Open Access Article
Open Access ArticleHydrogel particles-on-chip (HyPoC): a fluorescence micro-sensor array for IgG immunoassay†
Alessandra
De Masi‡
ac,
Pasqualina Liana
Scognamiglio‡§
*a,
Edmondo
Battista¶
ab,
Paolo Antonio
Netti
abc and
Filippo
Causa
 *abc
*abc
aCenter for Advanced Biomaterials for Healthcare@CRIB, Istituto Italiano di Tecnologia (IIT), Largo Barsanti e Matteucci 53, 80125 Naples, Italy. E-mail: pasqualina.scognamiglio@unibas.it; causa@unina.it
bInterdisciplinary Research Centre on Biomaterials (CRIB), University “Federico II”, Piazzale Tecchio 80, 80125 Naples, Italy
cDipartimento di Ingegneria Chimica del Materiali e della Produzione Industriale (DICMAPI), University “Federico II”, Piazzale Tecchio 80, 80125 Naples, Italy
First published on 14th April 2023
Abstract
Novel microparticles have generated growing interest in diagnostics for potential sensitivity and specificity in biomolecule detection and for the possibility to be integrated in a micro-system array as a lab-on-chip. Indeed, bead-based technologies integrated in microfluidics could speed up incubation steps, reduce reagent consumption and improve accessibility of diagnostic devices to non-expert users. To limit non-specific interactions with interfering molecules and to exploit the whole particle volume for bioconjugation, hydrogel microparticles, particularly polyethylene glycol-based, have emerged as promising materials to develop high-performing biosensors since their network can be functionalized to concentrate the target and improve detection. However, the limitations in positioning, trapping and mainly fine manipulation of a precise number of particles in microfluidics have largely impaired point-of-care applications. Herein, we developed an on-chip sandwich immunoassay for the detection of human immunoglobulin G in biological fluids. The detection system is based on finely engineered cleavable PEG-based microparticles, functionalized with specific monoclonal antibodies. By changing the particle number, we demonstrated tuneable specificity and sensitivity (down to 3 pM) in serum and urine. Therefore, a controlled number of hydrogel particles have been integrated in a microfluidic device for on-chip detection (HyPoC) allowing for their precise positioning and fluid exchange for incubation, washing and target detection. HyPoC dramatically decreases incubation time from 180 minutes to one minute and reduces washing volumes from 3.5 ml to 90 μL, achieving a limit of detection of 0.07 nM (with a dynamic range of 0.07–1 nM). Thus, the developed approach represents a versatile, fast and easy point-of-care testing platform for immunoassays.
Introduction
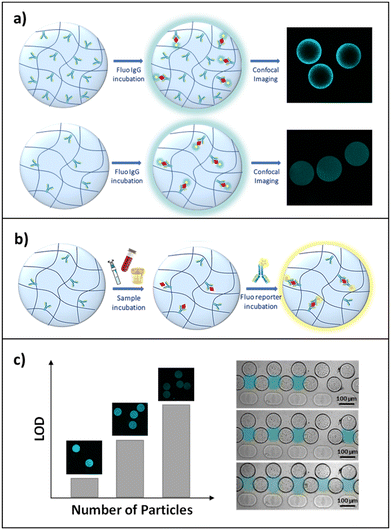
Advances in medical diagnostics and patient-tailored therapy require robust methods for the sensitive and rapid measurement of protein biomarkers. Traditionally, protein detection has been carried out through analytical techniques like time- and labour-intensive enzyme linked immunosorbent assays (ELISAs). Because of the great quantity of validated antibody pairs available, sandwich ELISAs are the most common implementation of this assay. This scheme has been applied to a number of strategies, including planar and particle arrays.1 Fixed design, long detection times (from 1.5 h to 4 hours) and low sensitivity (between 0.24 ng ml−1 and 1.2 μg ml−1)2–5 make, however, planar arrays unsuitable for rapid sample processing in diagnostic applications. For this reason, in applications where the concentrations involved are very low (a few ng ml−1), such as the detection of immunoglobulins in urine,6–8 it is vital to develop sensors that are both sufficiently sensitive and rapid. To overcome these issues, suspension bead-based microarrays have been developed in recent years as alternative platforms, providing more chemical flexibility through particle modification during or post-synthesis,9 greater sensitivity,10 the possibility to encode particles to perform multiplex assays11 and improved reaction kinetics due to solution-phase diffusion of biomolecules.12–14 Moreover, to reduce incubation times and limit the consumption of reagents, also favouring their mixing, the scientific community has been focusing its attention on the design of microfluidic devices for the positioning and trapping of particles, especially through serpentine and well geometries.15–17 These devices facilitate the fine manipulation of few particles, allowing the volume to be controlled in which the assay is performed through the number of particles trapped and ensuring a perfusive flow through porous particles. This allows the target to be efficiently detected even in small sample amounts, leading to shortened assay times and lower reagent consumption and cost. To further improve the performance of bead-based biosensors, there has been growing interest in hydrogel-based technologies since they represent a versatile tool to perform fast and accurate bioassays due to their hydrophilic, biocompatible and highly flexible properties.18–21 Furthermore, compared to non-porous beads, these three-dimensional hydrogel microparticles offer a larger effective surface area for capture probe immobilization,12,13,22–24 thereby providing enhanced binding capacity, and consequently, a higher sensitivity.25,26 In particular, poly(ethylene)glycol (PEG) has emerged as the perfect candidate to synthesise hydrogel microparticles with anti-fouling properties, preventing non-specific interactions between the gel and interfering biomolecules, allowing selective binding of the target even in complex fluids.27,28 The diffusion of biomacromolecules within the polymer network remains, however, a major challenge in the development and feasibility of hydrogel-based suspension arrays.29 As a result, numerous approaches have aimed to alter the hydrogel network porosity and/or the partition coefficient of the diffusing molecules, e.g. with addition of salts/PEG to the solution or hydrogel,30 or through applied cyclic mechanical forces.31 An increase in the pore size of the hydrogel may result in greater diffusivity but nevertheless may compromise the density of the embedded probe.32,33To overcome these challenges, we have recently implemented a simple preparation for the one-step generation of porous and reactive PEG-based hydrogel microparticles, through the addition of a specific cleavable cross-linker, N,N′-(1,2-dihydroxyethylene)bisacrylamide (DHEBA), to a PEGDA pre-polymer solution.34 Interestingly, this new strategy simplifies all synthetic steps, from the realization of large pores to bio-conjugation. In detail, the oxidative cleavage of DHEBA increases porosity, and simultaneously, for each new pore, two functional groups are generated, useful for probe functionalization under mild conditions and without any biomolecule pre-modification. We found that the addition of DHEBA induced a highly porous morphology in microparticles enhancing the accessibility to large proteins. The particles were optimized in terms of immobilized probe density, to ensure homogeneous target binding and, at the same time, a sufficiently high fluorescence signal. Afterwards, to demonstrate the suitability of this tool for a rapid and sensitive protein-based assay, here we applied these finely engineered microparticles to the development of an in-gel sandwich assay for human immunoglobulin G (IgG) detection in biological fluids (Fig. 1). The specificity and selectivity of our system were determined and confirmed by performing the assay in complex media, fetal bovine serum and synthetic urine, showing sensitivity in the low picomolar range. We also demonstrated that a reduction of the number of particles in which the assay is performed translates to a further signal amplification, lowering the detection limit and compressing the dynamic range of the micro-sensor, which allows for more accurate analysis.
However, manual manipulation of few micro-sized particles can be difficult and time-consuming,17 and although the increased accessibility of antibodies to the hydrogel network results in a more time-efficient analysis, the mass transport in these porous micro-substrates can still be limited by agitation and mixing of reagents. For these reasons, in order to make diagnostic tests more accessible and exploit the versatility of hydrogel microparticles to develop point-of-care (POC) diagnostics, our particles are trapped in designated positions inside a microfluidic device (HyPoC). This microsensor integrates micro-sized suspension arrays with fluidic channels to enable IgG-sample perfusion, improving reaction rates and reducing sample and washing volumes, while maintaining excellent sensitivity. Definitively, the HyPoC system has proved to be a sensitive, versatile and fast platform to perform IgG detection and quantification.
Materials and methods
Hydrogel microparticle synthesis
Polyethylene glycol diacrylate (PEGDA700 by Sigma-Aldrich) solutions at a concentration of 10% were prepared with 0.07% of N,N′-(1,2-dihydroxyethylene)bisacrylamide (DHEBA). The pre-polymer solution was vortexed for 1 min at room temperature, adding 0.5% v/v of Darocur1173 (Sigma-Aldrich) as a photo-initiator. The droplet generation was performed starting from the microfluidic emulsification of pre-polymer solution in a continuous phase (oil). A T-junction glass chip (Dolomite Microfluidics) was used to obtain 75 μm diameter droplets, using light mineral oil (LMO) with 5% (v/v) SPAN 80 as a continuous phase. The droplets were polymerized on flow by means of a UV lamp. After photo-polymerization, the particles were collected and washed. Regarding the cleavage of the crosslinker DHEBA, the reaction with sodium periodate (50 mM) was carried out at 50 °C and under stirring for 16 hours, as already optimized in our previous work.34 After cleavage, a small number of particles were collected, washed with a solution of 0.1 M sodium bicarbonate and then visualized under a microscope. The number of microparticles was evaluated by diluting the stock suspension and spotting 10 drops (10 μL each) on a thin glass slide. The drops were visualized with an inverted microscope (IX 71 Olympus) equipped with a 10× objective, and the microparticles in each drop were counted and mediated. Then, the mean number of particles in 10 μL of diluted suspension was multiplied for the dilution factor to evaluate the number of particles contained in 10 μL of the stock suspension.Labelling of antibodies
The human immunoglobulin type G (hIgG, Sigma Aldrich) and the monoclonal anti-human IgG-FAB specific antibody (anti-hIgG-FAB, Sigma Aldrich) were labelled with a fluorophore exploiting their amino groups. In detail, the reaction was carried out with 0.55 mg of hIgG (or anti-hIgG-FAB) and a solution (30 μM) of atto647 N-N-hydroxysuccinimide diluted in a final volume of 500 μL (PBS buffer). The mixture was vortexed for 30 min at room temperature. After the reaction, the conjugates (hIgG-atto647N and anti-hIgG-FABatto647N) were dialysed and characterized through a spectrophotometric measurement to calculate their concentration and degree of functionalization (Fig. S1†). The UV spectra were obtained on a JASCO J-815 CD spectropolarimeter (Jasco Inc., Easton, MD, USA). The solutions were prepared in TRIS buffer at a concentration of 0.1 mg mL−1. The spectra were acquired averaging three scans between 200 and 700 nm with a 2 nm bandwidth. The scan rate used was 100 nm min−1. All the spectra were corrected for background by subtracting a blank scan of buffer.Primary antibody optimization
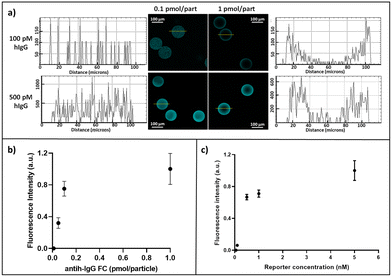
In order to optimize the capture antibody concentration for microparticle conjugation, different amounts (0.05, 0.1 and 1 pmol per particle) of monoclonal anti-human IgG-FC specific (anti-hIgG-FC, Sigma Aldrich) antibody have been explored, carrying out the reaction in the presence of 5% v/v of sodium cyanoborohydride (NaCNBH3, Sigma Aldrich) (Overnight, RT, 500 rpm). Then, TRIS buffer was added to block all unreacted aldehyde groups. After several washes with buffer solutions, hIgG-atto647N (100 and 500 pM) was added to microparticles and incubated for 90 minutes (37 °C, 500 rpm). After several washing steps to remove unbound hIgG-atto647N, images using a confocal microscope (CLSM Leica SP5, objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) were acquired and analysed using ImageJ35 software to evaluate the residual fluorescence intensity within the microparticles. The error is represented as the standard deviation.Reporter antibody optimization
Particles were conjugated with 0.1 pmol per particle of anti-hIgG-FC. After several washes with buffer solutions, a solution of hIgG (at a concentration of 1 nM) was added to microparticles and incubated for 90 minutes (37 °C, 500 rpm). The sample was finally split and then incubated with different concentrations (0.1, 0.2, 0.5, 1, and 5 nM) of anti-hIgG-FABatto647N (90 minutes). After washing, images using a confocal microscope (CLSM Leica SP5, objective 20× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) were acquired and analysed using ImageJ35 software to evaluate the fluorescence intensity and the error is represented as the standard deviation.Sandwich immunoassay sensitivity evaluation and modulation
To study the influence of the particle number on assay sensitivity, cleaved hydrogel microparticles were conjugated with 0.1 pmol per particle anti-hIgG-FC. Three different concentrations of hIgG-atto647N (0.1, 0.5 and 1 nM) were added to 10, 50 and 100 microparticles (total reaction volume 100 μL) and incubated for 90 minutes (37 °C, 500 rpm). After washing, images using a confocal microscope (CLSM Leica SP5, objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) were acquired and analysed with ImageJ.35 To perform the immunoassay, anti-hIgG-FC-conjugated microparticles were incubated for 90 minutes (at 37 °C and 500 rpm) with different concentrations of hIgG (0–1000 pM) in a total reaction volume of 100 μL (ten particles for each sample). After several washes, the microparticles were finally incubated for 90 minutes with anti-hIgG-FABatto647N. Confocal images (CLSM Leica SP5, objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) were recorded and analysed with ImageJ35 software to evaluate the mean fluorescence intensity in three independent experiments. In detail, the fluorescence intensity of a single particle is evaluated as the mean value in a circular region of interest (ROI) with the exact diameter of the particle. The average fluorescence intensity of the particles used for the experiments (about 10, 50 and 100 particles) has been mediated over the replicates and plotted, and the error is represented as the standard deviation over the triplicate experiments. The fluorescence emission of the functionalized particles was acquired as the background signal and its value was subtracted from the signal intensity of each point. Data points were fitted with a non-linear regression based on the Langmuir isotherm model using GraphPad Prism.The CV% inter-assay measures the consistency of replicate samples between experiments, while the CV% intra-assay measures the consistency of replicate samples in single experiments, both are calculated as follows:
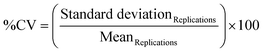 | (1) |
| LOD = 3σ0/S | (2) |
Cross-reactivity tests and sandwich immunoassay in complex fluids
Firstly, cleaved microparticles were conjugated by adding 0.1 pmol per particle of an anti-dioxin antibody (creative diagnostics), with an overnight incubation at room temperature, under stirring at 500 rpm. The conjugated microparticles were then washed and incubated for 90 minutes, at 37 °C and 500 rpm with different concentrations of hIgG. After several washes, the microparticles were finally incubated for 90 minutes with anti-hIgG-FABatto647N. Another specificity test is performed by firstly incubating anti-hIgG-FC-conjugated particles with different concentrations of HSA (90 minutes, 37 °C, 500 rpm) and secondly with anti-hIgG-FABatto647N (90 minutes, 37 °C, 500 rpm). All the experiments were carried out with ten particles per sample and the error is represented as SEM. Confocal images (CLSM Leica SP5, objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) were recorded and analysed with ImageJ35 software to evaluate the fluorescence intensity. The fluorescence emission of the functionalized microparticles was acquired as the background signal and its value was subtracted from the signal intensity of each point. To test the suitability of the in-gel immunoassay for direct use in biological samples, the biosensor performance in fetal bovine serum (FBS) and in synthetic urine was evaluated. In the first experiment, the antibody-conjugated microparticles were incubated with different concentrations of hIgG spiked in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 diluted FBS (90 minutes, 37 °C, 500 rpm). The particles were washed, incubated with antihIgG-FABatto647N (90 minutes, 37 °C, 500 rpm), washed again and finally visualized. Regarding the second experiment, the synthetic urine was prepared as an aqueous solution of salts and human serum albumin (HSA) in the following concentrations: CaCl 0.44 g L−1, MgCL2 × 2H2O 0.52 g L−1, urea 25 g L−1, NaCl 4.8 g L−1, Na2SO4 2.34 g L−1, KCl 1.5 g L−1, NH4Cl 1 g L−1, and HSA 10 g L−1. The cleaved particles were conjugated with the primary antibody, washed and incubated with different concentrations of human IgG spiked in the synthetic urine solution. After washing, the microparticles were finally incubated with anti-hIgG-FABatto647N. All the experiments were carried out with ten particles per sample, the error was represented as the standard deviation and the LOD was calculated using eqn (2). The confocal images were acquired using a CLSM Leica SP5 (objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) and analysed with ImageJ software to evaluate the fluorescence intensity. The fluorescence emission of the functionalized hydrogel microparticles was acquired as the background signal and its value was subtracted from the signal intensity of each point.
5 diluted FBS (90 minutes, 37 °C, 500 rpm). The particles were washed, incubated with antihIgG-FABatto647N (90 minutes, 37 °C, 500 rpm), washed again and finally visualized. Regarding the second experiment, the synthetic urine was prepared as an aqueous solution of salts and human serum albumin (HSA) in the following concentrations: CaCl 0.44 g L−1, MgCL2 × 2H2O 0.52 g L−1, urea 25 g L−1, NaCl 4.8 g L−1, Na2SO4 2.34 g L−1, KCl 1.5 g L−1, NH4Cl 1 g L−1, and HSA 10 g L−1. The cleaved particles were conjugated with the primary antibody, washed and incubated with different concentrations of human IgG spiked in the synthetic urine solution. After washing, the microparticles were finally incubated with anti-hIgG-FABatto647N. All the experiments were carried out with ten particles per sample, the error was represented as the standard deviation and the LOD was calculated using eqn (2). The confocal images were acquired using a CLSM Leica SP5 (objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633, emission wavelength 648–710 nm) and analysed with ImageJ software to evaluate the fluorescence intensity. The fluorescence emission of the functionalized hydrogel microparticles was acquired as the background signal and its value was subtracted from the signal intensity of each point.
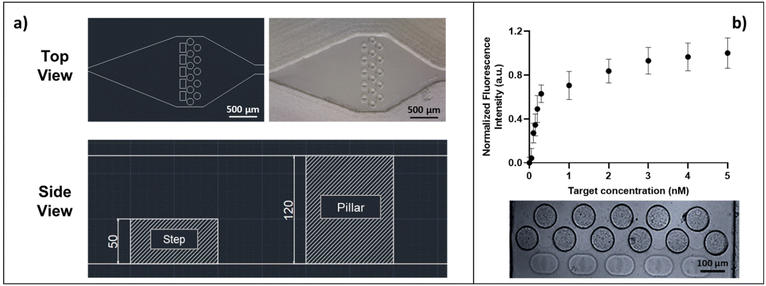
Microfluidic device for hydrodynamic particle trapping: design and realization
The microfluidic devices for hydrogel particle entrapment were made of polydimethylsiloxane (PDSM) via conventional replica molding following the fabrication process in Fig. S2.† All the microfluidic devices realized and tested were designed in 2D using Autocad and then transformed into three-dimensional files with Deskam. The CAM files were then loaded on a micro-milling machine (Mini-mill/GX, Minitech Machinery Corporation; end mills 0.1–0.5 mm TS-2-SR6, Performance Micro Tool), to obtain a negative mold of the device on a 1.2 mm thick polymethylmethacrylate (PMMA) plate. The molds were cleaned with ethanol, sonicated and finally used to replicate the microfluidic chip. The replicas were realized by pouring onto the mold a mixture of PDMS, in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio with a thermo-initiator, and Silwet 0.8% (Silwet l-77, Momentive). PDSM was cured in a stove at 90 °C for 2 hours, detached from the master, punched, cleaned and bonded on a thin glass slide via oxygen plasma treatment for 1 min with subsequent heat treatment at 90 °C for 1 hour. The slides used for bonding presented a high level of optical quality, even in fluorescence, an essential feature to ensure high sensitivity of the assay inside the device. After bonding, the microfluidic chip was connected to the flow through pipes and fittings to carry out loading tests. In order to test the capability of the microfluidic devices to trap hydrogel microparticles in well-defined positions, a highly diluted suspension of microparticles in buffer (10 particles per ml) was injected into the device in cycles of 100 μL each, using a 1 mL syringe. Images of the microparticles inside the trapping chamber were acquired every loading cycle with an inverted microscope (IX 71 Olympus) equipped with a 10× objective. On average, for each loading cycle, a single particle is trapped.
1 ratio with a thermo-initiator, and Silwet 0.8% (Silwet l-77, Momentive). PDSM was cured in a stove at 90 °C for 2 hours, detached from the master, punched, cleaned and bonded on a thin glass slide via oxygen plasma treatment for 1 min with subsequent heat treatment at 90 °C for 1 hour. The slides used for bonding presented a high level of optical quality, even in fluorescence, an essential feature to ensure high sensitivity of the assay inside the device. After bonding, the microfluidic chip was connected to the flow through pipes and fittings to carry out loading tests. In order to test the capability of the microfluidic devices to trap hydrogel microparticles in well-defined positions, a highly diluted suspension of microparticles in buffer (10 particles per ml) was injected into the device in cycles of 100 μL each, using a 1 mL syringe. Images of the microparticles inside the trapping chamber were acquired every loading cycle with an inverted microscope (IX 71 Olympus) equipped with a 10× objective. On average, for each loading cycle, a single particle is trapped.
Hydrogel particles on-chip (HyPoC): immunoassay optimization and dynamic range characterization
To optimize washing steps and target binding, the device was loaded with 5 functionalized particles (0.1 pmol per particle monoclonal anti-hIgG-FC). Then, 100 μL of a 1 μM solution of hIgG-atto647N was injected into the loaded device. After the perfusing time, the microparticles were washed by injecting TRIS buffer solutions (45 μL at a time). Confocal images of the hydrogel microparticles inside the device were acquired after binding with the target and at every washing step (CLSM Leica SP5, objective 10× DRY, scan speed of 400 Hz, excitation wavelength 633). The fluorescence signal was then evaluated using ImageJ35 software and the error was presented as SEM. To perform the assay within HyPoC, devices were loaded with five functionalized microparticles each (0.1 pmol per particle anti-hIgG-FC). Then, different amounts of hIgG (range 0–5 nM) were injected with a volume of 20 μL each into the loaded devices using a pipette. After the perfusing time (∼1 min), the hydrogel microparticles were washed with 90 μL (45 μL at a time) of TRIS buffer solution. Then, 100 μL of anti-hIgG-FABatto647N (1 nM) was injected into all the loaded devices. After a washing step, images of the hydrogel microparticles inside the device were acquired using an optical microscope Zeiss axio observer Z1 equipped with a Colibri 5 epifluorescence LED-light source, Hamamatsu Camera Orca Flash 4.0 and a 10× dry objective. The mean fluorescence signal was evaluated using ImageJ35 software and the error for each target concentration was evaluated by applying the rules of uncertainty propagation on the mean over four independent experiments, performed on different devices (40 devices) loaded with five microparticles each.Results and discussion
Primary and reporter antibody optimization
For point-of-care (POC) diagnostics, the timing of the assay becomes critical and slow kinetics cannot be circumvented by long incubation times. During a hydrogel-based assay, target molecules diffuse into the hydrogel and react with immobilized probe molecules, resulting in a reaction–diffusion process. The goal of any POC bioassay is to maximize the signal from the target captured within the hydrogel over the short assay time, which is related to the flux of the target into the hydrogel. Many bioassays are performed under conditions where the rate of reaction within the hydrogel is much faster than the rate of diffusion.32 The ratio of these rates is known as the Damköhler number (Da):36,37where ka represents the forward rate constant, P0 is the initial probe concentration, L is the particle's radius, and Dgel is the diffusivity of the target in the hydrogel matrix. The signal variation within the hydrogel is therefore dependent on the Da. Here we sought to apply this knowledge of flux into the hydrogels to maximize the response of a biological assay by increasing polymer porosity, through the realization of hydrogel microparticles based on the copolymerization of PEG-diacrylate and N,N′-(1,2-dihydroxyethylene)-bisacrylamide, a crosslinker that simultaneously produces large pores and reactive aldehydes for 3D capture probe bioconjugation. Previous results showed a great accessibility of these microparticles to antibodies and their complexes, without affecting their diffusion rate within their porous network.34 Thus, to prove the suitability of these particles as diagnostic tools, here we developed a highly sensitive and specific in-gel immunoassay for the detection of human IgG (hIgG). First, anti-hIgG-FC, an antibody able to recognize the FC region of human IgG, was chosen as the primary antibody and conjugated to the polymer network of microparticles. Based on the amount of aldehyde groups generated and calculated in our previous study,34 different ratios of anti-hIgG-FC were explored (0.05, 0.1 and 1 pmol per particle). The response of these differently conjugated microparticles was assessed after their incubation with two different concentrations of fluorescent target hIgG-atto647N (100 and 500 pM). After washing the particles to remove the unbound hIgG-atto647N, the residual fluorescence was evaluated. As shown in Fig. 2a, hydrogel microparticles functionalized with the higher concentration present a more inhomogeneous fluorescence distribution and profile than those functionalized with the lowest concentration of anti-hIgG-FC and this inhomogeneity becomes even more evident at a low target concentration.
This effect can be described by the Damköhler number (Da). In fact, in the case of 1 pmol per particle functionalized particles, the Da number is higher (Da ≫ 1) than that of microparticles with 0.1 pmol per particle, so the binding between anti-hIgG-FC and hIgG-atto647N is much faster than the diffusion of the target which reacts with immobilized anti-hIgG-FC before reaching the centre of the particle. Consequently, the signal is confined to the outer layer of the microparticle, resulting in an inhomogeneous fluorescence distribution inside the microparticle, as also reflected by the high standard deviation of the plotted data in Fig. 2b. As opposed, in the case of a low functionalization of the microparticle with the primary antibody, the number of binding sites is much lower than the product of the reaction volume and the equilibrium dissociation constant, and consequently the fractional occupancy is maximized.38 Therefore, the use of 0.1 pmol per particle anti-hIgG-FC is preferable for achieving a high degree of target binding while ensuring a homogeneous fluorescence distribution, as also suggested by Ekins' “ambient analyte” theory,39 which predicts that the detection limit of an immunoassay can be improved by reducing the amount of capture antibodies. After the primary antibody optimization, the concentration of anti-hIgG-FABatto647N, a labelled reporter antibody able to recognize the FAB region of hIgG, was optimized to perform the sandwich assay. The anti-hIgG-FC conjugated microparticles were incubated with the target hIgG (1 nM), and subsequently, different concentrations of anti-hIgG-FABatto647N have been explored. As Fig. 2c shows, the fluorescence intensity increases until a plateau is reached. Since the fluorescence signals corresponding to the 5 nM anti-hIgG-FABatto647N solution exhibit high inhomogeneity (high standard error), probably reflecting a steric hindrance and difficulties in removing the unbound reporter antibody during washing steps, the optimal reporter antibody concentration is fixed at 1 nM.
Sandwich immunoassay sensitivity evaluation and modulation
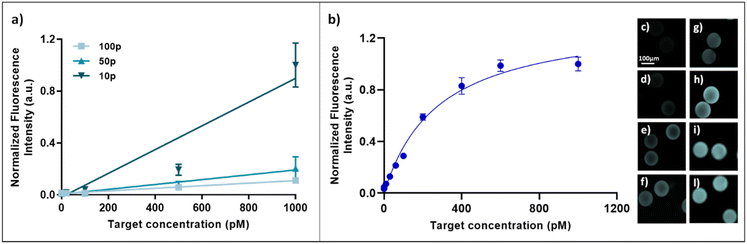
The number of particles can affect the responsivity of the assays, as the hydrogel microparticles have the ability to concentrate the analyte in a very small volume, thus amplifying the signal. For this reason, the capture of the fluorescently labelled hIgG target in the presence of different numbers of particles was studied. In detail, the primary antibody conjugated-microparticles were in numbers of 10, 50 and 100 (following appropriate dilutions of the stock) and were incubated with three different concentrations of hIgG-atto647N, exploring a range between 100 pM and 1 nM. As shown in Fig. 3a, for each target concentration, the fluorescence signal intensity increases by lowering the particle number, due to a reduction in total capture volume. The lower the particle number in which hIgG-atto647N is confined, the more intense the fluorescence signal is. Again, the ambient analyte theory is confirmed, indicating the achievement of an optimal sensitivity by maximizing the fractional occupancy of the capture area. Consequently, as the capture volume decreased from approximately 6.9 × 10−5 (100 particles) to 6.9 × 10−6 (10 particles) cm3 in this particle dilution assay, the limit of detection (LOD) approximately decreased from 50 pM to 6 pM (Fig. S3†). Thus, all the further experiments were performed with 10 particles. These optimized hydrogel microparticles were finally applied to a sandwich immunoassay for the detection of hIgG in TRIS buffer at different concentrations, exploring a range from 0.1 to 1000 pM. In detail, to construct a calibration curve, 10 anti-hIgG-FC conjugated microparticles were incubated with hIgG for 90 minutes, as optimized by performing binding kinetic studies (details in ESI,† Fig. S4). After washing steps, the microparticles were incubated with the labelled reporter antibody (anti-hIgG-FABatto647N). The fluorescence emission versus the hIgG target concentration is plotted (Fig. 3b), showing a specific dose–response binding curve, where the fluorescence signal increases with the target concentration until a plateau is reached. The linear fraction of the curve was used to calculate the LOD value, as reported in the Materials and methods section, which was found to be 4.2 pM (0.63 ng ml−1). Moreover, we calculated a coefficient of variation (CV%) inter-assay of 9.72%. This result shows that our assay has a confidence level comparable to that of the well-established ELISAs (CV% inter-assay about 10–17%).40–42 To further improve the micro-sensor sensitivity, the assay was performed using only five particles (Fig. S5†). As previously demonstrated in Fig. 3a, lowering the number of particles potentially both increases the fluorescence signal and decreases the LOD of the immunosensor; in fact, the limit of detection with five particles decreases up to 3 pM, corresponding to 0.45 ng ml−1. In addition to the limit of detection, another prominent aspect of effectiveness used to compare biosensors is the dynamic range. A fixed dynamic range complicates (or even precludes) the use of biosensors in many applications. Therefore, the development of biosensors with a suitably and easily tuned dynamic range is a valid strategy.43 Here, the dynamic range is scalable by more than an order of magnitude by simply tuning the number of particles and, in particular, it widens from 3–20 pM (5 particles) to 4.2–400 pM (10 particles).Cross-reactivity tests and sandwich immunoassay in complex fluids
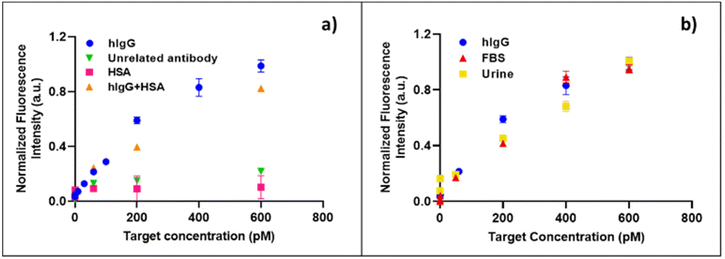
One of the major difficulties in immunoassays is related to the interference with the fluorescence signal either due to non-specific adsorption within microparticles or non-specific interaction with capture and reporter antibodies. In order to investigate the specificity of the assay, several experiments were performed. Firstly, the primary antibody was replaced with an unrelated antibody (anti-dioxin antibody), which was conjugated to the microparticles under the same conditions used for anti-hIgG-FC. After incubation with the hIgG target and the reporter antibody anti-hIgG-FABatto647N, the fluorescence intensities from the particles were recorded and quantified. In this case, the fluorescence intensity did not decrease with increasing target concentration (Fig. 4a), demonstrating that the hIgG target is specifically recognized by the primary antibody anti-hIgG-FC conjugated within microparticles, without any target non-specific adsorption or its physical entrapment within the material. A second control test was performed by replacing the hIgG target with the most abundant plasma protein, the human serum albumin (HSA). The anti-hIgG-FC conjugated microparticles were firstly incubated with different concentrations of HSA, exploring the same range used for the detection of hIgG, and secondly, with the fluorescent reporter antibody. The HSA is not captured by the particles, confirming the high specificity of the conjugated primary antibody and the anti-adhesive properties of the material (Fig. 4a). In order to investigate the selectivity of the developed micro-sensor, the IgG detection in the presence of a great excess of HSA as an interfering molecule was performed. The conjugated hydrogel microparticles were incubated for 90 minutes with different mixture samples, containing a fixed HSA concentration (3 nM) and different concentrations of hIgG (from 0 to 600 pM). After several washes, the microparticles were finally incubated with the fluorescent reporter antibody. The experimental data (Fig. 4a) demonstrate that our system can detect selectively hIgG even in the presence of a high concentration of interfering molecules, as happens physiologically in biological fluids (human serum and/or urine). To confirm the high performance of our assay in the presence of a great variety of interfering molecules, the detection of hIgG is analysed, spiking the target in two different biological fluids. In detail, the anti-hIgG-FC immobilized microparticles were incubated with different concentrations of hIgG diluted in fetal bovine serum (FBS) and synthetic urine. The data shown in Fig. 4b demonstrate that our system is still able to detect the target in FBS medium and in a very similar way when it is present in a simple buffered solution, showing a detection limit of 13 pM (1.95 ng ml−1). The same result is obtained when synthetic urine is used as a complex medium (Fig. 4b), and a comparable LOD of 23 pM (3.45 ng ml−1) is calculated. A large number of studies demonstrated how altered levels of IgG in urine are strongly related to renal injuries such as diabetic kidney disease (DKD)44,45 and post-transplant kidney dysfunctions.46,47 In the case of kidney damage, its filtration barrier does not work properly thus causing the presence of proteins in urine (proteinuria). Depending on the severity of the renal injury, low molecular weight proteins, such as albumin and transferrin, or higher molecular weight proteins such as IgG may be present in the patient's urine. Active monitoring of urinary IgG levels, together with transferrin, IgM, cystatin C, podocytes and other biomarkers, can help to predict the course of the disease since its earliest stages, in order to prevent kidney failure.48,49 In this clinical scenario, the capability of our system to detect IgG in urine in the picomolar range could represent a powerful tool for diagnosis and monitoring of kidney dysfunctions. Compared with a commercial ELISA kit (Invitrogen) to quantify total IgG,4 our in-gel immunoassay definitely shows improved assay time and sensitivity. Furthermore, compared with other bead-based assays,50,51 our hydrogel particle based (HyP) immunoassay exhibits a significant ∼100-fold reduction in detection limit.HyPoC: microfluidic device for particle trapping and sandwich immunoassay on-chip
For point-of-care (POC) diagnostics, the timing of the assay becomes critical and slow kinetics cannot be circumvented by long incubation times. The immunoassay implementation inside the microfluidic device is therefore extremely helpful, as the flux of the target into the hydrogel is forced. For this purpose, we designed and fabricated a microfluidic device for hydrodynamic entrapment of particles, which can reduce both the assay time and reagent consumption and facilitate the handling of few particles (fabrication process in ESI† Fig. S2). The microfluidic chip design was based on the optimization of several parameters: i) stable trapping of the microparticles, ii) absence of fluid and particle backflow, iii) homogeneous distribution of the sample inside all the particles loaded into the chip, iv) no need to use a syringe or pressure pump to load or use the device, v) high transparency to the fluorescence signal (atto647N), and vi) simple design, without involving any expensive materials and/or complex techniques for the production. The trapping chamber (depth 120 μm, width 1.07 mm) can host up to five particles as shown in Fig. 5a. The optimal design provides the presence of five traps (each trap consisting of three pillars and one step), sized to block soft particles with diameters between 100 and 110 μm. The space between pillars is minimized to ensure good entrapment stability, thus preventing the squeezing of soft microparticles during injections at high pressure. Furthermore, the traps are positioned in a single line to guarantee a homogeneous distribution of the target samples into all trapped hydrogel microparticles. The device allows the hydrodynamic entrapment of soft particles in an efficient way, avoiding their backflow, and thus representing a valid alternative to the classical serpentines and wells widely used in the literature to block particles and encapsulate cells.17,52,53 In detail, during the loading of the devices with a highly diluted suspension of particles, a resistance to the flow is observed due to the reduced channel size compared to the particle size, which causes a squeeze of the particles flowing along the inlet channel with a lower flow rate, improving the handling of single particles as widely reported in the literature.15,54 In our case, when a microparticle is close to an empty trap, it is forced by the flow to squeeze over the entering step (side view, Fig. 5a) and is stably confined between the pillars by hydrodynamic forces. For each loading cycle, on average, a single particle enters the device and is trapped into the chamber. Once a trap is filled by a microparticle, the flow resistance increases so that the next particle is guided towards an empty trap, until all five particles are trapped (Fig. S6†). Preliminary tests were carried out to confirm the HyPoC micro-sensor ability to speed up the incubation and washing steps, as detailed in the ESI.† These experiments (Fig. S7†) demonstrate that the incubation time dramatically decreases, from 180 minutes, for the classic off-chip assay, to less than one minute when the assay is performed inside the microfluidic device. Furthermore, the washing volumes are also reduced, passing from about 3.5 ml off-chip to 90 μL on-chip. To characterize the HyPoC performance in hIgG detection, the sandwich assay on chip was carried out. The devices were firstly loaded with five antibody-conjugated microparticles each, and then different target amounts were flushed inside the chamber with a pipette. After washing steps, a solution of the anti-hIgG-FABatto647N reporter was injected into each device and, after washing again, the fluorescence emission from the microparticles was registered and measured. The results obtained from four independent experiments for each target concentration (Table S1†) are averaged and shown in Fig. 5b. The fluorescence emission versus the hIgG target concentration is plotted, showing a specific dose–response binding curve, where the fluorescence signal increases with the target concentration until a plateau is reached, demonstrating that the target is efficiently captured from the microparticles and recognized by the reporter antibody. The linear fraction of the curve (Fig. S9†) was used to calculate the LOD value, which was found to be 0.07 nM, corresponding to a concentration of 10.5 ng ml−1. The CV% inter- and intra-assay values were calculated and were respectively equal to 6.1% and 1.5%. These values are adequate to ensure good repeatability and robustness of the assay. Interestingly, our microfluidic device holds great promise as it allows a sandwich test to be performed in under 10 minutes, like most FDA-approved POC technologies which take from 30 seconds to 1 hour.55 The combination between our system sensitivity and dynamic range (0.07–1 nM corresponding to 10.5–150 ng ml−1), with values comparable to other particle based systems,56 allows for rapid monitoring of IgG levels in urine. For this reason, our HyPoC system could represent a valid technology for early diagnosis of kidney diseases,7 where very low levels of immunoglobulins need to be detected. Furthermore, the modulation of the sensitivity and dynamic range through the number of particles makes our system extremely adaptable to different clinical applications. In particular, a wide dynamic range (appreciable with a high particle number) is required to detect IgG variations in serum which are key proteins to follow the course of inflammation and evaluate appropriate pharmacological therapies.57–59 Indeed, as a future perspective, a trapping device for a greater number of particles could be designed to monitor IgG in blood, whose physiological levels are much higher (7000–18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg ml−1 (ref. 60)) than those in urine. In this scenario, according to the results obtained, our HyPoC micro-sensor could represent a versatile platform to perform rapid and easy quantification of IgG in serum and urine, without any sample pre-treatment.
000 μg ml−1 (ref. 60)) than those in urine. In this scenario, according to the results obtained, our HyPoC micro-sensor could represent a versatile platform to perform rapid and easy quantification of IgG in serum and urine, without any sample pre-treatment.
Conclusions
Recent years have witnessed an explosion in the development of antibody arrays for immunoassays. Suspension array platforms have provided faster reaction kinetics, flexibility, and high-throughput quantification. In this study, we demonstrate the ability to adapt a traditional sandwich immunoassay to a rapid particle-based assay for human-IgG detection, as a proof of concept. The hydrogel microparticles proved high sensitivity, specificity and selectivity for the target, features that confirm their applicability for direct use in complex fluids without any sample purification. After primary antibody and reporter optimization, the micro-sensor was tested in buffer, fetal bovine serum and synthetic urine, showing a limit of detection in the picomolar range, as a result of the combined effects of PEG's anti-fouling properties and highly porous network. We further showed that this simple microparticle-based hIgG detection system provided a similar detection time and significantly higher sensitivity compared to conventional immunoassays, including ELISA and 2D bead-based assays. Since the chemical properties of particles can be easily tuned and optimized through post-synthesis reactions, these cleavable hydrogel microparticles could be an ideal platform for the detection of a great variety of molecules with different sizes. Moreover, in this work we successfully realized a portable HyPoC micro-sensor for hIgG quantification, in which a controlled number of particles are trapped. We demonstrated the system sensitivity (0.07 nM) and its capability to speed up the sample incubation (less than a minute) and reduce reagent consumption during washing steps (less than one hundred microliters). Moreover, the tuneability of sensitivity and the dynamic range, associated with the modulation of the number of particles, proved the great versatility of our system, which is easily adaptable to different applications, representing a great improvement in the field of portable analytical devices for biodetection.Author contributions
A. D. M. and P. L. S. designed and performed all the experiments. P. L. S and F. C. conceived the idea, E. B. and P. N. contributed to the discussion and F. C. coordinated the activities. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.References
- B. Schweitzer, S. Roberts, B. Grimwade, W. Shao, M. Wang, Q. Fu, Q. Shu, I. Laroche, Z. Zhou, V. T. Tchernev, J. Christiansen and S. F. Kingsmore, Nat. Biotechnol., 2002, 20, 359–365 CrossRef CAS PubMed.
- Human IgG (Total) ELISA Kit (ARG80135) - arigo Biolaboratories, https://www.arigobio.com/Human-IgG-Total-ELISA-Kit-ARG80135.html, (accessed 5 February 2023).
- IgG (Total) Human ELISA Kit - Invitrogen, https://www.thermofisher.com/elisa/product/IgG-Total-Human-ELISA-Kit/BMS2091, (accessed 5 February 2023).
- N. America, Human IgG total Ready-SET-Go! ® Human IgG total Ready-SET-Go! ELISA Europe/International* Human IgG total ELISA Ready-SET-Go!® Catalog Number: 88–50550 RUO: For Research Use Only, vol. 642.
- Human IgG, Total, ELISA kit, https://www.cygnustechnologies.com/human-igg-total-elisa-kit.html, (accessed 7 June 2021).
- F. Nagaoka, T. Yamazaki, S. Akashi-Takamura and M. Itoh, Vaccines, 2021, 9, 778 CrossRef CAS PubMed.
- S. Mohandas, S. Balan and D. Mourya, Indian J. Med. Res., 2022, 155, 11–21 CrossRef CAS PubMed.
- C. Eamudomkarn, P. Sithithaworn, C. Kamamia, A. Yakovleva, J. Sithithaworn, S. Kaewkes, A. Techasen, W. Loilome, P. Yongvanit, C. Wangboon, P. Saichua, M. Itoh and J. M. Bethony, PLoS One, 2018, 13(7) DOI:10.1371/JOURNAL.PONE.0192598.
- H. J. Lee, Y. H. Roh, H. U. Kim, S. M. Kim and K. W. Bong, Lab Chip, 2018, 19, 111–119 RSC.
- Y. Wang, V. Shah, A. Lu, E. Pachler, B. Cheng and D. Di Carlo, Lab Chip, 2021, 21, 3438–3448 RSC.
- Y. H. Roh, H. J. Lee, J. Y. Kim, H. U. Kim, S. M. Kim and K. W. Bong, Lab Chip, 2020, 20, 2841–2850 RSC.
- M. A. Al-Ameen and G. Ghosh, Biosens. Bioelectron., 2013, 49, 105–110 CrossRef CAS PubMed.
- S. C. Chapin, D. C. Pregibon and P. S. Doyle, Lab Chip, 2009, 9, 3100–3109 RSC.
- B. He, J. S. Sang and B. L. Sang, Anal. Chem., 2007, 79, 5257–5263 CrossRef CAS PubMed.
- L. Chen, J. J. Kim and P. S. Doyle, Biomicrofluidics, 2018, 12, 024102 CrossRef PubMed.
- J. J. Kim, L. Chen and P. S. Doyle, Lab Chip, 2017, 17, 3120–3128 RSC.
- Z. Zhao, M. A. Al-Ameen, K. Duan, G. Ghosh and J. Fujiou Lo, Biosens. Bioelectron., 2015, 74, 305–312 CrossRef CAS PubMed.
- R. Langer and D. A. Tirrell, Nature, 2004, 428, 487–492 CrossRef CAS PubMed.
- A. Y. Rubina, A. Kolchinsky, A. A. Makarov and A. S. Zasedatelev, Proteomics, 2008, 8, 817–831 CrossRef CAS PubMed.
- M. E. Helgeson, S. C. Chapin and P. S. Doyle, Curr. Opin. Colloid Interface Sci., 2011, 16, 106–117 CrossRef CAS PubMed.
- A. S. Hoffman, Adv. Drug Delivery Rev., 2012, 64, 18–23 CrossRef.
- D. C. Appleyard, S. C. Chapin and P. S. Doyle, Anal. Chem., 2011, 83, 193–199 CrossRef CAS PubMed.
- J. Moorthy, R. Burgess, A. Yethiraj and D. Beebe, Anal. Chem., 2007, 79, 5322–5327 CrossRef CAS PubMed.
- J. Kim, J. Heo and R. M. Crooks, Langmuir, 2006, 22, 10130–10134 CrossRef CAS PubMed.
- H. Kawaguchi, Prog. Polym. Sci., 2000, 25, 1171–1210 CrossRef CAS.
- J. Chou, J. Wong, N. Christodoulides, P. N. Floriano, X. Sanchez and J. McDevitt, Sensors, 2012, 12, 15467–15499 CrossRef PubMed.
- N. C. Padmavathi and P. R. Chatterji, Macromolecules, 1996, 29, 1976–1979 CrossRef CAS.
- D. Hutanu, Mod. Chem. Appl., 2014, 02, 2–7 Search PubMed.
- T. M. Squires, R. J. Messinger and S. R. Manalis, Nat. Biotechnol., 2008, 26, 417–426 CrossRef CAS PubMed.
- S. H. Gehrke, L. H. Uhden and J. F. McBride, J. Controlled Release, 1998, 55, 21–33 CrossRef CAS PubMed.
- M. B. Albro, N. O. Chahine, R. Li, K. Yeager, C. T. Hung and G. A. Ateshian, J. Biomech., 2008, 41, 3152–3157 CrossRef PubMed.
- D. C. Pregibon and P. S. Doyle, Anal. Chem., 2009, 81, 4873–4881 CrossRef CAS PubMed.
- N. W. Choi, J. Kim, S. C. Chapin, T. Duong, E. Donohue, P. Pandey, W. Broom, W. A. Hill and P. S. Doyle, Anal. Chem., 2012, 84, 9370–9378 CrossRef CAS PubMed.
- A. De Masi, P. L. Scognamiglio, E. Battista, P. A. Netti and F. Causa, J. Mater. Chem. B, 2022, 10, 1980–1990 RSC.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- (PDF) ANALYSIS OF TRANSPORT PHENOMENA | Ritu Ranjan ch18m020 - Academia.edu, https://www.academia.edu/39057895/ANALYSIS_OF_TRANSPORT_PHENOMENA, (accessed 20 June 2022).
- E. Battista, F. Causa and P. Netti, Gels, 2017, 3, 20 CrossRef PubMed.
- Z. A. Parpia and D. M. Kelso, Anal. Biochem., 2010, 401, 1–6 CrossRef CAS PubMed.
- D. Wild and E. Kodak, The Immunoassay Handbook, Elsevier Ltd, 2013 Search PubMed.
- W. Lexmond, J. van der Mee, F. Ruiter, B. Platzer, G. Stary, E. H. Yen, E. Dehlink, S. Nurko and E. Fiebiger, J. Immunol. Methods, 2011, 373, 192–199 CrossRef CAS PubMed.
- M. Kelly and B. DeSilva, AAPS J., 2007, 9, E156–E163 CrossRef PubMed.
- J. W. Lee, V. Devanarayan, Y. C. Barrett, R. Weiner, J. Allinson, S. Fountain, S. Keller, I. Weinryb, M. Green, L. Duan, J. A. Rogers, R. Millham, P. J. O'Brien, J. Sailstad, M. Khan, C. Ray and J. A. Wagner, Pharm. Res., 2006, 23, 312–328 CrossRef CAS PubMed.
- A. Vallée-Bélisle, F. Ricci and K. W. Plaxco, J. Am. Chem. Soc., 2012, 134, 2876–2879 CrossRef PubMed.
- T. Narita, M. Hosoba, M. Kakei and S. Ito, Diabetes Care, 2006, 29, 142–144 CrossRef CAS PubMed.
- Y. Qin, S. Zhang, S. Cui, X. Shen, J. Wang, X. Cui, M. Zuo, Z. Gao, J. Zhang, J. Yang, H. Zhu and B. Chang, J. Endocrinol. Invest., 2021, 44, 1981–1988 CrossRef CAS PubMed.
- H. Amer, J. C. Lieske, A. D. Rule, W. K. Kremers, T. S. Larson, C. R. F. Palacios, M. D. Stegall and F. G. Cosio, Am. J. Transplant., 2013, 13, 676–684 CrossRef CAS PubMed.
- E. N. Broeders, K. M. Wissing, M. Hazzan, L. Ghisdal, A. D. Hoang, C. Noel, F. Mascart and D. Abramowicz, Transplant Int., 2008, 21, 57–64 CAS.
- C. Wang, C. Li, W. Gong and T. Lou, New urinary biomarkers for diabetic kidney disease, 2013 Search PubMed.
- A. Zyłka, P. Dumnicka, B. Kuśnierz-Cabala, A. Gala-Błądzińska, P. Ceranowicz, J. Kucharz, A. Ząbek-Adamska, B. Maziarz, R. Drozdz and M. Kuźniewski, Mediators Inflammation, 2018, 2018, 7659243 Search PubMed.
- F. Lacharme, C. Vandevyver and M. A. M. Gijs, Microfluid. Nanofluid., 2009, 7, 479–487 CrossRef.
- C. T. Lin, S. H. Kuo, P. H. Lin, P. H. Chiang, W. H. Lin, C. H. Chang, P. H. Tsou and B. R. Li, Sens. Actuators, B, 2020, 316, 128003 CrossRef CAS.
- C. Kim, J. H. Bang, Y. E. Kim, J. H. Lee and J. Y. Kang, Sens. Actuators, B, 2012, 166–167, 859–869 CrossRef CAS.
- A. Forget, A. L. S. Burzava, B. Delalat, K. Vasilev, F. J. Harding, A. Blencowe and N. H. Voelcker, Biomater. Sci., 2017, 5, 828–836 RSC.
- L. Romita, S. Thompson and D. K. Hwang, Sci. Rep., 2020, 10, 15687 CrossRef CAS PubMed.
- S. Sharma, J. Zapatero-Rodríguez, P. Estrela and R. O'Kennedy, Biosensors, 2015, 5, 577–601 CrossRef PubMed.
- B. Della Ventura, M. Gelzo, E. Battista, A. Alabastri, A. Schirato, G. Castaldo, G. Corso, F. Gentile and R. Velotta, ACS Appl. Mater. Interfaces, 2019, 11, 3753–3762 CrossRef CAS PubMed.
- H. M. Dijstelbloem, C. G. M. Kallenberg and J. G. J. Van De Winkel, Trends Immunol., 2001, 22, 510–516 CrossRef CAS PubMed.
- M. Novokmet, E. Lukić, F. Vučković, Ž. Durić, T. Keser, K. Rajšl, D. Remondini, G. Castellani, H. Gašparović, O. Gornik and G. Lauc, Sci. Rep., 2014, 4, 1–10 Search PubMed.
- H. Zhang, P. Li, D. Wu, D. Xu, Y. Hou, Q. Wang, M. Li, Y. Li, X. Zeng, F. Zhang and Q. Shi, Medicine, 2015, 94, e387 CrossRef CAS PubMed.
- A. Gonzalez-Quintela, R. Alende, F. Gude, J. Campos, J. Rey, L. M. Meijide, C. Fernandez-Merino and C. Vidal, Clin. Exp. Immunol., 2008, 151, 42 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2lc01080a |
| ‡ These authors contributed equally to this work. |
| § Current address: Dipartimento di Scienze, Università degli Studi della Basilicata, via Nazario Sauro, 85, 85100, Potenza, Italy. |
| ¶ Current address: Department of Innovative Technologies in Medicine & Dentistry, University “G. d'Annunzio” Chieti-Pescara, Via dei Vestini, 31, 66100 Chieti, Italy. |
| This journal is © The Royal Society of Chemistry 2023 |