Surface modification for improving immunoassay sensitivity
Xiaohu
Zhou
 and
Bo
Zheng
and
Bo
Zheng
 *
*
Institute for Cell Analysis, Shenzhen Bay Laboratory, Shenzhen 518132, China. E-mail: bozheng@szbl.ac.cn
First published on 13th January 2023
Abstract
Immunoassays are widely performed in many fields such as biomarker discovery, proteomics, drug development, and clinical diagnosis. There is a growing need for high sensitivity of immunoassays to detect low abundance analytes. As a result, great effort has been made to improve the quality of surfaces, on which the immunoassay is performed. In this review article, we summarize the recent progress in surface modification strategies for improving the sensitivity of immunoassays. The surface modification strategies can be categorized into two groups: antifouling coatings to reduce background noise and nanostructured surfaces to amplify the signals. The first part of the review summarizes the common antifouling coating techniques to prevent nonspecific binding and reduce background noise. The techniques include hydrophilic polymer based self-assembled monomers, polymer brushes, and surface attached hydrogels, and omniphobicity based perfluorinated surfaces. In the second part, some common nanostructured surfaces to amplify the specific detection signals are introduced, including nanoparticle functionalized surfaces, two dimensional (2D) nanoarrays, and 2D nanomaterial coatings. The third part discusses the surface modification techniques for digital immunoassays. In the end, the challenges and the future perspectives of the surface modification techniques for immunoassays are presented.
1. Introduction
Immunoassays are highly selective bio-affinity testing to detect the presence or measure the concentration of an analyte of interest by utilizing a biorecognition agent with a specific affinity to the target analyte.1 The first immunoassay was introduced in 1959 with radioactive labels to detect plasma insulin.2 Since then, immunoassays have become an essential method in many fields such as biomarker discovery, proteomics, drug development, and clinical diagnosis.1 Today, approximately 66% of clinical decisions are based on the results of in vitro diagnosis (IVD) testing in modern clinical practice,3 while more than 1/3 of the IVD testing is based on immunoassays, especially for diagnosing infectious diseases and cancers.4,5Great efforts have been made to improve the specificity, sensitivity, and reproducibility of immunoassays.1,6–10 In this review, we will focus on the sensitivity issue of immunoassays.1,6,11 The sensitivity of immunoassays could be evaluated by the signal-to-noise ratio. The strategy of improving the sensitivity of the immunoassay therefore involves either amplifying the signals or reducing the background noise. To amplify the signals, labeling the affinity agents with enzymes is the most effective strategy, because the enzyme label could generate many detectable signal molecules for a single binding event. With this strategy, the enzyme-linked immunosorbent assay (ELISA) is the most commonly used immunoassay technique.7,8 Various nanomaterials are also developed and utilized to amplify the signals in immunoassays.12–14 Nanomaterials with high surface-to-volume ratios could conjugate to multiple labels, such as enzymes or fluorophores. Additionally, quantum dots or polymer dots with high fluorescence efficiency and photostability could be directly used as optical labels.12 Another signal-enhanced immunoassay is the nucleic acid-based immunoassay.15,16 By utilizing the amplification power of polymerase chain reaction (PCR), immuno-PCR was introduced to detect proteins with greater sensitivity than conventional immunoassays.15–17 To reduce the background noise, the most effective strategy is decreasing the nonspecific binding, which is the major source of background noise.11,18–20 The most popular practice is to use various blocking agents to quench or block the unreacted active site on the substrate surface after the immobilization of the capture agents.
There are now many techniques to improve the sensitivity of immunoassays, and the current review focuses on the recent development of surface modification strategies to increase the sensitivity of immunoassays (Fig. 1). Immunoassays are typically performed on surfaces, such as the surface of microplates, glass slides, and microbeads. The biorecognition agent in the immunoassay is immobilized on a surface as the probe to specifically capture the target molecule, which ranges from molecules to virus particles. The surface's chemical and physical properties determine the quality and quantity of the immobilized probe and affect the specificity and sensitivity of the assay. According to the working mechanism, the surface modification strategies can be categorized into two groups: the antifouling coating strategy for background noise reduction and the nanostructured surface for signal amplification.1,6,11,20
 | ||
| Fig. 1 The scheme of the surface modification strategies for signal amplification and background noise reduction of immunoassays. | ||
This review article consists of three parts. The first part summarizes the common antifouling coating techniques to prevent nonspecific binding and reduce background noise, including hydrophilic polymer based self-assembled monomers (SAMs), polymer brushes, and surface attached hydrogels, and omniphobicity based perfluorinated surfaces. In the second part, some common nanostructured surfaces to amplify the specific detection signals are introduced, including nanoparticle functionalized surfaces, two dimensional (2D) nanoarrays, and 2D nanomaterial coatings. The third part discusses the surface modification techniques for digital immunoassays. In the end, the challenges and the future perspectives of the surface modification techniques for immunoassays are presented.
2. Antifouling coatings for background noise reduction
Proteins with a variety of amino-acid residues and conformation flexibility could adsorb onto nearly any surface via non-covalent interactions, such as hydrogen bonds, electrostatic and ionic interactions, and hydrophobic interactions.18,32,33 Researchers have developed many types of blocking agents to prevent nonspecific binding, including natural proteins such as skim milk powder and BSA, synthetic polymers such as polyethylene glycol (PEG) and poly(vinyl alcohol) (PVA), and surfactants such as Tween 20 and Triton-X100.18,34 However, there is a lack of standardized guidance to choose the suitable blocking buffer and strategy. As a result, the appropriate blocking strategies are mostly chosen empirically.35,36 Unlike the blocking strategy which aims to block the unreacted active site after capture agent immobilization, antifouling coating aims to modify the substrate surface to prevent nonspecific binding before the affinity agent immobilization, followed by an additional activated step to immobilize the capture agent to the antifouling surface. After the capture agent immobilization, the additional blocking or deactivation step is taken to restore the activated surface to be antifouling.20,33,37,38According to the molecular systems and architectures of the coating materials, we could generally divide the antifouling coating strategy into two classes: (1) the hydrated layer-based antifouling coating, including forming self-assembled monolayers (SAMs), polymer brushes, and hydrogels; and (2) the omniphobicity based perfluorinated surface (Table 1).
| Antifouling strategies | Principle | Advantages | Drawbacks | Achievement |
|---|---|---|---|---|
| ALCAM: activated leukocyte cell adhesion molecule; AFP: α-fetoprotein; AFB1: aflatoxinB1; BNP: B-type natriuretic peptide; CEA: carcinoembryonic antigen; CRP: C-reactive protein; HBsAg: hepatitis B surface antigen; hCG: human chorionic gonadotropin; HER2: human epidermal growth factor receptor 2; IL-6: interleukin 6; IL-10: interleukin 10; OPG: human osteoprotegerin; OTA: ochratoxin A; PSA: prostate-specific antigen; TNFα: tumor necrosis factor α; TSH: thyroid stimulating hormone; tTG: tissue transglutaminase; ZEN: zearalenone. | ||||
| Self-assembled monolayers (SAMs) | Hydration layer based physical and energy barrier to prevent fouling | Good biocompatibility; effective antifouling properties; convenient fabrication procedures | Antifouling effect susceptible to solvents; some SAMs susceptible to oxidative damage or hydrolysis; limited antifouling effects on complex biological milieu | α-Synuclein antibody,50 B7-H6 tumor biomarker,55 CRP,51,52 glucagon,48 insulin,48,49 IgG,53 paclitaxel and cyclophosphamide,54 somatostatin48 |
| Polymer brushes | Same as above | Good biocompatibility; effective antifouling properties on complex biological milieu | Antifouling effect susceptible to solvents; some coatings susceptible to oxidative damage or hydrolysis; complex fabrication procedures | ALCAM,67,68 AFP,60,65,66 AFB1,65 BNP,60 CEA,64,66 COVID-19,62 Ebola,61E. coli,69 HBsAg,64 hCG,68 HIV,60 IL-6,59,60 Leptin,60 OPG,59 OTA,65 PSA,60,66 Salmonella,69 SARS-COV-2 variants,63 TNFα,60 TSH,68 ZEN65 |
| Hydrogel coating | Same as above | Good biocompatibility; effective antifouling properties on complex biological milieu; 3D network for high protein loading capacity | Same as above | HER2,77 IgG,73 TSH,75 tTG78 |
| Perfluorinated surfaces | Omniphobicity | Effective antifouling properties; good stability | Limited strategy to modify limited data for immunoassay application | IL-6 and IL-10 (ref. 81–83 and 86) |
The most commonly used strategy to fabricate the substrate surface with antifouling properties is to make the surface strongly hydrated.21,32,39,40 The hydration layer would form a physical and energy barrier to prevent molecule adsorption. A wide range of molecule systems with hydrophilic or ionic groups which can form strongly hydrated layers have been employed to fabricate antifouling substrate surfaces, such as PEG and its derivatives, zwitterionic materials, peptides, polysaccharides, and other polymers.11,21,33
PEG and its derivatives are the most widely used antifouling materials with the advantages of high water solubility, superior biocompatibility, and non-toxicity. Since the 1970s, PEG and its derivatives have been employed for deposition on surfaces to endow the surfaces with antifouling properties.21,22,41 However, the major drawback is that PEG and its derivatives would be auto-oxidized when oxygen and transition metals are present, which would make the PEGylated surfaces unstable and lose the antifouling properties.42,43 In the past decades, because of their high oxidative resistance and hydrolytic stability, zwitterionic materials such as carboxybetaine (CB), sulfobetaine (SB), and phosphorylcholine (PC) have been investigated extensively as promising substitutes to PEG in developing antifouling surfaces.11,21,22 Zwitterionic polymer materials contain equally oppositely charged moieties, and these charged moieties could form a more stable and thicker hydration layer on the surface via electrostatic interactions, which could provide a comparable or even better antifouling effect than PEGylated surfaces.44,45
Practically, these coating polymers could be deposited on surfaces either by “grafting to” or “grafting from” methods.11,21 Polymer coatings could be grafted to the surface either by physisorption or chemisorption.46–49 Although the “grafting to” strategy is a more convenient fabrication method and could provide a moderate antifouling effect, it is difficult to graft polymer coatings with high density and thickness due to steric hindrance between polymer chains, which could weaken the antifouling properties of the polymer layer for the incomplete cover.33,50 Alternatively, the “grafting from” strategy is a bottom-up method, and the polymer chains are grown via surface-initiated controlled radical polymerization from a functionalized surface.24,51 In this way, the composition, architecture, and overall properties of the polymer brush could be precisely controlled, so that the grafting density, coating thickness, and hydrophilicity of the polymer can be enhanced to present better antifouling properties.
In addition to the hydrated layer-based antifouling coatings, omniphobic perfluoropolymers provide another type of antifouling coating with a different working mechanism.21,22 More specifically, the hydrated antifouling surfaces exhibit fouling-resistant properties with the hydrated water layers on top of the surfaces resisting the foulant to attach, while the perfluorinated surface shows fouling-release properties with minimum interaction between the foulants and surfaces, and the attached foulants could be easily released from the surfaces.22,52
2.1 Self-assembled monolayers (SAMs)
Self-assembled monolayers (SAMs) are highly ordered molecular assemblies formed spontaneously on surfaces. Whitesides and co-workers performed a systematic study on the antifouling properties of structure–property relationships with various SAMs, and proposed the “Whitesides rules” of antifouling SAMs: 1) the presence of hydrophilic groups, 2) the presence of hydrogen-bond acceptors but not hydrogen-bond donors, 3) overall electrically neutral.41,48,53–55 Oligo(ethylene glycol)-terminated SAMs (OEG-SAM)56–60 and zwitterionic SAMs61–63 are the most widely used antifouling SAMs for immunoassays.Recently, Castiello et al. introduced a multiplex surface plasmon resonance (SPR) immunoassay based on PEG-based SAMs to simultaneously quantify three human pancreatic islet hormones: insulin, glucagon, and somatostatin.56 Their work utilized the mixed self-assembly monolayer of thiolated polyethylene glycol (CH3O-PEG-SH) and 16-mercaptohexadecanoic acid (MHDA) with a molar ratio of 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 as the substrate surface for the immunoassay. The thiolated PEG served as the antifouling agent to prevent nonspecific adsorption, while the MHDA with the carboxyl end group was used for hormone immobilization (Fig. 2A). The antibody functionalized spots on the SAM showed high specificity to the corresponding hormones without notable interference from other nonspecific proteins due to the excellent antifouling properties, and the limit of detection (LOD) of the hormones in multiplex mode could reach 5.81 ng mL−1 (1 nM) for insulin, 13.93 ng mL−1 (4 nM) for glucagon, and 402.7 ng mL−1 (246 nM) for somatostatin (Fig. 2A).56 Davis and co-workers reported a series of PEG-based SAMs to construct ultrasensitive electrochemical and label-free biosensors to quantify insulin,57 the α-synuclein antibody,58 and C-reactive protein.59,60 Thiolated PEG (HS-(CH2)11-(EG)3-OCH2-COOH) was self-assembled as the supporting substrate surface and simultaneously provided the antifouling effect and the antibody anchoring layer. All of these OEG-SAM based biosensors showed picomolar sensitivity to quantify target proteins.57–60 For example, the biosensor for insulin quantification exhibited a clinically relevant linear range from 29.04 pg mL−1 (5 pM) to 290.4 pn mL−1 (50 nM) with an LOD of 6.97 pg mL−1 (1.2 pM) in 50% serum,57 while the biosensor for α-synuclein antibody quantification showed a dynamic linear range of 8.0–160 ng mL−1 (0.5–10 nM) and an LOD of 0.88 ng mL−1 (55 pM).58 Furthermore, the SAM-based biosensor can be regenerated and reused by immersion in buffer to disassociate the antibody–antigen complex.57,58
2 as the substrate surface for the immunoassay. The thiolated PEG served as the antifouling agent to prevent nonspecific adsorption, while the MHDA with the carboxyl end group was used for hormone immobilization (Fig. 2A). The antibody functionalized spots on the SAM showed high specificity to the corresponding hormones without notable interference from other nonspecific proteins due to the excellent antifouling properties, and the limit of detection (LOD) of the hormones in multiplex mode could reach 5.81 ng mL−1 (1 nM) for insulin, 13.93 ng mL−1 (4 nM) for glucagon, and 402.7 ng mL−1 (246 nM) for somatostatin (Fig. 2A).56 Davis and co-workers reported a series of PEG-based SAMs to construct ultrasensitive electrochemical and label-free biosensors to quantify insulin,57 the α-synuclein antibody,58 and C-reactive protein.59,60 Thiolated PEG (HS-(CH2)11-(EG)3-OCH2-COOH) was self-assembled as the supporting substrate surface and simultaneously provided the antifouling effect and the antibody anchoring layer. All of these OEG-SAM based biosensors showed picomolar sensitivity to quantify target proteins.57–60 For example, the biosensor for insulin quantification exhibited a clinically relevant linear range from 29.04 pg mL−1 (5 pM) to 290.4 pn mL−1 (50 nM) with an LOD of 6.97 pg mL−1 (1.2 pM) in 50% serum,57 while the biosensor for α-synuclein antibody quantification showed a dynamic linear range of 8.0–160 ng mL−1 (0.5–10 nM) and an LOD of 0.88 ng mL−1 (55 pM).58 Furthermore, the SAM-based biosensor can be regenerated and reused by immersion in buffer to disassociate the antibody–antigen complex.57,58
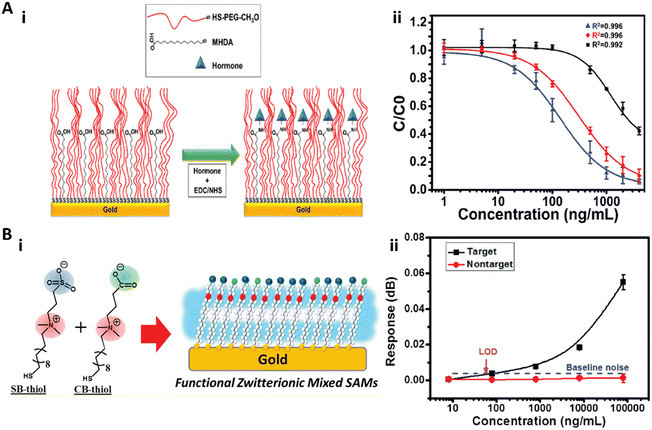 | ||
| Fig. 2 A-i) The schematic illustration of the mixed SAM formed on the Au film. The thiolated polyethylene glycol (CH3O-PEG-SH) in the mixed SAM served as the spacer and the antifouling agent to prevent the nonspecific adsorption, while the 16-mercaptohexadecanoic acid (MHDA) was used for hormone immobilization. A-ii) Multiplex hormone calibration curves of the SPR sensor for insulin (blue), glucagon (red), and somatostatin (black). Reproduced from ref. 56, Copyright 2018 American Chemical Society. B-i) The schematic illustration of the functional zwitterionic mixed SAMs with sulfobetaine-thiol (SB-thiol) and carboxybetaine-thiol (CB-thiol). B-ii) Calibration curve for detection of the target and nontarget secondary antibodies with the SPR biosensor. Reproduced from ref. 61, Copyright 2019 American Chemical Society. | ||
Huang and co-workers reported a mixed zwitterionic antifouling SAM with SB-thiols and CB-thiols for the immunoassay in an SPR biosensor (Fig. 2B).61 They demonstrated that the SB-thiol SAM with higher surface hydrophilicity and packing density exhibited an overall better antifouling effect than the CB-thiol SAM due to the ionic association of CB moieties, while CB-thiol with a carboxylate group was more suitable for protein immobilization. Therefore, the zwitterionic mixed SAM with 10% CB-thiol in mole fraction was utilized to detect the rabbit polyclonal IgG on an SPR biosensor. This biosensor exhibited high sensitivity with a LOD of 55.8 ng mL−1 and high specificity when the nontarget chicken polyclonal IgG was present in the sample (Fig. 2B).61
2.2 Polymer brushes
Although SAMs have shown good antifouling effects with buffer solutions or diluted complex media, the low surface density and thickness of the SAMs limit their antifouling performance with the complex biological milieu.33 Polymer brushes provide an alternative option, which are thin polymeric films of tightly packed polymer chains with one end attached to a surface. Polymer brushes fabricated by the method of “grafting from” a surface have been most commonly used in recent years to obtain dense and thick polymer brushes for the antifouling effect.11,24A variety of polymer brushes have been synthesized to construct antifouling surfaces, PEG-based poly(oligo(ethylene glycol) methyl ether methacrylate) (POEGMA), hydroxy-containing poly(N-(2-hydroxypropyl)methacrylamide) (PHPMA) and zwitterionic polymer brushes (PSBMA, PCBMA, PCBAA, etc.) are most commonly used brushes to construct antifouling surfaces for immunoassays.11,24,64,65
Chilkoti and co-workers first introduced POEGMA brushes as antifouling surfaces to construct antibody microarrays to quantify proteins in serum and blood with femtomolar sensitivity.66,67 Hucknall et al. invented a simple noncovalent immobilization strategy to fabricate antibody microarrays by inkjet printing the antibodies onto the dried POEGMA brush.67 Then, based on the simple strategy to fabricate antibody microarrays, Chilkoti and co-workers developed the POEGMA brush-based self-contained immunoassay (the “D4 assay”) to convert the sandwich immunoassay to a point-of-care test (POCT).68–71 In the D4 assay chip, the POEGMA brushes were grown from glass by surface-initiated atom transfer radical polymerization (SI-ATRP). The capture antibodies were then immobilized by noncontact inkjet printing on the dried POEGMA brushes, while the fluorophore-labelled detection antibodies were printed on the hydrated brushes near the spots of the capture antibody.68 Recently, based on the D4 assay platform, Heggestad et al. developed a multiplexed portable COVID-19 serological assay with the double-antigen (DA) bridging format to quantify the antibody response to three different SARS-CoV-2 antigens (Fig. 3). The nucleocapsid (N), spike S1 domain (S1), and the receptor binding domain (RBD) of S1 were printed as the stable capture antigens, and the fluorescence conjugated S1 and N-terminal domain (NTD) of N were printed nearby on the trehalose pad as the dissolvable detection antigens. When a sample is dispensed, the detection antigens would be re-dissolved in the sample solution. The antibodies in the sample specifically bind to the capture and detection antigens and form the sandwich complex with the fluorescence signals (Fig. 3A). The authors validated the DA-D4 assay platform with two formats, open format DA-D4 (Fig. 3B and E) and microfluidic DA-D4 (Fig. 3C, F, and G). With the homemade fluorescence detector D4Scope (Fig. 3D), the microfluidic DA-D4 could quantify the SARS-CoV-2 antibodies in 60 min.70
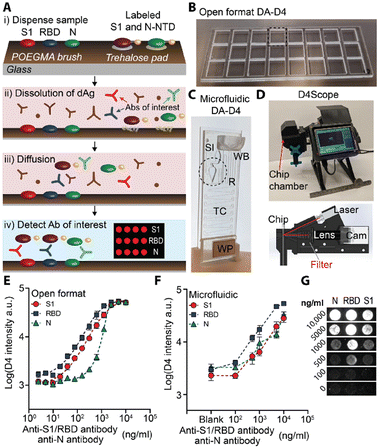 | ||
| Fig. 3 A) Schematic illustration of the DA-D4 assay. B and C) Photographs of (B) the open format DA-D4 assay with 24 individual assays and (C) microfluidic DA-D4. SI: Sample inlet; WB: wash buffer; TC: timing channel; WP: wicking pad. D) D4Scope and the schematic illustration of the optical path of D4Scope. E and F) Dose-response curves to the SARS-CoV-2 antibody from € the open format DA-D4 assay and (F) microfluidic DA-D4 with antibody spiked undiluted human serum. G) Fluorescence images with concentration of the SARS-CoV-2 antibody from the microfluidic DA-D4 assay with D4Scope. Reproduced from ref. 70, Copyright 2021 American Association for the Advancement of Science. | ||
Li and co-workers fabricated a series of ultrasensitive copolymer brush-based immunoassays by copolymerizing the glycidyl methacrylate (GMA) monomer with oligo(ethylene glycol) methacrylate (OEGMA).72–74 GMA in the POEGMA-co-GMA brush would provide anchor sites to immobilize proteins and enhance the loading stability and capacity.72 Recently, Hu et al. fabricated a hierarchically nanostructured substrate for an ultrasensitive immunoassay by growing the POEGMA-co-GMA brush on a zinc oxide (ZnO) nanorod array (Fig. 4A-i).74 The capture antibodies could be immobilized on the copolymer brushes with high stability and density due to the GMA moiety while preventing the nonspecific binding because of the PEG chains. Furthermore, the underlying ZnO nanorods could largely amplify the fluorescence signals. To validate this hybrid substrate for the sensitive immunoassay, the cancer biomarker CEA in human serum was quantified and the LOD could reach as low as 100 fg mL−1 (Fig. 4A-ii).74
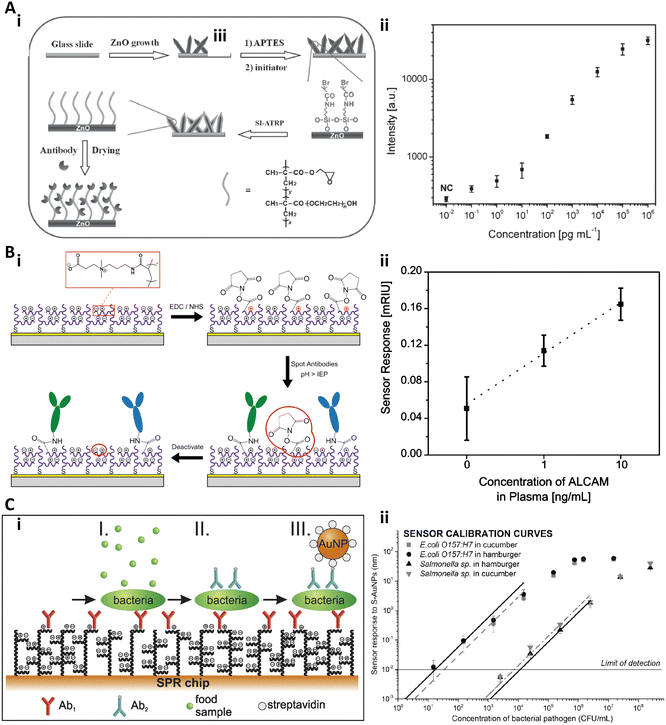 | ||
| Fig. 4 A-i) Schematic illustration of the fabrication of the ZnO@POEGMA-co-GMA substrate with the antifouling and fluorescence enhancement properties; A-ii) Dose responses of CEA quantification with the immunoassay on the ZnO@POEGMA-co-GMA substrate. Reproduced from ref. 74, Copyright 2015 Wiley. B-i) Schematic illustration of the immobilization strategy on pCB brushes. B-ii) Dose responsive curve for ALCAM spiked in undiluted human plasma. Reproduced from ref. 76, Copyright 2013 American Chemical Society. C-i) Schematic illustration of the three-step assay to detect bacterial pathogens in crude food samples. C-ii) Calibration curves of E. coli O 157:H7 and Salmonella sp. in cucumber and in hamburger. Reproduced from ref. 77, Copyright 2016 Elsevier. | ||
Jiang and co-workers systematically studied the antifouling properties of some zwitterionic polymer brushes and developed a series of zwitterionic brush based biosensors.44,49,75,76 Brault et al. reported a pCB brush based SPR imaging sensor for multiplexed profiling of protein targets (Fig. 4B).76 The pCB brushes were grafted from a gold substrate via SI-ATRP. Then, the antibody against the activated leukocyte cell adhesion molecule (anti-ALCAM), the antibody against thyroid stimulating hormone (anti-TSH), and the antibody against β-human chorionic gonadotropin (anti-hCG) were spotted on the pCB brushes via microcontact printing with the conventional coupling chemistry (Fig. 4B-i). The authors found that the functionalization and detection performance of such a substrate increased with the pH and antibody concentration, and the pCB brushes exhibited an excellent antifouling effect both before and after antibody immobilization. The LOD of this pCB platform to quantify ALCAM in undiluted human plasma was 1 ng mL−1 (Fig. 4B-ii).76 Homola and co-workers presented another SPR biosensor based on antifouling pCBAA polymer brushes to detect bacterial pathogens in crude food samples.77 The pCBAA brushes were grown on a gold substrate via SI-ATRP and then the capture antibodies were immobilized on the brushes with NHS/EDC coupling chemistry. The bacterial pathogen detection included three steps: bacteria were captured by the immobilized anti-bacteria antibodies (I), followed by labelling with biotinylated secondary antibodies (II) and conjugating with streptavidin-coated spherical gold nanoparticles (III) (Fig. 4C-i). The LOD was 57 colony-forming unit (CFU) mL−1 in cucumber and 17 CFU mL−1 in hamburger for E. coli O 157:H7, while 7.4 × 103 CFU mL−1 and 11.7 × 103 CFU mL−1 for Salmonella (Fig. 4C-ii).77
2.3 Hydrogel coating
Coating surfaces by a hydrogel layer is an effective strategy to achieve protein resistance in biologically complex fluids. Hydrogels are crosslinked hydrophilic polymer networks that can absorb large amounts of water. With the intrinsic biological inert and antifouling properties, hydrogels have been widely used for biosensors.78–80 Similar to the SAMs and polymer brushes, the PEG and zwitterionic based hydrogels are commonly used for immunoassays.81–86Yang and co-workers reported a crosslinked cylinder gel array based 3D immunoassay with an antifouling polymer brush as a background (Fig. 5A).81 First, poly(ethylene glycol) methyl methacrylate (PEGMMA) polymer brushes were grafted onto the surface to construct an antifouling background, and then the functional cylinder arrays with reactive epoxy groups were crosslinking copolymerized with glycidyl methacrylate (GMA) and polyethylene glycol diacrylate (PEGDA). The abundant reactive epoxy groups in the cylinder arrays could efficiently immobilize antibodies with high capacity. Then, the functionalized 3D cylinder gel arrays on the antifouling PEG brushes could serve as the immunoassay substrate to quantify protein targets (Fig. 5A-i). The authors demonstrated that this gel on the polymer brush substrate could detect and quantify the IgG in human plasma (Fig. 5A-ii–iv).81 Recently, by utilizing the C–H-insertion reaction-based strategy, Scherag et al. developed a blocking-free and substrate-independent serological microarray immunoassay based on a surface-attached hydrogel.86 This antifouling hydrogel based blocking-free microarray showed 2.5-fold higher sensitivity than the BSA-blocked surface.86
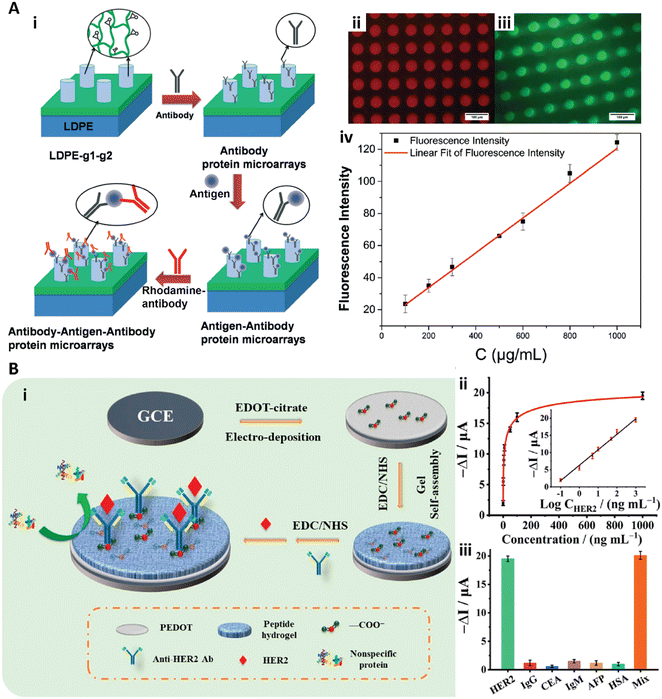 | ||
| Fig. 5 A-i) Schematic illustration of the sandwich immunoassay to detect IgG in human serum with the cylinder PEGDA-GMA gel array. A-ii, iii) Fluorescence images of RBITC labeled anti IgG (ii) and FITC-labeled anti IgG (iii). A-iv) Calibration curve with different IgG concentrations. Reproduced from ref. 81, Copyright 2014 Royal Society of Chemistry. B-i) Schematic illustration of the peptide hydrogel-based biosensor to detect HER2. B-ii) Dose response signal change of the peptide hydrogel biosensor to HER2 and the calibration curve (inset). B-iii) Response of the biosensor to HER2 with 0.1 mg mL−1 interference protein of IgG, CEA, IgM, AFP, and HSA, and a mixed solution (mix). Reproduced from ref. 85, Copyright 2021 American Chemical Society. | ||
Recently, Wang et al. introduced antifouling zwitterionic peptide hydrogel based electrochemical biosensors for ultra-sensitive quantification of human epidermal growth factor receptor 2 (HER2) in human blood (Fig. 5B).85 In their work, the antifouling peptide hydrogel was fabricated on the conducting polymer poly(3,4-ethylene dioxythiophene) (PEDOT) film. The PEDOT film served as a stable and conducting substrate for signal transduce. The peptide hydrogel consisted of the short peptide of Phe-Glu-Lys-Phe functionalized with a fluorene methoxycarbonyl group (Fmoc-FEKF) and the hydrophilicity of the hydrogel provided the antifouling properties. The anti-HER2 antibodies were immobilized on the peptide hydrogels with the EDC/NHS coupling chemistry for HER2 detection (Fig. 5B-i). This peptide hydrogel based biosensor exhibited high sensitivity and selectivity and quantified HER2 with a large linear range from 0.1 ng mL−1 to 1.0 μg mL−1 and the LOD of 45 pg mL−1 (Fig. 5B-ii and iii).85
2.4 Perfluorinated surfaces
Unlike the antifouling hydration layer, the perfluorinated surface is both hydrophobic and oleophobic, making it inert to most compounds. The perfluorinated coating can be produced by the combination of oxygen plasma and perfluorosilane or by CYTOF™ treatment.26–28,30,31,87 The perfluorinated surfaces is antifouling due to the low affinity interactions between most molecules and the surface. We expect the perfluorinated coating to be stable for the same reason. In contrast, the hydrophilic material-based coating strategies, including the SAMs, polymer brushes, and hydrogel coating, utilize the hydration layer as the physical and energy barrier to prevent fouling. The hydration layer-based coatings are sensitive to the aqueous solvent, which could alter the antifouling behaviour of the coatings. Furthermore, some hydration layer-based coatings may detach and degraft from the surfaces due to the hydrolysis of siloxane, amide, or ester bonds, or cleavage of Au–S in the hydration layer. Additionally, some hydrophilic polymers, such as PEG and peptide-based materials, would suffer from oxidative damage.11,21–24 Fluorine-based coatings have been used as antifouling materials for many years in industry applications.22,30 However, due to the difficulty of modification, the perfluorinated surfaces were not considered as the substrate for sensing until recently.25–28The situation of perfluorinated surfaces changed with the introduction of polydopamine (PDA) as a versatile polymer for surface modification.88–90 Due to their unique properties such as extraordinary adhesiveness even to the perfluoropolymer surface, excellent biocompatibility and convenient synthesis and coating conditions, PDA coatings are widely used in many chemical and biomedical applications.25–29,91,92
Recently, our group developed a series of perfluorinated surface-based ultralow background substrates for immunoassays.25–28 First, a PDA microspot array was formed on a perfluorinated surface by incubating the perfluorinated surface in dopamine solution with microfluidic technologies. The polymerization of the dopamine to form the PDA spots involves the oxidation of the dopamine to 5,6-dihydroxyindole, and the self-assembly of the dopamine and 5,6-dihydroxyindole through hydrogen bonding and π-stacking.88,91 Thereafter, the as-formed PDA patterned perfluorinated surface could be applied as an ultralow background substrate for standard immunoassays (Fig. 6A).26 The capture antibody could be immobilized on the PDA spot via Michael addition or Schiff-base reactions, while the omniphobic fluorinated surface would prevent the nonspecific protein adsorption. After protein immobilization, a blocking buffer was utilized to block the unreacted anchor sites on the PDA spots. The PDA patterned perfluoropolymer-based substrate was validated to quantify cytokine with the sandwich immunoassay format (Fig. 6A).26 Additionally, we have found that the protein immobilization density could be increased by tuning the morphology and roughness of the PDA spots, thus improving the sensitivity of the immunoassays.26
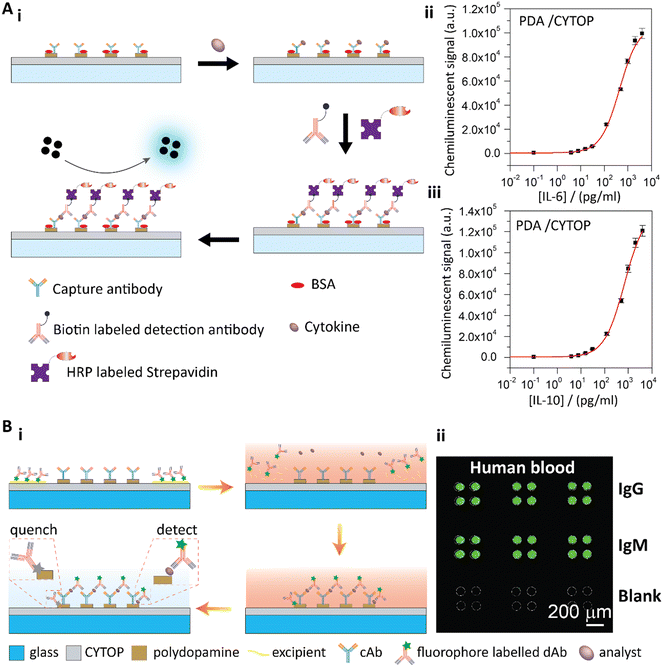 | ||
| Fig. 6 A-i) Schematic illustration of the sandwich cytokine immunoassay. A-ii and iii) Standard curves of IL-6 (ii) and IL-10 (iii) detection with the PDA patterned perfluorinated substrates. Reproduced from ref. 26, Copyright 2019 Elsevier. B-i) Schematic illustration of the PDA patterned perfluorinated surface-based self-contained blocking-free immunoassay platform for one-step POCT. B-ii) One-step testing of IgG and IgM from human blood on the self-contained, blocking-free POCT platform. Reproduced from ref. 27, Copyright 2020 Elsevier. | ||
To further utilize the unique properties of PDA and the perfluoropolymer, we developed a blocking-free and self-contained immunoassay platform with one-step POCT ability (Fig. 6B).27 In our work, we used a fluorophore-labelled detection antibody in a sandwich immunoassay format to generate the fluorescence signals on PDA. We observed that PDA effectively quenched the fluorescence93 of the nonspecifically adsorbed detection antibody, while the fluorescence of the detection antibody from the sandwich immunoassay was not affected due to the distance between the antibody and the PDA surface. Therefore, the conventional blocking step was no longer necessary. Furthermore, the perfluoropolymer surface showed unique fouling-release properties due to the ultralow surface energy of the perfluoropolymer molecules. Therefore, the detection antibody with other reagents could be temporally stored on the perfluoropolymer surface. When a sample solution was added to the surface, the temporally stored reagents would re-dissolve in the sample solution to conjugate the target molecules, and then the conjugated complexes would be captured by the immobilized capture antibodies. After a simple washing step to remove the remaining sample solution, the fluorescence signals of the sandwich immunoassay could be detected (Fig. 6B-i). By combining the PDA coating and perfluorinated surface, we achieved a blocking-free and self-contained immunoassay for one-step POCT.27 To demonstrate this blocking-free and self-contained immunoassay platform for one-step POCT, IgG and IgM solutions spiked into PBS and FBS solutions were quantitatively analyzed. The detection limit was 0.57 ng mL−1 for IgG and 0.83 ng mL−1 for IgM in PBS solutions on the blocking-free substrates, and the dynamic ranges were in three orders of magnitude from about several ng mL−1 to several μg mL−1. The blocking-free and self-contained substrate showed the best sensitivity compared to the BSA blocked substrates and the commercial microplates. With the excellent antifouling properties of the perfluorinated surface, this blocking-free, self-contained and one-step POCT immunoassay platform could directly detect the analytes from human blood (Fig. 6B-ii).
3. Nanostructured substrate surfaces for signal enhancement
With the development of nanomaterials and nanoengineering techniques in the past decades, nanostructured surfaces have been widely used in biosensors for highly sensitive sensing.14,94–103 Generally, the nanostructured surface would enhance the sensing sensitivity through two ways. First, the nanostructured surface would provide more anchor sites for capture agent immobilization with high capacity, which will enhance the capture efficiency.14,104–109 For example, Zhang et al. designed a microfluidic chip with a 3D herringbone nanopatterned surface to detect tumor-associated exosomes in plasma.109 This nanostructured surface in the microchip significantly increased the detection sensitivity to 10 exosomes per μL in plasma by not only increasing the surface area and probe density but also promoting microscale mass transfer.109 Secondly, the nanostructured surface would enhance the binding associated optical or electric signals due to the surface plasmonic properties of the nanomaterials. For example, localized surface plasmon resonance (LSPR)110 greatly enhances the electric fields near the surface and generates a maximum optical absorption at the plasmon resonance frequency.110 Furthermore, these enhanced electric fields could dramatically amplify the external spectroscopic and fluorescence signals of the immunoassays, leading to surface-enhanced Raman scattering (SERS),96,111 surface-enhanced infrared absorption spectroscopy (SEIRAS),103 and surface-enhanced fluorescence (SEF).98,112According to the structure, we classify the signal-enhanced nanostructured surfaces into three types: the nanoparticle functionalized surfaces,113–117 the surfaces with ordered 2D nanoarrays,118–129 and the surfaces coated with 2D nanomaterials.130–133
3.1 Nanoparticle functionalized surfaces
The functionalization of nanoparticles on a surface is extensively used for highly sensitive immunoassays.113–116,134–136 Sen and co-workers developed a silver nanoparticle (AgNP) functionalized surface based LSPR biosensor for detection of the dengue NS1 antigen in whole blood.113 A randomly arranged AgNP array was fabricated by thermal annealing of a thin silver film on a silicone substrate (Fig. 7A-i) and the as prepared biosensor showed a refractive index (RI) sensitivity of 10−3, which could be used to detect protein binding. By integrating a blood-plasma separation membrane to form a microfluidic chip, the biosensor could rapidly quantify the dengue NS1 antigen from only 10 μL of whole blood in 30 min. This biosensor for dengue NS1 antigen detection gave a linear range from 0.5 μg mL−1 to 50 μg mL−1 with a LOD of 0.06 μg mL−1 (Fig. 7A-ii).113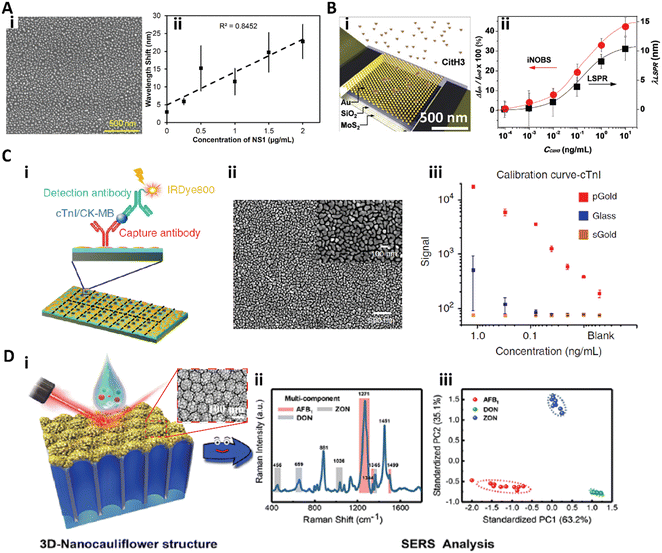 | ||
| Fig. 7 A-i) SEM image of the silver nanoparticle functionalized surface; A-ii) calibration curve for dengue NS1 antigen detection in whole human blood. Reproduced from ref. 113, Copyright 2019 Elsevier. B-i) Scheme of the iNOBS architecture; B-ii) calibration curves for CitH3 detection from the iNOBS and the LSPR spectroscopy setup. Reproduced from ref. 114, Copyright 2019 Wiley. C-i) Scheme of antigen (cTnI: cardiac troponin I; CK-MB: creatine kinase isoenzyme MB) detection on the plasmonic gold nano-island (pGold) chip. C-ii) SEM image of the pGold chip. C-iii) Calibration curves of cTnI quantification with pGold (red), glass (blue), and sputter gold (sGold, yellow) chips. Reproduced from ref. 115, Copyright 2020 Springer Nature. D-i) Scheme of SERS measurement with the 3D-nanocauliflower substrate. D-ii) SERS spectra of multiplex detection of three mycotoxins (AFB1, ZON, and DON) in maize. D-iii) Principal component analysis (PCA) plot computed from the SERS spectra (iii). Reproduced from ref. 116, Copyright 2016 American Chemical Society. | ||
Park et al. developed an integrated nano-optoelectronic biosensor (iNOBS) to detect citrullinated histone H3 (CitH3), an infection biomarker accompanying cell death in neutrophils (Fig. 7B).114 The iNOBS combined the nanoscale plasmonic and photoelectronic effects, which consisted of a hexagonal array of hemispherical gold nanoparticles (AuNPs) on a thin silicon dioxide film as the nanoplasmonic optical filter, a multilayer MoS2 photoconductive channel under the nanostructured film, and a microfluidic well on the top (Fig. 7B-i). The iNOBS could rapidly detect CitH3 in 20 min with only 2.5 μL sample, providing a large dynamic range from 10−4 to 10 ng mL−1 with a LOD of 0.87 pg mL−1 (56 fM), which is 250-fold higher sensitivity than that of the commercial ELISA (Fig. 7B-ii).114
Xu et al. introduced a gold nanostructure functionalized plasmonic chip for diagnosis and monitoring of myocardial infarction (Fig. 7C).115 The plasmonic gold nano-island (pGold) consists of discontinuous gold nano-islands with a size of 80–200 nm and a pitch of 10–30 nm (Fig. 7C-ii). The pGold chip could enhance the near-infrared fluorescence up to 130-fold in fluorescence immunoassay detection due to the enhanced SPR produced by the gold nano-islands, while the continuous gold film (sGold) would quench the fluorescence. For the detection of cardiac troponin I (cTnI), a biomarker of myocardial infarction, the pGold chip provided a dynamic range of 0.01–1.20 ng mL−1 with a LOD of 0.0100 ng mL−1 and a limit-of-quantification (LOQ) of 0.057 ng mL−1 (Fig. 7C-iii). For clinical practice, the diagnostic sensitivity and specificity of the pGold chip assay for myocardial infarction diagnosis were significantly better than those of the standard chemiluminescence assay.115
To further improve the density and intensity of the plasmonic hot spots, Han and co-workers fabricated a 3D-nanocauliflower SERS substrate for multiplex mycotoxin detection (Fig. 7D).116 This 3D-nanocauliflower substrate was fabricated by sputtering gold nanoparticles on a polydimethylsiloxane coated anodic aluminium oxide complex substrate (Fig. 7D-i), and the as-prepared 3D nanostructured surface provided tremendous contact area and hot spots. This 3D-nanocauliflower SERS substrate was successful in simultaneously detecting three common mycotoxins in maize, aflatoxin B1 (AFB1), deoxynivalenol (DON), and zearalenone (ZON), and provided a LOD of 1.8, 47.7, and 24.8 ng mL−1, respectively (Fig. 7D-ii and iii).116
3.2 2D nanoarray
Nanopatterns with repeated units arranged in an ordered array, such as a nanopillar array, nanoantenna array, nanohole array, and nanoslit array, are known as nanoarrays or nanolattices.118,119 The ordered nanopatterns with specifically designed sizes and shapes could generate a localized surface plasmon and surface-enhanced field, which could provide ultrasensitive immunosensing.118–120,122–129Zhou and co-workers fabricated a nanopillar array based nanostructured substrate for sensitive detection of prostate specific antigen (PSA) (Fig. 8A).128 This nanopillar array with a size of 140 nm × 140 nm and a gap of 400 nm was fabricated by nanoimprinting onto a glass substrate (Fig. 8A-i), which could serve as the LSPR generator to enhance the fluorescence intensity of the fluorophore within an optimal distance of 10–15 nm. Therefore, a sandwich immunoassay with quantum dots (QDs) as fluorescent labels was employed on this nanostructured substrate to detect the PSA (Fig. 8A-ii). The QD bioassay on the nanopillar array for PSA detection could reach an LOD of 10 pg mL−1 with 100 ms integration time (Fig. 8A-iii). Interestingly, at the same time, the authors also fabricated a nanohole array based substrate with similar size for PSA detection, while the same QD bioassay on this nanohole array based substrate showed worse detection sensitivity due to the lower plasmonic field intensity and less surface and volume coverage of the QDs.128 Zang et al. reported a nanoantenna array based biosensor which detected Ebola virus with a notable LOD of 220 fg mL−1.
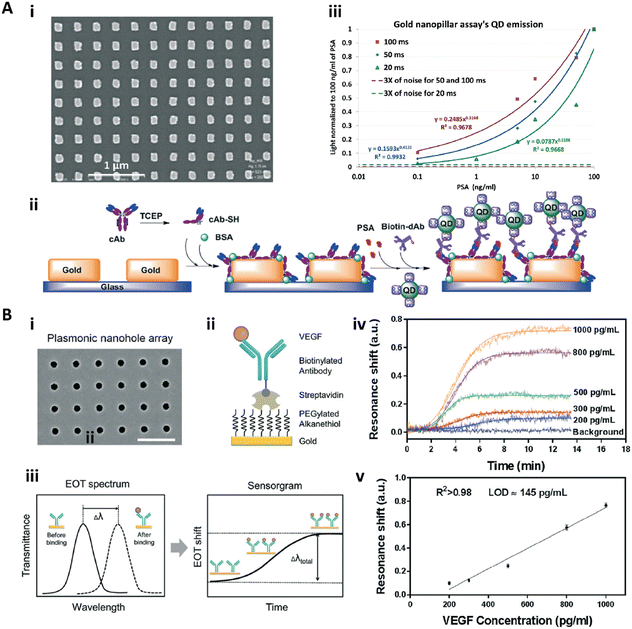 | ||
| Fig. 8 A-i) SEM image of the gold nanopillar array; A-ii) schematic illustration of the sandwich immunoassay for PSA detection with quantum dots (QDs) as the labelling fluorophore. A-iii) Calibration curves of the immunoassay on gold nanopillar arrays with different integration times. Reproduced from ref. 128, Copyright 2015 Royal Society of Chemistry. B-i) SEM image of the gold nanohole array; B-ii) schematic illustration for specific VEGF detection; B-iii) scheme of the sensing principle of the real-time plasmonic detection (EOT: extraordinary optical transmission); B-iv) the sensorgrams of VEGF detection with the concentration from 200–1000 pg mL−1. B-v) Calibration curve for VEGF detection. Reproduced from ref. 125, Copyright 2017 Royal Society of Chemistry. | ||
Altug and co-workers presented a series of gold nanohole array based substrate for immunoassays.123–127 In particular, Li et al. fabricated a nanohole array based biosensor for real-time analysis of live cell secretion by directly detecting the vascular endothelial growth factor (VEGF) in complex cell media.125 The gold nanohole array was fabricated by a deep-UV lithography method, and the as-fabricated nanohole was 200 nm in diameter with a pitch of 600 nm (Fig. 8B-i). Due to the plasmonic properties of the gold nanohole array, the extraordinary optical transmission spectra137 could be used to detect and quantify the biomolecular binding (Fig. 8B-iii and iv). To further improve the sensitivity, the antifouling PEG-based SAM was formed on the surface to immobilize the antibody complexes (Fig. 8B-ii). The biosensor achieved a VEGF sensitivity in complex cell media of 145 pg mL−1 with a linear dynamic range from 200 pg mL−1 to 1 ng mL−1 (Fig. 8B-v).125 This nanohole array based biosensor was then applied for high throughput analysis and POCT.123,126,127
3.3 2D nanomaterial coating
2D nanomaterials are sheet-like materials with the thickness of a single atom or a few atoms but a lateral size larger than 100 nm.130,131 The enhanced electronic, optical, and electrochemical properties of such a structure make the 2D nanomaterials extremely valuable for biosensing application.130–133 For example, a high surface to volume ratio provides more active site and increases the detection efficiency. Additionally, the SPR signal intensity is strongly dependent on the thickness and conductivity of the functional layer, and the 2D nanomaterial based SPR signal could be significantly enhanced.Graphene and its derivatives are the most commonly used 2D nanomaterials for immunosensing.133,138–145 Singh et al. presented a homogeneous single graphene layer based SPR immunosensor with sensitivity enhancement.140 The homogeneous single graphene layer was grown on a thin gold film by chemical vapor deposition (CVD), and then the biotinylated cholera toxin antigens were immobilized on the graphene via copper coordinated nitrilotriacetic acid (NTA), which was attached to graphene via π-stacking (Fig. 9A-i). Due to the amplification of the SPR signal by the graphene, the as-prepared immunosensor could specifically detect the anticholera toxin with an LOD of 4 pg mL−1 (Fig. 9A-ii).140 Xie et al. reported a graphene oxide (GO) enhanced fluorescence immunoassay to detect human IgG, which displayed a wide linear dynamic range of 0.01–800 ng mL−1 with an LOD of 0.006 ng mL−1, about 25-fold higher sensitive than that without GO.146 Recently, Jiang and co-workers developed a GO-based ultrasensitive SERS substrate for CEA detection.147 By utilizing gold nanorods as immunoprobes, this GO enhanced SERS platform provided a dynamic linear range from 10 pg mL−1 to 1 μg mL−1 with an LOD of 3.01 pg mL−1 for CEA detection.147
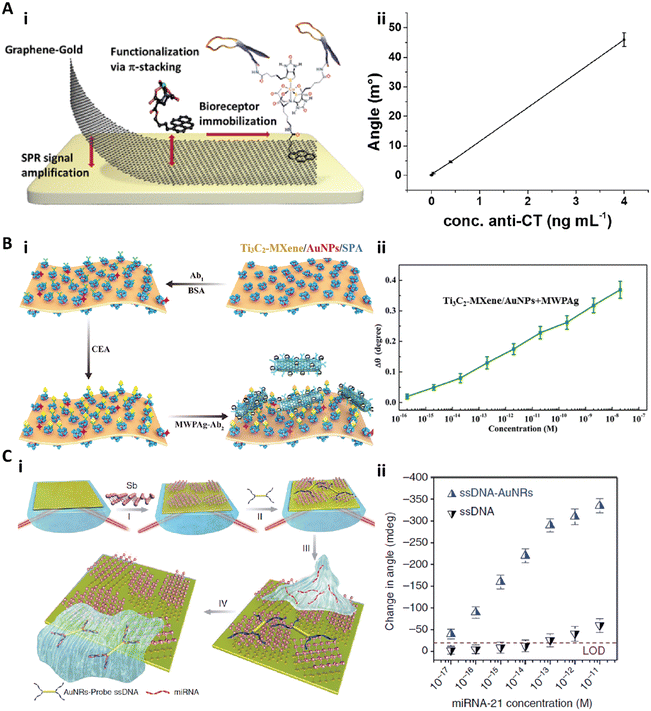 | ||
| Fig. 9 A-i) Schematic illustration of functionalized monolayer graphene for an enhanced SPR immunosensor. A-ii) Calibration curve of the graphene-based SPR immunosensor for anticholera toxin (anti-CT). Reproduced from ref. 140, Copyright 2015 American Chemical Society. B-i) Schematic illustration of the fabrication and detection procedure of the Ti3C2-MXene based SPR biosensor. B-ii) Relationship between the SPR signal shift and the concentration of CEA ranging from 10−16 to 10−8 M. Reproduced from ref. 148, Copyright 2019 Elsevier. C-i) Schematic illustration of fabrication of the antimonene nanosheet based miRNA biosensor. C-ii) The relationship between the SPR angle and miRNA concentration on the antimonene nanosheet based biosensor with or without gold nanorods. Reproduced from ref. 152, Copyright 2019 Springer Nature. | ||
Besides the graphene based 2D materials, other newly developed 2D nanomaterials are also employed as functional coatings to enhance the sensitivity of SPR biosensors.131,148–153 Recently, Wu et al. introduced an ultrathin Ti3C2-MXene nanosheet based SPR immunosensor for CEA detection.148 MXenes are a class of 2D transition metal carbides, nitrides, or carbonitrides with a few atoms thick.150 The Ti3C2-MXene nanosheets were fabricated by etching bulk Ti3AlC2, and then decorated with AuNPs. The anti-CEA antibody (Ab1) was immobilized on the Ti3C2-MXene/AuNP composites via staphylococcal protein A (SPA). To detect the CEA with high sensitivity, the detection anti-CEA antibody (Ab2) was tagged with multiwall carbon nanotubes–polydopamine–silver nanoparticles (MWPAg) as the signal enhancer (Fig. 9B-i). This Ti3C2-MXene based biosensor achieved a CEA assay sensitivity of 12.6 fg mL−1 (0.07 fM) with a dynamic range of 0.36 fg mL−1 (2 aM) to 3.6 μg mL−1 (20 nM) (Fig. 9B-ii).148 Xue et al. presented another antimonene nanosheet based SPR sensor for miRNA detection.152 This biosensor was created by assembling the antimonene nanosheets on the gold surface, followed by adsorption of gold nanorod–ssDNA complexes (AuNR-ssDNAs) as a probe on the antimonene. When the target miRNA was applied, the adsorbed AuNR-ssDNAs with a complementary sequence to the miRNA would be released, resulting in a significant decrease of the SPR angle (Fig. 9C-i). Due to the strong interaction between the ssDNA and antimonene and the signal enhancement by AuNRs, this biosensor could achieve a miRNA sensitivity of 0.13 fg mL−1 (10 aM) (Fig. 9C-ii).152
4. Surface modification for digital immunoassay
The ultimate sensitive immunoassay is to detect and quantify the target molecules at a single molecule resolution, which could be termed as the “digital immunoassay”.6,154 To make the single specific biorecognition distinct and detectable, the common strategy for the digital immunoassay is to stochastically compartmentalize the target molecules in the assay solution into femtoliter compartments in the “one-or-none” pattern, and the specific signal could be strongly amplified in the confined volume for distinguished detection.6,154 Therefore, most of the digital assays are homogeneous bead and microdroplet-based immunoassays by employing the beads as the capture substrates confined with target molecules in femtoliter microdroplets.87,155–160 However, such digital assays suffer from the low loading efficiency of the beads and the complex preparation process of the beads.87,155,159–162The heterogeneous solid phase based digital immunoassay could address the problems of the bead based digital assay. Single molecule fluorescence microscopy techniques, such as confocal microscopy and total internal reflection microscopy (TIRF), have been applied to develop the heterogeneous digital assay because of their ability to detect signals in a thin optical section with low background noise.163–169 Since the fluorescence signal from the single specific biorecognition is limited by the quantum yield of the fluorescing molecule, reducing background noise becomes essential in digital assays.6,165–169 The heterogeneous solid phase based digital immunoassay requires spatially isolating each single binding with others for distinguished counting, and surface modification with higher resolution is required.6,28,165–169 The Ha group pioneered the heterogeneous solid phase based digital assay, the single molecule pull down assay, by combining the pull down assay with TIRF microscopy.165 Here, the mixed polymer brush of PEG and biotinylated PEG was utilized to immobilize the capture antibody and to prevent nonspecific binding.165 Similarly, Zhang et al. reported a split aptamer based digital ATP assay with the mixed polymer brush of poly(L-lysine)-poly(ethylene glycol)-biotin (PLL-PEG-biotin) and PLL-PEG as the modified sensing surface.168 Weng et al. developed an aptasensor for small molecule detection on mPEG-succinimidyl valerate and biotin-PEG-succinimidyl valerate coated surfaces.169
Recently, our group developed a heterogeneous digital assay based on a PDA patterned perfluorinated surface (Fig. 10A).28 The PDA microspots with sub-micron resolution were fabricated on the antifouling perfluorinated surface via negative microcontact printing.170 Then, the capture antibody could be immobilized on the PDA spots with high stability and capacity.25,26 After the standard sandwich immunoassay was performed on the PDA patterned perfluorinated surface, a femtoliter droplet array of the substrate solution could be generated on the PDA microspot array via discontinuous dewetting with air flow. After incubation of the droplet array in air for 90 min, the detection results could be counted (Fig. 10A-i). This bead-free digital assay for IL-6 detection provided a dynamic range from 25.1 fg mL−1 (1.2 fM) to over 8.4 pg mL−1 (400 fM) with an LOD of 25.1 fg mL−1 (1.2 fM), and showed 30-fold higher sensitivity than the commercial ELISA (Fig. 10A-ii).28
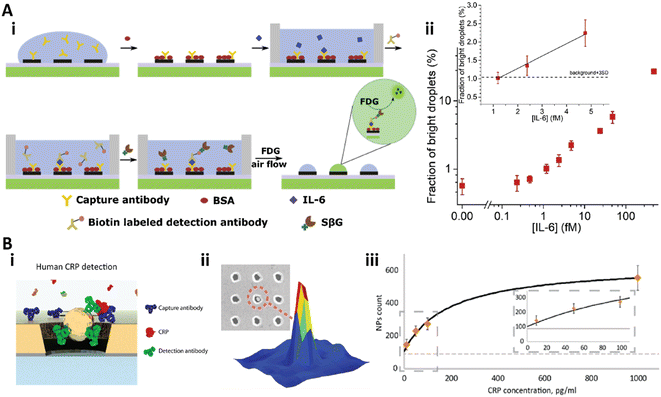 | ||
| Fig. 10 A) Bead-free digital immunoassay on a PDA patterned perfluorinated surface. i) Schematic illustration of the sandwich digital immunoassay to detect IL-6; ii) the relationship between the bright droplets and the concentration of IL-6 in log–log space. Inset: Linear relationship between the bright droplets and IL-6 at low concentration. Reproduced from ref. 28, Copyright 2021 Elsevier. B) The nanohole array based plasmonic digital assay. i) Scheme of the sandwich digital immunoassay on the gold nanohole array; ii) corresponding signal of the detection; iii) calibration curve of the CRP detection. Reproduced from ref. 173, Copyright 2018 American Chemical Society. | ||
Plasmonic nanomaterials are also utilized for digital assays with plasmon induced signal amplification.171–174 Recently, Belushkin et al. developed a gold nanohole array based digital assay for C-reactive protein (CRP) detection (Fig. 10B).173 The anti-CRP capture antibody was immobilized on the nanohole array. Then, the analyte solution was applied and washed, followed by the solution of Au NP tagged detection anti-CRP antibody (Fig. 10B-i). The Au NPs could be detected under bright-field imaging when the NPs were close to or in a nanohole, which would decrease the extraordinary optical transmission spectra (Fig. 10B-ii).137 This digital assay achieved an LOD of 29 pg mL−1 (Fig. 10B-iii).173
5. Conclusions
In this review, we have summarized the recent development in surface modification strategies to improve the sensitivity of immunoassays. According to the working mechanism and the effect of the modified surface, the surface modification can be categorized into two groups: antifouling coatings to reduce background noise and nanostructured surfaces to amplify the signals. However, the two groups of surface modification are not mutually exclusive, i.e., the same surface modification strategy may affect both signal and noise levels, and multiple surface modification strategies could be applied simultaneously. For example, the antifouling polymer brushes not only prevent nonspecific binding for background noise reduction, but also provide more active or activatable anchor sites for high protein loading capacity to enhance the signal. Applying the two groups of surface modification strategies to the same substrate might further improve the immunoassay sensitivity by simultaneously amplifying the signals and reducing the background noise. However, due to the complex modification procedures and limited consistency, most studies focus on only some specific modification strategies at one time. For example, routine blocking procedures were usually utilized when the research work was to develop a new nanostructured surface for signal amplification (Fig. 7A–C, 8A, 9B, and 10B). Recently, with the development of the surface modification techniques, some studies have successfully combined the two surface modification strategies to further improve the sensitivity (Fig. 4A and 8B).74,125Highly sensitive immunoassays are important to meet the growing need for the detection of low abundance analytes. High quality surfaces play an important role in achieving high sensitivity. Most of the modified surfaces for immunoassays are currently still limited to laboratory research, while only a few have been successfully commercialized.11,68 To improve the situation, the techniques of surface modification need to be simpler and yield more consistent performance, and these two kinds of surface modification strategies should be utilized synergistically to bring better sensitivity.
With the ongoing development of materials science and nanotechnology, we envision that more novel surface modification strategies will be developed towards the immunoassay of single or few molecules,28,87,154,155 which is the ultimate goal of ultrasensitive immunoassays.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Shenzhen Bay Laboratory (B. Z.) and the National Natural Science Foundation of China (Grant No. 21903055, X. Z.).Notes and references
- L. Cohen and D. R. Walt, Chem. Rev., 2019, 119, 293–321 CrossRef CAS PubMed.
- R. S. Yalow and S. A. Berson, Nature, 1959, 184, 1648–1649 CrossRef CAS PubMed.
- U. P. Rohr, C. Binder, T. Dieterle, F. Giusti, C. G. M. Messina, E. Toerien, H. Moch and H. Hendrikschäfer, PLoS One, 2016, 11, e0149856 CrossRef PubMed.
- B. D. Kevadiya, J. Machhi, J. Herskovitz, M. D. Oleynikov, W. R. Blomberg, N. Bajwa, D. Soni, S. Das, M. Hasan, M. Patel, A. M. Senan, S. Gorantla, J. E. McMillan, B. Edagwa, R. Eisenberg, C. B. Gurumurthy, S. P. M. Reid, C. Punyadeera, L. Chang and H. E. Gendelman, Nat. Mater., 2021, 20, 593–605 CrossRef CAS PubMed.
- M. Uhlén, E. Björling, C. Agaton, C. A. K. Szigyarto, B. Amini, E. Andersen, A. C. Andersson, P. Angelidou, A. Asplund, C. Asplund, L. Berglund, K. Bergström, H. Brumer, D. Cerjan, M. Ekström, A. Elobeid, C. Eriksson, L. Fagerberg, R. Falk, J. Fall, M. Forsberg, M. G. Björklund, K. Gumbel, A. Halimi, I. Hallin, C. Hamsten, M. Hansson, M. Hedhammar, G. Hercules, C. Kampf, K. Larsson, M. Lindskog, W. Lodewyckx, J. Lund, J. Lundeberg, K. Magnusson, E. Malm, P. Nilsson, J. Ödling, P. Oksvold, I. Olsson, E. Öster, J. Ottosson, L. Paavilainen, A. Persson, R. Rimini, J. Rockberg, M. Runeson, Å. Sivertsson, A. Sköllermo, J. Steen, M. Stenvall, F. Sterky, S. Strömberg, M. Sundberg, H. Tegel, S. Tourle, E. Wahlund, A. Waldén, J. Wan, H. Wernérus, J. Westberg, K. Wester, U. Wrethagen, L. L. Xu, S. Hober and F. Pontén, Mol. Cell. Proteomics, 2005, 4, 1920–1932 CrossRef PubMed.
- Z. Farka, M. J. Mickert, M. Pastucha, Z. Mikušová, P. Skládal and H. H. Gorris, Angew. Chem., 2020, 59, 10746–10773 CrossRef CAS PubMed.
- S. D. Gan and K. R. Patel, J. Invest. Dermatol., 2013, 133, 1–3 CrossRef PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CrossRef CAS PubMed.
- M. F. Templin, D. Stoll, M. Schrenk, P. C. Traub, C. F. Vöhringer and T. O. Joos, Trends Biotechnol., 2002, 20, 160–166 CrossRef CAS PubMed.
- A. H. C. Ng, U. Uddayasankar and A. R. Wheeler, Anal. Bioanal. Chem., 2010, 397, 991–1007 CrossRef CAS PubMed.
- J. T. Heggestad, C. M. Fontes, D. Y. Joh, A. M. Hucknall and A. Chilkoti, Adv. Mater., 2020, 32, 1903285 CrossRef CAS PubMed.
- Z. Farka, T. Juřík, D. Kovář, L. Trnková and P. Skládal, Chem. Rev., 2017, 117, 9973–10042 CrossRef CAS PubMed.
- M. Liang and X. Yan, Acc. Chem. Res., 2019, 52, 2190–2200 CrossRef CAS PubMed.
- E. C. Welch, J. M. Powell, T. B. Clevinger, A. E. Fairman and A. Shukla, Adv. Funct. Mater., 2021, 31, 2104126 CrossRef CAS.
- T. Sano, C. L. Smith and C. R. Cantor, Science, 1992, 258, 120–122 CrossRef CAS PubMed.
- K. Lind and M. Kubista, J. Immunol. Methods, 2005, 304, 107–116 CrossRef CAS PubMed.
- S. Fredriksson, M. Gullberg, J. Jarvius, C. Olsson, K. Pietras, S. M. Gústafsdóttir, A. Östman and U. Landegren, Nat. Biotechnol., 2002, 20, 473–477 CrossRef CAS PubMed.
- A. Frutiger, A. Tanno, S. Hwu, R. F. Tiefenauer, J. Vörös and N. Nakatsuka, Chem. Rev., 2021, 121, 8095–8160 CrossRef CAS PubMed.
- G. Ward, A. Simpson, L. Boscato and P. E. Hickman, Clin. Biochem., 2017, 50, 1306–1311 CrossRef CAS PubMed.
- W. J. Brittain, T. Brandsetter, O. Prucker and J. Rühe, ACS Appl. Mater. Interfaces, 2019, 11, 39397–39409 CrossRef CAS PubMed.
- C. Jiang, G. Wang, R. Hein, N. Liu, X. Luo and J. J. Davis, Chem. Rev., 2020, 120, 3852–3889 CrossRef CAS PubMed.
- A. M. C. Maan, A. H. Hofman, W. M. de Vos and M. Kamperman, Adv. Funct. Mater., 2020, 30, 2000936 CrossRef CAS.
- Z. Ding, C. Chen, Y. Yu and S. de Beer, J. Mater. Chem. B, 2022, 10, 2430–2443 RSC.
- J. O. Zoppe, N. C. Ataman, P. Mocny, J. Wang, J. Moraes and H. A. Klok, Chem. Rev., 2017, 117, 1105–1318 CrossRef CAS PubMed.
- H. Feng, Q. Zhang, H. Ma and B. Zheng, Analyst, 2015, 140, 5627–5633 RSC.
- Q. Liu, X. Zhou, H. Wu, L. Wu and B. Zheng, Sens. Actuators, B, 2019, 287, 306–311 CrossRef CAS.
- Q. Liu, X. Zhou, H. Wu and B. Zheng, Biosens. Bioelectron., 2020, 165, 112394 CrossRef CAS PubMed.
- S. Qian, H. Wu, B. Huang, Q. Liu, Y. Chen and B. Zheng, Sens. Actuators, B, 2021, 345, 130341 CrossRef CAS.
- B. Shen, B. Xiong and H. Wu, Biomicrofluidics, 2015, 9, 044111 CrossRef PubMed.
- M. Lejars, A. Margaillan and C. Bressy, Chem. Rev., 2012, 112, 4347–4390 CrossRef CAS PubMed.
- H. Peng, Polym. Rev., 2019, 59, 739–757 CrossRef CAS.
- Q. Wei, T. Becherer, S. Angioletti-Uberti, J. Dzubiella, C. Wischke, A. T. Neffe, A. Lendlein, M. Ballauff and R. Haag, Angew. Chem., Int. Ed., 2014, 53, 8004–8031 CrossRef CAS PubMed.
- H. Vaisocherová, E. Brynda and J. Homola, Anal. Bioanal. Chem., 2015, 407, 3927–3953 CrossRef PubMed.
- R. V. Vogt, D. L. Phillips, L. Omar Henderson, W. Whitfield and F. W. Spierto, J. Immunol. Methods, 1987, 101, 43–50 CrossRef CAS PubMed.
- M. E. Gibbs and J. Kennebunk, ELISA Tech. Bull., 2001, 1–6 Search PubMed.
- R. Ahirwar, S. Bariar, A. Balakrishnan and P. Nahar, RSC Adv., 2015, 5, 100077–100083 RSC.
- I. Víšová, M. Houska and H. Vaisocherová-Lísalová, Analyst, 2022, 147, 2597–2614 RSC.
- D. Kim and A. E. Herr, Biomicrofluidics, 2013, 7, 041501 CrossRef PubMed.
- S. Chen, L. Li, C. Zhao and J. Zheng, Polymer, 2010, 51, 5283–5293 CrossRef CAS.
- G. Liu, X. Sun, X. Li and Z. Wang, Polymer, 2022, 14, 826 CAS.
- E. Ostuni, R. G. Chapman, R. E. Holmlin, S. Takayama and G. M. Whitesides, Langmuir, 2001, 17, 5605–5620 CrossRef CAS.
- S. P. Lyu and D. Untereker, Int. J. Mol. Sci., 2009, 10, 4033–4065 CrossRef CAS PubMed.
- Y. Du, J. Gao, T. Chen, C. Zhang, J. Ji and Z. K. Xu, Langmuir, 2017, 33, 7298–7304 CrossRef CAS PubMed.
- J. Ladd, Z. Zhang, S. Chen, J. C. Hower and S. Jiang, Biomacromolecules, 2008, 9, 1357–1361 CrossRef CAS PubMed.
- F. Surman, T. Riedel, M. Bruns, N. Y. Kostina, Z. Sedláková and C. Rodriguez-Emmenegger, Macromol. Biosci., 2015, 15, 636–646 CrossRef CAS PubMed.
- J. H. Lee, J. Kopecek and J. D. Andrade, J. Biomed. Mater. Res., 1989, 23, 351–368 CrossRef CAS PubMed.
- N. Xia, Y. Hu, D. W. Grainger and D. G. Castner, Langmuir, 2002, 18, 3255–3256 CrossRef CAS.
- K. L. Prime and G. M. Whitesides, J. Am. Chem. Soc., 1993, 115, 10714–10721 CrossRef CAS.
- F. Sun, K. Wu, H. C. Hung, P. Zhang, X. Che, J. Smith, X. Lin, B. Li, P. Jain, Q. Yu and S. Jiang, Anal. Chem., 2017, 89, 10999–11004 CrossRef CAS PubMed.
- C. R. Emmenegger, E. Brynda, T. Riedel, Z. Sedlakova, M. Houska and A. B. Alles, Langmuir, 2009, 25, 6328–6333 CrossRef CAS PubMed.
- M. Badoux, M. Billing and H.-A. Klok, Polym. Chem., 2019, 10, 2925–2951 RSC.
- M. S. Selim, M. A. Shenashen, S. A. El-Safty, S. A. Higazy, M. M. Selim, H. Isago and A. Elmarakbi, Prog. Mater. Sci., 2017, 87, 1–32 CrossRef CAS.
- K. L. Prime and G. M. Whitesides, Science, 1991, 252, 1164–1167 CrossRef CAS PubMed.
- R. E. Holmlin, X. Chen, R. G. Chapman, S. Takayama and G. M. Whitesides, Langmuir, 2001, 17, 2841–2850 CrossRef CAS PubMed.
- R. G. Chapman, E. Ostuni, R. Shuichi Takayama, A. Erik Holmlin, Lin Yan and G. M. Whitesides, J. Am. Chem. Soc., 2000, 122, 8303–8304 CrossRef CAS.
- F. R. Castiello and M. Tabrizian, Anal. Chem., 2018, 90, 3132–3139 CrossRef CAS PubMed.
- M. Xu, X. Luo and J. J. Davis, Biosens. Bioelectron., 2013, 39, 21–25 CrossRef CAS PubMed.
- T. Bryan, X. Luo, L. Forsgren, L. A. Morozova-Roche and J. J. Davis, Chem. Sci., 2012, 3, 3468–3473 RSC.
- A. V. Patil, F. C. Bedatty Fernandes, P. R. Bueno and J. J. Davis, Anal. Chem., 2015, 87, 944–950 CrossRef CAS PubMed.
- T. Bryan, X. Luo, P. R. Bueno and J. J. Davis, Biosens. Bioelectron., 2013, 39, 94–98 CrossRef CAS PubMed.
- Y. S. Wang, S. Yau, L. K. Chau, A. Mohamed and C. J. Huang, Langmuir, 2019, 35, 1652–1661 CrossRef CAS PubMed.
- S. S. Panikar, G. Ramírez-García, S. Sidhik, T. Lopez-Luke, C. Rodriguez-Gonzalez, I. H. Ciapara, P. S. Castillo, T. Camacho-Villegas and E. De La Rosa, Anal. Chem., 2019, 91, 2100–2111 CrossRef CAS PubMed.
- S. S. Panikar, N. Banu, J. Haramati, G. Y. Gutierrez-Silerio, B. E. Bastidas-Ramirez, M. C. Tellez-Bañuelos, T. A. Camacho-Villegas, S. Del Toro-Arreola and E. De la Rosa, Anal. Chim. Acta, 2020, 1138, 110–122 CrossRef CAS PubMed.
- M. Krishnamoorthy, S. Hakobyan, M. Ramstedt and J. E. Gautrot, Chem. Rev., 2014, 114, 10976–11026 CrossRef CAS PubMed.
- Z. Lei, J. Gao, X. Liu, D. Liu and Z. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 10174–10182 CrossRef CAS PubMed.
- H. Ma, J. Hyun, P. Stiller and A. Chilkoti, Adv. Mater., 2004, 16, 338–341 CrossRef CAS.
- A. Hucknall, D. H. Kim, S. Rangarajan, R. T. Hill, W. M. Reichert and A. Chilkoti, Adv. Mater., 2009, 21, 1968–1971 CrossRef CAS PubMed.
- D. Y. Joh, A. M. Hucknall, Q. Wei, K. A. Mason, M. L. Lund, C. M. Fontes, R. T. Hill, R. Blair, Z. Zimmers, R. K. Achar, D. Tseng, R. Gordan, M. Freemark, A. Ozcan and A. Chilkoti, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 7054–7062 CrossRef PubMed.
- C. M. Fontes, B. D. Lipes, J. Liu, K. N. Agans, A. Yan, P. Shi, D. F. Cruz, G. Kelly, K. M. Luginbuhl, D. Y. Joh, S. L. Foster, J. Heggestad, A. Hucknall, M. H. Mikkelsen, C. F. Pieper, R. W. Horstmeyer, T. W. Geisbert, M. D. Gunn and A. Chilkoti, Sci. Transl. Med., 2021, 13, 9696 CrossRef PubMed.
- J. T. Heggestad, D. S. Kinnamon, L. B. Olson, J. Liu, G. Kelly, S. A. Wall, S. Oshabaheebwa, Z. Quinn, C. M. Fontes, D. Y. Joh, A. M. Hucknall, C. Pieper, J. G. Anderson, I. A. Naqvi, L. Chen, L. G. Que, T. Oguin, S. K. Nair, B. A. Sullenger, C. W. Woods, T. W. Burke, G. D. Sempowski, B. D. Kraft and A. Chilkoti, Sci. Adv., 2021, 7, 4901–4926 CrossRef PubMed.
- J. T. Heggestad, R. J. Britton, D. S. Kinnamon, S. A. Wall, D. Y. Joh, A. M. Hucknall, L. B. Olson, J. G. Anderson, A. Mazur, C. R. Wolfe, T. H. Oguin, B. A. Sullenger, T. W. Burke, B. D. Kraft, G. D. Sempowski, C. W. Woods and A. Chilkoti, Sci. Adv., 2021, 7, 7682 CrossRef PubMed.
- W. Hu, Y. Liu, Z. Lu and C. M. Li, Adv. Funct. Mater., 2010, 20, 3497–3503 CrossRef CAS.
- W. Hu, X. Li, G. He, Z. Zhang, X. Zheng, P. Li and C. M. Li, Biosens. Bioelectron., 2013, 50, 338–344 CrossRef CAS PubMed.
- W. Hu, Y. Liu, T. Chen, Y. Liu and C. M. Li, Adv. Mater., 2015, 27, 181–185 CrossRef CAS PubMed.
- H. Vaisocherová, W. Yang, Z. Zhang, Z. Cao, G. Cheng, M. Piliarik, J. Homola and S. Jiang, Anal. Chem., 2008, 80, 7894–7901 CrossRef PubMed.
- N. D. Brault, A. D. White, A. D. Taylor, Q. Yu and S. Jiang, Anal. Chem., 2013, 85, 1447–1453 CrossRef CAS PubMed.
- H. Vaisocherová-Lísalová, I. Víšová, M. L. Ermini, T. Špringer, X. C. Song, J. Mrázek, J. Lamačová, N. Scott Lynn, P. Šedivák and J. Homola, Biosens. Bioelectron., 2016, 80, 84–90 CrossRef PubMed.
- G. C. Le Goff, R. L. Srinivas, W. A. Hill and P. S. Doyle, Eur. Polym. J., 2015, 72, 386–412 CrossRef CAS PubMed.
- I. Y. Jung, J. S. Kim, B. R. Choi, K. Lee and H. Lee, Adv. Healthcare Mater., 2017, 6, 1601475 CrossRef PubMed.
- D. Buenger, F. Topuz and J. Groll, Prog. Polym. Sci., 2012, 37, 1678–1719 CrossRef CAS.
- Z. Lin, Y. Ma, C. Zhao, R. Chen, X. Zhu, L. Zhang, X. Yan and W. Yang, Lab Chip, 2014, 14, 2505–2514 RSC.
- Y. H. Roh, J. Seo, J. Y. Kim, H. U. Kim, S. J. Mun, J. H. Seo and K. W. Bong, Analyst, 2020, 145, 5482–5490 RSC.
- Y. N. Chou, F. Sun, H. C. Hung, P. Jain, A. Sinclair, P. Zhang, T. Bai, Y. Chang, T. C. Wen, Q. Yu and S. Jiang, Acta Biomater., 2016, 40, 31–37 CrossRef CAS PubMed.
- R. Randriantsilefisoa, J. L. Cuellar-Camacho, M. S. Chowdhury, P. Dey, U. Schedler and R. Haag, J. Mater. Chem. B, 2019, 7, 3220–3231 RSC.
- W. Wang, R. Han, M. Chen and X. Luo, Anal. Chem., 2021, 93, 7355–7361 CrossRef CAS PubMed.
- F. D. Scherag, A. Mader, M. Zinggeler, N. Birsner, R. E. Kneusel, T. Brandstetter and J. Rühe, Biomacromolecules, 2018, 19, 4641–4649 CrossRef CAS PubMed.
- S. H. Kim, S. Iwai, S. Araki, S. Sakakihara, R. Iino and H. Noji, Lab Chip, 2012, 12, 4986–4991 RSC.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Science, 2007, 318, 426–430 CrossRef CAS PubMed.
- H. Lee, J. Rho and P. B. Messersmith, Adv. Mater., 2009, 21, 431–434 CrossRef CAS PubMed.
- S. M. Kang, N. S. Hwang, J. Yeom, S. Y. Park, P. B. Messersmith, I. S. Choi, R. Langer, D. G. Anderson and H. Lee, Adv. Funct. Mater., 2012, 22, 2949–2955 CrossRef CAS PubMed.
- D. R. Dreyer, D. J. Miller, B. D. Freeman, D. R. Paul and C. W. Bielawski, Chem. Sci., 2013, 4, 3796 RSC.
- L. Wu, H. Feng, D. Guo and B. Zheng, RSC Adv., 2014, 4, 60002–60006 RSC.
- W. Qiang, W. Li, X. Li, X. Chen and D. Xu, Chem. Sci., 2014, 5, 3018–3024 RSC.
- H. Altug, S. H. Oh, S. A. Maier and J. Homola, Nat. Nanotechnol., 2022, 17, 5–16 CrossRef CAS PubMed.
- S. Zeng, D. Baillargeat, H. P. Ho and K. T. Yong, Chem. Soc. Rev., 2014, 43, 3426–3452 RSC.
- X. Liu, J. Guo, Y. Li, B. Wang, S. Yang, W. Chen, X. Wu, J. Guo and X. Ma, J. Mater. Chem. B, 2021, 9, 8378–8388 RSC.
- A. Minopoli, A. Acunzo, B. Della Ventura and R. Velotta, Adv. Mater. Interfaces, 2022, 9, 2101133 CrossRef CAS.
- D. Semeniak, D. F. Cruz, A. Chilkoti and M. H. Mikkelsen, Adv. Mater., 2022, 2107986 CrossRef PubMed.
- B. Jia, J. Chen, J. Zhou, Y. Zeng, H.-P. Ho and Y. Shao, Nano Res., 2022, 15, 8367–8388 CrossRef.
- A. M. Shrivastav, U. Cvelbar and I. Abdulhalim, Commun. Biol., 2021, 4, 1–12 CrossRef PubMed.
- S. Y. Ding, J. Yi, J. F. Li, B. Ren, D. Y. Wu, R. Panneerselvam and Z. Q. Tian, Nat. Rev. Mater., 2016, 1, 1–16 Search PubMed.
- B. Mu, J. Zhang, T. P. McNicholas, N. F. Reuel, S. Kruss and M. S. Strano, Acc. Chem. Res., 2014, 47, 979–988 CrossRef CAS PubMed.
- F. Neubrech, C. Huck, K. Weber, A. Pucci and H. Giessen, Chem. Rev., 2017, 117, 5110–5145 CrossRef CAS PubMed.
- L. Zheng, Y. Wan, P. Qi, Y. Sun, D. Zhang and L. Yu, Talanta, 2017, 167, 600–606 CrossRef CAS PubMed.
- L. Wang, W. Lu, W. Zhu, H. Wu, F. Wang and X. Xu, Sens. Actuators, B, 2020, 304, 127330 CrossRef CAS.
- E. Borberg, M. Zverzhinetsky, A. Krivitsky, A. Kosloff, O. Heifler, G. Degabli, H. P. Soroka, R. S. Fainaro, L. Burstein, S. Reuveni, H. Diamant, V. Krivitsky and F. Patolsky, Nano Lett., 2019, 19, 5868–5878 CrossRef CAS PubMed.
- S. Hou, J. F. Chen, M. Song, Y. Zhu, Y. J. Jan, S. H. Chen, T. H. Weng, D. A. Ling, S. F. Chen, T. Ro, A. J. Liang, T. Lee, H. Jin, M. Li, L. Liu, Y. S. Hsiao, P. Chen, H. H. Yu, M. S. Tsai, M. D. Pisarska, A. Chen, L. C. Chen and H. R. Tseng, ACS Nano, 2017, 11, 8167–8177 CrossRef CAS PubMed.
- P. Zhang, M. He and Y. Zeng, Lab Chip, 2016, 16, 3033–3042 RSC.
- P. Zhang, X. Zhou, M. He, Y. Shang, A. L. Tetlow, A. K. Godwin and Y. Zeng, Nat. Biomed. Eng., 2019, 3, 438–451 CrossRef CAS PubMed.
- J. Cao, T. Sun and K. T. V. Grattan, Sens. Actuators, B, 2014, 195, 332–351 CrossRef CAS.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CrossRef CAS PubMed.
- Y. Jeong, Y. M. Kook, K. Lee and W. G. Koh, Biosens. Bioelectron., 2018, 111, 102–116 CrossRef CAS PubMed.
- P. P. Austin Suthanthiraraj and A. K. Sen, Biosens. Bioelectron., 2019, 132, 38–46 CrossRef CAS PubMed.
- Y. Park, B. Ryu, Q. Deng, B. Pan, Y. Song, Y. Tian, H. B. Alam, Y. Li, X. Liang and K. Kurabayashi, Small, 2020, 16, 1905611 CrossRef CAS PubMed.
- W. Xu, L. Wang, R. Zhang, X. Sun, L. Huang, H. Su, X. Wei, C. C. Chen, J. Lou, H. Dai and K. Qian, Nat. Commun., 2020, 11, 1654 CrossRef CAS PubMed.
- J. Li, H. Yan, X. Tan, Z. Lu and H. Han, Anal. Chem., 2019, 91, 3885–3892 CrossRef CAS PubMed.
- B.-B. Fu, X.-D. Tian, J.-J. Song, B.-Y. Wen, Y.-J. Zhang, P.-P. Fang and J.-F. Li, Anal. Chem., 2022, 94, 9578–9585 CrossRef CAS PubMed.
- V. G. Kravets, A. V. Kabashin, W. L. Barnes and A. N. Grigorenko, Chem. Rev., 2018, 118, 5912–5951 CrossRef CAS PubMed.
- J. Jiang, X. Wang, S. Li, F. Ding, N. Li, S. Meng, R. Li, J. Qi, Q. Liu and G. L. Liu, NANO, 2018, 7, 1517–1531 CAS.
- J. S. Pang, I. G. Theodorou, A. Centeno, P. K. Petrov, N. M. Alford, M. P. Ryan and F. Xie, ACS Appl. Mater. Interfaces, 2019, 11, 23083–23092 CrossRef CAS PubMed.
- C. Zhu, G. Meng, P. Zheng, Q. Huang, Z. Li, X. Hu, X. Wang, Z. Huang, F. Li and N. Wu, Adv. Mater., 2016, 28, 4871–4876 CrossRef CAS PubMed.
- T. I. Wong, S. Han, L. Wu, Y. Wang, J. Deng, C. Y. L. Tan, P. Bai, Y. C. Loke, X. Da Yang, M. S. Tse, S. H. Ng and X. Zhou, Lab Chip, 2013, 13, 2405–2413 RSC.
- N. Fabri-Faja, O. Calvo-Lozano, P. Dey, R. A. Terborg, M. C. Estevez, A. Belushkin, F. Yesilköy, L. Duempelmann, H. Altug, V. Pruneri and L. M. Lechuga, Anal. Chim. Acta, 2019, 1077, 232–242 CrossRef CAS PubMed.
- F. Yesilkoy, R. A. Terborg, J. Pello, A. A. Belushkin, Y. Jahani, V. Pruneri and H. Altug, Light: Sci. Appl., 2018, 7, 17152 CrossRef PubMed.
- X. Li, M. Soler, C. I. Özdemir, A. Belushkin, F. Yesilköy and H. Altug, Lab Chip, 2017, 17, 2208–2217 RSC.
- P. Dey, N. Fabri-Faja, O. Calvo-Lozano, R. A. Terborg, A. Belushkin, F. Yesilkoy, A. Fàbrega, J. C. Ruiz-Rodriguez, R. Ferrer, J. J. González-López, M. C. Estévez, H. Altug, V. Pruneri and L. M. Lechuga, ACS Sens., 2019, 4, 52–60 CrossRef CAS PubMed.
- X. Li, M. Soler, C. Szydzik, K. Khoshmanesh, J. Schmidt, G. Coukos, A. Mitchell and H. Altug, Small, 2018, 14, 1800698 CrossRef PubMed.
- H. Y. Song, T. I. Wong, A. Sadovoy, L. Wu, P. Bai, J. Deng, S. Guo, Y. Wang, W. Knoll and X. Zhou, Lab Chip, 2015, 15, 253–263 RSC.
- F. Zang, Z. Su, L. Zhou, K. Konduru, G. Kaplan and S. Y. Chou, Adv. Mater., 2019, 31, 1902331 CrossRef PubMed.
- C. Tan, X. Cao, X. J. Wu, Q. He, J. Yang, X. Zhang, J. Chen, W. Zhao, S. Han, G. H. Nam, M. Sindoro and H. Zhang, Chem. Rev., 2017, 117, 6225–6331 CrossRef CAS PubMed.
- N. Rohaizad, C. C. Mayorga-Martinez, M. Fojtů, N. M. Latiff and M. Pumera, Chem. Soc. Rev., 2021, 50, 619–657 RSC.
- D. T. Nurrohman, Y. H. Wang and N. F. Chiu, Front. Chem., 2020, 8, 728 CrossRef CAS PubMed.
- D. T. Nurrohman and N. F. Chiu, Nanomaterials, 2021, 11, 1–30 CrossRef PubMed.
- J. Kim, S. Y. Oh, S. Shukla, S. B. Hong, N. S. Heo, V. K. Bajpai, H. S. Chun, C. H. Jo, B. G. Choi, Y. S. Huh and Y. K. Han, Biosens. Bioelectron., 2018, 107, 118–122 CrossRef CAS PubMed.
- L. Cui, Y. Li, M. Lu, B. Tang and C. Y. Zhang, Biosens. Bioelectron., 2018, 99, 1–7 CrossRef CAS PubMed.
- X. Ren, X. Meng, D. Chen, F. Tang and J. Jiao, Biosens. Bioelectron., 2005, 21, 433–437 CrossRef CAS PubMed.
- T. W. Ebbesen, H. J. Lezec, H. F. Ghaemi, T. Thio and P. A. Wolff, Nature, 1998, 391, 667–669 CrossRef CAS.
- P. O. Patil, G. R. Pandey, A. G. Patil, V. B. Borse, P. K. Deshmukh, D. R. Patil, R. S. Tade, S. N. Nangare, Z. G. Khan, A. M. Patil, M. P. More, M. Veerapandian and S. B. Bari, Biosens. Bioelectron., 2019, 139, 111324 CrossRef CAS PubMed.
- W. Wei, J. Nong, Y. Mei, C. Zhong, G. Lan and W. Hu, Sens. Actuators, B, 2018, 273, 1548–1555 CrossRef CAS.
- M. Singh, M. Holzinger, M. Tabrizian, S. Winters, N. C. Berner, S. Cosnier and G. S. Duesberg, J. Am. Chem. Soc., 2015, 137, 2800–2803 CrossRef CAS PubMed.
- L. He, Q. Pagneux, I. Larroulet, A. Y. Serrano, A. Pesquera, A. Zurutuza, D. Mandler, R. Boukherroub and S. Szunerits, Biosens. Bioelectron., 2017, 89, 606–611 CrossRef CAS PubMed.
- N. F. Chiu, S. Y. Fan, C. Du Yang and T. Y. Huang, Biosens. Bioelectron., 2017, 89, 370–376 CrossRef CAS PubMed.
- X. Xiong, Y. Chen, H. Wang, S. Hu, Y. Luo, J. Dong, W. Zhu, W. Qiu, H. Guan, H. Lu, J. Yu, J. Zhang and Z. Chen, ACS Appl. Mater. Interfaces, 2018, 10, 34916–34923 CrossRef CAS PubMed.
- N. F. Chiu, T. L. Lin and C. T. Kuo, Sens. Actuators, B, 2018, 265, 264–272 CrossRef CAS.
- S. Taniselass, M. K. M. Arshad and S. C. B. Gopinath, Biosens. Bioelectron., 2019, 130, 276–292 CrossRef CAS PubMed.
- K. X. Xie, S. H. Cao, Z. C. Wang, Y. H. Weng, S. X. Huo, Y. Y. Zhai, M. Chen, X. H. Pan and Y. Q. Li, Sens. Actuators, B, 2017, 253, 804–808 CrossRef CAS.
- Y. Chen, H. Liu, J. Jiang, C. Gu, Z. Zhao and T. Jiang, ACS Appl. Bio Mater., 2020, 3, 8012–8022 CrossRef CAS PubMed.
- Q. Wu, N. Li, Y. Wang, Y. Liu, Y. Xu, S. Wei, J. Wu, G. Jia, X. Fang, F. Chen and X. Cui, Biosens. Bioelectron., 2019, 144, 111697 CrossRef CAS PubMed.
- Q. Wu, N. Li, Y. Wang, Y. Xu, J. Wu, G. Jia, F. Ji, X. Fang, F. Chen and X. Cui, Anal. Chem., 2020, 92, 3354–3360 CrossRef CAS PubMed.
- X. Zhu, Y. Zhang, M. Liu and Y. Liu, Biosens. Bioelectron., 2021, 171, 112730 CrossRef CAS PubMed.
- H. Zhang, Z. Wang, Q. Zhang, F. Wang and Y. Liu, Biosens. Bioelectron., 2019, 124–125, 184–190 CrossRef CAS PubMed.
- T. Xue, W. Liang, Y. Li, Y. Sun, Y. Xiang, Y. Zhang, Z. Dai, Y. Duo, L. Wu, K. Qi, B. N. Shivananju, L. Zhang, X. Cui, H. Zhang and Q. Bao, Nat. Commun., 2019, 10, 1–9 CrossRef PubMed.
- S. Barua, H. S. Dutta, S. Gogoi, R. Devi and R. Khan, ACS Appl. Nano Mater., 2018, 1, 2–25 CrossRef CAS.
- H. Noji, Y. Minagawa and H. Ueno, Lab Chip, 2022, 22, 3092–3109 RSC.
- D. M. Rissin, C. W. Kan, T. G. Campbell, S. C. Howes, D. R. Fournier, L. Song, T. Piech, P. P. Patel, L. Chang, A. J. Rivnak, E. P. Ferrell, J. D. Randall, G. K. Provuncher, D. R. Walt and D. C. Duffy, Nat. Biotechnol., 2010, 28, 595–599 CrossRef CAS PubMed.
- X. Wang, L. Cohen, J. Wang and D. R. Walt, J. Am. Chem. Soc., 2018, 140, 18132–18139 CrossRef CAS PubMed.
- Y. Obayashi, R. Iino and H. Noji, Analyst, 2015, 140, 5065–5073 RSC.
- J. U. Shim, R. T. Ranasinghe, C. A. Smith, S. M. Ibrahim, F. Hollfelder, W. T. S. Huck, D. Klenerman and C. Abell, ACS Nano, 2013, 7, 5955–5964 CrossRef CAS PubMed.
- A. M. Maley, P. M. Garden and D. R. Walt, ACS Sens., 2020, 5, 3037–3042 CrossRef CAS PubMed.
- K. Akama, N. Iwanaga, K. Yamawaki, M. Okuda, K. Jain, H. Ueno, N. Soga, Y. Minagawa and H. Noji, ACS Nano, 2019, 13, 13116–13126 CrossRef CAS PubMed.
- D. Decrop, G. Pardon, L. Brancato, D. Kil, R. Zandi Shafagh, T. Kokalj, T. Haraldsson, R. Puers, W. Van Der Wijngaart and J. Lammertyn, ACS Appl. Mater. Interfaces, 2017, 9, 10418–10426 CrossRef CAS PubMed.
- A. H. Kunding, L. L. Busk, H. Webb, H. W. Klafki, M. Otto, J. P. Kutter and M. Dufva, Lab Chip, 2018, 18, 2797–2805 RSC.
- W. E. Moerner and D. P. Fromm, Rev. Sci. Instrum., 2003, 74, 3597–3619 CrossRef CAS.
- S. Weiss, Science, 1999, 283, 1676–1683 CrossRef CAS PubMed.
- A. Jain, R. Liu, B. Ramani, E. Arauz, Y. Ishitsuka, K. Ragunathan, J. Park, J. Chen, Y. K. Xiang and T. Ha, Nature, 2011, 473, 484–488 CrossRef CAS PubMed.
- E. Burgin, A. Salehi-Reyhani, M. Barclay, A. Brown, J. Kaplinsky, M. Novakova, M. A. A. Neil, O. Ces, K. R. Willison and D. R. Klug, Analyst, 2014, 139, 3235–3244 RSC.
- A. Salehi-Reyhani, S. Sharma, E. Burgin, M. Barclay, A. Cass, M. A. A. Neil, O. Ces, K. R. Willison and D. R. Klug, Lab Chip, 2013, 13, 2066–2074 RSC.
- H. Zhang, Y. Liu, K. Zhang, J. Ji, J. Liu and B. Liu, Anal. Chem., 2018, 90, 9315–9321 CrossRef CAS PubMed.
- R. Weng, S. Lou, L. Li, Y. Zhang, J. Qiu, X. Su, Y. Qian and N. G. Walter, Anal. Chem., 2019, 91, 1424–1431 CrossRef CAS PubMed.
- H. Wu, L. Wu, X. Zhou, B. Liu and B. Zheng, Small, 2018, 14, 1802128 CrossRef PubMed.
- A. B. Taylor and P. Zijlstra, ACS Sens., 2017, 2, 1103–1122 CrossRef CAS PubMed.
- D. Sevenler, G. G. Daaboul, F. Ekiz Kanik, N. L. Ünlü and M. S. Ünlü, ACS Nano, 2018, 12, 5880–5887 CrossRef CAS PubMed.
- A. Belushkin, F. Yesilkoy and H. Altug, ACS Nano, 2018, 12, 4453–4461 CrossRef CAS PubMed.
- Y. Wang, Y. Yang, C. Chen, S. Wang, H. Wang, W. Jing and N. Tao, ACS Sens., 2020, 5, 1126–1131 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |
