Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Label-free microfluidic cell sorting and detection for rapid blood analysis
Nan
Lu
 ab,
Hui Min
Tay
ab,
Hui Min
Tay
 a,
Chayakorn
Petchakup
a,
Chayakorn
Petchakup
 a,
Linwei
He
a,
Lingyan
Gong
a,
Kay Khine
Maw
a,
Linwei
He
a,
Lingyan
Gong
a,
Kay Khine
Maw
 a,
Sheng Yuan
Leong
a,
Wan Wei
Lok
a,
Hong Boon
Ong
a,
Ruya
Guo
c,
King Ho Holden
Li
a,
Sheng Yuan
Leong
a,
Wan Wei
Lok
a,
Hong Boon
Ong
a,
Ruya
Guo
c,
King Ho Holden
Li
 ab and
Han Wei
Hou
ab and
Han Wei
Hou
 *abd
*abd
aSchool of Mechanical and Aerospace Engineering, Nanyang Technological University, 50 Nanyang Avenue, Blk N3, Level 2, Room 86 (N3-02c-86), 639798, Singapore. E-mail: hwhou@ntu.edu.sg; Tel: +65 67904950
bHP-NTU Digital Manufacturing Corporate Lab, Nanyang Technological University, 65 Nanyang Drive, Block N3, 637460, Singapore
cKey Laboratory of Agricultural Information Acquisition Technology, Ministry of Agriculture and Rural Affairs, China Agricultural University, Beijing, 100083, China
dLee Kong Chian School of Medicine, Nanyang Technological University, 11 Mandalay Road, Clinical Sciences Building, 308232, Singapore
First published on 19th January 2023
Abstract
Blood tests are considered as standard clinical procedures to screen for markers of diseases and health conditions. However, the complex cellular background (>99.9% RBCs) and biomolecular composition often pose significant technical challenges for accurate blood analysis. An emerging approach for point-of-care blood diagnostics is utilizing “label-free” microfluidic technologies that rely on intrinsic cell properties for blood fractionation and disease detection without any antibody binding. A growing body of clinical evidence has also reported that cellular dysfunction and their biophysical phenotypes are complementary to standard hematoanalyzer analysis (complete blood count) and can provide a more comprehensive health profiling. In this review, we will summarize recent advances in microfluidic label-free separation of different blood cell components including circulating tumor cells, leukocytes, platelets and nanoscale extracellular vesicles. Label-free single cell analysis of intrinsic cell morphology, spectrochemical properties, dielectric parameters and biophysical characteristics as novel blood-based biomarkers will also be presented. Next, we will highlight research efforts that combine label-free microfluidics with machine learning approaches to enhance detection sensitivity and specificity in clinical studies, as well as innovative microfluidic solutions which are capable of fully integrated and label-free blood cell sorting and analysis. Lastly, we will envisage the current challenges and future outlook of label-free microfluidics platforms for high throughput multi-dimensional blood cell analysis to identify non-traditional circulating biomarkers for clinical diagnostics.
Introduction
Blood is the most complex biofluid in our body and consists of a diverse range of circulating cell types (red blood cells (RBCs), white blood cells (WBCs), platelets, etc.) and biomolecules (lipids, proteins and nuclei acids) that reflect our health and disease status.1 Routine blood testing is considered a standard clinical procedure in the diagnosis of many diseases. A typical clinical blood test measures your complete blood count (CBC), metabolic and lipid profiles, and hormone levels to detect pathological conditions including diabetes, cancer and anemia.2,3 While CBC using hematoanalyzers can detect the presence of infections based on cell enumeration or the neutrophil–lymphocyte ratio,4 increasing clinical evidence has shown that probing of cellular functions,5,6 biophysical phenotypes,7,8 and non-traditional cellular markers (e.g. monocyte–platelet aggregates9) is more comprehensive and invaluable in disease detection (e.g. sepsis and COVID-19 (ref. 10–12)). Platelet activation is also observed in viral infections such as human immunodeficiency virus (HIV),13,14 hepatitis C virus (HCV)15 and dengue,16,17 with thrombocytopenia as a clinical hallmark in dengue patients. Besides immune cells, diseased cells such as circulating tumor cells (CTCs) are present in blood during cancer metastasis which are crucial for cancer prognosis, diagnosis and treatment monitoring.18,19Apart from cellular components, there is increasing interest in studying cell-derived extracellular vesicles (EVs) and their roles in cell–cell communication and disease pathophysiology.20 Circulating EVs (exosomes and microvesicles) in blood often contain disease-specific information from their host cells, making them ideal biomarkers for non-invasive liquid biopsy21 in inflammation and injury,22–24 diabetes,21 CVD,25 cancer26 and COVID-19.27 Recent technological advances have also demonstrated EV-based diagnosis for cancer,28–30 Alzheimer's disease31 and Parkinson's disease32 with superior detection sensitivity to conventional testing methods. Due to their nanoscale sizes (∼50 nm–1 μm),33 EV isolation remains a major bottleneck and researchers are actively developing novel tools and assays to study EV biology and improve their clinical adoption.
While blood possesses numerous cell types and components which makes it a rich source of biomarkers, these cells and components can in turn increase the complexity of analysis as the high content of cellular components (∼50% v/v) and RBC abundance (>99% of all cells) pose significant technical challenges for many blood tests and diagnostics. For example, the scarcity of circulating tumor cells (CTCs) in blood (1–100 CTCs among billions of RBCs)34,35 is a major bottleneck for CTC isolation and detection. This is further aggravated by pre-treatment steps such as centrifugation and RBC lysis which could cause cell loss.36 Conventional sample preparation steps (e.g. centrifugation, density gradient centrifugation) are time consuming, laborious and could also cause unnecessary activation of sensitive immune cells,6 platelet activation37 and shear-induced microparticle shedding.38 Other approaches such as immunocapture using antibodies could alter cell function or phenotype which complicates downstream analysis.34 Therefore, fast and efficient blood separation methods are highly sought after to gain deeper insights into their biophysiological functions and empower clinicians with more comprehensive blood analysis. Microfluidics has revolutionized biomedical research with unprecedented sensitivity and efficiency by enabling small blood volume handling and precise control of the cell microenvironment. In particular, label-free microfluidic approaches which utilize intrinsic cell properties for cell separation and detection (without using antibodies) are extremely attractive for low-cost and rapid diagnostics. Recently, a plethora of technologies have been reported for label-free blood cell sorting based on cell size, shape, density and deformability. As these cellular biophysical properties are commonly linked to cell biology and pathology,39 they can be further exploited for label-free cell phenotyping to identify cell diseases and cellular states40 including lymphocyte activation,41 sepsis,42 diabetes43 and renal diseases.44
While several reviews have been published on microfluidic cell-sorting techniques,45,46 and single cell analysis,47–49 in this review, we aim to provide a comprehensive overview on recent microfluidics technologies (selecting publications after 2015) for label-free blood cell separation and detection. We will first focus on various blood fractionation techniques for microscale (WBCs, platelets) and nanoscale (EVs) blood components (Fig. 1). We will next summarize the applications of single cell analysis for label-free detection (optical, mechanical and electrical) of cellular properties as novel blood-based biomarkers in diseases (Fig. 8). Thirdly, we will discuss machine learning approaches used for multi-parametric cell profiling, and showcase integrated microfluidic platforms that achieve complete label-free blood cell sorting and analysis. Lastly, we will offer our perspectives on the challenges and future directions of microfluidic-based label-free blood diagnostics.
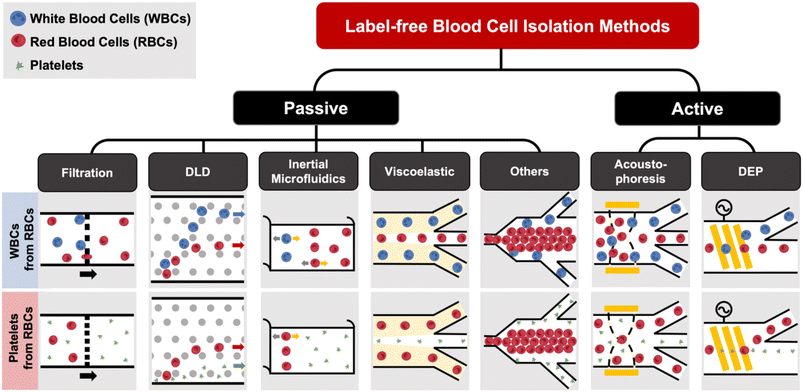
Label-free blood fractionation
Conventional cell sorting methods include physical filtration,50,51 density gradient centrifugation (e.g. Ficoll or Percoll),52 and addition of labels to identify target cells prior to isolation.53 The current gold standard is fluorescence activated cell sorting (FACS)51,54 which uses flow cytometry to detect fluorescently-labelled cells and sort them into different populations based on surface marker expression. Another popular approach is magnetic activated cell sorting (MACS)53–55 that uses antibodies conjugated with magnetic labels for positive or negative cell selection. Despite the high specificity and separation efficiency of these techniques, they are mostly limited for research use due to laborious labeling,51,56 high cost (e.g. antibodies),1 and sorting performance being dependent on the operators.53 The current gold standard for EV isolation is differential ultracentrifugation (UC),57 which is not suitable for clinical testing as it is time-consuming (∼4–5 h) and prone to EV losses.58 Other commercial approaches such as immunoaffinity might affect EV functionality59 or result in high protein contamination.60 Size-exclusion chromatography (SEC), which achieves size-based EV separation through a resin column, has been gaining traction in recent years.61,62 A common issue for the aforementioned EV isolation methods is that they all require pre-processed blood (plasma) as the starting sample, which itself involves several manual centrifugation steps. In this section, we will discuss recent progress in microfluidic label-free sorting of different blood cellular constituents using active (acoustophoresis, dielectrophoresis) and passive (viscoelastic, inertial focusing, deterministic lateral displacement (DLD), filtration, biomimetic cell margination) methods (Fig. 1). The representative microfluidic techniques based on different cell types for blood fractionation and their performance metrics are summarized in Tables 1–3.| Principles | Sample type | Throughput | Efficiency/recovery | Purity/sensitivity/enrichment | Ref. |
|---|---|---|---|---|---|
| Filtration | Spiked blood | 10 mL h−1 | >90% recovery | N.A. | 63 |
| Diluted blood | 10 mL h−1 | 77.7% efficiency | 17.44% purity | 64 | |
| Whole blood | 2.5 mL h−1 | 99% efficiency | N.A. | 65 | |
| Spiked blood | 1 mL h−1 | 95% efficiency | 99% purity | 69 | |
| Whole blood | 37.5 μL min−1 | 72.1% recovery | 15.1% purity | 236 | |
| 232-Fold enrichment | |||||
| Diluted blood | 40 μL min−1 | N.A. | 78 ± 14% purity | 237 | |
| Diluted blood | 1 mL min−1 | ∼94% efficiency | ∼4000-Fold enrichment | 70 | |
| DLD | Whole blood | 0.5 mL h−1 | ∼91.8% recovery for large CTC clusters | N.A. | 79 |
| ∼48% recovery for small CTC clusters | |||||
| Whole blood | 10 μL min−1 | ∼80% recovery | N.A. | 81 | |
| Whole blood | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per s 000 cells per s |
N.A. | 0.91 sensitivity | 83 | |
| Spiked RBC | 110 s 100 mL−1 | 95% recovery | N.A. | 86 | |
| Inertial microfluidics | Whole blood | 720 μL min−1 | 88.1% efficiency | N.A. | 95 |
| Whole blood | 0.4 mL min−1 | >80% efficiency | N.A. | 93 | |
| Whole blood | 80 mL h−1 | 90% recovery | N.A. | 238 | |
| Viscoelastic focusing | Whole blood | 200 μL min−1 | 67% recovery | N.A. | 100 |
| Whole blood | 12 μL min−1 | 98% efficiency | N.A. | 101 | |
| Whole blood | 30 μL h−1 | 76% efficiency | N.A. | 102 | |
| DEP | Lysed blood | 0.4 μL min−1 | N.A. | N.A. | 124 |
| PBMCs | 35 μL min−1 | 53–70% efficiency | N.A. | 121 | |
| Spiked blood | 6 μL h−1 | 100% recovery | 81% purity | 122 | |
| PBMC | 4–7 μL h−1 | N.A. | N.A. | 123 | |
| Acoustophoresis | Diluted blood | 1.0 μL min−1 | N.A. | 2500-Fold enrichment | 108 |
| Spiked phosphate buffer solution | 0.3 μL min−1 | 90% ± 2.4% efficiency | N.A. | 109 | |
| Lysed blood | 6 mL h−1 | 91.8 ± 1.0% efficiency for breast CTCs | 20-Fold enrichment for breast CTCs | 110 | |
| 84.1 ± 2.1% efficiency for prostate CTCs | 20-Fold enrichment for prostate CTCs | ||||
| Lysed blood | 2.5 mL h−1 | 87% recovery | 0.11% purity | 111 | |
| 162-Fold enrichment | |||||
| Lysed blood | 100 μL min−1 | 85.0% efficiency for prostate CTCs | 53 ± 27-fold enrichment for prostate CTCs | 112 | |
| 89.8% efficiency for breast CTCs | 84 ± 30-fold enrichment for prostate CTCs | ||||
| Lysed blood | 7.5 mL h−1 | 86% efficiency | N.A. | 239 | |
| Others | Diluted blood | 3 mL min−1 | 96.6% efficiency | N.A. | 133 |
| Whole blood | 0.5–5 μL min−1 | >80% efficiency | 12-Fold enrichment | 5 | |
| Whole blood | 3 mL min−1 | 95.9 ± 3.1% recovery | N.A. | 152 | |
| Whole blood | 6.75 mL h−1 | 89.8% efficiency | N.A. | 134 | |
| Diluted blood | 800 μL min−1 | 83% efficiency | N.A. | 174 | |
| WBCs | 1.2 mL h−1 | 82.2% efficiency | N.A. | 145 |
| Principles | Sample type | Throughput | Efficiency/recovery | Purity/sensitivity/enrichment | Ref. |
|---|---|---|---|---|---|
| Filtration | Whole blood | 1 mL min−1 | >80% recovery | N.A. | 71 |
| Whole blood | 600 nL min−1 | 97% recovery | ∼70% purity | 66 | |
| 60-Fold enrichment | |||||
| Whole blood | 100 ± 30 mL min−1 | 30% efficiency | N.A. | 72 | |
| DLD | Spiked blood | DLD1: 100–600 μL min−1 | DLD1: 50% recovery | N.A. | 84 |
| DLD2: 7–50 μL min−1 | DLD2: 25% recovery | ||||
| DEP | 1000× diluted plasma | 2 μL, 2 min | N.A. | N.A. | 127 |
| Whole blood | 10 μL min−1 | ∼78% efficiency | N.A. | 125 | |
| 20× diluted blood | 18 μL h−1 | 97% efficiency | N.A. | 126 | |
| Viscoelastic focusing | Whole blood | 10–32 μL min−1 | N.A. | N.A. | 103 |
| Acoustophoresis | Undiluted whole blood | 20 mL min−1 | 87.3% efficiency | 82.9% purity | 240 |
| Whole blood | 1.5 μL min−1 | 86% efficiency | 7.7-Fold enrichment | 114 | |
| Undiluted whole blood | 10 mL min−1 | >85% efficiency | N.A. | 241 | |
| Diluted whole blood | 80 μL min−1 | 90% efficiency | 103-Fold enrichment | 242 | |
| Culture media | 5 mL min−1 | N.A. | N.A. | 116 | |
| Others | Whole blood | 0.66 μL min−1 | N.A. | 99.9% purity | 142 |
| Whole blood | 1.5 min for plasma, 15 min for buffy coat | N.A. | 100% purity | 141 |
| Principles | Sample type | Throughput | Efficiency/recovery | Purity/sensitivity/enrichment | Ref. |
|---|---|---|---|---|---|
| Filtration | Diluted plasma | 50 μL min−1 | >97% efficiency | N.A. | 75 |
| >80% recovery | |||||
| Undiluted serum | 90 μL min−1 | N.A. | N.A. | 73 | |
| Plasma | 5–10 μL min−1 | N.A. | N.A. | 74 | |
| DLD | Serum | 900 μL h−1 | ∼70% recovery | 2.6–3.0-Fold enrichment | 77 |
| Inertial microfluidics | Whole blood | 80 μL min−1 | ∼15% efficiency | N.A. | 97 |
| Viscoelastic focusing | Adenocarcinoma human alveolar | 200 μL h−1 | >80% recovery | >90% purity | 99 |
| Basal epithelial cells | |||||
| DEP | Culture media | 1.5 μL min−1 | 81% recovery | 95% purity | 130 |
| Plasma | 3–5 μL min−1 | N.A. | N.A. | 128 | |
| Acoustophoresis | Plasma | 0.5 μL min−1 | N.A. | N.A. | 243 |
| Undiluted whole blood | 4 μL min−1 | 82.4% efficiency | 98.4% purity | 244 |
Filtration
Microfluidic filtration separate cells based on size and can be broadly divided into three categories: membrane filtration,63,64 pillar filtration,65 and tangential flow filtration.66 While readers are highly encouraged to learn more about the various filtration mechanisms from other excellent review articles,45,67,68 this section focuses on their applications to isolate CTCs,63–65,69 leukocytes,70 platelets,66,71 bacteria,72 and EVs73–75 from whole blood. In general, key advantages of filtration methods include scalable throughput, low-cost and simple operation. However, most blood filtration devices are affected by clogging and bubble formation issues that may affect the sorting efficacy. Selection of membrane size cut-off and flow parameters is also an important consideration as larger cells may be trapped or deform through pores under high pressure which results in lower cell recovery.Membrane filtration generally uses commercial membranes with through holes to trap or isolate larger cells. Kang et al.64 improved the membrane with unique tapered-slits to increase the sample throughput for CTC isolation (Fig. 2A). This tapered-slit filter fabricated with a photosensitive polymer was connected to a syringe to achieve a high CTC capture rate of 77.7% and viability of 80.6%. Besides single CTC isolation, Sarioglu et al.65 developed a cluster-chip based on pillar filtration to isolate CTC clusters from whole blood directly (Fig. 2B). A set of triangular pillars were designed to capture CTC clusters which successfully identified CTC clusters in 30–40% of patients with metastatic breast or prostate cancer or with melanoma. Another pillar-based device was used for continuous separation of platelets from whole blood in a radial sample flow direction (Fig. 2C).66 The multiple radial parallel paths and self-generating cross flow greatly minimize clogging to achieve a ∼60× platelet enrichment.
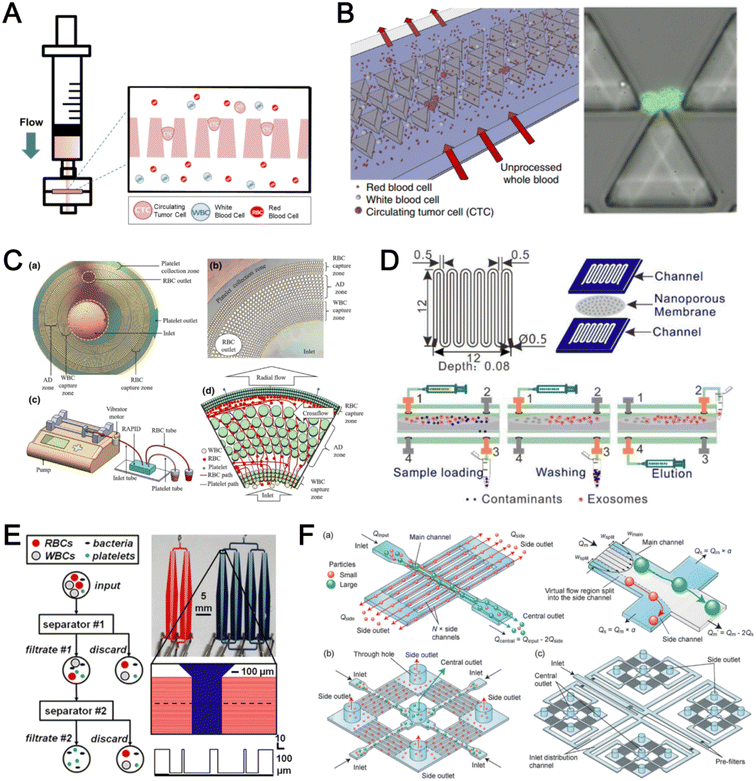 | ||
| Fig. 2 Microfluidic filtration cell sorting. (A) CTC isolation using a tapered-slit filter fabricated with a photosensitive polymer device. Reproduced from ref. 64 with permission from Ivyspring International Publisher. (B) Schematic of the cluster-chip with triangular pillars for CTC cluster isolation. Reproduced from ref. 65 with permission from Springer Nature. (C) High throughput radial pillar device for platelet enrichment. (a) Microscope image of the device; (b) magnified image of a sector of the device shows the varying sizes of the pillars and the pillar gaps in each of the three zones; (c) schematic of the experimental set-up for platelet enrichment from whole blood; (d) schematic of the radial, and the cross flows. Reproduced from ref. 66 with permission from Springer Science + Business Media. (D) Microfluidic tangential flow filtration, or cross-flow filtration device for exosome isolation and purification. Reproduced from ref. 75 with permission from Elsevier. (E) Microfluidic cross-flow device for bacteria separation from diluted whole blood. Reproduced from ref. 72 with permission from Elsevier. (F) Schematic of the numbering-up chip for direct leukocyte and erythrocyte separation. (a) A separation unit for particle sorting and particle behaviors at a branch point; (b) a square, integrated structure composed of 4 separation units; (c) four arranged structures shown in image (b) connected to a symmetrically branched inlet distribution channel. Reproduced from ref. 70 with permission from the Royal Society of Chemistry. | ||
Another form of filtration is tangential flow filtration, also commonly known as cross-flow filtration. In this approach, the sample will flow close and parallel (“pinched”) to the filtration structure using an additional sheath flow, while a pressure gradient is established to promote cells passing through small gaps perpendicular to the flow direction. Han et al.75 fabricated a microfluidic chip by bonding two polymethyl methacrylate (PMMA) layers with a nanoporous membrane to isolate and purify exosomes from human blood with a higher recovery rate (>80%) (Fig. 2D). Raub et al.72 designed a microfluidic device using cross-flow filtration for separation of bacteria from blood cells (Fig. 2E). This chip could remove 97–98% of RBCs and retain 30% of bacteria from undiluted whole blood with a higher relative bacterial abundance of 8200%. A new hydrodynamic filtration multiplexed method was reported to separate leukocytes from diluted blood samples at an ultra-high throughput of 15 mL min−1 and a leukocyte recovery ratio of ∼94% (Fig. 2F).70
Deterministic lateral displacement (DLD)
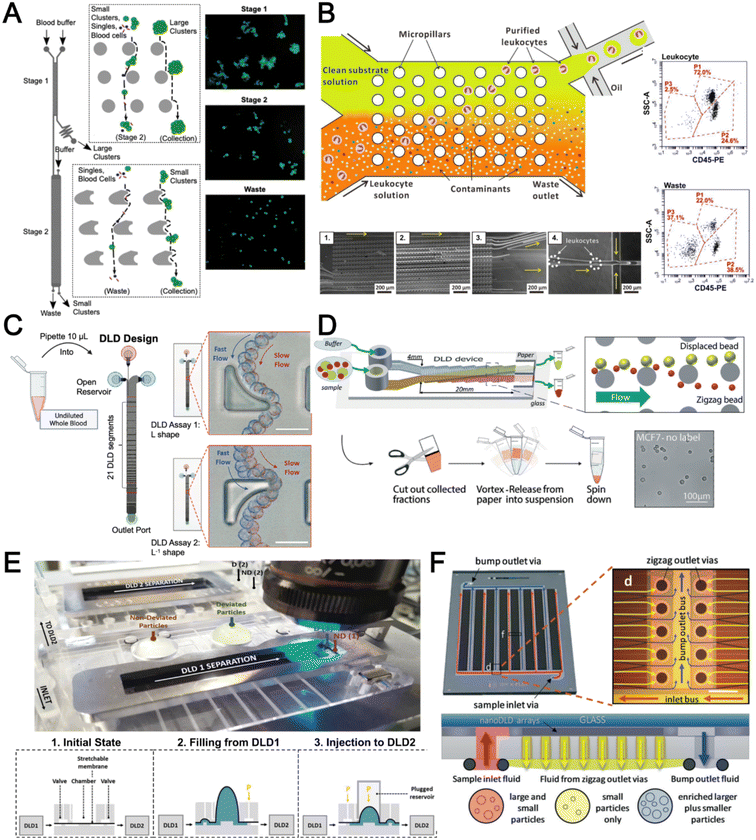
Deterministic lateral displacement (DLD) is an established microscale separation technique that uses an array of micropillars to displace larger particles laterally76 in a bumping migratory flow trajectory.76–78 By carefully designing the critical sorting diameter (Dc) based on the formula: Dc = 1.4Ge0.48, where G is the spacing between pillars and e is the row shift fraction (e = tan![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) q) when q is the tilt angle,79,80 DLD can be exploited for blood processing to separate CTCs,81–83 leukocytes,84 bacteria77 and EVs (Fig. 3).77 Since the first DLD work reported in 2004,85 many research groups have explored different sorting applications and improved the technique in terms of scalability and separation resolution with different pillar designs. Recent advances in blood fractionation include the use of a novel cascaded DLD approach to isolate CTC clusters from whole blood with a recovery yield of 91.8% and 48% for large and small CTC clusters, respectively.79 In contrast to conventional single-stage DLD devices, Au et al. integrated asymmetrical pillars and channel height restriction in a dual-stage DLD chip to achieve high CTC yield and integrity with a lower shear stress rate (Fig. 3A).79 Besides particle separation, efforts have also been made to incorporate different functionalities in DLD systems. An example is the integration of a droplet generator with DLD to encapsulate single DLD-purified leukocytes in droplets for protease analysis (Fig. 3B).81 Another work demonstrated the application of a label-free and rapid (15 min) combinational DLD assay to measure the biophysical properties (cell deformability) of immune cell population from 20 mL of whole blood for point-of-care testing (Fig. 3C).83 These measurements were correlated to patient clusters with different levels of immune severity, thus facilitating early detection of asymptomatic-infected individuals before the condition exacerbates into sepsis.
q) when q is the tilt angle,79,80 DLD can be exploited for blood processing to separate CTCs,81–83 leukocytes,84 bacteria77 and EVs (Fig. 3).77 Since the first DLD work reported in 2004,85 many research groups have explored different sorting applications and improved the technique in terms of scalability and separation resolution with different pillar designs. Recent advances in blood fractionation include the use of a novel cascaded DLD approach to isolate CTC clusters from whole blood with a recovery yield of 91.8% and 48% for large and small CTC clusters, respectively.79 In contrast to conventional single-stage DLD devices, Au et al. integrated asymmetrical pillars and channel height restriction in a dual-stage DLD chip to achieve high CTC yield and integrity with a lower shear stress rate (Fig. 3A).79 Besides particle separation, efforts have also been made to incorporate different functionalities in DLD systems. An example is the integration of a droplet generator with DLD to encapsulate single DLD-purified leukocytes in droplets for protease analysis (Fig. 3B).81 Another work demonstrated the application of a label-free and rapid (15 min) combinational DLD assay to measure the biophysical properties (cell deformability) of immune cell population from 20 mL of whole blood for point-of-care testing (Fig. 3C).83 These measurements were correlated to patient clusters with different levels of immune severity, thus facilitating early detection of asymptomatic-infected individuals before the condition exacerbates into sepsis.
 | ||
| Fig. 3 Deterministic lateral displacement (DLD). (A) Schematic representation of a cascaded DLD array device for isolation of large and small CTCs clusters. Reproduced from ref. 79 with permission from Nature Research. (B) An integrated continuous-flow microfluidic assay to separate leukocytes from lysed blood solution and resuspension in clean substrate solution for subsequent droplet encapsulation. Reproduced from ref. 81 with permission from the American Chemical Society. (C) Unconventional L- and inverse-L shaped pillar arrays for sorting and biophysical measurement of immune cell population from whole blood directly. Reproduced from ref. 83 with permission from Wiley. (D) Capillary-driven based DLD platform to isolate MCF-7 from RBCs. Reproduced from ref. 86 with permission from Royal Society of Chemistry. (E) A two-stage cascaded DLD platform for isolating E. coli from whole blood. Reproduced from ref. 84 with permission from Nature Research. (F) NanoDLD arrays on a silicon–glass microfluidic chip to separate EVs from serum samples. Reproduced from ref. 77 with permission from the Royal Society of Chemistry. | ||
To improve portability, a capillary paper pump-based DLD method was recently reported to isolate MCF-7 breast cancer cells (>90% viability) from RBCs (Fig. 3D).86 The capillary pumps rely on a negative pressure imposed by the downstream capillary action to initiate fluid flow over a prewetted or hydrophilic DLD array surface. Finally, researchers have also successfully scaled down the DLD cell sorting technique for smaller (sub-micron) bacteria and EV isolation.77,84 One study highlighted E. coli isolation from whole blood using a two-stage cascaded DLD approach to achieve a recovery yield of 25–50% (Fig. 3E).84 Smith et al. also reported a microfluidic silicon–glass nanoDLD array to selectively enrich large EVs from serum using a pillar gap distance of 225 nm. This resulted in the highest yield of ∼50% with a significantly shorter processing time (60 min) as compared to ultracentrifugation (Fig. 3F).77 While DLD is a highly versatile size-based blood separation technique, it should be noted that the sorting performance can be influenced by other factors including particle shape and deformability, and the presence of cell aggregates.76 Clogging issues and non-specific cell binding must be minimized during blood processing which can otherwise directly impact cell movements within the pillar array.
Inertial microfluidics
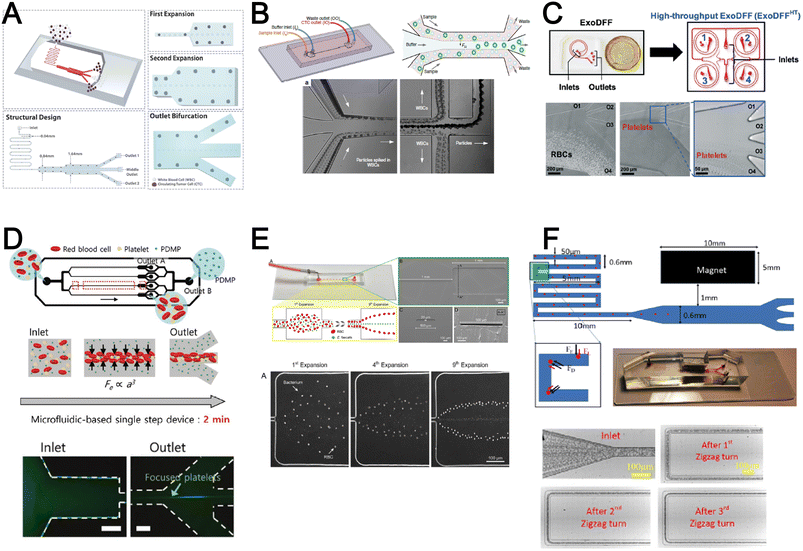
Inertial microfluidics is a hydrodynamic cell focusing method based on the interplay between inertial lift forces and Dean drag forces acting on particles under Newtonian flow. Several review articles have recently discussed the fundamentals and applications of inertial microfluidics.87–89 As these forces are particle size dependent, different cells are focused on distinct equilibrium positions to achieve continuous and high throughput (∼mL min−1) cell separation for different bio-fluids including blood, urine,90 and semen samples.91 When designing inertial cell sorters, it is important to consider both target and non-target cell sizes, starting sample concentration and volume, fluid properties, and channel dimensions for optimum focusing.92Abdulla et al. reported a zigzag microchannel for label-free separation of CTCs from whole blood.93 The zigzag structure was used to promote size-based particle migration and help stabilize the focused particles and larger CTCs (>10 μm) to the channel center (Fig. 4A). Zhou et al. used a multi-flow straight microfluidic device to demonstrate the separation of CTCs directly from whole blood (Fig. 4B). Using a 2-inlet system, the larger CTCs migrated to the channel center into a clean buffer flow with a high purity (>87%) and recovery (>93%) rate.94 Spiral inertial microfluidics is another popular cell sorting design due to its larger channel dimensions which minimizes clogging issues during blood processing. Zhu et al. reported a novel inertial microfluidic (IM) cube which was integrated with four subunits of spiral channels and stacked in multiple layers to extract WBCs from 1.3 mL of whole blood within 2 min.95 The design is based on passive secondary flow mixing and inertial sorting to achieve complete RBC lysis and a white blood cell extraction efficiency of 88.1%.
 | ||
| Fig. 4 Inertial microfluidics and viscoelastic focusing for blood separation. (A) Illustration of cell distribution positions and force analysis in a zigzag structured with a straight expansion channel based inertial microfluidic chip. Reproduced from ref. 93 with permission from the American Chemical Society. (B) Size-based differential migration of cells towards buffer flow in the middle of the main channel. Reproduced from ref. 94 with permission from Springer Nature. (C) Images of a spiral shaped single and multiplexed ExoDFF device for isolation of circulating EVs. Reproduced from ref. 97 with permission from the Royal Society of Chemistry. (D) Schematic illustration of platelet-derived micro-particle (PDMP) separation in a straight microfluidic device using viscoelastic non-Newtonian fluid. Reproduced from ref. 100 with permission from Wiley. (E) Schematic representation of red blood cell and bacteria sorting in a cascaded contraction–expansion microchannels. Reproduced from ref. 101 with permission from Elsevier. (F) Illustration of particle migration at the U-turn zigzag channel. Reproduced from ref. 103 with permission from Wiley. | ||
A major bottleneck in inertial microfluidics is the inability to separate small particles as they experience insufficient inertial forces, or require excessively long channel lengths to migrate to their equilibrium positions (large pressure drop). To address this issue, Tay et al. reported a novel transient (non-equilibrium) cell focusing method in spiral channels based on subtle differences in the particle innermost distance (Dinner) from the channel inner wall to separate sub-micron bacteria and nanoparticles.96 This was subsequently adapted to isolate EVs from whole blood directly (ExoDFF) and multiplexed to process 5 mL of whole blood within an hour with the same EV separation performance (Fig. 4C).97
Viscoelastic focusing
Another passive microfluidic cell sorting method gaining in popularity is using viscoelastic focusing effects. Briefly, the elasticity of dilute polymer solutions causes the lateral migration of single particles to focus on the channel center due to a non-uniform distribution of the normal stress between the centerline and the walls of the microchannel.98 Besides optimizing flow rates or channel dimensions, one can also tune the concentration of the polymer (poly(ethylene oxide), PEO) to control viscoelastic forces and particle separation resolution at lower flow rates. Liu et al. reported a straight microchannel design to separate exosomes from other larger EVs with high separation purity (>90%) and recovery (>80%) by adding a small amount of PEO (0.1 wt%) into the cell culture medium.99Recently, a sheathless focusing device using viscoelastic fluid was reported to isolate platelet-derived micro-particles (PDMPs ∼ 0.3–2 μm) from RBCs with a 4.8-fold increase in yield as compared to the centrifugation within 2 min (Fig. 4D).100 Similarly, Bilican et al. also developed a cascaded expanding-contracting straight microchannel for sheathless separation of Enterococcus faecalis (∼0.5–2 μm) in viscoelastic fluid with 98% separation efficiency (Fig. 4E).101 As single inlet devices are simple to operate, there are several other interesting sheathless cell sorting devices using viscoelastic focusing to achieve undiluted whole blood separation,102 integration with positive magnetophoresis (Fig. 4F),103 and a scaled-up double spiral design.104
Although both inertial and viscoelastic focusing microfluidic technologies are useful for separating nano to microscale particles from blood, there are still challenges to improve the separation dynamic range in a single chip for complete blood fractionation. The use of viscoelastic or polymer solution can enhance cell focusing but may affect downstream analysis (e.g. mass spectrometry) with additional chemical components or rare cell isolation. Future studies are warranted to investigate cell biocompatibility and functionalities after prolonged exposures (∼days). More details on viscoelastic microfluidic mechanisms and applications can be found in a review by Papautsky et al.105
Acoustophoresis
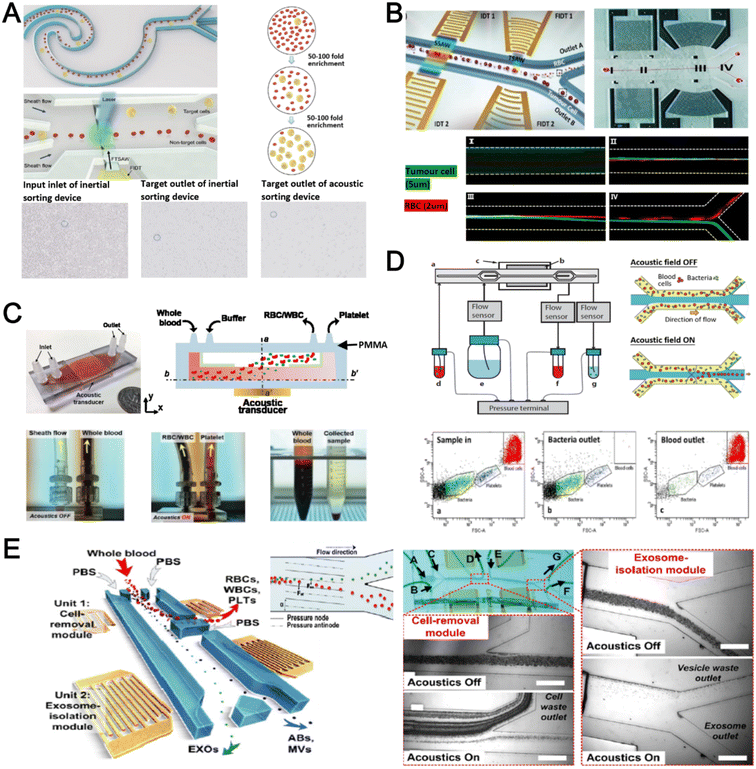
Acoustophoresis (or acoustofluidics) is an active method which uses an external acoustics field to manipulate particles. In general, microfluidic devices consist of interdigitated transducers (IDTs) using a piezoelectric material to generate bulk acoustic waves (BAWs) and surface acoustic waves (SAWs). This leads to the formation of pressure and anti-pressure nodes for size or density-based particle separation in a contactless and label-free manner.106,107 With recent advances in improving biocompatibility, cascaded designs, multiplexed cell sorting and scalable manufacturing, acoustofluidic technologies have demonstrated their potential for point-of-care blood-based clinical applications.Using a hybrid platform, Zhou et al. combined passive (inertia sorting) and active (acoustic sorting) methods to isolate CTCs from 10× diluted whole blood (Fig. 5A).108 The first section consists of a reverse ‘S’-curved microchannel design to deplete RBCs while focusing the larger CTCs to the acoustic sorter to sort fluorescence-labelled MCF-7 cells. In another work, Wang et al. utilized a pair of IDTs to generate standing SAWs (SSAW) to focus CTCs and RBCs on a single pressure node at the channel centre. Next, larger particles (>5 μm) would be deflected laterally due to the larger acoustic radiation induced by travelling pulsed SAWs (TSAW).109 The authors applied this to sort U87 glioma cells from the RBCs without the need for sample dilution (Fig. 5B). Similar CTC acoustic sorting devices were also reported by other groups with improved throughput110,111 or combined with negative selection of WBCs.112
 | ||
| Fig. 5 Acoustofluidics based blood fractionation. (A) CTC isolation and enrichment using a hybrid (inertia and acoustic) microfluidic sorting device. Reproduced from ref. 108 with permission from the Royal Society of Chemistry. (B) Tumour cells isolated by a multi-stage surface acoustic wave (SAW) device. Reproduced from ref. 109 with permission from Elsevier. (C) Platelet isolation using a PMMA plastic-based device. Reproduced from ref. 113 with permission from the Royal Society of Chemistry. (D) Clinical-scale automated isolation of bacteria. Reproduced from ref. 115 with permission from Nature Research. (E) Exosome isolation by taSSAW (tilted-angle standing SAW) using a dual-stage acoustofluidic device. Reproduced from ref. 117 with permission from United States National Academy of Sciences. | ||
Key advantages of acoustofluidic platforms include the tunability of the acoustics field strength for smaller particles and scalability for large sample volume processing. For example, Gu et al. isolated platelets (2–3 μm) using a PMMA disposable device which is suitable for rapid fabrication and clinical use (Fig. 5C).113 By producing a pressure node near the top layer with a quarter-wavelength resonator, blood cells were deflected vertically to the top layer to achieve a platelet recovery rate of 87.3% at an ultra-high throughput of 20 mL min−1. Another scaled up platelet separation device fabricated by the lithography method was reported using SSAW.114 Besides platelets, bacterial separation (>90% recovery) from whole blood is also reported based on bulk acoustophoresis (Fig. 5D).115 Another work was reported by Devendran et al. to separate bacteria at 5 mL min−1 by utilizing diffractive-acoustic SAW (DASAW) to focus particles along a 90-degree angled serpentine channel with only a single travelling wave.116
For sub-micron particles, Wu et al. developed a dual-modular acoustofluidic platform with a microscale cell-removal module followed by an nanoscale exosome isolation module.117 Both sections use tilted-angle standing SAW (taSSAW) fields to filter out blood components larger than 1 μm (>99.9% blood cell removal) in the first stage, and a higher frequency of 40 MHz to separate EVs and apoptotic bodies with a purity rate of 98.4% at the 2nd stage (Fig. 5E). Taken together, acoustics-based sorters are scalable, versatile and have excellent sorting performances. Due to frequent bubble formation and high setup costs, future work will focus on improving the device robustness and their translation for clinical use.
Dielectrophoresis
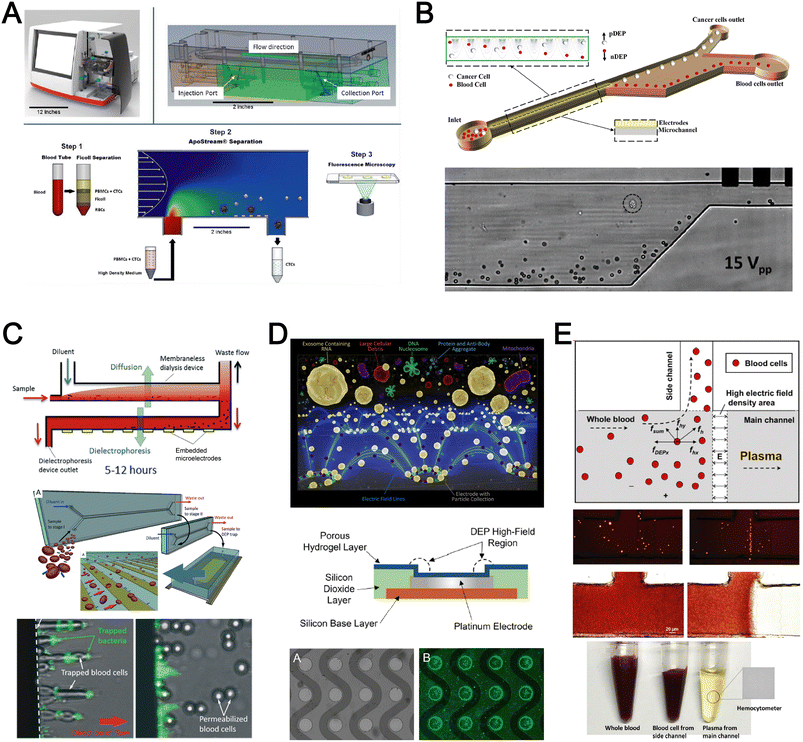
Dielectrophoresis (DEP) is another established active label-free cell sorting method based on electrical properties of particles and medium. Under a non-uniform electric field, particles or cells are polarised and a net dipole moment is induced on particles' surfaces, thereby allowing them to migrate along or against the electric field depending on the excitation frequency, particle size, and electrode configuration.118 In most devices, the electrodes are orientated to exert DEP forces perpendicular to the flow direction for lateral particle sorting into different channel outlets.119,120For DEP-based cell sorting, it is often combined with flow fractionation to take advantage of the parabolic velocity profile and laminar flow in microchannels. The ApoStream® system is a commercial product that separates CTCs from peripheral blood mononuclear cells (PBMCs) using DEP-based flow fractionation (Fig. 6A).121 Briefly, CTCs introduced at the channel bottom experienced stronger DEP forces towards the bottom outlet while smaller PBMCs are repelled vertically to the channel center (faster flow) and eluted as waste. Similarly, Alazzam et al. applied lateral DEP to separate MDA-MB-231 cancer cells from blood cells with 100% recovery and 81% purity (Fig. 6B) using a low conductivity sucrose–dextrose medium.122 To enhance CTC sorting, Dudaie et al. integrated DEP with image-based CTC discrimination using interferometric phase microscopy to achieve 98% classification success and 69% sorting accuracy.123 Besides sorting, DEP can also be applied for leukocyte activation profiling in a sepsis murine model since activated leukocytes are larger in size and thus experience stronger DEP forces to deflect to different channel positions.124
 | ||
| Fig. 6 Dielectrophoresis (DEP) based blood fractionation. (A) ApoStream® system for separating CTCs from peripheral blood mononuclear cells (PBMCs) under continuous flow. Reproduced from ref. 121 with permission from Public Library of Science. (B) Continuous sorting of human breast cancer cells from blood cells in low conductivity sucrose–dextrose medium. Reproduced from ref. 122 with permission from Wiley. (C) Combined microdialysis and DEP for bacterial isolation from blood. Reproduced from ref. 125 with permission from the Royal Society of Chemistry. (D) Rapid exosome isolation from whole blood using an alternating current electrokinetic (ACE) microarray. Reproduced from ref. 128 with permission from the American Chemical Society. (E) Continuous cell-free plasma extraction from undiluted blood. Reproduced from ref. 131 with permission from Wiley. | ||
Instead of DEP-induced particle lateral deflection under continuous flow, smaller bacteria and exosomes can be attracted and trapped at the electrodes by DEP forces to facilitate detection. D'Amico et al. combined microdialysis and dielectrophoresis to deplete permeabilized red blood cells before trapping bacteria on electrodes to achieve ∼79% separation efficiency using spiked blood samples (103–105 CFU mL−1) (Fig. 6C).125 Numerous DEP studies also reported higher bacteria separation efficiency126 or detection limit127 using diluted blood or plasma. For exosome isolation, Ibsen et al. reported a rapid EV isolation using an alternating current electrokinetic (ACE) microarray from 50 μL of undiluted blood, followed by on-chip immunofluorescence detection of EV proteins within 30 min (Fig. 6D).128 It was further used to distinguish pancreatic ductal adenocarcinoma patients from healthy subjects with 99% sensitivity and 82% specificity.129 Submicron particle separation was also demonstrated using combined DEP with acoustophoresis to separate exosomes (<200 nm) from microvesicles (>300 nm) in culture media with 95% purity and 81% recovery.130 Yang et al. reported a microfluidic DEP device for direct plasma extraction from undiluted blood which can be automated to replace conventional centrifugation (Fig. 6E).131
Overall, DEP is a tunable blood fractionation method and can complement passive size-based sorting methods to fractionate cells of similar sizes but different electrical properties. However, issues including Joule heating, limited throughput, and the need for a low conductivity buffer need to be addressed to facilitate the translation of DEP technologies for clinical testing.132
Other label-free blood fractionation methods
Aside from the aforementioned techniques, several non-conventional methods have also been reported for blood sorting applications. Tay et al. reported a microfluidic device to separate WBCs from undiluted whole blood based on the effect of biomimetic cell margination (Fig. 7A), a microcirculatory phenomenon whereby deformable RBCs migrate laterally toward the axial centre (Fahraeus effect) and result in an RBC-free layer (containing WBCs) at the vessel periphery.5 In another work, Mutlu et al. devised a non-equilibrium inertial separation array (NISA) that utilizes inertial lift forces to separate WBCs from blood. Unlike DLD, NISA adopts longer rectangular islands that use size-dependent inertial lift forces to push cells away from channel walls (Fig. 7B).133 The authors reported a sample throughput of ∼3 mL min−1 with 96.6% WBCs yield and 0.0059% RBC contamination, making NISA suitable for large blood volume processing applications such as banked blood cleansing and rare cell enrichment.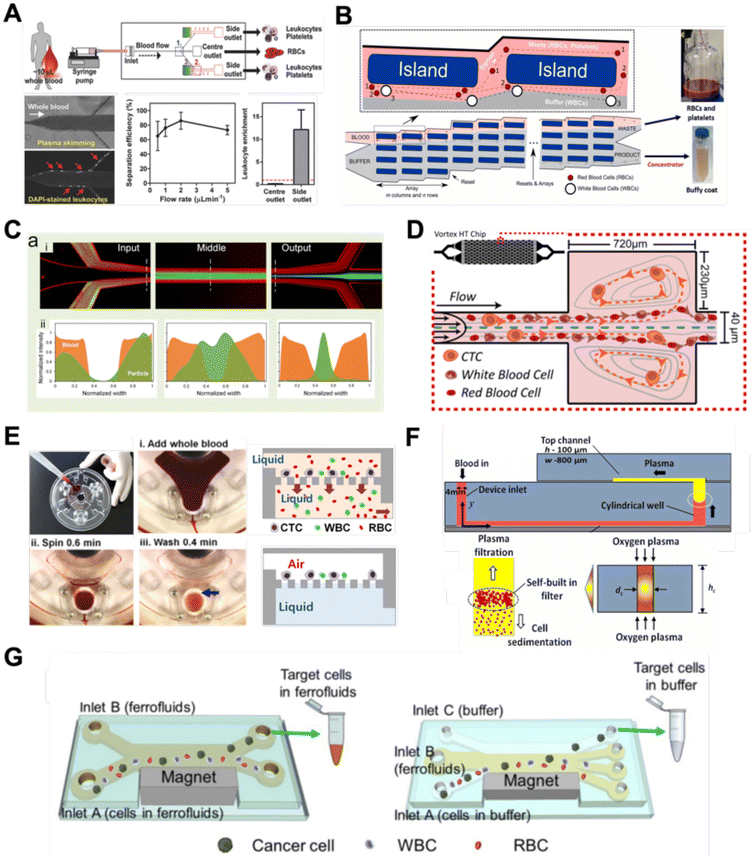 | ||
| Fig. 7 Other label-free fractionation methods. (A) Isolation of WBCs from a drop of blood using the biomimetic cell margination effect. Reproduced from ref. 5 with permission from Wiley. (B) Non-equilibrium inertial separation array (NISA) for isolation of WBCs from whole blood by passive inertial lift forces exerted by an island array. Reproduced from ref. 133 with permission from Nature Research. (C) Shear-induced diffusion (SID) used to separate 18.7 μm fluorescent particles (green) from 2× diluted whole blood (red). (i) Overlapped pseudocolored images indicating a high-efficiency separation; (ii) corresponding intensity profiles across the dashed lines in part (i). Reproduced from ref. 134 with permission from Springer Nature. (D) High Throughput Vortex Chip (Vortex HT) that traps larger cells in microvortices developed in reservoirs, while allowing smaller cells to pass through to achieve separation. Reproduced from ref. 137 with permission from Impact Journals. (E) Lab-on-a-disc system equipped with fluid-assisted separation technology (FAST) for CTC isolation from whole blood. Reproduced from ref. 152 with permission from the American Chemical Society. (F) Microfluidic device using a combination of capillary flow, differential wetting and sedimentation effects for the separation of plasma. Reproduced from ref. 142 with permission from Nature Research. (G) Negative magnetophoresis separation of cancer cells using ferrofluid. Reproduced from ref. 145 with permission from Royal Society of Chemistry. | ||
Another interesting label-free blood separation technique is shear-induced diffusion (SID), used for isolating CTCs94,134 and leukocytes135 from whole blood. The set-up involves a sandwiched buffer solution co-flowed with two sample streams, where target cells migrate from the side into the cell-free central stream under the influence of shear-induced diffusion and inertial forces. While the underlying mechanism of SID has yet to be fully elucidated,135 larger cells (WBCs, CTCs) in the blood samples are observed to migrate into the buffer stream faster than smaller cells (RBCs) due to size-dependency of SID and inertial forces.134 This size-based migration and focusing has also been attributed to the downstream velocity of particles,135 which scales inversely with particle size. Using this strategy, Zhou et al. demonstrated the separation of beads (18.7 μm) and HepG2 cancer cells from whole blood at a high throughput of 6.75 mL h−1 (106–107 cells per s) and an efficiency of 89.8% (Fig. 7C).134 As a proof-of-concept, the authors also applied SID for the isolation of CTCs from hepatocarcinoma subjects. While they were able to detect one CTC from 1 mL of blood via immunostaining, the purity of the sorted samples was affected by large WBCs. A low shear rate at the flow interface must be carefully maintained to minimize RBC diffusion.136 Nevertheless, the ability of SID to process highly concentrated samples and achieve higher throughput than typical inertial microfluidics shows great potential as a label-free cell isolation method. For CTC isolation, several studies have reported the use of microvortices to trap CTCs by designing a series of narrow channels with expanded reservoirs.137–140 Rapid flow in the narrow channel generates inertial forces to trap larger cells into the vortices in the reservoirs while smaller cells are allowed to pass through. After separation, the trapped CTCs can be “released” from the reservoirs using a slower flow rate and eluted into a small volume for off-chip processing. Using this phenomenon, Che et al. developed and clinically validated a high throughput vortex chip for size-based CTC enrichment with a capture efficiency of 83% at a throughput of 800 μL min−1 of whole blood (Fig. 7D).137 In a study by Dhar et al., they integrated vortex trapping of CTCs with single-cell encapsulation in a single platform termed SPEC (size-based purification and encapsulation of cells) by using the reservoirs as mini reaction chambers for the downstream study of rare cell secretion at the single-cell level.138
Using a different strategy, Kim et al. reported a lab-on-a-disc platform with an integrated fluid-assisted separation technology (FAST) to selectively capture CTCs on the polyethylene membrane filter (8 μm pore size) based on centrifugal effects. The novelty of this technique is the addition of a stably-held liquid throughout the size-based CTC isolation process, which reduced clogging and increased the recovery rate significantly from 54.0 ± 21.0% to 95.9 ± 3.1% (Fig. 7E). Another interesting work based on centrifugal force is the development of an ultra low-cost (<20 cents), human-powered paper centrifuge inspired by a whirligig toy, aptly termed ‘paperfuge’.141 A high rotational speed of 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 r.p.m. (∼30
000 r.p.m. (∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g) was reported which can separate plasma from whole blood in <1.5 min when blood-filled capillary tubes are mounted on the discs. By modifying channel surface properties, Maria et al. proposed a capillary flow-driven, double layered PDMS device with a wettability gradient for plasma extraction (Fig. 7F).142 As blood enters the bottom microchannel via capillary action and moves up the vertical well, the hydrophobic region enhances plasma separation by exploiting the velocity differences between plasma and cells. Combined with sedimentation effects, 2 μL of plasma was extracted from <10 μL whole blood in 15 min for blood glucose measurements.
000 g) was reported which can separate plasma from whole blood in <1.5 min when blood-filled capillary tubes are mounted on the discs. By modifying channel surface properties, Maria et al. proposed a capillary flow-driven, double layered PDMS device with a wettability gradient for plasma extraction (Fig. 7F).142 As blood enters the bottom microchannel via capillary action and moves up the vertical well, the hydrophobic region enhances plasma separation by exploiting the velocity differences between plasma and cells. Combined with sedimentation effects, 2 μL of plasma was extracted from <10 μL whole blood in 15 min for blood glucose measurements.
Lastly, while magnetic isolation is typically affinity-based using antibodies, an interesting label-free approach is negative magnetophoresis using ferrofluids.143–146 Ferrofluids are stable colloid suspensions of ferromagnetic nanoparticles such as magnetite or maghemite147 dispersed in a non-magnetic carrier fluid such as water or organic solvent.148 Cells and particles suspended in ferrofluids behave as “magnetic voids”149 which are deflected to distinct size-based flow trajectories when an external magnetic field is applied to attract magnetic nanoparticles.145,150 As most commercial ferrofluids are not biocompatible,143 careful selection of the ferrofluid material, pH and surfactant is crucial for cell manipulation.147 Zhao et al.145 developed a biocompatible approach using a ferrofluid to isolate cancer cells at low concentrations (∼100 cells per mL) from WBCs at a throughput of 1.2 mL h−1 and an average efficiency of 82.2% (Fig. 7G). A custom-made biocompatible ferrofluid flow is flanked by two buffer streams, and the blood sample is introduced through one of the buffer inlets. An external magnetic force pushes the target cancer cells into the opposite buffer flow, thereby minimizing the residency time in the ferrofluid stream and increasing the cell viability (94.4 ± 1.3%) when tested with 7 cancer cell lines. It should be noted that the adoption of ferrofluids for cell separation is limited by hindrance in optical imaging due to light diffraction of ferrofluids.148,151 Hence, the nanoscopic properties of ferrofluids have to be further optimized to develop new bio-applications.
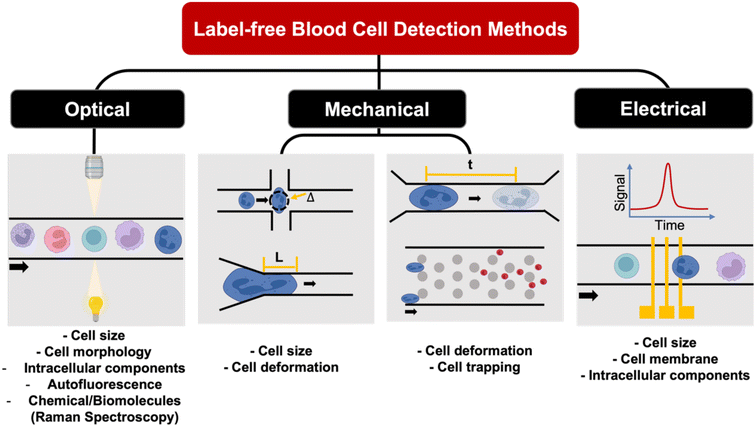
Label-free single cell analysis
Unlike conventional biological assays which provide average (bulk) readout, single cell analysis of a large cell population is a powerful method to probe cell heterogeneity. Label-free single cell analysis enabled by microfluidics offers alluring prospects including minimal sample preparation time, cost-effectivity and usefulness in diseases with no known biomarkers. These approaches mostly analyze intrinsic cellular properties such as cell morphology, spectrochemical properties, dielectric parameters and mechanical characteristics for non-invasive liquid biopsy153 and point-of-care diagnostics.154 Herein, we summarize recent innovations on microfluidic label-free blood cell analysis broadly classified as optical imaging, biomechanical analysis, and electrical profiling of single cells (Fig. 8 and Table 4).| Method | Sample type | Key results | Ref. |
|---|---|---|---|
| Optical properties | |||
| Digital holographic microscopy | Plasma | Discriminate healthy and pathological samples; classify leukemias | 159 |
| QPI (quantitative phase imaging) flow cytometry | Culture medium | Ultra high-throughput (>10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 leukemic cells per s); flow (2.3 m s−1) 000 leukemic cells per s); flow (2.3 m s−1) |
245 |
| Fluorescence lifetime imaging microscopy (FLIM) | Diluted whole blood | Combination of single-cell trapping array separation with real-time FLIM imaging of leukemia cells | 163 |
| Light-scattering | Culture medium | Distinguish and count CD4+ and CD8+ cells (∼79% accuracy) | 155 |
| Stimulated Raman scattering | Culture medium | Discriminate non-differentiated and differentiated 3T3-L1 cells | 168 |
| Stimulated Raman scattering | Whole blood cells, PBMCs, Jurkat cells, HT29 cells | Characterization and classification of cancer cells in blood (∼140 cells per s) | 169 |
| Optical diffraction tomography | Culture medium | Identification of individual lymphocytes (B, CD4+ T, CD8+ T cells) by 3D-RI tomograms | 160 |
| Interferometric plasmonic microscopy | Culture medium | Imaging, sizing, tracking single exosomes | 161 |
| Electrical properties | |||
| Impedance cytometry (multi-frequency) | Diluted whole blood | Accurate classification and quantification of 3 subtypes of leukocytes | 190 |
| Impedance cytometry (multi-frequency) | Whole blood | Label-free discrimination of platelets, erythrocytes, monocytes, granulocytes and lymphocytes (∼3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells in 45 min) 000 cells in 45 min) |
193 |
| Impedance cytometry (single-frequency) | Whole blood (RBCs and PBMCs) | Distinguish 1) normal RBCs and GA treated RBCs; and 2) lymphocytes and monocytes (throughput: 20 μL h−1) | 194 |
| Impedance cytometry (multi-shell model) | Malaria culture | Plasmodium falciparum infection characterisation based on membrane capacitance and cytoplasmic conductivity derived from impedance signal | 195 |
| Impedance spectroscopy | Whole blood | Isolation of CTCs and WBCs from whole blood, and classification of CTCs and WBCs | 197 |
| Impedance spectroscopy | Whole blood | Distinguish sickle red blood cells and normal cells from whole blood using microfluidic chip with oxygen control | 199 |
| Mechanical properties | |||
| Optical quantify cell circularity | Blood cells/differentiated blood cells | Plotted mechanical phenotypes for different types of blood cells | 171 |
| Showed mechanical properties sensitivity to cytoskeleton change | |||
| Optical quantify cell geometry | Diluted whole blood | Enriched CTCs from whole blood and performed cell mechano-phenotyping on the same device | 174 |
| Optical quantify cell geometrical change during deformation | HL-60 | Measured cell Young's modulus and fluidity | 177 |
| Electrical quantify cell geometry change | WBCs/HL-60 | Used electrical signatures to represent cell geometry and quantified cell geometrical change in a label-free and optics-free setup | 178 |
| Electrical quantify cell transit time | CTCs | Used electrical signals to represented cell transition time in constriction channel and characterized cell mechanical properties with transition time | 182 |
Optical properties
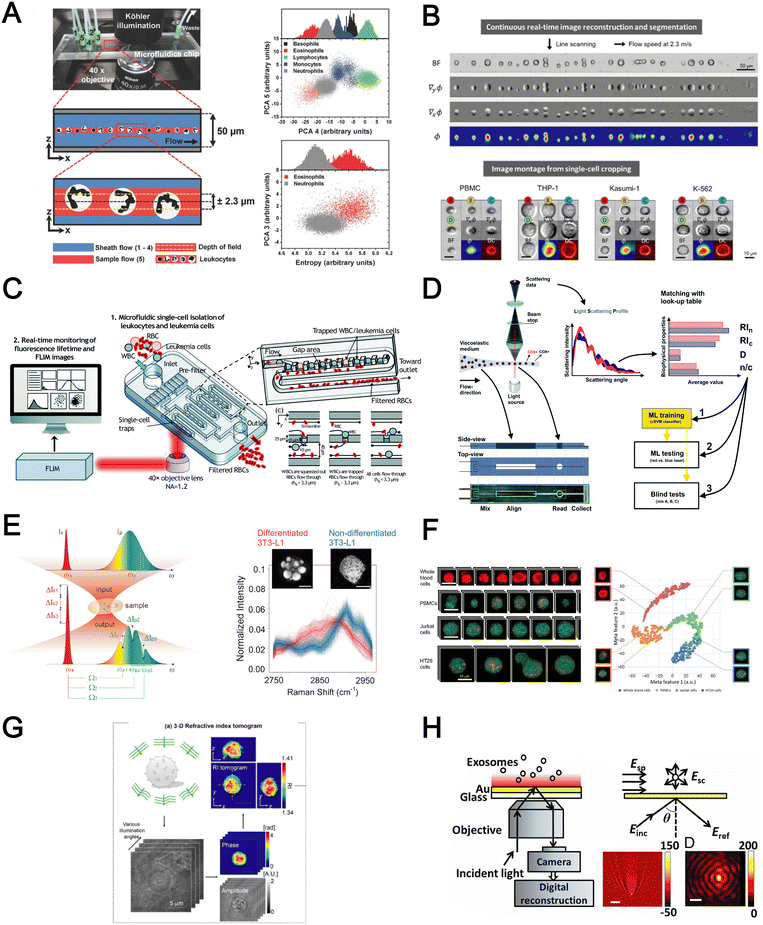
Optical detection methods for blood analysis can be based on imaging of cell morphology,48 or using spectrometry (absorption, scattering),155,156 surface plasmon resonance,157,158 refraction159,160 and interferometric plasmonic microscopy161 to characterize cellular spectrochemical properties. For whole blood analysis, single cell imaging is typically used for blood cell counting or resolving its size or morphology. Ugele et al. reported differential digital holographic (DH) microscopy for native leukocyte detection.159 The in-flow imaging of a 2D hydrodynamically focused leukocyte stream was performed by using a fixed focal height and moderate depth of field (±2.3 μm) (Fig. 9A). Based on principal component analysis (PCA) of the morphological parameters of the reconstructed images, the differentiation of nine leukocyte subtypes in healthy and pathological samples was achieved. Another interesting work is a high-throughput single leukemic cell imaging platform by utilizing ultrafast quantitative phase imaging (QPI), aptly termed as multiplexed asymmetric-detection time-stretch optical microscopy (multi-ATOM).162 The multi-ATOM flow cytometer system allows 12-dimensional single-cell biophysical phenotyping at an ultra-large-scale (>1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per s) (Fig. 9B). Such a combination of high throughput and cellular content provides sufficient statistics to distinguish multiple types of leukemia cells from the blood with high accuracy (∼92–97%). An integrated platform combining a cell trapping array and phasor-fluorescence lifetime imaging microscopy (phasor-FLIM) for leukemia cell identification was also reported for leukemia cell screening.163 The array was designed with 1600 packed single-cell traps to filter out RBCs and capture WBC/leukemia cells (Fig. 9C). The trapped single leukemia cells (THP-1, Jurkat and K562 cells) were then distinguished from WBCs in the phasor-FLIM lifetime map as they exhibited significant shift towards shorter fluorescence lifetime. The authors highlighted that this metabolic characterization indicating a higher ratio of free/bound NADH of leukemia cells was due to their stronger glycolysis for rapid proliferation. The identification of T-lymphocyte subclasses (CD4+ and CD8+) by a light scattering approach was recently proposed by Rossi et al. (Fig. 9D).155
000 cells per s) (Fig. 9B). Such a combination of high throughput and cellular content provides sufficient statistics to distinguish multiple types of leukemia cells from the blood with high accuracy (∼92–97%). An integrated platform combining a cell trapping array and phasor-fluorescence lifetime imaging microscopy (phasor-FLIM) for leukemia cell identification was also reported for leukemia cell screening.163 The array was designed with 1600 packed single-cell traps to filter out RBCs and capture WBC/leukemia cells (Fig. 9C). The trapped single leukemia cells (THP-1, Jurkat and K562 cells) were then distinguished from WBCs in the phasor-FLIM lifetime map as they exhibited significant shift towards shorter fluorescence lifetime. The authors highlighted that this metabolic characterization indicating a higher ratio of free/bound NADH of leukemia cells was due to their stronger glycolysis for rapid proliferation. The identification of T-lymphocyte subclasses (CD4+ and CD8+) by a light scattering approach was recently proposed by Rossi et al. (Fig. 9D).155
 | ||
| Fig. 9 Label free cell detection based on optical properties. (A) Microfluidic cell presentation and five-part DIFF of leukocytes. Reproduced from ref. 159 with permission from Wiley. (B) Multi-contrast single-cell images captured in the multi-ATOM flow cytometer. Reproduced from ref. 162 with permission from Wiley. (C) Isolation and identification of single leukemia cells from blood based on single-cell trapping and phaser-FLIM imaging. Reproduced from ref. 163 with permission from the Royal Society of Chemistry. (D) Identification of CD4+ and CD8+ T-lymphocytes using light scattering and machine learning. Reproduced from ref. 155 with permission from the Royal Society of Chemistry. (E) Multiplexed stimulated Raman scattering process and SRS spectra of two cell populations. Reproduced from ref. 168 with permission from Optica Publishing Group. (F) SRS images of whole blood cells, PBMCs, Jurkat cells, and HT29 cells. Classification of cancer cells in blood using the t-SNE plot. Reproduced from ref. 169 with permission from the National Academy of Sciences. (G) Identification of individual lymphocytes using optical diffraction tomography. Reproduced from ref. 160 with permission from Nature Research. (H) iPM system for single exosome detection based on interferometric scattering. Images of a 100 nm silica nanoparticle without (left) and with (right) an image-reconstruction process (scale bar: 300 nm). Reproduced from ref. 161 with permission from United States National Academy of Sciences. | ||
Raman spectroscopy (also commonly termed as Raman flow cytometry) is another powerful optical-based technology which directly probes characteristic intracellular molecular vibrations by measuring the inelastic scattering of incident photons.164 This is particularly useful to measure metabolites or biomolecules (e.g. unsaturated fatty acids, carotenoids and polysaccharides) in live cells which are difficult to detect using fluorescent labelling methods. While spontaneous Raman scattering limits the detection throughput, newer alternatives such as stimulated Raman scattering (SRS) and coherent anti-Stokes Raman scattering (CARS) significantly improve light–sample interaction for high throughput chemical fingerprinting of flowing cells165,166 or Raman-activated cell sorting.167 Zhang et al. reported a multiplexed SRS flow cytometry platform based on broadband laser excitation and multiplex spectral detection.168 The system can measure the chemical contents of single particles at high throughput (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 spectra per s and 11
000 spectra per s and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 particles per s) and help discriminate non-differentiated and differentiated 3T3-L1 cells by quantification of different chemical compositions accumulated in single cells (Fig. 9E). A more recent study reported label-free multi-colour SRS microscopy of fast-flowing cells on a 3D acoustic focusing microfluidic device to study cell metabolic heterogeneity in microalgae, blood cells and cancer cells.169 The setup was based on a fast pulse pair-resolved wavelength-switchable Stokes laser and a galvanometric scanner, achieving a high flow speed of 2 cm s−1 and a high image acquisition speed of 24k lines per s for 4-color SRS signal acquisition. With the help of machine learning to analyse large cell populations (∼10
000 particles per s) and help discriminate non-differentiated and differentiated 3T3-L1 cells by quantification of different chemical compositions accumulated in single cells (Fig. 9E). A more recent study reported label-free multi-colour SRS microscopy of fast-flowing cells on a 3D acoustic focusing microfluidic device to study cell metabolic heterogeneity in microalgae, blood cells and cancer cells.169 The setup was based on a fast pulse pair-resolved wavelength-switchable Stokes laser and a galvanometric scanner, achieving a high flow speed of 2 cm s−1 and a high image acquisition speed of 24k lines per s for 4-color SRS signal acquisition. With the help of machine learning to analyse large cell populations (∼10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells), the authors demonstrated high-precision characterization and classification of cancer cells in blood at an unprecedented throughput of ∼140 cells per s (Fig. 9F).
000 cells), the authors demonstrated high-precision characterization and classification of cancer cells in blood at an unprecedented throughput of ∼140 cells per s (Fig. 9F).
Four biophysical properties including cell dimension, nucleus-to-cytosol ratio, refractive index of the nucleus and cytosol for each cell were converted from the light-scattering profile and combined with machine learning for automatic counting and characterization of CD4+ versus CD8+ with 79% accuracy. In another work, Yoon et al. used 3D refractive index (RI) tomography to identify lymphocyte cell types based on quantitative morphological and biochemical properties of individual lymphocytes (Fig. 9G).160 The novelty of this work is the combination of RI tomography with machine learning to enable the identification of lymphocyte cell types (B, CD4+ T and CD8+ T cells) with high test accuracy (75.93%).
Besides cell imaging, label-free imaging of single exosomes was reported by Yang et al. using interferometric plasmonic microscopy (iPM).161 This was achieved by monitoring the real-time adsorption of exosomes on a modified Au surface and recording the iPM intensity to determine the size distribution by image reconstruction (Fig. 9H). In summary, label-free optical approaches offer numerous advantages in terms of throughput, information-rich cell images, and easier coupling to machine learning for automated blood analysis. The challenges need to be addressed include the precise focusing of cells, efficient removal of bulk RBCs prior to analysis and high setup cost.
Mechanical properties
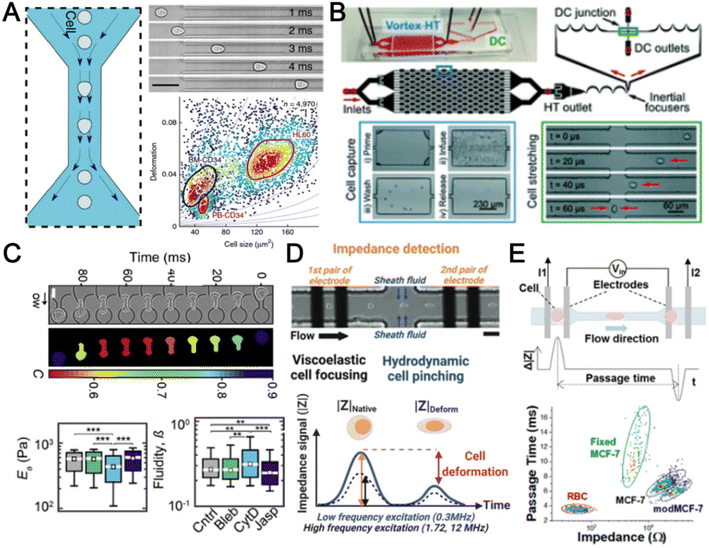
The cytoskeleton is an interconnected network of protein filaments that provides cell shape, determines cell resistance to deformation, and interacts with the extracellular tissue environment. As it is a dynamic and adaptive structure that is affected by cellular activities, accessing cell mechanics is important to study mechanobiology and pathophysiology.170 In this section, we highlight recent advances in microfluidics deformability cytometers for label-free blood cell profiling.The most direct approach to quantify cell deformability is to image single cell geometrical changes under mechanical stimuli such as shear force,171 hydrodynamic force,172 and electric force.173 Otto et al. reported a real-time deformability cytometry (RT-DC) platform that quantifies cell circularity change under shear force using high-speed imaging (Fig. 10A).171 Cell mechanical phenotypes (cell size, deformability) and their sensitivity to cytoskeleton changes were measured and used as biomarkers to identify PBMCs, RBCs, granulocytes, and platelets from whole blood directly. To analyse rare cell events, Che et al. reported a vortex-mediated deformability cytometry (VDC) platform that performed on-chip circulating tumour cell (CTC) enrichment and deformability assessment directly from whole blood (Fig. 10B).174 Their results showed a better CTC detection rate (93.8%) using the biophysical phenotypes (cell size, deformability) as compared to affinity-based immunofluorescence (71.4%). Besides cell profiling for drug treatment study175 or point-of-care diagnostics,8,176 cell mechanical properties such as Young's modulus and fluidity can also be determined if calibrated with known reference materials. For example, Nyberg et al. applied quantitative deformability cytometry (q-DC) to optically track cell geometrical changes to study cell creeping (Fig. 10C).177 After calibration with agarose gel particles, a power-law rheology (PLR) model was used to represent cell creeping in the constriction channel to extract quantitative information on cell elasticity and fluidity of leukaemia cells (HL-60). To avoid the use of expensive high-speed cameras, researchers have developed impedance-based detection methods to profile cell biophysical properties by quantifying the cell geometrical change,178 or cell creeping process179 in an optics-free and label-free manner. A recent study was reported by Petchakup et al. who defined a novel “electrical deformability index” for single cell electro-mechano-phenotyping (Fig. 10D).178 By measuring electrical signals at different frequencies (0.3 MHz, 1.72 MHz and 12 MHz), multiple biophysical parameters of single cells such as cell size, deformability, membrane opacity and nucleus opacity are simultaneously measured to study neutrophil dysfunction.
 | ||
| Fig. 10 Label free cell detection based on mechanical properties. (A) Real-time deformability cytometry (RT-DC) that optically quantified cell geometrical change under shear force. Reproduced from ref. 171 with permission from Springer Nature. (B) Vortex-mediated deformability cytometry (VDC) that enriched CTCs and hydrodynamically deformed cells on the same device. Reproduced from ref. 174 with permission from the Royal Society of Chemistry. (C) Quantitative deformability cytometry (q-DC) that utilized creeping energy to quantitatively measure cell intrinsic properties. Reproduced from ref. 177 with permission from Cell Press. (D) Impedance deformability cytometry that represented cell deformation with impedance signatures. Reproduced from ref. 178 with permission from Wiley. (E) Impedance-based deformability cytometry that quantified cell transition time in the constriction channel with impedance signatures. Reproduced from ref. 182 with permission from the American Chemical Society. | ||
Besides quantifying changes in cell shape, another approach to analyse cell deformability is by measuring cell transit time in constriction channels using imaging,180 suspended microchannel resonators (SMRs),181 and impedance readout.182 For example, impedance signals are detected when a cell passes through electrodes at the start and end of the channel to calculate cell transition time (Fig. 10E).182 Clear differences between RBCs and cancer cells (MCF-7 and modified MCF-t (softer)) were observed by comparing the cell impedance magnitude and cell deformability. However, major challenges include clogging issues and the requirement for low working sample concentration to avoid multiple cells entering the constricted channel. For RBC analysis, microfluidics deformability studies are typically performed in batch analysis to determine an average deformability score based on batch deforming pressure,183 relative lateral displacement,184 and filtration gap size.185 In summary, label-free biophysical profiling of WBCs and RBCs is highly promising for low-cost blood phenotyping. Future work will focus on extending the mechanical cellular assessment to small particles such as platelets and EVs.
Electrical properties
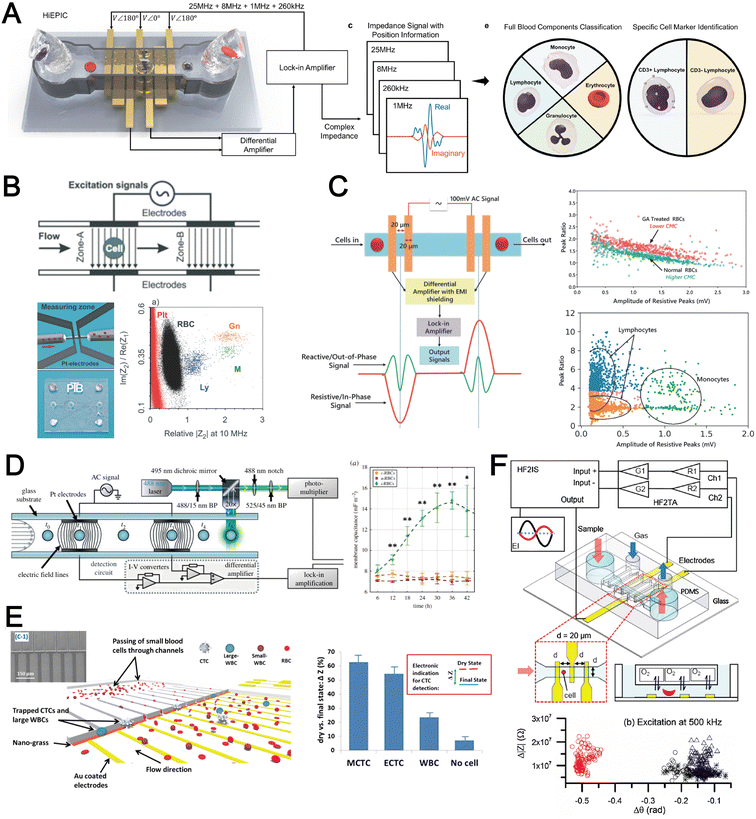
The cell membrane and intracellular content exhibit distinct dielectric and electrical properties for different cell types which can be exploited for label-free single cell detection. A powerful electrical biosensing modality is impedance cytometry which takes advantage of the dielectric dispersion effect under alternating electric field at varying frequencies to characterize different cellular components. It is widely applied for single cell analysis including cell classification and counting,186,187 and cell status monitoring.188,189The impedance magnitude and opacity (ratio of impedance magnitudes at different frequencies) are the most frequently used signatures for cell characterization. Conventional microfluidic impedance cytometers utilize 2D coplanar electrodes for impedance measurement,186,187 but the signal is affected by the vertical position of the particles in the channel. Zhong et al. proposed a high throughput (1000 cells per second) and position-insensitive coplanar electrode-based cytometry platform with a double differential electrode configuration (Fig. 11A). The additional position information provided by this novel electrode configuration contributes to a higher accuracy of cell detection to classify different leukocyte subtypes.190,191 A complicated electrode configuration with two pairs of parallel (top and bottom) electrodes and three additional pairs of co-planar electrodes is also proposed to minimise the effect of particle position on the signal magnitude by reducing the cross current flow into neighbouring detection electrodes.192 This work demonstrated successful classification and numeration of different leukocytes from human blood after erythrocyte lysis. Another strategy is using a 3D parallel electrode configuration to differentiate platelets, erythrocytes, monocytes, granulocytes and lymphocytes in blood by two pairs of top and bottom parallel electrodes (Fig. 11B).193 In addition to impedance-based cell detection, simultaneous capture of cell size and membrane property at single low frequency was reported by Mahesh et al. (Fig. 11C).194 They showed the relationship between the cell membrane capacitance and the ‘double peak’ profile in the out-of-phase signal within low frequencies (beta dispersion regime) to distinguish normal RBCs and glutaraldehyde-treated RBCs based on a single frequency signal measurement.
 | ||
| Fig. 11 Label free cell detection based on electrical properties. (A) Top and bottom parallel electrode configuration for label-free discrimination of platelets, erythrocytes, monocytes, granulocytes and lymphocytes. Reproduced from ref. 190 with permission from Elsevier. (B) Copland electrodes for cell classification using single frequency. Reproduced from ref. 193 with permission from the Royal Society of Chemistry. (C) Microfluidic impedance cytometry for dielectric characterization of Plasmodium falciparum-infected red blood cells. Reproduced from ref. 194 with permission from the Royal Society of Chemistry. (D) Microfluidic impedance spectroscopy device with trapping design for diagnosis of CTCs. Reproduced from ref. 195 with permission from the Royal Society. (E) Impedance microfluidic cytometer with oxygen control for detection of sickle red blood cells. Reproduced from ref. 197 with permission from Wiley. (F) HiEPIC system for blood cell classification. Reproduced from ref. 199 with permission from Elsevier. | ||
Besides blood profiling, impedance cytometry is also used for blood-based disease detection. For example, malaria-infected RBCs (Plasmodium falciparum) have higher membrane capacitance and cytoplasmic conductivity which would be useful for pre-enrichment of infected RBCs for malaria diagnosis (Fig. 11D).195 Application of impedance cytometry for CTC detection in whole blood has also been reported with integrated CTC trapping (Fig. 11E)196,197 or under continuous flow.198 Liu et al. developed a microfluidic chip allowing oxygen control for the detection of sickle red blood cells (Fig. 11F).199 Interestingly, significant differences in impedance profile were revealed between normal cells and sickle cells under normoxia, and between sickle cells under normoxia and hypoxia.
In summary, impedance-based detection is a promising label-free blood cell analysis technique which does not require expensive high-speed camera imaging. Although the signal is generally dependent on particle position in microchannels, recent advances have achieved higher sensing sensitivity through better electrode configurations and channel structures, as well as developing new signal processing strategies. Future work will focus on improving electrode fabrication and the calibration using reference materials for large-scale clinical testing.
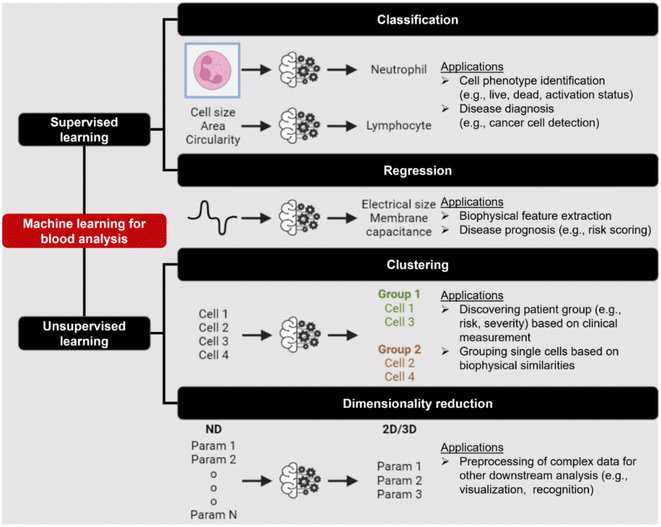
Label-free cell analysis using machine learning approaches
Machine learning (ML) is a field of computer algorithms that can learn relations of data without explicit programming or human intervention. With enhanced computing power and data storage, ML has gained increasing attention for a wide variety of biomedical applications especially in biomarker discovery200 and clinical diagnostics.201 Emerging applications of ML to synergize microfluidics include optimizing device design, operation and data processing/analysis. ML can be used to construct microfluidic design models based on experimental performance metrics (e.g. sorting efficiency, yield) and channel features (e.g., channel geometry, flow rate) to improve design performance or predict the behavior of new samples. This can potentially ease the iterative device optimization process and lower manufacturing and testing costs.127 Microfluidic devices integrated with imaging/sensing modalities (e.g., camera, electrodes, flow/pressure sensors) can incorporate ML to analyze real-time information (e.g., particle trajectory, particle properties, flow rate, pressure drop) “on-the-fly” and provide feedback to pumps/actuators for automated flow manipulation or resolving operational issues (e.g. channel clogging). These aspects have been extensively covered in a recent review by McIntyre et al.202 Lastly, ML can enhance the data analysis pipeline in preprocessing steps such as feature extraction or denoising, as well as in postprocessing steps including developing a prediction model and compressing multi-dimensional data for visualization.As label-free cell characterization may lack specificity and sensitivity compared to antibody-based detection, multi-parametric analysis of intrinsic cell properties combined with advanced data analysis can potentially improve the assay accuracy. In this section, we will highlight recent work that utilize ML for microfluidics label-free blood cell phenotyping in two broad classifications, namely 1) supervised learning and 2) unsupervised learning (Fig. 12 and Table 5). For more detailed information, the readers are highly encouraged to refer to other excellent reviews for microfluidic applications203–205 and neural networks/deep learning.206,207
| Category | Technique | Samples | Features | Applications | Ref. |
|---|---|---|---|---|---|
| Classification or regression | SVM with RFE | WBC, MCF-7, HL-60 | Features extracted from brightfield images from deformability cytometer | Classification of spiked cancers with WBC with <5% error in classification rate for differently mixed samples | 246 |
| Logistic regression | WBC, PLT and PLT aggregate | Morphological features from brightfield image | Classification with specificity of 96.6% | 247 | |
| Decision tree | PBMCs, MCF-7 and MDA-MB231 | Cell size, maximum intensity, and mean intensity | Cell classification with AUROC >0.9 for all features | 217 | |
| Decision tree, SVM, LDC and k-NN | RBC (discocyte, echinocyte and spherocyte) | Features from DIHM (12D) | Comparison of classification algorithms and decision tree achieved the best accuracy (98.18%) | 248 | |
| CNN | WBC, PLT and PLT aggregate | Brightfield image | Classification of PLT aggregate with specificity of 99% and selectivity 82% | 166 | |
| N/A | PBMC/THP-1, Kasumi-1 and K-562 | Features extracted from QPI (12D) | Classification with 94% accuracy. AUROC for one-versus-all classification were 0.975/0.920, 0.95 and 0.952 for PBMC, THP-1, K-562 and Kasumi-1, respectively | 249 | |
| CNN (autoencoder) | K562, K562/ADM cells and RBC | Brightfield image | Quantification of drug-induced morphological changes and segmentation of RBC and cancer cells from the images with 86% pixel-wise accuracy | 210 | |
| CNN | Whole blood cells, PBMCs, Jurkat cells, and HT29 cells | Stimulated Raman scattering images | Classification accuracy of >93% | 250 | |
| RNN (LSTM-NN) | RBCs and beads | Impedance signal measured in horizontal and vertical direction | Extraction of diameter velocity and positions of cells | 213 | |
| NN | PBMC, small cell lung cancer (NSCLC) | 80 optofluidic features from brightfield and quantitative phase imaging | Detection of spiked NSCLC (H2170) PBMC with 98% accuracy | 219 | |
| SVM | WBC, SW480 and SW620 | Optical phased delay features | Classification sensitivity, specificity, and accuracy of 97.8%, 98.9% and 99.6%, respectively | 251 | |
| SVM | CD4 and CD8 T-lymphocytes | Features from light scattering profile (size, nucleus-to-cytosol ratio, refractive index of nucleus and cytosol) | Classification with 70.31% and 87.10% for unstimulated and stimulated cells, respectively | 252 | |
| NN | Neutrophils, RBCs | Images from RT-FDC | Classification of neutrophils with 95.5% accuracy | 253 | |
| CNN | WBC subtypes | Brightfield images from RT-DC | Classification of blood cells and classification of T-cells and B-cells (transfer learning from Nitta et al.166) | 209 | |
| SVM | RBC | Features extracted from trained CNN (AlexNET) | Classification of different kinds of anemia (SCD, THAL and HS) | 227 | |
| SVM with radial basis function | WBC from healthy and sepsis patients | 38 biophysical markers quantified from DLD-devices at different flow rates and designs | Prediction of sepsis infection with AUROC of 0.97 | 218 | |
| CNN | RBC | Brightfield images | Classification of deformable and non-deformable sickle RBC and non-sickle RBC with accuracy of 96% ± 0.3% | 254 | |
| CNN | RBC | Images | Classification of RBC from different outlets and determine RBC rigidity score | 255 | |
| CNN | Bead, RBC and ghost RBC | Impedance images (stacked impedance signal at 8 frequencies) | Classification, calibration of measurement using bead properties and extraction of electrical parameters (cell size, membrane capacitance and cytoplasm permittivity and conductivity) | 212 | |
| NN | WBC | Impedance parameters (cell size, membrane capacitance, cytoplasmic conductivity) | Classification with 93.5% accuracy | 216 | |
| NN | WBC including eosinophils and basophils | Impedance parameters (cell size, membrane capacitance, cytoplasmic conductivity) | Classification rate of 80.8% for neutrophil vs. eosinophil, 77.7% for neutrophil vs. basophil and 59.3% for neutrophil vs. basophil | 215 | |
| NN | MCF-7, A549, HeLa, HL60 and GM12878 | Impedance peaks at four different frequencies | Extraction of electrical properties and classification with 91.5% accuracy | 256 | |
| CNN | WBC | Brightfield images | Classification with 81.4% accuracy | 257 | |
| MASK-RCNN, CNN–GRU | HL-60 (untreated, cytochalasin D and nocodazole) | Brightfield images | Segmentation using Mask-RCNN and classification of image sequence using CNN–GRU with accuracy of over 90% | 211 | |
| Clustering | DBSCAN | WBC and bead | Impedance parameters (cell size and opacity) | Extraction of cell cluster from heterogenous population | 189 |
| GMM | HL-60 cells and MG-63 | Area and deformability from RT-DC | Cluster cell populations in mixtures | 224 | |
| Hierarchical clustering | WBC from healthy and sepsis patients | 38 biophysical markers quantified from DLD-devices at different flow rates and designs | Group patients into 8 groups | 218 | |
| Dimensionality reduction | t-SNE | PBMC/THP-1, Kasumi-1 and K-562 | Features extracted from QPI (12D) | Visualization of cell type distribution in 2D t-SNE | 249 |
| PCA | PBMC, small cell lung cancer (NSCLC) | 80 optofluidic features from brightfield and quantitative phase imaging | Visualization of spiked H2170 distribution in 3 dimensions-PCA | 219 | |
| PCA | WBC, SW480 and SW620 | Optical phased delay features | Visualization cell distribution in 3 dimensions-PCA | 251 | |
| PCA | WBC from healthy and sepsis patients | 38 biophysical markers quantified from DLD-devices at different flow rates and designs | Visualization of healthy and sepsis patient data | 218 | |
| Pre-trained CNN (AlexNET) | RBC | Brightfield image | Extraction of latent features from brightfield image | 227 | |
| UMAP | Neutrophils (untreated, apoptosis, necrosis and NETosis) | Impedance-deformability parameters (10D) | Visualization of different neutrophil phenotypes in 2D UMAP | 178 |
Supervised learning
Supervised learning aims to establish connections between example inputs (e.g., image, signal, and data) and their corresponding outputs such as labels (cell types, pathological phenotypes) or single cell features (single cell properties, shape) for classification or regression problems, respectively. Traditionally, classification pipelines include preprocessing (filtering) and feature extraction to extract single cell features (e.g., single cell properties) followed by training a classifier to recognize labels from extracted features.206 The choice of classifier can vary based on applications and data complexity. Popular classifiers mentioned in the literature are tree-based model (e.g., decision tree, random forest), support vector machine (SVM) and neural networks. With the emergence of deep learning,207 the classification problem is greatly simplified as deep neural networks are capable of learning crucial representative features for classification directly from raw inputs without the need for preprocessing or feature extraction. To improve the performance of the model built with small-size data, one can also make use of transfer learning in which a pretrained model from one task can be used as a building block for another model for a similar task.208 Here we list the utilities of supervised learning based on input types (image, signal and data) as shown in Fig. 13.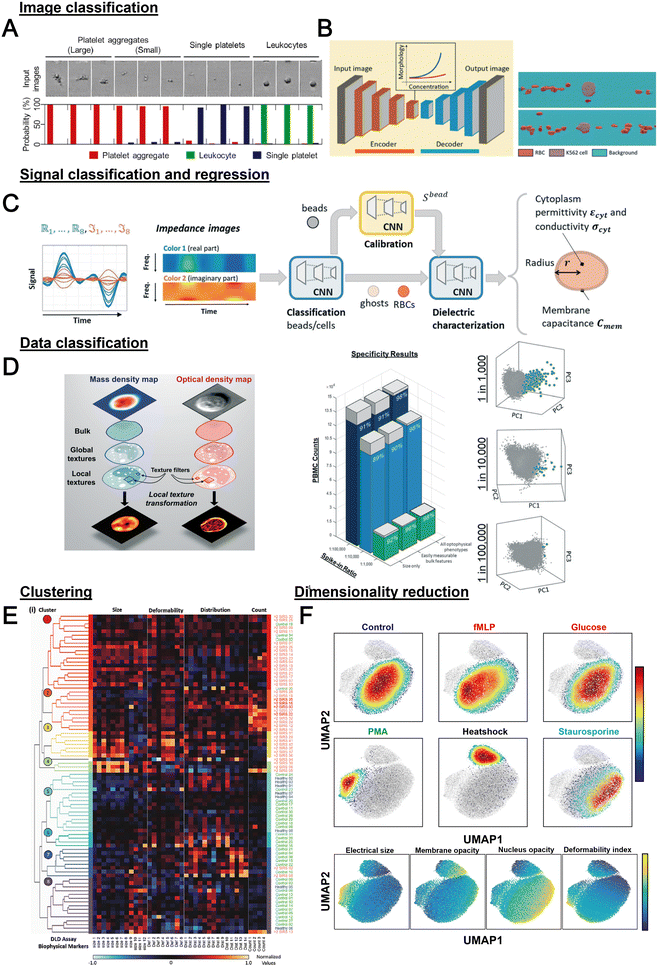 | ||
| Fig. 13 Machine learning for blood analysis. (A) Image classification of leukocytes and platelet aggregates from brightfield images captured from an intelligent flow cytometer. Reproduced from ref. 166 with permission from Elsevier. (B) Convolutional autoencoders for cancer cell and RBC segmentation and assessment of drug-induced morphological changes using maximum mean discrepancy (MMD). Reproduced from ref. 210 with permission from the Royal Society of Chemistry. (C) Integrated framework for multifrequency impedance signal processing for classification of beads, RBCs and ghost RBCs and extraction of electrical properties of single cells. Reproduced from ref. 212 with permission from the Royal Society of Chemistry. (D) Classification of spiked NSCLS in PBMC samples using opto-physical features from QPI and brightfield images. Reproduced from ref. 219 with permission from the Royal Society of Chemistry. (E) Hierarchical clustering for grouping of DLD extracted measurement for sepsis analysis. Reproduced from ref. 218 with permission from Wiley. (F) Dimensionality reduction using UMAP to compress electrical-based biophysical parameters for visualization of different neutrophil phenotypes. Reproduced from ref. 178 with permission from Wiley. | ||
Many ML models are viewed as a black box model as it can be difficult to comprehend how models reach their final outcomes which may fail to attract trust and acceptance for clinical uses.220 To improve interpretability and explainability of a model, certain approaches can be done including feature importance (e.g., Shapley value,221 permutation importance222) to understand the contribution/weight of features on a final prediction, or attention mechanism (e.g., class activation map (CAM)223) to visualize which part of an image that influences a class prediction.
Unsupervised learning
In contrast to supervised learning, unsupervised learning can learn patterns/structures from unlabeled data and be categorized as either clustering or dimensionality reduction (Fig. 14). Clustering involves grouping of unlabeled data/features into several groups based on certain similarity metrics (e.g., Euclidean distance, cosine similarity) or distributions. For example, density-based spatial clustering of applications with noise (DBSCAN) can be used to group single cell electrical features and extract a target cell population for quantification.189 Herbig et al. employed Gaussian mixture models (GMM) to cluster HL-60 cells and MG-63 from cell mixtures based on the area and deformability extracted from cell deformability measurements.224 Hierarchical clustering was also applied to group patients based on the similarities of their DLD biophysical parameters and certain cluster groups were shown to correlate with long hospitalization stay (Fig. 13E).218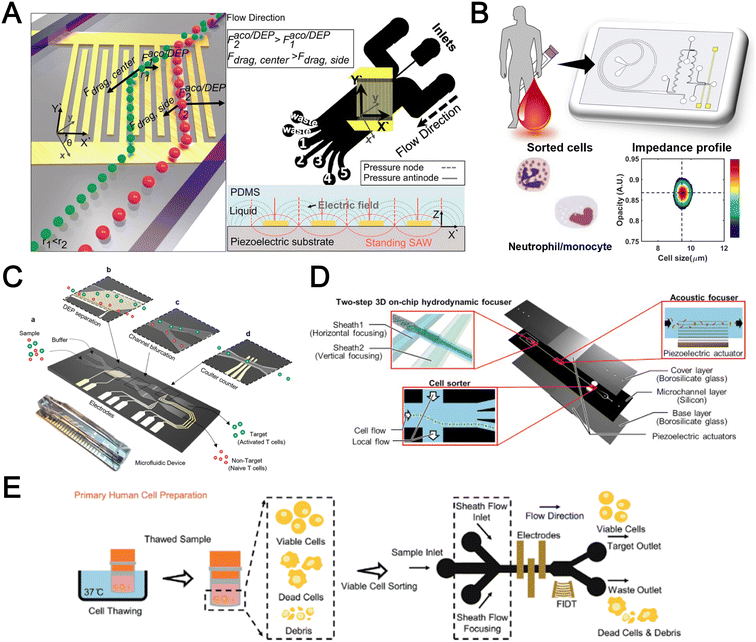 | ||
| Fig. 14 Integrated microfluidic platforms for blood applications. (A) Integrated acoustic and DEP for exosome separation. Reproduced from ref. 130 with permission from the American Chemical Society. (B) Integrated inertial impedance cytometer. Reproduced from ref. 189 with permission from the Royal Society of Chemistry. (C) DEP-impedance cytometer for differential counting of activated lymphocytes. Reproduced from ref. 233 with permission from the American Chemical Society. (D) Intelligent imaging flow cytometer. Reproduced from ref. 166 with permission from Elsevier. (E) Impedance activated surface acoustic wave cell sorter. Reproduced from ref. 235 with permission from Wiley. | ||
Dimensionality reduction simplifies high dimensional data into low-dimensional data (usually 2, 3 dimensions) for data visualization and analysis. In some scenario, having numerous features does not always guarantee an increase in predictive performance and can even deteriorate performance of certain models. This is because adding more features makes feature space more sparse and difficult for models to generalize (curse of dimensionality). Introducing dimensionality reduction as an intermediate step prior to other ML techniques can alleviate dimensionality issues. Techniques such as principal component analysis (PCA), t-distributed stochastic neighbor embedding (t-SNE) and uniform manifold approximation and projection (UMAP) are widely used in flow cytometry analysis and biological studies.225,226 Inspired by these methods, Petchakup et al. utilized UMAP to visualize biophysical features of different neutrophil phenotypes (Fig. 13F).178 Another way to harness the power of transfer learning is to exploit pre-trained CNN for feature extractor. As pre-trained CNNs (e.g., AlexNET) are trained from a large-scale image dataset, it has already captured all important features or patterns. By passing image input (high-dimensional data) through a pre-trained network, it can output the latent features (low-dimensional data) which can be used in the subsequent task.227 They used trained CNN to extract features from brightfield images of RBCs and subsequently SVM for classification of different RBC phenotypes such as sickle cell disease (SCD), thalassemia syndromes (THAL) and hereditary spherocytosis (HS).227
Taken together, these exciting studies have shown that ML can enhance blood analysis. It is possible to develop an intelligent microfluidic system to provide fast, automated, and accurate analysis without the need for clinical expertise or mechanistic biological knowledge. It should be noted that to produce a model with good generalization, large data collection is sometimes required, and comes with the cost of additional time and resources (biological samples, donor samples). Implementing ML for analysis usually incurs additional computational resources (processors) which may compromise the overall system setup (footprint). As ML exponentially evolves, other novel ML utilities have also been reported for biomedical image analysis including “virtual staining”228 in which CNNs were designed to understand the associations between morphological features of label-free images (e.g., autofluorescence, brightfield) and stained images (histological, antibody stained) for image-to-image translation to produce stained images without the need for laborious staining. A potential use for blood cell analysis is to construct a virtual staining model of subcellular components such as the nucleus and mitochondria in single immune cells for label-free quantification applications. It is also possible to use generative models to synthesize more training data (e.g., biomedical images) to improve predictive model generalization.229 This can be readily adopted to produce more events of rare cells especially CTCs from microfluidic assays to improve a predictive model.
Integrated label-free blood cell sorting and analysis
A powerful aspect of microfluidics is to miniaturize and integrate sample preparation and biological assays on a single chip to automate medical analysis. Herein, we highlight several integrated microfluidic approaches for complete label-free blood analysis (Fig. 14).Multiparametric sorting or sensing
To achieve higher separation efficiency, one can combine multiple sorting mechanisms to enhance particle separation. For example, Tayebi et al. combined acoustic and DEP forces for exosome separation (Fig. 14A).130 The ability to interrogate various single cell features is crucial for single cell analysis for elucidating immune cellular functions or disease pathology using multi-colour flow cytometry.230 As intrinsic or biophysical features obtained from single measurement might be limited, multi-parametric single cell measurement is important to enhance assay sensitivity or for ML applications. A common pairing is mechanical measurement with optical or impedance detection as changes in cell mechanics are often reflected in cell morphology which can be quantified using image or impedance readout. For example, Petchakup et al. demonstrated single cell electro-mechano-phenotyping (14 biophysical parameters of single cells) to study different neutrophil phenotypes during activation and apoptosis.178Integrated sample processing
As conventional RBC lysis or density centrifugation steps can be time-consuming and laborious, integrating a sample preprocessing module will facilitate direct blood processing and detection of target cells. An integrated spiral microfluidic chip with an impedance cytometer was used for removal of small blood components (e.g., lymphocytes, platelets and RBCs) to allow electrical interrogation of monocytes, neutrophils or activated lymphocytes from diluted blood or PBMCs (Fig. 14B).189,231 Raillon et al. utilized inertial vortex induced CTC trapping prior to impedance cytometry.232 Han et al. applied DEP for pre-sorting of large, activated-lymphocytes from small native-lymphocytes for subsequent differential impedance counting (Fig. 14C).233 Another interesting integration is to increase detection specificity by differential counting of target cells before and after traversing a cell capturing chamber functionalized with capture antibodies.234Integrated active sorting
To elucidate associations between label-free features and biological functions, an actuated cell sorter can be added to select cells based on label-free features (impedance, optical, mechanical). A great example is reported by Nitta et al. in which they incorporated various microfluidic modalities such as an acoustic focuser and dual membrane push–pull cell sorter to complement high-speed imaging for platelet aggregate sorting based on morphological features (Fig. 14D).166 Zhong et al. demonstrated acoustic sorting using impedance signatures for live cell sorting from thawed PBMC samples (Fig. 14E).235Conclusions and outlook
In summary, microfluidics or lab-on-a-chip technologies provide numerous advantages including reduced preparation time, small sample and reagent volume requirements, precise control over the cell microenvironment and ease of integration. As blood is routinely sampled and contains a rich source of cellular and molecular biomarkers, this review highlights the recent advancements in label-free microfluidic blood cell sorting and analysis for clinical diagnostics. By eliminating the need for expensive antibodies or chemicals, label-free microfluidic assays not only provide valuable insights for intrinsic single cell multi-parameter analysis, but are also essential for identifying cell types and understanding their biological functions. It is especially powerful for blood-based diagnostics as it requires minimal sample preparation and is more cost effective and scalable for clinical testing. Label-free cell sorting often exploits biophysical differences in different cell types to achieve separation and there is a variety of passive or active microfluidic methods reported for blood fractionation. Label-free cell detection approaches include optical measurement of cell morphology and intracellular components, impedance cytometry to detect cellular and sub-cellular electrical properties at multiple frequencies, as well as quantifying cellular mechanics (e.g. cell deformability) which can be indicative of cell dysfunction in diseases. While label-free cell detection may lack specificity and sensitivity as compared to affinity-based (e.g. antibodies) methods, recent studies have greatly improved their detection accuracy and clinical utilities by enabling multi-parametric cell analysis and high dimensional data processing. Coupled with machine learning approaches, they are highly promising for automated high-throughput blood analysis and biomarker discovery in biomedical research.Numerous technological advancements have been demonstrated for both label-free blood cell sorting and detection. Besides improving sample throughput and separation performance, integrated and user-friendly microfluidic systems will be key to automate and scale up for high throughput clinical samples testing. Especially for label-free separation approaches, multi-physics or multi-stage cell sorting can potentially refine resolution separation with the trade-off of increased device complexity. For example, isolation of immune cells from whole blood can be initial RBC bulk removal by cell size differences using inertial focusing or ferrofluids, followed by 2nd step fractionation of leukocyte subtypes (e.g., neutrophils and monocytes) based on other biophysical properties (e.g. cell density) by acoustophoresis. To achieve a complete label-free “sample-in-answer-out” microfluidic platform, multiple functionalities (sorting, sensing and analytics) can be integrated to enable high-content and high throughput analysis of multiple blood components. Nevertheless, microfluidic integration is a non-trivial task. Besides having increased complexity in chip design and instrumentation, researchers have to consider flow rate disparity between sorting and detection modules governed by different working principles. Several studies have shown that it is useful to include an additional flow focuser to align cells and siphon excess particle-free medium to reduce the flow rate. Secondly, incorporation of active sorting elements (DEP, acoustic) requires additional equipment and it is important to consider their miniaturisation for point-of-care testing. Lastly, certain techniques such as DEP and viscoelastic flow use specific medium conditions (e.g., conductivity, viscosity) by adding chemicals to blood samples. Combining upstream buffer exchange or particle sorting can help alleviate this issue. We envision that ML-powered microfluidics will improve device operation and robustness and significantly leverage label-free biophysical/chemical features of circulating cellular targets as novel blood-based biomarkers.
Taken together, label-free microfluidics technologies enable new assay development to identify novel biomarkers and non-traditional risk factors in disease diagnostics. By offering a complete label-free blood sample preparation and analysis workflow (cell sorting, cell detection, machine learning methods), we are optimistic that this will reduce barriers to adoption and better guide the readers to develop new clinical applications in liquid biopsy.
Author contributions
Nan Lu: conceptualization, literature search, writing – original draft, review and editing. Hui Min Tay: conceptualization, literature search, writing – original draft, review and editing. Chayakorn Petchakup: literature search, writing – original draft. Linwei He: literature search, writing – original draft. Lingyan Gong: literature search, writing – original draft. Kay Khine Maw: literature search, writing – original draft. Sheng Yuan Leong: literature search, writing – original draft. Wan Wei Lok: literature search, writing – original draft. Hong Boon Ong: literature search, writing – original draft. Ruya Guo: literature search, writing – original draft. King Ho Holden Li: funding acquisition, writing – review & editing. Han Wei Hou: conceptualization, funding acquisition, project administration, supervision, writing – review & editing.Conflicts of interest
The authors have declared that no conflict of interest exists.Acknowledgements
The authors would like to acknowledge support from Singapore Ministry of Education Academic Research Fund (MOE AcRF) Tier 2 (MOE-T2EP30120-0004) and under the RIE2020 Industry Alignment Fund – Industry Collaboration Projects (IAF-ICP) Funding Initiative, as well as cash and in-kind contributions from the industry partner, HP Inc., through the HP-NTU Digital Manufacturing Corporate Lab. Schematics were created with https://BioRender.com.References
- Z. T. Yu, K. M. Aw Yong and J. Fu, Small, 2014, 10, 1687–1703 CrossRef CAS PubMed.
- H. N. Iskandar and M. A. Ciorba, Transl. Res., 2012, 159, 313–325 CrossRef CAS PubMed.
- J. M. Rhea and R. J. Molinaro, MLO Med. Lab. Obs., 2011, 43, 10–12 Search PubMed , 16, 18; quiz 20, 22.
- S. A. Jamali, M. T. Turnbull, T. Kanekiyo, P. Vishnu, A. C. Zubair, C. C. Raper, R. G. Tawk and W. D. Freeman, J. Stroke Cerebrovasc. Dis., 2020, 29, 104631 CrossRef PubMed.
- H. M. Tay, R. Dalan, K. H. H. Li, B. O. Boehm and H. W. Hou, Small, 2018, 14, 1702832 CrossRef PubMed.
- H. W. Hou, C. Petchakup, H. M. Tay, Z. Y. Tam, R. Dalan, D. E. K. Chew, K. H. H. Li and B. O. Boehm, Sci. Rep., 2016, 6, 29410 CrossRef CAS PubMed.
- M. Kubánková, B. Hohberger, J. Hoffmanns, J. Fürst, M. Herrmann, J. Guck and M. Kräter, Biophys. J., 2021, 120, 2838–2847 CrossRef PubMed.
- L. Guillou, R. Sheybani, A. E. Jensen, D. Di Carlo, T. S. Caffery, C. B. Thomas, A. M. Shah, H. T. K. Tse and H. R. O'Neal, Jr., PLoS One, 2021, 16, e0246980 CrossRef CAS PubMed.
- H. M. Tay, W. H. Yeap, R. Dalan, S. C. Wong and H. W. Hou, Anal. Chem., 2018, 90, 14535–14542 CrossRef CAS PubMed.
- L. Tan, Q. Wang, D. Zhang, J. Ding, Q. Huang, Y.-Q. Tang, Q. Wang and H. Miao, Signal Transduction Targeted Ther., 2020, 5, 33 CrossRef CAS PubMed.
- Y. Mei, S. E. Weinberg, L. Zhao, A. Frink, C. Qi, A. Behdad and P. Ji, EClinicalMedicine, 2020, 26, 100475 CrossRef PubMed.
- D. Zhang, R. Guo, L. Lei, H. Liu, Y. Wang, Y. Wang, H. Qian, T. Dai, T. Zhang, Y. Lai, J. Wang, Z. Liu, T. Chen, A. He, M. O'Dwyer and J. Hu, J. Leukocyte Biol., 2021, 109, 13–22 CrossRef CAS PubMed.
- B. B. Nkambule, V. Mxinwa, Z. Mkandla, T. Mutize, K. Mokgalaboni, T. M. Nyambuya and P. V. Dludla, BMC Med., 2020, 18, 357 CrossRef PubMed.
- E. C. Mesquita, E. D. Hottz, R. T. Amancio, A. B. Carneiro, L. Palhinha, L. E. Coelho, B. Grinsztejn, G. A. Zimmerman, M. T. Rondina, A. S. Weyrich, P. T. Bozza and F. A. Bozza, Sci. Rep., 2018, 8, 14999 CrossRef PubMed.
- F. Scudiero, R. Valenti, R. Marcucci, G. D. Sanna, A. M. Gori, A. Migliorini, R. Vitale, B. Giusti, E. D. Vito, G. Corda, R. Paniccia, D. Zirolia, M. E. Canonico and G. Parodi, J. Am. Heart Assoc., 2020, 9, e016441 CrossRef CAS PubMed.
- C.-H. Chao, W.-C. Wu, Y.-C. Lai, P.-J. Tsai, G.-C. Perng, Y.-S. Lin and T.-M. Yeh, PLoS Pathog., 2019, 15, e1007625 CrossRef CAS PubMed.
- A. Ojha, D. Nandi, H. Batra, R. Singhal, G. K. Annarapu, S. Bhattacharyya, T. Seth, L. Dar, G. R. Medigeshi, S. Vrati, N. K. Vikram and P. Guchhait, Sci. Rep., 2017, 7, 41697 CrossRef CAS PubMed.
- M. L. Jones, J. Siddiqui, K. J. Pienta and R. H. Getzenberg, Prostate, 2013, 73, 176–181 CrossRef CAS PubMed.
- M. C. Miller, G. V. Doyle and L. W. Terstappen, J. Oncol., 2010, 2010, 617421 Search PubMed.
- X. Zhang, X. Yuan, H. Shi, L. Wu, H. Qian and W. Xu, J. Hematol. Oncol., 2015, 8, 83 CrossRef PubMed.
- C. Castaño, A. Novials and M. Párrizas, Diabetes/Metab. Res. Rev., 2019, 35, e3107 CrossRef PubMed.
- C. Lässer, S. E. O'Neil, G. V. Shelke, C. Sihlbom, S. F. Hansson, Y. S. Gho, B. Lundbäck and J. Lötvall, J. Transl. Med., 2016, 14, 181 CrossRef PubMed.
- M. J. Martinez-Bravo, C. J. Wahlund, K. R. Qazi, R. Moulder, A. Lukic, O. Rådmark, R. Lahesmaa, J. Grunewald, A. Eklund and S. Gabrielsson, J. Allergy Clin. Immunol., 2017, 139, 1186–1194 CrossRef CAS PubMed.
- Z. Lanyu and H. Feilong, Biomed. Pharmacother., 2019, 113, 108748 CrossRef PubMed.
- D. W. Freeman, N. Noren Hooten, E. Eitan, J. Green, N. A. Mode, M. Bodogai, Y. Zhang, E. Lehrmann, A. B. Zonderman, A. Biragyn, J. Egan, K. G. Becker, M. P. Mattson, N. Ejiogu and M. K. Evans, Diabetes, 2018, 67, 2377–2388 CrossRef CAS PubMed.
- S. A. Melo, L. B. Luecke, C. Kahlert, A. F. Fernandez, S. T. Gammon, J. Kaye, V. S. LeBleu, E. A. Mittendorf, J. Weitz, N. Rahbari, C. Reissfelder, C. Pilarsky, M. F. Fraga, D. Piwnica-Worms and R. Kalluri, Nature, 2015, 523, 177–182 CrossRef CAS PubMed.
- J. Inal, Br. J. Haematol., 2020, 190, e218–e220 CrossRef CAS PubMed.
- S. Cheng, Y. Li, H. Yan, Y. Wen, X. Zhou, L. Friedman and Y. Zeng, Lab Chip, 2021, 21, 3219–3243 RSC.
- K. S. Yang, H. Im, S. Hong, I. Pergolini, A. F. Del Castillo, R. Wang, S. Clardy, C. H. Huang, C. Pille, S. Ferrone, R. Yang, C. M. Castro, H. Lee, C. F. Del Castillo and R. Weissleder, Sci. Transl. Med., 2017, 9, eaal3226 CrossRef PubMed.
- K. Liang, F. Liu, J. Fan, D. Sun, C. Liu, C. J. Lyon, D. W. Bernard, Y. Li, K. Yokoi, M. H. Katz, E. J. Koay, Z. Zhao and Y. Hu, Nat. Biomed. Eng., 2017, 1, 0021 CrossRef CAS PubMed.
- C. Z. J. Lim, Y. Zhang, Y. Chen, H. Zhao, M. C. Stephenson, N. R. Y. Ho, Y. Chen, J. Chung, A. Reilhac, T. P. Loh, C. L. H. Chen and H. Shao, Nat. Commun., 2019, 10, 1144 CrossRef PubMed.
- M. Guo, J. Wang, Y. Zhao, Y. Feng, S. Han, Q. Dong, M. Cui and K. Tieu, Brain, 2020, 143, 1476–1497 CrossRef PubMed.
- Y. Kwon and J. Park, Micro Nano Syst. Lett., 2022, 10, 14 CrossRef.
- J. Lee, O. Sul and S. B. Lee, Micromachines, 2020, 11, 481 CrossRef PubMed.
- Y. Song, T. Tian, Y. Shi, W. Liu, Y. Zou, T. Khajvand, S. Wang, Z. Zhu and C. Yang, Chem. Sci., 2017, 8, 1736–1751 RSC.
- P. Bankó, S. Y. Lee, V. Nagygyörgy, M. Zrínyi, C. H. Chae, D. H. Cho and A. Telekes, J. Hematol. Oncol., 2019, 12, 48 CrossRef PubMed.
- A. C. Söderström, M. Nybo, C. Nielsen and P. J. Vinholt, Clin. Chem. Lab. Med., 2016, 54, 1913–1920 Search PubMed.
- C.-y. Chiang and C. Chen, J. Biomed. Sci., 2019, 26, 9 CrossRef PubMed.
- I. V. Pivkin, Z. Peng, G. E. Karniadakis, P. A. Buffet, M. Dao and S. Suresh, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 7804–7809 CrossRef CAS PubMed.
- D. Di Carlo, J. Lab. Autom., 2012, 17, 32–42 CrossRef CAS PubMed.
- N. Kang, Q. Guo, E. Islamzada, H. Ma and M. D. Scott, Integr. Biol., 2018, 10, 207–217 CrossRef CAS PubMed.
- K. Crawford, A. DeWitt, S. Brierre, T. Caffery, T. Jagneaux, C. Thomas, M. Macdonald, H. Tse, A. Shah, D. Di Carlo and H. R. O'Neal, Am. J. Respir. Crit. Care Med., 2018, 198, 280–282 CrossRef CAS PubMed.
- C. Petchakup, H. Yang, L. Gong, L. He, H. M. Tay, R. Dalan, A. J. Chung, K. H. H. Li and H. W. Hou, Small, 2022, 18, e2104822 CrossRef PubMed.
- V. Faustino, R. O. Rodrigues, D. Pinho, E. Costa, A. Santos-Silva, V. Miranda, J. S. Amaral and R. Lima, Micromachines, 2019, 10, 645 CrossRef PubMed.
- S. Zhu, F. Jiang, Y. Han, N. Xiang and Z. Ni, Analyst, 2020, 145, 7103–7124 RSC.
- Y. Shen, Y. Yalikun and Y. Tanaka, Sens. Actuators, B, 2019, 282, 268–281 CrossRef CAS.
- T. W. Murphy, Q. Zhang, L. B. Naler, S. Ma and C. Lu, Analyst, 2017, 143, 60–80 RSC.
- T. R. Carey, K. L. Cotner, B. Li and L. L. Sohn, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2019, 11, e1529 Search PubMed.
- T. Luo, L. Fan, R. Zhu and D. Sun, Micromachines, 2019, 10, 104 CrossRef PubMed.
- X. Lu, M. Tayebi and Y. Ai, Electrophoresis, 2021, 42, 2281–2292 CrossRef CAS PubMed.
- N. Liu, C. Petchakup, H. M. Tay, K. H. H. Li and H. W. Hou, in Applications of Microfluidic Systems in Biology and Medicine, ed. M. Tokeshi, Springer Singapore, Singapore, 2019, pp. 99–150, DOI:10.1007/978-981-13-6229-3_5.
- Y. Sun and P. Sethu, Biomed. Microdevices, 2018, 20, 77 CrossRef PubMed.
- S. Pandey, N. Mehendale and D. Paul, in Handbook of Single-Cell Technologies, ed. T. S. Santra and F.-G. Tseng, Springer Singapore, Singapore, 2022, pp. 207–234, DOI:10.1007/978-981-10-8953-4_6.
- B. A. Sutermaster and E. M. Darling, Sci. Rep., 2019, 9, 227 CrossRef PubMed.
- C. W. T. Shields, C. D. Reyes and G. P. Lopez, Lab Chip, 2015, 15, 1230–1249 RSC.
- S. A. Faraghat, K. F. Hoettges, M. K. Steinbach, D. R. van der Veen, W. J. Brackenbury, E. A. Henslee, F. H. Labeed and M. P. Hughes, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 4591–4596 CrossRef CAS PubMed.
- J. Stam, S. Bartel, R. Bischoff and J. C. Wolters, J. Chromatogr., B, 2021, 1169, 122604 CrossRef CAS PubMed.
- M. Li, A. J. Rai, G. J. DeCastro, E. Zeringer, T. Barta, S. Magdaleno, R. Setterquist and A. V. Vlassov, Methods, 2015, 87, 26–30 CrossRef CAS PubMed.
- B. J. Tauro, D. W. Greening, R. A. Mathias, H. Ji, S. Mathivanan, A. M. Scott and R. J. Simpson, Methods, 2012, 56, 293–304 CrossRef CAS PubMed.
- T. Soares Martins, J. Catita, I. Martins Rosa, A. B. da Cruz E Silva Odete and A. G. Henriques, PLoS One, 2018, 13, e0198820 CrossRef PubMed.
- L. S. Gaspar, M. M. Santana, C. Henriques, M. M. Pinto, T. M. Ribeiro-Rodrigues, H. Girão, R. J. Nobre and L. Pereira de Almeida, Mol. Ther.--Methods Clin. Dev., 2020, 18, 723–737 CrossRef CAS PubMed.
- K. Sidhom, P. O. Obi and A. Saleem, Int. J. Mol. Sci., 2020, 21, 6466 CrossRef CAS PubMed.
- X. Fan, C. Jia, J. Yang, G. Li, H. Mao, Q. Jin and J. Zhao, Biosens. Bioelectron., 2015, 71, 380–386 CrossRef CAS PubMed.
- Y. T. Kang, I. Doh, J. Byun, H. J. Chang and Y. H. Cho, Theranostics, 2017, 7, 3179–3191 CrossRef CAS PubMed.
- A. F. Sarioglu, N. Aceto, N. Kojic, M. C. Donaldson, M. Zeinali, B. Hamza, A. Engstrom, H. Zhu, T. K. Sundaresan, D. T. Miyamoto, X. Luo, A. Bardia, B. S. Wittner, S. Ramaswamy, T. Shioda, D. T. Ting, S. L. Stott, R. Kapur, S. Maheswaran, D. A. Haber and M. Toner, Nat. Methods, 2015, 12, 685–691 CrossRef CAS PubMed.
- N. Mehendale, O. Sharma, S. Pandey and D. Paul, Biomed. Microdevices, 2018, 20, 75 CrossRef PubMed.
- A. Dalili, E. Samiei and M. Hoorfar, Analyst, 2018, 144, 87–113 RSC.
- W. Su, H. Li, W. Chen and J. Qin, TrAC, Trends Anal. Chem., 2019, 118, 686–698 CrossRef CAS.
- S. W. Lee, K. A. Hyun, S. I. Kim, J. Y. Kang and H. I. Jung, J. Chromatogr. A, 2015, 1377, 100–105 CrossRef CAS PubMed.
- R. Ozawa, H. Iwadate, H. Toyoda, M. Yamada and M. Seki, Lab Chip, 2019, 19, 1828–1837 RSC.
- H. Xia, B. C. Strachan, S. C. Gifford and S. S. Shevkoplyas, Sci. Rep., 2016, 6, 35943 CrossRef CAS PubMed.
- C. B. Raub, C. Lee and E. Kartalov, Sens. Actuators, B, 2015, 210, 120–123 CrossRef CAS.
- M. Dehghani, K. Lucas, J. Flax, J. McGrath and T. Gaborski, Adv. Mater. Technol., 2019, 4, 1900539 CrossRef CAS PubMed.
- V. Riazanski, G. Mauleon, K. Lucas, S. Walker, A. M. Zimnicka, J. L. McGrath and D. J. Nelson, Commun. Biol., 2022, 5, 13 CrossRef CAS PubMed.
- Z. Han, C. Peng, J. Yi, D. Zhang, X. Xiang, X. Peng, B. Su, B. Liu, Y. Shen and L. Qiao, Sens. Actuators, B, 2021, 333, 129563 CrossRef CAS.
- A. Hochstetter, R. Vernekar, R. H. Austin, H. Becker, J. P. Beech, D. A. Fedosov, G. Gompper, S. C. Kim, J. T. Smith, G. Stolovitzky, J. O. Tegenfeldt, B. H. Wunsch, K. K. Zeming, T. Kruger and D. W. Inglis, ACS Nano, 2020, 14, 10784–10795 CrossRef CAS PubMed.
- J. T. Smith, B. H. Wunsch, N. Dogra, M. E. Ahsen, K. Lee, K. K. Yadav, R. Weil, M. A. Pereira, J. V. Patel, E. A. Duch, J. M. Papalia, M. F. Lofaro, M. Gupta, A. K. Tewari, C. Cordon-Cardo, G. Stolovitzky and S. M. Gifford, Lab Chip, 2018, 18, 3913–3925 RSC.
- K. K. Zeming, T. Salafi, C. H. Chen and Y. Zhang, Sci. Rep., 2016, 6, 22934 CrossRef CAS PubMed.
- S. H. Au, J. Edd, A. E. Stoddard, K. H. K. Wong, F. Fachin, S. Maheswaran, D. A. Haber, S. L. Stott, R. Kapur and M. Toner, Sci. Rep., 2017, 7, 2433 CrossRef PubMed.
- H. Okano, T. Konishi, T. Suzuki, T. Suzuki, S. Ariyasu, S. Aoki, R. Abe and M. Hayase, Biomed. Microdevices, 2015, 17, 9964 CrossRef PubMed.
- T. Jing, Z. Lai, L. Wu, J. Han, C. T. Lim and C. H. Chen, Anal. Chem., 2016, 88, 11750–11757 CrossRef CAS PubMed.
- N. Tottori and T. Nisisako, Anal. Chem., 2019, 91, 3093–3100 CrossRef CAS PubMed.
- K. K. Zeming, R. Vernekar, M. T. Chua, K. Y. Quek, G. Sutton, T. Kruger, W. S. Kuan and J. Han, Small, 2021, 17, e2006123 CrossRef PubMed.
- E. Pariset, C. Parent, Y. Fouillet, B. Francois, N. Verplanck, F. Revol-Cavalier, A. Thuaire and V. Agache, Sci. Rep., 2018, 8, 17762 CrossRef CAS PubMed.
- L. R. Huang, E. C. Cox, R. H. Austin and J. C. Sturm, Science, 2004, 304, 987–990 CrossRef CAS PubMed.
- T. S. H. Tran, B. D. Ho, J. P. Beech and J. O. Tegenfeldt, Lab Chip, 2017, 17, 3592–3600 RSC.
- D. Huang, J. Man, D. Jiang, J. Zhao and N. Xiang, Electrophoresis, 2020, 41, 2166–2187 CrossRef CAS PubMed.
- Y. Gou, Y. Jia, P. Wang and C. Sun, Sensors, 2018, 18, 1762 CrossRef PubMed.
- J. Zhang, S. Yan, D. Yuan, G. Alici, N. T. Nguyen, M. Ebrahimi Warkiani and W. Li, Lab Chip, 2016, 16, 10–34 RSC.
- A. S. Rzhevskiy, S. Razavi Bazaz, L. Ding, A. Kapitannikova, N. Sayyadi, D. Campbell, B. Walsh, D. Gillatt, M. Ebrahimi Warkiani and A. V. Zvyagin, Cancers, 2020, 12, 81 CrossRef CAS PubMed.
- A. Rahi, M. Kazemi, E. Pishbin, S. Karimi and H. Nazarian, Analyst, 2021, 146, 7230–7239 RSC.
- J. M. Martel and M. Toner, Annu. Rev. Biomed. Eng., 2014, 16, 371–396 CrossRef CAS PubMed.
- A. Abdulla, T. Zhang, K. Z. Ahmad, S. Li, J. Lou and X. Ding, Anal. Chem., 2020, 92, 16170–16179 CrossRef CAS PubMed.
- J. Zhou, A. Kulasinghe, A. Bogseth, K. O'Byrne, C. Punyadeera and I. Papautsky, Microsyst. Nanoeng., 2019, 5, 8 CrossRef PubMed.
- S. Zhu, D. Wu, Y. Han, C. Wang, N. Xiang and Z. Ni, Lab Chip, 2020, 20, 244–252 RSC.
- H. M. Tay, S. Kharel, R. Dalan, Z. J. Chen, K. K. Tan, B. O. Boehm, S. C. J. Loo and H. W. Hou, NPG Asia Mater., 2017, 9, e434 CrossRef.
- H. M. Tay, S. Y. Leong, X. Xu, F. Kong, M. Upadya, R. Dalan, C. Y. Tay, M. Dao, S. Suresh and H. W. Hou, Lab Chip, 2021, 21, 2511–2523 RSC.
- B. Kim, S. S. Lee, T. H. Yoo and J. M. Kim, Electrophoresis, 2021, 42, 2238–2245 CrossRef CAS PubMed.
- C. Liu, J. Guo, F. Tian, N. Yang, F. Yan, Y. Ding, J. Wei, G. Hu, G. Nie and J. Sun, ACS Nano, 2017, 11, 6968–6976 CrossRef CAS PubMed.
- J. Nam, J. Yoon, H. Jee, W. S. Jang and C. S. Lim, Adv. Mater. Technol., 2020, 5, 2000612 CrossRef CAS.
- I. Bilican, J. Chromatogr. A, 2021, 1652, 462366 CrossRef CAS PubMed.
- M. A. Faridi, H. Ramachandraiah, I. Banerjee, S. Ardabili, S. Zelenin and A. Russom, J. Nanobiotechnol., 2017, 15, 3 CrossRef PubMed.
- D. Yuan, S. Yadav, H. T. Ta, H. Fallahi, H. An, N. Kashaninejad, C. H. Ooi, N.-T. Nguyen and J. Zhang, Electrophoresis, 2021, 42, 2230–2237 CrossRef CAS PubMed.
- T. Kumar, H. Ramachandraiah, S. N. Iyengar, I. Banerjee, G. Mårtensson and A. Russom, Sci. Rep., 2021, 11, 8467 CrossRef CAS PubMed.
- J. Zhou and I. Papautsky, Microsyst. Nanoeng., 2020, 6, 113 CrossRef CAS PubMed.
- M. Wu, A. Ozcelik, J. Rufo, Z. Wang, R. Fang and T. Jun Huang, Microsyst. Nanoeng., 2019, 5, 1–18 CrossRef PubMed.
- J. Rufo, F. Cai, J. Friend, M. Wiklund and T. J. Huang, Nat. Rev. Dis. Primers, 2022, 2, 1–21 CrossRef.
- Y. Zhou, Z. Ma and Y. Ai, RSC Adv., 2019, 9, 31186–31195 RSC.
- K. Wang, W. Zhou, Z. Lin, F. Cai, F. Li, J. Wu, L. Meng, L. Niu and H. Zheng, Sens. Actuators, B, 2018, 258, 1174–1183 CrossRef CAS.
- M. Antfolk, C. Magnusson, P. Augustsson, H. Lilja and T. Laurell, Anal. Chem., 2015, 87, 9322–9328 CrossRef CAS PubMed.
- C. Magnusson, P. Augustsson, A. Lenshof, Y. Ceder, T. Laurell and H. Lilja, Anal. Chem., 2017, 89, 11954–11961 CrossRef CAS PubMed.
- K. Cushing, E. Undvall, Y. Ceder, H. Lilja and T. Laurell, Anal. Chim. Acta, 2018, 1000, 256–264 CrossRef CAS PubMed.
- Y. Gu, C. Chen, Z. Wang, P. H. Huang, H. Fu, L. Wang, M. Wu, Y. Chen, T. Gao, J. Gong, J. Kwun, G. M. Arepally and T. J. Huang, Lab Chip, 2019, 19, 394–402 RSC.
- C. Richard, A. Fakhfouri, M. Colditz, F. Striggow, R. Kronstein-Wiedemann, T. Tonn, M. Medina-Sánchez, O. G. Schmidt, T. Gemming and A. Winkler, Lab Chip, 2019, 19, 4043–4051 RSC.
- P. Ohlsson, K. Petersson, P. Augustsson and T. Laurell, Sci. Rep., 2018, 9, 9156 CrossRef PubMed.
- C. Devendran, K. Choi, J. Han, Y. Ai, A. Neild and D. J. Collins, Lab Chip, 2020, 20, 2674–2688 RSC.
- M. Wu, Y. Ouyang, Z. Wang, R. Zhang, P. H. Huang, C. Chen, H. Li, P. Li, D. Quinn, M. Dao, S. Suresh, Y. Sadovsky and T. J. Huang, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10584–10589 CrossRef CAS PubMed.
- L. Velmanickam, V. Jayasooriya, M. S. Vemuri, U. R. Tida and D. Nawarathna, Electrophoresis, 2022, 43, 212–231 CrossRef CAS PubMed.
- B. Sarno, D. Heineck, M. J. Heller and S. D. Ibsen, Electrophoresis, 2021, 42, 539–564 CrossRef CAS PubMed.
- G. I. Russo, N. Musso, A. Romano, G. Caruso, S. Petralia, L. Lanzanò, G. Broggi and M. Camarda, Cancers, 2021, 14, 198 CrossRef PubMed.
- P. Balasubramanian, R. J. Kinders, S. Kummar, V. Gupta, D. Hasegawa, A. Menachery, S. M. Lawrence, L. Wang, K. Ferry-Galow and D. Davis, PLoS One, 2017, 12, e0175414 CrossRef PubMed.
- A. Alazzam, B. Mathew and F. Alhammadi, J. Sep. Sci., 2017, 40, 1193–1200 CrossRef CAS PubMed.
- M. Dudaie, N. Nissim, I. Barnea, T. Gerling, C. Duschl, M. Kirschbaum and N. T. Shaked, J. Biophotonics, 2020, 13, e202000151 CrossRef CAS PubMed.
- J. L. Prieto, H.-W. Su, H. W. Hou, M. P. Vera, B. D. Levy, R. M. Baron, J. Han and J. Voldman, Lab Chip, 2016, 16, 4333–4340 RSC.
- L. D'Amico, N. Ajami, J. Adachi, P. Gascoyne and J. Petrosino, Lab Chip, 2017, 17, 1340–1348 RSC.
- E. Bisceglia, M. Cubizolles, C. I. Trainito, J. Berthier, C. Pudda, O. Français, F. Mallard and B. Le Pioufle, Sens. Actuators, B, 2015, 212, 335–343 CrossRef CAS.
- K.-H. Chen, S.-H. Lee, L.-C. Kok, T.-O. Ishdorj, H.-Y. Chang and F.-G. Tseng, Biosens. Bioelectron., 2022, 197, 113740 CrossRef CAS PubMed.
- S. D. Ibsen, J. Wright, J. M. Lewis, S. Kim, S.-Y. Ko, J. Ong, S. Manouchehri, A. Vyas, J. Akers and C. C. Chen, ACS Nano, 2017, 11, 6641–6651 CrossRef CAS PubMed.
- J. M. Lewis, A. D. Vyas, Y. Qiu, K. S. Messer, R. White and M. J. Heller, ACS Nano, 2018, 12, 3311–3320 CrossRef CAS PubMed.
- M. Tayebi, D. Yang, D. J. Collins and Y. Ai, Nano Lett., 2021, 21, 6835–6842 CrossRef CAS PubMed.
- F. Yang, Y. Zhang, X. Cui, Y. Fan, Y. Xue, H. Miao and G. Li, Biotechnol. J., 2019, 14, 1800181 CrossRef PubMed.
- Y. Li, Y. Wang, K. Wan, M. Wu, L. Guo, X. Liu and G. Wei, Nanoscale, 2021, 13, 4330–4358 RSC.
- B. R. Mutlu, K. C. Smith, J. F. Edd, P. Nadar, M. Dlamini, R. Kapur and M. Toner, Sci. Rep., 2017, 7, 9915 CrossRef PubMed.
- J. Zhou, C. Tu, Y. Liang, B. Huang, Y. Fang, X. Liang, I. Papautsky and X. Ye, Sci. Rep., 2018, 8, 9411 CrossRef PubMed.
- J. Zhou and I. Papautsky, Lab Chip, 2019, 19, 3416–3426 RSC.
- M. Li, C. Ge, Y. Yang, M. Gan, Y. Xu, L. Chen and S. Li, Anal. Bioanal. Chem., 2022, 414, 7683–7694 CrossRef CAS PubMed.
- J. Che, V. Yu, M. Dhar, C. Renier, M. Matsumoto, K. Heirich, E. B. Garon, J. Goldman, J. Rao, G. W. Sledge, M. D. Pegram, S. Sheth, S. S. Jeffrey, R. P. Kulkarni, E. Sollier and D. Di Carlo, Oncotarget, 2016, 7, 12748–12760 CrossRef PubMed.
- M. Dhar, J. N. Lam, T. Walser, S. M. Dubinett, M. B. Rettig and D. Di Carlo, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 9986–9991 CrossRef CAS PubMed.
- M. Dhar, J. Wong, A. Karimi, J. Che, C. Renier, M. Matsumoto, M. Triboulet, E. B. Garon, J. W. Goldman, M. B. Rettig, S. S. Jeffrey, R. P. Kulkarni, E. Sollier and D. Di Carlo, Biomicrofluidics, 2015, 9, 064116 CrossRef PubMed.
- E. Sollier, D. E. Go, J. Che, D. R. Gossett, S. O'Byrne, W. M. Weaver, N. Kummer, M. Rettig, J. Goldman, N. Nickols, S. McCloskey, R. P. Kulkarni and D. Di Carlo, Lab Chip, 2014, 14, 63–77 RSC.
- M. S. Bhamla, B. Benson, C. Chai, G. Katsikis, A. Johri and M. Prakash, Nat. Biomed. Eng., 2017, 1, 0009 CrossRef CAS.
- M. S. Maria, P. E. Rakesh, T. S. Chandra and A. K. Sen, Sci. Rep., 2017, 7, 43457 CrossRef PubMed.
- A. Shamloo and M. Besanjideh, IEEE Trans. Biomed. Eng., 2020, 67, 372–378 Search PubMed.
- Y. Liu, W. Zhao, R. Cheng, B. N. Harris, J. R. Murrow, J. Hodgson, M. Egan, A. Bankey, P. G. Nikolinakos, T. Laver, K. Meichner and L. Mao, Lab Chip, 2021, 21, 1706–1723 RSC.
- W. Zhao, R. Cheng, S. H. Lim, J. R. Miller, W. Zhang, W. Tang, J. Xie and L. Mao, Lab Chip, 2017, 17, 2243–2255 RSC.
- W. Zhao, T. Zhu, R. Cheng, Y. Liu, J. He, H. Qiu, L. Wang, T. Nagy, T. D. Querec, E. R. Unger and L. Mao, Adv. Funct. Mater., 2016, 26, 3990–3998 CrossRef CAS PubMed.
- W. Zhao, R. Cheng, J. R. Miller and L. Mao, Adv. Funct. Mater., 2016, 26, 3916–3932 CrossRef CAS PubMed.
- M. Frenea-Robin and J. Marchalot, Magnetochemistry, 2022, 8, 11 CrossRef CAS.
- J. Zeng, Y. Deng, P. Vedantam, T.-R. Tzeng and X. Xuan, J. Magn. Magn. Mater., 2013, 346, 118–123 CrossRef CAS.
- A. R. Kose, B. Fischer, L. Mao and H. Koser, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21478–21483 CrossRef CAS PubMed.
- C.-D. Xue, J.-M. Zhao, Z.-P. Sun, J.-T. Na, Y.-J. Li and K.-R. Qin, Microfluid. Nanofluid., 2021, 25, 97 CrossRef.
- T. H. Kim, M. Lim, J. Park, J. M. Oh, H. Kim, H. Jeong, S. J. Lee, H. C. Park, S. Jung, B. C. Kim, K. Lee, M. H. Kim, D. Y. Park, G. H. Kim and Y. K. Cho, Anal. Chem., 2017, 89, 1155–1162 CrossRef CAS PubMed.
- H. Pei, L. Li, Z. Han, Y. Wang and B. Tang, Lab Chip, 2020, 20, 3854–3875 RSC.
- L. Guillou, R. Sheybani, A. E. Jensen, D. Di Carlo, T. S. Caffery, C. B. Thomas, A. M. Shah, H. T. K. Tse and H. R. O'Neal, Jr., PLoS One, 2021, 16, e0246980 CrossRef CAS PubMed.
- D. Rossi, D. Dannhauser, M. Telesco, P. A. Netti and F. Causa, Lab Chip, 2019, 19, 3888–3898 RSC.
- I. W. Schie, J. Ruger, A. S. Mondol, A. Ramoji, U. Neugebauer, C. Krafft and J. Popp, Anal. Chem., 2018, 90, 2023–2030 CrossRef CAS PubMed.
- A. A. Sina, R. Vaidyanathan, S. Dey, L. G. Carrascosa, M. J. Shiddiky and M. Trau, Sci. Rep., 2016, 6, 30460 CrossRef CAS PubMed.
- J. Su, ACS Photonics, 2015, 2, 1241–1245 CrossRef CAS.
- M. Ugele, M. Weniger, M. Stanzel, M. Bassler, S. W. Krause, O. Friedrich, O. Hayden and L. Richter, Adv. Sci., 2018, 5, 1800761 CrossRef PubMed.
- J. Yoon, Y. Jo, M. H. Kim, K. Kim, S. Lee, S. J. Kang and Y. Park, Sci. Rep., 2017, 7, 6654 CrossRef PubMed.
- Y. Yang, G. Shen, H. Wang, H. Li, T. Zhang, N. Tao, X. Ding and H. Yu, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10275–10280 CrossRef CAS PubMed.
- K. C. M. Lee, M. Wang, K. S. E. Cheah, G. C. F. Chan, H. K. H. So, K. K. Y. Wong and K. K. Tsia, Cytometry, 2019, 95, 510–520 CrossRef PubMed.
- D. H. Lee, X. Li, N. Ma, M. A. Digman and A. P. Lee, Lab Chip, 2018, 18, 1349–1358 RSC.
- J. Gala de Pablo, M. Lindley, K. Hiramatsu and K. Goda, Acc. Chem. Res., 2021, 54, 2132–2143 CrossRef CAS PubMed.
- H. W. Wang, N. Bao, T. L. Le, C. Lu and J. X. Cheng, Opt. Express, 2008, 16, 5782–5789 CrossRef CAS PubMed.
- N. Nitta, T. Sugimura, A. Isozaki, H. Mikami, K. Hiraki, S. Sakuma, T. Iino, F. Arai, T. Endo, Y. Fujiwaki, H. Fukuzawa, M. Hase, T. Hayakawa, K. Hiramatsu, Y. Hoshino, M. Inaba, T. Ito, H. Karakawa, Y. Kasai, K. Koizumi, S. Lee, C. Lei, M. Li, T. Maeno, S. Matsusaka, D. Murakami, A. Nakagawa, Y. Oguchi, M. Oikawa, T. Ota, K. Shiba, H. Shintaku, Y. Shirasaki, K. Suga, Y. Suzuki, N. Suzuki, Y. Tanaka, H. Tezuka, C. Toyokawa, Y. Yalikun, M. Yamada, M. Yamagishi, T. Yamano, A. Yasumoto, Y. Yatomi, M. Yazawa, D. Di Carlo, Y. Hosokawa, S. Uemura, Y. Ozeki and K. Goda, Cell, 2018, 175, 266–276 CrossRef CAS PubMed , e213.
- M. Lindley, J. Gala de Pablo, W. Peterson, A. Isozaki, K. Hiramatsu and K. Goda, Adv. Mater. Technol., 2022, 7, 2101567 CrossRef CAS.
- C. Zhang, K.-C. Huang, B. Rajwa, J. Li, S. Yang, H. Lin, C.-s. Liao, G. Eakins, S. Kuang, V. Patsekin, J. P. Robinson and J.-X. Cheng, Optica, 2017, 4, 103–109 CrossRef CAS.
- Y. Suzuki, K. Kobayashi, Y. Wakisaka, D. Deng, S. Tanaka, C. J. Huang, C. Lei, C. W. Sun, H. Liu, Y. Fujiwaki, S. Lee, A. Isozaki, Y. Kasai, T. Hayakawa, S. Sakuma, F. Arai, K. Koizumi, H. Tezuka, M. Inaba, K. Hiraki, T. Ito, M. Hase, S. Matsusaka, K. Shiba, K. Suga, M. Nishikawa, M. Jona, Y. Yatomi, Y. Yalikun, Y. Tanaka, T. Sugimura, N. Nitta, K. Goda and Y. Ozeki, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 15842–15848 CrossRef CAS PubMed.
- C. Rianna and M. Radmacher, AIP Conf. Proc., 2016, 1760, 020057 CrossRef.
- O. Otto, P. Rosendahl, A. Mietke, S. Golfier, C. Herold, D. Klaue, S. Girardo, S. A.-O. Pagliara, A. Ekpenyong, A. Jacobi, M. Wobus, N. Töpfner, U. F. Keyser, J. Mansfeld, E. Fischer-Friedrich and J. Guck, Nat. Methods, 2015, 12, 199–202 CrossRef CAS PubMed.
- Y. Deng, S. P. Davis, F. Yang, K. S. Paulsen, M. Kumar, R. Sinnott DeVaux, X. Wang, D. S. Conklin, A. Oberai, J. I. Herschkowitz and A. J. Chung, Small, 2017, 13, 1700705 CrossRef PubMed.
- A. Menachery, J. Sapudom, A. Vembadi, A. Alatoom, J. Teo and M. A. Qasaimeh, BioTechniques, 2020, 70, 29–36 CrossRef PubMed.
- J. Che, V. Yu, E. B. Garon, J. W. Goldman and D. Di Carlo, Lab Chip, 2017, 17, 1452–1461 RSC.
- M. Urbanska, H. E. Muñoz, J. Shaw Bagnall, O. Otto, S. R. Manalis, D. Di Carlo and J. Guck, Nat. Methods, 2020, 17, 587–593 CrossRef CAS PubMed.
- M. Kubánková, B. Hohberger, J. Hoffmanns, J. Fürst, M. Herrmann, J. Guck and M. Kräter, Biophys. J., 2021, 120, 2838–2847 CrossRef PubMed.
- K. D. Nyberg, K. H. Hu, S. H. Kleinman, D. B. Khismatullin, M. J. Butte and A. C. Rowat, Biophys. J., 2017, 113, 1574–1584 CrossRef CAS PubMed.
- C. Petchakup, H. Yang, L. Gong, L. He, H. M. Tay, R. Dalan, A. J. Chung, K. H. H. Li and H. W. Hou, Small, 2022, 18, 2104822 CrossRef CAS PubMed.
- Y. Feng, H. Chai, W. He, F. Liang, Z. Cheng and W. Wang, Small Methods, 2022, 2200325 CrossRef CAS PubMed.
- A. A.-O. Ekpenyong, N. Toepfner, C. Fiddler, M. Herbig, W. Li, G. Cojoc, C. Summers, J. Guck and E. A.-O. Chilvers, Sci. Adv., 2017, 3, 2375–2548 Search PubMed.
- J. Shaw Bagnall, S. Byun, S. Begum, D. T. Miyamoto, V. C. Hecht, S. Maheswaran, S. L. Stott, M. Toner, R. O. Hynes and S. R. Manalis, Sci. Rep., 2015, 5, 18542 CrossRef CAS PubMed.
- Y. Zhou, D. Yang, Y. Zhou, B. L. Khoo, J. Han and Y. Ai, Anal. Chem., 2018, 90, 912–919 CrossRef CAS PubMed.
- M.-E. Myrand-Lapierre, X. Deng, R. R. Ang, K. Matthews, A. T. Santoso and H. Ma, Lab Chip, 2015, 15, 159–167 RSC.
- Q. Guo, S. P. Duffy, K. Matthews, E. Islamzada and H. Ma, Sci. Rep., 2017, 7, 6627 CrossRef PubMed.
- Y. Man, D. Maji, R. An, S. P. Ahuja, J. A. Little, M. A. Suster, P. Mohseni and U. A. Gurkan, Lab Chip, 2021, 21, 1036–1048 RSC.
- D. Holmes, D. Pettigrew, C. H. Reccius, J. D. Gwyer, C. van Berkel, J. Holloway, D. E. Davies and H. Morgan, Lab Chip, 2009, 9, 2881–2889 RSC.
- A. De Ninno, R. Reale, A. Giovinazzo, F. R. Bertani, L. Businaro, P. Bisegna, C. Matteucci and F. Caselli, Biosens. Bioelectron., 2020, 150, 111887 CrossRef CAS PubMed.
- C. Petchakup, H. M. Tay, W. H. Yeap, R. Dalan, S. C. Wong, K. H. H. Li and H. W. Hou, Biosens. Bioelectron., 2018, 118, 195–203 CrossRef CAS PubMed.
- C. Petchakup, H. M. Tay, K. H. H. Li and H. W. Hou, Lab Chip, 2019, 19, 1736–1746 RSC.
- J. Zhong, Q. Tang, M. Liang and Y. Ai, Sens. Actuators, B, 2022, 367, 132068 CrossRef CAS.
- J. Zhong, M. Liang and Y. Ai, Lab Chip, 2021, 21, 2869–2880 RSC.
- S. Subramaniam, U. Ahmed, S. Benerjee, P. Jain and P. Chowdhury, 2019 11th International Conference on Communication Systems & Networks (COMSNETS), Bengaluru, India, 2019, pp. 829–832 Search PubMed.
- P. Simon, M. Frankowski, N. Bock and J. Neukammer, Lab Chip, 2016, 16, 2326–2338 RSC.
- K. Mahesh, M. Varma and P. Sen, Lab Chip, 2020, 20, 4296–4309 RSC.
- C. Honrado, L. Ciuffreda, D. Spencer, L. Ranford-Cartwright and H. Morgan, J. R. Soc., Interface, 2018, 15, 20180416 CrossRef PubMed.
- S.-I. Han and K.-H. Han, Anal. Chem., 2015, 87, 10585–10592 CrossRef CAS PubMed.
- S. A. Hosseini, M. Abdolahad, S. Zanganeh, M. Dahmardeh, M. Gharooni, H. Abiri, A. Alikhani, S. Mohajerzadeh and O. Mashinchian, Small, 2016, 12, 883–891 CrossRef CAS PubMed.
- C. Raillon, J. Che, S. Thill, M. Duchamp, B. X. E. Desbiolles, A. Millet, E. Sollier and P. Renaud, Cytometry, Part A, 2019, 95, 1085–1095 CrossRef CAS PubMed.
- J. Liu, Y. Qiang, O. Alvarez and E. Du, Sens. Actuators, B, 2018, 255, 2392–2398 CrossRef CAS PubMed.
- M. Leclercq, B. Vittrant, M. L. Martin-Magniette, M. P. Scott Boyer, O. Perin, A. Bergeron, Y. Fradet and A. Droit, Front. Genet., 2019, 10, 452 CrossRef CAS PubMed.
- P. Rajpurkar, E. Chen, O. Banerjee and E. J. Topol, Nat. Med., 2022, 28, 31–38 CrossRef CAS PubMed.
- D. McIntyre, A. Lashkaripour, P. Fordyce and D. Densmore, Lab Chip, 2022, 22, 2925–2937 RSC.
- J. Zheng, T. Cole, Y. Zhang, J. Kim and S.-Y. Tang, Biosens. Bioelectron., 2021, 194, 113666 CrossRef CAS PubMed.
- A. Isozaki, J. Harmon, Y. Zhou, S. Li, Y. Nakagawa, M. Hayashi, H. Mikami, C. Lei and K. Goda, Lab Chip, 2020, 20, 3074–3090 RSC.
- K. C. M. Lee, J. Guck, K. Goda and K. K. Tsia, Trends Biotechnol., 2021, 39, 1249–1262 CrossRef CAS PubMed.
- L. Alzubaidi, J. Zhang, A. J. Humaidi, A. Al-Dujaili, Y. Duan, O. Al-Shamma, J. Santamaría, M. A. Fadhel, M. Al-Amidie and L. Farhan, J. Big Data, 2021, 8, 53 CrossRef PubMed.
- Y. LeCun, Y. Bengio and G. Hinton, Nature, 2015, 521, 436–444 CrossRef CAS PubMed.
- J. J. Bird, J. Kobylarz, D. R. Faria, A. Ekárt and E. P. Ribeiro, IEEE Access, 2020, 8, 54789–54801 Search PubMed.
- M. Kräter, S. Abuhattum, D. Soteriou, A. Jacobi, T. Krüger, J. Guck and M. Herbig, Adv. Sci., 2021, 8, 2003743 CrossRef PubMed.
- H. Kobayashi, C. Lei, Y. Wu, C.-J. Huang, A. Yasumoto, M. Jona, W. Li, Y. Wu, Y. Yalikun, Y. Jiang, B. Guo, C.-W. Sun, Y. Tanaka, M. Yamada, Y. Yatomi and K. Goda, Lab Chip, 2019, 19, 2688–2698 RSC.
- C. Combs, D. D. Seith, M. J. Bovyn, S. P. Gross, X. Xie and Z. S. Siwy, Biomicrofluidics, 2022, 16, 014104 CrossRef CAS.
- F. Caselli, R. Reale, A. De Ninno, D. Spencer, H. Morgan and P. Bisegna, Lab Chip, 2022, 22, 1714–1722 RSC.
- C. Honrado, J. S. McGrath, R. Reale, P. Bisegna, N. S. Swami and F. Caselli, Anal. Bioanal. Chem., 2020, 412, 3835–3845 CrossRef CAS PubMed.
- N. Wang, R. Liu, N. Asmare, C.-H. Chu and A. F. Sarioglu, Lab Chip, 2019, 19, 3292–3304 RSC.
- M. Wang, J. Zhang, H. Tan, D. Chen, Y. Lei, Y. Li, J. Wang and J. Chen, Cytometry, 2022, 101, 639–647 CrossRef CAS PubMed.
- H. Tan, M. Wang, Y. Zhang, X. Huang, D. Chen, Y. Li, M.-H. Wu, K. Wang, J. Wang and J. Chen, Cytometry, 2022, 101, 630–638 CrossRef CAS PubMed.
- D. K. Singh, C. C. Ahrens, W. Li and S. A. Vanapalli, Lab Chip, 2017, 17, 2920–2932 RSC.
- K. K. Zeming, R. Vernekar, M. T. Chua, K. Y. Quek, G. Sutton, T. Krüger, W. S. Kuan and J. Han, Small, 2021, 17, 2006123 CrossRef CAS PubMed.
- D. M. D. Siu, K. C. M. Lee, M. C. K. Lo, S. V. Stassen, M. Wang, I. Z. Q. Zhang, H. K. H. So, G. C. F. Chan, K. S. E. Cheah, K. K. Y. Wong, M. K. Y. Hsin, J. C. M. Ho and K. K. Tsia, Lab Chip, 2020, 20, 3696–3708 RSC.
- J. Amann, A. Blasimme, E. Vayena, D. Frey, V. I. Madai and Q. c. the Precise, BMC Med. Inf. Decis. Making, 2020, 20, 310 CrossRef PubMed.
- B. Rozemberczki, L. Watson, P. Bayer, H.-T. Yang, O. Kiss, S. Nilsson and R. Sarkar, arXiv, 2022, preprint, arXiv:2202.05594 Search PubMed.
- A. Altmann, L. Toloşi, O. Sander and T. Lengauer, Bioinformatics, 2010, 26, 1340–1347 CrossRef CAS PubMed.
- R. R. Selvaraju, M. Cogswell, A. Das, R. Vedantam, D. Parikh and D. Batra, Proceedings of the IEEE International Conference on Computer Vision (ICCV), 2017, pp. 618–626 Search PubMed.
- M. Herbig, A. Mietke, P. Müller and O. Otto, Biomicrofluidics, 2018, 12, 042214 CrossRef CAS PubMed.
- D. Kobak and P. Berens, Nat. Commun., 2019, 10, 5416 CrossRef PubMed.
- M. W. Dorrity, L. M. Saunders, C. Queitsch, S. Fields and C. Trapnell, Nat. Commun., 2020, 11, 1537 CrossRef CAS PubMed.
- V. Rizzuto, A. Mencattini, B. Álvarez-González, D. Di Giuseppe, E. Martinelli, D. Beneitez-Pastor, M. D. M. Mañú-Pereira, M. J. Lopez-Martinez and J. Samitier, Sci. Rep., 2021, 11, 13553 CrossRef CAS PubMed.
- Y. Rivenson, H. Wang, Z. Wei, K. de Haan, Y. Zhang, Y. Wu, H. Günaydın, J. E. Zuckerman, T. Chong, A. E. Sisk, L. M. Westbrook, W. D. Wallace and A. Ozcan, Nat. Biomed. Eng., 2019, 3, 466–477 CrossRef CAS PubMed.
- X. Yi, E. Walia and P. Babyn, Med. Image Anal., 2019, 58, 101552 CrossRef PubMed.
- P. F. Virgo and G. J. Gibbs, Ann. Clin. Biochem., 2012, 49, 17–28 CrossRef CAS PubMed.
- C. Petchakup, P. E. Hutchinson, H. M. Tay, S. Y. Leong, K. H. H. Li and H. W. Hou, Sens. Actuators, B, 2021, 339, 129864 CrossRef CAS.
- C. Raillon, J. Che, S. Thill, M. Duchamp, B. X. E. Desbiolles, A. Millet, E. Sollier and P. Renaud, Cytometry, Part A, 2019, 95, 1085–1095 CrossRef CAS PubMed.
- P. Han, S. Yosinski, Z. A. Kobos, R. Chaudhury, J. S. Lee, T. M. Fahmy and M. A. Reed, ACS Nano, 2020, 14, 8646–8657 CrossRef CAS PubMed.
- U. Hassan, N. N. Watkins, B. Reddy, G. Damhorst and R. Bashir, Nat. Protoc., 2016, 11, 714–726 CrossRef CAS PubMed.
- J. Zhong, P. Li, M. Liang and Y. Ai, Adv. Mater. Technol., 2022, 7, 2100906 CrossRef CAS.
- Y. Cheng, X. Ye, Z. Ma, S. Xie and W. Wang, Biomicrofluidics, 2016, 10, 014118 CrossRef PubMed.
- M. Yamada, W. Seko, T. Yanai, K. Ninomiya and M. Seki, Lab Chip, 2017, 17, 304–314 RSC.
- H. Chen, Sci. Rep., 2018, 8, 4042 CrossRef PubMed.
- M. Wu, P. H. Huang, R. Zhang, Z. Mao, C. Chen, G. Kemeny, P. Li, A. V. Lee, R. Gyanchandani and A. J. Armstrong, Small, 2018, 14, 1801131 CrossRef PubMed.
- Y. Gu, C. Chen, Z. Wang, P.-H. Huang, H. Fu, L. Wang, M. Wu, Y. Chen, T. Gao and J. Gong, Lab Chip, 2019, 19, 394–402 RSC.
- Y. Chen, M. Wu, L. Ren, J. Liu, P. H. Whitley, L. Wang and T. J. Huang, Lab Chip, 2016, 16, 3466–3472 RSC.
- P. Ohlsson, K. Petersson, P. Augustsson and T. Laurell, Sci. Rep., 2018, 8, 1–11 CAS.
- M. Wu, C. Chen, Z. Wang, H. Bachman, Y. Ouyang, P.-H. Huang, Y. Sadovsky and T. J. Huang, Lab Chip, 2019, 19, 1174–1182 RSC.
- M. Wu, Y. Ouyang, Z. Wang, R. Zhang, P.-H. Huang, C. Chen, H. Li, P. Li, D. Quinn and M. Dao, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 10584–10589 CrossRef CAS PubMed.
- S. M. Azarin, J. Yi, R. M. Gower, B. A. Aguado, M. E. Sullivan, A. G. Goodman, E. J. Jiang, S. S. Rao, Y. Ren, S. L. Tucker, V. Backman, J. S. Jeruss and L. D. Shea, Nat. Commun., 2015, 6, 8094 CrossRef PubMed.
- M. Masaeli, D. Gupta, S. O'Byrne, H. T. K. Tse, D. R. Gossett, P. Tseng, A. S. Utada, H.-J. Jung, S. Young, A. T. Clark and D. Di Carlo, Sci. Rep., 2016, 6, 37863 CrossRef CAS PubMed.
- Y. Jiang, C. Lei, A. Yasumoto, H. Kobayashi, Y. Aisaka, T. Ito, B. Guo, N. Nitta, N. Kutsuna, Y. Ozeki, A. Nakagawa, Y. Yatomi and K. Goda, Lab Chip, 2017, 17, 2426–2434 RSC.
- T. Go, H. Byeon and S. J. Lee, Biosens. Bioelectron., 2018, 103, 12–18 CrossRef CAS PubMed.
- K. C. M. Lee, M. Wang, K. S. E. Cheah, G. C. F. Chan, H. K. H. So, K. K. Y. Wong and K. K. Tsia, Cytometry, Part A, 2019, 95, 510–520 CrossRef.
- A. A. Nawaz, M. Urbanska, M. Herbig, M. Nötzel, M. Kräter, P. Rosendahl, C. Herold, N. Toepfner, M. Kubánková, R. Goswami, S. Abuhattum, F. Reichel, P. Müller, A. Taubenberger, S. Girardo, A. Jacobi and J. Guck, Nat. Methods, 2020, 17, 595–599 CrossRef CAS PubMed.
- M. Dudaie, N. Nissim, I. Barnea, T. Gerling, C. Duschl, M. Kirschbaum and N. T. Shaked, J. Biophotonics, 2020, 13, e202000151 CrossRef CAS PubMed.
- D. Rossi, D. Dannhauser, M. Telesco, P. A. Netti and F. Causa, Lab Chip, 2019, 19, 3888–3898 RSC.
- A. A. Nawaz, M. Urbanska, M. Herbig, M. Nötzel, M. Kräter, P. Rosendahl, C. Herold, N. Toepfner, M. Kubánková, R. Goswami, S. Abuhattum, F. Reichel, P. Müller, A. Taubenberger, S. Girardo, A. Jacobi and J. Guck, Nat. Methods, 2020, 17, 595–599 CrossRef CAS PubMed.
- N. Praljak, S. Iram, U. Goreke, G. Singh, A. Hill, U. A. Gurkan and M. Hinczewski, PLoS Comput. Biol., 2021, 17, e1008946 CrossRef CAS PubMed.
- E. S. Lamoureux, E. Islamzada, M. V. J. Wiens, K. Matthews, S. P. Duffy and H. Ma, Lab Chip, 2022, 22, 26–39 RSC.
- Y. Feng, Z. Cheng, H. Chai, W. He, L. Huang and W. Wang, Lab Chip, 2022, 22, 240–249 RSC.
- H. Jeon, M. Wei, X. Huang, J. Yao, W. Han, R. Wang, X. Xu, J. Chen, L. Sun and J. Han, Anal. Chem., 2022, 94, 6394–6402 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |