Open Access Article
Open Access ArticleBioinspired human stomach-on-a-chip with in vivo like function and architecture†
Daniel A.
Ferreira
 abc,
João P.
Conde
de,
Mario
Rothbauer
fg,
Peter
Ertl
abc,
João P.
Conde
de,
Mario
Rothbauer
fg,
Peter
Ertl
 h,
Pedro L.
Granja‡
h,
Pedro L.
Granja‡
 *ab and
Carla
Oliveira‡
*ab and
Carla
Oliveira‡
 *aij
*aij
ai3S – Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Rua Alfredo Allen, 208, 4200-135 Porto, Portugal. E-mail: pgranja@i3s.up.pt
bINEB – Instituto de Engenharia Biomédica, Universidade do Porto, Rua Alfredo Allen, 208, 4200-135 Porto, Portugal
cICBAS – Instituto de Ciências Biomédicas Abel Salazar, Universidade do Porto, Rua Jorge de Viterbo Ferreira, 228, 4050-313 Porto, Portugal
dDepartment of Bioengineering, Instituto Superior Técnico, Universidade de Lisboa, Av. Rovisco Pais, 1, 1049-001 Lisboa, Portugal
eInstituto de Engenharia de Sistemas e Computadores – Microsistemas e Nanotecnologia (INESC MN), Rua Alves Redol, 9, 1000-029 Lisboa, Portugal
fDepartment of Orthopedics and Trauma Surgery, Karl Chiari Lab for Orthopaedic Biology, Orthopedic Microsystems, Medical University of Vienna, Währinger Gürtel 18-20, 1090 Vienna, Austria
gInstitute of Applied Synthetic Chemistry, Cell Chip Group, Vienna University of Technology (TUW), Getreidmarkt, 9/163, 1060 Vienna, Austria
hFaculty of Technical Chemistry, Vienna University of Technology (TUW), Getreidemarkt 9, 1060 Vienna, Austria
iIpatimup – Institute of Molecular Pathology and Immunology, Universidade do Porto, Rua Alfredo Allen, 208, 4200-135 Porto, Portugal. E-mail: carlaol@ipatimup.pt
jDepartment of Pathology, Faculty of Medicine, University of Porto, Alameda Prof. Hernâni Monteiro, 4200-319 Porto, Portugal
First published on 9th January 2023
Abstract
The lack of biomimetic in vitro models capable of reproducing the complex architecture and the dynamic environment of the gastric mucosa, delay the development of diagnostic and therapeutic tools. Recent advances in microengineering made possible the fabrication of bioinspired microdevices capable of replicating the physiological properties of an organ, inside a microfluidics chip. Herein, a bioinspired stomach-on-a-chip (SoC) device is described, supporting peristalsis-like motion and reconstituting organ-level epithelial architecture and function. The device simulates the upper epithelial interface, representing the three innermost layers of the gastric mucosa, namely the epithelial barrier, the basement membrane and the lamina propria. The dynamic environment imparted by mechanical actuation of the flexible on-chip cell culture substrate, was the main driver in the development of epithelial polarization and differentiation traits characteristic of the native gastric mucosa, and allowed partial recapitulation of gastric barrier function. These traits were not affected by the addition of a mesenchymal population to the system, which was able to remodel the surrounding extracellular matrix, nor by the potential epithelial–mesenchymal cross-talk. The engineered platform highlights the importance of addressing the mechanical microenvironment of the native organ, to potentiate an organ-level response of the artificial tissue. The proposed SoC represents an appealing tool in personalized medicine, with bio-relevance for the study of gastric diseases and an alternative to current animal models.
1. Introduction
The stomach plays a central role in human physiology, and gastric dysfunction poses serious health risks with associated non-neglectable mortality and morbidity rates.1–3 For instance, gastric cancer, represented the fourth leading cause of estimated cancer related deaths in 2020.4 The demand for better study models accurately recapitulating human gastric physiology is high, mainly to improve the way therapies for gastric-associated diseases are currently tested.Two-dimensional (2D) cell culture models, lacking the complexity of human tissue architecture and physiology, are commonly used for early stages of drug development but, as expected, their simplified nature has a very low predictive power in drug efficacy.5,6 Three dimensional (3D) models, engineered with bio-inspired matrices replicating the chemistry, assembly, multicellular nature and architecture of living organs, provide a better simulation of physiological conditions.7,8 Similarly, addition of primary and stem cells, was key to the development of increasingly complex in vitro gastric models.9–12 Notwithstanding, these models were only developed for non-human cells. Recently, a long-lived, air-liquid interface gastric mucosoid culture model of human origin, was developed, closely resembling the phenotypical features of normal human gastric epithelium.13 However, it represented the gastric epithelial layer only, limiting the evaluation of physiological responses. We previously reported a 3D two-layered model, mimicking the gastric mucosa and lamina propria.14 The augmented 3D complexity reflected more closely the architecture of the native organ, while the fibroblastic components provided bioactive signaling to the epithelial layer. Despite notable progress, even the most advanced 3D culture models are limited and still lack the dynamic nature of the gastric mucosa, which is known to be extremely important for normal gastrointestinal physiology.15,16In vivo, cells are continuously sensing their vicinity and mechanical cues can, to a large extent, modulate the cellular response at the biochemical level.17,18 The contribution of biomechanical forces is particularly relevant in tissues such as the gut, where cells are repeatedly exposed to strain.19–22 Recent developments in soft-lithography, in particular in the fabrication of microactuators, enabled the creation of organ-on-a-chip devices, consisting in microfluidic platforms with capability to replicate a given functional sub-unit of a human organ.21–25 This bioinspired technology enabled the application of precisely defined amounts of strain to cell culture substrates, thus allowing to understand, to a greater level of detail, the implications of mechanotransduction at the cellular and molecular level.19,26
Recently, Lee and colleagues reported preliminary developments on a stomach-on-a-chip (SoC) model, where gut organoids were encased within a microfluidic device and exposed to cyclic peristalsis-like stretching.27 While presenting a promising model to address the effects of dynamic actuation on a differentiated gastric mucosa, the system used a matrigel-derived supporting gel that poorly recapitulated the human gastric extracellular matrix (ECM) composition and architecture. A more physiologically relevant environment, resembling the native gastric ECM, could be a critical asset when addressing system level response within these devices. Applications focusing on pathological ECM remodeling, as is the case during tumor invasion, could particularly benefit from such a model, since these processes are largely mediated by alterations in ECM components.28,29
In the current work, we propose a tissue engineering approach to design and fabricate a human-derived biomimetic SoC microdevice, experiencing peristalsis-like movement and intraluminal flow, with architecture and function emulating the native organ. The currently developed model was engineered to represent the three innermost layers of the gastric mucosa, namely the epithelial barrier (characterized by a tight cellular monolayer), the basement membrane (a layer mediating the adhesion and communication between the epithelial layer and the adjacent ECM), and an analogue of the lamina propria (consisting of a collagen type I gel with embedded normal gastric fibroblasts, able to potentially provide paracrine crosstalk between the lamina propria analogue and the epithelial layer and remodel the surrounding ECM). We then characterized the cellular and molecular alterations induced by the dynamic environment of the SoC, to assess its ability to promote functional and molecular traits characteristic of the native gastric mucosa.
The developed SoC represents a human-derived experimental in vitro asset for pre-clinical studies on host–pathogen interactions, drug–drug interactions, drug absorption or targeted delivery of compounds and nanoparticles, among many other possibilities.
2. Results and discussion
2.1. Design and fabrication of a multilayered stomach-on-a-chip (SoC) device
The SoC was designed and fabricated using a rapid prototyping method that we previously described.25To represent the three innermost layers of the stomach wall (epithelial barrier, basement membrane and supporting lamina propria), the SoC was devised as 9-layered device (Fig. 1a). The top layer was produced from thick PDMS, fabricated by gravity casting. The fluidic network was produced from 500 μm thick PDMS sheets and consisting of 3 fluid routing structures, intercalated by a 16 μm thick polyethylene terephthalate (PET) membrane (Fig. 1a and b). The proposed 3-tiered fluidic structure was designed to enable the replication of an epithelial barrier architecture on the top chamber and importantly, to provide the efficient incorporation of a lamina propria analogue, in the middle culture chamber. This allowed ECM-embedded fibroblasts to be adequately fed by flowing cell culture media above and underneath it. The microactuator was bonded directly below the fluidic network and is composed of a 2-part sub-assembly, namely a 250 μm thick PDMS membrane juxtaposed on a rectangular vacuum chamber of the same thickness (Fig. 1c). The entire biochip was sealed against a microscope glass slide.
2.2. On-chip emulation of stomach epithelium dynamics and architecture
To model the dynamic stomach microenvironment, peristalsis-like motion was imparted to the cell culture substrate by applying vacuum to the microactuator at cyclical intervals. The stretching pattern was modeled as a sinusoidal wave of 0.15 Hz frequency, which closely resembles the peristalsis pattern of the gut,22,30,31 at a maximum vacuum pressure of −100 mbar. We have previously shown that under these conditions, the integrated microactuator could produce surface stretching of approximately 10%, which is within the physiological range of strain that gut cells experience in vivo.25 Furthermore, as the microactuator was placed directly underneath the cell culture chamber, membrane flexing was effected from below, resulting in a tri-axial displacement of the cell culture membrane.Cell culture media was perfused over the epithelial layer at a flow rate of 2 μL min−1. As shear stress is not a dominant force on the stomach lumen, the fluidic routing structures were designed to keep shear stress at a low level. Taking in consideration a channel profile, 0.20 cm wide and 0.05 cm high, and a volumetric flow rate of 3.3 × 10−5 cm3 s−1, we estimated the shear stress over the epithelial cell culture to be about 0.003 dyne per cm2. This value is in line with the physiological shear stress experienced by gut cells in vivo, which is expected to range between 0.002 and 0.080 dyne per cm2.22,32
Emulation of the stomach epithelium was achieved on the two upper fluidic channels. The top channel encased the basement membrane analogue, consisting of a thin 50 μg mL−1 fibronectin gel coating the PET membrane, on top of which an epithelial monolayer was seeded (Fig. 1d). To mimic the selective permeability of the gastric mucosa it is important that the epithelial layer forms a cohesive, tightly packed monolayer, expressing fully developed cell-to-cell contacts. Here, we used a monolayer culture of MKN74 epithelial gastric cells (Table S1†). This cell line displays moderate differentiation, a typical epithelial cobblestone-like morphology and exhibits several characteristics of its epithelial origin. It displays fully developed and correctly localized tight junctions that mediate paracellular traffic across the epithelial barrier, as shown by staining of tight junction partners ZO-1 and occludin (Fig. S1a†). It also shows functional E-cadherin mediated cell-to-cell contacts, via adherens-junctions, as observed by co-immunofluorescence against E-cadherin and adherens-junction partner α-catenin (Fig. S1b†). This adhesion complex mediates tight monolayer integrity, by establishing cell-to-cell contacts and promotes an indirect link between the actin cytoskeleton of adjacent cells. The ability of cells to form an effective barrier was assessed by measuring the transepithelial electrical resistance (TEER). After day 2 post seeding, the cell line already exhibited TEER values above 100 Ω cm2, characteristic of a fully formed barrier, reaching an average of about 157 Ω cm2 after 5 days in culture (Fig. S1c†).
Taken together, these observations suggest that the MKN74 cell line was well suited to emulate the gastric barrier in the current SoC model.
To mimic the lamina propria, we used a 3 mg mL−1 collagen type I gel, since it is the most prevalent ECM protein in the human gastric lamina propria.33 The collagen gel was then embedded with normal gastric NST-20 fibroblasts (Table S1†), potentially introducing physiologically relevant ECM remodeling of the lamina propria analogue. The ECM was directly injected in the central fluidic channel, while the top and bottom perfusion channels acted as a direct feeding layer to the ECM embedded fibroblasts (Fig. 1d).
2.3. On-chip conditions did not affect cellular homeostasis of the epithelial barrier
Although PDMS-based devices are permeable to air, it is unlikely that this exchange is sufficient to ensure an efficient buffering capacity of the atmospheric 5% CO2, used in conventional cell culture incubators, particularly in pressurized perfusion systems, such as the one used here. To ensure that cells growing under CO2 depletion and cells growing under conventional cell culture atmosphere had the same basal readings, we compared their phenotype, proliferation, metabolic activity and doubling time, when fed with CO2-independent culture medium in a CO2-depleted atmosphere or with RPMI 1640 in the presence of CO2. No morphological differences were registered between the two groups (Fig. 2a). Furthermore, Ki67 staining and quantification at 48 h of growth showed mostly proliferative populations (Fig. 2b and c), and both exhibited a comparable doubling rate (Fig. 2d). Interestingly, we observed that cells growing under standard conditions, displayed a higher metabolic rate at early time points, compared to those grown in a CO2-depleted atmosphere. However, this difference equalized after ca. 8 days in culture (Fig. 2e). This observation could suggest that cells growing under CO2-depleted conditions, and fed with buffered medium, may need a longer period to attach and stabilize onto the substrate and start the replicative process, until they reach a similar doubling rate compared to cells growing in standard conditions.2.4. Impact of embedding stomach fibroblasts within a collagen type I gel
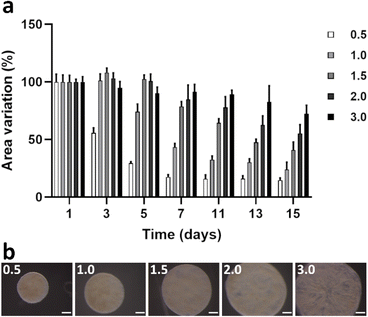
To properly emulate the lamina propria of the stomach wall, it is important that the designed model replicates, at least in part, the cellular milieu of its native counterpart. Here, we used a normal stomach fibroblast cell line (NST-20) embedded in a collagen type I gel. We aimed to potentiate the crucial native ECM remodeling. In this context, it was important to assess if ECM embedded fibroblasts were metabolically stable and growing in optimal conditions within the hydrogel gel.We first studied the off-chip behavior of gastric NST-20 fibroblasts, seeded at an initial density of 2 × 105 cells per mL, within a collagen type I gel. Observations were carried out over an increasing range of collagen type I densities from 0.5 to 3.0 mg mL−1. Metabolic activity of ECM-embedded fibroblasts was assessed over a period of 17 days and quantified by resazurin assay.
Our observations showed that the metabolic activity increased proportionally with increasing matrix density (Fig. 3a). For 0.5 mg mL−1 gels, metabolic activity peaked at day 5 and reached a stable plateau until the end of the experimental period. For any other gel density, metabolic activity increased steadily, reaching a peak at day 11. The same conditions displayed a small reduction in metabolic activity after day 11, which stabilized around day 15–17 in culture. Our results suggested that fibroblasts growing in a 3.0 mg mL−1 collagen gel needed a longer time to stabilize and start the replicative process (Fig. 3a). This was most likely due to the stiffer environment in which cells were initially seeded. However, once cells stabilized and developed full anchoring, the stiffer conditions seemed more advantageous for cell metabolism. To address the hypothesis that the high metabolic rates observed for higher density gels could result from cells growing selectively on the surface of the hydrogel, and thus not within a truly 3D environment, we performed a live–dead assay at day 13 post-seeding to assess qualitatively the viability of the embedded fibroblasts and imaged the entire hydrogel volume by z-stack imaging (Fig. S2†). Our results showed that cells remained viable within the hydrogel, with no appreciable differences in viability from 1.0 to 3.0 mg mL−1. In contrast, the 0.5 mg mL−1 gel exhibited a high number of dead cells in the core of the collagen discs (Fig. 3b). We have observed that the lower density gels potentiated very fast cell growth and migration and within 3 days, the collagen gel was completely occupied by the embedded fibroblasts. In turn, this could potentiate a higher density of cells in the core of the hydrogel, which could ultimately lead to a nutrient-depleted region within the core of the gel. Calcein staining also allowed morphological characterization of the stomach fibroblasts within the 3D matrices. Cells displayed a typical elongated spindle shape, characteristic of a mesenchymal phenotype. Protrusions establishing contact between adjacent cells could also be observed, suggesting that cell-to-cell signaling was being promoted within the collagen gel (Fig. 3b). Finally, evidence of ECM remodeling was also observed, as shown by immunocytochemistry against collagen type IV and fibronectin, both components of the native gastric lamina propria. De novo expression of collagen type IV could be observed as early as day 1 post-seeding (Fig. 3c), with extensive deposition of both collagen type IV and fibronectin matrix proteins after 11 days in culture (Fig. 3c). Of note, deposition of collagen type IV seemed to co-localize with thick bundles of collagen type I near the vicinity of fibroblasts, also suggesting de novo deposition of collagen type I (Fig. 3d). These results pointed to a robust remodeling of the lamina propria by the fibroblastic population, thus increasing the biomimetic dimensionality of the lamina propria analogue. Interestingly, remodeling of collagen type IV has been previously associated with malignant transformation in gastric tumors.34De novo expression of accessory ECM molecules such as collagen type IV within our model, could be effectively explored to elucidate its role in the malignant transformation leading to gastric cancer. Taken together these observations suggested that NST-20 fibroblasts were not adversely affected by the embedding within the collagen type I hydrogel. Under these circumstances, they can reliably be used to mimic the environment of the gastric lamina propria.
It is well understood that fibroblasts anchored to a collagen substrate exert contractile forces that can reshape the collagen fibers into a higher density structure with concomitant loss of volume.35,36 However, integrating on-chip a fibroblast-laden collagen hydrogel requires the hydrogel to withstand continued mechanical distension and retain, as much as possible, its shape in order to fill-in the culture chamber completely, thus avoiding disruption of fluid flow. Therefore, we next conducted a gel contraction assay, using the same range of collagen gel densities described above. We monitored fibroblast-laden hydrogel volume contraction every other day, for a period of 15 days. We observed a decrease of gel area proportional to decreasing hydrogel density, with lower density gels contracting significantly more. A 0.5 mg mL−1 gel contracted on average ca. 86% of its original size by day 15 in culture. In sharp contrast, a 3.0 mg mL−1 gel retained 77% of its original size over the same period (Fig. 4a and b).
Taking these observations into consideration, along with the fact that 3.0 mg mL−1 hydrogels could sustain mechanical actuation for extended periods of time without loss of integrity (Fig. S3†), we decided to conduct further experimentation using a fibroblast laden 3.0 mg mL−1 collagen type I gel.
2.5. A fibronectin thin gel mimicked the basement membrane
In vivo, the basement membrane anchors the epithelial layer, assisting in the maintenance of epithelial integrity and playing a crucial role in tissue compartmentalization.37,38 Given its preponderant role in tissue maintenance and disease,39,40 we aimed to reconstitute an anchoring substrate mimicking the in vivo gastric basement membrane. Here, we tested on a single-channel microfluidic chip several coating formulations, consisting of one of the major structural components of the gastric basement membrane, collagen type IV (at 4 mg mL−1) and one accessory molecule, fibronectin (at 50 μg mL−1),41 that functions as a bridging molecule between the most abundant structural molecules, directly linking the ECM to the epithelial cells. Namely we tested a pure collagen type IV coating, mixtures of collagen type IV and fibronectin at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 ratios and a pure fibronectin coating. Our results showed that MKN74 cells seeded over a thin coating of collagen type IV displayed an atypical phenotype, with a rounded shape. Over time, cells tended to remain isolated and exhibited cellular protrusions (Fig. 5a, panel 1). This suggested that cells were attempting to establish typical epithelial cell-to-cell contacts, but were unable to form a sheet-like epithelial monolayer. In contrast, when a mixture of higher fibronectin concentration was used as surface coating, MKN74 cells started exhibiting the typical epithelial phenotype, with cells displaying a cobblestone-like appearance and occupying the entire culture chamber surface (Fig. 5a, panel 2 and 3). This observation was more prominent when cells were seeded in a gel of pure fibronectin (Fig. 5a, panel 4), where cells displayed a normal phenotype, comparable to that of cells grown routinely in cell cultured-treated plastic vessels. We also quantified the number of cells showing a fully adherent phenotype vs. cells displaying an aberrant loosely adherent phenotype, 4 h post-seeding (Fig. 5b). Our results showed that, on average, about 82% of the cells seeded over a fibronectin gel were fully adherent and displayed an epithelial phenotype. In contrast, when seeded in any of the other coating formulations, the percentage of adherent cells was reduced drastically. Populations seeded on pure collagen type IV and a 4
4 ratios and a pure fibronectin coating. Our results showed that MKN74 cells seeded over a thin coating of collagen type IV displayed an atypical phenotype, with a rounded shape. Over time, cells tended to remain isolated and exhibited cellular protrusions (Fig. 5a, panel 1). This suggested that cells were attempting to establish typical epithelial cell-to-cell contacts, but were unable to form a sheet-like epithelial monolayer. In contrast, when a mixture of higher fibronectin concentration was used as surface coating, MKN74 cells started exhibiting the typical epithelial phenotype, with cells displaying a cobblestone-like appearance and occupying the entire culture chamber surface (Fig. 5a, panel 2 and 3). This observation was more prominent when cells were seeded in a gel of pure fibronectin (Fig. 5a, panel 4), where cells displayed a normal phenotype, comparable to that of cells grown routinely in cell cultured-treated plastic vessels. We also quantified the number of cells showing a fully adherent phenotype vs. cells displaying an aberrant loosely adherent phenotype, 4 h post-seeding (Fig. 5b). Our results showed that, on average, about 82% of the cells seeded over a fibronectin gel were fully adherent and displayed an epithelial phenotype. In contrast, when seeded in any of the other coating formulations, the percentage of adherent cells was reduced drastically. Populations seeded on pure collagen type IV and a 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of collagen type IV and fibronectin displayed ca. 90% of the population with a non-adherent rounded phenotype. Cells seeded on a 1
1 mixture of collagen type IV and fibronectin displayed ca. 90% of the population with a non-adherent rounded phenotype. Cells seeded on a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 mixture of collagen type IV and fibronectin performed slightly better, with around 77% of the population remaining non adherent. Increasing the fibronectin proportion did show a gradual, even if not statistically significant, increase in the number of loosely adherent cells, displaying protrusions (Fig. 5b). It was also evident that the pure fibronectin anchoring substrate was well suited for the rapid development of a confluent monolayer under fluid flow conditions. Cells started adhering to the basement membrane as early as 30 min post-seeding forming an 80% confluent monolayer within 24 h (Fig. 5b), similarly to what was observed in Transwell inserts. Taking these observations in consideration, we opted to use a pure thin fibronectin coating as a basement membrane analogue, sacrificing the biomimetics of a mixed gel of collagen type IV and fibronectin, for a more functional adhesion substrate capable of performing well under flow conditions.
4 mixture of collagen type IV and fibronectin performed slightly better, with around 77% of the population remaining non adherent. Increasing the fibronectin proportion did show a gradual, even if not statistically significant, increase in the number of loosely adherent cells, displaying protrusions (Fig. 5b). It was also evident that the pure fibronectin anchoring substrate was well suited for the rapid development of a confluent monolayer under fluid flow conditions. Cells started adhering to the basement membrane as early as 30 min post-seeding forming an 80% confluent monolayer within 24 h (Fig. 5b), similarly to what was observed in Transwell inserts. Taking these observations in consideration, we opted to use a pure thin fibronectin coating as a basement membrane analogue, sacrificing the biomimetics of a mixed gel of collagen type IV and fibronectin, for a more functional adhesion substrate capable of performing well under flow conditions.
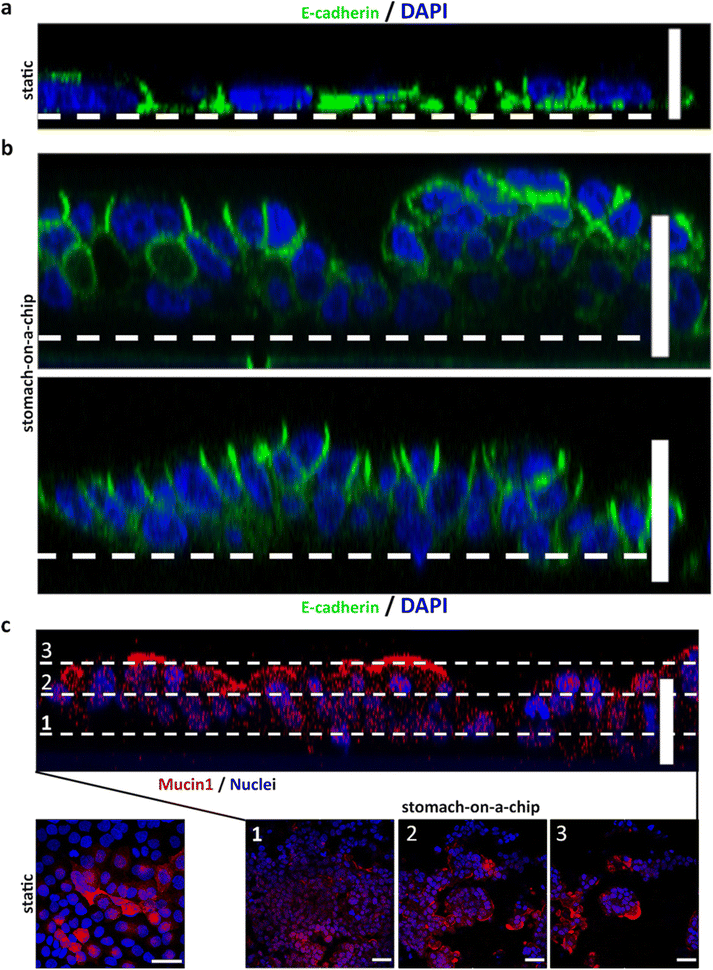
2.6. Stomach-on-a-chip device replicates the gastric epithelial barrier
The engineered stomach epithelium was housed within the upper portion of the biochip structure. The perforated membrane allowed communication between the adjacent fluidic structures, while ensuring that cells were kept within their designated confines, as was observed by staining the epithelial cells and the fibroblasts with cell tracker (Fig. 6a, upper panel). Epithelial cells formed a tightly knit epithelium once confluency was reached (Fig. 6a, lower panel). Importantly, barrier function did not appear to be affected by on-chip culture, since we observed that on-chip, MKN74 cells correctly expressed and localized at the cell membrane, adherens-junctions partner E-cadherin and tight-junctions partner ZO-1 (Fig. 6b). To understand the contribution of the dynamic conditions, towards the establishment of in vitro gastric function, we quantified pepsin activity at day 7 in culture, under static vs. dynamic conditions (n = 3). Comparison with the Transwell static system (S), showed that a regimen of flow alone (F), was not sufficient to induce an increase in pepsin activity. On the other hand, when fluid flow was coupled with biomechanical actuation (F + MA) an increase in pepsin activity was observed (Fig. 6c). Here, we showed that mechanical stretching of the epithelial population induced a significant increase in pepsin activity, suggesting that mechanical actuation might be promoting cytodifferentiation, similar to that observed in the native organ. Finally, as the fibroblast-laden hydrogel is at the inner core of the chip, we also qualitatively assessed the viability of the fibroblastic population growing on-chip by the live–dead assay. No major deviation was observed compared to our previous results obtained on off-chip collagen discs, with most of the fibroblastic population being viable within the hydrogel (Fig. 6d). This confirmed the efficiency of the lower perfusion compartment in providing correct feeding to the embedded fibroblasts.2.7. Stomach-on-a-chip replicates gastric epithelial barrier-function and selective permeability
Selective permeability to small molecules and drugs is a characteristic of epithelial interfaces, such as that of the human stomach. Therefore, we compared transport of a small 40 kDa molecule, namely fluorescein isothiocyanate (FITC)-labelled dextran, across the artificial epithelium to assess the impact of dynamic conditions, as well as of the engineered 3D architecture on paracellular transport across the epithelial barrier. Experimental conditions were measured on-chip for epithelial cells with culture media flow only (F), flow and mechanical actuation (F + MA), or the fully assembled SoC containing the epithelial and the fibroblastic populations and subjected to flow and mechanical actuation (SoC + F + MA). All of these conditions were compared to cells growing in static Transwell conditions (S). With this setup, we aimed to assess independently the contribution of each factor, namely the effect of flow, mechanical actuation and the on-chip emulation of the lamina propria, towards the establishment of organ level function within the chip. Here, a solution of FITC-labelled dextran was either added to the apical side of the epithelium in the Transwell static condition or perfused over the apical side of the SoC at a flow rate of 2 μL min−1. Samples were collected from the basal side chamber every 30 min, for a total period of 180 min. The Transwell inserts were slowly stirred for 5 s after each sample collection, to counteract the static nature of the Transwell system and to avoid the saturation of the epithelial barrier surface with FITC-labelled dextran molecules, which could limit diffusion across the barrier. Our observations showed a marked difference in paracellular permeability for cells growing under static conditions compared to dynamic conditions, with a significant difference already observable at 30 min (Fig. 7a). Flow alone (F) was sufficient to induce a marked decrease in the permeability of the gastric epithelium, in agreement to what we have previously reported, as flow alone was sufficient to induce alterations in the phenotype and molecular signature of these cells on a simplified microfluidic model of the gastric epithelium.25 A similar permeability profile was observed for MKN74 cells under flow and mechanical actuation (F + MA), as well as for the fully assembled SoC (SoC + F + MA). In all conditions tested, the gastric layer was tightly bound and fully formed, as observed by E-cadherin staining (Fig. 7b). Overall, our results suggest that dynamic conditions play a significant role in the establishment of a tightly knit epithelium, displaying highly selective permeability across the epithelial barrier when compared to a static model. This is a relevant observation, as conventional gastric static layouts, used as diffusion models across the epithelial barrier, may be underestimating the contribution of paracellular selectivity across the barrier.Finally, we aimed to characterize the phenotypic and molecular traits developed by the SoC gastric epithelium under dynamic conditions. As reported above, MKN74 cells formed a selective barrier after 2 days in culture when seeded at a density of 3.0 × 105 cells per mL. Taking this in consideration, the MKN74 cell population was seeded on-chip and allowed to reach confluency over a period of 3 days, followed by 3 days under flow regime and mechanical stretching of the cell culture substrate. Upon application of mechanical stretching and fluid flow, the SoC epithelium thickened considerably, reaching an average thickness of 38.5 μm and globular-like structures developed across its surface (Fig. 8a, panel 2 and 3). This is in sharp contrast to what was observed in static MKN74 cultures grown in Transwell inserts, where cells formed a flattened monolayer similar to a squamous epithelium (Fig. 8a, panel 1). Accordingly, the bioengineered SoC epithelium acquired a polarized state, with cells displaying a columnar shape with elongated nuclei located at the basal side and E-cadherin expression exclusively across the basolateral region. This phenotype closely resembled a pseudostratified columnar epithelium, similar in many aspects to its in vivo counterpart. Concurrently, expression of mucin-1 could be observed across the apical surface of the globular structures (Fig. 8b). This is in accordance to what can be observed in the native stomach42 and as previously reported for other MKN74 3D gastric models.25,43,44 Overall, our observations showed that the bioengineered device closely mimicked the gastric architecture and microenvironment by reproducing the three innermost-layers of the gastric epithelium. Furthermore, the peristalsis-like actuation and intra-luminal flow simulation seemed capable of producing physiologically relevant stimuli, inducing the MKN74 cell line, an otherwise moderately differentiated cell line that typically forms a squamous-like epithelium, to undergo further differentiation and acquire phenotypical characteristics typical of the normal gastric epithelium. Importantly, the introduction of the mesenchymal population to the stomach-on-a-chip device and the concomitant epithelial–mesenchymal cross-talk, did not affect the development of the differentiated state of the artificial gastric epithelium. The engineered SoC was not only capable of replicating the gastric mucosa architecture, by displaying a degree of differentiation and polarization characteristic of the native organ, but allowed also the partial reproduction of gastric function, as shown by enhanced barrier function. Here, we chose to represent the fibroblastic population, typically found within the gastric lamina propria and essential for tissue homeostasis. However, the lamina propria's cellular and molecular milieu is diverse and other aspects could be easily studied. Components of the immune system could be added to study response to induced inflammation or to tumor initiation. Likewise, an endothelial interface could be established on the lower perfusion channel, thus increasing the complexity of the system to represent the vasculature of the lamina propria, a model that would be well suited to study tumor invasion and intravasation.
3. Concluding remarks
Organ-on-a-chip devices enable a faithful replication of the tissue microenvironment, reproducing both architecture and in vivo dynamic conditions in one single device to potentiate the development of organotypic function. In the present work, we designed and fabricated a stomach-on-a-chip (SoC) device replicating both architectural and functional traits of the native organ. Our model could sustain a long-term 3D co-culture of epithelial and mesenchymal cells, without deleterious effects on cell homeostasis and viability. Importantly, we have demonstrated its capability for eliciting organ-level response, where cells developed phenotypic, molecular and functional traits characteristic of the normal gastric mucosa, that are usually not observable in 2D cultures. These observations outline the relevance of faithfully replicating in vitro the microenvironment of the gastric mucosa and, in particular, the importance of mimicking the dynamic nature of the native organ, in order to engineer a biomimetic analogue. Unlike the organoid-on-a-chip gastric system previously reported by Lee and colleagues, our stratified design allows easy access to both the apical and the basal side of the gastric epithelium, presenting a versatile model that can be used to study or simulate both luminal and systemic infection of the gastric mucosa. A further advantage of the currently proposed system is the use of a biomimetic supporting extracellular matrix (ECM), as a lamina propria analogue.The presently reported SoC constitutes a valuable tool to study gastric absorption dynamics, as well as host-pathogen interplay and directed-targeting therapies.
In summary, the low cost of the fabrication method coupled with the robust physiological response that the system elicits, makes the present SoC device a functional microfluidic platform to model the dynamic gastric microenvironment.
4. Experimental
Design
The conceived stomach-on-a-chip (SoC) consisted of 9 intercalated parts (Fig. 1a). From top to bottom, the top 6 layers formed the fluidic portion of the biochip. Layers 7 and 8 composed the microactuator and the bottom layer was a microscope slide. Polydimethylsiloxane (PDMS) microchannels were designed with a 10 mm (length) × 2 mm (width) main rectangular chamber and access holes measuring 2 mm in diameter (Fig. 1b). Each fluidic layer was separated by a 16 μm thick perforated polyethylene terephthalate (PET: it4ip) membrane with pore size of 8 μm and pore density of 6 × 104 cm2. The microactuator was a two-part assembly, composed of a 250 μm flexible PDMS membrane on top of a vacuum chamber. The vacuum chamber was 250 μm-thick and 11 mm (length) × 3.5 mm (width). The vacuum chamber was larger than the fluidic channels, thus ensuring proper actuation of the entire cell culture surface (Fig. 1c). The whole structure was assembled on top of a microscope glass slide and, for that reason, each structure was centered on a 26 mm square PDMS laminate, to match the width of the microscope slide.Fabrication
The SoC microdevice was fabricated from pre-cured PDMS laminated sheets and PET porous membranes using a method previously developed by our group.25 Briefly, the device was designed using computer aided design (CAD) software, exported to a cutting plotter (model CAMM-1 GS-24; Roland DG) and machined onto pre-cured 915 mm wide PDMS sheets (MVQ silicones). The top layer was fabricated by gravity casting. Prior to assembly, every material was thoroughly cleaned. Machined PDMS plates were washed with isopropanol (IPA) and air dried with compressed air. PET membranes were immersed in IPA, incubated in an ultra-sound bath for 15 min and dried with pressurized air. Microscope slides were incubated sequentially (5 min by ultrasound) in 2% (v/v) Hellmanex III (Helma Analytics) solution in double distilled (DD) water, followed by acetone and a final rinse in DD water. Clean slides were dried with compressed air and clean materials were either used immediately or stored in a dust-free environment.Assembly of the chip was performed sequentially from top to bottom (Fig. 1a). PDMS plates and glass slides were bonded by exposure to O2 plasma for 1 min (Zepto Plasma Laboratory Unit, Diener). Structures were then brought into conformal contact, placed between two glass slides and pressure was applied overnight. The outline of the microchannels in each layer was used as a guide to puncture access inlet and outlet holes through the top layer, with a 20 ga luer stub (Instech). To integrate the PET membranes, silanization of the PET surface was required. Following the cleaning procedure, both sides of the membrane were exposed to O2 plasma and immersed in 2% (v/v) bis-amino silane, 1% (v/v) DD water in IPA solution (20 min at 80 °C), followed by thorough cleaning with IPA and cured for 30 min at 70 °C. Membranes were then immersed for 30 min in 70% ethanol (v/v). Prior to bonding, both sides of the PDMS structures were exposed to O2 plasma and the wetted membrane was immediately sandwiched between them. This sub-assembly was further cured for 1 h at 70 °C.
Cell culture
The human-derived gastric cancer cell lines AGS and MKN74 were available at i3S/Ipatimup's cell line repository. For the SoC, the human-derived adenocarcinoma epithelial cell line, MKN74 was used. The human stomach normal fibroblast cell line NST-20 was a kind gift of Dr. Luis Filipe Santos Silva (i3S/Ipatimup, Portugal).Cell lines were routinely maintained at 37 °C and 5% CO2, under a humidified atmosphere and fed with RPMI 1640 (Gibco) medium, supplemented with 10% (v/v) fetal bovine serum (FBS; Biowest) and 1% (v/v) penicillin/streptomycin (pen/strep; Biowest). Cells were reseeded when confluency reached about 80% and tested for the presence of mycoplasma when thawed and every month when in culture.
Before on-chip seeding, MKN74 and NST-20 cells were conditioned for at least 48 h in CO2-independent cell culture medium (Gibco), supplemented with 4 mM of L-glutamine (Gibco), 10% (v/v) FBS (Biowest) and 1% (v/v) pen/strep (Biowest) in a humidified normal atmosphere (i.e., without CO2 buffering) at 37 °C.
Chip priming was started by sterilization with UV radiation (20 min), followed by flushing with 70% ethanol (20 min). Finally, the devices were rinsed with phosphate buffered saline (PBS; 3 × 15 min; Fisher BioReagents). Following sterilization, the fluidic portion was incubated with a 50 μg mL−1 fibronectin solution (Sigma) for 2 h at room temperature (RT).
NST-20 fibroblasts were embedded in a collagen type I gel. For that purpose, cells were harvested by trypsinization and suspended in a solution of 3 mg mL−1 collagen type I to a final density of 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per mL. The resulting suspension was carefully injected into the middle-tier microchannel and incubated for 30 min at 37 °C to allow gelification. The device was then coupled to a piezoelectric controller via polyether ether ketone (PEEK) tubing (Idex) and CO2-independent culture medium was perfused at a flow rate of 2 μL min−1. The system was allowed to equilibrate overnight, with the microactuator set to −50 mbar constant vacuum pressure to allow degassing of potential microbubbles present in the culture.25
000 cells per mL. The resulting suspension was carefully injected into the middle-tier microchannel and incubated for 30 min at 37 °C to allow gelification. The device was then coupled to a piezoelectric controller via polyether ether ketone (PEEK) tubing (Idex) and CO2-independent culture medium was perfused at a flow rate of 2 μL min−1. The system was allowed to equilibrate overnight, with the microactuator set to −50 mbar constant vacuum pressure to allow degassing of potential microbubbles present in the culture.25
To replicate the epithelial barrier, MKN74 cells were collected by trypsinization and resuspended to a density of 3.0 × 105 cells per mL. This suspension was then injected via a 4-way PEEK valve onto the upper fluidic channel. Cell seeding density was controlled under a phase contrast microscope and culture media flow was reinstated in both top and bottom fluidic channels at a flow rate of 2 μL min−1, 1 h after cell seeding. Cells were allowed a 72 h growth and stabilization period after which mechanical stretching was effected by application of cyclic vacuum. To emulate peristalsis motion experienced by gut cells in vivo, the stretching pattern was modelled as a sinusoidal wave of 0.15 Hz frequency and maximum vacuum pressure at −100 mbar.
Cell tracker staining
To monitor cell loading and compartmentalization on the SoC, MKN74 epithelial cells and NST-20 fibroblasts were stained with CellTracker™ green CMFDA and CellTracker™ red CMPTX dyes, respectively. Staining was performed according to the manufacturer's instructions. Briefly, cells were harvested and suspended in CellTracker staining solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution) for 30 min at 37 °C. Staining solution was then removed and cell seeding and/or embedding was performed as described above. Epithelial layer was grown to confluency after which the chip was imaged under a confocal microscope (model SP5; Leica Microsystems).
1000 dilution) for 30 min at 37 °C. Staining solution was then removed and cell seeding and/or embedding was performed as described above. Epithelial layer was grown to confluency after which the chip was imaged under a confocal microscope (model SP5; Leica Microsystems).
Morphological characterization
For immunofluorescence imaging, cells were washed in PBS with 0.05 mg mL−1 of CaCl2 (3 × 5 min), fixed with 4% (v/v) paraformaldehyde suspended in PBS (20 min at RT) and thoroughly washed with PBS (3 × 5 min). Cells were further incubated in 50 mM NH4Cl (10 min), followed by permeabilization of cellular membranes with 0.2% (v/v) Triton X-100 in PBS (5 min). Blocking was achieved with 5% (v/v) bovine serum albumin (BSA; Sigma) solution in PBS. Primary antibodies used were rabbit anti-E-cadherin (24E10; dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; Cell Signaling), rabbit anti-ZO1 (dilution 1
100; Cell Signaling), rabbit anti-ZO1 (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; ThermoFisher Scientific), rabbit anti-ocludin (dilution 1
200; ThermoFisher Scientific), rabbit anti-ocludin (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50; Santa Cruz), rabbit anti-collagen I (dilution 1
50; Santa Cruz), rabbit anti-collagen I (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; Rockland), mouse anti-collagen IV (dilution 1
100; Rockland), mouse anti-collagen IV (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; Dako), and rabbit anti-fibronectin (dilution 1
100; Dako), and rabbit anti-fibronectin (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150; Sigma). Primary antibodies were diluted in 5% (v/v) BSA suspended in PBS and incubated at 4 °C overnight. Secondary antibodies were diluted in 5% (v/v) BSA diluted in PBS and incubation was performed in the dark for 2 h at RT. Antibodies used were anti-rabbit Alexa Fluor 488 and/or anti-mouse Alexa Fluor 594 (dilution 1
150; Sigma). Primary antibodies were diluted in 5% (v/v) BSA suspended in PBS and incubated at 4 °C overnight. Secondary antibodies were diluted in 5% (v/v) BSA diluted in PBS and incubation was performed in the dark for 2 h at RT. Antibodies used were anti-rabbit Alexa Fluor 488 and/or anti-mouse Alexa Fluor 594 (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500; Thermo Scientific). Cellular preparations were mounted in Vectashield containing 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) for nuclei counterstaining. Image acquisition was performed with a confocal microscope as before.
500; Thermo Scientific). Cellular preparations were mounted in Vectashield containing 2-(4-amidinophenyl)-1H-indole-6-carboxamidine (DAPI) for nuclei counterstaining. Image acquisition was performed with a confocal microscope as before.
Characterization of a lamina propria analogue and analysis of ECM deposition
To emulate the lamina propria, a collagen type I (5 mg mL−1; Ibidi) hydrogel embedded with normal stomach fibroblasts (NST-20 cell line) was used. A range of collagen type I densities was tested, namely 0.5, 1.0, 1.5, 2.0 and 3.0 mg mL−1. Cell embedding and gelification of hydrogels was performed according to the manufacturer's recommendations. Briefly, NST-20 cells were trypsinized and resuspended to a final cellular density of 2 × 105 cells per mL. After combining the fibroblasts with the collagen type I, 30 μL per gel of the resulting suspension were plated on the bottom of a flat-faced well of a 24-well plate well (Corning). To trigger gelification and ensure cell viability, fibroblast-embedded hydrogels were incubated for 30 min at 37 °C under a humidified, normal (without CO2) atmosphere. After gelification, 1 mL of CO2-independent medium (supplemented as described above) was added to each well and gel contraction was monitored for 15 days. Discs were photographed every other day under a stereomicroscope (Olympus SZX10) coupled with a camera (Olympus EP50) and disc perimeter was measured using image analysis software (Fiji). The results were plotted as average percent of size difference over time, normalized to initial disc size (day 0). ECM remodeling was studied by immunofluorescence. Fibroblast-embedded discs were collected at day 11 post-seeding and stained with primary antibodies against collagen type I, collagen type IV or fibronectin. Image acquisition was done in a confocal microscope as before.Metabolic activity of collagen-embedded fibroblasts
Metabolic activity was assessed by the resazurin assay. NST-20 cells were embedded in collagen type I discs as described before. Metabolic activity was assessed after the first 24 h in culture and every 48 h for 15 days, for which a stock solution of 0.1 mg mL−1 resazurin (Sigma) was diluted to 20% (v/v; resazurin/cell culture media). Culture media was replaced with 500 μL of this solution and incubated at 37 °C for 2 h. 200 μL of the resulting supernatant were transferred to an opaque 96-well plate with clear bottom (Greiner) and the fluorescence signal was measured (excitation 530 nm/emission 590 nm) in a multiwell plate reader (model Sinergy MX HM550; Biotek Instruments). Metabolic activity was expressed as average relative fluorescence units (RFUs) ± standard deviation (SD).Cell viability assay
To assess viability of the NST-20 fibroblasts embedded on a collagen type I gel, a live dead assay was performed. Briefly, a staining solution containing 2 μg mL−1 of calcein and 2.5 μg mL−1 of propidium iodide was prepared. Cells were washed thrice with unsupplemented culture medium, followed by a 20 min incubation at 37 °C with staining solution. Finally, cells were washed twice with unsupplemented culture medium and immediately imaged under a confocal microscope as before. Z-stacks were acquired with a 2 μm z-step and post-processed using image analysis software (Fiji).45Epithelium characterization
The on-chip formed epithelium was fixed and stained with an antibody against E-cadherin as described above. Z-stacked fluorescence images were acquired with a 2 μm z-step value, using a confocal microscope as before. Z-stacks were post-processed and epithelium height measured using image analysis software (Fiji). For 3D rendering of the stomach epithelium, z-stacks were processed using Fiji's 3D viewer plugin.Paracellular permeability
To measure paracellular permeability, transport of fluorescein isothiocyanate (FITC)-labelled dextran (40 kDa; Sigma) across the epithelium was assessed over time. For on-chip measurements, cells were grown to confluency and cultured under cell culture media flow (2 μL min−1) with or without exposure to cyclic mechanical actuation for 3 days, at a 0.15 Hz frequency. FITC-dextran solution (1 mg mL−1) was perfused through the top channel and 30 μL samples were collected at the lower channel outlet every 30 min, for a period of 3 h. Samples were analyzed in a multiwell plate reader as before at 488 nm excitation and 520 nm emission. The conditions flow (F) and flow coupled with actuation (F + A) were assessed in a double channel chip, without the addition of the lamina propria analogue. Thus, the contribution of each condition was estimated independently. Static conditions were studied using Transwell inserts (Corning), coated with fibronectin (50 μg mL−1). Cells were allowed to reach confluency and grown for 3 additional days. Permeability was assessed by adding 200 μL of 1 mg mL−1 FITC-dextran solution to the apical side of the Transwell and collecting 50 μL samples from the basal side, while replenishing the same volume with fresh medium. Samples were collected up to 3 h and analyzed in a multiwell plate reader as before at 488 nm excitation and 520 nm emission. In both dynamic and static conditions, permeability was assessed as the relative transport of small molecules across the epithelial barrier and expressed as the fluorescence value collected at the basal channel, normalized to the fluorescence signal of the solution added to the apical side. Results were plotted as average percentage of FITC-dextran release against elapsed time (n = 3).Pepsin/pepsinogen activity
Gastric epithelial function was measured by quantifying the specific activity of pepsin/pepsinogen. A pepsin/pepsinogen activity assay kit (Biovision) was used. The procedure was performed according to the manufacturer's instructions. Briefly, cells were grown for 3 days post-confluency, either in Transwell inserts (static conditions) or on-chip with culture media flow and in the presence or absence of cyclic mechanical stretching (0.15 Hz frequency and −100 mbar vacuum pressure). Samples (100 μL) were collected from the basal side and 20 μL were added to 80 μL of pepsin assay buffer. A pepsin substrate working solution (pepsin substrate stock solution and pepsin assay buffer at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (v/v)), was then added to the previous mixture to start the reaction. Fluorescence was measured in a multiwell plate reader as before at 328 nm excitation and 418 nm emission in kinetic mode for 1 h at 37 °C. Sample pepsin activity was calculated according to the following equation:
10 (v/v)), was then added to the previous mixture to start the reaction. Fluorescence was measured in a multiwell plate reader as before at 328 nm excitation and 418 nm emission in kinetic mode for 1 h at 37 °C. Sample pepsin activity was calculated according to the following equation:Sample![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pepsin pepsin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) activity = B/(ΔT × P) activity = B/(ΔT × P) |
Statistical analysis
Data was compiled and analyzed using the software Graphpad Prism (Graphpad software). Data plotted in graphical format is presented as mean value ± SD. Cell culture doubling time and metabolic rates (n = 6) were compared using a two-way ANOVA statistical test. The latter were analyzed in conjunction with a Sidak's post hoc multiple comparison test. Relative transport across the paracellular route and cell adhesion assay, were analyzed using a two-way ANOVA in conjunction with Tukey's multiple comparison test. Pepsin activity mean values were compared using a non-parametric Mann–Whitney U test (n = 3). Statistical significance was defined as p < 0.05 (*), p < 0.01 (**), p < 0.0001 (****).Author contributions
Daniel A. Ferreira: conceptualization, investigation, writing – original draft, methodology. João P. Conde: writing – review & editing, funding acquisition, supervision. Mario Rothbauer: writing – review & editing, methodology. Peter Ertl: writing – review & editing, funding acquisition, supervision. Pedro L. Granja: writing – review & editing, funding acquisition, project administration, supervision, conceptualization. Carla Oliveira: writing – review & editing, funding acquisition, project administration, supervision, conceptualization.Conflicts of interest
Authors declare no conflict of interest.Acknowledgements
This work was supported by Fundação para a Ciência e Tecnologia (FCT) and BiotechHealth Programme (Doctoral Programme on Cellular and Molecular Biotechnology Applied to Health Sciences; ref. PD/00016/2012), by Programa Operacional Potencial Humano (POPH), and by SkinChip project (PTDC/BBB-BIO/1889/2014). The work has been also financed by: Fundo Europeu de Desenvolvimento (FEDER) Regional funds through the COMPETE 2020 – Operational Programme for Competitiveness and Internationalization (POCI), Portugal 2020, and by Portuguese funds through FCT/Ministério da Ciência, Tecnologia e Inovação in the framework of the projects: (1) 3DChroMe (PTDC/BTM-TEC/30164/2017), (2) LEGOH project (PTDC/BTM-TEC/6706/2020), (3) DETONATE project (2022.02951.PTDC); Norte Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF) for project NORTE-01-0145-FEDER-072678 - Consórcio PORTO.CCC – Porto.Comprehensive Cancer Center. DAF acknowledges FCT for their support through a FCT/BiotechHealth PhD programme grant, ref. PD/BD/105976/2014 and salary support from the LEGOH project (PTDC/BTM-TEC/6706/2020). JPC acknowledges FCT funding through INESC MN (Unidade ID 5367).The authors would also like to thank: Jorge Ferreira (Epithelial Polarity and Cell Division Group, i3S/IBMC) for granting access to the plasma cleaner equipment and for the insightful scientific support; i3S Scientific Platform (Biointerfaces and Nanotechnology core facility, i3S/INEB), member of the national infrastructure PPBI – Portuguese Platform of Bioimaging (PPBI-POCI-01-0145-FEDER-022122), in particular Maria Lázaro for support and access to the SP5 confocal microscope; Aureliana Sousa (Biofabrication Group, i3S) for scientific support and discussion; Sara C. Neves (Biofabrication Group, i3S) for support with the dynamic viscosity measurements; Celso Reis (Glycobiology in Cancer group, i3S) for kindly providing the antibody against Mucin-1; Ana Rita Matos (Expression Regulation in Cancer Group, i3S) for support with Biorender software and the layout of the graphical abstract.
Notes and references
- R. H. Hunt, M. Camilleri, S. E. Crowe, E. M. El-Omar, J. G. Fox, E. J. Kuipers, P. Malfertheiner, K. E. McColl, D. M. Pritchard, M. Rugge, A. Sonnenberg, K. Sugano and J. Tack, Gut, 2015, 64, 1650–1668 CrossRef CAS.
- M. H. Ross and W. Pawlina, Histology : a text and atlas, Lippincott Williams & Wilkins, Philadelphia, Pa., London, 6th edn, 2011 Search PubMed.
- P. J. Kumar and M. L. Clark, Kumar & Clark's clinical medicine, Saunders Elsevier, Edinburgh, 7th edn, 2009 Search PubMed.
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209–249 CrossRef.
- S. A. Langhans, Front. Pharmacol., 2018, 9, 6 CrossRef PubMed.
- J. Arrowsmith and P. Miller, Nat. Rev. Drug Discovery, 2013, 12, 569 CrossRef CAS PubMed.
- F. Pampaloni, E. G. Reynaud and E. H. Stelzer, Nat. Rev. Mol. Cell Biol., 2007, 8, 839–845 CrossRef CAS.
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. J. Yang, Assay Drug Dev. Technol., 2014, 12, 207–218 CrossRef CAS.
- N. Barker, M. Huch, P. Kujala, M. van de Wetering, H. J. Snippert, J. H. van Es, T. Sato, D. E. Stange, H. Begthel, M. van den Born, E. Danenberg, S. van den Brink, J. Korving, A. Abo, P. J. Peters, N. Wright, R. Poulsom and H. Clevers, Cell Stem Cell, 2010, 6, 25–36 CrossRef CAS.
- T. Grikscheit, A. Srinivasan and J. P. Vacanti, J. Pediatr. Surg., 2003, 38, 1305–1309 CrossRef.
- T. Sato, R. G. Vries, H. J. Snippert, M. van de Wetering, N. Barker, D. E. Stange, J. H. van Es, A. Abo, P. Kujala, P. J. Peters and H. Clevers, Nature, 2009, 459, 262–265 CrossRef CAS PubMed.
- A. L. Speer, F. G. Sala, J. A. Matthews and T. C. Grikscheit, J. Surg. Res., 2011, 171, 6–14 CrossRef CAS PubMed.
- F. Boccellato, S. Woelffling, A. Imai-Matsushima, G. Sanchez, C. Goosmann, M. Schmid, H. Berger, P. Morey, C. Denecke, J. Ordemann and T. F. Meyer, Gut, 2019, 68, 400–413 CrossRef CAS.
- B. N. Lourenco, T. Dos Santos, C. Oliveira, C. C. Barrias and P. L. Granja, Acta Biomater., 2018, 82, 68–78 CrossRef CAS.
- M. Bashashati and R. A. Hejazi, Dig. Dis. Sci., 2018, 63, 3164–3166 CrossRef PubMed.
- I. R. Sanderson, Am. J. Clin. Nutr., 1999, 69, 1028S–1034S CrossRef CAS PubMed.
- T. Mammoto, A. Mammoto and D. E. Ingber, Annu. Rev. Cell Dev. Biol., 2013, 29, 27–61 CrossRef PubMed.
- C. T. Lim, A. Bershadsky and M. P. Sheetz, J. R. Soc., Interface, 2010, 7, S291–S293 CrossRef PubMed.
- O. T. Guenat and F. Berthiaume, Biomicrofluidics, 2018, 12, 042207 CrossRef.
- C. Moraes, M. Likhitpanichkul, C. J. Lam, B. M. Beca, Y. Sun and C. A. Simmons, Integr. Biol., 2013, 5, 673–680 CrossRef PubMed.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Lab Chip, 2012, 12, 2165–2174 RSC.
- A. O. Stucki, J. D. Stucki, S. R. Hall, M. Felder, Y. Mermoud, R. A. Schmid, T. Geiser and O. T. Guenat, Lab Chip, 2015, 15, 1302–1310 RSC.
- M. Wufuer, G. Lee, W. Hur, B. Jeon, B. J. Kim, T. H. Choi and S. Lee, Sci. Rep., 2016, 6, 37471 CrossRef CAS PubMed.
- D. A. Ferreira, M. Rothbauer, J. P. Conde, P. Ertl, C. Oliveira and P. L. Granja, Adv. Sci., 2021, 8, 2003273 CrossRef CAS PubMed.
- E. Ergir, B. Bachmann, H. Redl, G. Forte and P. Ertl, Front. Physiol., 2018, 9, 1417 CrossRef.
- K. K. Lee, H. A. McCauley, T. R. Broda, M. J. Kofron, J. M. Wells and C. I. Hong, Lab Chip, 2018, 18, 3079–3085 RSC.
- X. He, B. Lee and Y. Jiang, Adv. Exp. Med. Biol., 2016, 936, 73–91 CrossRef CAS.
- W. P. Daley, S. B. Peters and M. Larsen, J. Cell Sci., 2008, 121, 255–264 CrossRef CAS.
- C. P. Gayer and M. D. Basson, Cell. Signalling, 2009, 21, 1237–1244 CrossRef CAS PubMed.
- M. D. Basson, Digestion, 2003, 68, 217–225 Search PubMed.
- T. Ishikawa, T. Sato, G. Mohit, Y. Imai and T. Yamaguchi, J. Theor. Biol., 2011, 279, 63–73 CrossRef CAS PubMed.
- A. Gillessen, B. Voss, J. Rauterberg and W. Domschke, Scand. J. Gastroenterol., 1993, 28, 688–689 CrossRef CAS PubMed.
- Z. N. Wang and H. M. Xu, World J. Gastroenterol., 2000, 6, 438–439 CrossRef.
- E. Bell, B. Ivarsson and C. Merrill, Proc. Natl. Acad. Sci. U. S. A., 1979, 76, 1274–1278 CrossRef CAS.
- T. Zhang, J. H. Day, X. Su, A. G. Guadarrama, N. K. Sandbo, S. Esnault, L. C. Denlinger, E. Berthier and A. B. Theberge, Front. Bioeng. Biotechnol., 2019, 7, 196 CrossRef PubMed.
- A. Pozzi, P. D. Yurchenco and R. V. Iozzo, Matrix Biol., 2017, 57–58, 1–11 CrossRef CAS.
- T. Rozario and D. W. DeSimone, Dev. Biol., 2010, 341, 126–140 CrossRef CAS PubMed.
- R. Sekiguchi and K. M. Yamada, Curr. Top. Dev. Biol., 2018, 130, 143–191 CAS.
- A. M. Moreira, J. Pereira, S. Melo, M. S. Fernandes, P. Carneiro, R. Seruca and J. Figueiredo, Cells, 2020, 9, 394 CrossRef CAS PubMed.
- A. C. Y. Li and R. P. H. Thompson, J. Clin. Pathol., 2003, 56, 885–887 CrossRef CAS PubMed.
- D. Boltin and Y. Niv, J. Gastrointest. Dig. Syst., 2013, 3, 15519 Search PubMed.
- S. Rocha, J. Carvalho, P. Oliveira, M. Voglstaetter, D. Schvartz, A. R. Thomsen, N. Walter, R. Khanduri, J. C. Sanchez, A. Keller, C. Oliveira and I. Nazarenko, Adv. Sci., 2019, 6, 1800948 CrossRef.
- M. Balmana, S. Mereiter, F. Diniz, T. Feijao, C. C. Barrias and C. A. Reis, Molecules, 2018, 23, 2815 CrossRef.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2lc01132h |
| ‡ Joint senior authorship. |
| This journal is © The Royal Society of Chemistry 2023 |