Open Access Article
Open Access ArticleVascularized human brain organoid on-chip
Sin Yen
Tan
a,
Xiaohan
Feng
 a,
Lily Kwan Wai
Cheng
b and
Angela Ruohao
Wu
a,
Lily Kwan Wai
Cheng
b and
Angela Ruohao
Wu
 *abcde
*abcde
aDepartment of Chemical and Biological Engineering, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong. E-mail: angelawu@ust.hk
bDivision of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong
cState Key Laboratory of Molecular Neuroscience, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong S.A.R, China
dHong Kong Branch of Guangdong Southern Marine Science and Engineering Laboratory (Guangzhou), The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong S.A.R, China
eCenter for Aging Science, The Hong Kong University of Science and Technology, Clear Water Bay, Kowloon, Hong Kong S.A.R, China
First published on 31st May 2023
Abstract
Modelling the human brain in vitro has been extremely challenging due to the brain's intricate cellular composition and specific structural architecture. The recent emergence of brain organoids that recapitulate many key features of human brain development has thus piqued the interest of many to further develop and apply this in vitro model for various physiological and pathological investigations. Despite ongoing efforts, the existing brain organoids demonstrate several limitations, such as the lack of a functional human vasculature with perfusion capability. Microfluidics is suited to enhance such brain organoid models by enabling vascular perfusion and a curated blood–brain barrier microenvironment. In this review, we first provide an introduction to in vivo human brain development and present the state-of-the-art in vitro human brain models. We further elaborate on different strategies to improve the vascularized human brain organoid microenvironment using microfluidic devices, while discussing the current obstacles and future directions in this field.
1. Introduction
The age-old question of what makes us human has intrigued neuroscientists to extensively study the human brain physiology and diseases for over a century, but paradoxically the brain remains the most poorly understood human organ.1 This is primarily due to its diverse yet highly regulated cellular interactions involving roughly 86 billion neuronal cells temporally and spatially coordinated with 85 billion glial cells, together with specialized vascular cells, to form and maintain a neuronal circuit consisting of an estimated 7000 synapses.2–4 Further adding to this complexity is its structural and functional heterogeneities and specificities across multiple brain regions.5 Indeed, development of various brain regions requires not only intricately orchestrated cellular organization but also a multitude of biochemical and mechanical signals, making the human brain an extraordinarily complicated organ to be explored.6 In addition to the brain's innate complexity, the inaccessibility of live human brain tissues and the inadequacy of human specific features in other model organisms have prompted the search for a more physiologically relevant in vitro human brain model.7Recently, the emergence of brain organoids, which are in vitro 3D human brain-like tissues derived from embryonic stem cells (ESCs) or pluripotent stem cells (iPSCs), represents a significant achievement in mimicking the complex cellular features and functionality of the human developing brain.8 They contain multiple brain-specific cell types that spatially organize into layers with specialized lineage commitment.8,9 More strikingly, they recently form a functional human neural circuitry through integration with their host's circuitry.10 Although promising, brain organoids did not accurately replicate all features of the human brain.11–15 Their tissue architecture only displays early stage structural organisation; therefore, they are only capable of mimicking the human brain development at the prenatal stage, which precludes their uses in exploring the aging brain and aging-associated neurodegenerative diseases.16 Furthermore, they lack some important cell types such as immune cells and vascular cells. Without vascular cells to maintain their metabolic needs, they eventually develop necrotic cores and cease to mature further.16–18 Another often overlooked issue is the organoid variability.19,20 There are variations in sizes and cell types between different batches, and even between individual organoids from the same batch. Various cell lines and bioreactors used in the organoid generation workflow introduce even larger disparities between organoids from different laboratories.19
Microfluidic technology is a promising solution for some of the abovementioned limitations. First, microfluidics facilitates the construction of a physiologically similar 3D microenvironment, which is largely missing in the current brain organoid models. This microenvironment includes proper spatial and temporal distribution of non-neuronal cells and signalling molecules surrounding the brain, as well as controllable induction of mechanical stimuli such as physiological fluid flow and shear stress.21 Second, microfluidics potentially allows standardization of brain organoids through the application of physical constraints, such as micropillars on-chip to control and reduce their size variation.22,23 Third, microfluidics enables the generation of a perfusable vascular network for potential brain organoid vascularization.24–26 Notably, accessible vascular lumens indicate the capability to allow delivery of substances, such as drugs or immune cells into the brain organoid. Given these advantages, there has been great interest in integrating microfluidics and organoid technologies.
Recently, several articles have provided valuable insights into the integration of brain organoids and on-chip technology albeit without much emphasis given to vascularization.27–30 Another review paper has discussed in detail about the vascular engineering approaches that are possibly applicable to vascularize various organoids mimicking different human organs.31 Although brain organoids and vasculature have been widely discussed, review papers specifically targeting the vascularization of brain organoids on-chip are inadequate. In this review, we aim to primarily focus on the vascularization of the human brain, in particular the curation of an in vitro vascularized brain organoid using microfluidics. We first briefly review the in vivo biological complexity of the human brain development and further highlight the existing advanced human brain models. We discuss the use of microfluidics to generate a functional vascularized human brain organoid on-chip and the current approaches to vascularize organoids. We also discuss various model design considerations including inclusion of relevant cell types, interfacing with the extracellular matrix as well as generating the relevant mechanical cues. We end by highlighting the remaining challenges and proposing future directions to inspire new solutions to overcome these shortcomings in the emerging field of organoids and microfluidics.
2. In vivo biology of the human brain and animal models
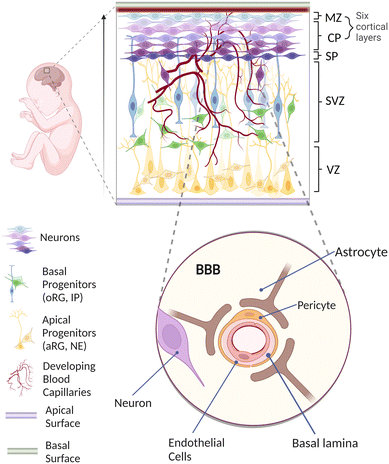
Understanding the pivotal stages of human brain development is of utmost importance for generating a physiologically realistic in vitro vascularized brain organoid on-chip.32,33In vivo, all the cortical neurons are derived from neuroepithelial (NE) cells through neurogenesis involving multiple sequential migration and differentiation throughout different layers of proliferative zones (Fig. 1). Initially, NE cells located at the ventricular zone (VZ) elongate to become apical radial glial (aRG) cells that eventually produce not only more aRG cells but also more differentiated cells, including basal progenitors (BPs) and neurons.34–38 BPs such as outer radial glial (oRG) cells and intermediate progenitors (IPs) subsequently migrate basally and dominate a newly formed subventricular zone (SVZ).39 While differentiating to become matured neurons, these cells continue to migrate outward and arrange themselves to occupy the cortical plate (CP), marginal zone (MZ) and subplate (SP) in an inside-out manner.40–45 Overall, the neurons have migrated approximately a few millimeters from the VZ to reach the final layer, resulting in a well-organized formation of six cortical layers.46In parallel with neurogenesis, vascularization of the human brain occurs mainly through angiogenic invasion from a mesoderm-derived vascular network into the developing brain.56–60 This invasion, followed by the constant vascular sprouting and remodelling, allows close interactions between vascular cells and various neuronal cells to establish a specialized organotypic feature known as the blood–brain barrier (BBB).59,61,62 As shown in the enlarged schematic in Fig. 1, the BBB is a highly selective border comprising endothelial cells (ECs), pericytes (PCs) and astrocytes (ACs) to maintain brain homeostasis via regulating solute exchange between the circulating blood and the brain.63,64 More evidence has implied that BBB dysfunction is one of the factors causing neurological diseases including multiple sclerosis (MS), brain ischemia and Alzheimer's disease (AD). For example, in MS, vascular dysfunction triggers immune cells to pass through the BBB from the blood circulation, resulting in neuronal damage through demyelination and chronic neuroinflammation.65 Brain ischemia is also associated with the disruption of the BBB, in which inadequate blood supply leads to the degradation of tight junctions and accumulation of cytokines,66 causing further break down in the BBB that enables immune cells to extravasate into the brain parenchyma.67,68 Similarly, in AD, the leakage of blood vessels causes BBB breakdown with the accumulation of a high level of metabolic solute such as amyloid beta plaques.69,70
At present, researchers most often employ animal models to study the BBB because there are few alternatives, and while animal models have provided very significant insight into the biological mechanisms involving the BBB, the intrinsic genetic and cytoarchitecture differences of animal brains to the human brain often result in failures to translate meaningful results to the clinic.71–75Table 1 shows a brief summary of the significant differences between the human and animal brains.
| Major differences | Human brain | Animal brain |
|---|---|---|
| Neo-anatomical | • More gyrification (cortical wrinkles)• Much larger brain size compared to rodents47 | • Less gyrification in primates, and absent in rodents49• Smaller brain size compared to humans • Cortical thickness: varies, 0.4 mm in mice, 1–2 mm in macaque48 |
| Cellular components | • More neurons compared to rodents and other primates• More complex neural networks50 • More expanded neural proliferative layers with subzones such as the appearances of outer SVZ during neurogenesis51 • More astrocytes to process 10× more GFAP+ than rodents52 |
• Chimpanzees have 2× less neurons compared to humans47• Mice have 10× less neurons compared to humans50 • Neural proliferative layer such as outer SVZ is absent in rodents during neurogenesis51,53 |
| Brain developmental timeline | • Human gestation period requires around 40 weeks54• Human neurogenesis requires around 27 weeks55 | • Mouse gestation period requires around 20 days• Mouse neurogenesis takes arounds 2 weeks55 |
3. Advanced in vitro human brain models
Advancements of in vitro human brain models raise the prospect of utilizing them to complement animal models as they can replicate key features of the human brain in a controlled laboratory setting.76 It is important to note that current in vitro human brain models are still impossible to exactly recapitulate the complexity of the human brain, and further validation tests using animal models and humans are necessary. Nonetheless, effective modelling of the human brain, especially with the tightly controlled BBB, remains one of the major bottlenecks for drug discovery and for understanding human brain disorders.77 Due to the diverse cell types involved, specific structure, inherent complex biochemical factors and dynamic cerebral blood flow, the human brain is difficult to accurately recreate in vitro.78 One of the earliest methods involved culturing human-derived brain cells on culture dishes, which could be either monoculture with a specific brain cell type or cocultures with multiple interacting cells.79,80 Although this method is reproducible, scalable, and easy to manipulate with widely available standardized operating protocols, it does not recapitulate the transport system of the BBB for functional tests such as permeability, cell migration and diffusion assays.The Transwell system, consisting of two compartments separated by a porous membrane, is a simplified model of the human BBB that has been widely applied to replicate the barrier and transport properties of the BBB for small molecules and drug permeability studies.81 Typically, ECs are cultured on top of the membrane while other brain parenchymal cells are either attached onto the opposite side of the membrane or at the bottom of the well. Several components of the Transwell system, such as the materials and pore sizes of the membrane could be designed and selected to customize for different experimental conditions.82 However, due to their static environment, they do not account for the dynamic properties of the BBB, such as the supply of nutrients and oxygen as well as the flow of cerebrospinal fluid.
Microfluidics could overcome the shortcomings of the Transwell system and potentially supplement in vivo studies of the human brain by allowing heterotypic human derived cell interaction while enabling realistic mimicry of human blood flow using perfusion. Indeed, these two features are crucial for recapitulating the human BBB environment. For instance, Park et al. demonstrated the advantages of having these features by separating ACs, PCs and ECs into top and bottom microfluidic compartments while allowing them to interact with each other via a porous membrane under physiological fluid flow (Fig. 2A).83 Importantly, this system allows the integration of trans-epithelial electrical resistance (TEER) electrodes for monitoring and quantitating the barrier function in a real-time manner.83,84 These yielded BBB functions similar to those of in vivo.83,85 However, the top-bottom configuration might obstruct the simultaneous real-time imaging of all BBB cell types. To address this issue, side-by-side chambers such as the SyM-BBB model by Prabhakarpandian et al. have been designed by using microposts or micropillars.86 Similarly, Deosarkar et al. designed a circular microfluidic compartment for culturing neonatal brain cells along with the vascular cells in the side channels separated by defined pores.87 However, they only included two BBB cell types (ACs and ECs) originating from rats instead of humans. As discussed earlier, the cells from rats bear different phenotypes compared to those of humans. Nonetheless, this assay has been proven to be useful in delineating the molecular mechanism of protein kinase for sepsis-induced brain inflammation and assessing antibody movement across the blood–brain barriers.88,89 While 2D BBB on-chips capture the important elements of the cell biology and provide many significant insights into BBB permeability, they often lack a functional 3D brain tissue environment, making this approach less ideal for the investigation of tissue level biological processes and systems.
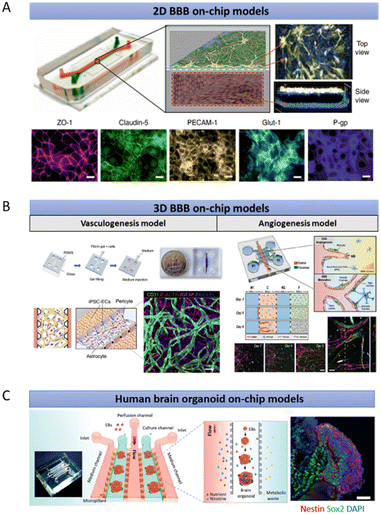 | ||
| Fig. 2 Existing microfluidic-based human brain models. (A) A representative 2D model of a microfluidic chip built upon the classical design by Huh et al., consisting of two channels with an upper channel containing PCs and ACs, separated from a lower channel containing ECs, by a porous membrane with pores that are at least 1 μm in diameter.125 (B) Representative models of 3D vasculogenesis and angiogenesis models to mimic the BBB with microposts to separate side-by-side channels. The inter-post open regions allow the opening of the vascular lumens through a thick hydrogel. (C) Brain organoid on-chip models to investigate the nicotine exposure effects towards the human brain. The figures are reproduced with permission/under Creative Commons license.83,90,91,126 | ||
3D BBB on-chips represent an alternative in vitro model that better mimics their counterparts in vivo in terms of biochemical heterogeneity, barrier functionality, and structural arrangement. Most of these 3D systems rely on the polydimethylsiloxane (PDMS)-based photolithography technique to create interspaced microposts for the formation of vascular open lumens, as depicted in Fig. 2B. These 3D luminal vascular networks, culturing alongside neuronal and perivascular cells, allow either dextran or bead perfusion for the measurement of the BBB integrity. For example, Campisi et al. reconstituted the human BBB environment by developing a 3D vasculogenesis-based vascular network that interacts directly with PCs and ACs.90 This results in low vascular permeability comparable to that of the in vivo BBB. By adopting a similar microfluidic design that features interspaced microposts, Lee et al. generated brain angiogenic sprouts with lumens to interact with both the brain and perivascular cells because barrier formation and maturation are often associated with angiogenesis (Fig. 2B).91 Brown et al. mixed 2D cultures of ECs, ACs and PCs with 3D cultures of human neurons in a BBB model termed neurovascular unit (NVU).92 They not only included all the crucial BBB cell types but also established flow with appropriate shear stress in their microfluidic model. They further demonstrated the applications of the NVU chip for drug study and metabolite analysis, which are challenging to achieve using a limited number of cells isolated from the microfluidic device.93,94 Additionally, the NVU chip was functionally coupled with other micro-physiological systems for analysing the penetration of metabolites through the BBB, in which the results were further validated in human studies.95,96 Although these models are capable of simulating direct cellular interactions in a 3D microenvironment, they lack in vivo cytoarchitectural structures and only involve limited cell types. Furthermore, the lifespan of these culture systems is short, rendering them unreliable to mimic many BBB related diseases, such as the long-term developing vascular dysfunction in AD.97
Alternatively, brain organoids containing diverse cell types might better recapitulate the human brain physiology with increased complexity, longer lifespan, and appropriate developmental timing.98 Typically, the first step of generating brain organoids is to form embryoid bodies (EBs) from ESCs or iPSCs using either suspension culture, hanging drop or microwell methods. Suspension culture is one of the earliest developed approaches that culture adherent cells using a non-adherent dish to force the cells to aggregate to form EBs. Random aggregation of the cells using this method often leads to large variability of EB sizes.99,100 Hanging drop is another method that allows the generation of EBs through simple inversion of the well plate containing tiny drops of cell suspension to subsequently allow EBs to form at the bottom of the droplet.101 While being straightforward, this requires careful handling and manipulation of the liquid to minimize cell loss. Microwell approach is another alternative that enables cells to aggregate and grow in a non-adherent microwell until they are limited by the growth space in the well. This creates homogeneous EBs with consistent sizes, making this method widely adopted for the current brain organoid culturing approaches.100,102–105
Generally, there are two widely applicable approaches for culturing brain organoids, each has its own pros and cons. The first approach uses an unguided approach, which takes advantage of the intrinsic capability of ESCs or iPSCs to self-organize and form neuroepithelial cells that adopt the neuroectodermal fate.98 The neuroepithelial cells self-organize into multiple neural rosettes that look like the neural tube, and the addition of a supporting matrix into the system such as 3D Matrigel further improves the growth of the neural rosettes. This subsequently results in the formation of multiple interdependent brain regions such as the dorsal cortex, hippocampus, and choroid plexus.98,105–107 However, these multiple brain regions formed in the organoid position themselves randomly without a proper and organized arrangement as their counterpart in vivo.108 Although this self-patterning of the cerebral organoid allows researchers to analyse and understand how discrete human brain regions develop and interact with each other, they suffer from significant batch-to-batch and organoid-to-organoid variability.98,109 In the second approach, extrinsic factors such as signalling molecules are often added into the system to direct the development and patterning of the neuroepithelial cells to become a specific brain region such as the forebrain and cortex.110–112 This strategy allows not only a detailed deconstruction of the organogenesis process but also a more reproducible and consistent organoid production system. However, not all the signals involved in producing each region of the human brain are known. This subsequently restricts the capability of forming multiple regions of the human brain in an individual organoid as well as the studies of the interaction between multiple regions. There are ongoing efforts to address this limitation by generating assembloids through the fusion of multiple brain-region specific organoids.113,114
It is important to note that while the brain organoid and brain spheroid are both in 3D shape and these terms are often interchangable, they are very different in terms of size, complexity, maturity, and reproducibility.98,115 The brain spheroid typically refers to aggregation of any brain cells into a brain tissue, while the brain organoid is structurally more complex and has organotypic distinct brain regions with heterotypic interacting cells that are absent in the spheroid; therefore, the brain organoid is usually larger in size.116 Compared to the organoid, the brain spheroid is easier to be generated and reproduced due to the availability of highly standardized protocols that allow researchers to easily form homogeneous spherical aggregates to replicate the simple tissue structure.117
A recent review paper discussed in depth about various brain organoid assays for investigating neurological phenotypes and brain diseases.54 Although brain organoids currently have broad applications, when they increase in size, limited nutrient supply to their inner cores causes necrotic cell death. The necrotic cells further release lysates that cause endoplasmic reticulum stress to the surviving cells at the outer layer of the organoid.118 All of these prevent the current brain organoids from maturing beyond the embryonic stage. Incorporating functional vasculatures may address this and possibly improve their overall lifespan and maturation level through the continuous supply of oxygen and nutrients. Additionally, an ideal vascularized brain organoid on-chip with better recapitulation of the organotypic phenotype and features of the BBB would act as a versatile and cost-effective in vitro platform for high-throughput screening of drug efficacy or toxicity in the therapeutic discovery of neurological diseases to restore BBB integrity.119
To date, efforts to generate functional vascularized human brain organoids show varying degrees of success, as summarized in Table 2. The main reason is that the vasculature generated using these approaches remains non-perfusable as these models do not possess any accessible sites to allow entry into the vasculature. To address the aforementioned limitations, a recent focus has shifted to the potential of integrating organoid technology and bioengineering.120 Several groups employed on-chip technologies to culture brain organoids. Karzburn et al. cultured a brain organoid inside a confined compartment of a microfluidic device to investigate the mechanism of brain wrinkling.121 By utilizing the closed compartment to constrain the height of the organoid, they could easily perform in situ whole-organoid fluorescence real-time imaging, which is challenging to achieve using a traditional dish model. Microfluidics has also been utilized to improve the reproducibility and reduce the size variation of brain organoids. For example, Ao et al. devised a one-stop assembly approach for culturing brain organoids from the beginning to the end within a single microfluidic chip without too much disturbance.122 They not only constrained the brain organoids to ensure their sizes to consistently remain at 2 μm but also exposed them to atmospheric oxygen to prevent necrotic core formation. Additionally, the microfluidic device features a bottom layer perfusable chamber to supply the medium to the upper layer brain organoids through a polytetrafluoroethylene-coated wire mesh. Although this hydrophobic wire mesh allowed the EBs to form without adhering to the surface, it might obstruct the real-time imaging of the brain organoids in the device.122 Seiler et al. developed an automated cell feeding on-chip platform to control the flow rate and feeding schedule for maintaining the brain organoid culture, and to minimize the effect of uncontrolled variables while changing the medium.123 To further improve nutrient absorption and allow the formation of longer neuroepithelial-like zones, Romero-Morales et al. designed a miniaturized spinner named Spin∞ that allows long term culture of brain organoids.124
| Approaches to vascularize brain organoids | Novelty | Limitations | Ref. |
|---|---|---|---|
| Note: These papers are chosen because they are the pioneers in establishing novel strategies for vascularizing the brain organoid. | |||
| Transplantation into animals | Establishment of vascular perfusion using the host vasculature | Vascular system is not entirely of human origin | 132 |
| Potential host cell contamination in the brain organoid | |||
| Inherent differences between the vasculatures of humans and animals | |||
| Direct incorporation of ECs into brain organoids | Brain organoids that harboured vessel-like structures | Lack of functional vascular perfusion | 133 |
| HUVECs are not brain-specific ECs | |||
| Gene overexpression | Brain organoids contain vessel-like structures that have BBB characteristics | Lack of functional vascular perfusion | 134 |
| Fusion with blood vessel organoids | Simultaneous establishment of vasculatures and microglia-like cells in the brain organoid | Lack of functional vascular perfusion | 135 |
In another example, Wang et al. investigated the effect of prenatal nicotine exposure on a brain organoid via perfusion flow (Fig. 2C).126 However, they only characterized the maturity and functionality of the brain organoid on-chip around one month old that recapitulates the early foetal brain development in which the neurons are still not fully matured and the oligodendrocytes are still largely missing.127,128 The effect of perfusion flow on neuronal activities such as synchronized bursts and spikes could only be detected in greater than two month old organoids.127 Indeed, most of the established brain organoid protocols allow the brain organoids to further mature for up until one year to mimic the later stage of foetal brain development.98,104,105 The significance of the culture period has also been reviewed and discussed by Gopurappilly et al.129 Similarly, Ao et al. examined the infiltration of young and old monocytes into a 45 day old brain organoid using a 3D printed microdevice.130 A prolonged culture of the brain organoid to reflect the aged brain senescent phenotypes would be necessary to improve our understanding of the brain aging. In the same study, they confined the brain organoid in their platform to allow it to develop into a pancake-shape structure to reduce the inner core necrosis. Nevertheless, the question of whether perfusion flow could overcome the problem of late-stage brain organoid necrosis remains unanswered. Most importantly, none of these models has vasculature in their system.
4. Strategies for creating a vascularized brain organoid with a physiological microenvironment on-chip
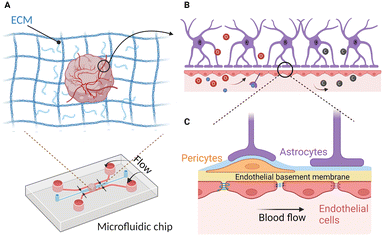
Many existing brain on-chips could reconstitute the human BBB environment at the cellular level, while the brain organoid could mimic the human brain at the organ level albeit without the existence of the BBB. Although the brain organoid is structurally different from the human BBB on-chip, these two technologies share many similar technical and biological challenges and could be potentially combined to develop an on-chip model of the vascularized brain organoid. Importantly, vascularization of the brain organoids could be potentially achieved through the creation of a BBB microenvironment on-chip. Therefore, various strategies that are pivotal for recreating a physiologically relevant brain microenvironment will be discussed in this section.Successful vascularization of organoids using on-chip technologies has been demonstrated for kidney and liver organoids. By subjecting flow over the top of the kidney organoid in a macrofluidic system, Homan et al. developed vascularized kidney organoids that exhibit enhanced maturity, as reflected by the increase in the adult gene expression level compared to the non-vascularized organoid.136 However, their vasculature is not connected to any external circulatory system for the establishment of a functional perfusable vascular network. Jin et al. employed microfluidics to generate 3D vascularized liver organoids in the presence of fluidic flow to enhance their oxygen and nutrient supply.137 These studies demonstrated the feasibility of using microfluidics to improve vascularization of various organoids. Likewise, vascularization of the brain organoids could be further enhanced by integrating them with a perfusable vascular network on-chip to allow the delivery of substances into them (Fig. 3). In this section, we discuss and highlight various controllable components that can be used to develop a realistic vascularized brain organoid on-chip.
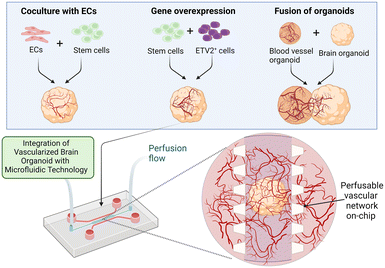 | ||
| Fig. 3 Enhanced vascularization of the brain organoid using microfluidics. Vascularization of the brain organoid has been thus far attained via three major approaches: the coculture of ECs with stem cells, the overexpression of specific genes and the fusion of brain and blood vessel organoids. These approaches, as summarized in Table 2, could be potentially combined with microfluidics to generate a perfusable brain vascular network. Perfusion flow could also be incorporated into the system to mimic the physiological environment.25,131 Image created with https://biorender.com. | ||
4.1 Bioengineering approaches for achieving anastomosis of the perfusable vascular network and brain organoid
Vascularization of the brain organoid could potentially be achieved using a self-assembly method by allowing the perfusable vascular network to further sprout into the organoid through angiogenesis. A proof of concept has been demonstrated in numerous studies in which perfusable vascularization was achieved for various spheroids through vascular invasion.138–140 One of the earliest models developed by Sobrino et al. allow micro-tumors to interact with a self-assembled vascular network to form vascularized micro-tumors.141 Recently, Straehla et al. created a soft lithography-based microfluidic model of vascularized glioblastoma with a self-organized perfusable vascular network surrounded by PCs and ACs to mimic the human BBB.140 By using a stereolithography-based 3D printed microfluidic chip, Salmon et al. applied a similar vascularization strategy by coculturing a self-assembled vascular network and PCs at circular channels flanking a middle brain organoid chamber. This spatial arrangement eventually led to the invasion of the vascular sprouts into the brain organoid.142 However, long term perfusion culture of the brain organoid through perfusable vascular network remains to be investigated. Nonetheless, this self-assembly approach is not only highly similar to the in vivo vasculature formation processes, but also comparable to the unguided brain organoid formation processes that are largely dependent on the self-organizing ability of the cells.9Another method for generating a perfusable vascular network is predesigned patterning. Normally, a scaffold or mold is used to create the hollow channel, followed by EC seeding into the respective channel. For instance, based on the work pioneered by Janigro and his colleagues,143 Cucullo et al. developed a dynamic in vitro (DIV)-BBB model, where ECs are grown on the luminal surface of hollow fibers while ACs are juxtaposed on the abluminal surface of ECs to simulate the cyto-architecture of a blood vessel under flow for not only studying EC–AC interaction but also remodeling perfusion and trans-endothelial migration of immune cells.144,145 Although this DIV-BBB model physiologically replicated the BBB characteristics in a long-term culture setting, it has limitations such as macro-scale setup with large hollow fibers (>100 μm in diameter) that inaccurately represent the brain capillaries (7–10 μm) and the use of polypropylene hollow fibers that are not inherently biocompatible for cell attachment.146
The latest method of predesigned patterning involves 3D printing techniques. This combination of techniques can directly print the specific type of cell at designated positions. For example, Kolesky et al. developed a 3D vascularized tissue that can be perfused on-chip for more than 6 weeks. By using multiple inks, they bio-printed an integration of mesenchymal stem cells (MSCs) and fibroblasts into a 3D thick tissue embedded with a vascular channel lined with ECs on a perfusion chip. Interestingly, they further differentiated the MSCs into an osteogenic tissue by perfusing the cells with a differentiation medium containing relevant growth factors.147 Later, the same research group generated a vascularized cerebral organoid by constructing a scaffold for a perfusable vascular network via both 3D bioprinting and patterning of the sacrificial ink containing ECs in densely packed tissues.148 The vasculature generated using this approach has lumen sizes ranging from 400 μm to 1000 μm, rendering it more suitable for mimicking larger vessels such as arteries and veins instead of the brain capillaries (7–10 μm).149–152 Although this method allows the formation of perfusable vessels with specific geometries and diameters, the development of intricate network structures of the vasculatures is technically challenging compared to the self-assembly method. Regardless of the methods applied to vascularize the brain organoid, while the presence of short-term perfusable vasculature may improve nutrient and oxygen supply to the organoids and further increase their lifespan and maturity level, the major challenge is to achieve a long-term functional perfusion as well as to maintain the structure and maturity of the vasculature inside the organoids.
In addition to creating a vascular network, bioengineering approaches such as the incorporation of fiber microfilaments as scaffolds have been applied to increase the surface area of the brain organoid, which resulted in an improved efficiency of neural induction.153 Such an advanced engineered brain organoid with improved complexity and functionality could be further vascularized and cultured in the microfluidic device for perfusion flow.
4.2 Recapitulation of the BBB environment for the vascularized brain organoid
To truly generate an in vitro BBB microenvironment, one of the major challenges is the incorporation of various cell types with their defined structural arrangement and accurate developmental time points. The anatomical reconstruction of the in vitro BBB microenvironment involves the accurate representation of ECs lining together to form the endothelium that is covered by PCs in proximity and governed by neuronal cells in distance.154,155In terms of their functions, ECs are specific in the human brain compared to those in other organs because they possess prominent characteristics that are instrumental to the barrier function, such as increased expression of junction proteins between adjacent ECs and reduced permeability of their network to allow passages of solutes.156,157 Their significant functions for protecting the barrier integrity suggest that the choice of the EC type to be added into the in vitro models needs to be carefully weighed. In the past decade, ECs from rodents were widely applied in in vitro human BBB models. Due to their non-human origin, human ECs have been used to replace the rodent ECs, including the use of HUVECs,158–161 brain-specific microvascular ECs such as HBMECs162 and HCMEC/D3.163 Despite these commercially available ECs have the advantages of being reproducible with stable performance when it comes to incorporating ECs into organoids, they have limited passage numbers and eventually show senescent phenotypes. Also, HBMECs have been shown to lose their in vivo phenotype for prolonged culture.164 These phenotype changes might render them inappropriate for vascularizing brain organoids that typically require a long culture period. Alternatively, stem cell derived ECs such as ESCs or iPSCs could be a more relevant source for building the vascularized brain organoid on-chip because derived ECs share the same origin as the brain organoids that are also largely derived from stem cells. Lippmann et al. developed a series of protocols for differentiating stem cells into BBB ECs and neural cells with the minimal uses of exogenous factors.165–167 These differentiation protocols were further improved by Hollmann et al. and Neal et al. to have a shorter derivation time168 and reduced batch effect caused by the variation in composition and quality of serum.169 These stem cell derived ECs were proven to be useful for many BBB models and could be potentially incorporated into the in vitro vascularized brain organoid models.
Furthermore, it is necessary to consider other co-existing cells in supporting the vascularization of brain organoids and contributing to the barrier function and homeostasis in the brain. Mural cells such as PCs mainly regulate the blood vessel diameter and support the endothelium growth. In vivo, the coverage of PCs around micro-vessels is much greater in the brain than that of other tissues,170 which suggests the indispensable role of PCs in supporting growth of cerebral microvasculature, as well as contributing to the BBB. However, most of the traditional in vitro BBB models fail to include PCs at a correct spatial arrangement.171–174 Rather than having the PCs wrapping around the ECs like the cellular arrangement in vivo, conventional cultures often involve transwell co-cultures and direct-contact mixtures of ECs with PCs without the vascular network formation.175 In the microfluidic setting, increasing studies in co-culturing pericytes with ECs showed an overall enhancement in the barrier function. Kim et al. demonstrated that inclusion of pericytes into their vasculature platform increased numbers of junctions and branches yet greatly decreased vascular permeability as well as the vascular diameter as opposed to the EC monoculture.176 This suggests the importance of the synergistic effect of ECs and PCs towards the BBB functions.
Other important cells involved in the BBB are ACs that regulate the contraction and relaxation of microvasculature and neurons that regulate contractility of ACs to PCs in response to the neuronal metabolic demands.177–180 In the physiological state, the distance between a neuron and a capillary is within 10–30 μm.181 However, in traditional BBB models such as co-cultures using the Transwell system, the spatial distance between neural cells and ECs is much greater than those of in vivo.182–184 Microfluidics can overcome this by offering a smaller culture distance between neural cells and ECs, allowing the vascular–neural interaction to be more accurately reflected. For example, Brown et al. established a microfluidic model of the human brain where ECs are positioned in a controlled manner relevant to the physiological distance to ACs.185 By further applying physiological shear stress, ECs formed vascular lumens with BBB characteristics. This co-culture model demonstrated proper spatial patterning of both cells to allow the mimicry of ACs' end-feet protruding towards the vascular network. In addition, microfluidics enables sequential introduction of heterotypic cells into the device, which is difficult to achieve in traditional in vitro models. For instance, Shin et al. modelled BBB dysfunction through sequential culturing of neuronal cells and ECs in a microfluidic chip following their respective maturation period to prevent them from interacting with each other before maturation.186
In general, strategies to incorporate PCs and ECs into brain organoids on-chip could be categorised into direct and indirect approaches.21 Direct approaches are relatively straightforward with the addition of both cell types into the same channel as the brain organoid, whereas indirect approaches often involve more complicated processes such as co-differentiation with stem cells or via fusion with blood vessel organoids as discussed earlier.187 Since both ACs and neurons originate from neural stem cells and can be found in matured human brain organoids, incorporation of ACs and neurons into the vascularized brain organoids is not necessary. However, one should be cognizant of the fact that the ACs and neurons generated in brain organoids using the current methods resemble cells from the mid-gestational brain development stage. Generation of more functional, matured neurons and ACs in vascularized brain organoid models is still essential for the development of fully functional neuronal circuits.
4.3 Extracellular matrix
The extracellular matrix (ECM) is another important factor in determining the success of brain organoid vascularization because it dictates much of the cellular and organoid behavior on-chip, which mainly depends on the ECM sources and various fabrication methods. Although the natural brain ECM is largely composed of proteoglycans and hyaluronic acid, brain organoid culture systems often utilize Matrigel, which mainly consists of four major basement membrane ECM proteins such as laminin, collagen IV, entactin, and heparin sulfate proteoglycan perlecan.188 A high similarity of the protein composition of the brain organoid ECM to the brain ECM in vivo would be ideal for supporting the structural integrity of vascularized brain organoids.In general, the ECM can be classified into natural, synthetic, or decellularized hydrogels, depending on the sources and preparation methods. To construct the perfusable vascular network, collagen or fibrin gels from natural sources are often the choices because their interaction with ECs often results in vascular lumen formation.189–191 In the unguided brain organoid forming procedures, Matrigel is used to support the expansion of the neuroepithelial buds. Pham et al. used Matrigel to embed EC-coated brain organoids, which then led to robust vascularization.192 However, Matrigel alone is inadequate to support the vascular lumens since the perfusion within the Matrigel-coated vascularized brain organoid is not achievable. In this case, various ECM components could be incorporated into microfluidic channels for culturing different mural cells to induce vascularization with lumen formation.
Alternatively, synthetic hydrogels can also be applied. Polyethylene glycol (PEG) has been widely employed as a synthetic scaffold to promote the growth of various cell types due to its hydrophilic and biocompatible properties.188,193,194 For example, Ranga et al. developed a PEG-based hydrogel to recapitulate the key features of neural morphogenesis during brain organoid generation.195 Another group used the PEG-based synthetic gel to investigate EC sprouting by embedding the ECs in hydrogel spheres.196 Gelatin methacrylate (GelMA) is another type of versatile and bioinert synthetic gel that enables various chemical and physical modifications to improve the growth of cells and their interactions with the scaffold.197 O'Grady et al. demonstrated this by modifying the GelMA with an N-cadherin extracellular peptide epitope, which subsequently enhanced the growth and maturity of the neurons to form a synaptically connected neuronal network.198 Although synthetic gels allow us to control their chemical and physical properties and tailor important ECM components for different purposes, the gel materials inherently lack many valuable ECM proteins and cell-friendly components. Due to this reason, cell-friendly peptides such as arginylglycylaspartic acid (RGD) peptides and matrix metalloproteinase (MMP)-cleavable peptides are commonly added into the customized synthetic gel for vascularization.199,200 Importantly, it was recently demonstrated that adjusting the concentrations of RGD peptides would significantly affect the development of vascular lumens in the microfluidic device.200
Additionally, the ECM can also be prepared through the decellularization of the whole brain tissue. Cho et al. demonstrated that the decellularized human brain tissue ECM improves brain organoid growth in a microfluidic device.201 Although the decellularized ECM can promote organoid maturation and vascularization, there are still issues related to experimental reproducibility, ECM component inconsistency, and potential ECM protein loss due to intensive preparation steps.202 Since the Young's modulus of the human brain is usually less than 2.4 kPa,203 a natural or synthetic hydrogel with similar stiffness and viscosity will be more suitable for mimicking the brain microenvironment.
Regardless of their sources, all the currently available hydrogels have non-negligible drawbacks, such as short-term durability and elasticity. This makes it difficult to completely recapitulate the ECM microenvironment on-chip long term. In order to construct elastic and long-lasting hydrogels, it is crucial to optimize and modify the recipe of various ECM components inside the gel to improve their overall performance. For in-depth understanding of engineered matrices for various types of organoids, we recommend a review paper by Kratochvil et al.204
4.4 Mechanical stimulation
In addition, vascularized brain organoids on-chip can be improved by adding biophysical cues, such as the shear stress induced by transmural flow and the interstitial flow from the chip. Shear stress, as a result of blood flow, is one of the essential mechanical factors affecting ECs' luminal surface and further influencing specific gene expression to produce biochemical factors for the penetration of blood vessels into the brain organoid. ECs react to shear stress via the regulation of gene expression as well as the cytoskeletal remodeling and cellular alignment towards the flow direction, which further affect the adherens junction complexes and cell proliferation. Thus, fluid shear stress could affect the barrier functionalities of the interface between the perfusable vascular network and the brain organoid. For example, pulsatile flow and high shear stress have been shown to cause changes in the phenotype of the brain ECs and barrier impairment.205 Under shear stress with laminar flow, ECs elongate and form tight junctions with reduced vascular permeability. In contrast, under shear stress with turbulent flow, ECs experience weakened tight junctions with higher permeability and proinflammatory expression levels.206 Since the shear stress could be adjusted by the flow rate, engineers should properly address this important factor while designing a perfusion flow system for the vascularized brain organoid on-chip.205Interstitial flow, which has a flow rate of 0.1–10 μm s−1 through the ECM, plays an essential role in vascularizing the brain organoid in vitro. This important mechanical factor has been demonstrated to be not only critical in affecting the vascular network and lymphatic endothelium but also effective in improving brain organoid and spheroid maturation.207–209 Winkelman et al. investigated the effects of interstitial flow towards the brain microvascular network formation and found that the interstitial flow allows the ECs to form a perfusable vascular network with improved BBB characteristics compared to the static culture. In another example, Park et al. cultured neuro-spheroids on-chip at different levels of interstitial flows and discovered that the brain spheroids under interstitial flow could form higher neuronal network activities.210 Wang et al. utilized a mechanical syringe pump to perfuse their brain organoid cultures on-chip, in which the short-term cultured brain organoids expressed maturation markers under flow perfusion.126 This suggests that the implementation of interstitial flow would direct the growth of the vascularized brain organoid on-chip toward more physiologically relevant conditions.
Hydrogel stiffness is another mechanical aspect that can also influence brain organoid vascularization. The stiffness of various hydrogels can be tuned by modifying their density, which subsequently affects the gel degradability and pore sizes. Zhang et al. showed that the ECM stiffness could modulate synapse connectivity and transmission in neuronal networks, suggesting that hydrogel stiffness is important for neuronal activities.211 Although numerous studies evaluated the hydrogel stiffness for promoting vascular network formation, the optimal stiffness to support the generation of vascular open lumens has not yet been determined.212,213
In summary, the duration, frequency, and amplitude of mechanical forces are critical factors for mimicking physiological mechanical forces. Although the effects of the mechanical stimulation towards vascularized brain organoids have not been figured out, existing studies of how these factors influence brain organoids hint at their importance in constructing fully functional vascularized brain organoids-on-chip (Fig. 4).
5. Challenges and future directions
We have provided an overview of the existing research and potential strategies to generate a physiologically relevant vascularized brain organoid on-chip model. Although there is great potential to create an advanced in vitro model for the understanding of various mechanisms related to vascular-brain interactions, there remains a large gap between the current organoid-on-chip technology and brain development in vivo. Future research can focus more on refining the presented existing models or integrating the advantages from different studies (Fig. 5). The vascularized brain organoid on-chip model is the convergence of microfluidic and organoid technology that requires collaborative work from stem cell experts and biomedical engineers. Therefore, considerations must be equally made for the recapitulation of the functions and structures of the human brain as well as the precise control of the cell microenvironment using microfluidic chips. We conclude by proposing challenges that might be encountered while developing vascularized brain organoid on-chip models.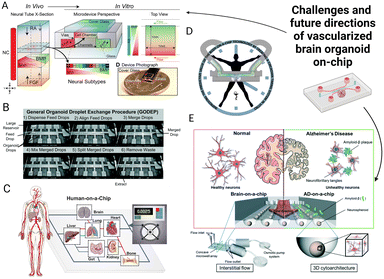 | ||
| Fig. 5 Challenges and future directions of the vascularized brain organoid-on-chip. (A) The microfluidic chip forms biochemical cue gradient by diffusion.232 (B) The advanced organoid-on-chip system could potentially reduce variability.233 (C) Human-on-chip systems can mimic organ interactions in vivo.231 (D) Rhythm on a chip can mimic circadian rhythms by constant flow and periodic agents.224 (E) A microfluidic model to investigate Alzheimer's disease mechanisms.210 The figures are reproduced with permission. | ||
5.1 Precise regulation of the microenvironment
The most common challenge is to precisely regulate a balanced microenvironment for the brain organoid and the perfusable vascular network. Biochemical cues should be specifically designed and optimized for the integration of both different cultures on-chip. For instance, certain growth factors that are beneficial for vascularization might be detrimental to the brain organoid at certain stages. For such, an optimal and balanced medium composition for the vascularized brain organoid on-chip would need to be formulated. Recently, Singh et al. developed a system named Microformulator that can test and trace concentration changes of the component in a medium over a long period of time. This platform could be suitable for optimizing the medium formulation for culturing vascularized brain organoids as well as stem cell differentiation.214 Also, the spatial–temporal distribution of growth factors could potentially improve the cellular organization of the brain organoid as well as promote vascularization towards the inner core of the organoid.5.2 Integrated sensors on-chip
Integrated sensors such as TEER and oxygen sensors on-chip could rapidly monitor physiological and biochemical changes within the microfluidic microenvironment. Recently, a commercial product named microfluidic OrganoTEER from MIMETAS achieved high-throughput TEER measurement for 3D tissue models on-chip using built-in electrodes and impedance spectroscopy.215 Although Cakir et al. also demonstrated the possibility to conduct the TEER measurement by directly inserting micro-electrodes on different regions of vascularized brain organoids,218 it is still technically very challenging due to the large size of the brain organoid.217 Quantification of the barrier permeability using a fluorescently labelled compound and confocal microscope has been demonstrated for BBB organoids.218,219 Such a method could also be potentially applied for vascularized brain organoids. Furthermore, an oxygen sensor could be incorporated into a microfluidic system for monitoring the hypoxic conditions experienced by the brain organoid. For example, a recent study developed an open-top microfluidic chip with an integrated oxygen sensor to analyse the changes of oxygen metabolism in vascularized cancer organoids.220,2215.3 Technical stability
To achieve a high-throughput vascularized brain organoid culture on-chip, technical stability must be taken into consideration. For neuroscientists, it could be challenging to use a microfluidic device for culturing cells. A simple factor such as undetected bubble formation in the microfluidic channels may potentially ruin the entire experiment. More creative inventions such as the bubble trap are required to improve the operation stability and efficiency to culture organoids on-chip.222 To enhance the reproducibility of organoid cultures, engineers should also streamline the mechanical automated process on-chip without relying too much on manual operation.5.4 Mimicking physiological circadian rhythms
To date, most brain organoid research studies often overlook a crucial part of the human brain, which is the circadian rhythms that regulate the behavioural and physiological rhythms of the brain. A rhythm on-chip has been built by introducing chemical messengers with hormonal regulation,223 and this could be potentially integrated with the vascularized brain organoid on-chip model to achieve persistent circadian behaviours. This would improve the predictive value of the whole system for clinical applications.224 Additionally, the Microformulator described previously in section 5.1 could also potentially help in controlling the hormone circadian rhythm on-chip by offering the advantage of automation instead of the manual changing of the medium at a specified interval over time.2145.5 Personalized medicine
Other than the purpose of mimicking diseases, there is great potential to use a vascularized brain organoid model for regenerative medicine. Novel strategies could be developed to generate neural stem cells or produce organoids for transplantation, which might aid functional brain recovery via neural circuit integration and motor function improvement. However, these approaches are still distant from implementation, mainly due to the low efficiency in transplantation as well as the ethical and safety concerns.225,226 A vascularized brain organoid-on-chip may potentially improve engraftment and functional recovery with complex physiological features and an optimal microenvironment for organoid expansion, which will lead to a revolution in regenerative medicine. Additionally, many neurological drugs failed to cross the BBB at the in vivo preclinical drug testing stage. Vascularized brain organoids on-chip are possibly useful for this purpose due to the existence of the blood–brain interface. Given the cost-effectiveness compared to animals and the reduced contamination risk of microfluidic devices, this advanced model could also be used in the personalized medicine field by using patient-source stem cells, which allow personalized drug screening for different individuals. For instance, Pham et al. demonstrated a successful attempt in creating a vascularized brain organoid model with patient derived iPSCs and ECs. This has paved a way for vascularized brain organoids to be applicable in the personalized medicine field.227 However, the cost for generating robust patient-derived brain organoids is still higher than normal cell line therapy. The cost for generating a batch of organoids is currently estimated to be around USD 1000 to 5000.228 This is mainly due to the long culturing period that might take years, as well as the use of expensive materials such as Matrigel and the medium. Thus, there have been ongoing efforts attempting to find better alternatives to reduce the time and cost for producing brain organoids. For example, the cheaper synthetic hydrogel could possibly replace the relatively more expensive Matrigel for culturing the brain organoid.2295.6 Human organs-on-chips
Recently, researchers constructed linked microfluidic chips that coculture different organoids to simulate multi-organ interactions.230 Although a single organoid-on-chip provides a powerful platform for modelling individual organs separately, a linked multi-organoid-on-chip is important to precisely assess the multi-organ interactions.231 Furthermore, multi-organoid chips would greatly benefit from standardized automatic instruments and high-resolution imaging systems to acquire vigorous quantitative readouts. Overall, vascularized brain organoid-on-chip models could be applied in many different fields. We believe that a multi-organoid microfluidic chip containing the vascularized brain organoid and other vascularized organoids while being linked together by the perfusable vascular network would be invaluable for various clinical applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding: Hong Kong Research Grant Council [16209820]; the Innovation and Technology Commission Hong Kong [ITCPD/17-9]; the Hong Kong University of Science and Technology Center for Aging Science; the Lo Ka Chung Foundation through the Hong Kong Epigenomics Project; the Chau Hoi Shuen Foundation.Notes and references
- W. Penfield, Proc. R. Soc. Med., 1968, 61, 831–840 CAS.
- A. Araque and M. Navarrete, Philos. Trans. R. Soc., B, 2010, 365, 2375–2381 CrossRef PubMed.
- J. C. Silbereis, S. Pochareddy, Y. Zhu, M. Li and N. Sestan, Neuron, 2016, 89, 248–268 Search PubMed.
- S. Herculano-Houzel, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 10661–10668 CrossRef CAS PubMed.
- L. N. Miterko, E. P. Lackey, D. H. Heck and R. V. Sillitoe, Front. Neural Circuits, 2018, 12, 1–12 CrossRef PubMed.
- X. Jiang and J. Nardelli, Neurobiol. Dis., 2016, 92, 3–17 CrossRef CAS PubMed.
- P. R. Manger, J. Cort, N. Ebrahim, A. Goodman, J. Henning, M. Karolia, S. L. Rodrigues and G. Štrkalj, Front. Neuroanat., 2008, 2, 1–7 Search PubMed.
- M. A. Lancaster, M. Renner, C.-A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Nature, 2013, 501, 373–379 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Nat. Protoc., 2014, 9, 2329–2340 CrossRef CAS PubMed.
- O. Revah, F. Gore, K. W. Kelley, J. Andersen, N. Sakai, X. Chen, M.-Y. Li, F. Birey, X. Yang, N. L. Saw, S. W. Baker, N. D. Amin, S. Kulkarni, R. Mudipalli, B. Cui, S. Nishino, G. A. Grant, J. K. Knowles, M. Shamloo, J. R. Huguenard, K. Deisseroth and S. P. Paşca, Nature, 2022, 610, 319–326 Search PubMed.
- N. Sun, X. Meng, Y. Liu, D. Song, C. Jiang and J. Cai, J. Biomed. Sci., 2021, 28, 1–16 CrossRef PubMed.
- J. G. Camp, F. Badsha, M. Florio, S. Kanton, T. Gerber, M. Wilsch-Bräuninger, E. Lewitus, A. Sykes, W. Hevers, M. Lancaster, J. A. Knoblich, R. Lachmann, S. Pääbo, W. B. Huttner and B. Treutlein, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 15672–15677 CrossRef CAS PubMed.
- G. Quadrato, T. Nguyen, E. Z. Macosko, J. L. Sherwood, S. M. Yang, D. R. Berger, N. Maria, J. Scholvin, M. Goldman, J. P. Kinney, E. S. Boyden, J. W. Lichtman, Z. M. Williams, S. A. McCarroll and P. Arlotta, Nature, 2017, 545, 48–53 CrossRef CAS PubMed.
- A. E. Trevino, N. Sinnott-Armstrong, J. Andersen, S. J. Yoon, N. Huber, J. K. Pritchard, H. Y. Chang, W. J. Greenleaf and S. P. Paşca, Science, 2020, 367(6476), eaay1645 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Science, 2014, 345, 1247125 Search PubMed.
- M. G. Andrews and A. R. Kriegstein, Annu. Rev. Neurosci., 2022, 45, 23–39 CrossRef PubMed.
- J. J. Marshall and J. O. Mason, Brain Res., 2019, 1724, 146427 CrossRef CAS PubMed.
- G. T. Norris and J. Kipnis, J. Exp. Med., 2019, 216, 60–70 CrossRef CAS PubMed.
- N. De Souza, Nat. Methods, 2017, 14, 655 CrossRef CAS.
- D. Hernández, L. A. Rooney, M. Daniszewski, L. Gulluyan, H. H. Liang, A. L. Cook, A. W. Hewitt and A. Pébay, Stem Cell Rev. Rep., 2022, 18, 718–731 Search PubMed.
- S. Y. Tan, Z. Leung and A. R. Wu, Small, 2020, 16, 1905055 CrossRef CAS PubMed.
- F. Yu, W. Hunziker and D. Choudhury, Micromachines, 2019, 10, 165 CrossRef PubMed.
- Y. Zhu, L. Wang, H. Yu, F. Yin, Y. Wang, H. Liu, L. Jiang and J. Qin, Lab Chip, 2017, 17, 2941–2950 RSC.
- S. Kim, H. Lee, M. Chung and N. L. Jeon, Lab Chip, 2013, 13, 1489–1500 RSC.
- M. B. Chen, J. A. Whisler, J. Fröse, C. Yu, Y. Shin and R. D. Kamm, Nat. Protoc., 2017, 12, 865–880 CrossRef CAS PubMed.
- S. Y. Tan, Q. Jing, Z. Leung, Y. Xu, L. K. W. Cheng, S. S. T. Tam and A. R. Wu, Lab Chip, 2022, 1037–1043 Search PubMed.
- M. Hofer and M. P. Lutolf, Nat. Rev. Mater., 2021, 6, 402–420 CrossRef CAS PubMed.
- H. Castiglione, P.-A. Vigneron, C. Baquerre, F. Yates, J. Rontard and T. Honegger, Pharmaceutics, 2022, 14, 2301 CrossRef CAS PubMed.
- J. Song, S. Bang, N. Choi and H. N. Kim, Biomicrofluidics, 2022, 16, 061301 CrossRef CAS PubMed.
- G. Saorin, I. Caligiuri and F. Rizzolio, Semin. Cell Dev. Biol., 2023, 144, 41–54 CrossRef CAS PubMed.
- S. Grebenyuk and A. Ranga, Front. Bioeng. Biotechnol., 2019, 7, 1–12 Search PubMed.
- B. Martynoga, D. Drechsel and F. Guillemot, Cold Spring Harbor Perspect. Biol., 2012, 4(10) DOI:10.1101/cshperspect.a008359.
- J. C. Silbereis, S. Pochareddy, Y. Zhu, M. Li and N. Sestan, Neuron, 2016, 89, 248–268 CrossRef CAS PubMed.
- S. Ackerman, in The Development and Shaping of the Brain, National Academies Press (US), Washington (DC), 1992, vol. 6, pp. vii–viii Search PubMed.
- P. Malatesta, E. Hartfuss and M. Götz, Development, 2000, 127, 5253–5263 CrossRef CAS PubMed.
- B. M. Howard, Z. Mo, R. Filipovic, A. R. Moore, S. D. Antic and N. Zecevic, Neuroscientist, 2008, 14, 459–473 CrossRef PubMed.
- J. Stiles and T. L. Jernigan, Neuropsychol. Rev., 2010, 20, 327–348 Search PubMed.
- M. Renner, M. A. Lancaster, S. Bian, H. Choi, T. Ku, A. Peer, K. Chung and J. A. Knoblich, EMBO J., 2017, 36, 1316–1329 Search PubMed.
- D. A. Lim and A. Alvarez-Buylla, Cold Spring Harbor Perspect. Biol., 2016, 8, 1–33 CAS.
- I. Bystron, C. Blakemore and P. Rakic, Nat. Rev. Neurosci., 2008, 9, 110–122 Search PubMed.
- L. C. Greig, M. B. Woodworth, M. J. Galazo, H. Padmanabhan and J. D. Macklis, Nat. Rev. Neurosci., 2013, 14, 755–769 CrossRef CAS PubMed.
- M. Berry and A. W. Rogers, J. Anat., 1965, 99, 691–709 CAS.
- J. B. Angevine and R. L. Sidman, Nature, 1961, 192, 766–768 CrossRef PubMed.
- X. Tan and S. H. Shi, Wiley Interdiscip. Rev.: Dev. Biol., 2013, 2, 443–459 CrossRef CAS PubMed.
- C. G. Silva, E. Peyre and L. Nguyen, Nat. Rev. Neurosci., 2019, 20, 318–329 CrossRef CAS PubMed.
- S. M. Williams, D. Purves, G. J. Augustine, D. Fitzpatrick, L. C. Katz, A.-S. LaMantia and J. O. McNamara, Neuroscience, Sinauer Associates, Sunderland (MA), 2nd edn, 2001 Search PubMed.
- U. Dicke and G. Roth, Philos. Trans. R. Soc., B, 2016, 371, 20150180 CrossRef PubMed.
- T. Hayashi, Y. Hou, M. F. Glasser, J. A. Autio, K. Knoblauch, M. Inoue-Murayama, T. Coalson, E. Yacoub, S. Smith, H. Kennedy and D. C. van Essen, NeuroImage, 2021, 229, 117726 CrossRef CAS PubMed.
- B. D. Semple, K. Blomgren, K. Gimlin, D. M. Ferriero and L. J. Noble-Haeusslein, Prog. Neurobiol., 2013, 106–107, 1–16 CrossRef PubMed.
- S. Loomba, J. Straehle, V. Gangadharan, N. Heike, A. Khalifa, A. Motta, N. Ju, M. Sievers, J. Gempt, H. S. Meyer and M. Helmstaedter, Science, 2022, 377(6602), eabo0924 CrossRef CAS PubMed.
- B. K. Stepien, S. Vaid and W. B. Huttner, Front. Cell Dev. Biol., 2021, 9, 1–20 Search PubMed.
- N. A. Oberheim, T. Takano, X. Han, W. He, J. H. C. Lin, F. Wang, Q. Xu, J. D. Wyatt, W. Pilcher, J. G. Ojemann, B. R. Ransom, S. A. Goldman and M. Nedergaard, J. Neurosci., 2009, 29, 3276–3287 CrossRef CAS PubMed.
- D. E. Eigenmann, G. Xue, K. S. Kim, A. V. Moses, M. Hamburger and M. Oufir, Fluids Barriers CNS, 2013, 10, 33 CrossRef PubMed.
- J. Sidhaye and J. A. Knoblich, Cell Death Differ., 2021, 28, 52–67 CrossRef PubMed.
- K. Toma, T. C. Wang and C. Hanashima, Dev., Growth Differ., 2016, 58, 59–72 CrossRef PubMed.
- K. H. Plate, J. Neuropathol. Exp. Neurol., 1999, 58, 313–320 CrossRef CAS PubMed.
- M. R. Harrigan, N. Ole-Schmidt, P. M. L. Black, M. Laurans, M. Gunel, J. T. Rutka and A. T. Parsa, Neurosurgery, 2003, 53, 639–661 Search PubMed.
- M. Marín-Padilla and D. S. Knopman, J. Neuropathol. Exp. Neurol., 2011, 70, 1060–1069 CrossRef PubMed.
- M. Tata and C. Ruhrberg, Neuronal Signaling, 2018, 2, 1–13 CrossRef PubMed.
- M. Tata, C. Ruhrberg and A. Fantin, Mech. Dev., 2015, 138, 26–36 CrossRef CAS PubMed.
- P. A. Stewart and M. J. Wiley, Dev. Biol., 1981, 84, 183–192 CrossRef CAS PubMed.
- H. C. Bauer, I. A. Krizbai, H. Bauer and A. Traweger, Front. Neurosci., 2014, 8, 1–21 Search PubMed.
- R. Daneman and A. Prat, Cold Spring Harbor Perspect. Biol., 2015, 7, a020412 CrossRef PubMed.
- K. Shah and T. Abbruscato, Conn's Translational Neuroscience, 2016, pp. 141–146 Search PubMed.
- M. D'haeseleer, M. Cambron, L. Vanopdenbosch and J. de Keyser, Lancet Neurol., 2011, 10, 657–666 CrossRef PubMed.
- W. Abdullahi, D. Tripathi and P. T. Ronaldson, Am. J. Physiol., 2018, 315, C343–C356 CrossRef CAS PubMed.
- A. Kassner and Z. Merali, Stroke, 2015, 46, 3310–3315 CrossRef PubMed.
- M. Gelderblom, F. Leypoldt, K. Steinbach, D. Behrens, C.-U. Choe, D. A. Siler, T. V. Arumugam, E. Orthey, C. Gerloff, E. Tolosa and T. Magnus, Stroke, 2009, 40, 1849–1857 CrossRef PubMed.
- J. Klohs, Neurodegener. Dis., 2019, 19, 109–127 CrossRef PubMed.
- M. Nedergaard, Science, 2013, 340, 1529–1530 CrossRef CAS PubMed.
- B. K. Harvey, C. T. Richie, B. J. Hoffer and M. Airavaara, J. Neural Transm., 2011, 118, 27–45 CrossRef PubMed.
- H. B. van der Worp, D. W. Howells, E. S. Sena, M. J. Porritt, S. Rewell, V. O'Collins and M. R. Macleod, PLoS Med., 2010, 7, e1000245 CrossRef PubMed.
- J. P. Garner, ILAR J., 2014, 55, 438–456 CrossRef CAS PubMed.
- A. Akhtar, Camb. Q. Healthc. Ethics, 2015, 24, 407–419 CrossRef PubMed.
- P. Mukherjee, S. Roy, D. Ghosh and S. K. Nandi, Lab. Anim. Res., 2022, 38, 1–17 Search PubMed.
- R. D. Kamm, APL Bioeng., 2021, 5, 10–12 Search PubMed.
- W. M. Pardridge, NeuroRx, 2005, 2, 3–14 CrossRef PubMed.
- W. A. Banks, Biochim. Biophys. Acta, Mol. Basis Dis., 2010, 1802, 881–888 CrossRef CAS PubMed.
- J. Rauh, J. Meyer, C. Beuckmann and H. J. Galla, Prog. Brain Res., 1992, 91, 117–121 CAS.
- P. D. Bowman, S. R. Ennis, K. E. Rarey, A. L. Betz and G. W. Goldstein, Ann. Neurol., 1983, 14(4), 396–402 CrossRef CAS PubMed.
- S. Nakagawa, M. A. Deli, H. Kawaguchi, T. Shimizudani, T. Shimono, Á. Kittel, K. Tanaka and M. Niwa, Neurochem. Int., 2009, 54, 253–263 CrossRef CAS PubMed.
- L. L. Bischel, P. N. Coneski, J. G. Lundin, P. K. Wu, C. B. Giller, J. Wynne, B. R. Ringeisen and R. K. Pirlo, J. Biomed. Mater. Res., Part A, 2016, 104, 901–909 CrossRef CAS PubMed.
- T. E. Park, N. Mustafaoglu, A. Herland, R. Hasselkus, R. Mannix, E. A. FitzGerald, R. Prantil-Baun, A. Watters, O. Henry, M. Benz, H. Sanchez, H. J. McCrea, L. C. Goumnerova, H. W. Song, S. P. Palecek, E. Shusta and D. E. Ingber, Nat. Commun., 2019, 10, 1–12 CrossRef PubMed.
- O. Y. F. Henry, R. Villenave, M. J. Cronce, W. D. Leineweber, M. A. Benz and D. E. Ingber, Lab Chip, 2017, 17, 2264–2271 Search PubMed.
- B. Srinivasan, A. R. Kolli, M. B. Esch, H. E. Abaci, M. L. Shuler and J. J. Hickman, J. Lab. Autom., 2015, 20, 107–126 CrossRef CAS PubMed.
- B. Prabhakarpandian, M. C. Shen, J. B. Nichols, I. R. Mills, M. Sidoryk-Wegrzynowicz, M. Aschner and K. Pant, Lab Chip, 2013, 13, 1093–1101 RSC.
- S. P. Deosarkar, B. Prabhakarpandian, B. Wang, J. B. Sheffield, B. Krynska and M. F. Kiani, PLoS One, 2015, 10, 1–21 CrossRef PubMed.
- T. B. Terrell-Hall, M. I. Nounou, F. El-Amrawy, J. I. G. Griffith and P. R. Lockman, Oncotarget, 2017, 8, 83734–83744 CrossRef PubMed.
- Y. Tang, F. Soroush, S. Sun, E. Liverani, J. C. Langston, Q. Yang, L. E. Kilpatrick and M. F. Kiani, J. Neuroinflammation, 2018, 15, 1–12 CrossRef CAS PubMed.
- M. Campisi, Y. Shin, T. Osaki, C. Hajal, V. Chiono and R. D. Kamm, Biomaterials, 2018, 180, 117–129 CrossRef CAS PubMed.
- S. Lee, M. Chung, S. Lee and N. L. Jeon, Biotechnol. Bioeng., 2020, 117, 748–762 CrossRef CAS PubMed.
- J. A. Brown, V. Pensabene, D. A. Markov, V. Allwardt, M. D. Neely, M. Shi, C. M. Britt, O. S. Hoilett, Q. Yang, B. M. Brewer, P. C. Samson, L. J. McCawley, J. M. May, D. J. Webb, D. Li, A. B. Bowman, R. S. Reiserer and J. P. Wikswo, Biomicrofluidics, 2015, 9, 054124 CrossRef PubMed.
- J. A. Brown, S. L. Faley, Y. Shi, K. M. Hillgren, G. A. Sawada, T. K. Baker, J. P. Wikswo and E. S. Lippmann, Fluids Barriers CNS, 2020, 1–12 Search PubMed.
- J. A. Brown, S. G. Codreanu, M. Shi, S. D. Sherrod, D. A. Markov, M. D. Neely, C. M. Britt, O. S. Hoilett, R. S. Reiserer, P. C. Samson, L. J. Mccawley, D. J. Webb, A. B. Bowman and J. A. Mclean, J. Neuroinflammation, 2016, 1–17 Search PubMed.
- L. Vernetti, A. Gough, N. Baetz, S. Blutt, J. R. Broughman, J. A. Brown, J. Foulke-abel, N. Hasan, J. In, E. Kelly, O. Kovbasnjuk, J. Repper, N. Senutovitch, J. Stabb, C. Yeung, N. C. Zachos, M. Donowitz and M. Estes, Sci. Rep., 2017, 7(1), 42296 CrossRef CAS PubMed.
- D. Del Rio, F. Zimetti, P. Caffarra, M. Tassotti, F. Bernini, F. Brighenti, A. Zini and I. Zanotti, Nutrients, 2017, 9(10), 1053 CrossRef PubMed.
- C. Hajal, B. Le Roi, R. D. Kamm and B. M. Maoz, Annu. Rev. Biomed. Eng., 2021, 23, 359–384 CrossRef CAS PubMed.
- M. A. Lancaster, M. Renner, C. A. Martin, D. Wenzel, L. S. Bicknell, M. E. Hurles, T. Homfray, J. M. Penninger, A. P. Jackson and J. A. Knoblich, Nature, 2013, 501, 373–379 CrossRef CAS PubMed.
- E. K. Hollmann, A. K. Bailey, A. V. Potharazu, M. D. Neely, A. B. Bowman and E. S. Lippmann, Fluids Barriers CNS, 2017, 14, 9 Search PubMed.
- D. P. Spelke, D. Ortmann, A. Khademhosseini, L. Ferreira and J. M. Karp, Methods Mol. Biol., 2011, 690, 151–162 CrossRef CAS PubMed.
- C. Cerdan, S. H. Hong and M. Bhatia, Current Protocols in Stem Cell Biology, 2007, 3, 1D.2 CrossRef PubMed.
- Y. S. Hwang, G. C. Bong, D. Ortmann, N. Hattori, H. C. Moeller and A. Khademhosseinia, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 16978–16983 Search PubMed.
- G. Pettinato, X. Wen and N. Zhang, Sci. Rep., 2014, 4, 7402 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Nat. Protoc., 2014, 9, 2329–2340 Search PubMed.
- S. L. Giandomenico, M. Sutcliffe and M. A. Lancaster, Nat. Protoc., 2021, 16, 579–602 CrossRef CAS PubMed.
- J. G. Camp, F. Badsha, M. Florio, S. Kanton, T. Gerber, M. Wilsch-Bräuninger, E. Lewitus, A. Sykes, W. Hevers, M. Lancaster, J. A. Knoblich, R. Lachmann, S. Pääbo, W. B. Huttner and B. Treutlein, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 15672–15677 CrossRef CAS PubMed.
- M. A. Lancaster and J. A. Knoblich, Nat. Protoc., 2014, 9, 2329–2340 CrossRef CAS PubMed.
- I. Kelava and M. A. Lancaster, Dev. Biol., 2016, 420, 199–209 CrossRef CAS PubMed.
- M. Renner, M. A. Lancaster, S. Bian, H. Choi, T. Ku, A. Peer, K. Chung and J. A. Knoblich, EMBO J., 2017, 36, 1316–1329 CrossRef CAS PubMed.
- X. Qian, H. N. Nguyen, M. M. Song, C. Hadiono, S. C. Ogden, C. Hammack, B. Yao, G. R. Hamersky, F. Jacob, C. Zhong, K. Yoon, W. Jeang, L. Lin, Y. Li, J. Thakor, D. A. Berg, C. Zhang, E. Kang, M. Chickering, D. Nauen, C.-Y. Ho, Z. Wen, K. M. Christian, P.-Y. Shi, B. J. Maher, H. Wu, P. Jin, H. Tang, H. Song and G. Ming, Cell, 2016, 165, 1238–1254 CrossRef CAS PubMed.
- P. J. Susaimanickam, F. R. Kiral and I. H. Park, Int. J. Stem Cells, 2022, 15, 26–40 CrossRef PubMed.
- F. Birey, J. Andersen, C. D. Makinson, S. Islam, W. Wei, N. Huber, H. C. Fan, K. R. C. Metzler, G. Panagiotakos, N. Thom, N. A. O'Rourke, L. M. Steinmetz, J. A. Bernstein, J. Hallmayer, J. R. Huguenard and S. P. Pasca, Nature, 2017, 545, 54–59 CrossRef CAS PubMed.
- Y. Miura, M. Y. Li, F. Birey, K. Ikeda, O. Revah, M. V. Thete, J. Y. Park, A. Puno, S. H. Lee, M. H. Porteus and S. P. Paşca, Nat. Biotechnol., 2020, 38, 1421–1430 CrossRef CAS PubMed.
- J. Andersen, O. Revah, Y. Miura, N. Thom, N. D. Amin, K. W. Kelley, M. Singh, X. Chen, M. V. Thete, E. M. Walczak, H. Vogel, H. C. Fan and S. P. Paşca, Cell, 2020, 183, 1913–1929 CrossRef CAS PubMed , e26.
- L. Song, X. Yuan, Z. Jones, K. Griffin, Y. Zhou, T. Ma and Y. Li, Sci. Rep., 2019, 9, 1–16 CrossRef PubMed.
- E. Fennema, N. Rivron, J. Rouwkema, C. van Blitterswijk and J. de Boer, Trends Biotechnol., 2013, 31, 108–115 CrossRef CAS PubMed.
- P. E. C. Leite, M. R. Pereira, G. Harris, D. Pamies, L. M. G. dos Santos, J. M. Granjeiro, H. T. Hogberg, T. Hartung and L. Smirnova, Part. Fibre Toxicol., 2019, 16, 22 CrossRef PubMed.
- P. Rohne, S. Wolf, C. Dörr, J. Ringen, A. Holtz, R. Gollan, B. Renner, H. Prochnow, M. Baiersdörfer and C. Koch-Brandt, Cell Stress Chaperones, 2018, 23, 77–88 CrossRef CAS PubMed.
- W. Neuhaus, Neural Regener. Res., 2017, 12, 1607 Search PubMed.
- E. Garreta, R. D. Kamm, S. M. Chuva de Sousa Lopes, M. A. Lancaster, R. Weiss, X. Trepat, I. Hyun and N. Montserrat, Nat. Mater., 2021, 20, 145–155 CrossRef CAS PubMed.
- E. Karzburn, A. Kshirsagar, S. R. Cohen, J. H. Hanna and O. Reiner, Nat. Phys., 2018, 14, 515–522 Search PubMed.
- Z. Ao, H. Cai, D. J. Havert, Z. Wu, Z. Gong, J. M. Beggs, K. Mackie and F. Guo, Anal. Chem., 2020, 92, 4630–4638 CrossRef CAS PubMed.
- S. T. Seiler, G. L. Mantalas, J. Selberg, S. Cordero, S. Torres-Montoya, P. V. Baudin, V. T. Ly, F. Amend, L. Tran, R. N. Hoffman, M. Rolandi, R. E. Green, D. Haussler, S. R. Salama and M. Teodorescu, Sci. Rep., 2022, 12, 1–12 CrossRef PubMed.
- A. I. Romero-Morales, B. J. O'Grady, K. M. Balotin, L. M. Bellan, E. S. Lippmann and V. Gama, HardwareX, 2019, 6, e00084 CrossRef PubMed.
- D. Huh, H. J. Kim, J. P. Fraser, D. E. Shea, M. Khan, A. Bahinski, G. A. Hamilton and D. E. Ingber, Nat. Protoc., 2013, 8, 2135–2157 CrossRef CAS PubMed.
- Y. Wang, L. Wang, Y. Zhu and J. Qin, Lab Chip, 2018, 18, 851–860 RSC.
- L. O. Porciúncula, L. Goto-Silva, P. F. Ledur and S. K. Rehen, Front. Neurosci., 2021, 15 DOI:10.3389/fnins.2021.674563.
- P. S. Cheah, J. O. Mason and K. H. Ling, Neuroscience Research Notes, 2019, 2, 1–6 CrossRef.
- R. Gopurappilly and R. Pal, Bioengineering of brain organoids: Advancements and challenges, Elsevier Inc., 2022 Search PubMed.
- Z. Ao, S. Song, C. Tian, H. Cai, X. Li, Y. Miao, Z. Wu, J. Krzesniak, B. Ning, M. Gu, L. P. Lee and F. Guo, Adv. Sci., 2022, 9, 1–10 Search PubMed.
- S. Kim, H. Lee, M. Chung and N. L. Jeon, Lab Chip, 2013, 13, 1489–1500 RSC.
- A. A. Mansour, J. T. Gonçalves, C. W. Bloyd, H. Li, S. Fernandes, D. Quang, S. Johnston, S. L. Parylak, X. Jin and F. H. Gage, Nat. Biotechnol., 2018, 36, 432–441 CrossRef CAS PubMed.
- Y. Shi, L. Sun, M. Wang, J. Liu, S. Zhong, R. Li, P. Li, L. Guo, A. Fang, R. Chen, W. P. Ge, Q. Wu and X. Wang, PLoS Biol., 2020, 18, 1–29 Search PubMed.
- B. Cakir, Y. Xiang, Y. Tanaka, M. H. Kural, M. Parent, Y. J. Kang, K. Chapeton, B. Patterson, Y. Yuan, C. S. He, M. S. B. Raredon, J. Dengelegi, K. Y. Kim, P. Sun, M. Zhong, S. Lee, P. Patra, F. Hyder, L. E. Niklason, S. H. Lee, Y. S. Yoon and I. H. Park, Nat. Methods, 2019, 16, 1169–1175 CrossRef CAS PubMed.
- X.-Y. Sun, X.-C. Ju, Y. Li, P.-M. Zeng, J. Wu, Y.-Y. Zhou, L.-B. Shen, J. Dong, Y.-J. Chen and Z.-G. Luo, eLife, 2022, 11, 1–28 Search PubMed.
- K. A. Homan, N. Gupta, K. T. Kroll, D. B. Kolesky, M. Skylar-Scott, T. Miyoshi, D. Mau, M. T. Valerius, T. Ferrante, J. V. Bonventre, J. A. Lewis and R. Morizane, Nat. Methods, 2019, 16, 255–262 CrossRef CAS PubMed.
- Y. Jin, J. Kim, J. S. Lee, S. Min, S. Kim, D. H. Ahn, Y. G. Kim and S. W. Cho, Adv. Funct. Mater., 2018, 28, 1–15 Search PubMed.
- Y. Nashimoto, R. Okada, S. Hanada, Y. Arima, K. Nishiyama, T. Miura and R. Yokokawa, Biomaterials, 2020, 229, 119547 CrossRef CAS PubMed.
- Y. Nashimoto, T. Hayashi, I. Kunita, A. Nakamasu, Y.-S. Torisawa, M. Nakayama, H. Takigawa-Imamura, H. Kotera, K. Nishiyama, T. Miura and R. Yokokawa, Integr. Biol., 2017, 9, 506–518 CrossRef PubMed.
- J. P. Straehla, C. Hajal, H. C. Safford, G. S. Offeddu, N. Boehnke, T. G. Dacoba, J. Wyckoff, R. D. Kamm and P. T. Hammond, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2118697119 CrossRef CAS PubMed.
- A. Sobrino, D. T. T. Phan, R. Datta, X. Wang, S. J. Hachey, M. Romero-López, E. Gratton, A. P. Lee, S. C. George and C. C. W. Hughes, Sci. Rep., 2016, 6, 31589 CrossRef CAS PubMed.
- I. Salmon, S. Grebenyuk, A. R. Abdel Fattah, G. Rustandi, T. Pilkington, C. Verfaillie and A. Ranga, Lab Chip, 2022, 22, 1615–1629 RSC.
- D. Janigro, K. A. Stanness, C. Soderland and G. A. Grant, Curr. Protoc. Toxicol., 2000, 3, 1–11 Search PubMed.
- L. Cucullo, N. Marchi, M. Hossain and D. Janigro, J. Cereb. Blood Flow Metab., 2011, 31, 767–777 CrossRef CAS PubMed.
- L. Cucullo, M. S. McAllister, K. Kight, L. Krizanac-Bengez, M. Marroni, M. R. Mayberg, K. A. Stanness and D. Janigro, Brain Res., 2002, 951, 243–254 CrossRef CAS PubMed.
- A. Williams-Medina, M. Deblock and D. Janigro, Front. Med. Technol., 2021, 2 DOI:10.3389/fmedt.2020.623950.
- D. B. Kolesky, K. A. Homan, M. A. Skylar-Scott and J. A. Lewis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3179–3184 CrossRef CAS PubMed.
- M. A. Skylar-Scott, S. G. M. Uzel, L. L. Nam, J. H. Ahrens, R. L. Truby, S. Damaraju and J. A. Lewis, Sci. Adv., 2019, 5(9), eaaw2459 Search PubMed.
- A. D. Wong, M. Ye, A. F. Levy, J. D. Rothstein, D. E. Bergles and P. C. Searson, Front. Neuroeng., 2013, 6, 1–22 Search PubMed.
- C. Nicholson, Rep. Prog. Phys., 2001, 64, 815–884 CrossRef CAS.
- H. Duvernoy, S. Delon and J. L. Vannson, Brain Res. Bull., 1983, 11, 419–480 CrossRef CAS PubMed.
- R. Saiga, M. Uesugi, A. Takeuchi, K. Uesugi, Y. Suzuki, S. Takekoshi, C. Inomoto, N. Nakamura, Y. Torii, I. Kushima, S. Iritani, N. Ozaki, K. Oshima, M. Itokawa, M. Arai and R. Mizutani, Sci. Rep., 2021, 11, 1–9 CrossRef PubMed.
- S. L. Giandomenico, M. Sutcliffe and M. A. Lancaster, Nat. Protoc., 2021, 16, 579–602 CrossRef CAS PubMed.
- M. D. Sweeney, S. Ayyadurai and B. V. Zlokovic, Nat. Neurosci., 2016, 19, 771–783 CrossRef CAS PubMed.
- E. Kang and J. W. Shin, Int. J. Nanomed., 2016, 11, 2397–2406 CrossRef CAS PubMed.
- U. Kniesel and H. Wolburg, Cell. Mol. Neurobiol., 2000, 20, 57–76 CrossRef CAS PubMed.
- N. J. Abbott and P. A. Revest, Cerebrovascular and Brain Metabolism Reviews, 1991, 3, 39–72 CAS.
- J. H. Yeon, D. Na, K. Choi, S.-W. Ryu, C. Choi and J.-K. Park, Biomed. Microdevices, 2012, 14, 1141–1148 CrossRef CAS PubMed.
- C. Hajal, G. S. Offeddu, Y. Shin, S. Zhang, O. Morozova, D. Hickman, C. G. Knutson and R. D. Kamm, Nat. Protoc., 2022, 17, 95–128 CrossRef CAS PubMed.
- G. N. Grifno, A. M. Farrell, R. M. Linville, D. Arevalo, J. H. Kim, L. Gu and P. C. Searson, Sci. Rep., 2019, 9, 13957 CrossRef PubMed.
- P. P. Partyka, G. A. Godsey, J. R. Galie, M. C. Kosciuk, N. K. Acharya, R. G. Nagele and P. A. Galie, Biomaterials, 2017, 115, 30–39 CrossRef CAS PubMed.
- D. E. Eigenmann, G. Xue, K. S. Kim, A. V. Moses, M. Hamburger and M. Oufir, Fluids Barriers CNS, 2013, 10, 33 CrossRef PubMed.
- B. Weksler, I. A. Romero and P.-O. Couraud, Fluids Barriers CNS, 2013, 10, 16 CrossRef PubMed.
- T. Qian, S. E. Maguire, S. G. Canfield, X. Bao, W. R. Olson, E. V. Shusta and S. P. Palecek, Sci. Adv., 2017, 3(11), e1701679 CrossRef PubMed.
- E. S. Lippmann, S. M. Azarin, J. E. Kay, R. A. Nessler, H. K. Wilson, A. Al-Ahmad, S. P. Palecek and E. V. Shusta, Nat. Biotechnol., 2012, 30, 783–791 CrossRef CAS PubMed.
- E. S. Lippmann, M. C. Estevez-Silva and R. S. Ashton, Stem Cells, 2014, 32, 1032–1042 Search PubMed.
- E. S. Lippmann, A. Al-Ahmad, S. P. Palecek and E. V. Shusta, Fluids Barriers CNS, 2013, 10, 1–14 CrossRef PubMed.
- E. K. Hollmann, A. K. Bailey, A. V. Potharazu, M. D. Neely, A. B. Bowman and E. S. Lippmann, Fluids Barriers CNS, 2017, 14, 9 CrossRef PubMed.
- E. H. Neal, N. A. Marinelli, Y. Shi, P. M. McClatchey, K. M. Balotin, D. R. Gullett, K. A. Hagerla, A. B. Bowman, K. C. Ess, J. P. Wikswo and E. S. Lippmann, Stem Cell Rep., 2019, 12, 1380–1388 CrossRef CAS PubMed.
- D. Shepro and N. M. L. Morel, FASEB J., 1993, 7, 1031–1038 CrossRef CAS PubMed.
- U. Tigges, J. V. Welser-Alves, A. Boroujerdi and R. Milner, Microvasc. Res., 2012, 84, 74–80 CrossRef CAS PubMed.
- Y. He, Y. Yao, S. E. Tsirka and Y. Cao, Stroke, 2014, 45, 2514–2526 CrossRef PubMed.
- C. D. Anfuso, G. Lupo, L. Romeo, G. Giurdanella, C. Motta, A. Pascale, C. Tirolo, B. Marchetti and M. Alberghina, J. Lipid Res., 2007, 48, 782–793 CrossRef CAS PubMed.
- S. Tarallo, E. Beltramo, E. Berrone and M. Porta, Acta Diabetol., 2012, 49, 141–151 CrossRef CAS PubMed.
- J. A. Kim, N. D. Tran, Z. Li, F. Yang, W. Zhou and M. J. Fisher, J. Cereb. Blood Flow Metab., 2006, 26, 209–217 Search PubMed.
- J. Kim, M. Chung, S. Kim, D. H. Jo, J. H. Kim and N. L. Jeon, PLoS One, 2015, 10, e0133880 CrossRef PubMed.
- S. Takahashi, Neuropathology, 2020, 40, 121–137 CrossRef CAS PubMed.
- N. J. Abbott, L. Rönnbäck and E. Hansson, Nat. Rev. Neurosci., 2006, 7, 41–53 CrossRef CAS PubMed.
- Y. Xia, Q. He, Y. Li, S. Chen, M. Huang, Y. Wang, Y. Gao, Y. Huang, M. Wang, L. Mao and B. Hu, PLoS One, 2013, 8, e68891 CrossRef CAS PubMed.
- Y. Igarashi, H. Utsumi, H. Chiba, Y. Yamada-Sasamori, H. Tobioka, Y. Kamimura, K. Furuuchi, Y. Kokai, T. Nakagawa, M. Mori and N. Sawada, Biochem. Biophys. Res. Commun., 1999, 261, 108–112 CrossRef CAS PubMed.
- T. A. Lovick, L. A. Brown and B. J. Key, Neuroscience, 1999, 92, 47–60 CrossRef CAS PubMed.
- K. Luyts, D. Napierska, D. Dinsdale, S. G. Klein, T. Serchi and P. H. M. Hoet, Toxicol. In Vitro, 2015, 29, 234–241 CrossRef CAS PubMed.
- E. M. Vedula, J. L. Alonso, M. A. Arnaout and J. L. Charest, PLoS One, 2017, 12, e0184330 Search PubMed.
- R. Kumar, S. Harris-Hooker, R. Kumar and G. Sanford, Vasc. Cell, 2011, 3, 27 Search PubMed.
- T. D. Brown, M. Nowak, A. V. Bayles, B. Prabhakarpandian, P. Karande, J. Lahann, M. E. Helgeson and S. Mitragotri, Bioeng. Transl. Med., 2019, 4, e10126 CrossRef PubMed.
- Y. Shin, S. H. Choi, E. Kim, E. Bylykbashi, J. A. Kim, S. Chung, D. Y. Kim, R. D. Kamm and R. E. Tanzi, Adv. Sci., 2019, 6, 1900962 CrossRef CAS PubMed.
- X.-Y. Sun, X.-C. Ju, Y. Li, P.-M. Zeng, J. Wu, Y.-Y. Zhou, L.-B. Shen, J. Dong, Y.-J. Chen and Z.-G. Luo, eLife, 2022, 11, 1–28 Search PubMed.
- E. A. Aisenbrey and W. L. Murphy, Nat. Rev. Mater., 2020, 5, 539–551 CrossRef CAS PubMed.
- M. B. Chen, J. A. Whisler, J. Fröse, C. Yu, Y. Shin and R. D. Kamm, Nat. Protoc., 2017, 12, 865–880 CrossRef CAS PubMed.
- S. Kim, H. Lee, M. Chung and N. L. Jeon, Lab Chip, 2013, 13, 1489–1500 Search PubMed.
- S. Y. Tan, Q. Jing, Z. Leung, Y. Xu, L. K. W. Cheng, S. S. T. Tam and A. R. Wu, Lab Chip, 2022, 1037–1043 Search PubMed.
- M. T. Pham, K. M. Pollock, M. D. Rose, W. A. Cary, H. R. Stewart, P. Zhou, J. A. Nolta and B. Waldau, NeuroReport, 2018, 29, 588–593 CrossRef PubMed.
- M. B. Mellott, K. Searcy and M. V. Pishko, Biomaterials, 2001, 22, 929–941 CrossRef CAS PubMed.
- X. bin Kong, Q. Y. Tang, X. Y. Chen, Y. Tu, S. Z. Sun and Z. L. Sun, Neural Regener. Res., 2017, 12, 1003–1008 CrossRef PubMed.
- A. Ranga, M. Girgin, A. Meinhardt, D. Eberle, M. Caiazzo, E. M. Tanaka and M. P. Lutolf, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, E6831–E6839 Search PubMed.
- D. G. Belair, M. P. Schwartz, T. Knudsen and W. L. Murphy, Acta Biomater., 2016, 39, 12–24 CrossRef CAS PubMed.
- A. I. van den Bulcke, B. Bogdanov, N. de Rooze, E. H. Schacht, M. Cornelissen and H. Berghmans, Biomacromolecules, 2000, 1, 31–38 CrossRef CAS PubMed.
- B. J. O'Grady, K. M. Balotin, A. M. Bosworth, P. M. McClatchey, R. M. Weinstein, M. Gupta, K. S. Poole, L. M. Bellan and E. S. Lippmann, ACS Biomater. Sci. Eng., 2020, 6, 5811–5822 Search PubMed.
- M. P. Lutolf and J. A. Hubbell, Nat. Biotechnol., 2005, 23, 47–55 CrossRef CAS PubMed.
- J. Liu, H. Long, D. Zeuschner, A. F. B. Räder, W. J. Polacheck, H. Kessler, L. Sorokin and B. Trappmann, Nat. Commun., 2021, 12, 3402 CrossRef CAS PubMed.
- A.-N. Cho, Y. Jin, Y. An, J. Kim, Y. S. Choi, J. S. Lee, J. Kim, W.-Y. Choi, D.-J. Koo, W. Yu, G.-E. Chang, D.-Y. Kim, S.-H. Jo, J. Kim, S.-Y. Kim, Y.-G. Kim, J. Y. Kim, N. Choi, E. Cheong, Y.-J. Kim, H. S. Je, H.-C. Kang and S.-W. Cho, Nat. Commun., 2021, 12, 4730 CrossRef CAS PubMed.
- A. Fritschen and A. Blaeser, Biomaterials, 2021, 268, 120556 CrossRef CAS PubMed.
- S. Budday, R. Nay, R. de Rooij, P. Steinmann, T. Wyrobek, T. C. Ovaert and E. Kuhl, J. Mech. Behav. Biomed. Mater., 2015, 46, 318–330 CrossRef PubMed.
- M. J. Kratochvil, A. J. Seymour, T. L. Li, S. P. Paşca, C. J. Kuo and S. C. Heilshorn, Nat. Rev. Mater., 2019, 4, 606–622 CrossRef CAS PubMed.
- F. Garcia-Polite, J. Martorell, P. del Rey-Puech, P. Melgar-Lesmes, C. C. O'Brien, J. Roquer, A. Ois, A. Principe, E. R. Edelman and M. Balcells, J. Cereb. Blood Flow Metab., 2017, 37, 2614–2625 CrossRef CAS PubMed.
- C. J. Mandrycky, C. C. Howard, S. G. Rayner, Y. J. Shin and Y. Zheng, J. Mol. Cell. Cardiol., 2021, 159, 1–13 CrossRef CAS PubMed.
- M. A. Winkelman, D. Y. Kim, S. Kakarla, A. Grath, N. Silvia and G. Dai, Lab Chip, 2022, 22, 170–192 Search PubMed.
- A. A. Tomei, S. Siegert, M. R. Britschgi, S. A. Luther and M. A. Swartz, J. Immunol., 2009, 183, 4273–4283 CrossRef CAS PubMed.
- D. O. Miteva, J. M. Rutkowski, J. B. Dixon, W. Kilarski, J. D. Shields and M. A. Swartz, Circ. Res., 2010, 106, 920–931 CrossRef CAS PubMed.
- J. Park, B. K. Lee, G. S. Jeong, J. K. Hyun, C. J. Lee and S.-H. Lee, Lab Chip, 2015, 15, 141–150 RSC.
- Q. Y. Zhang, Y. Y. Zhang, J. Xie, C. X. Li, W. Y. Chen, B. L. Liu, X. A. Wu, S. N. Li, B. Huo, L. H. Jiang and H. C. Zhao, Sci. Rep., 2014, 4, 12–15 Search PubMed.
- A. J. Berger, K. M. Linsmeier, P. K. Kreeger and K. S. Masters, Biomaterials, 2017, 141, 125–135 CrossRef CAS PubMed.
- B. N. Mason, A. Starchenko, R. M. Williams, L. J. Bonassar and C. A. Reinhart-King, Acta Biomater., 2013, 9, 4635–4644 CrossRef CAS PubMed.
- D. Singh, S. P. Deosarkar, E. Cadogan, V. Flemington, A. Bray, J. Zhang, R. S. Reiserer, D. K. Schaffer, G. B. Gerken, C. M. Britt, E. M. Werner, F. D. Gibbons, T. Kostrzewski, C. E. Chambers, E. J. Davies, A. R. Montoya, J. H. L. Fok, D. Hughes, K. Fabre, M. P. Wagoner, J. P. Wikswo and C. W. Scott, PLoS Biol., 2022, 20, e3001624 CrossRef CAS PubMed.
- A. Nicolas, F. Schavemaker, K. Kosim, D. Kurek, M. Haarmans, M. Bulst, K. Lee, S. Wegner, T. Hankemeier, J. Joore, K. Domansky, H. L. Lanz, P. Vulto and S. J. Trietsch, Lab Chip, 2021, 21, 1676–1685 RSC.
- B. Cakir, Y. Xiang, Y. Tanaka, M. H. Kural, M. Parent, Y.-J. Kang, K. Chapeton, B. Patterson, Y. Yuan, C.-S. He, M. S. B. Raredon, J. Dengelegi, K.-Y. Kim, P. Sun, M. Zhong, S. Lee, P. Patra, F. Hyder, L. E. Niklason, S.-H. Lee, Y.-S. Yoon and I.-H. Park, Nat. Methods, 2019, 16, 1169–1175 CrossRef CAS PubMed.
- M. Odijk, A. D. van der Meer, D. Levner, H. J. Kim, M. W. van der Helm, L. I. Segerink, J.-P. Frimat, G. A. Hamilton, D. E. Ingber and A. van den Berg, Lab Chip, 2015, 15, 745–752 RSC.
- G.-A. Kim, N. J. Ginga and S. Takayama, Cell. Mol. Gastroenterol. Hepatol., 2018, 6, 123–131 CrossRef PubMed , e1.
- S. Bergmann, S. E. Lawler, Y. Qu, C. M. Fadzen, J. M. Wolfe, M. S. Regan, B. L. Pentelute, N. Y. R. Agar and C. Cho, Nat. Protoc., 2018, 13, 2827–2843 CrossRef CAS PubMed.
- C. Bouquerel, W. César, L. Barthod, S. Arrak, A. Battistella, G. Gropplero, F. Mechta-Grigoriou, G. Zalcman, M. C. Parrini, M. Verhulsel and S. Descroix, Lab Chip, 2022, 22, 4443–4455 RSC.
- Y. Nashimoto, R. Mukomoto, T. Imaizumi, T. Terai, S. Shishido, K. Ino, R. Yokokawa, T. Miura, K. Onuma, M. Inoue and H. Shiku, Biosens. Bioelectron., 2023, 219, 114808 CrossRef CAS PubMed.
- J. D. Stucki and O. T. Guenat, Lab Chip, 2015, 15, 4393–4397 RSC.
- K. J. Cyr, O. M. Avaldi and J. P. Wikswo, Exp. Biol. Med., 2017, 242, 1714–1731 CrossRef CAS PubMed.
- J.-M. Fustin, M. Li, B. Gao, Q. Chen, T. Cheng and A. G. Stewart, Curr. Opin. Pharmacol., 2019, 48, 127–136 CrossRef CAS PubMed.
- Z. Wang, S.-N. Wang, T.-Y. Xu, C. Hong, M.-H. Cheng, P.-X. Zhu, J.-S. Lin, D.-F. Su and C.-Y. Miao, CNS Neurosci. Ther., 2020, 26, 682–697 CrossRef CAS PubMed.
- O. Revah, F. Gore, K. W. Kelley, J. Andersen, N. Sakai, X. Chen, M.-Y. Li, F. Birey, X. Yang, N. L. Saw, S. W. Baker, N. D. Amin, S. Kulkarni, R. Mudipalli, B. Cui, S. Nishino, G. A. Grant, J. K. Knowles, M. Shamloo, J. R. Huguenard, K. Deisseroth and S. P. Paşca, Nature, 2022, 610, 319–326 CrossRef CAS PubMed.
- M. T. Pham, K. M. Pollock, M. D. Rose, W. A. Cary, H. R. Stewart, P. Zhou, J. A. Nolta and B. Waldau, NeuroReport, 2018, 29, 588–593 CrossRef PubMed.
- H. K. Hopkins, E. M. Traverse and K. L. Barr, Pathogens, 2021, 10, 1510 CrossRef CAS PubMed.
- S. Bose, H. Clevers and X. Shen, Med, 2021, 2, 1011–1026 CrossRef PubMed.
- L. J. Y. Ong, T. Ching, L. H. Chong, S. Arora, H. Li, M. Hashimoto, R. Dasgupta, P. K. Yuen and Y. C. Toh, Lab Chip, 2019, 19, 2178–2191 RSC.
- N. Picollet-D'hahan, A. Zuchowska, I. Lemeunier and S. le Gac, Trends Biotechnol., 2021, 39, 788–810 CrossRef PubMed.
- C. J. Demers, P. Soundararajan, P. Chennampally, G. A. Cox, J. Briscoe, S. D. Collins and R. L. Smith, Development, 2016, 143, 1884–1892 CrossRef CAS.
- S. H. Au, M. D. Chamberlain, S. Mahesh, M. V. Sefton and A. R. Wheeler, Lab Chip, 2014, 14, 3290–3299 RSC.
| This journal is © The Royal Society of Chemistry 2023 |