Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Efficient full-length IgG secretion and sorting from single yeast clones in droplet picoreactors†
Esteban
Lebrun
 ab,
Vasily
Shenshin
ab,
Vasily
Shenshin
 a,
Cécile
Plaire
a,
Cécile
Plaire
 a,
Vincent
Vigneres
c,
Théo
Pizette
a,
Bruno
Dumas
a,
Jean-Marc
Nicaud‡
a,
Vincent
Vigneres
c,
Théo
Pizette
a,
Bruno
Dumas
a,
Jean-Marc
Nicaud‡
 b and
Guillaume
Mottet‡
b and
Guillaume
Mottet‡
 *a
*a
aLarge Molecules Research, Sanofi, 94400 Vitry-Sur-Seine, France. E-mail: guillaume.mottet@sanofi.com
bUniversité Paris-Saclay, INRAE, AgroParisTech, Micalis Institute, 78350 Jouy-en-Josas, France
cArcale, 31100 Toulouse, France
First published on 6th July 2023
Abstract
The search for new antibodies is a major field of pharmaceutical research that remains lengthy and costly due to the need for successive library screenings. Existing in vitro and in vivo antibody discovery processes require that libraries are repeatedly subcloned to switch the antibody format or the secretory host, a resource-intensive process. There is an urgent need for an antibody identification platform capable of screening large antibody libraries in their final soluble format. Previous attempts to develop such a platform have struggled to combine large antibody libraries with screening of high specificity, while retaining sufficient library diversity coverage (ability to detect rare events). Here, we describe a new antibody screening platform based on the encapsulation of antibody secreting yeast cells into picoreactor droplets. We developed and optimized a Yarrowia lipolytica yeast strain capable of growing and secreting full-length human IgGs in picoreactors, and applied a microfluidics-based high-throughput screening approach to sort and recover target-specific antibody-secreting yeasts. Critically, the direct recovery of secretory yeasts allows for downstream screening and antibody characterization, without the need to reformat or subclone the coding sequences. We successfully increased the diversity coverage of sorting the antibody library without compromising sorting specificity by developing a new fluorescence signal processing methodology. By combining this drastically enhanced sorting efficiency with the high-throughput capability of droplet microfluidics, and the rapid growth of Y. lipolytica, our new platform is capable of screening millions of antibodies per day and enriching for target-specific ones in 4 days. This platform will enable the efficient screening of antibody libraries in a variety of contexts, including primary screening of synthetic libraries, affinity maturation, and identification of multi-specific or cross-reactive antibodies.
Introduction
Over the past decades, the identification of new monoclonal antibodies has become a major field of research in the pharmaceutical industry1–3 due to their strong therapeutic potential for various diseases.4 However, the process of antibody discovery remains lengthy and costly because of multiple selection steps, which do not systematically lead to the identification of potential pharmaceuticals.The usual in vitro antibody discovery process includes successive screenings of immune or synthetic libraries, starting with a selection based on display technologies (e.g., phage display, yeast display).5,6 These technologies enable the rapid testing of deep libraries but must be formatted as displayable antibody fragments such as single-chain variable-fragment (scFv) or fragment antigen-binding (Fab) to be produced and displayed efficiently by non-mammalian cells. After this first selection, the sequences of the binder antibody fragments need to be recovered, as DNA fragments or by sequencing, to be reformatted as full-length immunoglobulin (IgG).5–7 This smaller IgGs library can then be secreted by mammalian cells to perform characterization or secondary screenings based on the antibody functionality. In the case of single B cell screening, the first selection step can be performed directly with full-length IgG, but the reformatting step is still required to change the mouse antibody backbone to a human one, and because B cells cannot be grown efficiently after screening. In these processes, the reformatting step is time-consuming and subject to yield variation.
More importantly, none of these methods can be used to perform functional assays on soluble agonistic antibodies, as the latter must be in the final secreted IgG format to be fully functional. Recent studies have shown that more affine antibodies are not necessarily more functional, demonstrating the value of performing an initial functional assay as early in the process as possible, before investing significant resources in reformatting or producing binder antibodies.8–10 Thus, there is a need for a high-throughput platform for the primary screening of secreted full-length IgGs, allowing a selection as closely as possible to the final antibody format and the immediate availability of the secretory cells for further antibody characterization (e.g., ELISA, SPR, functional assay, …).
The greatest challenge of antibody library screening is to maintain the genotype–phenotype linkage during and after the selection process. Display technologies manage to keep the linkage at high throughputs, while secretion-based technologies rely on clonal isolation and sequestration, generally restricted to the use of multiple microwell plates or commercial devices with a few thousand nanowells.11 However, recent advances in droplet microfluidics opened the way to gathering confined environments and high-throughput screening into one single technology.12 Several in-droplet immunoassays have been developed during the past decade to screen antibody-secreting mammalian cells,13–15 hybridomas,16–20 or B lymphocytes.19,21 Although they have demonstrated their ability to isolate specific and even functional IgGs, these screening platforms are limited by the cell types they rely one. B lymphocytes cannot be grown after sorting, which requires the recovery of antibody sequences before further characterization; the production of a hybridoma library takes several months and cannot cover the diversity of the original immune library; and the transfectability of mammalian cells is too low to generate primary libraries as diverse as other synthetic libraries.5,6,22,23
Due to their robustness, fast growth, and engineerability, yeasts are emerging as promising alternatives to replace mammalian cells in a droplet-based antibody screening platform.24 Their good transformability allows the generation of relatively large gene libraries, which can be further enriched by yeast mating.25 Furthermore, yeasts are already established in the antibody discovery process5,6,23 and are known to be efficient hosts for producing complex molecules,26,27 including full-length IgGs.28–30 Some have even developed a switchable yeast-based system for IgG display and secretion, allowing rapid binding screening with the displayed format followed by characterization of the secreted antibodies in wells.25,31,32
Several attempts were performed to use yeasts as secretory cells for droplet-based high-throughput screening. These studies used yeasts to secrete enzymes,33 interleukins,34 and even antibodies.35 From an antibody screening perspective, the study of Fang et al. was promising, but inherent limitations in the gel droplet technology (e.g., polydisperse drop size, drop-to-drop antibody contamination, low encapsulation yield) led to numerous false positives and poor diversity coverage, making the process unreliable for deep library screening. These limitations shall be overcome by the rational generation of droplet emulsions coupled with direct droplet reading and sorting.
Herein, we report a novel approach for the high-throughput screening of secreted full-length human IgGs based on encapsulating secretory yeasts in picoliter reactors. To this end, the yeast Yarrowia lipolytica was chosen as chassis because of its great bioproduction and secretion abilities36–39 and its capability to grow in picoreactors.33 In this study, we describe the engineering of Y. lipolytica to achieve full-length human IgG secretion. It includes a deep screening of secretion signal peptides to build a robust antibody expression cassette and investigating several strain optimizations to increase antibody production. Then, building on prior work,21,40 we present an in-drop high-throughput immunoassay demonstrating the first efficient identification and enrichment of antibody-secreting yeasts. Finally, we validate the recovery of sorted yeasts for further antibody production and characterization, with no need for antibody reformatting.
Materials and methods
Vector and strain construction
Growth media and culture conditions
The E. coli strains were grown at 37 °C in lysogeny broth (LB) medium supplemented with either kanamycin sulfate (50 μg mL−1) or ampicillin (100 μg mL−1). The Y. lipolytica strains were grown at 28 °C in rich yeast extract–peptone–dextrose medium (YPD) or minimal yeast nitrogen base glucose medium (YNB), prepared as described below. The YPD medium contained 10 g L−1 of yeast extract (Difco, Paris, France), 10 g L−1 of peptone (Difco), and 10 g L−1 of glucose (Sigma Aldrich, Saint-Quentin Fallavier, France). The YNB medium contained 1.7 g L−1 of yeast nitrogen base without amino acids or ammonium sulfate (YNBww; Difco), 5.0 g L−1 of NH4Cl, and 50 mM phosphate buffer (pH 6.8). It was supplemented with either 10 g L−1 or 5 g L−1 of glucose, respectively for transformant selection on plate and droplet experiments. To meet the auxotrophic requirement, lysine (0.8 g L−1) or uracil (0.1 g L−1) were added to the culture medium when necessary. Solid media were created by adding 1.5% agar. IgG production was induced by adding 2.5 g L−1 of erythritol.Quantification of antibodies by biolayer interferometry (BLI)
The BLI was performed with an Octet QK System (Sartorius, NY, USA) and Protein A biosensors (Sartorius, #18-5012). Ninety microliters of culture supernatant was mixed with 10 μL of HBS 10× into a half-area 96-well plate (Greiner Bio-One, Les Ulis, France, #675076). Column 11 of the plate was filled with 100 μL per well of 10 mM glycine pH 1.5 for the regeneration of the sensors. Column 12 of the plate was filled with 100 μL per well of YDP for the neutralization of the sensors. In parallel, a standard made of several concentrations of purified antibodies (0, 5, 10, 20, 40, 60, 80, 100 μg mL−1) in YDP was prepared. The biosensors were rehydrated with YPD for 10 minutes before the dosage. Each sample was measured by BLI following the manufacturer's instructions and its antibody concentration was calculated based on the standard curve.Quantification of antibodies by enzyme-linked immunosorbent assay (ELISA)
An ELISA 96-well plate (Thermo Fisher Scientific, #442404) was coated overnight with 100 μL per well of solution A (16 mM Na2CO3 + 34 mM NaHCO3, pH 9.6) complemented with 2.5 μg mL−1 of anti-FC antibody (Jackson ImmunoResearch #109-005-098) and rinsed twice with 300 μL per well of solution B (PBS 1× + 0.05% Tween20). The plate was saturated for 1 hour with 200 μL per well of solution C (PBS 1× + 0.05% Tween20 + 1% BSA) before being rinsed twice with 300 μL per well of solution B. Each sample was diluted in solution C, and 100 μL of the diluted sample was placed into a well of the plate (raw supernatants were diluted by 1000; purified supernatants were diluted by 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). In parallel, a standard was prepared with a purified antibody (IgG1-kappa) ranging from 2 mg L−1 to 0.5 ng L−1 (serial dilution). The plate was incubated at room temperature for 1 hour before being rinsed 5 times with 300 μL per well of solution B. 100 μL of peroxidase-labeled anti-kappa antibody (Merck, Darmstadt, Germany, #A7164) diluted by 2000 in solution C was added into each well. The plate was incubated at room temperature for 1 hour before being rinsed 5 times with 300 μL per well of solution B. 100 μL of TMB (peroxidase substrate, Thermo Fischer Scientific, #34028) was added to each well for 15 minutes, and the reaction was stopped with 100 μL of hydrochloric acid. The OD450nm was measured in each well.
000). In parallel, a standard was prepared with a purified antibody (IgG1-kappa) ranging from 2 mg L−1 to 0.5 ng L−1 (serial dilution). The plate was incubated at room temperature for 1 hour before being rinsed 5 times with 300 μL per well of solution B. 100 μL of peroxidase-labeled anti-kappa antibody (Merck, Darmstadt, Germany, #A7164) diluted by 2000 in solution C was added into each well. The plate was incubated at room temperature for 1 hour before being rinsed 5 times with 300 μL per well of solution B. 100 μL of TMB (peroxidase substrate, Thermo Fischer Scientific, #34028) was added to each well for 15 minutes, and the reaction was stopped with 100 μL of hydrochloric acid. The OD450nm was measured in each well.
Quantification of antibody fragments by capillary electrophoresis
JMY8652 was grown in 10 mL of YPD into a leaned glass tube shaken at 160 rpm for 24 hours at 28 °C. This preculture was used for sowing 2 L baffled Erlenmeyer flasks at an OD600nm of 0.1 in 400 mL of YPD complemented with 2.5 g L−1 of erythritol. The flask was incubated for 48 hours at 28 °C with shaking at 100 rpm. After cultivation, the cultures were centrifuged to pellet the cells and the supernatants were filtered at 0.2 μm. Supernatant antibody fragments were purified on an OPUS column (Repligen, MA, USA) filled with 0.5 mL of MabSelect PrismA (Cytiva, MA, USA). Concentrated antibody fragments were eluted in 1 mL of acetate buffer pH 2.8 (acetate + sodium acetate in water) and desalted using a MidiTrap G25 (Cytiva) to resuspend them in PBS 1×. The final concentration of antibody fragments was estimated by measuring the OD280nm and measured by ELISA. The quality of produced antibodies was measured by capillary electrophoresis using the LabChip GX Automated Electrophoresis Systems (PerkinElmer, MA, USA) according to the manufacturer's instructions.Droplet microfluidic platform
The platform was based on the principle of fluorescence-activated droplet sorting (FADS) proposed by Baret et al.43 The platform was divided into five parts: (I) fluidic system, (II) excitation system, (III) microscope, (IV) sensor/calculator/actuator system, and (V) supervisor system. (I) The fluidic system enabled to control the flows inside the microfluidic chip. It was based on syringe pumps (neMESYS Low Pressure, Cetoni, Korbußen, Germany) and a pressure controller (LineUP Flow EZ, Fluigent, Le Kremlin-Bicêtre, France). (II) The excitation system was made with a four-laser box (405 nm, 488 nm, 561 nm, and 647 nm, Oxxius, Lannion, France) which was coupled to an optical fiber and a Powell lens to transform the laser dots into laser lines. (III) The RAAM microscope (ASI, OR, USA) was equipped with a 40× objective (ELWD 40XC O.N 0.6, Nikon, Tokyo, Japan) and an optomechanical part: Semrock for the filter (IDEX Health & Science, WA, USA) and Thorlabs (NJ, USA) for the mechanical components. (IV) The optomechanical part enabled connecting the optical fiber to bring the lasers to the objective and irradiating the microfluidic chip's channel. Similarly, it brought the fluorescent signal through different filters to the PMTs (Hamamatsu Photonics K.K., Shizuoka, Japan, #H10723). Fluorescent signals detected by the PMTs were converted into a voltage proportional to the photon flow. The analogic voltage signal emitted by the PMTs was digitized and analyzed with the fast electronic chips USB-7856R FPGA Kintex-7 160T (NI, TX, USA) and a 16 bits digital analogic convertor (DAC) with a conversion time of 1 μs per channel. Droplet detection was done by threshold crossing of one designated PMT leader at 200 kHz (average of 5 acquisitions for noise reduction). While the PMT leader was above the defined threshold, information about extreme values and derivative sign changes was memorized. The information was then interpreted when the PMT leader passed under the threshold. When the user-defined requirements were met for the droplet, a signal was generated by the DAC and passed through the high voltage amplificator TREK 2220-CE (Advanced Energy, CO, USA) a few microseconds after the droplet detection. This caused the positive droplets to be deviated in the sorting channel by dielectrophoresis. (V) The dedicated Labview human-machine interface allowed controlling PMTs supply, visualizing PMTs signals, displaying detected droplets data in 3D graphs, and setting all requirements on droplet detection and sorting.Microfluidic chips
The microfluidic chips were made with the standard process: PDMS structures obtained by a casting on a SU-8 mold were bounded on a glass slide.44 We used two kinds of microfluidic chips. The first one was a droplet maker with three entrances: two for the aqueous phases and one for the HFE oil (Novec 7500, 3M, MN, USA) mixed with 2% of surfactant (RAN Biotechnologies, MA, USA). One outlet enabled collecting the emulsion in a dedicated tube. The second microfluidic chip was made for droplet sorting. One entrance was dedicated to the emulsion and another enabled droplet spacing by oil injection. Two outlets allowed us to separate sorted drops from discarded ones (waste channel). Three other channels were made to build the electrodes by melted indium injection. Both microfluidic chips were manufactured with a channel thickness of 50 μm.Droplet production, collection, and sorting
The first step consisted of aspirating aqueous phases into two 200 μL tips filled with HFE oil at 1500 μl h−1. The two tips were then plugged in the PDMS chip. The droplet production was made by pushing the aqueous phases at 400 μL h−1 and the oil phase at 1200 μL h−1. It generated 30 pL droplets at 5000 Hz. The emulsion was collected into a 1.5 mL tube filled with oil and closed by a PDMS plug with two entrances: one bringing the emulsion and another removing the oil. The sorting was done by pushing the emulsion out of the collection tube at 100 μL h−1 and injecting oil at 300 mBar to space the drops, resulting in a flow rate of approximately 500 drops per second. When a drop of interest was detected, an electric field was applied to sort it (300 V, 10 kHz, 150 μs).In-droplet immunoassay
A bioassay solution containing 20 nM of Alexa647 anti-huFc F(ab′)2 (Jackson ImmunoResearch, PA, USA, #309-606-008), 10 nM of proprietary His-tagged CD86, 12 nM of Alexa 488 anti-His IgG (Cell Signaling Technology, MA, USA, #14930S), and 2.5 g L−1 of erythritol in YNB was centrifuged for 1 h at 4 °C/21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g to pellet fluorescent aggregates. Streptavidin-coated magnetic nanoparticles (Ademtech, Pessac, France, #3231) were incubated for 30 min in DPBS supplemented with saturating concentrations of biotinylated anti-huKappa VHH (Thermo Fisher Scientific). After washing with DPBS, the nanoparticles were incubated for 20 minutes in Pluronic. Finally, the nanoparticles were resuspended into the bioassay solution. In parallel, overnight yeast cultures were rinsed twice with DPBS, filtered at 5 μm to avoid yeasts aggregates, and diluted to reach a cell density between 10 and 30 million cells per milliliter, according to the targeted Poisson distribution. The bioassay solution and the yeasts were co-encapsulated as separated aqueous flows. Recorded data were analyzed afterward using FlowJo software.
000 g to pellet fluorescent aggregates. Streptavidin-coated magnetic nanoparticles (Ademtech, Pessac, France, #3231) were incubated for 30 min in DPBS supplemented with saturating concentrations of biotinylated anti-huKappa VHH (Thermo Fisher Scientific). After washing with DPBS, the nanoparticles were incubated for 20 minutes in Pluronic. Finally, the nanoparticles were resuspended into the bioassay solution. In parallel, overnight yeast cultures were rinsed twice with DPBS, filtered at 5 μm to avoid yeasts aggregates, and diluted to reach a cell density between 10 and 30 million cells per milliliter, according to the targeted Poisson distribution. The bioassay solution and the yeasts were co-encapsulated as separated aqueous flows. Recorded data were analyzed afterward using FlowJo software.
Statistics
Statistics were calculated with Prism software. When the aim was to determine the mean value of biological replicates, the Standard Error of the Mean (SEM) was indicated. When the aim was to determine the variability of experimental replicates, the Standard Deviation (SD) was indicated. An unpaired t-test was used when comparing two independent experimental conditions. Ordinary one-way ANOVA was used when comparing an experimental control condition with several treatments/mutations.Results and discussion
Evaluating a set of signal peptides combinations for antibody production
Regarding the production of complex molecules such as antibodies, the secretion pathway is a significant bottleneck that can lead to cell toxicity, protein aggregation when it is overwhelmed, and ultimately no secretion. Aside from strain secretion efficiency, protein secretability is strongly dependent on the signal peptide (SP) that leads it through the cell, and the SP's efficiency is itself dependent on the following protein sequence. To find the best SPs for the antibody heavy chain (HC) and light chain (LC), we have designed a cloning strategy based on a Y. lipolytica's integrative vector through which multiple antibody's variable sequences and SPs can be tested in parallel. This vector (GGE429; Fig. S1†) contains the constant sequences of the human IgG1 heavy chain (CH) and the human kappa light chain (CL), along with Golden Gate cloning sites for the integration of the variable sequences of the light chain (VL) and the heavy chain (VH), the two SPs, and the promoter (pEYK). The final structure of the assembled plasmid is presented in Fig. 1a. pEYK stands for the previously described hybrid promoter pEYK450-5AB and was chosen because it is erythritol-inducible and conducts bidirectional transcription.45 These characteristics allow to control IgG production initiation, improve the genetic stability of the IgG expression cassette,46 and allow the co-expression of both the HC and the LC. The use of a bidirectional promoter also facilitates the transfer of the expression cassette from a yeast secretory strain to a mammalian cell line while maintaining the pairing of the antibody heavy and light chains.47 Moreover, pEYK exhibits a strength ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 between the two transcriptional directions, which is known to be beneficial for antibody production.48 We selected 13 SPs (Y. lipolytica's SP1 to 10 as described by Celińska et al.;49 SSAF, the S. cerevisiae's pre-pro SP of the mating pheromone alpha-factor;50 SSLP, Y. lipolytica's pre-pro SP of the triacylglycerol lipase LIP2;51 and SSAEP, Y. lipolytica's pre-pro SP of the alkaline extracellular protease XPR2 (ref. 52)) and performed pooled assemblies to generate a theoretical set of 169 IgG expression cassettes, each with a unique combination of SPs along with the variable sequences of the anti-CD3 antibody muromonab-CD3 (OKT3 (ref. 53)) (Fig. S1†). During secretion, SPs were cleaved by the cellular machinery to release unaltered IgGs in the medium. For further reference, the SPs combinations are named by giving first the LC SP and then the HC SP (e.g., SP1-SP2).
3 between the two transcriptional directions, which is known to be beneficial for antibody production.48 We selected 13 SPs (Y. lipolytica's SP1 to 10 as described by Celińska et al.;49 SSAF, the S. cerevisiae's pre-pro SP of the mating pheromone alpha-factor;50 SSLP, Y. lipolytica's pre-pro SP of the triacylglycerol lipase LIP2;51 and SSAEP, Y. lipolytica's pre-pro SP of the alkaline extracellular protease XPR2 (ref. 52)) and performed pooled assemblies to generate a theoretical set of 169 IgG expression cassettes, each with a unique combination of SPs along with the variable sequences of the anti-CD3 antibody muromonab-CD3 (OKT3 (ref. 53)) (Fig. S1†). During secretion, SPs were cleaved by the cellular machinery to release unaltered IgGs in the medium. For further reference, the SPs combinations are named by giving first the LC SP and then the HC SP (e.g., SP1-SP2).
Up to four clones expressing each unique antibody expression cassette were grown for two days with erythritol to induce the production of OKT3. Then, supernatants' antibody concentrations were measured by biolayer interferometry (BLI) (Fig. S2†) and enzyme-linked immunosorbent assay (ELISA) (Fig. 1b).
The BLI method is based on the affinity between the bacterial protein A and the fragment crystallizable region (Fc) of several antibodies, such as the human IgG1. Thus, the BLI provided information on the concentration of secreted Fc fragments and unassembled HC in the supernatant, which was helpful in combination with ELISA to determine the quality of secreted IgGs. However, it did not give information on antibodies' folding nor the presence of the LC. By contrast, the ELISA method is based on a double binding of both the HC and the LC. Thus, the ELISA provided us with accurate information on secreted full-length antibodies and half-antibodies concentrations. This dosage revealed that we were able to produce up to 11 nM of OKT3 in yeast supernatant with substantial variations depending on the SPs combination used (Fig. 1b). Thirty-five combinations led to almost no detectable IgG production, and only 20% led to a concentration greater than 6.5 nM, which we consider a threshold for in-droplet IgG and IgG-antigen binding detections (Fig. S3†). Additionally, several SPs appeared less efficient than others in producing IgGs, regardless of their associated combination: IgG production was negatively affected when SP2, SP7, and SP9 were used to secrete the LC and when SP3 and SP8 were used to secrete the HC. We assume this chain-dependent impact resulted from protein–protein interactions preventing the cell machinery from recognizing the SP. On the other hand, every SPs combination containing SP10, SSAF, SSLP, or SSAEP led to weak IgG secretion, regardless of their linked IgG chain.
BLI and ELISA measurements were mostly correlated (Fig. S4†) but interestingly, the concentration measured by ELISA was on average 24 times and up to hundreds of times lower than the concentration measured by BLI, depending on the SP combination used. It suggests that a large proportion of HC and LC produced by the yeasts did not correctly assemble into antibodies. For the 12 best SPs combinations (SP1-SP4, SP1-SP6, SP3-SP1, SP3-SP4, SP3-SP6, SP4-SP1, SP4-SP7, SP5-SP1, SP5-SP4, SP6-SP4, SP8-SP6, and SP6-SP6), the estimated proportion of full-length IgGs in the antibody fragments mixture was between 4.1% and 8.3%. This hypothesis was validated by purifying the antibody fragments from the SP4-SP1 supernatant and performing capillary electrophoresis (Fig. 1c). Capillary electrophoresis results are difficult to quantify, but we estimated that 4.9% of supernatants antibody fragments were detectable in ELISA (at least one HC and one LC), and only 1.3% were full IgGs.
Although the proportion of assembled IgGs in supernatants was relatively weak, it demonstrated the first full-length IgG secretion by Y. lipolytica.
Testing the robustness of SPs combinations for antibody production
To select the best SPs combination for producing a large variety of antibodies, we tested the robustness of the 12 combinations that have led to the highest OKT3 productions. These SPs combinations were assembled into GGE429 with the variable sequences of five other antibodies: three anti-CD3, one anti-CD86, and one anti-TNP. Among the six tested antibodies, the VL sequence variability was up to 51%, and the VH variability was up to 45%. Up to eight clones expressing each antibody expression cassette were cultivated for two days with erythritol, and the antibody fragments concentrations in the filtered supernatants were measured by ELISA (Fig. 1d).Interestingly, the concentrations of supernatant antibodies were similar for each IgG across the different SPs combinations, but we identified strong variations from one IgG to another, ranging from an average of 0.3 nM for anti-TNP to up to 35 nM for anti-CD3 Blinatumomab. Thus, for the selected SP combinations, the IgG production efficiency mainly depended on the IgG variable sequences and less on the SPs. It suggests that each of these SPs combinations could be used independently in the context of library secretion. For further experiments, we kept the combination SP4-SP1 for FUN-1 (anti-CD86) and OKT3 (anti-CD3), and SP3-SP6 for the anti-TNP IgG because other combinations led to a barely detectable concentration for this IgG. With these SPs combinations, the three antibodies were produced at 1.1, 10.6, and 0.9 nM, respectively.
Optimizing Yarrowia lipolytica for antibody secretion in droplets
Given the specific context of in-droplet secretion, we needed to tune our yeast strain to allow its encapsulation and maximize the concentration of secreted IgGs.Y. lipolytica can form hyphae when exposed to various stressful conditions, including high cell density. This feature would have been a significant obstacle for single-cell encapsulation and subsequent droplet integrity, with the risk of drop-to-drop yeast contamination. Thus, we needed to control and block this morphological form of our yeast strain to achieve efficient encapsulation. We used CRISPR-Cas9 to perform a deletion of MHY1 (YALI1_B28150g), the responsible gene for Y. lipolytica's filamentous growth54 (Fig. 1e). In order to assess the effect of MHY1 deletion on IgG secretion, JMY9320 (OKT3_SP4-SP1) and JMY8652 (OKT3_SP4-SP1 Δmhy1) strains were grown for two days with erythritol, and antibody fragments concentrations in the filtered supernatants were measured by ELISA (Fig. 1f). The deletion of MHY1 caused a 14.2% (± 5.8%) decrease in OKT3 supernatant concentration, but the secretion efficiency remained higher than the theoretical in-drop detection limit (i.e., 6.5 nM; Fig. S3†).
To increase the supernatant concentration of secreted IgGs and decrease the number of unassembled IgG fragments, we drew on previous studies29,55 to optimize the yeast strain by over-expressing five molecular chaperones: the Y. lipolytica's protein disulphide isomerase (ylPDI), the human protein disulphide isomerase (huPDI), the Y. lipolytica's endoplasmic reticulum (ER) oxidoreductin (ERO1), the Y. lipolytica's binding immunoglobulin protein (BiP), and the Y. lipolytica's nucleotide exchange factor SLS1. PDI and ERO1 are ER-resident enzymes that catalyze disulfide bond formation within proteins. BiP binds newly synthesized proteins during ER translocation and maintains them in a competent state for subsequent folding and oligomerization. SLS1 is an ER-resident protein interacting with BiP for protein translocation. We also tested the overexpression of HAC1, a Y. lipolytica's transcriptional activator involved in the unfolded protein response that activates the production of ER chaperones such as PDI and BiP.56 The effect of these optimizations on IgG secretion was assessed, and we observed an increase in secreted IgGs concentrations by 7% with ylPDI and 25% with huPDI (Fig. S5†). It constitutes the first step for improving IgG secretion by Y. lipolytica. However, due to experiment parallelization, this improvement was not included in the following experiments.
Additionally, the previously mentioned capillary electrophoresis performed on the yeast supernatant revealed that 80% of secreted LC and 84% of secreted HC were glycosylated (Fig. S6†). Because of significant differences between human and yeast glycosylations, which can be much more ramified than human ones, we assumed that the Y. lipolytica's glycosylations could be responsible for the weak proportion of properly folded IgGs in the supernatant. Moreover, the VH of OKT3 contains one free cysteine, which could also have disturbed its folding. Thus, with the same objective to increase the purity of secreted IgGs, we mutated the sequence of OKT3 to remove both glycosylation sites and the free cysteine. However, no significative improvement in secreted IgGs concentration could be achieved (Fig. S7†).
Assessing Yarrowia lipolytica's secretion efficiency in droplets
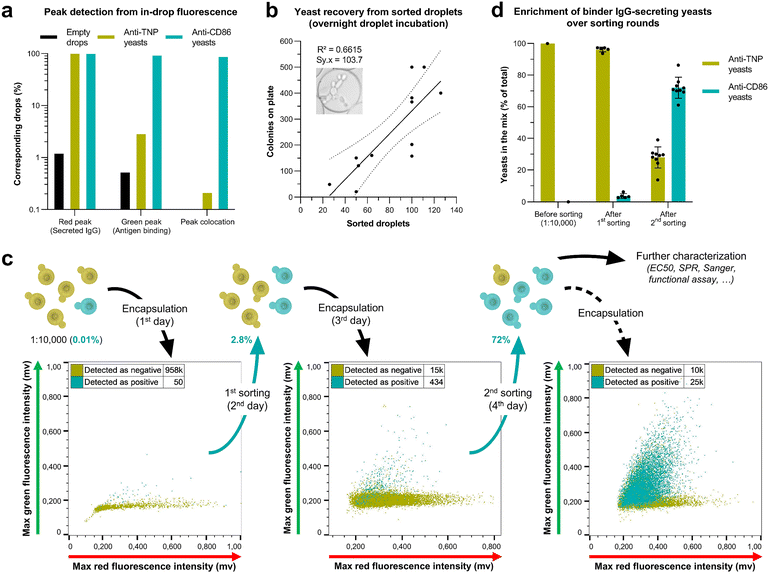
To evaluate the ability of Y. lipolytica to secrete IgGs in picoliter droplets, we adapted the fluorescence-based in-droplet single-cell bioassay developed by Eyer et al.21,40 to encapsulate IgG-secreting yeasts and detect the secretion of the anti-CD86 IgG FUN-1 (Fig. 2). The method developed by Eyer et al. is based on the use of magnetic nanoparticles capable of forming a bead line under the effect of a magnetic field. The huge advantage of using these microline-forming nanoparticles instead of microparticles or antigen-exposing cells is that they eliminate the limitations of a double Poisson distribution. Indeed, Poisson's law still applies when encapsulating the nanoparticles, but each drop of the emulsion ultimately contains a single bead line, which allows estimating the amount of IgGs secreted in each drop as well as their avidity. Our new immunoassay rests on magnetic nanoparticles coated with anti-kappa heavy-chain antibodies (VHH), whose function is to cluster secreted IgGs onto the bead line. When IgGs are secreted in the drop, they are also bound by Alexa647-fused anti-Fc F(ab′)2, which leads to aggregation of red fluorescence on the bead line. If secreted IgGs can bind to the antigen, here Alexa488-fused CD86, green fluorescence is also aggregated on the bead line. Thus, droplets containing binder IgGs can be sorted by reading their internal fluorescence, and secretory yeasts can be recovered for further antibody characterization.To assess the efficacy of this method, we used a droplet-maker microfluidic chip to generate picoliter drops containing a single anti-CD86-secreting yeast (positive control) or anti-TNP-secreting yeast (negative control). To ensure single-cell encapsulations, yeasts were diluted in accordance with the Poisson distribution (λ = 0.1; 10% of yeast-containing drops). Drops were incubated overnight at room temperature for yeasts to grow and produce IgGs. Then, the drops were re-injected into a sorting microfluidic chip, where they were scanned with overlaid laser lines, and their internal fluorescence distribution was read using photomultiplier tubes (PMTs). Furthermore, the fluorescence distribution was also visualized by fluorescence microscopy. As expected, drops containing anti-CD86-secreting yeasts exhibited red and green fluorescence aggregation on the bead line, while those containing anti-TNP-secreting yeasts had a diffuse green fluorescence and only exhibited aggregation of red fluorescence on the bead line (Fig. 3a; Video S1†). It translates into either overlapping green and red peaks or one red peak alone when reading the drops by laser scanning. Interestingly, fluorescence intensities displayed by the bead line were much higher than expected based on IgG concentrations measured by ELISA in yeast supernatants. By reading maximum red and green fluorescence intensities in droplets containing various concentrations of purified IgGs, we assessed that the smallest detectable IgG concentration with our microfluidic platform was around 6.5 nM (Fig. S3†). Measured fluorescence intensities in yeasts-containing drops, together with their discriminability from empty drops, revealed that in-drop yeasts-secreted IgGs concentrations were equivalent to this threshold. It suggests a higher quality of IgGs secreted at the single-cell scale than those secreted in batch cultures, potentially due to less cell stress.
In the context of antibody library screening, most of the secreting cells must be able to secrete IgGs at a detectable scale. Thus, we wanted to determine the proportion of encapsulated yeasts that secreted IgGs. To this end, the antibody-secreting strains were transformed with a yellow fluorescent protein (YFP) expression cassette, which acted as a cell tracer to discriminate between empty drops and encapsulated yeasts (Fig. 3b). It pointed out a decrease in size from yeast-containing droplets during overnight incubation (Fig. 3c and S8†). Although unexpected, this phenomenon was described by Hofmann et al. as a result of an osmotic pressure decrease in yeast-containing drops, leading to water transfer from them to empty drops.57 We also hypothesize that it could result from increased drop density while the medium is converted into cells. This fortunate discovery provided a way to isolate yeast-containing drops from empty ones without needing an extra cell tracer, thus releasing one fluorescence channel for additional usage.
Then, maximum red fluorescence in drops containing IgG-secreting yeasts was compared to that in empty drops and drops containing non-secreting yeasts. In doing so, we demonstrated that both the anti-TNP and the anti-CD86 strains could secrete detectable IgGs with 99% efficacy, revealing the remarkable survival of encapsulated yeasts (Fig. 3d). Importantly, this tremendous efficacy was only observed when the emulsion was incubated into a large volume of HFE oil (Fig. S9†), which acted as an essential oxygen supply for the yeasts.
Furthermore, to test the ability of the method to discriminate between binder and non-binder IgGs, we compared the maximum green fluorescence of droplets containing the anti-TNP and anti-CD86 yeasts. By setting the proper threshold of green fluorescence, we were able to isolate 86.5% of anti-CD86 drops, with very few false positives coming from anti-TNP drops (0.2%) (Fig. 3e). When manually examining the droplets' fluorescence profiles, we realized that green peaks were present in positive drops harboring a weak green fluorescence signal. However, the maximum green fluorescence reached by these little peaks was sometimes lower than the green fluorescence background in negative drops. This phenomenon is noticeable in the dot plots of Fig. 4c. Thus, the discrimination between anti-TNP and anti-CD86 yeasts has been further improved by replacing the maximum fluorescence reading with a real-time detection of fluorescence peaks inside the drops. Then, to minimize the number of false positive events coming from the detection of false peaks by the algorithm, drops were sorted when harboring a colocation of a read peak and a green peak. Through the use of fluorescence peak detection, we managed to increase the detection of positive events while preserving the sorting specificity (Fig. 4a and S10†). Thereby, we demonstrated the ability of encapsulated Y. lipolytica to secrete full-length IgGs, and we validated the efficacy of the new in-drop bioassay to identify a large proportion of binder IgG-secreting yeasts with few false positive contaminations.
Sorting and recovering binder antibodies in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 000 dilution
000 dilution
One assumed benefit of using yeasts instead of mammalian cells was their high robustness, which should have allowed the efficient and rapid recovery of ready-to-secrete cells after the screening process. To confirm this assumption, we sorted yeast-containing drops and evaluated the recoverability of sorted yeasts by spreading the emulsion on YPD-agarose plates and counting the number of colonies after overnight incubation. By sorting drops containing only one yeast each (after 1 hour drop incubation), we demonstrated that we could recover up to 100% of sorted yeasts, provided that at least a hundred drops were sorted (Fig. S11†). More interestingly, when sorting drops containing several yeasts (after overnight drop incubation), we recovered many more colonies on the plates, indicating droplet breaking and yeasts release during the spreading (Fig. 4b). It allowed the sorting of very few drops (at least 30), representing around one nanoliter of emulsion. This result demonstrated the ability of our process to isolate and recover very rare elements from a potential library.
Once the immunoassay efficiency, in-drop antibody secretion, and yeast recovery from sorted drops were validated, we mimicked an antibody library screening by mixing anti-CD86 and anti-TNP yeasts at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ratio (Fig. 4c). This mix was encapsulated into picoliter droplets at λ = 0.5 to maximize the proportion of yeast-containing drops (≃40%), thus reducing the time needed to sort enough positive drops. After overnight incubation, drops were read at 500 Hz, and those showing colocation of green and red fluorescence peaks were sorted (average of 1.6 positive events per minute) and poured into 10 mL of YPD. After a second overnight incubation, the sorted yeasts were encapsulated for a second sorting round at λ = 0.1 to minimize co-encapsulation of anti-TNP and anti-CD86 yeasts into the same drops. On the next day, drops with peak colocation were sorted and poured into 10 mL of YPD to recover the yeasts. Finally, after a last overnight incubation, yeasts were encapsulated again to evaluate the final anti-CD86 yeasts abundance in the cell mixture. The microfluidic measurements of anti-TNP and anti-CD86 yeasts abundance after the first and the second sortings were confirmed by Sanger sequencing the IgG expression cassette of hundreds of yeasts isolated from the mixture. With this two-round selection procedure, we were able to enrich the yeast population that secreted antigen-specific IgGs by an average of 7200-fold within four days. From a 1
000 ratio (Fig. 4c). This mix was encapsulated into picoliter droplets at λ = 0.5 to maximize the proportion of yeast-containing drops (≃40%), thus reducing the time needed to sort enough positive drops. After overnight incubation, drops were read at 500 Hz, and those showing colocation of green and red fluorescence peaks were sorted (average of 1.6 positive events per minute) and poured into 10 mL of YPD. After a second overnight incubation, the sorted yeasts were encapsulated for a second sorting round at λ = 0.1 to minimize co-encapsulation of anti-TNP and anti-CD86 yeasts into the same drops. On the next day, drops with peak colocation were sorted and poured into 10 mL of YPD to recover the yeasts. Finally, after a last overnight incubation, yeasts were encapsulated again to evaluate the final anti-CD86 yeasts abundance in the cell mixture. The microfluidic measurements of anti-TNP and anti-CD86 yeasts abundance after the first and the second sortings were confirmed by Sanger sequencing the IgG expression cassette of hundreds of yeasts isolated from the mixture. With this two-round selection procedure, we were able to enrich the yeast population that secreted antigen-specific IgGs by an average of 7200-fold within four days. From a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution of anti-CD86 yeasts, we brought them to 2.8% (± 1.5%) after the first sorting and 72.1% (± 6.7%) after a second enrichment (Fig. 4d).
000 dilution of anti-CD86 yeasts, we brought them to 2.8% (± 1.5%) after the first sorting and 72.1% (± 6.7%) after a second enrichment (Fig. 4d).
General discussion
In this study, we faced three major challenges: to develop a yeast strain capable of secreting full-length IgGs; to validate this secretion at the single-cell scale in picoliter reactors; and to detect and discriminate the secreted IgGs with high sensitivity. Over the last decade, very few examples of full-length IgG-secreting yeasts have been described. For this reason, we turned to the non-conventional yeast specie Y. lipolytica. This yeast has shown a good ability to secrete complex proteins, and a first proof of concept has demonstrated its compatibility with droplet microfluidics.33Knowing that it would be challenging to successfully secrete proteins as complex as IgGs, we screened 132 IgG expression cassettes with unique SPs combinations. This screening resulted in 80% of unusable IgG expression cassettes, demonstrating the relevance of this extensive SP screening strategy. Thus, we have demonstrated the first use of the yeast Y. lipolytica for the secretion of full-length human IgGs. Following these encouraging results, we have tested the robustness of the 12 best SPs combinations through the secretion of several IgGs and demonstrated that secretion efficiency depended more on the IgG sequence than on the SPs combination. It suggests that our secretion system would remain efficient for the secretion of most other IgGs with no need to find a suitable SPs combination again, which is essential from the perspective of antibody library screening. Regarding the promoter, the ability to induce IgG production with erythritol avoided IgG contamination of the medium prior to encapsulation into droplets, as seen in analogous studies.34,35 Additionally, we deleted the gene MHY1, responsible for Y. lipolytica's filamentous growth, to optimize the strain for encapsulation into picoliter droplets, avoid drop-to-drop yeast contamination (as seen previously33), and improve sorted yeasts recovery. Although Y. lipolytica could secrete IgGs, most HC and LC remained unassembled into the supernatant. To solve this issue, we made Y. lipolytica express several chaperones and removed glycosylation sites from secreted antibody sequences, but none of these attempts significantly increased supernatant IgG concentration. Nevertheless, the 25% increase in IgG concentration obtained by overexpressing huPDI is a first step towards the optimization of Y. lipolytica for IgG secretion at high concentration levels. Since Y. lipolytica is known to secrete various proteases, we believe that deleting some of them would be an efficient approach to further improve secreted IgGs concentration.
Droplet microfluidics is a powerful technology able to maintain the genotype–phenotype linkage for secretory applications without sacrificing the number of parallelized experiments. Besides, the miniaturization of single-cell experiments allows high-throughput screening of secreted libraries and drastically reduces the need for reagent volume. Herein, we demonstrated the ability of yeasts to grow and secrete full-length IgGs in picoliter reactors. Despite the low IgG concentrations produced by yeasts compared to mammalian cells, we have shown that yeast-secreted IgGs were highly detectable and distinguishable by our in-drop immunoassay. This statement was even true for the anti-CD86 and anti-TNP IgGs, which were the least secreted ones in our tests. Although we were successful in producing IgGs, the issue of nutrient and oxygen availability and metabolic waste accumulation is generally critical in a closed picoliter reactor. We quickly found that nutrients and waste were not a problem for Y. lipolytica's growth in picoreactors, but, as expected,58 oxygenation was a limitation in several experiments. Fortunately, one of the characteristics of fluorinated oil is that oxygen is highly soluble in it.59,60 Thus, by increasing the volume of fluorinated oil in the emulsion manifold, we directly increased the oxygen supply available to the yeast and allowed good IgG production. This approach should be transferable to reduce cell stress and increase cell productivity in any other experiment involving encapsulated yeasts.33,61
There are several advantages of having successfully produced IgGs in closed picoreactors. First, it is a one-step bioassay, which does not require washing or reagents addition during the process. Secondly, having picoreactors in emulsion form instead of agarose beads prevents contamination of antibodies from drop to drop (with the fluorinated oil and surfactant acting as a hydrophobic barrier60), allows rapid monodisperse encapsulations, and ensure low agglomeration and high transparency of reactors. These are significant improvements over previous agarose bead-based studies.34,35 Moreover, yeasts' ability to survive, divide and produce in picoreactors is a major asset for the screening process. Indeed, these abilities allowed yeast recovery from very few sorted droplets (equivalent to less than one nanoliter of emulsion), which means that our sorting platform is capable of isolating rare events. Furthermore, the great survival of yeasts after the sorting process and their ability to grow rapidly allowed us to reuse them in further screenings. It is therefore possible to run a series of enrichment cycles very quickly, or even to screen antibodies according to other criteria. This is a major improvement over display technologies or single B-cell screening, which require subcloning of antibody sequences to reformat them or modify the secretory host.
Another success of this study comes from our fast numerical analysis of the droplets' fluorescent signal. We have shown that basic signal analysis based on maximum intensities induces both false positives (low specificity) and false negatives (low diversity coverage). Previous studies have successfully enriched specific antibodies using a strict sorting threshold to increase the specificity of their method at the expense of the diversity coverage.34,35 This makes those platforms incompatible with antibody library screening under realistic conditions. Using an elementary algorithm based on a derivative calculation to detect fluorescence peaks and map the internal fluorescence of the drops, we increased our signal-to-noise ratio and thus increased the diversity coverage without decreasing its specificity. With this strategy, we demonstrated for the first time the ability of an IgGs screening platform to enrich target-specific antibodies by 7200-fold while capturing nearly 90% of positive events.
Conclusions
We have developed a new pipeline for the high-throughput screening of antibodies which combines the benefits of yeast surface display (easily editable and handleable yeasts and big antibody libraries) and mammalian cells libraries (secreted full-length IgGs). Yeast survivability and fast growth permitted successive sorting rounds to enrich the yeast population that secreted antigen-specific IgGs. Thus, we could use a permissive λ = 0.5 (≃40% occupancy) for the first round to increase sorting speed and, by extension, the maximum number of interrogated drops, and a stringent λ = 0.1 (≃10% occupancy, clonal distribution) for the second round to clean the sorted population from false positives. During a screening step, the microfluidic platform was able to read 1.8 million drops per hour (500 Hz), corresponding to 720k yeasts per hour (at λ = 0.5) and 3.6 million yeasts per run (5 hours of sorting). Thus, this new pipeline offers the potential to screen several million candidates within a few days with more reliability and reproducibility than previously described methods. Additionally, the antibody-secreting cells obtained after the enrichment can be used directly to produce IgGs for further functional assays without needing an antibody reformatting step. This pipeline is suitable for screening antibody libraries in a variety of contexts, such as primary screening of synthetic libraries, affinity maturation, and identification of multi-specific or cross-reactive antibodies. In the near future, it could also be used to screen immune libraries by pre-formatting with in-drop overlap extension PCR,62,63 or for primary screening based on a soluble agonistic functional assay. Furthermore, some could adapt it for high-throughput screening of any yeast strain and any protein whose secretion is necessary for its function, making it a universal platform for protein and strain selection in directed evolution.Author contributions
E. L. conducted the project, designed and performed the experiments, and analyzed the data. V. S. performed in-drop experiments and replicated the enrichment. C. P. determined optimal in-drop bioassay parameters. V. V. developed the LabVIEW software and the real-time peak detection algorithm. T. P. purified supernatant IgG fragments prior to capillary electrophoresis. G. M., J. M. N., and B. D. conceptualized and designed the project. G. M. and J. M. N. supervised the project. E. L. and G. M. wrote the manuscript. All authors reviewed and approved the final manuscript.Conflicts of interest
J.-M. Nicaud is a co-inventor listed on the EP patent application (EP17305575.7) filed on May 18, 2017, and on the EP patent PCT application (PCT/EP2018/062600; WO2018210867A1) filed on May 15, 2018, by the Université de Liege (ULG), Université Libre de Bruxelles (ULB) and Institut National de la Recherche Agronomique (INRA). These patents describe applications for the inducible promoter in gene expression and synthetic biology. There are no other conflicts to declare.Acknowledgements
Esteban Lebrun received a PhD scholarship funded by a CIFRE grant from the Sanofi R&D Department (n° 2019/1206). We thank Dana Brunner, Radi Khodr, and Solène Ludwig for their assistance during their internship, Evelyne Deschamps for her support with BLI experiments, Nathalie Couteault and Laurent Maton for their support with ELISA experiments, Léa Vidal for providing the chaperones CDS, Emmanuelle Vigne for her suggestions regarding the experimental strategy, and Thomas Bouquin for sponsoring the projetc. We thank Tristan Rossignol, Klervi Desrumeaux, Thomas Bouquin, Bruno Teste, and Caitlin Gillis for reviewing the final manuscript.References
- D. M. Ecker, S. D. Jones and H. L. Levine, mAbs, 2015, 7, 9–14 CrossRef CAS PubMed.
- A. L. Grilo and A. Mantalaris, Trends Biotechnol., 2019, 37, 9–16 CrossRef CAS PubMed.
- H. Kaplon, M. Muralidharan, Z. Schneider and J. M. Reichert, mAbs, 2020, 12, 1703531 CrossRef PubMed.
- S. Singh, N. K. Kumar, P. Dwiwedi, J. Charan, R. Kaur, P. Sidhu and V. K. Chugh, Curr. Clin. Pharmacol., 2018, 13, 85–99 CrossRef PubMed.
- P. J. Kennedy, C. Oliveira, P. L. Granja and B. Sarmento, Crit. Rev. Biotechnol., 2018, 38, 394–408 CrossRef CAS PubMed.
- J. Ministro, A. M. Manuel and J. Goncalves, in Current Applications of Pharmaceutical Biotechnology, ed. A. C. Silva, J. N. Moreira, J. M. S. Lobo and H. Almeida, Springer International Publishing, Cham, 2019, vol. 171, pp. 55–86 Search PubMed.
- R. H. Reader, R. G. Workman, B. C. Maddison and K. C. Gough, Mol. Biotechnol., 2019, 61, 801–815 CrossRef CAS PubMed.
- X. Yu, C. M. Orr, H. T. C. Chan, S. James, C. A. Penfold, J. Kim, T. Inzhelevskaya, C. I. Mockridge, K. L. Cox, J. W. Essex, I. Tews, M. J. Glennie and M. S. Cragg, Nature, 2023, 614, 539–547 CrossRef CAS PubMed.
- P. A. Mayes, K. W. Hance and A. Hoos, Nat. Rev. Drug Discovery, 2018, 17, 509–527 CrossRef CAS PubMed.
- J. S. Schardt, H. S. Jhajj, R. L. O'Meara, T. S. Lwo, M. D. Smith and P. M. Tessier, Drug Discovery Today, 2022, 27, 31–48 CrossRef CAS PubMed.
- M. Jorgolli, T. Nevill, A. Winters, I. Chen, S. Chong, F.-F. Lin, M. Mock, C. Chen, K. Le, C. Tan, P. Jess, H. Xu, A. Hamburger, J. Stevens, T. Munro, M. Wu, P. Tagari and L. P. Miranda, Biotechnol. Bioeng., 2019, 116, 2393–2411 CrossRef CAS PubMed.
- L. Weng and J. E. Spoonamore, Micromachines, 2019, 10, 734 CrossRef PubMed.
- Y. Wang, R. Jin, B. Shen, N. Li, H. Zhou, W. Wang, Y. Zhao, M. Huang, P. Fang, S. Wang, P. Mary, R. Wang, P. Ma, R. Li, Y. Tian, Y. Cao, F. Li, L. Schweizer and H. Zhang, Sci. Adv., 2021, 7, eabe3839 CrossRef CAS PubMed.
- W. N. Lin, M. Z. Tay, J. X. E. Wong, C. Y. Lee, S.-W. Fong, C.-I. Wang, L. F. P. Ng, L. Renia, C.-H. Chen and L. F. Cheow, Lab Chip, 2022, 22, 2578–2589 RSC.
- R. Gaa, K. Kumari, H. M. Mayer, D. Yanakieva, S.-P. Tsai, S. Joshi, R. Guenther and A. Doerner, Artif. Cells, Nanomed., Biotechnol., 2023, 51, 74–82 CrossRef CAS PubMed.
- B. E. Debs, R. Utharala, I. V. Balyasnikova, A. D. Griffiths and C. A. Merten, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 11570–11575 CrossRef PubMed.
- L. Mazutis, J. Gilbert, W. L. Ung, D. A. Weitz, A. D. Griffiths and J. A. Heyman, Nat. Protoc., 2013, 8, 870–891 CrossRef CAS PubMed.
- N. Shembekar, H. Hu, D. Eustace and C. A. Merten, Cell Rep., 2018, 22, 2206–2215 CrossRef CAS PubMed.
- R. Ding, K.-C. Hung, A. Mitra, L. W. Ung, D. Lightwood, R. Tu, D. Starkie, L. Cai, L. Mazutis, S. Chong, D. A. Weitz and J. A. Heyman, RSC Adv., 2020, 10, 27006–27013 RSC.
- J. Rutkauskaite, S. Berger, S. Stavrakis, O. Dressler, J. Heyman, X. Casadevall i Solvas, A. deMello and L. Mazutis, iScience, 2022, 25, 104515 CrossRef CAS PubMed.
- A. Gérard, A. Woolfe, G. Mottet, M. Reichen, C. Castrillon, V. Menrath, S. Ellouze, A. Poitou, R. Doineau, L. Briseno-Roa, P. Canales-Herrerias, P. Mary, G. Rose, C. Ortega, M. Delincé, S. Essono, B. Jia, B. Iannascoli, O. Richard-Le Goff, R. Kumar, S. N. Stewart, Y. Pousse, B. Shen, K. Grosselin, B. Saudemont, A. Sautel-Caillé, A. Godina, S. McNamara, K. Eyer, G. A. Millot, J. Baudry, P. England, C. Nizak, A. Jensen, A. D. Griffiths, P. Bruhns and C. Brenan, Nat. Biotechnol., 2020, 38, 715–721 CrossRef PubMed.
- A. Pedrioli and A. Oxenius, Trends Immunol., 2021, 42, 1143–1158 CrossRef CAS PubMed.
- H. R. Hoogenboom, Nat. Biotechnol., 2005, 23, 1105–1116 CrossRef CAS PubMed.
- A. Patel, U. Rova, P. Christakopoulos and L. Matsakas, Bioengineering, 2022, 9, 751 CrossRef PubMed.
- L. Rhiel, S. Krah, R. Günther, S. Becker, H. Kolmar and B. Hock, PLoS One, 2014, 9, e114887 CrossRef PubMed.
- R. Fleer, Curr. Opin. Biotechnol., 1992, 3, 486–496 CrossRef CAS PubMed.
- Recombinant Protein Production in Yeast, ed. B. Gasser and D. Mattanovich, Springer New York, New York, NY, 2019, vol. 1923 Search PubMed.
- A. H. Horwitz, C. P. Chang, M. Better, K. E. Hellstrom and R. R. Robinson, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 8678–8682 CrossRef CAS PubMed.
- T. Suzuki, S. Baba, M. Ono, K. Nonaka, K. Ichikawa, M. Yabuta, R. Ito and Y. Chiba, J. Biosci. Bioeng., 2017, 124, 156–163 CrossRef CAS PubMed.
- T. I. Potgieter, M. Cukan, J. E. Drummond, N. R. Houston-Cummings, Y. Jiang, F. Li, H. Lynaugh, M. Mallem, T. W. McKelvey, T. Mitchell, A. Nylen, A. Rittenhour, T. A. Stadheim, D. Zha and M. d'Anjou, J. Biotechnol., 2009, 139, 318–325 CrossRef CAS PubMed.
- J. A. Rakestraw, D. Aird, P. M. Aha, B. M. Baynes and D. Lipovsek, Protein Eng., Des. Sel., 2011, 24, 525–530 CrossRef CAS PubMed.
- H. Rouha, A. Badarau, Z. C. Visram, M. B. Battles, B. Prinz, Z. Magyarics, G. Nagy, I. Mirkina, L. Stulik, M. Zerbs, M. Jägerhofer, B. Maierhofer, A. Teubenbacher, I. Dolezilkova, K. Gross, S. Banerjee, G. Zauner, S. Malafa, J. Zmajkovic, S. Maier, R. Mabry, E. Krauland, K. D. Wittrup, T. U. Gerngross and E. Nagy, mAbs, 2015, 7, 243–254 CrossRef CAS PubMed.
- T. Beneyton, S. Thomas, A. D. Griffiths, J.-M. Nicaud, A. Drevelle and T. Rossignol, Microb. Cell Fact., 2017, 16, 18 CrossRef PubMed.
- D. Yanakieva, A. Elter, J. Bratsch, K. Friedrich, S. Becker and H. Kolmar, Sci. Rep., 2020, 10, 10182 CrossRef CAS PubMed.
- Y. Fang, T. H. Chu, M. E. Ackerman and K. E. Griswold, mAbs, 2017, 9, 1253–1261 CrossRef CAS PubMed.
- E. Celińska and J.-M. Nicaud, Appl. Microbiol. Biotechnol., 2019, 103, 39–52 CrossRef PubMed.
- J. Ma, Y. Gu, M. Marsafari and P. Xu, J. Ind. Microbiol. Biotechnol., 2020, 47, 845–862 CrossRef CAS PubMed.
- C. Madzak, Mol. Biotechnol., 2018, 60, 621–635 CrossRef CAS PubMed.
- C. Madzak, J. Fungi, 2021, 7, 548 CrossRef CAS PubMed.
- K. Eyer, R. C. L. Doineau, C. E. Castrillon, L. Briseño-Roa, V. Menrath, G. Mottet, P. England, A. Godina, E. Brient-Litzler, C. Nizak, A. Jensen, A. D. Griffiths, J. Bibette, P. Bruhns and J. Baudry, Nat. Biotechnol., 2017, 35, 977–982 CrossRef CAS PubMed.
- M. Larroude, Y. Park, P. Soudier, M. Kubiak, J. Nicaud and T. Rossignol, Microb. Biotechnol., 2019, 12, 1249–1259 CrossRef CAS PubMed.
- M. Larroude, H. Trabelsi, J.-M. Nicaud and T. Rossignol, Biotechnol. Lett., 2020, 42, 773–785 CrossRef CAS PubMed.
- J.-C. Baret, O. J. Miller, V. Taly, M. Ryckelynck, A. El-Harrak, L. Frenz, C. Rick, M. L. Samuels, J. B. Hutchison, J. J. Agresti, D. R. Link, D. A. Weitz and A. D. Griffiths, Lab Chip, 2009, 9, 1850 RSC.
- J. C. McDonald and G. M. Whitesides, Acc. Chem. Res., 2002, 35, 491–499 CrossRef CAS PubMed.
- L. Vidal, E. Lebrun, Y.-K. Park, G. Mottet and J.-M. Nicaud, Microb. Cell Fact., 2023, 22, 7 CrossRef CAS PubMed.
- D. Fiebig, J. P. Bogen, S. C. Carrara, L. Deweid, S. Zielonka, J. Grzeschik, B. Hock and H. Kolmar, Front. Bioeng. Biotechnol., 2022, 10, 794389 CrossRef PubMed.
- S. Schlatter, S. H. Stansfield and D. M. Dinnis, et al. , Biotechnol. Prog., 2005, 21(1), 122–133 CrossRef CAS PubMed.
- S. Schlatter, S. H. Stansfield, D. M. Dinnis, A. J. Racher, J. R. Birch and D. C. James, Biotechnol. Prog., 2005, 21, 122–133 CrossRef CAS PubMed.
- E. Celińska, M. Borkowska, W. Białas, P. Korpys and J. M. Nicaud, Appl. Microbiol. Biotechnol., 2018, 102, 5221–5233 CrossRef PubMed.
- D. Stotzler and W. Duntze, Eur. J. Biochem., 1976, 65, 257–262 CrossRef CAS PubMed.
- G. Pignède, H. Wang, F. Fudalej, C. Gaillardin, M. Seman and J.-M. Nicaud, J. Bacteriol., 2000, 182, 2802–2810 CrossRef PubMed.
- S. Matoba, J. Fukayama, R. A. Wing and D. M. Ogrydziak, Mol. Cell. Biol., 1988, 8, 4904–4916 CAS.
- C. Emmons and L. G. Hunsicker, Iowa Med., 1987, 77, 78–82 CAS.
- O. Konzock and J. Norbeck, PLoS One, 2020, 15, 1–11 CrossRef PubMed.
- B. Gasser, M. Maurer, J. Gach, R. Kunert and D. Mattanovich, Biotechnol. Bioeng., 2006, 94, 353–361 CrossRef CAS PubMed.
- M. H. Oh, S. A. Cheon, H. A. Kang and J.-Y. Kim, Yeast, 2010, 27, 443–452 CrossRef CAS PubMed.
- T. W. Hofmann, S. Hänselmann, J. Janiesch and C. H. J. Böhm, Impact of Osmosis on Micro-Droplets - A New Route to Novel Sensors, Seattle, Washington, USA, 2011 Search PubMed.
- M. Gorczyca, J. Kaźmierczak, S. Steels, P. Fickers and E. Celińska, Yeast, 2020, 37, 559–568 CrossRef CAS PubMed.
- M. A. Hamza, G. Serratrice, M. J. Stebe and J. J. Delpuech, J. Am. Chem. Soc., 1981, 103, 3733–3738 CrossRef CAS.
- P. Gruner, B. Riechers, L. A. Chacòn Orellana, Q. Brosseau, F. Maes, T. Beneyton, D. Pekin and J.-C. Baret, Curr. Opin. Colloid Interface Sci., 2015, 20, 183–191 CrossRef CAS.
- G. Woronoff, P. Nghe, J. Baudry, L. Boitard, E. Braun, A. D. Griffiths and J. Bibette, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10660–10666 CrossRef CAS PubMed.
- A. S. Adler, R. A. Mizrahi, M. J. Spindler, M. S. Adams, M. A. Asensio, R. C. Edgar, J. Leong, R. Leong and D. S. Johnson, mAbs, 2017, 9, 1270–1281 CrossRef CAS PubMed.
- B. Wang, B. J. DeKosky, M. R. Timm, J. Lee, E. Normandin, J. Misasi, R. Kong, J. R. McDaniel, G. Delidakis, K. E. Leigh, T. Niezold, C. W. Choi, E. G. Viox, A. Fahad, A. Cagigi, A. Ploquin, K. Leung, E. S. Yang, W.-P. Kong, W. N. Voss, A. G. Schmidt, M. A. Moody, D. R. Ambrozak, A. R. Henry, F. Laboune, J. E. Ledgerwood, B. S. Graham, M. Connors, D. C. Douek, N. J. Sullivan, A. D. Ellington, J. R. Mascola and G. Georgiou, Nat. Biotechnol., 2018, 36, 152–155 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00403a |
| ‡ These authors jointly supervised the work. |
| This journal is © The Royal Society of Chemistry 2023 |