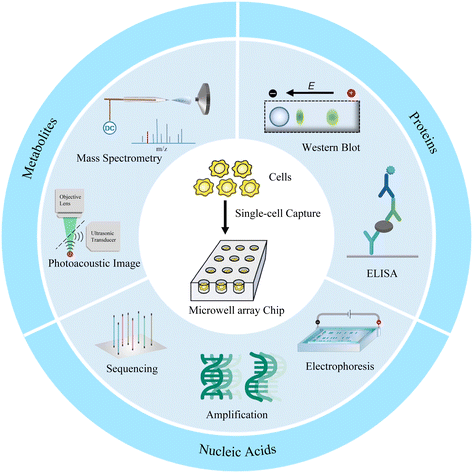
Microwell array chip-based single-cell analysis
Jin
Zhang
a,
Jing
Xue
a,
Ningfeng
Luo
a,
Feng
Chen
 a,
Badong
Chen
*b and
Yongxi
Zhao
a,
Badong
Chen
*b and
Yongxi
Zhao
 *a
*a
aInstitute of Analytical Chemistry and Instrument for Life Science, The Key Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi'an Jiaotong University, Xi'an, Shaanxi 710049, P. R. China. E-mail: yxzhao@mail.xjtu.edu.cn
bInstitute of Artificial Intelligence and Robotics and the College of Artificial Intelligence, Xi'an Jiaotong University, Xi'an, Shaanxi 710049, P. R. China. E-mail: chenbd@mail.xjtu.edu.cn
First published on 10th January 2023
Abstract
Single-cell profiling is key to uncover the cellular heterogeneity and drives deep understanding of cell fate. In recent years, microfluidics has become an ideal tool for single-cell profiling owing to its benefits of high throughput and automation. Among various microfluidic platforms, microwell has the advantages of simple operation and easy integration with in situ analysis ability, making it an ideal technique for single-cell studies. Herein, recent advances of single-cell analysis based on microwell array chips are summarized. We first introduce the design and preparation of different microwell chips. Then microwell-based cell capture and lysis strategies are discussed. We finally focus on advanced microwell-based analysis of single-cell proteins, nucleic acids, and metabolites. The challenges and opportunities for the development of microwell-based single-cell analysis are also presented.
1. Introduction
Recent advances in single-cell analysis have illustrated that even cells of the same type showed obvious heterogeneity, including fate, morphology, and differential expression of biomolecules, due to differences in their microenvironment.1,2 Traditional bulk analysis masks cellular heterogeneity, leading to the missing of important biological information. Hence, analysing cells at the single-cell level helps us gain a more comprehensive cognition of life processes.3,4In contrast to bulk assay, single-cell analysis requires manipulation of a sample volume as small as a picoliter to investigate individual cells. Early single-cell analysis methods rely mainly on well plates5,6 and flow cytometry.7,8 Among them, plate-based methods often require manual separation of individual cells into the plate, which is inefficient and labor-intensive.9 Although flow cytometry enables automated single-cell sorting, it is bulky, mechanically complex, expensive, and can only be used for cell analysis at fixed time points.10 In recent years, microfluidics has proven to be an ideal tool for efficient single-cell studies. Compared with traditional techniques, microfluidics can limit reaction volumes from nanoliters to picoliters, enabling highly sensitive analysis. The structure and function of microfluidic chips can be optionally changed to meet the needs of single-cell profiling; the chips mainly include droplet microfluidic chips,11 microvalve-based chips,12,13 and microwell-based chips.14 Droplet microfluidics typically utilizes oil phase shearing of the aqueous phase to generate massive droplets within a short time, enabling separate compartments to reduce cross-contamination. It is a powerful tool for ultrahigh-throughput single-cell capture and the droplets enable precise manipulation such as sorting, fusion, and splitting. However, to ensure that each droplet encapsulates no more than one cell, a Poisson distribution-based limiting dilution method is required, which results in a large number of empty droplets.15 Microvalve-based chips utilize pressure-controlled valves to manipulate the flow/blocking of fluids within microchannels. Valves can be switched on and off, enabling precise manipulation of fluids in time and space. However, commonly used valve-based devices are constructed by multi-layer soft lithography, which requires complex structural design and fabrication.16 Microwell-based chips consist of picoliter containers and use the cell's own gravity to capture individual cells. They are usually obtained by standard soft lithography and have the advantages of simple design, easy microfabrication, and convenient operation. Compared with droplet or microvalve chips, microwell-based chips can provide a more stable microenvironment and have been applied to high-throughput single-cell culture17 and in situ analysis.18,19
Microwell has been proven to be a very cogent platform for single-cell research. In this review, we first describe the design and preparation of microwell-based chips, including shape design and material selection, and their advantages as well as limitations. We then discuss the appropriate single-cell isolation approaches and lysis strategies on the basis of targets and microwell chips. Finally, advanced single-cell analyses of proteins, nucleic acids, and metabolites based on microwell chips are presented (Fig. 1). We also provide perspectives on the challenges and opportunities of microwell-based single-cell analysis.
2. Microwell array chip preparation and manipulation of single cells
2.1 Microwell array chip preparation
The selection of microwell chips is critical and depends on the application. Different analysis methods and targets require microwells of different materials and/or geometries. In the following section, we summarize the preparatory work of microwell array chips, including material selection, shape and size design, and fabrication.The earliest materials for microfluidic devices are glass and silicon wafers. Walt et al. designed a microwell array based on optical imaging fibers to enable real-time observation of the response of living cells to environmental changes.21 The microwell chip was fabricated by etching techniques combined with fluorescence analysis techniques to allow the study of optically detectable events. However, even a single wafer requires long processing time and expensive equipment and consumables, not to mention mass production. Therefore, a low-cost and easy-to-handle material is needed to fabricate microfluidic chips. Since Whiteside's research group established soft lithography protocols as a microfabrication technique in 1998,22 polydimethylsiloxane (PDMS) has become the most commonly used material. From the manufacturing point of view, PDMS has the advantages of high replication fidelity, good mechanical strength, and easy integration with glass. In addition, PDMS exhibits good biocompatibility, favourable permeability and low autofluorescence, making it promising for applications in biomedical engineering. However, PDMS also has disadvantages. It swells in many common solvents, especially hydrocarbon-based solvents.23 Hydrogels are also frequently used materials for microwell chips. Since tuning the overall material properties of the device can be achieved by adjusting the molecular weight or ratio of monomers, hydrogels offer special advantages.24 Microwells of polyethylene glycol are prepared using soft lithography and are used for the fusion of single-cell pairs. The yield of correct cell fusion hybridization is greatly improved with the help of PEG microwells.25 Microwell chips based on agarose and polyacrylamide have been developed for single-cell electrophoresis experiments.26 Meanwhile, microwell arrays based on polyacrylamide were designed to perform three-dimensional cell cultures.27 Polymers have attracted much attention in recent years due to their diverse properties. Many polymers have been used to prepare microwell chips, such as polystyrene,28 PMMA,29 epoxy resin,30 and fluorocarbon polymers.31
Circles, squares and triangles are the most commonly used microwell shapes. Since most cells are spherical in shape, microwells are usually designed to be circular.32 Compared with round microwells, square microwells can generate higher local cytoskeletal tension at the corners, and the increase in cytoskeletal tension promotes the local assembly of focal adhesion and stress fibers. Thus, human mesenchymal stem cells in square wells tend to proliferate in situ and differentiate into osteoblasts.33 Gravity-based capture of cells is common in circular and square microwells, and triangles are validated to be the optimal shape for hydrodynamics-based single-cell capture after five different shapes of microwells were tested in simulations.32 The continuous flow of the aqueous phase creates a strong backflow in the triangular microwell, which effectively traps cells. Once cells are captured, they change flow patterns in the microwells, preventing other cells from being captured. Thus, triangular wells prefer to capture single cells, while square or round ones have more possibility to isolate multiple cells (Fig. 2a). In the design of the 3D microwell culture device, the concave bottom structure is most commonly used, because such a bottom is conducive to the growth of individual aggregates in the microwells.34,35 In conical microwells, cell aggregates of uniform size tend to form in each microwell.36,37 Microwells of different shapes can also be used for coding. Eng et al. provided a broad compositional and geometric tunability of shape-encoded microwell patterns. Using precisely aligned hydrogel shapes, they studied the migration patterns of human mesenchymal stem cells and endothelial cells (Fig. 2c).38
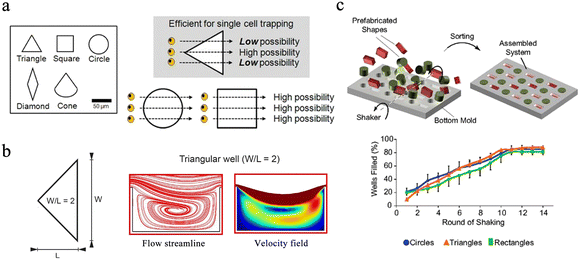 | ||
| Fig. 2 Different designs of microwell chips for single-cell analysis. (a) The equilateral triangle has been proven to be an ideal well shape. Scale bar: 50 μm. Reproduced with permission.32 Copyright 2009, Springer Nature Publishing. (b) Triangular microwell with W/L = 2 has the best trapping capability because of the enhanced recirculation vortex. Reproduced with permission.39 Copyright 2019, Springer Nature Publishing. (c) Extensive compositional and geometric tunability of shape-coded microwell patterns. Reproduced with permission.38 Copyright 2013, National Academy of Sciences. | ||
The size of the well is also the key factor during the design of the microchips. Ideally, the width and height of the well should match a single particle and not accommodate multiple targets. In hydrodynamics-based single-cell capture chips, the ratio of the width (W) and length (L) of the triangular microwell will significantly affect the capture efficiency of single cells. Microwells with a W/L ratio of 2 are experimentally proven to have better capture capability owing to the enhanced recirculation vortex (Fig. 2b).39 Nevertheless, when performing 3D microwell-based cell culture, the aspect ratio (diameter/height) of the microwell determines its performance. A high aspect ratio favors long-term cell culture,40 while a low aspect ratio with a shallow microwell favors spheroid formation.41
2.2 Manipulation of single cells
Gravity-induced single-cell capture is the simplest and most direct method to isolate single cells. Typically, the cell suspension is placed over the well chip, and after the cells are settled, the chip is washed to remove any uncaptured cells (Fig. 3a).50 However, gravity-based single-cell isolation requires a long time, resulting in inefficient single-cell trapping.51 To ensure that cells are captured individually, the cell suspension is usually diluted, resulting in a single-cell capture efficiency of about 10%. In order to improve the single-cell occupancy efficiency, Huang et al. designed a truncated cone-shaped microwell array chip and deposited the cells into the microwells under the action of centrifugal force, realizing the occupancy rate of single cells up to about 90% in a few seconds (Fig. 3b).52 This approach has been applied to investigate the dynamic reactions of individual lung cancer cells to pharmacological treatments.53
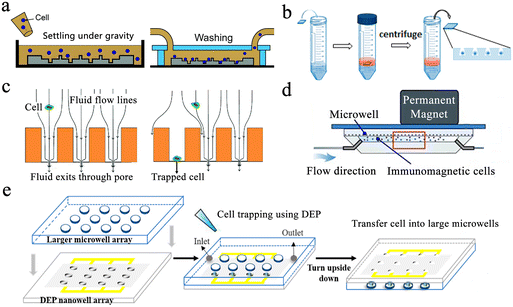 | ||
| Fig. 3 Single-cell capture strategies in microwell chips. (a) Gravity-induced single-cell seeding. Reproduced with permission.50 Copyright 2019, Royal Society of Chemistry. (b) Centrifugation-based single-cell capture. Reproduced with permission.52 Copyright 2015, American Chemical Society. (c) Hydrodynamic-assisted single-cell isolation. Reproduced with permission.54 Copyright 2015, Royal Society of Chemistry. (d) Immunomagnetic single-cell trapping. Reproduced with permission.55 Copyright 2019, Springer Nature Publishing. (e) Dielectrophoresis (DEP)-trapping approach to perform cell capture. Reproduced with permission.56 Copyright 2020, American Chemical Society. | ||
Hydrodynamic-assisted single-cell isolation exploits fluid streamlines within microwells, where cell motion trajectories follow these streamlines. At the same time, under the action of gravity, the cells progressively settle, lowering the streamline as a result. The generation of strong vortex flow lines in triangular microwells facilitates the capture of individual prostate tumor cells in the microwells, which can achieve a single-cell occupancy of 62%.32 Swennenhuis et al. employed microwells with pores at the bottom to capture single cells with the help of negative pressure (Fig. 3c).54 When negative pressure is applied, fluid can pass through the pores, creating hydrodynamic forces that drag individual cells toward the pores in the center of the microwell bottom. Cells are larger in size than pores, so they remain in the microwell and do not pass through. When a cell lands on a pore, the flow of sample through that particular well is stopped, and no other cells enter the same well, thus achieving a single-cell trapping rate of 67%. In the process of cell capture, target cells can be screened and captured by external forces, and the most commonly used external forces include magnetic and electric fields. Integrating magnetic fields with microfluidic systems can be applied to capture rare target cells coupled to magnetic beads. Huang et al. described a microfluidic chip based on a microwell array with integrated permanent magnets for immunomagnetic single-cell capture (Fig. 3d).55 With this method, they achieved high-purity capture of human acute monocytic leukemia cells and realized a capture efficiency of 62%. Precise separation of captured target cells from mixed samples is an advantage of this method. However, the binding of magnetic beads and applied magnetic field forces may negatively affect cell viability. Electro-manipulation techniques use the force generated by an electric field to handle individual cells. Cells are polarized due to exposure to an electric field, and manipulation of cells is achieved by moving them to regions of strong electric fields using dielectrophoretic forces. Using a sub-Poisson method, dielectrophoresis-trapping-nanowell-transfer (dTNT) methodology was achieved with a single-cell capture efficiency of 91.84% (Fig. 3e).56
After the cell lysis is complete, reducing cross-contamination between wells is a key factor for accurate single-cell analysis. Microwell-seq was a strategy for sequencing the transcriptomes of thousands of single cells in open microwell arrays.63 The method utilized chemical lysis and self-made barcoded beads to capture single-cell transcripts for transcriptome analysis. Cell doublets need to be removed manually under a microscope, which was labour-intensive and complicated. Seqwell utilized a semipermeable membrane with nanopores to seal microwells.64 After cell lysis, the semipermeable membrane traps biomacromolecules in the well, reducing cross-contamination. However, semipermeable membranes are unable to retain small molecules from cell lysates within the wells. Burak et al. presented scFTD-seq, a single-cell RNA-sequencing platform based on freeze–thaw lysed cells and slide-sealed microwells.61 This slide-sealing-based approach reduces cross-contamination between wells, but still requires manual handling of slides. Then, a microwell-based single-cell RNA-sequencing platform was developed by Yuan and Sims.65 After cells and barcoded beads are captured in pairs, lysis buffer was introduced, and the microwells were immediately sealed with fluorine oil. This approach ensures extremely low cross-contamination with the aid of an automated flow delivery system.
3. Single-cell analysis
Single-cell analysis is the most efficient way to conduct comprehensive heterogeneity studies from cellular phenotype to gene expression. Microwell array chips have become a popular choice to conduct single-cell studies owing to their advantages of easy fabrication, low cost, and simple operation. In this section, we summarize recent advances in microwell-based single-cell analysis, including proteins, nucleic acids, and metabolites.3.1 Single-cell protein analysis
Protein analysis plays an essential role in cellular biochemical process monitoring as well as in clinical diagnostic research. Accurate measurements of single-cell protein are vital to obtaining a comprehensive view of the cells. The microwell-based platform isolates single cells and confines the target to a small volume, resulting in high local concentrations and enabling more precise protein detection.Enzyme-linked immunosorbent assay (ELISA) is one of the most widely used methods for detecting and quantifying proteins. Traditional ELISA-based methods for hybridoma screening take a long time and cannot use multiple antibodies in a single screen. To address this issue, a microengraving-based assay for single-cell secreted factors was developed. Using antigen-modified slides and microwell arrays containing single cells, this method enabled the determination of various secretions from large numbers of single cells in a semi-quantitative manner (Fig. 4a).66 In order to increase the sensitivity of the platform, a hybridization chain reaction was added to amplify the signals produced by sandwich immunoassays for the simultaneous detection of three released proteins.67 However, this method had difficulty detecting low-concentration proteins. To detect low-abundance proteins in blood, digital ELISA methods were developed.68 This method loaded beads with specific antibody modifications into femtoliter-sized microwells, resulting in single-molecule arrays (SiMoAs). SiMoAs ensured that the concentration of the fluorescence signal molecule was high enough for detection. This strategy enabled detection of enzyme concentrations as low as one single molecule. Through the continuous improvement of SiMoAs, researchers have realized the detection of small molecules,69 proteins,70–72 cytokines73 and their simultaneous detection.74 However, this method was unable to detect multiple low-abundance proteins simultaneously in a single cell. To accomplish multiplexed protein detection, a beads-on-barcode antibody microarray (BOBarray) has been developed (Fig. 4b). It gave each protein a pair of unique identifiers (bead size and fluorescent color) and up to 12 different proteins can be encoded using four different bead sizes and three different colors. The chip of 60 pL BOBarray enables the integration of single-cell capture lysis and subsequent protein detection.75
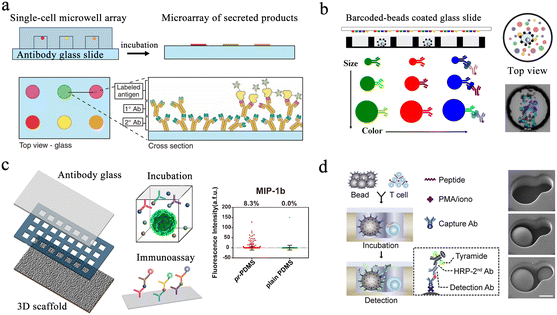 | ||
| Fig. 4 Microwell-based single-cell protein analysis. (a) Schematic of microengraving method. Single-cell microwell arrays were clamped and cultured with antibody modified slides to obtain single-cell secretion arrays. Reproduced with permission.66 Copyright 2006, Springer Nature Publishing. (b) Overview of beads-on-barcode antibody microarray (BOBarray). The size and color of beads are used as identifiers in combination with single-cell microwell array to achieve multiplex protein detection. Reproduced with permission.75 Copyright 2016, American Chemical Society. (c) Schematic of paper-based 3D microwell chip. Using 3D scaffolds to mimic extracellular physical substrates, the effect of 3D microenvironment on cellular protein secretion was investigated. Reproduced with permission.76 Copyright 2018, American Chemical Society. (d). Scheme of hierarchical loading microwell chip (HL-Chip). This chip realized efficient pairing of multiple single cells with functionalized beads according to the size difference, and performed secretion analysis of single T cells. Reproduced with permission.77 Copyright 2020, Elsevier. | ||
Microengraving-based assays confine cells to small spaces and can affect the physiological state of cells. The modification and capture of the bead in the SiMoA method have greater randomness. Single-cell analysis based on these two platforms is still a two-dimensional culture of cells, which has difficulty restoring the real microenvironment in vivo. Therefore, a method integrating three-dimensional cell culture and cancer cell co-culture was developed to address these issues (Fig. 4c).76 The platform cultured tumor cells with macrophages within the same microwell, supporting single-cell cultures on a 3D matrix using a high-density micropillar array. Scatter plots in Fig. 4c show the profile of MIP-1b protein detection from U87 single cells on pr-PDMS and plain PDMS. None of the above methods can be used for cell-to-cell interaction analysis. Then, a hierarchical loading microwell chip (HL-Chip) was developed.77 The platform enabled high-precision dispensing of single cells/beads based on size differences, achieving 91.8% pairing efficiency of bead–cell (Fig. 4d). High-throughput secretion of individual T cells and tumor–T cell interactions were investigated using this platform.
Western blotting (WB) is an alternative protein detection technique applied in cellular and molecular biology. Researchers can distinguish particular proteins from complicated protein mixtures isolated from cells using WB. However, the average value of cell populations required by traditional blotting covers the abundant single-cell behavior in complicated populations. Single-cell western blot (scWB) is a single-cell protein analysis technique that provides an approach to further deepen our cognition of cell-to-cell differentiation in protein-related cellular characteristics.26 Specifically, the scWestern array consisted of thousands of microwells that are templated on a photosensitive polyacrylamide gel and placed on glass microscope slides. The scWB analysis is divided into six steps (Fig. 5a).78 Although antibody probe cross-reactivity and fixation errors continue to be confusing variables, immunoassays are the accepted standard for identifying subcellular protein localization in single cells. To improve selectivity, Herr et al. introduced a subcellular western blotting technique that enables direct measurement of nucleoplasmic protein distribution in single cells, providing more accurate protein signals in heterogeneous cell populations (Fig. 5b).79 After isolating single cells in multi-layered microwells, the researchers sequentially lysed the cell membrane and nuclear membrane and performed western blotting on the lysates, respectively. Six targets have been identified by subcellular single-cell western blots, including spliceosome-associated proteins that were released from big protein and RNA complexes as well as compact-sized proteins (7 kDa difference). However, cell separation disrupts multiple proteins when analyzing cell suspensions for proteins. Analysis of adherent cells reduces disturbance and sample loss. Therefore, in situ scWB was developed for the protein analysis of individual attached cells.80 To reduce the influence during cell culture, the microwell was functionalized with fibronectin (Fig. 5c). Following the reattachment, cells were lysed and western blotting was performed. This platform provided a solution for single-cell detection of losable proteins.
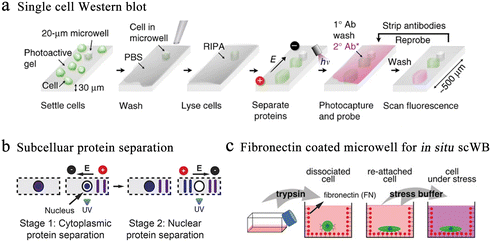 | ||
| Fig. 5 Microwell-based single-cell western blot analysis. (a) Working flow of scWestern analysis. Polyacrylamide gels are selected as the material for the microwells, and individual cells are deposited into the microwells for lysis, electrophoresis, protein photocapturing and antibody detection. Reproduced with permission.26 Copyright 2014, Springer Nature Publishing. (b) Subcellular scWestern blotting. On the basis of scWestern, two-dimensional electrophoresis was introduced to separate and detect the cytoplasm (stage 1) from the nucleus (stage 2), realizing subcellular protein localization in individual cells. Reproduced with permission.79 Copyright 2017, Springer Nature Publishing. (c) In situ single-cell western blot detects protein expression in adherent cells. Fibronectin-coated microwells allow cell cultures to be combined with scWestern, reducing interference during sample preparation. Reproduced with permission.80 Copyright 2019, Wiley. | ||
To analyze the phenotypic relationship between parental and offspring cells, a translocation and secretion assay (TransSeA) was established.81 Using TransSeA, researchers transferred the daughter cells individually to a new plate for culturing. In addition, single-cell extracellular vesicle (EV) analysis based on immunological methods has the characteristics of simple operation and rapid analysis. However, this class of methods is limited by the proteomic parameters of EV analysis per cell. Therefore, it is not sufficient to comprehensively analyze the heterogeneity of EV secretion. The method for multiplexed profiling of single-cell EVs is also necessary. Lu et al. combined spatially patterned antibody barcodes with microwell chips to achieve multiplex analysis of EV secretion from more than 1000 single cells. With the help of barcoded antibodies, multiple EV phenotypes were detected simultaneously.82 In addition, multiplexed analysis of single-cell secretion helps to understand the cellular communication more comprehensively.
3.2 Single-cell nucleic acid analysis
Nucleic acid is responsible for the storage, transfer and expression of genetic information in organisms. Single-cell nucleic acid analysis has significantly improved our recognition of cellular heterogeneity.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 single cells simultaneously on a small chip, achieving 100 times that of conventional throughput.87 Due to the complicated form of comets, each one must be examined separately. This means extensive and potential bias will appear when detecting each comet. To solve these problems, HaloChip was developed.88 Cells were neatly embedded in a thin layer of agarose gel, followed by DNA treatment. DNA fragments from the damage diffused out of the nucleus, forming a circular halo. Single-cell DNA damage can be accurately detected based on the shape and size of the circles. At the same time, the chip can also be used for the assessment of nucleic acid damage repair. In X-ray-induced damage repair experiments, LNCaP was observed to possess stronger repair ability than HeLa cells and MG-63 cells. The HaloChip is considered as a variant of the comet chip experiment.
000 single cells simultaneously on a small chip, achieving 100 times that of conventional throughput.87 Due to the complicated form of comets, each one must be examined separately. This means extensive and potential bias will appear when detecting each comet. To solve these problems, HaloChip was developed.88 Cells were neatly embedded in a thin layer of agarose gel, followed by DNA treatment. DNA fragments from the damage diffused out of the nucleus, forming a circular halo. Single-cell DNA damage can be accurately detected based on the shape and size of the circles. At the same time, the chip can also be used for the assessment of nucleic acid damage repair. In X-ray-induced damage repair experiments, LNCaP was observed to possess stronger repair ability than HeLa cells and MG-63 cells. The HaloChip is considered as a variant of the comet chip experiment.
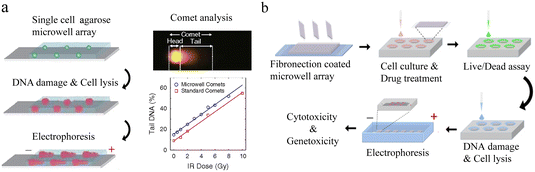 | ||
| Fig. 6 Microwell-based single-cell nucleic acid electrophoresis analysis. (a) Schematic of comet chip assay. Agarose microwell arrays are designed to capture large numbers of individual cells and subsequently characterize DNA damage by gel electrophoresis. By dividing the head/tail of the comet and calculating the parameters, the high sensitivity and high precision of DNA damage analysis of single cells were achieved. Reproduced with permission.83 Copyright 2010, National Academy of Sciences. (b) Working flow of single-cell cytotoxicity and genetoxicity analysis. Multiplex analysis can be performed in combination with cell culturing. Reproduced with permission.85 Copyright 2016, American Chemical Society. | ||
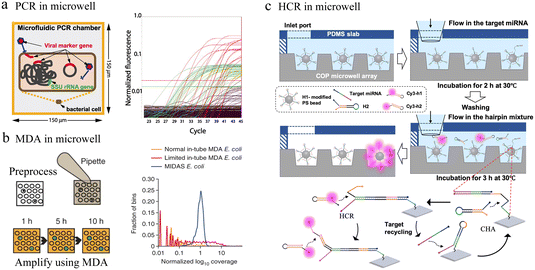 | ||
| Fig. 7 Microwell-based single-cell nucleic acid amplification analysis. (a) Microwell-based digital PCR for detection of single bacteria for viruses. Possible genuine host–virus associations were detectable by this assay. Reproduced with permission.90 Copyright 2011, The American Association for the Advancement of Science. (b) Schematic of microwell displacement amplification system. The pretreatment process includes cell lysis, degeneration and neutralization. Reproduced with permission.91 Copyright 2013, Springer Nature Publishing. (c) Schematic illustration of the cascaded DNA circuit process in the microwell-bead array. The cascaded DNA circuits consist of catalytic hairpin assembly and hybrid chain reaction. Reproduced with permission.92 Copyright 2021, American Chemical Society. | ||
Typically, different temperatures are involved in PCR cycling, whereas isothermal amplification can be carried out at a constant temperature. In microfluidic chips, single-cell nucleic acid amplification analysis using isothermal amplification instead of PCR can significantly reduce the complexity of the system. Multiple displacement amplification (MDA) is one of the most commonly applied isothermal amplification strategies. To reduce amplification bias, the microwell displacement amplification system (MIDAS) performs single-cell MDA in a tiny volume down to 12 nL (Fig. 7b).91 Compared with MDA results in a test tube, MIDAS had a markedly reduced level of amplification bias.
Enzyme-based amplification methods all involve multiple primers and non-specific extensions, which greatly limit the accuracy of ultra-low copy nucleic acid detection in single cells. To address this issue, a bead array analysis method based on an enzyme-free amplification was developed. This method used the cascaded DNA circuits of hybrid chain reaction (HCR) and catalytic hairpin assembly (CHA) for signal amplification and successfully identified single-base mutant miRNAs with good specificity in the presence of high concentrations of interfering nucleic acids (Fig. 7c).92 Any aptamer or probe sequence was compatible with the programmable DNA circuits, which would significantly lower the costs and boost accuracy as well as throughput.
Single-cell transcriptome analysis reflects the expression of genes under certain conditions, which is closely related to cell differentiation and variation. Fan et al. performed pairing of single cells with barcoded beads in microwells, allowing downstream operations.93 Barcode information was added to the cDNA for post target single-cell transcriptional profiling and single-molecule analysis. Yet this unsealed microwell-based platform suffers from inaccurate single-cell information due to the molecular diffusion among wells. To reduce cross-contamination, several sealing strategies were applied to microwell-based single-cell sequencing. The Seq-well64 platform used a semi-permeable polycarbonate membrane to seal the microwells, obtaining high-quality single-cell resolution sequencing data (Fig. 8a). This membrane allowed small molecules to pass through while preventing the passage of mRNA-like macromolecules, critically avoiding cross-contamination. However, the complex chemical modification of the well array and the exacting attachment impeded its broad application.
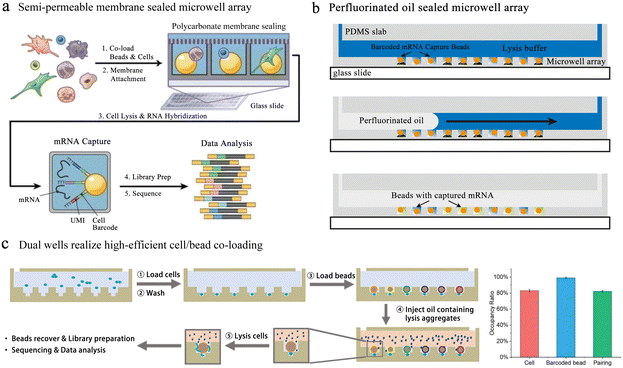 | ||
| Fig. 8 Microwell-based single-cell sequencing. (a) Seq-well: a platform using a semi-permeable polycarbonate membrane to seal the microwells. The application of semi-permeable membranes has greatly reduced the cross-contamination between microwells. Reproduced with permission.64 Copyright 2017, Springer Nature Publishing. (b) An oil-sealing microwell chip for single-cell RNA sequencing. Oil seal technology advances the automation of high-throughput single-cell sequencing platforms based on microwells. Reproduced with permission.94 Copyright 2015, BioMed Central Ltd. (c) Well-paired-seq: using dual wells to realize efficient cell/bead co-loading. Reproduced with permission.97 Copyright 2022, Wiley. | ||
In addition to semi-permeable membranes, oils are also used for microwell sealing. Bose et al. fabricated a microwell array in a thin PDMS layer on top of a glass slide with a microfluidic flow channel above.94 To ensure the array sealing, oil with lysis buffer was quickly injected into the flow channel to replace the liquid. Cell lysis and mRNA capture were performed in the well under oil seal conditions (Fig. 8b). This method can be used for unbiased RNA sequencing of a single cell, but the throughput was relatively low. Thus, a large amount of cells can be sequenced when the platform is improved to be automated.95 However, this design restricts buffer exchange and demands integrated temperature, which has an influence on usability and portability. Therefore, a convenient, low-cost platform for high-throughput scRNA-seq is attractive. Microwell-seq used agarose as material to fabricate well chips, reducing the cost of single-cell sequencing.63 Using this strategy, researchers successfully mapped mammalian adult adrenal gland hierarchy across a species and mouse cell atlas. The absence of temperature controllers and pumps also made Microwell-seq easier to use in individual laboratories. Subsequently, the process was accelerated with costs reduced through system optimization of TaqMan qPCR in Microwell-seq, which was known as Microwell-seq 2.0.96
In the above-mentioned methods, to ensure the accuracy of cell sequencing data and reduce doublets, the capture of cells is based on Poisson distribution, which means that the pairing rate of cells and barcode beads is quite low. Well-paired-seq utilized dual-microwell chips to achieve an extremely high cell/bead pairing ratio (Fig. 8c).97 This chip consisted of a flow channel and dual wells, and the dual well included a bead trap and a cell trap. When laminar fluid passed through the flow channel, the velocity at which the beads located below the main flow capture the fluid in the well was significantly reduced. The cell trap below the bead trap had a lower flow rate, which allowed for more efficient capture of single cells and a higher pairing rate with the barcoded bead. Cell lysis was performed in a confined space by mixing lysis reagents in mineral oil, effectively reducing cross-contamination among wells. Transcript expression of peripheral blood mononuclear cells (PBMCs) and drug-treated cells was profiled through Well-paired-seq, revealing the heterogeneity of single cells.
Linking single-cell omics signatures with imaging informative features may help to unravel the molecular mechanisms driving the growth of cell subsets. Microwell chip-based assays have evolved to enable integrated analysis of single-cell omics sequencing data with multiple phenotypic information. SCOPE-seq used a barcoded bead containing a second barcode to link live-cell images with single-cell transcriptome sequencing data.65 After co-loading cells with barcode beads into the microwell chip, the second barcode in each well can be optically decoded. Thus, images obtained by microscopy from individual cells can be directly linked to genome-wide expression profiles. However, this method had limited throughput, low mRNA capture efficiency, and required complex procedures to link the two barcodes. In SCOPE-seq 2, the introduction of error-correcting codes addressed these issues well and also improved the throughput and linking accuracy.98 Zhang et al. used specific coordinate oligonucleotides to encode the position of each well, which were identified together with mRNA during library preparation.99 These coordinate oligonucleotides allowed each cell barcode to be traced back to its origin well, thus linking it to the optical data collected for that cell. In contrast to SCOPE-seq, this strategy did not require complex optical decoding. In addition, microwell chip-based sequencing and imaging linking analysis have enabled photophysiological analysis of microalgae100 and integration of the phenotype and transcriptome of patient-derived tumor organoids.101
3.3 Single-cell metabolite analysis
Significant changes in cellular metabolic characteristics are also manifestations of cellular heterogeneity. The detection of single-cell metabolites is of great significance for the diagnosis of cancer and the comprehensive understanding of key processes such as embryonic development.Mass spectrometry is one of the common methods for single-cell metabolite analysis due to its ability to perform accurate and sensitive detection of large numbers of chemical molecules. Zhang's group proposed a single-cell metabolite quantification method combining microwell arrays and droplet microextraction mass spectrometry.102 Microwells were able to confine single cells in a certain space and avoided the random diffusion of target molecules during the microextraction process, which greatly improves the precision and accuracy of detection. The glucose phosphate calibration curve measured by this method varied linearly from aM to fM range. Liu's team designed and fabricated a microwell array-based chip combined with matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS).103 Using this platform, they detected eight phospholipids from one single cell and characterized the structures by MS/MS spectroscopy.
The oxygen consumption rate (OCR) of a cell is related to the metabolism of the cell directly. The distribution of single-cell OCR is an important parameter of metabolic heterogeneity. Lidstrom et al. designed a glass microwell array containing immobilized luminescence sensors to quantitatively measure oxygen consumption by individual cells at fM per min resolution.104 However, this method needed to embed a large number of micro-oxygen sensor arrays in the microwell chip, which made the preparation process of this chip very complicated. To address this issue, Wang et al. combined photoacoustic microscopy with a microwell array chip to measure OCR.105 This method separated single cells by microwells and then combined photoacoustic imaging to measure the change of hemoglobin oxygen in each microwell, realizing label-free metabolic detection of tumor cells.106
4. Conclusions and future outlook
In this review, we have discussed the design and fabrication of microwell-based chips and their applications in single-cell nucleic acid, protein, and small molecule analysis. Microfluidics, including droplet, microwell and other technologies, has been proved to be a powerful tool for single-cell analysis. These technologies have their own features. A huge amount of droplets can be produced in a short time to achieve ultrahigh-throughput single-cell encapsulation. During the process of producing droplets, a large number of droplets without cells are generated due to the Poisson distribution. These empty droplets can be screened by sorting techniques or combined with methods such as inertial focusing to obtain droplets with high single-cell rates. Meanwhile, droplet operations, including sorting, fusion and splitting, realize the precise control of high-throughput single droplets. Therefore, droplet technology has become the most commercialized platform. In contrast, microwell-based chips can achieve high-throughput single-cell isolation with hyper-Poisson distribution through simple size design. Microwell chips have various structures with flexible operations; for instance, roofless microwell chips permit direct sample loading without a pump. Using a modified glass slide as the roof enables microwell chip secretion capture and detection. In addition, the fixed spatial location of each well can be used to connect multiple discrete measurements, so the microwell platform is easy to integrate with methods such as single-cell culture and in situ analysis. The ability to link single-cell omics features with cellular phenotypic features, especially time-varying phenotypic changes, with the help of microwell chips can provide insight into the molecular basis of cellular function. However, the overall throughput capability of microwells is limited by chip size, supporting less precision in environmental control than valve-based systems. Furthermore, multi-omics analysis is one of the future directions of single-cell profiling, so it is crucial to accurately capture and detect multiple targets in single cells. Although microwells can be sealed through adding a roof, such as a glass slide or a semipermeable membrane, it is difficult to achieve complete isolation among microwells. Therefore, cross-contamination between microwells remains a tough problem. As microwell technologies continue to mature, these obstacles will be overcome associated with deeper understanding of single-cell biology. Overall, we envision that the microwell platform will provide multi-dimensional analysis methods for oncology, immunology and drug screening.Author contributions
Supervision: Y. X. Zhao, B. D. Chen; funding acquisition: Y. X. Zhao; investigation and visualization: F. Chen, J. Zhang; writing – original draft: J. Zhang; writing – review & editing: J. Zhang, J. Xue, N. F. Luo; F. Chen.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (grant number 22125404).Notes and references
- T. Masuda, R. Sankowski, O. Staszewski, C. Böttcher, L. Amann, Sagar, C. Scheiwe, S. Nessler, P. Kunz, G. van Loo, V. A. Coenen, P. C. Reinacher, A. Michel, U. Sure, R. Gold, D. Grün, J. Priller, C. Stadelmann and M. Prinz, Nature, 2019, 566, 388–392 CrossRef PubMed.
- E. Papalexi and R. Satija, Nat. Rev. Immunol., 2018, 18, 35–45 CrossRef PubMed.
- M. H. Rohban, H. S. Abbasi, S. Singh and A. E. Carpenter, Nat. Commun., 2019, 10, 1–6 CrossRef PubMed.
- F. Buettner, K. N. Natarajan, F. P. Casale, V. Proserpio, A. Scialdone, F. J. Theis, S. A. Teichmann, J. C. Marioni and O. Stegle, Nat. Biotechnol., 2015, 33, 155–160 CrossRef PubMed.
- H. Shi, A. Colavin, T. K. Lee and K. C. Huang, Nat. Protoc., 2017, 12, 429–438 CrossRef PubMed.
- A.-C. Villani, R. Satija, G. Reynolds, S. Sarkizova, K. Shekhar, J. Fletcher, M. Griesbeck, A. Butler, S. Zheng, S. Lazo, L. Jardine, D. Dixon, E. Stephenson, E. Nilsson, I. Grundberg, D. McDonald, A. Filby, W. Li, P. L. De Jager, O. Rozenblatt-Rosen, A. A. Lane, M. Haniffa, A. Regev and N. Hacohen, Science, 2017, 356, eaah4573 CrossRef PubMed.
- P. O. Krutzik, J. M. Crane, M. R. Clutter and G. P. Nolan, Nat. Chem. Biol., 2008, 4, 132–142 CrossRef CAS.
- F. Porichis, M. G. Hart, M. Griesbeck, H. L. Everett, M. Hassan, A. E. Baxter, M. Lindqvist, S. M. Miller, D. Z. Soghoian and D. G. Kavanagh, Nat. Commun., 2014, 5, 5641 CrossRef CAS.
- D. P. Ivanov and A. M. Grabowska, Sci. Rep., 2017, 7, 41160 CrossRef CAS PubMed.
- S. M. Prakadan, A. K. Shalek and D. A. Weitz, Nat. Rev. Genet., 2017, 18, 345–361 CrossRef CAS.
- S. Y. Teh, R. Lin, L. H. Hung and A. P. Lee, Lab Chip, 2008, 8, 198–220 RSC.
- S. B. Im, M. J. Uddin, G. J. Jin and J. S. Shim, Lab Chip, 2018, 18, 1310–1319 RSC.
- Y. Chen, Z. Fang, B. Merritt, D. Strack, J. Xu and S. Lee, Lab Chip, 2016, 16, 3024–3032 RSC.
- S. Lindstrm, M. Hammond, H. Brismar, H. A. Svahn and A. Ahmadian, Lab Chip, 2009, 9, 3465–3471 RSC.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed.
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, 1903905 CrossRef CAS.
- S. Wiedenmann, M. Breunig, J. Merkle, C. von Toerne, T. Georgiev, M. Moussus, L. Schulte, T. Seufferlein, M. Sterr, H. Lickert, S. E. Weissinger, P. Möller, S. M. Hauck, M. Hohwieler, A. Kleger and M. Meier, Nat. Biomed. Eng., 2021, 5, 897–913 CrossRef CAS PubMed.
- S. Bian, Y. Zhou, Y. Hu, J. Cheng, X. Chen, Y. Xu and P. Liu, Sci. Rep., 2017, 7, 42512 CrossRef CAS PubMed.
- Y. Hu, X. Sui, F. Song, Y. Li and P. Liu, Nat. Commun., 2021, 12, 1–14 CrossRef.
- X. Hou, Y. S. Zhang, T. D. Santiago, M. M. Alvarez, J. Ribas, S. J. Jonas, P. S. Weiss, A. M. Andrews, J. Aizenberg and A. Khademhosseini, Nat. Rev. Mater., 2017, 2, 17016 CrossRef CAS.
- L. C. Taylor and D. R. Walt, Anal. Biochem., 2000, 278, 132–142 CrossRef CAS PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- R. M. Kiran and S. Chakraborty, J. Appl. Polym. Sci., 2020, 137, 48958 CrossRef.
- A. B. Bernard, C. C. Lin and K. S. Anseth, Tissue Eng., Part C, 2012, 18, 583–592 CrossRef PubMed.
- L. Huang, Y. Chen, W. Huang and H. Wu, Lab Chip, 2018, 18, 1113–1120 RSC.
- A. J. Hughes, D. P. Spelke, Z. Xu, C. C. Kang and A. E. Herr, Nat. Methods, 2014, 11, 749–755 CrossRef PubMed.
- A. R. Thomsen, C. Aldrian, P. Bronsert, Y. Thomann, N. Nanko, N. Melin, G. Rücker, M. Follo, A. L. Grosu, G. Niedermann, P. G. Layer, A. Heselich and P. G. Lund, Lab Chip, 2018, 18, 179–189 RSC.
- M. R. Dusseiller, D. Schlaepfer, M. Koch, R. Kroschewski and M. Textor, Biomaterials, 2005, 26, 5917–5925 CrossRef PubMed.
- Y. Xu, F. Xie, T. Qiu, L. Xie, W. Xing and J. Cheng, Biomicrofluidics, 2012, 6, 016504 CrossRef PubMed.
- M. Kobayashi, S. H. Kim, H. Nakamura, S. Kaneda and T. Fujii, PLoS One, 2015, 10, e0139980 CrossRef PubMed.
- Y. Tokimitsu, H. Kishi, S. Kondo, R. Honda, K. Tajiri, K. Motoki, T. Ozawa, S. Kadowaki, T. Obata and S. Fujiki, Cytometry, Part A, 2010, 71, 1003–1010 Search PubMed.
- J. Y. Park, M. Morgan, A. N. Sachs, J. Samorezov, R. Teller, Y. Shen, K. J. Pienta and S. Takayama, Microfluid. Nanofluid., 2010, 8, 263–268 CrossRef CAS.
- X. Xu, W. Wang, K. Kratz, L. Fang, Z. Li, A. Kurtz, N. Ma and A. Lendlein, Adv. Healthcare Mater., 2014, 3, 1991–2003 CrossRef CAS.
- Y. C. Chen, X. Lou, Z. Zhang, P. Ingram and E. Yoon, Sci. Rep., 2015, 5, 12175 CrossRef PubMed.
- K. C. Hribar, D. Finlay, X. Ma, X. Qu, M. G. Ondeck, P. H. Chung, F. Zanella, A. J. Engler, F. Sheikh, K. Vuori and S. C. Chen, Lab Chip, 2015, 15, 2412–2418 RSC.
- Š. Selimović, F. Piraino, H. Bae, M. Rasponi, A. Redaelli and A. Khademhosseini, Lab Chip, 2011, 11, 2325–2332 RSC.
- Y. Y. Choi, B. G. Chung, D. H. Lee, A. Khademhosseini, J. H. Kim and S. H. Lee, Biomaterials, 2010, 31, 4296–4303 CrossRef CAS PubMed.
- G. Eng, B. W. Lee, H. Parsa, C. D. Chin, J. Schneider, G. Linkov, S. K. Sia and G. Vunjak-Novakovic, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4551–4556 CrossRef CAS.
- R. L. Lai and N. T. Huang, Microfluid. Nanofluid., 2019, 23, 121 CrossRef.
- A. Napolitano, D. Dean, A. Man, J. Youssef, D. Ho, A. Rago, M. Lech and J. Morgan, BioTechniques, 2007, 43, 500–500 CrossRef PubMed.
- J. Dahlmann, G. Kensah, H. Kempf, D. Skvorc, A. Gawol, D. Elliott, G. Dräger, R. Zweigerdt, U. Martin and I. Gruh, Biomaterials, 2013, 34, 2463–2471 CrossRef CAS PubMed.
- W. Liu, J. Wang, X. Xu, C. Zhao, X. Xu and P. S. Weiss, ACS Nano, 2021, 15, 12180–12188 CrossRef CAS.
- S. Lindstrm, R. Larsson and H. A. Svahn, Electrophoresis, 2008, 29, 1219–1227 CrossRef PubMed.
- J. M. Cha, H. Bae, N. Sadr, S. Manoucheri, F. Edalat, K. Kim, S. B. Kim, I. K. Kwon, Y. S. Hwang and A. Khademhosseini, Macromol. Res., 2015, 23, 245–255 CrossRef CAS.
- H. H. Jeong, J. H. Lee, Y. M. Noh and Y. Kim, Macromol. Res., 2013, 21, 534–540 CrossRef CAS.
- H. Tekin, M. Anaya, M. Brigham, C. Nauman, R. Langer and A. Khademhosseini, Lab Chip, 2010, 10, 2411–2418 RSC.
- J. J. Kim, K. W. Bong, E. Reátegui, D. Irimia and P. S. Doyle, Nat. Mater., 2017, 16, 139–146 CrossRef.
- J. Garra, T. Long, J. Currie, T. Schneider, R. White and M. Paranjape, J. Vac. Sci. Technol., A, 2002, 20, 975–982 CrossRef.
- M. Holl, T. Ray, S. Bhushan, D. R. Meldrum and H. Zhu, J. Micromech. Microeng., 2015, 19, 65013–65020(65018) Search PubMed.
- C. Wang, L. Ren, W. Liu, Q. Wei, M. Tan and Y. Yu, Analyst, 2019, 144, 2811–2819 RSC.
- G. Dan, J. Feng, Z. Min and Y. Jiang, Analyst, 2019, 144, 766–781 RSC.
- L. Huang, Y. Chen, Y. Chen and H. Wu, Anal. Chem., 2015, 87, 12169 CrossRef CAS PubMed.
- S. M. Park, J. Y. Lee, S. Hong, S. H. Lee, I. K. Dimov, H. Lee, S. Suh, Q. Pan, K. Y. Li, A. M. Wu, S. M. Mumenthaler, P. Mallick and L. P. Lee, Lab Chip, 2016, 16, 3682–3688 RSC.
- J. F. Swennenhuis, A. G. Tibbe, M. Stevens, M. R. Katika, J. Van Dalum, H. D. Tong, C. J. Van Rijn and L. W. Terstappen, Lab Chip, 2015, 15, 3039–3046 RSC.
- N. T. Huang, Y. J. Hwong and R. L. Lai, Microfluid. Nanofluid., 2018, 22, 16 CrossRef.
- Z. Bai, Y. Deng, D. Kim, Z. Chen, Y. Xiao and R. Fan, ACS Nano, 2020, 14, 7412–7424 CrossRef CAS.
- Q. Huang, S. Mao, M. Khan and J.-M. Lin, Analyst, 2019, 144, 808–823 RSC.
- L. Nan, Z. Jiang and X. Wei, Lab Chip, 2014, 14, 1060–1073 RSC.
- F. Stumpf, J. Schoendube, A. Gross, C. Rath, S. Niekrawietz, P. Koltay and G. Roth, Biosens. Bioelectron., 2015, 69, 301–306 CrossRef CAS PubMed.
- K. Leung, H. Zahn, T. Leaver, K. M. Konwar, N. W. Hanson, A. P. Page, C. C. Lo, P. S. Chain, S. J. Hallam and C. L. Hansen, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7665–7670 CrossRef CAS.
- D. Burak, C. Jin-Young, K. Zhang, D. William, T. Durga, B. Marcus, C. Joe and R. Fan, Nucleic Acids Res., 2018, 3, e16 Search PubMed.
- W. Gao, X. Zhang, H. Yuan, Y. Wang and J. Zhao, Biosens. Bioelectron., 2019, 139, 111326 CrossRef CAS.
- X. Han, R. Wang, Y. Zhou, L. Fei, H. Sun, S. Lai, A. Saadatpour, Z. Zhou, H. Chen, F. Ye, D. Huang, Y. Xu, W. Huang, M. Jiang, X. Jiang, J. Mao, Y. Chen, C. Lu, J. Xie, Q. Fang, Y. Wang, R. Yue, T. Li, H. Huang, S. H. Orkin, G.-C. Yuan, M. Chen and G. Guo, Cell, 2018, 172, 1091–1107 CrossRef CAS PubMed.
- T. M. Gierahn, M. H. W. Ii, T. K. Hughes, B. D. Bryson, A. Butler, R. Satija, S. Fortune, J. C. Love and A. K. Shalek, Nat. Methods, 2017, 14, 395–398 CrossRef CAS.
- J. Yuan, J. Sheng and P. A. Sims, Genome Biol., 2018, 19, 1–5 CrossRef.
- J. C. Love, J. L. Ronan, G. M. Grotenbreg, D. Van and H. L. Ploegh, Nat. Biotechnol., 2006, 24, 703 CrossRef CAS.
- J. Choi, K. Routenberg Love, Y. Gong, T. M. Gierahn and J. C. Love, Anal. Chem., 2011, 83, 6890 CrossRef.
- D. M. Rissin, C. W. Kan, T. G. Campbell, S. C. Howes, D. R. Fournier, L. Song, T. Piech, P. P. Patel, L. Chang, A. J. Rivnak, E. P. Ferrell, J. D. Randall, G. K. Provuncher, D. R. Walt and D. C. Duffy, Nat. Biotechnol., 2010, 28, 595–599 CrossRef.
- X. Wang, L. Cohen, J. Wang and D. R. Walt, J. Am. Chem. Soc., 2018, 140, 18132–18139 CrossRef PubMed.
- S. M. Schubert, L. M. Arendt, W. Zhou, S. Baig and D. R. Walt, Sci. Rep., 2015, 5, 11034 CrossRef PubMed.
- X. Wang, A. F. Ogata and D. R. Walt, J. Am. Chem. Soc., 2020, 142, 15098–15106 CrossRef PubMed.
- C. Wu, P. M. Garden and D. R. Walt, J. Am. Chem. Soc., 2020, 142, 12314–12323 CrossRef PubMed.
- D. Wu, M. D. Milutinovic and D. R. Walt, Analyst, 2015, 140, 6277–6282 RSC.
- X. Wang and D. R. Walt, Chem. Sci., 2020, 11, 7896–7903 RSC.
- L. Yang, Z. Wang, Y. Deng, Y. Li, W. Wei and Q. Shi, Anal. Chem., 2016, 88, 11077–11083 CrossRef CAS PubMed.
- R. Bai, L. Li, M. Liu, S. Yan, C. Miao, R. Li, Y. Luo, T. Liu, B. Lin, Y. Ji and Y. Lu, Anal. Chem., 2018, 90, 5825–5932 CrossRef CAS.
- Y. Zhou, N. Shao, R. Castro, P. Zhang and L. Qin, Cell Rep., 2020, 31, 107574 CrossRef CAS PubMed.
- C. C. Kang, K. A. Yamauchi, J. Vlassakis, E. Sinkala, T. A. Duncombe and A. E. Herr, Nat. Protoc., 2016, 11, 1508–1530 CrossRef PubMed.
- K. A. Yamauchi and A. E. Herr, Microsyst. Nanoeng., 2017, 3, 16079 CrossRef CAS.
- Y. Zhang, I. Naguro and A. E. Herr, Angew. Chem., Int. Ed., 2019, 58, 13929–13934 CrossRef CAS.
- W. Cai, Y.-J. Chiu, V. Ramakrishnan, Y. Tsai, C. Chen and Y.-H. Lo, Lab Chip, 2018, 18, 3154–3162 RSC.
- Y. Ji, D. Qi, L. Li, H. Su, X. Li, Y. Luo, B. Sun, F. Zhang, B. Lin, T. Liu and Y. Lu, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 5979–5984 CrossRef CAS.
- D. K. Wood, D. M. Weingeist, S. N. Bhatia and B. P. Engelward, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10008–10013 CrossRef CAS.
- C. Watson, J. Ge, J. Cohen, G. Pyrgiotakis, B. P. Engelward and P. Demokritou, ACS Nano, 2014, 8, 2118–2133 CrossRef CAS PubMed.
- L. Li, W. Wang, M. Ding, G. Luo and Q. Liang, Anal. Chem., 2016, 88, 6734–6742 CrossRef CAS.
- C. Chao, P. N. Le and B. P. Engelward, ACS Biomater. Sci. Eng., 2020, 6, 2427–2439 CrossRef CAS.
- Y. Li, X. Feng, D. Wei, L. Ying and B. Liu, Anal. Chem., 2013, 85, 4066–4073 CrossRef CAS PubMed.
- Y. Qiao, C. Wang, M. Su and L. Ma, Anal. Chem., 2012, 84, 1112 CrossRef CAS.
- W. Liu, Z. Li, Y. Liu, Q. Wei, Y. Liu, L. Ren, C. W. Ang and Y. Yu, RSC Adv., 2019, 9, 2865–2869 RSC.
- A. D. Tadmor, E. A. Ottesen, J. R. Leadbetter and R. Phillips, Science, 2011, 333, 58–62 CrossRef CAS.
- J. Gole, A. Gore, A. Richards, Y. J. Chiu and K. Zhang, Nat. Biotechnol., 2013, 31, 1126 CrossRef CAS.
- F. Jin and D. Xu, Anal. Chem., 2021, 93, 11617–11625 CrossRef PubMed.
- H. C. Fan, G. K. Fu and S. Fodor, Science, 2015, 347, 1258367 CrossRef PubMed.
- S. Bose, Z. Wan, A. Carr, A. H. Rizvi, G. Vieira, D. Pe'er and P. A. Sims, Genome Biol., 2015, 16, 120 CrossRef PubMed.
- J. Yuan and P. A. Sims, Sci. Rep., 2016, 6, 33883 CrossRef PubMed.
- H. Chen, Y. Liao, G. Zhang, Z. Sun, L. Yang, X. Fang, H. Sun, L. Ma, Y. Fu, J. Li, Q. Guo, X. Han and G. Guo, Cell Discovery, 2021, 7, 107 CrossRef PubMed.
- K. Yin, M. Zhao, L. Lin, Y. Chen, S. Huang, C. Zhu, X. Liang, F. Lin, H. Wei, H. Zeng, Z. Zhu, J. Song and C. Yang, Small Methods, 2022, 2200341 CrossRef PubMed.
- Z. Liu, J. Yuan, A. Lasorella, A. Iavarone and P. A. Sims, Sci. Rep., 2020, 10, 1–15 CrossRef.
- J. Q. Zhang, C. A. Siltanen, L. Liu, K. C. Chang and A. R. Abate, Genome Biol., 2020, 21, 1–11 CrossRef PubMed.
- L. Behrendt, M. M. Salek, E. L. Trampe, V. I. Fernandez, K. S. Lee, M. Kühl and R. Stocker, Sci. Adv., 2020, 6, eabb2754 CrossRef CAS PubMed.
- Y. Wu, K. Li, Y. Li, T. Sun, C. Liu, C. Dong, T. Zhao, D. Tang, X. Chen, X. Chen and P. Liu, Nucleic Acids Res., 2021, 50, e28 CrossRef.
- J. Feng, X. Zhang, L. Huang, H. Yao, C. Yang, X. Ma, S. Zhang and X. Zhang, Anal. Chem., 2019, 91, 5613–5620 CrossRef CAS PubMed.
- W. Xie, D. Gao, F. Jin, Y. Jiang and H. Liu, Anal. Chem., 2015, 87, 7052–7059 CrossRef CAS PubMed.
- T. W. Molter, S. C. Mcquaide, M. T. Suchorolski, T. J. Strovas, L. W. Burgess, D. R. Meldrum and M. E. Lidstrom, Sens. Actuators, B, 2009, 135, 678–686 CrossRef CAS PubMed.
- P. Hai, T. Imai, S. Xu, R. Zhang, R. L. Aft, J. Zou and L. V. Wang, Nat. Biomed. Eng., 2019, 3, 381–391 CrossRef CAS PubMed.
- A. J. Walsh, J. T. Sharick and M. C. Skala, Nat. Biomed. Eng., 2019, 3, 333–334 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |