Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interfacing centrifugal microfluidics with linear-oriented 8-tube strips and multichannel pipettes for increased throughput of digital assays†
Yu-Kai
Lai
 ab,
Yu-Ting
Kao
ab,
Yu-Ting
Kao
 b,
Jacob Friedrich
Hess
b,
Jacob Friedrich
Hess
 ab,
Silvia
Calabrese
ab,
Silvia
Calabrese
 a,
Felix
von Stetten
a,
Felix
von Stetten
 ab and
Nils
Paust
ab and
Nils
Paust
 *ab
*ab
aHahn-Schickard, Georges-Koehler-Allee 103, 79110 Freiburg, Germany. E-mail: Nils.Paust@Hahn-Schickard.de; Tel: +49 761 203 73245
bLaboratory for MEMS Applications, IMTEK – Department of Microsystems Engineering, University of Freiburg, Georges-Koehler-Allee 103, 79110 Freiburg, Germany
First published on 3rd May 2023
Abstract
We present a centrifugal microfluidic cartridge for the eight-fold parallel generation of monodisperse water-in-oil droplets using standard laboratory equipment. The key element is interfacing centrifugal microfluidics with its design based on polar coordinates to the linear structures of standard high-throughput laboratory automation. Centrifugal step emulsification is used to simultaneously generate droplets from eight samples directly into standard 200 μl PCR 8-tube strips. To ensure minimal manual liquid handling, the design of the inlets allows the user to load the samples and the oil via a standard multichannel pipette. Simulation-based design of the cartridge ensures that the performance is consistent in each droplet generation unit despite the varying radial positions that originate from the interface to the linear oriented PCR 8-tube strip and from the integration of linear oriented inlet holes for the multichannel pipettes. Within 10 minutes, sample volumes of 50 μl per droplet generation unit are emulsified at a fixed rotation speed of 960 rpm into 1.47 × 105 monodisperse droplets with a mean diameter of 86 μm. The overall coefficient of variation (CV) of the droplet diameter was below 4%. Feasibility is demonstrated by an exemplary digital droplet polymerase chain reaction (ddPCR) assay which showed high linearity (R2 ≥ 0.999) across all of the eight tubes of the strip.
Introduction
Digital assays allow for precise and absolute quantification of target molecules in certain dynamic ranges without the need for reference standards.1 They are widely used for the quantification of nucleic acids,2 the detection of low levels of proteins,3,4 measurements of enzymatic activities5 and for single cell phenotypic analysis.6 In order to detect single molecules, the target molecules are split into thousands or even millions of small partitions.7,8 Each partition carries a discrete number of target molecules and the distribution follows Poisson statistics. Absolute quantification of the target molecules is assessed by counting the ratio of partitions that contain a target molecule. From this binary “yes” or “no” answer, the term “digital” is derived.In recent years, emulsion-based approaches, which utilize water-in-oil (W/O) droplets as partitions, have become popular due to the possibility of high-throughput partitioning and flexible handling of the droplets.9 The droplets are generated by microfluidic devices10 based on different strategies, such as T-junction emulsification,11 flow-focusing,12 or step-emulsification.13,14 In addition to this classification by the droplet generation methods, microfluidic devices can be categorized into on-chip15–18 and off-chip19–27 systems. On-chip systems automate the complete workflow within a single microfluidic device; off-chip systems allow for dispensing of generated droplets into reaction tubes. Whereas integrated on-chip systems are typically limited in the degree of parallelization and require complex and costly instrumentation,28 off-chip systems allow for processing of subsequent workflows such as droplet visualization or thermal cycling29–31 with high flexibility and theoretically in a highly parallel manner. If the workflows can be performed on standard laboratory equipment, a cost-efficient automation strategy that does not require additional lab space is provided.
To date, several off-chip systems for droplet generation in reaction tubes have been developed.19–27 Some require syringe pumps for flow control,19–22 which makes it difficult to parallelize droplet generation of multiple samples. Moreover, microfluidic setups (e.g., syringe pumps and tubing) often require special knowledge for operation, thus, adopting them to different laboratories is challenging. Commercial instruments, which can be easily applied in different laboratories, often come at high costs (e.g., QX 200™ ∼80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–100
000–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 USD, Bio-Rad, USA). Off-chip systems based on centrifugal microfluidics can generate droplets on standard centrifuges without the need for specialized equipment.23–27 Current solutions typically consist of a cartridge or a syringe needle27 that is placed into a reaction tube for droplet generation, which allows the user to emulsify one sample per tube. Schulz et al.23 presented a cartridge using centrifugal step-emulsification,32 which enables the generation of monodisperse droplets in 2 ml reaction tubes on standard centrifuges. However, similar to other off-chip solutions, this solution is not intended for increased throughput sample processing. Each cartridge must be loaded manually in the tubes, which results in an increase of manual pipetting steps with the rising number of samples.
000 USD, Bio-Rad, USA). Off-chip systems based on centrifugal microfluidics can generate droplets on standard centrifuges without the need for specialized equipment.23–27 Current solutions typically consist of a cartridge or a syringe needle27 that is placed into a reaction tube for droplet generation, which allows the user to emulsify one sample per tube. Schulz et al.23 presented a cartridge using centrifugal step-emulsification,32 which enables the generation of monodisperse droplets in 2 ml reaction tubes on standard centrifuges. However, similar to other off-chip solutions, this solution is not intended for increased throughput sample processing. Each cartridge must be loaded manually in the tubes, which results in an increase of manual pipetting steps with the rising number of samples.
This paper describes a novel “off-chip” cartridge design strategy that seamlessly adapts linear-oriented laboratory tools to centrifugal microfluidics, enabling increased throughput of digital assays by employing centrifugal step emulsification. The microfluidic cartridge, specifically designed according to this strategy, supplies droplets to a standard 200 μl PCR 8-tube strip, not only to enhance the throughput of sample processing but also to fit to the format of standard laboratory instrumentation. Up to 50 μl aqueous sample in each droplet generation unit (DGU) can be emulsified in fluorinated oil. A multichannel pipette supplies the oil and the sample. We developed a simulation-supported fluidic cartridge design strategy that connects the centrifugal microfluidic system in polar coordinates to both the linear-oriented 8-tube strip and the linear-oriented multichannel pipette while maintaining similar flow conditions in each DGU to ensure the supply of monodisperse droplets to each of the eight tubes. The cartridge design strategy is validated through experimental evaluation of droplet generation and assay performance. The cartridge was manufactured by hot embossing. It is amenable to injection molding and insertion into the centrifuge is very simple. Thus, we introduce a cost-efficient and convenient tool for laboratory automation of digital assays.
Results and discussion
Automated workflow
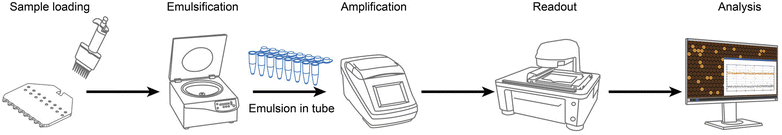
The laboratory workflow based on the new multi-sample emulsification cartridge is depicted in Fig. 1. First, the sample and oil are loaded into the droplet generation cartridge by a standard multichannel pipette. The cartridge is installed on the centrifuge for emulsification directly into the standard PCR 8-tube strip. After PCR amplification in a thermal cycler, the emulsion is ready for downstream processing, such as image-based droplet analysis as demonstrated in this work, or demulsification within a sequencing workflow. PCR amplification in droplets has been adopted for amplifying rare target in sequencing workflows, e.g., single-cell whole-genome sequencing,30,31,33 offering advantages like reduced competition and bias. The cartridge presented in this work is compatible with the aforementioned sequencing workflow, making it potentially suitable for sequencing applications as well. However, investigating this aspect is beyond the scope of this work.Working principle
For emulsification, the following steps must be carried out: (Fig. 2a–d):1. Install two sets of 8-tube strips and the cartridges on the customized rotor.
2. Put on the adaptors and screw on tight.
3. Load oil and sample into the inlets by a multichannel pipette.
4. Centrifuge 4 minutes for 25 μl or 7 minutes for 50 μl sample at 960 rpm.
5. Remove adaptors and cartridges.
6. Collected emulsions at the bottom of 8-tube strips are ready for further steps (e.g., thermal cycling).
The dimension of the customized rotor resembled the centrifugal radius of the commercial rotor F45-48-PCR rotor (Eppendorf AG, Hamburg, Germany).34 In our current design, two cartridges (16 samples) are processed simultaneously, which balances the mass. A slightly adjusted design of the holder would allow processing of three cartridges (24 samples) in parallel. An asymmetric jigsaw-fit structure would provide a quick and fail-safe way to attach the cartridge to the rotor.
The use of multichannel pipettes reduces the pipetting steps from 16 to 2 per 8 samples. The pipetting order of sample and oil does not make any difference in our workflow. On the contrary, the previously reported cartridges24–27 usually require the user to load the oil, insert the cartridge, and load the sample one after the other. The presented system can thus save time and reduce potential manual errors. In addition, the pipetting steps can be fully automated with a robotic pipetting station thanks to the compatibility of the cartridge with standard laboratory consumables.
The microfluidic cartridge consists of a single hot-embossed plastic part and a sealing film. The dimensions of the cartridge are 76 × 74 × 3.5 mm3 (l·w·h). As shown in Fig. 2e, the eight DGUs are implemented with a constant pitch of 9 mm, which is the pitch dimension of standard multichannel pipettes and of 8-tube strips. The diameter of the inlet holes is 3 mm, which allows proper insertion of the pipette tips into the inlets for correct loading and reduces the risk of leakage. During centrifugation at a fixed rotational speed, each unit undergoes various centrifugal acceleration (see Table 1). The detailed fluidic design of a single DGU is described in the following chapter. Each DGU consists of an array of 8 nozzles, which are oriented towards the center of rotation. After droplet generation, the droplets are spun out of the cartridge via the outlet (Fig. 2f). Each DGU provides an extended outlet feature for droplet transfer on the backside of the cartridge that reaches into the tubes to ensure that generated emulsions from each DGUs are completely conveyed into the corresponding tube (Fig. 2g). The distance between the outlet and the inner surface of the tube strip is designed to be ∼1.5 mm. The short distance allows for a smooth transfer of the emulsions to the tube thus avoiding a coalescence of droplets as they contact the inner surface of the tube. After centrifugation, generated emulsions sit at the bottom of the tube strip, ready for the next step.
| DGU case | Δxnzl (mm) | Δynzl (mm) | r nzl (mm) | RCF (×g) |
|---|---|---|---|---|
| 1 | 70 | 31.5 | 77.0 | 79.5 |
| 2 | 70 | 22.5 | 73.5 | 75.9 |
| 3 | 70 | 13.5 | 71.5 | 73.8 |
| 4 | 70 | 4.5 | 70.0 | 72.8 |
| 5 | 70 | 4.5 | 70.0 | 72.8 |
| 6 | 70 | 13.5 | 71.5 | 73.8 |
| 7 | 70 | 22.5 | 73.5 | 75.9 |
| 8 | 70 | 31.5 | 77.0 | 79.5 |
Fluidic design
Each DGU consists of the following geometric components: an inlet for oil, an inlet for the sample, an oil channel, a sample channel, an array of eight step nozzles, a chamber for droplet generation, and an outlet (see Fig. 3a). The fluidic concept of a single DGU was described previously.23 Since the single DGU from the previous work had shown excellent droplet monodispersity, we aimed to apply the same flow conditions on our system. Here, the main challenge was to provide the same flow conditions to each DGU. Since forces on a rotating system are oriented radially outwards, the design in centrifugal microfluidics is based on polar coordinates. To fit the linear structures to the design, different radial positions of the inlets and outlets and thus varying forces acting on these features cannot be avoided. To provide similar flow conditions to each DGU and thus monodisperse droplets, a simulation-supported design strategy has been applied that consists of the following steps. First, the array of eight nozzles and chambers are positioned towards the center of rotation to ensure that the direction of the centrifugal force is the same in each DGU. As illustrated in Fig. 2e, sample and oil inlets are placed at the same distance of x-direction for fitting the dimension of the multichannel pipette. The same applies for the outlets, to provide the interface to the 8-tube strip. Sample channel and oil channels are then derived by simulation to ensure that the desired flowrates from previous work23 is realized in each DGU. | ||
| Fig. 3 (a) Fluidic design using DGU1 as an example. The origin of polar coordinates is set at the center of rotation. The values of Δx, Δy and rnzl are listed in Table 1. (b) Model block representation of the fluidic network of single DGU in (a). The model consists of volume chamber, channel junction, and isoradial and radial channels. Ground symbol represents a connection to the atmospheric pressure. Numbers indicate the equivalent fluidic component shown in (a). The color boxes represent the filled fluid in the channel. Blue: dispersed phase. Yellow: oil phase. Green: emulsion phase. (c) Measured mean sample flowrate and validation of simulation model from individual DGUs. The dash line marks the mean flowrate in a case of ideal design. Error bars denote standard deviation from three performed cartridges. | ||
Network simulation model
A MATLAB based fluidic network simulation with lumped elements was set up to predict flowrates and optimize channel lengths (Fig. 3b). In a network simulation describing centrifugal microfluidic structures, the fluidic components (see Fig. 3a), can be translated to basic elements such as radially or iso-radially oriented channels, volume chambers or T-junctions as illustrated in Fig. 3b and detailed by Schwarz et al.35 Each fluidic component is represented by a model block with corresponding physical functions. The sample and oil inlets are represented by volume chambers with fill level function r(Vspl) and r(Voil), which are radial position functions in relation to the current fluid volumes inside the chambers. Channels with arbitrary orientation can be simplified to radial and isoradial channels.35 Each element consists of a physical function that derives the total pressure difference across the element and the flowrate Q through the element. At constant rotational speed, the predominant pressure difference is the sum of centrifugal force (Δpc) and viscous dissipation (Δpv). The equations are given as follows: | (1) |
| Δpc = −RQ | (2) |
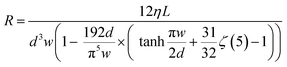 | (3) |
 , where Δx and Δy are the x-distance and y-distance between the point of the interest and the center of rotation (see Fig. 3a for an example). The hydraulic resistances of the sample and oil channels, Rspl and Roil, are adjusted for each individual DGU to match the mean flowrates of all DGUs. To simplify the optimization and manufacturing of the cartridges, we fixed the cross-section dimension of the sample channel, oil channel and outlet channel among all DGUs, and only altered channel lengths (dimensions are listed in ESI,† Table S2). The fluidic properties, density and viscosity of the three phases, sample, oil and mixture phases (highlighted by color boxes in Fig. 3b), must be known to run the simulation. While the fluidic properties of the sample and the oil phase are known, the density and viscosity of the generated emulsion are not evident. Due to the conservation of mass and the assumption of incompressible fluids, the density of the emulsion can simply be calculated using the volume fraction:
, where Δx and Δy are the x-distance and y-distance between the point of the interest and the center of rotation (see Fig. 3a for an example). The hydraulic resistances of the sample and oil channels, Rspl and Roil, are adjusted for each individual DGU to match the mean flowrates of all DGUs. To simplify the optimization and manufacturing of the cartridges, we fixed the cross-section dimension of the sample channel, oil channel and outlet channel among all DGUs, and only altered channel lengths (dimensions are listed in ESI,† Table S2). The fluidic properties, density and viscosity of the three phases, sample, oil and mixture phases (highlighted by color boxes in Fig. 3b), must be known to run the simulation. While the fluidic properties of the sample and the oil phase are known, the density and viscosity of the generated emulsion are not evident. Due to the conservation of mass and the assumption of incompressible fluids, the density of the emulsion can simply be calculated using the volume fraction:| ρeml = φρspl + (1 − φ)ρoil | (4) |
![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl/(
spl/(![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl +
spl + ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) oil).
oil). ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl and
spl and ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) oil are the mean sample and mean oil flowrates, respectively. The effective viscosity for calculating the flow resistance of emulsions in microchannels, however, cannot be calculated analytically as, in addition to the volume fractions, the viscosity depends on complex flow patterns influenced by interfacial forces and by the size of the droplets. It has been observed in previous studies that the presence of droplets in a channel contributes to an additional pressure drop,36 resulting in an effective viscosity higher than an average viscosity derived from volume fractions. Effective viscosities have extensively been studied37–39 and implemented in network simulations40–42 previously. However, the derived values are not suitable for the scenario in our system in which droplets are much more compact and squeezed in the outlet channel (see ESI,† Fig. S1). To account for the additional pressure drop due to the emulsion's transport through the outlet channel, we estimated the effective viscosity by fitting parameters using experimental data from the previous study22 on which the current design is based (see ESI,† S2). The determination procedure of effective viscosity is described in detail in ESI,† S2. As a result, an effective viscosity of ηeml = 10.5 mPa s was used in this work. The mean flowrates of
oil are the mean sample and mean oil flowrates, respectively. The effective viscosity for calculating the flow resistance of emulsions in microchannels, however, cannot be calculated analytically as, in addition to the volume fractions, the viscosity depends on complex flow patterns influenced by interfacial forces and by the size of the droplets. It has been observed in previous studies that the presence of droplets in a channel contributes to an additional pressure drop,36 resulting in an effective viscosity higher than an average viscosity derived from volume fractions. Effective viscosities have extensively been studied37–39 and implemented in network simulations40–42 previously. However, the derived values are not suitable for the scenario in our system in which droplets are much more compact and squeezed in the outlet channel (see ESI,† Fig. S1). To account for the additional pressure drop due to the emulsion's transport through the outlet channel, we estimated the effective viscosity by fitting parameters using experimental data from the previous study22 on which the current design is based (see ESI,† S2). The determination procedure of effective viscosity is described in detail in ESI,† S2. As a result, an effective viscosity of ηeml = 10.5 mPa s was used in this work. The mean flowrates of ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl = 0.13 μl s−1 and
spl = 0.13 μl s−1 and ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) oil = 0.09 μl s−1 for sample and oil flow, respectively, were chosen as design values for all DGUs with φ = 0.591 and ρeml = 1251.1 kg m−3 (eqn (4)).
oil = 0.09 μl s−1 for sample and oil flow, respectively, were chosen as design values for all DGUs with φ = 0.591 and ρeml = 1251.1 kg m−3 (eqn (4)).
The result of the simulation study is a parameter set for the sample and oil channel length for each DGU as depicted in Table 2, while the cross-section areas of the sample and the oil channel are 62 × 58 μm2 and 49 × 37 μm2, respectively.
| DGU case | L spl (mm) | L oil (mm) |
|---|---|---|
| 1 | 15.28 | 16.14 |
| 2 | 15.87 | 15.75 |
| 3 | 16.11 | 14.75 |
| 4 | 16.28 | 14.91 |
| 5 | 16.40 | 14.39 |
| 6 | 16.72 | 13.96 |
| 7 | 16.84 | 13.34 |
| 8 | 17.03 | 13.19 |
Validation of the simulation model and the manufactured cartridge
The manufactured cartridge turned out to have some deviations from the ideal design as listed in ESI,† S4. For the comparison of the simulation results with the experiments, the dimensions of the manufactured cartridge were measured and applied to the simulation.Then, we experimentally evaluated the mean sample flowrate ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl and compared it with the results of the proposed simulation model to validate our design strategy. 50 μl of PCR buffer (1×) and 50 μl of oil were supplied to each DGU in the cartridge and then emulsified. The emulsification time from each DGU was determined from video sequences. We obtained
spl and compared it with the results of the proposed simulation model to validate our design strategy. 50 μl of PCR buffer (1×) and 50 μl of oil were supplied to each DGU in the cartridge and then emulsified. The emulsification time from each DGU was determined from video sequences. We obtained ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl by dividing the sample volume of 50 μl in each DGU by the measured emulsification time. The depth and width of the sample, oil, nozzles and outlet channels were measured by optical profilometry (the values are listed in ESI,† S4). The measured dimension parameters are used in the simulation model. In this case, the flowrate conditions are unknown and thus ρeml is unknown (eqn (4)). To obtain the correct results in which the solved mean flowrates
spl by dividing the sample volume of 50 μl in each DGU by the measured emulsification time. The depth and width of the sample, oil, nozzles and outlet channels were measured by optical profilometry (the values are listed in ESI,† S4). The measured dimension parameters are used in the simulation model. In this case, the flowrate conditions are unknown and thus ρeml is unknown (eqn (4)). To obtain the correct results in which the solved mean flowrates ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl and
spl and ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) oil, and ρeml fit eqn (4), one can use an iterative method: start a simulation with an arbitrary φ, e.g., 0.5, as an initial guess and then utilize the solved
oil, and ρeml fit eqn (4), one can use an iterative method: start a simulation with an arbitrary φ, e.g., 0.5, as an initial guess and then utilize the solved ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl and
spl and ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) oil as an input for the next simulation run. After two runs, the predetermined emulsion density matched the outcome of eqn (4) with a difference less than 0.001 kg m−3. After the integration of all manufacturing variances to the simulation, the model using manufactured dimensions (red circle, Fig. 3c) showed a good agreement with the experimental results (black square, Fig. 3c). The maximum variance within the experimental
oil as an input for the next simulation run. After two runs, the predetermined emulsion density matched the outcome of eqn (4) with a difference less than 0.001 kg m−3. After the integration of all manufacturing variances to the simulation, the model using manufactured dimensions (red circle, Fig. 3c) showed a good agreement with the experimental results (black square, Fig. 3c). The maximum variance within the experimental ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl and compensated
spl and compensated ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl is ∼2.2% in the case of DGU3. The accordance of the simulated and experimental results suggests that the variance of measured
spl is ∼2.2% in the case of DGU3. The accordance of the simulated and experimental results suggests that the variance of measured ![[Q with combining macron]](https://www.rsc.org/images/entities/i_char_0051_0304.gif) spl is mainly attributed to tolerances of the manufacturing technology. In addition, the experimental result also supports our simulation-based design strategy, which integrates linearly arranged fluidic elements on the centrifugal microfluidic platform.
spl is mainly attributed to tolerances of the manufacturing technology. In addition, the experimental result also supports our simulation-based design strategy, which integrates linearly arranged fluidic elements on the centrifugal microfluidic platform.
Droplet characterization
We investigated the performance of the cartridge by examining the diameter of the generated droplets. The PCR buffer (1×) was used as sample phase for droplet characterization. 50 μl of PCR buffer and oil per DGU were pipetted into the cartridge and then emulsified. After 7 minutes, all reaction mixes were completely emulsified, and no residual liquid was visible, neither in the sample inlets or channels, nor on the inner surface of the tube strip. As shown in Fig. 4a, the mean of the droplet diameter from each DGU has a slight variance. The largest mean droplet diameter (88.9 μm) from DGU8 is about 1.07-fold the smallest mean droplet diameter (82.7 μm) from DGU1. This variance originates from tolerances of the applied manufacturing technology. The cartridge has a relatively large area (76 × 74 mm2) compared, for example, to the previously presented microfluidic cartridges from Schulz et al.23 (33.6 × 7.8 mm2). Higher milling tolerance is usually expected as the work piece is larger.We investigated how manufacturing variance affected droplet diameter variation. First, the mean nozzle depth from each DGU was obtained by optical profilometry (the values are listed in the ESI,† S4). Dividing the mean droplet diameter by the mean nozzle depth of the corresponding DGU (red dots in Fig. 4a), the variance of the mean droplet diameter decreased to 1.03-fold between the largest value and the smallest value (DGU2 and DGU8). The overall nozzle dimension has a mean depth of 25.6 ± 0.81 μm with CV < 3.5% (64 nozzles among 8 DGUs). Despite the slight differences of droplet diameters among DGUs, the overall droplet diameter distribution (Fig. 4b) revealed good droplet monodispersity (CV < 5%). Hence, we can consider that the droplet diameters from different DGUs are identical and can be used for digital droplet PCR (d = 86.5 μm) with estimated 1.47 × 105 generated droplets in each DGU.
ddPCR assay
To demonstrate compatibility of the cartridge to biochemical assays, exemplary ddPCRs for the detection of the human cystic fibrosis transmembrane conductance regulator (CFTR) gene were performed in serial dilutions. Triplicates were run in each DGU for each dilution step and for every run, a fresh cartridge was used. Cartridges were loaded and processed as explained in the workflow description. The ddPCR was evaluated using an automated fluorescence microscope and image data analysis software. The concentration was calculated by applying Poisson statistics.43 Evaluation of the ddPCR showed homogeneous, monodisperse droplet images with a clear differentiation between positive and negative droplets (see Fig. 5a). By plotting expected values against measured values, a very good concordance of template concentrations could be observed for each serial dilution per DGU with R2 values between R2 ≥ 0.998 and R2 ≥ 0.999 (Fig. 5b, ESI,† S6) as well as for the overall experimental performance (red line, Fig. 5c) with R2 ≥ 0.999.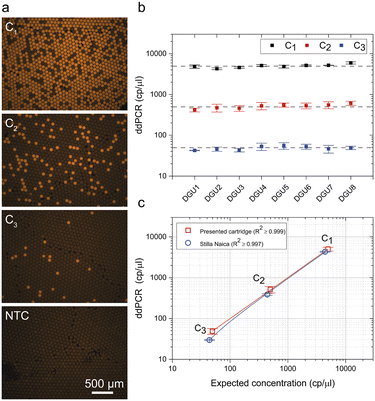 | ||
| Fig. 5 Analysis of the digital droplet polymerase chain reaction (ddPCR). (a) Exemplary microscopic fluorescence images of droplet monolayers after amplification. Serial dilution of target template (C1–C3) and a non-template control (NTC) are shown (using the presented cartridge). (b) Measured template concentrations per DGU. The dashed line represents the expected concentration. Error bars represent standard deviations of three repetitions. (c) Comparison of presented cartridge and commercial instrument is performed by plotting overall concentration against the expected concentration. Each data point represents the mean of concentration of three repetitions of the eight DGUs for the presented cartridge and four repetitions for the Stilla Naica system. The linear fit curve is represented by the solid line. RAW data are presented in the ESI,† S6. | ||
Additional experiments were performed separately to verify quantitative accuracy against commercial ddPCR platform (Naica system, Stilla Technologies, Villejuif, France). These experiments showed comparable results in terms of linearity, with an R2 value of ≥0.997 (Fig. 5c).
The good concordance between template concentration in the single DGU as well as for the overall set-up demonstrates that the cartridge performance did not interfere with introduced biomolecules nor impair assay performance. Therefore, it is suitable for increased throughput biochemical assays.
The presented cartridge is an open system without any lid during sample emulsification. Usually low DNA-concentrations are applied before amplification, and the risk of cross contamination can be considered low, similar to other common methods of DNA extraction such as the use of spin-columns. The accordance of the ddPCR result also supports our assumption.
Materials and methods
Microfluidic cartridge design and manufacturing
The microfluidic structure was designed in a computer-aided-design software (Dassault Systèmes SE, SolidWorks, Vélizy-Villacoublay, France) and MATLAB based fluidic networking simulation (MathWorks Corp., Matlab, Natick, MA, USA) and manufactured by the Hahn-Schickard Lab-on-a-Chip Foundry.44 The PMMA (Evonik AG, Essen, Germany) master was milled by a CNC milling machine (KERN Evo mill, KERN Microtechnik GmbH, Eschenlohe, Germany). The milled surface was measured by confocal microscopy (DUO Vario, Confovis GmbH, Jena, Germany). Then, the positive stamp was produced. Polydimethylsiloxane (PDMS) (mixture of elastosil 607 and elastosil 675, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, with monomer/crosslinker ratios of 9
1, with monomer/crosslinker ratios of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 respectively) was poured onto the milled master, and centrifuged (Zentrifuge Rotanta 460 R, Hettich GmbH, Kirchlengern, Germany). The positive PDMS stamp was used to create replicates by hot embossing, transferring structures onto cyclic olefin copolymer substrate (COC) (TOPAS COC 5013, TOPAS Advanced Polymers GmbH, Raunheim, Germany). Finally, the cartridge was sealed manually using a pressure sensitive adhesive film (9795R diagnostic tape, 3M Corp., Saint Paul, MN, USA).
1 respectively) was poured onto the milled master, and centrifuged (Zentrifuge Rotanta 460 R, Hettich GmbH, Kirchlengern, Germany). The positive PDMS stamp was used to create replicates by hot embossing, transferring structures onto cyclic olefin copolymer substrate (COC) (TOPAS COC 5013, TOPAS Advanced Polymers GmbH, Raunheim, Germany). Finally, the cartridge was sealed manually using a pressure sensitive adhesive film (9795R diagnostic tape, 3M Corp., Saint Paul, MN, USA).
Centrifuge and customized rotor
The rotor was designed in Solidworks and printed by a commercially available 3D printer (Original Prusa i3, Prusa Research sro, Czech Republic) using polyethylene terephthalate glycol filament (PETG, Prusa Research sro, Czech Republic).To perform and observe droplet generation, a customized centrifuge was used (Euler player 2nd generation, BioFluidix GmbH, Freiburg im Breisgau, Germany).
Droplet generation and readout
For all experiments, fluorinated oil with surfactant dSurf (4% (w/w) in Novec™ 7500, Fluigent, France) was used as a continuous phase. The PCR buffer (1×) was used as a dispersed phase in this paper with a measured viscosity of 1.65 mPa s (see ESI,† S1) and was prepared by mixing 2× ddPCR Supermix for probes (Bio-Rad Laboratories Inc., Hercules, CA, USA) with distilled water (UltraPure DNase/RNase free, Invitrogen Corp., Carlsbad, CA, USA). Pipetting was performed using an Eppendorf Research Plus 8-channel pipette (Eppendorf AG, Hamburg, Germany) and epT.I.P.S. Standard 2–200 μL pipette tips (Eppendorf AG, Hamburg, Germany). No leakage was observed through all experiments. Droplet diameters were evaluated by the following procedure: bright field images were recorded by microscope Observer Z1 (Zeiss GmbH, Jena, Germany) and then further analyzed using MATLAB based build-in circle detection algorithm (see ESI,† S5).ddPCR
The ddPCR experiment on the presented cartridge was set-up according to Schulz et al.23 25 μl reaction mixes were pipetted into each DGU of the cartridge with a multichannel pipette (Eppendorf AG, Hamburg, Germany). After droplet generation, the emulsion in the 8-tube strip (Biozym Scientific GmbH, Hessisch Oldendorf, Germany) was covered with 15 μl mineral oil (Sigma Aldrich, St. Louis, MO, USA) and transferred to a thermal cycler (T 100 Thermal Cycler, Bio-Rad Laboratories Inc., Hercules, CA, USA). The cycling protocol was as follows: 5 min at 94 °C, 45 cycles (15 s at 94 °C and 60 s at 58 °C), and holding at 23 °C until readout. After cycling, droplets were transferred into cell counting chamber slides (Countess™, Invitrogen Corp., Carlsbad, CA, USA), where the droplets aligned in a monolayer. Positive and negative droplets (N ≈ 2000 per sample) were counted using a fluorescence microscope (Lionheart LX, BioTek Instruments GmbH, Bad Friedrichshall, Germany) and the corresponding image analysis software package (Gen5).A comparative experiment on a commercial ddPCR platform was performed using the Stilla Naica system (Stilla Technologies, Villejuif, France). The same assay was used with a minor adjustment: 5× PerfeCTa qPCR ToughMix (QuantaBio, Beverly, MA, USA) was used in the reaction mix instead of 2× ddPCR Supermix for probes. 25 μl reaction mixes were pipetted into sample inlets on Sapphire Chips (Stilla Technologies, Villejuif, France). The same thermal cycling protocol was applied after an additional step of partitioning droplets (40 °C for 12 min). 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 27
000 to 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets were evaluated after thermal cycling using the corresponding readout instrument and software (Prism 6, Crystal Miner).
000 droplets were evaluated after thermal cycling using the corresponding readout instrument and software (Prism 6, Crystal Miner).
Conclusions
We present a microfluidic cartridge comprising eight “droplet generation units”, DGUs, for increased throughput sample emulsification as a part of a parallelized and automated laboratory workflow. The automated workflow is suitable for digital droplet assay and can be adapted in any laboratory as it requires no specialized instruments. The user-friendly installation simplifies cartridge set up with minimal manual steps within a short period of time. Compared to existing solutions,23–27 the use of eight channel pipettes reduces pipetting steps for loading sample and oil from 16 to 2. Using our simulation-based strategy, we derived an in silico design that provides consistent flow conditions for each DGU despite the challenges of interfacing the linearly oriented eight channel pipette and the eight tube strips to the centrifugal microfluidic chip designed in polar coordinates. We demonstrated that the cartridge enables the user to emulsify eight samples for ddPCR with good reproducibility, regardless of the DGU position. The presented cartridge design is tailored to the Eppendorf rotor for 6× PCR 8-tube strips. By applying the introduced design strategy, however, adaptation of the cartridge design to rotors with different dimensions is straightforward. One critical parameter is the distance between the rotor's center and the installed PCR 8-tube strip, as this affects the orientation and radial position of each DGU on the cartridge (parameters shown in Fig. 3(a)). The second parameter is the rotor radius required to hold the cartridge. In the presented cartridge, the length (x-direction) of the DGU is 40 mm, and additional length is necessary to achieve sufficiently large hydrostatic pressures at the start of centrifugation. Consequently, the minimum rotor radius should be no less than 50 mm. This also determines the centrifuge models compatible with the presented cartridge.The current 3D printed rotor allows for the operation of 16 samples per run by processing two cartridges simultaneously. By adapting the rotor design, one can easily integrate at least three cartridges. It is possible that higher throughput can be achieved by using a customized rotor for 4 × 8 DGUs or by using a 96-spin plate rotor with stacked DGU units on top of a 96-well PCR plate (12 × 8 DGUs, not shown in this study). Furthermore, the current design required adaptors to fix the cartridges to the rotor and therefore, a customized centrifuge was used. Once the cartridge is transferred to injection molding, a cartridge can be designed to fit directly to the commercially available rotor and thereby become a powerful tool for cost-effective digital assay implementations.
Finally, we demonstrate the interfacing of centrifugal microfluidics with linear-oriented standard equipment. The design strategy is not limited to droplet generation. Arranging fluidic elements in a linear-oriented manner enables compact fluidic design and reduces the footprint, resulting in increased throughput. This approach could further extend parallelization for various applications in diagnostics and bioanalytical processes, including lateral flow strips,45 immunoassays46 and nucleic acid assays.47 This streamlining could offer a way to make the most of the major advantages of centrifugal microfluidics when handling small volumes in a highly precise manner.48,49 Efficiency is further enhanced by the excellent technical benefits of linear-oriented laboratory equipment for high-throughput laboratory automation.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the team at Hahn-Schickard Lab-on-a-Chip Foundry for manufacture of cartridges, engineer Moritz Boesenberg and Daniel Frejek for 3D printing, and Dr. Martin Schulz for discussion at an early stage of development. The authors also thank Tobias Lange and Tobias Gross for providing samples. The scientific work was funded by Federal Ministry of Education and Research (BMBF) within the projects FREEDOM (project number 13N15288).References
- A. S. Basu, Digital Assays Part I: Partitioning Statistics and Digital PCR, SAGE Publications Inc, SLAS Technol., 2017, 22, 369–386 CrossRef PubMed.
- E. A. Ottesen, J. W. Hong, S. R. Quake and J. R. Leadbetter, Microfluidic Digital PCR Enables Multigene Analysis of Individual Environmental Bacteria, Science, 2006, 314, 1464–1467 CrossRef CAS PubMed.
- L. Cohen, N. Cui, Y. Cai, P. M. Garden, X. Li, D. A. Weitz and D. R. Walt, Single Molecule Protein Detection with Attomolar Sensitivity Using Droplet Digital Enzyme-Linked Immunosorbent Assay, ACS Nano, 2020, 14, 9491–9501 CrossRef CAS PubMed.
- J. U. Shim, R. T. Ranasinghe, C. A. Smith, S. M. Ibrahim, F. Hollfelder, W. T. S. Huck, D. Klenerman and C. Abell, Ultrarapid generation of femtoliter microfluidic droplets for single-molecule-counting immunoassays, ACS Nano, 2013, 7, 5955–5964 CrossRef CAS PubMed.
- Y. Obayashi, R. Iino and H. Noji, A single-molecule digital enzyme assay using alkaline phosphatase with a cumarin-based fluorogenic substrate, Analyst, 2015, 140, 5065–5073 RSC.
- Y. Zeng, R. Novak, J. Shuga, M. T. Smith and R. A. Mathies, High-performance single cell genetic analysis using microfluidic emulsion generator arrays, Anal. Chem., 2010, 82, 3183–3190 CrossRef CAS PubMed.
- D. Witters, B. Sun, S. Begolo, J. Rodriguez-Manzano, W. Robles and R. F. Ismagilov, Digital biology and chemistry, Lab Chip, 2014, 14, 3225–3232 RSC.
- B. Vogelstein and K. W. Kinzler, Digital PCR, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef CAS PubMed.
- M. T. Guo, A. Rotem, J. A. Heyman and D. A. Weitz, Droplet microfluidics for high-throughput biological assays, Lab Chip, 2012, 12, 2146–2155 RSC.
- K. S. Elvira, F. Gielen, S. S. H. Tsai and A. M. Nightingale, Materials and methods for droplet microfluidic device fabrication, Lab Chip, 2022, 22, 859–875 RSC.
- B. J. Hindson, K. D. Ness, D. A. Masquelier, P. Belgrader, N. J. Heredia, A. J. Makarewicz, I. J. Bright, M. Y. Lucero, A. L. Hiddessen, T. C. Legler, T. K. Kitano, M. R. Hodel, J. F. Petersen, P. W. Wyatt, E. R. Steenblock, P. H. Shah, L. J. Bousse, C. B. Troup, J. C. Mellen, D. K. Wittmann, N. G. Erndt, T. H. Cauley, R. T. Koehler, A. P. So, S. Dube, K. A. Rose, L. Montesclaros, S. Wang, D. P. Stumbo, S. P. Hodges, S. Romine, F. P. Milanovich, H. E. White, J. F. Regan, G. A. Karlin-Neumann, C. M. Hindson, S. Saxonov and B. W. Colston, High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number, Anal. Chem., 2011, 83(22), 8604–8610 CrossRef CAS PubMed.
- L. Yobas, S. Martens, W.-L. Ong and N. Ranganathan, High-performance flow-focusing geometry for spontaneous generation of monodispersed droplets, Lab Chip, 2006, 6, 1073–1079 RSC.
- F. Schuler, M. Trotter, M. Geltman, F. Schwemmer, S. Wadle, E. Domínguez-Garrido, M. López, C. Cervera-Acedo, P. Santibáñez, F. von Stetten, R. Zengerle and N. Paust, Digital droplet PCR on disk, Lab Chip, 2016, 16, 208–216 RSC.
- M. Schulz, F. von Stetten, R. Zengerle and N. Paust, Centrifugal step emulsification: How buoyancy enables high generation rates of monodisperse droplets, Langmuir, 2019, 35, 9809–9815 CrossRef CAS PubMed.
- W. Lyu, J. Zhang, Y. Yu, L. Xu and F. Shen, Slip formation of a high-density droplet array for nucleic acid quantification by digital LAMP with a random-access system, Lab Chip, 2021, 21, 3086–3093 RSC.
- J. J. Agresti, E. Antipov, A. R. Abate, K. Ahn, A. C. Rowat, J.-C. Baret, M. Marquez, A. M. Klibanov, A. D. Griffiths and D. A. Weitz, Ultrahigh-throughput screening in drop-based microfluidics for directed evolution, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 4004–4009 CrossRef CAS PubMed.
- V. Yelleswarapu, J. R. Buser, M. Haber, J. Baron, E. Inapuri and D. Issadore, Mobile platform for rapid sub–picogram-per-milliliter, multiplexed, digital droplet detection of proteins, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4489–4495 CrossRef CAS PubMed.
- M. Nie, M. Zheng, C. Li, F. Shen, M. Liu, H. Luo, X. Song, Y. Lan, J.-Z. Pan and W. Du, Assembled Step Emulsification Device for Multiplex Droplet Digital Polymerase Chain Reaction, Anal. Chem., 2019, 91, 1779–1784 CrossRef CAS PubMed.
- B. Shi, W. Di, Y. Jiang, J. An and W. Wenming, Off-Chip Vertical Step Emulsification Droplets Preparation Device Applied for Droplet Digital PCR, Adv. Mater. Interfaces, 2020, 7, 2001074 CrossRef CAS.
- S. Zhao, Z. Zhang, F. Hu, J. Wu and N. Peng, Massive droplet generation for digital PCR via a smart step emulsification chip integrated in a reaction tube, Analyst, 2021, 146, 1559–1568 RSC.
- Y. Li, H. Cherukury, L. Labanieh, W. Zhao and D.-K. Kang, Rapid Detection of β-Lactamase-Producing Bacteria Using the Integrated Comprehensive Droplet Digital Detection (IC 3D) System, Sensors, 2020, 20, 4667 CrossRef CAS PubMed.
- T. J. Abram, H. Cherukury, C.-Y. Ou, T. Vu, M. Toledano, Y. Li, J. T. Grunwald, M. N. Toosky, D. F. Tifrea, A. Slepenkin, J. Chong, L. Kong, D. V. Del Pozo, K. T. La, L. Labanieh, J. Zimak, B. Shen, S. S. Huang, E. Gratton, E. M. Peterson and W. Zhao, Rapid bacterial detection and antibiotic susceptibility testing in whole blood using one-step, high throughput blood digital PCR, Lab Chip, 2020, 20, 477–489 RSC.
- M. Schulz, S. Probst, S. Calabrese, A. R. Homann, N. Borst, M. Weiss, F. von Stetten, R. Zengerle and N. Paust, Versatile Tool for Droplet Generation in Standard Reaction Tubes by Centrifugal Step Emulsification, Molecules, 2020, 25, 1914 CrossRef CAS PubMed.
- Z. Chen, P. Liao, F. Zhang, M. Jiang, Y. Zhu and Y. Huang, Centrifugal micro-channel array droplet generation for highly parallel digital PCR, Lab Chip, 2017, 17, 235–240 RSC.
- H. Yamashita, M. Morita, H. Sugiura, K. Fujiwara, H. Onoe and M. Takinoue, Generation of monodisperse cell-sized microdroplets using a centrifuge-based axisymmetric co-flowing microfluidic device, J. Biosci. Bioeng., 2015, 119, 492–495 CrossRef CAS PubMed.
- D. C. Shin, Y. Morimoto, J. Sawayama, S. Miura and S. Takeuchi, Centrifuge-based step emulsification device for simple and fast generation of monodisperse picoliter droplets, Sens. Actuators, B, 2019, 301, 127164 CrossRef CAS.
- W. Zhou, C. Liu, T. Zhang, K. Jiang, H. Li, Z. Zhang and Y. Tang, Low Cost, Easily-Assembled Centrifugal Buoyancy-Based Emulsification and Digital PCR, Micromachines, 2022, 13, 171 CrossRef PubMed.
- J. Madic, A. Zocevic, V. Senlis, E. Fradet, B. Andre, S. Muller, R. Dangla and M. E. Droniou, Three-color crystal digital PCR, Biomol. Detect. Quantif., 2016, 10, 34–46 CrossRef CAS PubMed.
- D. Pekin, Y. Skhiri, J.-C. Baret, D. Le Corre, L. Mazutis, C. B. Salem, F. Millot, A. El Harrak, J. Brian Hutchison, J. W. Larson, D. R. Link, P. Laurent-Puig, A. D. Griffiths and V. Taly, Quantitative and sensitive detection of rare mutations using droplet-based microfluidics, Lab Chip, 2011, 11, 2156–2166 RSC.
- Y. Fu, F. Zhang, X. Zhang, J. Yin, M. Du, M. Jiang, L. Liu, J. Li, Y. Huang and J. Wang, High-throughput single-cell whole-genome amplification through centrifugal emulsification and eMDA, Commun. Biol., 2019, 2, 1–10 CrossRef PubMed.
- Y. Fu, C. Li, S. Lu, W. Zhou, F. Tang, X. Sunney Xie and Y. Huang, Uniform and accurate single-cell sequencing based on emulsion whole-genome amplification, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 11923–11928 CrossRef CAS PubMed.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. von Stetten and N. Paust, Centrifugal step emulsification applied for absolute quantification of nucleic acids by digital droplet RPA, Lab Chip, 2015, 15, 2759–2766 RSC.
- X. Li, D. Zhang, W. Ruan, W. Liu, K. Yin, T. Tian, Y. Bi, Q. Ruan, Y. Zhao, Z. Zhu and C. Yang, Centrifugal-Driven Droplet Generation Method with Minimal Waste for Single-Cell Whole Genome Amplification, Anal. Chem., 2019, 91, 13611–13619 CrossRef CAS PubMed.
- Eppendorf AG, Original instructions. Centrifuge 5804/5804 R Centrifuge 5810/5810 R, Rotor F-45-48-PCR 11.1.9, 2019, https://www.eppendorf.com/product-media/doc/en/330816/Centrifugation_Operating-manual_Centrifuge-5804-5804-R_Centrifuge-5810-5810-R.pdf.
- I. Schwarz, S. Zehnle, T. Hutzenlaub, R. Zengerle and N. Paust, System-level network simulation for robust centrifugal-microfluidic lab-on-a-chip systems, Lab Chip, 2016, 16, 1873–1885 RSC.
- C. N. Baroud, F. Gallaire and R. Dangla, Dynamics of microfluidic droplets, Lab Chip, 2010, 10, 2032 RSC.
- B. J. Adzima and S. S. Velankar, Pressure drops for droplet flows in microfluidic channels, J. Micromech. Microeng., 2006, 16, 1504–1510 CrossRef.
- V. Labrot, M. Schindler, P. Guillot, A. Colin and M. Joanicot, Extracting the hydrodynamic resistance of droplets from their behavior in microchannel networks, Biomicrofluidics, 2009, 3, 12804 CrossRef PubMed.
- D. A. Sessoms, M. Belloul, W. Engl, M. Roche, L. Courbin and P. Panizza, Droplet motion in microfluidic networks: Hydrodynamic interactions and pressure-drop measurements, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2009, 80, 16317 CrossRef CAS PubMed.
- A. Grimmer, M. Hamidović, W. Haselmayr and R. Wille, Advanced Simulation of Droplet Microfluidics, ACM J. Emerg. Technol. Comput. Syst., 2019, 15, 1–16 CrossRef.
- T. Glawdel and C. L. Ren, Global network design for robust operation of microfluidic droplet generators with pressure-driven flow, Microfluid. Nanofluid., 2012, 13, 469–480 CrossRef CAS.
- G. Tetradis-Meris, D. Rossetti, C. Pulido de Torres, R. Cao, G. Lian and R. Janes, Novel Parallel Integration of Microfluidic Device Network for Emulsion Formation, Ind. Eng. Chem. Res., 2009, 48, 8881–8889 CrossRef CAS.
- S. Dube, J. Qin, R. Ramakrishnan and X. Wu, Mathematical Analysis of Copy Number Variation in a DNA Sample Using Digital PCR on a Nanofluidic Device, PLoS One, 2008, 3, e2876 CrossRef PubMed.
- Lab-on-a-Chip Foundry, https://www.hahn-schickard.de/en/service-portfolio/production/lab-on-a-chip-foundry.
- D. M. Kainz, S. M. Früh, T. Hutzenlaub, R. Zengerle and N. Paust, Flow control for lateral flow strips with centrifugal microfluidics, Lab Chip, 2019, 19, 2718–2727 RSC.
- J. Park, V. Sunkara, T.-H. Kim, H. Hwang and Y.-K. Cho, Lab-on-a-Disc for Fully Integrated Multiplex Immunoassays, Anal. Chem., 2012, 84, 2133–2140 CrossRef CAS PubMed.
- Y. Chen, N. Zong, F. Ye, Y. Mei, J. Qu and X. Jiang, Dual-CRISPR/Cas12a-Assisted RT-RAA for Ultrasensitive SARS-CoV-2 Detection on Automated Centrifugal Microfluidics, Anal. Chem., 2022, 94, 9603–9609 CrossRef CAS PubMed.
- P. Juelg, M. Specht, E. Kipf, M. Lehnert, C. Eckert, M. Keller, T. Hutzenlaub, F. von Stetten, R. Zengerle and N. Paust, Automated serial dilutions for high-dynamic-range assays enabled by fill-level-coupled valving in centrifugal microfluidics, Lab Chip, 2019, 19, 2205–2219 RSC.
- J.-N. Klatt, M. Depke, N. Goswami, N. Paust, R. Zengerle, F. Schmidt and T. Hutzenlaub, Tryptic digestion of human serum for proteomic mass spectrometry automated by centrifugal microfluidics, Lab Chip, 2020, 20, 2937–2946 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3lc00339f |
| This journal is © The Royal Society of Chemistry 2023 |