Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Pump-less, recirculating organ-on-a-chip (rOoC) platform†‡
Mathias
Busek§
 *ab,
Aleksandra
Aizenshtadt§
a,
Timo
Koch
c,
Anna
Frank
ade,
Ludivine
Delon
b,
Mikel Amirola
Martinez
a,
Alexey
Golovin
b,
Clotilde
Dumas
a,
Justyna
Stokowiec
a,
Stefan
Gruenzner
f,
Espen
Melum
adegh and
Stefan
Krauss
ab
*ab,
Aleksandra
Aizenshtadt§
a,
Timo
Koch
c,
Anna
Frank
ade,
Ludivine
Delon
b,
Mikel Amirola
Martinez
a,
Alexey
Golovin
b,
Clotilde
Dumas
a,
Justyna
Stokowiec
a,
Stefan
Gruenzner
f,
Espen
Melum
adegh and
Stefan
Krauss
ab
aHybrid Technology Hub – Centre of Excellence, Institute of Basic Medical Science, University of Oslo, P.O. Box 1110, 0317 Oslo, Norway
bDep. of Immunology and Transfusion Medicine, Oslo University Hospital, P.O. Box 4950, 0424 Oslo, Norway. E-mail: mathias.busek@medisin.uio.no
cDep. of Mathematics, University of Oslo, P.O. Box 1083, 0316 Oslo, Norway
dNorwegian PSC Research Center, Division of Surgery, Inflammatory Diseases and Transplantation, Oslo University Hospital, Oslo, Norway
eResearch Institute of Internal Medicine, Division of Surgery, Inflammatory Diseases and Transplantation, Oslo University Hospital, Oslo, Norway
fChair of Microsystems, Technische Universität Dresden, 01069 Dresden, Germany
gInstitute of Clinical Medicine, Faculty of Medicine, University of Oslo, P.O. Box 1110, 0317 Oslo, Norway
hSection of Gastroenterology, Department of Transplantation Medicine, Division of Surgery, Inflammatory Diseases and Transplantation, Oslo University Hospital, Oslo, Norway
First published on 20th December 2022
Abstract
We developed a novel, pump-less directional flow recirculating organ-on-a-chip (rOoC) platform that creates controlled unidirectional gravity-driven flow by a combination of a 3D-tilting system and an optimized microfluidic layout. The rOoC platform was assembled utilizing a layer-to-layer fabrication technology based on thermoplastic materials. It features two organoid compartments supported by two independent perfusion channels and separated by a hydrogel barrier. We developed a computational model to predict wall shear stress values and then measured the flow rate in the microfluidic channels with micro-Particle-Image-Velocimetry (μPIV). The suitability of the rOoC for functional culture of endothelial cells was tested using HUVECs seeded in the perfusion channels. HUVECs aligned in response to the directional flow, formed a barrier and were able to sprout into the organoid compartments. Next, we demonstrated the viability of human stem-cell derived liver organoids in the organoid compartments. Finally, we show the possibility to circulate immune cells in the microfluidic channels that retain viability without being trapped or activated. The rOoC platform allows growing and connecting of two or more tissue or organ representations on-chip with the possibility of applying gradients, endothelial barriers, microvasculature and circulating cells independent of external tubing and support systems.
Introduction
Organ-on-a-Chip (OoC) technology combines microfluidics and cell biology to mimic tissue or organ functionality in vitro. OoCs are on the verge of widespread use as new models with significant advantages for studying diseases and drug response/toxicity in both academia and the pharmaceutical industry.1 A variety of platforms were developed from 2010 onwards when the first complex OoC, a Lung-on-a-Chip model comprising a co-cultivation of lung alveolar and endothelial cells (ECs) on the two sides of a porous membrane, was presented.2 A major driver for OoC development is the pharmaceutical industry.3 Bringing a new drug to the market can cost more than 1 billion US$ (ref. 4) and OoC technology is expected to reduce R&D costs by 10–30%.5 Another important aspect of OoC technology is in vitro disease modelling6 as human physiology frequently differs from laboratory animal physiology. In addition, pathological processes may be controlled and monitored on-chip. Finally, OoC technology is an important step toward reducing animal testing in line with the 3R initiative.7However, after more than 10 years of development, OoC technology is still limited in its applicability and scalability. This might be caused by the necessity of most formats to use expensive and complicated driving/pumping systems thus limiting long-term stability and usability for smaller laboratories. Beside usability and scalability issues connected with pump-driven systems, the necessary tubing might cause unwanted adsorption of drugs, metabolites and cytokines thus limiting the usability of OoCs.8 This is not only due to the materials used for tubes (often silicone) but also because of the increased dead volume and surface area. At the same time, widely used and easy to scale platforms with gravity-driven flow provide limited functionality and/or not always physiologically relevant flow characteristics. For instance, conventional platforms on rockers generate bidirectional flow, opposite to unidirectional flow – optimal for functional endothelial cells.9
Bidirectional flow characteristics have been demonstrated to be present at atherosclerosis-prone areas10 and are associated with oxidative stress, metabolic disruption and activation of pro-inflammatory signalling in endothelial cells.9,11 In contrast, unidirectional flow creates conditions that are beneficial for homeostasis and quiescence of endothelial cells.12,13 Therefore, there is a need for a microfluidic platform that combines controlled, unidirectional flow, commonly seen in pump-driven platforms, with scalability and applicability, being a feature of devices based on gravity-driven flow.
Recently, we surveyed peers in academia to understand the major limitations, needs and advantages of OoC technology.14 We identified major problems in terms of usability, and robustness which is in line with surveys conducted by others.15 Moreover, we found that many scientists aim for more complex in vitro models for their studies than those that are currently available on user-friendly platforms. Hence, important features of new OoCs generation include among others the incorporation of blood vessel-like structures, 3D culture models and immune cells. Existing OoCs tend to combine these sought-after features at the cost of low scalability and usability.16–19
Here we present a novel, scalable, and easy-to-use OoC platform, termed rOoC that not only creates a directed flow without the need for pumps and tubing but also allows the implementation of biological complexity on-chip addressing several of the concerns raised above. The rOoC platform supports on-chip vascularization, growth factor and nutrient gradients, 3D organoids culture and the incorporation of circulating cells. We present the layout and fabrication of the rOoC platform, its technical characterization, and a mathematical model to predict the flow in the device under different conditions. Next, we demonstrate stable EC culture over 2 weeks of directional flow-dependent endothelial alignment and sprouting of microvessels into the organoid compartment (OC) of the chip. We then show the long-term viability and functionality of 3D liver organoids on-chip. Finally, we demonstrate that the rOoC platform is a suitable platform also for circulating immune cells. The device can support several perfusion and OCs that are separated by an extracellular matrix (ECM) barrier, a layout that can in principle be applied to a plethora of single and combined organ models.
Results
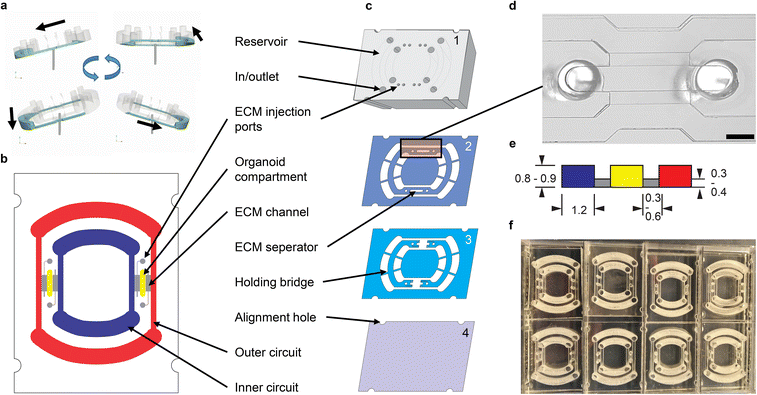
Platform design and fabrication
Adapted from common gravity-driven perfusion systems placed on a 2D-tilting platform,20 we introduce a novel concept of using a 3D-tilting/rotating platform (Fig. 1a) to produce a directed recirculating flow on our rOoC platform without using pumps. To generate unidirectional flows, we propose the usage of elongated and high reservoirs (Fig. 1 and 2a). By connecting two reservoirs with microchannels, a fluidic ring circuit is produced and allows the cyclic filling and emptying of the reservoirs hence producing a directed flow as shown in the Video S1.† The main effect responsible for the directionality of the flow is that the liquid level in the reservoirs equalizes during tilting thus causing the inlet to run dry and thus preventing backflow. For actuation, we use a standard laboratory rotator (3D rotator waver, VWR Avantor, USA). The flow rate in the rOoC depends on the volume (both maximum and filled with liquid) and geometry of the reservoirs, the perfusion channel (PC) dimensions, as well as the tilt and rotation speed of the platform.The PC dimensions are based on the sizes of smaller arteries in the body.21 We use a cross-section of 1.2 × 0.8 mm2. To have a media-to-cell ratio comparable to standard cell culture plastics (96/48 well plates), we selected a reservoir volume of 200 μL for the inner and 300 μL for the outer circuit. The reservoir shape was designed half-round as this geometry is less prone to trapping circulating cells. The general chip layout (Fig. 1b) follows the commonly used three-lane concept as this is especially useful for vascularization studies.22 Our unique actuation principle gives us the possibility to place the two PCs within each other with two OCs positioned in between (Fig. 1b). We selected a length of 4 mm, a width between 0.5 and 1.2 mm and a height of 0.8/0.9 mm for the OCs, as this is big enough to house several organoids (between 200–500 μm each) and to allow endothelial sprouting into the OC. For loading, we mixed cells or organoids with ECM and injected the mixture into the OC through the two ECM injection ports (diameter: 1 mm, accessible by a 10 μL-pipette tip). When the ECM solidifies, the organoids are fixed in place.
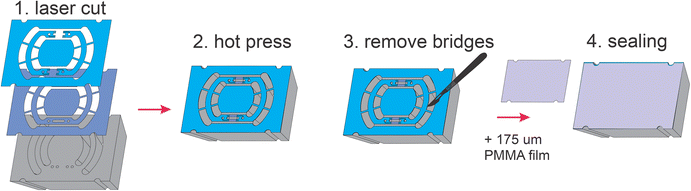
The rOoC is built up of 4 layers of poly(methyl methacrylate) (PMMA) (Fig. 1c). The layers are designed in a CAD program, laser-cut, precisely stacked and thermally bonded together.23,24 The process is detailed in the methods part.
The 4 layers are:
1. The top layer (6 mm) includes laser-engraved reservoirs and ECM injection ports.
2. Layer 2 is 0.5 mm thick and includes the reservoirs, PCs, OCs and connecting holes for the ECM-cell mixture.
3. Layer 3 includes PCs and OCs and is 0.4 mm thick. In contrast to layer 2, a connection between the PC and OCs is included that can be filled with ECM.
4. The rOoC is sealed with a thin film of PMMA (0.175 mm).
Together, layers 2 and 3 form a stepped channel layout (Fig. 1d) that prevents ECM from entering the PCs.25 Due to capillary forces, the liquid ECM is retained in the region where the channel height is abruptly increased. By using this layout, a biodegradable separation layer26 is formed between the OCs and the PCs allowing for spatial defining of the cell composition25 and studying cellular interaction such as angiogenesis.27 The connection area is 0.3–0.6 mm wide so that ECs can sprout into the OC while ensuring that ECM is not accidentally blocking the PCs. Additional separate ECM channels can be used to fill the ECM (possibly mixed with other cells) independently from the OCs. For on-chip confocal imaging, only PMMA films with superior optical clarity were chosen. To improve scalability, up to 8 rOoCs can be placed in a standard rectangular 8 well plate (Thermo Scientific, catalogue no. 267062) and covered with a lid (Fig. 1e). In this configuration, the evaporation was measured (by weighing) to be 31 ± 16 μL per day for a total filling volume of 500 μL (n = 6).
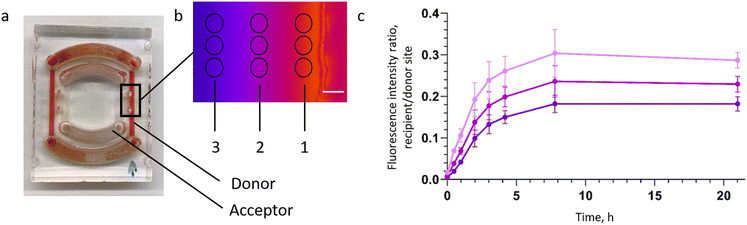
The configuration of the rOoC with two nested circuits of PCs separated by ECM channels and OCs allows filling the two circuits with different media to create a gradient in the OC, which can be maintained in the ECM by dosing reagents in either the outer or the inner circuit. In many biological processes, including differentiation, vascularization and functional subdivisions such as liver zonation, growth factor-, nutrient- and gas gradients play a crucial role and need to be included when representing tissues or organs. To test the capability of the rOoC device to establish and maintain gradients, 70 kDa fluorescein-labelled dextran was filled in the inner PC and its gradient within the OC was observed under a fluorescence microscope. The detailed setup and experimental data are given in Appendix A. The dextran gradient was produced and assessed to be stable over 6 hours in the rOoC. Note that the gradient formation and characteristics will differ between molecules and used ECM due to different molecular weights and interactions between the ECM and the permeating substances (binding to ECM).
Flow characterisation and modelling
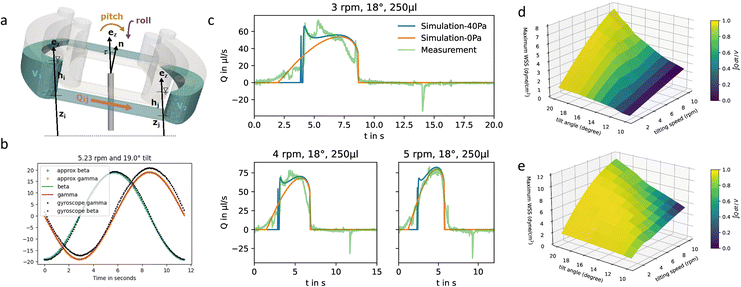
For ECs cultivated at the PC walls, the wall shear stress (WSS) is an important parameter for cell functionality. We have developed a mathematical model to predict flow rates and WSS values in the rOoC and cross-validated it with micro-particle velocimetry measurements (μPIV). From the flow velocity v, the WSS can be calculated from the gradient of the tangential velocity in the normal direction to the wall: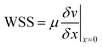 | (1) |
![[n with combining right harpoon above (vector)]](https://www.rsc.org/images/entities/i_char_006e_20d1.gif) that is tilted by an angle of α to the z-axis (Fig. 2a). This motion leads to a time-dependent pitch and roll (β, γ) of the platform. We measured both values with a gyroscope (Arduino 9-Axis Motion Shield coupled to an Arduino Uno, Arduino, Ivrea, Italy – source code see Code S1‡). The data was recorded on a computer and saved as CSV-File. A plot of β and γ is given in Fig. 2b. Mathematically, β and γ can be described using trigonometric functions and the rotation function ϑ(t) = 2πft (a detailed explanation is given in the ESI Text†):
that is tilted by an angle of α to the z-axis (Fig. 2a). This motion leads to a time-dependent pitch and roll (β, γ) of the platform. We measured both values with a gyroscope (Arduino 9-Axis Motion Shield coupled to an Arduino Uno, Arduino, Ivrea, Italy – source code see Code S1‡). The data was recorded on a computer and saved as CSV-File. A plot of β and γ is given in Fig. 2b. Mathematically, β and γ can be described using trigonometric functions and the rotation function ϑ(t) = 2πft (a detailed explanation is given in the ESI Text†):γ(ϑ(t)) = sin−1(−sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) τ τ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ϑ) ≈ −α ϑ) ≈ −α![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ϑ, ϑ, | (2) |
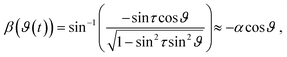 | (3) |
The computational model (detailed in the methods part, source code in Code S3‡) allows for calculating the flow rates dynamically for different reservoir geometries, tilt angle, angular velocities, and filling volumes. Changing channel geometry requires the re-computation of the channel transmissibility and relations between flow rate and velocity or WSS. The following parameters have been used for the computational model:
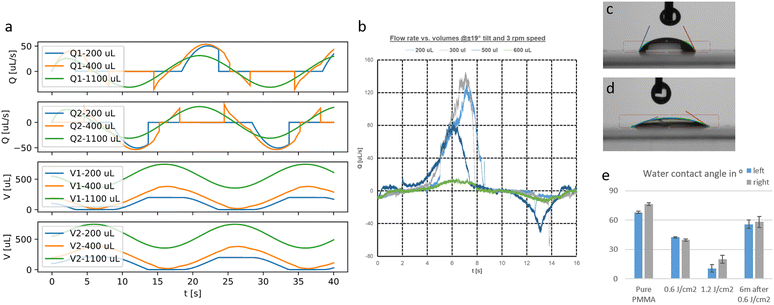
The calculation yields the flow-time curve given in Fig. 2c that aligns well with the measured flow-time curves (flow measurement described in the materials and methods part). We calculated the flow-time curve with two critical pressure values (0 and 40 Pa) to investigate the influence of the surface tension on the flow characteristics. It seems that for low tilting speeds (<4 rpm) a better fitting to the measured data can be obtained for 40 Pa critical pressure whereas for speeds >5 rpm, surface effects can be neglected and the curve aligns very well with the prediction using 0 Pa critical pressure.
The calculated flow profile (see ESI Text†) indicates that the flow velocity is maximum in the middle of the channel and reaches zero near the walls. In contrast, the WSS (plot see ESI Text†) is maximum at the middle of each wall but zero at the corners. We next calculated the maximum WSS with the parameters given in Table 1 (viscosity μ = 0.8 mPa s for T = 37 °C) for different tilts and rotation speeds for both the outer and the inner PC (Vres = 200 μL, lc = 12 mm) and plotted these values in Fig. 2d and e. The colour in the plots indicates whether the whole reservoir volume is transported thus indicating if the flow is unidirectional 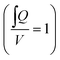 . If this quotient becomes <1 a backflow occurs and if this quotient equals zero, the flow is fully bidirectional similar to 2D tilting systems (no net flow in one direction). In the rOoC, this is for instance the case for low tilts (<10°), high rotations speed (>10 rpm) and predominantly in the inner PC. Moreover, bidirectional flow occurs for higher filling volumes (Appendix B).
. If this quotient becomes <1 a backflow occurs and if this quotient equals zero, the flow is fully bidirectional similar to 2D tilting systems (no net flow in one direction). In the rOoC, this is for instance the case for low tilts (<10°), high rotations speed (>10 rpm) and predominantly in the inner PC. Moreover, bidirectional flow occurs for higher filling volumes (Appendix B).
| Expression | b c | w c | l C,out | V max | p c | α | f | μ | P |
|---|---|---|---|---|---|---|---|---|---|
| Name | PC width | PC height | PC length | Reservoir volume | Critical pressure | Tilt | Tilting speed | Fluid viscosity | Fluid density |
| Unit | mm | mm | mm | μL | Pa | ° | rpm | mPa s | kg m−3 |
| Value | 1.2 | 0.8 | 16.4 | 300 | 0/40 | 10–20 | 2–10 | 1 | 1001 |
Endothelial lining and sprouting on-chip
Vasculature provides oxygen and nutrients for most tissue and supports organ function and physiology. The lining of the luminal surface of the vasculature depends on the substrate as well as on the mechanical features, also called haemodynamic forces, such as flow, pressure, speed and direction that translate into shear stress on the EC membrane. Hence, WSS is a key parameter influencing the morphology and physiology of ECs as demonstrated in different studies.28,29To evaluate the behaviour of ECs in the rOoC platform, we first tested the viability and morphology of human umbilical vein endothelial cells (HUVECs) cultured over 2 weeks in the inner and outer PCs (3 independent experiments with 3 rOoCs each). For this, the OCs were filled with ECM (Geltrex), and subsequently, both outer and inner PCs of the rOoC chip were coated with a mixture of collagen I type and Geltrex in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio. HUVECs were plated in a concentration of 2 × 106 cells per mL and left to attach for 1 h without tilting. Then, perfusion was performed by placing the rOoC on the 3D-tilting platform. HUVECs formed a confluent monolayer covering the walls and the bottom of the PCs 24 h after plating. The confluent monolayer was preserved for at least 14 days of culture under both high and low WSS (Fig. S1†). Elongation and alignment of ECs are the main morphological changes that can be observed within hours of exposure to flow stimulation.30–32 Previously, it has been demonstrated that the alignment of ECs in vivo is characteristic of large blood vessels with unidirectional and laminar flow. Correspondingly, the ECs alignment in vitro is stronger for a unidirectional flow compared to a bidirectional flow.33 We used the following settings to investigate alignment and sprouting (WSS values from Fig. 2d and e for inner/outer PC):
1 ratio. HUVECs were plated in a concentration of 2 × 106 cells per mL and left to attach for 1 h without tilting. Then, perfusion was performed by placing the rOoC on the 3D-tilting platform. HUVECs formed a confluent monolayer covering the walls and the bottom of the PCs 24 h after plating. The confluent monolayer was preserved for at least 14 days of culture under both high and low WSS (Fig. S1†). Elongation and alignment of ECs are the main morphological changes that can be observed within hours of exposure to flow stimulation.30–32 Previously, it has been demonstrated that the alignment of ECs in vivo is characteristic of large blood vessels with unidirectional and laminar flow. Correspondingly, the ECs alignment in vitro is stronger for a unidirectional flow compared to a bidirectional flow.33 We used the following settings to investigate alignment and sprouting (WSS values from Fig. 2d and e for inner/outer PC):
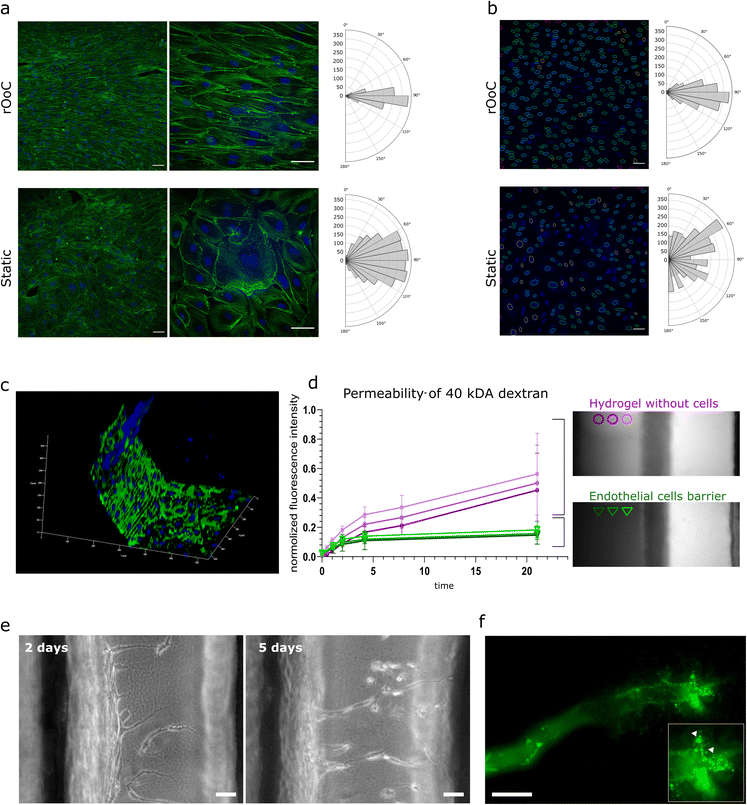
The cells were cultured for 4 days in the outer and inner PCs using the high-WSS setting, fixed and subsequently stained with FITC-conjugated phalloidin and DAPI to visualize the actin cytoskeleton and the nuclei respectively. Representative confocal images in Fig. 3a illustrate a clear difference between cells cultured under static conditions, and directional flow. This is not only apparent in the alignment of the actin fibres as shown in the magnified images (Fig. 3a, right side), but also in the alignment of nuclei (Fig. 3b). To quantify HUVECs alignment to the flow direction, orientation histograms were created for both actin filaments (Fig. 3a) and nuclei (Fig. 3b). The analysis was expanded to different flow settings, and a comparison between inner and outer circuits (fluorescence images and alignment histograms Fig. S2†).
Obtained results demonstrated stronger alignment in the outer PC compared to the inner PC and for increased tilting speeds (high-WSS vs. medium-WSS). This corresponds with the WSS values for both PCs and settings given in Table 2.
| Setting | Tilt [°] | Speed [rpm] | WSSinner [dyne per cm2] | WSSouter [dyne per cm2] |
|---|---|---|---|---|
| High-WSS | 15 | 2 | 1.5 | 3 |
| Medium-WSS | 15 | 1 | 0.8 | 1.5 |
| Low-WSS | 10 | 1 | 0.3 | 1.2 |
Although others have reported a positive impact of unidirectional flow in contrast to bidirectional flow on endothelial cells, we wanted to study the impact of these different flow conditions in the rOoC. For this, we modified the system and replaced the 2 U-shaped reservoirs by 4 round reservoirs (5 mm diameter each) to produce a bidirectional flow with the same amplitude and frequency when tilted on a conventional 2d-rocker (Mimetas OrganoFlow®). We plated HUVECs in both devices (2 each) and cultivated them for 2 days with the medium-WSS setting. Next, we performed a live/dead staining and counted the ratio between dead and living cells as described in the methods part. We demonstrated not only a decreased alignment but also a significantly increased rate of dead cells under bidirectional perfusion – 0.3 compared to 0.08 under unidirectional perfusion. The results of this analysis are summarized in Fig. S7.†
Next, we evaluated the formation of an endothelial barrier in the rOoC. As mentioned above, ECs formed a confluent monolayer not only on the bottom but also on the walls of the PCs (Fig. 3c). The barrier formation in the outer channel was evaluated at the interface with hydrogel by the ratio of fluorescence between the gel region and the outer PC (Fig. 3d). For this, EGM2 medium was supplemented with 40 kDa FITC-conjugated dextran (250 μg ml−1) and introduced into the outer PC with or without HUVECs similarly to the experiment done in Appendix A. The permeability assay confirmed the establishment of an effective barrier preventing dextran leakage (Fig. 3d).
The sprouting of new microvessels from a monolayer of ECs defines the initiation of angiogenesis and is an important process in physiological and pathological cascades. To test ECs sprouting on the rOoC, we plated HUVECs in the outer PC as described above (2 independent experiments with 3 parallels each) and performed perfusion at the low-WSS setting (Table 2). This setting has been empirically optimised and defined as an optimal setting for sprouting. The first tip cells in the ECM were observed after 48 h of culture in the HUVEC endothelialized rOoC. To stabilize newly formed microvessels and promote their growth, we generated a gradient of angiogenic factors in the ECM compartment, as described in the methods part. From day 5 onwards, we observed the formation of microvessels with discernible lumens that extended towards the inner PC. Using GFP-labelled microbeads that were added to the endothelialized PC on day 6 after sprouting, we demonstrated that the newly formed microvessels are perfusable as beads were detected at the tip of these microvessels (Fig. 3e).
Liver organoids on-chip
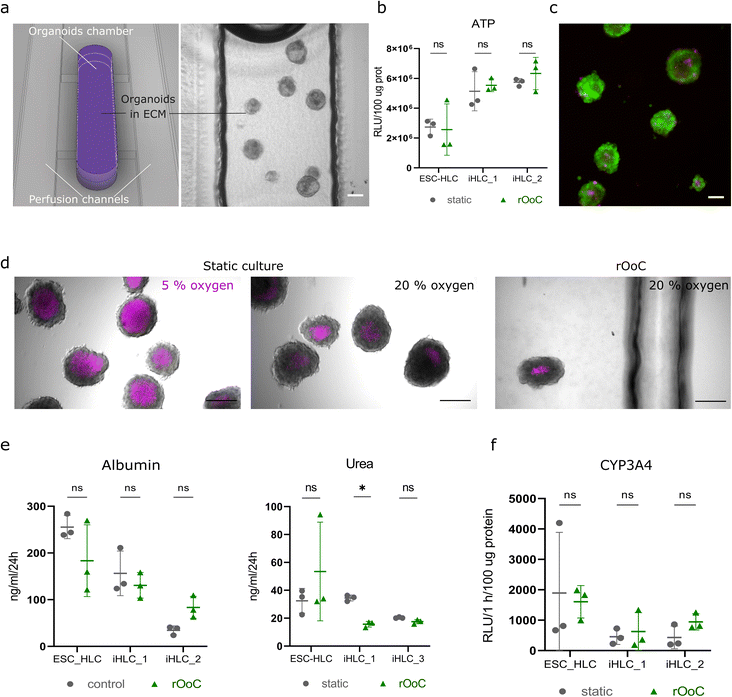
Next, we tested whether the rOoC platform supports the viability and functionality of 3D liver organoids cultured in the OC. For this, we differentiated human embryonic stem cells (H1 line) and induced pluripotent stem cells (WTC-11 and WTSIi013-A cell lines) towards hepatocyte-like cells grown as 3D organoids (ESC-HLC, iHLC_1 and iHLC_2, correspondingly) together termed as HLC organoids. Protocol and characterization of the liver organoids are shown in Fig. S4.† After 24 days of differentiation, HLC organoids were mixed with Geltrex in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio and introduced into the OCs (5–20 organoids each). HLC organoids were cultured in the rOoC platform for up to 2 weeks (3 different stem-cell derived HLCs used with 3 parallel rOoCs in each experiment).
2 ratio and introduced into the OCs (5–20 organoids each). HLC organoids were cultured in the rOoC platform for up to 2 weeks (3 different stem-cell derived HLCs used with 3 parallel rOoCs in each experiment).
During this time HLC organoids preserved their overall morphological characteristics (Fig. 4a), however, some organoids exhibited sprouting of non-hepatic cells, due to the presence of an endothelial lineage in the HLC organoids. After 2 weeks, organoids in the rOoC had the same level of viability as in standard 3D static cell culture, without significant changes in ATP content (N = 3 PSC lines) (Fig. 4b). Live/dead staining after 2 weeks of culture was also performed to visualize cell viability in the HLC organoids (Fig. 4c). To further characterize the culture conditions in the OC, we stained the HLC organoids after 48 h of on-chip culture in the standard incubator (20% (v/v) oxygen, 5% (v/v) CO2, 370 °C) with Image-iT™ Red Hypoxia Reagent (Thermo Fisher Scientific, Fig. 4d). As a static positive control, we used free-floating organoids cultured in a hypoxia incubator (5% (v/v) oxygen), and as static negative controls free-floating HLC organoids placed in a standard incubator. Unlike the positive controls, organoids cultured on-chip showed a similar number of hypoxic cells as the negative control, mostly restricted to the organoids core as commonly seen. However, when more than 20 organoids were cultured in each OC, HLC organoids experienced hypoxic conditions, especially in the centre of the OC indicating a reduction of oxygen supply in these areas.
To test the functionality of HLC organoids, we compared the albumin and urea production of HLC organoids cultured under static conditions in an ultra-low attachment plate, or cultured in the rOoC (Fig. 4e). HLC organoids in the rOoC exhibited similar levels of albumin and urea secretion as those cultured under static conditions. Next, we tested CYP3A4 enzymatic activity, essential for the biotransformation of more than 30% of all prescribed drugs in the liver, in HLC organoids using a luciferase-based assay based on Luciferin IPA (Promega, Sweden). The luciferase-based assay (Fig. 4f) indicated the same level of CYP3A4 activity in HLC organoids cultured in the rOoC platform and under static conditions.
Taken together, we demonstrated that the rOoC is suitable for the culture of 3D organoids, taking the example of HLC organoids, and supports their viability and functionality over at least 2 weeks of culture.
Circulating immune cells
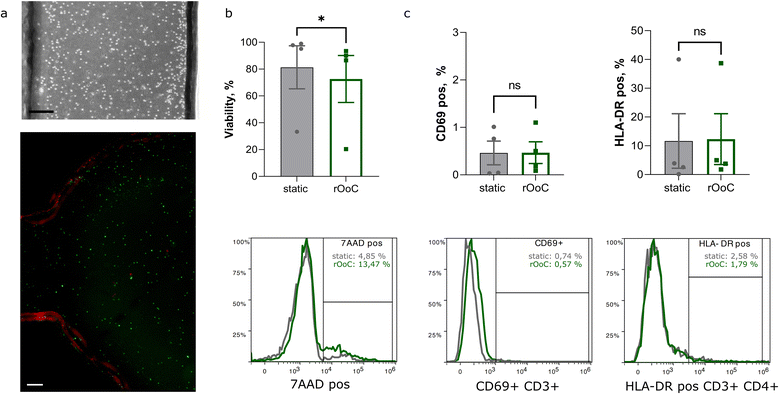
Immune cells, both tissue-resident and circulating, play a fundamental role in nearly all diseases. While tissue-resident immune cells may be included in an organoid, immune cell circulation and homing are more challenging to reproduce in vitro, requiring either a Transwell® or a microfluidic setting.34 Maintaining circulating immune cells in a microfluidic system can be difficult due to immune cell trapping and activation. Trapping may occur in tubing, reservoirs and internal pumps, reducing the number of cells that can interact with stationary tissue models. Unwanted activation of immune cells may occur due to mechanical stress35e.g. in areas with high shear forces or, in the case of internal pumps, by moving parts. Activation of immune cells changes the characteristics of the cells and may superimpose an effect that is alien to the intended physiological or disease model.To determine viability as well as potential trapping and activation of immune cells in the rOoC platform, we isolated peripheral blood mononuclear cells (PBMCs) from 4 healthy donors (see Table 3) as described in the methods part. Next, PBMCs were introduced in the outer PC of the rOoC platform at a density of 3 × 105 cells per mL in RPMI medium and the device was perfused on the platform to produce the previously defined high-WSS condition (Table 2) in the outer PCs (for each of the 4 independent experiments, 3 parallel rOoCs were used). The movement of the PBMCs on-chip was tracked using a miniaturized digital microscope (Dino-Lite Edge 3.0, Dino-Lite Europe B.V., Netherlands). As shown in the Video S2,† PBMCs followed the general flow in the PCs without any visible trapping at any point of the chip. After the culture in the rOoC and under static conditions, we stained the cells with the dead-marker 7-aminoactinomycin D (7-AAD) and labelled them with monoclonal antibodies to determine cellular subsets and possible activation by flow cytometry. As seen in Fig. 5, as shown by representative bright field image of cells in the channel and LIVE/DEAD labelled cells in the reservoirs, PBMC were evenly distributed in the device (Fig. 5a). PBMC cultured in the rOoC maintained high viability (above 85%) after 24 h of culture, except for the samples from one donor, which was characterized by low viability in both static conditions and the rOoC (Fig. 5b). Importantly, we did not observe a significant change in percentages of T-cells populations with early (CD69) or late (HLA-DR) activation markers.
| Gender | Age | |
|---|---|---|
| Donor 1 | Female | 32 |
| Donor 2 | Male | 36 |
| Donor 3 | Male | 37 |
| Donor 4 | Female | 35 |
Experimental
Device fabrication
The rOoC chips are made of several laser-machined PMMA sheets (Hesaglas®, Topacryl AG, Switzerland) thermally bonded together. First, each layer was designed in Autodesk Inventor® 2019 (Autodesk Inc., USA) and saved as individual DXF files for laser machining. The top part (6 mm thick) containing the reservoirs was processed with a conventional CO2 laser cutter (Beambox®, Flux Inc., Taiwan). Reservoirs were laser-engraved using a laser power of 12 W and engraving speed of 25 mm s−1 and holes and the outline was laser-cut with 24 W and 6 mm s−1. Thin PMMA films (0.3/0.5 mm) containing the PCs and OCs are laser micro-structured using an RDX-500 (Pulsar Photonics GmbH, Germany) equipped with a UV (343 nm) femtosecond laser (Pharos PH2–10 W, Light Conversion, Lithuania). The films were cut with a speed of 500 mm s−1 and a peak fluence of 11.03 J cm−2.After structuring, the films were cleaned in an ultrasonic bath (USC100T, VWR International, USA) for 15 min, sprayed with a nitrogen gun, and dried in an oven at 65 °C for 15 min. Next, the substrates were UV-activated using an Excimer lamp with a 172 nm emission wavelength (ExciJet172 55-130, Ushio GmbH, Germany). The distance to the lamp was adjusted to 1 mm and the substrates were exposed to a dose of 0.6 J cm−2. Finally, the parts were aligned on an aluminium plate using alignment pins and transferred to a pneumatic hot-press (AirPress-0302, Across International LLC, USA). For equal pressure distribution and control, cleanroom paper was put on top of the stack and an electronic pressure regulator (ITV1030, SMC Corporation, Japan) was used to control the bonding pressure. After UV activation, PMMA can be thermally bonded below its glass transition temperature (Tg = 100 °C). We used a bonding pressure of 2.4 MPa, a temperature of 84 °C, and a bonding time of 15 min. In a previous study, test devices were fabricated with this technology that withstood loads >40 N with a thermal-induced channel collapse of less than 6 μm for a 200 mm2 large chamber.23
To prevent freestanding structures (e.g. in layer 2 or 3) from falling apart, small bridges (0.5 mm wide) were included that were later removed using a knife. This made it necessary to first bond the top parts of the device separately and later seal them in a second step (see Fig. 6). For on-chip confocal imaging, the devices were sealed with a 175 μm thick PMMA film (Merck Sigma, USA).
Flow measurement
The flow was measured by micro-Particle Image Velocimetry with a setup as described earlier.36 We used dyed 10 μm polystyrene microparticles (Merck Sigma, USA) in a concentration between 0.1% and 1% (m/v) as tracers. Images were acquired in transmission by using a digital microscope (H800-96S-ES3, United Scope LLC, USA) coupled with a high-speed camera (acA1300-200uc, Basler AG, Germany). The microscope was mounted to a customized stand including a LED backlight illumination as shown in Fig. S5.† The stand was assembled using laser-cut PMMA sheets and mounted on the moving platform. Particle movement was observed in a field of view of 592 × 100 Px (1 Px = 5.33 μm) at 2270 fps and 50 μs acquisition time. Later, image stacks of 10.000 frames were evaluated using an optical flow algorithm previously applied to calculate the motion of beating cardiomyocytes.37 The setup is suitable to detect flow velocities up to 100 mm s−1. The measurement was calibrated by applying defined flow rates to channels with a similar cross-section using a syringe pump as a pressure source (Aladdin AL-1000, World Precision Instruments LLC, USA). A calibration factor of 1.84 ± 0.4 was determined to calculate the flow rate Q from the measured velocity vm and the channel cross section (b, w):| Q ≈ 1.84·b·w·vm | (4) |
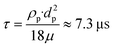 | (5) |
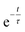 ). This value is much lower than the recording frequency of 2270 fps thus, it can be stated that the particles follow the flow to a good extent.
). This value is much lower than the recording frequency of 2270 fps thus, it can be stated that the particles follow the flow to a good extent.
Computational model
The flow in the rOoC is driven by the pressure difference between the inlet and outlet:| Δp = ρg(zi + hi − (zj + hj)), | (6) |
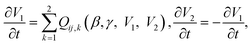 | (7) |
Based on these equations, we first developed a simplified model to understand the basic physics involved in the system (Appendix B). As it turned out, several additional effects needed to be taken into account:
• Due to the curved geometry of the reservoir, the water table height h non-linearly depends on the water volume and current tilt angles. To determine the liquid level in the reservoirs, we intersect the tilted reservoir geometry with the horizontal plane such that the intersected volume equals the current filling volume (determined with eqn (7)). This nonlinear problem is solved iteratively using Brent's method.
• Since the reservoirs have a fixed maximum volume (Vmax), the volume balance ensures that the volume never exceeds this value.
• The abrupt dimension change at the in-/outlets of the reservoirs acts as capillary-stop-valves.39 If the reservoirs run dry at one in-/outlet, the air–liquid interface will prevent backflow until a critical pressure pc is reached. This pressure is defined by the surface tension δ of the medium, the contact angle on the plastic surface θ and the estimated curvature radius of the liquid–air interface R:
pc = 2δ·cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ·R−1 θ·R−1 | (8) |
Endothelial cells culture
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was used as a 3D scaffold (bellow referred to as the extracellular matrix, ECM). A stock solution of 3 mg mL−1 rat tail collagen type I was neutralized with 1% 1 M NaOH (Sigma, S5761) and 10% 10× phosphate buffered saline, PBS (Thermo Fisher Scientific). The neutralized collagen was kept on ice until use and used within 10 min. The organ chamber was filled with 17 μL of ECM. After polymerization of ECM (incubated for 10 min at 37 °C), PCs were coated with 1% (v/v) of the same ECM diluted in a cold DMEM/F12 medium. Endothelial cells were dissociated by incubation with Trypsin/EDTA (Sigma, T3924-100ML), and re-suspended in EGM2-medium at a concentration of 2 × 106 cells per mL. 50 μL of the cell suspension was dispensed into the PCs and incubated for 1 h at 37 °C, with 5% CO2. For an even distribution of the cells to both walls, we first placed the rOoC on the left side and let the cells attach for 45 to 60 min. Next, a second aliquot of cell suspension was added to the PCs and the rOoC was placed on the right side for the same time. After the cells were attached to both walls, the rOoC was incubated for another 45 min to let cells attach to the bottom of the PCs. After that rOoC was placed on the 3D tilting platform. The following settings of the platform were used in the experiments with endothelial cells: speed 1–5 rpm, and tilt of 10–17°. The medium was changed daily.
1 was used as a 3D scaffold (bellow referred to as the extracellular matrix, ECM). A stock solution of 3 mg mL−1 rat tail collagen type I was neutralized with 1% 1 M NaOH (Sigma, S5761) and 10% 10× phosphate buffered saline, PBS (Thermo Fisher Scientific). The neutralized collagen was kept on ice until use and used within 10 min. The organ chamber was filled with 17 μL of ECM. After polymerization of ECM (incubated for 10 min at 37 °C), PCs were coated with 1% (v/v) of the same ECM diluted in a cold DMEM/F12 medium. Endothelial cells were dissociated by incubation with Trypsin/EDTA (Sigma, T3924-100ML), and re-suspended in EGM2-medium at a concentration of 2 × 106 cells per mL. 50 μL of the cell suspension was dispensed into the PCs and incubated for 1 h at 37 °C, with 5% CO2. For an even distribution of the cells to both walls, we first placed the rOoC on the left side and let the cells attach for 45 to 60 min. Next, a second aliquot of cell suspension was added to the PCs and the rOoC was placed on the right side for the same time. After the cells were attached to both walls, the rOoC was incubated for another 45 min to let cells attach to the bottom of the PCs. After that rOoC was placed on the 3D tilting platform. The following settings of the platform were used in the experiments with endothelial cells: speed 1–5 rpm, and tilt of 10–17°. The medium was changed daily.
Liver organoid culture
Hepatic differentiation was performed using a modification of previously published protocols.40 Briefly, iPSC/ESCs were differentiated toward definitive endoderm in IMDM/F12 media containing 1% (v/v) lipid concentrate (Thermo Fisher Scientific, catalogue no. 11905031), 100 μg ml−1 transferrin, 3 μM CHIR99021 (Tocris Bioscience, catalogue no. 4423), 50 nM PI-103 (Tocris Bioscience, catalogue no. 2930) and 100 ng ml−1 activin A (Peprotech, catalogue no. 120-14P) for 24 h and 100 ng ml−1 activin A for subsequent 48 h. The definitive endoderm cells were treated with 10 ng mL−1 FGF2 (Peprotech, catalogue no. 100-18B) and 20 ng mL−1 BMP4 (Peprotech, catalogue no. 120-05) in IMDM/F12 medium supplemented with 1% (v/v) N-2 (Thermo Fisher Scientific, catalogue no. 17502-048), 1% (v/v) B-27 minus vitamin A (Thermo Fisher Scientific, catalogue no. 12587010) and 1% (v/v) lipid concentrate, then with 5 μM A8301 (Stem Cell Technologies, catalogue no. 72022), 20 ng mL−1 HGF (Peprotech, catalogue no. 100-39H), 20 ng mL−1 BMP4, 1% (v/v) B-27 with vitamin A for 3 more days and with 25 ng mL−1 HGF, 1% (v/v) DMSO for another 5 days. At day 12, cells were detached by accutase and aggregated in the agarose U bottom microwells in the presence of 25 ng mL−1 HGF, 0.1 μM dexamethasone, 10 μM Y-27632, 0.5% (v/v) ITS, 0.1% (v/v) lipids concentrate, 100 μM Ascorbic acid-2 phosphate (AAP), 1% (v/v) B-27 (without vitamin A) and 1% (v/v) N-2. After formation of spheroids at day 13, media was replaced with William's E media, supplemented with 5% (v/v) FBS, 20 ng ml−1 HGF and 10 ng ml−1 oncostatin M (Peprotech, catalogue no. 300-10), 1% (v/v) ITS, 100 μM AAP, 0.1 μM dexamethasone. For further maturation, organoids were cultured in microwells in William's E media, supplemented with 1% (v/v) ITS, 0.1 μM Dexamethasone, 20 ng ml−1 oncostatin M and 1% (v/v) MEM non-essential amino acids solution (Thermo Fisher Scientific, catalogue no. 11140050), 10 uM DAPT for another 10 days with the replacement of media every 48 h. On day 18, organoids were additionally incubated for 1 h in the same medium supplemented with 5% (v/v) of Geltrex.
Immunofluorescence staining and microscopy
Cell cultures in rOoC were fixed in 4% (w/v) PFA for 20 min on the orbital shaker. Three washing steps (each 10 min) in DPBS using an orbital shaking followed each step. Permeabilization and blocking was performed by incubation in PBS with 1% (m/v) BSA (Sigma Aldrich), 0.2% (v/v) Triton-X100 (Sigma Aldrich) and 0.5% (v/v) DMSO at RT for 2 h on the orbital shaker. Staining with primary antibodies was performed for 24 h (at 4 °C) with subsequently 2 h incubation with secondary antibodies (Jackson ImmunoResearch, West Grove, PA) diluted with 1% BSA, 0.1% Triton-X100 in PBS. Primary antibodies (Ab) used in this study: rabbit polyclonal Ab to human serum albumin (Abcam, catalogue no. ab2406, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), rabbit polyclonal to VE-cadherin (Abcam, catalogue no. ab33168, 1
400), rabbit polyclonal to VE-cadherin (Abcam, catalogue no. ab33168, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), mouse monoclonal to human CD31 (Abcam, catalogue no. ab24590, 1
400), mouse monoclonal to human CD31 (Abcam, catalogue no. ab24590, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200). Secondary Ab used in this study: CY™3 AffiniPure Donkey Anti-Rabbit IgG (H + L) (catalogue no. 711-165-152, 1
200). Secondary Ab used in this study: CY™3 AffiniPure Donkey Anti-Rabbit IgG (H + L) (catalogue no. 711-165-152, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgG (H + L) (catalogue no. 715-605-150, 1
400), Alexa Fluor® 647 AffiniPure Donkey Anti-Mouse IgG (H + L) (catalogue no. 715-605-150, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400) (all secondary Ab are from Jackson ImmunoResearch). Nuclear counterstaining was performed with 1 μg mL−1 Hoechst 33258 (Sigma Aldrich). Confocal microscopy was performed on a Zeiss 700 laser scanning confocal microscope using standard filter sets and laser lines with a 40× oil immersion objective. Images were acquired using Zen software (Zeiss) as Z-stacks with 2 μm spacing between stacks. The confocal images were analyzed using Fiji software13 and are displayed as a resulting Z-stack.
400) (all secondary Ab are from Jackson ImmunoResearch). Nuclear counterstaining was performed with 1 μg mL−1 Hoechst 33258 (Sigma Aldrich). Confocal microscopy was performed on a Zeiss 700 laser scanning confocal microscope using standard filter sets and laser lines with a 40× oil immersion objective. Images were acquired using Zen software (Zeiss) as Z-stacks with 2 μm spacing between stacks. The confocal images were analyzed using Fiji software13 and are displayed as a resulting Z-stack.
RNA extraction and real-time polymerase chain reaction (PCR)
RNA was isolated using an RNeasy Micro kit (Qiagen, Germany) according to the manufacturer's protocol. cDNA was synthesized using a High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, catalogue no. 4368814). Gene expression analysis was performed using a TaqMan Universal mix on a TaqMan ViiA7 Real-Time PCR System. The primers used in this study were: ALB (Hs00609411_m1), HNF4A (Hs00230853_m1), CYP3A4 (Hs00604506_m1), A1AT (SERPINA, Hs01097800_m1) and RPL (Hs02338565_gH), all purchased from Thermo Fisher Scientific. RPL was used as endogenous control. The levels of expression of genes of interest were quantified by ddCt with normalization to undifferentiated stem cells (d 0) differentiated from the H1 line. Data represent three donor PHH samples, and HLC differentiated from 3 cell lines.PBMC isolation and culture
The whole blood samples were collected into 5 mL EDTA tubes from 4 healthy subjects:Human peripheral blood mononuclear cells (PBMC) were isolated by a density gradient technique using the Lymphoprep™ gradient (StemCell Technologies, 07851) using the vendor's protocol. PBMC were cultured for 24 h both in the rOoC and under static conditions, using an advanced RPMI medium (Thermo Fisher Scientific, catalogue no. 12633012), supplemented with 2% (v/v) FBS, 1% (v/v) penicillin–streptomycin and then analyzed by Flow cytometry.
Flow cytometry
PBMC were collected from the rOoC and static plates and centrifuged at 300 g for 5 minutes. Next, cells were washed once with FACS buffer (PBS, supplemented with 1% BSA and 0.05% sodium azide) and centrifuged again at 300g for 5 min. Staining for immune subsets and activation markers was performed for 30 min, followed by a wash step with FACS buffer and analysis at a “Gallios” flow cytometer (Beckman Coulter Life Sciences, United States). The following antibodies were used at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 in FACS buffer:
250 in FACS buffer:
• Anti-Human CD4 FITC (eBioscience 11-0049-42), Clone RPA-T4
• Anti-Human CD3 PE (BioLegend 300308), Clone HIT3a
• Anti-Human CD69 PE/Cy7 (Biolegend 310912), Clone FN50
• Anti-Human HLA-DR APC/Cy7 (Miltony Biotech 130-111-792), Clone REA805
• 7AAD ready-made solution (Sigma SML1633), dilution: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100
100
Image processing
The alignment of ECs was analysed with two different toolchains for the cytoskeleton and the nuclei. Image pre-processing was done in Fiji,41 cell/nuclei segmentation was performed with Cellpose42 and for post-processing, we used Python with the libraries Matplotlib and OpenCV. For actin-stained fluorescence/confocal and phase-contrast images the following process was applied:a) For confocal images only: Z-projection in Fiji.
b) Conversion in an 8 bit image.
c) Analysis of cell alignment using the OrientationJ Vector Field43 plugin from Fiji:
• Gradient: Riesz-Filters
• Local window: 10 Px
• Grid size: 40 Px
d) The generated vector field is saved as a CSV file and loaded in a Python script (Code S4‡) that extracts and plots the orientation histograms.
The orientation of the nuclei was evaluated as follows:
I. For confocal images only: Z-projection in Fiji.
II. Conversion in 8 bit image and application of a Gaussian-blur filter (sigma = 2).
III. Segmentation in Cellpose using the “nuclei method” and a size of 20–100 Px (depending on magnification) and saving as an ImageJ contour file.
IV. Reading the contour files in a Python script (Code S5‡) that fits ellipses to the contours and calculates nuclei orientation and aspect ratio.
V. Plotting of nuclei orientation histograms using the script from d).
Cell counting for immune cells and EC viability assay was performed in Fiji with following toolchain:
1. Convert to 8 bit.
2. Remove background (rolling ball = 10 Px).
3. Gaussian-blur filtering (sigma = 2).
4. Auto-threshold with OTSO method (LI for dead cells).
5. Remove channel walls if necessary.
6. Analyse-particle macro (minimum size = 5 Px).
Statistics
Statistical analyses and graph generation were performed using GraphPad PRISM 9 (GraphPad Software Inc.). Unless specifically stated, a two-tailed, paired t-test (with unequal variances) was applied for the comparison of the two groups. If not stated otherwise, a one-way ANOVA analysis was applied. The data are presented as mean ± SD. Statistical significance was assigned as not significant (NS) P > 0.05; or significant with *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.Conclusions
We present a novel, pump-less rOoC platform capable to cultivate different types of cells or organ representations in a closed microfluidic circuit with a unidirectional flow. The device is built of laser-structured thermoplastic films that are thermally bonded to form a multi-layer device. One of the main advantages of this technology over the often used soft lithography process44 is that thermoplastic films do not adsorb small molecules to the extent to which soft lithography materials do (especially Polydimethylsiloxane (PDMS)45).Besides fabrication, our layout presents several advantages over pump-driven platforms: air bubbles are automatically trapped in the reservoirs avoiding channels blockages and cells death, cell/organoid loading and media exchange are simply done by pipetting and many systems can be run in parallel (8 rOoCs per plate, >10 plates per rotator). Compared to pump-driven systems that require controlling units and connecting tubes, the rOoC platform has higher scalability and lower cost46 and does not require the integration of flexible membranes23 for fluid manipulation. The integration of flexible membranes increases fabrication time, costs, and potentially influences the behaviour of biological material by the introduction of another material into the system and by the implementation of local high pressure, which will in particular affect circulating cells. Compared to other gravity-flow-driven systems, the rOoC platform generates a unidirectional instead of a bidirectional flow. In the rOoC platform, we see a strong and shear stress dependent alignment of endothelial cells (Fig. 3a, b and S2†) after several days of flow stimulation. Furthermore, we see a significant reduction in cell viability if endothelial cells are culture under bidirectional flow conditions (obtained by modifying the reservoir layout) compared to unidirectional flow perfusion with the rOoC reservoir geometry under the same tilting conditions in accordance with the previous reports.33
As organoids are the main building block in 3D cell culture, we demonstrated the suitability to embed and culture liver organoids in the rOoC. For this, we use a specific chamber layout with a stepped channel geometry that can be used to structure the ECM and provide a biologically relevant and biodegradable separation between PCs and OCs.27 We have shown the organoid functionality using stem-cell-derived liver organoids and proved that the organoids stay healthy, not showing a significant hypoxic core, secrete albumin and urea and show drug metabolizing activities at the same level as the static control. The advantage of an ECM-based separation layer instead of a separating polymer membrane is that this layout can be used to vascularize tissue representations. We have demonstrated that the platform allows EC to sprout into the OC. To achieve long-term stable perfusable microvessels, an interstitial flow47,48 will be added in a future iteration of the device. To do so, the layout can be adapted to generate a sufficient pressure gradient between both perfusion channels, e.g. by using bigger reservoirs or placing both perfusion circuits next to each other instead of one in the other. This will lead to an alternating and not parallel flow direction in both PCs thus increasing the pressure gradient compared to the current configuration. Finally, we showed that circulating immune cells could be cultured without trapping in external tubing, integrated pumps or accumulation in medium reservoirs. At the same time, the ECM separation layer allows for immune interaction with the tissue representations cultured in the OC e.g. to study immune cell homing or inflammatory processes. Significant sedimentation or trapping of immune cells did not occur for tilting speed of 1 rpm (lowest possible setting with the current platform) or more. Nevertheless, this does not mean that sedimentation might occur for lower speeds thus indicating a possible limitation of the current platform for low perfusion speeds.
Most importantly, we have thoroughly characterized the rOoC platform and developed a computational model to predict flow rates and WSS values. This will help us to conduct a model-driven design approach in the future to tune the platform to new biological questions. The flow characteristics of a fluidic system can be classified using the Reynolds (Re) and the Womersley (Wo) numbers (definition see ESI Text†). Re relates to the relationship between viscous to inertial forces in the liquid flow, whereby high values of Re lead to turbulences. Wo is used to describe if inertial effects due to unsteady pressure lead to a disturbed flow profile (oscillatory flow). The maximum Re number in the rOoC is 115, which is clearly within a laminar regime and the maximum Wormesley number in our system is Womax = 0.2 thus the flow can be considered undisturbed by transient effects (calculations and channel dimensions see ESI Text†). Given these flow conditions, we mimic healthy conditions for EC lining.30 The flow rate/WSS can be tuned either by design or by changing the driving conditions (tilt, speed, volume) thus we can model an arterial and a venous side (different WSS) in one device. For example, as described in Appendix B, a high reservoir volume will result in a bidirectional flow pattern that is associated with a bidirectional flow pattern resulting in loss of EC formation and is associated with inflammation and heart diseases.30
Moreover, we observed that surface tension effects influence the flow characteristics of the system, especially for low tilting speeds. Thus, the modelled curves align the measurements well when the critical pressure is taken into account whereas for higher speeds the effect seems to be not present. A possible explanation for this effect is evaporation. Due to the high-intensity LED illumination that is required for the μPIV measurement, the evaporation is significantly higher than in the incubator and might cause the liquid to evaporate fully in the reservoirs when they run dry. This is especially true for lower tilting speeds as the remaining liquid in the emptied reservoir has more time to evaporate.
In theory, different flow patterns (continuous, pulsatile and oscillatory) can be achieved by the rOoC platform. Strictly speaking, the generated flow-time curve depends on the reservoir layout, the pitch/roll of the platform (Fig. 2b) and the hydraulic conductivity (ESI Text†). Due to the relatively high channel cross-section and actuation frequencies (limited by the used rotation platform) used in this study, the flow rate is relatively high and pulsatile. Nevertheless, a modified version of the rotation platform driving a pump-less OoC system presented earlier49 might be suitable to obtain more control over the flow conditions. For instance, a programmable stepper motor can be used to perform the tilt of the reservoirs much faster than the fluid actuation from one reservoir to the other. Combining this with a reduced channel cross-section an almost square-shaped flow-velocity curve might be obtained. Using our developed computational model, we performed a flow prediction to prove this assumption (Fig. S8†). Even more control over the pitch and roll of the platform and thus the flow in the rOoC system might be possible by using a Stewart platform50 in the future.
In summary, the rOoC platform provides a robust and scalable new tool for building comprehensive in vitro models by incorporating three central components: long-term cultures of functional endothelial cells, ECM-embedded 3D organ representations and circulating immune cells. This, together with the versatility of the flow regulation should provide a valuable next-generation platform for disease modelling and interrogation.
Ethics
The regional Committees for Medical and Health Research Ethics of South East Norway (REK) approved all experiments with donor samples (reference number: 2012-286).Author contributions
M. B. developed the technology, designed and characterized the devices, prepared figures, and wrote the paper together with A. A. and S. K. Furthermore, A. A. designed the biological part of the paper, performed cell experiments and prepared figures. T. K. performed computational modelling and wrote the modelling part of the paper and the ESI Text.† A. F., L. D., M. A.-M., C. D. and J. S. performed cell experiments and biological readouts. A. G. and S. G. fabricated the microdevices. S. K. and E. M. supervised the work, provided laboratory space, and procured funding for this study. All authors have read and agreed to the published version of the manuscript.Data availability
The Arduino code to measure the tilt of the platform using a gyroscope (Code 1), Python script used to generate plots for pitch and roll in Fig. 2b (Code S2), Python script used to calculate the orientation of the cytoskeleton in Fig. 3 (Code S4), Python script used to calculate the orientation of the segmented nuclei in Fig. 3 (Code S5) and Python script used for the simplified flow model calculations in Appendix B (Code S6) are provided in our Github repository: https://github.com/MathiasBusek/Code-Tilting-Paper. The source code of the developed computational model based on DuMux51 (Code S3) is provided in our Github repository: https://github.com/timokoch/dumux-microfluidics.Conflicts of interest
M. B., A. A., M. A.-M., and S. K. have applied for a patent covering the main principle of fluid actuation and application and are planning to commercialize the technology via a start-up company.Appendices
Appendix A: Diffusion experiment
To test the capability of the rOoC device to establish and maintain gradients, the diffusion of fluorescein-labelled dextran (10, 40 and 70 kDa, Merck Sigma, USA) from one side of the OC to the other was observed. First, the organ chamber was filled with an ECM (Geltrex, Thermo Fisher Scientific, catalogue no. A1413201) to generate a flow barrier. Next, a dextran solution dissolved in PBS (concentration: 0.25 mg mL−1) was injected into the reservoirs either connected to the inner or outer PC (donor side). This setup is similar to the rOoC chip filled with red food colour in the inner circuit as shown in Fig. 7a. Now, the fluorescence intensity in three different regions of interest (ROIs) (1, 2, and 3 marked in Fig. 7b) was measured with a fluorescence microscope (Axio Vert. A1, Zeiss, Germany). Images were recorded every 5 min with a constant exposure time (28 ms) and excitation intensity. Next, the mean intensity in the FOW was evaluated using Fiji.41 Finally, the fluorescence signals were normalized to the maximum intensity and averaged over all ROIs. Fig. 7c shows the normalized fluorescence intensity for 70 kDa dextran depending on the distance from the PCs.Appendix B: Simple model & characterisation
In a rough approximation, the reservoirs can be simplified as rectangular (width bR, length lR). The height difference within the reservoirs (ΔhR) and between the in- and outlets of the PCs (Δhc) can then be calculated from the pitch and roll β, γ and lengths.ΔhR, Δhc = lR·sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β, lc·sin β, lc·sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) γ γ | (C1) |
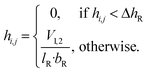 | (C2) |
| Expression | l C | b c | w c | l R | b R | α | f | μ | ρ |
|---|---|---|---|---|---|---|---|---|---|
| Unit | mm | mm | mm | mm | mm | ° | rpm | mPa s | kg m−3 |
| Value | 20 | 3.2 | 0.5 | 20 | 3 | 20 | 3 | 1 | 1001 |
The dynamic viscosity μ for the used media (EGM™-2 BulletKit™, Lonza, Switzerland) is not detailed reported but can be estimated from similar media like DMEM with FBS supplement.52 Given the FBS content of 2% for EGM-2, we approximate the viscosity to be 1 mPa s at 20 °C and 0.8 mPa s at 37 °C. Table 4 denotes a different PC cross-section than given in Fig. 1d. The reason behind this is that the optical setup used to measure the flow velocity has a very high focal length thus detecting almost all particles in the PC. The 3D component of the flow field will lead to a very high tracking error as particle patterns are overlapping each other. To overcome this problem, the PC height for the devices used for the flow studies has been reduced to 0.5 mm. To achieve a similar channel transmissibility σ (approximation error: 1.2%), the PC width was increased to 3.2 mm. In terms of flow rate, this geometry is equal to the cross-section used for the cell experiments but can be measured with the current optical setup without using light-sheet illumination or objectives with a high numerical aperture.
Fig. 8a shows the calculated flow rate and volumes for different initial volumes using the simplified model. Up to a filling volume of 300 μL (excluding the PC volume), the flow is unidirectional as the reservoirs run dry and the whole volume is exchanged. Above this value, a negative flow component is occurring due to liquid remaining on the other side of the tilted reservoir causing a reversed flow at certain positions of the platform. The transported volume is either defined by the amount filled in (V0) or the maximum tilt and the reservoir geometry:
ΔVmax = lc·sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) α·bR·lR ≈ 410 μL α·bR·lR ≈ 410 μL | (C3) |
To validate the developed model, we conducted flow measurements with the μPIV setup explained in the Materials and Methods section. The flow was measured in the middle between both reservoirs of the outer PC. A flow-time curve for different initial filling volumes is given in Fig. 8b. In contrast to the simplified model, the flow is in reality unidirectional even for higher filling volumes and rises much faster. Moreover, in contrast to Fig. 8a, the flow rate is much reduced for a filling volume of 600 μL compared to 500 μL.
Another important aspect is surface effects at the air–liquid interface. When the reservoirs run dry, an air–liquid interface is formed that will present a resistance to the flow according to eqn (6). To obtain a reasonable value for the critical pressure pc, the contact angle has to be measured. We used drops of deionized (DI) water on different substrates of PMMA (Contact Angle Goniometer, Ossila, UK). In Fig. 8c and d, the contact angle for a pure PMMA substrate and after exposure to a dose of 1.2 J cm−2 UV light is shown. The contact angle is reduced from 80° to 20° and the grade of hydrophilicity depends on the applied UV dose and fades away only slightly over one year (Fig. 8e). The critical pressure depends furthermore on the surface tension δ (for cell media values between 50 and 70 mN m−1 have been reported53,54) and the curvature of the liquid–air interface. As these values are difficult to define exactly, the critical pressure was measured experimentally for different in-/outlet geometries. The experimental setup was as follows:
The liquid level is increased until the receiving reservoir is filled. The needed value of Δp is defined as the critical pressure pc for this geometry and cell media. By using this method, we determined a critical pressure of 40 Pa.
Acknowledgements
Flow cytometry assays and quality control of iPSC lines were performed at The Norwegian Centre for Stem Cell Research, Oslo University Hospital. Contact angle measurements were done at the chair of Biomedical Engineering from Prof. Nikolaj Gadegaard at University of Glasgow. This work received funding from the Research Council of Norway through its Centers of Excellence scheme; project No. 262613, open project support No. 315399 and a Qualification grant No. 329001. Furthermore, financial resources were granted from the South-Eastern Norway Regional Health Authority Innovation project No. 30629 and the European Union's Horizon 2020 Research and Innovation program under the Marie Skłodowska-Curie Actions Grant, agreement No. 801133 (Scientia fellowship), PSC Partners Seeking a Cure and the Norwegian PSC Research Center.Notes and references
- L. A. Low, C. Mummery, B. R. Berridge, C. P. Austin and D. A. Tagle, Nat. Rev. Drug Discovery, 2021, 20, 345–361 CrossRef CAS PubMed.
- D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin and D. E. Ingber, Science, 2010, 328, 1662–1668 CrossRef CAS PubMed.
- P. Vulto and J. Joore, Nat. Rev. Drug Discovery, 2021, 1–2 Search PubMed.
- U. Marx, T. Akabane, T. B. Andersson, E. Baker, M. Beilmann, S. Beken, S. Brendler-Schwaab, M. Cirit, R. David, E.-M. Dehne, I. Durieux, L. Ewart, S. C. Fitzpatrick, O. Frey, F. Fuchs, L. G. Griffith, G. A. Hamilton, T. Hartung, J. Hoeng, H. Hogberg, D. J. Hughes, D. E. Ingber, A. Iskandar, T. Kanamori, H. Kojima, J. Kuehnl, M. Leist, B. Li, P. Loskill, D. L. Mendrick, T. Neumann, G. Pallocca, I. Rusyn, L. Smirnova, T. Steger-Hartmann, D. A. Tagle, A. Tonevitsky, S. Tsyb, M. Trapecar, B. van de Water, J. van den E. Raaij, P. Vulto, K. Watanabe, A. Wolf, X. Zhou and A. Roth, ALTEX, 2020, 37, 365–394 Search PubMed.
- N. Franzen, W. H. van Harten, V. P. Retèl, P. Loskill, J. van den Eijnden-van Raaij and M. IJzerman, Drug Discovery Today, 2019, 24, 1720–1724 CrossRef PubMed.
- M. A. Lancaster and M. Huch, Dis. Models Mech., 2019, 12, dmm039347 CrossRef CAS PubMed.
- R. C. Hubrecht and E. Carter, Animals, 2019, 9(10), 54 CrossRef PubMed.
- T. E. Winkler and A. Herland, ACS Appl. Mater. Interfaces, 2021, 13, 45161–45174 CrossRef CAS PubMed.
- N. Niu, S. Xu, Y. Xu, P. J. Little and Z.-G. Jin, Trends Pharmacol. Sci., 2019, 40, 253–266 CrossRef CAS PubMed.
- P. R. Myers and M. A. Tanner, Arterioscler., Thromb., Vasc. Biol., 1998, 18, 717–722 CrossRef CAS PubMed.
- Y. Yang, P. Fathi, G. Holland, D. Pan, N. S. Wang and M. B. Esch, Lab Chip, 2019, 19, 3212–3219 RSC.
- J.-J. Chiu and S. Chien, Physiol. Rev., 2011, 91, 327–387 CrossRef PubMed.
- J. Shin, S.-G. Hong, S. Y. Choi, M. E. Rath, J. Saredy, D. G. Jovin, J. Sayoc, H.-S. Park, S. Eguchi, V. Rizzo, R. Scalia, H. Wang, S. R. Houser and J.-Y. Park, Redox Biol., 2022, 50, 102252 CrossRef CAS PubMed.
- M. Busek, A. Aizenshtadt, M. A. Martinez, L. Delon and S. Krauss, Biosensors, 2022, 12(2), 126 CrossRef PubMed.
- A. Junaid, A. Mashaghi, T. Hankemeier and P. Vulto, Curr. Opin. Biomed. Eng., 2017, 1, 15–22 CrossRef.
- M. N. S. de Graaf, A. Vivas, A. D. van der Meer, C. L. Mummery and V. V. Orlova, Micromachines, 2022, 13, 1359 CrossRef PubMed.
- R. Novak, M. Ingram, S. Marquez, D. Das, A. Delahanty, A. Herland, B. M. Maoz, S. S. F. Jeanty, M. R. Somayaji, M. Burt, E. Calamari, A. Chalkiadaki, A. Cho, Y. Choe, D. B. Chou, M. Cronce, S. Dauth, T. Divic, J. Fernandez-Alcon, T. Ferrante, J. Ferrier, E. A. FitzGerald, R. Fleming, S. Jalili-Firoozinezhad, T. Grevesse, J. A. Goss, T. Hamkins-Indik, O. Henry, C. Hinojosa, T. Huffstater, K.-J. Jang, V. Kujala, L. Leng, R. Mannix, Y. Milton, J. Nawroth, B. A. Nestor, C. F. Ng, B. O'Connor, T.-E. Park, H. Sanchez, J. Sliz, A. Sontheimer-Phelps, B. Swenor, G. Thompson, G. J. Touloumes, Z. Tranchemontagne, N. Wen, M. Yadid, A. Bahinski, G. A. Hamilton, D. Levner, O. Levy, A. Przekwas, R. Prantil-Baun, K. K. Parker and D. E. Ingber, Nat. Biomed. Eng., 2020, 4, 407–420 CrossRef PubMed.
- A. Vivas, A. van den Berg, R. Passier, M. Odijk and A. D. van der Meer, Lab Chip, 2022, 22, 1231–1243 RSC.
- B. Zhang, A. Korolj, B. F. L. Lai and M. Radisic, Nat. Rev. Mater., 2018, 3, 257–278 CrossRef.
- S. J. Trietsch, E. Naumovska, D. Kurek, M. C. Setyawati, M. K. Vormann, K. J. Wilschut, H. L. Lanz, A. Nicolas, C. P. Ng, J. Joore, S. Kustermann, A. Roth, T. Hankemeier, A. Moisan and P. Vulto, Nat. Commun., 2017, 8, 262 CrossRef PubMed.
- E. Doutel, F. J. Galindo-Rosales and L. Campo-Deaño, Materials, 2021, 14, 7402 CrossRef CAS PubMed.
- Y. Wang, L. Wang, Y. Guo, Y. Zhu and J. Qin, RSC Adv., 2018, 8, 1677–1685 RSC.
- M. Busek, S. Nøvik, A. Aizenshtadt, M. Amirola-Martinez, T. Combriat, S. Grünzner and S. Krauss, Biosensors, 2021, 11, 162 CrossRef CAS PubMed.
- U. Klotzbach, F. Sonntag, S. Grünzner, M. Busek, F. Schmieder and V. Franke, Adv. Opt. Technol., 2014, 3, 515–521 CrossRef CAS.
- C. Su, Y. J. Chuah, H. B. Ong, H. M. Tay, R. Dalan and H. W. Hou, Biosensors, 2021, 11, 509 CrossRef CAS PubMed.
- P. Vulto, S. Podszun, P. Meyer, C. Hermann, A. Manz and G. A. Urban, Lab Chip, 2011, 11, 1596–1602 RSC.
- X. Zhao, Z. Xu, L. Xiao, T. Shi, H. Xiao, Y. Wang, Y. Li, F. Xue and W. Zeng, Front. Bioeng. Biotechnol., 2021, 9, 637048 CrossRef PubMed.
- N. Baeyens, C. Bandyopadhyay, B. G. Coon, S. Yun and M. A. Schwartz, J. Clin. Invest., 2016, 126, 821–828 CrossRef PubMed.
- E. Roux, P. Bougaran, P. Dufourcq and T. Couffinhal, Front. Physiol., 2020, 11, 861 CrossRef PubMed.
- C. A. Dessalles, C. Leclech, A. Castagnino and A. I. Barakat, Commun. Biol., 2021, 4, 1–15 CrossRef PubMed.
- S. Li, N. F. Huang and S. Hsu, J. Cell. Biochem., 2005, 96, 1110–1126 CrossRef CAS PubMed.
- R. Steward, D. Tambe, C. C. Hardin, R. Krishnan and J. J. Fredberg, Am. J. Physiol., 2015, 308, C657–C664 CrossRef CAS PubMed.
- Y. I. Wang and M. L. Shuler, Lab Chip, 2018, 18, 2563–2574 RSC.
- S. Shim, M. C. Belanger, A. R. Harris, J. M. Munson and R. R. Pompano, Lab Chip, 2019, 19, 1013–1026 RSC.
- J. M. Hope, J. A. Dombroski, R. S. Pereles, M. Lopez-Cavestany, J. D. Greenlee, S. C. Schwager, C. A. Reinhart-King and M. R. King, BMC Biol., 2022, 20, 61 CrossRef CAS PubMed.
- M. Busek, C. Polk, T. Albrecht, U. Marx, J. König and F. Sonntag, Biomed. Tech., 2012, 57, 927–930 Search PubMed.
- T. J. Kolanowski, M. Busek, M. Schubert, A. Dmitrieva, B. Binnewerg, J. Pöche, K. Fisher, F. Schmieder, S. Grünzner, S. Hansen, A. Richter, A. El-Armouche, F. Sonntag and K. Guan, Acta Biomater., 2020, 102, 273–286 CrossRef CAS PubMed.
- S. Burgmann, N. Van der Schoot, C. Asbach, J. Wartmann and R. Lindken, Houille Blanche, 2011, 97, 55–61 CrossRef.
- S. Wang, X. Zhang, C. Ma, S. Yan, D. Inglis and S. Feng, Biosensors, 2021, 11, 405 CrossRef CAS PubMed.
- G. Lee, H. Kim, J. Y. Park, G. Kim, J. Han, S. Chung, J. H. Yang, J. S. Jeon, D.-H. Woo, C. Han, S. K. Kim, H.-J. Park and J.-H. Kim, Biomaterials, 2021, 269, 120529 CrossRef CAS PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- C. Stringer and M. Pachitariu, Cellpose 2.0: how to train your own model, bioRxiv, 2022, preprint, DOI:10.1101/2022.04.01.486764.
- Z. Püspöki, M. Storath, D. Sage and M. Unser, in Focus on Bio-Image Informatics, ed. W. H. De Vos, S. Munck and J.-P. Timmermans, Springer International Publishing, Cham, 2016, pp. 69–93 Search PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- M. W. Toepke and D. J. Beebe, Lab Chip, 2006, 6, 1484–1486 RSC.
- I. Wagner, E.-M. Materne, S. Brincker, U. Süßbier, C. Frädrich, M. Busek, F. Sonntag, D. A. Sakharov, E. V. Trushkin, A. G. Tonevitsky, R. Lauster and U. Marx, Lab Chip, 2013, 13, 3538–3547 RSC.
- P. A. Galie, D.-H. T. Nguyen, C. K. Choi, D. M. Cohen, P. A. Janmey and C. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 7968–7973 CrossRef CAS PubMed.
- J. W. Song and L. L. Munn, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15342–15347 CrossRef CAS PubMed.
- P. Fathi and M. B. Esch, in Organ-on-a-Chip: Methods and Protocols, ed. M. Rasponi, Springer US, New York, NY, 2022, pp. 177–199 Search PubMed.
- B. Dasgupta and T. S. Mruthyunjaya, Mech. Mach. Theory, 2000, 35, 15–40 CrossRef.
- T. Koch, D. Gläser, K. Weishaupt, S. Ackermann, M. Beck, B. Becker, S. Burbulla, H. Class, E. Coltman, S. Emmert, T. Fetzer, C. Grüninger, K. Heck, J. Hommel, T. Kurz, M. Lipp, F. Mohammadi, S. Scherrer, M. Schneider, G. Seitz, L. Stadler, M. Utz, F. Weinhardt and B. Flemisch, Comput. Math. Appl., 2021, 81, 423–443 CrossRef.
- C. Poon, Measuring the density and viscosity of culture media for optimized computational fluid dynamics analysis of in vitro devices, bioRxiv, 2020, preprint, DOI:10.1101/2020.08.25.266221.
- S. Kebbouche-Gana, M. L. Gana, S. Khemili, F. Fazouane-Naimi, N. A. Bouanane, M. Penninckx and H. Hacene, J. Ind. Microbiol. Biotechnol., 2009, 36, 727–738 CrossRef CAS PubMed.
- M. Jokinen, K. Pittois, S. van den Akker, I. Gutschoven, T. Assmuth, T. Metz, H. Lehtilä and P. Alanne, Cytotechnology, 2020, 72, 271–282 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: For Fig. S1–S8, Video S1 and S2, ESI Table and ESI Text please see DOI: https://doi.org/10.1039/d2lc00919f |
| ‡ Code S1, Code S2 and Code S3–S6 available in GitHub repository: https://github.com/MathiasBusek/Code-Tilting-Paper. Code S3 available in GitHub repository: https://github.com/timokoch/dumux-microfluidics. |
| § Authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |