Sci. Rep., 2021, 11, 5287 CrossRef CAS PubMed.
- Y.-L. Fang, C.-H. Wang, Y.-S. Chen, C.-C. Chien, F.-C. Kuo, H.-L. You, M. S. Lee and G.-B. Lee, Lab Chip, 2021, 21, 113–121 RSC.
- M. Bernhardt, D. R. Pennell, L. S. Almer and R. Schell, J. Clin. Microbiol., 1991, 29, 422–425 CrossRef CAS PubMed.
- S. Kassegne, A. Khosla, D. Patel, N. Paramesh, N. Harwood and B. Arya, Microsyst. Technol., 2015, 21, 719–732 CrossRef CAS.
- D. J. Vitello, R. M. Ripper, M. R. Fettiplace, G. L. Weinberg and J. M. Vitello, J. Vet. Med., 2015, 2015, 152730 Search PubMed.
- M. Glynn, C. Nwankire, K. Lemass, D. J. Kinahan and J. Ducrée, Microsyst. Nanoeng., 2015, 1, 1–9 Search PubMed.
- K. Zhang, S. Qin, S. Wu, Y. Liang and J. Li, Chem. Sci., 2020, 11, 6352–6361 RSC.
- M. Alizadeh, R. L. Wood, C. M. Buchanan, C. G. Bledsoe, M. E. Wood, D. S. McClellan, R. Blanco, T. V. Ravsten, G. A. Husseini and C. L. Hickey, et al. , Colloids Surf., B, 2017, 154, 365–372 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00594a |
| ‡ These authors contributed equally to this work and are shared first authors. |
| This journal is © The Royal Society of Chemistry 2023 |